Abstract
Sarcospan is an integral membrane component of the dystrophin-glycoprotein complex (DGC) found at the sarcolemma of striated and smooth muscle. The DGC plays important roles in muscle function and viability as evidenced by defects in components of the DGC, which cause muscular dystrophy. Sarcospan is unique among the components of the complex in that it contains four transmembrane domains with intracellular N- and C-terminal domains and is a member of the tetraspan superfamily of proteins. Sarcospan is tightly linked to the sarcoglycans, and together these proteins form a subcomplex within the DGC. Stable expression of sarcospan at the sarcolemma is dependent upon expression of the sarcoglycans. Here we describe the generation and analysis of mice carrying a null mutation in the Sspn gene. Surprisingly, the Sspn-deficient muscle maintains expression of other components of the DGC at the sarcolemma, and no gross histological abnormalities of muscle from the mice are observed. The Sspn-deficient muscle maintains sarcolemmal integrity as determined by serum creatine kinase and Evans blue uptake assays, and the Sspn-deficient muscle maintains normal force and power generation capabilities. These data suggest either that sarcospan is not required for normal DGC function or that the Sspn-deficient muscle is compensating for the absence of sarcospan, perhaps by utilizing another protein to carry out its function.
The dystrophin-glycoprotein complex (DGC) is a multisubunit protein complex composed of integral membrane, peripheral membrane, and cytoplasmic proteins expressed at the sarcolemma of striated muscle fibers (see references 7, 41, 54, and 62 for reviews). Isolation and cloning of proteins within this complex have provided key insights into the function of the DGC and its role in normal muscle physiology. The skeletal muscle DGC is composed of dystrophin (9, 31); the syntrophins (22); α- and β-dystroglycan (33); α-, β-, γ-, and δ-sarcoglycan (3, 36, 42, 48, 50, 51, 55, 56); and sarcospan (14). Recently, the DGC within smooth muscle fibers has been characterized (64). This complex differs from the striated muscle DGC in at least two respects. First, ɛ-sarcoglycan replaces α-sarcoglycan within the sarcoglycan subcomplex, and second, differences occur in the glycosylation pattern of α-dystroglycan in the smooth muscle DGC.
One likely function of the DGC is to provide a structural link between the extracellular matrix and the actin cytoskeleton, thereby maintaining the stability of the sarcolemma under contractile forces (10, 21). This link occurs through dystrophin, which binds to filamentous actin (31, 58, 59), and dystroglycan—with its β-subunit binding to dystrophin (37) and its α-subunit interacting with the extracellular matrix component laminin-2 (24). The DGC likely plays other roles in normal muscle physiology by interacting with cell signaling molecules or other proteins at the sarcolemma. The skeletal muscle DGC has been the most fully characterized in this respect, and several laboratories have demonstrated interactions between DGC components and known signaling molecules (1, 4, 5, 11, 12, 25, 28, 34, 44, 60, 71). Additionally, the skeletal, cardiac, and smooth muscle-specific forms of the DGC may have different functions and specific transient binding partners in each tissue.
Sarcospan is a transmembrane protein found in skeletal, cardiac, and smooth muscle DGCs (14, 15, 64). It was identified as a component of the rabbit skeletal muscle DGC by peptide sequence analysis of the 25-kDa DGC component (14). Primary sequence analysis predicts that sarcospan contains four transmembrane domains with cytoplasmic NH2 and COOH termini (14, 29, 61). This makes sarcospan unique among other integral membrane components of the DGC, which contain single-transmembrane domains. Protein sequence analysis designates sarcospan as a member of the tetraspan superfamily, most closely related to Rom-1, peripherin, and uroplakin (14). The tetraspan proteins are thought to function as molecular facilitators, mediating interaction between proteins at the plasma membrane. This is an attractive function for a component of the DGC: to facilitate protein-protein interactions within the plane of the plasma membrane. The tetraspan proteins have also been implicated in cell signaling activities including cell adhesion, migration, and proliferation (45, 70). This suggests that sarcospan may be an important mediator of signaling functions within the DGC.
Sarcospan is tightly associated with the sarcoglycans to form a subcomplex within the DGC, the sarcoglycan-sarcospan (SG-SSPN) complex (15). In animal models for the limb-girdle muscular dystrophies, namely, in the α-, β-, and δ-sarcoglycan knockout mice (2, 13, 17, 18) and the cardiomyopathic hamster (63), there is a concomitant loss of sarcospan with the sarcoglycans at the sarcolemma. When the DGC has been successfully restored in Sgca-deficient mice or cardiomyopathic hamsters by adenoviral injections of either α-sarcoglycan or δ-sarcoglycan, respectively, sarcospan expression is also restored (15, 17). Biochemical evidence further demonstrates this association. The SG-SSPN complex cofractionates when the DGC is disrupted by alkaline treatment or in the mdx mouse, a dystrophin-deficient animal model for Duchenne muscular dystrophy. Additionally, sarcospan can be coimmunoprecipitated with the sarcoglycans in an in vitro cell expression system (15) and from skeletal muscle membrane preparations (C. S. Lebakken and K. P. Campbell, unpublished data). One function of the SG-SSPN complex is to stabilize α-dystroglycan at the membrane (32). Furthermore, the SG-SSPN complex may interact with other sarcolemma proteins and be involved in cell signaling processes.
Primary genetic defects in several components of the DGC lead to muscular dystrophies. Duchenne and Becker muscular dystrophies are caused by mutations in the dystrophin gene. Four forms of autosomal recessive limb-girdle muscular dystrophies (types 2C, 2D, 2E, and 2F) are caused by primary mutations in each of the four sarcoglycan genes (reviewed in references 8, 41, and 62). Additionally, mutations in the laminin-α2 chain cause a severe form of congenital muscular dystrophy. To date, no human mutations have been found in the dystroglycans, dystrobrevins, syntrophins, or sarcospan, nor have any unclassified muscular dystrophies been mapped to their chromosomal locations. Human-null mutations in the dystroglycan gene would likely lead to an early embryonic lethality. Recent data suggest that dystroglycan is important in basement membrane formation (30), and dystroglycan-null mice die at a very early embryonic stage (68). α-Dystrobrevin-deficient mice maintain expression of DGC components, however, and develop a mild muscular dystrophy (26). In contrast, disruption of the α1-syntrophin gene in mice does not result in muscle degeneration (38).
To address the consequences of sarcospan deficiency, we have generated a sarcospan-deficient mouse through standard techniques of homologous recombination in embryonic stem (ES) cells. In this study, we demonstrate that the Sspn-deficient mice do not show any hallmarks of a muscular dystrophy phenotype. Surprisingly, the other components of the DGC are maintained at the sarcolemma in the absence of sarcospan. The muscle appears normal by histological analysis, and it maintains sarcolemma integrity as determined by serum creatine kinase measurements and the absence of Evans blue dye uptake. Additionally, the muscle maintains normal force and power generation capacity. These data suggest either that sarcospan is not required for normal DGC function or, more likely, that the muscle compensates for the loss of sarcospan in some manner.
MATERIALS AND METHODS
Isolation of murine sarcospan genomic and cDNA clones.
The murine sarcospan genomic DNA sequence was obtained by screening a 129/SvJ mouse genomic library (Lambda FIX II; Stratagene, La Jolla, Calif.) using a mouse sarcospan expressed sequence tag (EST) (IMAGE clone 406138; obtained from Research Genetics, Inc., Huntsville, Ala.) containing a portion of the sarcospan 3′ untranslated region (UTR). Clones containing the sarcospan insert were analyzed by restriction enzyme mapping and DNA sequencing using dye terminator cycling on a 373 Stretch Fluorescent Automated sequencer (Applied Biosystems). Twenty-four kilobases of the sarcospan genomic DNA sequence was analyzed and found to contain exons 2 and 3 and flanking regions. Murine sarcospan cDNA was isolated by screening a mouse skeletal muscle library (a generous gift of Jeffrey Chamberlain, University of Michigan) with the same EST. Clones were analyzed by DNA sequencing as described above and are identical to that previously published by Scott and colleagues (61) (GenBank accession no. U02487).
Generation of the Sspn targeting plasmid.
The Sspn targeting plasmid was constructed using a modified version of the positive-negative selection vector pPNT (obtained from Richard Mulligan, Whitehead Institute, Cambridge, Mass.) (67). A neomycin cassette flanked by loxP sites replaced the neomycin cassette from the original pPNT vector. The short arm of homology in the targeting vector was a 2.1-kb BsaHI fragment containing a 3′ portion of intron 1. This fragment was inserted into the modified pPNT vector in the ClaI cloning site upstream of the phosphoglycerate kinase (PGK)-neomycin resistance cassette. Orientation of this fragment was confirmed by restriction enzyme digestion and sequencing. To generate the long-arm region of homology, a 10-kb NheI/NotI fragment containing a portion of the 3′ UTR, located in exon 3, and downstream sequences was isolated from a genomic clone and subcloned into pBluescript KS(+) (pBS). From this, a 6.5-kb BamHI fragment was subsequently isolated and inserted into the BamHI cloning site distal to the PGK-neomycin resistance cassette. Orientation of this fragment was also confirmed by restriction enzyme digestion and sequencing. The vector contained a thymidine kinase cassette distal to the long arm. The mutant gene therefore lacked 7.6 kb, which included 223 bp of intron 1, exon 2, intron 2, and 1.8 kb of exon 3 (which encompasses the entire coding region of exon 3).
Generation of Sspn null mice.
The targeting construct was linearized with NotI and introduced into 2 × 107 R1 ES cells by electroporation (Bio-Rad Gene Pulser; Hercules, Calif.). The ES cells were maintained on feeder layers under the selective pressure of G418 and ganciclovir. Colonies surviving the selection were isolated, expanded, and screened for targeting fidelity by Southern blot analysis (Fig. 1). Nine targeted clones were obtained from 236 analyzed. These clones were subsequently reconfirmed by Southern blot analysis by digestion with XbaI (X, Fig. 1A) and hybridization with a second probe (probe 2, Fig. 1A) to verify correct targeting. Finally, Southern blot analysis was performed with a PGK-neomycin resistance gene probe to confirm a single targeting event. Cells from two targeted clones (U104 and U202) were microinjected into C57BL/6J blastocysts and transferred into pseudopregnant recipients. Chimeric animals resulting from the microinjections were bred to C57BL/6J mice, and agouti pups were screened for germ line transmission of the mutant allele. The genotypes from these matings and all subsequent matings were determined by PCR on DNA from tail biopsy specimens (Fig. 1). The following oligonucleotide primers were used: SPNi1FA (primer 1), ACTCCCTGGAATACAGAGGAACT; SPNi2RA (primer 2), TACAAGGGGACAGACACTCAGAC; and NEOspnKO (primer 3), TTTCTCTTGATTCCCACTTTGTG. PCR conditions were as follows: denaturation at 94°C for 2 min, followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1.5 min at 72°C. Southern blot analysis was periodically performed to confirm the fidelity of the PCR. We have obtained identical results from both targeted lines (U104 and U202). Analysis thus far has been carried out on the hybrid C57BL/6–129-SV/J background. However, initial results analyzing Sspn-deficient mice on the 129-SV/J background are consistent with the data presented in this report. All animals were housed in the animal care unit of the University of Iowa College of Medicine according to animal care guidelines.
FIG. 1.
Generation of sarcospan-deficient mice by homologous recombination in ES cells. (A) Restriction map of the wild-type murine Sspn locus, the targeting construct, and the targeted locus. A 7.6-kb region including exon 2 (E2) and the coding region of exon 3 (E3) (black boxes) was deleted and replaced by a PGK-neomycin cassette (Neo) flanked by loxP sites (arrowhead). (B) Southern blot analysis. Using probe 1 (as illustrated in panel A), the wild-type locus contains a 16.2-kb EcoRV (RV) fragment, whereas the targeted allele contains a 7.5-kb EcoRV (RV) fragment. (C) Genotype analysis by PCR. Primer sites are shown in panel A, denoted by arrows; using primers 1 and 2, the wild-type allele (+/+) corresponds to an 840-bp PCR product; using primers 1 and 3, the null allele (−/−) corresponds to a 425-bp product. (D) Northern blot analysis of total RNA isolated from skeletal muscle of wild-type, heterozygous, and Sspn-deficient mice using a cDNA probe spanning the entire sarcospan coding region. A 4.5-kb transcript is detected in the wild type (+/+) and heterozygous (+/−) mice but is not detected in the Sspn-deficient mice (−/−). (E) Immunoblot analysis. KCl-washed skeletal muscle microsomes were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed for sarcospan using the polyclonal R256 antibody, generated against the N-terminal cytoplasmic domain of murine sarcospan. A 20-kDa band is detected in the wild-type and heterozygous mouse preparations (+/+ and +/−) but is not detected in the Sspn-deficient mouse preparations (−/−).
Antibodies.
Polyclonal antibodies against mouse sarcospan were generated by injecting New Zealand White rabbits at intramuscular and subcutaneous sites with a C-terminal sarcospan–glutathione-S-transferase fusion protein (amino acids 186 to 216 of mouse sarcospan) (R235) (15) or with an N-terminal sarcospan–glutathione-S-transferase fusion protein (amino acids 1 to 25 of mouse sarcospan: MGRKPSPRAQELPEEEARTCCGCRF) (R256). Affinity purification of sarcospan antibodies was accomplished using Immobilon-P (Millipore, Burlington, Mass.) strips containing the C-terminal or N-terminal sarcospan maltose binding protein-fusion proteins. Antibody specificity was verified for immunofluorescence and immunoblotting by competition experiments utilizing the corresponding fusion proteins and peptides synthesized to these regions. Polyclonal antibodies to α-sarcoglycan (rabbit 98) (55), β-sarcoglycan (goat 26) (17), γ-sarcoglycan (rabbit 245) (17), δ-sarcoglycan (rabbit 214) (17), ɛ-sarcoglycan (rabbit 232) (17), dystrophin (rabbit 31) (53), the laminin-α2 chain, β-dystroglycan (rabbit 83), and α-dystroglycan (affinity purified from goat 20) (33) were used as previously described (17). Monoclonal antibodies IIH6, against α-dystroglycan (20), and 8D5, against β-dystroglycan (42), were previously characterized. Monoclonal antibodies Ad1/20A6, against α-sarcoglycan; BSarc1/5B1, against β-sarcoglycan; and 35DAG/21B5, against γ-sarcoglycan, were generated in collaboration with L. V. B. Anderson (Newcastle General Hospital, Newcastle upon Tyne, United Kingdom).
RNA isolation and Northern blot analysis.
Total RNA from control, heterozygous, and homozygous-null skeletal muscle was extracted using RNAzol (Tel-Test, Friendswood, Tex.) according to manufacturer specifications. Twenty micrograms of total RNA was run on a 1.25% agarose gel containing 5% formaldehyde and transferred to a Hybond N+ membrane (Amersham Corp., Arlington Heights, Ill.). RNA was cross-linked to the membrane using a Stratagene UV cross-linker. Membranes were prehybridized and hybridized using standard methods. Washes were carried out at 65°C in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate)–1% sodium dodecyl sulfate (SDS) followed by 0.1× SSC–0.1% SDS.
Immunofluorescence and histological analysis.
Skeletal, cardiac, and smooth muscle from the small intestine was dissected from wild-type, heterozygous, and Sspn-deficient mice and rapidly frozen in liquid nitrogen-cooled isopentane. Seven-micrometer cryosections were analyzed by immunofluorescence as previously described (17) or by staining with hematoxylin and eosin (H&E).
Measurement of serum creatine kinase levels.
Serum creatine kinase levels were quantified using a kinetic determination of creatine kinase activity in serum obtained from the retro-orbital sinus. The creatine kinase assay was carried out on a Hitashi 917 spectrophotometer by the University of Iowa Department of Pathology.
Immunoblot analysis of membrane preparations.
KCl-washed membranes from skeletal muscle were prepared as previously described (52) with the addition of two protease inhibitors, calpeptin and calpain inhibitor I (17) (Calbiochem-Nova-Biochem Corp., San Diego, Calif.). Proteins were resolved by SDS-polyacrylamide gel electrophoresis on 3 to 15% linear gradient gels and transferred to nitrocellulose membranes. Immunoblot analysis was performed as previously described (52), except that the blots were developed using enhanced chemiluminescence (Supersignal; Pierce Chemical Co.).
Contractile properties.
Contractile properties of the extensor digitorum longus (EDL) and soleus muscles of wild-type, heterozygous, and Sspn-null mice (n = 9 for EDL and n = 10 for soleus muscles) were measured in vitro. Mice were anesthetized with avertin (13 to 17 μl/g of body weight). Muscles were isolated and removed carefully from the anesthetized mice and immersed in an oxygenated (95% O2 and 5% CO2) bath containing a buffered mammalian Ringer's solution, pH 7.4, which included curare. The solution was maintained at 25°C. The tendons of the muscles were tied securely to a force transducer and a fixed post. Muscles were stimulated directly by the current flow between two large platinum electrodes (6). The voltage of the stimulator was set to provide maximum twitch force, and the muscle length was set at optimum length for force development. With the muscle at optimum length, the frequency of stimulation was increased until force plateaued at maximum isometric tetanic force (Po). Each muscle was removed from the bath and blotted, and the tendons were trimmed from the muscle prior to weighing to obtain the muscle mass. Total fiber cross-sectional area and specific Po were calculated based on the direct measurements of muscle mass, muscle length, fiber length, and Po (6). The force (kilonewtons) was divided by the total fiber cross-sectional area (square meters) to obtain an estimate of the specific force (kilonewtons per square meter) of the EDL and soleus muscles. Each data set was analyzed by a two-way analysis of variance appropriate for unequal sample sizes (Statistical Analysis System, Cary, N.C.). Significance was set a priori at P < 0.05.
RESULTS
Generation of sarcospan-deficient mice.
To generate sarcospan-deficient mice, we first isolated genomic DNA clones from a 129/SvJ genomic library by hybridization screening with a mouse EST corresponding to a portion of the 3′ UTR of mouse sarcospan. Isolated clones were analyzed by restriction enzyme digestion and DNA sequencing. From these genomic clones, a targeting vector was designed to replace exon 2 and the coding sequence of exon 3 of sarcospan, which encode three of the four transmembrane domains, the extracellular loop, and the C-terminal intracellular domain of sarcospan (Fig. 1A). Southern blot analysis of 236 neomycin-resistant ES cells demonstrated homologous recombination in nine independent clones (shown for one clone in Fig. 1B). Two of these clones were used to generate chimeric founder mice. Heterozygous mice from the F1 generation were identified by PCR analysis and were crossed to obtain Sspn-deficient mice (PCR screen is demonstrated in Fig. 1C). Sspn-deficient mice are born from heterozygous matings in a predicted Mendelian inheritance frequency. Northern blot analysis of skeletal muscle RNA from wild-type, heterozygous, and homozygous Sspn-deficient mice revealed a 4.5-kb sarcospan-specific transcript in the wild-type and heterozygous mice which was absent in the Sspn-deficient skeletal muscle (Fig. 1D).
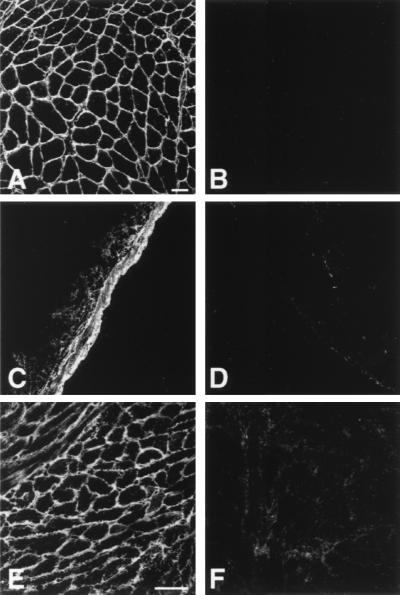
To verify that there was no sarcospan protein present in the Sspn-deficient mice, we performed immunoblot analysis of KCl-washed skeletal muscle microsomes from wild-type, heterozygous, and Sspn-deficient animals. A strong sarcospan-specific band at 20 kDa is detected in both the wild-type control and heterozygous mouse samples (Fig. 1E). This band is not detected in the preparations from the Sspn-deficient mice. Identical results were obtained when total skeletal muscle homogenates were immunoblotted for the sarcospan protein (data not shown). Transverse cryosections (7 μm) of quadriceps muscles from control or Sspn-deficient animals were stained with the sarcospan-specific antibodies. In control muscle, sarcospan is present at the sarcolemma and within neuromuscular junctions of skeletal muscle. Sarcospan protein is not detected in the Sspn-deficient muscle (Fig. 2A and B). Additionally, sarcospan protein was detected by immunofluorescence analysis in smooth muscle of the small intestine (Fig. 2C and D) and cardiac muscle (Fig. 2E and F) from the wild-type and heterozygous animals but was not detected in Sspn-deficient animals. Both the immunoblot analysis and immunofluorescence analysis were carried out using two separate antibodies generated against either the N-terminal or the C-terminal domain of sarcospan. Note that the N-terminal domain is encoded by exon 1, which was not within the deleted region of the gene. Failure to detect this epitope with the N-terminal antibody verifies that a null allele has been generated.
FIG. 2.
Sarcospan-deficient mice do not express sarcospan protein. Muscle cryosections (7 μm) were stained with the R256 polyclonal antibody generated against the N-terminal cytoplasmic domain of mouse sarcospan. Sarcospan is expressed in skeletal, cardiac, and smooth muscle of wild-type animals but is not detected in the Sspn-deficient tissues. Shown are results of immunofluorescence analysis of wild-type (A, C, and E) and Sspn-deficient (B, D, and E) quadriceps skeletal muscle (A and B), smooth muscle of the small intestine (C and D), and cardiac muscle (E and F). Bars, 50 μm.
Other components of the DGC are maintained at the sarcolemma in Sspn-deficient animals.
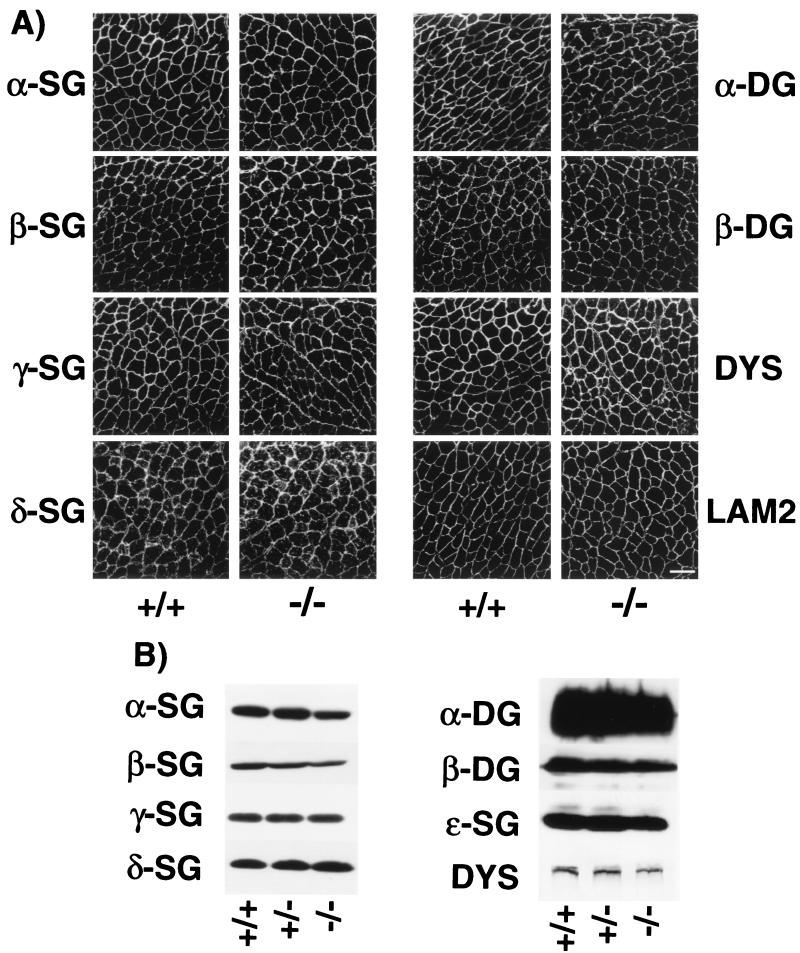
We have previously documented that sarcospan is tightly associated with the sarcoglycans to form a subcomplex within the DGC. From analysis of several animal models of sarcoglycanopathies, we have demonstrated that sarcospan expression at the sarcolemma is dependent upon expression of an intact sarcoglycan complex (15, 17, 63). To evaluate whether loss of sarcospan affects the expression of the sarcoglycans and other components of the DGC, immunofluorescence analysis was performed on cryosections of quadriceps muscle from control and Sspn-deficient mice. The sarcoglycans (α, β, γ, and δ), α- and β-dystroglycan, and dystrophin are all expressed at the sarcolemma in both control and Sspn-deficient mice (Fig. 3A). This demonstrates that although the localization of sarcospan to the sarcolemma is dependent on the sarcoglycans, the reverse is not the case. Namely, expression of the sarcoglycans is not dependent on sarcospan. These results were identical with mice ranging from 1 day to 8 months of age. To verify this result, immunoblot analysis was performed on KCl-washed microsomes obtained from wild-type, heterozygous, or Sspn-deficient skeletal muscle (Fig. 3B). Consistent with immunofluorescence analysis, the sarcoglycans (α, β, γ, δ, and ɛ), α- and β-dystroglycan, and dystrophin are expressed at similar levels in membrane preparations.
FIG. 3.
Components of the DGC are maintained at the sarcolemma in Sspn-deficient mice. (A) Immunofluorescence analysis of wild-type (+/+) and Sspn-deficient (−/−) quadriceps muscle for components of the DGC. Expression levels of the sarcoglycans (α-, β-, γ-, and δ-SG), α- and β-dystroglycan (α-, and β-DG), and dystrophin (DYS) as well as the laminin-α2 chain (LAM2) are unaffected in the Sspn−/− muscle. Several muscle groups in mice up to 8 months of age have been analyzed with identical results. Bar, 100 μm. (B) Immunoblot analysis of wild-type (+/+), heterozygous (+/−), and Sspn-deficient (−/−) mouse KCl-washed microsome preparations from skeletal muscle. Equal protein loading (100 μg) in each lane was verified by Coomassie blue staining of lanes that were identically loaded (data not shown). Expression levels of each of the tested proteins are unaltered in the Sspn-deficient mice. Abbreviations are defined in the legend to panel A.
We also analyzed expression of the DGC components in cardiac and smooth muscle from the Sspn-deficient mice. Consistent with the finding in skeletal muscle, expression of the sarcoglycans, dystroglycans, dystrophin, and utrophin is unaffected in the Sspn-deficient mice (data not shown).
Sarcospan-deficient animals do not develop a muscular dystrophy phenotype.
Although Sspn-deficient mice maintain proper expression and localization of the DGC at the sarcolemma, this does not demonstrate that the DGC remains functionally intact in these animals. If the DGC was not functionally intact, the animals might exhibit a phenotype similar to the loss of the complex (i.e., a muscular dystrophy) or a milder myopathy as is observed in the α-dystrobrevin-deficient mice (26). To test this, we first examined the skeletal and cardiac muscle morphology of Sspn-deficient mice. Cryosections (10 μm) of quadriceps, biceps, diaphragm, neck, and cardiac muscles were stained with H&E. The skeletal muscle from the Sspn-deficient mice appears normal and does not display any classical hallmarks of a muscular dystrophy phenotype such as necrosis, fibrosis, centrally located nuclei, calcification, or fat infiltration (shown for quadriceps, Fig. 4A and B). Analysis of the cardiac tissue also indicates that there are no observable dystrophic alterations in the heart. We have analyzed mice up to 8 months in age, and muscle from the Sspn-deficient animals is indistinguishable from control muscle.
FIG. 4.
Skeletal muscle from Sspn-deficient mice has normal morphology and maintains membrane integrity. The figure shows H&E staining of cryosections from the quadriceps of littermate, sex-matched wild-type (A) or Sspn-deficient (B) mice, Evans blue uptake in Sgca-deficient (C) or Sspn-deficient (D) skeletal muscle, and measurement of serum creatine kinase levels (E). The presence of the muscle-specific enzyme creatine kinase in the blood is another indication of membrane permeability. As a positive control, the serum creatine kinase levels of mdx mice were compared to those of wild-type (+/+), heterozygous (+/−), and Sspn-deficient (−/−) mice. No significant differences in the serum creatine kinase levels between the Sspn-deficient mice and their controls have been detected. However, the mdx mice exhibit very high serum creatine kinase levels. Error bars represent the standard errors of the means.
One diagnostic indication of muscular dystrophy is the evidence of an efflux of muscle-specific intracellular proteins into the serum as well as an influx of serum proteins into the muscle (16, 39, 57). This altered protein localization is likely reflective of altered membrane integrity in the dystrophic muscle. To test the sarcolemmal integrity of the Sspn-deficient muscles, we performed quantitative serum creatine kinase assays on age- and sex-matched wild-type, heterozygous, and Sspn-deficient mice (n = 6, 6, and 7, respectively). Wild-type, heterozygous, and Sspn-deficient animals all exhibited similar, low serum creatine kinase levels of 543 ± 116, 770 ± 403, and 362 ± 141 U/liter, respectively (Fig. 4E). As a control, serum from mdx mice was also analyzed for creatine kinase levels, and these were found to be extremely high (19,622 ± 3,786 U/liter) (n = 4).
To further analyze sarcolemma integrity in the Sspn-deficient animals, we performed Evans blue dye uptake experiments. Evans blue dye is a membrane-impermeant molecule that binds to serum albumin and enters into muscle fibers with damaged sarcolemmal membranes (46, 65). Evans blue dye was injected through the retro-orbital sinus, and the animals were sacrificed 3 h after injection. Upon gross examination, no Evans blue uptake was evident in muscles of either the Sspn-deficient or wild-type mice. In contrast, significant uptake was observed in skeletal muscle of mdx and α-sarcoglycan-deficient mice, which served as positive controls in this assay. Additionally, cryosections of skeletal muscle were analyzed microscopically for evidence of Evans blue uptake. There was no evidence of Evans blue accumulation in either the wild-type or the Sspn-deficient muscles (Fig. 4D). However, many Evans blue-positive fibers were found in mdx and Sgca-deficient mice (Fig. 4C).
Sspn-deficient muscle maintains normal force and power generation capabilities.
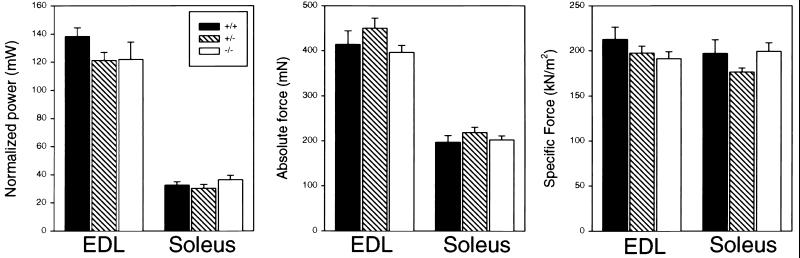
To examine if the Sspn-deficient mice had physiological muscle defects, we analyzed force and power generation capabilities of isolated EDL and soleus muscles. Total fiber cross-sectional area and specific Po were calculated based on the direct measurements of muscle mass, muscle length, fiber length, and Po (6). The force (kilonewtons) was divided by the total fiber cross-sectional area (square meters) to obtain an estimate of the specific force (kilonewtons per square meter) of the EDL and soleus muscles. We found no differences in the muscle mass, Po, or specific Po among the wild-type, heterozygous, or Sspn-deficient mice (Fig. 5).
FIG. 5.
Sarcospan-deficient muscle maintains normal power and force generation capabilities. Contractile properties of the EDL and soleus muscles of homozygous (+/+), heterozygous (+/−), and Sspn-deficient (−/−) male mice were measured in vitro. No significant differences in the normalized power, absolute force, or specific force generation between the groups of mice (n = 9 soleus muscles from each genotype and n = 10 EDL muscles from each genotype) were found. Error bars represent the standard errors of the means.
DISCUSSION
Sarcospan is a unique integral membrane component of the DGC found in skeletal, cardiac, and smooth muscle cells. We have generated sarcospan-deficient mice through standard homologous recombination within the Sspn gene. No sarcospan transcript or protein is detected in these animals. Surprisingly, the mice maintain expression of the other integral membrane components of the DGC and do not develop a muscular dystrophy phenotype. Analysis of skeletal and cardiac muscle from these mice demonstrates normal histological parameters. The sarcolemma of the Sspn-deficient mice is not perturbed, as demonstrated by normal levels of serum creatine kinase and the lack of Evans blue-positive fibers in the Evans blue uptake assay. Additionally, Sspn-deficient muscle maintains normal power and force generation capabilities. These data suggest that a functional DGC is formed and that either sarcospan is not required for normal DGC function or the Sspn-deficient muscle is compensating for the loss of sarcospan.
This is the first report describing a null mutation in an integral membrane component of the DGC in mice that does not result in either muscular dystrophy or, as in the case of dystroglycan, early embryonic lethality. Mice with null mutations in α-, β-, γ-, and δ-sarcoglycan have recently been described (2, 13, 17, 18, 27). Each of these mice develops a progressive muscular dystrophy with a dramatic reduction in the expression of the other sarcoglycans at the sarcolemma, as is observed in patients with sarcoglycan mutations (reviewed in references 7, 41, and 62). Additionally, in the α-, β-, and δ-sarcoglycan-deficient mice sarcospan expression is also dramatically reduced at the sarcolemma. The SG-SSPN complex is also lost in the smooth muscle of δ- and β-sarcoglycan-deficient mice. Disruption of the smooth muscle DGC accelerates the progression of the muscular dystrophy and leads to the cardiomyopathy observed in these animals (13, 18). Recently, the targeted disruption of the α1-syntrophin gene has been reported. As in the Sspn-deficient mice, there are no gross skeletal muscle defects observed in these animals, and presumably the rest of the DGC is maintained (38). In contrast, although the DGC is maintained at the sarcolemma in α-dystrobrevin-deficient mice, they do develop a mild muscular dystrophy and cardiomyopathy (26).
The finding that Sspn-deficient mice maintain normal muscle viability, integrity, and function is important and suggests that human muscle may also be able to compensate for the loss of sarcospan. This may explain why there have been no mutations found in the sarcospan gene which cause a muscular dystrophy phenotype. It is possible, however, that dominant mutations in the sarcospan gene that result in expression of a nonfunctional sarcospan protein might result in disruption of the complex and may be manifested in a muscular dystrophy phenotype, perhaps in a dominant form of limb-girdle muscular dystrophy.
Sarcospan is an integral component of the DGC by all tested criteria: (i) sarcospan expression is lost in Duchenne muscular dystrophy patients and in the mdx mouse, (ii) sarcospan copurifies with the DGC and additionally cofractionates with the complex on sucrose gradient columns (14), and (iii) sarcospan binds with the DGC to laminin affinity columns (R. H. Crosbie and K. P. Campbell, unpublished results). Although we hypothesize that there is a protein functionally replacing sarcospan within the DGC, we cannot rule out the possibility that sarcospan is not required for a functional complex. It is also possible that the role of sarcospan is to link the DGC to other proteins at the plasma membrane. Candidates for such interactions include but are not limited to the α5β1 and α7β1 integrins, both of which are important for normal muscle physiology (47, 66).
Sarcospan and the sarcoglycans are tightly associated and form a subcomplex within the DGC. In animal models for limb-girdle muscular dystrophy types 2D, 2B, and 2F, expression of sarcospan at the sarcolemma is severely reduced along with that of the sarcoglycans (2, 13, 17, 18). We have analyzed this tight association biochemically by demonstrating that sarcospan cofractionates with the sarcoglycans after they have been dissociated from the dystroglycans and dystrophin by alkaline treatment and by immunoprecipitation analysis (15). Given this tight association between sarcospan and the sarcoglycans, the finding that the sarcoglycans remain expressed in the Sspn-deficient mice is surprising. We hypothesize that there is a protein functionally replacing sarcospan within the DGC of the Sspn-deficient mice. The identity of this protein remains unknown. Perhaps a homologue of sarcospan, which has not been previously identified, is compensating for the loss of sarcospan. It is also interesting to speculate that an unrelated protein may be functionally replacing sarcospan within the complex.
Sarcospan is a member of the tetraspan superfamily of proteins, which is a large family of proteins expressed in nearly all cell types (45, 70). Each tetraspan protein has four conserved hydrophobic membrane-spanning domains and intracellular N- and C-terminal domains. Across species, the most conserved regions of sarcospan lie within its transmembrane domains, suggesting that these domains are functionally important. It is possible that another tetraspan family member is carrying out the function of sarcospan in its absence. Alternatively, many other four-transmembrane proteins have been demonstrated to mediate protein-protein interactions within the plasma membrane. Examples of such proteins include the connexins (43, 72, 73), which associate to form gap junctions; the claudins (23, 49), which mediate interactions within tight junctions; and the gamma subunits of voltage-gated calcium channels (19, 35, 40, 69), which associate with the α1, α2δ-, and β-subunits of the calcium channel. Future work may lead us to the identification of a protein compensating for the loss of sarcospan in the Sspn-deficient muscle.
ACKNOWLEDGMENTS
We are indebted to members of the Campbell laboratory for valuable discussions, critical review of the manuscript, and reagents. Thanks go to Karmen Munson, Bridget Ruff, Clare Gerstein, Sahar Rezayazdi, and David Thomsen for technical assistance.
We also thank the University of Iowa Diabetes and Endocrinology Research Center (NIH DK25295), the University of Iowa DNA Sequencing Core Facility, and the Contractility Core of the Nathan Shock Center (PO1AG-13283). C. S. Lebakken is supported by the Iowa Cardiovascular Interdisciplinary Research Fellowship (HL07121). This work was also supported by a grant from the Muscular Dystrophy Association to K. P. Campbell. K. P. Campbell is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Anderson J T, Rogers R P, Jarrett H W. Ca2+-calmodulin binds to the carboxyl-terminal domain of dystrophin. J Biol Chem. 1996;271:6605–6610. doi: 10.1074/jbc.271.12.6605. [DOI] [PubMed] [Google Scholar]
- 2.Araishi K, Sasaoka T, Imamura M, Noguchi S, Hama H, Wakabayashi E, Yoshida M, Hori T, Ozawa E. Loss of the sarcoglycan complex and sarcospan leads to muscular dystrophy in beta-sarcoglycan-deficient mice. Hum Mol Genet. 1999;8:1589–1598. doi: 10.1093/hmg/8.9.1589. [DOI] [PubMed] [Google Scholar]
- 3.Bönnemann C G, Modi R, Noguchi S, Mizuno Y, Yoshida M, Gussoni E, McNally E M, Duggan D J, Angelini C, Hoffman E P. Beta-sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat Genet. 1995;11:266–273. doi: 10.1038/ng1195-266. [DOI] [PubMed] [Google Scholar]
- 4.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Wu Z, Huang F, Xia H, Peters M F, Froehner S C, Bredt D S. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 5.Brenman J E, Chao D S, Xia H, Aldape K, Bredt D S. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- 6.Brooks S V, Faulkner J A. Contractile properties of skeletal muscles from young, adult and aged mice. J Physiol (London) 1988;404:71–82. doi: 10.1113/jphysiol.1988.sp017279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushby K M. The limb-girdle muscular dystrophies—multiple genes, multiple mechanisms. Hum Mol Genet. 1999;8:1875–1882. doi: 10.1093/hmg/8.10.1875. [DOI] [PubMed] [Google Scholar]
- 8.Bushby K M. Making sense of the limb-girdle muscular dystrophies. Brain. 1999;122:1403–1420. doi: 10.1093/brain/122.8.1403. [DOI] [PubMed] [Google Scholar]
- 9.Byers T J, Kunkel L M, Watkins S C. The subcellular distribution of dystrophin in mouse skeletal, cardiac, and smooth muscle. J Cell Biol. 1991;115:411–421. doi: 10.1083/jcb.115.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell K P. Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 11.Cartaud A, Coutant S, Petrucci T C, Cartaud J. Evidence for in situ and in vitro association between beta-dystroglycan and the subsynaptic 43K rapsyn protein. Consequence for acetylcholine receptor clustering at the synapse. J Biol Chem. 1998;273:11321–11326. doi: 10.1074/jbc.273.18.11321. [DOI] [PubMed] [Google Scholar]
- 12.Chao D S, Gorospe J R, Brenman J E, Rafael J A, Peters M F, Froehner S C, Hoffman E P, Chamberlain J S, Bredt D S. Selective loss of sarcolemmal nitric oxide synthase in Becker muscular dystrophy. J Exp Med. 1996;184:609–618. doi: 10.1084/jem.184.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coral-Vazquez R, Cohn R D, Moore S A, Hill J A, Weiss R M, Davisson R L, Straub V, Barresi R, Bansal D, Hrstka R F, Williamson R, Campbell K P. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: a novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98:465–474. doi: 10.1016/s0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- 14.Crosbie R H, Heighway J, Venzke D P, Lee J C, Campbell K P. Sarcospan, the 25-kDa transmembrane component of the dystrophin-glycoprotein complex. J Biol Chem. 1997;272:31221–31224. doi: 10.1074/jbc.272.50.31221. [DOI] [PubMed] [Google Scholar]
- 15.Crosbie R H, Lebakken C S, Holt K H, Venzke D P, Straub V, Lee J C, Grady R M, Chamberlain J S, Sanes J R, Campbell K P. Membrane targeting and stabilization of sarcospan is mediated by the sarcoglycan subcomplex. J Cell Biol. 1999;145:153–165. doi: 10.1083/jcb.145.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Amore P A, Brown R H, Jr, Ku P T, Hoffman E P, Watanabe H, Arahata K, Ishihara T, Folkman J. Elevated basic fibroblast growth factor in the serum of patients with Duchenne muscular dystrophy. Ann Neurol. 1994;35:362–365. doi: 10.1002/ana.410350320. [DOI] [PubMed] [Google Scholar]
- 17.Duclos F, Straub V, Moore S A, Venzke D P, Hrstka R F, Crosbie R H, Durbeej M, Lebakken C S, Ettinger A J, van der Meulen J, Holt K H, Lim L E, Sanes J R, Davidson B L, Faulkner J A, Williamson R, Campbell K P. Progressive muscular dystrophy in alpha-sarcoglycan-deficient mice. J Cell Biol. 1998;142:1461–1471. doi: 10.1083/jcb.142.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durbeej, M., R. D. Cohn, R. F. Hrstka, S. A. Moore, V. Allamand, B. L. Davidson, R. A. Williamson, and K. P. Campbell. Disruption of the β-sarcoglycan gene reveals pathogenetic complexity of limb-girdle muscular dystrophy type 2E. Mol. Cell, in press. [DOI] [PubMed]
- 19.Eberst R, Dai S, Klugbauer N, Hofmann F. Identification and functional characterization of a calcium channel gamma subunit. Pfluegers Arch. 1997;433:633–637. doi: 10.1007/s004240050324. [DOI] [PubMed] [Google Scholar]
- 20.Ervasti J M, Campbell K P. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 21.Ervasti J M, Campbell K P. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froehner S C, Adams M E, Peters M F, Gee S H. Syntrophins: modular adapter proteins at the neuromuscular junction and the sarcolemma. Soc Gen Physiol Ser. 1997;52:197–207. [PubMed] [Google Scholar]
- 23.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gee S H, Blacher R W, Douville P J, Provost P R, Yurchenco P D, Carbonetto S. Laminin-binding protein 120 from brain is closely related to the dystrophin-associated glycoprotein, dystroglycan, and binds with high affinity to the major heparin binding domain of laminin. J Biol Chem. 1993;268:14972–14980. [PubMed] [Google Scholar]
- 25.Gee S H, Madhavan R, Levinson S R, Caldwell J H, Sealock R, Froehner S C. Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins. J Neurosci. 1998;18:128–137. doi: 10.1523/JNEUROSCI.18-01-00128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grady R M, Grange R W, Lau K S, Maimone M M, Nichol M C, Stull J T, Sanes J R. Role of α-dystrobrevin in the pathogenesis of dystrophin-dependent muscular dystrophies. Nat Cell Biol. 1999;1:215–220. doi: 10.1038/12034. [DOI] [PubMed] [Google Scholar]
- 27.Hack A A, Ly C T, Jiang F, Clendenin C J, Sigrist K S, Wollmann R L, McNally E M. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J Cell Biol. 1998;142:1279–1287. doi: 10.1083/jcb.142.5.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasegawa M, Cuenda A, Spillantini M G, Thomas G M, Buee-Scherrer V, Cohen P, Goedert M. Stress-activated protein kinase-3 interacts with the PDZ domain of alpha1-syntrophin. A mechanism for specific substrate recognition. J Biol Chem. 1999;274:12626–12631. doi: 10.1074/jbc.274.18.12626. [DOI] [PubMed] [Google Scholar]
- 29.Heighway J, Betticher D C, Hoban P R, Altermatt H J, Cowen R. Coamplification in tumors of KRAS2, type 2 inositol 1,4,5 triphosphate receptor gene, and a novel human gene, KRAG. Genomics. 1996;35:207–214. doi: 10.1006/geno.1996.0340. [DOI] [PubMed] [Google Scholar]
- 30.Henry M D, Campbell K P. A role for dystroglycan in basement membrane assembly. Cell. 1998;95:859–870. doi: 10.1016/s0092-8674(00)81708-0. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman E P, Brown R H, Jr, Kunkel L M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 32.Holt K H, Campbell K P. Assembly of the sarcoglycan complex. Insights for muscular dystrophy. J Biol Chem. 1998;273:34667–34670. doi: 10.1074/jbc.273.52.34667. [DOI] [PubMed] [Google Scholar]
- 33.Ibraghimov-Beskrovnaya O, Ervasti J M, Leveille C J, Slaughter C A, Sernett S W, Campbell K P. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 34.Iwata Y, Pan Y, Yoshida T, Hanada H, Shigekawa M. Alpha1-syntrophin has distinct binding sites for actin and calmodulin. FEBS Lett. 1998;423:173–177. doi: 10.1016/s0014-5793(98)00085-4. [DOI] [PubMed] [Google Scholar]
- 35.Jay S D, Ellis S B, McCue A F, Williams M E, Vedvick T S, Harpold M M, Campbell K P. Primary structure of the gamma subunit of the DHP-sensitive calcium channel from skeletal muscle. Science. 1990;248:490–492. doi: 10.1126/science.2158672. [DOI] [PubMed] [Google Scholar]
- 36.Jung D, Duclos F, Apostol B, Straub V, Lee J C, Allamand V, Venzke D P, Sunada Y, Moomaw C R, Leveille C J, Slaughter C A, Crawford T O, McPherson J D, Campbell K P. Characterization of delta-sarcoglycan, a novel component of the oligomeric sarcoglycan complex involved in limb-girdle muscular dystrophy. J Biol Chem. 1996;271:32321–32329. doi: 10.1074/jbc.271.50.32321. [DOI] [PubMed] [Google Scholar]
- 37.Jung D, Yang B, Meyer J, Chamberlain J S, Campbell K P. Identification and characterization of the dystrophin anchoring site on beta-dystroglycan. J Biol Chem. 1995;270:27305–27310. doi: 10.1074/jbc.270.45.27305. [DOI] [PubMed] [Google Scholar]
- 38.Kameya S, Miyagoe Y, Nonaka I, Ikemoto T, Endo M, Hanaoka K, Nabeshima Y, Takeda S. Alpha1-syntrophin gene disruption results in the absence of neuronal-type nitric-oxide synthase at the sarcolemma but does not induce muscle degeneration. J Biol Chem. 1999;274:2193–2200. doi: 10.1074/jbc.274.4.2193. [DOI] [PubMed] [Google Scholar]
- 39.Kaye D, Pimental D, Prasad S, Maki T, Berger H J, McNeil P L, Smith T W, Kelly R A. Role of transiently altered sarcolemmal membrane permeability and basic fibroblast growth factor release in the hypertrophic response of adult rat ventricular myocytes to increased mechanical activity in vitro. J Clin Investig. 1996;97:281–291. doi: 10.1172/JCI118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letts V A, Felix R, Biddlecome G H, Arikkath J, Mahaffey C L, Valenzuela A, Bartlett F S, 2nd, Mori Y, Campbell K P, Frankel W N. The mouse stargazer gene encodes a neuronal Ca2+-channel gamma subunit. Nat Genet. 1998;19:340–347. doi: 10.1038/1228. [DOI] [PubMed] [Google Scholar]
- 41.Lim L E, Campbell K P. The sarcoglycan complex in limb-girdle muscular dystrophy. Curr Opin Neurol. 1998;11:443–452. doi: 10.1097/00019052-199810000-00006. [DOI] [PubMed] [Google Scholar]
- 42.Lim L E, Duclos F, Broux O, Bourg N, Sunada Y, Allamand V, Meyer J, Richard I, Moomaw C, Slaughter C, et al. Beta-sarcoglycan: characterization and role in limb-girdle muscular dystrophy linked to 4q12. Nat Genet. 1995;11:257–265. doi: 10.1038/ng1195-257. [DOI] [PubMed] [Google Scholar]
- 43.Lo C W. Genes, gene knockouts, and mutations in the analysis of gap junctions. Dev Genet. 1999;24:1–4. doi: 10.1002/(SICI)1520-6408(1999)24:1/2<1::AID-DVG1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Lumeng C, Phelps S, Crawford G E, Walden P D, Barald K, Chamberlain J S. Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat Neurosci. 1999;2:611–617. doi: 10.1038/10165. [DOI] [PubMed] [Google Scholar]
- 45.Maecker H T, Todd S C, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 46.Matsuda R, Nishikawa A, Tanaka H. Visualization of dystrophic muscle fibers in mdx mouse by vital staining with Evans blue: evidence of apoptosis in dystrophin-deficient muscle. J Biochem (Tokyo) 1995;118:959–964. doi: 10.1093/jb/118.5.959. [DOI] [PubMed] [Google Scholar]
- 47.Mayer U, Saher G, Fassler R, Bornemann A, Echtermeyer F, von der Mark H, Miosge N, Poschl E, von der Mark K. Absence of integrin alpha 7 causes a novel form of muscular dystrophy. Nat Genet. 1997;17:318–323. doi: 10.1038/ng1197-318. [DOI] [PubMed] [Google Scholar]
- 48.McNally E M, Yoshida M, Mizuno Y, Ozawa E, Kunkel L M. Human adhalin is alternatively spliced and the gene is located on chromosome 17q21. Proc Natl Acad Sci USA. 1994;91:9690–9694. doi: 10.1073/pnas.91.21.9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nigro V, Piluso G, Belsito A, Politano L, Puca A A, Papparella S, Rossi E, Viglietto G, Esposito M G, Abbondanza C, Medici N, Molinari A M, Nigro G, Puca G A. Identification of a novel sarcoglycan gene at 5q33 encoding a sarcolemmal 35 kDa glycoprotein. Hum Mol Genet. 1996;5:1179–1186. doi: 10.1093/hmg/5.8.1179. [DOI] [PubMed] [Google Scholar]
- 51.Noguchi S, McNally E M, Ben Othmane K, Hagiwara Y, Mizuno Y, Yoshida M, Yamamoto H, Bonnemann C G, Gussoni E, Denton P H, et al. Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science. 1995;270:819–822. doi: 10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- 52.Ohlendieck K, Campbell K P. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J Cell Biol. 1991;115:1685–1694. doi: 10.1083/jcb.115.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohlendieck K, Ervasti J M, Snook J B, Campbell K P. Dystrophin-glycoprotein complex is highly enriched in isolated skeletal muscle sarcolemma. J Cell Biol. 1991;112:135–148. doi: 10.1083/jcb.112.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozawa E, Noguchi S, Mizuno Y, Hagiwara Y, Yoshida M. From dystrophinopathy to sarcoglycanopathy: evolution of a concept of muscular dystrophy. Muscle Nerve. 1998;21:421–438. doi: 10.1002/(sici)1097-4598(199804)21:4<421::aid-mus1>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 55.Roberds S L, Anderson R D, Ibraghimov-Beskrovnaya O, Campbell K P. Primary structure and muscle-specific expression of the 50-kDa dystrophin-associated glycoprotein (adhalin) J Biol Chem. 1993;268:23739–23742. [PubMed] [Google Scholar]
- 56.Roberds S L, Leturcq F, Allamand V, Piccolo F, Jeanpierre M, Anderson R D, Lim L E, Lee J C, Tomé F M, Romero N B, et al. Missense mutations in the adhalin gene linked to autosomal recessive muscular dystrophy. Cell. 1994;78:625–633. doi: 10.1016/0092-8674(94)90527-4. [DOI] [PubMed] [Google Scholar]
- 57.Rosalki S B. Serum enzymes in disease of skeletal muscle. Clin Lab Med. 1989;9:767–781. [PubMed] [Google Scholar]
- 58.Rybakova I N, Amann K J, Ervasti J M. A new model for the interaction of dystrophin with F-actin. J Cell Biol. 1996;135:661–672. doi: 10.1083/jcb.135.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rybakova I N, Ervasti J M. Dystrophin-glycoprotein complex is monomeric and stabilizes actin filaments in vitro through a lateral association. J Biol Chem. 1997;272:28771–28778. doi: 10.1074/jbc.272.45.28771. [DOI] [PubMed] [Google Scholar]
- 60.Schultz J, Hoffmuller U, Krause G, Ashurst J, Macias M J, Schmieder P, Schneider-Mergener J, Oschkinat H. Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels. Nat Struct Biol. 1998;5:19–24. doi: 10.1038/nsb0198-19. [DOI] [PubMed] [Google Scholar]
- 61.Scott A F, Elizaga A, Morrell J, Bergen A, Penno M B. Characterization of a gene coamplified with Ki-ras in Y1 murine adrenal carcinoma cells that codes for a putative membrane protein. Genomics. 1994;20:227–230. doi: 10.1006/geno.1994.1157. [DOI] [PubMed] [Google Scholar]
- 62.Straub V, Campbell K P. Muscular dystrophies and the dystrophin-glycoprotein complex. Curr Opin Neurol. 1997;10:168–175. doi: 10.1097/00019052-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 63.Straub V, Duclos F, Venzke D P, Lee J C, Cutshall S, Leveille C J, Campbell K P. Molecular pathogenesis of muscle degeneration in the delta-sarcoglycan-deficient hamster. Am J Pathol. 1998;153:1623–1630. doi: 10.1016/s0002-9440(10)65751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Straub V, Ettinger A J, Durbeej M, Venzke D P, Cutshall S, Sanes J R, Campbell K P. Epsilon-sarcoglycan replaces alpha-sarcoglycan in smooth muscle to form a unique dystrophin-glycoprotein complex. J Biol Chem. 1999;274:27989–27996. doi: 10.1074/jbc.274.39.27989. [DOI] [PubMed] [Google Scholar]
- 65.Straub V, Rafael J A, Chamberlain J S, Campbell K P. Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J Cell Biol. 1997;139:375–385. doi: 10.1083/jcb.139.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taverna D, Disatnik M H, Rayburn H, Bronson R T, Yang J, Rando T A, Hynes R O. Dystrophic muscle in mice chimeric for expression of alpha5 integrin. J Cell Biol. 1998;143:849–859. doi: 10.1083/jcb.143.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 68.Williamson R A, Henry M D, Daniels K J, Hrstka R F, Lee J C, Sunada Y, Ibraghimov-Beskrovnaya O, Campbell K P. Dystroglycan is essential for early embryonic development: disruption of Reichert's membrane in Dag1-null mice. Hum Mol Genet. 1997;6:831–841. doi: 10.1093/hmg/6.6.831. [DOI] [PubMed] [Google Scholar]
- 69.Wissenbach U, Bosse-Doenecke E, Freise D, Ludwig A, Murakami M, Hofmann F, Flockerzi V. The structure of the murine calcium channel gamma-subunit gene and protein. Biol Chem. 1998;379:45–50. doi: 10.1515/bchm.1998.379.1.45. [DOI] [PubMed] [Google Scholar]
- 70.Wright M D, Tomlinson M G. The ins and outs of the transmembrane 4 superfamily. Immunol Today. 1994;15:588–594. doi: 10.1016/0167-5699(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 71.Yang B, Jung D, Motto D, Meyer J, Koretzky G, Campbell K P. SH3 domain-mediated interaction of dystroglycan and Grb2. J Biol Chem. 1995;270:11711–11714. doi: 10.1074/jbc.270.20.11711. [DOI] [PubMed] [Google Scholar]
- 72.Yeager M. Structure of cardiac gap junction intercellular channels. J Struct Biol. 1998;121:231–245. doi: 10.1006/jsbi.1998.3972. [DOI] [PubMed] [Google Scholar]
- 73.Yeager M, Unger V M, Falk M M. Synthesis, assembly and structure of gap junction intercellular channels. Curr Opin Struct Biol. 1998;8:517–524. doi: 10.1016/s0959-440x(98)80131-0. [DOI] [PubMed] [Google Scholar]