ABSTRACT
Fluoroquinolones and cephalosporins are critically important antimicrobial classes for both human and veterinary medicine. We previously found a drastic increase in enrofloxacin resistance in clinical Escherichia coli isolates collected from diseased pigs from the United States over 10 years (2006 to 2016). However, the genetic determinants responsible for this increase have yet to be determined. The aim of the present study was to identify and characterize the genetic basis of resistance against fluoroquinolones (enrofloxacin) and extended-spectrum cephalosporins (ceftiofur) in swine E. coli isolates using whole-genome sequencing (WGS). blaCMY-2 (carried by IncA/C2, IncI1, and IncI2 plasmids), blaCTX-M (carried by IncF, IncHI2, and IncN plasmids), and blaSHV-12 (carried by IncHI2 plasmids) genes were present in 87 (82.1%), 19 (17.9%), and 3 (2.83%) of the 106 ceftiofur-resistant isolates, respectively. Of the 110 enrofloxacin-resistant isolates, 90 (81.8%) had chromosomal mutations in gyrA, gyrB, parA, and parC genes. Plasmid-mediated quinolone resistance genes [qnrB77, qnrB2, qnrS1, qnrS2, and aac-(6)-lb′-cr] borne on ColE, IncQ2, IncN, IncF, and IncHI2 plasmids were present in 24 (21.8%) of the enrofloxacin-resistant isolates. Virulent IncF plasmids present in swine E. coli isolates were highly similar to epidemic plasmids identified globally. High-risk E. coli clones, such as ST744, ST457, ST131, ST69, ST10, ST73, ST410, ST12, ST127, ST167, ST58, ST88, ST617, ST23, etc., were also found in the U.S. swine population. Additionally, the colistin resistance gene (mcr-9) was present in several isolates. This study adds valuable information regarding resistance to critical antimicrobials with implications for both animal and human health.
IMPORTANCE Understanding the genetic mechanisms conferring resistance is critical to design informed control and preventive measures, particularly when involving critically important antimicrobial classes such as extended-spectrum cephalosporins and fluoroquinolones. The genetic determinants of extended-spectrum cephalosporin and fluoroquinolone resistance were highly diverse, with multiple plasmids, insertion sequences, and genes playing key roles in mediating resistance in swine Escherichia coli. Plasmids assembled in this study are known to be disseminated globally in both human and animal populations and environmental samples, and E. coli in pigs might be part of a global reservoir of key antimicrobial resistance (AMR) elements. Virulent plasmids found in this study have been shown to confer fitness advantages to pathogenic E. coli strains. The presence of international, high-risk zoonotic clones provides worrisome evidence that resistance in swine isolates may have indirect public health implications, and the swine population as a reservoir for these high-risk clones should be continuously monitored.
KEYWORDS: plasmids, ESBLs, swine, USA, PMQR, WGS, cephalosporin, fluoroquinolone, antimicrobial resistance, PacBio, long-read sequencing, high-risk clones, epidemic plasmids
INTRODUCTION
Antimicrobial resistance has emerged as an issue of grave concern in both human and veterinary medicine. Food animals are considered potential reservoirs of antimicrobial-resistant and zoonotic pathogens such as Escherichia coli, although the extent of spread of resistant bacteria via the food chain is still under debate (1). Critically important antimicrobials for human medicine such as cephalosporins and fluoroquinolones are still used in many parts of the world to treat diseased food animals, including swine in the United States (2–4). Furthermore, certain genetic determinants responsible for resistance to antimicrobials approved for use in animals (such as ceftiofur and enrofloxacin) and those used in human medicine (such as cefoxitin and ciprofloxacin) are the same (5, 6). It is therefore important to monitor the circulation of genes responsible for resistance to such critically important antimicrobials in bacteria present in humans and animals to develop better source attribution models and targeted interventions in both human and veterinary medicine (7). A recent ban on colistin use in animal agriculture in China is an example of surveillance of antimicrobial resistance genes leading to actual policy changes (8) and a decrease in colistin resistance (9).
Resistance to extended-spectrum cephalosporins is complex and mediated by extended-spectrum beta-lactamases (ESBLs) (commonly encoded by the blaTEM, blaSHV, and blaCTX-M genes), carbapenemases (encoded by blaKPC, blaNDM, blaOXA-48, etc.), plasmidic AmpC (pAmpC; commonly encoded by the blaCMY genes), and mutations in AmpC promoter regions in the chromosome (10, 11). These bla genes may be inserted on bacterial chromosomes but are usually present on plasmids with the potential to disseminate horizontally to other bacterial strains (12). blaCTX-M genes are reported as the most prevalent ESBL genes worldwide in humans and animals (13). However, blaCMY-2 genes were primarily responsible for extended-spectrum cephalosporin resistance in bacteria of food animal origin in North America, while other ESBL-encoding genes were not reported until the late 2000s (14). Nevertheless, recent reports have also suggested the emergence of ESBL genes in bacteria of food animal origin in teh United States over the last decade (15). So far, blaCMY-2 genes have been found to be present on IncA/C2 and IncI1 plasmids in farm animals both globally (16–19) and in the United States (20, 21). On the other hand, blaCTX-M genes have been found to be present on IncF, IncI1, and IncN in farm animals in the United States (15, 22, 23).
Resistance to fluoroquinolones is mainly mediated by multiple chromosomal mutations in certain genes (gyrA, gyrB, parE, and parC). Additionally, plasmid-mediated quinolone resistance genes (such as qnr) and upregulation of efflux pumps confer variable levels of resistance to this antimicrobial family (24). qnr genes encoded in plasmids were also found in Salmonella isolates collected from retail pork, cecal samples from healthy pigs, and clinical samples from diseased pigs in the same period, suggesting a likely role in the increase in phenotypic resistance (25–27). An increase in fluoroquinolone resistance was recently reported in Salmonella enterica isolates from diseased pigs in Minnesota between 2006 and 2015 (2). A similar increase in phenotypic resistance to a fluoroquinolone (enrofloxacin) was also reported for the same time frame in swine E. coli clinical isolates (28), though the genetic determinants mediating this increase have not been determined yet.
Selective pressure due to exposure to antimicrobials or other chemicals can lead to a quick emergence of resistant bacterial strains (29). Clones of these resistant strains can quickly disseminate through the populations if they have key fitness advantages over nonresistant strains (29). In some instances, the presence of additional virulence factors can also confer additional fitness advantage on these bacterial clones. A hallmark example of clonal dissemination of a dominant E. coli clone is the global emergence of the highly pathogenic E. coli ST131 lineage that has been associated with the acquisition of mutations in quinolone resistance-determining genes or virulent IncF epidemic plasmids carrying blaCTX-M genes (30).
Although increasing information on the prevalence of phenotypic resistance in bacteria (including E. coli) of animal origin is generated by national antimicrobial resistance (AMR) monitoring programs such as NARMS (31), there is limited information on the genetic backbone mediating these resistance phenotypes. This may be of particular importance in the case of critically important antimicrobials such as fluoroquinolones, cephalosporins, or carbapenems. The objective of this study was to characterize the genetic basis of fluoroquinolone and extended-spectrum cephalosporin resistance in phenotypically resistant E. coli isolates collected from diseased pigs in the United States between 2014 and 2015 using both short-read (Illumina) and long-read (PacBio) whole-genome sequencing (WGS).
RESULTS
Genetic determinants conferring extended-spectrum cephalosporin and fluoroquinolone resistance.
Of 106 ceftiofur-resistant isolates, 87 (82.1%) carried blaCMY-2 genes (Fig. 1). These genes were not present in the remaining 105 ceftiofur-susceptible isolates. Isolates carrying this gene belonged to 24 different sequence types (STs), with ST12 (n = 21) and ST101 (n = 10) being the dominant STs (Fig. 1). Nineteen isolates of 11 different STs carried blaCTX-M genes (Fig. 1). All of the 19 blaCTX-M-carrying isolates were ceftiofur resistant. Five isolates of 3 different STs carried the blaSHV-12 gene, and 2 of these five isolates were ceftiofur susceptible. Twenty-two of the 106 ceftiofur-resistant isolates carried blaCTX-M or blaSHV-12 genes, whereas only 2 of the 105 ceftiofur-susceptible isolates carried blaSHV-12 genes. Four isolates carried combinations of blaCMY-2-blaCTX-M or blaCMY-2-blaSHV-12 genes.
FIG 1.

Maximum likelihood tree constructed using the core gene alignment of Escherichia coli isolates collected from diseased pigs at UMN-VDL between 2014 and 2015. This unrooted tree was created by mapping raw reads on E. coli K-12 substrain MG1655 (accession NZ_AJGD00000000.1), followed by extraction of SNPs and phylogenetic tree construction (GTR plus gamma substitution model, 1,000 bootstrap replicates) using RAxML version 8.0. Raw reads mapped onto 84.1% to 94.9% of the reference sequence E. coli K-12 substrain MG1655. Phylogenetic tree was constructed using 100,569 recombinant-free sites. Ceftiofur and enrofloxacin MIC values (in μg/ml) are labeled in red and blue to denote resistant and susceptible isolates, respectively. Heat map shows presence of chromosomal mutations in quinolone resistance-determining regions (QRDRs) (green squares), plasmid-mediated quinolone resistance (PMQR) genes (blue stars), ESBL/pAmpC genes (red circles), and virulotypes (APEC, ExPEC, UPEC, ETEC, and STEC) (purple triangles). Median pairwise SNP distances (MPDs) were estimated using ST-specific references. Colored clusters represent groups of isolates with an SNP distance to the next closest isolate of less than 100. Unknown, plasmids/chromosomes carrying these genes were not identified for these isolates.
Multiple fluoroquinolone resistance-associated genes and mutations were detected in 106 of the 110 enrofloxacin-resistant E. coli isolates (Table 1; see also Table S1 in the supplemental material), while only four of the 101 susceptible isolates presented any of them (specifically, single mutations in the gyrA gene [S83L or D87Y]). Isolates resistant to enrofloxacin belonged to 30 different STs.
TABLE 1.
Pattern of genetic determinants of enrofloxacin resistance in E. coli clinical isolates of swine origin
| MIC (μg/ml) | Pattern of genetic determinants (no. of isolates) | ST type(s) (no. of isolates) |
|---|---|---|
| >2 | gyrA(S83L) + gyrA(D87Y or D87N or D87G) + parC(S80I or S80R) ± other genetic determinantsa (49) | 744 (11), 100 (10), 224 (4), 410 (3), 10 (2), 457 (2), 617 (2), 4981 (2), 88 (1), 93 (1), 167 (1), 977 (1), 1585 (1), 2161 (1), 3901 (1) |
| 2 | gyrA(S83L) + parC(S80I or S80R) (23) | 100 (21), 58 (1), 90 (1) |
| qnrB19 (2) | 361 (1), 2496 (1) | |
| No genetic determinants (1) | 5926 (1) | |
| 1 | gyrA(S83L) + parC(S80I or S80R) (7) | 100 (6), 69 (1) |
| gyrA(S83L) only (1) | 6234 (1) | |
| gyrA(D87G) + qnrB2 (1) | 10 (1) | |
| aac(6′)-Ib-cr + qnrB2 (1) | 540 (1) | |
| Single PMQR (qnrB19, qnrS1, qnrS2, qnrB2, or qnrB77) (8) | 10 (4), 101 (1), 641 (1), 847 (1), 5759 (1) | |
| 0.25–0.5 | gyrA(S83L) ± aac(6′)-Ib-cr (6) | 6234 (2), 10 (1), 58 (1), 101 (1), 410 (1) |
| Single PMQR (qnrB19, qnrS1, qnrS2, qnrB2) (5) | 10 (3), 93 (1), 1112 (1) | |
| gyrA (D87G or D87Y) (3) | 10 (1), 88 (1), 641 (1) | |
| aac(6′)-Ib-cr + qnrB2 (1) | 641 (1) | |
| No genetic determinants (2) | 641 (1), 3057 (1) | |
| ≤0.125 | gyrA(S83L) (3) | 10 (1), 847 (1), unknown (1) |
| gyrA(D87Y) (1) | 90 (1) |
This genetic determinant might or might not be present in isolates with that particular MIC value. Other determinants were parC(A56T or E84G), parE(S458A or L416F), and single PMQR [aac(6′)-Ib-cr, qnrB77, or qnrB19].
Detailed virulence, antimicrobial resistance, and typing metadata of all isolates generated in this study. Download Table S1, XLSX file, 0.2 MB (213.1KB, xlsx) .
Copyright © 2020 Hayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Six different types of plasmid-mediated quinolone resistance (PMQR) genes were identified in a total of 25 isolates spread across 7 states (Fig. 1; Table S1). These 25 isolates belonged to 16 different STs (Table 1, Fig. 1, and Table S1). Enrofloxacin MIC values for isolates with a single PMQR gene, two PMQR genes, and one PMQR gene plus a chromosomal mutation (gyrA-S83L, D87G, or parE-D476A) ranged between 0.5 and 1.0 μg/ml, with the exception of two isolates that carried only qnrB19 but had an enrofloxacin MIC value of 2 μg/ml.
Description of assembled plasmids carrying PMQR and ESBL genes.
We assembled complete E. coli chromosome and plasmid sequences using both long and short reads from 10 isolates (six isolates carrying only blaCTX-M genes, two carrying blaSHV-12 genes, one carrying blaCTX-M and qnrB77 genes, and one carrying blaCTX-M and blaCMY-2 genes) (Table 2). In seven of the isolates, blaCTX-M genes were present on IncFII (blaCTX-M-14, -15, -27) and IncHI2 (blaCTX-M-55) plasmids, with sizes ranging between 69 and 240 kbp. blaCTX-M genes were present in regions flanked by IS26, ISEcp1, IS5, IS6, and Tn3 family transposases, which were often truncated (Table 2; Fig. 2, 3, and 4). In one isolate, blaCTX-M-15 was present on the E. coli chromosome, flanked by transposases similar to those surrounding blaCTX-M-15 in the IncFII plasmids. Plasmids with blaCTX-M-14 or blaCTX-M-27 carried only blaCTX-M or one other AMR gene [erm(B), a macrolide resistance gene], whereas the plasmids carrying blaCTX-M-15 and blaCTX-M-55 also bore genes which can confer resistance to aminoglycosides, penicillins, macrolides, or trimethoprim (Table 2; Fig. 2 to 4). Additionally, some of these blaCTX-M-15 and blaCTX-M-55 plasmids also carried genes that can cause resistance to sulfonamides, phenicols, or tetracyclines (Table 2; Fig. 2 and 4). Two of the blaCTX-M-15-carrying IncFII plasmids also harbored the aac(6′)-Ib-cr gene which can confer resistance to both aminoglycosides and fluoroquinolones (Table 2; Fig. 4).
TABLE 2.
Characteristics of plasmids assembled in this study
| Plasmid (GenBank accession no.) | Gene of interest | Size of plasmid (kbp) | Replicon type (pMLST) | ST | Other AMR gene(s) [drug resistance]a | Virulence gene(s) |
|---|---|---|---|---|---|---|
| p77 (MT077889) | bla CTX-M-14 | 76 | IncF (F2:A-:B-) | 10 | traT | |
| p37 (MT077885) | bla CTX-M-27 | 75 | IncF (F2:A-:B-) | 744 | erm(B) [MA] | traT |
| p62 (MT077887) | bla CTX-M-27 | 69 | IncF (F2:A-:B-) | 10 | traT | |
| p1 (MT077880) | bla CTX-M-15 | 171 | IncF (F31:F36:A4:B1) | 617 | aadA5, aac(3)-IIa, aac(6′)-Ib-cr [AM]; blaOXA-1 [PE]; mph(A) [MA]; sul1, dfrA17 [TS]; catB3 [PH]; and tet(B) [TE] | traT, sitA, iucC, iutA |
| p2 (MT077881) | bla CTX-M-15 | 168 | IncF (F31:F36:A4:B1) | 58 | aac(6′)-Ib-cr [AM]; blaOXA-1 [PE]; mph(A) [MA]; dfrA17 [TS]; catB3 [PH]; and tet(B) [TE] | traT, sitA, iucC, iutA |
| p4 (MT077882) | bla CTX-M-15 | 115 | IncF (F48:A1:B49) | 744 | aac(3)-IIa [AM]; blaTEM-1b [PE]; mph(A) [MA]; and dfrA17 [TS] | traT |
| p65 (MT077888) | bla CTX-M-55 | 241 | IncHI2 (ST-2) | 165 | aac(3)-IId, aadA2, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id [AM]; blaTEM-1b [PE]; mph(A) [MA]; sul1, dfrA12 [TS]; and tet(M) [TE] | terC |
| p33 (MT077884) | bla SHV-12 | 302 | IncHI2 (ST-1) | 641 | aac(6′)-Ib3, aac(6′)-IIc, aph(6′)-Id, aph(3′)-Ib, aadA2, aac(6′)-Ib-cr [AM]; blaTEM-1b [PE]; qnrB2 [FL]; ere(A) [MA]; sul1, sul2, dfrA19 [TS]; tet(D) [TE]; and mcr-9 [CO] | terC |
| p39 (MT077886) | bla SHV-12 | 289 | IncHI2 (ST-1) | 1112 | aph(3″)-Ib, aph(6′)-Id, aph(3′)-Ia, aac(6′)-IIc [AM]; blaTEM-1b [PE]; ere(A) [MA]; sul1 [TS]; catA2 [PH]; tet(D) [TE]; and mcr-9 [CO] | terC |
| pCMY (MT816498) | bla CMY-2 | 168 | IncA/C2 (ST-3) | 10 | aac(3)-VIa, aadA24, aph(3″)-Ib, aph(6)-Id [PE]; sul1, sul2 [TS]; floR [PH]; and tet(A) [TE] | |
| p23 (MT077883) | qnrB77 | 59 | IncN (unknown) | 4981 | aac(3)-VIa, aadA1 [AM]; and dfrA15 [TS] |
AM, aminoglycosides; PE, penicillins; FL, fluoroquinolones; MA, macrolides; TS, trimethoprim/sulfonamide; PH, phenicols; TE, tetracyclines; CO, colistin.
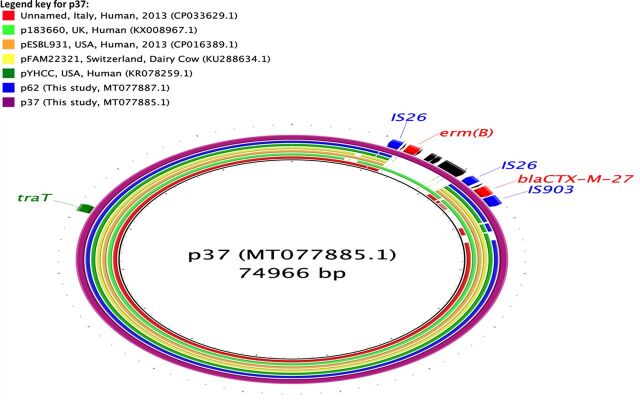
FIG 2.
Circular maps representing comparisons of blaCTX-M-14 (p77)- and blaCTX-M-55 (p65)-carrying plasmids available at GenBank and plasmids assembled in this study. The innermost rings (not colored black) represent the top plasmids with high nucleotide identity and coverage with respect to reference plasmids (p77 and p65). The legends at the top left present plasmid name, country, animal species/human, and year of isolation, where available. Areas of the plasmids carrying AMR genes are presented in the outermost rings. AMR genes, genes associated with mobile elements, and virulence genes are colored and labeled in red, blue, and green, respectively. Truncated genes are presented with Δ as a prefix.
FIG 3.
Circular map representing comparison of blaCTX-M-27 (p37 and p62)-carrying plasmids available at GenBank and plasmids assembled in this study. The innermost rings (not colored black) represent the top plasmids with high nucleotide identity and coverage with respect to reference plasmid (p37). The legend at the top left presents plasmid name, country, animal species/human, and year of isolation, where available. Area of the plasmid carrying AMR genes is presented in the outermost ring. AMR genes, genes associated with mobile elements, and virulence genes are colored and labeled in red, blue, and green, respectively.
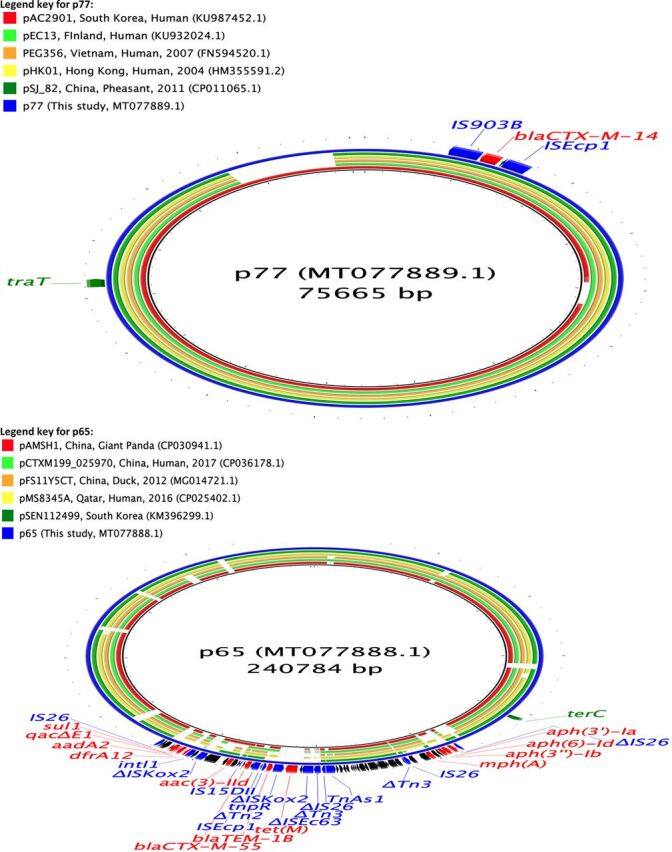
FIG 4.
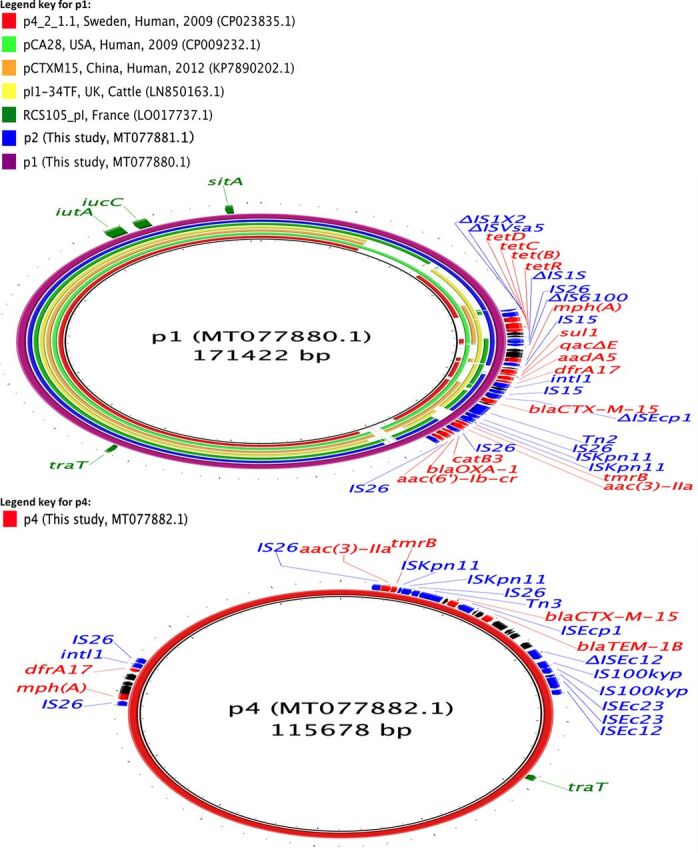
Circular maps representing comparisons of blaCTX-M-15 (p1, p2, and p4)-carrying plasmids available at GenBank and plasmids assembled in this study. The innermost rings (not colored black) represent the top plasmids with high nucleotide identity and coverage with respect to reference plasmids (p1). There were no plasmids similar to p4. The legends at the top left present plasmid name, country, animal species/human, and year of isolation, where available. Areas of the plasmids carrying AMR genes are presented in outermost rings. AMR genes, genes associated with mobile elements, and virulence genes are colored and labeled in red, blue, and green, respectively. Truncated genes are presented with Δ as a prefix.
The two plasmids carrying blaSHV-12 genes assembled were of large IncHI2-type plasmids (approximately 287 to 300 kbp) and carried genes for resistance to aminoglycosides, sulfonamides, trimethoprim, tetracyclines, penicillins, phenicols (only p39), and macrolides (Table 2; Fig. 5). blaSHV-12 genes were present in a region flanked by intact IS6 family transposases. One of these plasmids also carried genes for resistance to fluoroquinolones [qnrB2, aac(6′)-Ib-cr] and both of these plasmids also carried a colistin resistance gene (mcr-9) (Table 2; Fig. 5).
FIG 5.
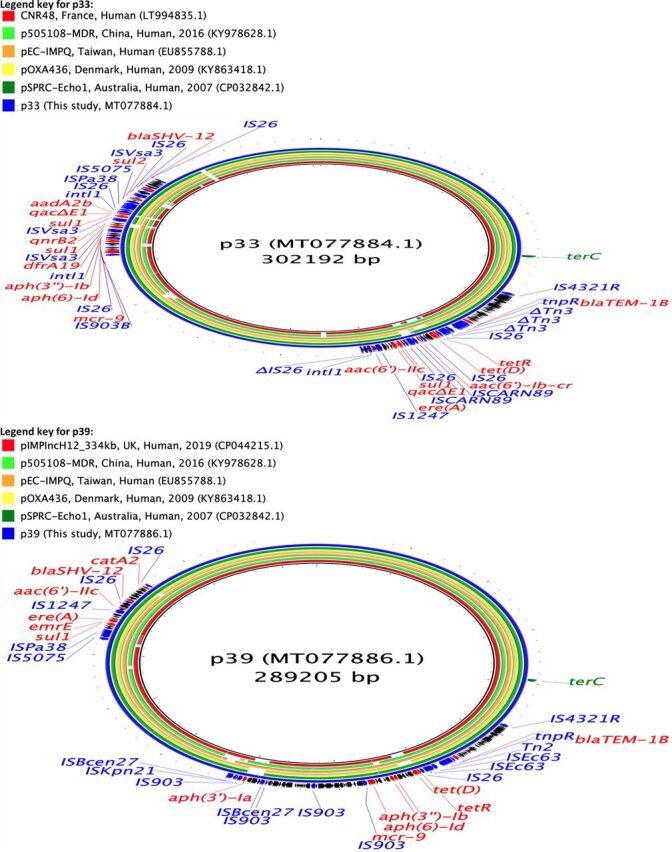
Circular maps representing comparisons of blaSHV-12 (p33 and p39)-carrying plasmids available at GenBank and plasmids assembled in this study. The innermost rings (not colored black) represent the top plasmids with high nucleotide identity and coverage with respect to reference plasmids (p33 and p39). The legends at the top left present plasmid name, country, animal species/human, and year of isolation, where available. Areas of the plasmids carrying AMR genes are presented in the outermost rings. AMR genes, genes associated with mobile elements, and virulence genes are colored and labeled in red, blue, and green, respectively.
The plasmid carrying blaCMY-2 (pCMY) assembled here was present alongside a blaCTX-M-27-carrying plasmid IncF (F2:A-:B-) in an E. coli ST10 isolate. This plasmid was of IncA/C2-ST3 type, 168 kbp in size, and carried resistance genes to aminoglycosides, sulfonamides, phenicols, and tetracyclines (Table 2; Fig. 6). The blaCMY-2 gene was flanked by IS1380 transposase on one side and blc-sugE on the other, and this genetic environment is commonly associated with the presence and dissemination of blaCMY-2 genes universally. This plasmid (pCMY) had 92% coverage and 100% nucleotide identity with a blaCMY-2-carrying plasmid assembled recently from Salmonella enterica isolates from diseased pigs in the United States (accession number MK191845.1). Similarly, pCMY had 93% coverage and 99.97% nucleotide identity with respect to pUMNK88 (accession number HQ023862.1), which was among the first blaCMY-2-carrying plasmids to be isolated and assembled from E. coli isolates collected from diseased pigs in the United States in 2008. This plasmid was highly similar (97% coverage, 99% nucleotide identity) to another plasmid isolated from cow in the United States in 2002 (accession number FJ621588.1), suggesting that blaCMY-2-carrying IncA/C2 plasmids isolated from farm animals in the United States have remained relatively conserved over a long duration with the exception of a gain or loss of some AMR genes.
FIG 6.
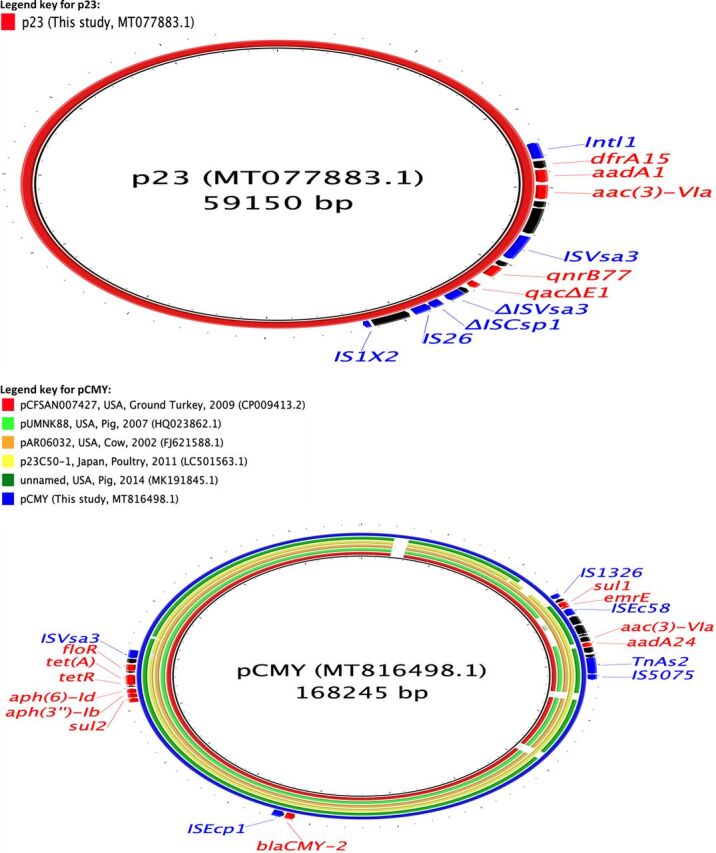
Circular maps representing comparisons of qnrB77 (p23)- and blaCMY-2 (pCMY)-carrying plasmids available at GenBank and plasmids assembled in this study. The innermost rings (not colored black) represent the top plasmids with high nucleotide identity and coverage with respect to reference plasmids (pCMY). There were no plasmids similar to p23. The legends at the top left present plasmid name, country, animal species/human, and year of isolation, where available. Areas of the plasmids carrying AMR genes are presented in the outermost rings. AMR genes and genes associated with mobile elements are colored and labeled in red and blue, respectively. Truncated genes are presented with Δ as a prefix.
In addition to these ESBL-encoding plasmids, we also assembled a 59-kbp IncN plasmid carrying a qnrB77 gene (Table 2; Fig. 6). This plasmid was present in a ST4981 isolate which also carried an ESBL-encoding gene (blaCTX-M-15) chromosomally. This plasmid also carried resistance genes to trimethoprim and aminoglycosides. The qnrB77 gene was flanked by a complete and a truncated transposase of the IS91 family of transposases (Table 2; Fig. 6).
Some of these plasmids (p1, p23, p33, and p65) also carried genes (qacΔE) that determine resistance to quaternary ammonium compounds. pCMY also carried the sugE gene that modulates resistance to quaternary ammonium compounds. Genes related to heavy metal resistance such as mercury (merCDEPTR), arsenic (arsHB), copper (pcoES), and tellurium (terABCDWX) resistance were also present on plasmids carrying blaSHV-12 and pCMY (merCDEPTR operon only). Plasmids carrying blaCTX-M-55 genes also carried tellurium resistance genes (terABCDWX). Additionally, all the plasmids assembled in this study carried mobility genes (tra set of genes) and genes that can aid in plasmid maintenance and stability. All of the IncFII and IncHI2 plasmids carried genes coding for at least one toxin-antitoxin system, e.g., the IncFII plasmids carried pemI-pemK genes, and all the IncHI2 and IncA/C2 plasmids carried higA-higB genes. Similarly, the qnrB77-carrying IncN plasmid also carried mobility genes (tra) and genes encoding proteins that aid in plasmid stability (stbB-stbC genes), antirestriction systems (ardA-ardB genes), and mutagenesis (mucA-mucB genes).
The comparison of these assembled plasmids with the PLSDB database resulted in the identification of several previously described plasmids with a high similarity (>80% coverage, and >98% nucleotide identity). To summarize, most of the plasmids carrying ESBL-encoding genes assembled in this study were similar to plasmids harbored on various Enterobacteriaceae and collected from various sources (animals, humans, and the environment) across different continents and shared the same molecular context around the genes of interest (qnr and bla genes) (Fig. 2 and 6). In contrast, we were not able to identify similar plasmids to the blaCTX-M-15-carrying IncFII (plasmid multilocus sequence type [pMLST], F48:A1:B49) and the qnrB77-carrying IncN plasmids found in this study.
We were also able to assemble 61 putative plasmids using these assembled plasmids as references (Table 3). Additionally, 1 blaCTX-M-15 was identified to be present on the chromosome of a ST4981 isolate. We also identified reference plasmids by conducting nucleotide BLAST searches in the GenBank database using contigs on which resistance genes were present. By mapping short reads onto these reference plasmids, we further identified that blaCMY-2 genes were present on a variety of IncI1-ST65, -12, -2, -20, -23, and -266 and IncI2 plasmids (Table 3). We also identified putative blaCTX-M-27-IncF- (F24:A-:B1), blaCTX-M-55-IncF- (F18:A-:B1), and blaCTX-M-55-IncI1-ST16 plasmids in our study (Table 3). Finally, we identified 6 qnrB19-ColE plasmids (sizes ranging between 2.8 and 3.2 kbp) and 2 qnrS2-IncQ2 plasmids (approximately 7.7 kbp in size) present as contigs in the draft E. coli isolates assembled (Table 3). Overall, by combining these different strategies, we were able to identify the AMR genes-plasmid/chromosome combinations for 85.1%, 91.7%, and 81.5% of the blaCMY-2, blaCTX-M/SHV-12, and PMQR genes, respectively.
TABLE 3.
Characteristics of putative plasmids assembled in this study
| Gene | Reference sequence GenBank accession no. (size [kbp]) | pMLST | E. coli ST (no. of isolates) | Median coveragea (range) | Median SNP difference (range) | Virulence gene(s) |
|---|---|---|---|---|---|---|
| bla CMY-2 | MT816498.1 (168) | IncA/C2-ST3 (n = 53) | ST12 (20), ST100 (8), ST101 (6), ST10 (2), ST23 (2), ST410 (2), ST58 (2), ST744 (2), ST90 (2), ST127 (1), ST224 (1), ST6234 (1), ST69 (1), ST88 (1), ST93 (1), ST977 (1) | 89.6 (72.3–97.2) | 2 (0–18) | |
| LC501512.1 (80) | IncI1-ST65 (n = 6) | ST10 (4), ST100 (1), ST23 (1) | 95.5 (94.2–97.0) | 10 (5–40) | cia | |
| MK191846.1 (99) | IncI1-ST12 (n = 5) | ST58 (2), ST75 (2), ST744 (1) | 99.2 (95.5–99.9) | 2 (0–157) | cib | |
| CP023356.1 (95) | IncI1-ST2 (n = 3) | ST101 (1), ST410 (1), ST58 (1) | 98.8 (97.1–100) | 3 (2–4) | cia | |
| CP027535.1 (101) | IncI1-ST20 (n = 1) | ST2025 (1) | 92.9 | 5 | cib | |
| CP009566.1 (95) | IncI1-ST23 (n = 1) | ST10 (1) | 94.7 | 2 | cia | |
| CP029976.1 (34) | IncI1-ST266 (n = 1) | ST766 (1) | 99.9 | 14 | ||
| CP043196.1 (65) | IncI2 (n = 3) | ST154 (1), ST23 (1), ST58 (1) | 98.5 (97.9–98.6) | 4 (4–10) | ||
| NAb | Unknownc (n = 13) | ST10 (4), ST101 (2), ST12 (1), ST3057 (1), ST369 (1), ST410 (1), ST4373 (1), ST73 (1), ST90 (1) | NA | NA | NA | |
| bla CTX-M-14 | MT077889.1 (76) | IncF (F2:A-:B-) (n = 1) | ST10 (1) | 91.7 | 0 | traT |
| bla CTX-M-15 | MT077880.1 (171) | IncF (F31:36:A4:B1) (n = 2) | ST617 (1), ST167 (1) | 74.0–91.5 | 3–24 | traT, sitA, iucC, iutA |
| MT077882.1 (115) | IncF (F48:A1:B49) (n = 1) | ST744 (1) | 96.9 | 0 | traT | |
| NA | Unknown (n = 1) | ST410 (1) | NA | NA | NA | |
| bla CTX-M-27 | AP017621.1 (117) | IncF (F24:A-:B1) (n = 1) | ST1585 (1) | 80.7 | 48 | hlyF, ompT, sitA |
| bla CTX-M-55 | MN158989.1 (128) | IncF (F18:A-:B1) (n = 3) | ST457 (2), ST744 (1) | 90.1 (89.1–90.6) | 5 (1–48) | cma, cvaC, hlyF, iucC, iutA, ompT, sitA, traT |
| KX246268.1 (86) | Inc1-ST16 (n = 1) | ST101 (1) | 98.9 | 0 | ||
| bla SHV-12 | MT077886.1 (289) | IncHI2A-ST1 (n = 2) | ST10 (2) | 95.1–95.2 | 8–9 | terC |
| NA | Unknown (n = 1) | ST641 (1) | NA | NA | NA | |
| qnrB19 | KY991369.1 (3) | ColE (n = 6) | ST10 (1), ST2161 (1), ST2496 (1), ST361 (1), ST5759 (1), ST93 (1) | 95.8 (91.6–100) | 0 | |
| qnrB2 | MT077886.1 (289) | IncHI2-ST1 (n = 2) | ST10 (2) | 95.1–95.2 | 8–9 | terC |
| NA | Unknown (n = 2) | ST10 (1), ST540 (1) | NA | NA | NA | |
| qnrB77 | MT077883.1 (59) | IncN unknown (n = 2) | ST10 (1), ST100 (1) | 90.7–91.9 | 2–4 | |
| qnrS1 | NA | Unknown (n = 2) | ST10 (1), ST641 (1) | NA | NA | NA |
| qnrS2 | KT896500.1 (7.7) | IncQ2 (n = 2) | ST10 (1), ST101 (1) | 99–100 | 0 | |
| NA | Unknown (n = 2) | ST10 (1), ST847 (1) | NA | NA | NA | |
| aac-(6′)-lb-cr | MT077880.1 (170) | IncF (F31:36:A4:B1) (n = 2) | ST617 (1), ST167 (1) | 74.0–91.5 | 3–24 | traT, sitA, iucC, iutA |
| NA | Unknown (n = 1) | ST410 (1) | NA | NA | NA |
Coverage was estimated using following formula: (size of reference − size of putative plasmid)/size of reference × 100.
NA, not applicable.
Unknown, plasmids/chromosomes carrying these genes were not identified for these isolates.
Clonal and horizontal transmission of AMR.
The spread of ceftiofur and enrofloxacin resistance was bimodal. Both widespread dissemination of clones with specific E. coli-plasmid combinations as well as horizontal transmission of plasmids between genetically unrelated E. coli STs contributed to successful spread of AMR in the United States.
Fifty-four E. coli isolates belonging to 16 different STs harbored blaCMY-IncA/C3-ST3 plasmids (Table 3), which is an indicator of successful horizontal dissemination of these plasmids across multiple E. coli strains. Similarly, 6 E. coli-blaCMY-2-IncI1-ST65 isolates were of 3 different STs (ST10, ST23, and ST100) and 5 E. coli-blaCMY-2-IncI1-ST12 isolates were also of 3 different STs (ST58, ST75, and ST744) (Table 3). Similar to that for blaCMY-2 genes, blaCTX-M genes were also present on a wide variety of plasmids and E. coli STs. For example, blaCTX-M-55 genes were present on 3 different plasmid types and present in 5 E. coli isolates of 4 different STs (ST165, ST457, ST744, and ST101) (Tables 3). qnrB19 was the most prevalent PMQR gene and was located on ColE plasmids in 6 E. coli isolates, all of which were of different STs (Table 3). The details of the prevalence of blaCTX-M, blaSHV-12, and PMQR genes and their association with E. coli STs and plasmids are presented in Table 3.
There were also some clusters of genetically similar isolates having same ST-AMR genetic mechanism-plasmid type combinations, indicating clonal transmission of certain resistant bacterial clones throughout swine populations in the United States. Clusters of isolates with genetic distance between the next closest isolate being <100 single nucleotide polymorphisms (SNPs) and carrying genetic mechanisms of resistance relevant to this study are indicated in Fig. 1. The biggest examples of clonal transmission in our data set are ST100 E. coli isolates, 36 of which had chromosomal mutations in gyrA83 and parC80 and had a means median pairwise SNP distance (MPD) of 36 SNPs (range, 4 to 63 SNPs) (Fig. 1). Seventeen ST744 isolates had chromosomal mutations in gyrA83, gyrA87, parC56, and parC80, with an MPD of 115 SNPs (range, 10 to 203 SNPs) (Fig. 1). Twenty-two of the 23 ST12 and 6 of the 12 ST101 E. coli isolates carried blaCMY-2-IncA/C2-ST3 (MPD between these 22 ST12 isolates, 66 SNPs [range, 32 to 164 SNPs]; MPD between the 6 ST101 isolates, 70 SNPs [range, 31 to 87 SNPs]) (Fig. 1). It should be noted that some very close isolates harbored different AMR genes and plasmids. For example, in related ST10 isolates (MPD, 101 SNPs [range, 20 to 121 SNPs]), blaCMY-2 genes were present on IncA/C2-ST3, IncI1-ST23, and IncI1-ST65 plasmids (Fig. 1).
Comparison with other isolates available on Enterobase.
Some of the isolates of ST1581, -58, -457, -4981, and -744 in our study were found to be within 20 allelic differences of isolates collected from humans globally based on core genome multilocus sequence type (cgMLST) (Fig. 7). ST744 E. coli similar to those in this study were isolated from humans, pet animals, wild animals, farm animals, and environmental samples globally (Fig. 7). Specifically, these ST744 isolates were genetically similar (within 20 allelic differences) to those isolated from diseased and nondiseased humans in the Philippines, the Netherlands, the United States, Russia, Germany, the United Kingdom, Colombia, Denmark, and Switzerland. ST744 isolates from our study and the Enterobase collection consistently had mutations in quinolone resistance-determining regions (QRDRs) of gyrA and parC genes. Moreover, 2 ST744 isolates from our study were similar to isolates collected from diseased humans in the United States and the Netherlands (39 to 83 SNP differences) and also harbored blaCTX-M-15-carrying IncF (F48:A1:B49) plasmids, which have been described to be novel in this study (Fig. 7). Some of these ST744 isolates (including one from this study) were classified as avian pathogenic E. coli (APEC).
FIG 7.

Maximum likelihood tree constructed using the core gene alignment of selected Escherichia coli isolates collected in this study and isolates available at Enterobase. E. coli isolates from Enterobase were selected by identifying those that were within 20 allelic differences (same HC20) of the isolates assembled in our study. This unrooted tree was created by mapping raw reads on E. coli K-12 substrain MG1655 (accession NZ_AJGD00000000.1), followed by extraction of SNPs and phylogenetic tree construction (GTR plus gamma substitution model, 1,000 bootstrap replicates) using RAxML version 8.0. Raw reads mapped onto 84.2% to 94.2% of the reference sequence E. coli K-12 substrain MG1655. Phylogenetic tree was constructed using 43,503 recombinant-free sites. Heat map shows presence of chromosomal mutations in quinolone resistance-determining regions (QRDRs) (green squares), plasmid-mediated quinolone resistance (PMQR) genes (blue stars), ESBL/pAmpC genes (red circles), and virulotypes (APEC, ExPEC, UPEC, and ETEC) (purple triangles). Median pairwise SNP distances (MPDs) were estimated using ST-specific references. Colored clusters represent groups of isolates with an SNP distance to the next closest isolate of less than 100. Unknown, plasmids/chromosomes carrying these genes were not identified for these isolates.
ST1581, ST457, and ST4981 isolates from our study also carried the same AMR genes, mutations, and plasmids as present in similar human isolates (Fig. 7). The ST457 isolate from this study was not classified into a virulotype, but other ST457 isolates in the Enterobase were classified as extraintestinal pathogenic E. coli (ExPEC). One of the APEC ST58 isolates was genetically similar to another isolate collected from a diseased human in the United Kingdom (SNP difference, 60).
Several other E. coli isolates were found to be genetically similar to those isolated from animals only. These consisted of ST10, ST23, ST224, ST847, and ST12. ST10, ST12, and ST847 were similar to those isolated from pork or healthy pigs in the United States, and ST10 and ST12 isolates from this study and the Enterobase collection had same genetic determinants of ceftiofur and enrofloxacin resistance. In this comparative analysis, 26 of 28 ST12 isolates were classified as ExPEC and uropathogenic E. coli (UPEC).
Resistance determinants to other critical antimicrobials.
No carbapenem resistance genes were identified in our collection, but the mcr-9 gene was present in 7 isolates belonging to 6 different STs. These isolates carried both the mcr-9 gene and either a pAmpC, an ESBL, or a PMQR gene (Table 4). Descriptions of these isolates are presented briefly in Table 4. mcr-9 was also present in two of the ESBL plasmids assembled in this study (Table 2).
TABLE 4.
Characteristics of isolates carrying mcr-9 genes
| Isolate SRA accession no. | ST (serotype, phylotype) | Other AMR genes in the isolate [drug resistance]a | Virulotype | Plasmid replicons (pMLST results) |
|---|---|---|---|---|
| SRR11048580 | 540 (O9:H10, A) | aac(6′)-IIc, aadA2b, aac(6′)-Ib3, aph(3″)-Ib, aph(6)-Id, aac(6′)Ib-cr [AM]; blaTEM-1b [PE]; qnrB2 [FL]; ere(A), mdf(A) [MA]; sul1, sul2, sul3, dfrA12, dfrA19 [TS]; and tet(A), tet(M) [TE] | IncHI2A (ST1), IncI1 (ST266), IncX1 | |
| SRR11045321 | 641 (O121:H10, B1) | aac(6′)-Ib3, aac(6′)-IIc, aph(6′)-Id, aph(3″)-Ib, aadA2, aac(6′)-Ib-cr [AM]; blaTEM-1b [PE]; qnrB2 [FL]; ere(A), mdf(A) [MA]; sul1, sul2, dfrA19 [TS]; tet(D), tet(B) [TE]; blaSHV-12 [EC] | ETEC | IncFII (F13:F29:A-:B-), IncHI2A (ST1), p011 |
| SRR11046298 | 1112 (O142:H27, A) | aph(3″)-Ib, aph(6′)-Id, aph(3′)-Ia, aac(6′)-IIc, aadA2 [AM]; blaTEM-1b [PE]; qnrB2 [FL]; ere(A), mdf(A) [MA]; sul1, dfrA19 [TS]; catA2 [PH]; tet(B), tet(D) [TE]; and blaSHV-12 [EC] | ExPEC | IncFIB, IncFII (F2:A-:B25), IncHI2A (ST1) |
| SRR11047778 | 90 (O149:H19, C) | aac(3)-VIa, aadA1, aadA2, aadA5, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, armA [AM]; blaTEM-1b [PE]; mph(E), msr(E), mdf(A) [MA]; sul2, dfrA1 [TS]; floR [PH]; tet(A), tet(B) [TE]; and blaCMY-2 [EC] | APEC | Col156, IncA/C2, IncFIB, IncFIC, IncFII (F108:A-:B42), IncHI2A (ST unknown), IncI1 (ST12) |
| SRR11074163 | 101 (O82:H8, B1) | aac(3)-VIa, aadA1, aph(3″)-Ib, aph(6)-Id [AM]; mdf(A) [MA]; sul1, sul2 [TS]; floR [PH]; tet(A) [TE]; and blaCMY-2, blaCTX-M-55 [EC] | APEC | IncFIB, IncFII (F24:A-:B1), IncHI2A (ST unknown), IncI1 (ST16), IncX1 |
| SRR11074700 | 10 (O141:H4, A) | aac(6′)-IIc, aadA2b, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id [AM]; blaTEM-1b [PE]; qnrB2 [FL]; ere(A), mdf(A) [MA]; dfrA19, sul1, sul2 [TS]; tet(B), tet(D) [TE]; and blaCMY-2, blaSHV-12 [EC] | ETEC, STEC | IncFIB, IncFII (F-:A-:B42), IncHI2A (ST1), IncI1 (ST65), IncI2, p011 |
| SRR11074696 | 10 (O141:H4, A) | aac(6′)-IIc, aadA2b, aph(3′)-Ib, aph(3′)-Ia, aph(6)-Id [AM]; blaTEM-1b [PE]; qnrB2 [FL]; ere(A), mdf(A) [MA]; dfrA19, sul1, sul2 [TS]; tet(B), tet(D) [TE]; and blaCMY-2, blaSHV-12 [EC] | ETEC, STEC | IncFIB, IncFII (F-:A-:B42), IncHI2A (ST1), IncI1 (ST65), IncI2, p011 |
AM, aminoglycosides; PE, penicillins; FL, fluoroquinolones; MA, macrolides; TS, trimethoprim/sulfonamide; PH, phenicols; TE, tetracyclines; CO, colistin; EC, extended-spectrum cephalosporin.
DISCUSSION
Whole-genome sequencing (WGS) of enrofloxacin- and ceftiofur-resistant E. coli revealed multiple determinants conferring resistance to these critical antimicrobials, which were present on a wide spectrum of STs recovered from the major swine-producing states in the United States. The use of both long- and short-read WGS technologies identified the genetic context of these resistance determinants for several isolates, suggesting determinants by which resistance may be spreading, such as plasmids carrying blaCMY-2, which previously established in Salmonella and E. coli populations circulating in food animals in the United States (14). We also assembled plasmids not previously described in isolates from swine or other food animals or retail meat in the United States.
Nearly 82% of the ceftiofur-resistant E. coli isolates carried a blaCMY-2 gene, which is consistent with findings in ceftiofur-resistant Salmonella isolates from diseased pigs collected during the same study period (26). However, 24 E. coli isolates in this study (including 2 isolates nonresistant to ceftiofur) carried blaCTX-M or blaSHV-12 genes, indicating a much higher prevalence (18%) of blaCTX-M in our isolates compared to that in ceftiofur-resistant Salmonella of swine origin (26). Still, our data suggest a more limited distribution of blaCTX-M genes compared with reports in extended-spectrum cephalosporin-resistant E. coli isolates retrieved from swine in other upper-income countries in Europe and Asia such as Belgium (97.5%) and Hong Kong (87.5%) (32, 33). ESBL genes are responsible for extended-spectrum cephalosporin resistance globally in food animals (13). However, until the late 2000s, these genes were not found in food animal isolates collected in North America (34). In a study on E. coli isolates collected from diseased pigs at the University of Minnesota Veterinary Diagnostic Laboratory (UMN-VDL) in 2008, all ceftiofur-resistant isolates carried blaCMY-2 genes (35), whereas blaCTX-M-carrying E. coli in finishing pigs in the United States were first identified in 2011 (36). Since then, more recent studies have also reported the sporadic occurrence of blaCTX-M genes in Enterobacteriaceae isolates of swine origin (including pork) in the United States (37, 38). Our study reinforces the results that ESBL genes might have been introduced in E. coli collected from pigs during the late 2000s and early 2010s (34).
Similar to that for ESBLs, the presence of PMQR genes [qnr, aac(6′)-Ib-cr] in food animal isolates in the United States had not been reported until recently (25, 26, 39, 40). There has also been an increase in PMQR genes in clinical Salmonella isolates from humans in the United States, and animal sources have been postulated to contribute to this surge (40). In this study, the presence of PMQR genes without additional QRDR mutations was sufficient to yield MIC values to the intermediate susceptibility levels (0.25 to 1 μg/ml) but not above (with the exception of 2 qnrB19-carrying isolates). This is consistent with previous reports suggesting PMQR genes such as qnrB and qnrS confer only lower level resistance to quinolones by inhibiting the binding of quinolones to DNA gyrase (41). However, these PMQR genes are known to supplement resistance caused by other determinants such as altered target enzymes (DNA gyrase), efflux pump activities, and deficiencies in outer membrane porin channels (42). The presence of PMQR genes in zoonotic bacteria and their clinical impact on both human and animal health should therefore be continuously monitored.
To the best of our knowledge, this is the first study to describe completely assembled plasmids carrying blaCTX-M-14, -15, -27, and -55, blaSHV-12, and qnrB77 in E. coli isolates of swine origin in the United States. However, the close identities between some plasmids in this study and those already described in humans and animals globally indicate that the presence of ESBL genes in this isolate collection could be part of the pandemic expansion of ESBLs (13). blaCTX-M-15 and blaCTX-M-14 are considered the predominant ESBL genes in humans globally (13) and have also been identified in food animals, including pigs, worldwide (43–46). The plasmids carrying blaCTX-M-15 identified in our study were highly similar (98% coverage, >99% nucleotide identity) to other plasmids found in human E. coli isolates collected in the United States between 2009 and 2010 (47) (GenBank accession number CP009232), which were also described to have the same plasmid backbone as other ESBL gene-carrying plasmids reported worldwide (47). blaCTX-M-14-carrying plasmids identical to those found here were previously reported in human isolates in Hong Kong and characterized as an epidemic plasmid type (pHK01) (48) which has spread globally to other Asian (China, Vietnam, and South Korea) and European (Finland) countries (unpublished; GenBank accession numbers NC_013727.1, KU932024.1, KU987452.1, NC_013542.1, and NZ_CP018973.1). Families of insertion sequences (IS26, ISEcp9, and IS6) that were part of the above-mentioned genetic contexts have also been demonstrated to be involved in transposing ESBL-encoding genes across plasmids and bacterial chromosomes (49).
IncA/C2 and IncI1 plasmids were the carriers of blaCMY-2 genes, which is consistent with previous studies conducted on Salmonella enterica and Escherichia coli from both farm animals and humans in the United States (20, 21, 26, 50, 51). The blaCMY-2-carrying IncI1-ST12, IncI1-ST65, and IncA/C2-ST3 plasmids assembled here were highly similar (>99% nucleotide identity) to those isolated from broiler E. coli in Japan (which, in turn, were highly similar to plasmids from European poultry) (52). This suggests that closely related blaCMY-2-carrying plasmids are disseminated globally in livestock.
In contrast to ESBL and pAmpC genes, qnr genes were also present on short (∼3 kbp) ColE and medium-sized (∼7 kbp) IncQ2 plasmids. ColE plasmids harboring qnrB19 genes are among the most commonly isolated PMQR-plasmid combinations globally, and these have been isolated from bacteria of family Enterobacteriaceae from several animals, meat, and humans globally (25, 53–57). qnrS2-harboring IncQ2 plasmids have largely been found from aquatic sources such as a wastewater treatment plant in Israel (58) and rivers and aquaculture facilities in China (59), indicative of potential exchange of resistant bacteria between environmental sources and swine herds (21, 60).
It has been widely believed that the presence of plasmids in the absence of selective pressure imposes a metabolic fitness cost to the bacterial host (61). However, the fitness cost imposed due to plasmid carriage depends on the plasmid-bacterial host combination (62–64). There are several plasmid characteristics that facilitate plasmid stability in bacterial hosts: for example, IncF and IncI1-pMLST12 plasmids similar to those assembled here have a narrow host range and carry factors such as toxin-antitoxin systems which help maintain their stability in bacterial hosts in the absence of antimicrobial pressure (52, 65, 66). Similarly, IncHI2 plasmids similar to those assembled here carry genes which confer resistance to heavy metals, mutagenesis induction systems, etc., which can also contribute to their stability (67). IncA/C2, IncHI2, IncN, and IncQ2 plasmids have a broad host range and can survive in multiple bacterial species, including bacteria present in the environment, which can aid in their persistence and dissemination outside animal hosts (68, 69). In a study using blaCMY-2-carrying E. coli isolated from pigs in the United States, larger plasmids (IncI1 and IncA/C2 plasmids similar to the one assembled in this study) were shown to coexist without imposing metabolic costs to the bacterial host (62). Epidemic plasmids identical to those found in our study such as pHK01-like plasmids have been demonstrated to be conjugative in vitro (70). Hence, it can be postulated that these plasmids might aid in the successful establishment of ESBL and PMQR genes and persistence of AmpC beta-lactamases as mechanisms of cephalosporin and quinolone resistance in swine E. coli in the United States.
In addition to the above-mentioned properties, IncF plasmids also possessed virulence genes which can contribute to the fitness of bacterial clones inside mammalian hosts. For example, the traT gene related to serum resistance was found on every blaCTX-M-harboring IncF plasmid, except for IncF (F24:A-:B1), and this gene has been consistently associated with urinary tract infections and sepsis in humans (71–74). Similarly, hlyF (hemolysin), iutA (iron uptake), and ompT (outer membrane protease) genes present on some of the blaCTX-M-IncF plasmids are very commonly found in avian pathogenic E. coli (75). iucC (aerobactin synthesis), iutA, and ompT genes also play key roles in the pathogenesis of severe extraintestinal infections in humans (76–78). The presence of virulence factors and antimicrobial resistance genes on epidemic IncF plasmids have previously contributed to global domination of E. coli ST131 clones (30) and can potentially aid in the successful dissemination of emerging E. coli lineages identified in this study.
The main enrofloxacin-resistant swine-specific ST identified in this study was ST100, which is associated with porcine enterotoxigenic infections and has spread clonally throughout the U.S. swine population (79). Enrofloxacin was approved to treat swine enteric infections in the United States in 2012 (2), and the association of enterotoxigenic ST100 E. coli with enrofloxacin resistance might be of concern for swine health because of the potential decrease in clinical efficacy of enrofloxacin in treating scours due to bacterial resistance.
Many of the isolates in this study can be considered to be “high-risk” clones. These clones are characterized by global dissemination, ease of transmission from host to host, disease-causing abilities, and acquisition of genetic characteristics that provide a competitive advantage over other bacterial clones, such as virulence factors, epidemic plasmids, and antimicrobial resistance genes (30). One outstanding example of a high-risk clone in our database is the fluoroquinolone-resistant ST744 isolates which were also closely related to isolates collected globally from multiple host species and the environment as well as from diseased humans. ST744 isolates belonged to phylogroup A and E. coli in this phylogroup are not as virulent as those belonging to phylogroups B2 and D2 (80). Nonetheless, ST744 has been sporadically associated as a disease-causing agent carrying resistance to critical antimicrobials such as colistin and carbapenem from human patients worldwide (81–83). The global distribution of fluoroquinolone-resistant ST744 clones should be worrisome, as fluoroquinolones are critical antimicrobials for treating systemic infections in humans (84). Fluoroquinolone-resistant ST410 was also present in our data set. Recently, Roer et al. described the emergence of ST410 (phylogroup A) high-risk clones globally, indicating that the clones of lowly virulent E. coli are also capable of widespread dissemination and carriage of antimicrobial resistance genes (85). More experiments and clinical studies are needed to determine the true pathogenicity and fitness of ST410 and ST744 high-risk clones.
ESBLs have been associated with pandemic ST131 E. coli in humans (86). However, in this study, only one ST131 isolate was identified, and it was susceptible to both antimicrobial classes under study but was classified as ExPEC and APEC. Manges et al. (87) recently published a review of the most prevalent global ExPEC lineages, and 12 of the top 20 ExPEC E. coli STs listed in this review were present in the U.S. swine E. coli isolates (ST131, ST69, ST10, ST73, ST410, ST12, ST127, ST167, ST58, ST88, ST617, and ST23) (87). In a study conducted on clinical E. coli isolates from a north California community, 47% of ExPEC strains consisted of ST127, -73, -69, -10, -12, and -88, which were all present in our study (88). Isolates of several STs in this study carried blaCTX-M-epidemic plasmids similar to IncF (F31:F36:A4:B1) and IncF (F2:A-:B-), had mutations in QRDRs conferring fluoroquinolone resistance, and were classified into several virulotypes of public health concern (Shiga toxin-producing E. coli [STEC], ExPEC, APEC, and UPEC), presenting further evidence of the presence of potentially high-risk zoonotic clones in the swine population in the United States.
The colistin resistance gene (mcr-9) was recently described for the first time in an S. enterica serovar Typhimurium isolate collected from a human patient in Washington state and was able to confer colistin resistance to E. coli isolates cloned with this gene (89). Tyson et al. further evaluated the presence of this gene in Salmonella and E. coli collected from animal meat in the United States and found that this gene was present on large IncHI2 plasmids similar to those found here or integrated into bacterial chromosomes (90). Tyson et al. also found that mcr-9-carrying bacterial isolates were all susceptible to colistin (90); hence, the clinical relevance of this gene on human health is still debatable. Regardless of clinical impact, colistin has never been used in swine in the United States; therefore, the presence of the mcr-9 gene in swine could be an indicator of the complex transmission dynamics of resistant determinants across different ecosystems and/or a coselection of resistant determinants due to the use of other unrelated antimicrobials.
Several considerations must be accounted for when interpreting these results. An association between antimicrobial use and presence of these resistance genes cannot be established due to the lack of information on the use of antimicrobials. Also, the public health implications of our findings could be limited by the removal of diseased pigs, such as the ones from which these resistant and potentially zoonotic STs were retrieved, from the food chain.
Conclusions.
We have identified and characterized a wide range of genetic determinants of resistance to some critically important antimicrobial classes in swine clinical E. coli isolates, some of which had never been described in isolates of animal origin in the United States. We also highlighted the presence of high-risk clones and epidemic plasmids in swine E. coli with a potential to negatively impact human and animal health.
MATERIALS AND METHODS
Description of isolates.
A total of 211 E. coli isolates recovered from diseased pigs at the University of Minnesota Veterinary Diagnostic Laboratory (UMN-VDL) between 2014 and 2015 were included in this study. E. coli isolates available at the UMN-VDL infectious agent repository were classified as ceftiofur non-wild type (MICs ≥ 2 μg/ml) and enrofloxacin non-wild type (MIC ≥ 0.25 μg/ml) (91). These MIC values were routinely estimated during the processing of diagnostic submissions and were based on the results of broth microdilution tests performed using Clinical and Laboratory Standards Institute guidelines (92). For ease of interpretation, “non-wild-type” and “wild-type” isolates are referred to as “resistant” and “susceptible,” respectively. All the ceftiofur- and/or enrofloxacin-resistant isolates available at the UMN-VDL infectious disease repository collected between 2014 and 2015 were selected for this study, and a random selection of susceptible isolates from the same period was used for comparative purposes. Of these 211 isolates, 110 were enrofloxacin resistant and 106 were ceftiofur resistant, with 41 isolates being resistant to both ceftiofur and enrofloxacin. Forty-six isolates susceptible to both antimicrobials were added to assess the presence of resistance genes and chromosomal mutations in susceptible isolates. Only one isolate per farm was selected in order to avoid duplicity of potentially identical clones circulating in the same farm.
Short-read sequencing and in silico typing of E. coli isolates.
Isolates were first subjected to short-read sequencing using Illumina HiSeq 2500 (2 × 125 bp). The mean phred scores of raw reads was greater than 30 for all isolates. Raw reads were then trimmed using Trimmomatic version 0.39 (settings: sliding window mode; number of bases to average across, 4; average quality required, 20) (93). The raw reads were uploaded to and assembled using the QAssembly version 3.61 pipeline provided by the Enterobase webserver (94). This pipeline assembles draft genomes using Spades version 3.9.0 (95). The assemblies were further polished using BWA version 0.7.12 (96) to align reads back onto the assemblies, and these assemblies were polished using consensus bases or indels using bcftools version 1.2 (97). The assemblies were then passed for downstream analyses only if they meet the following criteria: number of bases, 3.7 to 6.4 Mbp; N50, >20 kb; number of contigs, <800; proportion of N’s, <3%; species assignment using Kraken, >70% contigs. The assembly statistics are as follows: average coverage, 107.3× (range, 52 to 405×); average N50, 135,520.4 bp (range, 40,154 to 340,030 bp); average size, 5.26 Mbp (range, 4.62 to 5.94 Mbp); and average number of contigs (>200 bp size), 209.4 (range, 71 to 392).
Draft genomes, assembly statistics, serotyping, phylotyping, and cgMLST results for these E. coli isolates were downloaded from the Enterobase webserver. Draft genomes were uploaded to the Center for Genomic Epidemiology (CGE) webserver to identify multilocus sequence type (MLST version 2.0.4) (98), acquired resistance genes and chromosomal mutations in quinolone resistance-determining regions (QRDRs) of gyrA, gyrB, parC, and parE genes (ResFinder version 3.2) (99), plasmid multilocus sequence type (pMLST version 0.1.0) (100), plasmid replicon type (Plasmid Finder version 2.0.1) (100), and virulence factors (virulence finder version 2.0) (101). A minimum nucleotide identity of 90% and minimum length of 60% compared to reference sequences were used as thresholds for classifying virulence and resistance genes. Draft genomes were annotated using PROKKA (version 1.13) (102).
Isolates were classified into virulotypes based on the virulence genes present as follows: (a) enterotoxigenic E. coli (ETEC) if any of the heat stable toxin genes (sta, stb, or astA) or heat-labile toxin gene (ltcA) were present, (b) Shiga toxin-producing E. coli if Shiga toxin genes (stx) were present, (c) extraintestinal pathogenic E. coli (ExPEC) if two or more of the genes papA-papC, sfa-focDE, afa/draBC, kpsMII, or iutA were present (103), (d) avian pathogenic E. coli (APEC) if all of the following genes, iutA, hlyF, iss, iroN, and ompT, were present (75), and (e) uropathogenic E. coli if three or more of the genes chuA, fyuA, vat, and yfcV were present (104).
Phylogenetic analysis.
For the phylogenetic analysis, raw reads were first mapped to a reference genome (E. coli strain K-12 substrain MG1655; accession NZ_AJGD00000000.1), and full gene alignments were assembled using snippy (default values: minimum mapping quality, 60; minimum coverage, 10; version 4.4.5) (105). ClonalframeML (default values, version 1.12) was used to detect recombinant regions from these full gene alignments (106), which were subsequently masked by using maskrc-svg (version 0.5) (107). Single nucleotide polymorphisms (SNPs) were extracted from these recombination-masked alignments using snp-sites (version 2.5.1) (108), and pairwise SNP distances were estimated using snp-dists (version 0.7.0) (108). Maximum likelihood trees were then built using a general time-reversible (GTR) with gamma substitution model through RAxML (version 8.0) (109). Support for nodes on trees was assessed using 1,000 bootstrap replicates, and the phylogenetic tree was made and genomic features were annotated using iTOL (version 4.0) (110). Based on this analysis, we identified STs with SNP distances of less than 100 between at least 2 isolates and ran an ST-specific phylogenetic analysis for these STs using the following reference sequences: ST10 (NZ_AJGD00000000.1), ST12 (CP010151.1), ST23 (CP007491.1), ST58 (CP043744.1), ST88 (CP031546.1), ST90 (CP020520.1), ST100 (CP002729.1), ST101 (CP024821.1), ST224 (CP035339.1), ST410 (CP031231.1), ST457 (CP024826.1), ST641 (CP046000.1), ST744 (GCF_001682305), ST847 (CP010344.1), and ST4981 (CP017980.1). For each ST-specific analysis, the chromosomal sequence with maximum genome coverage available at the RefSeq database was selected as a reference. The steps for SNP estimation and measurement of SNP distances were repeated for each ST-specific analysis as mentioned above for E. coli strain K-12 substrain MG1655 as the reference. In the text and figures, median pairwise SNP distances (MPDs) and the ranges of SNPs mentioned are based on these ST-specific analyses.
We downloaded raw reads of all isolates available at Enterobase which belonged to the same cgMLST clusters (defined by a maximum difference of 20 alleles [“HC20”]) as the ceftiofur- or enrofloxacin-resistant isolates in our study. Phylogenetic analyses using E. coli K-12 substrain MG1655 and ST-specific references and typing (virulence genes, AMR genes, phylotyping, pMLST, plasmid analysis, and serotyping) were repeated using the same steps as mentioned above for comparing our isolates with similar isolates (same HC20) available at Enterobase.
Detailed results of serotyping, phylotyping, MLST, cgMLST, virulence factors, antimicrobial resistance genes, plasmid replicon typing, and associated metadata are available in Tables S1 and S2 in the supplemental material.
Detailed virulence, antimicrobial resistance, and typing metadata of isolates available on Enterobase that were within 20 allele differences (as per cgMLST schema provided at Enterobase) of isolates in this study. Download Table S2, XLSX file, 0.1 MB (127.8KB, xlsx) .
Copyright © 2020 Hayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assembly of plasmids using long-and short-read sequencing.
Additionally, long-read sequencing was performed on a subset of isolates carrying blaSHV-12, blaCTX-M, blaCMY-2, and qnrB77 genes in the analysis described above using Pacific Biosciences (PacBio) RSII technology (SMRT Cell 1M v3). This subset was selected on the basis of presence of ESBL genes to represent all genomic contexts around these genes (available from contigs assembled on short reads). Long reads were first corrected for errors using LoRDEC (version 0.9) (111). Unicycler (version 0.4.7) (112) was used to obtain de novo hybrid assemblies of these isolates using both long and short reads, and assemblies were visualized using Bandage (version 0.8.1) (113). Complete plasmid genomes (here referred to as “assembled plasmids”) were uploaded to the ISsaga webserver (114) for identification of insertion sequences and to the CGE webserver to perform analyses as mentioned above. The assembled plasmids were also analyzed against a blast database of reference plasmids available at the PLSDB webserver (115) to identify closely related plasmids also carrying antimicrobial resistance genes of interest (ESBL and PMQR). Plasmid sequences with a query coverage of >80% and nucleotide identity >90% were downloaded, and the top five closely related plasmids and genomes to each of the ones found here were visually compared using BRIG (version 0.95) (116).
Assembly of putative plasmids.
“Putative plasmids” were also assembled using these assembled plasmids. This was performed by mapping short reads of isolates carrying the same ESBL, PMQR, or pAmpC gene as the assembled plasmids carrying the corresponding genes. The mapping was conducted using snippy (version 4.4.5) with the values mentioned above. Pileups were generated after mapping short reads using SAMtools (version 1.10) with mapping quality capped at 60 (97). Pileups were then converted to fasta format using Galaxy tools 1.0.2 (117).
In the cases where raw reads did not sufficiently map to the assembled plasmids (query coverage of putative plasmid <70% compared to assembled plasmids), we identified the closest “reference plasmids” by doing a BLASTN search of the contigs carrying AMR genes on the NCBI server and assembled putative plasmids by mapping short reads onto these reference plasmids. Contigs from our study had >90% query coverage compared to these reference plasmids, with a nucleotide identity of >99%. We also confirmed that plasmids in our study were indeed similar to these reference plasmids by comparing the pMLST results for these reference plasmid sequences, putative plasmids assembled, draft E. coli genomes, and the contigs carrying these AMR gene in draft genomes. Analysis of pMLSTs for reference plasmids, putative plasmids, and draft E. coli sequences provided complete information on the sequence type of the plasmids. The pMLST results for just the contigs carrying AMR genes provided only partial matches, with the exception of short plasmids. These assembled and putative plasmids were then annotated, and genomic features such as virulence factors and insertion sequences were identified as mentioned above. Putative plasmids were also assembled for the isolates downloaded from Enterobase using same methods as described above.
Data availability.
Short reads generated during this project have been submitted at NCBI GenBank under BioProject accessions PRJNA605257, PRJNA605064, and PRJNA604903. Complete plasmid sequences have been submitted at GenBank under accession numbers MT077880, MT077881, MT077882, MT077883, MT077884, MT077885, MT077886, MT077887, MT077888, MT077889, and MT816498.
ACKNOWLEDGMENTS
This work was supported by the USDA National Institute of Food and Agriculture (Animal Health Formula Fund project MIN-62-091), Swine Disease Eradication Center fund, and the Rapid Agricultural Response Fund (RARF) at the University of Minnesota.
REFERENCES
- 1.Aarestrup FM. 2015. The livestock reservoir for antimicrobial resistance: a personal view on changing patterns of risks, effects of interventions and the way forward. Philos Trans R Soc Lond B Biol Sci 370:20140085. doi: 10.1098/rstb.2014.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong S, Rovira A, Davies P, Ahlstrom C, Muellner P, Rendahl A, Olsen K, Bender JB, Wells S, Perez A, Alvarez J. 2016. Serotypes and antimicrobial resistance in Salmonella enterica recovered from clinical samples from cattle and swine in Minnesota, 2006 to 2015. PLoS One 11:e0168016. doi: 10.1371/journal.pone.0168016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmo LP, Nielsen LR, Alban L, Müntener CR, Schüpbach-Regula G, Magouras I. 2017. Comparison of antimicrobial consumption patterns in the Swiss and Danish cattle and swine Production (2007–2013). Front Vet Sci 4:26. doi: 10.3389/fvets.2017.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Food and Drug Administration. 2017. Summary report on antimicrobials sold or distributed for use in food-producing animals. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 5.Neuert S, Nair S, Day MR, Doumith M, Ashton PM, Mellor KC, Jenkins C, Hopkins KL, Woodford N, de Pinna E, Godbole G, Dallman TJ. 2018. Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Front Microbiol 9:592. doi: 10.3389/fmicb.2018.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edirmanasinghe R, Finley R, Parmley EJ, Avery BP, Carson C, Bekal S, Golding G, Mulvey MR. 2017. A whole-genome sequencing approach to study cefoxitin-resistant Salmonella enterica serovar Heidelberg isolates from various sources. Antimicrob Agents Chemother 61:e01919-16. doi: 10.1128/AAC.01919-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magouras I, Carmo LP, Stärk KDC, Schüpbach-Regula G. 2017. Antimicrobial usage and -resistance in livestock: where should we focus? Front Vet Sci 4:148. doi: 10.3389/fvets.2017.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh TR, Wu Y. 2016. China bans colistin as a feed additive for animals. Lancet Infect Dis 16:1102–1103. doi: 10.1016/S1473-3099(16)30329-2. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Xu C, Zhang R, Chen Y, Shen Y, Hu F, Liu D, Lu J, Guo Y, Xia X, Jiang J, Wang X, Fu Y, Yang L, Wang J, Li J, Cai C, Yin D, Che J, Fan R, Wang Y, Qing Y, Li Y, Liao K, Chen H, Zou M, Liang L, Tang J, Shen Z, Wang S, Yang X, Wu C, Xu S, Walsh TR, Shen J. 2020. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect Dis 20:1161–1171. doi: 10.1016/S1473-3099(20)30149-3. [DOI] [PubMed] [Google Scholar]
- 10.Lalak A, Wasyl D, Zając M, Skarżyńska M, Hoszowski A, Samcik I, Woźniakowski G, Szulowski K. 2016. Mechanisms of cephalosporin resistance in indicator Escherichia coli isolated from food animals. Vet Microbiol 194:69–73. doi: 10.1016/j.vetmic.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Caroff N, Espaze E, Gautreau D, Richet H, Reynaud A. 2000. Analysis of the effects of -42 and -32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J Antimicrob Chemother 45:783–788. doi: 10.1093/jac/45.6.783. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez I, Thomas K, Van Essen A, Schink A-K, Day M, Chattaway M, Wu G, Mevius D, Helmuth R, Guerra B, SAFEFOODERA-ESBL consortium. 2014. Chromosomal location of blaCTX-M genes in clinical isolates of Escherichia coli from Germany, The Netherlands and the UK. Int J Antimicrob Agents 43:553–557. doi: 10.1016/j.ijantimicag.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-Mβ-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 14.Frye JG, Jackson CR. 2013. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals Front Microbiol 4:135. doi: 10.3389/fmicb.2013.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wittum TE, Mollenkopf DF, Erdman MM. 2012. Detection of Salmonella enterica isolates producing CTX-M cephalosporinase in U.S. livestock populations. Appl Environ Microbiol 78:7487–7491. doi: 10.1128/AEM.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y-F, Zhang W-H, Ren S-Q, Yang L, Lü D-H, Zeng Z-L, Liu Y-H, Jiang H-X. 2014. IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS One 9:e96738. doi: 10.1371/journal.pone.0096738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri Junior A, Darini ALC. 2017. Diversity of plasmids harboring blaCMY-2 in multidrug-resistant Escherichia coli isolated from poultry in Brazil. Diagn Microbiol Infect Dis 88:361–364. doi: 10.1016/j.diagmicrobio.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Castellanos LR, Donado-Godoy P, León M, Clavijo V, Arevalo A, Bernal JF, Timmerman AJ, Mevius DJ, Wagenaar JA, Hordijk J. 2017. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the Colombian poultry chain. PLoS One 12:e0170777. doi: 10.1371/journal.pone.0170777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castellanos LR, van der Graaf-van Bloois L, Donado-Godoy P, Mevius DJ, Wagenaar JA, Hordijk J, Zomer AL. 2019. Phylogenomic Investigation of IncI1-Iγ plasmids harboring blaCMY-2 and blaSHV-12 in Salmonella enterica and Escherichia coli in multiple countries. Antimicrob Agents Chemother 63:e02546-18. doi: 10.1128/AAC.02546-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernández-Alarcón C, Singer RS, Johnson TJ. 2011. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6:e23415. doi: 10.1371/journal.pone.0023415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pornsukarom S, Thakur S. 2017. Horizontal dissemination of antimicrobial resistance determinants in multiple Salmonella serotypes following isolation from the commercial swine operation environment after manure application. Appl Environ Microbiol 83:e01503-17. doi: 10.1128/AEM.01503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afema JA, Ahmed S, Besser TE, Jones LP, Sischo WM, Davis MA. 2018. Molecular epidemiology of dairy cattle-associated Escherichia coli carrying blaCTX-M genes in Washington state. Appl Environ Microbiol 84:e02430-17. doi: 10.1128/AEM.02430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittum TE, Mollenkopf DF, Daniels JB, Parkinson AE, Mathews JL, Fry PR, Abley MJ, Gebreyes WA. 2010. CTX-M-type extended-spectrum β-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog Dis 7:1575–1579. doi: 10.1089/fpd.2010.0615. [DOI] [PubMed] [Google Scholar]
- 24.Holmes AH, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, Guerin PJ, Piddock LJV. 2016. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 25.Tyson GH, Tate HP, Zhao S, Li C, Dessai U, Simmons M, McDermott PF. 2017. Identification of plasmid-mediated quinolone resistance in Salmonella isolated from swine ceca and retail pork chops in the United States. Antimicrob Agents Chemother 61:e01318-17. doi: 10.1128/AAC.01318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elnekave E, Hong SL, Lim S, Hayer SS, Boxrud D, Taylor AJ, Lappi V, Noyes N, Johnson TJ, Rovira A, Davies P, Perez A, Alvarez J. 2019. Circulation of plasmids harboring resistance genes to quinolones and/or extended-spectrum cephalosporins in multiple Salmonella enterica Serotypes from swine in the United States. Antimicrob Agents Chemother 63:e02602-18. doi: 10.1128/AAC.02602-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elnekave E, Hong S, Mather AE, Boxrud D, Taylor AJ, Lappi V, Johnson TJ, Vannucci F, Davies P, Hedberg C, Perez A, Alvarez J. 2018. Salmonella enterica serotype 4,[5],12:i:- in swine in the United States Midwest: an emerging multidrug-resistant clade. Clin Infect Dis 66:877–885. doi: 10.1093/cid/cix909. [DOI] [PubMed] [Google Scholar]
- 28.Hayer SS, Rovira A, Olsen K, Johnson TJ, Vannucci F, Rendahl A, Perez A, Alvarez J. 25 February 2020. Prevalence and trend analysis of antimicrobial resistance in clinical Escherichia coli isolates collected from diseased pigs in the USA between 2006 and 2016. Transbound Emerg Dis doi: 10.1111/tbed.13528. [DOI] [PubMed] [Google Scholar]
- 29.Baker S, Thomson N, Weill F-X, Holt KE. 2018. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 360:733–738. doi: 10.1126/science.aar3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathers AJ, Peirano G, Pitout JDD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Food and Drug Administration. 2019. NARMS now. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 32.Ho PL, Chow KH, Lai EL, Lo WU, Yeung MK, Chan J, Chan PY, Yuen KY. 2011. Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to “critically important” antibiotics among food animals in Hong Kong, 2008-10. J Antimicrob Chemother 66:765–768. doi: 10.1093/jac/dkq539. [DOI] [PubMed] [Google Scholar]
- 33.Van Damme I, Garcia-Graells C, Biasino W, Gowda T, Botteldoorn N, de Zutter L. 2017. High abundance and diversity of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in faeces and tonsils of pigs at slaughter. Vet Microbiol 208:190–194. doi: 10.1016/j.vetmic.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Roach S, Wallinga D. 2013. Commentary on genetic mechanisms of antimicrobial resistance in bacteria from U.S. food animals: ESBLs are here. Front Microbiol 4:214. doi: 10.3389/fmicb.2013.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson TJ, Shepard SM, Rivet B, Danzeisen JL, Carattoli A. 2011. Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid 66:144–151. doi: 10.1016/j.plasmid.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Mollenkopf DF, Mirecki JM, Daniels JB, Funk JA, Henry SC, Hansen GE, Davies PR, Donovan TS, Wittum TE. 2013. Escherichia coli and Klebsiella pneumoniae producing CTX-M cephalosporinase from swine finishing barns and their association with antimicrobial use. Appl Environ Microbiol 79:1052–1054. doi: 10.1128/AEM.03169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tadesse DA, Li C, Mukherjee S, Hsu C-H, Bodeis Jones S, Gaines SA, Kabera C, Loneragan GH, Torrence M, Harhay DM, McDermott PF, Zhao S. 2018. Whole-genome sequence analysis of CTX-M containing Escherichia coli isolates from retail meats and cattle in the United States. Microb Drug Resist 24:939–948. doi: 10.1089/mdr.2018.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang F, Wu Z, Zheng Y, Frana TS, Sahin O, Zhang Q, Li G. 2019. Genotypes and antimicrobial susceptibility profiles of hemolytic Escherichia coli from diarrheic piglets. Foodborne Pathog Dis 16:94–103. doi: 10.1089/fpd.2018.2480. [DOI] [PubMed] [Google Scholar]
- 39.Awosile B, McClure J, Sanchez J, Rodriguez-Lecompte JC, Keefe G, Heider LC. 2018. Salmonella enterica and extended-spectrum cephalosporin-resistant Escherichia coli recovered from Holstein dairy calves from 8 farms in New Brunswick, Canada. J Dairy Sci 101:3271–3284. doi: 10.3168/jds.2017-13277. [DOI] [PubMed] [Google Scholar]
- 40.Cummings KJ, Rodriguez‐Rivera LD, Norman KN, Ohta N, Scott HM. 2017. Identification of a plasmid-mediated quinolone resistance gene in Salmonella isolates from Texas dairy farm environmental samples. Zoonoses Public Health 64:305–307. doi: 10.1111/zph.12318. [DOI] [PubMed] [Google Scholar]
- 41.Tran JH, Jacoby GA, Hooper DC. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother 49:118–125. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martínez-Martínez L, Pascual A, García I, Tran J, Jacoby GA. 2003. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother 51:1037–1039. doi: 10.1093/jac/dkg157. [DOI] [PubMed] [Google Scholar]
- 43.Liu B-T, Yang Q-E, Li L, Sun J, Liao X-P, Fang L-X, Yang S-S, Deng H, Liu Y-H. 2013. Dissemination and characterization of plasmids carrying oqxAB-blaCTX-M genes in Escherichia coli Isolates from Food-Producing Animals. PLoS One 8:e73947. doi: 10.1371/journal.pone.0073947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norizuki C, Kawamura K, Wachino J-I, Suzuki M, Nagano N, Kondo T, Arakawa Y. 2018. Detection of Escherichia coli producing CTX-M-1-group extended-spectrum beta-lactamases from pigs in Aichi Prefecture, Japan, between 2015 and 2016. Jpn J Infect Dis 71:33–38. doi: 10.7883/yoken.JJID.2017.206. [DOI] [PubMed] [Google Scholar]
- 45.Horton RA, Randall LP, Snary EL, Cockrem H, Lotz S, Wearing H, Duncan D, Rabie A, McLaren I, Watson E, La Ragione RM, Coldham NG. 2011. Fecal carriage and shedding density of CTX-M extended-spectrum β-lactamase-producing Escherichia coli in cattle, chickens, and pigs: implications for environmental contamination and food production. Appl Environ Microbiol 77:3715–3719. doi: 10.1128/AEM.02831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian GB, Wang HN, Zhang AY, Zhang Y, Fan WQ, Xu CW, Zeng B, Guan ZB, Zou LK. 2012. Detection of clinically important β-lactamases in commensal Escherichia coli of human and swine origin in western China. J Med Microbiol 61:233–238. doi: 10.1099/jmm.0.036806-0. [DOI] [PubMed] [Google Scholar]
- 47.Li J-J, Spychala CN, Hu F, Sheng J-F, Doi Y. 2015. Complete nucleotide sequences of blaCTX-M-harboring IncF plasmids from community-associated Escherichia coli strains in the United States. Antimicrob Agents Chemother 59:3002–3007. doi: 10.1128/AAC.04772-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho PL, Lo WU, Wong RCW, Yeung MK, Chow KH, Que TL, Tong AHY, Bao JYJ, Lok S, Wong SSY. 2011. Complete sequencing of the FII plasmid pHK01, encoding CTX-M-14, and molecular analysis of its variants among Escherichia coli from Hong Kong. J Antimicrob Chemother 66:752–756. doi: 10.1093/jac/dkr010. [DOI] [PubMed] [Google Scholar]
- 49.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Folster JP, Pecic G, Bolcen S, Theobald L, Hise K, Carattoli A, Zhao S, McDermott PF, Whichard JM. 2010. Characterization of extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg isolated from humans in the United States. Foodborne Pathog Dis 7:181–187. doi: 10.1089/fpd.2009.0376. [DOI] [PubMed] [Google Scholar]
- 51.Folster JP, Pecic G, McCullough A, Rickert R, Whichard JM. 2011. Characterization of blaCMY-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog Dis 8:1289–1294. doi: 10.1089/fpd.2011.0944. [DOI] [PubMed] [Google Scholar]
- 52.Shirakawa T, Sekizuka T, Kuroda M, Suzuki S, Ozawa M, Abo H, Furuya Y, Akama R, Matsuda M, Shimazaki Y, Kijima M, Kawanishi M. 2020. Comparative genomic analysis of third-generation-cephalosporin-resistant Escherichia coli harboring the blaCMY-2-positive IncI1 group, IncB/O/K/Z, and IncC plasmids isolated from healthy broilers in Japan. Antimicrob Agents Chemother 64:e02385-19. doi: 10.1128/AAC.02385-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran T, Andres P, Petroni A, Soler-Bistué A, Albornoz E, Zorreguieta A, Reyes-Lamothe R, Sherratt DJ, Corso A, Tolmasky ME. 2012. Small plasmids harboring qnrB19: a model for plasmid evolution mediated by site-specific recombination at oriT and Xer sites. Antimicrob Agents Chemother 56:1821–1827. doi: 10.1128/AAC.06036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammerl JA, Beutlich J, Hertwig S, Mevius D, Threlfall EJ, Helmuth R, Guerra B. 2010. pSGI15, a small ColE-like qnrB19 plasmid of a Salmonella enterica serovar Typhimurium strain carrying Salmonella genomic island 1 (SGI1). J Antimicrob Chemother 65:173–175. doi: 10.1093/jac/dkp383. [DOI] [PubMed] [Google Scholar]
- 55.Pallecchi L, Riccobono E, Sennati S, Mantella A, Bartalesi F, Trigoso C, Gotuzzo E, Bartoloni A, Rossolini GM. 2010. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob Agents Chemother 54:678–682. doi: 10.1128/AAC.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunha MPV, Lincopan N, Cerdeira L, Esposito F, Dropa M, Franco LS, Moreno AM, Knöbl T. 2017. Coexistence of CTX-M-2, CTX-M-55, CMY-2, FosA3, and QnrB19 in extraintestinal pathogenic Escherichia coli from poultry in Brazil. Antimicrob Agents Chemother 61:e02474-16. doi: 10.1128/AAC.02474-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyson GH, Li C, Hsu C-H, Bodeis-Jones S, McDermott PF. 2019. Diverse fluoroquinolone resistance plasmids from retail meat E. coli in the United States. Front Microbiol 10:2826. doi: 10.3389/fmicb.2019.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manageiro V, Romão R, Moura IB, Sampaio DA, Vieira L, Ferreira E, Caniça M, Network EuSCAPE-Portugal. 2018. Molecular epidemiology and risk factors of carbapenemase-producing Enterobacteriaceae isolates in Portuguese hospitals: results from European survey on carbapenemase-producing Enterobacteriaceae (EuSCAPE). Front Microbiol 9:2843. doi: 10.3389/fmicb.2018.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen Y, Pu X, Zheng W, Hu G. 2016. High prevalence of plasmid-mediated quinolone resistance and IncQ plasmids carrying qnrS2 gene in bacteria from rivers near hospitals and aquaculture in China. PLoS One 11:e0159418. doi: 10.1371/journal.pone.0159418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sapkota AR, Curriero FC, Gibson KE, Schwab KJ. 2007. Antibiotic-resistant enterococci and fecal indicators in surface water and groundwater impacted by a concentrated swine feeding operation. Environ Health Perspect 115:1040–1045. doi: 10.1289/ehp.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.San Millan A, MacLean RC. 2017. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr 5:MTBP-0016-2017. doi: 10.1128/microbiolspec.MTBP-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson TJ, Singer RS, Isaacson RE, Danzeisen JL, Lang K, Kobluk K, Rivet B, Borewicz K, Frye JG, Englen M, Anderson J, Davies PR. 2015. In vivo transmission of an IncA/C plasmid in Escherichia coli depends on tetracycline concentration, and acquisition of the plasmid results in a variable cost of fitness. Appl Environ Microbiol 81:3561–3570. doi: 10.1128/AEM.04193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buckner MMC, Saw HTH, Osagie RN, McNally A, Ricci V, Wand ME, Woodford N, Ivens A, Webber MA, Piddock LJV. 2018. Clinically relevant plasmid-host interactions indicate that transcriptional and not genomic modifications ameliorate fitness costs of Klebsiella pneumoniae carbapenemase-carrying plasmids. mBio 9:e02303-17. doi: 10.1128/mBio.02303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schaufler K, Semmler T, Pickard DJ, de Toro M, de la Cruz F, Wieler LH, Ewers C, Guenther S. 2016. Carriage of extended-spectrum beta-lactamase-plasmids does not reduce fitness but enhances virulence in some strains of pandemic E. coli lineages Front Microbiol 7:336. doi: 10.3389/fmicb.2016.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 66.Smith H, Bossers A, Harders F, Wu G, Woodford N, Schwarz S, Guerra B, Rodríguez I, van Essen-Zandbergen A, Brouwer M, Mevius D. 2015. Characterization of epidemic IncI1-Iγ plasmids harboring ambler class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob Agents Chemother 59:5357–5365. doi: 10.1128/AAC.05006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.García-Fernández A, Carattoli A. 2010. Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum β-lactamase and quinolone resistance genes. J Antimicrob Chemother 65:1155–1161. doi: 10.1093/jac/dkq101. [DOI] [PubMed] [Google Scholar]
- 68.Shintani M, Sanchez ZK, Kimbara K. 2015. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol 6:242. doi: 10.3389/fmicb.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 70.Ho PL, Lo WU, Yeung MK, Li Z, Chan J, Chow KH, Yam WC, Tong AHY, Bao JYJ, Lin CH, Lok S, Chiu SS. 2012. Dissemination of pHK01-like incompatibility group IncFII plasmids encoding CTX-M-14 in Escherichia coli from human and animal sources. Vet Microbiol 158:172–179. doi: 10.1016/j.vetmic.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 71.Abd El-Baky RM, Ibrahim RA, Mohamed DS, Ahmed EF, Hashem ZS. 2020. Prevalence of virulence genes and their association with antimicrobial resistance among pathogenic E. coli isolated from Egyptian patients with different clinical infections. Infect Drug Resist 13:1221–1236. doi: 10.2147/IDR.S241073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daga AP, Koga VL, Soncini JGM, de Matos CM, Perugini MRE, Pelisson M, Kobayashi RKT, Vespero EC. 2019. Escherichia coli bloodstream infections in patients at a university hospital: virulence factors and clinical characteristics. Front Cell Infect Microbiol 9:191. doi: 10.3389/fcimb.2019.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nojoomi F, Ghasemian A. 2019. The relation of phylogroups, serogroups, virulence factors and resistance pattern of Escherichia coli isolated from children with septicemia. New Microbes New Infect 29:100517. doi: 10.1016/j.nmni.2019.100517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hung W-T, Cheng M-F, Tseng F-C, Chen Y-S, Shin-Jung Lee S, Chang T-H, Lin H-H, Hung C-H, Wang J-L. 2019. Bloodstream infection with extended-spectrum beta-lactamase–producing Escherichia coli: the role of virulence genes. J Microbiol Immunol Infect 52:947–955. doi: 10.1016/j.jmii.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 75.Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol 46:3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bakhshi M, Zandi H, Bafghi MF, Astani A, Ranjbar VR, Vakili M. 2020. A survey for phylogenetic relationship; presence of virulence genes and antibiotic resistance patterns of avian pathogenic and uropathogenic Escherichia coli isolated from poultry and humans in Yazd, Iran Gene Rep 20:100725. doi: 10.1016/j.genrep.2020.100725. [DOI] [Google Scholar]
- 77.Najafi S, Rahimi M, Nikousefat Z. 2019. Extra-intestinal pathogenic Escherichia coli from human and avian origin: detection of the most common virulence-encoding genes. Vet Res Forum 10:43–49. doi: 10.30466/vrf.2019.34307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Desloges I, Taylor JA, Leclerc J-M, Brannon JR, Portt A, Spencer JD, Dewar K, Marczynski GT, Manges A, Gruenheid S, Moual HL, Thomassin J-L. 2019. Identification and characterization of OmpT-like proteases in uropathogenic Escherichia coli clinical isolates. MicrobiologyOpen 8:e915. doi: 10.1002/mbo3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abraham S, Jordan D, Wong HS, Johnson JR, Toleman MA, Wakeham DL, Gordon DM, Turnidge JD, Mollinger JL, Gibson JS, Trott DJ. 2015. First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J Glob Antimicrob Resist 3:273–277. doi: 10.1016/j.jgar.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Micenková L, Bosák J, Vrba M, Ševčíková A, Šmajs D. 2016. Human extraintestinal pathogenic Escherichia coli strains differ in prevalence of virulence factors, phylogroups, and bacteriocin determinants. BMC Microbiol 16:218. doi: 10.1186/s12866-016-0835-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perdigão Neto LV, Corscadden L, Martins RCR, Nagano DS, Cunha MPV, Neves PR, Franco LAM, Moura MLN, Rizek CF, Guimarães T, Boszczowski Í, Rossi F, Levin AS, Stabler RA, Costa SF. 2019. Simultaneous colonization by Escherichia coli and Klebsiella pneumoniae harboring mcr-1 in Brazil. Infection 47:661–664. doi: 10.1007/s15010-019-01309-2. [DOI] [PubMed] [Google Scholar]
- 82.Tacão M, Tavares R dos S, Teixeira P, Roxo I, Ramalheira E, Ferreira S, Henriques I. 2017. mcr-1 and blaKPC-3 in Escherichia coli sequence type 744 after meropenem and colistin therapy, Portugal. Emerg Infect Dis 23:1419–1421. doi: 10.3201/eid2308.170162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:e30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085. [DOI] [PubMed] [Google Scholar]
- 84.Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. 2009. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin Infect Dis 49:132–141. doi: 10.1086/599374. [DOI] [PubMed] [Google Scholar]
- 85.Roer L, Overballe-Petersen S, Hansen F, Schønning K, Wang M, Røder BL, Hansen DS, Justesen US, Andersen LP, Fulgsang-Damgaard D, Hopkins KL, Woodford N, Falgenhauer L, Chakraborty T, Samuelsen Ø, Sjöström K, Johannesen TB, Ng K, Nielsen J, Ethelberg S, Stegger M, Hammerum AM, Hasman H. 2018. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e00337-18. doi: 10.1128/mSphere.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pitout JDD, DeVinney R. 2017. Escherichia coli ST131: a multidrug-resistant clone primed for global domination. F1000Res 6:195. doi: 10.12688/f1000research.10609.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. 2019. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev 32:e00135-18. doi: 10.1128/CMR.00135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamaji R, Rubin J, Thys E, Friedman CR, Riley LW. 2018. Persistent pandemic lineages of uropathogenic Escherichia coli in a college community from 1999 to 2017. J Clin Microbiol 56:e01834-17. doi: 10.1128/JCM.01834-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. 2019. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tyson GH, Li C, Hsu C-H, Ayers S, Borenstein S, Mukherjee S, Tran T-T, McDermott PF, Zhao S. 2020. The mcr-9 gene of Salmonella and Escherichia coli is not associated with colistin resistance in the United States. Antimicrob Agents Chemother 64:e00573-20. doi: 10.1128/AAC.00573-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.EUCAST. 2019. MIC and zone distributions and ECOFFs. https://www.eucast.org/mic_distributions_and_ecoffs/.
- 92.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimcirobial disk and dilution susceptibility tests for bacteria isolated from animals VET08, 4th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 93.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou Z, Alikhan N-F, Mohamed K, Fan Y, Achtman M, Agama Study Group. 2020. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res 30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 13033997 [q-bio.GN]. https://arxiv.org/abs/1303.3997.
- 97.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 103.Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. 2012. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun 80:1554–1562. doi: 10.1128/IAI.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Merino I, Porter SB, Johnston B, Clabots C, Thuras P, Ruiz-Garbajosa P, Cantón R, Johnson JR. 2020. Molecularly defined extraintestinal pathogenic Escherichia coli status predicts virulence in a murine sepsis model better than does virotype, individual virulence genes, or clonal subset among E. coli ST131 isolates. Virulence 11:327–336. doi: 10.1080/21505594.2020.1747799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seemann T. 2020. snippy: fast bacterial variant calling from NGS reads. https://github.com/tseemann/snippy.
- 106.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kwong J, Seemann T. 2019. maskrc-svg. Httpsgithubcomkwongjmaskrc-Svg. https://github.com/kwongj/maskrc-svg.
- 108.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane A, Harris SR. 2016. SNP-sites: rapid efficient extraction of SNPs from multi- FASTA alignments. Microb Genom 5:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Salmela L, Rivals E. 2014. LoRDEC: accurate and efficient long read error correction. Bioinformatics 30:3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. 2011. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol 12:R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Galata V, Fehlmann T, Backes C, Keller A. 2019. PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res 47:D195–D202. doi: 10.1093/nar/gky1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed virulence, antimicrobial resistance, and typing metadata of all isolates generated in this study. Download Table S1, XLSX file, 0.2 MB (213.1KB, xlsx) .
Copyright © 2020 Hayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed virulence, antimicrobial resistance, and typing metadata of isolates available on Enterobase that were within 20 allele differences (as per cgMLST schema provided at Enterobase) of isolates in this study. Download Table S2, XLSX file, 0.1 MB (127.8KB, xlsx) .
Copyright © 2020 Hayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Short reads generated during this project have been submitted at NCBI GenBank under BioProject accessions PRJNA605257, PRJNA605064, and PRJNA604903. Complete plasmid sequences have been submitted at GenBank under accession numbers MT077880, MT077881, MT077882, MT077883, MT077884, MT077885, MT077886, MT077887, MT077888, MT077889, and MT816498.