Abstract
The regulation of cap-independent translation directed by the internal ribosome entry sites (IRESs) present in some viral and cellular RNAs is poorly understood. Polypyrimidine-tract binding protein (PTB) binds specifically to several viral IRESs. IRES-directed translation may be reduced in cell-free systems that are depleted of PTB and restored by reconstitution of lysates with recombinant PTB. However, there are no data concerning the effects of PTB on IRES-directed translation in vivo. We transfected cells with plasmids expressing dicistronic transcripts in which the upstream cistron encoded PTB or PTB deletion mutants (including a null mutant lacking amino acid residues 87 to 531). The downstream cistron encoded a reporter protein (chloramphenicol acetyltransferase [CAT]) under translational control of the poliovirus IRES which was placed within the intercistronic space. In transfected BS-C-1 cells, transcripts expressing wild-type PTB produced 12-fold more reporter protein than similar transcripts encoding the PTB null mutant. There was a 2.4-fold difference in CAT produced from these transcripts in HeLa cells, which contain a greater natural abundance of PTB. PTB similarly stimulated CAT production from transcripts containing the IRES of hepatitis A virus or hepatitis C virus in BS-C-1 cells and Huh-7 cells (37- to 44-fold increase and 5 to 5.3-fold increase, respectively). Since PTB had no quantitative or qualitative effect on transcription from these plasmids, we conclude that PTB stimulates translation of representative picornaviral and flaviviral RNAs in vivo. This is likely to reflect the stabilization of higher ordered RNA structures within the IRES and was not observed with PTB mutants lacking RNA recognition motifs located in the C-terminal third of the molecule.
The cap-independent initiation of translation on the plus-sense genomic RNAs of picornaviruses and some flaviviruses requires binding of the small ribosome subunit to the RNA at a site located hundreds of nucleotides downstream of its 5′ terminus (34, 54, 75). This interaction is controlled by highly structured, cis-acting RNA elements located within the 5′ nontranslated region (5′NTR), the internal ribosome entry site (IRES). IRES elements were first identified within viral RNAs but have been increasingly recognized within the lengthy 5′NTRs of cellular mRNAs encoding certain critical mammalian growth factors, including fibroblast growth factor 2 (76), human insulin-like growth factor II (74), and platelet-derived growth factor B (4), as well as eukaryotic translation initiation factor 4G (eIF4G) (19). In directing the internal initiation of translation, these viral and cellular IRES elements function in concert with both canonical and possibly noncanonical trans-acting cellular translation initiation factors. Factors influencing the efficiency of internal ribosome entry directed by IRESs are likely to play important roles in determining the cellular tropisms of some viruses or the posttranscriptional regulation of proteins translated under the control of a cellular IRES. However, these factors remain incompletely characterized.
Like all picornaviruses, poliovirus and hepatitis A virus (HAV) contain functional IRES elements within their 5′NTRs (11, 54). However, these two picornaviral IRESs share negligible primary sequence homology and have distinctly different predicted secondary RNA structures (11, 60). They can be considered representative of two major structural classes of IRES elements that are found within the Picornaviridae (11, 59, 60, 79). Hepatitis C virus (HCV) is another positive-stranded RNA virus which is classified within the family Flaviviridae (16). The 5′NTR of HCV also contains an IRES which directs the translation of the HCV polyprotein in a cap-independent fashion (30, 63, 75). Although HCV and HAV both replicate predominantly if not exclusively within hepatocytes of infected humans, the IRES elements of these viruses have no sequences in common, nor have they been shown to contain conserved higher-ordered structures (11, 13). The HCV IRES is thus representative of a third structural class of viral IRES elements (40).
Relatively little is known of the specific mechanisms by which IRES elements direct the internal entry of the 40S ribosome subunit on their RNAs, although it is apparent that higher-ordered RNA structures within viral IRESs are essential to this process. Some requirements may also exist for short conserved primary nucleotide sequences among both picornaviral and flaviviral IRESs (29, 61), but the secondary and tertiary RNA structure (i.e., conformation) of the IRES generally appears to be more important (11, 29, 59, 60). It is also clear that IRES elements require trans-acting cellular factors to direct translation. Each of the canonical translation initiation factors, with the probable exception of intact eIF4G, appear necessary, at least for picornaviral IRESs (41, 53, 67). However, in vitro studies also suggest important roles for one or more cellular proteins that do not typically participate in the cap-dependent translation of cellular mRNAs (6–8, 28, 31, 32, 37, 48–50). Such trans-acting cellular factors presumably interact specifically with the cis-acting RNA structures within the IRES (47, 50, 57, 81). Since translation of the viral polyprotein is an essential step in viral replication, the intracellular abundance of these factors may be an important determinant of the extent to which specific cell types are permissive for virus replication.
Efficient and accurate translation directed by the poliovirus (or closely related rhinovirus) IRES in rabbit reticulocyte lysate is dependent on supplementation with cellular extracts or ribosomal salt washes prepared from poliovirus-permissive HeLa cells (10). Similarly, an as yet unconfirmed report suggests that translation mediated by the HAV IRES in reticulocyte lysates may be enhanced by the addition of cytoplasmic extracts from mouse liver (23). In addition, available data strongly support the existence of one or more cell-type-specific proteins which facilitate (or impede) cap-independent translation directed by the HAV IRES in cultured cells (17, 18, 69). Mutations which confer cell-type-specific differences in viral translation and replication have also been described within the poliovirus IRES (70). However, the cell-type-specific translation factors responsible for these differences are unknown.
A 52-kDa protein, La, is present at relatively high levels within HeLa cell extracts and binds specifically to the poliovirus 5′NTR (47). It may be the cellular factor responsible for the correction of aberrant translation of poliovirus RNA in reticulocyte lysates (47, 48). La is a predominantly nuclear protein. However, it is redirected to the cytoplasm, the site of virus replication, in poliovirus-infected cells (48). Poly(rC) binding protein 2 (PCBP2) also binds specifically to the poliovirus IRES and appears to be an essential factor required for efficient translation of poliovirus RNA in HeLa cells (6). In addition to La and PCBP2, the 57-kDa polypyrimidine-tract binding protein (PTB; also known as hnRNP I) binds specifically to the IRES sequences of several picornaviruses, including poliovirus and HAV (15, 27, 44, 80). A specific interaction with the HCV IRES has also been claimed (2), although PTB has greater affinity for the 3′NTR as well as a segment within the 5′-proximal open reading frame of HCV RNA (33). Like La, PTB is a predominantly nuclear protein, although it is present in lower abundance in the cytoplasm of many cells. PTB preferentially binds RNAs containing pyrimidine-rich tracts and binds with greatest affinity to pyrimidine-rich RNA tracts containing the core sequence UCUU or UCUUC (20, 52, 56, 71). Its normal cellular functions are incompletely defined, but there is increasing evidence that PTB plays a role in 3′ splice site selection, spliceosome assembly on pre-mRNAs, and the regulation of pre-mRNA splicing (22, 24, 42, 43, 52, 73, 77). Despite evidence that PTB binds to pyrimidine-rich segments of viral IRESs, its functional involvement in viral translation has been controversial.
The depletion of PTB from translationally active cell lysates may impair translation directed by both picornaviral and flaviviral IRESs but generally does not hinder translation of cellular mRNAs (2, 8, 28, 37, 49). However, both the IRES of Theiler's murine encephalitis virus, which is closely related both in structure and sequence to the IRES of encephalomyocarditis virus (EMCV) (59), another picornavirus, and the IRES of HCV retained good translational activity in PTB-depleted reticulocyte lysates while the EMCV IRES did not (37). More recently, PTB has been shown to stimulate translation directed by a variant EMCV IRES sequence with minimally altered secondary RNA structure (38). Two groups of investigators noted that IRES-dependent translation could not be restored by the addition of recombinant PTB to lysates which had been depleted of PTB by immunoadsorption (2, 28). These latter results suggest that PTB had been removed as a complex with an unrelated but essential translation factor(s). In contrast, when lysates which were depleted of PTB by RNA affinity procedures were reconstituted with recombinant PTB, there was good restoration of IRES-directed translational activity (37, 49). The complexity of cellular protein interactions with IRES elements is further evidenced by the fact that unr, a cytoplasmic RNA binding protein, may interact synergistically with PTB in stimulating translation directed by the rhinovirus, but not the poliovirus, IRES in vitro (31). Nonetheless, an overriding concern is that no data have yet been presented which support a role for PTB or any other noncanonical translation factor in cap-independent translation directed by a viral or cellular IRES in vivo.
In this study, we used a novel system to demonstrate that the transient expression of PTB from the upstream cistron of dicistronic transcripts stimulates cap-independent translation of a downstream cistron directed by picornaviral (poliovirus and HAV) as well as flaviviral (HCV) IRES elements placed within the intercistronic space. We show that the enhancement of IRES-dependent translation is dependent on the C-terminal one-third of the PTB molecule, which contains two putative RNA recognition motifs (RRMs). The results suggest that there are quantitative differences in the requirements of various viral IRESs for PTB in vivo and are consistent with the possibility that PTB and possibly other cellular factors may be important determinants of the host range of these positive-strand RNA viruses. Since IRES elements also exist in some cellular mRNAs (4, 19, 46, 51, 66, 74, 76), these observations may also be relevant to posttranscriptional control of the expression of certain cellular genes.
MATERIALS AND METHODS
Plasmids.
pRc/CMV-PTB contains the complete PTB coding region preceded by the cytomegalovirus (CMV) immediate-early (IE) and T7 promoters. This was constructed by inserting the 1.6-kb EcoRI fragment comprising the human PTB coding sequence from pGemPTB (22) (generously provided by Mariano Garcia-Blanco, Duke University), ligated to HindIII linkers, into the HindIII site of the mammalian expression vector pRc/CMV (Invitrogen, San Diego, Calif.). pPwt/AC, pPwt/SC, and pPwt/CC contain dicistronic transcriptional units in which an intact upstream PTB coding sequence is separated from a downstream chloramphenicol acetyltransferase (CAT) reporter sequence by insertion of the entire 5′NTR of either HAV (nucleotides [nt] 1 to 734 of HM175/P16 virus) or Sabin type 1 poliovirus (nt 1 to 742) (Fig. 1A) or the 5′NTR and 5′-proximal 8 nt of the open reading frame of HCV (Fig. 1B), respectively. These plasmids were constructed by inserting the HindIII-NotI fragments of pHAV-CAT1 or pSAB-CAT, respectively, into pRc/CMV-PTB. The construction of plasmids pHAV-CAT1 and pSAB-CAT has been described previously (69, 78). pPwt/CC is a discistronic plasmid that contains the 5′ 353 nt of the HCV-N strain of HCV fused in-frame with the CAT sequence downstream of the PTB sequence. It was constructed by inserting the NheI-BamHI fragment of pHCV-N2 (30) into pwtCAT (64). The 5′NTR-CAT sequence in the resulting construct (HindIII-BamHI fragment) was then inserted into pRc/CMV-PTB as described above.
FIG. 1.
(A) Organization of dicistronic transcripts expressing human PTB by cap-dependent translation and expressing CAT under the translational control of an IRES within the intercistronic space. Plasmid designations appear at the left; in each, “x” is “S” for constructs containing the Sabin type 1 poliovirus 5′NTR in the intercistronic space or “A” for constructs containing the HAV 5′NTR. Transcription is under control of a composite CMV-T7 promoter. Intact PTB is expressed by pPwt/SC or pPwt/AC and contains a total of 531 amino acid residues. In pΔ87-118/xC, 32 amino acid residues have been removed from the PTB sequence by an in-frame deletion. Frameshift mutations in pΔ87-531/xC (null mutant) and pΔ361-531/xC result in termination of PTB translation at Thr86 and Val360, respectively. At the bottom is a graphical representation of the PTB molecule showing locations of the four RRMs with respect to these deletion mutations. (B) Organization of dicistronic transcripts encoding the wt or null mutant PTB upstream of the HCV IRES and a downstream CAT reporter protein. The upstream sequences are identical to those shown for the related transcripts in panel A, but the intercistronic space contains the complete HCV 5′NTR fused naturally to the 5′-most 8 nt of the HCV open reading frame (Δ Core) and in-frame CAT coding sequence.
Additional dicistronic plasmids contain mutated PTB sequences (Fig. 1) in which an internal in-frame deletion removes 32 amino acid residues (pΔ87-118/AC or pΔ87-118/SC) or frameshift mutations lead to termination of translation at residue 86 (pΔ87-531/AC, pΔ87-531/SC, and pΔ87-531/CC) or 360 (pΔ361-531/SC and pΔ361-531/AC) of the PTB sequence. pΔ87-118/AC was constructed by digestion of pPwt/AC with BstEII, removal of a 96-bp fragment (nt 268 to 363), and religation. pΔ87-531/AC was created by blunt ending the BstEII digest with Klenow enzyme prior to religation. For the construction of pΔ361-531/AC, a 0.95-kb BstXI-EcoNI PTB fragment was subcloned into pΔ355-532 (78). The resulting plasmid was digested by AccI, blunt ended with Klenow enzyme, and religated, generating a frameshift mutation at nt 1094 within the PTB coding sequence. The modified BstXI-EcoNI fragment was reinserted into pPwt/AC to create pΔ361-531/AC. pPTB-Awt-RLuc and pΔ87-531-Awt-RLuc are plasmids derived from pPwt/AC and pΔ87-531/AC, respectively, in which the wild-type (wt) HAV IRES (69) directs the translation of renilla luciferase from the downstream cistron. Plasmids pΔ87-118/SC, pΔ87-531/SC, and pΔ361-531/SC (Fig. 1A) and pΔ87-531/CC (Fig. 1B) were similarly constructed from pPwt/SC and pPwt/CC, respectively.
pCMV-2A contains the poliovirus ΔVP3-VP1-2Apro coding sequence preceded by the CMV IE and T7 promoters. The His20 residue of poliovirus 2Apro is replaced by Asn in pCMV-2A(H20N), resulting in an inactive protease (82). These plasmids were constructed by removing the 1.5-kb BamHI-HindIII fragments of plasmids pEP2A and pEP2AH20N (82) and blunt ending with Klenow enzyme. Following ligation to HindIII linkers, these fragments were inserted into the HindIII site of pRc/CMV to create pCMV-2A and pCMV-2A(H20N), respectively. All plasmid constructions were validated by restriction endonuclease analysis and/or DNA sequencing.
Cells.
The BS-C-1 cells used in these experiments were obtained from David Anderson (MacFarlane-Burnett Centre, Melbourne, Australia), as these proved more readily transfected than BS-C-1 cells that had been obtained directly from the American Type Culture Collection. Both BS-C-1 and HeLa cells were grown in 1× minimum essential medium supplemented with Earles' salts (Life Technologies Inc., Grand Island, N.Y.), glutamine, antibiotics, and 5 and 10% fetal bovine serum, respectively. Huh-7 cells were grown in Dulbecco's modified Eagle's medium (high glucose) supplemented with glutamine, antibiotics, and 10% fetal bovine serum.
In vitro transcription and translation reactions.
Plasmid DNAs were linearized by digestion with NotI. RNA transcription was carried out with T7 RNA polymerase in 100-μl reaction mixtures containing 2.5 μg of template DNA and Riboprobe Gemini System II reagents (Promega, Madison, Wis.) at 37°C for 1 h. Template DNA was removed by the addition of 2.5 U of RQ1 DNase to each reaction mixture, followed by incubation at 37°C for 30 min. RNA content was determined by spectrophotometric analysis and RNA integrity confirmed by sodium dodecyl sulfate (SDS)-agarose gel electrophoresis. Micrococcal nuclease-treated rabbit reticulocyte lysates (Promega) were programmed for translation by the addition of 1 μg of RNA per 25-μl reaction mixture containing [35S] methionine and then incubated at 30°C for 1 h. Translation products were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel and autoradiography. Translation products were quantitated by PhosphorImager analysis with ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
DNA transfections and reporter gene assays.
Cells were transfected by electroporation with a Gene Pulser apparatus (Bio-Rad, Richmond, Calif.) at a setting of 1.4 kV and 25 μF, with the pulse controller unit set at maximum resistance and two pulses. Cells (0.5 ml containing 5 × 106 cells) were transfected with 20 μg of DNA and placed at room temperature or on ice (BS-C-1 cells) for 10 min. Cells were subsequently plated into 60-mm-diameter culture dishes and placed in a 5% CO2 environment at 37°C. Alternatively, some DNA transfections were carried out by a liposome-mediated method as noted. For these transfections, nearly confluent cells grown in 60-mm-diameter plastic dishes were transfected with plasmid DNA mixed with FuGENE 6 (Boehringer Mannheim); 100 μl of OptiMEM (Gibco-BRL) and 6 μl of FuGENE reagent were incubated for 10 min at room temperature prior to the addition of plasmid DNA (2 μg). The mixture was incubated for an additional 15 min at room temperature, and 100 μl was added directly to cells fed previously with 2 ml of growth medium.
Cells were assayed for reporter gene activity approximately 48 h following transfection. For CAT assays, the cell culture medium was removed by aspiration, the cells were washed five times with phosphate-buffered saline, pH 7.5 (PBS), and 1 ml of TEN buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl) was added. After 5 min, the cells were mechanically removed from the plastic surface, and the cell suspension was transferred to a microcentrifuge tube. Cells were pelleted at 4°C for 10 min in a microcentrifuge, resuspended in 150 μl of 250 mM Tris-HCl (pH 8.0), and lysed by freeze-thawing. CAT activity in the cell lysates was determined by a phase extraction assay which quantitates butyrylated [14C]chloramphenicol products by liquid scintillation counting following xylene extraction (Promega). In the standard assay, cell lysates were incubated with CAT substrate for 18 h prior to phase extraction. Where noted, an alternative short incubation assay involved a reduced incubation period (60 min) prior to extraction. These two assays generate different absolute values for the abundance of butyrylated [14C]chloramphenicol product but comparable estimates of the relative CAT activities of cell lysates generated in individual experiments. The protein content of cell lysates was established by dotMETRIC assay (Research Products International Corp., Mount Prospect, Il.).
RNase protection assay.
RNase protection assays were carried out using reagents and protocols supplied with a lysate RNase protection kit (United States Biochemicals, Cleveland, Ohio). Transfected cell monolayers in 60-mm-diameter dishes were lysed by addition of 200 μl of a solution containing 4 M guanidine thiocyanate, 25 mM sodium citrate, and 0.5% sarcosyl. Lysates were stored at −70°C until use in hybridization reactions. The RNA probe was a 0.22-kb 32P-labeled antisense CAT probe transcribed from pRC/CMV-CAT with SP6 RNA polymerase. Reactions also included a 0.14-kb 32P-labeled human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antisense probe to monitor the amount of total cellular RNA in each lysate. These antisense probes produce protected RNA fragments of 0.18 and 0.10 kb, respectively. Probes (106 cpm) were added to lysates and incubated at 37°C for 17 h, diluted, and extensively digested with RNases and proteinase K as directed by the manufacturer. Protected RNA fragments were precipitated with isopropanol, separated on 6% denaturing polyacrylamide gels, visualized by autoradiography, and quantitated by PhosphorImager (Molecular Dynamics) analysis.
Northern blot analysis of reporter transcripts.
Total RNA was extracted from BS-C-1 cells 48 h posttransfection with FuGENE (see above), using RNAqueous (Ambion, Austin, Tex.) as recommended by the manufacturer. The poly(A)+ RNA fraction was purified from total RNA on Oligotex spin columns (Oligotex mRNA; Qiagen, Santa Clarita, Calif.), separated by electrophoresis in a formaldehyde-agarose gel, and blotted onto a BrightStar-Plus nylon membrane (NorthernMax, Ambion). The membrane was subsequently hybridized with a mixture of two different CAT-specific, [32P]CTP-labeled riboprobes, which were complementary to the 3′ 125 nt or the 300-nt EcoRI-NcoI segment of the CAT sequence, respectively. The hybridized blots were imaged on a PhosphorImager (Molecular Dynamics).
Immunofluorescence detection of PTB.
The intracellular abundance of PTB was monitored in normal and transfected cells by indirect immunofluorescence using a polyclonal rabbit antibody to a recombinant glutathione S-transferase (GST)–PTB fusion protein (generously provided by James G. Patton, Vanderbilt University). Cells were allowed to adhere to eight-chamber tissue culture chamber slides, fixed with 4% paraformaldehyde for 20 min at room temperature, and permeabilized with 0.2% Triton × 100 in PBS for 15 min at room temperature (56). Cells were incubated with a 1:100 dilution of the primary antibody for 2 h at room temperature. After extensive washing with PBS, cells were incubated with tetramethyl rhodamine isothiocyanate (TRITC)-labeled anti-rabbit immunoglobulin (Dako, Inc., Carpintiera, Calif.), diluted 1:400, for 1 h at room temperature. Following additional washes, slides were mounted in Vectashield fluid (Vector Laboratories, Burlingame, Calif.) and examined for specific fluorescence with a Nikon Eclipse E800 microscope.
Cell fractionation and immunoblot analysis.
Forty-eight hours following DNA transfection of BS-C-1 or Huh-7 cells, cells were mechanically collected, centrifuged at 500 × g, and resuspended in NP-40 lysis buffer A (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% [vol/vol] NP-40). The resuspended cells were vortexed for 10 s, and then kept on ice for 10 min. After removal of the nuclei by centrifugation, the supernatant fluid was collected as the cytoplasmic fraction. The nuclear pellet was washed in NP-40 lysis buffer once, followed by centrifugation at 500 × g to prepare a nuclear fraction. For immunoblot analysis, 10-μg aliquots of the cytoplasmic extract and corresponding nuclear fraction were separated by SDS-PAGE (12.5% gel). Following electrotransfer to polyvinylidene difluoride membranes at 100 V for 2 h, and membranes were blocked with 5% skim milk in 0.1% Tween–PBS for 1 h. Following two washes with the same buffer, membranes were probed with polyclonal rabbit anti-PTB antibody (Intronn) at a concentration of 3.2 μg/ml for 1 h. The membranes were washed twice and incubated with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G for 40 min. After thorough washing of the membranes, proteins were visualized with an enhanced chemiluminescence reagent kit (Amersham International Plc.) according to the manufacturer's recommended procedure.
RESULTS
Dicistronic reporter transcripts which encode PTB in the upstream cistron.
The cap-independent, internal initiation of translation of the picornaviral polyprotein is dependent on the interaction of trans-acting cellular factors with cis-acting RNA sequences within the viral IRES. To determine whether PTB plays a functional role in this process, we constructed a series of plasmids containing dicistronic transcriptional units in which the upstream cistron encodes PTB (or various PTB deletion mutants) and the downstream cistron encodes the reporter protein, CAT, under the translational control of the IRES of either Sabin type 1 poliovirus or a cell culture-adapted HAV (Fig. 1A). We reasoned that the cap-dependent translation of human PTB from the upstream cistron of pPwt/SC and pPwt/AC transcripts (Fig. 1A) would facilitate an analysis of its effects on the immediately adjacent IRES in transient expression assays. This approach eliminated the need for plasmid cotransfections and ensured that the expressed PTB would be present within the microenvironment of the IRES, thus enhancing the likelihood of detecting either a positive or negative effect on viral translation.
The PTB mutants encoded by these plasmids included a frameshift/deletion mutation that terminated translation at amino acid residue 86 of PTB (Δ87-531, null mutant), which normally comprises 531 amino acid residues (Fig. 1A). The small N-terminal segment of PTB that is expressed by the Δ87-531 mutant contains the nuclear localization signal of PTB but only part of the first of four putative RRMs (21, 39, 56). Because it is unlikely to have any RNA binding activity, and thus any influence on IRES-directed translation, plasmids encoding this null mutant served to establish the basal level of IRES activity in transfected cells. Other PTB mutants included a frameshift mutation leading to the termination of translation at residue 360 (Δ361-531) and an in-frame deletion mutation removing residues 87 to 118 from PTB (Δ87-118) (Fig. 1A).
To confirm the effects of these mutations on the PTB products expressed from the constructs depicted in Fig. 1A, T7 transcripts synthesized in vitro were used to program translation in rabbit reticulocyte lysates. PTB normally migrates in SDS-PAGE as a doublet band with an apparent molecular mass of ∼57 kDa (20, 52). This product, along with the ∼24-kDa CAT protein produced from the downstream cistron under control of the poliovirus IRES, was readily apparent in the products of translation reactions programmed with pPwt/SC transcripts which encode wild-type PTB (Fig. 2, lane 2). In contrast, the mutant PTB produced in reactions programmed with pΔ87-118/SC transcripts migrated as a doublet band with an apparent molecular mass slightly less than that of wild-type PTB (Fig. 2, lane 3), consistent with the internal deletion of 32 amino acid residues. The mutant PTB produced from pΔ361-531/SC transcripts also migrated as a doublet band but with an apparent molecular mass of only ∼38 kDa (Fig. 2, lane 5). This result is consistent with the deletion of the C-terminal one-third of the PTB molecule that results from the frameshift mutation in these transcripts. No PTB product was evident in reactions programmed with pΔ87-531/SC transcripts (null mutant), which contains a frameshift mutation resulting in the production of a very abbreviated polypeptide of only 86 amino acid residues (Fig. 2, lane 4). The translation products from transcripts containing the HAV IRES were identical to those shown in Fig. 2 (data not shown).
FIG. 2.
Products of cell-free translation reactions programmed with synthetic dicistronic RNA transcripts containing the poliovirus 5′NTR within the intercistronic space (Fig. 1A). Reaction mixes (25 μl) were programmed with 1 μg of the indicated T7 transcripts (lanes 2 to 5) or no RNA (lane 1). Intact PTB appears as a doublet band with an apparent molecular mass of ∼57 kDa (lane 2), while mutant PTBs appear as more rapidly migrating doublet bands (lanes 3 and 5). No PTB product is evident from translation of the null mutant, pΔ87-531/SC (lane 4), consistent with the small predicted size of this product (∼10.7 kDa). CAT is produced from all four transcripts and migrates with an apparent molecular mass of ∼24 kDa (lanes 2 to 5).
To determine whether the production of human PTB in reticulocyte lysates programmed with pPwt/SC or pPwt/AC influenced the efficiency of CAT translation directed by the picornavirus IRESs, we quantified the amounts of CAT protein produced in these reactions by PhosphorImager analysis. For each IRES, results were standardized to the amount of CAT produced in lysates programmed with transcripts encoding the PTB null mutant, Δ87-531. The results of this experiment are shown in Table 1. The quantities of CAT produced in these reactions did not vary by more than ∼2-fold for each IRES. Furthermore, when transcripts containing the poliovirus and HAV IRESs were compared, there was no correlation between the quantity of CAT produced and the type of PTB molecule expressed by individual constructs. Thus, these results suggest that either the quantity of endogenous PTB is not limiting for IRES-directed translation in reticulocyte lysates or the quantity of newly synthesized PTB produced in these lysates is insufficient to influence the translational activity of the relatively large quantity of RNA (1 μg) used to program each reaction.
TABLE 1.
PhosphorImager quantitation of CAT produced under control of the poliovirus or HAV IRES in rabbit reticulocyte lysates programmed with dicistronic RNA
Amount of CAT protein produced in each reaction in a representative experiment, normalized to that produced in reactions programmed with RNAs encoding the Δ87-531 null mutant, assigned a value of 1. In reactions performed with no RNA, CAT production was not detected.
pPwt constructs (Fig. 1A).
PTB enhances poliovirus IRES-mediated translation in vivo.
In contrast to rabbit reticulocyte and HeLa cell lysates that contain abundant PTB, cytoplasmic extracts prepared from BS-C-1 cells contain a substantially lower abundance of PTB (15). The lower endogenous level of PTB expression in BS-C-1 cells should enhance the probability of detecting an effect of PTB overexpression on translation directed by the poliovirus or HAV IRES elements in vivo. Figure 3A summarizes results obtained in replicate experiments in which BS-C-1 cells were transfected with plasmids containing the poliovirus IRES. Transcription from these plasmids in vivo was driven by the CMV IE promoter; thus, PTB was translated by a typical cap-dependent process. In contrast, translation of CAT from the downstream cistron of the dicistronic transcripts was dependent on internal ribosome entry mediated by the viral IRES. Reporter protein activity was measured in cell lysates approximately 48 h following transfection.
FIG. 3.
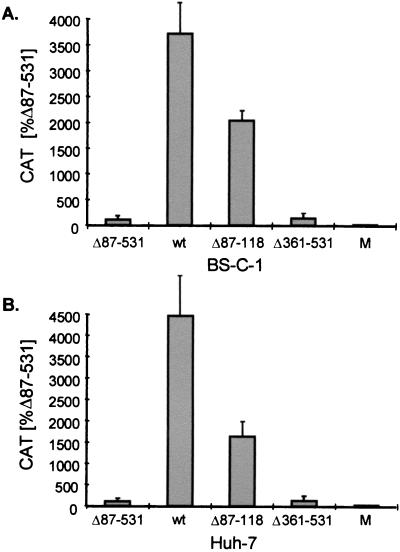
Impact of PTB transient expression on levels of CAT reporter protein activity in cultured mammalian cells which were transfected with plasmids containing the poliovirus 5′NTR within the intercistronic space (Fig. 1A). CAT activities were measured 46 to 50 h following transfection and were normalized to those obtained following transfection of the null mutant pΔ87-531/SC (100%). Error bars indicate the standard deviations of results obtained in two separate experiments, each involving two replicate transfections (total of four transfections). (A) Relative CAT activities following DNA transfection of BS-C-1 cells which contain a low cytoplasmic abundance of PTB. Transfection with the null mutant, pΔ87-531/SC, generated a mean CAT activity value of 777 cpm, while CAT activity in mock-transfected cells (m) was 69 cpm. (B) Relative CAT activities in transfected H1-HeLa cells which contain a greater natural abundance of PTB than BS-C-1 cells. Transfection with the null mutant, pΔ87-531/SC, generated a mean CAT activity value of 22,715 cpm, while CAT activity in mock-transfected cells was 67 cpm.
Compared with cells transfected with the null mutant, pΔ87-531/SC (Fig. 1A), BS-C-1 cells transfected with pPwt/SC (expressing intact PTB) or pΔ87-118/SC (expressing PTB with a small 32-residue internal deletion) produced 11- to 12-fold more CAT activity. In contrast, no increase in CAT expression over that produced by the null mutant was observed following transfection with pΔ361-531/SC, which encodes a truncated PTB mutant lacking the C-terminal one-third of the molecule. Since the increased CAT activity observed in cells transfected with pPwt/SC or pΔ87-118/SC could reflect either stimulation of the poliovirus IRES by PTB or increased CMV promoter-directed transcription or mRNA stability in BS-C-1 cells, we assessed the levels of RNA transcripts in transfected BS-C-1 cells by an RNase protection assay employing an RNA probe complementary to the CAT sequence. Protected CAT-specific RNA fragments were normalized to levels of GAPDH transcripts detected by RNase protection. These results demonstrated that differences in reporter protein activity were not due to differences in the abundance of CAT transcripts, as PhosphorImager analysis indicated that RNA transcript levels varied from 1.0-fold (pΔ87-118/SC) to 2.2-fold (pPwt/SC) that of the null transcript (data not shown). In addition, the protein concentrations of these lysates were similar. Thus, we conclude that the transient overexpression of PTB significantly enhances translation of the CAT reporter protein from these dicistronic transcripts in BS-C-1 cells. This enhancement is absolutely dependent on the presence of the PTB sequence downstream of residue 360, which includes most of RRM-3 and all of RRM-4 (21, 39, 56).
Compared to BS-C-1 cells, HeLa cells contain a much higher natural abundance of cytoplasmic PTB (15). Thus, we anticipated that the effect of transient PTB overexpression on translation of the reporter protein might be less in HeLa cells than in BS-C-1 cells. This proved to be the case, as shown in Fig. 3B. Transient expression of wt PTB or the pΔ87-118/SC mutant had only a modest (2.4-fold) stimulatory effect on CAT expression directed by the poliovirus IRES in transfected HeLa cells. This did not reflect poor transfection efficiency or low transcriptional activity in HeLa cells, as the basal levels of translation in cells transfected with the null mutant were considerably higher than in BS-C-1 cells. As in BS-C-1 cells, no enhancement of CAT activity was observed with the large frameshift deletion mutant, pΔ361-531/SC (Fig. 3B). Thus, the natural cytoplasmic abundance of PTB in HeLa cells appears to be almost sufficient for optimal poliovirus IRES-mediated initiation of translation.
Stimulation of HAV IRES-directed translation by PTB.
The HAV and poliovirus 5′NTRs contain IRES elements with distinctly different RNA secondary structures that are representative of two general structural classes of IRESs found in picornaviruses (11, 60). Compared with the poliovirus IRES, the HAV IRES has exceptionally low intrinsic translational activity both in vitro and in vivo (12, 69, 78). Thus, it was of interest to see whether PTB was also capable of stimulating the translation of a reporter protein placed downstream of the HAV IRES in a dicistronic transcript. Strikingly, when dicistronic plasmids containing the HAV 5′NTR (Fig. 1A) were transfected into BS-C-1 cells, the stimulatory effect of PTB on translation of the reporter protein was substantially greater than its effect on the poliovirus IRES. CAT activity was approximately 37-fold greater in cells transfected with pPwt/AC, which encodes wt PTB, than in cells transfected with the related null mutant, pΔ87-531/AC (Fig. 4A).
FIG. 4.
Impact of PTB transient expression on levels of CAT reporter protein activity in cells which were transfected with plasmids (Fig. 1A) containing the HAV 5′NTR within the intercistronic space (results shown are means of four separate transfections from a total of two experiments; see the legend to Fig. 3). (A) Relative CAT activities following DNA transfection of BS-C-1 cells. Transfection with the null mutant, pΔ87-531/AC, generated a mean CAT activity value of 2531 cpm, while CAT activity in mock-transfected cells (M) was 60 cpm. (B) Relative CAT activities in transfected Huh-7 cells, which are derived from a human hepatocellular carcinoma. Transfection with the null mutant, pΔ87-531/AC, generated a mean CAT activity value of 536 cpm, while CAT activity in mock-transfected cells was 65 cpm. CAT assay incubation times were extended compared to those used in the experiments shown in Fig. 4 because of the expected low basal rate of HAV translation (78).
As with the poliovirus IRES (Fig. 3), deletion of amino acid residues downstream of residue 360 of PTB abrogated the enhancement in translation of the reporter protein that was observed with pPwt/AC (Fig. 4A). However, in contrast to results obtained with the poliovirus IRES (Fig. 3), the deletion of amino acid residues 87 to 118 in mutant pΔ87-118/AC resulted in an almost 50% reduction of the enhancement of translation observed with wt PTB. Similar results were obtained in multiple transfection experiments. Thus, an intact RRM-1 must be present in the PTB molecule for maximum stimulation of translation from transcripts containing the HAV IRES, while this does not appear to be the case with the poliovirus IRES. As with the poliovirus constructs, RNase protection assays confirmed that the differences in CAT activity observed in BS-C-1 cells transfected with plasmids containing the HAV IRES were not due to differences in the abundance of CAT-specific transcripts. RNase protection assays indicated that the levels of transcripts varied from only 0.8- to 1.4-fold that of the null transcript (data not shown).
Consistent with previous studies (69, 78) the HAV IRES demonstrated much lower intrinsic translation-initiating activity than the poliovirus IRES within the context of otherwise identical dicistronic transcripts. To eliminate interexperimental differences in the absolute values obtained in the CAT assay (reflecting differences in the length of the incubation step in the enzymatic assay; see the legend to Fig. 4), frozen cell lysates collected in multiple experiments were reassayed for CAT activity in a single assay (Table 2). In the absence of expression of a functional PTB molecule from the upstream cistron (pΔ87-531 null mutants), the level of CAT activity expressed under translational control of the HAV IRES was approximately 19-fold less than that expressed under control of the poliovirus IRES in BS-C-1 cells (Table 2), despite similar levels of CAT-specific RNA transcripts detected by RNase protection assays (data not shown). However, the difference between the translational activities of HAV and the poliovirus IRES elements decreased to approximately sixfold when IRES activities were compared in the context of transcripts expressing wt PTB (pPwt/AC vs. pPwt/SC) (Table 2). These results indicate that the transient overexpression of PTB partially compensates for the intrinsically poor activity of the HAV IRES, compared with the poliovirus IRES, in vivo.
TABLE 2.
CAT activities in lysates of BS-C-1 cells transfected with dicistronic plasmids containing the poliovirus or HAV IRES and expressing either intact PTB or the Δ87-531 null mutant
| PTB segment | Mean cpm ± SDa
|
|
|---|---|---|
| Poliovirus 5′NTR | HAV 5′NTR | |
| Δ87-531 | 2,408 ± 30.6 | 129 ± 12.1 |
| wtb | 28,652 ± 1,167 | 4,886 ± 202 |
Frozen lysates from multiple experiments were retested in a single CAT assay with 125-μl reaction volumes containing 10 μl of lysate incubated at 37°C for 1 h. The values shown were obtained with cell lysates from four separate transfections carried out in two independent experiments.
pPwt constructs (Fig. 1A).
To determine the extent to which PTB might mediate enhancement of translation directed by the HAV IRES in a human hepatocyte-derived cell line, plasmids containing the HAV IRES were transfected into Huh-7 cells. The results of these experiments closely paralleled those obtained in BS-C-1 cells. A 44-fold-greater level of CAT activity was obtained following transfection of Huh-7 cells with pPwt/AC compared to the null mutant, pΔ87-531/AC (Fig. 4B). As in BS-C-1 cells, the deletion of amino acid residues 87 to 118 substantially reduced the ability of PTB to stimulate HAV-directed translation (resulting in ∼30% of the stimulation observed with intact PTB), while deletion of residues downstream of residue 360 completely eliminated the translation-enhancing activity. We conclude from these experiments that transient expression of PTB in either BS-C-1 or Huh-7 cells enhances translation directed by the IRES of HAV. The 37- to 44-fold enhancement of HAV translational activity was significantly greater than the 12-fold enhancement observed with the poliovirus IRES in BS-C-1 cells (Fig. 3A) and much greater than the 2.4-fold increase observed with poliovirus in HeLa cells (Fig. 3B).
Enhanced CAT expression from dicistronic RNAs encoding PTB is not due to altered nuclear processing of transcripts.
Since we found that PTB overexpression did not influence the abundance of CAT transcripts in transfected cells, the results described above strongly suggest that PTB enhances the efficiency of translation directed by different picornaviral IRES elements in vivo. However, because of growing evidence that PTB may regulate exon selection during pre-mRNA splicing (14, 24, 43, 55, 73), we considered the possibility that overexpression of PTB might result in selection of a cryptic splice site within these dicistronic transcripts. This could alter the primary structure of the transcripts and possibly the nature of the process by which the translation of the reporter protein is initiated. However, it would seem to be a very unlikely explanation for the translational enhancement observed in these experiments, since splicing events would have to have occurred fortuitously in both the HAV and poliovirus sequences in pPwt/AC and pPwt/SC transcripts, respectively, in order to remove the viral IRES and render the reporter protein subject to a conventional translation initiation mechanism. Nonetheless, to formally exclude this possibility, we carried out a Northern analysis of RNA transcripts produced in BS-C-1 cells following transfection with plasmids containing the HAV IRES, pPwt/AC, and the related null mutant, pΔ87-531/AC (Fig. 5). The probe for these experiments was a mixture of labeled RNAs complementary to the CAT sequence. These results demonstrated a single transcript of appropriate size in BS-C-1 cells that had been transfected with either plasmid (Fig. 5, lanes 1 and 2). Consistent with earlier RNase protection studies (data not shown), the abundance of this transcript was equivalent in cells transfected with either plasmid, despite the significantly greater expression of CAT in cells transfected with pPwt/AC. No transcript was detected in mock-transfected cells (lane 3). These experiments provide further proof that the overexpression of PTB results in enhanced internal initiation of translation directed by representative picornaviral IRES elements in vivo.
FIG. 5.
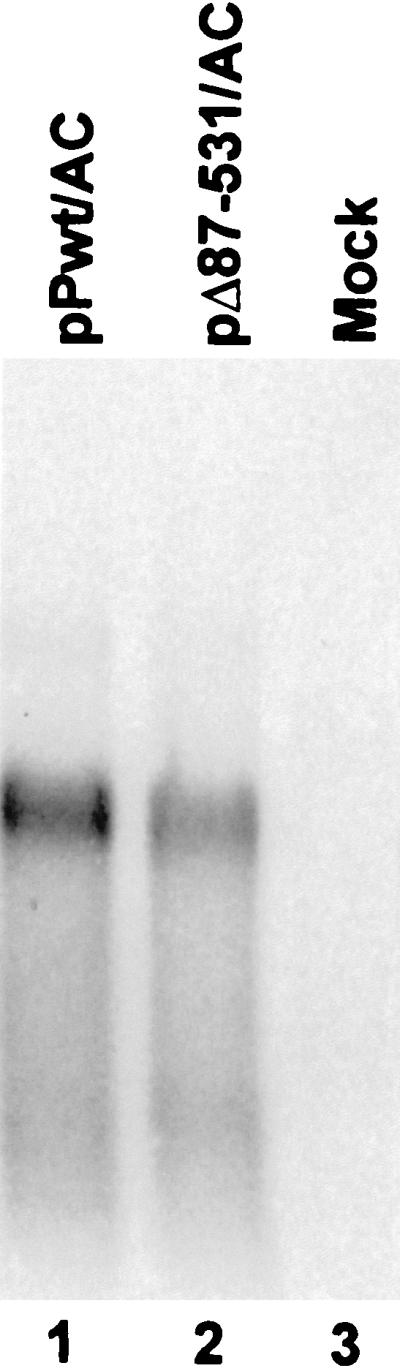
Northern blot analysis of the poly(A) fraction of RNA extracted from BS-C-1 cells following DNA transfection with constructs containing the HAV IRES, pPwt/AC (lane 1), or its related null mutant pΔ87-531/AC (lane 2). Lane 3 was loaded with RNA from mock-transfected cells. The probe for hybridization was complementary to the CAT sequence.
The enhanced translational activity of the IRES in dicistronic transcripts encoding PTB is dependent on translation of the upstream PTB reading frame.
The dicistronic RNA transcripts encoding the PTB mutants shown in Fig. 1 differ minimally from the nucleotide sequence of the related wt PTB construct, despite large differences in the nature of the PTB proteins expressed from the upstream reading frames. Furthermore, the frameshift and deletion mutations which alter the expression of PTB are located far upstream of the IRES sequence (Fig. 1A). Thus, the differences that we observed in the translational activities of these transcripts are unlikely to reflect altered RNA structure in the region of the IRES. However, to formally exclude this possibility, we determined whether the greater translational activity of the poliovirus IRES within the context of the dicistronic pPwt/SC and pΔ87-118/SC transcripts (Fig. 3) was dependent on expression of PTB from the first cistron.
BS-C-1 cells were cotransfected with a dicistronic plasmid (pPwt/SC, pΔ87-118/SC, or the null mutant pΔ87-531/SC) (Fig. 1A) and a plasmid expressing either the wt protein or a proteolytically inactive form of the poliovirus 2Apro protease under transcriptional control of the CMV promoter [pCMV-2A or pCMV-2A(H20N), respectively] (Fig. 6A). In addition to directing the cis-active primary cleavage of the poliovirus polyprotein, 2Apro directs the proteolytic cleavage of eIF4G, an essential component of the cellular cap-binding complex (3, 72). It may also have a direct, trans-activating effect on the poliovirus IRES (25, 45). Thus, the expression of 2Apro should inhibit the cap-dependent initiation of PTB translation from the upstream cistron of the dicistronic transcripts but should not hinder (and could actually enhance) the cap-independent translation of the reporter protein which is under control of the poliovirus IRES. As shown in Fig. 6B, the PTB-mediated enhancement of IRES activity observed with pPwt/SC was almost completely eliminated by coexpression of the wt 2Apro but not the proteolytically inactive 2Apro(H20N) in which His20 of 2Apro is substituted with Asn (82). These results confirm that the enhanced activity of the poliovirus IRES in the pPwt/SC and pΔ87-118/SC transcripts is dependent on translation of the upstream cistron (PTB) and is not a function of intrinsic transcript sequence or higher-ordered RNA structure within the region of the IRES. It is important to note that in this experiment, CAT activities were substantially increased in cells expressing the proteolytically active form of 2Apro compared to that in cells expressing the proteolytically inactive 2Apro(H20N). The increase in IRES-directed translation in the presence of 2Apro is likely due to the shutdown of cap-dependent translation, and greater availability of ribosomes or protein translation factors, in addition to any direct effects of 2Apro on the IRES (25, 45).
FIG. 6.
Enhancement of the translational activity of the poliovirus IRES within dicistronic transcripts is dependent on the cap-dependent translation of PTB from the upstream cistron. (A) Organization of plasmids encoding protease 2A (2Apro) of poliovirus under control of the CMV IE promoter. pCMV-2A expresses wt 2Apro, while pCMV-2A(H20N) expresses a mutant 2Apro which lacks proteolytic activity. (B) BS-C-1 cells were cotransfected with dicistronic plasmid DNAs plus either pCMV-2A (grey bars) or pCMV-2A(H20N) (black bars). Results shown represent mean values obtained in a total of four transfections of plasmid DNAs ± standard deviation, normalized for each series to that obtained with the null mutant, pΔ87-531/SC (100%). Absolute CAT activities following transfection of the null mutant, pΔ87-531/SC, were 28,908 cpm with pCMV-2A cotransfection and 2,726 cpm with pCMV-2A(H20N) cotransfection, compared to 60 cpm for mock transfection.
Indirect immunofluorescence detection of PTB in normal and transfected cells.
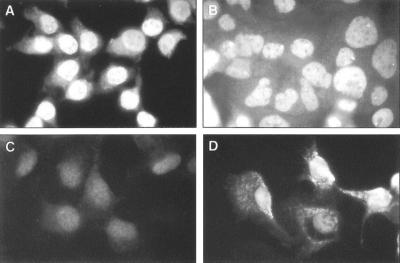
An important consideration in interpreting the enhancement of IRES-directed translation due to overexpression of PTB is the extent to which the abundance of cytoplasmic PTB has been increased beyond the physiologic range. Thus, we assessed the intracellular abundance and subcellular localization of PTB in both normal and transfected cells by an indirect immunofluorescence method using a primary rabbit polyclonal antibody raised to a GST-PTB fusion protein (56) (Fig. 7). Specificity of the immunofluorescence (TRITC) staining was controlled by similarly staining cells with normal rabbit serum (data not shown). In normal HeLa cells (Fig. 7A), there was brilliant PTB-specific nuclear staining. This was located predominantly in the nucleoplasm, sparing the nucleoli but with two or more intense perinucleolar deposits present within the majority of cells, as described previously (21, 56). In contrast, normal BS-C-1 cells demonstrated only very weak specific nuclear staining for PTB, with none of the bright perinucleolar deposits that were noted in HeLa cells (Fig. 7C). Nonspecific background fluorescence within the cytoplasm of BS-C-1 cells often rivaled the intensity of the nuclear staining. These results are consistent with the lower PTB content of BS-C-1 cells noted previously by Chang et al. (15). Normal Huh-7 cells demonstrated a level of specific nuclear PTB staining that was intermediate between that of BS-C-1 and HeLa cells (Fig. 7B). Some Huh-7 cells contained intense perinucleolar deposits of PTB similar to those observed in HeLa cells but with less frequency than in HeLa cells.
FIG. 7.
Indirect immunofluorescence detection of PTB in normal HeLa (A) Huh-7 (B), and BS-C-1 cells (C) and BS-C-1 cells 24 h following electroporation of the pPwt/AC plasmid (D). The primary antibody for these studies was rabbit anti-GST-PTB, while the secondary antibody was TRITC-labeled swine antibody to rabbit immunoglobulin.
Following transfection with pPwt/AC, the majority of BS-C-1 cells demonstrated brilliant PTB-specific staining (Fig. 7D), present as a granular cytoplasmic fluorescence with conspicuous perinuclear accumulation. In many cells there was prominent, diffuse staining of the nucleoplasm without evidence of the intense perinucleolar deposits observed in normal HeLa and Huh-7 cells. In general, the intensity of PTB staining in transfected BS-C-1 cells was similar to that in normal HeLa cells (Fig. 7A). Thus, the overall level of PTB expression required for stimulation of IRES-directed translation in BS-C-1 cells appears to be within what is the physiologic range for HeLa cells, although it is likely that the cytoplasmic levels of this predominantly nuclear protein were increased substantially above what is normally present in BS-C-1 cells.
Immunoblot detection of PTB in normal and transfected cells.
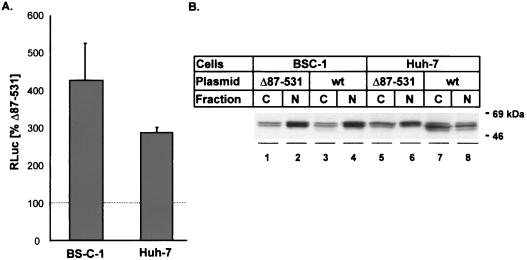
To further assess the increase in the abundance of PTB in transfected cells in relation to the increase observed in IRES-directed translation, we used an immunoblot assay to monitor the amounts of PTB present in cytoplasmic and nuclear fractions (Fig. 8). First, we compared IRES activities in cells transfected with dicistronic plasmids that express (i) either wt PTB or the PTB null mutant from the upstream cistron and (ii) renilla luciferase from the downstream cistron under the control of the wt HAV IRES (pPTB-Awt-RLuc and pΔ87-531-Awt-RLuc, respectively). The HAV IRES in these constructs differs at three bases from the HM175/P16 HAV IRES used in the preceding experiments, and we have shown it to be significantly less active in BS-C-1 cells (69). Transfection was via a liposome-mediated method rather than by electroporation. Despite these differences, the results were similar to those obtained in the preceding experiments. BS-C-1 cells transfected with pPTB-Awt-RLuc expressed over fourfold more renilla luciferase than control cells transfected with the pΔ87-531-Awt-RLuc null mutant, while there was approximately a threefold difference in Huh-7 cells (Fig. 8A). The lesser degree of translational enhancement compared with that shown in Fig. 4 may reflect the lower efficiency of liposome-mediated transfection compared with electroporation. This was particularly evident with the BS-C-1 cells and may also reflect a greater level of constitutive expression of PTB in these particular cells (see below). These results show that the increased IRES activity observed with PTB overexpression is not dependent on the reporter protein, which was renilla luciferase in these experiments and CAT in the experiments shown in Fig. 3 and 4.
FIG. 8.
Immunoblot detection of PTB in transfected BS-C-1 and Huh-7 cells in comparison to increases in translation directed by the wt IRES of HAV. (A) Relative expression of renilla luciferase (RLuc) following DNA transfection of cells with a dicistronic plasmid expressing wt PTB from the upstream cistron, compared with a matched control plasmid encoding the PTB null mutant, Δ87-531, in the upstream cistron. RLuc was translated from the downstream cistron by a cap-independent process under control of the wt 5′NTR of HAV. Transfection was by a liposome-mediated procedure. (B) Immunoblot analysis of BS-C-1 and Huh-7 cells following DNA transfection under conditions identical to those used in panel A with monocistronic plasmids expressing either wt PTB or the null mutant, Δ87-531. C, cytoplasmic fraction; N, nuclear fraction.
To assess the level of PTB overexpression under these conditions, we carried out immunoblot assays of cells transfected under identical conditions with a related set of monocistronic plasmids expressing PTB and the null mutant, Δ87-531 (Fig. 8B). PTB was visualized as a doublet band migrating with an apparent molecular mass of ∼52 kDa. In both BS-C-1 and Huh-7 cells transfected with the null mutant, the nuclear fraction contained a significantly greater abundance of PTB than the cytoplasmic fraction. The PTB content of the Huh-7 cells (particularly the cytoplasmic fraction) was greater than that of the BS-C-1 cells (compare lanes 5 and 1 in Fig. 8B). These results are generally consistent with the immunofluorescence results shown in Fig. 7. However, the abundance of PTB in the cytoplasmic fraction of the BS-C-1 cells used in these particular experiments appears to have been greater than that we observed in previous experiments (15). Thus, it is possible that an increase in the constitutive level of expression of PTB by these BS-C-1 cells may have contributed to the lesser magnitude of translational enhancement observed with PTB overexpression in these cells (compare Fig. 8A with Fig. 4). Transfection with the wt PTB-expressing plasmid resulted in little apparent increase in the total cellular PTB content of BS-C-1 cells (compare lanes 3 and 4 with 1 and 2 in Fig. 8B) and only a moderate increase in the PTB content of Huh-7 cells (compare lanes 7 and 8 with 5 and 6). Interestingly, however, transfection with this plasmid resulted in the appearance of a new minor PTB species migrating with a slightly smaller apparent mass than the major doublet bands (best visualized in lanes 3, 7, and 8). The increase in PTB content appeared to be predominantly nuclear in BS-C-1 cells, but there were significant increases in the cytoplasmic abundance of PTB in transfected Huh-7 cells. The results shown in Fig. 8 confirm that relatively small increases in the cytoplasmic abundance of PTB may be associated with significant increases in HAV IRES activity, particularly in BS-C-1 cells, and they argue strongly for a physiologically relevant role for PTB in the regulation of viral IRES elements in vivo.
PTB also stimulates cap-independent translation directed by the HCV IRES.
In addition to picornaviruses, IRES elements have been demonstrated within the 5′NTRs of certain flaviviruses (HCV, GBV-B, and the pestiviruses bovine viral diarrhea virus and classical swine fever viruses) (29, 30, 62, 63, 63a, 65, 75;. These flaviviral IRESs contain common RNA secondary structures and share limited primary nucleotide sequence identity, but they show no relatedness to any picornaviral IRES and thus can be considered to comprise a wholly different class of viral IRES elements (13, 26, 29, 40). Ali and Siddiqui (2) reported that PTB binds to the IRES of HCV and that the immunodepletion of PTB from reticulocyte lysates substantially reduced the ability of the HCV IRES to direct translation. In addition, Ito and Lai (33) have suggested that PTB regulates HCV translation in vitro through interactions with RNA sequences in the 5′ and 3′ NTRs as well as the 5′-proximal open reading frame. Nonetheless, Kaminski et al. (37) found that reticulocyte lysates depleted of PTB on an RNA affinity column were able to support a normal level of HCV-directed translation. Thus, the available in vitro data bearing on the role of PTB in HCV translation are contradictory. To determine whether transient expression of PTB in vivo would enhance translation directed by the IRES of HCV in a manner similar to that observed with picornaviral IRESs, we constructed dicistronic plasmids similar to those shown in Fig. 1A but containing (in lieu of the picornaviral IRES) the HCV 5′NTR and the first 8 nt of the HCV open reading frame fused in frame to the CAT coding sequence and either the wt or null mutant PTB in the upstream cistron (pPwt/CC and pΔ87-571/CC respectively [Fig. 1B]).
When these constructs were transfected into BS-C-1 or Huh-7 cells, we observed an approximately fivefold increase in CAT expression from the construct encoding wt PTB relative to that encoding the null mutant (Fig. 9). As was the case with cells transfected with dicistronic constructs containing the HCV IRES, Northern analysis demonstrated only a single transcript of equal size in BS-C-1 cells transfected with either of the HCV constructs (data not shown). There were no consistent differences in the abundance of this transcript in cells transfected with the null mutant, pΔ87-571/CC, and that encoding wt PTB. We conclude from these results that cap-independent translation directed by the IRES of HCV is also stimulated by transient expression of PTB in either BS-C-1 or Huh-7 cells, although the extent of translational enhancement was considerably less than that observed with the HAV IRES in these cell types (Fig. 4). Thus, the translational activity of the HCV IRES appears to be less affected by the abundance of cellular PTB than either the poliovirus or HAV IRES.
FIG. 9.
PTB stimulates the HCV IRES following transfection of mammalian cells with plasmids containing dicistronic transcriptional units (see Fig. 1B and the legend to Fig. 3). (A) CAT activities in lysates collected following DNA transfection of BS-C-1 cells with the null mutant, pΔ87-531/CC, or pPwt/CC. Transfection was by a cationic liposome-mediated method (see Materials and Methods) and with the null mutant, pΔ87-531/CL, generated a mean CAT activity of 22,907 cpm. Because of variation in transfection efficiency was greater with liposome-mediated transfection than with electroporation as in the experiments shown in Fig. 3 and 4, the results are shown as the ratio of CAT activities produced by the wt versus null mutant expression vectors. The data shown represent the mean values obtained in seven separate transfection experiments ± standard deviation. (B) CAT activities following DNA transfection of Huh-7 cells with the same plasmids. Transfection with the null mutant, pΔ87-531/CL, generated a mean CAT activity of 30,170 cpm. Results shown represent the mean values obtained in two separate transfection ± range.
DISCUSSION
We sought to develop direct evidence for a role for PTB in IRES-directed translation in vivo by determining whether the transient overexpression of PTB enhances or otherwise alters the translational activity of representative picornaviral and flaviviral IRES elements. Toward this end, we used a novel expression system involving the cap-dependent translation of PTB from the upstream cistron of dicistronic transcripts in which an intercistronic IRES controlled translation of a downstream reporter protein sequence. We studied the translational activity of these transcripts in several cell types, including BS-C-1 cells which normally express PTB at relatively low levels (Fig. 7C) (15). We found that the expression of PTB significantly enhanced the translational activity of two structurally different IRES elements from distantly related picornaviruses, poliovirus and HAV (Fig. 3 and 4), as well as that of a third, structurally distinct IRES which is present within the 5′NTR of a flavivirus, HCV (Fig. 9). While our data do not specifically exclude a general increase in cellular translation due to PTB expression (i.e., both cap-dependent and IRES-directed translation), such a phenomenon would not account for the wide variance in the degree to which different IRES elements were transactivated by PTB. Moreover, we saw no increase in β-galactosidase expression when we cotransfected β-galactosidase expression vectors with PTB constructs (data not shown). The data presented here thus provide an important in vivo confirmation of prior reports suggesting that PTB may function as a noncanonical internal initiation factor in cell-free translation systems (2, 8, 28, 37, 38, 49).
The approach taken to demonstrate this role for PTB in IRES-directed translation was unique in that PTB or PTB variants were expressed from the same dicistronic mRNA that contained the viral IRES and downstream reporter protein-coding sequence. This obviated the need for cotransfection of multiple plasmids. It also ensured that PTB was produced in every cell that expressed the reporter protein under control of the IRES and within the microenvironment of the IRES. The use of BS-C-1 cells that normally contain a relatively low level of cytoplasmic PTB (15) (Fig. 7C) allowed us to demonstrate that stimulation of IRES-directed translation occurs with levels of PTB expression that are similar to or only modestly higher than those levels that are normally present in HeLa cells (Fig. 7A and D). RNase protection assays and Northern analysis of transfected cells (Fig. 5) demonstrated that the increase observed in IRES-directed translation was not due to quantitative or qualitative differences in transcription from transfected plasmids. Furthermore, experiments involving the coexpression of the poliovirus 2Apro protease (Fig. 6) demonstrated that the increase in IRES-directed translation required the cap-dependent translation of PTB from the upstream cistron of the expressed transcripts, indicating that the increase in translation was not due to altered folding of transcripts containing mutated PTB segments.
The results obtained with dicistronic plasmids encoding different PTB mutants (Fig. 1A) provide insight into the functional domains of PTB that are required for stimulation of IRES activity in vivo. Previous analyses have suggested that PTB may contain up to four RRMs, each of which comprise about 80 amino acid residues (21, 39, 56). Only part of the first of these four RRMs is present in the 86-residue N-terminal fragment of PTB expressed from the null mutant, Δ87-531 (Fig. 1A). This small protein fragment is unlikely to have any RNA binding activity, although it does contain the nuclear localization signal of PTB (56). This suggests that the Δ87-531 product was not functioning as a dominant negative mutant in these transient expression experiments and supports the use of dicistronic plasmids encoding the null mutant to establish basal levels of IRES activity in the absence of PTB overexpression. In contrast, the large C-terminal deletion in the pΔ361-531 frameshift mutant eliminates most of RRM-3 and all of RRM-4 but leaves RRM-1 and RRM-2 intact (Fig. 1A). The resulting PTB fragment did not stimulate translation directed by either picornaviral IRES (Fig. 3 and 4). This is consistent with previous observations which indicate that RRM-3 and RRM-4 play a critical role in the RNA binding activity of both murine and human forms of PTB, although they are not required for dimerization of the molecule (9, 56). It is also consistent with previous results obtained in a cell-free translation system by Kaminski et al. (37).
The deletion of residues 87 to 118 from the pΔ87-118 mutant eliminates the conserved core residues of the RRM-1 motif. Previous studies indicate that this domain is not essential for either dimerization or RNA binding activities of PTB (9, 56). We found that this PTB mutant was capable of fully stimulating translation directed by the poliovirus IRES (Fig. 3). However, it was able to only partially stimulate the translational activity of the HAV IRES in either BS-C-1 or Huh-7 cells (Fig. 4). These results indicate a subtle difference between these picornaviral IRES elements in their requirements for this noncanonical translation initiation factor. However, detailed studies of the RNA binding activities of the Δ87-118 mutant will be required to understand the molecular basis underlying this difference.
The mechanism by which PTB stimulates the translational activities of picornaviral IRESs is unknown. However, available evidence suggests that the internal initiation of HAV and HCV translation, like that of EMCV, involves the binding of the 40S ribosome subunit to the viral RNA at a position close to the initiator AUG codon, rather than at an upstream site with subsequent 3′ scanning to the AUG codon as occurs with the poliovirus IRES (1, 11, 12, 29, 35, 36). Thus, it is likely that PTB facilitates the initial interaction of the 40S subunit with these viral RNAs, rather than its subsequent 3′ movement along the RNA (50). This initial interaction between the 40S subunit and the viral RNA is highly dependent on proper folding of the IRES, suggesting the possibility that the translation-enhancing activity of PTB may result from a stabilizing action on higher-ordered RNA structures. The notion that PTB might serve as an RNA chaperone (37) is strengthened by binding studies which indicate that PTB specifically interacts with more than one pyrimidine-rich RNA segment within each IRES (15, 27, 44, 80). There are also multiple RRMs within the PTB molecule, which exists in solution predominantly as a homodimer (21, 56). Thus, it is likely that the PTB dimer makes multiple contacts with the viral RNA. This could facilitate its putative role as a protein which stabilizes RNA structure.
However, an alternative mechanism by which PTB might stimulate IRES-directed translation is suggested by studies that identified a specific interaction between the cellular glycolytic enzyme GAPDH and the IRES of HAV (68). GAPDH, through its NAD+ binding groove, binds specifically to multiple pyrimidine-rich segments within the 5′NTRs of HAV and EMCV (15, 68). Circular dichroic spectropolarimetry demonstrated that the interaction of GAPDH with a stem-loop located within the IRES of HAV results in the destabilization of this RNA structure, an effect which would likely reduce the translational competence of the IRES (68). Since PTB competes strongly with GAPDH for binding to this RNA segment (68), it could stimulate HAV IRES activity by preventing the binding of GAPDH to the RNA. Since translation is an essential step in viral replication, the relative abundance of these proteins within the cytoplasm of infected cells could represent an important host determinant of the extent to which a cell is permissive for replication of a virus.
In comparison with HAV-directed translation, there was only a modest enhancement of HCV translation following the transient overexpression of PTB in either BS-C-1 or Huh-7 cells (e.g., compare Fig. 4 and 7). Nonetheless, the fivefold enhancement of HCV translation that we observed from transcripts encoding the intact PTB is consistent with prior studies that have suggested that PTB stimulated IRES-dependent translation in vitro (2). However, our observations should not be construed as indicating that PTB is absolutely required for the activity of the HCV (or either picornaviral) IRES, and it is noteworthy that other studies have shown that HCV translation is not impaired in PTB-depleted rabbit reticulocyte lysates (37). It is not clear why there is such a great difference in the extent to which the efficiencies of the HAV and HCV IRES elements are enhanced by PTB. It may be that the HCV IRES binds PTB with only low affinity (33, 37). The poliovirus IRES also appears to have relatively low affinity for PTB (57), and it was stimulated to an intermediate degree by PTB in BS-C-1 cells (Fig. 3A) (we did not assess poliovirus translation in Huh-7 cells). It is also possible that the lesser increase in translational efficiency that was observed with the HCV IRES reflects greater intrinsic stability of the essential higher-ordered RNA structures within the HCV IRES than in picornaviral IRESs or that the interaction of PTB with the HCV IRES does not enhance the stability of these structures. In support of this argument is the observation that the HCV IRES, but not picornaviral IRESs, can form binary complexes with the 40S ribosome particle in the absence of any additional cellular proteins (58). Finally, it may be that the HCV IRES has lower affinity for cellular helix-destabilizing proteins such as GAPDH (68). Further experimentation will be required to distinguish between these possibilities.
IRES elements are not restricted to viral RNAs, and they have been increasingly recognized in a small subset of cellular mRNAs (46, 66). Interestingly, these include mRNAs which direct the translation of important mammalian regulatory proteins, such as fibroblast growth factor 2, insulin-like growth factor II, and platelet-derived growth factor B (4, 74, 76). The activity of these cellular IRES elements may vary according to the differentiation state of the cell (4, 5), adding an additional layer of posttranscriptional control to the regulation of these important genes. It is reasonable to suspect that cellular proteins which influence the translational activity of viral IRES elements, such as PTB, may also play a role in regulating the translational activity of cellular IRESs. Thus, the characterization of such proteins is of potentially broad significance to a better understanding of cellular mechanisms for posttranscriptional regulation of gene expression.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (RO1-AI32599 and U19-AI40035), the Advanced Technology Program of the Texas Higher Education Coordinating Board, and the Swiss National Science Foundation (FK 364/95 to R.G.).
We are grateful to Mariano A. Garcia-Blanco and James G. Patton for freely sharing PTB reagents and data in advance of publication.
REFERENCES
- 1.Agol V I. The 5′-untranslated region of picornaviral genomes. Adv Virus Res. 1991;40:103–180. doi: 10.1016/S0065-3527(08)60278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvey J C, Wyckoff E E, Yu S F, Lloyd R, Ehrenfeld E. cis- and trans-cleavage activities of poliovirus 2A protease expressed in Escherichia coli. J Virol. 1991;65:6077–6083. doi: 10.1128/jvi.65.11.6077-6083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein J, Sella O, Le S-Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein J, Shefler I, Elroy-Stein O. The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA leader is relieved during megakaryocytic differentiation. J Biol Chem. 1995;270:10559–10565. doi: 10.1074/jbc.270.18.10559. [DOI] [PubMed] [Google Scholar]
- 6.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borman A, Howell M T, Patton J G, Jackson R J. The involvement of a splicesome component in internal initiation of human rhinovirus RNA translation. J Gen Virol. 1993;74:1775–1788. doi: 10.1099/0022-1317-74-9-1775. [DOI] [PubMed] [Google Scholar]
- 8.Borovjagin A, Pestova T, Shatsky I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994;351:299–302. doi: 10.1016/0014-5793(94)00848-5. [DOI] [PubMed] [Google Scholar]
- 9.Bothwell A L, Ballard D W, Philbrick W M, Lindwall G, Maher S E, Bridgett M M, Jamison S F, Garcia-Blanco M A. Murine polypyrimidine tract binding protein: purification, cloning, and mapping of the RNA binding domain. J Biol Chem. 1991;266:24657–24663. [PubMed] [Google Scholar]
- 10.Brown B A, Ehrenfeld E. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396–405. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- 11.Brown E A, Day S P, Jansen R W, Lemon S M. The 5′ nontranslated region of hepatitis A virus: secondary structure and elements required for translation in vitro. J Virol. 1991;65:5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown E A, Zajac A J, Lemon S M. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J Virol. 1994;68:1066–1074. doi: 10.1128/jvi.68.2.1066-1074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown E A, Zhang H, Ping L-H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan R C, Black D L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang K H, Brown E A, Lemon S M. Cell type-specific proteins which interact with the 5′ nontranslated region of hepatitis A virus RNA. J Virol. 1993;67:6716–6725. doi: 10.1128/jvi.67.11.6716-6725.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Day S P, Murphy P, Brown E A, Lemon S M. Mutations within the 5′ nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J Virol. 1992;66:6533–6540. doi: 10.1128/jvi.66.11.6533-6540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funkhouser A W, Schultz D E, Lemon S M, Purcell R H, Emerson S U. Hepatitis A virus translation is rate-limiting for virus replication in MRC-5 cells. Virology. 1999;254:268–278. doi: 10.1006/viro.1998.9548. [DOI] [PubMed] [Google Scholar]
- 19.Gan W N, Rhoads R E. Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 20.García-Blanco M A, Jamison S F, Sharp P A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 21.Ghetti A, Pinol-Roma S, Michael W M, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil A, Sharp P A, Jamison S F, Garcia-Blanco M A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 23.Glass M J, Summers D F. Identification of a trans-acting activity from liver that stimulates hepatitis A virus translation in vitro. Virology. 1993;193:1047–1050. doi: 10.1006/viro.1993.1225. [DOI] [PubMed] [Google Scholar]
- 24.Gooding C, Roberts G C, Smith C W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 25.Hambidge S J, Sarnow P. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc Natl Acad Sci USA. 1992;89:10272–10276. doi: 10.1073/pnas.89.21.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q-L, Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellen C U T, Pestova T V, Litterst M, Wimmer E. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J Virol. 1994;68:941–950. doi: 10.1128/jvi.68.2.941-950.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellen C U T, Witherell G W, Schmid M, Shin S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 30.Honda M, Ping L-H, Rijnbrand R C A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosomal entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 31.Hunt S L, Hsuan J J, Totty N, Jackson R J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunt S L, Jackson R J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito T, Lai M M. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 34.Jang S K, Krausslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E C. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang S K, Pestova T V, Hellen C U T, Witherell G W, Wimmer E. Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme. 1990;44:292–309. doi: 10.1159/000468766. [DOI] [PubMed] [Google Scholar]
- 36.Kaminski A, Howell M T, Jackson R J. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 38.Kaminski A, Jackson R J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 40.Lemon S M, Honda M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 41.Liebig H D, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lampher B, Rhoads R E, Kuechler E, Skern T. Purification of two picornaviral 2A proteinases: interaction with eIF-4 gamma and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 42.Lin C H, Patton J G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 43.Lou H, Helfman D M, Gagel R F, Berget S M. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luz N, Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1991;65:6486–6494. doi: 10.1128/jvi.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macadam A J, Ferguson G, Fleming T, Stone D M, Almond J W, Minor P D. Role for poliovirus protease 2A in cap independent translation. EMBO J. 1994;13:924–927. doi: 10.1002/j.1460-2075.1994.tb06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 47.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;3:1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 48.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysates. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niepmann M. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 1996;388:39–42. doi: 10.1016/0014-5793(96)00509-1. [DOI] [PubMed] [Google Scholar]
- 50.Niepmann M, Petersen A, Meyer K, Beck E. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J Virol. 1997;71:8330–8339. doi: 10.1128/jvi.71.11.8330-8339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh S-K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 52.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1993;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 53.Pause A, Methot N, Svitkin Y, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 55.Perez I, Lin C H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 56.Perez I, McAfee J G, Patton J G. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- 57.Pestova T V, Hellen C U T, Wimmer E. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pilipenko E V, Blinov V M, Chernov B K, Dmitrieva T M, Agol V I. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pilipenko E V, Blinov V M, Romanova L I, Sinyakov A N, Maslova S V, Agol V I. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- 61.Pilipenko E V, Gmyl A P, Maslova S V, Svitkin Y V, Sinyakov A N, Agol V I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 62.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 63.Reynolds J E, Kaminiski A, Kettinen H J, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63a.Rijnbrand R, Abell G, Lemon S M. Mutational analysis of the GB virus B interval ribosome entry site. J Virol. 2000;74:773–783. doi: 10.1128/jvi.74.2.773-783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 65.Rijnbrand R, van der Stratten T, Van Rijn P A, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarnow P. Translation of glucose-regulated protein 7S/immunoglobulin heavy-chain binding protein mRNA is increased in poliovirus-infected cells at a time when cap-dependent translation of cellular mRNAs is inhibited. Proc Natl Acad Sci USA. 1989;87:5795–5799. doi: 10.1073/pnas.86.15.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scheper G C, Voorma H O, Thomas A A. Eukaryotic initiation factors-4E and -4F stimulate 5′ cap-dependent as well as internal initiation of protein synthesis. J Biol Chem. 1992;267:7269–7274. [PubMed] [Google Scholar]
- 68.Schultz D E, Hardin C C, Lemon S M. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′ nontranslated RNA of hepatitis A virus. J Biol Chem. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 69.Schultz D E, Honda M, Whetter L E, McKnight K L, Lemon S M. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J Virol. 1996;70:1041–1049. doi: 10.1128/jvi.70.2.1041-1049.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shiroki K, Ishii T, Aoki T, Ota Y, Yang W-X, Komatsu T, Ami Y, Arita M, Abe S, Hashizume S, Nomoto A. Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J Virol. 1996;71:1–8. doi: 10.1128/jvi.71.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 72.Sommergruber W, Ahorn H, Klump H, Seipelt J, Zoephel A, Fessl F, Krystek E, Blaas D, Kuechler E, Liebig H-D, Skern T. 2A proteinases of coxsackie- and rhinovirus cleave peptides derived from eIF-4G via a common recognition motif. Virology. 1994;198:741–745. doi: 10.1006/viro.1994.1089. [DOI] [PubMed] [Google Scholar]
- 73.Southby J, Gooding C, Smith C W. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutually exclusive exons. Mol Cell Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teerink H, Vorma H O, Thomas A A M. The human insulin-like growth factor II leader I contains an internal ribosomal entry site. Biochim Biophys Acta. 1995;1264:405–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- 75.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vagner S, Gensac M-C, Marlet A, Bayard F, Amalric F, Prats H, Prats A-C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Pederson T. A 62,000 molecular weight spliceosome protein crosslinks to the intron polypyrimidine tract. Nucleic Acids Res. 1990;18:5995–6001. doi: 10.1093/nar/18.20.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whetter L E, Day S P, Elroy-Stein O, Brown E A, Lemon S M. Low efficiency of the 5′ nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J Virol. 1994;68:5253–5263. doi: 10.1128/jvi.68.8.5253-5263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 80.Witherell G W, Gil A, Wimmer E. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry. 1993;32:8268–8275. doi: 10.1021/bi00083a030. [DOI] [PubMed] [Google Scholar]
- 81.Witherell G W, Wimmer E. Encephalomyocarditis virus internal ribosomal entry site RNA-protein interactions. J Virol. 1994;68:3183–3192. doi: 10.1128/jvi.68.5.3183-3192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu S F, Lloyd R E. Characterization of the roles of conserved cysteine and histidine residues in poliovirus 2A protease. Virology. 1992;186:725–735. doi: 10.1016/0042-6822(92)90039-r. [DOI] [PubMed] [Google Scholar]