Abstract
Meat quality data can only be obtained after slaughter when selection decisions about the live animal are already too late. Carcass estimated breeding values present major precision problems due to low accuracy, and by the time an informed decision on the genetic merit for meat quality is made, the animal is already dead. We report for the first time, a targeted next-generation sequencing (NGS) of single nucleotide polymorphisms (SNP) of lipid metabolism genes in Tattykeel Australian White (TAW) sheep of the MARGRA lamb brand, utilizing an innovative and minimally invasive muscle biopsy sampling technique for directly quantifying the genetic worth of live lambs for health-beneficial omega-3 long-chain polyunsaturated fatty acids (n-3 LC-PUFA), intramuscular fat (IMF), and fat melting point (FMP). NGS of stearoyl-CoA desaturase (SCD), fatty acid binding protein-4 (FABP4), and fatty acid synthase (FASN) genes identified functional SNP with unique DNA marker signatures for TAW genetics. The SCD g.23881050T>C locus was significantly associated with IMF, C22:6n-3, and C22:5n-3; FASN g.12323864A>G locus with FMP, C18:3n-3, C18:1n-9, C18:0, C16:0, MUFA, and FABP4 g.62829478A>T locus with IMF. These add new knowledge, precision, and reliability in directly making early and informed decisions on live sheep selection and breeding for health-beneficial n-3 LC-PUFA, FMP, IMF and superior meat-eating quality at the farmgate level. The findings provide evidence that significant associations exist between SNP of lipid metabolism genes and n-3 LC-PUFA, IMF, and FMP, thus underpinning potential marker-assisted selection for meat-eating quality traits in TAW lambs.
Keywords: SNP, FASN, SCD, FABP4, IMF, FMP, eating quality, TAW MARGRA lamb, biopsy, n-3 LC-PUFA
1. Introduction
Eating quality is the single largest determinant of consumer acceptability and satisfaction with meat products. The eating and nutritional quality of lamb is influenced by intramuscular fat (IMF) content [1], fat melting point (FMP), tenderness, juiciness, flavor [2], and health-promoting omega-3 long-chain polyunsaturated fatty acids (n-3 LC-PUFA) that optimize retinal, maternal, and childhood brain functions while minimizing the risks associated with cardiovascular and chronic diseases [3,4].
In a recent review of the development, calibration, and validation of objective measurement technologies for carcass composition, lean, fat, and meat-eating quality traits in the Australian and New Zealand livestock industries, Gardner et al. [5] highlighted the inherent difficulties associated with the poor measurement of meat-eating quality and lean meat yield. Attempts to predict IMF [6,7,8], intramuscular connective tissue [9], composition and quality characteristics [10], tenderness, ultimate pH, and IMF content [11,12,13] from near infra-red based regression equations were characterized by low accuracy, inconsistency, and divergence between calibration and validation data. Such inaccuracies lead to lamb inefficiencies and an estimated annual value-chain wastage costs of $130 million to the Australian beef industry [5].
However, meat quality data can only be obtained after slaughter when selection decisions about the live animal are already too late. Carcass estimated breeding values [14,15], visual marbling score and meat imaging camera marbling systems [16], and dual X-ray absorptiometry scanner based computed tomography determined fat, lean muscle, and bone compositions of lamb carcasses [17] are all useful technological advancements, but still present precision problems due to low accuracy, and by the time an informed decision on the genetic merit for meat quality is made, the animal is already dead. In a study of associations of sire estimated breeding values and objective meat quality measurements with sensory scores in Australian lamb, Pannier et al. [18] confirmed the growing concerns that selecting for lean meat yield would reduce consumer eating quality and concluded that careful monitoring of selection programs is needed to maintain lamb eating quality. In an experimental trial to understand the impact of sire lean meat yield breeding value on carcass composition, meat quality, nutrient, and mineral content of Australian lamb, Knight et al. [14] concluded that to avoid deterioration in meat quality, the nutritional content of lamb and fresh meat color, Australian sheep producers will need to incorporate other aspects of meat quality when selecting sires with increased lean meat yield. To date, the use of conventional laboratory-based fat extraction, ‘slip point’ and gas chromatography methods still remain the most accurate techniques for not only measuring IMF, FMP, and n-3 LC-PUFA, but also for predicting consumer acceptance of beef and sheep meat [19]. Herein, we report for the first time, a combination of an innovative and minimally invasive longissimus dorsi thoracis et lumborum muscle biopsy sampling of Tattykeel Australian White (TAW) sheep exclusive to MARGRA lamb brand, laboratory-based IMF, FMP, and fatty acid analyses, and advanced genomics technique of next-generation sequencing (NGS) of single nucleotide polymorphisms (SNP) of lipid metabolism genes for directly quantifying the genetic worth of live lambs for health-beneficial n-3 LC-PUFA, IMF, and FMP. The primary objective was to conduct a NGS of stearoyl-CoA desaturase (SCD), fatty acid binding protein-4 (FABP4), and fatty acid synthase (FASN) lipogenic genes to identify functional SNP that provide unique DNA marker signatures for TAW genetics, breeding, and selection programs for meat-eating quality. The hypothesis tested was that significant associations exist between SNP of lipid metabolism genes and n-3 LC-PUFA, FMP, and FMP underpinning potential marker-assisted selection for meat-eating quality traits in TAW lambs.
2. Materials and Methods
2.1. Animals and Experimental Design
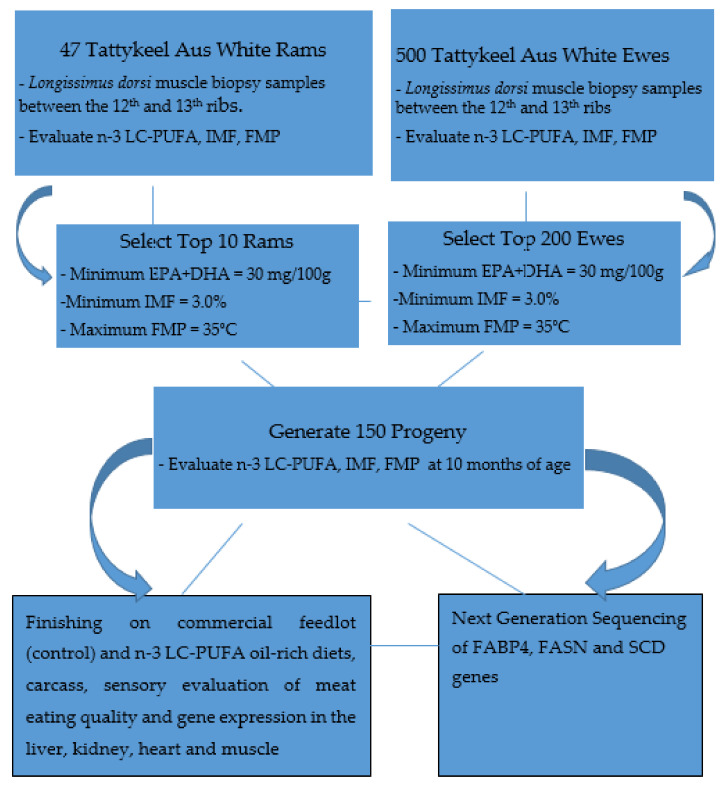
The experimental design for the selection, breeding, and evaluation of n-3 LC-PUFA, IMF, and FMP in Tattykeel Australian White (TAW) sheep is shown in Figure 1.
Figure 1.
Experimental design for the selection, breeding, and evaluation of n-3 LC-PUFA, IMF, and FMP in Tattykeel Australian White sheep.
Three composite generations—parental, first (F1), and second (F2) composite generations of lambs were bred, raised, and maintained under the same management at the Tattykeel Australian White Stud in Black Springs, Oberon, New South Wales, Australia. The parental composite generation comprised 47 rams mated to 500 ewes after evaluating their longissimus dorsi thoracis et lumborum muscle biopsy samples for health-beneficial n-3 LC-PUFA, IMF, and FMP with minimum thresholds set at 30 mg/100 g, 3.0%, and 35 °C, respectively. The top 10 rams and 200 ewes were selected and mated to generate 150 progeny whose muscle biopsy samples were laboratory tested for n-3 LC-PUFA, IMF, FMP, and genomic DNA sequenced at 10 months of age prior to being finished at a commercial feedlot. The Poll Dorset and Texel were used as positive control and the Rambouillet as the negative control in assessing extracted genomic DNA, polymerase chain reaction products, and next-generation sequencing procedures in the laboratory. Details of the muscle biopsy procedure and laboratory analyses of IMF, FMP, and fatty acid composition had already been published [2] and are only briefly summarized below.
2.2. Muscle Biopsy Sampling Procedure
The biopsy procedure for sampling the Longissimus dorsi muscle from the 12th–13th ribs was first described in cattle [20] and modified in sheep [2]. Pewan et al. [2] published the details of the biopsy procedure in sheep, and these will not be repeated here.
2.3. Determination of Intramuscular Fat
Details of the procedures for laboratory analysis of intramuscular fat were published by Pewan et al. [2], Holman et al. [21], and Flakemore et al. [22] and will not be repeated here.
2.4. Determination of Fat Melting Point
Details of the laboratory analysis of fat melting point were published by Pewan et al. [2], Holman et al. [21], and Flakemore et al. [22] and needless to repeat herein.
2.5. Determination of Fatty Acid Composition
Fatty acid composition including n-3 LC-PUFA analysis of Longissimus dorsi muscle biopsy samples was analyzed by means of gas chromatography–mass spectrophotometry procedure described in detail by Malau-Aduli et al. [23] based on modified Bligh and Dyer [24], Miller et al. [25], and Clayton [26] methods. Details were published by Pewan et al. [2].
2.6. Blood Collection and Genomic DNA Extraction
About 10 mL of blood was collected from Tattykeel Australian White, Poll Dorset, and Texel (positive control) lambs of the same age and under the same management conditions by jugular venipuncture into vacutainers containing EDTA. Blood samples were stored at −80 °C until ready for genomic DNA (gDNA) extraction. gDNA was extracted from 2 mL of blood using NucleoSpin Blood Kits (Macherey-Nagel GmbH and Co. KG, Neumann-Neander-Str. 6-8. 52355 Duren, Germany) according to the manufacturer’s protocol. gDNA yield was quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop, Thermo Fisher Scientific Australia Pty Ltd., Scoresby, Victoria, Australia).
2.7. Primer Design
2.7.1. FASN, FABP4, and SCD Primers
All primers were designed using Geneious Prime Software Program 2020 v.2.2 (http://www.geneious.com). A targeted candidate gene approach of lipid metabolism genes (FASN, FABP4, and SCD) was utilized. Single coding sequences of each gene deposited in the National Center for Biotechnology Information (NCBI) database (Genbank) of FASN, FABP4, and SCD of Ovis aries breed were used as reference points. In order to amplify the 18 kb of the FASN gene (Accession Number: NC_040262.1), a long-range PCR approach was used to split the gene sequence into 3 overlapping fragments of 8.5 kb each (FASN1, FASN2, and FASN3), comprising approximately 91% of the total gene sequence. For the 4 kb FABP4 (NC_040260.1) and 12 kb SCD (NC_040273.1) gene fragments, a single primer set was designed as shown in Table 1. All primers were synthesized at Integrated DNA Technologies Pte. Ltd., Melbourne, Australia (Itddna.Com (accessed on 12 June 2021)).
2.7.2. Long-Range PCR
Due to the different fragment lengths and DNA composition, it was necessary to use 3 different long-range PCR approaches to amplify the FASN, FABP4, and SCD genes. During optimization, all 3 approaches were tested for all 3 genes, but only the best performing combinations were utilized.
2.7.3. FASN Gene
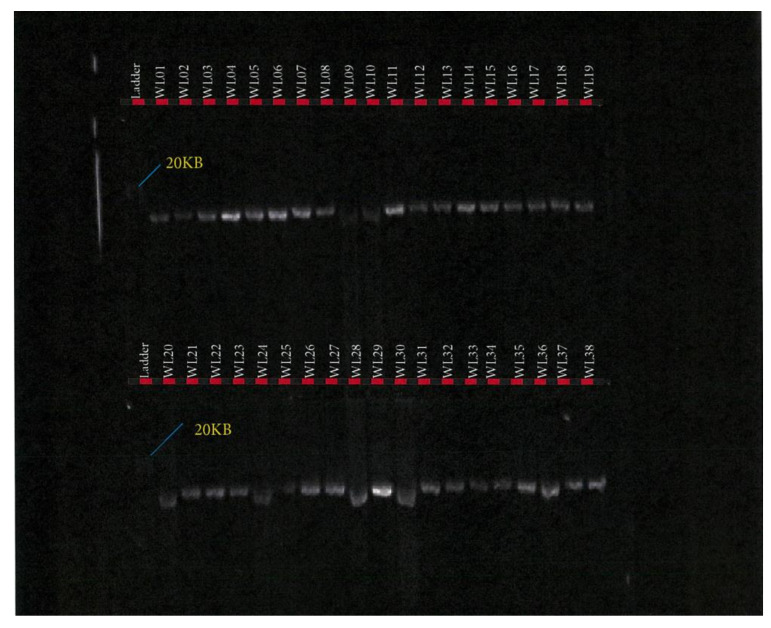
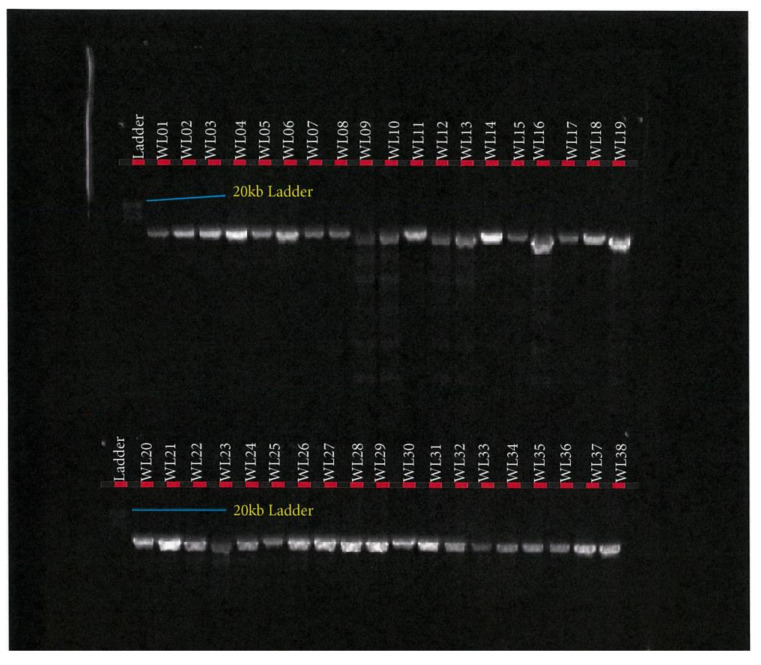
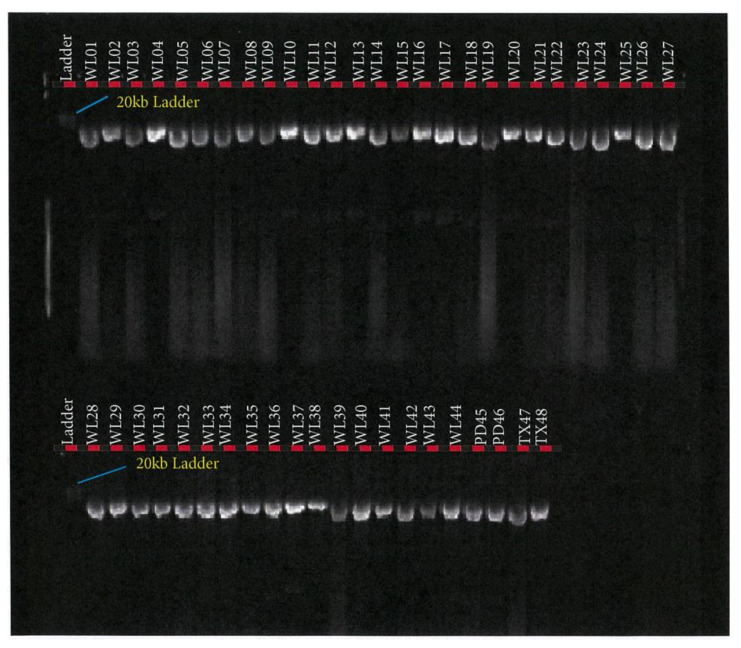
FASN PCR amplification assay was performed using the TakaRa PrimeSTAR GXL Master Mix (TaKaRa Bio Inc., Kusatsu, Shiga, Japan). PCR reaction assay was set up in a total volume of 50 µL containing 10 µL of 5× TakaRa PrimeSTAR GXL Buffer, 200 μM of TaKaRa dNTP Mixture, 1.25 units of TaKaRa PrimeSTAR GXL DNA Polymerase, 0.2 μM of each primer (IDT, Melbourne, Australia), and 100 ng of DNA template. PCR was performed in a SimpliAmp™ Thermal Cycler (Thermofisher Scientific, Melbourne, Australia), in a 2-step protocol using the following conditions: 98 °C initial denaturation for 1 min (1 cycle); 98 °C denaturation for 10 s; 68 °C annealing/extension for 10 min for 30 cycles. PCR success was checked in 0.8% agarose gel electrophoresis as depicted in Figure 2, Figure 3 and Figure 4.
Figure 2.
FASN fragment 1 PCR product in Tattykeel Australian White (WL), Poll Dorset (PD), and Texel (TX) lambs.
Figure 3.
FASN fragment 2 PCR product in Tattykeel Australian White (WL), Poll Dorset (PD), and Texel (TX) lambs.
Figure 4.
FASN fragment 3 PCR product in Tattykeel Australian White (WL), Poll Dorset (PD), and Texel (TX) lambs.
2.7.4. FABP4 and SCD
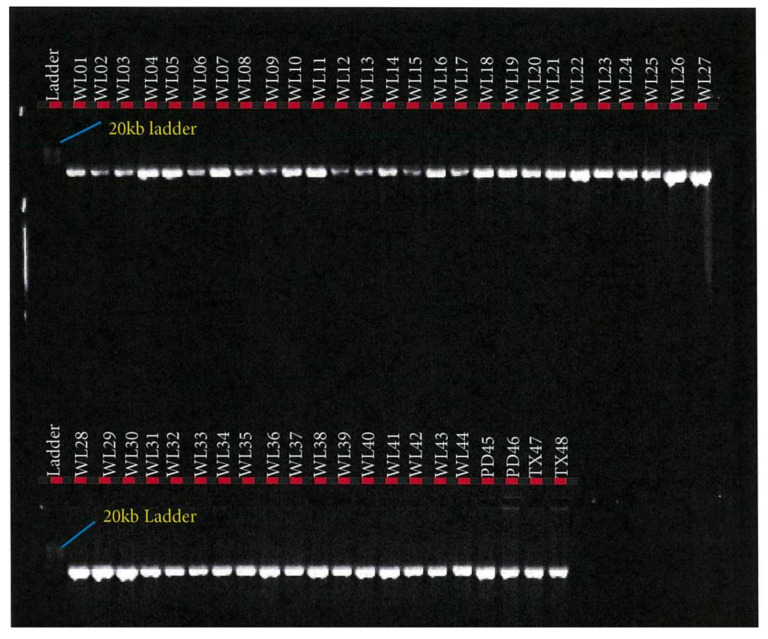
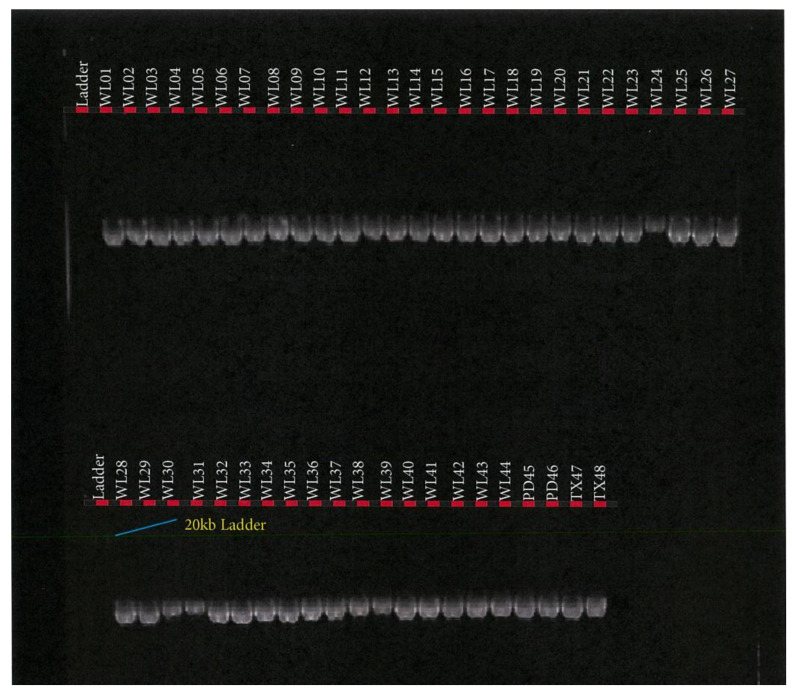
For the FAPB4 gene, Platinum™ SuperFi™ II PCR Master Mix (Thermofisher Scientific, Australia) was used, while for the SCD gene, Hot Start II High-Fidelity PCR Master Mix (Thermofisher Scientific, Australia) was used under the same PCR conditions. The amplification reactions were performed in a total volume of 50 µL containing 25 µL of 2× Platinum™ SuperFi™ II PCR Master Mix or Phusion Hot Start II High-Fidelity PCR Master Mix (Thermofisher Scientific, Australia), 0.5 µM of each primer (IDT, Australia), and 100 ng of DNA template. PCR was performed in a SimpliAmp™ Thermal Cycler (Thermofisher Scientific, Australia), in a 3-step protocol, using the following conditions: 98 °C initial denaturation in 1 min (1 cycle); 98 °C for denaturation 15 s; 60 °C (FABP4)/and 65 °C (SCD) annealing for 15 s; 72 °C extension for 9 min; 72 °C final extension for 9 min; 4 °C hold for 35 cycles. PCR success was checked in 0.8% agarose gel electrophoresis, as depicted in Figure 5 and Figure 6.
Figure 5.
SCD PCR product in Tattykeel Australian White (WL), Poll Dorset (PD), and Texel (TX) lambs.
Figure 6.
FABP4 PCR product of Tattykeel Australian White (WL), Poll Dorset (PD), and Texel (TX) lambs.
2.8. PCR Clean-Up
Sera-Mag™ SpeedBeads was prepared according to Faircloth et al. [27] and used to clean the PCR products using a Zephyr NGS Workstation (Caliper Lifesciences, Perkin-Elmer) and quantified using a Promega dsDNA Quantifluor System Kit (Ref: E2670, 00002484139) on an Enspire Workstation (Perkin-Elmer). The 5 different PCR products were pooled at approximately 0.4 nM to ensure even coverage during sequencing using Quantifluor dsDNA System (Promega, Madison, WI, USA). The products were normalized to 2 ng/µL using 10 mM Tris-HCl (pH 8.0). Final dilution to 0.2 ng/µL with 10 mM Tris-HCl (pH 8.0) was conducted in preparation for library preparation and final accuracy checks using the Illumina NexteraXT DNA.
2.9. Library Preparation, Quantification, Normalization, and Sequencing
Libraries were prepared using Nextera XT DNA Library Prep kit (Illumina, CA, USA) in accordance with the manufacturer’s protocols using the recommended input of 5 µL of 0.2 ng/µL gDNA per sample. This was followed by Sera-Mag™ SpeedBeads purification using 0.6× beads and 2 washes using 80% ethanol to select fragments > 250 bp and remove unincorporated adapters. Each DNA library fragment size and concentration was determined using Agilent High Sensitivity D5000 reagents and ScreenTape on the Tape Station 4200 Instrument (Agilent Technologies, Santa Clara, CA, USA) according to the Agilent assay quick guide. Additionally, all individual libraries were quantified using QuantiFluor® dsDNA System (Promega, Madison, WI, USA) to give an additional concentration estimate. The resultant size and concentration data from Tape Station and Quantifluor system were used to normalize each library to 4 nM by diluting with 10 mM Tris-HCl (pH 8.5) prior to pooling. An equal volume of 5 µL was pooled and sequenced on an Illumina MiSeq benchtop sequencer, using a 500-cycle MiSeq Reagent Nano Kit v2 with a 10 pM input and 10% PhiX spike-in.
2.10. Bioinformatics and Next Generation Sequencing Data Analysis
Genomic data analysis was performed using commercial bioinformatics program Geneious Prime software program 2020 v.2.24 (http://www.geneious.com (accessed on 12 June 2021)) to analyze the sequences. The following reference sequences deposited in the NCBI database were used for comparative analysis: NC_040262.1, NC_040260.1, and NC_040273.1 for FASN, FABP4, and SCD genes, respectively. Next Generation Sequenced data were retrieved from Illumina Dashboard-BaseSpace Sequence Hub (https://basespace.illumina.com/dashboard (accessed on 15 July 2021)) as paired read data in 2 separate forward and reverse read lists in FASTQ format. The retrieved raw reads were subjected to quality control measures. Reads were trimmed and adapters removed using the BBDuk trimmer in Geneious Prime 2020 v.2.2 with the default setting for paired-end reads. The Quality (Q) value of Phred score was set at 20 to improve sequenced data and increase the likelihood of calling true SNPs to 99%. Short reads with a minimum length of 20 bp were discarded, resulting in clean reads. Regions of low coverage were excluded when calling SNPs using the Annotate and Predict → Find Low/High Coverage. The reads were mapped to reference in Geneious. The reference sequences were retrieved from NCBI database (Genbank) of FASN, FABP4, and SCD of Ovis aries breed. The Sensitivity was set on the Medium Sensitivity/Fast and Fine-Tuning (iterate up to 5 times) option selected to improve the results by aligning reads to each other in addition to the reference sequence. Major allele frequencies from the next-generation sequence data based on observed and expected genotypes were computed using the Hardy–Weinberg equilibrium principle as described by Graffelman et al. [28].
2.11. Statistical Analyses
All statistical analyses of the associations between detected SNP of the 3 genes and meat-eating quality traits were performed using R statistical software version 3.6.3 [29]. Linkage disequilibrium as an index of non-random association between alleles of different loci, was estimated as the difference between the frequency of gametes carrying the pair of alleles A and B at two loci (pAB) and the product of the frequencies of those alleles (pA and pB), DAB = pAB − pApB, where the allele pair AB is a haplotype and pAB is the haplotype frequency [30]. Major and minor allele frequencies were computed, and the Hardy–Weinberg Equilibrium was tested using the chi-square test. Pearson’s residual correlation analysis was carried out to examine the relationships between genomic variants and meat quality traits (FA, FMP, and IMF). Linear mixed models procedure was used to investigate differences in FMP, IMF, and fatty acid profiles of the TAW lambs due to FABP4, SCD, and FASN variants fitting the fixed effect of allele substitution for individual SNP and random effect of animal (for pedigree) accounting for composite generation effects. Functional allele mutations at the coding regions of identified FABP4, SCD, and FASN loci were statistically analyzed for association with FMP, IMF, and fatty acids. Least-square means were compared using the Tukey-adjusted multiple comparisons test. The full statistical model was:
where Yij = dependent variable (FMP, IMF, FA) of jth TAW of ith composite generation, µ = overall mean, αi = effect of the ith composite generation, FA = the genotype FASN (AA, GA and GG), SC = the genotype SCD (CC, CT and TT), SK = the genotype FABP4 (GG, GA and AA), γ = effect of the genotype, and eij = residual error.
3. Results
This study of SCD, FASN, and FABP4 lipogenic genes SNP in TAW lamb muscle biopsy samples bred, selected, and evaluated as per the experimental design shown in Figure 1, was based on the Geneious-designed primers whose sequences are presented in Table 1 and successful polymerase chain reactions (PCR) products are presented in Figure 2, Figure 3, Figure 4, Figure 5 and Figure 6.
Table 1.
Primer sequences for FABP4, FASN, and SCD polymerase chain reaction assays #.
| Gene | Sequence | Length (bp) | Ta (°C) | Fragment Length (bp) | |
|---|---|---|---|---|---|
| FASN 1 | Forward | CCTACTTTCCCATGCTCAGAGAA | 23 | 68 | 7890 |
| Reverse | CTACGTTGCTGAGGAAGAACTCTA | 24 | 68 | ||
| FASN 2 | Forward | ACCGTCTCTCCTTCTTCTTTGAC | 23 | 68 | 8798 |
| Reverse | GAAGTTGAGGGAGGCGTAATAGAT | 24 | 68 | ||
| FASN 3 | Forward | CTAGAGTTCTTCCTCAGCAACGTA | 24 | 68 | 9288 |
| Reverse | GCCAGGGAGCTGTGAATAATACTA | 24 | 68 | ||
| FABP4 | Forward | TTGTTGAATGGCTGGGCTTATAAC | 24 | 60 | 4107 |
| Reverse | TAAGAAAATACTTCCTGGGGCACA | 24 | 60 | ||
| SCD | Forward | CAAACTTAGGTCTGCAACTTTCGT | 24 | 65 | 11,545 |
| Reverse | TTTCCCACTTCAACTCACCCTATT | 24 | 65 |
#FASN, Fatty Acid Synthase; FABP4, Fatty Acid Binding Protein 4; SCD, Stearoyl-CoA Desaturase; Ta, annealing temperature.
3.1. SCD, FASN, and FABP4 Gene SNP Variants and Genotypes
Using the Poll Dorset and Texel as positive controls, and Rambouillet as negative controls, eight SCD gene SNP loci (g.23880613A>G; g.23881050T>C; g.23883280G>A; g.23885910C>A; g.23887165A>G; g.23888763C>T; g.23889346T>G; g.23890209T>C) with major allele frequencies ranging from 0.53 to 0.93 were identified as depicted in Table 2. It was evident from Table 2 that TAW lambs were all heterozygous at three loci (g.23881050T>C, g.23883280G>A g.23885910C>A) in the parental, first, and second composite generations, thereby presenting a genetic divergence from the homozygous variants seen in the Poll Dorset, Texel and Rambouillet controls.
Table 2.
SCD gene SNP (major allele frequency) in TAW 1, Poll Dorset (+control), and Rambouillet (−control) lambs.
| Lamb breed, generation, type of control and genotypes (major allele frequencies in brackets) Parental composites 1st (F1) and 2nd (F2) composites Positive (+) and negative (−) controls | ||||||
| SNP Locus |
TAW Parents
(n = 147) |
TAW F1
(n = 75) |
TAW F2
(n = 75) |
Poll Dorset
(+n = 2) |
Texel
(+n = 2) |
Rambouillet
(−n = 2) |
| g.23880613A>G | GG (0.82) | GG (0.93) | GG (0.73) | GG | GG | AA |
| g.23881050T>C | CT (0.58) | CT (0.54) | CT (0.90) | CC | CC | TT |
| g.23883280G>A | AG (0.53) | AG (0.71) | AG (0.60) | AA | AA | GG |
| g.23885910C>A | AC (0.57) | AC (0.71) | AC (0.53) | CC | CC | CC |
| g.23887165A>G | GA (0.69) | GG (0.82) | GG (0.70) | GG | GG | AA |
| g.23888763C>T | TC (0.58) | TC (0.54) | CC (0.93) | CC | CC | CC |
| g.23889346T>G | GT (0.68) | GG (0.82) | GG (0.70) | GG | GG | TT |
| g.23890209T>C | CT (0.67) | CC (0.82) | CC (0.70) | CC | CC | TT |
1 TAW, Tattykeel Australian White.
As depicted in Table 3, nine functional SNP covering 91% of the FASN gene sequence were identified. The genotypes at the nine loci were all the same in TAW, indicating a consistent heredity pattern from the composite TAW parents to the first and second generations, which were all distinguishable from the Rambouillet negative control breed. For the FABP4 gene, three SNP loci were genotyped with major allele frequencies ranging from 0.50 to 0.97 (Table 4).
Table 3.
FASN gene SNP (major allele frequency) in TAW 1, Poll Dorset (+control) and Rambouillet (−control) lambs.
| Lamb breed, generation, type of control, and genotypes (major allele frequencies in brackets) Parental composite 1st (F1) and 2nd (F2) composites, Positive (+) and negative (−) controls | ||||||
| SNP Locus |
TAW Parents
(n = 147) |
TAW F1
(n = 75) |
TAW F2
(n = 75) |
Poll Dorset
(+n = 2) |
Texel
(+n = 2) |
Rambouillet
(−n = 2) |
| g.12316077T>G | GG (0.89) | GG (0.86) | GG (0.95) | GG | GG | TT |
| g.12318491A>G | GG (0.89) | GG (0.86) | GG (0.95) | GG | GG | AA |
| g.12320583T>C | CC (0.89) | CC (0.86) | CC (0.97) | CC | CC | TT |
| g.12321671T>C | CC (0.89) | CC (0.86) | CC (0.97) | CC | CC | TT |
| g.12323864A>G | GA (0.70) | GA (0.69) | GA (0.70) | GG | GG | AA |
| g.12324288G>A | AG (0.69) | AG (0.68) | AG (0.69) | AA | AA | GG |
| g.12326992T>C | CC (0.88) | CC (0.79) | CC (0.90) | CC | CC | TT |
| g.12327084->CT | CT (0.50) | CT (0.50) | CT (0.50) | CT | CT | TT |
| g.12328120T>C | CC (0.89) | CC (0.86) | CC (0.97) | CC | CC | TT |
1 TAW, Tattykeel Australian White.
Table 4.
FABP4 gene SNP (major allele frequency) in TAW 1, Poll Dorset (+control) and Rambouillet (−control) lambs.
| Lamb breed, generation, type of control, and genotypes (major allele frequencies in brackets) Parental composites 1st (F1) and 2nd (F2) composites Positive (+) and negative (−) controls | ||||||
| SNP Locus |
TAW Parents
(n = 147) |
TAW F1
(n = 75) |
TAW F2
(n = 75) |
Poll Dorset
(+n = 2) |
Texel
(+n = 2) |
Rambouillet
(−n = 2) |
| g.62826961T>C | CT (0.61) | TT (0.64) | CT (0.60) | TT | TT | TT |
| g.62826965C>G | GC (0.61) | GC (0.57) | GC (0.60) | GG | GG | CC |
| g.62829478A>T | AT (0.55) | AT (0.61) | AT (0.53) | AA | AA | AA |
1 TAW, Tattykeel Australian White.
3.2. Correlations between SCD, FASN, and FABP4 Gene SNP, FMP, IMF, and Fatty Acids
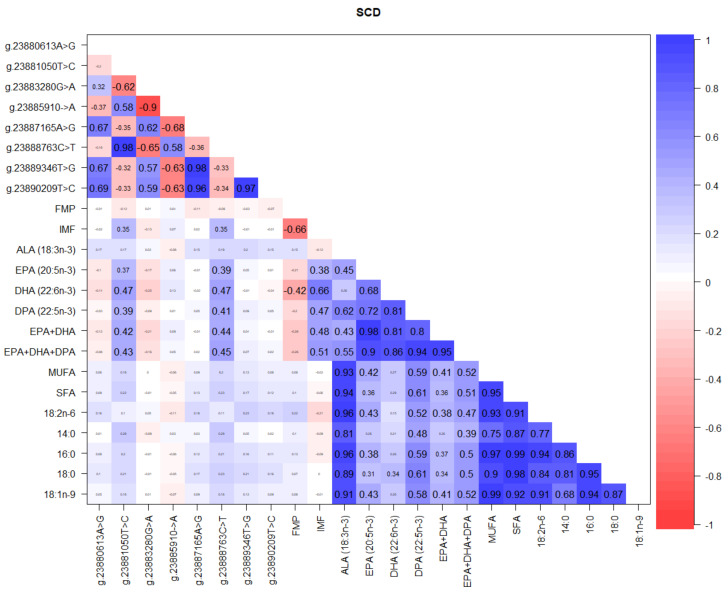
Figure 7 shows significant correlations between detected SCD SNP loci, several fatty acids and other meat-eating quality traits. Among SCD SNP loci, the highest correlations of 0.98 were observed between g.23888763C>T and g.23881050T>C; g.23889346T>G and g.23887165A>G. Moderate correlations between health-promoting n-3 LC-PUFA (EPA, DHA, and DPA), and g.23888763C>T and g.23881050T>C loci ranging from 0.37 to 0.47 were observed. IMF was moderately to highly correlated with n-3 LC-PUFA (0.38–0.66), while FMP was negatively correlated with IMF (−0.66) and DHA (−0.42). Among the different fatty acids and their summations, very high correlations of up to 0.99 were evident (Figure 7).
Figure 7.
Correlations between SCD gene SNP loci, IMF, FMP, and fatty acids in TAW lambs.
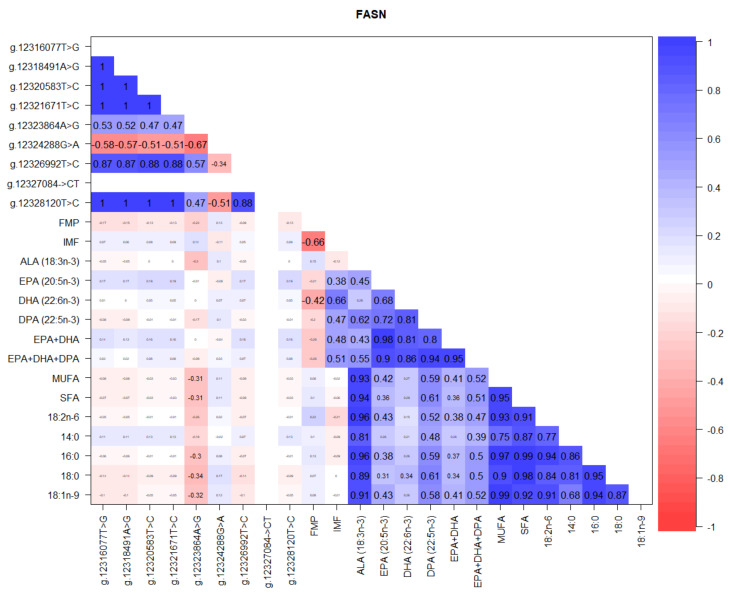
Figure 8 shows that among FASN gene SNP, there were highly significant correlations between the loci, while correlations between the g.12323864A>G locus and most fatty acids were negative, ranging from −0.3 to −0.34. Negative correlations between IMF and FMP (−0.66) and DHA (−0.42) were also observed, while the highest positive correlations were between the various fatty acids (Figure 8).
Figure 8.
Correlations between FASN gene SNP loci, IMF, FMP, and fatty acids in TAW lambs.
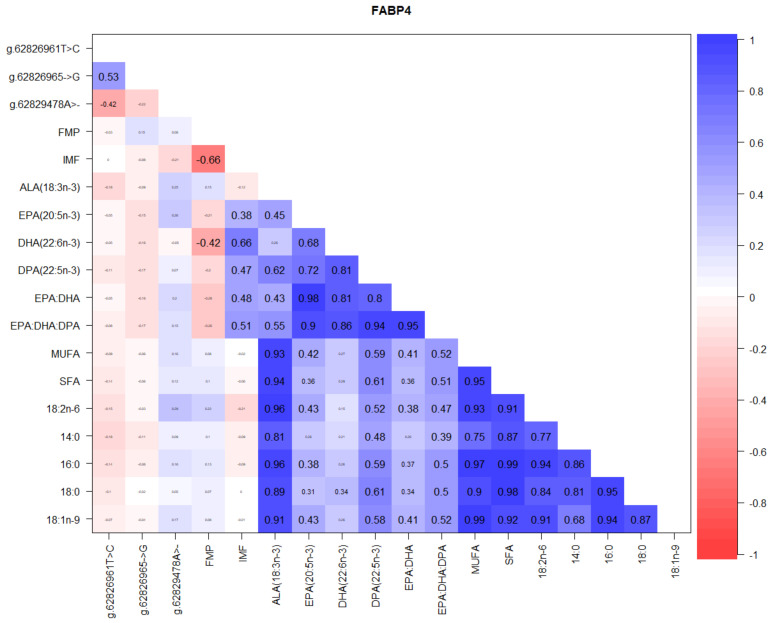
Figure 9 shows that among FABP4 gene SNP, the highest correlation of 0.53 was between the loci g.62826965C>G and g.62826961T>C, while a negative correlation of −0.42 was observed between g.62826965C>G and g.62829478A>T. Consistently positive correlations between IMF and n-3 LC-PUFA of up to 0.66 with DHA, 0.47 with DPA, and 0.38 with EPA were also observed, while the highest positive correlations were among the various fatty acids and their summations (Figure 9).
Figure 9.
Correlations between FABP4 gene SNP loci, IMF, FMP, and fatty acids in TAW lambs.
3.3. Associations between SCD, FASN and FABP4 SNP, FMP, IMF, and Fatty Acids
Descriptive statistics of mean, standard deviation, and coefficient of variation of the meat quality traits and full suite of fatty acids breakdown are presented in Table 5. FMP had a mean of 33.65 °C with a standard deviation of 2.74 and coefficient of variation of 8.14%, while IMF averaged 4.43% with a standard deviation of 1.31 and coefficient of variation of 29.58%.
Table 5.
Associations between SNP mutations and FMP, IMF, and fatty acids in TAW lambs #.
| SNP effect (p-values) SCD FABP4 FASN | ||||||
| Variable | Mean | SD | CV (%) | g.23881050T>C | g.62829478A>T | g.12323864A>G |
| FMP (°C) | 33.65 | 2.74 | 8.14 | 0.2700 | 0.6115 | 0.0544 * |
| IMF (%) | 4.43 | 1.31 | 29.58 | 0.0089 ** | 0.0539 * | 0.1915 |
| Fatty acids (mg/100 g) | ||||||
| ALA (C18:3n-3) | 163.03 | 192.27 | 117.94 | 0.7755 | 0.1419 | 0.0033 ** |
| EPA (C20:5n-3) | 25.20 | 11.62 | 46.10 | 0.7683 | 0.1023 | 0.9810 |
| DHA (C22:6n-3) | 8.43 | 4.16 | 49.27 | 0.0111 * | 0.2145 | 0.9480 |
| DPA (C22:5n-3) | 23.85 | 13.70 | 57.44 | 0.0532 * | 0.3894 | 0.0927 |
| EPA + DHA | 33.64 | 14.75 | 43.84 | 0.2036 | 0.4794 | 0.9915 |
| EPA + DHA + DPA | 57.49 | 26.97 | 46.92 | 0.0728 | 0.8958 | 0.2004 |
| MUFA | 3694.70 | 4099.08 | 110.94 | 0.6824 | 0.3949 | 0.0025 ** |
| SFA | 4392.18 | 5238.81 | 119.28 | 0.4000 | 0.5472 | 0.0029 ** |
| C18:2n-6 | 253.68 | 247.70 | 97.64 | 0.6781 | 0.0647 | 0.0138 * |
| C14:0 | 287.92 | 437.58 | 151.98 | 0.0632 | 0.7354 | 0.1190 |
| C16:0 | 2076.17 | 2419.46 | 116.53 | 0.5414 | 0.3751 | 0.0039 ** |
| C18:0 | 1683.83 | 2065.71 | 122.68 | 0.3891 | 0.9125 | 0.0012 ** |
| C18:1n-9 | 2901.10 | 3212.65 | 110.74 | 0.8555 | 0.3696 | 0.0023 ** |
#* p < 0.05, ** p < 0.01; SFA, Saturated fatty acids; MUFA, Monounsaturated fatty acids; SD, Standard Deviation; CV, Coefficient of variation.
Table 5 also shows that the SCD g.23881050T>C SNP was significantly associated with IMF (p < 0.0089) and DHA (p < 0.0111), while FABP4 g.62829478A>G SNP was associated with only IMF (p < 0.0539). The FASN g.12323864A>G SNP was associated with FMP (p < 0.0544), ALA (p < 0.0033), MUFA (p < 0.0025), SFA (p < 0.0025), C18:2n-6 (p < 0.0138), C16:0 (p < 0.0039), C18:0 (p < 0.0012) and C18:1n-9 (p < 0.0023) fatty acids (Table 5).
3.4. Tukey-Adjusted Multiple Comparison Tests for Significant SNP, FMP, IMF, and Fatty Acids
As depicted in Table 6, Tukey-adjusted multiple genotype comparison tests at the SCD g.23881050T>C SNP locus confirmed significant differences where the homozygous TT genotype had the highest DHA (11.00 ± 2.34 mg/100 g), IMF (5.43 ± 0.516%), and DPA (27.1 ± 3.26 mg/100 g) compared to the CC genotype with the lowest DHA (7.00 ± 2.11 mg/100 g), IMF (3.98 ± 0.312%), and DPA (17.9 ± 6.81 mg/100 g). The heterozygous genotype CT had intermediate DPA (7.64 ± 2.09 mg/100 g), IMF (4.39 ± 0.287%), and DPA (19.4 ± 6.74 mg/100 g) that were in-between the highest and lowest values (Table 6).
Table 6.
Tukey-adjusted multiple comparisons between SNP mutations and FMP, IMF, and fatty acids in TAW lambs #.
| Multiple Genotype Comparisons | ||||||
| SNP Locus | Variable | Mean ± SE | Genotypes | Difference ± SE | p-Value | |
| SCD g.23881050T>C | DHA (C22:6n-3) (mg/100 g) | |||||
| CC | 7.00 ± 2.11 | CC vs. | CT | −0.639 ± 0.834 | 0.7247 | |
| CT | 7.64 ± 2.09 | CC vs. | TT | −3.998 ± 1.334 | 0.0105 * | |
| TT | 11.00 ± 2.34 | CT vs. | TT | −3.359 ± 1.235 | 0.0223 * | |
| IMF (%) | ||||||
| CC | 3.98 ± 0.312 | CC vs. | CT | −0.407 ± 0.323 | 0.4224 | |
| CT | 4.39 ± 0.287 | CC vs. | TT | −1.446 ± 0.532 | 0.0222 * | |
| TT | 5.43 ± 0.516 | CT vs. | TT | −1.038 ± 0.502 | 0.1041 | |
| DPA (C22:5n-3) (mg/100 g) | ||||||
| CC | 17.9 ± 6.81 | CC vs. | CT | −1.56 ± 2.65 | 0.8270 | |
| CT | 19.4 ± 6.74 | CC vs. | TT | −9.19 ± 4.25 | 0.0850 | |
| TT | 27.1 ± 3.26 | CT vs. | TT | −7.63 ± 3.93 | 0.0356 * | |
| FASN g.12323864A>G | FMP (°C) | |||||
| GG | 34.2 ± 0.4 | GG vs. | GA | 0.81 ± 0.64 | 0.4201 | |
| GA | 33.4 ± 0.3 | GG vs. | AA | 2.98 ± 1.61 | 0.0536 * | |
| AA | 31.5 ± 1.5 | GA vs. | AA | 2.16 ± 1.60 | 0.3685 | |
| ALA (C18:3n-3) (mg/100 g) | ||||||
| GG | 188.7 ± 67.6 | GG vs. | GA | 114.7 ± 39.9 | 0.0149 * | |
| GA | 74.0 ± 66.7 | GG vs. | AA | 147.2 ± 100.1 | 0.3115 | |
| AA | 41.5 ± 113.7 | GA vs. | AA | 32.6 ± 99.8 | 0.9430 | |
| MUFA (mg/100 g) | ||||||
| GG | 4524 ± 1384 | GG vs. | GA | 2617 ± 867 | 0.0099 ** | |
| GA | 1907 ± 1361 | GG vs. | AA | 3089 ± 2175 | 0.3363 | |
| AA | 1436 ± 2415 | GA vs. | AA | 472 ± 2168 | 0.9742 | |
| SFA (mg/100 g) | ||||||
| GG | 5479 ± 1715 | GG vs. | GA | 3270 ± 1121 | 0.0132 * | |
| GA | 2208 ± 1684 | GG vs. | AA | 4162 ± 2812 | 0.3068 | |
| AA | 1317 ± 3086 | GA vs. | AA | 892 ± 2803 | 0.9458 | |
| C18:2n-6 (mg/100 g) | ||||||
| GG | 281 ± 84.8 | GG vs. | GA | 142.5 ± 52.2 | 0.0216 * | |
| GA | 139 ± 83.4 | GG vs. | AA | 105.8 ± 130.8 | 0.6988 | |
| AA | 175 ± 146.4 | GA vs. | AA | −36.7 ± 130.4 | 0.9573 | |
| C16:0 (mg/100 g) | ||||||
| GG | 2539 ± 800 | GG vs. | GA | 1475 ± 518 | 0.0158 * | |
| GA | 1063 ± 786 | GG vs. | AA | 1826 ± 1298 | 0.3433 | |
| AA | 713 ± 1429 | GA vs. | AA | 350 ± 1294 | 0.9604 | |
| C18:0 (mg/100 g) | ||||||
| GG | 2227 ± 658 | GG vs. | GA | 1419 ± 441 | 0.0056 ** | |
| GA | 809 ± 646 | GG vs. | AA | 1711 ± 1106 | 0.2756 | |
| AA | 516 ± 1205 | GA vs. | AA | 292 ± 1102 | 0.9620 | |
| C18:1n-9 (mg/100 g) | ||||||
| GG | 3589 ± 1078 | GG vs. | GA | 2103 ± 679 | 0.0080 ** | |
| GA | 1486 ± 1060 | GG vs. | AA | 2353 ± 1704 | 0.3566 | |
| AA | 1236 ± 1892 | GA vs. | AA | 250 ± 1698 | 0.9882 | |
| FABP4 g.62829478A>T | IMF (%) | |||||
| A | 4.57 ± 0.39 | A vs. | AA | 0.07 ± 0.344 | 0.0556 | |
| AA | 3.92 ± 0.39 | |||||
#* p < 0.05, ** p < 0.01; SFA, Saturated fatty acids; MUFA, Monounsaturated fatty acids; SD, Standard Deviation; CV, Coefficient of variation.
There were many more significant genotype variations at the FASN g.12323864A>G SNP mutation that was associated with FMP, ALA, MUFA, SFA, C18:2n-6, C18:1n-9, C18:0, and C16:0, in which the homozygous genotype GG had the highest values compared to the lowest values in AA genotype for all variables, with the exception of C18:2n-6 that was lowest in the heterozygous GA genotype (Table 6). In contrast, at the FABP4 g.62829478A>G SNP locus, only IMF variation tended towards significance between the genotypes (p < 0.06).
4. Discussion
It is well-established that DNA-based inheritance enables the transmission of selected phenotypes across generations either without changes in the DNA sequence through epigenetic inheritance [31] or through functional mutations involving changes in only one base pair (single nucleotide polymorphisms—SNP). Through next-generation sequencing, SNP are valuable for detecting genetic variability and genomic prediction in sheep breeding programs [32], developing breed-specific DNA markers for breed identification [33,34], animal productivity [35], parentage assignment [36,37], forensics [38], and prediction of meat quality traits [39,40,41].
The prediction of meat-eating quality traits is highly challenging due to the hurdles associated with low accuracy of estimated breeding values, inconsistency in technical ease of measurement in live animals, non-repeatable reproducibility of carcass data, and high costs of rapid generation of data from large scale consumer sensory panels [19]. While the n-3 LC-PUFA profile of lamb and beef can be nutritionally enhanced using rumen-protected dietary supplements and pasture-based feeding [42,43,44], several research findings [2,4] emphasized the need for the more permanent and cumulative genetic selection route for meat sheep producers to guarantee the consistency of their lamb products in order to meet consumer preferences and adapt to the dynamics of purchasing decisions based on meat-eating quality. Consumers prefer meat with low FMP, moderate IMF, and fatty acid composition with proportionately more of the health-promoting n-3 LC-PUFA [45,46]. Since humans and other vertebrates lack Δ15 desaturase enzyme to synthesize n-3 LC-PUFA, they must obtain these from dietary intake sources in order to meet their daily requirement of 500 mg of n-3 LC-PUFA [4]. Therefore, lamb producers can tap into the omega-3 functional meat market niche through novel strategies for developing healthy meat products and reducing saturated fats [47] by matching their sheep breeding and production system to meet this health-conscious consumer preference [2,4].
4.1. SCD Gene Polymorphism
The SCD gene increases the desaturation of stearic acid to oleic acid and a functional variant in the SCD gene promoter affects fattening performance, carcass traits, meat quality, blood metabolites, and gene expression in ovine muscle [48,49]. Our results herein showing that TAW lambs were all heterozygous at three SCD SNP loci g.23881050T>C, g.23883280G>A and g.23885910C>A in the parental, first, and second composite generations (Table 2), presents a hereditary pattern and genetic divergence from the homozygous variants seen in the Poll Dorset, Texel, and Rambouillet controls that can be used as molecular markers for breed-specific identification. The significant correlations (Figure 7) and associations (Table 5 and Table 6) between detected SCD SNP loci, several fatty acids, and other meat-eating quality traits in TAW sheep is in consonance with other studies in Bashby × Argali [50], Rasa Aragonesa [49], Iranian fat- and thin-tailed [51], Poll Dorset × Border Leicester × Merino [52], Spanish, French, Egyptian, and Israeli sheep breeds [53] and Spanish goats [54]. In a comprehensive review of the genetics of n-3 LC-PUFA metabolism and meat-eating quality in TAW lambs [4], it was reported that although they were renowned for an outstanding low fat melting point (28–39 °C), high n-3 LC-PUFA EPA + DHA content (33–69 mg/100 g), marbling (3.4–8.2%), tenderness (20.0–38.5 N), and overall consumer liking (7.9–8.5), correlations between n-3 LC-PUFA profile, SCD, FABP4, FASN, and other lipogenic genes and meat quality traits presented major knowledge gaps. Therefore, the significant differences and associations observed at the SCD g.23881050T>C SNP locus in the present study where TAW lambs with the TT genotype had the highest DHA, IMF, and DPA compared to CC and CT genotypes (Table 6), have not only filled these knowledge gaps, but also equip lamb producers at the farmgate level to use this locus as a molecular marker for selection and breeding targeted at improving marbling and health-beneficial n-3 LC-PUFA. Since IMF in lamb has a moderately high heritability of 0.32–0.48 [55], has a direct relationship with tenderness, juiciness, and flavor [56] and surpasses the minimum acceptable consumer satisfaction threshold of 4% [57], the TAW lamb is well-positioned for a rapid genetic improvement for these meat-eating quality traits using the SCD gene g.23881050T>C SNP locus for identifying lambs at an early age.
4.2. FASN Gene Polymorphism
FASN catalyzes the synthesis of fatty acids such as palmitate from acetyl-CoA and malonyl-CoA, in the presence of NADPH, into long-chain saturated fatty acids, hence its involvement with fat deposition and fatty acid synthesis [58]. While novel genetic polymorphisms and gene expressions associated with carcass traits in Texel [59] and Rasa Aragonesa [60] sheep have been published, Sanz et al. [61] reported that only few studies focused on genetic variation in 5′ regulatory regions of genes involved in fat synthesis and metabolism pathways that could be good candidate genes. They went on to identify FASN gene polymorphisms and the potential use of these variants as markers associated with fat-related traits in Assaf, Roja Mallorquina, and Rasa Aragonesa sheep breeds [61]. In TAW sheep, our current study was the first to report significant genotype variations at the FASN g.12323864A>G SNP locus associated with FMP, ALA, MUFA, SFA, C18:2n-6, C18:1n-9, C18:0, and C16:0, in which the homozygous genotype GG had the highest values compared to the lowest values in AA genotype, with the exception of C18:2n-6 that was lowest in the heterozygous GA genotype (Figure 8, Table 5 and Table 6). This finding fills in a significant knowledge gap in sheep where very little has been reported on FASN gene, in stark contrast to many publications in cattle [62,63,64,65] and pigs [66,67,68,69]. Since fatty acid compositions determine the melting point and quality of fat and are closely related to the nutrition and meat-eating quality of lambs [70], our findings will assist TAW lamb producers to select the FASN genotypes best suited to their environments, market specifications, and processing needs in achieving efficiency in their management operations aimed at meeting consumer demand for healthy and nutritious lamb eating quality.
4.3. FABP Gene Polymorphism
The proteins of the FABP4 family are small molecular-weight proteins that have a high binding affinity for long-chain fatty acids, participate in fatty-acid transportation from the plasma membrane to the sites of β-oxidation, triacylglycerol, and phospholipid synthesis, and variation in FABP4 gene has been reported to affect fat deposition, growth, and meat production in sheep [71,72]. Several other research findings in sheep have demonstrated that dietary manipulation of omega-3 fatty acids can influence intramuscular fat deposition, growth, milk, wool, and meat quality [42,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90], while only a handful of studies [52,91,92,93] have validated independent associations of carcass quality, shear force, intramuscular fat percentage, and omega-3 polyunsaturated fatty acid content with gene markers or the expression of genes encoding enzymes regulating fat metabolism in Australian lamb. Therefore, our current findings at the FABP4 g.62829478A>- SNP locus showing consistently positive correlations between IMF and n-3 LC-PUFA of up to 0.66 with DHA, 0.47 with DPA, and 0.38 with EPA (Figure 9 and Table 5 and Table 6) provides a novel molecular marker for TAW sheep producers to select and breed lambs that are not only of high meat-eating quality, but also provide a healthy product for brain growth and development. This stems from the fact that IMF provides the needed marbling for taste, juiciness, and tenderness, while DHA being the major prevalent fatty acid in the brain membrane, is vital for the maintenance of healthy and functional brain development in infants and adults [94]. In pigs, Shang et al. [95] identified 3 FABP gene SNP and demonstrated that the genotype C-1375G was associated with fat deposition, while Gao et al. [96] reported that an association analysis of FABP SNP indicated that the polymorphism had a significant effect on marbling, in which pigs with the DD genotype had higher marbling than CD and CC genotypes, but the difference between CD and CC genotypes was not significant. They also reported that this FABP SNP had a highly significant effect on intramuscular fat content (p < 0.01). Our current study is the first report in TAW, which provides foundational data for the selection and breeding of lambs for marbling and healthy n-3 LC-PUFA using the identified SNP herein.
5. Conclusions
This study has provided novel insights into the shared genetic control of the fat melting point, intramuscular fat content, and health-beneficial omega-3 long-chain fatty acid composition traits that are helpful in designing breeding strategies to genetically improve meat-eating quality traits in TAW lambs while they are still alive. The early decision making utilizing this innovative and minimally invasive longissimus dorsi thoracis et lumborum muscle biopsy sampling technique for directly quantifying the genetic worth of live lambs overcomes the problem of waiting to collect meat quality data after slaughter when selection decisions about the live animal are already too late. As the present data are laboratory-tested, personalized, and customized to actual individual lamb performance and not based on estimated breeding values, precision problems due to low accuracy are minimized. The identified SNP of these lipid metabolism genes can also be used for breed-specific identification and marker-assisted selection of Tattykeel Australian White (TAW) sheep exclusive to MARGRA lamb brand for high-end meat-eating quality. Next-generation sequencing of the FABP4, FASN, and SCD genes also provides foundational data underpinning their roles in fatty acid metabolism unique to the TAW breed.
Acknowledgments
The authors gratefully acknowledge James Cook University Postgraduate Research Scholarship Program and College of Public Health, Medical and Veterinary Sciences (for PhD scholarship awarded to S.B.P.), the Australian Commonwealth Department of Industry’s Innovations Connections, CSIRO SIEF Ross Metcalf STEM Business Industrial Research Fellowship (awarded to J.R.O.) and Tattykeel Australian White Pty Ltd. (for research funding, access to flock and farm resources), CSIRO Marine and Atmosphere Lab, Hobart (for fatty acid analysis) and the National Veterinary Research Institute Vom, Nigeria (study leave approval for S.B.P.). We are also grateful to Sandra Infante Villamil, staff and management of the Gundagai Meat Processing plant for their assistance with abattoir meat sampling, and technical support at the Molecular and Evolutionary Ecology Laboratory, James Cook University, Townsville, Queensland.
Author Contributions
Conceptualization, A.E.O.M.-A.; methodology, A.E.O.M.-A., J.R.O., S.B.P., F.W.M., A.M.B.; R.H., R.C.E., B.W.B.H., M.L.E.H.; O.A.A. and R.T.K.; software, A.E.O.M.-A.; validation, A.E.O.M.-A.,S.B.P., J.R.O., formal analysis, S.B.P.; O.A.A.; investigation, A.E.O.M.-A., J.R.O., S.B.P., F.W.M., A.M.B.; R.H., R.C.E., B.W.B.H., M.L.E.H.; O.A.A. and R.T.K.; resources, A.E.O.M.-A., R.T.K. and O.A.A.; data curation, writing—original draft preparation, S.B.P.; writing—reviewing and editing, A.E.O.M.-A., R.T.K. and O.A.A.; supervision, A.E.O.M.-A., R.T.K. and O.A.A.; project administration, A.E.O.M.-A.; funding acquisition, A.E.O.M.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Innovations Connections Research Grant from the Australian Commonwealth Government’s Department of Industry, Commonwealth Scientific and Industrial Research Organisation (CSIRO) Science and Industry Endowment Fund (SIEF) Ross Metcalf STEM Business Industrial Research Fellowship (awarded to J.R.O.), Tattykeel Australian White Pty Ltd. and a PhD Research Scholarship from the James Cook University, Townsville, Queensland, Australia (awarded to S.B.P.).
Institutional Review Board Statement
Ethical approval for the use of animals and other procedures described herein was granted by the James Cook University Animal Ethics Committee (Permit No. A0015657) in compliance with the Australian Code for Care and Use of Animals for Scientific Purposes (Eighth edition, 2013).
Data Availability Statement
Data available on request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thomas E.M., Roden J.A., Haresign W., Richardson R., Lambe N.R., Clelland N., Gardner G.E., Scollan N.D. Meat eating and nutritional quality of lambs sired by high and low muscle density rams. Animal. 2021;15:100136. doi: 10.1016/j.animal.2020.100136. [DOI] [PubMed] [Google Scholar]
- 2.Pewan S.B., Otto J.R., Kinobe R.T., Adegboye O.A., Malau-Aduli A.E.O. MARGRA lamb eating quality and human health-promoting omega-3 long-chain polyunsaturated fatty acid profiles of Tattykeel Australian White Sheep: Linebreeding and gender effects. Antioxidants. 2020;9:1118. doi: 10.3390/antiox9111118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heck R.T., Lorenzo J.M., Dos Santos B.A., Cichoski A.J., de Menezes C.R., Campagnol P.C.B. Microencapsulation of healthier oils: An efficient strategy to improve the lipid profile of meat products. Curr. Opin. Food Sci. 2021;40:6–12. doi: 10.1016/j.cofs.2020.04.010. [DOI] [Google Scholar]
- 4.Pewan S.B., Otto J.R., Huerlimann R., Budd A.M., Mwangi F.W., Edmunds R.C., Holman B.W.B., Henry M.L.E., Kinobe R.T., Adegboye O.A., et al. Genetics of omega-3 long-chain polyunsaturated fatty acid metabolism and meat eating quality in Tattykeel Australian White lambs. Genes. 2020;11:587. doi: 10.3390/genes11050587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner G.E., Apps R., McColl R., Craigie C.R. Objective measurement technologies for transforming the Australian and New Zealand livestock industries. Meat Sci. 2021;179:108556. doi: 10.1016/j.meatsci.2021.108556. [DOI] [PubMed] [Google Scholar]
- 6.Fowler S.M., Wheeler D., Morris S., Mortimer S.I., Hopkins D.L. Partial least squares and machine learning for the prediction of intramuscular fat content of lamb loin. Meat Sci. 2021;177:108505. doi: 10.1016/j.meatsci.2021.108505. [DOI] [PubMed] [Google Scholar]
- 7.Lambe N.R., Clelland N., Draper J., Smith E.M., Yates J., Bunger L. Prediction of intramuscular fat in lamb by visible and near-infrared spectroscopy in an abattoir environment. Meat Sci. 2021;171:108286. doi: 10.1016/j.meatsci.2020.108286. [DOI] [PubMed] [Google Scholar]
- 8.Alvarenga T.I.R.C., Hopkins D.L., Morris S., McGilchrist P., Fowler S.M. Intramuscular fat prediction of the semimembranosus muscle in hot lamb carcases using NIR. Meat Sci. 2021;181:108404. doi: 10.1016/j.meatsci.2020.108404. [DOI] [PubMed] [Google Scholar]
- 9.Andueza D., Picard F., Hocquette J.F., Listrat A. Prediction of the intramuscular connective tissue components of fresh and freeze-dried samples by near infrared spectroscopy. Meat Sci. 2021;179:108537. doi: 10.1016/j.meatsci.2021.108537. [DOI] [PubMed] [Google Scholar]
- 10.Patel N., Toledo-Alvarado H., Bittante G. Performance of different portable and hand-held near-infrared spectrometers for predicting beef composition and quality characteristics in the abattoir without meat sampling. Meat Sci. 2021;178:108518. doi: 10.1016/j.meatsci.2021.108518. [DOI] [PubMed] [Google Scholar]
- 11.Dixit Y., Pham H.Q., Realini C.E., Agnew M.P., Craigie C.R., Reis M.M. Evaluating the performance of a miniaturized NIR spectrophotometer for predicting intramuscular fat in lamb: A comparison with benchtop and hand-held Vis-NIR spectrophotometers. Meat Sci. 2020;162:108026020. doi: 10.1016/j.meatsci.2019.108026. [DOI] [PubMed] [Google Scholar]
- 12.Dixit Y., Hitchman S., Hicks T.M., Lim P., Wong C.K., Holibar L., Gordon K.C., Loeffen M., Farouk M.M., Craigie C.R., et al. Non-invasive spectroscopic and imaging systems for prediction of beef quality in a meat processing pilot plant. Meat Sci. 2021;181:108410. doi: 10.1016/j.meatsci.2020.108410. [DOI] [PubMed] [Google Scholar]
- 13.Knight M.I., Linden N., Ponnampalam E.N., Kerr M.G., Brown W.G., Hopkins D.L., Baud S., Ball A.J., Borggaard C., Wesley I. Development of VISNIR predictive regression models for ultimate pH, meat tenderness (shear force) and intramuscular fat content of Australian lamb. Meat Sci. 2019;155:102–108. doi: 10.1016/j.meatsci.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Knight M.I., Butler K.L., Linden N.P., Burnett V.F., Ball A.J., McDonagh M.B., Behrendt R. Understanding the impact of sire lean meat yield breeding value on carcass composition, meat quality, nutrient and mineral content of Australian lamb. Meat Sci. 2020;170:108236. doi: 10.1016/j.meatsci.2020.108236. [DOI] [PubMed] [Google Scholar]
- 15.Anderson F., Williams A., Pannier L., Pethick D.W., Gardner G.E. Sire carcass breeding values affect body composition in lambs—2. Effects on fat and bone weight and their distribution within the carcass as measured by computed tomography. Meat Sci. 2016;116:243–252. doi: 10.1016/j.meatsci.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Stewart S.M., Gardner G.E., Williams A., Pethick D.W., McGilchrist P., Kuchida K. Association between visual marbling score and chemical intramuscular fat with camera marbling percentage in Australian beef carcasses. Meat Sci. 2021;181:108369. doi: 10.1016/j.meatsci.2020.108369. [DOI] [PubMed] [Google Scholar]
- 17.Connaughton S.L., Williams A., Anderson F., Kelman K.R., Peterse J., Gardner G.E. Dual energy X-ray absorptiometry predicts lamb carcass composition at abattoir chain speed with high repeatability across varying processing factors. Meat Sci. 2021;181:108413. doi: 10.1016/j.meatsci.2020.108413. [DOI] [PubMed] [Google Scholar]
- 18.Pannier L., Gardner G.E., Pearce K.L., McDonagh M., Ball A.J., Jacob R.H., Pethick D.W. Associations of sire estimated breeding values and objective meat quality measurements with sensory scores in Australian lamb. Meat Sci. 2014;96:1076–1087. doi: 10.1016/j.meatsci.2013.07.037. [DOI] [PubMed] [Google Scholar]
- 19.Holman B.W.B., Hopkins D.L. The use of conventional laboratory-based methods to predict consumer acceptance of beef and sheep meat: A review. Meat Sci. 2021;181:108586. doi: 10.1016/j.meatsci.2021.108586. [DOI] [PubMed] [Google Scholar]
- 20.Malau-Aduli A.E.O., Siebert B.D., Bottema C.D.K., Pitchford W.S. Breed comparison of the fatty acid composition of muscle phospholipids in Jersey and Limousin cattle. J. Anim. Sci. 1998;76:766–773. doi: 10.2527/1998.763766x. [DOI] [PubMed] [Google Scholar]
- 21.Holman B., Flakemore A., Kashan I.A., Malau-Aduli A.E.O. Spirulina supplementation, sire breed, sex and basal diet effects on lamb intramuscular fat percentage and fat melting points. Int. J. Vet. Med. Res. Rep. 2014;2014:1–9. doi: 10.5171/2014.263951. [DOI] [Google Scholar]
- 22.Flakemore A.R., Balogun R.O., McEvoy P.D., Malau-Aduli B.S., Nichols P., Malau-Aduli A.E.O. Genetic variation in intramuscular fat of prime lambs supplemented with varying concentrations of degummed crude canola oil. Int. J. Nutr. Food Sci. 2014;3:203–209. doi: 10.11648/j.ijnfs.20140303.22. [DOI] [Google Scholar]
- 23.Malau-Aduli A.E.O., Holman B.W.B., Kashani A., Nichols P.D. Sire breed and sex effects on the fatty acid composition and content of heart, kidney, liver, adipose and muscle tissues of purebred and first-cross prime lambs. Anim. Prod. Sci. 2016;56:2122–2132. doi: 10.1071/AN14906. [DOI] [Google Scholar]
- 24.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Phys. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Miller M.R., Nichols P.D., Barnes J., Davies N.W., Peacock E.J., Carter C.G. Regiospecificity profiles of storage and membrane lipids from the gill and muscle tissue of atlantic salmon (Salmo salar L.) grown at elevated temperature. Lipids. 2006;41:865–876. doi: 10.1007/s11745-006-5042-5. [DOI] [PubMed] [Google Scholar]
- 26.Clayton E. In: Graham Centre Monograph No. 4: Long-Chain Omega-3 Polyunsaturated Fatty Acids in Ruminant Nutrition: Benefits to Animals and Humans. Nugent T., Nicholls C., editors. Charles Sturt University; Wagga-Wagga, NSW, Australia: 2014. [Google Scholar]
- 27.Faircloth B.C., Glenn T.C., White N.D. Illumina Library Prep Protocol. Release 2.1 23 February 2014. [(accessed on 8 June 2021)]. Available online: https://buildmedia.readthedocs.org/media/pdf/protocols-libprep/latest/protocols-libprep.pdf.
- 28.Graffelman J., Jain D., Weir B. A genome-wide study of Hardy–Weinberg equilibrium with next generation sequence data. Hum. Genet. 2017;136:727–741. doi: 10.1007/s00439-017-1786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2021. [(accessed on 10 August 2021)]. Rstudio Version 1.3.1056. Available online: http://www.R-project.org/ [Google Scholar]
- 30.Slatkin M. Linkage disequilibrium—Understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khatib H. Transgenerational epigenetic inheritance in farm animals: How substantial is the evidence? Livest. Sci. 2021;250:104557. doi: 10.1016/j.livsci.2021.104557. [DOI] [Google Scholar]
- 32.Sharifi R.S., Noshahr F.A., Seifdavati J., Evrigh N.H., Cipriano-Salazar M., Mariezcurrena-Berasain M.A. Comparison of haplotype method using for genomic prediction versus single SNP genotypes in sheep breeding programs. Small Rum. Res. 2021;199:106380. doi: 10.1016/j.smallrumres.2021.106380. [DOI] [Google Scholar]
- 33.Deus A.R.S.D., Silva G.R., Sena L.S., Britto F.B., Carvalho D.A.D., Freitas J.V.G.D., Sarmento J.L.R. Comparison of kinship estimates in Santa Inês sheep using microsatellite and genome-wide SNP markers. Small Rum. Res. 2021;201:106399. doi: 10.1016/j.smallrumres.2021.106399. [DOI] [Google Scholar]
- 34.Xu Z., Diao S., Teng J., Chen Z., Feng X., Cai X., Yuan X., Zhang H., Li J., Zhang Z. Breed identification of meat using machine learning and breed tag SNPs. Food Control. 2021;125:107971. doi: 10.1016/j.foodcont.2021.107971. [DOI] [Google Scholar]
- 35.Krivoruchko A., Sermyagin A., Saprikina T., Golovanova N., Kvochko A., Yatsyk O. Genome wide associations study of single nucleotide polymorphisms with productivity parameters in Jalgin merino for identification of new candidate genes. Gene Rep. 2021;23:101065. doi: 10.1016/j.genrep.2021.101065. [DOI] [Google Scholar]
- 36.Kumar H., Panigrahi M., Saravanan K.A., Parida S., Bhushan B., Gaur G.K., Dutt T., Mishra B.P., Singh R.K. SNPs with intermediate minor allele frequencies facilitate accurate breed assignment of Indian Tharparkar cattle. Gene. 2021;777:145473. doi: 10.1016/j.gene.2021.145473. [DOI] [PubMed] [Google Scholar]
- 37.Long J. Parentage analysis using genome-wide high-density SNP microarray. Gene. 2021;785:145605. doi: 10.1016/j.gene.2021.145605. [DOI] [PubMed] [Google Scholar]
- 38.Tao R., Wang S., Chen A., Xia R., Zhang X., Yang Q., Qu Y., Zhang S., Li C. Parallel sequencing of 87 STR and 294 SNP markers using the prototype of the SifaMPS panel on the MiSeq FGx™ system. Forensic Sci. Int. Genet. 2021;52:102490. doi: 10.1016/j.fsigen.2021.102490. [DOI] [PubMed] [Google Scholar]
- 39.Grochowska E., Lisiak D., Akram M.Z., Adeniyi O.O., Luhken G., Borys B. Association of a polymorphism in exon 3 of the IGF1R gene with growth, body size, slaughter and meat quality traits in Colored Polish Merino sheep. Meat Sci. 2021;172:108314. doi: 10.1016/j.meatsci.2020.108314. [DOI] [PubMed] [Google Scholar]
- 40.Lopes F.B., Baldi F., Passafaro T.L., Brunes L.C., Costa M.F.O., Eifert E.C., Narciso M.G., Rosa G.J.M., Lobo R.B., Magnabosco C.U. Genome-enabled prediction of meat and carcass traits using Bayesian regression, single-step genomic best linear unbiased prediction and blending methods in Nelore cattle. Animal. 2021;15:100006. doi: 10.1016/j.animal.2020.100006. [DOI] [PubMed] [Google Scholar]
- 41.Marín-Garzón N.A., Magalhães A.F.B., Mota L.F.M., Fonseca L.F.S., Chardulo L.A.L., Albuquerque L.G. Genome-wide association study identified genomic regions and putative candidate genes affecting meat color traits in Nellore cattle. Meat Sci. 2021;171:108288. doi: 10.1016/j.meatsci.2020.108288. [DOI] [PubMed] [Google Scholar]
- 42.Perkins L., Cantwell M., Hows S., Scollan N. The impact of nutrition on intramuscular omega-3 fatty acid composition of lamb meat: A systematic review and meta-analysis. Anim. Sci. Proc. 2021;12:28. doi: 10.1016/j.anscip.2021.03.039. [DOI] [Google Scholar]
- 43.Maggiolino A., Bragaglio A., Salzano A., Rufrano D., Claps S., Sepe L., Damiano S., Ciarcia R., Dinardo F.R., Hopkins D.L., et al. Dietary supplementation of suckling lambs with anthocyanins: Effects on growth, carcass, oxidative and meat quality traits. Anim. Feed Sci. Technol. 2021;276:114925. doi: 10.1016/j.anifeedsci.2021.114925. [DOI] [Google Scholar]
- 44.Moloney A.P., O’Riordan E.G., McGee M., Carberry C.M., Moran L., Menamin K.M., Monahan F.J. Growth, efficiency and the fatty acid composition of blood and muscle from previously grazed late-maturing bulls fed rumen protected fish oil in a high concentrate finishing ration. Livest. Sci. 2021;244:104344. doi: 10.1016/j.livsci.2020.104344. [DOI] [Google Scholar]
- 45.Realini C.E., Pavan E., Johnson P.L., Font-i-Furnols M., Jacob N., Agnew M., Craigie C.R., Moon C.D. Consumer liking of M. longissimus lumborum from New Zealand pasture-finished lamb is influenced by intramuscular fat. Meat Sci. 2021;173:108380. doi: 10.1016/j.meatsci.2020.108380. [DOI] [PubMed] [Google Scholar]
- 46.Stampa E., Schipmann-Schwarze C., Hamm U. Consumer perceptions, preferences, and behavior regarding pasture-raised livestock products: A review. Food Qual. Prefer. 2020;82:103872. doi: 10.1016/j.foodqual.2020.103872. [DOI] [Google Scholar]
- 47.López-Pedrouso M., Lorenzo J.M., Gullón B., Campagnol P.C.B., Franco D. Novel strategy for developing healthy meat products replacing saturated fat with oleogels. Curr. Opin. Food Sci. 2021;40:40–45. doi: 10.1016/j.cofs.2020.06.003. [DOI] [Google Scholar]
- 48.Liu G., Ding Y., Chen Y., Yang Y. Effect of energy intake and L-carnitine on fattening performance, carcass traits, meat quality, blood metabolites, and gene expression of lamb. Small Rum. Res. 2020;183:106025. doi: 10.1016/j.smallrumres.2019.106025. [DOI] [Google Scholar]
- 49.Calvo J.H., González-Calvo L., Dervishi E., Blanco M., Iguácel L.P., Sarto P., Pérez-Campo F.M., Serrano M., Bolado-Carrancio A., Rodríguez-Rey J.C., et al. A functional variant in the stearoyl-CoA desaturase (SCD) gene promoter affects gene expression in ovine muscle. Livest. Sci. 2019;219:62–70. doi: 10.1016/j.livsci.2018.11.015. [DOI] [Google Scholar]
- 50.Wang X., Fang C., He H., Cao H., Liu L., Jiang L., Ma Y., Liu W. Identification of key genes in sheep fat tail evolution based on RNA-seq. Gene. 2021;781:145492. doi: 10.1016/j.gene.2021.145492. [DOI] [PubMed] [Google Scholar]
- 51.Aali M., Moradi-Shahrbabak H., Moradi-Shahrbabak M., Sadeghi M., Kohram H. Polymorphism in the SCD gene is associated with meat quality and fatty acid composition in Iranian fat- and thin-tailed sheep breeds. Livest. Sci. 2016;188:81–90. doi: 10.1016/j.livsci.2016.04.003. [DOI] [Google Scholar]
- 52.Alvarenga T.I.R.C., Chen Y., Lewandowski P., Ponnampalam E.N., Sadiq S., Clayton E.H., van de Ven R.J., Perez J.R.O., Hopkins D.L. The expression of genes encoding enzymes regulating fat metabolism is affected by maternal nutrition when lambs are fed algae high in omega-3. Livest. Sci. 2016;187:53–60. doi: 10.1016/j.livsci.2016.02.013. [DOI] [Google Scholar]
- 53.García-Fernández M., Gutiérrez-Gil B., García-Gámez E., Arranz J.J. Genetic variability of the Stearoyl-CoA desaturase gene in sheep. Mol. Cell. Probes. 2009;23:107–111. doi: 10.1016/j.mcp.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 54.Avilés C., Horcada A., Polvillo O., Membrillo A., Anaya G., Molina A., Alcalde M.J., Panea B. Association study between variability in the SCD gene and the fatty acid profile in perirenal and intramuscular fat deposits from Spanish goat populations. Small Rum. Res. 2016;136:127–131. doi: 10.1016/j.smallrumres.2016.01.008. [DOI] [Google Scholar]
- 55.Mortimer S., Van der Werf J., Jacob R.H., Hopkins D., Pannier L., Pearce K., Gardner G.E., Warner R.D., Geesink G.H., Hocking Edwards J.E., et al. Genetic parameters for meat quality traits of Australian lamb meat. Meat Sci. 2014;96:1016–1024. doi: 10.1016/j.meatsci.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 56.Liu J., Chriki S., Ellies-Oury M.-P., Legrand I., Pogorzelski G., Wierzbicki J., Farmer L., Troy D., Polkinghorne R., Hocquette J.-F., et al. European conformation and fat scores of bovine carcasses are not good indicators of marbling. Meat Sci. 2020;170:108233. doi: 10.1016/j.meatsci.2020.108233. [DOI] [PubMed] [Google Scholar]
- 57.Pannier L., Pethick D., Geesink G., Ball A., Jacob R., Gardner G. Intramuscular fat in the longissimus muscle is reduced in lambs from sires selected for leanness. Meat Sci. 2014;96:1068–1075. doi: 10.1016/j.meatsci.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 58.Raza S.H.A., Gui L., Khan R., Schreurs N.M., Xiaoyu W., Wu S., Mei C., Wang L., Ma X., Wei D., et al. Association between FASN gene polymorphisms, ultrasound carcass traits and intramuscular fat in Qinchuan cattle. Gene. 2018;645:55–59. doi: 10.1016/j.gene.2017.12.034. [DOI] [PubMed] [Google Scholar]
- 59.Armstrong E., Ciappesoni G., Iriarte W., Da Silva C., Macedo F., Navajas E.A., Brito G., San Julián R., Gimeno D., Postiglioni A. Novel genetic polymorphisms associated with carcass traits in grazing Texel sheep. Meat Sci. 2018;145:202–208. doi: 10.1016/j.meatsci.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 60.Dervishi E., Serrano C., Joy M., Serrano M., Rodellar C., Calvo J.H. The effect of feeding system in the expression of genes related with fat metabolism in semitendinous muscle in sheep. Meat Sci. 2011;89:91–97. doi: 10.1016/j.meatsci.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Sanz A., Serrano C., Ranera B., Dervishi E., Zaragoza P., Calvo J.H., Rodellar C. Novel polymorphisms in the 5′UTR of FASN, GPAM, MC4R and PLIN1 ovine candidate genes: Relationship with gene expression and diet. Small Rum. Res. 2015;123:70–74. doi: 10.1016/j.smallrumres.2014.10.010. [DOI] [Google Scholar]
- 62.Fang X., Zhao Z., Jiang P., Haibin X., Hang Y., Yang R. Identification of the bovine HSL gene expression profiles and its association with fatty acid composition and fat deposition traits. Meat Sci. 2017;131:107–118. doi: 10.1016/j.meatsci.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Papaleo-Mazzucco J., Goszczynski D.E., Ripoli M.V., Meluccia L.M., Pardo A.M., Colatto E., Rogberg-Muñoz A., Mezzadra C.A., Depetris G.J., Giovambattista G., et al. Growth, carcass and meat quality traits in beef from Angus, Hereford and cross-breed grazing steers, and their association with SNPs in genes related to fat deposition metabolism. Meat Sci. 2016;114:121–129. doi: 10.1016/j.meatsci.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 64.Barton L., Bures D., Kott T., Rehak D. Associations of polymorphisms in bovine DGAT1, FABP4, FASN, and PPARGC1A genes with intramuscular fat content and the fatty acid composition of muscle and subcutaneous fat in Fleckvieh bulls. Meat Sci. 2016;114:18–23. doi: 10.1016/j.meatsci.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 65.Yeon S.H., Lee S.H., Choi B.H., Lee H.J., Jang G.W., Lee K.T., Kim K.H., Lee J.H., Chung H.Y. Genetic variation of FASN is associated with fatty acid composition of Hanwoo. Meat Sci. 2013;94:133–138. doi: 10.1016/j.meatsci.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 66.Zappaterra M., Luis D., Zambonelli P., Mele M., Serra A., Costa L.N., Davoli R. Association study between backfat fatty acid composition and SNPs in candidate genes highlights the effect of FASN polymorphism in large white pigs. Meat Sci. 2019;156:75–84. doi: 10.1016/j.meatsci.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y., Zhang J., Gong H., Cui L., Zhang W., Ma J., Chen C., Ai H., Xiao S., Huang L., et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Sci. 2019;150:47–55. doi: 10.1016/j.meatsci.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 68.Renaville B., Bacciu N., Lanzoni M., Mossa F., Piasentier E. Association of single nucleotide polymorphisms in fat metabolism candidate genes with fatty acid profiles of muscle and subcutaneous fat in heavy pigs. Meat Sci. 2018;139:220–227. doi: 10.1016/j.meatsci.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 69.Zappaterra M., Deserti M., Mazza R., Braglia S., Zambonelli P., Davoli R. A gene and protein expression study on four porcine genes related to intramuscular fat deposition. Meat Sci. 2016;121:27–32. doi: 10.1016/j.meatsci.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 70.Ates S., Keles G., Demirci U., Dogan S., Kirbas M., Filley S.J., Parker N.B. The effects of feeding system and breed on the performance and meat quality of weaned lambs. Small Rum. Res. 2020;192:106225. doi: 10.1016/j.smallrumres.2020.106225. [DOI] [Google Scholar]
- 71.Yan W., Zhou H., Hu J., Luo Y., Hickford J.G.H. Variation in the FABP4 gene affects carcass and growth traits in sheep. Meat Sci. 2018;145:334–339. doi: 10.1016/j.meatsci.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 72.Bai J.L., Xu H.W., Zang R.X., He H.J., Cai Y., Cao X., Peng F.J., Han J., Wu J.P., Yang J.T. Cloning of the heart fatty acid-binding protein (H-FABP) gene and its tissue-specific expression profile in the Lanzhou fat-tailed sheep, Ovis aries. Small Rum. Res. 2013;112:114–122. doi: 10.1016/j.smallrumres.2012.12.016. [DOI] [Google Scholar]
- 73.Malau-Aduli A.E.O., Nguyen D.V., Le V.H., Nguyen Q.V., Otto J.R., Malau-Aduli B.S., Nichols P.D. Correlations between growth and wool quality traits of genetically divergent Australian lambs in response to canola or flaxseed oil supplementation. PLoS ONE. 2019;14:e0208229. doi: 10.1371/journal.pone.0208229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le H.V., Nguyen D.V., Nguyen Q.V., Malau-Aduli B.S., Nichols P.D., Malau-Aduli A.E.O. Fatty acid profiles of muscle, liver, heart and kidney of Australian prime lambs fed different polyunsaturated fatty acids enriched pellets in a feedlot system. Sci. Rep. 2019;9:1238. doi: 10.1038/s41598-018-37956-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen Q.V., Malau-Aduli B.S., Cavalieri J., Nichols P.D., Malau-Aduli A.E.O. Enhancing omega-3 long-chain polyunsaturated fatty acid content of dairy-derived foods for human consumption. Nutrients. 2019;11:743. doi: 10.3390/nu11040743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nguyen Q.V., Le H.V., Nguyen D.V., Malau-Aduli B.S., Nichols P.D., Malau-Aduli A.E.O. Enhancement of dairy sheep cheese eating quality with increased omega-3 long-chain polyunsaturated fatty acids. J. Dairy Sci. 2019;102:211–222. doi: 10.3168/jds.2018-15215. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen Q.V., Le V.H., Nguyen D.V., Malau-Aduli B.S., Nichols P.D., Malau-Aduli A.E.O. Supplementing grazing dairy ewes with oil and rumen-protected EPA + DHA pellets enhances health-beneficial n-3 long-chain polyunsaturated fatty acids in sheep milk. Eur. J. Lipid Sci. Technol. 2018;120:1700256. doi: 10.1002/ejlt.201700256. [DOI] [Google Scholar]
- 78.Nguyen Q.V., Le H.V., Nguyen D.V., Nish P., Otto J.R., Malau-Aduli B.S., Nichols P.D., Malau-Aduli A.E.O. Supplementing dairy ewes grazing low quality pastures with plant-derived and rumen-protected oils containing eicosapentaenoic acid and docosahexaenoic acid pellets increases body condition score and milk, fat, and protein yields. Animals. 2018;8:241. doi: 10.3390/ani8120241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen D.V., Malau-Aduli B.S., Cavalieri J., Nichols P.D., Malau-Aduli A.E.O. Supplementation with plant-derived oils rich in omega-3 polyunsaturated fatty acids for lamb production. Vet. Anim. Sci. 2018;6:29–40. doi: 10.1016/j.vas.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen D.V., Le V.H., Nguyen Q.V., Malau-Aduli B.S., Nichols P.D., Malau-Aduli A.E.O. Omega–3 long-chain fatty acids in the heart, kidney, liver and plasma metabolite profiles of Australian prime lambs supplemented with pelleted canola and flaxseed oils. Nutrients. 2017;9:893. doi: 10.3390/nu9080893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen D.V., Flakemore A.R., Otto J.R., Ives S.W., Smith R.W., Nichols P.D., Malau-Aduli A.E.O. Nutritional value and sensory characteristics of meat eating quality of Australian prime lambs supplemented with pelleted canola and flaxseed oils: Fatty acid profiles of muscle and adipose tissues. Intern. Med. Rev. 2017;3:1–21. doi: 10.18103/imr.v3i3.295. [DOI] [Google Scholar]
- 82.Flakemore A.R., Malau-Aduli B.S., Nichols P.D., Malau-Aduli A.E.O. Omega-3 fatty acids, nutrient retention values, and sensory meat eating quality in cooked and raw Australian lamb. Meat Sci. 2017;123:79–87. doi: 10.1016/j.meatsci.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 83.Fowler S.M., Morris S., Hopkins D.L. Nutritional composition of lamb retail cuts from the carcases of extensively finished lambs. Meat Sci. 2019;154:126–132. doi: 10.1016/j.meatsci.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 84.Cardoso D.B., Medeiros G.R., Guim A., Azevedo P.S., Suassuna D.M.L., Jr., Maciel M.V., Costa C.A., Lopes L.A., Silva J.L., Véras A.S.C., et al. Growth performance, carcass traits and meat quality of lambs fed with increasing levels of spineless cactus. Anim. Feed Sci. Technol. 2021;272:114788. doi: 10.1016/j.anifeedsci.2020.114788. [DOI] [Google Scholar]
- 85.Díaz M.T., Pérez C., Sánchez C.I., Lauzurica S., Cañeque V., González C., De La Fuente J. Feeding microalgae increases omega 3 fatty acids of fat deposits and muscles in light lambs. J. Food Compos. Anal. 2017;56:115–123. doi: 10.1016/j.jfca.2016.12.009. [DOI] [Google Scholar]
- 86.Ponnampalam E.N., Butler K.L., Jacob R.H., Pethick D.W., Ball A.J., Hocking Edwards J.E., Geesink G., Hopkins D.L. Health beneficial long chain omega-3 fatty acid levels in Australian lamb managed under extensive finishing systems. Meat Sci. 2014;96:1104–1110. doi: 10.1016/j.meatsci.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 87.Ponnampalam E.N., Butler K.L., Pearce K.M., Mortimer S.I., Pethick D.W., Ball A.J., Hopkins D.L. Sources of variation of health claimable long chain omega-3 fatty acids in meat from Australian lamb slaughtered at similar weights. Meat Sci. 2014;96:1095–1103. doi: 10.1016/j.meatsci.2012.11.039. [DOI] [PubMed] [Google Scholar]
- 88.Ponnampalam E.N., Sinclair A.J., Egan A.R., Ferrier G.R., Leury B.J. Dietary manipulation of muscle long-chain omega-3 and omega-6 fatty acids and sensory properties of lamb meat. Meat Sci. 2002;60:125–132. doi: 10.1016/S0309-1740(01)00113-9. [DOI] [PubMed] [Google Scholar]
- 89.Ponnampalam E.N., Trout G.R., Sinclair A.J., Egan A.R., Leury B.J. Comparison of the color stability and lipid oxidative stability of fresh and vacuum packaged lamb muscle containing elevated omega-3 and omega-6 fatty acid levels from dietary manipulation. Meat Sci. 2001;58:151–161. doi: 10.1016/S0309-1740(00)00143-1. [DOI] [PubMed] [Google Scholar]
- 90.Álvarez-Rodríguez J., Ripoll G., Lobón S., Sanz A., Blanco M., Joy M. Alfalfa but not milk in lamb’s diet improves meat fatty acid profile and α-tocopherol content. Food Res. Int. 2018;107:708–716. doi: 10.1016/j.foodres.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 91.Knight M.I., Daetwyler H.D., Hayes B.J., Hayden M.J., Ball A.J., Pethick D.W., McDonagh M.B. An independent validation association study of carcass quality, shear force, intramuscular fat percentage and omega-3 polyunsaturated fatty acid content with gene markers in Australian lamb. Meat Sci. 2014;96:1025–1033. doi: 10.1016/j.meatsci.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 92.Malau-Aduli A.E.O., Kashani A. Molecular genetics-nutrition interactions in the expression of AANAT, ADRB3, BTG2 and FASN genes in the heart, kidney and liver of Australian lambs supplemented with Spirulina (Arthrospira platensis) Genes Genom. 2015;37:633–644. doi: 10.1007/s13258-015-0294-1. [DOI] [Google Scholar]
- 93.Kashani A., Holman B.W.B., Malau-Aduli A.E.O. Single nucleotide polymorphisms of the ovine ADRB3 gene in crossbred Australian sheep supplemented with Spirulina (Arthrospira platensis) cyanobacterial microalgae. Biomed. J. Sci. Tech. Res. 2017;1:462–467. doi: 10.26717/BJSTR.2017.01.000218. [DOI] [Google Scholar]
- 94.Mallick R., Basak S., Duttaroy A.K. Docosahexaenoic acid, 22:6n-3: Its roles in the structure and function of the brain. Int. J. Dev. Neurosci. 2019;79:21–31. doi: 10.1016/j.ijdevneu.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 95.Shang P., Zhang B., Zhang J., Duan M., Wu L., Gong X., Tang K., Zhang H., Chamba Y. Expression and single-nucleotide polymorphisms of the H-FABP gene in pigs. Gene. 2019;710:156–160. doi: 10.1016/j.gene.2019.05.061. [DOI] [PubMed] [Google Scholar]
- 96.Gao Y., Zhang Y.H., Zhang S., Li F., Wang S., Dai L., Jiang H., Xia S., Liu D., Sun B., et al. Association of A-FABP gene polymorphism in intron 1 with meat quality traits in Junmu No. 1 white swine. Gene. 2011;487:170–173. doi: 10.1016/j.gene.2011.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available on request.