Abstract
Paramecium internal eliminated sequences (IESs) are short AT-rich DNA elements that are precisely eliminated from the germ line genome during development of the somatic macronucleus. They are flanked by one 5′-TA-3′ dinucleotide on each side, a single copy of which remains at the donor site after excision. The timing of their excision was examined in synchronized conjugating cells by quantitative PCR. Significant amplification of the germ line genome was observed prior to IES excision, which starts 12 to 14 h after initiation of conjugation and extends over a 2- to 4-h period. Following excision, two IESs were shown to form extrachromosomal circles that can be readily detected on Southern blots of genomic DNA from cells undergoing macronuclear development. On these circular molecules, covalently joined IES ends are separated by one copy of the flanking 5′-TA-3′ repeat. The similar structures of the junctions formed on the excised and donor molecules point to a central role for this dinucleotide in IES excision.
DNA rearrangements have been found in a wide range of living organisms (3) but have reached a remarkable extent in ciliates: deletion of internal sequences, DNA scrambling, and chromosomal fragmentation are developmentally programmed during the sexual phase of their life cycle (33). These unicellular eukaryotes are characterized by the presence of two functionally different nuclei within the same cytoplasm. The polyploid macronucleus, also called the somatic nucleus, is transcribed during vegetative growth and governs the cell phenotype but progressively degenerates during sexual reproduction. The diploid micronucleus is transcriptionally silent during vegetative growth and can be viewed as the germ line nucleus, since it is able to undergo meiosis. In Paramecium aurelia, two successive divisions of the zygotic nucleus take place after fertilization and produce four identical diploid nuclei, two of which become the new micronuclei while the other two differentiate into macronuclei. Macronuclear development extends over two cell cycles (4) and is accompanied by intensive replication to reach the final ploidy of the mature macronucleus, estimated to be between 800 and 1,700N according to previous studies (33, 38). It involves extensive rearrangements of the germ line genome: fragmentation of micronuclear chromosomes coupled to telomere addition to form the macronuclear chromosome ends (13) and precise deletion of internal eliminated sequences (IESs) that interrupt coding and non-coding DNA (reviewed in reference 22).
Paramecium IESs are single-copy sequences of AT-rich noncoding DNA and are flanked by two 5′-TA-3′ repeats, one of which is retained in the macronuclear genome after excision (1, 7, 9, 12, 21, 24, 28, 32, 37, 40, 44). Since they were initially identified by sequence comparison of macronuclear and micronuclear versions of a given locus, their size—ranging between 26 and 882 bp—represents, by convention, the whole length of the deleted sequence and formally includes one copy of the flanking 5′-TA-3′. Based on an extrapolation of their frequency within sequenced regions of the genome, their number was estimated to be around 50,000 per haploid germ line genome (12). Paramecium IESs belong to the family of ciliate TA IESs also found in the hypotrichs Euplotes and Oxytricha (18). A statistical analysis led to the identification of a loosely conserved 8-bp consensus (5′-TAYAGYNR-3′), present as inverted repeats at the ends of Paramecium IESs and including the flanking 5′-TA-3′ (21). This consensus bears similarities to the ends of Tc1/mariner transposons and of transposon-like Tec elements and some TA IESs of Euplotes (18). It was therefore proposed that IESs have evolved from ancestral transposons but have lost their coding capacity and rely on cellular functions for their excision (22). Genetic studies demonstrated that the terminal 8 bp of IES ends, including the flanking 5′-TA-3′, plays a functional role in excision (9, 25, 26). Furthermore, Paramecium IES excision is subjected to a homology-dependent epigenetic control since the presence of an IES in the parental macronucleus was shown in some cases to inhibit excision of the corresponding sequence during development of the macronucleus of the next sexual generation (11, 12).
Several lines of evidence have indicated that excision of TA IESs and transposon-like elements in Oxytricha and Euplotes is accompanied by the formation of extrachromosomal circular molecules, detectable in significant amounts in cells undergoing macronuclear development (19, 23, 46). Under the assumption that such molecules are direct excision products, double-stranded DNA cutting was proposed to initiate the reaction at one (46) or both (19, 23) ends of the excised sequence. In Tetrahymena, a ciliate more closely related to Paramecium than hypotrichs, IESs have also been found, but they differ significantly from TA IESs (22). Extrachromosomal forms of these elements were not detected on Southern blots of DNA from developing macronuclei (2), although putative circularized forms could be amplified by extensive PCR (36, 47). It was therefore proposed that IES excision in Tetrahymena essentially produces unstable excised molecules and that circles are secondary, dead-end products of the reaction. Transient DNA cuts were detected at only one end of these sequences during macronuclear development, indicating that excision might proceed via a transposition-like pathway, including successive DNA transesterification steps, and result primarily in the formation of linear excised molecules (35).
As a first step in investigating the DNA transactions and the putative enzymatic machinery participating in Paramecium IES excision, we used a quantitative PCR approach to study the timing of IES elimination. Our data indicate that it starts after three rounds of DNA replication have taken place in the developing macronuclei (anlagen) and is completed by the time of the first cell division (the karyonidal division), when the two anlagen segregate into daughter cells. These experiments allowed the concomitant detection of abundant, circularized forms of two IESs. The structure of the circle junctions suggests that Paramecium IES excision may involve DNA cutting at the flanking 5′-TA-3′ dinucleotide.
MATERIALS AND METHODS
Paramecium strains and growth conditions.
P. tetraurelia stocks 51 and d4.2 are entirely homozygous strains carrying the A51 and A29 alleles, respectively, of the gene encoding surface antigen A (39). Both carry the G51 allele of the paralogous gene encoding surface antigen G. Culture conditions were as described previously (11). Unless otherwise stated, cells were grown at 27°C. Under starvation conditions, macronuclear development was induced by conjugation of reactive cells of complementary mating types or through a self-fertilization process called autogamy. Autogamy was monitored by staining cells with a 20:1 (vol/vol) mix of carmine red (0.5% in 45% acetic acid) and fast green (1% in ethanol) or by 4′,6-diamidino-2-phenylindole (DAPI) staining.
The availability of a micronuclear DNA library from stock 51 (40) prompted us to focus our study on the IESs identified in this strain. However, the original stock 51 available in our laboratory proved to be inadequate for the standardization of conjugation experiments, mainly because of its relatively slow growth (three to four doublings every 24 h for stock 51 versus four to five doublings for d4.2). We therefore crossed stocks 51 and d4.2 and selected a fast-growing mating-type E F2 clone homozygous for the A51 allele. This clone was designated speedy, and a mating-type O revertant was isolated for subsequent conjugation experiments.
Synchronization of macronuclear development.
Reactive cells were mixed and incubated at room temperature for 1 h 45 min to allow the first conjugating pairs to form. Rich medium (2 volumes) was added, and the cells were incubated at 27°C for 45 min to stop further conjugation: only 30% of the cells were involved in the formation of stable synchronous pairs. These were hand sorted and transferred into rich medium. A fixed number of 60 cells were picked at the indicated time points from the time of exconjugant separation until the completion of karyonidal division and treated with 1% Nonidet P-40 (NP-40) for 10 min at 65°C before being subjected to PCR analysis (26).
Quantitative PCR analysis of total genomic DNA.
Aliquots (10 μl) of NP-40-lysed cells (representing 10 cells) were subjected to PCR amplification in a Perkin-Elmer Cetus DNA thermal cycler, using 1 U of Tfl DNA polymerase (Promega) and 10 to 100 pmol of each primer in a final volume of 25 μl of commercial buffer (pH 9) (Promega) supplemented with 1.25 mM MgSO4. For IES 51A2591, 100 pmol of primer 51A3 and 10 pmol of primer 51A-3′ (Table 1) were used in the following amplification program: 6 cycles of 60 s at 92°C, 60 s at 54°C, and 60 s at 74°C, and 20 cycles of 20 s at 92°C, 20 s at 54°C, and 60 s at 74°C. For IES 51G4404, 10 pmol of each primer (51G3 and 51G6) (Table 1) was used and the program was 6 cycles of 60 s at 92°C, 60 s at 63°C, and 60 s at 74°C, followed by 30 cycles of 20 s at 92°C, and 75 s at 63°C; a final termination cycle (3 min at 72°C) was included for both IESs. To ensure that amplification yields were proportional to the input DNA concentration, serial dilutions of total genomic DNA extracted from a 400-ml vegetative culture of P. tetraurelia stock speedy (mating-type E) were amplified with the desired set of PCR primers. The resulting products were analyzed by Southern blot hybridization with a 32P-labeled oligonucleotide and quantified using a Fuji phosphorimager. The number of amplification cycles for each primer set was chosen to give a good signal/input proportionality within a 1- to 15-fold range of input DNA concentrations, starting from the equivalent of the mean micronuclear signal obtained from two independent NP-40-treated samples of 10 vegetative cells (because of the lack of synchrony of vegetative cultures, this signal may vary within a 2-fold range between two different control samples). These conditions gave a perfectly proportional signal for IES 51A2591, while saturation was observed for IES 51G4404 at input DNA concentrations above 15 equivalents of the vegetative micronuclear signal. This could lead to an underestimation of the highest amplification factors obtained for IES 51G4404.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence (5′ to 3′) | Positions |
|---|---|---|
| 51G3 | CAACATGTGCTGCATATAATGTAGG | 7325–7349a |
| 51G5 | GATGGTATTATAAATCGAATATTAATTCTG | 7366–7395a |
| 51G6 | CAGCCGGTATTATTAAGTAACTTTAAAAAG | 7601–7572a |
| 51G7 | TTTTGAAATATTTTCAAGTTTTTGGACTAC | 7509–7480a |
| 51G8 | ACAATATATATTTACTTGATAATATTTTCC | 7510–7539a |
| 51A1 | AGATTTATATCTTTTTTCTCAAATTCAGC | 4052–4080b |
| 51A3 | ATTCTAAAATTAACAAAAATACATTATTTTAC | 3800–3831b |
| 51A4 | TTGAGAAGTTAAGAAATAAAATGATGG | 4168–4142b |
| 51A9 | CAATATTATACATCTAGAACTTATAGTTAG | 4051–4022b |
| 51A-3′ | TTTTATGGCATTAAGCTTGTGTCAT | 4222–4198b |
| 51AΔ28 | GTTATAGTGATTATTAAAATACTTGATTCAT | 3917–3947b |
PCR detection of IES circular junctions.
Primer sets 51G7 plus 51G8 and 51A1 plus 51A9 (Table 1) were used for the detection of the junctions between the ends of IES 51G4404 and 51A2591, respectively (Table 1). For 10 μl of NP-40 cell lysates, the PCR conditions were 6 cycles of 60 s at 92°C, 60 s at 54°C, and 60 s at 72°C followed by 36 cycles of 20 s at 92°C, 20 s at 54°C, and 60 s at 72°C, in a final volume of 25 μl. When 1 μl of total genomic DNA was used as a template, the initial 6 cycles were followed by 32 cycles of 20 s at 92°C, 20 s at 54°C, and 60 s at 72°C. For all PCR amplifications, 10 pmol of each primer was used and a final termination step of 3 min at 72°C was included.
DNA manipulations.
λ51Amic and λ51Gmic phages harbor the micronuclear A51 and G51 genes, respectively (12). Plasmid pRA593 is a pBR322 derivative carrying a minitransposon derived from bacterial insertion sequence IS911. In the presence of IS911 transposase, the minitransposon is excised in vivo as a 411-bp covalently closed supercoiled minicircle (31). A 1-kb DNA ladder (Gibco BRL) was used as a linear size standard. All oligonucleotides (Eurobio or Eurogentec) are listed in Table 1.
For large-scale preparations, Paramecium genomic DNA was extracted from 100- to 400-ml cultures grown to a cell density of 500 to 1,000 cells/ml (11). Restriction or DNA modification enzymes (New England Biolabs or Boehringer Mannheim) were used as specified by the suppliers.
DNA fragments used as hybridization probes were labeled by random priming as described previously (12) with [α-32P]dATP (3,000 Ci/mmol; Amersham). Oligonucleotide probes were labeled with [γ-32P]ATP (5,000 Ci/mmol; Amersham) and T4 polynucleotide kinase (New England Biolabs).
PCR-mediated sequencing of DNA fragments purified by the QIAquick PCR purification kit procedure (Qiagen) was performed with oligonucleotide primers labeled with [γ-33P]ATP (2,500 Ci/mmol; Amersham), using the fmol sequencing kit (Promega).
Southern blotting.
For the identification of extrachromosomal IESs, equivalents of 1 μl of a total genomic DNA preparation at an optical density at 260 nm of 1,200 were treated with 20 μg of Rnase A per ml and loaded on a 1.5% agarose (Appligene)–1.5% NuSieve (FMC) gel in 1× Tris-borate-EDTA (TBE) buffer. PCR amplification products were analyzed on 3% NuSieve (FMC) or 1.5% agarose gels (Appligene) in 1× TBE buffer. Alkaline transfer onto Hybond N+ membranes (Amersham), hybridization with 32P-labeled DNA fragments, and washing conditions were as described previously (12), while hybridization with 32P-labeled oligonucleotides was carried out for 1 h at 60°C followed by slow cooling to 30°C and washing at 35°C in 2× SSC (0.3 M NaCl, 30 mM sodium citrate)–0.1% sodium dodecyl sulfate.
RESULTS
Timing of IES excision.
In Paramecium, the formation of a new macronucleus in rich medium takes approximately 22 h from the time the zygotic nucleus has divided twice, and extensive DNA amplification was reported to occur in the developing macronucleus during this period (4). To investigate precisely the timing of IES excision relative to DNA amplification, macronuclear development in strain speedy was synchronized through conjugation of reactive cells of complementary mating types. After cell mixing, early-conjugating pairs that formed within 1 h 45 min were picked and transferred into rich medium. Separation of exconjugants occurred within a 1-h interval about 6 h 30 min after mixing, and karyonidal division took place within a 4-h period around 18 h after mixing. Since fragments of the degenerating parental macronucleus coexist with the anlagen over the entire period of macronuclear development (4), it was impossible, in total-DNA preparations, to distinguish the new macronuclear junctions generated by IES excision from the sequences of the parental macronucleus. Instead, we analyzed the micronuclear junctions between IESs and their flanking macronuclear-destined sequences.
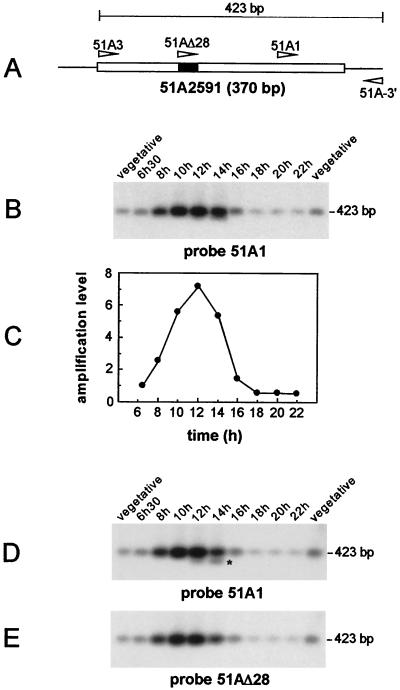
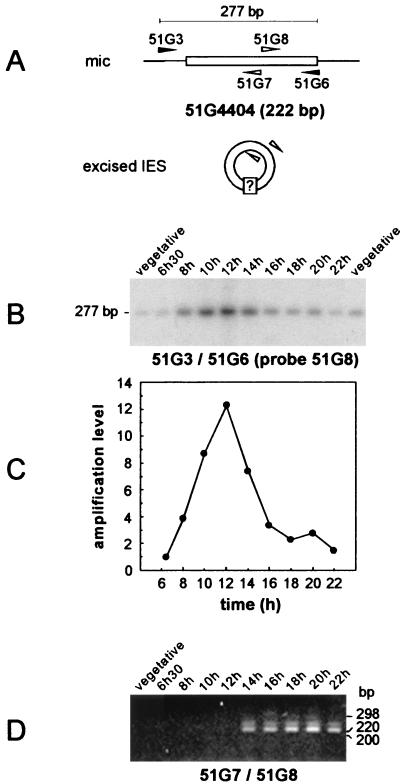
Our study focused on two IESs, 51A2591 (370 bp) (Fig. 1A) and 51G4404 (222 bp) (Fig. 2A), identified in the micronuclear versions of P. tetraurelia surface antigen genes A51 and G51, respectively (28, 32). The physical association of the two IESs with their flanking sequences was monitored in the same time course experiment, using synchronized cell samples taken every 2 h from the time of exconjugant separation. For each IES, semiquantitative PCR (see Materials and Methods) was performed on 10-cell aliquots at each time point, with one primer inside the IES and the other within adjacent macronuclear DNA: primers 51A3 and 51A-3′ were expected to yield a 423-bp micronucleus-specific band for IES 51A2591 (Fig. 1A), while a 277-bp product was expected for IES 51G4404 amplified with primers 51G3 and 51G6 (Fig. 2A). PCR products were revealed by Southern blot hybridization with internal oligonucleotide probes, 51A1 for IES 51A2591 (Fig. 1B) and 51G8 for IES 51G4404 (Fig. 2B). Bands of the expected sizes were detected for both IESs, and their intensities varied in a similar way, starting from a background level characteristic of the vegetative micronuclear signal at the time of exconjugant separation (Fig. 1B and 2B, compare the bands obtained at 6 h 30 min with the vegetative controls), reaching a peak 12 h after cell mixing, and returning to the background level at around 16 h. Quantification of the radioactive signal at each time point indicated that for both IESs, the micronuclear signal increased over the first 6 h of macronuclear development, up to 7-fold for IES 51A2591 (Fig. 1C) and up to 12-fold for IES 51G4404 (Fig. 2C). A sharp decrease in its intensity was observed from 12 to 14 h after the start of conjugation, suggesting that most copies of each IES are excised at this time. After karyonidal division, the signal was essentially similar to that obtained from control vegetative cells and could be attributable to the micronuclei (22-h time point in Fig. 1B and 2B).
FIG. 1.
Excision of IES 51A2591 and of the internal 28-bp IES in synchronized exconjugant cells. (A) Schematic diagram of IES 51A2591, with the internal 28-bp IES indicated by a solid box. The oligonucleotides used in the study are represented by arrowheads. (B) Southern blot analysis of the PCR products amplified from synchronized exconjugant cells with primers 51A3 and 51A-3′. Samples were run on a 1.5% agarose gel and revealed with 32P-labeled oligonucleotide 51A1. Indicated time points refer to the time following the mixing of reactive cells. Two independent samples of 10 vegetative cells were used as controls for the micronuclear signal. (C) Quantitative analysis of the excision timing of IES 51A2591. The signals shown in panel B were quantified and normalized relative to the value obtained for the 6 h 30 min time point. (D) Prolonged electrophoresis of the samples shown in panel B on a 1.5% agarose gel. IES 51A2591-derived fragments were revealed by Southern blot hybridization with oligonucleotide 51A1. The 400-bp band lacking the 28-bp internal IES is marked by an asterisk. (E) Same blot as in panel D, but hybridized with oligonucleotide 51AΔ28.
FIG. 2.
Timing of IES 51G4404 excision and detection of joined IES ends in synchronized exconjugant cells. (A) Diagram of the PCR strategies used for the amplification of chromosomal IES 51G4404 prior to its excision (primers 51G3 and 51G6 [black arrowheads]) and for the detection of putative IES circular forms (primers 51G7 and 51G8 [white arrowheads]). (B) Southern blot analysis of the PCR products amplified from synchronized exconjugant cells with primers 51G3 and 51G6. Samples were revealed with 32P-labeled oligonucleotide 51G8 after electrophoresis on a 3% NuSieve gel. Indicated time points refer to the time following the mixing of reactive cells. Two independent samples of 10 vegetative cells were used as controls for the micronuclear signal. (C) Quantitative analysis of the timing of IES 51G4404 excision. The signals shown in panel B were quantified and normalized relative to the value obtained for the 6 h 30 min time point. (D) PCR amplification from the same NP-40-treated synchronized exconjugant cells as in panel B, using divergent primers 51G7 and 51G8 (see Materials and Methods). PCR products were run on a 3% NuSieve gel and revealed by ethidium bromide staining. The lane order is as in panel B, except that only one vegetative control was used.
The above data demonstrate that Paramecium IES amplification takes place early during macronuclear development, before excision occurs. They also indicate that IESs are excised in a defined time interval during the first cell cycle following zygotic-nucleus formation.
Excision of a 28-bp IES internal to IES 51A2591.
Prolonged electrophoresis of the PCR products amplified with primers 51A3 and 51A-3′ in the experiment described above (Fig. 1B) revealed an approximately 400-bp fragment migrating ahead of the major 423-bp species in the 12- and 14-h samples and hybridizing with probe 51A1 (Fig. 1D). Excision of a 28-bp sequence located within IES 51A2591 (Fig. 1A) and exhibiting the characteristics of an IES has previously been observed when excision of IES 51A2591 was abolished by a point mutation at one end (26) or when it was epigenetically inhibited by the parental macronucleus (12). The absence of the 28-bp sequence from the 400-bp PCR product in Fig. 1D was assayed by hybridization with oligonucleotide 51AΔ28, which includes the 28-bp sequence and three upstream adjacent nucleotides (Table 1). This failed to reveal the 400-bp fragment (Fig. 1E), suggesting that this additional band arises through excision of the internal 28-bp IES in the normal course of macronuclear development.
Joining of the ends of excised IESs.
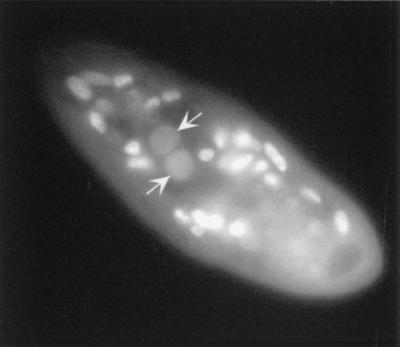
While the previous experiments allowed the identification of the time interval during which IESs are excised, it was of interest to investigate whether putative excision products could be detected. Following a strategy already described for other ciliates (22), divergent primers 51G7 and 51G8 hybridizing within IES 51G4404 were used to selectively amplify molecules in which both IES ends would have been covalently linked, as expected for a circular form of the excised sequence (Fig. 2A). PCR amplification products obtained from the same synchronized cell lysates as in the previous experiment were revealed by ethidium bromide staining (Fig. 2D). While no PCR product was detected from the vegetative control sample or at early time points (6 h 30 min to 12 h), a major product migrating just above the 220-bp size marker, hence exhibiting the expected size for IES 51G4404, was amplified from the 14-h sample and at all later time points (16 to 22 h). A species with a slightly higher molecular weight was also apparent: although its structure remains unclear, it most probably arose from the saturating PCR conditions used and accumulated with increasing numbers of amplification cycles (data not shown). Strikingly, IES excision (Fig. 2B) and the joining of ends (Fig. 2D) appeared to be closely linked in time, since both started to be detected between 12 and 14 h after cell mixing. DAPI staining of 14-h exconjugant cells revealed that anlagen were already clearly visible at this stage (Fig. 3).
FIG. 3.
DAPI staining of a 14-h exconjugant cell. The anlagen (arrows) are surrounded by the parental macronuclear fragments.
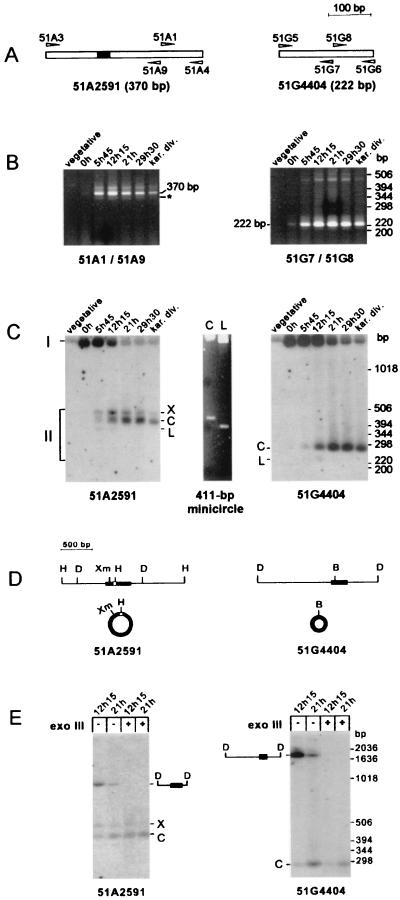
Since a PCR signal was detected during an 8-h period, at least in cells synchronized through conjugation, the same approach was applied to genomic DNA preparations from starved autogamous cells, in spite of the loss in synchrony generally observed for macronuclear development in large-scale autogamous cultures. Strain speedy was grown for 30 vegetative divisions before the cells were allowed to starve to induce autogamy, and macronuclear development was monitored by DAPI staining (data not shown). The t = 0 time point was arbitrarily defined as the time when 97% of the cell population exhibited a fragmented macronucleus. At 5 h 45 min, 100% of the cells were autogamous and harbored round fragments of the parental macronucleus; at 12 h 15 min, macronuclear fragments were located mainly around the cell periphery but the developing macronuclei were not yet visible. The anlagen were apparent in 50% of the cells at 21 h: at this time, the culture was divided in half and one aliquot was fed with rich medium. Both aliquots were further incubated for 8 h 30 min at 27°C. At 29 h 30 min, two anlagen were visible in all cells of the starved sample while karyonidal division had taken place in a fraction of the cells fed with rich medium. Total genomic DNA was extracted from aliquots of the culture taken at each of these time points and assayed for the presence of putative excision products by using suitable pairs of divergent PCR primers (Fig. 4A, primers 51A1 and 51A9 for IES 51A2591 and primers 51G7 and 51G8 for IES 51G4404). After agarose gel electrophoresis, a major product of the size of each IES was present in samples showing 100% autogamous cells (from 5 h 45 min to karyonidal division in both panels of Fig. 4B) and absent from vegetative controls (Fig. 4B). Low yields of this product were obtained in the t = 0 sample (Fig. 4B, both panels); this could reflect either early DNA rearrangements or some asynchrony in the autogamous cell population. Additional minor PCR products were observed for both IESs. For IES 51A2591, two additional bands were visible: a slower-migrating one, which could be of the same nature as the minor species described in Fig. 2C for IES 51G4404, and an approximately 340-bp fragment (Fig. 4B, left panel), which is described below. For IES 51G4404, a minor species of approximately 450 bp was amplified, but its structure was not elucidated.
FIG. 4.
Direct detection of circular forms of the excised IESs in autogamous cells. (A) Diagram of IESs 51A2591 and 51G4404, with the corresponding oligonucleotide primers represented by arrowheads. (B) PCR amplification of circular junctions from autogamous cells. Primer sets 51A1 plus 51A9 and 51G7 plus 51G8 were used for the detection of a junction between the ends of IES 51A2591 (left panel) and IES 51G4404 (right panel), respectively (see Materials and Methods). The t = 0 time point was arbitrarily defined as stated in the text, and other time points refer to later stages of autogamy (see the text for details), up to karyonidal division (kar. div.). Electrophoresis was carried out on a 3% NuSieve gel. The major amplification products are indicated by their sizes, and the 342-bp minor band corresponding to the form lacking the 28-bp IES internal to IES 51A2591 is indicated by an asterisk. (C) Southern blot analysis of uncut genomic DNA from autogamous cells. Electrophoresis was carried out on a 1.5% agarose–1.5% NuSieve gel. Chromosomal (I) and extrachromosomal (II) forms of IES 51A2591 (left panel) were revealed after hybridization with a 370-bp PCR fragment specific for the IES sequence and amplified with primers 51A3 and 51A4. A 236-bp fragment, specific for IES 51G4404 and amplified with primers 51G5 and 51G6, was used as a probe in the right panel. The central panel shows ethidium bromide staining of supercoiled (C) or linear (L) forms of a 411-bp control minicircle (see Materials and Methods), electrophoresed on the same gel. (D) Restriction maps of the micronuclear regions around IES 51A2591 (left) and IES 51G4404 (right). Chromosomal IESs are drawn as black boxes, and the 28-bp IES inside IES 51A2591 is shown as a white square. The corresponding circular IES molecules are not drawn to scale. Restriction enzymes: B, BsaI; D, DdeI; H, HinfI; Xm, XmnI. (E) DdeI and exonuclease III (exo III) treatment of total genomic DNA from 100% autogamous cells. DNA extracted from autogamous cells at t = 12 h 15 min and t = 21 h was treated with DdeI and, where indicated, incubated for 2 h at 37°C with 200 U of exonuclease III. Electrophoresis and hybridization probes were as in panel C. IESs are shown as black boxes on the diagrams of their corresponding micronuclear DdeI fragments.
The detection of a signal amplified with divergent PCR primers from synchronized exconjugants or from starved autogamous cells indicates that molecules carrying abutted IES ends are produced during macronuclear development in Paramecium.
Circular forms of the excised IESs.
The molecules responsible for the amplification signal obtained with divergent PCR primers (Fig. 2D and 4B) could be circular forms or direct tandem repeats of the IESs. To distinguish between these two alternatives, total unrestricted DNA extracted from cells at different stages of autogamy was directly analyzed by Southern blot hybridization with DNA probes corresponding to IESs 51A2591 or 51G4404 (Fig. 4C).
Two groups of bands were detected for each IES. High-molecular-weight band I migrates with the bulk of chromosomal DNA and corresponds to the germ line sequences present in the micronuclei and in the developing macronuclei. This was confirmed by restriction with DdeI, which does not cut within either IES (Fig. 4D). Band I was converted to a unique species exhibiting, for each IES, the size of the corresponding micronuclear fragment (exonuclease III-untreated samples in Fig. 4E). As expected from our previous quantitative analysis of synchronized macronuclear development, the intensity of band I first increased in young autogamous cells (t = 0 h and 5 h 45 min in Fig. 4C) and then decreased until the start of karyonidal division (t = 12 h 15 min and later in Fig. 4C). The second group of hybridizing bands (II in Fig. 4C) corresponds to extrachromosomal forms of the IESs, as indicated by their low molecular weight. They are specific for cells undergoing macronuclear development (t = 5 h 45 min and later time points in Fig. 4C), and their appearance is concomitant with the decrease of the chromosomal signal.
In the experiment in Fig. 4C, two major group II species (X and C) were detected for IES 51A2591 (Fig. 4C, left panel) and only one (C) was detected for IES 51G4404 (right panel). Their apparent molecular masses were higher than expected for a linear form of each IES. Interestingly, the major extrachromosomal form of IES 51A2591 (band C) has an apparent molecular mass that varies relative to linear size standards, according to the electrophoresis conditions used: from 310 bp on a 1.2% agarose gel (data not shown) to 432 bp on a 1.5% agarose–1.5% NuSieve gel (Fig. 4C, left panel). In young autogamous cells (t = 5 h 45 min and 12 h 15 min in Fig. 4C), a faint band (L) was also present at the position expected for a linear form of each IES. The nature of bands X and C was first assayed with restriction enzymes cutting once within each IES (Fig. 4D): IES 51A2591 bands X and C were converted to a single 370-bp species after restriction with XmnI or HinfI, while BsaAI treatment converted IES 51G4404 band C into one band of the size of the linear IES (data not shown). A similar mobility shift was observed after the linearization of a 411-bp double-stranded control minicircle (Fig. 4C). To confirm that the excised IESs were circular, DdeI-restricted DNA preparations were treated with E. coli exonuclease III, which preferentially uses blunt or recessed 3′ ends (such as DdeI-generated ends) as substrates. A rather complex pattern was observed for IES 51A2591: no degradation of band C was detected, while band X was partially sensitive to exonuclease III and the chromosomal DdeI fragment was completely degraded (Fig. 4E, left panel). IES 51G4404 band C was essentially resistant to the nuclease (right panel). In conclusion, their electrophoretic properties and the patterns obtained after restriction enzyme and exonuclease III treatment of bands C suggest that they represent double-stranded circular excised forms of IESs 51A2591 and 51G4404. The structure of IES 51A2591 band X is unclear; it could be a nicked or relaxed form of band C, supposing that C is a supercoiled circle.
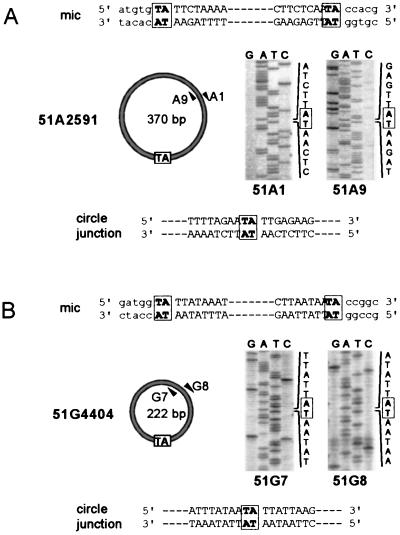
Sequence of the junctions on IES circles.
To determine the nucleotide sequence of the junctions between circularized IES ends, the PCR fragments in Fig. 2D and 4B were sequenced on both strands with the primers used for their amplification. For each IES, total PCR products from synchronized exconjugants (Fig. 2D, t = 14 to 22 h) or autogamous cells (Fig. 4B, t = 5 h 45 min to karyonidal division) were pooled and purified from residual unlabeled primers. En masse PCR-mediated sequencing allowed the identification of a unique and unambiguous sequence for each IES (Fig. 5A for IES 51A2591 and Fig. 5B for IES 51G4404). In both cases, IES ends were precisely joined, with a single copy of the flanking 5′-TA-3′ repeat at the junction. Sequencing of the gel-purified major amplification product from IES 51A2591 (the 370-bp species in Fig. 4B, left panel) revealed that it carried the 28-bp internal IES (data not shown), in agreement with its observed size on agarose gels (Fig. 4B). The very faint PCR band migrating ahead of this major 370-bp species (Fig. 4B, left panel) was also gel purified and sequenced with the same primers; it was shown to correspond to a 342-bp molecule with the same junction between the ends of IES 51A2591 but from which the internal 28-bp IES had been excised, leaving one 5′-TA-3′ dinucleotide at the new junction (data not shown).
FIG. 5.
Sequence of IES circles. Sequences of the ends (capital letters) and their flanking macronucleus-destined regions (lowercase letters) of IES 51A2591 (A) and IES 51G4404 (B) are shown at the top of each panel. The 5′-TA-3′ dinucleotide between joined IES ends is boxed in the diagram representing the circularized form of each IES. For purposes of clarity, only the structure of the major circular species observed for IES 51A2591 is shown in panel A (see the text for details). Arrowheads represent the divergent primers used for PCR amplification and sequencing of the circle junctions. Sequencing reactions were performed on both strands, and portions of the sequencing gels encompassing the joined ends of each IES are shown on the right. The nucleotide sequence of each junction is displayed at the bottom.
DISCUSSION
While the timing of IES excision in other ciliates has been known for a long time (2, 42, 45), this type of study has been delayed for Paramecium because of the difficulty in obtaining large amounts of cells synchronized for macronuclear development. We have overcome this problem by using manual sorting of early-conjugating pairs coupled to quantitative PCR. For the IESs studied here, excision was shown to start 12 to 14 h after the mixing of reactive cells and was essentially completed at 16 h. Beginning 14 h after initiation of conjugation, excised IES circles were formed, with the joined IES ends separated by a single copy of the flanking 5′-TA-3′ dinucleotide. A chromosomal junction resulting from the excision of a short IES inserted within a larger one was also transiently detected during the same time interval.
IES excision and DNA replication.
IES excision in Paramecium seems to take place during a specific period of the first cell cycle following micronuclear meiosis. This is based on the observations that the same elimination timing (12 to 14 h after initiation of conjugation) was observed for two IESs within different surface antigen genes and for a third, short IES (28 bp). Previous cytological analysis of macronuclear development following conjugation indicated that three doublings of the anlage DNA content take place before the 12-h time point (4). This would correspond to an 8-fold amplification of the IESs, in agreement with our observation that IESs 51A2591 and 51G4404 are amplified 7- and 12-fold, respectively, prior to their excision. Care must be taken, however, in the quantitative comparison of our data with those of Berger (4) because the specific contribution of the developing macronuclei cannot be distinguished from that of the new micronuclei in our experiments and because no information is available on the timing of micronuclear replication during the first cell cycle following conjugation. In addition, Berger's work provided an estimation of the overall DNA doubling rate within the anlage, but particular genomic loci could be amplified at different rates. On the other hand, we cannot exclude that early but rare IES excision events take place before the 12-h time point, which would decrease the apparent rate of IES replication. The results presented here nevertheless imply that up to 16 copies of each IES may be present in one anlage before their excision. This could allow significant variability in the excision products obtained from a single germ line sequence within the same macronucleus. Alternative rearrangement patterns have indeed been reported for several IESs in vegetative clones arising from unique macronuclear differentiation events (1, 9, 12).
A striking correlation exists between IES excision and DNA replication in the developing macronuclei of ciliates. In Paramecium, four discontinuous synchronous rounds of DNA synthesis were detected during the cell cycle preceding karyonidal division (4). Our data indicate that IES excision takes place during the third peak of DNA synthesis. In Tetrahymena, internal micronucleus-specific sequences are excised within a time interval during which the anlage DNA content increases from 4C to 8C (2). In Euplotes, two discrete periods of anlage DNA synthesis were observed during the first half of macronuclear development: IESs are excised at the end of the second period, after three to four doublings of the developing macronuclear genome have taken place (14, 42). The concomitant occurrence of DNA synthesis and IES excision may indicate that common factors participate in the control of both processes: recent reports have suggested that transcription factor-induced modifications of chromatin structure in eukaryotic cells can modulate the initiation of replication (17) and could regulate DNA rearrangements (29). A mechanistic link may also exist between DNA replication and IES excision: proteins involved in replication could take part in excision directly (14, 15), or, more indirectly, replication could induce DNA topological changes and chromatin remodelling (10), which might increase accessibility to the excision machinery. Whatever the case may be, IESs are always excised after a few replication cycles have taken place in the anlage. This could indicate that all requirements for excision are not met at the beginning of macronuclear development and that transcription or developmental activation of specific factors is needed.
Formation of excised IES circles.
This study demonstrates the developmentally programmed formation of excised double-stranded circular forms for two Paramecium IESs, which appear to result from the excision reaction. Indeed, their timing of appearance coincides with the dissociation of IESs from their chromosomal flanking sequences. Also, they are quite abundant in developing macronuclei, as suggested by their detection on Southern blots of DNA from autogamous cells. This point distinguishes Paramecium IESs from those of Tetrahymena, for which putative circularized forms could hardly be amplified by PCR (36, 47) and were not detected on Southern blots of DNA from developing macronuclei (2). As was proposed for Euplotes IESs (23, 41), Paramecium IES circles could be directly generated by the excision reaction. It should be noted, however, that trace amounts of molecules with the electrophoretic mobility expected for linear forms of the IESs are detected early during autogamy. Although the exact structure of these minor species remains to be established, this raises the possibility that IES excision primarily produces linear molecules that could be precursors to the circles. The relative abundance of the circles and their homogeneous nucleotide sequence would then imply that circularization is an efficient and precise reaction, involving little or no degradation of linear DNA ends. In a first attempt to investigate whether circular molecules are direct products of the excision reaction, the radioactive signals detected on the Southern blots in Fig. 4C were quantified to monitor the conversion of the chromosomal signal into the extrachromosomal signal. This indicated that the amount of extrachromosomal circles remained smaller than the maximum amplification level of the chromosomal IES (data not shown). However, it is worth pointing out that IES circles are not amplified in the final macronucleus, as indicated by their absence from vegetative cells. Therefore, since no information is available concerning the stability of the circular molecules (i.e., whether they are actively degraded or simply diluted out during subsequent cell divisions), one should be cautious in interpreting this result in terms of the circles being (or not being) primary excision products.
A majority of Paramecium IESs are shorter than the average 140-bp persistence length of B-DNA, which reflects the intrinsic flexibility of a DNA fragment (16, 43). It is difficult to imagine that sequences as short as 26 bp can form covalently closed double-stranded circular molecules. This could be in favor of the hypothesis that IES circles are not the primary products of the excision process. Alternatively, two mechanisms could exist for IES excision, depending on their capacity to circularize. In support of the latter hypothesis is the existence of a subgroup of short IESs in Euplotes, which are excised later during macronuclear development and could be excised via a pathway related to chromosome fragmentation (20). No evidence has been obtained for different excision timings of long and short IESs in Paramecium, but the only short IES examined here is internal to IES 51A2591. Identification of extrachromosomal forms of IES 51A2591 that still carry the 28-bp internal sequence shows that correct excision of the larger IES does not depend on prior excision of the internal one. Furthermore, the transient formation of a chromosomal junction resulting from excision of the 28-bp IES confirms that it can be excised from the germ line genome before the larger one (12, 26). Additional work is required to investigate the timing of short IES excision in more detail.
Mechanism of IES excision.
The relatively easy detection of extrachromosomal Paramecium IESs on Southern blots will be very helpful in further studies of the molecular mechanism of excision and in the identification of the different intermediates of the reaction. The present study has focused on the analysis of the abundant circular forms of the excised IESs. IES excision in Paramecium may formally be viewed as a recombination between short DNA repeats, in which one copy of the repeated 5′-TA-3′ dinucleotide is found between joined IES ends on the excised circle while the other is retained at the new junction formed on the macronuclear chromosome (hereafter referred to as the donor molecule, by analogy to other DNA excision systems, such as those involved in cut-and-paste transposition). Similar joining of the donor and excised molecules was previously reported only for TBE1 transposon-like elements in Oxytricha (46). TBE1 circle junctions carry a single copy of the 3-bp target site duplication, joined on each side to the terminal 3′ dG of each end. However, recent data have indicated that the nucleotide facing this dG in the duplex circle is not always its complement, which can result in a mispaired position on both sides of the 3-bp target sequence (K. Williams, T. Doak, and G. Herrick, personal communication). The double-stranded DNA junctions created during IES excision in Paramecium therefore appear to be unprecedented in ciliates. More generally, transposase-induced excision of DNA transposons moving via a cut-and-paste pathway (reviewed in reference 34) generates double-stranded breaks on the donor molecule, which is either lost, imprecisely repaired to give characteristic DNA footprints, or repaired by gene conversion with a homologous sequence as a template. In these systems, the transposon is generally excised as a linear molecule. In the particular case of Tc1/mariner transposable elements, the integrated copies of the transposon are flanked by repeated 5′-TA-3′ dinucleotides but the excised linear form does not contain any part of these repeats (30). Transposase-induced formation of a DNA minicircle, in which the joined ends of the excised sequence are separated by one copy of the 3-bp repeat initially flanking the element, was demonstrated for bacterial insertion sequence IS911, but the fate of the donor molecule is unclear (31). Finally, in mammalian V(D)J recombination, both the circular excised molecule and the resealed donor molecule have been analyzed (reviewed in reference 5). While the signal joints on the excised molecules result from the precise fusion of conserved heptamer sequences, rejoining of the coding ends on the donor molecule is accompanied by nucleotide addition or removal.
It has been proposed that Paramecium and Euplotes TA IESs have evolved from transposons of the Tc1/mariner family (22). This proposal is essentially based on the sequence conservation of the terminal 8 bp of these elements, including the flanking 5′-TA-3′ dinucleotides (21), and on the observation that the developmental excision of the transposon-like Tec elements of Euplotes, which are members of this family (8), produces circles with a junction structurally similar to those of Euplotes TA IESs (19, 23). Interesting differences exist at the nucleotide level between the excision products generated in the two ciliate species. The junctions formed between the ends of Paramecium circular IESs include a single copy of the flanking 5′-TA-3′ repeat with no additional nucleotide. In contrast, the circularized junctions of Euplotes elements include two copies of the 5′-TA-3′ dinucleotide, separated by 10 bp originating from the left and right flanking macronucleus-destined sequences, 6 bp of which apparently form a partial heteroduplex (19, 23). Provided that the excised circles are direct products of excision, a possible model for IES and Tec excision in this organism includes symmetrical double-stranded staggered cuts at the ends of the eliminated fragment, at the level of the 5′-TA-3′ dinucleotide on one side and further into the flanking DNA on the other. The precise location of initial DNA cuts distinguishes this putative mechanism from that proposed for the excision step of Tc1/mariner transposition (30), suggesting that the enzymes participating in the reactions differ to some extent. In addition, the enzymatic machinery responsible for the excision of TA IESs seems to have evolved more rapidly than the nucleotide constraints imposed on their ends since different circle junctions are found in Paramecium and Euplotes: ciliates could have independently developed different strategies to solve the problem of IES excision.
The characteristic junctions formed during IES excision in Paramecium hint at a specific role for the 5′-TA-3′ dinucleotide, as already inferred from statistical and genetic analyses (9, 21, 25), and suggest that it might be a target for single- or double-stranded DNA cutting at each IES end. This dinucleotide was also found at chromosomal junctions of developmental deletions induced by homology-dependent epigenetic effects (27) or by other developmentally programmed rearrangements of the germ line genome (6). The remarkable flexibility of the TA step (48) could be linked to the preferential use of this dinucleotide as a target site for DNA recombination. In the absence of any data concerning the molecular mechanism involved, it is tempting to speculate that a general TA-specific cleavage activity has been recruited for IES excision in Paramecium.
ACKNOWLEDGMENTS
We thank Kevin Williams, Tom Doak, and Glenn Herrick for communicating their results prior to publication; Janine Beisson, Anne-Marie Keller, and Françoise Ruiz for their advice in Paramecium culture handling; Rémi Dillys and Anne Turbé for their participation in the sequencing of IES circular junctions; and Robert Alazard for the gift of plasmid pRA593. Many thanks go to all members of the group for stimulating discussions and critical reading of the manuscript.
This work was supported by the Association pour la Recherche sur le Cancer (grant 1374), the Centre National de la Recherche Scientifique (Programme Génome), and the Ministère de l'Education Nationale de la Recherche et de la Technologie (Programme de Recherche fondamentale en Microbiologie et Maladies infectieuses et parasitaires). S.D. was a recipient of doctoral fellowships from the Association pour la Recherche sur le Cancer and from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Amar L. Chromosome end formation and internal sequence elimination as alternative genomic rearrangements in the ciliate Paramecium. J Mol Biol. 1994;236:421–426. doi: 10.1006/jmbi.1994.1154. [DOI] [PubMed] [Google Scholar]
- 2.Austerberry C F, Allis C D, Yao M C. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci USA. 1984;81:7383–7387. doi: 10.1073/pnas.81.23.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. [Google Scholar]
- 4.Berger J D. Nuclear differentiation and nucleic acid synthesis in well-fed exconjugants of Paramecium aurelia. Chromosoma. 1973;42:247–268. doi: 10.1007/BF00284774. [DOI] [PubMed] [Google Scholar]
- 5.Bogue M, Roth D B. Mechanism of V(D)J recombination. Curr Opin Immunol. 1996;8:175–180. doi: 10.1016/s0952-7915(96)80055-0. [DOI] [PubMed] [Google Scholar]
- 6.Bourgain-Guglielmetti F M, Caron F M. Molecular characterization of the D surface protein gene subfamily in Paramecium primaurelia. J Eukaryot Microbiol. 1996;43:303–313. doi: 10.1111/j.1550-7408.1996.tb03993.x. [DOI] [PubMed] [Google Scholar]
- 7.Breuer M, Schulte G, Schwegmann K J, Schmidt H J. Molecular characterization of the D surface protein gene subfamily in Paramecium tetraurelia. J Eukaryot Microbiol. 1996;43:314–322. doi: 10.1111/j.1550-7408.1996.tb03994.x. [DOI] [PubMed] [Google Scholar]
- 8.Doak T G, Doerder F P, Jahn C L, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubrana K, Le Mouël A, Amar L. Deletion endpoint allele-specificity in the developmentally regulated elimination of an internal sequence (IES) in Paramecium. Nucleic Acids Res. 1997;25:2448–2454. doi: 10.1093/nar/25.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duguet M. When helicase and topoisomerase meet! J Cell Sci. 1997;110:1345–1350. doi: 10.1242/jcs.110.12.1345. [DOI] [PubMed] [Google Scholar]
- 11.Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence in Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- 12.Duharcourt S, Keller A M, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forney J D, Blackburn E H. Developmentally controlled telomere addition in wild-type and mutant paramecia. Mol Cell Biol. 1988;8:251–258. doi: 10.1128/mcb.8.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frels J S, Jahn C L. DNA rearrangements in Euplotes crassus coincide with discrete periods of DNA replication during the polytene chromosome stage of macronuclear development. Mol Cell Biol. 1995;15:6488–6495. doi: 10.1128/mcb.15.12.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frels J S, Tebeau C M, Doktor S Z, Jahn C L. Differential replication and DNA elimination in the polytene chromosomes of Euplotes crassus. Mol Biol Cell. 1996;7:755–768. doi: 10.1091/mbc.7.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagerman P J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y F, Hao Z L, Li R. Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev. 1999;13:637–642. doi: 10.1101/gad.13.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs M E, Klobutcher L A. The long and the short of developmental DNA deletion in Euplotes crassus. J Eukaryot Microbiol. 1996;43:442–452. doi: 10.1111/j.1550-7408.1996.tb04503.x. [DOI] [PubMed] [Google Scholar]
- 19.Jaraczewski J W, Jahn C L. Elimination of Tec elements involves a novel excision process. Genes Dev. 1993;7:95–105. doi: 10.1101/gad.7.1.95. [DOI] [PubMed] [Google Scholar]
- 20.Klobutcher L A. Developmentally excised DNA sequences in Euplotes crassus capable of forming G quartets. Proc Natl Acad Sci USA. 1995;92:1979–1983. doi: 10.1073/pnas.92.6.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klobutcher L A, Herrick G. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 1995;23:2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klobutcher L A, Herrick G. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog Nucleic Acid Res Mol Biol. 1997;56:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- 23.Klobutcher L A, Turner L R, LaPlante J. Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev. 1993;7:84–94. doi: 10.1101/gad.7.1.84. [DOI] [PubMed] [Google Scholar]
- 24.Ling K Y, Vaillant B, Haynes W J, Saimi Y, Kung C. A comparison of internal eliminated sequences in the genes that encode two K+-channel isoforms in Paramecium tetraurelia. J Eukaryot Microbiol. 1998;45:459–465. doi: 10.1111/j.1550-7408.1998.tb05100.x. [DOI] [PubMed] [Google Scholar]
- 25.Mayer K M, Forney J D. A mutation in the flanking 5′-TA-3′ dinucleotide prevents excision of an internal eliminated sequence from the Paramecium tetraurelia genome. Genetics. 1999;151:597–604. doi: 10.1093/genetics/151.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer K M, Mikami K, Forney J D. A mutation in Paramecium tetraurelia reveals function and structural features of developmentally excised DNA elements. Genetics. 1998;148:139–149. doi: 10.1093/genetics/148.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer E, Butler A, Dubrana K, Duharcourt S, Caron F. Sequence-specific epigenetic effects of the maternal somatic genome on developmental rearrangements of the zygotic genome in Paramecium primaurelia. Mol Cell Biol. 1997;17:3589–3599. doi: 10.1128/mcb.17.7.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer E, Keller A M. A mendelian mutation affecting mating-type determination also affects developmental genomic rearrangements in Paramecium tetraurelia. Genetics. 1996;143:191–202. doi: 10.1093/genetics/143.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolas A. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc Natl Acad Sci USA. 1998;95:87–89. doi: 10.1073/pnas.95.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plasterk R H. The Tc1/mariner transposon family. Curr Top Microbiol Immunol. 1996;204:125–143. doi: 10.1007/978-3-642-79795-8_6. [DOI] [PubMed] [Google Scholar]
- 31.Polard P, Prere M F, Fayet O, Chandler M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992;11:5079–5090. doi: 10.1002/j.1460-2075.1992.tb05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preer L B, Hamilton G, Preer J R. Micronuclear DNA from Paramecium tetraurelia: serotype 51 A gene has internally eliminated sequences. J Protozool. 1992;39:678–682. doi: 10.1111/j.1550-7408.1992.tb04448.x. [DOI] [PubMed] [Google Scholar]
- 33.Prescott D M. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saedler H, Gierl A, editors. Current topics in microbiology and immunology. 204. Transposable elements. Berlin-Heidelberg, Germany: Springer-Verlag KG; 1996. [Google Scholar]
- 35.Saveliev S V, Cox M M. Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. EMBO J. 1996;15:2858–2869. [PMC free article] [PubMed] [Google Scholar]
- 36.Saveliev S V, Cox M M. The fate of deleted DNA produced during programmed genomic deletion events in Tetrahymena thermophila. Nucleic Acids Res. 1994;22:5695–5701. doi: 10.1093/nar/22.25.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott J, Leeck C, Forney J. Analysis of the micronuclear B type surface protein gene in Paramecium tetraurelia. Nucleic Acids Res. 1994;22:5079–5084. doi: 10.1093/nar/22.23.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soldo A T, Godoy G A. The kinetic complexity of Paramecium macronuclear deoxyribonucleic acid. J Protozool. 1972;19:673–678. doi: 10.1111/j.1550-7408.1972.tb03558.x. [DOI] [PubMed] [Google Scholar]
- 39.Sonneborn T M. Paramecium aurelia. In: King R C, editor. Handbook of genetics: plants, plant viruses and protists. Vol. 2. New York, N.Y: Plenum Press; 1974. pp. 469–594. [Google Scholar]
- 40.Steele C J, Barkocy-Gallagher G A, Preer L B, Preer J R. Developmentally excised sequences in micronuclear DNA of Paramecium. Proc Natl Acad Sci USA. 1994;91:2255–2259. doi: 10.1073/pnas.91.6.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tausta S L, Klobutcher L A. Detection of circular forms of eliminated DNA during macronuclear development in E. crassus. Cell. 1989;59:1019–1026. doi: 10.1016/0092-8674(89)90758-7. [DOI] [PubMed] [Google Scholar]
- 42.Tausta S L, Klobutcher L A. Internal eliminated sequences are removed prior to chromosome fragmentation during development in Euplotes crassus. Nucleic Acids Res. 1990;18:845–853. doi: 10.1093/nar/18.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Travers A A, Ner S S, Churchill M E. DNA chaperones: a solution to a persistence problem? Cell. 1994;77:167–169. doi: 10.1016/0092-8674(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 44.Vayssié L, Sperling L, Madeddu L. Characterization of multigene families in the micronuclear genome of Paramecium tetraurelia reveals a germline specific sequence in an intron of a centrin gene. Nucleic Acids Res. 1997;25:1036–1041. doi: 10.1093/nar/25.5.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen J, Maercker C, Lipps H J. Sequential excision of internal eliminated DNA sequences in the differentiating macronucleus of the hypotrichous ciliate Stylonichia lemnae. Nucleic Acids Res. 1996;24:4415–4419. doi: 10.1093/nar/24.22.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams K, Doak T G, Herrick G. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 1993;12:4593–4601. doi: 10.1002/j.1460-2075.1993.tb06148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao M C, Yao C H. Detection of circular excised DNA deletion elements in Tetrahymena thermophila during development. Nucleic Acids Res. 1994;22:5702–5708. doi: 10.1093/nar/22.25.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zakrzewska K. Static and dynamic conformational properties of AT sequences in B-DNA. J Biomol Struct Dyn. 1992;9:681–693. doi: 10.1080/07391102.1992.10507948. [DOI] [PubMed] [Google Scholar]