Abstract
CREB-binding protein (CBP) serves as a transcriptional coactivator in multiple signal transduction pathways. The Drosophila homologue of CBP, dCBP, interacts with the transcription factors Cubitus interruptus (CI), MAD, and Dorsal (DL) and functions as a coactivator in several signaling pathways during Drosophila development, including the hedgehog (hh), decapentaplegic (dpp), and Toll pathways. Although dCBP is required for the expression of the hh target genes, wingless (wg) and patched (ptc) in vivo, and potentiates ci-mediated transcriptional activation in vitro, it is not known that ci absolutely requires dCBP for its activity. We used a yeast genetic screen to identify several ci point mutations that disrupt CI-dCBP interactions. These mutant proteins are unable to transactivate a reporter gene regulated by ci binding sites and have a lower dCBP-stimulated activity than wild-type CI. When expressed exogenously in embryos, the CI point mutants cannot activate endogenous wg expression. Furthermore, a CI mutant protein that lacks the entire dCBP interaction domain functions as a negative competitor for wild-type CI activity, and the expression of dCBP antisense RNAs can suppress CI transactivation in Kc cells. Taken together, our data suggest that dCBP function is necessary for ci-mediated transactivation of wg during Drosophila embryogenesis.
CREB-binding protein (CBP) and p300 were identified through their interactions with phosphorylated CREB and the adenoviral protein E1A, respectively (10, 12, 20). Since the initial cloning of CBP/p300 and the characterization of their coactivator functions in CREB-mediated gene activation and E1A-mediated transformation, CBP/p300 have been shown to be the coactivators for many transcription factors and coactivators, including nuclear hormone receptors, p65, Stat, c-Myb, c-jun, Sap-1b, c-fos, Myo-D, p53, SRC-1/NCoA-1, TIF2/GRIP1, and p/CIP (4; for reviews, see references 31 and 36). These interactions suggest a role for CBP/p300 in a variety of cellular processes, including differentiation, cellular proliferation, immune response, homeostasis, tumorigenesis, and organogenesis. CBP/p300 are also the target for several other viral proteins such as simian virus 40 large T antigen and the human T-cell leukemia virus Tax-1 (13, 18). The current model for transcriptional activation proposes that CBP/p300 serve as bridging molecules between DNA-binding transcription factors and the basal transcriptional machinery. The finding that CBP/p300 interact with components of the basal transcriptional machinery, such as TFIIB, RNA polymerase, and RNA helicase A, supports this model as well (15, 19, 23).
Recently, CBP has been shown to possess intrinsic histone acetyltransferase (HAT) activity, suggesting that its ability to activate transcription involves the modification of chromatin (24). Consistent with this idea is the observation that CBP interacts synergistically with other HAT proteins that are thought to be actively involved in chromatin remodeling, such as p/CAF, p/CIP, and SRC-1/NCoA-1 (32, 33, 37, 40). All of these studies suggest that CBP integrates signals from different second messenger pathways into specific patterns of gene expression.
Haploinsufficiency of CBP causes Rubinstein-Taybi syndrome in humans, a genetic abnormality characterized by mental retardation, abnormal skeletal development, and susceptibility to cancers (28). Mice lacking either the CBP or p300 gene die early in embryogenesis. Disrupting one copy of CBP in mice results in abnormal skeletal development and a decrease in BMP-7 expression, while heterozygous p300 mutant mice show significant embryonic lethality (35, 41). The CBP/p300 transheterozygotes are lethal (41). Besides embryonic lethality, these knockout animals show multiple defects in neurogenesis, bone and heart development, and growth progress, suggesting that CBP/p300 are involved in many developmental processes governing cellular proliferation and differentiation (35, 41). Thus, CBP knockouts in mammalian systems are exceedingly complex and have not allowed an analysis of the requirement of CBP/p300 in the regulation of specific target genes.
We have isolated a Drosophila homologue of CBP (dCBP). Flies that are heterozygous for dCBP mutations develop normally; however, embryos that are homozygous for dCBP mutations die early in development and have severe defects, such as a twisted germ band, loss of head structures, and cuticular defects (1). dCBP has been shown to act as a coactivator for dorsal (dl) (2), mad (39), and cubitus interruptus (ci) (1), implicating dCBP as a positive activator of at least three signaling pathways, the Toll, decapentaplegic (dpp), and the hedgehog (hh) pathways. The dl target gene, twist (twi), is not expressed in dCBP-mutant embryos and when tested in tissue culture, dCBP can potentiate DL-mediated transcriptional activation (2). Certain dpp-regulated enhancers are not activated in dCBP-mutant embryos, and in vitro dCBP can interact with MAD, the transcription factor that mediates the activation of these enhancers (39). The loss of dCBP function results in the loss of hh target gene expression; wingless (wg) and patched (ptc) are induced but not maintained in dCBP mutant embryos, and dCBP mutant clones in the wing do not express ptc. Furthermore, in cell culture experiments, dCBP potentiates the transcriptional activity of cubitus interruptus (CI) (1). These results suggest but do not prove that dCBP is required for the activity of the transcription factors that mediate the various signaling cascades.
ci encodes a transcription factor responsible for transducing the hh signal into the nucleus (3, 11, 25, 27). It is both a transcriptional repressor and activator. The repression domain is in the N terminus, and the activation domain maps to the C terminus, which includes the dCBP interacting domain. There are five highly conserved zinc fingers that are responsible for DNA-binding activity. A proteolytic cleavage site lies C-terminal to the zinc finger domain between amino acids (aa) 650 to 700 (5, 21). In the absence of a hh signal, a substantial amount of CI is proteolysed, generating a 75-kDa protein containing the zinc finger domain and the N-terminal repressor domain (5). Presumably, this form of CI migrates into nucleus, binds to the CI binding sites in the target gene promoters, and functions as a repressor. When cells receive an hh signal, this proteolysis is inhibited and, through an unknown mechanism, the activator form of CI transactivates the hh target genes, presumably by recruiting its coactivator dCBP (1, 5).
To determine whether dCBP is absolutely required for CI activity and to define the CI-dCBP interaction more precisely, we performed a genetic screen to isolate point mutations in CI that disrupt its interaction with dCBP. We show that these mutant CI proteins cannot activate transcription in cell culture and cannot support wg expression in embryos. Furthermore, we demonstrate that the expression of the dCBP antisense RNAs can suppress CI transcriptional activity in cell culture. Taken together, these results demonstrate that dCBP is required for ci-mediated transcriptional activation of CI enhancers in cell culture and wg expression in embryos.
MATERIALS AND METHODS
Plasmid vectors.
For cell culture, we used pPac5c (kindly provided by M. Krasnow, Stanford University School of Medicine, and K. Thummel, University of Utah), which has the actin 5C proximal promoter to drive protein expression, as our expression vector in Kc cells. The cloning of pPac-luciferase, pPac-PKA, pPac-PKI, pPac-CI (wild type and m1-4), and pPac-CI protein kinase A (PKA) mutants was as described previously (8; note that m1-4 is the same as the “null” in this reference). The ADHCAT and ADHCAT/GLI6BS (ADHCAT with six GLI binding sites fused upstream of ADH promoter) reporter vectors have been described by Akimaru et al. (1). Briefly, the reporter gene ADHCAT/GLI6BS was constructed by inserting six copies of the GLI-binding site (5′-GCGTGGACCACCCAAGACGAAATT-3′) (16) into the BglII site of pAdhCAT (1, 17). Thus, the chloramphenicol acetyltransferase (CAT) expression is driven by 6-GLI enhancers and a minimal Adh promoter. The internal deletion of CI [CI(Δ)] was generated by the following procedure. The ci DNA fragments containing aa 918 to 1020 and aa 1160 to 1349 were PCR amplified, ligated with an engineered internal BglII site, and fused into the ci cDNA at the aa 918 HindIII site and the aa 1349 SmaI site. PCR sequences were confirmed by sequencing using Sequenase (version 2.0, U.S. Biochemical [USB]). The NotI fragment containing the dCBP cDNA was blunt ended and cloned into the pPac5c BamHI site to generate pPac-dCBP. The pPac-dCBP antisense vector was constructed by inserting the 4.7-kb EcoRV fragment of dCBP that encodes the 5′ coding region of dCBP (from aa 110 to 1655) into the pPac BamHI site in the reversed orientation. Hemagglutinin (HA)- or FLAG-tagged CI constructs were made by inserting oligonucleotides encoding HA or FLAG sequences into the MluI site at the fifth amino acid in CI. For yeast transformation, the pACT-CI(wt, 2.1) and pACT-CI(m1-4, 2.1) vectors were made by ligating the HincII-EcoRI fragment containing CI from aa 685 to 1377 with the pACT-2 (Clontech) digested with SmaI and EcoRI. These constructs are in frame with the GAL4 activation domain in pACT-2. The pACT-CI(Δwt, 2.1) and pACT-CI(Δm1-4, 2.1) vectors were made adopting the same strategy by using a CI fragment (HincII-EcoRI) that has aa 1020 to 1160 deleted. The CI-C vector (aa 984 to 1377) was originally detected in a two-hybrid yeast screen using dCBP (aa 835 to 1043) as the “bait” (1). This same dCBP bait was used in these studies. For Drosophila transformation, the pUAST-CI wild-type and mutant constructs were made by inserting the BamHI/NotI fragment containing full-length ci into the BglII and NotI sites of the pUAST vector (6). The CI(m1-3) mutant refers to a CI protein that is mutant for the three consensus RRxS PKA phosphorylation sites found in the CI activation domain. The mutants 103 and 459 are the two mutant CI proteins that cannot interact with dCBP.
Tissue culture and transfection.
The Kc cell line is an immortalized Drosophila embryonic cell line that is possibly a derivative of Drosophila hematopoietic cells (9). Kc cells were maintained, transfected, and assayed for CAT and luciferase activities as described elsewhere (8). For immunoprecipitations, cells were grown and transfected as described earlier (8).
Yeast two-hybrid and split-hybrid assays.
The yeast two-hybrid assay was performed as described earlier (38) with 25 mM 3-aminotriazole (3-AT) to select for strong interactions. The yeast split-hybrid assay was performed as described by Shih et al. (30). The construction of the yeast split-hybrid strain has been described elsewhere (30), and the genotype is MATa/MATα, his3Δ200/his3Δ200 trp1-901/trp1-901 leu2-3,112/leu2-3,112 ade2/ade2 URA3::(LexA operator)8-TetR LYS2::(Tet operator)2-HIS3. The “bait” was the same as the one used in the yeast two-hybrid screen. The library was made by fusing a CI fragment (aa 984 to 1377) to the GAL4 activation domain (pACT-2; Clontech). The DNA sequence ACG, which encodes aa 1161 in CI, was mutated to ACC by PCR to generate a SacII site without changing the amino acid sequence. Random mutagenesis was performed as described elsewhere (30) with primers flanking the NcoI site in the pACT-2 and the engineered SacII site in CI. The mutagenized PCR fragments were digested with NcoI and SacII and ligated into pACT-CI (aa 984 to 1377). The resulting mixture of plasmids was transformed into DH5α. The transformants were pooled together, and DNA was purified by CsCl gradient centrifugation. The split-hybrid screen was carried out in the presence of 15 mM 3-AT and absence of tetracycline to select for noninteractors. The noninteractors from the plates were subjected to a secondary liquid screen to identify true noninteractors. The clones from the secondary screen were cured on minimal glucose plates, and the library plasmids were recovered. These plasmids were digested with BglII to select for full-length CI fragment, and the full-length plasmids were transformed into the yeast two-hybrid system with LexA-dCBD as bait to confirm that they do not interact with dCBD in the presence of 20 mM 3-AT. The true noninteractors were sequenced using the USB system according to their protocol.
Western analysis and immunoprecipitation.
Kc cells were washed twice with phosphate-buffered saline, scraped off the plate, and resuspended in lysis buffer containing 100 mM potassium phosphate (pH 7.8) supplemented with 1 mM dithiothreitol. Cell extracts were made by use of three freeze-thaw cycles, followed by centrifugation at 5,000 rpm. Equal amounts of supernatant were mixed with loading buffer, heat denatured, and loaded on a 5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Fractionated cell extracts were then electronically transferred to an Immobilon-P membrane (Millipore) by using a semidry transfer apparatus and reacted to antibody according to the protocol provided by ECL Kits (Amersham). The anti-CI C-terminal antibody was provided by R. Holmgren (Northwestern University) and used at a concentration of 1:10. Immunoprecipitations were performed as described elsewhere (8).
Germ line transformation and whole-mount embryo in situ hybridization and immunostaining.
The HACI(wt), HACI(m1-3), HACI(m1-3*103), and HACI(m1-3*459) pUAST transgenic fly lines were generated as described earlier (29, 34). At least four independent transformants were generated for each HACI construct and tested for viability and expression with the prd-GAL4 line RG1 (42), kindly provided by D. Kalderon, Columbia University. The dual immunohistochemistry and in situ hybridizations of whole-mount embryos were carried out as described earlier (7).
RESULTS
A CI protein that lacks the dCBP interaction domain is a competitive inhibitor of wild-type CI activity in cell culture and the expression of antisense dCBP RNAs suppresses wild-type CI activity.
We have previously shown that CI interacts with dCBP both in vitro and in vivo (1) but have not demonstrated that this interaction is required for the expression of any particular target genes. CI binds dCBP in yeast two-hybrid and glutathione S-transferase (GST) pull-down assays. The domain in CI that interacts with dCBP has been mapped at between aa 1020 and 1160, and the domain in dCBP that interacts with CI is the CREB interacting domain (dCBD) that lies between aa 835 and 1043. A schematic representation of CI and dCBP is presented in Fig. 1. dCBP potentiates CI-mediated transcriptional activation in S2 cells and has been implicated as the coactivator of ci in vivo. To determine whether ci absolutely requires dCBP for its activation function, we used several approaches. The first approach was to make a CI mutant that has the dCBP interaction domain (between aa 1020 and 1160) deleted [CI(Δ)] and then to assess its activity. Because the dCBP interacting domain is located near the C-terminal end of CI and thus C terminal to the DNA-binding domain of CI, it is unlikely that a deletion of the dCBP interacting domain would affect the more medial CI-DNA binding activity (Fig. 1).
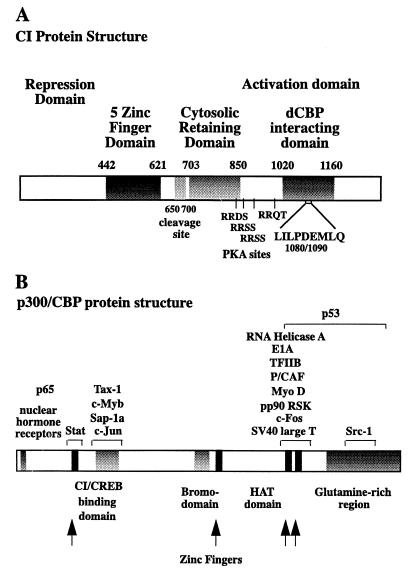
FIG. 1.
Schematic representations of CI and dCBP. (A) CI. The repressor domain extends from the N terminus to aa 442. The DNA binding domain is defined by a five-zinc-finger region from aa 442 to 621. The proteolytic cleavage site is in the residues from aa 650 to 700, and the domain required to retain CI in the cytoplasm is defined by aa 703 to 850. The four PKA phosphorylation sites are located at S838, S856, S892, and T1006. The dCBP interaction domain lies between aa 1020 and 1160, and the critical residues for the interaction are between aa 1080 and 1090. The CI activation domain extends from aa 970 to the C terminus. (B) dCBP. The CI interaction domain is identical to the CREB binding domain of CBP and extends from aa 835 to 1043. The Bromodomain is found between aa 1700 and 1943, and the activation domain is in the region of aa 2278 to the C terminus at aa 3276.
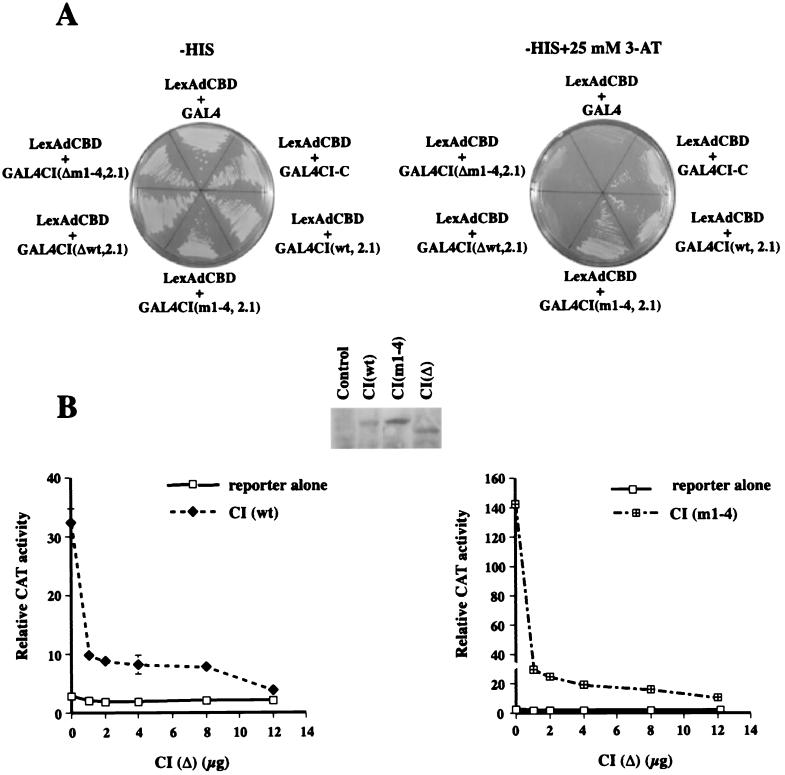
To ensure that the deletion mutant does not interact with dCBP, we performed binding assays in the yeast two-hybrid system. We fused the GAL4 transactivation domain to the CI fragment that encodes aa 685 to 1377 and includes the dCBP binding domain [CI(wt, 2.1)]. We also generated Gal4 fusion proteins with the CI aa 685 to 1377 fragment in which the serine-threonine residues in the consensus RRxS/T PKA sites had been mutated to alanine (Ser-838, Ser-856, Ser-892, and Thr-1006) [CI(m1-4, 2.1)]. In cell culture, CI proteins that carry these mutations are not proteolysed, and their activities are not regulated by PKA (7, 8). We then made deletions of the dCBP binding domain (aa 1020 to 1160) in the CI(wt 2.1) and CI(m1-4, 2.1) constructs, CI(Δwt, 2.1) and CI(Δm1-4, 2.1). All of these vectors encoding GAL4 fusion proteins were transformed into the L40 yeast two-hybrid strain that has the LexA-dCBP (aa 835 to 1043) vector as bait and LexA-driven HIS3 and lacZ as reporter genes. We also transformed L40 with the GAL4CI-C (aa 918 to 1377) construct, which was originally identified in a yeast two-hybrid screen to detect proteins that interact with dCBP, as a positive control. As shown in Fig. 2A, both CI(wt, 2.1) and CI(m1-4, 2.1) interact with dCBP (aa 835 to 1043) in the presence of 25 mM 3-AT, while the deletion mutants, CI(Δwt, 2.1) and CI(Δm1-4, 2.1), do not.
FIG. 2.
CI deleted for the dCBP interaction domain does not bind dCBP and is a negative competitor for wild-type CI activity. (A) The domain of dCBP from aa 825 to 1043 that binds to CI fused to the LexA DNA binding domain (the “bait”) and the “prey” [pACT-2, pACT-CI(wt, 2.1), pACT-CI(m1-4, 2.1), pACT-CI(Δwt,2.1) pACT-CI(Δm1-4,2.1), or pACT-CI-C (CI aa 984 to 1377)] were cotransformed into the yeast strain L40. The transformed yeast were plated on selective media in the presence or absence of 25 mM 3-AT (see Materials and Methods). (B) A total of 100 ng of pPac-luciferase, 5 μg of ADHCAT/GLI6BS, 1 μg of full-length pPac-CI(wt), or 1 μg of pPac-CI(m1-4) and increasing amounts of pPac-CI(Δ) were transfected into Kc cells. CAT activities were normalized to the corresponding luciferase activities. The data represent the means ± the standard error of the mean. (Inset) Western blot showing that the CI constructs used in panel B are expressed in Kc cells.
We then tested whether a full-length CI(Δ) could act as a competitive inhibitor of CI(wt) and CI(m1-4) in Kc cells. As shown in Fig. 2B, CI(Δ) has very little effect on the basal promoter activity within the concentration range tested. However, when increasing amounts of CI(Δ) are cotransfected with 1 μg of CI(wt) or CI(m1-4), CI(Δ) effectively suppresses the CI activity in a dose-dependent manner. CI(Δ) is effectively expressed in our Kc cell system as determined by Western analysis with an antibody against the C terminus of CI protein (Fig. 2B, inset). One explanation for this result is that the CI(Δ) binds to the CI-DNA binding site and competes with wild-type CI for DNA binding. Because the CI(Δ) does not have the dCBP interaction domain, dCBP is not recruited to the promoter region and we do not observe transcriptional activation. However, this experiment does not rule out the possibility that the deletion of 140 amino acids from the CI activation domain disrupts the structure of CI to such an extent that the protein can no longer interact with other required proteins or the basal transcriptional machinery. To assess whether dCBP is required for CI activity, we performed a dCBP antisense experiment.
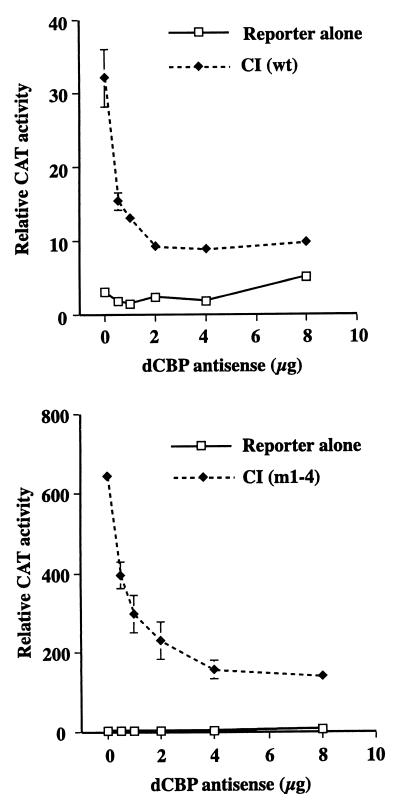
As shown in Fig. 3, transfecting up to 8 μg of an antisense dCBP vector that encodes antisense message for aa 110 to 1655 has a negligible effect on the basal promoter activity. However, in the presence of CI(wt), dCBP antisense effectively suppresses wild-type CI activity. Antisense dCBP RNAs were also able to suppress CI(m1-4) activity.
FIG. 3.
Expression of dCBP antisense RNA inhibits CI-mediated transcriptional activation. A total of 100 ng of pPac-luciferase, 5 μg of ADHCAT/GLI6BS, 1 μg of full-length pPac-CI(wt), or 1 μg of pPac-CI(m1-4) and increasing amounts of pPac-dCBP antisense vector were transfected into Kc cells. CAT activities were assayed 5 days after transfection and processed as described in Fig. 1B.
Identification of CI point mutations that disrupt the CI-dCBP interaction.
The deletion and antisense experiments support the hypothesis that dCBP is required for CI-mediated transcriptional activation. However, they cannot rule out the possibility that antisense dCBP has a secondary effect on unknown factors that are important for CI transcriptional activity. Ideally, a mutation in CI that affected only its ability to interact with dCBP would demonstrate the requirement of dCBP for CI function. To generate point mutations in CI that disrupt the CI-dCBP interaction, we performed a yeast genetic screen termed the split-hybrid assay (30). The split-hybrid system is a two-component reporter system that converts the disruption of a protein-protein interaction into a positive selection. The first reporter is the TetR gene that is driven by LexA binding sites. The second reporter is the HIS3 gene that is driven by the TetR operator sites. When a LexA binding domain is fused to a protein that interacts with another protein fused to the VP16 activation domain (or the GAL4 activation domain), TetR is expressed. The TetR then binds to the TetR operator sites upstream of HIS3 and suppresses the expression of HIS3. Thus, these yeast strains that contain interacting proteins cannot grow on plates lacking histidine. Mutations that disrupt the binding of the two interacting proteins, inhibit the activation of TetR. The lower doses of TetR allow the expression of HIS3, and yeast strains carrying these mutant proteins will grow in the absence of histidine. Tetracycline can be used to control LexA fusions that have intrinsic transcriptional activity and 3-AT, an inhibitor of the histidine pathway, can be used to select for weak protein-protein interactions.
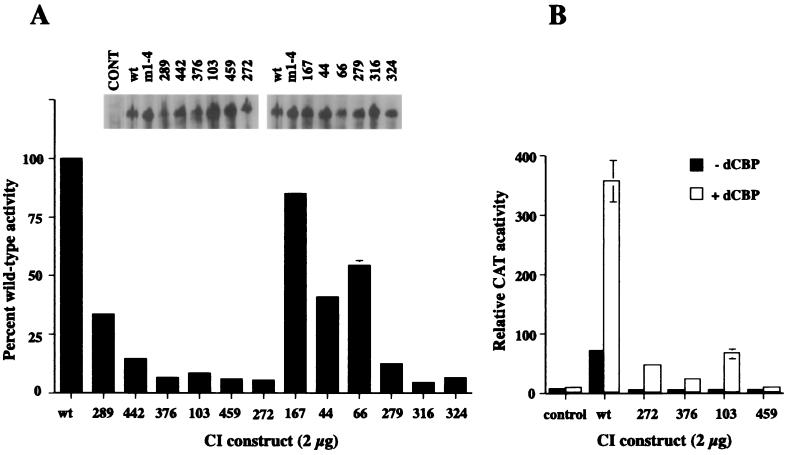
To generate mutations in CI that would disrupt the dCBP-CI interaction, we randomly mutagenized the dCBP interacting domain in CI (aa 1020 to 1160) and cloned these mutant fragments into pACT-CI that has the GAL4 activation domain fused to the C terminus of CI from aa 984 to 1377. This randomly mutagenized CI library was transformed into the split-hybrid yeast strain that carries a vector expressing a fusion protein between the LexA DNA binding domain and the CI-binding domain of dCBP (aa 835 to 1043). We screened 20,000 yeast transformants and isolated 500 colonies that grew in the presence of 3-AT on HIS-selective plates. A total of 365 of the 500 colonies grew well in liquid medium in the presence of different concentrations of 3-AT. Of these 365 yeast clones, 146 contain the full-length CI fragment, and 94 of them produce proteins that do not interact with dCBP when tested in a yeast two-hybrid system. The majority of these 94 clones are frameshift mutations (50), nonsense mutations (18), and multiple mutations (9). Only 10 are single or double mutations. Seven wild-type clones escaped the screen. The 12 clones containing the single, double, or triple mutations were cloned into full-length ci, and the vectors were transfected into Kc cells. We determined the transcriptional activities of these mutant proteins and the ability of dCBP to augment these activities. The results of these studies are shown in Fig. 4A and summarized in Table 1. A graphic representation of the effects of dCBP on the activities of four proteins with single mutations is illustrated in Fig. 4B.
FIG. 4.
Activities of the CI mutants in Kc cells. (A) A total of 100 ng of pPac-luciferase, 5 μg of ADHCAT/GLI6BS, and either 2 μg of pPac-FLAGCI(wt) or pPac-FLAGCI(split-hybrid mutant) were transfected into Kc cells. CAT activities were analyzed as for Fig. 1B. (Inset) Western blot showing that the CI mutant constructs are expressed in Kc cells. (B) CAT activities of 2 μg of the single mutation CI constructs in the presence or absence of 1 μg of pPac-dCBP. A summary of the results in panels A and B is presented in Table 1.
TABLE 1.
Summary of the split-hybrid mutant activities in Kc cellsa
| Clone no. | Mutation(s) | % CI(wt) activity | % CI(wt)-dCBP activity |
|---|---|---|---|
| 289 | I1040T | 33.4 ± 0.6 | ND |
| 442 | F1054L | 14.5 ± 1.0 | ND |
| 376 | L1082H | 6.1 ± 0.5 | 5.2 ± 0.2 |
| 103 | E1087G | 8.5 ± 0.3 | 10.0 ± 0.7 |
| 459 | M1088T | 5.7 ± 0.3 | 1.6 ± 0.1 |
| 272 | L1089P | 5.5 ± 0.2 | 16.1 ± 0.4 |
| 167 | D1145G | 84.6 ± 0.5 | 120 ± 8.1 |
| 44 | P1010A, H1123R | 40.7 ± 0.5 | 31.8 ± 0.3 |
| 66 | D1047N, K1151R | 54.2 ± 2.2 | 55.2 ± 3.1 |
| 279 | D1077G, K1151R | 12.4 ± 0.7 | 27.8 ± 4.7 |
| 316 | I1083T, L1089P, I1146M | 4.4 ± 0.3 | 29.4 ± 4.6 |
| 324 | E1087G, K1096R, D1098G | 6.5 ± 0.3 | 22.5 ± 1.0 |
A total of 2 μg of different CI mutant constructs compared to 2 μg of wild-type CI activity in the absence or presence of 1 μg of dCBP, with ADHCAT/GLI6BS as reporter and pac-luciferase as an internal control. ND, not determined.
When 2 μg of DNA was used to transfect Kc cells, 10 of the 12 mutants that did not interact with dCBP in yeast had significantly decreased activity in Kc cells; two preserved more than 50% wild-type CI activity. To determine whether the decrease in activity was due to insufficient protein expression, we performed a Western blot analysis of cells transfected with the different CI mutants. All of the transfected cells expressed full-length protein at levels comparable to or higher than cells expressing wild-type CI (Fig. 4A, inset). Four of the seven mutants with single amino acid changes had less than 10% of the wild-type activity. Two of the mutations, 272 and 103, involve residue changes to the known helix-disrupting residues proline and glycine, respectively. The two mutations 376 and 459 are relatively benign; mutation 376 changes a leucine to a ring histidine, and mutation 459 changes a methionine to threonine. We chose one disruption mutation, 103, and one benign mutation, 459, to further characterize the CI-dCBP interaction.
Construction and analysis of CI*103 and CI*459 double mutants that are independent of PKA regulation.
The purpose of the yeast screen and tissue culture experiments was to provide reagents to test the hypothesis that dCBP is required for CI function in vivo. We expected that the overexpression of CI carrying these hypomorphic mutations in flies would be of little consequence and would not affect endogenous wg expression. Certainly, these mutants would have less effect on wg expression than the effect observed when wild-type CI is overexpressed. We generated UAS-CI transgenic fly lines and crossed them with the prd-GAL4 line, in which the expression of the GAL4 activator is under the control of the paired (prd) promoter, to study the effect of overexpressing wild-type CI on endogenous wg expression (7). We do not observe any abnormalities or changes in endogenous wg expression when the wild-type CI is overexpressed in the prd domain (Fig. 6A and reference 7). This suggests that, like the endogenous wild-type CI, the exogenous CI protein is proteolyzed to the repressor form of the protein in the cells of the prd domain that do not receive a hh signal. To determine whether the hypomorphic CI*103 and CI*459 mutants could activate wg expression in the Drosophila embryo, we needed to assess the activity of CI*103 and CI*459 in forms that could not be proteolyzed and would thus be in an active form in the cells of the prd domain that do not receive a hh signal.
FIG. 6.
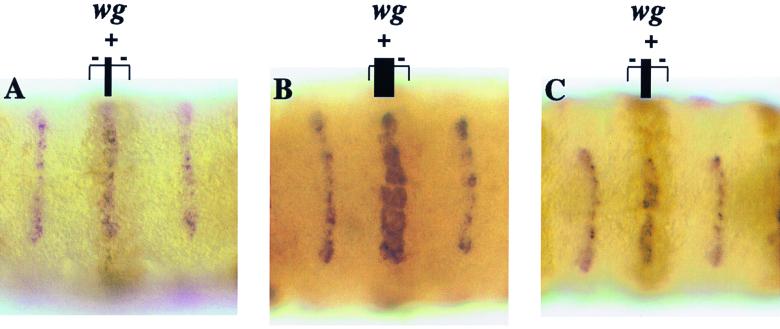
CI mutant for the dCBP binding cannot transactivate wg in the embryo. The brackets delineate the prd domain, and the vertical black bars represent the domains of wg expression. The cells posterior to the domain of wg expression are not competent to express wg (14). (A) UAS-HACI/+; prd-GAL4/+ embryos have a wild-type pattern of wg expression. Although overexpressed in cells that do not receive a HH signal, wild-type CI is regulated; it is proteolyzed in these cells to the repressor form. The exogenous wild-type CI is not proteolyzed in the cells abutting the anterior-posterior boundary that receive an HH signal, and the presumably full-length activated form ensures the activation of wg. (B) UAS-HACI(m1-3)/+; prd-GAL4/+ embryos. wg is misexpressed in the anterior cells of the prd domain that express the mutant CI and do not receive an HH signal. The mutant CI cannot be phosphorylated and thus is not proteolyzed in cells that do not receive a HH signal. The increased amount of activated CI is sufficient to ensure wg expression in these cells. (C) In UAS-HACI(m1-3*459)/+; prd-GAL4/+ embryos, the exogenous expression of the mutant CI should stimulate wg expression in the cells that do not receive an HH signal because it cannot be proteolyzed to the repressor form of CI. However, this is not the case. The overexpression of this mutant CI cannot activate wg to the degree seen in panel B because the ability of the mutant CI to interact with dCBP is reduced. This phenotype is identical to that of the UAS-HACI(m1-3*103)/+; prd-GAL4/+ embryos. In each panel, embryos are stained with an anti-HA antibody to detect exogenous CI and a wg antisense RNA probe. Anterior is left and ventral is forward.
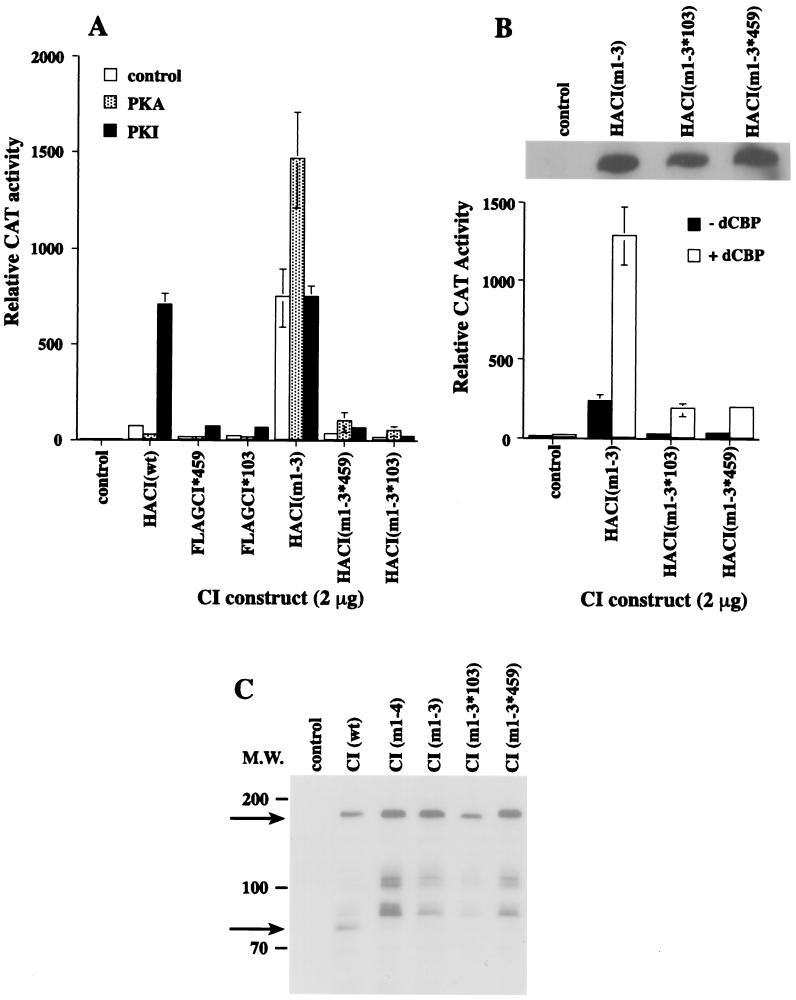
We generated double-mutant CI constructs that have the consensus RRxS PKA sites at Ser-838, Ser-856, and Ser-892 mutated to alanines in addition to the point mutations in CI*103 and CI*459 [CI(m1-3*103) and CI(m1-3*459)]. We have previously shown that when the four consensus PKA sites are mutated, CI is no longer proteolyzed, regulated by PKA, or regulated by hedgehog signaling to activate wg (7, 8). To use these two double-mutant forms of CI, it was necessary to ensure that PKA did not negatively regulate the activity of CI(m1-3), that dCBP could potentiate the activity of CI(m1-3), and that the double-mutant proteins were not proteolyzed. As shown in Fig. 5A, CI(m1-3) is 11-fold more active than CI(wt) in Kc cells, and this activity is not affected by the addition of the PKA inhibitor PKI. CI(wt) activity increases 10-fold in the presence of PKI. When additional PKA is introduced, the CI(wt) activity is suppressed threefold, while the activity of CI(m1-3) increases almost twofold. Thus, CI(m1-3), like CI(m1-4) (8), is no longer subject to the negative regulation of PKA.
FIG. 5.
CI that is mutant for the three consensus PKA sites and for which the dCBP binding domain is hypomorphic is independent of negative PKA regulation and minimally augmented by additional dCBP. (A) A total of 100 ng of pPac-luciferase, 5 μg of ADHCAT/GLI6BS, and 2 μg of pPac-HACI(wt), pPac-FLAGCI(split-hybrid mutant), pPac-HACI(m1-3), or pPac-HACI(m1-3 split-hybrid mutant), plus 4 μg of pPac, pPac-PKA, or pPac-PKI were transfected into Kc cells. CAT activities were analyzed as in Fig. 1B. (B) CAT activities of 2 μg of the PKA mutant CI construct or double-mutant CI constructs in the presence or absence of 1 μg of pPac-dCBP. (Inset) Western blot showing that the CI constructs used in the transfection assay were expressed in Kc cells. (C) A total of 10 μg of each pPac-HA CI constructs was transfected into Kc cells, immunoprecipitated, and probed with HA antibody. The arrow shows the position of the 75-kDa repressor form of CI. The larger fragments are presumed to be breakdown products because they are variable from gel to gel (for comparison, see references 7 and 8).
The maximal activities of CI(m1-3*103) and CI(m1-3*459) are reduced and represent approximately 20% of maximal CI(wt) activity and only 4% of maximal CI(m1-3) activity. These activities are similar to those of the single CI*103 and CI*459 mutants that also have approximately 20% the maximal wild-type activity. In the presence of PKI, the activities of the CI*103 and CI*459 mutants increase fivefold and represent only 10% of the PKI-stimulated wild-type CI activity. The activities of the double mutants do not increase significantly with the addition of PKI. When PKA is added to the double mutants, their activities are enhanced by three- to fourfold. The addition of dCBP augments the activities of CI(m1-3*103) and CI(m1-3*459), but these activities are sevenfold less than the dCBP-enhanced activity of CI(m1-3). The decreased activities observed for the CI double mutants are not due to low expression levels of the proteins. A Western blot probed with an anti-CI antibody (Fig. 5B, inset) demonstrates that the Kc cells express high levels of the mutant proteins. Although the CI double mutants are not proteolyzed to the repressor form in the cells (Fig. 5C), their activities are less potent compared to the transactivation achieved with the same dose of CI(wt).
The CI double mutants cannot activate endogenous wg expression in the Drosophila embryo.
We made upstream activation sequence (UAS)-HACI(m1-3), UAS-HACI(m1-3*103), and UAS-HACI(m1-3*459) transgenic fly lines to investigate the consequences of overexpressing the CI mutants that are defective in dCBP binding in vivo. UAS-HACI(m1-3), UAS-HACI(m1-3*103), and UAS-HACI(m1-3*459) lines were crossed to the prd-GAL4 line to generate flies that ectopically express HA-tagged CI in the paired (prd) expression domain. Figure 6 illustrates embryos that are double stained for the HA epitope and wg message. Our previous findings demonstrated that inhibition of PKA phosphorylation at Ser-838, Ser-856, and Ser-892 abolishes CI proteolysis and increases CI transcriptional activity and that, when the four consensus PKA sites are mutant, CI is able to activate wg in the absence of a HH signal (7, 8). In agreement with these studies, the ectopic expression of HACI(m1-3) results in the ectopic expression of wg in the anteriormost cells of the prd expression domain (Fig. 6B). In 97.8% (266 of 272) of the embryos counted, wg expression is expanded in the anterior cells of the prd domains. In contrast, the overexpression of HACI(m1-3*103) or HACI(m1-3*459) does not result in the ectopic expression of wg. In 94.5% (357 of 378) of the HACI(m1-3*103) and 93% (185 of 198) of the HACI(m1-3*459) embryos, the levels of wg message in the prd domains are nearly wild type compared to the neighboring segments that express endogenous CI (Fig. 6C). While animals expressing CI(m1-3) in the prd domains are virtually lethal (10% survival), those expressing HACI(m1-3*103) or HACI(m1-3*459) in the prd domains are viable.
DISCUSSION
The results of cell culture experiments have produced a model of CBP action in which the coactivator stimulates basal transcription by forming a molecular bridge between signal-responsive transcriptional activators and the basal transcriptional machinery (15, 19, 23, 31, 36). The discovery that CBP has an intrinsic acetyltransferase activity suggests that by acetylating histones, CBP might open the nucleosomes and allow activator access to the promoters (24). The fact that CBP can bind additional acetyltransferases further suggests that CBP is part of chromatin remodeling complexes (32, 33, 37, 40). Although these studies have defined some of the signaling pathways and types of transcriptional activities that involve CBP, it is not yet certain how CBP and its associated proteins function in vivo. The identification of the CBP homologue in Drosophila and the generation of mutations in the dCBP gene have identified some of the signaling pathways that use dCBP in flies. dCBP mutant animals do not express twist, the target gene of the Toll pathway (2), wg or ptc, targets of the hh pathway (1), or certain dpp-responsive enhancers (39). Although dCBP has been shown to interact with the transcription factors that activate these targets and in some cases to augment their activities, it has not been demonstrated whether dCBP is absolutely required for the activation of these factors.
We provide here evidence that the CI-dCBP interaction is required for the CI activation of a CI reporter in cell culture and for wg expression in embryos. The expression of antisense dCBP RNAs can suppress CI activity in cell culture. This suppression is detected after 4 days, demonstrating that dCBP has a long half-life in these cells. This result is consistent with the fact that maternal dCBP RNAs can rescue the lack of zygotic dCBP through embryonic development (1). To rule out the possibility that the loss of dCBP was having a secondary effect on proteins required for CI activity, we generated mutations in CI that could not interact with dCBP and assessed their abilities to transactivate CI targets in cell culture and the wg target gene in embryos.
The first mutant we generated was a CI deleted for the dCBP interaction domain. This protein cannot transactivate a CI reporter gene and acts as an inhibitor of wild-type CI in Kc cells. The dCBP interaction domain was defined by GST pull-down experiments and the fact that it interacts with a dCBP “bait” in yeast two-hybrid screens (1). While other regions of CI may also interact with dCBP, these interactions may not be sufficient to rescue the loss of the characterized interaction domain. An alternate explanation of these results may be that, in addition to deleting the dCBP interaction domain, this deletion ablates regions of CI that are needed to interact with the basal transcriptional machinery. In this case, the interaction with dCBP may only be required to enhance the activity. To differentiate these possibilities, we used the yeast split-hybrid system to generate point mutations in CI that would destabilize the CI-dCBP interaction. We analyzed mutants with single amino acid changes that are predicted either to maintain the overall structure of the CI activation domain or to disrupt the helical structure of the CBP interaction domain. The single site mutations should minimize the possibility that the CI mutants would not interact with other factors required for CI activity.
The four single site mutations that disrupt the CI-dCBP interaction and have the lowest activity fall in the amino acid sequence LILPDEMLQY. Mutation 376 is a lesion in the first L residue, while mutations 103, 459, and 272 are lesions in the E, M, and third L residues, respectively. These residues are highly conserved in the CBP binding domain of SREBP, a transcription factor that binds to and is activated by CBP in a phosphorylation-independent manner (22, 26). The first L, the M, and third L are exactly conserved, while the two similarly charged D and E residues are switched. This comparison suggests that this motif will be important for protein binding to CBP.
The CI mutation CI*459 is a change from methionine to threonine and is least likely to disrupt the conformation of the CI activation domain. In contrast, CI*103, changes a charged glutamic acid residue to a glycine. This mutation may interfere with dCBP binding either by changing a critical charge interaction or by disrupting the helical structure of the dCBP binding domain. Although neither protein binds dCBP in the yeast two-hybrid assay, both behave as hypomorphs in transient-transfection assays and have a maximal activity that is 20% of the maximal wild-type CI activity. That these mutations do not result in a complete loss of CI function suggests that they destabilize the CI-dCBP interaction and lower the affinity of CI for dCBP or that CI interacts with other dCBP residues and this secondary interaction(s) can allow some mutant CI activity. It is possible that the 103 and 459 mutations disrupt the interaction between CI and a factor other than dCBP and that this interaction is more critical to CI function than the CI-dCBP interaction. If so, one would expect that increases in the dosage of dCBP would not affect the mutant protein activity. This is not the case. Increasing the amount of dCBP can augment the activities of CI*103 and CI*459, although these activities are 10-fold less than those seen when dCBP coactivates wild-type CI. The results of these studies do not rule out the possibility that the dCBP interaction domain interacts with other factors, but they do support the hypothesis that the dCBP interaction with CI is required for wild-type CI function.
We next wanted to determine whether these mutant proteins could activate an endogenous CI promoter. In the cell culture assays we utilize an artificial promoter with multiple CI binding sites and assess the ability of the bound CI to recruit a specific coactivator. The CI-dCBP interaction may be crucial to the transactivation of these artificial promoters but not be essential on an endogenous promoter. The CI mutants behave as hypomorphs, and we would not expect to see a loss of activity in a background of wild-type CI. Therefore, we needed to assess the activities of the CI mutants in cells where wild-type CI is inactive. We took advantage of our finding that mutations in the four consensus RRxS/T PKA sites prevent proteolysis of CI and result in the transactivation of wg in cells that do not receive an HH signal (7, 8). We thus generated CI double mutants that carry the 103 and 459 lesions in the dCBP interaction domain and mutations in the three PKA phosphorylation sites.
The CI(m1-3) mutant behaves in the same manner as the CI(m1-4) mutant; its activity is not affected by the addition of the PKA inhibitor PKI and is enhanced twofold by additional PKA. This twofold activation suggests that there is a secondary, positive effect of PKA on CI activity; however, it is not significant in cell culture and cannot be detected in vivo (7). As with CI(m1-4), CI(m1-3) can transactivate the endogenous wg promoter in the anterior cells of the prd domain that do not receive an HH signal.
The CI double mutants behave in a manner consistent with their lesions. Their activities are severely reduced, are not affected by the presence of PKI, and are only mildly enhanced by additional PKA. When expressed in the prd domain, the CI double mutants cannot misexpress endogenous wg in the anterior cells that do not receive a HH signal. Thus, the CI-dCBP interaction is required for the CI-mediated activation of endogenous wg expression. Obviously, we will want to assess the role the CI-dCBP interaction at other CI target genes in various tissues during development. To determine the role of dCBP in signaling, it will be important to define which factors and promoters require dCBP and which are augmented by dCBP function.
ACKNOWLEDGMENT
This work was supported by a grant from the National Institutes of Health (DK4Y239).
REFERENCES
- 1.Akimaru H, Chen Y, Dai P, Hou D-X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signaling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 2.Akimaru H, Hou D, Ishii S. Drosophila CBP is required for dorsal-dependent twist gene expression. Nat Genet. 1997;17:211–214. doi: 10.1038/ng1097-211. [DOI] [PubMed] [Google Scholar]
- 3.Alexandre C, Jacinto A, Ingham P W. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- 4.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 5.Aza-Blanc P, Ramirez-Weber F-A, Kornberg T B. Proteolysis that is inhibited by hedgehog targets cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 6.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Cardinaux J-R, Goodman R H, Smolik S M. Mutants of cubitus interruptus that are independent of PKA regulation are independent of hedgehog signaling. Development. 1999;126:3607–3616. doi: 10.1242/dev.126.16.3607. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Gallaher N, Goodman R H, Smolik S M. Protein kinase A directly regulates the activity and proteolysis of cubitus interruptus. Proc Natl Acad Sci USA. 1998;95:2349–2354. doi: 10.1073/pnas.95.5.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherbas L, Moss R, Cherbas P. Transformation techniques for Drosophila cell lines. In: Goldstein L S B, Fyrberg E A, editors. Drosophila melanogaster: practical uses in cell and molecular biology. Vol. 44. New York, N.Y: Academic Press; 1994. pp. 161–179. [DOI] [PubMed] [Google Scholar]
- 10.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montiminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez M, Brunner M, Hafen E, Basler K. Sending and receiving the hedgehog signal: control by the Drosophila Gli protein cubitus interruptus. Science. 1996;272:1621–1625. doi: 10.1126/science.272.5268.1621. [DOI] [PubMed] [Google Scholar]
- 12.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 13.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingham P W. Localized hedgehog activity controls spatial limits of wingless transcription in the Drosophila embryo. Nature. 1993;366:560–562. doi: 10.1038/366560a0. [DOI] [PubMed] [Google Scholar]
- 15.Kee B L, Arias J, Montminy M R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 16.Kinzler K W, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krasnow M A, Saffman E E, Kornfeld K, Hogness D S. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell. 1989;57:1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- 18.Kwok R P S, Laurance M E, Lundblad J R, Goldman P S, Shih H-M, Connor L M, Marriott S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 19.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 20.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 21.Methot N, Basler C. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- 22.Naar A M, Beaurang P A, Robinson K M, Oliner J D, Avizonis D, Scheek S, Zwicker J, Kadonaga J T, Tjian R. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima T, Uchida C, Anderson S F, Lee C-G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 24.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 25.Ohlen T V, Lessing D, Nusse R, Hooper J E. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc Natl Acad Sci USA. 1997;94:2404–2409. doi: 10.1073/pnas.94.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliner J D, Anderson J M, Hansen S K, Zhou S, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 27.Orenic T V, Slusarski D C, Kroll K L, Holmgren R A. Cloning and characterization of the segment polarity gene cubitus interruptus Dominant of Drosophila. Genes Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- 28.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C M, Masuno M, Tommerup N, Ommen G-J B V, Goodman R H, Peters D J M, Breuning M H. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 29.Rose R E, Gallaher N M, Andrew D J, Goodman R H, Smolik S M. The CRE binding protein dCREB-A is required for Drosophila embryonic development. Genetics. 1997;146:595–606. doi: 10.1093/genetics/146.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shih H-M, Goldman P S, DeMaggio A J, Hollenberg S M, Goodman R H, Hoekstra M F. A positive genetic selection for disrupting protein-protein interaction: identification of CREB mutations that prevent association with the coactivator CBP. Proc Natl Acad Sci USA. 1996;93:13896–13901. doi: 10.1073/pnas.93.24.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shikama N, Lyon J, Thangue N B L. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 32.Smith C L, Onate S A, Tsai M-J, O'Malley B W. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 34.Spradling A. P element-mediated transformation. In: Roberts D B, editor. Drosophila: a practical approach. New York, N.Y: IRL Press; 1986. pp. 175–198. [Google Scholar]
- 35.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single CBP allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 37.Torchia J, Rose D W, Inostroza J, Kamel Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 38.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 39.Waltzer L, Bienz M. A function of CBP as a transcriptional co-activator during Dpp signalling. EMBO J. 1999;18:1630–1641. doi: 10.1093/emboj/18.6.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 41.Yao T-P, Oh S P, Fuchs M, Zhou N-D, Chng L-E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 42.Yoffe K B, Manoukian A S, Wilder E L, Brand A H, Perrimon N. Evidence for engrailed-independent wingless autoregulation in Drosophila. Dev Biol. 1995;170:636–650. doi: 10.1006/dbio.1995.1243. [DOI] [PubMed] [Google Scholar]