Abstract
Cyclic AMP (cAMP) stimulates the expression of numerous genes via the protein kinase A (PKA)-mediated phosphorylation of CREB at Ser133. Ser133 phosphorylation, in turn, promotes recruitment of the coactivator CREB binding protein and its paralog p300, histone acetyltransferases (HATs) that have been proposed to mediate target gene activation, in part, by destabilizing promoter bound nucleosomes and thereby allowing assembly of the transcriptional apparatus. Here we show that although histone deacetylase (HDAC) inhibitors potentiate target gene activation via cAMP, they do not stimulate transcription over the early burst phase, during which CREB phosphorylation and CBP/p300 recruitment are maximal. Rather, HDAC inhibitors augment CREB activity during the late attenuation phase by prolonging CREB phosphorylation on chromosomal but, remarkably, not on extrachromosomal templates. In reconstitution studies, assembly of periodic nucleosomal arrays on a cAMP-responsive promoter template potently inhibited CREB phosphorylation by PKA, and acetylation of these template-bound nucleosomes by p300 partially rescued CREB phosphorylation by PKA. Our results suggest a novel regulatory mechanism by which cellular HATs and HDACs modulate the phosphorylation status of nuclear activators in response to cellular signals.
Most signaling pathways promote cellular gene expression with burst attenuation kinetics; maximal rates of transcription are typically achieved within 30 min of stimulation, returning to baseline after 2 to 4 h (24). Transcriptional activation via the second messenger cyclic AMP (cAMP), for example, is rate limited by nuclear entry of protein kinase A (PKA) catalytic subunit, a passive process that plateaus after 15 to 30 min, coinciding with peak levels of CREB Ser133 phosphorylation and target gene activation (8, 9). Over the subsequent 2- to 4-hour attenuation phase, transcription rates return to prestimulus levels, reflecting, in part, the protein phosphatase 1 (PP-1)-mediated dephosphorylation of CREB at Ser133 (8).
The paralogous coactivators CREB binding protein (CBP) and p300 have been proposed to mediate target gene activation during the burst phase by acetylating promoter-bound nucleosomes and thereby allowing productive assembly of the transcriptional apparatus (3, 22). In cellular microinjection experiments where endogenous CBP activity is sequestered with anti-CBP antiserum, for example, histone acetyltransferase (HAT)-defective forms of CBP are unable to rescue target gene activation via CREB (14). Indeed, recent studies in other signaling systems have reinforced the notion that chromatin remodeling is a prerequisite for induction of signal dependent genes. p300 is capable of promoting target gene activation via the estrogen receptor in vitro, for example, on chromatin assembled but not on nonchromatinized templates (15). Stimulation of the beta interferon promoter in vivo, moreover, is accompanied by nucleosome acetylation over the promoter, and mutations in promoter-bound factors that abrogate recruitment of CBP correspondingly inhibit both nucleosome acetylation and target gene activation (23).
In addition to its effects on nucleosome remodeling, CBP has also been found to promote target gene expression via an association with RNA polymerase II complexes (12, 13, 19, 20). Such CBP-RNA polymerase II complexes appear competent to mediate target gene activation via Ser133-phosphorylated CREB [phospho-(Ser133)-CREB] comparably on naked DNA and nucleosome-assembled templates, suggesting that chromatin derepression, per se, may not be a prerequisite for target gene activation in response to cAMP (17).
Here we evaluate the importance of cellular HAT activities for transcriptional activation via CREB. Our studies reveal that although histone deacetylase (HDAC) inhibitors cooperate with cAMP signals on chromosomal templates, they do not potentiate target gene activation during the expected early burst phase, where CBP/p300 recruitment to the promoter is maximal. Rather, HDAC inhibitors promote transcription from cAMP-responsive genes during the attenuation phase, by prolonging Ser133 phosphorylation and thereby extending the ability of CREB to engage the transcriptional machinery via its association with CBP/p300. Our results suggest that chromatin-bound activators may be differentially phosphorylated in response to cellular signals depending, in part, on local chromatin structure.
MATERIALS AND METHODS
Cell culture.
The stable NIH 3T3 cell line D5, containing rat somatostatin gene sequences from 750 bp upstream of the promoter to 3 kb downstream of the coding region (18), was maintained in Dulbecco's minimal essential medium with 10% calf serum plus 200 μg of G418/ml.
Plasmids and transfections.
The dominant negative inhibitor A-CREB has been described previously (1). Approximately 4.4 × 105 cells per 100-mm-diameter dish were plated for transient transfection by the calcium phosphate coprecipitation technique. For each 100-mm-diameter dish, 8 μg of each construct was independently coprecipitated with 8 μg of pCA-GFP (green fluorescent protein) to select for transfected cells. D5 cells (107 per construct) were subjected to fluorescence-activated cell sorting (FACS) to obtain an average of 95% pure population of transfected cells. The sorted cells were then treated with various combinations of 10 μM forskolin, trichostatin A (TSA; 100 ng/ml; BIOMOL Research Laboratories), 15 mM sodium butyrate, or 100 nM trapoxin. For transient transfection assays, 2 × 105 cells were plated into six-well dishes. Each transfection contained 1 μg of CRE-CAT (chloramphenicol acetyltransferase) reporter plasmid and 50 ng of the Rous sarcoma virus (RSV)-based plasmid RSV-βGAL. CAT and β-galactosidase assays were performed as described previously. GAL4-CREB(1-283) plasmid (8 μg/100-mm-diameter dish) was transfected into D5 cells by calcium phosphate transfection as described above.
Northern blot analysis.
Total RNA was isolated from untreated cells or from cells treated with forskolin, TSA (100 ng/ml), 15 mM sodium butyrate, or 100 nM trapoxin. Northern blot analysis was performed as described previously, using either an antisense RNA probe to rat somatostatin or an α-tubulin cDNA probe (18).
Immunoblotting and immunocytochemistry.
D5 cells were treated with activators as described for various lengths of time and were lysed with sodium dodecyl sulfate (SDS)-urea lysis buffer. Whole-cell lysates (20 μg) were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% gel. Duplicate blots were probed with an anti-CREB antibody (244) or a phospho-Ser133-specific antiserum (5322) (9). Immune complexes were detected by chemiluminescence. Data for phospho-CREB are representative of a minimum of five independent experiments. Immunofluorescence assays were performed as previously described (2).
Chromatin immunoprecipitation assays (ChIPs).
D5 cells were grown to near confluence in 15-cm-diameter dishes and then treated for 30 min in the absence or presence of forskolin (10 μM) and/or TSA (100 ng/ml). Cellular histone proteins were cross-linked to chromatinized DNA for 10 min at 37°C by addition of 1% formaldehyde to the medium. Crude nuclei were isolated by Triton X-100 lysis (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 6.5]), resuspended in 0.5 ml of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.0]), and sonicated to reduce the chromatin DNA length to approximately 200 to 2,000 bp. The chromatin solution was diluted 10-fold in immunoprecipitation dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.0], 165 mM NaCl) and incubated with 4 μl of anti-acetylated H4 antiserum (gift from C. D. Allis, University of Virginia) (16); 2 μg of poly(dI-dC) was added to each reaction, and immune complexes were collected with protein A-agarose beads. Following extensive washing and elution in 1% SDS–0.1 M NaHCO3, DNA-histone protein cross-links were reversed by incubation at 65°C for 4 h. Released DNA was purified by proteinase K digestion, phenol extraction, and ethanol precipitation. Immunoprecipitated DNA was immobilized onto Zeta-Probe membrane (Bio-Rad) by slot blotting, and somatostatin gene sequences were detected by hybridization with an antisense RNA probe (4). The data are representative of three independent experiments.
Nuclear run-on transcription assay.
D5 cells were stimulated for various times (30 min to 4 h) with 10 mM forskolin and/or 100 ng of TSA per ml. Nuclear run-on transcription was performed as previously described. The data are representative of three independent experiments at the 30-min time point and two independent experiments for the time course (8).
Chromatin assembly and analysis.
Plasmid 3× CRE-MLP (20), containing three cAMP response element (CRE) sites inserted upstream of the adenovirus major late promoter, was used in assembly reactions. Drosophila S190 extract was preincubated with purified Drosophila core histones for 30 min at room temperature (15); 500 ng of 3× CRE-MLP plasmid (1.3 nM, final concentration) was added to the assembly mixture along with an ATP-generating system, and the reaction mixture was incubated for 3.5 h at 27°C. Following assembly, reactions were divided for either DNA supercoiling, micrococcal nuclease digestion, or cAMP-responsive element modulator (CREM) phosphorylation assays. For phosphorylation experiments, 3.9 nM CREM was incubated with the assembly reaction for 30 min at 27°C, and approximately 12 nM PKA catalytic subunit was added to each reaction. In time course experiments, aliquots of the reactions were stopped by mixing with 2× SDS loading buffer. Phospho-CREM levels were assessed by immunoblotting with phosphospecific antiserum.
Cellular PKA activation assay.
Methods for determining cellular PKA activity have been described elsewhere (6). D5 cells were stimulated for various times with 10 μM forskolin with or without TSA. Cells were then collected and lysed in 100 μl of HP (10 mM potassium phosphate [pH 6.8], 1 mM β-mercaptoethanol, 10 μg of leupeptin/ml, 10 mM magnesium acetate, 10 μM ATP containing 5 × 105 cpm of [γ-32P-ATP [3,000 Ci/mmol], 30 μg of Kemptide substrate). Background was determined from reactions lacking Kemptide substrate, and total PKA activity was estimated in reactions containing 20 μM dibutyryl-cAMP. Reaction mixtures were incubated for 5 min at 30°C, aliquots were spotted onto Whatman P-81 paper, and the filters were washed in 75 mM phosphoric acid twice for 1 min each time. 32P incorporation was determined by liquid scintillation counting. Data are representative of three independent experiments.
RESULTS
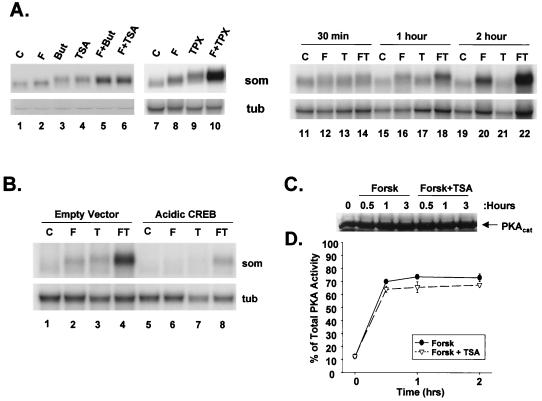
To test the importance of cellular HAT activities in promoting expression of cAMP-responsive genes, we used the HDAC inhibitors butyrate, TSA, and trapoxin. When added to cultures of D5 cells, all three inhibitors markedly potentiated somatostatin mRNA accumulation in cells costimulated with the cAMP agonist forskolin for 4 h (Fig. 1A, compare lanes 1, 2, 5, 6, 7, 8, and 10) but had minimal effects on target gene expression by themselves (compare lanes 11, 13, 15, 17, 19, and 21). Cooperativity between cAMP and HDAC inhibitor was most apparent at later times; a time course analysis of D5 cells revealed optimal synergism between these inducers at 2 to 4 h (Fig. 1A, compare lanes 14, 18, and 22). Arguing against integration-site- or cell-type-selective effects, HDAC inhibitors also potentiated chromosomal stomatostatin gene and endogenous c-fos gene expression in other NIH 3T3 lines as well as in clonal isolates of PC12 cells (not shown).
FIG. 1.
HDAC inhibitors act synergistically with cAMP agonist to promote somatostatin mRNA accumulation. (A) Lanes 1 to 10, Northern blot assays of total RNA from D5 cells expressing chromosomal copies of the rat somatostatin gene. Cells were treated with control vehicle (C), 10 μM forskolin (F), 15 mM sodium butyrate (But), 100 ng of TSA per ml, 100 nM trapoxin (TPX), or combinations of these reagents, as indicated, for 4 h. RNA was hybridized with a 32P-labeled somatostatin riboprobe (som) or α-tubulin cDNA probe (tub), as indicated. Lanes 11 to 22, time course analysis of somatostatin mRNA levels in D5 cells treated with forskolin (F), TSA (T), or both agents (FT) for times listed above the lanes. (B) CREB is required for synergistic effects of cAMP agonist and HDAC inhibitors on somatostatin gene expression, as determined by Northern blot assay of total RNA from D5 cells that were transfected with dominant negative A-CREB CMV expression vector (Acidic CREB) or CMV expression vector without insert (Empty Vector) plus RSV-GFP marker. After FACS sorting for transfected cells, GFP-positive cells were treated either without (C) or with 10 μM forskolin (F) and/or TSA (T). (C and D) HDAC inhibitors do not modulate cellular PKA activity or catalytic subunit expression levels. (C) Western blot assay of PKA catalytic subunit levels in D5 cells following treatment with forskolin or forskolin plus TSA for times listed above the lanes; (D) cellular PKA activation assay performed on extracts from D5 cells treated either with forskolin (Forsk) or forskolin plus TSA for various times. The line graph shows percentage of total cellular PKA activated by each treatment as measured by phosphorylation of synthetic Kemptide fragment. Total cellular PKA activity in each sample was estimated by incubating samples with 20 μM dibutyryl-cAMP to dissociate residual PKA holoenzyme.
cAMP has been shown to stimulate somatostatin gene expression via the PKA-mediated phosphorylation of CREB at Ser133 (7). To determine whether HDAC inhibitors promote somatostatin gene expression via a CREB-dependent mechanism, we used a dominant negative A-CREB cytomegalovirus (CMV) expression vector. The A-CREB polypeptide potently inhibited somatostatin mRNA accumulation in response to both forskolin and TSA but had no effect on α-tubulin mRNA levels in the same cells (Fig. 1B, compare lanes 4 and 8). These results indicate that CREB is required in order for HDAC inhibitors to augment somatostatin gene expression.
To rule out nonspecific effects of HDAC inhibitors on the cAMP pathway, we performed cellular PKA activation assays using synthetic Kemptide fragment as the phosphorylation substrate. Forskolin treatment induced 70% of total cellular PKA activity within 30 min of stimulation, and this effect persisted throughout the 2-h time course of treatment (Fig. 1C and D). Costimulation with TSA did not potentiate cellular PKA activity either alone (not shown) or in combination with forskolin stimulus (Fig. 1C and D). Indeed, total levels of PKA catalytic subunit remained constant in TSA-forskolin-stimulated cells by Western blot assay, indicating that HDAC inhibitors do not enhance CREB activity indirectly by stimulating PKA (Fig. 1C).
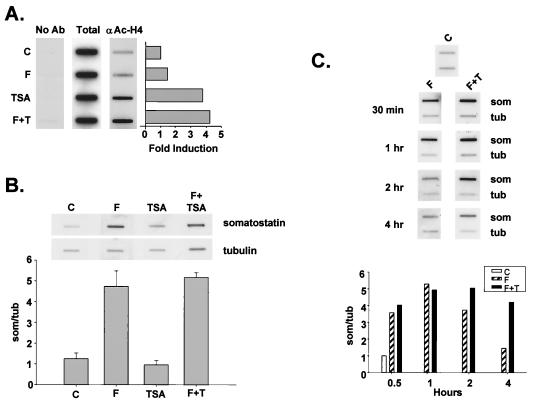
Following its recruitment to the promoter via phospho-Ser133 CREB, the coactivator CBP has been hypothesized to mediate target gene activation, in part, by acetylating and disrupting promoter-bound nucleosomes (3, 22). To determine whether HDAC inhibitors potentiate CREB activity by enhancing nucleosome acetylation over the somatostatin promoter during the burst phase, we performed ChIPs. Forskolin treatment had a small though reproducible effect on histone acetylation over the somatostatin promoter after 30 min (Fig. 2A). By contrast, TSA induced a robust fourfold increase in nucleosomal acetylation during the same time frame, and costimulation with cAMP had no greater effect than TSA alone (Fig. 2A).
FIG. 2.
HDAC inhibitors stimulate CREB activity during the late attenuation phase of transcription in response to cAMP. (A) ChIPs of untreated D5 cells (C) or D5 cells treated with forskolin (F) or TSA (T), either separately or in combination. Following a 30-minute stimulation, cells were treated with formaldehyde, and the cross-linked histone-DNA complexes were immunoprecipitated with anti-acetylated H4-specific antiserum. DNA recovered from each immunoprecipitation was analyzed for somatostatin gene sequences by slot blot hybridization with 32P-labeled somatostatin cDNA. No Ab, no primary antibody during immunoprecipitation; Total, 10% of total somatostatin hybridizing sequences applied to the immunoprecipitation reaction; αAc-H4, somatostatin hybridizing sequences that coimmunoprecipitate with somatostatin promoter sequences compared to control cells as shown in the bar graph. (B) Nuclear run-on transcription assay of D5 cells treated without (C) or with forskolin (F), TSA, or forskolin plus TSA for 30 min. 32P-labeled transcripts were hybridized to slot-blotted cDNAs encoding either somatostatin (som) or α-tubulin (tub). The bar graph shows mean ± standard error (n = 3) for each treatment. (C) Representative run-on transcription assay (n = 2) showing time course of somatostatin transcription in response to forskolin and HDAC inhibitor. D5 cells were treated with control vehicle (C), forskolin (F), or forskolin plus TSA (F+T) for times shown, and cells were harvested for run-on assay as described above. The bar graph shows relative somatostatin transcription rate in control and treated D5 cells normalized to tubulin.
To evaluate whether the TSA-dependent acetylation of nucleosomes over the somatostatin promoter during the burst phase is sufficient to promote somatostatin gene transcription in D5 cells costimulated with cAMP agonist, we performed run-on transcription assays. Following treatment with forskolin alone, somatostatin transcription rates increased four- to fivefold at 30 min (Fig. 2B). Remarkably, TSA had no effect on somatostatin gene expression at 30 min, either alone or in combination with forskolin (Fig. 2B). The lack of cooperativity between TSA and forskolin at this time point does not appear to reflect variable sensitivity of D5 cells to either inducer; TSA promoted H4 acetylation in greater than 60% of cells by immunofluorescence analysis, and forskolin stimulated Ser133 phosphorylation of CREB in about 90% of cells, suggesting that a majority of cells are responsive to both reagents (not shown).
The delayed effects of HDAC inhibitors on somatostatin mRNA accumulation in forskolin-treated D5 cells (Fig. 1A) prompted us evaluate somatostatin transcription rates at later times after stimulation. Following treatment with forskolin alone, somatostatin transcription rates returned to prestimulus levels by 4 h (Fig. 2C). By contrast, somatostatin transcription rates remained elevated throughout the attenuation phase in cells costimulated with forskolin plus TSA, suggesting that the underlying cooperativity between these inducers reflects a late effect of HDAC inhibitor on CREB activity (Fig. 2C).
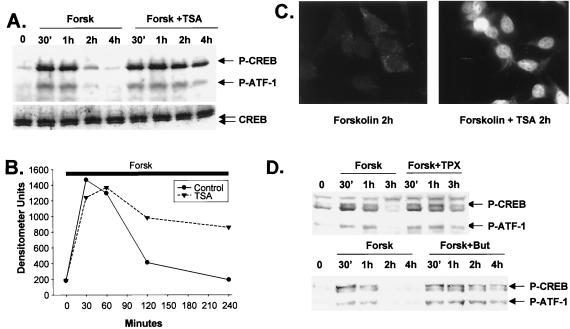
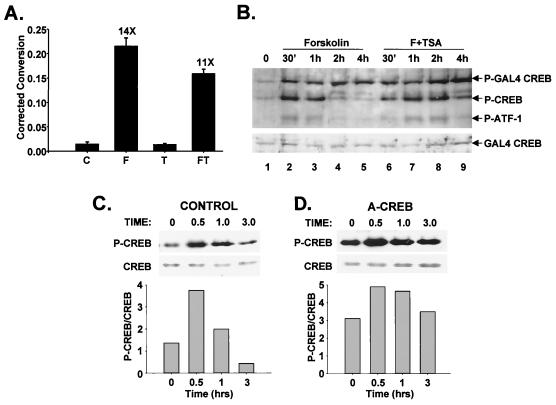
The importance of Ser133 phosphorylation for transcriptional induction via CREB prompted us to evaluate levels of phospho-Ser133 CREB in cells stimulated with cAMP agonist and/or HDAC inhibitor. Following treatment of D5 cells with forskolin, both CREB and its mammalian paralog ATF-1 were phosphorylated to maximal levels after 30 min (Fig. 3A). Reflecting the previously noted action of the Ser/Thr phosphatase PP-1 on both activators, levels of phospho-ATF-1 and phospho-CREB were diminished in parallel with target gene expression rates during the subsequent attenuation phase (Fig. 3A and B). TSA alone had no effect on CREB Ser133 phosphorylation during the 4-h assay period (not shown); when added in combination with forskolin, however, TSA strongly enhanced both CREB and ATF-1 phosphorylation in cells after 2 to 4 h (Fig. 3A). Similar results were observed by immunofluorescence analysis of D5 cells with phospho-CREB-specific antiserum. Nuclear phospho-Ser133) CREB staining was virtually undetectable after 2 h stimulation with forskolin alone, but phospho-CREB staining remained elevated in cells exposed to both forskolin plus TSA (Fig. 3C). Indeed, other HDAC inhibitors had similar effects: butyrate (15 mM) and trapoxin (100 nM) enhanced Ser133 phosphorylation of CREB and Ser64 phosphorylation of ATF-1 2 to 4 h following costimulation with cAMP agonist (Fig. 3D).
FIG. 3.
HDAC inhibitors potentiate target gene expression by prolonging Ser133 phosphorylation of CREB in cells costimulated with cAMP agonist. (A) Western blot assay of phospho-Ser133 CREB and total CREB levels in D5 whole-cell extracts following stimulation with forskolin (Forsk) or forskolin plus TSA for times indicated. Arrows (top to bottom) indicate phospho-Ser133 CREB, phospho-Ser64 ATF-1, and total CREB levels. (B) Graph showing relative levels of phospho-Ser133 CREB in cells treated with forskolin or forskolin plus TSA over time. The thick bar shows continuous treatment with forskolin for the length (240 min) of the assay. (C) Nuclear localization of phospho-Ser133 CREB in cells costimulated with forskolin plus TSA, visualized by immunofluorescence microscopy using phospho-Ser133 CREB-specific antiserum 5322 on D5 cells following a 2-h stimulation with forskolin or forskolin plus TSA, as indicated. (D) Other HDAC inhibitors promote CREB phosphorylation at late times in cells costimulated with forskolin. The Western blot represents extracts from D5 cells treated with forskolin (F), forskolin plus trapoxin (TPX; 100 nM), or forskolin plus sodium butyrate (But; 15 mM) for the times indicated.
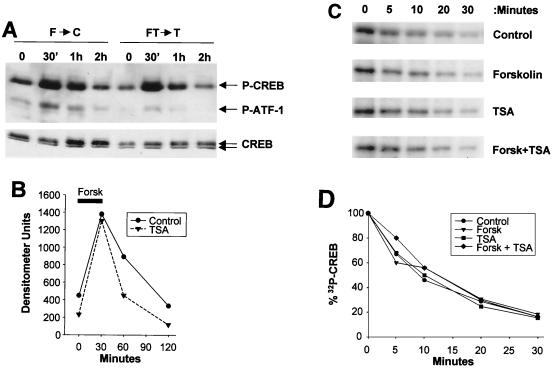
The importance of PP-1 in attenuating CREB activity at late times after cAMP induction prompted us to examine whether HDAC inhibitors act in part to block PP-1 activity. To test this model, we performed pulse-chase studies in which D5 cells were stimulated with forskolin or forskolin plus TSA for 30 min and then transferred either to control medium or to medium supplemented with TSA. Following removal of the forskolin stimulus, CREB was dephosphorylated with an estimated half-life of 30 min (Fig. 4A and B). TSA had no effect on cellular levels of phospho-Ser133 CREB during the chase period, indicating that PKA activity is continuously required for potentiation via HDAC inhibitors (Fig. 4A and B). CREB phosphatase activity in control and TSA-treated cells appeared comparable by in vitro phosphatase assay with phospho-Ser133 CREB, indicating that HDAC antagonists do not promote Ser133 phosphorylation by inhibiting cellular PP-1 activity (Fig. 4C and D).
FIG. 4.
cAMP agonist is continuously required for potentiation via HDAC inhibitors. (A) Western blot assay of D5 cells showing phospho-Ser133 CREB and phospho-Ser64 ATF-1 levels in control and TSA-treated D5 cells stimulated with forskolin (F) (2 μM) for 30 min and then transferred to control (F→C) medium or medium supplemented with TSA only (FT→T). Phospho-Ser133 CREB and total CREB levels in D5 whole-cell extracts were evaluated at different time points by Western blot assay. (B) Line graph showing relative levels of phospho-Ser133 CREB in control and TSA-treated cells in response to forskolin (Forsk) stimulation. The thick bar shows the time interval during which cells were exposed to forskolin (0 to 30 min). (C and D) HDAC inhibitors do not modulate CREB phosphatase activity in D5 cells. (C) In vitro CREB phosphatase assay using 32P-labeled phospho-Ser133 CREB plus nuclear extracts from control and treated D5 cells exposed to forskolin, TSA, or forskolin plus TSA for 2 h. Extracts (50 μg) were incubated with 32P-labeled CREB at 37°C for times indicated, and samples were visualized by SDS-PAGE. (D) Line graph comparing decay in phospho-Ser133 CREB levels, as determined following SDS-PAGE (C), from cells treated with HDAC inhibitor and/or forskolin compared to control.
Histone acetylation has been shown to enhance accessibility of nuclear factors and other regulatory factors to the mononucleosome (27). To evaluate whether acetylation of nucleosomes in response to HDAC inhibitors similarly increases accessibility of the signaling machinery to chromatin-bound CREB polypeptides, we compared CREB activities on chromosomal and plasmid templates, which are thought to assemble incompletely into chromatin structures and to promote transcription via chromatin-independent mechanisms (11). Following transient transfection into D5 cells, a somatostatin reporter plasmid was induced 14-fold by forskolin after 4 h. Remarkably, neither TSA (Fig. 5A), butyrate, nor trapoxin (not shown) had any effect on the extrachromosomal somatostatin template, either alone or in combination with cAMP agonist. Although we did not examine the chromatin state of the somatostatin reporter construct, the bulk of the evidence suggests that such templates do not recapitulate native genomic chromatin (25).
FIG. 5.
Persistent activation of non-chromatin-bound CREB polypeptides following stimulation with cAMP agonist suggests a chromatin-dependent mechanism for transcriptional attenuation. (A) HDAC inhibitors have no effect on cAMP-dependent induction of a nonchromosomal somatostatin gene in D5 cells. The bar graph shows CAT activity derived from a somatostatin promoter construct after normalizing to activity from cotransfected RSV-βGAL plasmid. Cells were stimulated with forskolin (F) and/or TSA (T), as indicated, for 4 h prior to harvesting. C, control. (B) Western blot assay of whole-cell extracts from D5 cells transfected with a GAL4-CREB(1-283) construct containing the GAL4 DNA binding domain but lacking the leucine zipper/DNA binding domain of CREB. At least 50% of cells were transfected, according to fluorescence from a cotransfected RSV-GFP expression vector. Cells were treated with forskolin alone or forskolin plus TSA (F+TSA) for times indicated, and levels of phospho-Ser133 CREB and phospho-Ser133 GAL-CREB(1-283) were evaluated by Western blot assay with phospho-Ser133-specific antiserum 5322. A Western blot with GAL4 antiserum to visualize total levels of GAL4-CREB in transfected cells is shown below. (C and D) Overexpression of dominant negative A-CREB polypeptide enhances phosphorylation of CREB in D5 cells stimulated with forskolin. Results show Western blot assay of D5 cells transfected with either A-fos (control) or A-CREB expression vector plus CMV-GFP fluorescence marker and then FACS sorted. A-fos- and A-CREB-expressing cells were stimulated for various times with forskolin, and whole-cell extracts were analyzed by Western blot assay with phospho-Ser133-specific antiserum or with nondiscriminating CREB antiserum (244). The bar graphs show ratio of phospho-CREB to total CREB at each time point, as estimated by densitometry.
The unresponsiveness of the extrachromosomal somatostatin template to HDAC inhibitor prompted us to examine whether the phosphorylation status of non-chromatin-bound CREB polypeptides might differ from that of the chromatin-bound population. Following transfection into D5 cells, a GAL4-CREB construct (amino acids 1 to 283) lacking the CREB DNA binding domain (amino acids 284 to 341), and therefore unable to bind to cellular CRE sites, was continuously phosphorylated in response to forskolin treatment with no discernible attenuation after 2 h (Fig. 5B, compare lanes 1 to 5). Costimulation of D5 cells with TSA had no effect on Ser133 phosphorylation of GAL4-CREB but potentiated Ser133 phosphorylation of endogenous CREB and Ser64 phosphorylation of the paralogous ATF-1 proteins in the same cells (Fig. 5B, compare lanes 4 and 8). These results suggest that HDAC inhibitors potentiate cellular gene expression via cAMP by enhancing phosphorylation of CREB and ATF-1 in a chromatin-dependent manner.
Based on the ability of A-CREB to heterodimerize with and block binding of CREB to cellular CRE sites (1), we examined whether overexpression of this dominant negative polypeptide would alter the status of Ser133 phosphorylation, in part, by liberating chromatin bound CREB protein and thereby increasing PKA accessibility. Following treatment with forskolin, CREB was maximally phosphorylated after 30 min, returning to prestimulus levels in control cells transfected with either empty expression vector or dominant negative control A-fos plasmid (Fig. 5C). By contrast, overexpression of A-CREB polypeptide markedly enhanced CREB Ser133 phosphorylation both under basal conditions and following forskolin treatment (Fig. 5D). Indeed, CREB remained heavily phosphorylated throughout the attenuation phase in A-CREB-expressing cells (1 to 3 h), underscoring the potential importance of chromatin localization for stimulus-appropriate regulation of CREB by PKA (Fig. 5C and D).
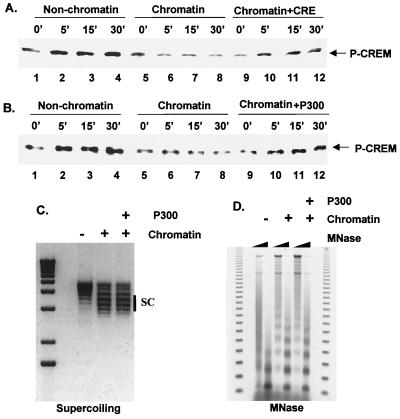
The ability of A-CREB to enhance CREB phosphorylation status led us to consider whether promoter-bound nucleosomes normally repress target gene transcription during the attenuation phase by blocking accessibility of PKA to its substrate. To test this model directly, we performed in vitro PKA phosphorylation assays on chromatin templates, using the CREB-related CREM protein as purified recombinant substrate. CREM was used in place of CREB for these assays due to its ease of purification from recombinant sources. Following assembly of periodic nucleosomal arrays on a cAMP-responsive template (3× CRE-MLP, 1.3 nM) with a Drosophila chromatin assembly extract (S190) (5), purified recombinant CREM protein (3.9 nM) was added to reactions. Under these conditions, CREM and CREB have been found to bind CRE sites on chromatin-assembled templates in a phosphorylation-independent manner (17). S190 extract induced low-level phosphorylation of CREM at Ser71 during chromatin assembly (Fig. 6A, lanes 1 and 2), but addition of PKA resulted in far higher levels of Ser71 phosphorylation in reactions lacking purified histones (Fig. 6A, compare lanes 2 and 5). PKA-mediated phosphorylation of CREM at Ser71 was severely attenuated on chromatin-assembled templates compared to templates lacking purified histones (Fig. 6A, compare lanes 2 to 5 and lanes 6 to 9). Supporting the notion that binding of CREM to chromatinized template is necessary to block accessibility to PKA, addition of CRE oligonucleotide in 100-fold molar excess (400 nM) relative to template rescued phosphorylation of CREM by PKA (Fig. 6A, compare lanes 6 to 9 and lanes 10 to 13).
FIG. 6.
Effect of nucleosomal templates on phosphorylation of CREM by PKA in vitro. (A) Western blot assay of phospho-Ser71 CREM and total CREM levels following addition of PKA to reactions containing chromatin assembled 3× CRE-MLP template. Addition of a 100-fold molar excess of CRE oligonucleotide competitor is indicated. (B) p300 partially rescues CREM Ser71 phosphorylation on chromatinized templates. Results show Western blot assay of phospho-Ser71 levels on chromatinized or nonchromatinized templates following incubation with PKA for times listed above the lanes. Addition of p300 (200 nM) plus TSA (100 ng/ml) to reactions is indicated. (C) One-dimensional chloroquine supercoiling (SC) gel from aliquots of the chromatin assembly reactions confirms nucleosome deposition on the 3× CRE-MLP template. (D) Micrococcal nuclease (MNase) digestion of the chromatin assembly reactions demonstrates the regularity of nucleosome spacing on the template. Increasing amounts of microccal nuclease are indicated by wedges.
The importance of CBP/p300 HAT activity for activation of cAMP-responsive genes prompted us to examine whether p300 could reverse the inhibitory effects of chromatin on PKA-mediated phosphorylation of CREM. When added to chromatin assembly reactions, p300 (200 nM) partially restored PKA-mediated phosphorylation of CREM, suggesting that nucleosome disruption may be sufficient to enhance accessibility of PKA to its target substrates (Fig. 6B, compare lanes 5 to 8 and lanes 9 to 12). Taken together, these observations support the notion that the phosphorylation status of a signal-dependent activator is in part determined by the configuration of the surrounding chromatin.
DISCUSSION
Nucleosome acetylation is thought to constitute an integral component in the process of target gene activation by extracellular stimuli. Our findings suggest that in addition to regulating assembly of the transcriptional machinery, promoter-bound nucleosomes may also modulate the phosphorylation status of nuclear activators by limiting their access to signal-dependent kinases.
Using a fibroblast line containing chromosomal copies of the somatostatin gene, we observed that three HDAC inhibitors (butyrate, TSA, and trapoxin) synergized with cAMP agonists to promote somatostatin gene expression on chromosomal but not extrachromosomal templates. Run-on transcription assays indicate that these inhibitors potentiate somatostatin transcription primarily during the late attenuation phase, by extending the time course over which CREB is phosphorylated in response to cAMP. Importantly, the initial rate and maximal amplitude of CREB phosphorylation were unaffected by HDAC inhibitors.
HDAC inhibitors do not appear to prolong CREB phosphorylation by either enhancing PKA or inhibiting PP-1 activities directly. Although we cannot rigorously exclude activation of other signaling pathways, our results point to a chromatin-dependent mechanism for these effects. In transient transfection assays, GAL4-CREB polypeptides lacking the CREB DNA binding domain and therefore unable to bind cellular CRE sites remained heavily phosphorylated throughout the attenuation phase and were unresponsive to HDAC inhibitor. Similarly, overexpression of a dominant negative A-CREB polypeptide that displaced CREB from its resident chromosomal sites strongly enhanced Ser133 phosphorylation during the late attenuation phase.
Imaging studies with fluorescence-tagged PKA suggest that the catalytic subunit enters the nucleus via passive diffusion in response to cAMP stimulation, equilibrating throughout the nuclear compartment, where it subsequently phosphorylates CREB (9, 10). Although Ser133 phosphorylation does not regulate CREB DNA binding activity per se, cross-linking and genomic footprinting studies support the notion that in some cases, occupancy of the CRE site by CREB on chromatin is modulated by cAMP agonist (21, 26). Evidence for an extrachromosomal pool of CREB that mobilizes to DNA upon PKA phosphorylation is lacking, but previous estimates of cellular CREB expression levels (5 × 104 copies per cell) support that notion (9).
Chromatin-bound CREB is likely to undergo several rounds of rephosphorylation during a 4-h stimulus, based on cellular pulse-chase experiments revealing that the half-life of phospho-Ser133 is 30 min. In view of their late effects, HDAC inhibitors may favor subsequent rounds of CREB phosphorylation either by promoting association of PKA with acetylated chromatin or, more likely, by enhancing access of chromatin bound CREB to PKA.
The inability of TSA to induce transcription from a transiently transfected template does not necessarily reflect a lack of chromatin structure on the reporter, but evidence from a number of laboratories supports that notion (25). Indeed, in vitro reconstitution studies with chromatin-assembled templates reveal that periodic nucleosomal arrays are capable of blocking the phosphorylation of CREB by PKA, and this effect can be partially reversed either by adding CRE oligonucleotide or by acetylating template-bound nucleosomes with p300. The inability of p300 to completely restore phosphorylation of CREM by PKA in these assays may reflect the less than stoichiometric acetylation of nucleosomes, owing perhaps to TSA-insensitive HDACs in Drosophila S190 extracts. Nevertheless, the low efficiency of CREB phosphorylation by PKA on unacetylated chromatin templates contrasts sharply with the high stoichiometry of cellular CREB phosphorylation observed during the initial phase of the cAMP response. In view of these results, a substantial pool of CREB in unstimulated cells may be targeted to acetylated chromatin or may actually be nonchromosomal, associating with chromatin subsequent to PKA phosphorylation and CBP recruitment. Studies focusing on the occupancy of CREB on these chromosomal sites following cAMP stimulus should provide new insights into this process.
ACKNOWLEDGMENTS
We thank Pamela Meluh for helpful advice with ChIPs assays, David Allis for providing acetylated histone-specific antisera, and Jackie Corbin for assistance with cellular PKA activation assays. We also thank Chuck Vinson for the gift of A-CREB plasmid.
This work was supported by NIH grants RO1-GM37828 and GM 19680.
REFERENCES
- 1.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty D, Vinson C. A dominant negative inhibitor of CREB reveals that it is a general mediator stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts A, Arias J, Hagiwara M, Montminy M, Feramisco J. Recombinant cyclic AMP response element binding protein (CREB) phosphorylated on Ser-133 is transcriptionally active upon its introduction into fibroblast nuclei. J Biol Chem. 1994;269:7623–7630. [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Braunstein M, Rose A, Holmes S, Allis D, Broach J. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1995;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 5.Bulger M, Kadonaga J. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:241–262. [Google Scholar]
- 6.Corbin J. Determination of the cAMP-dependent protein kinase activity ratio in intact tissues. Methods Enzymol. 1983;99:227–232. doi: 10.1016/0076-6879(83)99057-2. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 8.Hagiwara M, Alberts A, Brindle P, Meinkoth J, Feramisco J, Deng T, Karin M, Shenolikar S, Montminy M. Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell. 1992;70:105–113. doi: 10.1016/0092-8674(92)90537-m. [DOI] [PubMed] [Google Scholar]
- 9.Hagiwara M, Brindle P, Harootunian A, Armstrong R, Rivier J, Vale W, Tsien R, Montminy M R. Coupling of hormonal stimulation and transcription via cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13:4852–4859. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harootunian A, Adams S, Wen W, Meinkoth J, Taylor S, Tsien R. Movement of the free catalytic subunit of cAMP-dependent protein kinase into and out of the nucleus can be explained by diffusion. Mol Biol Cell. 1993;4:993–1002. doi: 10.1091/mbc.4.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong S, Stein A. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected COS-1 cells. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kee B, Arias J, Montminy M. Adaptor mediated recruitment of RNA polymerase II to a signal dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 13.Kim T, Maniatis T. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-b enhanceosome in vitro. Proc Natl Acad Sci USA. 1998;95:12191–12196. doi: 10.1073/pnas.95.21.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korzus E, Torchia J, Rose D, Xu L, Kurokawa R, McInerney E, Mullen T, Glass C, Rosenfeld M. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 15.Kraus W, Kadonaga J. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin R, Leone J, Cook R, Allis C. Antibodies raised to acetylated histones document the existence of deposition and transcription related histone acetylation in tetrahymena. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayall T, Sheridan P, Montminy M, Jones K. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 18.Montminy M R, Low M J, Tapia-Arancibia L, Reichlin S, Mandel G, Goodman R H. Cyclic AMP regulates somatostatin mRNA accumulation in primary diencephalic cultures and in transfected fibroblast cells. J Neurosci. 1986;6:803–813. doi: 10.1523/JNEUROSCI.06-04-01171.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakajima T, Uchida C, Anderson S, Lee C, Hurwitz J, Parvin J, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima T, Uchida C, Anderson S, Parvin J, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 21.Nichols M, Weih F, Schmid W, DeVack C, Kowenz-Leutz E, Luckow B, Boshart M, Schutz G. Phosphorylation of CREB affect its binding to high and low affinity sites: implications for cAMP induced gene transcription. EMBO J. 1992;11:3337–3346. doi: 10.1002/j.1460-2075.1992.tb05412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 23.Parekh B, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-b promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki K, Cripe T P, Koch S R, Andreone T L, Peterson D D, Beale E B, Granner K K. Multihormonal regulation of phosphoenolpyruvate carboxykinase gene transcription. J Biol Chem. 1984;259:15242–15251. [PubMed] [Google Scholar]
- 25.Smith C, Hager G. Transcriptional regulation of mammalian genes in vivo: a tale of two templates. J Biol Chem. 1997;272:27493–27496. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 26.Wolfl S, Martinez C, Majzoub J. Inducible binding of cyclic adenosine 3′,5′-monophosphate (cAMP) responsive element binding protein CREB to a cAMP responsive promoter in vivo. Mol Endocrinol. 1999;13:659–669. doi: 10.1210/mend.13.5.0282. [DOI] [PubMed] [Google Scholar]
- 27.Workman J, Kingston R. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]