Abstract
Repair of UV-induced DNA lesions in terminally differentiated human hNT neurons was compared to that in their repair-proficient precursor NT2 cells. Global genome repair of (6-4)pyrimidine-pyrimidone photoproducts was significantly slower in hNT neurons than in the precursor cells, and repair of cyclobutane pyrimidine dimers (CPDs) was not detected in the hNT neurons. This deficiency in global genome repair did not appear to be due to denser chromatin structure in hNT neurons. By contrast, CPDs were removed efficiently from both strands of transcribed genes in hNT neurons, with the nontranscribed strand being repaired unexpectedly well. Correlated with these changes in repair during neuronal differentiation were modifications in the expression of several repair genes, in particular an up-regulation of the two structure-specific nucleases XPG and XPF/ERCC1. These results have implications for neuronal dysfunction and aging.
The human genome is the target of DNA damage from multiple sources, both environmental, such as radiation or chemicals, and intracellular, such as the reactive products of cellular oxidative metabolism. DNA lesions are a threat to the organism for several reasons. First, they may arrest transcription, thereby preventing appropriate gene expression and potentially disturbing the cellular metabolism. Second, lesions encountered during DNA replication can result in the introduction of mutations, the accumulation of which may lead to cancer. Finally, DNA damage can prevent cell proliferation and kill cells, which can result in various growth and developmental defects (7).
Eucaryotic cells have evolved several systems, including nucleotide excision repair (NER), base excision repair, and mismatch repair, to repair inappropriate DNA alterations. Some are able to target transcribed genes specifically and repair them more rapidly than they can repair the global genome (10). Other mechanisms, involving the genes ATM and p53, can arrest the cell cycle until damage is repaired, and if this does not occur rapidly, they can trigger apoptosis (4). Finally, cells can tolerate some DNA lesions through translesion DNA synthesis and/or recombination.
However, cells that have undergone terminal differentiation, such as myotubes, adipocytes, or neurons, represent a special situation. (i) Since they no longer divide, apoptosis would result in an uncompensated cell loss for the organism, which would probably be a harmful situation. One would then expect that such cells would rely heavily on their repair systems to deal with lesions in their genomes. (ii) Recombinational repair most probably does not occur in cells that remain in interphase. (iii) Since these cells do not replicate their DNA, the risk of accumulating carcinogenic mutations is extremely low, as documented by the rarity of neuronal cancer in adults. Neurons and other postmitotic cells could thus, in principle, afford to repair only the portion of their genome that is really needed for their specialized functions, i.e., transcribed genes.
For these reasons, neurons constitute a particularly interesting system in which to study DNA repair, especially in conjunction with transcription. However, surprisingly little is known about the repair capabilities of neurons. It has been shown that mouse neuroblastoma cells become extremely UV sensitive after terminal differentiation (21) and that human neuroblastoma cells are deficient in removal of bulky adducts and exhibit low levels of unscheduled DNA synthesis, an indication of repair activity (15). Preliminary studies in our laboratory with rat pheochromocytoma PC12 cells differentiating into neuron-like cells have indicated a marked decrease in global genome repair but not in the repair of expressed genes (summarized in reference 11).
The present work makes use of the NT2-hNT system, a well-characterized human precursor/neuron cell system. It was undertaken to examine in detail the repair phenotype of human neurons and to try to understand how terminal differentiation might modulate DNA NER.
MATERIALS AND METHODS
Cell culture and treatments.
NT2 cells (Stratagene) were grown and differentiated as recommended by the supplier. Due to the poor quality of the commercially available cells, another batch was obtained directly from Peter Andrews (18). The cells were irradiated with 254-nm UV light using a Westinghouse SB-30 germicidal lamp at 1 W/m2. They were harvested by trypsinization, and DNA was prepared as described previously (25). In some cases, 6 h before irradiation, the cells were fed medium containing trichostatin A (ICN) dissolved at 1 mg/ml in ethanol.
T4 endo V sensitive-site assay.
DNA (200 ng) was digested with 0.07 μl of T4 endonuclease V (endo V) (activity, 2 × 1013 nicks/min/μl) (a kind gift from Stephen Lloyd) in TEV buffer (100 mM NaCl, 10 mM Tris [pH 7.5], 10 mM EDTA, 1 mg of bovine serum albumin per ml). The reaction mixture was loaded on 5 to 20% sucrose gradients containing 0.1 M NaOH and 10 mM EDTA, and centrifuged for 2 h at 30,000 rpm at 20°C in an SW-50.1 rotor (Beckman). Fractions (22 to 24 per gradient) were collected from the bottom, and 100 μl of each, mixed with 100 μl of 20× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), was slot blotted under vacuum on a Hybond N+ membrane (Amersham) and fixed for 20 min on 3MM paper (Whatman) soaked in 0.4 N NaOH. A 32P-labeled DNA probe was prepared from genomic DNA by using a nick translation labeling kit (Gibco). The membrane was prehybridized for 2 h at 42°C in 6× SSPE–5× Denhardt's solution–0.5% sodium dodecyl sulfate (SDS)–50% formamide–200 μg of denatured salmon sperm DNA per ml, hybridized overnight with the 32P-labeled probe at 42°C in 6× SSPE–0.5% SDS–50% formamide–100 mg of denatured salmon sperm DNA per ml, and washed for 5 min at 20°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% SDS, 15 min at 20°C in 2× SSC–0.1% SDS, 30 min at 37°C in 0.1× SSC–0.5% SDS, and 30 min at 65°C in 0.1× SSC–0.5% SDS, and rinsed in 0.1× SSC, and the 32P was quantified with a Bio-Rad GS-363 phosphorimager.
T4 endo V accessibility assay.
The T4 endo V accessibility assay was conducted as previously described (29). Briefly, cells were resuspended in a low-salt buffer (100 mM Tris [pH 8], 100 mM NaCl, 10 mM EDTA, 1 mg of bovine serum albumin per ml) and permeabilized by three cycles of freezing-thawing. Some samples were incubated in high-salt buffer (as above but with 2 M NaCl) for 15 minutes at room temperature before being diluted back to 100 mM NaCl. Samples containing 20,000 cells were digested for 20 min at 37°C with 0.1 μl of T4 endo V. The cells were then lysed in a layer of 1 N NaOH at the top of high-salt alkaline sucrose gradients (5 to 20% sucrose in 2 M NaCl–0.3 N NaOH–10 mM EDTA). The gradients were processed and DNA was detected in the fractions as described above.
Antibody assay.
Cellular DNA in Tris-EDTA (TE) was denatured by boiling. One volume of 2× SSPE was added, and samples were slot blotted in triplicate on a Hybond N+ membrane, using 60 ng of DNA for the CPD assay and 1 μg of DNA for the (6-4)PPs assay. The DNA was fixed as above, and the membranes were blocked overnight in phosphate-buffered saline (PBS)–0.2% Tween 20 (PBS-T) containing 5% (wt/vol) skim milk, washed in PBS-T, and incubated for 2 h at room temperature with monoclonal antibodies specific for cyclobutane pyrimidine dimer (CPD) or (6-4)pyrimidine-pyrimidone photoproducts [(6-4)PPs] (a kind gift from Toshio Mori [22]) diluted 1/2,000 in PBS. The membrane was washed as above and incubated for 1 h at room temperature with a peroxidase-labeled anti-mouse monoclonal antibody (Amersham) diluted 1/4,000 in PBS. After extensive washes with PBS-T, the membrane was assayed with an ECL chemiluminescence kit (Amersham). The signal was quantified using a Bio-Rad GS-363 phosphorimager.
TCR assay.
The transcription-coupled repair (TCR) assay was conducted essentially as described previously (25) except that the DNA could not be 3H labeled (since neurons do not replicate DNA). The CsCl gradient step was thus omitted, and after restriction, DNA was ethanol precipitated with 0.8 M LiCl, resuspended in TE, and subjected to T4 endo V digestion. The restriction enzymes were KpnI for the dihydrofolate reductase (DHFR) gene, SphI and HincII for the glucagon gene, EcoRI and BamHI for the NF-L gene, BclI and XhoI for the CK8 gene, and HindIII for the CK18 gene.
Quantitative RT-PCR.
RNA was prepared by resuspending trypsinized cells in 8 ml of GT buffer (4 M guanidinium thiocyanate, 0.1 M Tris [pH 7.5], 1% β-mercaptoethanol, 0.5% sodium lauroyl sarcosinate) and passing the lysate 10 times through a 20-gauge needle. It was then laid on a cushion of 1 ml of 40% (wt/vol) CsCl on top of 3 ml of 5.7 M CsCl in TE (pH 7.6) and centrifuged in an SW27 rotor for 22 h at 34,000 rpm and 20°C. The RNA pellet was dissolved in TE (pH 7.6), extracted with 1 volume of chloroform-butanol (1:1), ethanol (2 volumes) precipitated with 0.3 M sodium acetate (pH 5.2), and dissolved in nuclease-free water. RNA (3 μg) was reverse transcribed with 50 ng of random hexamers (Gibco) using a Superscript II kit (Gibco). PCR was performed using a touchdown method; 5 min at 94°C; then 10 cycles of 15 s at 94°C, 30 s at the calculated annealing temperature (Ta) plus 10°C, subtracting 1°C at each cycle, and 1 min at 72°C; then 18 to 25 cycles of 15 s at 94°C, 30 s at Ta, and 1 min at 72°C; followed by a final 5 min at 72°C. The reaction mixtures were assembled in a volume of 50 μl, using 400 nM each dATP, dGTP, and dTTP, 120 nM dCTP, 10 μCi of [α-32P]dCTP (Amersham), 1.5 mM MgCl2, and 1.25 U of Taq polymerase (Gibco). Three different dilutions of reverse transcription (RT) reaction mixture (up to 1/70 of the RT reaction mixture) were routinely used to verify that the band intensity was linearly dependent on the amount of template. The number of cycles and the respective amounts of primers for target and reference genes were adjusted in preliminary experiments to avoid saturation and to obtain bands of comparable intensity for both genes. A 5-μl sample of each reaction mixture was run on a 1.8% agarose gel, and the gels were dried and quantified with a Bio-Rad GS-363 phosphorimager. The intensity of each band was normalized to the number of cytidines in a given fragment.
Strand-specific RT-PCR.
Total cellular RNA was reverse transcribed as above, except that 50-ng portions of oligonucleotides specific for the sense or antisense strand were used instead of random hexamers as primers. In the case of the DHFR gene, RT was detected in the absence of any primer, probably due to self-priming of the mRNA. To overcome this problem, RT was first performed for 20 min in the absence of any primer, with 0.6 μl of the ddA mix from a sequencing kit (Promega), in a volume of 18 μl. Regular deoxynucleoside triphosphates (final concentration, 5 mM) and 50-ng portions of strand-specific primers were then added, and the reaction was continued as above for an additional 40 min.
RESULTS
Global genome repair is greatly attenuated upon neuronal differentiation.
The system used for our studies consisted of NT2 human neuroteratoma cells, which upon appropriate stimulation differentiate into neuron-like cells termed hNT neurons (18). This well-characterized system has the advantage that repair can be studied in the identical genetic background, with the two cell types differing only by the fact that the hNT neurons have attained terminal differentiation. We have used it to study the repair of UV-induced DNA damage. Short-wavelength UV radiation induces covalent bonds between adjacent pyrimidines on the same DNA strand. There are two main types of such dimers, depending upon where the bonds form: CPDs and (6-4)PPs. CPDs are induced to a higher level upon irradiation with 254-nm light (8), but (6-4)PPs are more rapidly repaired (31).
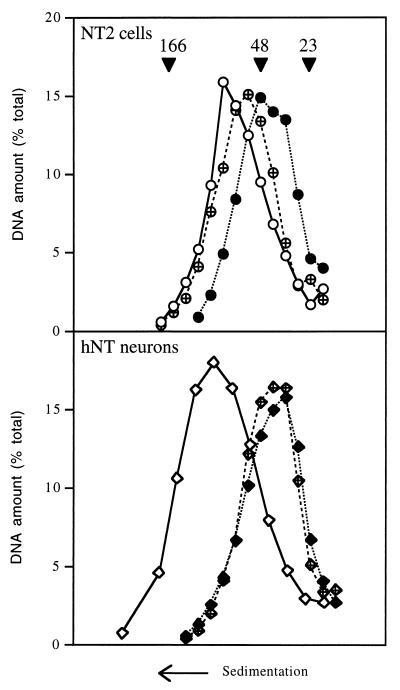
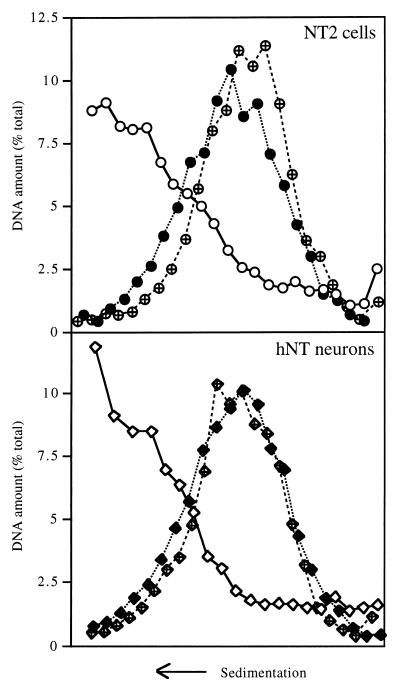
We first evaluated the status of the NER system at the global genome level in the two cell types by measuring the repair of CPDs. Cells were irradiated with UV light or not irradiated, and their DNA was extracted and digested with T4 endo V, an enzyme that nicks the DNA backbone wherever it contains a CPD. The DNA was then analyzed by sedimentation on alkaline sucrose gradients (Fig. 1). The nicked DNA sedimented more slowly than undamaged DNA did. When NT2 cells were allowed 24 h after irradiation to repair their DNA, the resulting peak shifted back toward the position of undamaged DNA, indicating that these cells repaired a significant fraction of the CPD lesions induced by UV irradiation. In striking contrast, no such shift was observed when the same experiment was carried out with hNT neurons. Calculations based upon the average size of the DNA fragments indicated that the average distance between CPDs was 25.6 kbp in NT2 cells immediately after irradiation and increased to 322.3 kbp within 24 h, which means that over 90% of the lesions were repaired. By contrast, the average distance between CPDs in hNT neurons was 14.7 kbp upon irradiation and 17.0 kbp after 24 h, which indicates that only 14% of the CPDs were removed.
FIG. 1.
Global genome repair of CPDs in NT2 precursor cells and hNT neurons. NT2 cells and hNT neurons were either not irradiated (open symbols) or irradiated with 254-nm UV light at 10 J/m2 and harvested immediately (solid symbols) or 24 h after irradiation (crossed symbols). Total DNA was digested with T4 endo V, fractionated on alkaline sucrose gradients, slot blotted, and detected with a 32P-labeled genomic probe. The arrowheads indicate the positions of the molecular size markers: bacteriophage T2 (166 kbp), bacteriophage λ (48 kbp), and the largest fragment of HindIII-digested phage λ (23 kbp).
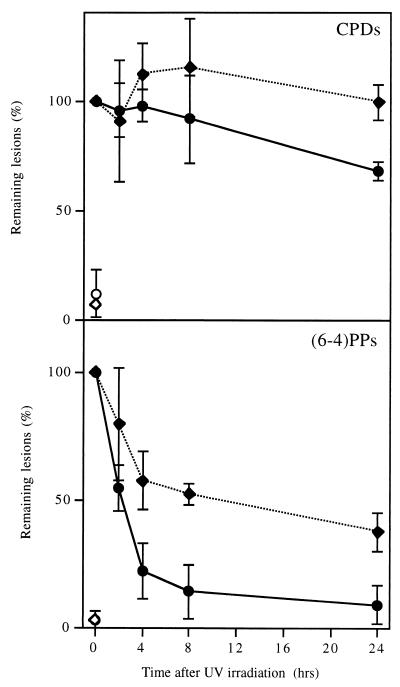
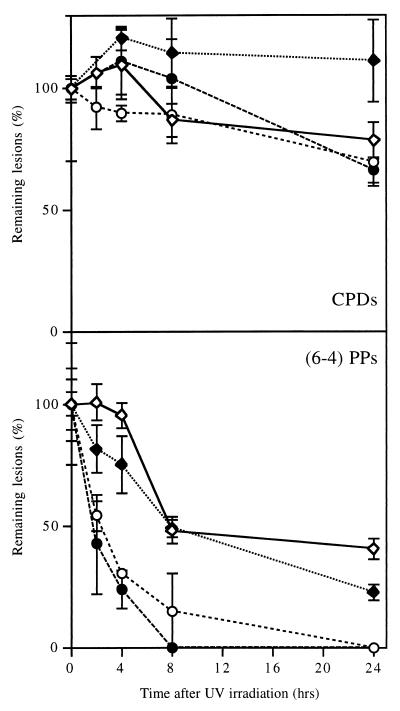
To determine whether the repair of CPDs and (6-4)PPs was equally affected by differentiation, cells were harvested at various times after irradiation with 254-nm UV light, and DNA was blotted on a nylon membrane and probed with monoclonal antibodies specific for CPDs or (6-4)PPs (22). Figure 2 shows that NT2 cells removed most of the (6-4)PPs from their DNA within a few hours whereas removal of CPDs took somewhat longer. This result is consistent with our previous observations on other cell types (5, 14), in that repair is somewhat underestimated with the anti-CPD antibody, probably due to the relatively weak specificity of this antibody. In this respect, the T4 endo V incision assay is more reliable, although it detects only CPDs. The antibody assay revealed that hNT neurons removed (6-4)PPs much more slowly than did NT2 cells and did not seem to repair CPDs at all, consistent with the results presented in Fig. 1. Experiments in which hNT neurons were allowed longer times for repair showed that (6-4)PPs were almost completely removed within 3 days whereas removal of CPDs was still not detectable by that time (data not shown). Thus, most domains of the genome did not appear to be totally inaccessible to repair enzymes, even though the repair of CPDs was severely attenuated.
FIG. 2.
Global genome repair of CPDs and (6-4)PPs in NT2 and hNT cells. DNA from NT2 cells (circles) and hNT neurons (diamonds) that were irradiated (solid symbols) or not (open symbols) was slot blotted on a nylon membrane and assayed with monoclonal antibodies specific for CPDs or (6-4)PPs. The results are the means of two to four experiments; the error bars indicate standard error of the mean.
Transcribed genes are efficiently repaired in hNT neurons.
Human cells generally repair transcribed genes with greater efficiency than the rest of their genome, a process known as TCR. The enhanced repair is confined to the transcribed strand of active genes, with the nontranscribed strand being only marginally better repaired than inactive DNA (10, 28).
To determine whether TCR was impaired in hNT neurons, genomic DNA was isolated from NT2 and hNT cells at various times after irradiation with UV light. The DNA was sequentially digested with appropriate restriction enzymes and with T4 endo V, run on a denaturing gel, subjected to Southern transfer, and hybridized with riboprobes specific for the transcribed or nontranscribed strand of the gene of interest. The treatment with T4 endo V resulted in nicking of the restriction fragments that contained at least one CPD. The percentage of intact fragments was determined by quantifying the full-size band and comparing it with a lane in which the T4 endo V treatment had been omitted. The Poisson expression was then used to derive the total number of CPDs present at various times after irradiation and hence the percentage that was repaired (25).
This analysis was performed on five different genes. The DHFR gene is transcribed in both cell lines, although transcription is twofold higher in NT2 than in hNT (see Fig. 7). The glucagon gene is not expressed in either cell type. The NF-L gene encodes the large subunit of the neurofilament complex; it is silent in NT2 cells but vigorously induced by differentiation. Both cytokeratin genes CK8 and CK18 are expressed in both cell types and at essentially similar levels.
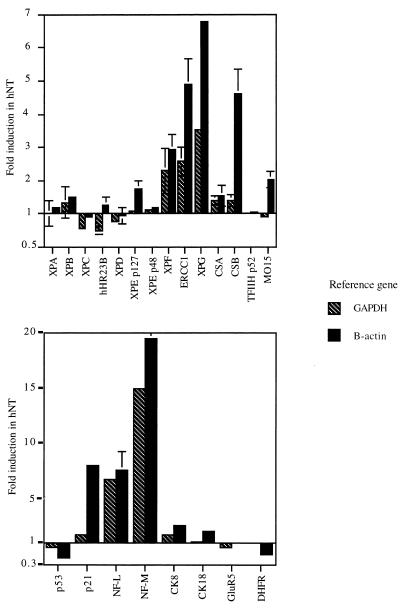
FIG. 7.
Effect of differentiation on gene expression. Total RNA from NT2 precursor cells and hNT neurons was reverse transcribed, and quantitative PCRs were performed for repair genes (top panel) or nonrepair genes (bottom panel) (note the change in scale). Normalization was achieved by coamplifying a reference gene, either the GAPDH or β-actin gene. Bars show the means of one to six measurements, depending on the gene; the error bars indicate standard error of the mean. Data are expressed as the ratio of expression in hNT neurons to that in NT2 cells.
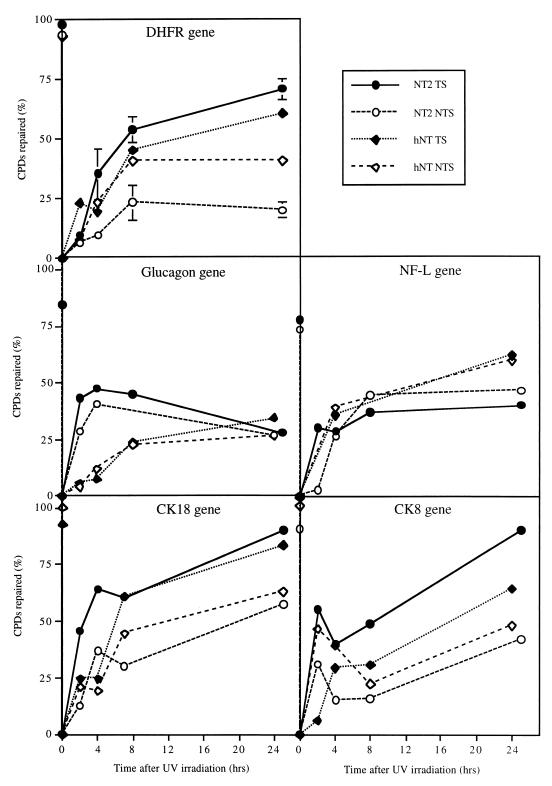
Figure 3 summarizes the results of our TCR analyses. It can be seen that although NT2 cells repaired both strands of active genes efficiently, the transcribed strand was better repaired than the nontranscribed one. This strand bias is typical of TCR and occurs because although both strands are subject to the global genome repair process, transcribed DNA strands are additionally repaired by the TCR machinery. This is illustrated by the two genes that are silent in NT2 cells, glucagon and NF-L, in which both strands were repaired with the same efficiency.
FIG. 3.
TCR in NT2 and hNT cells. DNA from UV-irradiated NT2 cells and hNT neurons was prepared at various times after irradiation, digested with restriction enzymes and then with T4 endo V, and subjected to a Southern analysis with riboprobes specific for the transcribed or nontranscribed strand of the gene of interest. Quantification of the fragment resistant to T4 endo V is an indication of the number of CPDs present at that time, from which the percentage of repair can be calculated. DNA from nonirradiated cells is included as a control; the corresponding points are reported on the y axis.
The situation was somewhat different for hNT neurons, which do not exhibit significant repair at the global genome level. As expected, for the silent glucagon gene, both strands were repaired poorly compared with NT2 cells: only about 25% of the lesions were eventually removed, which is consistent with the residual repair we observed at the global genome level (14% in Fig. 1). By contrast, in the four other genes that are transcribed in hNT neurons, the transcribed strand was repaired as well as in NT2 cells, indicating that TCR is proficient in these cells. The surprising finding was that the nontranscribed strand was also proficiently repaired in hNT neurons, at least to the same extent as in NT2 cells, and in some cases (i.e., DHFR and NF-L genes) was repaired as well as the transcribed strand. This lack of strand bias should not be mistaken for deficient TCR, however; in such a situation, both strands would be poorly repaired, as was the case for the silent glucagon gene. We observed the opposite for the expressed genes: both strands were repaired as efficiently as or even better than in NT2 cells. It is difficult to determine whether the gene-to-gene variations shown in Fig. 3 are due to heterogeneity in repair at the gene level or to experimental error, since the limited availability of hNT neurons did not allow us to accumulate enough data to run statistical tests. The data presented only allow us to conclude that the nontranscribed strand is proficiently repaired in active genes in hNT neurons and thus does not reflect the overall poor repair at the global genome level.
The nontranscribed strand does not become transcribed in hNT neurons.
One main concern, in view of our above results, is that there could be another transcription unit, in the opposite direction, downstream from the genes we studied. A tissue-specific activation of that unit in neurons would result in transcription of the previously nontranscribed strand, which could then account for the unusually efficient repair we observed.
To examine this possibility, we reverse transcribed RNA from NT2 or hNT cells, using primers specific for the DHFR or CK18 mRNA or for a putative antisense RNA that could result from the transcription of the nontranscribed strand in these genes. Semiquantitative PCR was then performed, with primers nested within the one used for RT. To our surprise, even in the absence of any primer, the RT yielded a detectable level of cDNA. This could have been due to secondary structures in the mRNA that would serve as self-primers or to impurities (e.g., oligonucleotides) in the preparation of reverse transcriptase. The first explanation seems more likely, since this phenomenon was much more marked with the DHFR gene than with the CK18 gene.
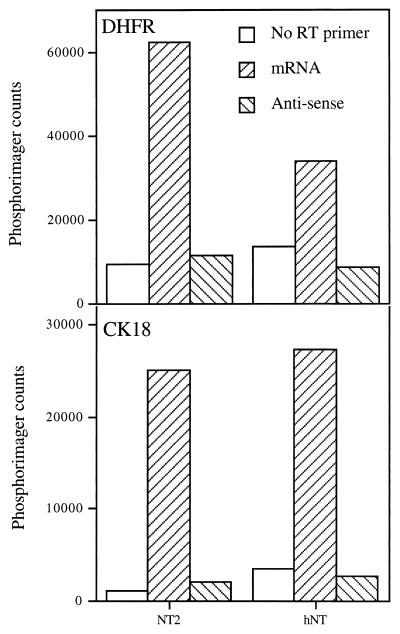
We were able to partially solve the problem by using a two-step approach. First, RT was performed in the absence of any primer, with a small amount of dideoxy-ATP. This was meant to quickly terminate the elongation of any nonspecific RT product. Then the dideoxy-ATP was diluted with an excess of normal deoxynucleotides, and RT was continued with the strand-specific primer of interest. Under these conditions, the background signal was reduced to manageable levels, and we detected no transcription products that could have originated from the nontranscribed strand of either the DHFR or the CK18 gene (Fig. 4).
FIG. 4.
Search for transcription products from either strand of the DHFR and CK18 genes. Total RNA from NT2 or hNT cells was reverse transcribed with primers specific for the DHFR or CK18 mRNA or for putative antisense RNA generated by transcription of the nontranscribed strand of these genes. As a control, RT reactions were performed with no primer. The resulting cDNA was then measured by semiquantitative PCR.
The defect in global genome repair is not due to a more compact chromatin structure.
To determine whether a more compact chromatin structure in hNT neurons could prevent repair enzymes from reaching the lesions, cells were UV irradiated, permeabilized, treated (or not) with 2 M NaCl to disrupt chromatin, and then digested with T4 endo V. The cells were then lysed directly at the top of alkaline sucrose gradients containing 2 M NaCl. After centrifugation, DNA was quantified in the fractions collected from the gradients. The sedimentation profiles obtained in this way reflect the number of UV-induced lesions that were incised by T4 endo V. As expected, the high-salt treatment unmasked a few lesions that were not previously accessible to the T4 endo V (Fig. 5). However, this unmasking effect was not any stronger in hNT neurons than in NT2 cells, indicating that chromatin was not more of an obstacle for T4 endo V accessibility to CPDs in hNT neurons than in the precursor cells.
FIG. 5.
Detection of CPDs by T4 endo V in NT2 and hNT cells. NT2 cells and hNT neurons were either not irradiated (open symbols) or irradiated (solid symbols) with 254-nm UV light at 10 J/m2, permeabilized, treated with T4 endo V, and lysed at the top of alkaline sucrose gradients. The DNA in gradient fractions was blotted and assayed with a 32P-labeled genomic probe. In some cases (crossed symbols), chromatin was disrupted by treating the cells with high salt prior to T4 endo V digestion.
Since it is conceivable that a small enzyme such as T4 endo V would not be impaired by chromatin structures that would be an obstacle for a huge “repairosome,” we cross-checked these results using a different approach. We made use of trichostatin A, a specific inhibitor of histone deacetylases, to destabilize chromatin and hopefully make neuronal DNA more accessible for repair enzymes. Preliminary experiments (results not shown) were conducted to determine the sublethal range of trichostatin A concentrations, and subsequent experiments were performed with trichostatin A at 25 to 50 μg/ml. In another pilot experiment, cells were treated with trichostatin A before being UV irradiated and subjected to the T4 endo V assay described in the previous paragraph. The resulting DNA profile (not shown) peaked in a position intermediate between that of DNA from untreated cells and that of DNA from cells treated with high salt. We took this as an indication that trichostatin A facilitates the access of T4 endonuclease V to the DNA, although not to the same extent as high salt does.
DNA from UV-irradiated cells pretreated with trichostatin A for 6 h or not pretreated was blotted onto a nylon membrane and assayed for the presence of CPDs and (6-4)PPs by using specific monoclonal antibodies (Fig. 6). Treatment with trichostatin A did not improve the repair of CPDs in either cell type. There was marginal improvement in the removal of (6-4)PPs after trichostatin A treatment, although one could argue whether this effect was significant. In any case, it was no more marked in hNT neurons than in NT2 precursor cells, a result that would not have been expected if chromatin structure changes had been responsible for the poor repair in neurons. In other words, destabilizing chromatin in hNT neurons did not cause their global genome repair capability to revert to what it had been prior to differentiation.
FIG. 6.
Effect of trichostatin A on the repair of CPDs and (6-4)PPs. DNA from NT2 cells (circles) and hNT neurons (diamonds) harvested at various times after UV irradiation was blotted onto a nylon membrane and assayed with monoclonal antibodies specific for CPDs or (6-4)PPs. For 6 h prior to irradiation, the cells were treated (solid symbols) or not treated (open symbols) with trichostatin A at 25 μg/ml for (6-4)PPs and 50 μg/ml for CPDs. Results are taken from two experiments performed in triplicate; the error bars indicate standard deviation.
Effect of differentiation on gene expression.
Quantitative RT-PCR experiments were performed to determine whether the changes in repair phenotype observed during differentiation might be explained by modulation in the expression of repair genes. The mRNA for the relevant genes was reverse transcribed and coamplified with a reference gene, either a glyceraldehyde phosphate dehydrogenase (GAPDH) or a β-actin gene, to allow precise comparisons between expression in NT2 and hNT cells. Figure 7 summarizes the results of these analyses for genes associated with repair (top panel) and other genes (bottom panel). It can be seen that expression of most repair genes remained unaffected by the differentiation process, with the main exceptions being the two NER incision factors, XPG (induced four- to sevenfold) and ERCC1/XPF (induced two- to fivefold). More modestly induced genes were CSB and MO15, although there was some discrepancy between the results obtained with GAPDH and β-actin as a control. In fact, in most cases, the use of β-actin as a control provided higher induction values than did the use of GAPDH. This is probably because the expression of one or both of these reference genes is affected by differentiation. Finally, because the XPC-hHR23B complex is necessary for the global genome repair subpathway, it is worth noting that both XPC and hHR23B were repressed to about 70% of the control values.
Among the genes not directly related to DNA repair, the neuron-specific neurofilament genes NF-L and NF-M were massively induced, as expected. The DHFR and p53 genes were repressed about twofold, whereas p21/WAF was induced two- to sevenfold.
DISCUSSION
The analysis of DNA metabolism in terminally differentiated cells poses quite a challenge to the experimenter since these cells do not grow in vitro. Resorting to primary cells is not easily done for human cells, particularly for neurons. The NT2-hNT cell system therefore represents a suitable compromise: the NT2 precursor cells, although neoplastic, are likely to present a reasonably normal phenotype, and the hNT neurons, resulting from the differentiation process, are very similar to normal neurons. In particular, hNT cells express neuron-specific genes (Fig. 7) and histological markers. They have been widely used for biochemical, cytological, and electrophysiological studies involving neurons (12, 18). The main drawback of this system is the low yield of hNT neurons, which limits the number and feasibility of some types of experiments.
Using the NT2-hNT model system for differentiating human neurons, we showed that upon differentiation, global genomic repair of CPDs was markedly decreased in hNT neurons (Fig. 1). A more detailed analysis, distinguishing CPDs from (6-4)PPs with specific monoclonal antibodies (Fig. 2), allowed us to detect low levels of repair for (6-4)PPs: nearly complete removal was achieved after 3 days in hNT neurons, whereas it was substantially completed within hours in NT2 cells. Since it is well known that (6-4)PPs are repaired faster than CPDs (31), it is conceivable that CPDs could be completely removed after several weeks. Unfortunately, this was impossible to assay since hNT cultures are never completely free of precursor cells: initially the proportion of NT2 cells is less than 5%, but because those cells replicate, they rapidly outnumber the hNT neurons. Another explanation for the residual repair of (6-4)PPs in hNT neurons is that they could be more accessible than CPDs to the repair complex, since they are concentrated in the linker regions between nucleosome cores (8).
Repair of transcribed genes, on the other hand, was fully functional in hNT neurons (Fig. 3). One could rationalize that since neurons do not need to replicate their DNA, they do not need to repair the bulk of their genome and therefore can concentrate on the genes that are expressed and essential for their function. In addition, it seems that hNT neurons display a peculiar transcription-coupled repair phenotype, in that they repair the nontranscribed strand better than would be expected for cells that are deficient in CPD repair in the global genome overall. A possible explanation for the latter observation is that a neuron-specific promoter triggers transcription of the opposite strand in hNT neurons. One can even envision that this could be a general strategy used by terminally differentiated cells to increase the repair of the nontranscribed strand in critical genes. We thus made use of a sensitive RT-PCR approach to try to detect such transcription products (Fig. 4). Under those conditions, we could not detect any transcription of the nontranscribed strand in hNT neurons.
By definition, TCR occurs only in the transcribed strand; the nontranscribed strand is supposedly dealt with by global genome repair (10). However, in hNT neurons, the global genome repair is greatly impaired whereas the repair of the nontranscribed strand is proficient in active genes, which calls for a different explanation. This phenomenon, which we tentatively have termed differentiation-associated repair (DAR), seems to be confined to transcribed genes, as judged by the poor repair of the silent glucagon gene in hNT neurons. Such a situation was previously encountered in two rodent model systems, rat myoblasts differentiating to myocytes (13) and rat PC12 pheochromocytoma cells that display a neuron-like phenotype upon differentiation (reviewed in reference 11). Human HL60 promyelocytic leukemia cells also showed an improved repair of the nontranscribed strand in the c-myc gene after differentiation (14). By contrast, different results were reported for Swiss mouse 3T3 cells, which, when differentiating into adipocytes, exhibited reduced unscheduled DNA synthesis (a measure of repair) (27) but also lost the capacity to selectively repair transcribed genes (2). However, strand-specific repair was not assayed in that study (2). To our knowledge, this is the first time DAR has been described in human neurons.
It is known that terminal differentiation in neurons is concomitant with important changes in the composition and structure of the chromatin, involving different histone variants and nucleosome repositioning (17). Thus, a possible explanation for our observations is that the chromatin structure in neurons is somewhat denser, which would make it more difficult for repair enzymes to access the lesions. By contrast, active genes would be exposed by the transcription process and both their strands could benefit from the increased availability of repair enzymes, relieved from their overall global genome repair commitment. However, we have not been able to validate such a model, even using two different approaches. First, the enzyme T4 endo V accessed CPDs in hNT neurons just as well as in NT2 cells (Fig. 5). Furthermore, a chromatin-destabilizing compound, trichostatin A, did not improve global genome repair in hNT neurons (Fig. 6). This suggests that the defect in global genome repair observed in hNT neurons is not due to a denser chromatin structure.
Another possibility is that differentiation affects the expression of one or more repair genes. For instance, a massive decrease in XPC protein could explain the poor global genome repair, mimicking the situation observed in XPC cells in which global genomic repair is defective but TCR is normal (30). It was not possible to measure protein levels directly for all the known repair enzymes because of the small amount of material that could be obtained from hNT cultures and because antibodies are not available for each enzyme. Therefore, we used a quantitative RT-PCR approach, keeping in mind the limitation that it would not detect any posttranscriptional regulation. Our measurements (Fig. 7) revealed only a small decrease in the mRNA levels of XPC and of its partner hHR23B. Such a modest change is not likely to be the cause of the massive impairment in global genome repair we have observed.
On the other hand, several repair genes were found to be up-regulated by differentiation, mainly the two NER structure-specific nucleases, XPG and XPF/ERCC1. The current view of the molecular mechanism for NER is that a bulky lesion in the DNA, whether a UV-induced dimer or a chemical adduct, is recognized by a complex of proteins involving XPC, XPA, possibly XPE, and the single-strand-DNA-binding protein RPA. A denaturation bubble is created around the lesion with the help of the helicases XPB and XPD, both of which are constituents of the TFIIH general transcription factor. XPG then incises the damaged strand 3′ to the bubble, and the heterodimer XPF/ERCC1 cuts it on the 5′ side, allowing the removal of a 27- to 29-nucleotide fragment containing the lesion. The resulting gap can now be sealed by DNA polymerase δ or ɛ and DNA ligase I. Although it is hard to imagine how an increase in the concentration of the two incision enzymes would result in a decrease in global genome repair, it could well contribute to the better repair of the nontranscribed strand in transcriptionally active genes. If this were true, it would imply that the limiting step for NER is not simply lesion recognition but, rather, incision of the DNA backbone. Indeed, it makes sense that lesion recognition would be very sensitive but poorly specific whereas incision, the first irreversible step, would occur only when the presence of a lesion has been verified.
It has been shown in this laboratory that p53-deficient cells exhibit an impairment in global genome repair of CPDs while retaining their capabilities for TCR (5, 6). This phenotype is similar to what we have observed in neurons, raising the possibility that p53 plays a role in DAR. Our RT-PCR quantification experiments with hNT neurons indeed revealed a 50% decrease in the level p53 mRNA but at the same time an increase in the level of p21/WAF, whose promoter is one of the targets of p53 (1). Since it is known that marginal amounts of p53 generally result in normal repair of (6-4)PPs (5), a twofold decrease is not likely to be responsible for the impaired global genome repair in hNT neurons. On the other hand, although p21/WAF can also be activated by a p53-independent pathway, its induction in hNT neurons strongly suggests a posttranscriptional activation of p53 that would stimulate the transcription of several target genes, including p21/WAF. A feedback mechanism, such as the known mdm2/p53 feedback loop (1), could be responsible for the subsequent transcriptional repression of p53. In fact, such a situation has been observed in several other terminally differentiated cell types such as myocytes and keratinocytes (20, 26, 32), in which p53 mRNA and, sometimes, protein levels are lower but the p53 activity is higher than that in precursor cells. Thus, it appears that p53 is generally activated posttranscriptionally upon differentiation. However, this activation is not expected to trigger the apoptosis pathway, since this would be a disastrous option for cells that do not multiply and cannot be replaced. Indeed, we have observed by microscopic examination or trypan blue counting that hNT neurons appear to be more resistant than NT2 cells to killing by UV radiation (data not shown).
We are currently investigating the possibility that this repair phenotype in differentiated neurons is not limited to NER and that other repair systems, such as base excision repair, are also affected. This would provide an alternative explanation for the spectacular increase in the level of XPG mRNA, which was even more striking than those of ERCC1 and XPF: coamplification of XPG with XPF indicated that the XPG/XPF ratio increases about twofold upon differentiation (data not shown). It has been recently shown that XPG, unlike other NER enzymes, is also involved in the removal of oxidative lesions that are traditionally thought to be repaired by base excision repair (although NER is also able to remove this type of damage [24]). Furthermore, XPG has been implicated in the TCR of such lesions (3, 23) and stimulates base excision repair in vitro (16). Neurons have a high cellular metabolism (9), and it is therefore likely that oxidative lesions are more of a challenge for them than are UV-induced lesions or bulky adducts. Stimulating the transcription of XPG and possibly other genes involved in base excision repair might be an important strategy to cope with a higher level of oxidative lesions. In this respect, the fact that about half the xeroderma pigmentosum group G patients also suffer from Cockayne syndrome, which includes severe neurological problems, emphasizes the important role of XPG in neurons.
Similarly, turning off repair at the global genome level to concentrate on the repair of transcribed genes may be the best strategy to deal with the continuing induction of damage in postmitotic cells. However, this strategy would result in poor repair of the nontranscribed strand in these genes, since this strand is repaired by the same mechanism as that for the bulk of the genome. Since the nontranscribed strand will be needed as a template for repairing the transcribed strand, it should not be allowed to accumulate damage for decades. If it were, there would be a significant probability that a lesion in the nontranscribed strand could interfere with the synthesis of a repair patch, within 29 nucleotides around a lesion in the transcribed strand. The NER machinery would then introduce a mutation in the transcribed strand when repairing it, a potentially disastrous situation. It therefore makes sense that a special mechanism, such as DAR, might ensure that the nontranscribed strand will be equally well repaired in terminally differentiated cells. It has often been suggested that neuron aging and some forms of dementia could be due to the accumulation of unrepaired DNA lesions, which would eventually interfere with neuronal function (9, 19). The mechanisms we suggest could be a way for normal neurons to delay, if not preclude, such events.
ACKNOWLEDGMENTS
We are indebted to Peter Andrews for providing us with NT2 cells and useful advice on how to grow them. We gratefully thank Toshio Mori for the gift of anti-CPD and anti-(6-4)PPs antibodies and Stephen Lloyd for the gift of T4 endonuclease V. We thank C. A. Smith and A. K. Ganesan for advice and helpful discussions.
This work was supported by grants from the Swiss National Science Foundation (823-046695) and from the Novartis Jubilaum Stiftung to T.N. and an Outstanding Investigator Grant, CA44349, from the National Cancer Institute to P.C.H.
REFERENCES
- 1.Bates S, Vousden K H. p53 in signalling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:12–19. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 2.Bill C A, Grochan B M, Meyn R E, Bohr V A, Tofilon P J. Loss of intragenomic DNA repair heterogeneity with cellular differentiation. J Biol Chem. 1991;266:21821–21826. [PubMed] [Google Scholar]
- 3.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from xeroderma pigmentosum group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 4.Enoch E, Norbury C. Cellular responses to DNA damage: cell-cycle checkpoints, apoptosis and the roles of p53 and ATM. Trends Biol Sci. 1995;20:426–430. doi: 10.1016/s0968-0004(00)89093-3. [DOI] [PubMed] [Google Scholar]
- 5.Ford J, Hanawalt P C. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 6.Ford J M, Hanawalt P C. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 8.Gale J M, Smerdon M J. UV induced (6-4) photoproducts are distributed differently than cyclobutane dimers in nucleosomes. Photochemi Photobiol. 1990;51:411–417. doi: 10.1111/j.1751-1097.1990.tb01732.x. [DOI] [PubMed] [Google Scholar]
- 9.Gensler H L, Bernstein H. DNA damage as the primary cause of aging. Q Rev Biol. 1981;56:279–303. doi: 10.1086/412317. [DOI] [PubMed] [Google Scholar]
- 10.Hanawalt P C. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 11.Hanawalt P C, Gee P, Ho L, Hsu R K, Kane C J M. Genomic heterogeneity of DNA repair. Role in aging? Ann N Y Acad Sci. 1992;663:17–25. doi: 10.1111/j.1749-6632.1992.tb38644.x. [DOI] [PubMed] [Google Scholar]
- 12.Hardy M, Younkin D, Tang C-M, Pleasure J, Shi Q-Y, Williams M, Pleasure D. Expression of non-NMDA glutamate receptor chanel genes by clonal human neurons. J Neurochem. 1994;63:482–489. doi: 10.1046/j.1471-4159.1994.63020482.x. [DOI] [PubMed] [Google Scholar]
- 13.Ho L, Hanawalt P C. Gene-specific DNA repair in terminally differentiating rat myoblasts. Mutat Res DNA Repair. 1991;255:124–141. doi: 10.1016/0921-8777(91)90047-s. [DOI] [PubMed] [Google Scholar]
- 14.Islas A, Hanawalt P C. DNA repair in the MYC and FMS proto-oncogenes in ultraviolet light-irradiated human HL60 promyelocytic cells during differentiation. Cancer Res. 1995;55:336–341. [PubMed] [Google Scholar]
- 15.Jensen L, Linn S. A reduced rate of bulky DNA adduct removal is coincident with differentiation of human neuroblastoma cells induced by nerve growth factor. Mol Cell Biol. 1988;8:3964–3968. doi: 10.1128/mcb.8.9.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klungland A, Höss M, Gunz D, Constantinou A, Clarkson S G, Doetsch P W, Bolton P H, Wood R D, Lindahl T. Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 17.Kuenzle C C, Heizmann C W, Hubscher U, Hobi R, Winkler G C, Jaeger A W, Morgenegg G. Chromatin changes accompanying neuronal differentiation. Cold Spring Harbor Symp Quant Biol. 1983;48:493–499. doi: 10.1101/sqb.1983.048.01.054. [DOI] [PubMed] [Google Scholar]
- 18.Lee V M Y, Andrews P W. Differentiation of NTERA-2 clonal human embryonal carcinoma cells into neurons involves the induction of all three neurofilament proteins. J Neurosci. 1986;6:514–521. doi: 10.1523/JNEUROSCI.06-02-00514.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 20.Lutzker S, Levine A. A functionally inactive p53 protein in teratocarcinoma cells is activated by either DNA damage or cellular differentiation. Nat Med. 1996;2:804–810. doi: 10.1038/nm0796-804. [DOI] [PubMed] [Google Scholar]
- 21.McCombe P, Lavin M, Kidson C. Control of DNA repair linked to neuroblastoma differentiation. Int J Radiat Biol Relat Stud Phys Chem Med. 1976;29:523–531. doi: 10.1080/09553007614550621. [DOI] [PubMed] [Google Scholar]
- 22.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4) photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 23.Nouspikel T, Lalle P, Leadon S A, Cooper P K, Clarkson S G. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc Natl Acad Sci USA. 1997;94:3116–3121. doi: 10.1073/pnas.94.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Scott A D, Neishabury M, Jones D H, Reed S H, Boiteux S, Waters R. Spontaneous mutation, oxidative DNA damage, and the roles of base and nucleotide excision repair in the yeast Saccharomyces cerevisiae. Yeast. 1999;15:205–218. doi: 10.1002/(SICI)1097-0061(199902)15:3<205::AID-YEA361>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 25.Spivak G, Hanawalt P C. Determination of damage and repair in specific DNA sequences. Gene Methods Companion Methods Enzymol. 1995;7:147–161. [Google Scholar]
- 26.Tamir Y, Bengal E. p53 protein is activated during muscle differentiation and participates with MyoD in the transcription of muscle creatine kinase gene. Oncogene. 1998;17:347–356. doi: 10.1038/sj.onc.1201929. [DOI] [PubMed] [Google Scholar]
- 27.Tofilon P J, Meyn R E. Influence of cellular differentiation on repair of ultraviolet-induced DNA damage in murine proadipocytes. Radiat Res. 1988;116:217–237. [PubMed] [Google Scholar]
- 28.van Hoffen A, Natarajan A T, Mayne L V, van Zeeland A A, Mullenders L H, Venema J. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Zeeland A A, Smith C A, Hanawalt P C. Sensitive determination of pyrimidine dimers in DNA of UV-irradiated mammalian cells. Introduction of T4 endonuclease V into frozen and thawed cells. Mutat Res. 1981;82:173–189. doi: 10.1016/0027-5107(81)90148-2. [DOI] [PubMed] [Google Scholar]
- 30.Venema J, van Hoffen A, Karcagi V, Natarajan A T, van Zeeland A A, Mullenders L H F. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol Cell Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vreeswijk M P, van Hoffen A, Westland B E, Vrieling H, van Zeeland A A, Mullenders L H. Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts in transcriptionally active and inactive genes in Chinese hamster cells. J Biol Chem. 1994;269:31858–31863. [PubMed] [Google Scholar]
- 32.Weinberg W, Azzoli C, Chapman K, Levine A, Yuspa S. p53-mediated transcriptional activity increases in differentiating epidermal keratinocytes in association with decreased p53 protein. Oncogene. 1995;10:2271–2279. [PubMed] [Google Scholar]