Abstract
Telomere length in human cells is controlled by a homeostasis mechanism that involves telomerase and the negative regulator of telomere length, TRF1 (TTAGGG repeat binding factor 1). Here we report that TRF2, a TRF1-related protein previously implicated in protection of chromosome ends, is a second negative regulator of telomere length. Overexpression of TRF2 results in the progressive shortening of telomere length, similar to the phenotype observed with TRF1. However, while induction of TRF1 could be maintained over more than 300 population doublings and resulted in stable, short telomeres, the expression of exogenous TRF2 was extinguished and the telomeres eventually regained their original length. Consistent with their role in measuring telomere length, indirect immunofluorescence indicated that both TRF1 and TRF2 bind to duplex telomeric DNA in vivo and are more abundant on telomeres with long TTAGGG repeat tracts. Neither TRF1 nor TRF2 affected the expression level of telomerase. Furthermore, the presence of TRF1 or TRF2 on a short linear telomerase substrate did not inhibit the enzymatic activity of telomerase in vitro. These findings are consistent with the recently proposed t loop model of telomere length homeostasis in which telomerase-dependent telomere elongation is blocked by sequestration of the 3′ telomere terminus in TRF1- and TRF2-induced telomeric loops.
The length of mammalian telomeres is governed by a homeostasis mechanism. Telomeres have a species-specific length setting (26) which is constant over the generations, despite high levels of telomerase activity in the germline (47). For instance, the telomeres of Mus musculus are maintained at 20 to 50 kb, while the closely related mouse species Mus spretus has telomeric tracts closer in length to human telomeres, usually ranging from 5 to 15 kb. Similarly, despite high levels of telomerase, the telomeres of many human tumor cell lines do not grow but are stably maintained at a setting characteristic for each individual cell line (12, 13, 44). Telomere length homeostasis is also evident when new telomeres are generated by transfection of short stretches of telomeric DNA into cultured cells. The transfected telomeric tracts are elongated, presumably by telomerase, until their length matches the other telomeres in the transfected cells (3, 23, 43). The new telomere presumably recruits telomere binding proteins and so attains all functions of telomeres, including the length regulation characteristic of a given cell. Regulated growth of a single telomere, as observed in this context, suggests that cells can measure and modulate the length of the telomeric repeat array at individual chromosome ends, implying a cis-acting regulatory mechanism.
Human telomeres contain two related TTAGGG repeat binding factors, TRF1 and TRF2 (7, 10, 11). Both TRF proteins have a Myb-like helix-turn-helix domain in their carboxy terminus and a central conserved domain that includes sequences responsible for the formation of homodimers. The two proteins do not heterodimerize, and they differ substantially at the N terminus, which is acidic in TRF1 but basic in TRF2 (10). TRF1 and -2 are most closely related in their Myb-type DNA binding domains (56% identity), and both proteins can bind duplex telomeric DNA in vitro. The DNA binding site of TRF1, as determined by systematic evolution of ligands by exponential enrichment (SELEX), is composed of two identical YTAGGGTTR half sites which each engage one Myb domain in the TRF1 homodimer (6). There is no constraint on the distance between the two half sites, and the sites can be bound in direct or inverted orientation. TRF1 has architectural features, including the ability to loop the sequence between two half sites (6) and the ability to pair two telomeric tracts (20). Although TRF2 displays a similar preference for duplex TTAGGG repeats in vitro (4, 10), its DNA binding features have not been established in detail.
In human cells, both TRF1 and TRF2 are predominantly located at chromosome ends where they contribute to the protection and maintenance of telomeric DNA. TRF1 has been demonstrated to regulate telomere length (44). Overexpression of TRF1 in the tetracycline-responsive human fibrosarcoma cell line HTC75 results in a gradual decline in telomere length at a rate of ∼10 bp/population doubling (PD). Conversely, the expression of a dominant negative allele of TRF1, which removes endogenous TRF1 from telomeres, leads to telomere elongation. In this system, TRF1 did not affect the activity of telomerase detectable in cell extracts, suggesting that TRF1 does not affect telomerase activity globally in the cell. Instead, we have proposed that TRF1 acts in cis as a negative length regulator at each individual telomere. According to the current model, an inappropriately long telomere would recruit a large amount of TRF1 protein, blocking telomerase-mediated elongation of that particular chromosome end and thus leading to a resetting of the telomere length in cis. A similar protein-counting model was proposed for telomere length homeostasis in yeast (31).
An important question is how TRF1, which binds along the length of the telomere, modulates telomerase, an enzyme that acts at telomere termini. One mechanism that could be considered in this context is that accessibility of a DNA end to telomerase is diminished by the presence of TRF1 on or near the telomere terminus. An alternative proposal has recently emerged from the finding that telomeres fold back, forming a large duplex lariat called the t loop (21). In the t loop, the 3′ single-stranded telomeric overhand of TTAGGG repeats is tucked into the duplex part of the telomeric repeat tract. The t loop is proposed to sequester telomeres from activities that might act on chromosome ends, including telomerase. In vitro studies suggest that telomerase requires an accessible 3′ overhang (28, 30, 46), a structure predicted to be absent from t loops. Therefore, t loops could control the action of telomerase at individual chromosome ends. Based on biochemical studies, the formation of t loops was proposed to involve both TRF1 and TRF2. TRF1 has the ability to induce bending, looping, and pairing of duplex telomeric DNA (5, 6, 20, 21), activities that could facilitate the folding back of the telomere. TRF2 was found to induce the invasion of the 3′ single-stranded TTAGGG repeat tail into duplex telomeric DNA, forming t loops in vitro (21; R. Stansel, T. de Lange, and J. D. Griffith, unpublished data). Thus, a t-loop-based mechanism for telomere length regulation would predict that both TRF1 and TRF2 are required for the length homeostasis of human telomeres. Specifically, the model predicts that, like TRF1, TRF2 acts as a negative regulator of telomere length.
The role of TRF2 in telomere length regulation cannot easily be examined through the inhibition of its function. Interference with the activity of TRF2 results in immediately deleterious phenotypes, possibly due to inappropriate exposure of unfolded telomere termini to DNA damage checkpoints and repair activities. For instance, cells forced to express a dominant negative allele of TRF2 rapidly initiate an apoptotic pathway (24), and telomeres lacking TRF2 lose their 3′ overhangs and undergo covalent fusions (45; A. Smogorzewska and T. de Lange, unpublished data). Therefore, we have addressed the role of TRF2 in telomere length regulation by overexpression of the full-length protein. The results demonstrate that TRF2 is a second negative regulator of telomere length in mammalian cells and are consistent with a t-loop-based mechanism for telomere length homeostasis.
MATERIALS AND METHODS
Cell lines.
Four clonal HTC75 cell lines (P4, P7, P12, and P33) expressing full-length TRF2 from a tetracyclin-repressed promoter were described previously (45). Two HTC75-derived clonal cell lines (J3 and J24) expressing an N-terminally FLAG-tagged deletion allele of TRF1 (TRF166-439) were generated by cotransfection of the pTetFLAGhTRF166-439 plasmid with a neomycin resistance marker. Inducible expression of TRF166-439 was tested by indirect immunofluorescence and immunoblotting with the FLAG-specific M2 monoclonal antibody (Kodak) on 24 independent clones isolated after selection in media containing 400 μg of G418 per ml. A control HTC75 cell line, B27, contained the empty vector (44). Cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with l-glutamine, penicillin-streptomycin, nonessential amino acids, 10% bovine calf serum (HyClone) and either hygromycin (90 μg/ml) or G418 (150 μg/ml). Drugs were alternated every other week. Cells were passaged at 1:16 when they reached 80% confluence. Noninduced and induced cells were grown in parallel with or without doxycycline (Sigma, 100 ng/ml). The cells from hamster cell lines AHL-1 (ATCC CCL 195), BHK-21 (ATCC CCL 8544), and CHO-K1 (ATCC CCL 61) were grown in DMEM supplemented as above.
Whole-cell extracts.
Cells grown in 10-cm-diameter dishes were harvested by scraping in 5 ml of cold phosphate-buffered saline (PBS) and were collected after centrifugation at 1,000 × g for 5 min. Cell pellets were resuspended in 4 to 6 volumes (50 to 200 μl) of ice-cold buffer C (20 mM HEPES-KOH [pH 7.9], 420 mM KCl, 25% glycerol, 0.1 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.2% Nonidet P-40, 1 μg of leupeptin, pepstatin, and aprotinin per ml) and were incubated for 30 min on ice and centrifuged for 10 min at 14,000 × g at 4°C. The supernatant was dialyzed for 2 h against buffer D (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.2 mM EGTA, 0.5 mM DTT, and 0.5 mM PMSF) at 4°C, was snap-frozen in liquid nitrogen, and was stored at −80°C. Protein concentrations were determined by using the Bradford assay (Bio-Rad, Hercules, Calif.) with bovine serum albumin as a standard.
Western blotting.
Proteins (30 μg) were separated on a sodium dodecyl sulfate (SDS)–9% polyacrylamide gel and were transferred to nitrocellulose by electroblotting. Membranes were stained with Ponceau S to verify that equal amounts of protein were fractionated in each lane. After blocking for 30 min in PBS containing 10% nonfat milk powder and 0.5% Tween 20, blots were incubated for 12 to 16 h at 4°C with the anti-TRF2 antibody 508 or 647 or with anti-TRF1 antibody 371 (44, 45; X.-D. Zhu and T. de Lange, unpublished data) or anti-cyclin D antibody (Santa Cruz), followed by three washes in PBS containing 0.1% nonfat dry milk powder and 0.1% Tween 20. Blots were incubated for 45 min with horseradish peroxidase-conjugated donkey anti-rabbit antibody (for 508, 647, and 371) or sheep anti-mouse antibody (for cyclin D) (Amersham) and were washed three times for 10 min. The secondary antibody was detected by using the ECL kit (Amersham).
Genomic blotting.
Cells from a subconfluent 15-cm-diameter dish (approximately 107 cells) were trypsinized, washed in cold PBS, and collected by centrifugation for 5 min at 1,000 × g, and cell pellets were frozen at −80°C. DNA was isolated as described (15) with inclusion of an RNase A digestion step, was cleaved with HinfI and RsaI, and was quantitated by fluorometry using Hoechst 33258. Three micrograms of DNA was size fractionated on a 0.7% agarose gel in 0.5× Tris-borate-EDTA (TBE). Gels were processed for genomic blotting and telomeric restriction fragments were detected with a TTAGGG probe as previously described (14). The median length of the telomeric restriction fragments was determined by using ImageQuant software after scanning with a PhosphorImager (44).
Transient expression of TRF166-439.
Logarithmically growing HeLa 1.2.11 cells (107 cells in 0.8 ml of HBS [21 mM HEPES, pH 7.05, 137 mM NaCl, 0.7 mM NaHPO4, 5 mM KCl, 6 mM glucose]) were electroporated (960 μF/320 mV) with 7.5 μg each of pTetFLAGhTRF166-439 and the tTA-expression vector pUHD15-1 (19), and the cells were grown on autoclaved coverslips in DMEM with 10% fetal bovine serum for 24 h before being processed for indirect immunofluorescence.
FISH on metaphase spreads.
Hamster cells were incubated with 0.5 μg of colcemid per ml of growth media at 37°C for 30 min. Cells were harvested by trypsinization and were incubated in 0.075 M KCl at 37°C for 7 min. Subsequently, cells were fixed in methanol-acetic acid (3:1) and were dropped onto water-wetted slides. Fluorescence in situ hybridization (FISH) was carried out as described before (27) with 0.5 μg of fluorescein isothiocyanate-conjugated (C3TA2)3 peptide nucleic acid (PNA) probe (Biotech BmpbH) per ml. The cells were embedded in a mixture of 90% glycerol and 10% PBS containing 1 mg of p-phenenylene diamine (Sigma) per ml with 0.2 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml.
Immunofluorescence and microscopy.
FLAG-tagged TRF166-439 was detected by using the anti-FLAG monoclonal antibody M2 (Sigma), endogenous full-length TRF1 was detected by using antibody 371C2 (44), and endogenous TRF2 was detected by using antibody 508 (45). M2 was detected with fluorescein isothiocyanate-conjugated sheep anti-mouse antibody (Jackson), and 371C2 and 508 were detected with tetramethyl rhodamine isocyanate-conjugated donkey anti-rabbit antibody (Jackson). Secondary antibodies did not cross-react, as shown by control experiments omitting either of the primary antibodies. DNA was counterstained with DAPI. Endogenous TRF1 and TRF2 on telomeres in hamster, HeLal.2.11, and HeLall cells (39, 45) were detected in parallel by using affinity-purified rabbit polyclonal antibodies 371 and 647, respectively (44; X.-D. Zhu and T. de Lange, unpublished data), and tetramethyl rhodamine isocyanate-conjugated donkey anti-rabbit secondary antibodies (Jackson). For analysis of the relative abundance of TRF1 and TRF2 on long and short HeLa telomeres, exposure times were 1.0 s for all photographs, and the images are presented without changes in settings. Photographs were taken with a Photometrix SenSyn camera installed on a Zeiss Axioplan2 microscope with IPLab software (Scanalytics, Inc.).
RNA isolation.
Total RNA was isolated as described (2). Briefly, cells were washed in PBS, lysed in 3 M LiCl-6 M urea, sonicated, and incubated on ice for 6 h. RNA was collected by centrifugation at 14,000 × g for 20 min at 4°C, was treated with proteinase K (50 μg/ml) in 200 μl of a solution containing 10 mM Tris-HCl [pH 7.5], 10 mM EDTA, 100 mM NaCl, and 1% SDS for 10 min at 37°C, was extracted with phenol-chloroform-isoamylalcohol (24:24:1), and was precipitated with isopropanol in the presence of 0.2 M Na-acetate, pH 5.5.
RNase protection.
The hTERT sequence was subcloned from pCl Neo-hEST2-HA (a gift from R. A. Weinberg) (32) into pcDNA3 (Invitrogen). A Styl fragment containing part of the hTERT sequence (nucleotide [nt] 3229 to 3452) and flanking vector sequences was purified and used in the MAXIscript in vitro transcription reaction (Ambion) with 5 μl of [α-32P]UTP (800 Ci/mmol, 10 mCi/ml). The template for the hTR RNase protection probe was similarly prepared by digesting pGRN78 (25) with XbaI, and the β-actin probe was obtained from Ambion. The β-actin probe was labeled to a specific activity 200-fold lower than that of the hTERT and hTR probes by diluting out the radiolabelled UTP with cold UTP. RNase protection reactions were performed with solutions from the Direct Protect Lysate Ribonuclease Protection Assay Kit (Ambion) with 20 μg of total RNA (isolated as described above) instead of cell lysates.
Partial purification of human telomerase.
Nuclear extracts from HeLa cells (16) were dialyzed against buffer A-100 (20 mM HEPES [pH 7.9], 1 mM EDTA, 1 mM EGTA, 10% glycerol, 100 mM KCl, 0.5 mM PMSF, 0.5 mM DTT), and were applied to a Spermine-agarose column (Sigma). The column was washed with 250 mM KCl, and 500 mM KCl in buffer A. Telomerase activity was eluted with buffer A containing 1 M KCl. The active fractions were pooled, dialyzed against buffer A-100, and loaded onto a Heparin Cl-6B column (Pharmacia) equilibrated in buffer A-100. The column was washed with buffer A containing 220 mM KCl, and active telomerase was step eluted with 500 mM KCl. Active fractions were pooled, dialyzed as described above, and applied to a MonoQ fast protein liquid chromatography column (Pharmacia), equilibrated with buffer A-100. The column was washed with buffer A containing 300 mM KCl, and telomerase was eluted with a linear gradient from 0.3 to 1 M salt in buffer A. The peak of active telomerase eluted at 500 mM KCl. Pooled active fractions showed a specific telomerase activity 100-fold higher than that of the input material as determined by the conventional telomerase assay (34).
Telomerase reactions.
Telomerase reactions were in a solution containing 25 mM Tris acetate (pH 8.2), 50 mM Na acetate (pH 8.3), 1 mM MgCl2, 1 mM EGTA, 5 mM dATP, 5 mM dTTP, 4.5 μM cold dGTP, 0.165 to 0.825 μM labelled dGTP (1 to 5 μl of [α32P]dGTP, 3,000 Ci/mmol, 10 mCi/ml; NEN), 0.1 μl of RNasin, and 1 mM spermidine (optional), in a volume of 20 μl, typically using 1 to 2 μl of telomerase fractions. Concentration of primer was 10 nM. Baculovirus-expressed TRF1 or TRF2 protein (5, 17), or bovine serum albumin (BSA) as a control, were incubated with the DNA in the telomerase reaction buffer for 30 min at room temperature. Subsequently, telomerase fractions were added, and reactions were incubated for 1 h at 30°C and stopped by the addition of 20 μl of H2O and 50 μl of 10 mM Tris (pH 8.0), 5 mM EDTA, and 0.1 mg of RNase/ml and incubation at 45°C for 10 min. Next, 50 μl of 10 mM Tris (pH 8.0) and 1% SDS containing 0.3 mg of proteinase K/ml were added, and the samples were incubated at 45°C for 10 min. Samples were extracted with 200 μl of phenol-chloroform-isoamyl alcohol and were precipitated by the addition of 50 μl of 3 M Na acetate (pH 5.2), 2.5 μg of tRNA, and 500 μl of ethanol and incubation on ice for 15 min. Samples were centrifuged at room temperature for 10 min, and the ethanol was removed. Pellets were dissolved in 4 μl of formamide loading dye, and all or half of the sample was loaded onto 6% polyacrylamide (19:1 acrylamide–bis-acrylamide) gels containing 7 M urea and 20% (vol/vol) formamide in 1× TBE. Gels were run at 40 to 55 W, at about 50 to 60°C external temperature. Oligonucleotides were gel purified on polyacrylamide–7 M urea gels and were annealed in a solution containing 10 mM Tris (pH 7.5), 1 mM EDTA, and 100 mM NaCl by boiling for 5 min and slow cooling (over a few hours) from 80°C to room temperature.
Gel shift analysis to monitor TRF1 and TRF2 binding to the telomerase substrates was performed by using end-labeled DNAs. Binding conditions were exactly the same as in telomerase assays except that deoxynucleoside triphosphates were omitted. Samples were run on 0.6% agarose, 0.1× TBE, at 130 V for about 45 min (4).
RESULTS
TRF2 affects telomere length.
To address the role of TRF2 in telomere length regulation, we employed a previously established tetracyclin-inducible expression system in the human fibrosarcoma cell line HTC75 (45). Although the HTC75 cells express telomerase, the length of their telomeres is stably maintained at a setting that is in part governed by TRF1 (44). Overexpression of TRF1 results in gradual and progressive loss of telomeric DNA. Here we explore the effect of long-term overexpression of TRF2. Four clonal cell lines (called P lines) that can be induced to express high levels of TRF2 by removal of doxycycline from the growth media were established. An initial, short-term (9-day) analysis of these cells revealed no striking phenotypes due to increased TRF2 expression (45), whereas the same study documented a rapid inhibition of cell proliferation by two truncated alleles of TRF2. Long-term growth of the P cell lines allowed us to assess the effect of elevated TRF2 expression on the steady-state length of the telomeres.
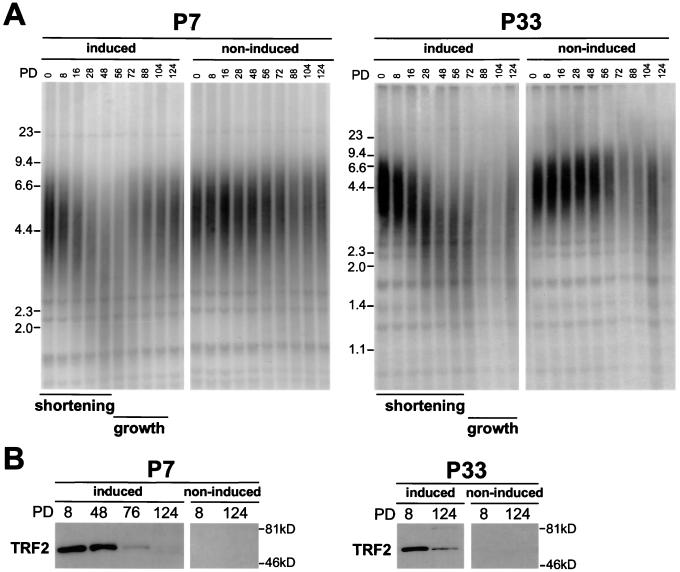
The cell lines were maintained with or without doxycycline in the media for 124 PDs, and their telomere length was assessed by genomic blotting. With one exception (P4), the cell lines showed consistent telomere length changes that were dependent on TRF2 induction (Fig. 1). The P4 cell line showed unanticipated fluctuations in telomere length that were not dependent on the presence of doxycycline in the medium (data not shown). The three other P cell lines showed a complex, biphasic pattern of telomere length changes in response to increased TRF2 levels. In the P7 cell line, the telomeres initially shortened at a rate of 82 bp per PD (calculated for the growth period from PD 0 to PD 28) when TRF2 was induced (Fig. 1A). Unexpectedly, around PD 48, the dynamics of the telomeres changed. At this point in the culture, the telomeres lengthened, reaching their original length setting by PD 88. The telomere length of the noninduced culture appeared relatively stable throughout the experiment. A similar course of events was seen in the P33 cell line, where the initial decline in telomere length was 47 bp per PD (growth window, PD 0 to PD 40) and was followed by modest telomere elongation that started at PD 72. The third cell line (P12) showed a telomere-shortening rate of 51 bp/PD in the first 8 PD after TRF2 induction, followed by a rapid increase in telomere length, reaching a final length setting that surpassed the starting value (data not shown). Overall, the overexpression of TRF2 led to an initial decline in telomere length and an eventual cessation of this decline followed by telomere elongation. These results indicate that TRF2, like TRF1, affects telomere maintenance.
FIG. 1.
Overexpression of TRF2 results in telomere shortening. (A) Two TRF2-expressing HTC75 cell lines (P7 and P33) were grown for 124 PD in media with (non-induced) or without (induced) doxycycline, and genomic DNA was isolated at the indicated PD. Protein-free DNA was digested with HinfI and RsaI, was size fractionated on an agarose gel, and was blotted. A telomere-specific TTAGGG repeat probe was used to detect the telomeric restriction fragments. Molecular weight standards are indicated to the left of the gels. Periods of telomere shortening and growth are highlighted below the lanes. (B) Immunoblotting analysis of the TRF2 expression in P7 and P33 cells under induced and noninduced conditions. TRF2 was detected with antibody 508 in immunoblots of equal amounts (30 μg) of whole-cell-extract proteins. Endogenous TRF2 is below detection limit under these conditions.
Complex telomere dynamics correlate with loss of TRF2 expression.
Although the biphasic nature of the telomere length changes in the P cell lines was unexpected, the fact that the pattern of telomere dynamics was similar in three independent cell lines suggested that this phenotype was an inherent property of HTC75 cells induced to express high levels of TRF2. To further understand the molecular basis of this phenotype, we analyzed the expression levels of TRF2. Immunoblotting showed that even though the initial expression of the exogenous TRF2 protein was very high, the levels of TRF2 declined in each of the cell lines with increasing PD (Fig. 1B, compare lanes at PDs 8 and 124 in the induced state) (data not shown for P12). The decline of the steady-state levels of TRF2 protein detectable by Western analysis was due to a decrease in number of cells that expressed TRF2 as well as in the level of expression in individual cells as deduced from the immunofluorescence staining. For instance, the percentage of immunofluorescence staining-positive cells dropped from 100% at PD 2 to 0% at PD 48 for P12 and from 100% at PD 2 to 21% at PD 76 for P7.
Loss of expression of a transfected gene during long-term growth of the transfected cells has been reported previously and is often ascribed to de novo methylation of the transgene. However, the rapid loss of TRF2 expression in comparison to the stable expression of TRF1 in the same system (see below) suggested that down regulation of TRF2 expression might confer a selective advantage on the cells. In this regard, we noted that the induced cultures contained many cells with an unusual nuclear morphology (multilobulated nuclei) (data not shown). The appearance of the aberrant nuclei tended to coincide with the loss of TRF2 expression. We also noted that each of the induced cultures grew somewhat slower than the uninduced controls. For instance, PD 40 was reached after about 40 days for the controls but 4 days later for the induced cultures. Although we cannot determine whether the altered nuclear morphology is relevant to the gradual disappearance of TRF2-expressing cells, the results are consistent with the possibility that overexpression of TRF2 (perhaps in conjunction with shortened telomeres) inhibits cell growth.
Long-term overexpression of full-length TRF1 and TRF166-439.
The complex telomere dynamics observed in cells overexpressing TRF2 and the apparent selection against high levels of TRF2 raised the question of whether similar events might be occurring during long-term overexpression of TRF1. Previous analysis of several HTC75 cell lines that overexpressed full-length TRF1 had revealed a gradual and progressive shortening of the telomeres, and no repression of the transfected TRF1 gene was noted (44). However, the initial analysis of these cell lines did not provide information on events after extensive telomere shortening. Thus, it was pertinent to analyze the long-term dynamics of telomeres in cells overexpressing TRF1.
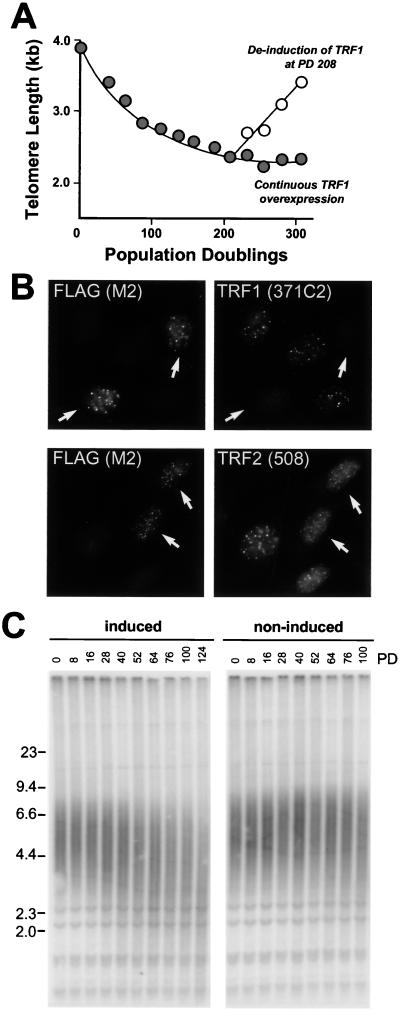
One clonal TRF1-expressing cell line, D4, was selected for this purpose, and its telomere length was monitored over 304 PD. As shown in Fig. 2A, this cell line showed the previously recorded gradual telomere shortening, and this shortening continued until approximately 1.5 kb of telomeric DNA was lost (at PD 200). At that stage, telomere length became stable, and no further changes were observed when the culture was grown for an additional 100 PD. No gross effects on growth rates were observed, and TRF1 expression was unaltered (data not shown). In fact, the overexpression of TRF1 was required to maintain the telomeres at a stable, short, length setting as demonstrated by the rapid telomere lengthening observed when doxycycline was added to the growth media (Fig. 2A).
FIG. 2.
Long-term overexpression of full-length TRF1 and TRF166-439 result in progressive telomere shortening. (A) Stabilization of telomeres at a short-length setting due to overexpression of TRF1. D4, a HTC75 cell line overexpressing TRF1 in absence of doxycycline (44), was maintained under inducing conditions for 304 PD, and telomere lengths were determined by genomic blotting as shown in Fig. 1. The plot represents mean telomere length values during induced growth (filled circles). At PD 208, doxycycline was added to a parallel culture and telomere length changes were monitored under noninduced conditions (open circles). (B) TRF166-439 localizes to telomeres, where it displaces TRF1 but not TRF2. TRF166-439, bearing an N-terminal FLAG tag, was transfected into HeLal.2.11 cells, and its subcellular localization was determined by using indirect immunofluorescence with the anti-FLAG antibody M2 (left panels). The effect of this TRF1 allele on endogenous TRF1 was determined with an antibody directed against the N terminus of TRF1 (371C2; right top panel) that does not detect the TRF166-439 mutant. The subnuclear localization of TRF2 was assessed with antibody 508 (right bottom panel). Arrows denote transfected cells. (C) TRF166-439 induces progressive telomere shortening. J24, an HTC75 cell line expressing TRF166-439, was maintained under induced and noninduced conditions, and changes in telomere length were evaluated as described in the legend of Fig. 1A.
In addition, we have not found a deleterious effect of overexpression of a TRF1 protein that lacks the NH2-terminal acidic domain (TRF166-439). This truncated protein localized to telomeres as seen by indirect immunofluorescence (Fig. 2B, left panels) and displaced the full-length endogenous TRF1 protein from telomeres as shown by the diminished immunofluorescence signal with an antibody specific to the acidic domain of TRF1 (371C2) (44) (Fig. 2B, top right panel). Overexpression of TRF166-439 had only a moderate effect on the relative abundance of TRF2 on telomeres, consistent with previous results (45) (Fig. 2B, bottom right panel). As was observed with full-length TRF1, cells expressing TRF166-439 displayed a gradual shortening of their telomeres. In the J24 cell line shown in Fig. 2C, the decline was 11 bp per PD over 124 PD, whereas a second cell line (J3) had more rapidly shortening telomeres (44 bp/PD) (data not shown). Neither of these cell lines showed the complex telomere dynamics observed in TRF2-overexpressing cell lines. Thus, the biphasic dynamics of telomeres and the rapid extinction of transgene expression appear to be exclusively associated with the overexpression of TRF2.
Accumulation of TRF1 and TRF2 at chromosome ends correlates with telomere length.
The telomere-shortening phenotype associated with high levels of TRF2 expression indicates a role for this factor in telomere length homeostasis. One possibility, similar to what was proposed for TRF1, is that TRF2 measures telomere length by binding to the duplex telomeric repeat tract. Indeed, according to band-shift analysis, TRF2 binds to duplex telomeric repeat arrays (4, 10). However, electron microscope analysis indicated that when TRF2 is associated with telomeric DNA in the t loop configuration, it is preferentially positioned on or near the t loop junction rather than along the telomeric tract (21). This preference for the t loop junction was not observed for TRF1 which displayed extensive binding along the duplex telomeric DNA even when the DNA was in the t loop configuration. It was therefore pertinent to establish whether TRF2 can bind to duplex telomeric DNA in vivo or whether the presence of this protein at chromosome ends reflects a preference for a structural feature of telomeres.
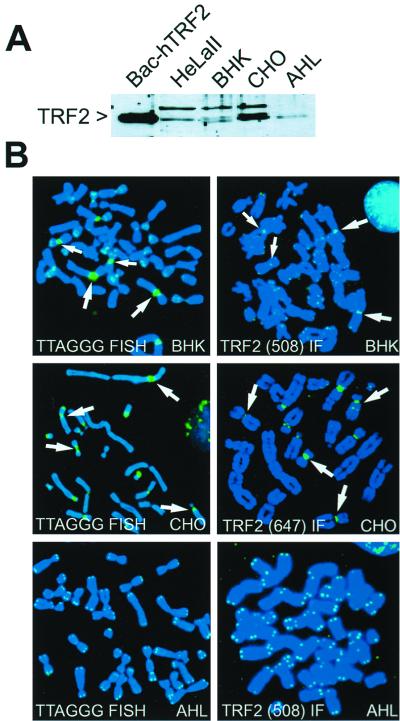
We addressed this question by asking whether TRF2 could associate with telomeric sequences at a chromosome-internal site. Such a situation is represented by certain hamster chromosomes which have sequences related to telomeric repeat DNA associated with their centromeres. Pericentromeric satellite sequences related to TTAGGG repeats are present in several chromosomes in the Syrian hamster cell line, BHK-21, and in the Chinese hamster cell line, CHO-K1, but not in any of the chromosomes of the Armenian hamster cell line, AHL-1 (1, 33) (Fig. 3). Since TRF2 is highly conserved (10), we were able to detect TRF2 in Syrian, Armenian, and Chinese hamster cells with a polyclonal antibody raised against an amino-terminal peptide of human TRF2 (508) (45) (Fig. 3A). By using this antibody in indirect immunofluorescence on metaphase chromosomes from BHK and AHL cells, we detected the expected telomeric staining pattern previously observed for TRF2 in other mammals. In addition, several of the BHK chromosomes showed prominent pericentric signals for TRF2. The use of a second independent polyclonal antibody (647) (X.-D. Zhu and T. de Lange, unpublished data) for indirect immunofluorescence on CHO chromosomes further verified the chromosome-internal accumulation of TRF2 at pericentric sites (Fig. 3B). Such interstitial signals were not seen in AHL cells, consistent with the lack of extensive pericentric telomere-related DNA in these cells. These data indicated that TRF2 has the ability to bind to chromosome-internal telomere-related sequences, suggesting that the binding of TRF2 is not dependent on the DNA being in a t loop configuration. Consistent with the poor conservation of TRF1 (9), we have not been able to detect hamster TRF1 with a polyclonal antibody raised against full-length human TRF1 or with a polyclonal antibody raised against a peptide of mouse Trf1. However, transfection of human TRF1 into BHK cells indicated that TRF1, like TRF2, can locate to the interstitial sites of telomere-related sequences (data not shown).
FIG. 3.
TRF2 can bind to interstitial telomeric repeat-related sequences. (A) Immunoblot of whole-cell extracts from HeLall cells and three hamster cell lines (BHK-21, CHO-K1, and AHL-1) showing expression of TRF2. TRF2 was detected with antibody 508. Baculovirus-derived human TRF2 (Bac-hTRF2) protein is used as a positive control (4). (B) Interstitial binding of TRF2 in BHK-21 and CHO-K1 chromosomes. Left panels: metaphase chromosomes from the indicated hamster cell lines were analyzed by FISH by using a PNA probe for TTAGGG repeats. Arrows indicate some of the BHK and CHO chromosomes with prominent interstitial telomere-related sequences. Right panels: indirect immunofluorescence with the TRF2 antibody 508 or 647 (as indicated). Arrows highlight TRF2 at pericentric interstitial sites in BHK and CHO chromosomes. No interstitial TTAGGG signals or chromosome-internal binding of TRF2 is observed in the AHL cells. DNA was stained with DAPI.
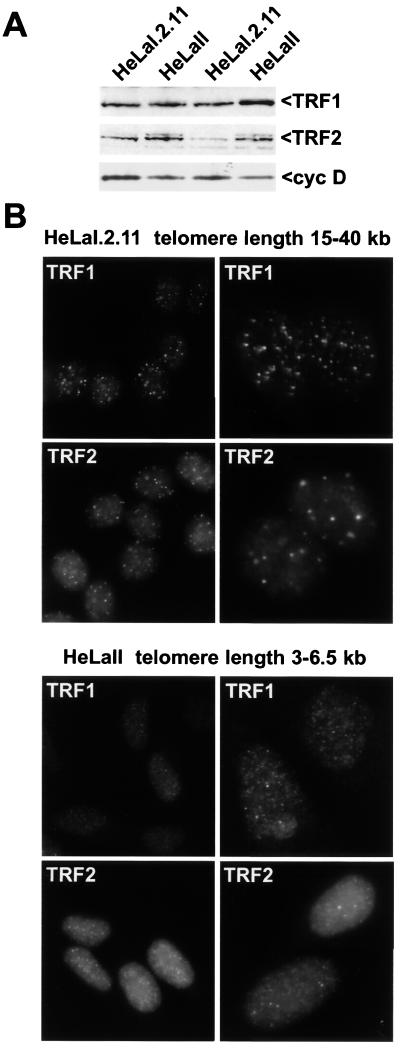
If the binding of TRF1 and TRF2 at telomeres occurs primarily on the duplex part of the telomere, it might be expected that longer telomeres recruit more protein. In contrast, if TRF2 is specifically bound to a structural feature of the t loop or in general requires the proximity of a DNA end for binding, its abundance at individual telomeres should not depend on their length. We therefore compared the TRF1 and TRF2 immunofluorescence signals in two closely related HeLa subclones that differ with regards to the length of the telomeric repeat tracts. HeLal.2.11 has telomeres of approximately 15 to 40 kb (21), whereas HeLall has shorter telomeres ranging from 3 to 6.5 kb (6 kb median) (39). Both cell lines express TRF1 and TRF2 as determined by immunoblotting (Fig. 4A). The abundance of TRF1 and TRF2 is slightly greater in the cells with shorter telomeres.
FIG. 4.
Greater relative abundance of TRF1 and TRF2 on long human telomeres. (A) Immunoblot showing expression of TRF1 and TRF2 in whole-cell extracts from two HeLa cell lines with different telomere lengths. TRF1 was detected with antibody 371; TRF2 was detected with antibody 647. Cyclin D is used as a loading control. In the first two lanes, proteins from equal numbers of cells were loaded. The second two lanes were normalized based on protein amounts (30 μg). (B) Indirect immunofluorescence signals for TRF1 and TRF2 in two HeLa cell lines with the indicated telomere lengths. Immunofluorescence staining of the two cell lines was executed in parallel and was processed identically, and images are presented without any adjustment. For each combination of cell line and antibody, nuclei are presented at two magnifications. DNA was stained with DAPI.
Indirect immunofluorescence was used to examine the apparent abundance of TRF1 and TRF2 at interphase telomeres. We chose to address this issue in interphase cells since the higher level of condensation of metaphase chromatin could result in misleading staining intensities. To detect TRF1, we employed an affinity-purified polyclonal antibody (371C2) that is specific for the N-terminal acidic domain (reference 44 and data not shown). Similarly, to detect TRF2, we employed an affinity-purified polyclonal serum directed against full-length TRF2 that is highly specific for TRF2 (647) (X.-D. Zhu and T. de Lange, unpublished data). Using these sera, we examined the two HeLa cell lines in parallel, and the immunofluorescence images were captured and reproduced under exactly the same conditions and settings (Fig. 4B). Comparison of the intensity of the telomeric dots observed with the TRF1-specific 371C2 serum revealed an obvious difference between the two HeLa cell lines. The cells with long telomeres (HeLal.2.11) showed a punctate pattern generally more pronounced than in the cells with shorter telomeres (HeLall). Although direct quantitative interpretation of immunofluorescence signals is not possible, the difference in intensity was very obvious and highly reproducible. Similarly, other cell lines bearing long telomeres have yielded brighter punctate patterns with our TRF1 sera than cells with short telomeres (data not shown). The striking difference in the immunofluorescence intensity of long versus short telomeres was also found when cells were stained for TRF2 (Fig. 4B). Although staining for TRF2 usually results in a higher overall nucleoplasmic signal, the correlation between the intensity of the punctate immunofluorescence signals and telomere length was still obvious. Furthermore, the TRF2 signal at telomeres of hamster metaphase chromosomes is greater in AHL cells which have more TTAGGG repeats at the chromosome ends than the other two hamster cell lines (Fig. 3B). These results indicate that TRF1 and TRF2 bind to the duplex part of the telomere and that their accumulation at chromosome ends is influenced by the length of the telomeric repeat tract.
Lack of direct effects of TRF1 and TRF2 on telomerase in vitro.
Previous studies indicated that overexpression or inhibition of TRF1 and TRF2 does not affect the expression level of telomerase as detected by the semiquantitative TRAP assay performed with extracts of cell lines expressing full-length or truncated proteins (44, 45). Similarly, we have failed to detect a change in the expression level of the hTERT mRNA or the hTR component of telomerase in cells overexpressing TRF2 (Fig. 5A). We also failed to detect an association between telomerase and TRF1 by immunoprecipitation or on glycerol gradients, and the addition of baculovirus-produced TRF1 protein did not affect telomerase activity in the TRAP assay (data not shown). Finally, no correlation was noted between expression of TRF1 or TRF2 and the expression of telomerase activity in immortalized human cell lines (data not shown).
FIG. 5.
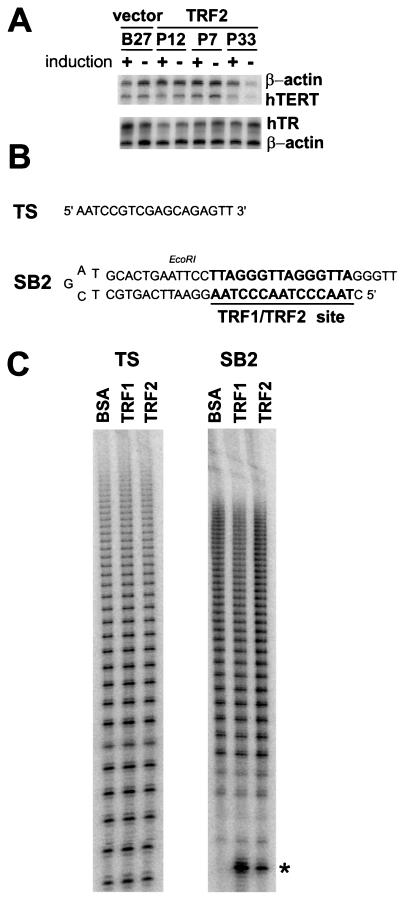
Lack of direct effects of TRF1 and TRF2 on telomerase. (A) Overexpression of TRF2 does not affect the expression of hTERT mRNA or hTR RNA. Total RNA was prepared from the indicated HTC75 cell lines after 9 days of induction with doxycycline. RNase protection assays were performed to determine the relative abundance of hTERT mRNA and hTR RNA. β-actin served as a control (as described in Materials and Methods). (B) Schematic of the telomerase substrates used in panel C. SB2 contains an optimal binding site for TRF1 and TRF2. TS does not bind TRF1 or TRF2. (C) Lack of effects of TRF1 and TRF2 on the in vitro activity of telomerase. Partially purified human telomerase (see Materials and Methods) was assayed with TS (25, 35) or SB2 as a substrate in the presence of excess baculovirus-derived TRF1 and TRF2, or BSA as a control. The assay uses incorporation of [α-32P]dGTP into telomerase products (conventional assay). The asterisk indicates a labeled form of the SB2 substrate that is only observed in the presence of the baculovirus-derived proteins.
These negative results would be expected if the regulation of telomere length by TRF1 and TRF2 occurs in cis. One cis-acting mechanism could be that telomeres containing TRF proteins are not an optimal substrate for telomerase. Accordingly, an in vitro system was developed to test whether the presence of TRF1 or TRF2 near a 3′ DNA end affects the action of telomerase on that terminus. The system employs a DNA that is a binding substrate for TRF1 and TRF2 and can also serve as a primer for telomerase-mediated addition of TTAGGG repeats. This DNA, called SB2, is a snapback oligonucleotide with a 3′ overhang of the sequence GGTT flanking a duplex segment with an optimal TRF1 binding site (TTAGGGTTAGGGTTAG [4, 6]) (Fig. 5B). Conditions were established in which the SB2 substrate was saturated with TRF1 or TRF2 protein using the buffer appropriate for a telomerase assay (data not shown). In parallel, control reactions were performed with TS, a single-stranded optimal substrate for telomerase (25, 35) that lacks a site for TRF1 and -2. Partially purified telomerase from HeLa cells was incubated with either TS or SB2 in the presence or absence of TRF1 or -2 (50 nM TRF1 and 35 nM TRF2, respectively), under conditions that were found to result in an approximately linear dependence of telomerase activity on primer concentration (data not shown). The enzymatic activity of telomerase was monitored based on the incorporation of [32P-α]dGTP into the typical 6-nt ladder of telomerase products (Fig. 5C). We engineered an EcoRI site into the duplex part of SB2, allowing us to verify that telomerase added TTAGGG repeats to the correctly folded substrate. Indeed, cleavage of the telomerase products with EcoRI and subsequent size fractionation on a denaturing gel indicated the expected downshift of the product ladder by approximately 40 nt (data not shown).
When TRF1 or TRF2 protein was added to the telomerase reactions with TS as a substrate, no effect on either the overall incorporation of labelled precursor or the size distribution of the products was noted (Fig. 5C). This result was consistent with the observation that the addition of TRF1 to TRAP assays had no effect on the activity of telomerase. Similarly, when SB2 was used as a substrate, the products appeared identical regardless of the presence of saturating amounts of TRF1 or TRF2 (BSA was used as a control). Because the elongation products were spread over a large range of molecular weights on the gel, and because of the presence of a substantial smearing from the unincorporated [32P-α]dGTP, quantitation of the telomerase products was not attempted. Nevertheless, repetitions of the experiment (n = 7) allowed us to rule out a major effect of either telomeric protein on telomerase activity under these conditions. Only minor variations from experiment to experiment were observed. Two overhangs, 5′GTTAGGGTT3′ and 5′GGTT3′, were tested in the assay with identical results. Telomerase assays were also performed in the presence of a 25-fold-higher amount of TRF1 and TRF2 (1.25 μM TRF1 and 0.877 μM TRF2). Even though the results showed greater variability under these conditions, presumably because of protein aggregation, no clear effect of either protein on telomerase was observed (data not shown). Thus, under these conditions, binding of TRF1 or TRF2 near the telomerase substrate site does not interfere with enzyme activity. Our experiments do not address other cis-acting mechanisms, and we cannot exclude at this stage that TRF1 or TRF2 affects telomere length maintenance in trans through affecting a limiting factor other than telomerase.
DISCUSSION
The maintenance of human telomeres by telomerase is regulated by a homeostasis mechanism. Previously, TRF1 was identified as a component of a negative feedback loop that limits telomere elongation and results in stable telomere length. Here we report that overexpression of a related telomere binding protein, TRF2, also results in telomere shortening, indicating that TRF2 is a second negative regulator of telomere length. The simplest explanation is that both TRF1 and TRF2 bind along the length of the duplex telomeric repeat array and function to measure telomere length. In agreement, we find that both proteins bind to double-stranded telomeric DNA in vivo and that their accumulation at telomeres is dependent on telomere length.
TRF1 and TRF2 vis-à-vis the protein-counting model of telomere length regulation.
Our observations can be interpreted within the context of a protein-counting model for telomere length regulation (31, 40, 44). According to this model, telomeres can exist in two states, an “open” state which allows telomerase to elongate the telomere and a “closed” state in which the enzyme cannot access or extend the telomere terminus. The switching between these two states is proposed to be governed by telomere binding proteins, TRF1 and TRF2 in human cells, which act as negative regulators through promoting the closed state. As telomeres are elongated by telomerase, they will bind a greater number of the negative regulators, increasing the chance of a switch to the closed state. After switching to the closed state, a telomere will gradually lose sequences with each cell division, leading to a smaller number of bound negative regulators and an improved chance of switching back to the open state. As a result, each individual telomere will approach a steady-state length determined by the activity of telomerase, the expression level of TRF1 and TRF2, and other regulatory factors (e.g., tankyrase [42]).
Several predictions of this model are tested here. First, the model predicts that the negative regulators are proteins that bind along the duplex part of the telomere. Although previous results demonstrated the association of TRF1 and TRF2 with human and mouse telomeres, it was not possible to distinguish between their binding to the duplex part of the telomere or to some other feature of the telomeric structure (e.g., the telomere terminus or a structural component of the t loop). By using indirect immunofluorescence, we discovered that TRF2 bound to telomere-related sequences that occur at chromosome-internal sites in some hamster chromosomes. Assuming that these interstitial sequences are present as duplex DNA, this result indicates that TRF2 can bind to duplex TTAGGG repeat tracts in vivo. Indirect evidence indicated that TRF1 also bound interstitial telomeric DNA.
A second prediction of the model is that the accumulation of TRF1 and TRF2 at telomeres reflects the length of the duplex telomeric tracts. Indeed, cells with long telomeres had more intense immunofluorescence signals for both proteins. Although a direct quantitative interpretation of immunofluorescence signals is not possible, the data are consistent with the view that longer telomeres attract a greater mass of TRF1 and TRF2, as predicted by the protein-counting model.
According to the protein-counting model, very short telomeres would not bind sufficient amounts of TRF1 or TRF2 to create the closed state. At this stage, the telomere elongation would again take place, preventing complete loss of the telomeric DNA and resulting in the stabilization of the telomeres at a short length. This scenario is consistent with the data on long-term overexpression of TRF1 presented here. After an initial period of telomere shortening, the telomeres stabilized at reduced length even though TRF1 levels remained high. In fact, the overexpression of TRF1 continued to dictate the short-length setting as demonstrated by rapid telomere elongation after repression of the exogenous TRF1. In sum, the telomere dynamics in HTC75 cells in response to changes in the levels of TRF1 and TRF2 are consistent with both proteins functioning as negative regulators of telomere length in a protein-counting mode for telomere length regulation.
A possible mechanism by which TRF1 and TRF2 regulate telomere length.
The protein-counting model of telomere length regulation does not define the nature of the open and closed states nor does it specify by what mechanism TRF1 and TRF2 affect the transition between these states. The only specific prediction of the model is that the events take place in cis, regulating the action of telomerase at individual chromosome ends. In agreement with this prediction, we find that neither the expression of telomerase components nor the global activity of the enzyme are affected by changes in expression of TRF1 or TRF2. We also did not find a direct effect of TRF1 or TRF2 on telomerase when the factors were positioned at or near the 3′ end of the substrate.
Recent findings on the structure of human telomeres suggest that t loops may represent the closed conformation. In t loops, the telomere terminus is tucked away in the duplex telomeric repeat array and is therefore likely to be inaccessible to telomerase. TRF2 can remodel telomeric DNA into t loops in vitro, and TRF1 has several biochemical activities that could be expected to promote t loop formation, suggesting that both proteins contribute to the remodelling of telomeres into t loops in vivo. The current data are consistent with this proposal, since both TRF1 and TRF2 act as negative regulators of telomere length as would be expected if both proteins promote the formation of a closed t loop state.
The emerging view, based on the current data, is that long telomeres recruit a larger amount of TRF1 and TRF2. The greater abundance of these proteins on longer telomeres is suggested to facilitate remodelling of the telomeres into t loops. In the t loop state, telomerase would no longer be able to elongate the telomere terminus, leading to loss of sequences with cell divisions. Eventually, this sequence loss would result in diminished binding of TRF1 and TRF2 to the telomere, which would consequently form t loops at a lower rate (or at a reduced frequency). The resulting (temporary) persistence of an unfolded telomere, a substrate for telomerase, would then again lead to telomere elongation. This model can be tested by assessing how TRF1 and TRF2 modulate the formation of t loops in vivo and by studying the interaction of telomerase with t loops in vitro and in vivo.
Comparison to telomere homeostasis in yeast.
A comprehensive view of telomere length control has emerged in the yeast Saccharomyces cerevisiae. Genetic, molecular genetic, and biochemical approaches have revealed the interplay between components of the telomeric chromatin (including Rap1p, Rif1p, Rif2p, and Cdc13p), trans-acting factors (such as telomerase [Est2p/TLC1]), and telomeric chromatin-associated proteins (Est1p and Est3p), as well as additional mediators like the Ku, the Rad50-Mre11-Xrs2 complex (8, 36, 38), Tel1p, and Stn1p (for reviews, see references 37 and 41). In comparison, the dissection of telomere length homeostasis in mammals is in its infancy, with only a few of the players available, most of which show no similarity to the yeast genes. For instance, TRF1 and TRF2 lack significant homology to Rap1p outside the Myb-type DNA binding fold. However, a strong common theme that has emerged is the likely involvement of a protein-counting mechanism in both systems (31, 44). It will be of interest to determine the possible structural similarities between the telomeric complex in yeast and mammals, in particular with regards to the presence of t loops. In this regard, Li and Lustig (29) raised the intriguing possibility of a transient loopback mechanism in combination with strand invasion to explain the rapid deletions they observed at yeast telomeres. Additional hints in favor of t loops in yeast emerged from studies of telomeric silencing (reviewed in reference 22), and Rap1p and TRF2 display intriguing similarities in biochemical activities that are relevant to telomere looping (17, 18, 21). Further parallel analysis of telomere length homeostasis in unicellular and multicellular organisms should reveal the diversity of solutions to this common regulatory problem.
ACKNOWLEDGMENTS
We thank members of the de Lange laboratory, especially Susan Smith and Jan Karlseder, for helpful discussions. Art Lustig is thanked for advice in the revision stage of this manuscript. Jason Lue is thanked for preparation of baculovirus-derived TRF1 and TRF2 proteins. R. A. Weinberg generously provided the hTERT cDNA. G.S. thanks K. Damm and A. Schnapp for useful discussions.
This work was supported by a grant to T.D.L. from the National Cancer Institute (CA76027) and an Irma T. Hirschl Career Scientist Award to T.D.L. T.D.L. is a recipient of the Burroughs Wellcome Fund Scholar Award in Toxicology. A.S. is supported by National Institutes of Health MSTP grant GM07739 to the Cornell/Rockefeller/Memorial Sloan-Kettering Tri-Institutional MD/PhD program. B.V.S. was supported by an HFSP fellowship, and A.B. was the recipient of a Comitato Promotore Telethon graduate fellowship.
REFERENCES
- 1.Ashley T, Ward D C. A “hot spot” of recombination coincides with an interstitial telomeric sequence in the Armenian hamster. Cytogenet Cell Genet. 1993;62:169–171. doi: 10.1159/000133464. [DOI] [PubMed] [Google Scholar]
- 2.Auffray C, Rougeon G. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 3.Barnett M, Buckle V, Evans E, Porter A, Rout D, Smith A, Brown W R A. Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res. 1993;21:27–36. doi: 10.1093/nar/21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi A. Characterization of DNA binding activities at vertebrate telomeres. Doctoral dissertation. New York, N.Y: The Rockefeller University; 1999. [Google Scholar]
- 5.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi A, Stansel R M, Fairall L, Griffith J D, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilaud T, Brun C, Ancelin K, Koering C E, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 8.Boulton S J, Jackson S P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broccoli D, Chong L, Oelmann S, Fernald A A, Marziliano N, van Steensel B, Kipling D, Le Beau M M, de Lange T. Comparison of the human and mouse genes encoding the telomeric protein, TRF1: chromosomal localization, expression, and conserved protein domains. Hum Mol Genet. 1997;6:69–76. doi: 10.1093/hmg/6.1.69. [DOI] [PubMed] [Google Scholar]
- 10.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 11.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 12.Counter C M, Avilion A A, LeFeuvre C E, Stewart N G, Greider C W, Harley C B, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells with express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Counter C M, Hirte H W, Bacchetti S, Harley C. Telomerase activity in human ovarian carcinoma. Proc Natl Acad Sci USA. 1994;91:2900–2904. doi: 10.1073/pnas.91.8.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lange T. Human telomeres are attached to the nuclear matrix. EMBO J. 1992;11:717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lange T, Shiue L, Myers R M, Cox D R, Naylor S L, Killery A M, Varmus H E. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam J P, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilson E, Laroche T, Gasser S M. Telomeres and the functional architecture of the nucleus. Trends Cell Biol. 1993;3:128–134. doi: 10.1016/0962-8924(93)90175-z. [DOI] [PubMed] [Google Scholar]
- 18.Gilson E, Muller T, Sogo J, Laroche T, Gasser S M. RAP1 stimulates single- to double-strand association of yeast telomeric DNA: implications for telomere-telomere interactions. Nucleic Acids Res. 1994;22:5310–5320. doi: 10.1093/nar/22.24.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracyclin-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffith J, Bianchi A, de Lange T. TRF1 promotes parallel pairing of telomeric tracts in vitro. J Mol Biol. 1998;278:79–88. doi: 10.1006/jmbi.1998.1686. [DOI] [PubMed] [Google Scholar]
- 21.Griffith J D, Comeau L, Rosenfield S, Stansel R M, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 22.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 23.Hanish J P, Yanowitz J, de Lange T. Stringent sequence requirements for telomere formation in human cells. Proc Natl Acad Sci USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 25.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L C, Coviello G M, Wright W E, Weinrich S L, Shay J W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 26.Kipling D, Cooke H J. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 27.Lansdorp P M, Verwoerd N P, van de Rijke F M, Dragowska V, Little M-T, Dirks R W, Raap A K, Tanke H J. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 28.Lee M S, Blackburn E H. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Lustig A J. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 30.Lingner J, Cech T R. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 32.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 33.Meyne J, Baker R J, Hobart H H, Hsu T C, Ryder O A, Ward O G, Wiley J E, Wurster-Hill D H, Yates T L, Moyzis R K. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990;99:3–10. doi: 10.1007/BF01737283. [DOI] [PubMed] [Google Scholar]
- 34.Morin G B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 35.Morin G B. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 36.Nugent C, Bosco G, Ross L, Evans S, Salinger A, Moore J, Haber J, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 37.Nugent C I, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 38.Polotnianka R, Li J, Lustig A. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- 39.Saltman D, Morgan R, Cleary M L, de Lange T. Telomeric structure in cells with chromosome end associations. Chromosoma. 1993;102:121–128. doi: 10.1007/BF00356029. [DOI] [PubMed] [Google Scholar]
- 40.Shore D. Telomerase and telomere-binding proteins: controlling the endgame. Trends Biochem Sci. 1997;22:233–235. doi: 10.1016/s0968-0004(97)01082-7. [DOI] [PubMed] [Google Scholar]
- 41.Shore D. Telomere length regulation: getting the measure of chromosome ends. Biol Chem. 1997;378:591–597. [PubMed] [Google Scholar]
- 42.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres [see comments] Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 43.Sprung C N, Reynolds G E, Jasin M, Murnane J P. Chromosome healing in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1999;96:6781–6786. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 45.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Blackburn E H. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–879. doi: 10.1093/emboj/16.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright W E, Piatyszek M A, Rainey W R, Byrd W, Shay J W. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]