Abstract
Among artisanal fermented beverages, kefir (fermented milk drink) and water kefir (fermented nondairy beverage) are of special interest because their grains can be considered natural reservoirs of safe and potentially probiotic strains. In the last years, several reports on Lacticaseibacillus paracasei (formerly Lactobacillus paracasei) isolated from both artisanal fermented beverages were published focusing on their health-promoting properties. Although this is not the predominant species in kefir or water kefir, it may contribute to the health benefits associated to the consumption of the fermented beverage. Since the classification of L. paracasei has been a difficult task, the selection of an adequate method for identification, which is essential to avoid mislabeling in products, publications, and some publicly available DNA sequences, is discussed in the present work. The last findings in health promoting properties of L. paracasei and the bioactive compounds are described and compared to strains isolated from kefir, providing a special focus on exopolysaccharides as effector molecules. The knowledge of the state of the art of Lacticaseibacillus paracasei from kefir and water kefir can help to understand the contribution of these microorganisms to the health benefits of artisanal beverages as well as to discover new probiotic strains for applications in food industry.
Keywords: Lacticaseibacillus paracasei, kefir, bioactive compounds, exopolysaccharide
1. Introduction
Fermented foods have played an important role in the human diet since the development of civilization and remain an integral part of local culture and traditions in many countries. A positive relationship between the consumption of fermented dairy products, health status and the intestinal microbiota has been reported. Fermented foods can contribute to health benefits by providing the consumer with beneficial microorganisms and nutritional benefits associated to changes performed in the food matrix during fermentation [1]. Some artisanal fermented foods contain several taxa and even different strains, with the population’s dynamic during processing being remarkably complex [2].
Among artisanal fermented beverages, kefir (fermented milk drink) and water kefir also known as “aquakefir” or “sugary kefir” (fermented non-dairy beverage) are of special interest because of their natural and artisanal production, which is compatible with sustainable technology. They have a common characteristic, the starter used for their production are kefir grains or water kefir grains that contain a complex microbiota immobilized in a polymeric matrix [3,4].
Due to their complex microbiota, kefir grains or water kefir grains can be considered as natural reservoirs of safe and potentially probiotic strains. Studies on kefir grain microbiota revealed that Lactobacillus kefiranofaciens, Lentilactobacillus kefiri and Lentilactobacillus parakefiri are the most representative species; however, also other species are described, including Lacticaseibacillus paracasei, Lactobacillus acidophilus, Lactobacillus delbrueckii subsp. bulgaricus, Lactiplantibacillus plantarum and Lactococcus lactis. More than 23 species of yeast have been identified as part of the kefir grain microbiota. These include Saccharomyces cerevisiae, S. unisporus, Candida kefyr and Kluyveromyces marxianus [5,6].
Water kefir grains have been less studied than those isolated from kefir, although they are also an important reservoir of highly competitive microorganisms. Some of the key microorganisms of water kefir fermentation are Lentilactobacillus hilgardii, Liquorilactobacillus nagelii, L. paracasei, Bifidobacterium aquikefiri, S cerevisiae and Dekkera bruxellensis [7].
Kefir and sugary kefir are described to exert beneficial effects on consumers’ health. The most relevant scientific findings on health benefits associated with kefir consumption were summarized by several authors [3,4,8,9]. The health promoting properties of artisanal fermented beverages could be attributed to the diversity of microorganisms that they contain [10].
Lacticaseibacillus paracasei (formerly Lactobacillus paracasei) is one of the species isolated from both artisanal fermented beverage’s starters. Although it is not the predominant species in kefir or water kefir, it may contribute to their health-promoting properties. It belongs to the genus Lacticaseibacillus (formerly Lactobacillus casei group (LCG)), which is composed of the closely related species L. casei, L. paracasei and L. rhamnosus, among others and contains several strains with a long history of apparently safe use in food and agricultural applications that have been studied for their health-promoting properties [11,12,13].
The present review will discuss some issues relating to the probiotic properties of L. paracasei and their effector molecules, focused on the role of exopolysaccharides (EPS). A special emphasis on strains isolated from artisanal fermented beverages will be performed highlighting the relevance of these products as sources for new probiotic strains for applications in food industry. Along the review, the taxonomic and genetic aspects of L. paracasei strains will be considered as well as the role of effector molecules associated with health-promoting properties.
2. Lacticaseibacillus paracasei: A Tour through the Evolution of Taxonomical Classification
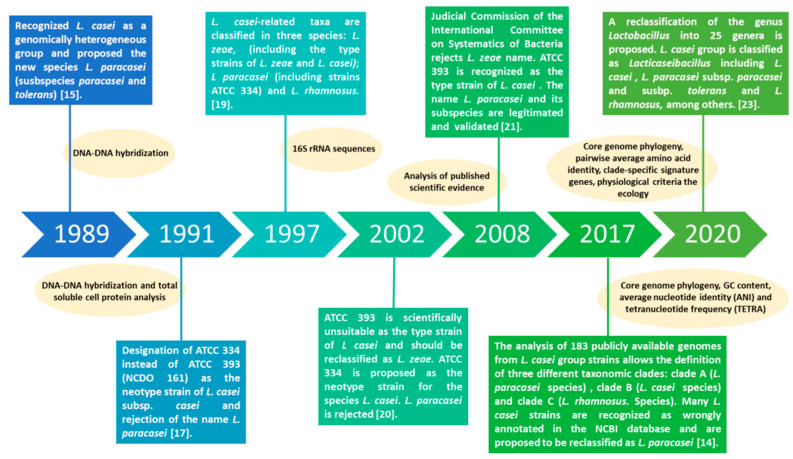
Due to the broad interest in L. casei group members for use in the food and pharmaceutical industries as probiotics, their genotypic and phenotypic properties have been extensively studied, and their classification and taxonomy have been discussed [14]. Unequivocally identification of L. casei group members has been a matter of extensive discussion since the initial description of the species by Collin et al. in 1989 [15]. Taking into consideration the heterogeneity in DNA–DNA hybridization assays and biochemical tests between different L. casei strains, the authors reclassified the group proposing a new species named L. paracasei with two subspecies L. paracasei subsp. paracasei and subsp. tolerans.
Since then, L. paracasei taxonomy has been convoluted together from the use of conventional phenotypic methods to the advance of molecular genotyping for determining evolution and phylogeny [14,16]. However, the consideration of L. paracasei as a novel species was first questioned on account of misunderstandings and discrepancies regarding the identification of the type strain [17,18,19,20]. Some authors suggested the rejection of the name L. paracasei and proposed that the strain designated as type strain L. casei ssp. casei ATCC 393 should, in fact, be reclassified as L. zeae and that L. paracasei strains should be reclassified under L. casei, with L. casei ATCC 334 as the neotype strain for this species [20].
However, these suggestions were not accepted by Judicial Commission of the International Committee on Systematics of Bacteria (2008) indicating that strain ATCC 393 was the type strain for L casei, rejecting the name L. zeae because it contravenes Rules 51b (1) and (2) of the International Code of Nomenclature of Bacteria and classifying ATCC 334 as a member of a different taxon named L. paracasei [21].
The advance in pan and core genome methods for determining evolutionary relationships provided new tools for taxonomy. An in-depth comparative genomic analysis of the L. casei group was performed by Wuyst et al. (2017) who proposed a classification of this group in three clades based on (a) differences in whole-genome Guanine and Cytosine (GC) content, (b) a core genome phylogenetic tree constructed on the alignment of 776 single-copy marker genes and (c) pairwise genome distances [14].
One clade represents the species of L. rhamnosus, including the type strain L. rhamnosus DSM 20021. The second clade contains L. casei and L. paracasei isolates and includes the L. paracasei type strain ATCC 334, while the third clade consist of L. casei and L. zeae isolates. However, since L. zeae isolates were reclassified as L. casei, this clade actually represents L. casei strains. Based on these results, most of the strains designated as either L. casei or L. paracasei subsp. paracasei in the literature are, in fact, in the same clade corresponding to the species named L. paracasei.
Salvetti et al. (2018) evaluated the relatedness of 269 species belonging primarily to the families Lactobacillaceae and Leuconostocaceae through the analysis of ribosomal proteins and housekeeping genes and the assessment of the average amino acid identity (AAI) and the percentage of conserved proteins (POCP) as the basis for reclassification [22].
By using a polyphasic approach (core genome phylogeny, (conserved) pairwise average amino acid identity, clade-specific signature genes, physiological criteria and the ecology of the organisms), Zheng et al. (2020) recently recommended the reclassification of the genus Lactobacillus into 25 genera that reflects the phylogenetic position of the microorganisms but also considers the ecological and metabolic properties. This new classification proposed the genus Lacticaseibacillus, which includes the species Lacticaseibacillus paracasei to name the formerly called Lactobacillus paracasei [23]. Figure 1 summarizes the evolution of classification and taxonomy of L. paracasei over the years with a non-exhaustive list of reports published.
Figure 1.
Timeline of the main events on evolution of classification and taxonomy of Lacticaseibacillus along with the methodology applied in each report. ATCC: American Type Culture Collection; NCDO. National Collection of Food Bacteria.
3. Identification and Classification of Lacticaseibacillus paracasei
Since the three species that belongs to the formerly called L. casei group are closely genotypically related, the selection of an appropriate methodology for their unequivocally classification is necessary [11].
The selection of an adequate method for identification is essential to avoid mislabeling in products, publications and some publicly available DNA sequences [11,24,25]. Heterogeneities found in the 16S rRNA genes of several strains of L. casei/paracasei was one of the difficulties encountered in obtaining clear-cut taxa within the L. casei/paracasei complex [26]. Furthermore, a response to niche adaptation led to loss of redundant genes followed by the acquisition of genes by horizontal gene transfer, which results in heterogeneity depending on the environment [24,27]. In fact, studies on L. paracasei strains isolated from cheese demonstrated that heterogeneities are also found even between strains isolated from the same niche [28].
Thus, several methods to unequivocally classify L. paracasei were proposed and were reviewed in detail [11,25]. Among them, phenotypic methods, such as a traditional fermentation test accompanied by a PCR (polymerase chain reaction) DNA-sequencing method for the analysis of 16S ribosomal RNA (16S rRNA) genes were some of the first methods proposed for classification of the L. casei group [19,29]. Iacumin et al. (2015) proposed the use of species-specific PCR and high-resolution melting (HRM) analysis. They applied these methods to evaluate 201 strains from international culture collections belonging to the L. casei group and found that 46 strains were different from their previous classification [30]. Huang and Lee (2011) proposed used dnaK sequencing to complement 16S rRNA for the classification of the L. casei group [31]. Multiplex PCR assay based on mutL was used to effectively distinguish L. casei, L. paracasei and L. rhamnosus [32].
Wuyst et al. (2017) performed a comparative analysis of 183 public available L. casei group genome assemblies and also found some inconsistencies in the taxonomy and reclassified all of the genomes accordingly [14]. Based on their results, the authors suggested the use of whole-genome ANI compared to L. casei group type strains to differentiate between the species L. casei and L. paracasei [14]. They also suggested that the use of the heme-dependent catalase or the superoxide dismutase gene as marker genes could be used for correct differentiation of these species if sequencing of the 16S rRNA gene leads to the identification of a member of the species L. casei/L. paracasei.
Huang and Huang (2018) successfully differentiated L. paracasei strains even into subspecies by using level matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDITOF MS) and an analytical in-house database (IHDB) accompanied with bioinformatics tools [33]. They used housekeeping gene sequencing and species-specific PCR to validate the MALDI-TOF MS platform. Housekeeping genes, such as the phenylalanyl t-RNA synthase alpha subunit (pheS) and RNA polymerase alpha subunit (rpoA), were also used to distinguish closely related species [34] as well as multilocus sequence analysis (MLSA) based on concatenated sequences of three housekeeping genes (dnaK, pheS and yycH) [35].
Silveraju et al. (2020) evaluated the pheS genes from a total of 266 type strains of lactobacilli for which genomes are publicly available. The extracted pheS sequences were compiled in a database (pheS-DB) to allow a comprehensive and confident characterization of the community diversity and structure of lactobacilli at the species level [36]. According to Huang et al. (2018) combining species- and subspecies-specific identification methods, housekeeping gene sequences and MALDI-TOF MS spectral pattern analysis resulted as quick and accurate methods for identifying strains in the L. casei group at the species and subspecies levels [25]. However, it should be taken into consideration that tests based on phenotypic markers should be performed under strictly controlled growth conditions.
Methods used for identification of L. paracasei strains isolated from traditional kefir or water kefir also varied. Most of them used 16S rRNA gene sequencing alone [37,38,39] or 16S rRNA gene sequencing accompanied by other methods, such as the use of species-specific primers [40,41] or ribosomal DNA restriction analysis (ARDRA), Repetitive Element Palindromic PCR (REP-PCR) fingerprint and Random Amplified Polymorphic DNA (RAPD) [42]. MALDI-TOF (Matrix-assisted laser desorption ionization time-of-flight) mass spectrometry was also used for the identification of strains isolated from kefir [43] as well as a polyphasic approach combining phenotypic and genotypic methods based on (GTG)5-R fingerprint and sequence analysis of the housekeeping gene encoding the α-subunit of bacterial phenylalanyl-tRNA synthase (pheS) [44]. Recently, Kim et al. (2020) performed a comparative genomic analysis of the pan genome and core genome of L. casei group species to designed species-specific primers for a differential identification of L. paracasei with real time PCR [45].
The diversity of methods developed for the identification of L. paracasei indicates that its differentiation from the closely related species is a difficult task and that a combined approach should be performed for an unequivocally classification. However, the advance in genome analysis contributes to the better understanding of L. paracasei phylogenetic relationship that would improve the selection of identification methods.
4. Lacticaseibacillus paracasei Health-Promoting Properties and the Effector Molecules Associated
The mechanisms related to the health effects displayed by probiotics (live microorganisms that, when administered in adequate amounts, confer a health benefit on the host [46]) are of interest among researchers. Probiotics can affect human/animal health in various ways, such as inhibiting intestinal pathogenic microorganisms, modulating the immune response, reducing serum cholesterol concentration, or exerting antioxidant activity, among others. Such effects may be due to the presence of the microorganism itself or to metabolites that it produces and that in certain cases releases into the environment (bacteriocins, exopolysaccharides and organic acids, for example).
Many are the benefits reported in the literature for L. paracasei strains [11,47]. These include antimicrobial and antibiofilm activity; immune system stimulation; anti-inflammatory, antioxidant, anti-obesity and anti-proliferative/proapoptotic, lipid metabolism improving, hypocholesterolemic and stress modulator effects; and the enhancement of intestinal bacterial microbiota; among others. Health promoting properties can be ascribed to the consumption of the viable microorganisms that generate molecules in situ that also contribute to well-being, as well as the metabolites produced during fermentation are associated with the changes made in the matrix of the foods that contain them.
Given that most effects of probiotics are strain-specific [48], the intrinsic high heterogeneity existing among L. paracasei strains makes this species an optimal source for the selection of novel candidate probiotic strains possessing unique technological and health-promoting traits [49]. Table 1 lists the most recent reports on health functional properties attributed to different strains isolated from different sources with a special focus on kefir and other traditional fermented products indicating if the study was performed with whole product, microorganisms or cell free supernatant.
Table 1.
Health functional properties attributed to Lacticaseibacillus paracasei strains isolated from different sources.
| L. paracasei Strain | Origin | Health Functional Properties | Biological Activity * | Reference |
|---|---|---|---|---|
| Lpc-37® | DuPont de Nemours, Inc. trademark | Stress, mood and well-being modulation | Reduction of perceived stress and improvement of biomarkers related to stress in a clinical trial. | [13] |
| GKS6 | Healthy infant feces | Antioxidant activity | Delay in the aging process in mice by enhancement of antioxidants activity, resulting in lower oxidative damage. | [50] |
| KBL 382 | Korean healthy feces | Anti-inflammatory activity—Microbiota modulation | Reduction of INF-γ, IL-4, IL-6, TNF and IL-17A levels and increase of anti-inflammatory cytokine IL-10 and CD4+CD25+Foxp3+ T regulatory cells in mesenteric lymph nodes levels. Improvement of cell tight junction and mucus thickness. Increase in bacterial diversity of fecal microbiota. | [51] |
| Ameliorates atopic dermatitis—Immunomodulatory activity—Microbiota modulation | Decrease in T helper cytokines and increase IL-10 and TGF-β production in skin tissue. Increase in the proportion of CD4+ CD25+ Foxp3+ T regulatory cells in mesenteric lymph nodes and changes in the composition of gut microbiota of oral treated mice. | [52] | ||
| 28.4 | Oral cavity of a caries-free individual | Immunomodulatory activity | Bacteria cells have antifungal activity against planktonic cells, biofilms and persisted cells of Candida auris. | [53] |
| Postbiotic elements (free-cell supernatant) inhibit C. auris in vitro and protect Galleria mellonella infected with C. auris enhancing its immune status. | ||||
| L1 | Sweet potato sour liquid | Microbiota modulation | Increase in the abundance of functions related to carbohydrate and protein metabolism and fatty acid biosynthesis in the intestinal microbiota. Improvement in the growth performance of chicken. | [54] |
| FZU103 | Traditional Hongqu rice wine | Improvement of lipid metabolism | Regulation of lipid metabolic pathways of pathogen free mice feed with high fat diet (fatty acid degradation, fatty acid elongation, unsaturated fatty acids biosynthesis, glycerolipid, glycerophospholipid and arachidonic acid metabolism, primary bile acid biosynthesis and riboflavin metabolism). Regulation of the expression of hepatic genes involved in lipid metabolism and bile acid homeostasis and promotion of fecal excretion of bile acids. | [55] |
| B-14 | Traditional fermented dairy product | Antiproliferative—Proapoptotic effects | Downregulation or upregulation of key genes in the cell proliferation, cell survival and intrinsic and extrinsic apoptosis pathways. | [56] |
| ZFM54 | New-born infant’s feces | Protection against foodborne pathogens | In vitro inhibition of Salmonella typhimurium, Micrococcus luteus and Listeria monocytogenes by production of a pore forming bacteriocin ZFM54. | [57] |
| M7 | Human breast milk | Hypocholesterolemic—Antioxidant activity—Protection against pathogens | Antibiofilm potential of EPS against several human pathogens. | [58] |
| NTU 101 | Human feces | Immunomodulatory—Antioxidant activity | Induction of pro-inflammatory molecules (nitric oxide, IL-6, TNFα and IL-1β) and phagocytic activity in murine macrophages Raw 264.7. Antioxidant activity (scavenging of 1,1-Diphenyl-2-picrylhydrazyl radicals, inhibition of linoleic acid peroxidation, reducing power, chelating ability on ferrous ions). | [59] |
| M5L | Kumiss | Antiproliferative activity | Apoptotic effect on human colorectal adenocarcinoma cell line HT-29 mediated by induction of oxidative stress and endoplasmic reticulum stress. | [60] |
| DG | Commercial product | Immunomodulatory activity | Induction of pro-inflammatory cytokines TNF α, IL-6 and the chemokines IL-8 and CCL20 in human monocytic cell line THP-1. | [49] |
| IJH SONE | Fig leaf | Anti-inflammatory activity | Anti-inflammatory effect mediated by inhibition of hyaluronidase activity. | [61] |
| Antiallergenic activity | Antiallergenic effect evidenced by oral and topic administration against contact dermatitis in mice. | [62] | ||
| LB-8 | Feces | Modulation of intestinal microbiota | Bifidogenic effect in vitro. | [63] |
| CIDCA 8339, 83120, 83121, 83123, 83124 | Kefir grains (Argentine) | Protection against pathogens | Adhesion to Caco-2 cells and prevention of Salmonella association/invasion in vitro depending on surface properties of the strain. | [64] |
| CIDCA83123, 83124, 8339 | Kefir grains (Argentine) | Immunomodulatory activity | Modulation of the intestinal epithelial innate immune response by viable whole cell. | [65] |
| Immunomodulatory activity | Fermented milk supernatants downregulate the induced innate immune response in intestinal and gastric cells, with lactate as the metabolite responsible of this effect. | [12,66] | ||
| CIDCA 83124, 8339 | Kefir grains (Argentine) | Microbiota modulation and changes in SCFA profile | EPS 8339 and EPS 83124 modify the microbiota by reducing the enterobacteria and increasing the production of propionic and butyric acid. | [67] |
| CIDCA 8339 | Kefir grains (Argentine) | Gastroprotection | Adhesion to AGS gastric cell line. The strain consumption shows a gastroprotective effect in an acute gastritis murine model. | [66] |
| Ž2 | Kefir grains (Tibet) | Immunomodulatory activity | Increase in the proportion of all T cells (CD3+), CD4+ lymphocytes and the ratio of CD4+:CD8+ cells in vivo and increase in the gene expression for mucins (MUC-1 and MUC-2) and IgA at intestinal level. | [43] |
| KL1-Liu | Kefir grains (Tibet) | Protection against pathogens | Mixed probiotic of L. paracasei KL1-Liu (EPS producer) and L. plantarum Zhang-LL reduces the mortality of pullorosis in chicks. | [68] |
| MRS59 | Kefir grains (Brazil) | Adhesion to intestinal epithelial cells | Adhesion to human Caco-2 epithelial cells, bacteriocin production. | [42] |
| Antimicrobial-Antioxidant activity | Antagonistic activity against food pathogens by bacteriocin-like inhibitory substance and antioxidative activity of cell extract. | |||
| FX6 | Kefir grains (Tibet) | Antimicrobial activity | Bacteriostatic effect on Pseudomonas putida due to the increase of bacterial membrane permeability and ability of the antimicrobial substance to affect the synthesis of protein and bind to genomic DNA. | [69] |
| Antimicrobial activity | Bacteriocin F1 with a wide antimicrobial spectrum. | [38] | ||
| Antimicrobial activity | Antibacterial effect on Serratia in chicken breast during refrigerated storage. | [70] | ||
| LAB2, LAB4 | Kefir grains (Iran) | Protection against pathogens | Neutralized cell-free supernatant inhibits the growth and the biofilm formation by uropathogenic E. coli. | [37] |
| SP5 | Kefir grains (Russia) | Antiproliferative activity | Reduction of cancer cell proliferation in vitro in a time- and concentration-dependent manner. | [40] |
| SP5 | Antioxidant activity | Fermentation metabolites produced by the breakdown of anthocyanins and other larger-in-size phenolic compounds present in chokeberry juice, leading to increased levels of total phenol content. | [71] | |
| AGR4 | Kefir grains (Greece) | Antiproliferative activity | Time- and dose-dependent antiproliferative activity of HT-29 cells and human melanoma cell line A375. | [41] |
| LMG R40086, LMG R39998, LMG R40122, LMG R40006 | Water kefir grains(Belgium) | Adhesion to intestinal epithelial cells | Adhesion ability to Caco-2 cells. | [64] |
| CT12 | Water kefir grains (Mexico) | Antimicrobial-Antioxidative activity | Antimicrobial, antifungal and antioxidant capacity of cell-free supernatant. | [39] |
* Colors indicates that assays were performed with whole L. paracasei cell (green), cell-free supernatant or non-viable cells (yellow), whole fermented product (light blue) and EPS fraction (pink).
Among L. paracasei that have been studied for their beneficial effects on consumer health, there are several isolates from kefir of Argentina, Brazil, Greece, Iran, Russia and Tibetan grains and water kefir from Mexico and Belgium. Most of the strains were obtained from kefir and only a few from water kefir. Protection against pathogens, immunomodulation and anti-inflammatory, prebiotic, antioxidant and antiproliferative activity are the beneficial properties that have been attributed to these microorganisms.
What is the mechanism by which L. paracasei confers health benefits on the host? What are the molecules involved in such effects? Although in recent years much effort has been made to study this, the answer is not conclusive, and there remains much to be elucidated. In some studies, a biological effect mediated by the viable microorganism was evidenced (indicated in green in Table 1), while, in other cases, the inanimate microorganisms and/or their components, known as postbiotics [72,73] as well as their metabolites, were responsible for the beneficial effect on health (indicated in yellow in Table 1). On certain occasions, an effect due to the whole fermented product (marked in light blue in Table 1) was described, and, in several reports, a beneficial effect of the EPS was demonstrated (marked in pink in Table 1).
As mentioned above, the probiotic effect of L. paracasei strains isolated from kefir could be associated with the presence of the microorganism. For example, viable whole cells of L. paracasei CIDCA 8339, CIDCA 83123 and CIDCA 83124 isolated from Argentinean kefir grains have the capacity to adhere to epithelial intestinal cells and protect against Salmonella invasion as well as immunomodulate and protect the gastric epithelium [64,66,74]. In the same direction, Karaffová et al. (2021) described that the strain I2 isolated from Tibetan kefir grains increased the proportion of all T cells (CD3+), CD4+ lymphocytes and the ratio of CD4+:CD8+ cells in vivo and increase the gene expression for mucins at the local intestinal level [43].
Otherwise, the probiotic effect of L. paracasei strains isolated from kefir could be associated with an effector molecule. The evidence provided in recent years has led to the evaluation of lactic acid as a health-promoting agent in fermented foods [75]. This acid, as the main product of the metabolism of lactic acid bacteria, influences its probiotic action, playing an important role both in the control of pathogens and in the immunomodulatory activity. It has been demonstrated that lactate can regulate critical functions of macrophages and dendritic cells and modulate the inflammatory activation of epithelial cells as well.
In addition to the organic acids consumed with the fermented foods, the acids produced by lactobacilli in situ can contribute to the probiotic action. It was described that the L. paracasei strains CIDCA 8339, 83123 and 83124 and isolated from kefir can adhere to Caco-2 cells and mucin after passage through simulated gastrointestinal tract [74] giving to the lactobacilli the possibility to produce lactate in the gut epithelium microenvironment. Intraluminal levels of lactate derived from fermentative metabolism of lactobacilli were shown to modulate the inflammatory response in the intestinal mucosa.
Furthermore, lactate can be used by the gut microbiota to produce acetate, propionate and butyrate; short chain fatty acids (SCFA) highly associated to gut health that have been proven to down regulate the pro-inflammatory responses in intestinal epithelial and myeloid cells [76,77]. Among the molecular mechanisms responsible for these functions, histone deacetylase dependent-modulation of gene expression and signaling through G-protein-coupled receptors have been described [77]. In addition, Bengoa et al. (2020) demonstrated that the in vivo gastroprotective effect exerted by L. paracasei CIDCA 8339 involves not only the direct interaction of the microorganism with the gastric mucosa but also the in situ production of other metabolites that could modulate the inflammatory response [66].
Regarding other effector molecules described for kefir isolated L. paracasei strains, exopolysaccharides, bacteriocins and other metabolites can be mentioned. Miao et al., 2014 reported a bacteriocin F1 produced by L. paracasei FX6 strain with a wide antimicrobial spectrum [38], and Leite et al. (2015) detected a bacteriocin-like inhibitory substance produced by L. paracasei MRS59 strain from Brazilian kefir grains [42].
Recently, Duan et al. (2020) found an antimicrobial substance from L. paracasei FX6 that was able to interfere with important cellular functions through affecting the synthesis of protein and binding to genomic DNA [69]. Even more, Ghane et al. (2020) showed that neutralized cell-free supernatant of L. paracasei LAB2 and LAB4 strains inhibited the growth and the biofilm formation by uropathogenic E. coli [37].
On the other hand, a relevant antimicrobial, antifungal and antioxidant capacity of cell-free supernatant of L. paracasei CT12, an isolate from Mexican water kefir grains, was described by Romero-Luna et al. (2020) [39]. Additionally, a significant reduction of cancer-cell proliferation in vitro was evidenced with the cell-free supernatant of L. paracasei SP5, isolated from Russian kefir grains [40] and L. paracasei AGR4 isolated from Greece kefir grain [41]. However, the nature of the compounds responsible for these effects has not been yet determined.
5. Exopolysaccharides as Effector Molecules Associated to Lacticaseibacillus paracasei Health Benefits
As mentioned above, one of the effectors molecules ascribed to the health-promoting properties of L. paracasei are extracellular polysaccharides, which can be secreted to the environment in the form of slime EPS or can be attached to the cell surface in the form of capsule (CPS) [78]. According to their structure, EPS are classified into two groups: homopolysaccharides (HoPS) and heteropolysaccharides (HePS) [79]. The first ones are composed of a single type of monosaccharide either glucose (α-glucans and β-glucans) or fructose (β-fructans).
Meanwhile, heteropolysaccharides, which constitute the majority of EPS produced by lactic acid bacteria (LAB), are composed of a backbone of repeating subunits branched or unbranched that consist of three to eight monosaccharides, commonly rhamnose, fructose, galactose or glucose. In some cases, HePS could contain some modifications, such as acetylations, pyruvylations and phosphorylations [80,81,82]. There are substantial differences in the number of enzymes and steps implied in the synthesis and export out of the cell between HoPS and HePS.
HoPS are synthetized completely outside the cell by the activity of an extracellular glycosyltransferase that uses sucrose as a substrate. Instead, HePS biosynthesis is a much more complex process. First, the cell must internalize monosaccharides and disaccharides from the growth media through the PEP-PTS system, although this can also occur by active transport. Once inside the cell, the sugar can be used in catabolic pathways (as sugar-6-phosphate) or for polysaccharide synthesis (as sugar-1-phosphate). Before polymerization begins, the sugar is activated by the addition of uridine diphosphate (e.g., UDP-glucose and UDP-galactose) or deoxithymidine diphosphate (e.g., dTDP-rhamnose), thus, forming sugar nucleotides.
Then, a priming glycosyltransferase (EpsE) binds the first sugar to a lipid carrier, followed by the action of other glycosyltransferases that continue with the saccharide unit formation. Finally, the undecaprenol-lipid bound with the repeated unit is translocated through the membrane by a flippase, the repeated units are joined by a polymerase, and other enzymes control the chain length and remove the lipid carrier [83].
Genes involved in EPS synthesis in different LAB strains were usually designated by alphabetical order of occurrence in a given locus [83]. In this way, genes codifying for different kind of enzymes were named equally leading to confusion in understanding the genes involved in EPS synthesis of different strains. Zeidan et al. (2017) described a general cluster structure for HePS synthesis in LAB, which was organized with five conserved genes at the 5′ end (epsABCDE) followed with a variable region that contains the flippase (wyx), polymerase (wzy) and glycosyltranfesases [83].
EPS clusters are conformed by four groups of genes that codify the proteins involved in assembly, modulation, glycosyltransferases and NDP-sugar biosynthesis. The enzymes involved in sugar nucleotides formation and monosaccharide decoration are not exclusively of polysaccharide synthesis pathway and can be codified within EPS cluster or not [83,84].
Smokvina et al. (2013) analyzed the pangenome, core genome and variable genome from L. paracasei by sequencing 34 L. paracasei strains from different niches and using complete genome sequences of three reference strains [27]. From this study, they found four different EPS clusters that were either chromosomally or plasmid encoded. EPS-1 or EPS-2 clusters were not detected simultaneously in the L. paracasei genomes studied. The third EPS cluster (EPS-3) was identified in most of the studied strains (29), and EPS-4 was detected in only one strain.
The presence of EPS-1 and EPS-3 suggests that rhamnose is an important constituent of EPS in L. paracasei strains since genes that encode rhamnosyltransferases and enzymes that catalyze the conversion of D-glucose-1-phosphate into dTDP-L-rhamnose are codified in those clusters [27]. Within the genome of L. paracasei DG, two EPS clusters were identified. Only one of them, EPS-a, was conserved in other five L. paracasei strains and L. casei Shirota. EPS-b, contained about 14 EPS-related genes, of which, 7 were only present in the DG strain and codified for putative glycosiltransferases, a flippase and a phosphotransferase [49].
Two EPS clusters were codified in the chromosome of L. paracasei IJH-SONE68 [61], while the LC2W strain presented only one EPS cluster with a similar structure to the cps cluster described in L. casei Shirota [85,86]. Although the EPS clusters of the mentioned L. paracasei strains presented certain differences in their genetic organization, most of them presented genes that encode enzymes that catalyze the conversion of D-glucose-1-phosphate into dTDP-L-rhamnose suggesting that rhamnose is an important constituent of EPS in these strains.
Regarding the L. paracasei strains isolated from kefir there is no information about the number of EPS clusters or the organization of the genes involved in EPS production. Up to the present, the complete genomic sequence of only one L. paracasei strain isolated from Tibetan kefir (submitted by Nanjing Agricultural University, Nanjing, Jiansu, China) named ZY-1 was available in the NCBI (National Center for Biotechnology Information) database and appears to be compound by the chromosome and four plasmids (pLPZ1-4).
Although the physiological role of EPS in the bacteria is not completely understood, it is thought to play an important role in the protection of microbial cells against harsh factors and conditions, such as toxic metals, innate immune factors, phage attack, osmotic stress and desiccation [81,82]. Moreover, this EPS layer could provide additional protection against low pH, bile salt, gastric and pancreatic enzymes, thus, guaranteeing the survival of orally administrated probiotics through the gastrointestinal tract [81].
In this context, it was demonstrated that the removal of the slightly attached EPS through successive washes with PBS led to a reduction in the tolerance of L. paracasei CIDCA 8339, CIDCA 83123 and CIDCA 83124 isolated from kefir grains to gastrointestinal conditions, supporting the protective role of EPS against stress conditions [65]. Similarly, Stack et al. (2010) demonstrated that the beta-glucan-producing L. paracasei NFBC 338 isolated from the human gastrointestinal tract showed higher tolerance to both technological and gastrointestinal stresses than the original non beta-glucan-producing strain [87].
Moreover, this surface polymer is involved in aggregation, biofilm formation and interaction of the lactobacilli with intestinal epithelial cells [81,88]. EPS from LAB are surface molecules that directly interact with intestinal mucosa exerting strain-dependent negative or positive effects in adhesion [89]. The removal of the EPS layer from L. paracasei DG did not significantly affect its adhesion to Caco-2 cells [49].
Otherwise, L. paracasei CIDCA 8339, CIDCA 83123 and CIDCA 83124 increased their ability to adhere to mucin and intestinal epithelial cells in vitro after the passage through gastrointestinal conditions that could be ascribed to induction of EPS synthesis. Proteomic analysis suggested that gastrointestinal stress induced changes in the expression of enzymes involve in EPS production, which could be responsible for the differences observed in the adhesion properties of lactobacilli [74].
Similarly, when comparing the adhesion ability of EPS-producing L. paracasei subsp. paracasei BGS J2-8 isolated from natural dairy product with its non-EPS producing mutant to Caco-2 and HT-29 intestinal cells, it was observed that the absence of EPS led to a significant reduction in the adhesion to both cells lines [88]. Probiotics, which are usually administered orally, must survive the gastrointestinal conditions in order to reach and colonize the gut. In this context, the production of EPS could imply an important advantage for potentially probiotic strains to display their health effects in situ.
In addition to their exceptional physicochemical properties, EPS present in foods constitute relevant bioactive metabolites with underlying biological activities that significantly contribute to the healthy properties of the final product [90]. The biological activities evidenced with L. paracasei EPS included antioxidative, anti-bacterial, cholesterol-lowering, immunoregulatory, anti-tumor, anti-inflammatory and prebiotic properties (Table 1).
High levels of certain blood lipids represent a risk factor for the development of cardiovascular diseases, one of the major causes of deaths worldwide [82]. In this context, the use of probiotics or probiotics’ metabolites, such as EPS emerge as an alternative to reduce blood cholesterol levels through diet. Bhat et al. (2019) demonstrated the cholesterol-reducing potential of EPS synthetized by L. paracasei M7, a strain isolated from breast milk.
From the hypocholesterolemic effect, this EPS displayed antioxidant and hydroxyl radical scavenging activity [58]. In the same direction, the EPS synthetized by L. paracasei subsp. paracasei NTU 101, a strain isolated from human feces, showed potential antioxidant activity evidenced by its ability to reduce linoleic acid peroxidation, its reducing power and its chelant and free radical scavenging activity [59].
On the other hand, the induction of oxidative stress and the consequent endoplasmic reticulum stress mediated by L. paracasei subsp. paracasei M5L EPS is responsible for the apoptotic effect attributed to this biomolecule on human colorectal adenocarcinoma cell line HT-29 [60]. Considering this, EPS that induced ROS production in malignant cells could eject a beneficial effect in patients that suffer from cancer diseases as a complement to traditional therapy.
The ability of EPS from L. paracasei strains to prevent adhesion to intestinal cells [88] and to reduce biofilm formation of pathogenic bacteria [58] has also been reported in the literature contributing to the treatment and prevention of infectious diseases produced by biofilm-forming pathogens [91].
Moreover, some EPS have the ability to interact with the immune system. However, the broad structural diversity among EPS may influence their recognition by receptors of the immune system resulting in different effects. On the other hand, Liu et al. (2011) demonstrated that L. paracasei subsp. paracasei NTU 101 EPS stimulated the growth of murine macrophages Raw 264.7 and increased the levels of pro-inflammatory molecules, such as IL-1 β, TNF α, IL-6 and nitric oxide, which consequently led to an induction of phagocytic activity [59].
Similarly, the rhamnose-rich EPS synthetized by L. paracasei DG enhanced the expression of pro-inflammatory cytokines TNF α, IL-6 and the chemokines IL-8 and CCL20 in human monocytic cell line THP-1, ejecting an immunostimulatory effect [49]. On the contrary, Noda et al. (2018) demonstrated the in vitro the anti-inflammatory effect of L. paracasei IJH SONE 68 EPS (isolated from fig leaf) mediated by the inhibition hyaluronidase [61].
Furthermore, the authors studied the potential used of this EPS as anti-allergenic against contact dermatitis using a picryl-chloride-induced delayed-type (type IV) allergy mice model. An anti-allergic effect was evidenced through oral administration as well as topic administration of this EPS, being the last effect likely mediated by the inhibition of the hyaluronidase activity but the mechanism is not described [62].
The prebiotic effect of EPS from L. paracasei strain has also been evidenced. Prebiotics are defined as “substrates that are selectively utilized by host microorganisms conferring a health benefit” [92]. Due to their complex structure, EPS produced by LAB can resist gastric and intestinal degradation reaching the intestinal level were they act as a substrate for commensal bacteria, stimulating the development of beneficial microorganisms and the production of bioactive metabolites, such as lactate and SCFA, which are involved in different cell mechanisms that play a key role in host health [81,82].
Traditionally, the targets of prebiotic action were mainly Lactobacillus and Bifidobacterium, genera with recognized probiotic properties [93]. Sarikaya et al. (2015) demonstrated that L. paracasei subsp. paracasei LB-8 EPS has the ability to modulate the intestinal microbiota by promoting the growth of Bifidobacterium breve in levels comparable to that of the known prebiotic inulin [63]. However, currently, the concept of prebiotics has expanded to include the modulation of other genera, in particular, those involved in the production of SCFA, such as Eubacterium, Propionibacterium, Roseburia and Akkermansia, among others [92,93].
The prebiotic potential of EPS synthetized by L. paracasei CIDCA 8339 and CIDCA 83124 has been studied in vitro evidencing that both polymers are metabolized by infant fecal microbiota leading to changes in the fecal microbiota profile and enhancing the production of SCFA, such as propionic and butyric acids [67]. In this context, EPS produced by LAB constitute an interesting alternative to establish a widely diverse intestinal microbiota usually associated with a healthy state. Figure 2 summarizes the health-promoting effects ascribed to EPS produced by different L. paracasei strains.
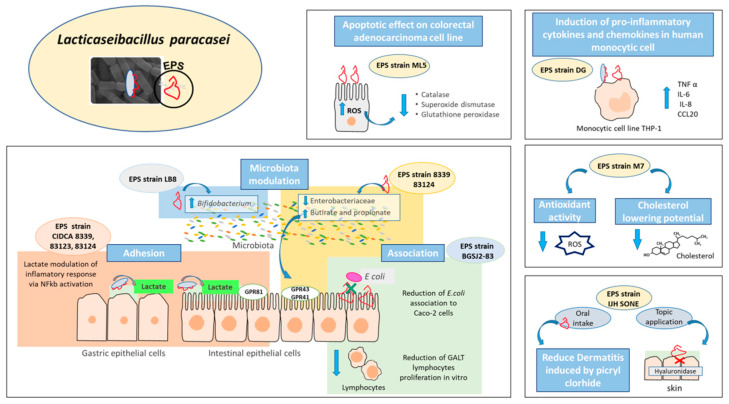
Figure 2.
Beneficial effects and mechanisms ascribed to EPS isolated from L. paracasei strains. At the gastrointestinal level, EPS produced by the BGSJ2-83 strain reduced the association of E. coli to Caco-2 cells and reduced GALT lymphocyte proliferation in vitro. EPS from the LB8 strain modulated the microbiota composition due to a bifidogenic effect; meanwhile, EPS from CIDCA 8339 and CIDCA 83124 strains reduced Enterobacteriaceae. In addition, EPS isolated from CIDCA 8339 and 83124 produced a change in the concentration of short chain fatty acids by increasing the butyrate and propionate levels. EPS is involved in the bacteria association to epithelial cells where the in situ production of lactate modulated the inflammatory response via NFkb activation. Additionally, EPS from MLS strain exerted an apoptotic effect on the human colorectal adenocarcinoma cell line HT-29. The oral intake of EPS produced by IJH-SONE reduced dermatitis induced by picryl-clorhide, and the topical application of EPS reduced the inflammation in mice. EPS produced by the DG strain induced pro-inflammatory cytokines and chemokines in the human monocytic cell line THP-1. EPS isolated from M7 strain presented anti-inflammatory activity and reduced cholesterol levels. ROS: reactive oxygen species.
As described above, many studies of EPS from L. paracasei strains are described in the literature. However, when it comes to strains isolated from kefir and water kefir, only a few reports have been published. These include the EPS-producing L. paracasei KL1-Liu strain isolated from Tibetan kefir that ejects a protective effect against Salmonella pullorum infection in chicks when administered in combination with L. plantarum Zhang-LL [68]. Nevertheless, it is not possible to attribute this health benefit exclusively to the strain or its EPS.
Although the EPS structure of this strain has been partly described [94], the EPS cluster genes organization has not been studied in detail yet. Similarly, L. paracasei CIDCA 8339, CIDCA 83120, CIDCA 83121, CIDCA 83123 and CIDCA 83124 (all EPS-producing strains isolated from kefir grains) significantly reduce the invasion of Salmonella to Caco-2/TC-7 intestinal epithelial cells. However, the role of EPS has not been elucidated.
Moreover, EPS from L. paracasei CIDCA 8339 and CIDCA 83124 have been characterized not only to define their biological activity (effect on the innate immune response and prebiotic effect) but also evidencing the role of the biopolymer in the protection of the probiotic against gastrointestinal conditions and its adhesion to intestinal cells. These properties are of great relevance for the maintenance of the viable microorganism close to the gastrointestinal epithelial cells to allow a deep cross talk between them producing in situ effectors molecules, such as lactate, bacteriocins and exopolysaccharides that may contribute to create a health-promoting microenvironment.
6. Conclusions
As was demonstrated by the literature reviewed, Lacticaseibacillus paracasei (formerly Lactobacillus paracasei) is one of the species isolated from artisanal fermented beverages, such as kefir (fermented milk drink) and water kefir (fermented non-dairy beverage), that is gaining attention to the scientific community as novel strains for application in the food industry. When a new isolate is obtained, the first challenge is the selection of an adequate method for identification to avoid mislabeling in products, scientific reports and the publication of DNA sequences. A special focus must be to unequivocally identify L. paracasei strains and differentiate from other species of the Lacticaseibacillus genus.
Recent advances in molecular data on the genes and their expression by using metagenomics, transcriptomics and metabolomics not only allow for a better understanding of the phylogeny of this group contributing to classification and identification but also the knowledge of the ability to synthesize molecules that could be involved in its probiotic function. The knowledge of the mechanisms involved in beneficial properties of L. paracasei strains from artisanal products, such as kefir or water kefir, would contribute to understand their role in the fermented beverage and to develop products with novel strains based on scientific evidence of their health-promoting effects.
As was described, biological effect may be due to the presence of either the viable bacteria non-viable bacteria/rest of cells (postbiotic), or metabolites released from L. paracasei into the environment. L. paracasei cell-free supernatants have immunomodulatory and gastroprotective properties ascribed to the presence of lactate, a common effector molecule of all fermented products.
In addition, the inhibitory properties against a range of pathogenic microorganisms could be exerted by the presence of lactate or owing to the production of bacteriocins by certain L. paracasei strains (strain-specific effectors). Health properties attributed to the presence of the viable microorganism could be associated to the molecules expressed on its surface. However, little information has been reported about the effector molecules associated with beneficial effects and/or the mechanisms of action.
Considering the revised reports, extracellular polysaccharides appear to be relevant effector surface molecules in L. paracasei. Although the study of EPS production genes and the biological activity from many L. paracasei strains is reported in the literature, it is noteworthy that little is known regarding the chemical characterization of these macromolecules as well as the relationship between their structure and technological and health functionality.
In light of the foregoing, it can be concluded that kefir grains and water kefir grains are significant sources for the isolation of new, safe L. paracasei strains. In particular, EPS-producing strains are very promising probiotics due to the double-benefit attributed to these polymers, both in helping the probiotic bacteria to colonize the gut and in acting as an effector molecule.
The present work emphasizes the relevance of isolating and investigating new EPS-producing L. paracasei strains from kefir and water kefir for their application in the development of new functional products, highlighting this as an emerging research field to explore in the future.
Author Contributions
Writing—original draft preparation, A.A.B., C.D., G.L.G. and A.G.A.; writing—review and editing, A.A.B., C.D., G.L.G. and A.G.A.; Investigation and figure design, A.A.B. and C.D.; Conceptualization, A.A.B., G.L.G. and A.G.A.; Supervision, A.G.A.; project administration and funding acquisition G.L.G. and A.G.A. All authors have read and agreed to the published version of the manuscript.
Funding
The present work was supported by CONICET, Universidad Nacional de La Plata (UNLP 18/X813, 2018–2021) and ANPCyT (PICT 2016-0639). AB is a postdoctoral fellows of Consejo Nacional de Ciencia y Tecnología (CONICET). CD is a postdoctoral fellow of Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT). GG and AA are members of Scientific Career of CONICET.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.González S., Fernández-Navarro T., Arboleya S., de los Reyes-Gavilán C.G., Salazar N., Gueimonde M. Fermented Dairy Foods: Impact on Intestinal Microbiota and Health-Linked Biomarkers. Front. Microbiol. 2019;10:1046. doi: 10.3389/fmicb.2019.01046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marco M.L., Heeney D., Binda S., Cifelli C.J., Cotter P.D., Foligné B., Gänzle M., Kort R., Pasin G., Pihlanto A., et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017;44:94–102. doi: 10.1016/j.copbio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Bengoa A.A., Iraporda C., Garrote G.L., Abraham A.G. Kefir micro-organisms: Their role in grain assembly and health properties of fermented milk. J. Appl. Microbiol. 2019;126:686–700. doi: 10.1111/jam.14107. [DOI] [PubMed] [Google Scholar]
- 4.Lynch K.M., Wilkinson S., Daenen L., Arendt E.K. An update on water kefir: Microbiology, composition and production. Int. J. Food Microbiol. 2021;345:109128. doi: 10.1016/j.ijfoodmicro.2021.109128. [DOI] [PubMed] [Google Scholar]
- 5.Garofalo C., Osimani A., Milanović V., Aquilanti L., De Filippis F., Stellato G., Di Mauro S., Turchetti B., Buzzini P., Ercolini D., et al. Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol. 2015;49:123–133. doi: 10.1016/j.fm.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Bourrie B.C.T., Willing B.P., Cotter P.D. The Microbiota and Health Promoting Characteristics of the Fermented Beverage Kefir. Front. Microbiol. 2016;7:647. doi: 10.3389/fmicb.2016.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh A.J., O’Sullivan O., Hill C., Ross R.P., Cotter P.D. Sequence-based analysis of the microbial composition of water kefir from multiple sources. FEMS Microbiol. Lett. 2013;348:79–85. doi: 10.1111/1574-6968.12248. [DOI] [PubMed] [Google Scholar]
- 8.Azizi N.F., Kumar M.R., Yeap S.K., Abdullah J.O., Khalid M., Omar A.R., Osman M.A., Mortadza S.A.S., Alitheen N.B. Kefir and Its Biological Activities. Foods. 2021;10:1210. doi: 10.3390/foods10061210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peluzio M.D.C.G., e Dias M.D.M., Martinez J.A., Milagro F.I. Kefir and Intestinal Microbiota Modulation: Implications in Human Health. Front. Nutr. 2021;8:638740. doi: 10.3389/fnut.2021.638740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slattery C., Cotter P.D., O’Toole P.W. Analysis of Health Benefits Conferred by Lactobacillus Species from Kefir. Nutrients. 2019;11:1252. doi: 10.3390/nu11061252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill D., Sugrue I., Tobin C., Hill C., Stanton C., Ross R.P. The Lactobacillus casei Group: History and Health Related Applications. Front. Microbiol. 2018;9:2107. doi: 10.3389/fmicb.2018.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bengoa A.A., Iraporda C., Acurcio L.B., de Cicco Sandes S.H., Costa K., Guimarães G.M., Arantes R.M.E., Neumann E., Nunes Á.C., Nicoli J.R., et al. Physicochemical, immunomodulatory and safety aspects of milks fermented with Lactobacillus paracasei isolated from kefir. Food Res. Int. 2019;123:48–55. doi: 10.1016/j.foodres.2019.04.041. [DOI] [PubMed] [Google Scholar]
- 13.Patterson E., Griffin S.M., Ibarra A., Ellsiepen E., Hellhammer J. Lacticaseibacillus paracasei Lpc-37® improves psychological and physiological markers of stress and anxiety in healthy adults: A randomized, double-blind, placebo-controlled and parallel clinical trial (the Sisu study) Neurobiol. Stress. 2020;13:100277. doi: 10.1016/j.ynstr.2020.100277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuyts S., Wittouck S., De Boeck I., Allonsius C.N., Pasolli E., Segata N., Lebeer S. Large-Scale Phylogenomics of the Lactobacillus casei Group Highlights Taxonomic Inconsistencies and Reveals Novel Clade-Associated Features. mSystems. 2017;2:e00061-17. doi: 10.1128/mSystems.00061-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins M.D., Phillips B.A., Zanoni P. Deoxyribonucleic Acid Homology Studies of Lactobacillus casei, Lactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 1989;39:105–108. doi: 10.1099/00207713-39-2-105. [DOI] [Google Scholar]
- 16.Wittouck S., Wuyts S., Lebeer S. Towards a Genome-Based Reclassification of the Genus Lactobacillus. Appl. Environ. Microbiol. 2019;85:e02155-18. doi: 10.1128/AEM.02155-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dellaglio F., Dicks L.M.T., DU Toit M., Torriani S. Designation of ATCC 334 in Place of ATCC 393 (NCDO 161) as the Neotype Strain of Lactobacillus casei subsp. casei and Rejection of the Name Lactobacillus paracasei (Collins et al., 1989): Request for an Opinion. Int. J. Syst. Evol. Microbiol. 1991;41:340–342. doi: 10.1099/00207713-41-2-340. [DOI] [Google Scholar]
- 18.Dicks L.M.T., Du Plessis E.M., Dellagl F., Lauer E. Reclassification of Lactobacillus casei subsp. casei ATCC 393 and Lactobacillus rhamnosus ATCC 15820 as Lactobacillus zeae norn. rev., designation of ATCC 334 as the neotype of L. casei subsp. casei, and Rejection of the Name Lactobacillus paracasei. Int. J. Syst. Evol. Microbiol. 1996;46:337–340. doi: 10.1099/00207713-46-1-337. [DOI] [PubMed] [Google Scholar]
- 19.Mori K., Yamazaki K., Ishiyama T., Katsumata M., Kobayashi K., Kawai Y., Inoue N., Shinano H. Comparative Sequence Analyses of the Genes Coding for 16S rRNA of Lactobacillus casei-Related Taxa. Int. J. Syst. Evol. Microbiol. 1997;47:54–57. doi: 10.1099/00207713-47-1-54. [DOI] [PubMed] [Google Scholar]
- 20.Dellaglio F., Felis G.E., Torriani S. The status of the species Lactobacillus casei (Orla-Jensen 1916) Hansen and Lessel 1971 and Lactobacillus paracasei Collins et al. 1989. Request for an opinion. Int. J. Syst. Evol. Microbiol. 2002;52:285–287. doi: 10.1099/00207713-52-1-285. [DOI] [PubMed] [Google Scholar]
- 21.Tindall B.J. The type strain of Lactobacillus casei is ATCC 393, ATCC 334 cannot serve as the type because it represents a different taxon, the name Lactobacillus paracasei and its subspecies names are not rejected and the revival of the name ‘Lactobacillus zeae’ contravenes Rules 51b (1) and (2) of the International Code of Nomenclature of Bacteria. Opinion 82. Int. J. Syst. Evol. Microbiol. 2008;58:1764–1765. doi: 10.1099/ijs.0.2008/005330-0. [DOI] [PubMed] [Google Scholar]
- 22.Salvetti E., Harris H.M.B., Felis G.E., O’Toole P.W. Comparative Genomics of the Genus Lactobacillus Reveals Robust Phylogroups That Provide the Basis for Reclassification. Appl. Environ. Microbiol. 2018;84:e00993-18. doi: 10.1128/aem.00993-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng J., Wittouck S., Salvetti E., Franz C.M.A.P., Harris H.M.B., Mattarelli P., O’Toole P.W., Pot B., Vandamme P., Walter J., et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 24.Fontana A., Zacconi C., Morelli L. Genetic Signatures of Dairy Lactobacillus casei Group. Front. Microbiol. 2018;9:2611. doi: 10.3389/fmicb.2018.02611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C.-H., Li S.-W., Huang L., Watanabe K. Identification and Classification for the Lactobacillus casei Group. Front. Microbiol. 2018;9:1974. doi: 10.3389/fmicb.2018.01974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vásquez A., Molin G., Pettersson B., Antonsson M., Ahrné S. DNA-based classification and sequence heterogeneities in the 16S rRNA genes of Lactobacillus casei/paracasei and related species. Syst. Appl. Microbiol. 2005;28:430–441. doi: 10.1016/j.syapm.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Smokvina T., Wels M., Polka J., Chervaux C., Brisse S., Boekhorst J., Van Vlieg J.E.T.H., Siezen R.J. Lactobacillus paracasei Comparative Genomics: Towards Species Pan-Genome Definition and Exploitation of Diversity. PLoS ONE. 2013;8:e68731. doi: 10.1371/journal.pone.0068731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefanovic E., McAuliffe O. Comparative genomic and metabolic analysis of three Lactobacillus paracasei cheese isolates reveals considerable genomic differences in strains from the same niche. BMC Genom. 2018;19:1–13. doi: 10.1186/s12864-018-4586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward L.J.H., Timmins M.J. Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett. Appl. Microbiol. 1999;29:90–92. doi: 10.1046/j.1365-2672.1999.00586.x. [DOI] [PubMed] [Google Scholar]
- 30.Iacumin L., Ginaldi F., Manzano M., Anastasi V., Reale A., Zotta T., Rossi F., Coppola R., Comi G. High resolution melting analysis (HRM) as a new tool for the identification of species belonging to the Lactobacillus casei group and comparison with species-specific PCRs and multiplex PCR. Food Microbiol. 2015;46:357–367. doi: 10.1016/j.fm.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Huang C.-H., Lee F.-L. The dnaK gene as a molecular marker for the classification and discrimination of the Lactobacillus casei group. Antonie Leeuwenhoek. 2011;99:319–327. doi: 10.1007/s10482-010-9493-6. [DOI] [PubMed] [Google Scholar]
- 32.Bottari B., Felis G.E., Salvetti E., Castioni A., Campedelli I., Torriani S., Bernini V., Gatti M. Effective identification of Lactobacillus casei group species: Genome-based selection of the gene mutL as the target of a novel multiplex PCR assay. Microbiology. 2017;163:950–960. doi: 10.1099/mic.0.000497. [DOI] [PubMed] [Google Scholar]
- 33.Huang C.-H., Huang L. Rapid species- and subspecies-specific level classification and identification of Lactobacillus casei group members using MALDI Biotyper combined with ClinProTools. J. Dairy Sci. 2018;101:979–991. doi: 10.3168/jds.2017-13642. [DOI] [PubMed] [Google Scholar]
- 34.Naser S.M., Dawyndt P., Hoste B., Gevers D., Vandemeulebroecke K., Cleenwerck I., Vancanneyt M., Swings J. Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int. J. Syst. Evol. Microbiol. 2007;57:2777–2789. doi: 10.1099/ijs.0.64711-0. [DOI] [PubMed] [Google Scholar]
- 35.Huang C.-H., Liou J.-S., Lee A.-Y., Tseng M., Miyashita M., Huang L., Watanabe K. Polyphasic characterization of a novel species in the Lactobacillus casei group from cow manure of Taiwan: Description of L. chiayiensis sp. nov. Syst. Appl. Microbiol. 2018;41:270–278. doi: 10.1016/j.syapm.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Silvaraju S., Menon N., Fan H., Lim K., Kittelmann S. Phylotype-Level Characterization of Complex Communities of Lactobacilli Using a High-Throughput, High-Resolution Phenylalanyl-tRNA Synthetase (pheS) Gene Amplicon Sequencing Approach. Appl. Environ. Microbiol. 2020;87:e02191-20. doi: 10.1128/AEM.02191-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghane M., Babaeekhou L., Ketabi S.S. Antibiofilm activity of kefir probiotic lactobacilli against uropathogenic Escherichia coli (UPEC) Avicenna J. Med. Biotechnol. 2020;12:221–229. [PMC free article] [PubMed] [Google Scholar]
- 38.Miao J., Guo H., Ou Y., Liu G., Fang X., Liao Z., Ke C., Chen Y., Zhao L., Cao Y. Purification and characterization of bacteriocin F1, a novel bacteriocin produced by Lactobacillus paracasei subsp. tolerans FX-6 from Tibetan kefir, a traditional fermented milk from Tibet, China. Food Control. 2014;42:48–53. doi: 10.1016/j.foodcont.2014.01.041. [DOI] [Google Scholar]
- 39.Romero-Luna H.E., Peredo-Lovillo A., Hernández-Mendoza A., Hernández-Sánchez H., Cauich-Sánchez P.I., Ribas-Aparicio R.M., Dávila-Ortiz G. Probiotic Potential of Lactobacillus paracasei CT12 Isolated from Water Kefir Grains (Tibicos) Curr. Microbiol. 2020;77:2584–2592. doi: 10.1007/s00284-020-02016-0. [DOI] [PubMed] [Google Scholar]
- 40.Mantzourani I., Chondrou P., Bontsidis C., Karolidou K., Terpou A., Alexopoulos A., Bezirtzoglou E., Galanis A., Plessas S. Assessment of the probiotic potential of lactic acid bacteria isolated from kefir grains: Evaluation of adhesion and antiproliferative properties in in vitro experimental systems. Ann. Microbiol. 2019;69:751–763. doi: 10.1007/s13213-019-01467-6. [DOI] [Google Scholar]
- 41.Plessas S., Kiousi D.E., Rathosi M., Alexopoulos A., Kourkoutas Y., Mantzourani I., Galanis A., Bezirtzoglou E. Isolation of a Lactobacillus paracasei Strain with Probiotic Attributes from Kefir Grains. Biomedicines. 2020;8:594. doi: 10.3390/biomedicines8120594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leite A.M.O., Miguel M.A.L., Peixoto R.S., Ruas-Madiedo P., Paschoalin V.M.F., Mayo B., Delgado S. Probiotic potential of selected lactic acid bacteria strains isolated from Brazilian kefir grains. J. Dairy Sci. 2015;98:3622–3632. doi: 10.3168/jds.2014-9265. [DOI] [PubMed] [Google Scholar]
- 43.Karaffová V., Mudroňová D., Mad’Ar M., Hrčková G., Faixová D., Gancarčíková S., Ševčíková Z., Nemcová R. Differences in Immune Response and Biochemical Parameters of Mice Fed by Kefir Milk and Lacticaseibacillus paracasei Isolated from the Kefir Grains. Microorganisms. 2021;9:831. doi: 10.3390/microorganisms9040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamet M.F., Londero A., Medrano M., Vercammen E., Van Hoorde K., Garrote G.L., Huys G., Vandamme P., Abraham A.G. Application of culture-dependent and culture-independent methods for the identification of Lactobacillus kefiranofaciens in microbial consortia present in kefir grains. Food Microbiol. 2013;36:327–334. doi: 10.1016/j.fm.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 45.Kim E., Yang S.-M., Cho E.-J., Kim H.-Y. Novel real-time PCR assay for Lactobacillus casei group species using comparative genomics. Food Microbiol. 2020;90:103485. doi: 10.1016/j.fm.2020.103485. [DOI] [PubMed] [Google Scholar]
- 46.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 47.Jones R.M. The Microbiota in Gastrointestinal Pathophysiology: Implications for Human Health, Prebiotics, Probiotics, and Dysbiosis. Academic Press; Cambridge, MA, USA: 2017. The Use of Lactobacillus casei and Lactobacillus paracasei in Clinical Trials for the Improvement of Human Health; pp. 99–108. [Google Scholar]
- 48.Azais-Braesco V., Bresson J.L., Guarner F., Corthier G. Not all lactic acid bacteria are probiotics, …but some are. Br. J. Nutr. 2010;103:1079–1081. doi: 10.1017/S0007114510000723. [DOI] [PubMed] [Google Scholar]
- 49.Balzaretti S., Taverniti V., Guglielmetti S., Fiore W., Minuzzo M., Ngo H.N., Ngere J.B., Sadiq S., Humphreys P.N., Laws A.P. A Novel Rhamnose-Rich Hetero-exopolysaccharide Isolated from Lactobacillus paracasei DG Activates THP-1 Human Monocytic Cells. Appl. Environ. Microbiol. 2017;83:2702–2718. doi: 10.1128/AEM.02702-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin S.-W., Tsai Y.-S., Chen Y.-L., Wang M.-F., Chen C.-C., Lin W.-H., Fang T.J. An Examination of Lactobacillus paracasei GKS6 and Bifidobacterium lactis GKK2 Isolated from Infant Feces in an Aged Mouse Model. Evid.-Based Complement. Altern. Med. 2021;2021:1–9. doi: 10.1155/2021/6692363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim W.-K., Jang Y.J., Seo B., Han D.H., Park S.J., Ko G.P. Administration of Lactobacillus paracasei strains improves immunomodulation and changes the composition of gut microbiota leading to improvement of colitis in mice. J. Funct. Foods. 2019;52:565–575. doi: 10.1016/j.jff.2018.11.035. [DOI] [Google Scholar]
- 52.Kim W.-K., Jang Y.J., Han D.H., Jeon K., Lee C., Han H.S., Ko G.P. Lactobacillus paracasei KBL382 administration attenuates atopic dermatitis by modulating immune response and gut microbiota. Gut Microbes. 2020;12:1819156. doi: 10.1080/19490976.2020.1819156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossoni R.D., De Barros P.P., do Carmo Mendonça I., Medina R.P., Silva D.H.S., Fuchs B.B., Junqueira J.C., Mylonakis E. The Postbiotic Activity of Lactobacillus paracasei 28.4 Against Candida auris. Front. Cell. Infect. Microbiol. 2020;10:397. doi: 10.3389/fcimb.2020.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y., Tian Y., Cao Y., Li J., Guo H., Su Y., Tian Y., Wang C., Wang T., Zhang L. Probiotic Properties of Lactobacillus paracasei subsp. paracasei L1 and Its Growth Performance-Promotion in Chicken by Improving the Intestinal Microflora. Front. Physiol. 2019;10:937. doi: 10.3389/fphys.2019.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lv X.-C., Chen M., Huang Z.-R., Guo W.-L., Ai L.-Z., Bai W.-D., Yu X.-D., Liu Y.-L., Rao P.-F., Ni L. Potential mechanisms underlying the ameliorative effect of Lactobacillus paracasei FZU103 on the lipid metabolism in hyperlipidemic mice fed a high-fat diet. Food Res. Int. 2021;139:109956. doi: 10.1016/j.foodres.2020.109956. [DOI] [PubMed] [Google Scholar]
- 56.Jam S.A.M., Talebi M., Alipour B., Khosroushahi A.Y. The therapeutic effect of potentially probiotic Lactobacillus paracasei on dimethylhydrazine induced colorectal cancer in rats. Food Biosci. 2021;41:101097. doi: 10.1016/j.fbio.2021.101097. [DOI] [Google Scholar]
- 57.Ye P., Wang J., Liu M., Li P., Gu Q. Purification and characterization of a novel bacteriocin from Lactobacillus paracasei ZFM54. LWT. 2021;143:111125. doi: 10.1016/j.lwt.2021.111125. [DOI] [Google Scholar]
- 58.Bhat B., Bajaj B.K. Hypocholesterolemic potential and bioactivity spectrum of an exopolysaccharide from a probiotic isolate Lactobacillus paracasei M7. Bioact. Carbohydr. Diet. Fibre. 2019;19:100191. doi: 10.1016/j.bcdf.2019.100191. [DOI] [Google Scholar]
- 59.Liu C.-F., Tseng K.-C., Chiang S.-S., Lee B.-H., Hsu W.-H., Pan T.-M. Immunomodulatory and antioxidant potential of Lactobacillus exopolysaccharides. J. Sci. Food Agric. 2011;91:2284–2291. doi: 10.1002/jsfa.4456. [DOI] [PubMed] [Google Scholar]
- 60.Song W., Hu P., Guo S., Hu J., Song C., Wang T., Gao Z., Yue T. Oxidative stress and endoplasmic reticulum stress contribute to L. paracasei subsp. paracasei M5L exopolysaccharide-induced apoptosis in HT-29 cells. Food Sci. Nutr. 2021;9:1676–1687. doi: 10.1002/fsn3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noda M., Sugimoto S., Hayashi I., Danshiitsoodol N., Fukamachi M., Sugiyama M. A novel structure of exopolysaccharide produced by a plant-derived lactic acid bacterium Lactobacillus paracasei IJH-SONE68. J. Biochem. 2018;164:87–92. doi: 10.1093/jb/mvy048. [DOI] [PubMed] [Google Scholar]
- 62.Noda M., Sultana N., Hayashi I., Fukamachi M., Sugiyama M. Exopolysaccharide Produced by Lactobacillus paracasei IJH-SONE68 Prevents and Improves the Picryl Chloride-Induced Contact Dermatitis. Molecules. 2019;24:2970. doi: 10.3390/molecules24162970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarikaya H., Aslim B., Yuksekdag Z. Assessment of anti-biofilm activity and bifidogenic growth stimulator (BGS) effect of lyophilized exopolysaccharides (l-EPSs) from Lactobacilli strains. Int. J. Food Prop. 2017;20:362–371. doi: 10.1080/10942912.2016.1160923. [DOI] [Google Scholar]
- 64.Zavala L., Golowczyc M.A., Van Hoorde K., Medrano M., Huys G., Vandamme P., Abraham A.G. Selected Lactobacillus strains isolated from sugary and milk kefir reduce Salmonella infection of epithelial cells in vitro. Benef. Microbes. 2016;7:585–595. doi: 10.3920/BM2015.0196. [DOI] [PubMed] [Google Scholar]
- 65.Bengoa A.A., Llamas M.G., Iraporda C., Dueñas M.T., Abraham A.G., Garrote G.L. Impact of growth temperature on exopolysaccharide production and probiotic properties of Lactobacillus paracasei strains isolated from kefir grains. Food Microbiol. 2018;69:212–218. doi: 10.1016/j.fm.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 66.Bengoa A.A., Errea A.J., Rumbo M., Abraham A.G., Garrote G.L. Modulatory properties of Lactobacillus paracasei fermented milks on gastric inflammatory conditions. Int. Dairy J. 2020;111:104839. doi: 10.1016/j.idairyj.2020.104839. [DOI] [Google Scholar]
- 67.Bengoa A.A., Dardis C., Gagliarini N., Garrote G.L., Abraham A.G. Exopolysaccharides from Lactobacillus paracasei Isolated from Kefir as Potential Bioactive Compounds for Microbiota Modulation. Front. Microbiol. 2020;11:583254. doi: 10.3389/fmicb.2020.583254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C., Li J., Zhang H., Xie Y., Xiong L., Liu H., Wang F. Effects of a probiotic on the growth performance, intestinal flora, and immune function of chicks infected with Salmonella pullorum. Poult. Sci. 2020;99:5316–5323. doi: 10.1016/j.psj.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duan X., Chen S., Duan S., Lan C., Yang Z., Cao Y., Miao J. Antibiotic activities of the natural antimicrobial substance produced by Lactobacillus paracasei FX-6 against Pseudomonas putida. LWT. 2020;123:109096. doi: 10.1016/j.lwt.2020.109096. [DOI] [Google Scholar]
- 70.Duan X., Duan S., Wang Q., Ji R., Cao Y., Miao J. Effects of the natural antimicrobial substance from Lactobacillus paracasei FX-6 on shelf life and microbial composition in chicken breast during refrigerated storage. Food Control. 2020;109:106906. doi: 10.1016/j.foodcont.2019.106906. [DOI] [Google Scholar]
- 71.Bontsidis C., Mallouchos A., Terpou A., Nikolaou A., Batra G., Mantzourani I., Alexopoulos A., Plessas S. Microbiological and Chemical Properties of Chokeberry Juice Fermented by Novel Lactic Acid Bacteria with Potential Probiotic Properties during Fermentation at 4 °C for 4 Weeks. Foods. 2021;10:768. doi: 10.3390/foods10040768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maehata H., Arai S., Iwabuchi N., Abe F. Immuno-modulation by heat-killed Lacticaseibacillus paracasei MCC1849 and its application to food products. Int. J. Immunopathol. Pharmacol. 2021;35:1–9. doi: 10.1177/20587384211008291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salminen S., Collado M.C., Endo A., Hill C., Lebeer S., Quigley E.M.M., Sanders M.E., Shamir R., Swann J.R., Szajewska H., et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021;18:649–667. doi: 10.1038/s41575-021-00440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bengoa A.A., Zavala L., Carasi P., Trejo S.A., Bronsoms S., de los Ángeles Serradell M., Garrote G.L., Abraham A.G. Simulated gastrointestinal conditions increase adhesion ability of Lactobacillus paracasei strains isolated from kefir to Caco-2 cells and mucin. Food Res. Int. 2018;103:462–467. doi: 10.1016/j.foodres.2017.09.093. [DOI] [PubMed] [Google Scholar]
- 75.Garrote G.L., Abraham A.G., Rumbo M. Is lactate an undervalued functional component of fermented food products? Front. Microbiol. 2015;6:629. doi: 10.3389/fmicb.2015.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iraporda C., Romanin D.E., Rumbo M., Garrote G.L., Abraham A.G. The role of lactate on the immunomodulatory properties of the nonbacterial fraction of kefir. Food Res. Int. 2014;62:247–253. doi: 10.1016/j.foodres.2014.03.003. [DOI] [Google Scholar]
- 77.Iraporda C., Errea A., Romanin D.E., Cayet D., Pereyra E., Pignataro O., Sirard J.-C., Garrote G.L., Abraham A.G., Rumbo M. Lactate and short chain fatty acids produced by microbial fermentation downregulate proinflammatory responses in intestinal epithelial cells and myeloid cells. Immunobiology. 2015;220:1161–1169. doi: 10.1016/j.imbio.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 78.Oleksy M., Klewicka E. Exopolysaccharides produced by Lactobacillus sp.: Biosynthesis and applications. Crit. Rev. Food Sci. Nutr. 2018;58:450–462. doi: 10.1080/10408398.2016.1187112. [DOI] [PubMed] [Google Scholar]
- 79.Rajoka M.S.R., Wu Y., Mehwish H.M., Bansal M., Zhao L. Lactobacillus exopolysaccharides: New perspectives on engineering strategies, physiochemical functions, and immunomodulatory effects on host health. Trends Food Sci. Technol. 2020;103:36–48. doi: 10.1016/j.tifs.2020.06.003. [DOI] [Google Scholar]
- 80.Ruas-Madiedo P., Abraham A.G., Mozzi F., de los Reyes-Gavilán C.G. Molecular Aspects of Lactic Acid Bacteria for Traditional and New Applications. Research Signpost; Kerala, India: 2008. Functionality of exopolysaccharides produced by lactic acid bacteria in an in vitro gastric system; pp. 137–166. [Google Scholar]
- 81.Ryan P.M., Ross R.P., Fitzgerald G.F., Caplice N.M., Stanton C. Sugar-coated: Exopolysaccharide producing lactic acid bacteria for food and human health applications. Food Funct. 2015;6:679–693. doi: 10.1039/C4FO00529E. [DOI] [PubMed] [Google Scholar]
- 82.Korcz E., Kerényi Z., Varga L. Dietary fibers, prebiotics, and exopolysaccharides produced by lactic acid bacteria: Potential health benefits with special regard to cholesterol-lowering effects. Food Funct. 2018;9:3057–3068. doi: 10.1039/C8FO00118A. [DOI] [PubMed] [Google Scholar]
- 83.Zeidan A.A., Poulsen V.K., Janzen T., Buldo P., Derkx P.M.F., Øregaard G., Neves A.R. Polysaccharide production by lactic acid bacteria: From genes to industrial applications. FEMS Microbiol. Rev. 2017;41:S168–S200. doi: 10.1093/femsre/fux017. [DOI] [PubMed] [Google Scholar]
- 84.Laws A., Gu Y., Marshall V. Biosynthesis, characterisation, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol. Adv. 2001;19:597–625. doi: 10.1016/S0734-9750(01)00084-2. [DOI] [PubMed] [Google Scholar]
- 85.Yasuda E., Serata M., Sako T. Suppressive Effect on Activation of Macrophages by Lactobacillus casei Strain Shirota Genes Determining the Synthesis of Cell Wall-Associated Polysaccharides. Appl. Environ. Microbiol. 2008;74:4746–4755. doi: 10.1128/AEM.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song X., Xiong Z.-Q., Kong L., Wang G., Ai L. Relationship between Putative eps Genes and Production of Exopolysaccharide in Lactobacillus casei LC2W. Front. Microbiol. 2018;9:1882. doi: 10.3389/fmicb.2018.01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stack H.M., Kearney N., Stanton C., Fitzgerald G.F., Ross R.P. Association of Beta-Glucan Endogenous Production with Increased Stress Tolerance of Intestinal Lactobacilli. Appl. Environ. Microbiol. 2010;76:500–507. doi: 10.1128/AEM.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Živković M., Miljković M.S., Ruas-Madiedo P., Markelić M.B., Veljović K., Tolinački M., Soković S., Korać A., Golić N. EPS-SJ Exopolisaccharide Produced by the Strain Lactobacillus paracasei subsp. paracasei BGSJ2-8 Is Involved in Adhesion to Epithelial Intestinal Cells and Decrease on E. coli Association to Caco-2 Cells. Front. Microbiol. 2016;7:286. doi: 10.3389/fmicb.2016.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Castro-Bravo N., Wells J.M., Margolles A., Ruas-Madiedo P. Interactions of Surface Exopolysaccharides from Bifidobacterium and Lactobacillus Within the Intestinal Environment. Front. Microbiol. 2018;9:2426. doi: 10.3389/fmicb.2018.02426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou Y., Cui Y., Qu X. Exopolysaccharides of lactic acid bacteria: Structure, bioactivity and associations: A review. Carbohydr. Polym. 2019;207:317–332. doi: 10.1016/j.carbpol.2018.11.093. [DOI] [PubMed] [Google Scholar]
- 91.Angelin J., Kavitha M. Exopolysaccharides from probiotic bacteria and their health potential. Int. J. Biol. Macromol. 2020;162:853–865. doi: 10.1016/j.ijbiomac.2020.06.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gibson G.R., Hutkins R., Sanders M.E., Prescott S.L., Reimer R.A., Salminen S.J., Scott K., Stanton C., Swanson K.S., Cani P.D., et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 93.Salazar N., Gueimonde M., de los Reyes-Gavilán C., Ruas-Madiedo P. Exopolysaccharides Produced by Lactic Acid Bacteria and Bifidobacteria as Fermentable Substrates by the Intestinal Microbiota. Crit. Rev. Food Sci. Nutr. 2016;56:1440–1453. doi: 10.1080/10408398.2013.770728. [DOI] [PubMed] [Google Scholar]
- 94.Liu H., Xie Y.H., Han T., Zhang H.X. Purification and Structure Study on Exopolysaccharides Produced by Lactobacillus paracasei KL1-Liu from Tibetan Kefir. Adv. Mater. Res. 2013;781–784:1513–1518. doi: 10.4028/www.scientific.net/AMR.781-784.1513. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.