Abstract
Phosphorylation of the transcription factor CREB leads to the recruitment of the coactivator, CREB binding protein (CBP). Recent studies have suggested that CBP recruitment is not sufficient for CREB function, however. We have identified a conserved protein-protein interaction motif within the CBP-binding domains of CREB and another transcription factor, SREBP (sterol-responsive element binding protein). In contrast to CREB, SREBP interacts with CBP in the absence of phosphorylation. We have exploited the conservation of this interaction motif to test whether CBP recruitment to CREB is sufficient for transcriptional activation. Substitution of six nonconserved amino acids from SREBP into the activation domain of CREB confers high-affinity, phosphorylation-independent CBP binding. The mutated CREB molecule, CREBDIEDML, activates transcription in F9 teratocarcinoma and PC12 cells even in the absence of protein kinase A (PKA). Addition of exogenous CBP augments the level of transcription mediated by CREBDIEDML, and adenovirus 12S E1A blocks transcription, implicating CBP in the activation process. Thus, recruitment of CBP to CREB is sufficient for transcriptional activation. Addition of PKA stimulates transcription induced by CREBDIEDML further, suggesting that a phosphorylation event downstream from CBP recruitment augments CREB signaling.
The signaling mechanism that activates genes through the cyclic AMP (cAMP)-regulated enhancer (CRE) (23) represents one of the most intensively studied transcriptional pathways. This pathway consists of protein kinase A (PKA), the transcription factor CREB, and the coactivator CREB binding protein (CBP) (3, 7, 18). CBP has been shown to participate in many additional transcriptional pathways as well (11), but the mechanism by which it activates gene expression remains uncertain. CBP and its homologue p300 interact with the basal transcription factors TFIID and TFIIB, as well as with the RNA polymerase II holoenzyme component, RNA helicase A (9, 24, 34), suggesting that one function of this coactivator is to stabilize the preinitiation complex. Other studies have argued that transcriptional activation through CBP/p300 occurs only in the context of chromatin, however (17). The involvement of chromatin in CBP function is consistent with the finding that CBP, and several associated proteins including P/CAF, steroid receptor coactivator 1, and p/CIP, have the ability to acetylate the amino-terminal tails of histone proteins (6, 31, 36, 38). These and other posttranslational modifications of chromatin components induced by the cAMP signaling cascade may stimulate transcription through nucleosome remodeling.
Of all transcription factor-CBP associations, only the interaction with phosphorylated CREB has been characterized in detail. Our lab has studied this association by using a fluorescence polarization binding assay and a genetic interaction assay in yeast (18, 30). These studies indicated that CREB phosphorylated at Ser133 binds to CBP with an affinity of approximately 350 nM and that this interaction depends on the phosphorylated Ser residue and several adjacent hydrophobic residues. Other laboratories have reached similar conclusions (26). Recently, the structure of the phosphorylated CREB-CBP complex has been solved by nuclear magnetic resonance spectroscopy (28). These studies demonstrated the importance of the phosphorylated serine in the CREB activation domain and showed that the interaction of phosphorylated CREB with CBP introduces structure into both components of the complex. Surprisingly, other transcription factors can interact with the CREB binding domain of CBP in the absence of phosphorylation. How this CBP domain participates in both phosphorylation-dependent and -independent interactions is unknown.
Although it was once believed that CREB phosphorylation was sufficient for gene activation, several studies have shown that these two events can be dissociated. The first indication that CREB phosphorylation might be insufficient for gene induction came from studies of the c-fos promoter (10). In both PC12 cells and primary neurons, depolarization activates the c-fos CRE in a manner that is blocked by inhibitors of PKA, despite the fact that CREB phosphorylation is maintained (16, 35). Precisely how PKA contributes to depolarization-induced gene activation is unknown, however. It is possible that PKA phosphorylates components of the transcriptional machinery downstream from CREB; conceivably, these other phosphorylation events could augment, or even be required for, the transcriptional response. Xu et al. (37) have suggested that PKA phosphorylates CBP directly, while Zanger et al. (39) have argued that PKA might affect a step downstream from CBP. Calcium-activated kinases, such as Ca2+/calmodulin kinase IV (CaMKIV), have also been proposed to augment transcription via CBP phosphorylation (5, 13, 14). Many of these experiments are problematic, however, because they are based on the analysis of GAL-CBP fusion proteins, several of which appear to activate transcription more potently than the full-length native protein (34). Thus, it is likely that the transcriptional activation function of these fragments is masked by the conformation of native CBP. Moreover, this conformation might not be recapitulated precisely in a molecule that is targeted to a promoter through a heterologous DNA binding domain at its amino terminus. Thus, how various kinases such as PKA contribute to CBP signaling remains uncertain.
By comparing the activation domain sequences of CREB and other transcription factors, we hoped to elucidate whether phosphorylation-dependent and -independent factors interact with CBP in a similar manner. We identified a region of homology between CREB and the sterol-responsive element binding protein (SREBP), a transcription factor that also interacts with the CREB-binding domain of CBP (25). SREBP is responsible for the transcriptional regulation of several key enzymes involved in cholesterol metabolism, including HMG-coenzyme A reductase, HMG coenzyme A synthase, and the low-density lipoprotein receptor (25).
By substituting amino acids from the activation domain of SREBP into CREB, we sought to develop a CREB molecule that could interact with CBP in the absence of phosphorylation. We then used this mutated CREB molecule to determine whether CBP recruitment alone was sufficient for gene activation. Studies from our lab and others have shown that PKA augments GAL-CBP function, but as indicated above, these experiments are problematic. We used the constitutively active CREB mutant to test whether PKA can facilitate transcription through a step downstream from CBP recruitment.
MATERIALS AND METHODS
Plasmids.
pGEX-3T4-SREBP1a1-50 was kindly provided by R. Tjian (University of California, Berkeley). Mutants mt1 to mt18 were derived directly from this vector by site-directed mutagenesis. pGEX-KG-CREB1-290 was constructed by inserting a PCR-generated fragment of rat CREB341 encoding amino acids 1 to 290 into the BamHI/HindIII sites of pGEX-KG. pRc/RSV-FLAG-CREB was constructed by inserting a synthetic, oligonucleotide-derived fragment encoding the FLAG epitope (DYKDDDDK) into the first NcoI site (amino acid position 3) of pRc/RSV-CREB341 (20). pRc/RSV-FLAG-CREBDIEDML was generated by site-directed mutagenesis of pRc/RSV-FLAG-CREB. The CRE-chloramphenicol acetyltransferase (CAT) reporter is the somatostatin-CAT fusion gene Δ (−71) described in Montminy et al. (23). pRc/RSV-GAL4-CREB was constructed by cloning GAL4-CREB4-283 (33) into pRc/RSV. GAL4-CREB4-283 encodes amino acids (aa) 1 to 147 of GAL4 fused to the N terminus of the CREB activation domain (aa 4 to 283). pRc/RSV-GAL4-CREBDIEDML was constructed as described above after site-directed mutagenesis. The GAL4-LUC (luciferase) reporter is 5XGAL4-TATA-Luciferase described by Sun et al. (32). pRSV-β-GAL contains the β-galactosidase gene driven by the Rous sarcoma virus (RSV) long terminal repeat. pRc/RSV-E1A-12S was described by Lundblad et al. (21), and pRc/RSV-CREB-M1 was described by Loriaux et al. (20). RSV-PKA was obtained from R. Maurer (Oregon Health Sciences University). Mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocols.
CBP binding assay.
HeLa cell nuclear extract was prepared as described elsewhere (4) from HeLa-S3 cells provided by the National Cell Culture Center, Minneapolis, Minn. Glutathione S-transferase (GST)–CREB1-290 was phosphorylated with PKA for 1 h at 30°C as described by Laurance et al. (19). Five micromolar GST, GST-CREB1-290, or GST-SREBP1a1-50 was incubated with 10 μl of glutathione-Sepharose beads (Pharmacia) in 150 μl of buffer A (50 mM HEPES [pH 7.6], 1 M NaCl, 0.2% NP-40, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]) for 1 h at 4°C. The beads were washed twice with 1 ml of buffer A and twice with 1 ml of HEG100 (20 mM HEPES [pH 7.6], 10% glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT). The GST-CREB bound beads were incubated with 900 μg of HeLa nuclear extract in 400 μl of HEG100 plus protease inhibitors (Complete; Boehringer Mannheim), 10 μM NaF, and 0.4 μM microcystin for 2 h at 4°C. The GST-SREBP-bound beads were incubated with 150 μg of HeLa nuclear extract in 400 μl of HEG100 plus protease inhibitors for 2 h at 25°C. The beads were washed three times with 1 ml of HEGN300 (20 mM HEPES [pH 7.6], 10% glycerol, 300 mM KCl, 0.1 mM EDTA, 0.1% NP-40, 1 mM DTT), resuspended in 15 μl of sodium dodecyl sulfate (SDS) loading buffer, heated to 95°C for 5 min, and electrophoresed on an SDS–6% polyacrylamide gel. After transfer to a polyvinylidene difluoride membrane, CBP was detected by Western blotting using a polyclonal antibody directed against CBP451-682. PKA was a gift from R. Maurer.
Protein expression and purification.
The CREB proteins used in the fluorescence polarization binding assays contain three Cys-Ser mutations (Cys300, Cys310, and Cys337) in the DNA-binding domain. Construction and expression of this vector (CREB/Ser) are described by Richards et al. (29). The serine mutations in CREB/Ser improve protein solubility but do not alter DNA binding, as demonstrated by the similar affinities of CREB/Ser and wild-type CREB to the CRE (29). CREB/Ser was phosphorylated stoichiometrically using the purified catalytic subunit of PKA in 50 mM morpholinepropanesulfonic acid (pH 6.8)–50 mM NaCl–2 mM MgCl2–1 mM DTT–1 mM ATP. The reaction mixture was incubated at 30°C for 20 min. Residual ATP was removed by dialysis against 10 mM Tris buffer (pH 8.0). Expression of the CBP fragment was as described previously (18).
Fluorescence polarization measurements.
Fluorescence polarization measurements were determined using a PanVera 2000 fluorescence polarization system. Samples were excited at 490 nm, and emission was measured at 510 nm. A 5′-fluoresceinated oligonucleotide corresponding to the top strand of the somatostatin CRE (5′ CCTGACGTCAGCCCCCTGACGTCAGG 3′) was purchased from Gibco BRL. This oligonucleotide was constructed to form a double-stranded CRE site (boldface) with a five-nucleotide hairpin loop. The binding reaction mixtures contained 1 nM fluoresceinated oligonucleotide in 25 mM Tris-HCl (pH 7.6)–50 mM NaCl–5 mM MgCl2–0.1 mM EDTA–1 mM DTT–5% glycerol–10 μg of poly(dI-dC)/ml–60 nM CREB. The reaction mixtures were titrated with successive additions of CBP (aa 451 to 682) from 0 to 2 μM. The stock concentration of CBP used for these titrations was high enough that the total volume did not have to be increased by more than 10%. Binding reactions were performed at 25°C, and samples were incubated for 2 min to achieve equilibrium of the complex formation. The binding curves were fit with a nonlinear least squares regression analysis. In addition to the rectangular hyperbolic binding function, the equation utilized for curve fit determination included a nonspecific component to account for the linear increase in polarization at higher protein concentrations.
Cell culture and transfection assays.
F9 teratocarcinoma cells were cultured on gelatin-coated plates in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, penicillin G (100 IU/ml), and streptomycin sulfate (100 mg/ml). PC12 cells were cultured on poly-l-lysine-coated plates in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated horse serum, 5% heat-inactivated fetal calf serum, penicillin G (100 IU/ml), and streptomycin sulfate (100 mg/ml). F9 cells were seeded at 4 × 105 per 100-mm-diameter plate, and PC12 cells were seeded onto eight 60-mm-diameter plates per confluent 75-mm2 flask 24 h prior to transfection. Supercoiled plasmid DNA was prepared by CsCl gradient centrifugation, and transfections were carried out using the Gibco BRL calcium phosphate transfection system (F9 cells) or the Qiagen SuperFect reagent (PC12 cells). Cells were washed and fed after 24 h (F9 cells) or 3 h (PC12 cells) and allowed to grow for an additional 24 h (F9 cells) or 45 h (PC12 cells) before harvesting. CRE-CAT and GAL4-LUC reporter activities were determined by CAT assay (20) or luciferase assay, respectively, and normalized to either total cellular protein (Bio-Rad protein assay) or β-galactosidase activity. All experiments were performed at least in triplicate.
RESULTS
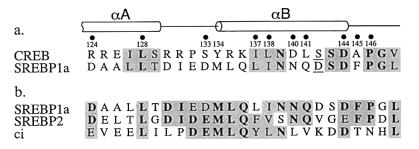
Nuclear magnetic resonance analysis of the phosphorylated CREB-CBP complex showed that after binding to CBP, CREB adopts a bihelical conformation with the helical axes approximately perpendicular to one another (28). The portion of SREBP1a that interacts with CBP resides within its amino-terminal 50 aa (25). Only a region of 25 residues within this portion is conserved in the functionally related protein SREBP2 (15), suggesting that this smaller domain is sufficient for CBP binding. Comparison of the primary sequences of CREB and the 25-aa domain of SREBP1a revealed some unexpected similarities, despite the fact that the two factors bind to CBP through phosphorylation-dependent and -independent mechanisms. Figure 1a aligns the phosphorylated Ser133 in CREB with an Asp in SREBP. Substitution of an Asp for Ser133 in CREB is not sufficient for CBP binding (8). A second Asp residue, which corresponds to Ser142 in CREB, may represent a conservative change, however. Mutation of CREB Ser142 to Asp has been shown to have no effect on binding of phosphorylated CREB to CBP (33). More importantly, the secondary structures of the CREB and SREBP regions are also likely to be conserved, as both the amino- and carboxy-terminal portions of the SREBP domain contain residues typically found in α helices. We hypothesized, therefore, that CREB and SREBP might contact CBP through related mechanisms.
FIG. 1.
(a) Comparison of CBP interaction domains of CREB and SREBP1a. Dots and numerals refer to residues in CREB known to be critical for CBP binding; shading denotes homologous residues, with identical positions in boldface. Underlined S and D residues represent possible additional conservative substitutions. α-Helical regions of CREB in the complex with CBP extend from aa 120 to 129 and aa 134 to 144 (28). (b) Comparison of CBP-interacting domains of SREBP1a, SREBP2, and Drosophila ci. Identical and conserved residues are indicated as above.
To test this hypothesis, we mutated residues in SREBP that corresponded to critical residues in CREB and measured the effects of these substitutions on CBP binding. These assays were performed by incubating HeLa cell nuclear extracts with GST or GST-SREBP1-50 fusion proteins immobilized on glutathione beads. The bound proteins were electrophoresed on SDS-polyacrylamide gels and immunoblotted using an antibody that recognizes the CREB binding domains of CBP and its homologue p300. Bacterially expressed fragments of CBP were also tested. These studies confirmed that the first 50 residues of SREBP1a bind to full-length CBP through a region previously identified as the CREB-binding domain (data not shown).
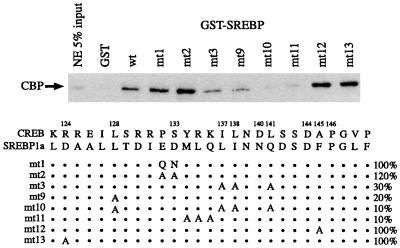
Hydrophobic residues Leu128, Ile137, Leu138, and Leu141 in CREB have been shown to be essential for CBP binding (26, 28, 30). These residues are represented by Leu, Leu, Ile, and Gln in SREBP1a (Fig. 1a). Molecular modeling suggested that each of the substitutions should be tolerated (unpublished observations), and alanine mutagenesis indicated that these residues all contribute to SREBP-CBP binding (Fig. 2). The combination of all four mutations (mt10) blocked binding by 90%, and mutation of the residue corresponding to CREB Leu128 was almost as effective (mt9). On the other hand, some interactions known to be important for phosphorylated CREB-CBP binding were not required for the SREBP interaction. Perhaps most unexpected was the relative unimportance of the Glu-Asp residues (mt1 and mt2), which were predicted to serve the same function in SREBP as the phosphorylated Ser in CREB. To our surprise, the negatively charged central portion of the SREBP interaction domain was not essential. In fact, change of these residues to Gln-Asn (mt1) or Ala-Ala (mt2) modestly increased the level of binding.
FIG. 2.
Binding of full-length CBP to GST-SREBP mutants. The schematic depicts mutants analyzed at the top. Numbers refer to amino acid positions in CREB; dots refer to retained SREBP residues. (For example, in mt1, the ED in SREBP is replaced by QN.) Percent binding (compared to wild-type SREBP) is indicated on the right. NE, nuclear extract.
Alignment of the CBP interaction domains of SREBP1a, SREBP2, and the Drosophila cubitus interruptus (ci) provided further insight into the determinants of coactivator binding (Fig. 1b). Drosophila ci is a particularly informative model because, in addition to the standard biochemical analyses, we have shown in genetic studies that the mutant phenotype caused by ectopic ci expression is suppressed by mutations in Drosophila CBP (1). Moreover, point mutations in ci that block association with Drosophila CBP also block the ability of the transcription factor to activate its endogenous target gene wingless (6a). As shown in Fig. 2, mutation of the Met-Leu-Gln residues in SREBP to Ala (mt11) completely blocks CBP binding. These residues are shared by SREBP1a, SREBP2, and ci.
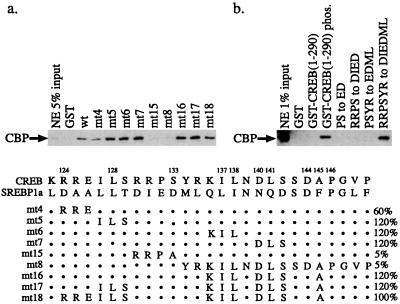
Having determined which residues in SREBP were required for CBP binding, we next tested whether amino acid sequences from CREB could be introduced into SREBP without disrupting the phosphorylation-independent CBP interaction (Fig. 3a). For these studies, we introduced blocks of amino acids from CREB into GST-SREBP1-50 and monitored binding of full-length CBP in HeLa nuclear extracts. Substitution of the Arg-Arg-Glu from CREB for the Asp-Ala-Ala of SREBP (mt4) reduced binding by about 40%. This result was somewhat unexpected because Arg124 of CREB contributes to CBP binding through an electrostatic interaction with Glu655 of CBP (28) and suggests that the binding mechanisms of phosphorylated CREB and SREBP to CBP are not identical. Other substitutions of CREB sequences into GST-SREBP had minimal effect, with the exception of mutations involving the midportion of the SREBP domain, Asp-Ile-Glu-Asp-Met-Leu (DIEDML; mt15 and mt8), which completely blocked binding. These studies indicated that only the central core of the SREBP sequence was required for phosphorylation-independent CBP binding. Therefore, a CREB protein containing this six-residue substitution might be expected to interact with CBP in a phosphorylation-independent manner.
FIG. 3.
Binding of full-length CBP to mutants of GST-SREBP1 (a) and GST-CREB (b). GST-CREB(1-290) phos. refers to GST fusion protein containing the CREB activation domain phosphorylated by PKA. Other mutations in panel b refer to substitutions of CREB residues with residues from SREBP. Only the DIEDML substitution conferred phosphorylation-independent binding. The schematic depicts substitution of CREB sequences into SREBP. Dots refer to SREBP residues left unchanged. Percent binding is indicated on the right. NE, nuclear extract.
To test this hypothesis, we constructed a GST-CREB fusion protein lacking the carboxy-terminal basic leucine zipper region. GST-CREB1-290 interacted with full-length CBP in a phosphorylation-dependent manner, as shown in Fig. 3b. These experiments were performed in the presence of phosphatase inhibitors to maintain phosphorylation of the Ser133 site. Western blots indicated that the PKA-treated GST-CREB1-290 remained phosphorylated despite the incubation with HeLa nuclear extracts (data not shown). CBP binding to phosphorylated GST-CREB1-290 was significantly weaker than to GST-SREBP, however. We next tested whether the sequential replacement of the central portion of the CREB activation domain containing the PKA recognition site (Arg-Arg-Pro-Ser-Tyr-Arg) with residues from SREBP would allow constitutive CBP binding. Substitution of the Pro-Ser sequence of CREB with Glu-Asp, the Arg-Arg-Pro-Ser with Asp-Ile-Glu-Asp, or the Pro-Ser-Tyr-Arg with Glu-Asp-Met-Leu did not allow phosphorylation-independent binding. However, when the entire Arg-Arg-Pro-Ser-Tyr-Arg of CREB was replaced with the DIEDML sequence of SREBP, binding was equivalent to that of phosphorylated GST-CREB1-290 (Fig. 3b). This mutant, designated CREBDIEDML, cannot be phosphorylated by PKA (data not shown).
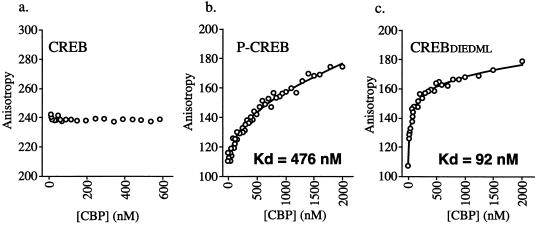
We used a fluorescence polarization binding assay to quantify the relative affinities of CBP for phosphorylated CREB and CREBDIEDML. Unlike the GST pull-down assays, this procedure can provide true equilibrium measurements of protein-DNA and protein-protein interactions. These experiments utilized a 5′-fluoresceinated oligonucleotide derived from the somatostatin CRE promoter sequence that was saturated with either CREBDIEDML, phosphorylated CREB, or unphosphorylated CREB. The affinities of the three CREB preparations for the CRE were identical (2 nM [data not shown]). CBP binding was assessed by measuring the association of a CBP fragment (aa 451 to 682) representing the CREB-binding domain. Complete binding to this preformed CREB-CRE complex was ascertained by monitoring the increases in fluorescence polarization values upon addition of CBP (Fig. 4). The average equilibrium dissociation constants for CBP binding were calculated to be 444 ± 27 nM in the presence of phosphorylated CREB and 96 ± 4 nM in the presence of CREBDIEDML. We detected no binding of CBP to nonphosphorylated CREB. These data demonstrate a significant increase in the affinity of CBP for CREBDIEDML compared to phosphorylated CREB and support the results of the GST-CREB1-290 binding studies presented above.
FIG. 4.
Fluorescence polarization binding curves of CBP (aa 451 to 682) for nonphosphorylated CREB-CRE (a), phosphorylated CREB-CRE (b), and CREBDIEDML-CRE (c) complexes. The change in anisotropy is plotted against the CBP protein concentration. The symbols represent the individual data points collected from successive additions of CBP to saturated CREB-CRE complexes. Each assay condition was performed in triplicate, and a representative binding curve is shown for each. No binding of CBP was detected to the complex containing nonphosphorylated CREB.
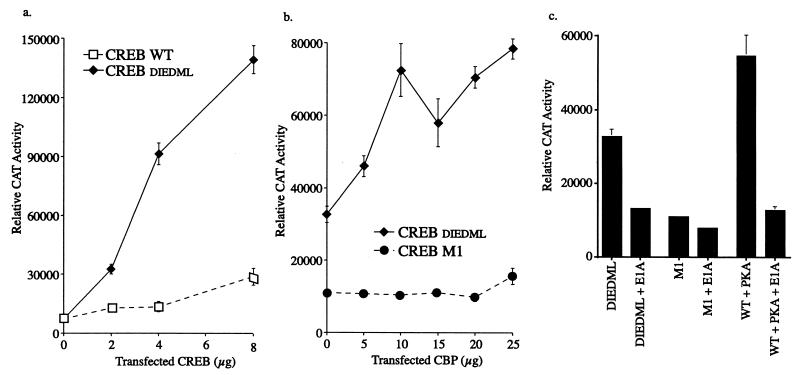
To test the functional consequences of the CREB mutations, plasmids encoding wild-type CREB and CREBDIEDML were introduced into F9 teratocarcinoma cells along with a CRE reporter. These cells contain low levels of CBP but normally require the addition of CREB and PKA to activate a CRE reporter (8). As shown in Fig. 5a, CREBDIEDML was approximately eightfold more active than the wild-type protein. To confirm that the activation mediated by CREBDIEDML is dependent on CBP, we cotransfected a CBP expression vector. We have shown previously that expression of exogenous CBP increases the transcriptional response to PKA-phosphorylated CREB but not to a mutant (CREB-M1) (8) that cannot be phosphorylated by PKA (18). Similarly, exogenous CBP augmented the response to CREBDIEDML but not to CREB-M1 (Fig. 5b) or wild-type CREB in the absence of PKA (data not shown). Activity mediated by CREBDIEDML alone is presumably due to the low level of endogenous CBP present in F9 teratocarcinoma cells. To confirm the involvement of endogenous CBP in CREBDIEDML function, we introduced an expression vector encoding 12S E1A along with wild-type or mutant CREB (Fig. 5c). Adenovirus 12S E1A blocks the activity of transcription factor pathways by interfering with CBP function (2, 21). 12S E1A blocked the activities of CREBDIEDML and wild-type CREB plus PKA but had no effect on CREB-M1. These data also support the hypothesis that CREBDIEDML activates transcription through CBP.
FIG. 5.
(a) Activation of CRE reporter in F9 teratocarcinoma cells as directed by wild-type (WT) CREB or CREBDIEDML. Values (mean ± standard error) are normalized for protein levels. (b) Effect of CBP on CREB-M1 and CREBDIEDML function. Cells were transfected with 6 μg of CREB plasmid and various amounts of CBP. (c) Effect of adenovirus 12S E1A on CREB function. DIEDML, M1, and WT refer to CREBDIEDML, CREB-M1, and wild-type CREB, respectively. Cells were transfected with 6 μg of CREB, 1 μg of E1A, and 4 μg of PKA, as indicated. The apparent increased activity of wild-type CREB in the presence of PKA, compared to CREBDIEDML, is due to PKA stimulation of the RSV promoter.
To test whether CREBDIEDML is active in other cell types, we introduced plasmids containing GAL4-CREB or GAL4-CREBDIEDML fusion genes, along with a GAL4-LUC reporter, into PC12 cells. GAL4-CREBDIEDML was almost five times more active than the GAL4–wild-type CREB fusion protein (Fig. 6). These data indicate that the transcriptional activity of CREBDIEDML is not limited to F9 cells. Rather, the DIEDML mutation may render CREB constitutively active in all cells that express CBP.
FIG. 6.
Activation of GAL4-LUC reporter by GAL4–wild-type (WT) CREB and GAL4-CREBDIEDML in PC12 cells. Cells were transfected with the indicated amounts of GAL4-CREB plasmids along with 4 μg each of GAL4-LUC reporter and pRSV-β-GAL. Relative luciferase (Luc) values were normalized to β-galactosidase activity.
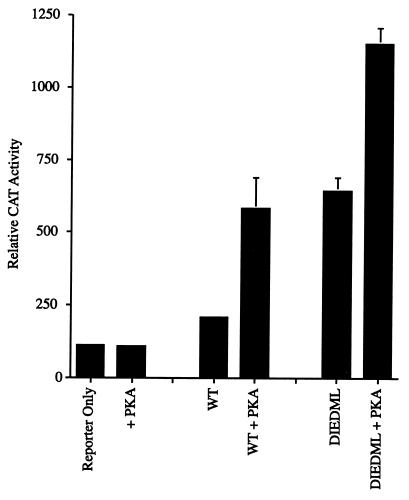
Addition of PKA to F9 teratocarcinoma cells expressing wild-type CREB increased reporter activity about fivefold, as previously reported (8) (Fig. 7). Surprisingly, PKA also increased activity of CREBDIEDML, despite the fact that CREBDIEDML is not a PKA target (Fig. 7 and data not shown). The activity of wild-type CREB plus PKA was about the same as that of CREBDIEDML minus PKA when the assays were normalized for β-galactosidase activity, although the mutant CREB binds to CBP somewhat better. On the other hand, activity of wild-type CREB was less than that seen with the mutant in the presence of PKA. These experiments indicate that the recruitment of CBP is sufficient for a significant portion of CREB activity, but full activation of the reporter requires the addition of PKA. Presumably, PKA also activates a component of the CREB-CBP cascade at a point downstream from CREB phosphorylation and CBP recruitment.
FIG. 7.
Activation of CRE reporter by CREB and CREBDIEDML in F9 teratocarcinoma cells in the presence and absence of PKA. WT designates wild-type CREB. Cells were transfected with 8 μg of CREB, 4 μg of PKA, and 6 μg of pRSV-β-GAL, as indicated. CAT activity was normalized for β-galactosidase activity.
DISCUSSION
Whether recruitment of CBP to CREB is sufficient for gene activation has been controversial. For example, recent studies have suggested that CBP cannot activate transcription in the absence of a calcium-dependent phosphorylation signal (13, 14). Characterization of these phosphorylation mechanisms in the context of the CREB-CBP pathway has been difficult, because CREB activation is also phosphorylation dependent. Thus, it has not been possible to separate kinases involved in CREB activation from those involved in CBP function. To address this issue, investigators have made use of GAL-CBP fusion genes, which target the coactivator to synthetic promoter sequences through a DNA-binding domain attached to the CBP amino terminus. Unfortunately, many of these GAL-CBP fusion proteins confer artifactually high or low levels of transcription.
We have used a different approach to address this question. By comparing the primary sequences of SREBP and other factors, we identified residues that have been conserved with the CBP interaction domain of CREB. Moreover, secondary structure predictions suggest that the CBP-binding region of SREBP is α helical, like that in the phosphorylated CREB-CBP complex. We showed by mutagenesis studies that the α-helical regions of the CBP interaction domains of phosphorylated CREB and SREBP are conserved functionally. On the other hand, the central, negatively charged portion of SREBP, which we initially hypothesized might correspond to the phosphoserine of CREB, was relatively unimportant. Replacing these residues with uncharged or smaller amino acids had no effect on binding, and replacing the PKA site in CREB with the negatively charged residues from SREBP did not promote phosphorylation-independent binding. Similarly, substitution of a single negatively charged residue for Ser133 fails to confer phosphorylation-independent CREB activation (12). Surprisingly, introduction of the 6-aa DIEDML sequence of SREBP was sufficient to allow phosphorylation-independent binding of CREB to CBP. Moreover, this sequence is conserved in SREBP2 and Drosophila ci, two other factors that interact with the CREB-binding domain of CBP.
While this paper was being prepared, a report by Parker et al. (27) described the properties of a CREB-Myb chimera which contained a 21-aa segment of c-Myb inserted into the activation domain of CREB. These workers found that the CREB-Myb chimera did not activate transcription significantly in the absence of PKA. Notably, the binding of c-Myb to CBP was determined to be 26-fold weaker than that of phosphorylated CREB. Thus, the chimeric CREB-Myb protein likely also interacts with CBP fairly weakly. It is also possible that the large c-Myb insert disrupts critical aspects of the native CREB structure. There are no obvious sequence similarities between the CBP interaction domains of c-Myb and SREBP. Our study indicates that the DIEDML mutation increases the affinity of CREB for CBP, compared to the phosphorylated wild-type protein. We propose that this difference in CBP binding contributes to the functional differences between the CREB-Myb and CREBDIEDML mutants. The DIEDML sequence, by itself, is not sufficient for the CBP interaction, however, as shown by the failure of the GST-SREBP mutants mt3, mt9, and mt10 (Fig. 2) to mediate wild-type binding.
The transcriptional activity of CREBDIEDML is consistent with the idea that CBP recruitment is sufficient for activation of CRE-containing promoters. Our data do not support the recent study by Hu et al. (14), however, who found that CBP was inactive in the absence of a supplemental Ca2+ signal. The differences between our results and those of Hu et al. could relate to their use of GAL-CBP fusion genes or the particular cell types that were examined. Hu et al. used primary E18 cortical neurons, while we utilized F9 teratocarcinoma cells. Endogenous PKA, CaMKII, and CaMKIV levels in F9 cells are insufficient for CREB-mediated gene activation (22), and so it is unlikely that the activation that we detect is due to the unrecognized stimulation of kinase pathways.
While CBP recruitment is sufficient to activate CREB-mediated transcription, PKA augments the response somewhat further. The lower activity of wild-type CREB than CREBDIEDML in the presence of PKA might relate to dynamic aspects of their interactions with CBP. Wild-type CREB probably binds to CBP only transiently, due to dephosphorylation, while the interaction of CBP with CREBDIEDML may be more sustained. Additionally, the moderately higher affinity of CREBDIEDML for CBP might contribute to its enhanced level of transcriptional activity. The modest stimulation of CREBDIEDML by PKA is also consistent with previous reports that CREB activation through Ca2+-stimulated kinase pathways can be blocked by PKA inhibitors. Whether PKA contributes to gene activation by phosphorylating CBP, as proposed by Xu et al. (37), or by modifying targets downstream from CBP is unknown.
ACKNOWLEDGMENTS
This work was supported by grants from the Swiss National Science Foundation Fellowship, the McKnight Foundation, and the National Institutes of Health.
We thank James Lundblad and Gail Mandel for thoughtful comments.
REFERENCES
- 1.Akimaru H, Chen Y, Dai P, Hou D X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signaling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 2.Arany Z, Newson D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 3.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 12.1.1–12.1.9. [Google Scholar]
- 5.Chawla S, Hardingham G E, Quinn D R, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 6a.Chen Y, Goodman R H, Smolik S M. Cabitus interruptus requires Drosophila CREB-binding protein to activate wingless expression in the Drosophila embryo. Mol Cell Biol. 2000;20:1616–1625. doi: 10.1128/mcb.20.5.1616-1625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chrivia J D, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 8.Dwarki V J, Montminy M, Verma I M. Both the basic region and the ‘leucine zipper’ domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 1990;9:225–232. doi: 10.1002/j.1460-2075.1990.tb08099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felzien L K, Farrell S, Betts J C, Mosavin R, Nabel G J. Specificity of cyclin E-Cdk2, TFIIB, and E1A interactions with a common domain of the p300 coactivator. Mol Cell Biol. 1999;19:4241–4246. doi: 10.1128/mcb.19.6.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginty D D, Glowacka D, Bader D S, Hidaka H, Wagner J A. Induction of immediate early genes by Ca2+ influx requires cAMP-dependent protein kinase in PC12 cells. J Biol Chem. 1991;266:17454–17458. [PubMed] [Google Scholar]
- 11.Goldman P S, Tran V K, Goodman R H. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Horm Res. 1996;52:103–119. [PubMed] [Google Scholar]
- 12.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 13.Hardingham G E, Chawla S, Cruzalegui F H, Bading H. Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron. 1999;22:789–798. doi: 10.1016/s0896-6273(00)80737-0. [DOI] [PubMed] [Google Scholar]
- 14.Hu S C, Chrivia J, Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- 15.Hua X, Yokoyama C, Wu J, Briggs M R, Brown M S, Goldstein J L, Wang X. SREBP-2, a second basic helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Impey S, Wayman G, Wu Z, Storm D R. Type I adenylyl cyclase functions as a coincidence detector for control of cyclic AMP response element-mediated transcription: synergistic regulation of transcription by Ca2+ and isoproterenol. Mol Cell Biol. 1994;14:8272–8281. doi: 10.1128/mcb.14.12.8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraus W L, Kadonaga J T. p300 and estrogen receptor cooperatively activate transcriptional via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bächinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–236. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 19.Laurance M E, Kwok R P S, Huang M S, Richards J P, Lundblad J R, Goodman R H. Differential activation of viral and cellular promoters by human T-cell lymphotropic virus-1 Tax and cAMP-responsive element modulator isoforms. J Biol Chem. 1997;272:2646–2651. doi: 10.1074/jbc.272.5.2646. [DOI] [PubMed] [Google Scholar]
- 20.Loriaux M M, Rehfuss R P, Brennan R G, Goodman R H. Engineered leucine zippers show that hemiphosphorylated CREB complexes are transcriptionally active. Proc Natl Acad Sci USA. 1993;90:9046–9050. doi: 10.1073/pnas.90.19.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 22.Masson N, Ellis M, Goodbourn S, Lee K A W. Cyclic AMP response element-binding protein and the catalytic subunit of protein kinase A are present in F9 embryonal carcinoma cells but are unable to activate the somatostatin promoter. Mol Cell Biol. 1992;12:1096–1106. doi: 10.1128/mcb.12.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montminy M R, Sevarino K A, Wagner J A, Mandel G, Goodman R H. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 25.Oliner J D, Andresen J M, Hansen S K, Zhou S, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB binding protein. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 26.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker D, River M, Zor T, Henrion-Caude A, Radhakrishnan I, Kumar A, Shapiro L H, Wright P E, Montminy M, Brindle P K. Role of secondary structure in discrimination between constitutive and inducible activators. Mol Cell Biol. 1999;19:5601–5607. doi: 10.1128/mcb.19.8.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 29.Richards J P, Bachinger H P, Goodman R H, Brennan R G. Analysis of the structural properties of cAMP-responsive element binding protein (CREB) and phosphorylated CREB. J Biol Chem. 1996;271:13716–13723. doi: 10.1074/jbc.271.23.13716. [DOI] [PubMed] [Google Scholar]
- 30.Shih H M, Goldman P S, DeMaggio A J, Hollenberg S M, Goodman R H, Hoekstra M F. A positive genetic selection for disrupting protein-protein interactions: identification of CREB mutations that prevent association with the coactivator CBP. Proc Natl Acad Sci USA. 1996;93:13896–13901. doi: 10.1073/pnas.93.24.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 32.Sun P, Enslen H, Myung P, Maurer R A. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- 33.Sun P, Maurer R A. An inactivating point mutation demonstrates that interaction of cAMP response element binding protein (CREB) with the CREB binding protein is not sufficient for transcriptional activation. J Biol Chem. 1995;270:7041–7044. doi: 10.1074/jbc.270.13.7041. [DOI] [PubMed] [Google Scholar]
- 34.Swope D L, Mueller C L, Chrivia J C. CREB binding protein activates transcription through multiple domains. J Biol Chem. 1996;21:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 35.Thompson M A, Ginty D D, Bonni A, Greenberg M E. L-type voltage-sensitive Ca2+ channel activation regulates c-fos transcription at multiple levels. J Biol Chem. 1995;270:4224–4235. doi: 10.1074/jbc.270.9.4224. [DOI] [PubMed] [Google Scholar]
- 36.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 37.Xu L, Lavinsky R M, Dasen J S, Flynn S E, McInerney E M, Mullen T M, Heinzel T, Szeto D, Korzus E, Kurokawa R, Aggarwal A K, Rose D W, Glass C K, Rosenfeld M G. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 38.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 39.Zanger K, Cohen L E, Hashimoto K, Radovick S, Wondisford F E. A novel mechanism for cyclic adenosine 3′,5′-monophosphate regulation of gene expression by CREB binding protein. Mol Endocrinol. 1999;13:268–275. doi: 10.1210/mend.13.2.0245. [DOI] [PubMed] [Google Scholar]