ABSTRACT
Shinkaia crosnieri is an invertebrate that inhabits an area around deep-sea hydrothermal vents in the Okinawa Trough in Japan by harboring episymbiotic microbes as the primary nutrition. To reveal physiology and phylogenetic composition of the active episymbiotic populations, metatranscriptomics is expected to be a powerful approach. However, this has been hindered by substantial perturbation (e.g., RNA degradation) during time-consuming retrieval from the deep sea. Here, we conducted direct metatranscriptomic analysis of S. crosnieri episymbionts by applying in situ RNA stabilization equipment. As expected, we obtained RNA expression profiles that were substantially different from those obtained by conventional metatranscriptomics (i.e., stabilization after retrieval). The episymbiotic community members were dominated by three orders, namely, Thiotrichales, Methylococcales, and Campylobacterales, and the Campylobacterales members were mostly dominated by the Sulfurovum genus. At a finer phylogenetic scale, the episymbiotic communities on different host individuals shared many species, indicating that the episymbionts on each host individual are not descendants of a few founder cells but are horizontally exchanged. Furthermore, our analysis revealed the key metabolisms of the community: two carbon fixation pathways, a formaldehyde assimilation pathway, and utilization of five electron donors (sulfide, thiosulfate, sulfur, methane, and ammonia) and two electron accepters (oxygen and nitrate/nitrite). Importantly, it was suggested that Thiotrichales episymbionts can utilize intercellular sulfur globules even when sulfur compounds are not usable, possibly also in a detached and free-living state.
IMPORTANCE Deep-sea hydrothermal vent ecosystems remain mysterious. To depict in detail the enigmatic life of chemosynthetic microbes, which are key primary producers in these ecosystems, metatranscriptomic analysis is expected to be a promising approach. However, this has been hindered by substantial perturbation (e.g., RNA degradation) during time-consuming retrieval from the deep sea. In this study, we conducted direct metatranscriptome analysis of microbial episymbionts of deep-sea squat lobsters (Shinkaia crosnieri) by applying in situ RNA stabilization equipment. Compared to conventional metatranscriptomics (i.e., RNA stabilization after retrieval), our method provided substantially different RNA expression profiles. Moreover, we discovered that S. crosnieri and its episymbiotic microbes constitute complex and resilient ecosystems, where closely related but various episymbionts are stably maintained by horizontal exchange and partly by their sulfur storage ability for survival even when sulfur compounds are not usable, likely also in a detached and free-living state.
KEYWORDS: chemosynthesis, deep sea, in situ analysis, metatranscriptome, symbiosis
INTRODUCTION
In environments without sunlight, ecosystems and geochemical cycles are often powered by chemosynthetic microbes that utilize reduced chemical compounds as energy sources (1–3). As a notable example, deep-sea hydrothermal vents can harbor an abundance of animals that actively proliferate by obtaining nutrient sources from their symbiotic chemosynthetic microbes (4). Beginning with the discovery of tubeworm Riftia pachyptila associated with microbes oxidizing reduced sulfur compounds in 1981 (5–7), symbioses with chemosynthetic microbes have been found for various invertebrate phyla, including Mollusca, Annelida, and Arthropoda, and their microbial symbionts are either inside (endosymbionts) or outside (episymbionts) the host bodies (8, 9). To obtain comprehensive and detailed knowledge on the taxonomic composition and metabolic activities of these microbes, a metatranscriptomic approach is expected to be promising (10); however, fundamental obstacles exist in the application of this approach to deep-sea environments (11).
Shinkaia crosnieri is a deep-sea invertebrate that predominantly (up to 465 individuals/m2) inhabits an area around hydrothermal vents in the Okinawa Trough in Japan (12). S. crosnieri utilizes episymbiotic microbes on the surface of its ventral setae as its primary nutrient source, and its symbiotic system is the most clearly characterized one among deep-sea episymbioses (13, 14). Those episymbionts were revealed to contain methane-utilizing and sulfur (thiosulfate and sulfide)-oxidizing chemolithotrophic bacteria (15, 16). Phylogenetically, 16S rRNA gene clone analysis reproducibly identified species belonging to the genus Sulfurovum in Epsilonproteobacteria (recently proposed to be renamed as Campylobacterota [17–19]) and the orders Thiotrichales and Methylococcales in Gammaproteobacteria as major episymbionts (20, 21). Of these episymbionts, fluorescence in situ hybridization coupled to nanoscale secondary ion mass spectrometry (FISH-NanoSIMS) revealed that the Sulfurovum episymbionts oxidize reduced sulfur compounds and fix inorganic carbon (15). The Thiotrichales and Methylococcales episymbionts were inferred to be thioautotrophs and methanotrophs, respectively, based on the physiology of related species and reverse transcription-PCR (RT-PCR)/phylogenetic analyses of pmoA, a key gene for methane oxidation (16). However, experimental investigation of the S. crosnieri episymbionts in laboratories is impeded because of difficulties in culturing these microbes and rearing the hosts by accurately simulating hydrothermal vent environments. Rearing S. crosnieri in a tank supplied with sulfide or methane was successfully conducted, but this process substantially changed the episymbiotic microbial community composition (22, 23).
Given these difficulties in laboratory experiments, metatranscriptomics by in situ RNA stabilization is expected to be promising for direct investigation of these episymbionts as mentioned above. However, in addition to the apparent difficulty of sampling from the deep sea itself, the time and environmental changes required by sampling processes constitute fundamental obstacles, although most of the metatranscriptomic studies of symbiotic and free-living deep-sea microbes stabilized RNA after onboard recovery (11, 24). During deep-sea sampling and onboard recovery, which usually takes more than an hour, the composition and activity of microbial communities can change considerably, while RNAs can be degraded (25), which means that true RNA profiles in the deep sea can be missed if experimental procedures are conducted after retrieval (26, 27). To solve these problems, in situ RNA stabilization (i.e., stabilization before retrieval by remote control) instead of onboard RNA stabilization (i.e., stabilization after retrieval) is desirable (16, 28–32); however, these two approaches have been neither systematically compared nor applied to metatranscriptomic analyses of deep-sea symbiotic microbial communities.
In this study, we applied in situ and onboard RNA stabilization methods to 16S rRNA and metatranscriptomic analyses of the S. crosnieri episymbiotic microbial communities to (i) delineate their taxonomic composition by increasing sensitivity and eliminating biases in PCR cloning, (ii) systematically estimate the effects of the sampling processes from deep-sea hydrothermal vents that are characterized by unusual chemical and physical conditions, and (iii) obtain comprehensive knowledge on their physiology.
RESULTS AND DISCUSSION
Total RNA (rRNA) analysis of S. crosnieri episymbiotic microbial communities.
S. crosnieri sampling was conducted at a 982-m-deep hydrothermal vent site in the Iheya North field in the Okinawa Trough in Japan (see Fig. S1 in the supplemental material) in 2015, and episymbiotic microbial community samples subjected to RNA stabilization either in situ or onboard were obtained from the host setae. Total RNA sequencing with Ion PGM System of six in situ and seven onboard RNA-stabilized samples from different host individuals yielded 5,430,057 and 4,847,064 reads, respectively (Table 1). After filtering short and low-quality reads, 619,694 and 695,480 rRNA reads, respectively, were used for taxonomic analyses.
TABLE 1.
Sequencing of total RNA
| Stabilization method | Sample ID | Specimen ID | No. of raw reads | No. of quality- controlled reads | Mean read length (bp) | No. of mapped reads |
|---|---|---|---|---|---|---|
| In situ | In situ total RNA 1 | 6 | 431,185 | 36,143 | 130.5 | 7,714 |
| In situ total RNA 2 | 8 | 1,276,721 | 143,663 | 134.4 | 27,636 | |
| In situ total RNA 3 | 9 | 1,109,657 | 133,618 | 142.4 | 25,482 | |
| In situ total RNA 4 | 10 | 695,387 | 129,258 | 158.7 | 38,269 | |
| In situ total RNA 5 | 11 | 623,241 | 83,526 | 142.8 | 27,014 | |
| In situ total RNA 6 | 12 | 1,293,866 | 93,486 | 129.4 | 25,424 | |
| Onboard | Onboard total RNA 1 | 1 | 426,120 | 79,465 | 155.5 | 16,378 |
| Onboard total RNA 2 | 2 | 759,925 | 66,155 | 128.8 | 15,304 | |
| Onboard total RNA 3 | 7 | 567,413 | 97,520 | 153.4 | 38,240 | |
| Onboard total RNA 4 | 8 | 607,935 | 100,741 | 152.2 | 23,636 | |
| Onboard total RNA 5 | 9 | 1,041,853 | 192,140 | 152.2 | 58,984 | |
| Onboard total RNA 6 | 10 | 776,235 | 58,905 | 123.2 | 16,047 | |
| Onboard total RNA 7 | 11 | 667,583 | 100,554 | 146.3 | 16,894 | |
Location of the Iheya North hydrothermal field, Okinawa Trough, in Japan. The figure is based on a map made with Natural Earth (naturalearthdata.com). Download FIG S1, EPS file, 1.1 MB (1.1MB, eps) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
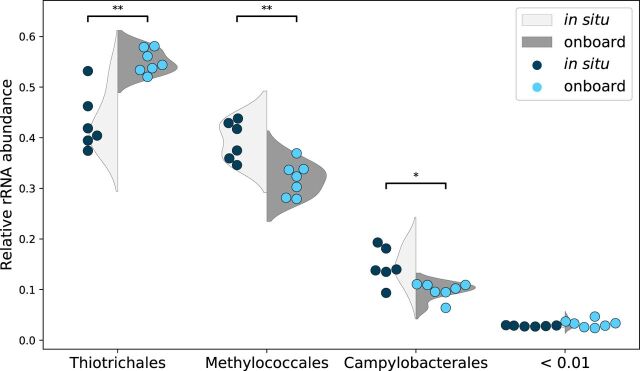
The relative abundances of the rRNA reads confirmed that the orders Thiotrichales, Methylococcales, and Campylobacterales were the major community members. These three major orders showed relative abundances of >5% in every sample and accounted for 95.3% to 97.6% in total (Fig. 1). At the genus level, Sulfurovum was particularly dominant and accounted for more than 99% of the Campylobacterales rRNA reads in all samples (see Table S1 in the supplemental material). Notably, Thiotrichales, Methylococcales, and Sulfurovum sequences were consistently found in a previous study that applied a PCR-based method to S. crosnieri episymbiotic microbial communities at the same site in 2007 (20). Therefore, Thiotrichales, Methylococcales, and Campylobacterales (or Sulfurovum) seemed to be stable as major members in S. crosnieri episymbiotic communities at least at this hydrothermal vent site, irrespective of possible PCR biases.
FIG 1.
Relative rRNA abundances at the order level. Three major orders for which the relative abundances of rRNA expression were >0.01 in all samples are shown. Each symbol shows the value for a sample (six in situ and seven onboard), and violin plots are presented for reference. Mann-Whitney U tests were performed (**, P < 0.01; *, P < 0.05).
Summary of rRNA reads assigned to genera within Campylobacterales (relative abundance [read numbers]). Download Table S1, DOCX file, 0.01 MB (14KB, docx) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
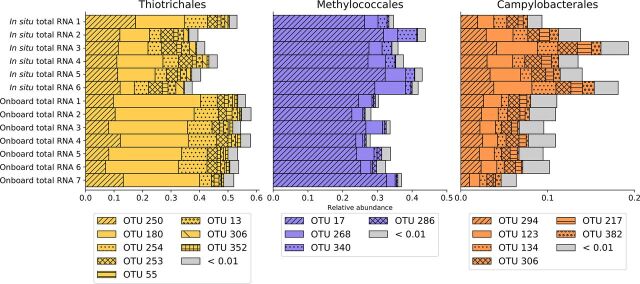
The Thiotrichales, Methylococcales, and Campylobacterales rRNA sequences contained 199, 132, and 337 SILVA identifiers, which were clustered to 82, 44, and 85 operational taxonomic units (OTUs), respectively, at a 95% similarity threshold (Fig. 2). These numbers suggest that each of the three S. crosnieri episymbiont taxa comprised dozens of species instead of a few specifically adapted species. The OTU compositions were highly similar among the different S. crosnieri individuals. The Thiotrichales and Methylococcales episymbionts contained two (OTUs 250 and 180) and one (OTU 17) abundant OTU, respectively. On the other hand, no specific dominant OTUs were observed in the Campylobacterales (Sulfurovum) episymbionts. The saturation of rarefaction curves for all samples suggested that the entire episymbiotic microbial communities were well sampled (Fig. S3a). Collectively, the large numbers of OTUs observed in each sample and the community composition similarities among the samples indicate that the episymbiotic community of each S. crosnieri individual is not composed of clonal descendants of a few “founder” cells that settled down after birth or molting of the host but is more complex than previously thought and maintained by horizontal exchange between host individuals around the hydrothermal vent.
FIG 2.
Relative rRNA abundances at the OTU level. OTUs clustered at the 95% identity threshold for which the relative abundances of rRNA expression were >0.01 in all samples are shown.
Episymbiotic microbial community comparisons between the in situ and onboard RNA stabilization methods.
While OTU richness and Faith’s phylogenetic diversity were not significantly different between the in situ and onboard RNA-stabilized samples (P > 0.05, Mann-Whitney U test; Fig. S3b and c), the in situ RNA-stabilized samples showed significantly larger Shannon diversity (P = 0.018, Mann-Whitney U test; Fig. S3d). Thus, while the OTU sets of the episymbiotic communities were not different between the sampling methods, skewness in their abundances decreased during the retrieval process from the deep sea. Weighted UniFrac and the principal coordinate analysis (PCoA) revealed that the in situ and onboard RNA-stabilized data were significantly different (P = 0.002, permutational multivariate analysis of variance [PERMANOVA], 999 permutations in each test, Fig. S4). It may be noted that the composition of the in situ total RNA 1 sample was exceptionally close to that of the onboard stabilized samples, but this sample was also characterized by the particularly small number of the mapped reads and OTUs, likely because of problems in sequencing (Table 1). We confirmed that exclusion of in situ total RNA 1 sample did not affect the results of the statistical tests of the OTU richness, Faith’s phylogenetic diversity, and Shannon diversity.
Comparison of the relative abundances between the in situ and onboard RNA stabilization methods showed relative increases in Thiotrichales (4.0% on average) and decreases in Methylococcales (11.9% on average) and Campylobacterales (6.5% on average) in the onboard stabilization samples, and these shifts were consistently observed regardless of OTUs among different host individuals (Fig. 2). Thus, as expected, the approximately 3-h sampling process without RNA stabilization likely affected the rRNA abundances of the major taxa in a systematic manner. In particular, Thiotrichales OTU 180 (corresponding to the genus Thiothrix) showed a substantial increase under onboard RNA stabilization (Table S2). Notably, Thiothrix was reported to store reduced sulfur compounds as intercellular sulfur granules and utilize them as energy sources by oxidization to sulfate (discussed later) (33–35).
Detected SILVA identifiers and OTUs. Download Table S2, DOCX file, 0.01 MB (13.2KB, docx) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metatranscriptomic analysis of S. crosnieri episymbiotic microbial communities.
rRNA-depleted RNA sequencing with Ion PGM System of two in situ and two onboard RNA-stabilized samples from different host individuals yielded 23,392,460 reads in total after quality control (Table 2). Transcriptomic assembly using all reads provided 146,985 contigs (N50, 597 bp; longest contig, 22,476 bp), among which 97,348 contigs were predicted to have 109,253 protein-coding sequences (CDSs). We conducted taxonomic and functional annotations of these CDSs, as well as 49,637 contigs on which no CDSs were predicted. Taxonomic annotations were given to 136,313 (85.8%) sequences (i.e., CDSs and contigs), among which 49,014, 27,321, and 26,141 were assigned to Thiotrichales, Methylococcales, and Sulfurovum, respectively. Functional annotations using the KEGG database annotated 78,748 (49.6%) sequences, which are regarded as genes hereinafter. Among those genes, 64,908 (82.4%) were taxonomically annotated to either of the three major taxa.
TABLE 2.
Sequencing and initial analysis of rRNA-depleted RNA
| Statistic |
In situ
|
Onboard |
||
|---|---|---|---|---|
| mRNA 1 | mRNA 2 | mRNA 1 | mRNA 2 | |
| Specimen ID | 4 | 8 | 2 | 10 |
| No. of raw reads | 6,611,280 | 4,229,368 | 6,941,648 | 7,117,406 |
| No. of quality-controlled reads | 5,820,455 | 4,126,616 | 6,572,709 | 6,872,680 |
| Mean read length (bp) | 111.2 | 157.6 | 140.5 | 140.9 |
| % Mapped effective reads | 74.7 | 72.1 | 72.7 | 74.7 |
| % Thiotrichales | 18.1 | 14.2 | 30.6 | 28.4 |
| % Methylococcales | 22.6 | 28.4 | 14.6 | 19.8 |
| % Sulfurovum | 18.7 | 16.8 | 5.3 | 10.3 |
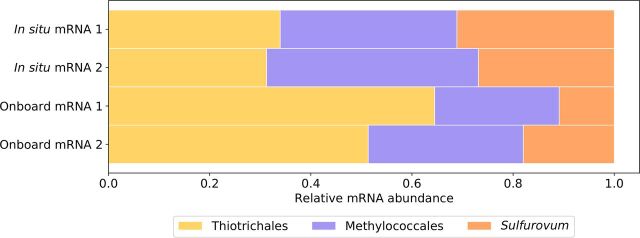
Transcript abundances were quantified by mapping the rRNA-depleted RNA sequences to the assembled contigs and calculating transcripts per million (TPM) values. Total TPM values of the transcripts that were taxonomically annotated to the three major taxa ranged from 58.8% to 69.6% depending on the samples (Table 2). These values were smaller than the rRNA (total RNA) abundances of the same three taxa, likely because of difficulties of taxonomic annotation of mRNA. The relative transcript abundances among the three major taxa are shown in Fig. 3. Compared to the relative rRNA abundances (Fig. 1), the Sulfurovum episymbionts showed larger mRNA abundances. This might be because the existence of their closely related genomes in the database enabled more sensitive annotation of Sulfurovum mRNA (although Sulfurovum had the fewest reference sequences: Thiotrichales, 513,398; Methylococcales, 268,111; and Sulfurovum, 53,394) and/or because the Sulfurovum episymbionts produce more mRNAs than the others.
FIG 3.
Relative mRNA abundances of the three major taxa. Relative abundances of total mRNA among Thiotrichales, Methylococcales, and Sulfurovum (dominating the Campylobacterales episymbionts) are shown. Each bar represents a sample (two in situ samples and two onboard samples).
Finally, comparison of the relative abundances between the in situ and onboard RNA stabilization methods showed that the two RNA stabilization methods similarly affected the rRNA and mRNA abundances (Fig. 1 and 3), i.e., an overall increase in the Thiotrichales profiles but an overall decrease in the Methylococcales and Campylobacterales (Sulfurovum) profiles.
Metatranscriptomic differences between the in situ and onboard RNA stabilization methods.
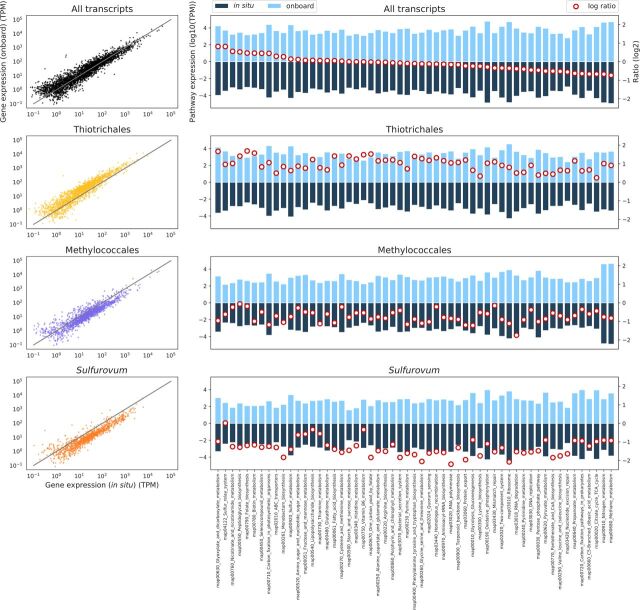
We then compared the transcriptomic profiles of the three major taxa, namely, Thiotrichales, Methylococcales, and Sulfurovum, in depth. Gene- and pathway-level relative transcript abundances were examined based on the functional KEGG Orthology (KO) annotations (Fig. 4, the left and right panels show abundances by genes and pathways, respectively). Those of the genes taxonomically assigned to the three major taxa are also shown in the second to fourth rows in the same figure. As described already, when the onboard RNA stabilization method was adopted, the relative abundances of Thiotrichales genes increased, and those of Methylococcales and Sulfurovum genes decreased. What was most notable here was that these shifts seemed to have been caused by transcriptome-wide changes that coincided with rRNA abundance changes but not with large changes in the abundance of specific genes (e.g., those required for metabolism at hydrothermal vents). It may also be notable that some genes and pathways went against the global trends between the two stabilization methods, either by chance or because of the physiological effects of sampling from the deep sea. However, as far as we investigated, we did not see any genes and pathways that exhibited concerted trends against the transcriptome-wide trends in all three major taxa, even among energy and carbon metabolism-related genes and pathways that were likely systematically affected by deep-sea sampling (Fig. 4). To compare mRNA abundance of each sample by cancelling out those transcriptome-wide effects in each taxon, the expression profiles were also visualized by normalizing per each taxon and sample (Fig. 5). First, the expression profiles of the same taxa from the replicate samples were most similar, showing the robustness of the results. Second, the expression profiles of the same taxa were most similar between the in situ and onboard RNA-stabilized samples when the transcriptome-wide effects were cancelled out. This also confirmed that the sample retrieval from the deep sea did not substantially affect expression of genes of specific pathways. Thus, we concluded that the transcriptome-wide shift due to deep-sea sampling was the major factor that affected the RNA expression profiles. Specifically, a reasonable explanation would be that the rRNA and mRNA of Methylococcales and Sulfurovum were globally degraded faster than those of Thiotrichales during deep-sea sampling, possibly because of global attenuation of biological activities by depletion of carbon and energy sources and temperature and hydrostatic pressure changes.
FIG 4.
mRNA abundances of genes and pathways compared by the RNA stabilization methods. The left panels show the expression abundance of each gene by the in situ (x axis) and onboard (y axis) RNA stabilization methods in TPM values. From the top to the bottom, data from all transcripts (black) and those assigned to Thiotrichales (yellow), Methylococcales (purple), and Sulfurovum (orange) are shown. Values from the two samples were averaged for each method. The gray line is x = y. The right panels show the total expression abundance of genes in each KEGG pathway. The upper and lower bars represent results from the onboard and in situ RNA stabilization methods, respectively, in log10 TPM values. Values from the two samples were averaged for each method. Only pathways with TPM values of >1,000 in at least one sample are shown. The red closed circles represent log ratios of TPM values obtained by both methods.
FIG 5.
mRNA abundance of pathways normalized for each sample and taxon. The heat map shows the total expression abundance of genes in each KEGG pathway, which was scaled by log10 and z-score normalized for each taxon and sample.
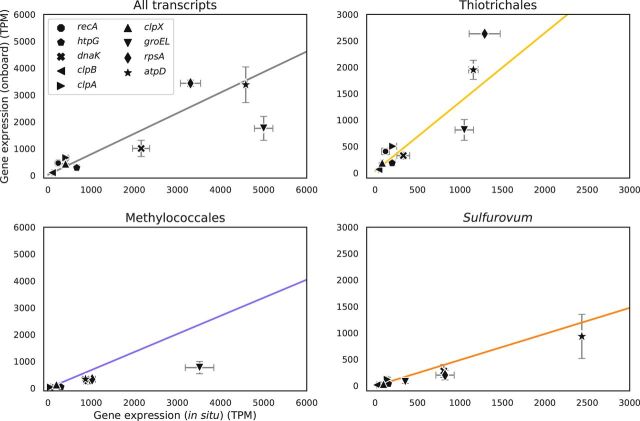
We note that previous studies reported gene-specific transcript abundance changes during deep-sea sampling. A study that applied quantitative PCR to analyze the transcript abundance of the Methylococcales pmoA gene, which is a key gene in methane oxidation, reported a 3.7-fold decrease in onboard RNA-stabilized samples of deep-sea S. crosnieri episymbionts compared to in situ RNA-stabilized samples (16). In this study, we observed a 1.82-fold decrease in the TPM values of the pmoA gene, and this could be in line with the 1.51-fold transcriptome-wide abundance decrease of Methylococcales (Fig. 3 and 4); therefore, the decrease in pmoA transcript abundance might actually be a part of the transcriptome-wide effects of deep-sea sampling. It was also reported that the expression of stress response genes (e.g., recA and hsp90) increases under onboard RNA stabilization (28, 30), although these studies dealt with neither S. crosnieri episymbionts nor deep-sea vent microbes. Thus, we analyzed nine genes (recA, dnaK, hsp90, clpB, clpA, clpX, groEL, rpsA, and atpD) for which the expression levels were expected to be increased by environmental stress. In our results, none of the nine genes showed an increase in all three major taxa but collectively followed the transcriptome-wide trends (Fig. 6). A previous study on endosymbionts of Ifremeria nautili, which inhabits deep-sea hydrothermal vents, reported that their 34 h of incubation with few or no electron donors increased the gene expression of the DnaK chaperone system (e.g., dnaK) and the Clp/Hsp100 family of ATP-dependent proteins (e.g., clpA and clpX) (36). However, there are fundamental differences between this study and the previous studies (e.g., 34-h incubation versus 171-min retrieval, endosymbionts versus episymbionts, and taxonomic compositions), and these differences might have led to different transcriptome-wide and gene-specific responses.
FIG 6.
mRNA abundances of stress-responsive genes by RNA stabilization methods. Expression abundance of each stress-responsive gene by the in situ (x axis) and onboard (y axis) RNA stabilization methods is shown. The four panels show data from all transcripts and those assigned to Thiotrichales, Methylococcales, and Sulfurovum. The error bars represent the larger and smaller values of the two samples for each method. The slope is a regression line calculated using the expression abundances of all genes in each major taxon or in total.
Different metatranscriptomic responses among the three major episymbiotic taxa.
We found that the rRNA and mRNA of Methylococcales and Sulfurovum were likely rapidly and globally degraded during deep-sea sampling, while those of Thiotrichales were kept relatively stable. We were then interested in why Thiotrichales episymbionts could keep their rRNA and mRNA more stable than those belonging to the other two major taxa.
We hypothesized that Thiotrichales episymbionts utilized intercellular sulfur globules as an energy source to escape the global attenuation of biological activities. Many sulfur-oxidizing bacteria belonging to Gammaproteobacteria, including Thiotrichales, form intracellular zero-valent sulfur globules as intermediates in sulfur oxidation (35, 37). The TusA and DsrAB proteins are known to be involved in the transfer and oxidization, respectively, of sulfur globules (38–40). In our metatranscriptomic results, we detected the expression of the tusA and dsrAB genes by Thiotrichales episymbionts (TPM values of more than 2,800 and 50, respectively, in all samples). Furthermore, a previous FISH-NanoSIMS analysis revealed that bicarbonate was incorporated into S. crosnieri episymbiotic microbial communities without electron donors and that bicarbonate was enriched in short and thin filamentous cells that resembled Thiotrichales cells (15). Therefore, the Thiotrichales episymbionts were assumed to be able to fix carbon even if electron donors are not supplied using intracellular sulfur globules, and this ability may help them survive even when sulfur compounds are not usable, likely also in a free-living state during horizontal exchange. Notably, the Sulfurovum episymbionts, which are also thioautotrophs but showed a decrease in mRNA abundances, expressed neither the tusA nor dsrAB genes.
Delineation of the metabolic activities of S. crosnieri episymbionts.
Finally, we investigated the physiology and ecology of the S. crosnieri episymbiotic community using the in situ RNA-stabilized metatranscriptomic data. Functional annotation using the KEGG database revealed the expression of genes of two carbon fixation pathways, genes of a formaldehyde assimilation pathway, and genes that exploit five electron donors (sulfide [HS−], thiosulfate [HS2O3−], sulfur [S0], methane [CH4], and ammonia [NH4+]) and two electron acceptors (oxygen [O2] and nitrate/nitrite [NO3−/NO2−]). We could not detect the expression of genes related to hydrogen oxidation or nitrogen fixation; these systems were recently observed in symbiotic chemolithotrophic microbes (41, 42).
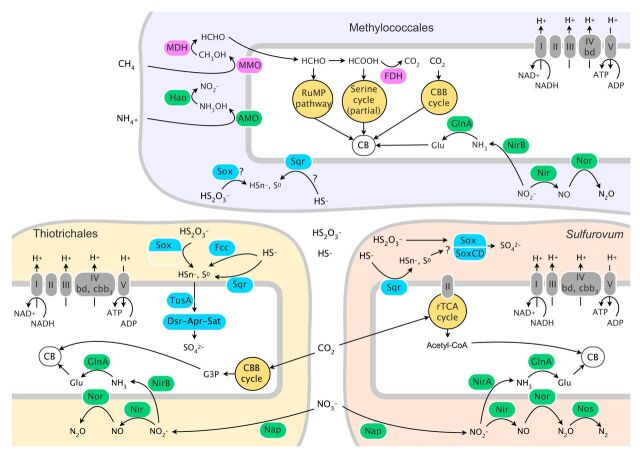
(i) Sulfur oxidation. Substantial expression of genes for oxidation of reduced sulfur compounds by the Thiotrichales, Methylococcales, and Sulfurovum episymbionts was observed (Fig. 7). Those genes included the Sox multiple-enzyme system (soxABCXYZ), sulfide:quinone oxidoreductase (sqr), flavocytochrome c (fccB), dissimilatory sulfite reductase (dsrAB, functioning in the reverse dissimilatory sulfate reduction pathway), adenylylsulfate reductase (aprAB), and sulfate adenylyltransferase (sat) genes (see Data Set S1 in the supplemental material). As other sulfur-oxidizing Gammaproteobacteria and Epsilonproteobacteria do (43, 44), the Thiotrichales episymbionts expressed all of these genes except soxC (i.e., the Sox system without SoxCD [37, 46]), and the Sulfurovum episymbionts expressed soxABCXYZ and sqr. These observations suggest that both bacteria can oxidize thiosulfate and sulfide to sulfate.
FIG 7.
Schematic model of the chemosynthetic metabolisms of the three major S. crosnieri episymbiont taxa. Symbols represent proteins involved in each metabolism (blue, sulfur metabolism; pink, methane metabolism; yellow, inorganic carbon fixation; green, nitrogen metabolism). AMO, ammonia monooxygenase; Apr, adenosine phosphosulfate reductase; CB, cellular biosynthesis; CBB cycle, Calvin-Benson-Bassham cycle; Dsr, reverse dissimilatory sulfite reductase; Fcc, flavocytochrome c; FDH, formate dehydrogenase; GlnA, glutamine synthetase; Hao, hydroxylamine; MDH, methanol dehydrogenase; MMO, methane monooxygenase; Nap, two membrane-bound periplasmic nitrate reductases; Nir, nitrite reductase; Nor, a set of membrane-bound nitric oxide reductases; Nos, a set of nitrous oxide reductases; rTCA cycle, reductive tricarboxylic acid cycle; RuMP pathway, ribulose monophosphate pathway; Sat, sulfate adenylyltransferase; Sox, sox enzyme system (Sox and SoxCD, representing the SoxABXYZ complex and the SoxCD complex, respectively); Sqr, sulfide–quinone reductase; TusA, sulfur carrier protein; I, NADH dehydrogenase; II, succinate dehydrogenase; III, complex III cytochrome bc1; IV, bd and cbb3, which are bd-type cytochrome c and cbb3-type cytochrome c, respectively; and V, complex V ATPase.
mRNA abundances of the genes shown in Fig. 7. Download Data Set S1, XLSX file, 0.05 MB (52.8KB, xlsx) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Methylococcales episymbionts expressed the soxBXY and sqr genes (TPM values of more than 40 in each of the two in situ RNA-stabilized samples), implying that they can also oxidize reduced sulfur compounds. Notably, this possibility of sulfur oxidization by the Methylococcales episymbionts of S. crosnieri has not been proposed thus far, whereas metagenomic studies of other microbial ecosystems containing methanotrophs (47, 48) and the genome sequences of Methylobacter whittenbury (49) and Methylomarinum vadi (50) indicated that methanotrophs in general can harbor genes for sulfur oxidization.
(ii) Methane metabolism by Methylococcales. Substantial expression of genes for methane metabolism, specifically for complete methane oxidation and formaldehyde (C1 compound) assimilation, by Methylococcales episymbionts was observed (Fig. 7 and Data Set S1). These genes included methane monooxygenase (pmoCAB), methanol dehydrogenase (mxaF), those for dehydrogenation of formaldehyde to formate (fae, mtdAB, mch, and fhc), formate dehydrogenase, and hexulose-6-phosphate synthase and hexulose-phosphate isomerase (hps and hpi) in the ribulose monophosphate (RuMP) pathway. Thus, the Methylococcales episymbionts were supposed to be able to oxidize methane to carbon dioxide and assimilate its intermediate, formaldehyde, as a carbon source. On the other hand, only weak expression of six out of nine serine pathway genes (another C1 compound assimilation pathway) and the ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) gene was observed. This implied that the Methylococcales episymbionts were mainly composed of type I Gammaproteobacteria methanotrophs, which do not possess RuBisCO and serine pathways, consistent with previous phylogenetic analysis of pmoA genes (16). Nonetheless, it should be noted that type X methanotrophs, which have RuBisCO and serine pathways (51), might also exist in the community. Expression of the hydroxylamine oxidoreductase gene was also observed, suggesting that Methylococcales episymbionts may oxidize ammonia with methane monooxygenase to nitrite via hydroxylamine.
(iii) Respiration and nitrogen metabolism. Expression of genes for both oxygen respiration and aerobic denitrification by all three major taxa was observed. Regarding oxygen respiration, the expression of cytochrome c oxidase cbb3-type subunit I (ccoNOPQ) and cytochrome c oxidase bd-type subunit I (cydAB) genes was detected in all three major taxa (Data Set S1). Regarding denitrification, the expression of the periplasmic nitrate reductase complex (napAB), two cytochrome cd-type nitrite reductases (nirK or nirS), nitric oxide reductase (nor), and nitrous oxide reductase (nosZ) genes was detected in the Thiotrichales and Sulfurovum episymbionts, while expression of only the nirK and norBC genes was detected in Methylococcales episymbionts. Thus, Methylococcales episymbionts seem to generate nitrous oxide (N2O), whereas Thiotrichales and Sulfurovum episymbionts seem to generate nitrogen (N2). The substantial expression of genes involved in denitrification suggests the possibility that the three major taxa utilize nitrate and nitrite as an alternative electron acceptor because of their competition for oxygen.
Expression of cytoplasmic nitrite reductase (nirBD or nirA) and nitrogen-assimilating glutamine synthetase (glnA) genes were detected for all three major taxa. Thus, we assume that nitrite in the periplasm would be transported to and assimilated in the cytoplasm.
(iv) Carbon fixation. Expression of the genes of two inorganic carbon fixation pathways among six pathways (43, 52) was observed in the Thiotrichales and Sulfurovum episymbionts. The Thiotrichales episymbionts expressed genes encoding the Calvin-Benson-Bassham (CBB) cycle, including the RuBisCO gene. The Sulfurovum episymbionts expressed genes encoding the reductive tricarboxylic acid (rTCA) cycle, including oor, frd, and acl genes. Thus, the CBB and rTCA cycles are likely utilized by the Thiotrichales and Sulfurovum episymbionts, respectively, which is consistent with the physiological characteristics of the cultured strains that belong to Gammaproteobacteria and Epsilonproteobacteria, respectively (43, 53–55).
Other notable observations would include that Thiotrichales showed relatively high expression of glyoxylate and dicarboxylate metabolism pathway genes and Methylococcales showed relatively high expression of the pentose phosphate pathway genes (Fig. 5).
Conclusion.
In this study, we performed direct metatranscriptome analysis of episymbionts of S. crosnieri by applying the in situ RNA stabilization equipment. To our knowledge, this is the first study that applied and systematically compared the different RNA stabilization approaches to deep-sea symbiotic microbial communities.
Comparison between the in situ and onboard RNA stabilization methods is highly demanded in applying the metatranscriptomic approach to deep-sea microbial communities, which can hardly be directly investigated by laboratory experiments, for three reasons. First, the onboard RNA stabilization method can give biased results that are different from those obtained by in situ RNA stabilization. Second, by doing so, the transcriptome-wide and gene-specific effects of deep-sea sampling can be discriminated. Third, by positively regarding such methodological differences as environmental perturbation, comparative metatranscriptomics may provide new insights into the physiology and ecology of microbial community members, as we did in this study. Last but not least, we note that these three factors would also be considered in metatranscriptomic analyses of other access-limited ecosystems.
The direct metatranscriptome analysis showed that methane utilization and sulfur oxidation are the main energy metabolisms of the S. crosnieri episymbionts. The dominant yet uncultured Thiotrichales, Methylococcales, and Sulfurovum expressed genes that encode almost all the components of the pathways for these chemoautotrophic systems. It may be notable that no expression of hydrogenase genes was detected in this study, while the genome of Sulfurovum sp. strain NBC37-1, which was isolated from an in situ sampler deployed on the same NBC hydrothermal vent site (i.e., not from the S. crosnieri body surface), carries four hydrogenase genes (55, 56). Whether the Sulfurovum episymbionts contribute to hydrogen flux will require further studies.
The results of the taxonomic clustering analysis indicated that the composition of the episymbiotic microbial populations is more complex than previously thought, where each of the three taxonomic clades likely contained diverse species. We anticipate that comparative analysis of metagenome-assembled genomes of the S. crosnieri episymbionts powered by long-read single-molecule sequencing technologies will help better understand their true biodiversity, immigration, and evolution within and among different hydrothermal vent habitats in the Okinawa Trough.
MATERIALS AND METHODS
Sample collection and RNA stabilization.
S. crosnieri individuals were collected from a single colony at the NBC hydrothermal vent site in the Iheya North field in the Okinawa Trough (27° 47.45′ N, 126° 53.80′ E, 982-m depth; see Fig. S1 in the supplemental material) during dive 1773 of the remotely operated vehicle (ROV) HyperDolphine on 18 January 2015. The details of the ROV are shown and described in Fig. S2. The seawater temperature of S. crosnieri habitats around the site was 5.7°C on average. All procedures were performed following the manufacturer’s instructions and with default settings unless otherwise noted.
Mounted equipment on the ROV HyperDolphine. The photograph shown in the left was taken by Masayuki Miyazaki. Download FIG S2, EPS file, 1.2 MB (1.2MB, eps) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ecological indices of the S. crosnieri episymbiotic microbial communities of the in situ and onboard RNA-stabilized samples. (a) Rarefaction curves. (b to d) Box-whisker plots of OTU richness (b), Faith’s phylogenetic diversity (c), and Shannon diversity (d). Points represent individual samples. The lines in the boxes represent medians. The boxes and whiskers represent ranges and interquartile ranges, respectively. Mann-Whitney U tests were performed (n.s., not significant; *, P < 0.05). Download FIG S3, EPS file, 0.1 MB (119.6KB, eps) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PCoA based on weighted UniFrac among the in situ and onboard RNA-stabilized samples. Variance explained by each PC is indicated on the axis. Plots represent results from individual samples of the in situ (dark blue) and onboard (light blue) RNA stabilization methods. Download FIG S4, EPS file, 0.04 MB (4.4KB, eps) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In situ RNA stabilization was conducted by following the procedure described by Watsuji et al. (16). Briefly, S. crosnieri individuals were collected in a sample container of the ROV using a suction sampler at the sampling site (Fig. S2). Then, the seawater in the container was replaced with aqueous sulfate salt solution (25 mM sodium citrate, 10 mM EDTA, and 700 g/liter ammonium sulfate; pH 5.2; colored with phenol red for visibility; prepared at 4°C before the ROV dive) by spontaneous diffusion in 9 min (sampling time, 11:46, stabilization time, 11:55 [Japan standard time {JST}]). The aqueous sulfate salt solution, which was proved to inhibit RNase activity (57), was used to provide a large amount of RNA stabilization solution. The S. crosnieri samples were then retrieved by a research vessel. The samples were washed three times using chilled aqueous sulfate salt solution to remove possibly contaminating bacteria that did not firmly attach to the S. crosnieri bodies. The setae covered with episymbionts were dissected from the host individuals and stored in RNAlater (Qiagen) at 4°C overnight and −80°C subsequently.
For onboard RNA stabilization, S. crosnieri individuals were collected in another sample container using another suction sampler during the same dive. To remove air and to keep the animals alive until retrieval by being kept in cold seawater, the container was prefilled with chilled filtered seawater and kept on −20°C ice packs before the dive. After collection, the S. crosnieri individuals were retrieved by a research vessel. The samples were washed three times using chilled sterile artificial seawater (25 g/liter NaCl, 4.2 g/liter MgCl2·6H2O, 3.4 g/liter MgSO4·7H2O, 0.5 g/liter KCl, 0.7 g/liter CaCl2·2H2O, and 14 mg/liter K2HPO4; pH 6.8) and submerged in aqueous sulfate salt solution. The duration from sample collection to submergence was 171 min (sampling time, 11:39; stabilization time, 14:30 [JST]). The setae covered with episymbionts were dissected from the host individuals and stored in RNAlater at 4°C overnight and −80°C subsequently. For this study, seta samples from seven S. crosnieri individuals each from the in situ and onboard RNA stabilization methods (i.e., from 14 individuals in total) were used. The carapace lengths and widths of the 14 individuals ranged from 30 to 48 mm and from 25 to 37 mm, respectively.
RNA extraction.
Total RNA extraction (for 16S rRNA analysis) and subsequent rRNA-depleted RNA preparation (for metatranscriptomic analysis) were conducted. Total RNA extraction was conducted using all 14 seta samples. RNA was extracted using an RNA PowerSoil Total RNA isolation kit (Mo Bio), treated using DNase I (Qiagen) for 10 min at room temperature, and recovered using an RNeasy minikit (Qiagen). The quantity and quality of the RNA were measured using a Qubit RNA HS assay kit with a Qubit 1.0 fluorometer (Invitrogen) and an Agilent RNA 6000 Pico kit with an Agilent 2100 Bioanalyzer (Agilent). We did not observe eukaryotic 18S and 28S rRNA peaks due to host RNA contamination using the Bioanalyzer. rRNA was depleted from four of the total RNA samples that had good quantities and qualities (two each from the in situ and onboard RNA stabilization methods). The samples were treated with the Ribo-Zero rRNA Removal kit for Gram-Negative Bacteria (Epicentre) and purified using RNA Clean & Concentrator (Zymo Research). Using a Bioanalyzer and Qubit, rRNA depletion was confirmed, and the RNA quantity was measured.
The RNA samples were kept at −80°C until sequencing.
Sequencing and initial quality check.
Sequencing of total RNA was conducted using 13 samples that comprised six in situ and seven onboard RNA-stabilized samples (one in situ sample was not used because of a quality problem). The RNA quantities and RNA integrity number (RIN) values ranged from 301.8 to 500.8 ng and from 5.6 to 6.7, respectively. Barcoded cDNA libraries were obtained using the Ion Total RNA-Seq kit v2 and Ion Xpress RNA-Seq Barcode 1–16 kit (Life Technologies). The quantities and library sizes of the cDNA libraries were measured using a Quant-iT dsDNA HS assay kit with a Qubit 2.0 fluorometer and an Agilent High Sensitivity DNA kit with an Agilent 2100 Bioanalyzer, respectively. Each cDNA library was diluted to 50 pM. Then, 5 μl of each solution was pooled to prepare two solutions so that each of the pooled solutions contained either the six in situ or seven onboard RNA-stabilized samples. From each of the two pooled solutions, 25 μl was loaded onto an Ion 318 v2BC Chip using an Ion PGM Hi-Q Chef kit with an Ion Chef system. Two sequencing runs were conducted using the Ion PGM Hi-Q Sequencing kit for 200-bp libraries with the Ion PGM System (Life Technologies). Sequencer-specific errors were corrected with Karect (58) (-celltype=haploid -matchtype=edit). Low-quality sequence regions and short reads were trimmed (-trim_qual_left 20 -trim_qual_right 20) and removed (-min_len 200), respectively, using PRINSEQ v0.20.4 (59).
Sequencing of the four rRNA-depleted RNA samples was conducted as follows. The RNA quantities and RIN values ranged from 316.8 to 436.8 ng and from 5.9 to 6.9, respectively. cDNA libraries were obtained using the Ion Total RNA-Seq kit v2. Quantities and library sizes were measured as described above. Each cDNA library was diluted to 20 pM. From each of the four solutions, 25 μl was loaded onto an Ion 318 v2BC Chip using an Ion PGM Hi-Q View Chef kit with an Ion Chef system. Four sequencing runs were conducted using the Ion PGM Hi-Q View Sequencing kit for 200-bp libraries with the Ion PGM System. Sequencer-specific errors were corrected with Karect (-celltype=haploid -matchtype=edit). Low-quality sequence regions and short reads were trimmed (-trim_qual_left 26 -trim_qual_right 26) and removed (-min_len 50), respectively, using PRINSEQ. Note that the low-quality filter was set at a stricter level here for subsequent metatranscriptome assembly.
rRNA analysis.
Reads generated by the two total RNA sequencing runs (i.e., those from 13 samples) were used for 16S rRNA analysis. Taxonomic annotation of each read was performed using BLASTn from BLAST+ 2.7.1 (60) against SILVA SSURef NR99 123 (61) (http://www.arb-silva.de/) and by retrieving the top hit that satisfied the criteria E value ≤ 1E−20, alignment length ≥ 180 bp, and identity ≥ 97%. If the top hit was attributed to multiple SILVA identifiers typically because a conserved region was sequenced, the most frequent identifier from the same stabilization method was chosen based on Occam’s razor. The relative abundance of each SILVA identifier was quantified by the assigned read numbers. Then, the SILVA sequences were clustered with a 95% identity threshold using UCLUST v1.2.22q (62, 63). Each cluster was designated an operational taxonomic unit (OTU). The abundance of each OTU was calculated as the sum of those of the contained SILVA identifiers. Alpha diversity (OTU richness, Shannon diversity, and Faith’s phylogenetic diversity) and beta diversity (weighted UniFrac) were calculated using skbio package v0.5.6 (scikit-bio.org). To calculate Faith’s phylogenetic diversity and weighted UniFrac, representative sequences of OTUs selected by UCLUST were aligned using MAFFT v7.470 (64) and phylogenetic trees were reconstructed using FastTree v2.1.11 (65) with the GTR+CAT model. Principal coordinate analysis (PCoA) was conducted based on weighted UniFrac using the prcomp package in R.
Metatranscriptomic analysis.
Reads from rRNA-depleted RNA sequencing were used for metatranscriptomic analysis. Because reference genomes of the episymbionts were not available, all reads generated by the four sequencing runs were merged and fed into the transcript assembly using Trinity v2.3.2 (45) (–min_contig_length 300). For quantification, we used RSEM v 1.2.30 (66) and Bowtie2 v 2.2.9 (67) through the align_and_estimate_abundance.pl script in Trinity (–est_method RSEM –aln_method bowtie2) to map the rRNA-depleted RNA sequencing reads to the assembled transcript contigs and quantify their abundances as transcripts per million (TPM) values.
Protein-coding sequences (CDSs) on each transcript contig were predicted by TransDecoder (http://transdecoder.github.io/). The taxonomic and functional annotations of the CDSs were conducted using BLASTp (E-value cutoff of 1E−5) against the NCBI nonredundant (NCBInr) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (68) (as of 12 March 2018) databases, respectively. Because the transcript contigs could contain partial genes that were missed by the CDS prediction, contigs without predicted CDSs were also queried against the NCBInr and KEGG databases for taxonomic and functional annotations, respectively, using BLASTx (E-value cutoff of 1E−5). For the taxonomic annotation using NCBInr, NCBI taxonomic identifiers (TaxIDs) were assigned to the BLAST hits by cross-referencing against the NCBI database (as of 26 March 2019; ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/accession2taxid). Then, the top hit in terms of E value (with bit scores in a tie) among the hits assigned to 1,751 TaxIDs that belonged to the order Thiotrichales or Methylococcales or genus Sulfurovum was retrieved (because >99% rRNA sequences assigned to the order Campylobacterales were assigned to Sulfurovum at the genus level [Fig. S1] and Sulfurovum did not belong to Campylobacterales but to unclassified Epsilonproteobacteria in NCBInr [as of 7 March 2018], the genus Sulfurovum was used instead of Campylobacterales for mRNA taxonomic assignment). For the functional annotation using KEGG, the KEGG Orthology (KO) number of the top hit was used if one was assigned.
KEGG orthology, module, and pathway analyses.
For each sample, a TPM value of each KO group was calculated as the sum of TPM values of CDSs and contigs that were functionally annotated to that KO. Likewise, the TPM value of each KEGG module and KEGG pathway was calculated as the sum of TPM values of KOs that belonged to each.
Analyses of the transcript abundances of the following genes were performed using the following KOs: pmoA by K10944, recA by K03553, rpsA by K02945, atpD by K02112, hsp90 by K04079, clpB by K03695, clpA by K03694, clpX by K03544, dnaK by K04043, groEL by K04077, and hupL by K06281. Analyses of the transcript abundances of the following functional categories were performed using the following KEGG modules (M numbers) and KOs: sulfur metabolism by M00176, M00596, M00595, K17229, K17230, and K17218; methane metabolism by M00174, M00346, M00345, M00344, M00140, K00122, K10713, K10714, and K01499; carbon fixation metabolism by M00165, M00173, M00579, M0376, M0375, M0374, M0377, and M0620; oxygen respiration by M00153, M00417, M00416, M00156, M00157, and M00159; nitrogen metabolism by M00175, M00531, M00530, M00529, and M00804; and adhesion mechanism by M00330 and K12549.
Ethics approval and consent to participate.
The locations for sample collection were not privately owned or protected in any way, and no specific permits were required for the described field studies and sample collection. The field studies did not involve any endangered or protected species.
Data availability.
The sequence data and assembled contigs were deposited in the DDBJ/ENA/GenBank database under BioProject identifier (ID) PRJDB9200. The bioinformatic scripts are available in github (https://github.com/kaorimo/MetatraAnalysis_mSystems2020).
ACKNOWLEDGMENTS
We thank the captain, crews, and onboard scientists of JAMSTEC R/V Natsushima, the operation team of ROV HyperDolphine, Takafumi Kasaya, Masahiro Yamamoto, and Thornton Blair for supporting sampling, Maki Tokuda and Miwako Tsuda for metatranscriptome sequencing, Ryoji Wani for supporting the project, and members of the Iwasaki Lab at the University of Tokyo for helpful discussions.
This research was supported by Japan Society for the Promotion of Science (grant 17J11518 to K.M. and grants 18H04136 and 19H05688 to W.I.).
We declare that we have no competing interests.
REFERENCES
- 1.Van Dover C. 2000. The ecology of deep-sea hydrothermal vents. Princeton University Press, Princeton, NJ. [Google Scholar]
- 2.Dick GJ. 2019. The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat Rev Microbiol 17:271–283. doi: 10.1038/s41579-019-0160-2. [DOI] [PubMed] [Google Scholar]
- 3.Takai K. 2019. Recent topics on deep-sea microbial communities in microbes and environments. Microbes Environ 34:345–346. doi: 10.1264/jsme2.ME3404rh. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanaugh CM, McKiness ZP, Newton ILG, Stewart FJ. 2006. Marine chemosynthetic symbioses, p 475–507. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), Prokaryotes. A handbook on the biology of bacteria: symbiotic associations, biotechnology, applied microbiology, 3rd ed, vol 1. Springer, New York, NY. [Google Scholar]
- 5.Cavanaugh CM, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB. 1981. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213:340–342. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- 6.Felbeck H. 1981. Chemoautotrophic potential of the hydrothermal vent tube worm. Science 213:336–338. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- 7.Rau GH. 1981. Hydrothermal vent clam and tube worm 13C/12C: further evidence of nonphotosynthetic food sources. Science 213:338–340. doi: 10.1126/science.213.4505.338. [DOI] [PubMed] [Google Scholar]
- 8.Dubilier N, Bergin C, Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 9.Goffredi SK. 2010. Indigenous ectosymbiotic bacteria associated with diverse hydrothermal vent invertebrates. Environ Microbiol Rep 2:479–488. doi: 10.1111/j.1758-2229.2010.00136.x. [DOI] [PubMed] [Google Scholar]
- 10.Baker BJ, Dick GJ. 2013. Omic approaches in microbial ecology: charting the unknown. Microbe 8:353–360. doi: 10.1128/microbe.8.353.1. [DOI] [Google Scholar]
- 11.Stewart FJ. 2013. Preparation of microbial community cDNA for metatranscriptomic analysis in marine plankton. Methods Enzymol 531:187−218. doi: 10.1016/B978-0-12-407863-5.00010-1. [DOI] [PubMed] [Google Scholar]
- 12.Thornton B, Bodenmann A, Pizarro O, Williams SB, Friedman A, Nakajima R, Takai K, Motoki K, Watsuji T, Hirayama H, Matsui Y, Watanabe H, Ura T. 2016. Biometric assessment of deep-sea vent megabenthic communities using multi-resolution 3D image reconstructions. Deep Sea Res Part I Oceanogr Res Pap 116:200−219. [Google Scholar]
- 13.Watsuji T, Yamamoto A, Motoki K, Ueda K, Hada E, Takaki Y, Kawagucci S, Takai K. 2015. Molecular evidence of digestion and absorption of epibiotic bacterial community by deep-sea crab Shinkaia crosnieri. ISME J 9:821–831. doi: 10.1038/ismej.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watsuji T, Tsubaki R, Chen C, Nagai Y, Nakagawa S, Yamamoto M, Nishiura D, Toyofuku T, Takai K. 2017. Cultivation mutualism between a deep-sea vent galatheid crab and its chemosynthetic epibionts. Deep Sea Res Part I Oceanogr Res Pap 127:13–20. doi: 10.1016/j.dsr.2017.04.012. [DOI] [Google Scholar]
- 15.Watsuji T, Nishizawa M, Morono Y, Hirayama H, Kawagucci S, Takahata N, Sano Y, Takai K. 2012. Cell-specific thioautotrophic productivity of epsilon-proteobacterial epibionts associated with Shinkaia crosnieri. PLoS One 7:e46282. doi: 10.1371/journal.pone.0046282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watsuji T, Yamamoto A, Takaki Y, Ueda K, Kawagucci S, Takai K. 2014. Diversity and methane oxidation of active epibiotic methanotrophs on live Shinkaia crosnieri. ISME J 8:1020–1031. doi: 10.1038/ismej.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 18.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, Sievert SM, Simon J, Campbell BJ, Hanson TE, Woyke T, Klotz MG, Hugenholtz P. 2017. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 8:682. doi: 10.3389/fmicb.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, Sievert SM, Simon J, Campbell BJ, Hanson TE, Woyke T, Klotz MG, Hugenholtz P. 2018. Erratum: Addendum: Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 9:772. doi: 10.3389/fmicb.2018.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watsuji T, Nakagawa S, Tsuchida S, Toki T, Hirota A, Tsunogai U, Takai K. 2010. Diversity and function of epibiotic microbial communities on the galatheid crab, Shinkaia crosnieri. Microbes Environ 25:288–294. doi: 10.1264/jsme2.me10135. [DOI] [PubMed] [Google Scholar]
- 21.Tsuchida S, Suzuki Y, Fujiwara Y, Kawato M, Uematsu K, Yamanaka T, Mizota C, Yamamoto H. 2011. Epibiotic association between filamentous bacteria and the vent-associated galatheid crab, Shinkaia crosnieri (Decapoda: Anomura). J Mar Biol Assoc UK 91:23–32. doi: 10.1017/S0025315410001827. [DOI] [Google Scholar]
- 22.Konishi M, Watsuji T, Nakagawa S, Hatada Y, Takai K, Toyofuku T. 2013. Effects of hydrogen sulfide on bacterial communities on the surface of galatheid crab, Shinkaia crosnieri, and in a bacterial mat cultured in rearing tanks. Microbes Environ 28:25–32. doi: 10.1264/jsme2.me12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watsuji T, Motoki K, Hada E, Nagai Y, Takaki Y, Yamamoto A, Ueda K, Toyofuku T, Yamamoto H, Takai K. 2018. Compositional and functional shifts in the epibiotic bacterial community of Shinkaia crosnieri Baba & Williams (a squat lobster from hydrothermal vents) during methane-fed rearing. Microbes Environ 33:348–356. doi: 10.1264/jsme2.ME18072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McQuillan JS, Robidart JC. 2017. Molecular-biological sensing in aquatic environments: recent developments and emerging capabilities. Curr Opin Biotechnol 45:43–50. doi: 10.1016/j.copbio.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 25.Takayama K, Kjelleberg S. 2000. The role of RNA stability during bacterial stress responses and starvation. Environ Microbiol 2:355–365. doi: 10.1046/j.1462-2920.2000.00119.x. [DOI] [PubMed] [Google Scholar]
- 26.La Cono V, Smedile F, La Spada G, Arcadi E, Genovese M, Ruggeri G, Genovese L, Giuliano L, Yakimov MM. 2015. Shifts in the meso- and bathypelagic archaea communities composition during recovery and short-term handling of decompressed deep-sea samples. Environ Microbiol Rep 7:450–459. doi: 10.1111/1758-2229.12272. [DOI] [PubMed] [Google Scholar]
- 27.Steiner PA, De Corte D, Geijo J, Mena C, Yokokawa T, Rattei T, Herndl GJ, Sintes E. 2019. Highly variable mRNA half-life time within marine bacterial taxa and functional genes. Environ Microbiol 21:3873–3884. doi: 10.1111/1462-2920.14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feike J, Jürgens K, Hollibaugh JT, Krüger S, Jost G, Labrenz M. 2012. Measuring unbiased metatranscriptomics in suboxic waters of the central Baltic Sea using a new in situ fixation system. ISME J 6:461–470. doi: 10.1038/ismej.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders JG, Beinart RA, Stewart FJ, Delong EF, Girguis PR. 2013. Metatranscriptomics reveal differences in in situ energy and nitrogen metabolism among hydrothermal vent snail symbionts. ISME J 7:1556–1567. doi: 10.1038/ismej.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgcomb VP, Taylor C, Pachiadaki MG, Honjo S, Engstrom I, Yakimov M. 2016. Comparison of Niskin vs. in situ approaches for analysis of gene expression in deep Mediterranean Sea water samples. Deep Sea Res Part II Top Stud Oceanogr 129:213–222. doi: 10.1016/j.dsr2.2014.10.020. [DOI] [Google Scholar]
- 31.Yum LK, Baumgarten S, Röthig T, Roder C, Roik A, Michell C, Voolstra CR. 2017. Transcriptomes and expression profiling of deep-sea corals from the Red Sea provide insight into the biology of azooxanthellate corals. Sci Rep 7:6442. doi: 10.1038/s41598-017-05572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyazaki J, Ikuta T, Watsuji T, Abe M, Yamamoto M, Nakagawa S, Takaki Y, Nakamura K, Takai K. 2020. Dual energy metabolism of the Campylobacterota endosymbiont in the chemosynthetic snail Alviniconcha marisindica. ISME J 14:1273–1289. doi: 10.1038/s41396-020-0605-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin JM, Shinabarger DL. 1983. Characterization of Thiothrix nivea. Int J Syst Bacteriol 33:841–846. doi: 10.1099/00207713-33-4-841. [DOI] [Google Scholar]
- 34.Williams TM, Unz RF. 1985. Filamentous sulfur bacteria of activated sludge: characterization of Thiothrix, Beggiatoa, and Eikelboom type 021N strains. Appl Environ Microbiol 49:887–898. doi: 10.1128/AEM.49.4.887-898.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maki JS. 2013. Bacterial intracellular sulfur globules: structure and function. J Mol Microbiol Biotechnol 23:270–280. doi: 10.1159/000351335. [DOI] [PubMed] [Google Scholar]
- 36.Seston SL, Beinart RA, Sarode N, Shockey AC, Ranjan P, Ganesh S, Girguis PR, Stewart FJ. 2016. Metatranscriptional response of chemoautotrophic Ifremeria nautilei endosymbionts to differing sulfur regimes. Front Microbiol 7:1074. doi: 10.3389/fmicb.2016.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frigaard NU, Dahl C. 2008. Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol 54:103−200. doi: 10.1016/S0065-2911(08)00002-7. [DOI] [PubMed] [Google Scholar]
- 38.Pott AS, Dahl C. 1998. Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144:1881–1894. doi: 10.1099/00221287-144-7-1881. [DOI] [PubMed] [Google Scholar]
- 39.Stockdreher Y, Sturm M, Josten M, Sahl HG, Dobler N, Zigann R, Dahl C. 2014. New proteins involved in sulfur trafficking in the cytoplasm of Allochromatium vinosum. J Biol Chem 289:12390–12403. doi: 10.1074/jbc.M113.536425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahl C. 2015. Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life 67:268–274. doi: 10.1002/iub.1371. [DOI] [PubMed] [Google Scholar]
- 41.Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, Hourdez S, Girguis PR, Wankel SD, Barbe V, Pelletier E, Fink D, Borowski C, Bach W, Dubilier N. 2011. Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476:176–180. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 42.Petersen JM, Kemper A, Gruber-Vodicka H, Cardini U, Van Der Geest M, Kleiner M, Bulgheresi S, Mußmann M, Herbold C, Seah BKB, Antony CP, Liu D, Belitz A, Weber M. 2017. Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat Microbiol 2:16195. doi: 10.1038/nmicrobiol.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakagawa S, Takai K. 2008. Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol Ecol 65:1–14. doi: 10.1111/j.1574-6941.2008.00502.x. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto M, Takai K. 2011. Sulfur metabolisms in epsilon-and gamma-proteobacteria in deep-sea hydrothermal fields. Front Microbiol 2:192. doi: 10.3389/fmicb.2011.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, Di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hensen D, Sperling D, Trüper HG, Brune DC, Dahl C. 2006. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol Microbiol 62:794–810. doi: 10.1111/j.1365-2958.2006.05408.x. [DOI] [PubMed] [Google Scholar]
- 47.Anantharaman K, Breier JA, Dick GJ. 2016. Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J 10:225–239. doi: 10.1038/ismej.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singleton CM, McCalley CK, Woodcroft BJ, Boyd JA, Evans PN, Hodgkins SB, Chanton JP, Frolking S, Crill PM, Saleska SR, Rich VI, Tyson GW. 2018. Methanotrophy across a natural permafrost thaw environment. ISME J 12:2544–2558. doi: 10.1038/s41396-018-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamilton R, Kits KD, Ramonovskaya VA, Rozova ON, Yurimoto H, Iguchi H, Khmelenina VN, Sakai Y, Dunfield PF, Klotz MG, Knief C, Op den Camp HJM, Jetten MSM, Bringel F, Vuilleumier S, Svenning MM, Shapiro N, Woyke T, Trotsenko YA, Stein LY, Kalyuzhnaya MG. 2015. Draft genomes of gammaproteobacterial methanotrophs isolated from terrestrial ecosystems. Genome Announc 3:e00515-15. doi: 10.1128/genomeA.00515-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flynn JD, Hirayama H, Sakai Y, Dunfield PF, Klotz MG, Knief C, Op den Camp HJM, Jetten MSM, Khmelenina VN, Trotsenko YA, Murrell JC, Semrau JD, Svenning MM, Stein LY, Kyrpides N, Shapiro N, Woyke T, Bringel F, Vuilleumier S, DiSpirito AA, Kalyuzhnaya MG. 2016. Draft genome sequences of gammaproteobacterial methanotrophs isolated from marine ecosystems. Genome Announc 4:e01629-15. doi: 10.1128/genomeA.01629-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol Rev 60:439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hügler M, Sievert SM. 2011. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu Rev Mar Sci 3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 53.Hügler M, Wirsen CO, Fuchs G, Taylor CD, Sievert SM. 2005. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ε subdivision of proteobacteria. J Bacteriol 187:3020–3027. doi: 10.1128/JB.187.9.3020-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takai K, Campbell BJ, Cary SC, Suzuki M, Oida H, Nunoura T, Hirayama H, Nakagawa S, Suzuki Y, Inagaki F, Horikoshi K. 2005. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl Environ Microbiol 71:7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakagawa S, Takaki Y, Shimamura S, Reysenbach A-L, Takai K, Horikoshi K. 2007. Deep-sea vent ε-proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci U S A 104:12146–12150. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takai K, Inagaki F, Nakagawa S, Hirayama H, Nunoura T, Sako Y, Nealson KH, Horikoshi K. 2003. Isolation and phylogenetic diversity of members of previously uncultivated ε-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol Lett 218:167–174. doi: 10.1111/j.1574-6968.2003.tb11514.x. [DOI] [PubMed] [Google Scholar]
- 57.Allewell NM, Sama A. 1974. The effect of ammonium sulfate on the activity of ribonuclease A. Biochim Biophys Acta 341:484–488. doi: 10.1016/0005-2744(74)90240-X. [DOI] [PubMed] [Google Scholar]
- 58.Allam A, Kalnis P, Solovyev V. 2015. Karect: accurate correction of substitution, insertion and deletion errors for next-generation sequencing data. Bioinformatics 31:3421–3428. doi: 10.1093/bioinformatics/btv415. [DOI] [PubMed] [Google Scholar]
- 59.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 63.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, Mcdonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location of the Iheya North hydrothermal field, Okinawa Trough, in Japan. The figure is based on a map made with Natural Earth (naturalearthdata.com). Download FIG S1, EPS file, 1.1 MB (1.1MB, eps) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of rRNA reads assigned to genera within Campylobacterales (relative abundance [read numbers]). Download Table S1, DOCX file, 0.01 MB (14KB, docx) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detected SILVA identifiers and OTUs. Download Table S2, DOCX file, 0.01 MB (13.2KB, docx) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
mRNA abundances of the genes shown in Fig. 7. Download Data Set S1, XLSX file, 0.05 MB (52.8KB, xlsx) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mounted equipment on the ROV HyperDolphine. The photograph shown in the left was taken by Masayuki Miyazaki. Download FIG S2, EPS file, 1.2 MB (1.2MB, eps) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ecological indices of the S. crosnieri episymbiotic microbial communities of the in situ and onboard RNA-stabilized samples. (a) Rarefaction curves. (b to d) Box-whisker plots of OTU richness (b), Faith’s phylogenetic diversity (c), and Shannon diversity (d). Points represent individual samples. The lines in the boxes represent medians. The boxes and whiskers represent ranges and interquartile ranges, respectively. Mann-Whitney U tests were performed (n.s., not significant; *, P < 0.05). Download FIG S3, EPS file, 0.1 MB (119.6KB, eps) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PCoA based on weighted UniFrac among the in situ and onboard RNA-stabilized samples. Variance explained by each PC is indicated on the axis. Plots represent results from individual samples of the in situ (dark blue) and onboard (light blue) RNA stabilization methods. Download FIG S4, EPS file, 0.04 MB (4.4KB, eps) .
Copyright © 2020 Motoki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The sequence data and assembled contigs were deposited in the DDBJ/ENA/GenBank database under BioProject identifier (ID) PRJDB9200. The bioinformatic scripts are available in github (https://github.com/kaorimo/MetatraAnalysis_mSystems2020).