Abstract
Vav works as a GDP/GTP exchange factor for Rac GTPases, thereby facilitating the transition of these proteins from the inactive (GDP-bound) into the active (GTP-bound) state. The stimulation of Vav exchange activity during cell signaling is mediated by tyrosine phosphorylation. To understand the roles of phosphorylation in the regulation of Vav activity, we have initiated the characterization of the residues of Vav that are phosphorylated during signal transduction. Here we show that a Y-to-F mutation in one of these residues, Y174, leads to the oncogenic activation of Vav and to the enhancement of other Vav-mediated signals such as those for cytoskeletal reorganization, JNK activation, and stimulation of the nuclear factor of activated T cells. The effect induced by the Y174F mutation is further accentuated by mutations in residue Y142 or Y160. The Y174F mutation has no effect on the exchange activity of Vav in vitro but results in higher levels of phosphorylation in vivo. Using a phosphospecific antibody, we found that Y174 is phosphorylated following stimulation of mitogenic and antigenic receptors. This phosphorylation event is conserved in Vav-2 and Vav-3, the other two members of the Vav family. These results identify a previously unknown mechanism for the oncogenic activation of Vav and suggest that the activity of this exchange factor is modulated by two antagonistic phosphorylation events, one involved in Vav activation and a second one implicated in Vav inactivation.
The Vav family is a novel group of signal transduction molecules with important roles in cell signaling and tumorigenesis (3). This family has four known members, three distributed in mammalian cells (Vav, Vav-2, and Vav-3) and one present in Caenorhabditis elegans (CelVav) (3, 23). Vav proteins are composed of seven different structural domains, including a calponin homology (CH) region, an acidic (Ac) domain, Dbl homology (DH) and pleckstrin homology (PH) regions, one zinc finger butterfly (ZF), and two SH3 regions flanking a single SH2 domain (3). The SH3 regions are not conserved in CelVav (31). At the biochemical level, Vav proteins promote the exchange of guanosine nucleotides in GTP-binding proteins of the Rho/Rac family, an action that facilitates the transition of these GTPases from their inactive (GDP-bound) to their active (GTP-bound) state (11, 13, 23, 27). Activation of these GTP-binding proteins leads in turn to both cytoskeletal and mitogenic changes in the cell, as demonstrated by the ability of Vav proteins to induce membrane ruffles and lamellipodia (24, 27), to activate JNK-1 (10, 11), and to stimulate Rho/Rac-responsive transcriptional factors such as the nuclear factor of activated T cells (NF-AT) and NF-κB (16, 22, 32). Despite their similar structures and biochemical activities, Vav family proteins show significant regulatory differences. Thus, while Vav is active preferentially on Rac-1, Rac-2, and RhoG, Vav-2 and Vav-3 target mainly RhoG and RhoA-like proteins (11, 23, 27). Moreover, whereas Vav is restricted to hematopoietic cells (5, 17), Vav-3 displays broader expression profiles (23) and Vav-2 is found ubiquitously (26).
To date, two mechanisms to promote the GDP/GTP exchange activity of Vav proteins in vitro and in vivo have been described. Under physiological conditions, the binding of extracellular stimuli to their specific membrane receptors triggers the phosphorylation of Vav on tyrosine residues (3), a posttranslational modification that activates the latent exchange activity of Vav proteins (11, 13, 23, 27). This phosphorylation is mediated by the Vav SH2, a structural domain that allows the interaction of Vav proteins with membrane and cytoplasmic tyrosine kinases (4, 12, 18, 21). The Vav PH region, via its binding to products of phosphatidylinositol-3-kinase, has been shown to cooperate in such activation, presumably by favoring higher phosphorylation levels of Vav (14). In addition to being stimulated by phosphorylation, Vav proteins become activated oncogenically by deletions that eliminate their N-terminal domains (9, 23, 27). Recent reports have shown that such hyperactivation appears to derive from the elimination of an intramolecular effect induced by the CH regions on Vav family members, because the enzyme activity of the truncated versions of these proteins becomes constitutively active independently of their phosphorylation status (23, 27).
Although the role of tyrosine phosphorylation in the activation of Vav proteins in vivo and in vitro is now well established, there are still pending questions whose resolution is important to fully understand the activation/deactivation cycle of Vav proteins. For example, we do not know yet the residue (or residues) whose phosphorylation determines the catalytic activation of Vav proteins nor do we have information about the structural changes that are behind such activation. Likewise, we do not know whether other Vav phosphorylation sites can have other regulatory functions, either by mediating intramolecular effects or by allowing the interaction of proteins capable of modulating the Vav signal output at different stages of cell stimulation.
In order to understand the mechanism of activation of Vav during cell signaling, we have initiated the characterization of the tyrosine residues of Vav that become phosphorylated upon incubation with Lck, a protein tyrosine kinase that activates the exchange activity of Vav in vitro and in vivo (11). These studies have identified so far 13 phosphorylation sites distributed throughout the entire Vav molecule, which are now being functionally characterized. Among those phosphorylation sites, we have identified three tyrosine residues in the Ac region of Vav (Tyr142, Tyr160, and Tyr174). To address the role of these phosphorylation sites in Vav regulation, we have created single and multiple combinations of tyrosine (Y)-to-phenylalanine (F) mutations at those positions and analyzed their effect on the biological activity of Vav. Here, we show that these changes constitute gain-of-function mutations for the full-length Vav protein, leading to enhanced Vav biological responses such as transformation, activation of JNK and NF-AT, and F-actin reorganization. However, these mutations were found to have no effect in the catalytic activity of Vav proteins towards Rac-1 and RhoG when tested in vitro. Based on these results, we propose that residue Y174 of Vav is involved in a feedback mechanism that downmodulates the activity of Vav during signal transduction processes.
MATERIALS AND METHODS
Antibodies, immunoprecipitation, and immunoblot analysis.
Monoclonal antibodies to hexahistidines, phosphotyrosine, and the CD3 molecule were from Sigma, Santa Cruz Biotechnology, and Dako, respectively. Antihemagglutinin (HA) antibodies were purchased from Babco. Anti-green fluorescent protein antibodies were obtained from Clontech. A polyclonal antiserum to the Vav DH domain that recognizes specifically Vav but not Vav-2 or Vav-3 was developed in rabbits using a chimeric maltose binding protein-Vav DH region as the immunogen (7). The generation of the antibody specific for the phosphorylated synthetic peptide containing residues 168 to 180 of mouse Vav was done at Research Genetics Inc. Immunoprecipitations and immunoblot analysis were conducted as previously described (7).
Plasmids and site-directed mutagenesis.
For expression in NIH 3T3 and Jurkat cells, all cDNAs were cloned in pMEX, a mammalian expression vector containing the mouse mammary tumor virus long terminal repeat. For the expression in cells containing the simian virus 40 (SV40) T antigen, the cDNAs were cloned in pcDNA3, a vector containing the cytomegalovirus promoter and the SV40 origin of replication (Invitrogen). The names of the vectors used in these studies and the description of their encoded proteins are found in the figures of this article. Plasmids for Vav-2 and Vav-3 have been described before (23, 27). Mutagenesis of Vav proteins was conducted using the Quickchange mutagenesis kit exactly as indicated by the commercial supplier (Stratagene). All constructs derived from PCR and/or site-directed mutagenesis were analyzed by automatic sequence analysis to eliminate the possibility of extra mutations. pcDNA-HA-JNK-1 and pNF-AT/luc were gifts from P. Crespo (Instituto de Investigaciones Biomédicas “Alberto Sols,” Madrid, Spain) and G. R. Crabtree (Departments of Developmental Biology and Pathology, Stanford University Medical School, Stanford, Calif.), respectively. Plasmids pFR-Luc and pFA2-cJun were obtained from Stratagene.
Cell stimulation, transfection assays, and immunofluorescence techniques.
NIH 3T3 cells and COS-1 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% calf serum (Hyclone). Jurkat cell lines (purchased from the American Tissue Culture Collection) were grown in RPMI-1640 supplemented with 15% fetal bovine serum (Hyclone). For focus formation assays, NIH 3T3 cells (1.5 × 105/10-cm-diameter plate) were transfected with the indicated plasmids using the calcium phosphate precipitation method (30). After transfections, cells were cultured for 15 days, fixed with formaldehyde, and stained with Giemsa for the final quantitation of foci. All these assays were repeated in duplicate at least four times. For morphological assays, NIH 3T3 cells were transfected using liposomes (Fugene-6; Roche Molecular Biochemicals) according to the manufacturer's recommendations. Cells were processed for immunofluorescence analysis as previously indicated (27). COS-1 cells were transfected using either the DEAE-dextran (for JNK-1 activation and expression experiments) or Fugene-6 (for the analysis of cytoskeletal changes) methods. For stimulation assays, exponentially growing COS-1 cells were serum starved for 24 h and then stimulated with 0.1 μg of epidermal growth factor (EGF; Gibco/BRL) · ml−1. Expression of proteins in hematopoietic cells was done by electroporation. To this end, exponentially growing Jurkat cells were harvested, resuspended in 400 μl of RPMI-1640 to a final concentration of 107 cell · ml−1, and electroporated (250 V and 960 μF) using a Bio-Rad apparatus. After electroporation, cells were left at room temperature for 10 min, diluted in RPMI-1640–10% fetal bovine serum, and cultured at 37°C and 5% CO2. After 48 h, cells were either left unstimulated or were stimulated for 8 h with anti-CD3 antibodies (10 μg · ml−1), washed with phosphate-buffered saline solution, and lysed in 25 mM Tris-HCl (pH 7.8)–2 mM dithiothreitol (DTT)–1 mM EDTA–1% Triton X-100–10% glycerol. Lysates were centrifuged to eliminate insoluble debris and stored at −70°C until further use. For the determination of luciferase activity, 30 μl of thawed cell lysates was mixed with 100 μl of luciferase buffer (20 mM Tricine [pH 7.8], 2.8 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 530 μM ATP, 270 μM coenzyme A [Sigma], 470 μM d-luciferin [Sigma]) and the luciferase activity in each sample was measured in a luminometer. For assays involving the detection of Vav phosphorylation, exponentially growing Jurkat cells were resuspended in serum-free RPMI-1640 at a concentration of 3 × 107 cell · ml−1 and stimulated for the periods of time indicated in the figures with anti-CD3 antibodies (10 μg · ml−1). For measuring the activation of JNK-1 in hematopoietic cells using immunocomplex kinase assays, we used a T-antigen-expressing Jurkat cell line (JMC-T) generously provided by H. Band (Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, Mass.) and D. Rothstein (Department of Medicine, Yale University, New Haven, Conn.).
Insect cell and bacterial expression vectors.
Baculoviruses were generated from pFASTBAC derivatives (Gibco/BRL) encoding Vav Y3xF (pHML5) and Vav Y174F (pHML16). All pFASTBAC constructs were then recombined in Escherichia coli with the baculovirus DNA using a helper phage, and successful recombinants were identified by lacZ gene inactivation and PCR. Baculovirus DNA was then transfected into Spodoptera frugiperda (Sf9) cells using liposomes (CellFECTIN; Gibco/BRL) to generate viral particles. The baculovirus encoding wild-type Vav was previously described (6). The different deletion mutants of the CH-plus-Ac region of Vav were cloned in pGEX-2T and purified from E. coli using affinity chromatography onto glutathione beads (Pharmacia/LKB) according to standard procedures. These constructs included portions of the Vav CH-plus-Ac region encompassed between residues 1 and 186 (pMLB21), 1 and 173 (pKES3), and 1 and 158 (pKES4). In addition, we used glutathione S-transferase (GST) fusion proteins containing the Vav CH-plus-Ac regions (residues 1 to 186) with the following point mutations: Y142F (pMAL51), Y160F (pMAL52), Y174F (pMAL53), Y142F plus Y174F (pMAL54), and Y160F plus Y174F (pMAL55). Rac-1 and RhoG proteins were induced in bacteria using pGEX derivatives (27).
Protein purification, GDP/GTP exchange, and in vitro kinase assays.
GST-Lck purified from Sf9 cells was generously provided by J. Fragnoli (Bristol-Myers Squibb Pharmaceutical Research Institute, Princeton, N.J.). Purification of Vav proteins from baculovirus-infected Sf9 cells and GTPases from E. coli cells was performed as described previously (27). The activity of GST-GTPases was demonstrated by [32P]GTP hydrolysis and was normalized for concentration using 35S-GTP-binding experiments (27). In vitro kinase reactions with Lck and [γ-32P]ATP were conducted as previously described (11). For γ-35S-GTP incorporation assays, the GTP-binding proteins (60 pmol) were incubated at room temperature for 30 min in loading buffer (20 mM Tris-HCl [pH 7.5], 5 μM GDP, 50 mM NaCl, 3 mM MgCl2, 0.1 mM DTT, 0.1 mM EDTA). Exchange reactions were then conducted at room temperature in 200 μl of exchange buffer (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 0.5 mM DTT, 100 mM NaCl, 0.5 mg of bovine serum albumin/ml, 5 μM γ-35S-GTP) containing 15 pmol of the GDP-loaded GTP-binding protein and, when appropriate, 3.5 pmol of the Vav and Vav Y174F proteins. At the times indicated in the figures, aliquots of the reaction mixture were removed in duplicate and passed through nitrocellulose filters (HAWPO25; Millipore). Filters were washed twice with 10 ml of an ice-cold solution containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 10 mM MgCl2. Filters were then transferred onto vials, solubilized by the addition of 1 ml of 2-methoxyethanol (Sigma), and counted with the aid of 10 ml of scintillation fluid per vial in a β counter. JNK-1 kinase assays were conducted in the presence of [γ-32P]ATP and GST–ATF-2 as the exogenous substrate, as indicated in the figure legends (10). Alternatively, JNK-1 activity was determined using a trans reporting method (Stratagene). In this case, Jurkat cells were electroporated with a reporter plasmid containing the luciferase gene under the regulation of Gal-4-binding sites (pFR-Luc; 5 μg) in combination with a plasmid (pFA2-cJun; 2 μg) encoding the c-Jun activation domain (AD) fused to the DNA binding domain (DBD; residues 1 to 147) of Gal-4. When required, transfections included the appropriate Vav-encoding vector (20 μg each). After the electroporation, the transactivation of the reporter gene was determined after 48 h using the luciferase assay described above. In this system, the transcriptional activity of the c-Jun AD–Gal-4 DBD is dependent on the activation of the endogenous JNK protein.
RESULTS
Oncogenic activation of full-length Vav by point mutations affecting phosphorylation sites present in the Ac region.
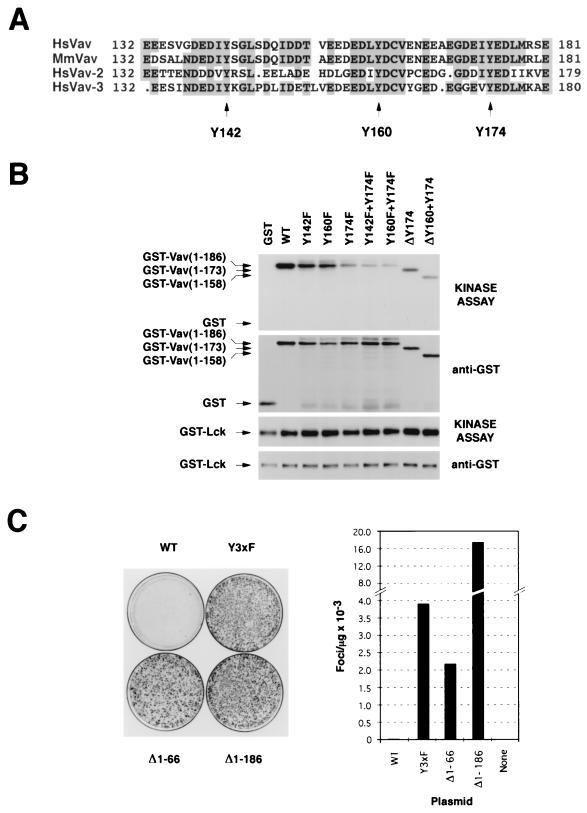
The primary structures of all known mammalian Vav proteins contain three conserved tyrosine residues located in their Ac regions (Fig. 1A). To test whether these residues are potential phosphorylation sites for protein tyrosine kinases, we purified from E. coli a collection of GST fusion proteins containing either deletions or point mutations at those three sites. Equal amounts of these chimeric proteins were then subjected to in vitro kinase assays in the presence of GST-Lck. As shown in Fig. 1B (top), the GST fusion protein containing the three phosphorylation sites of Vav was heavily phosphorylated after incubation with Lck. This phosphorylation decreased significantly when the Y174 residue was deleted from the fusion protein (ΔY174) or, alternatively, when it was mutated into phenylalanine (Fig. 1B, top). In contrast, the mutation of either Y142 or Y160 affected less dramatically the phosphorylation of the respective GST fusion protein by GST-Lck (Fig. 1B, top). The combination of the Y174F mutation with those affecting residues Y142 (Y142F plus Y174F) or Y160 (Y160F plus Y174F, ΔY160 plus Y174) further decreased, but did not abolish, the phosphorylation of the GST-Vav CH-plus-Ac fusion protein (Fig. 1B, top). GST-Lck could not phosphorylate the nonchimeric GST protein, demonstrating the specificity of the kinase reaction (Fig. 1B, top). Taken together, these results indicate that Lck recognizes preferentially the tyrosine residue of Vav located at position 174 and, with significantly less affinity, those present at positions 142 and 160. As a control, immunoblot analysis showed that all GST proteins used were at comparable concentrations (Fig. 1B, second from top). Similar amounts of kinase were also utilized, as determined by anti-GST immunoblotting and by the levels of autophosphorylated kinase present in each lane (Fig. 1B, lower two blots). Interestingly, a previous report demonstrated that synthetic peptides containing residue Y174 of Vav were phosphorylated with high affinity by another tyrosine kinase, Syk (1). This suggests that Y174 may be a target for different cytoplasmic tyrosine kinases, at least in vitro.
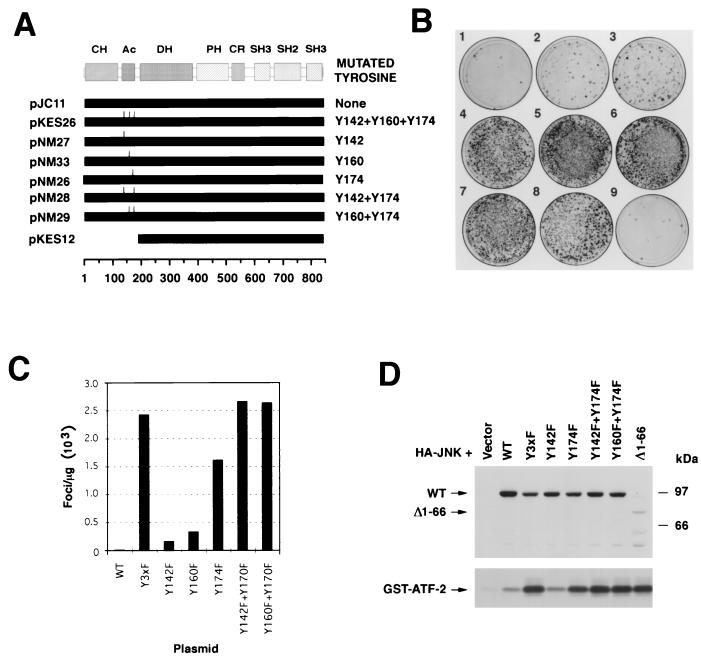
FIG. 1.
(A) Schematic representation of part of the Ac region of Vav family members. Identical residues are shaded. The tyrosine residues located at positions 142, 160, and 174 of Vav are indicated by arrows. Mm, Mus musculus; Hs, Homo sapiens. (B) In vitro kinase reaction conducted in the presence of GST-Lck and the indicated GST-Vav CH-plus-Ac fusion proteins (see Materials and Methods). Each fusion protein (0.5 μg) was incubated with 0.2 μg of GST-Lck and [γ-32P]ATP at room temperature. After 30 min, reactions were stopped by the addition of 2× sodium dodecyl sulfate sample buffer and the reaction mixtures were boiled, separated electrophoretically, and transferred onto nitrocellulose filters. After autoradiographic exposure (kinase assay), the filter was immunobloted with anti-GST antibodies to visualize the amount of protein present in each condition. WT, wild type. (C) Transforming activity of vav-containing constructs. NIH 3T3 cells were transfected with nonlinearized pJC11 (WT; 1 μg), pKES26 (Y3xF; 0.5 μg), pJC12 (Δ1-66; 0.5 μg), and pKES12 (Δ1-186; 0.1 μg) and then cultured for 15 days. Then cells were fixed and stained to visualize (left) and quantify (right) the foci of transformed cells.
To determine the biological role of these tyrosine residues, we generated a mammalian expression vector (pKES26) encoding a mutant Vav protein in which all three tyrosine residues (Y142, Y160, and Y174) were mutated to phenylalanine to avoid the appearance of cryptic phosphorylation sites. This mutant protein is referred to as Vav Y3xF. The transforming activity of the plasmid was then tested using focus formation assays with NIH 3T3 cells. For comparative purposes, these experiments included plasmids encoding either wild-type Vav (pJC11) or its two oncogenic versions, Vav (Δ1-66) (pJC12) and Vav (Δ1-186) (pKES12). To ensure that all these proteins were expressed with similar kinetics, all cDNAs were cloned in pMEX, a mammalian expression vector containing the mouse mammary tumor virus long terminal repeat. Consistent with previous results (9, 27), we found that the expression of wild-type Vav led to low levels of cellular transformation (20 to 30 foci/μg of transfected DNA) while the expression of Vav (Δ1-66) and (Δ1-186) resulted in moderate (≈2,000 foci/μg) and high (≈16,000 foci/μg) levels of oncogenic transformation, respectively (Fig. 1C). To our surprise, the Vav Y3xF construct showed also a high oncogenic potential under the same experimental conditions (≈4,000 foci/μg of transfected DNA; Fig. 1C). Vav Y3xF was approximately 130- and 2-fold more transforming than wild-type Vav and Vav (Δ1-66), respectively. However, Vav Y3xF showed lower transforming activity than the mutant Vav (Δ1-186) (Fig. 1C), a truncated protein lacking both the CH and Ac domains, whose exchange activity is independent of tyrosine phosphorylation (27).
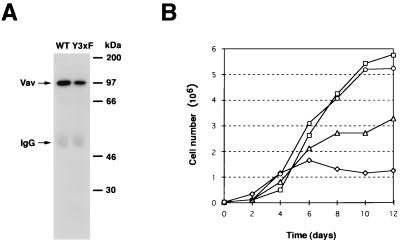
To confirm that this cellular transformation was derived from the expression of a full-length Vav protein in NIH 3T3 cells, we generated stable cell lines from several randomly picked foci of Vav- and Vav Y3xF-transformed cells. As shown in Fig. 2A, the expected size for the full-length Vav protein (97 kDa) was detected in both Vav- (B36-212 clone) and Vav Y3xF-transformed cells (X19-62 clone), confirming that the high transforming activity of Vav Y3xF is not due to the generation of a spurious truncated protein. We also observed consistently that the amount of Vav protein expressed by Vav Y3xF-transformed cells was lower than that expressed by Vav-transformed NIH 3T3 cells (Fig. 2A), ruling out the possibility that the transforming activity of this protein was due to higher levels of protein expression. Similar results were observed for three additional cell lines generated from independent foci (X19-61, X19-63, and X19-64; see Fig. 8A, right).
FIG. 2.
(A) Expression of Vav Y3xF in transformed cells. Exponentially growing NIH 3T3 cell clones obtained from randomly picked foci from Vav and Vav Y3xF transfections were lysed, immunoprecipitated with anti-Vav antibodies, and subjected to immunoblot analysis with anti-Vav antibodies. The mobilities of Vav and the immunoglobulin G (IgG) heavy chain are indicated by arrows. The migration of coelectrophoresed molecular weight markers is indicated on the right. WT, wild type. (B) Growth kinetics of parental NIH 3T3 cells (diamonds) and NIH 3T3 cell clones transformed by the expression of wild-type Vav (triangles), Vav (Δ1-186) (squares), and Vav Y3xF (circles). Exponentially growing cultures were trypsinized and seeded in duplicate in six-well plates (30,000 cells/well), and cells were counted at the indicated times. Values at each time point represent the mean of duplicate determinations performed in parallel. The value at day 0 represents the number of cells present 1 day after seeding to eliminate differences in cell growth due to variable plating efficiencies for the different cell clones.
FIG. 8.
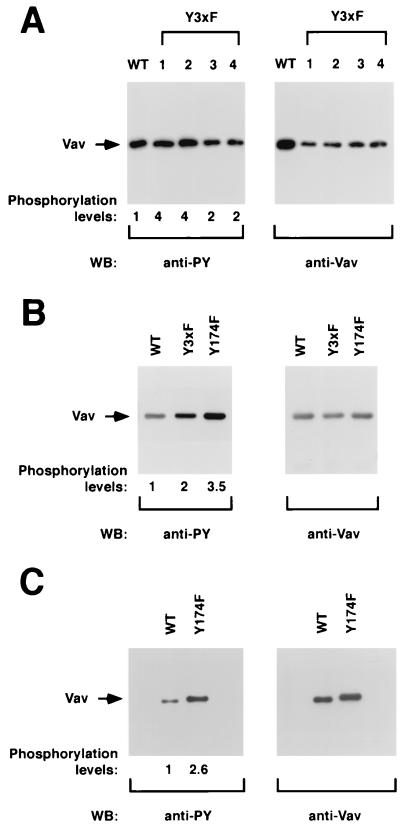
(A) Phosphorylation levels of Vav proteins in NIH 3T3 cells. One stable cell clone of NIH 3T3 cells expressing wild-type Vav (lane WT) and four independent cell clones expressing Vav Y3xF proteins (lanes 1 to 4) were lysed, immunoprecipitated with anti-Vav antibodies, and then subjected to Western blot (WB) analysis using either antiphosphotyrosine (anti-PY; left) or anti-Vav antibodies (right). (B) Phosphorylation of Vav proteins in COS-1 cells. Exponentially growing cells transfected with either wild-type Vav or the indicated Vav mutants were subjected to the same analysis as that described for panel A. (C) Phosphorylation levels of Vav proteins in Sf9 cells. Purified preparations of polyhistidine-tagged versions of wild-type and Vav Y174F proteins purified from Sf9 cells were separated electrophoretically and subjected to immunoblot analysis with the indicated antibodies. The mobilities of Vav proteins are indicated by arrows. For panel A, signals were developed either using 125I-labeled protein A (anti-Vav immunoblots) or by treatment with an anti-mouse immunoglobulin G antibody followed by incubations with 125I-labeled protein A (anti-PY). For panels B and C, signals were developed using the enhanced chemiluminescence method (ECL; Amersham). The overall levels of Vav phosphorylation (normalized by the relative amount of protein present in each case) are indicated underneath each panel. The levels of phosphorylation of wild-type Vav were given an arbitrary value of 1 in each case.
The establishment of stable cell lines expressing the Vav Y3xF protein allowed us to further study the proliferative and morphological changes induced by the expression of this mutant protein in NIH 3T3 cells. In agreement with the high oncogenicity of Vav Y3xF in focus formation assays, cells overexpressing this protein (X19-62 clone) showed the typical hallmarks of oncogenic transformation, including loss of contact inhibition and high proliferation rates in confluent cultures (Fig. 2B). Such behavior was observed in two additional cell lines expressing Vav Y3xF (X19-61 and X19-63) (data not shown). Likewise, cells transformed by the expression of the Vav (Δ1-66) and Vav (Δ1-186) deletion mutants displayed similar kinetics in culture (Fig. 2B and data not shown). All these cell lines were also capable of developing colonies in soft agar, indicating that their growth had become anchorage independent (data not shown). Taken together, these results indicate that the mutations in the phosphorylation sites of the Vav Ac region lead to the oncogenic activation of the Vav protein.
Expression of Y-to-F Vav mutants induces morphological change in NIH 3T3 cells.
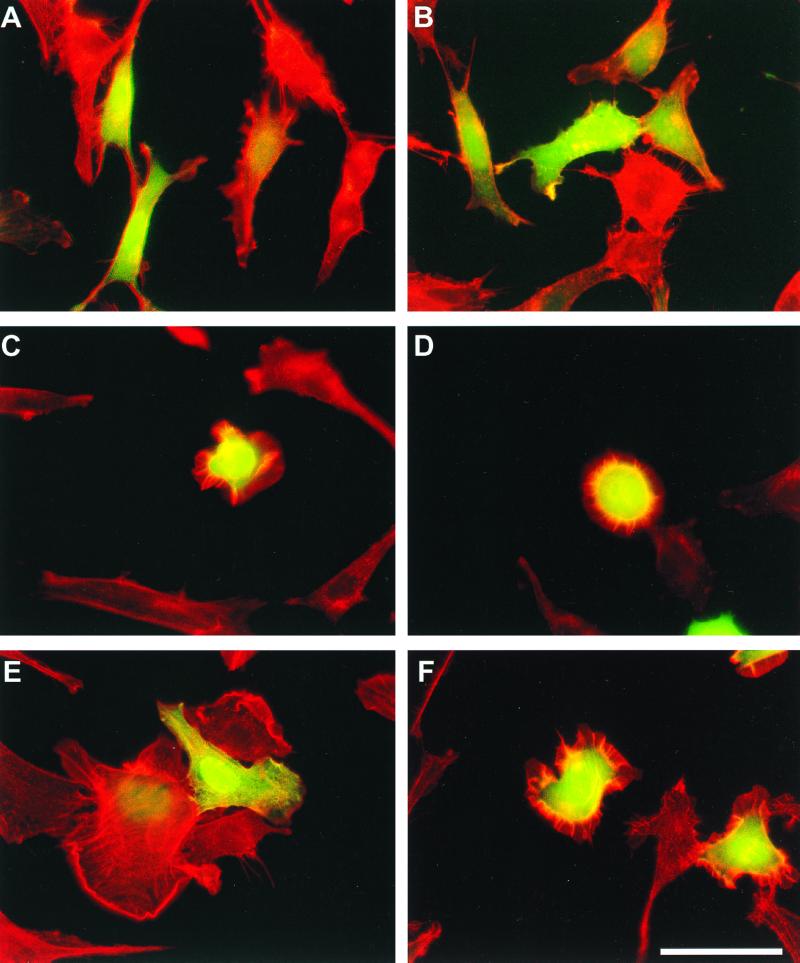
To further characterize the biological activity of Vav Y3xF, we analyzed the ability of this protein to induce cytoskeletal change in NIH 3T3 cells. To avoid any indirect epistatic effects derived from the oncogenic transformation of cells, we decided to evaluate the morphological change induced by this protein in transient transfections. To this end, we introduced in these cells the mammalian expression vectors indicated above (Fig. 1C). To facilitate the identification of the transfected cells, these plasmids (2 μg) were cotransfected with a mammalian expression vector (pEGFP-C1; 0.25 μg) containing an enhanced version of the green fluorescent protein (EGFP). Twenty-four hours after transfection, the cells were fixed, incubated with rhodamine-labeled phalloidin to visualize the actin filaments, and finally subjected to microscopy analysis. It was found that the expression of the EGFP either alone or in combination with wild-type Vav did not result in major morphological changes in rodent fibroblasts (Fig. 3A and B). By contrast, the expression of either Vav (Δ1-66) or Vav (Δ1-186) resulted in the induction of lamellipodia and membrane ruffles in the transfected cells (Fig. 3C and D). In addition, these proteins elicited the contraction of the actomyosin ring, leading to a pronounced retraction of the cell body (Fig. 3C and D). These morphological changes were previously described by us (27). In good agreement with the focus formation assays, the expression of Vav Y3xF caused marked F-actin reorganization in NIH 3T3 cells, resulting in the induction of lamellipodia and membrane ruffles (Fig. 3E and F). However, the formation of the actomyosin ring by Vav Y3xF-transfected cells was more heterogeneous, leading to the visualization of cells displaying either minor contractions of the cell body or a total absence of actomyosin contractility (Fig. 3E and F). Such variability was not observed in the mutants with the N terminus deleted (data not shown). Vav Y3xF was also capable of inducing extensive membrane ruffles upon transient expression in COS-1 cells (data not shown). Overall, these results indicate that the phosphorylation sites present in the Ac regions of Vav proteins play an important negative role in the regulation of Vav function.
FIG. 3.
Morphological change induced by transient expression of Vav proteins. NIH 3T3 cells were transfected with plasmids encoding EGFP either alone (A) or together with vectors encoding wild-type Vav (B), Vav (Δ1-66) (C), Vav (Δ1-186) (D), or Vav Y3xF (E and F). After 24 h, cells were fixed with paraformaldehyde, stained with rhodamine-phalloidin, and subjected to microscopy analysis to visualize the fluorescence derived from EGFP (green) and from rhodamine-phalloidin (red). Both images were superimposed using a computer program. Bar (F), 100 μm. Similar results were obtained in three independent experiments.
Residue Y174 is the main regulatory site for Vav transformation.
To narrow down the tyrosine residues of the Ac domain that were important for Vav oncogenic activation, we next tested the transforming activity of Vav proteins containing different combinations of Y-to-F mutations in those residues using focus formation assays (Fig. 4A). The expression in NIH 3T3 cells of Vav proteins with point mutation Y142F or Y160F resulted in a small, although significant, increase in the oncogenic potential of the respective proteins (Fig. 4B, plates 2 and 3). By contrast, the transfection of a Vav protein with a single point mutation at Y174 resulted in a strong transformation response (Fig. 4B, plate 4), although this effect was weaker than that observed after expression of either the Vav Y3xF or Vav (Δ1-186) mutants (Fig. 4B, plates 7 and 8). Interestingly, the expression of Vav with two Y-to-F mutations involving residues Y142 and Y174 or Y160 and Y174 triggered levels of cellular transformation similar to those found with Vav Y3xF (Fig. 4B, plates 5 and 6, and C).
FIG. 4.
(A) Schematic representation of the proteins used in these experiments. The names of plasmids are on the left. Solid bars, lengths of the proteins expressed; tick marks, mutated tyrosine residues in the proteins. Amino acid numbers are shown at the bottom. (B) Transforming activity of Vav Y-to-F mutants. NIH 3T3 cells were transfected using the calcium phosphate precipitation method with vectors encoding wild-type Vav (pJC11 plasmid; 1 μg; plate 1), Vav Y142F (pNM27 plasmid; 1 μg; plate 2), Vav Y160F (pNM33; 1 μg; plate 3), Vav Y174F (pNM26; 1 μg; plate 4), Vav Y142F+Y174F (pNM28; 1 μg; plate 5), Vav Y160F+Y174F (pNM29; 1 μg; plate 6), Vav Y3xF (pKES26; 1 μg; plate 7), and Vav (Δ1-186) (pKES12; 0.1 μg; plate 8). Cells were also mock transfected with no vector to be used as a negative control (plate 9). After 15 days of culture, cells were fixed and stained with Giemsa to visualize the foci of transformed cells. (C) Quantification of foci from a representative transfection of the plasmids indicated in panel B. WT, wild type. (D) (Bottom) Activation of JNK-1 by Y-to-F Vav mutants. COS-1 cells were transfected using the DEAE-dextran method with a vector encoding HA–JNK-1 (3 μg) together with empty vector or with plasmids encoding the indicated Vav mutants (5 μg). After 48 h, cells were washed and lysed, and the resulting cellular extracts were immunoprecipitated with anti-HA antibodies. Final immunocomplexes were subjected to in vitro kinase reactions in the presence of [γ-32P]ATP and GST–ATF-2 as the substrate. (Top) Expression of Vav proteins in this transfection as determined by anti-Vav immunoblotting of total cellular lysates. The mobilities of Vav proteins and the phosphorylated GST–ATF-2 are indicated by arrows. The migration of molecular weight markers is indicated on the right. This is a representative experiment of three independent kinase assays.
To check the activity of these proteins in a different biological readout, we measured the ability of the Y-to-F Vav mutants to promote activation of JNK-1, a well-known downstream element of the Rac-1 pathway that is stimulated by the expression of the Vav oncoprotein (10, 11). To this end, we overexpressed an HA-tagged version of JNK-1 in COS-1 cells either alone or in combination with the indicated (Fig. 4D) point mutants of Vav. After 48 h, the catalytic activity of this serine/threonine kinase was tested by immunocomplex kinase assays using a GST–ATF-2 fusion protein as the exogenous substrate. As shown in Fig. 4D (bottom), the coexpression of Vav Y3xF, Vav Y142F+Y174F, and Vav Y160F+Y174F resulted in levels of JNK-1 activation as high as those observed with one of the oncogenic versions of Vav (Δ1-66). The coexpression of Vav Y174F resulted also in marked JNK-1 activation, although such stimulation was slightly weaker than that found with the mutants described above (Fig. 4D, bottom). The cotransfection of HA–JNK-1 with either wild-type Vav or Vav Y142F resulted in only minor levels of activation of this serine/threonine kinase (Fig. 4D, bottom). Low levels of activation of wild-type Vav in this system were previously reported (10). Immunoblot analysis with anti-Vav antibodies of the total cellular lysates derived from the transfected COS-1 demonstrated that all Vav proteins were expressed at comparable levels in these experiments (Fig. 4D, top). The only exception was the oncogenic version of Vav that showed significantly lower levels of expression in the lysates. Further analysis demonstrated that this is due to the predominant presence of this version of Vav in the Triton X-100-insoluble fraction (data not shown). A similar distribution of Vav (Δ1-66) was recently demonstrated by others (19). Likewise, similar levels of HA–JNK-1 were detected in all transfections, as determined by immunoblot analysis of total cellular lysates with anti-HA antibodies (data not shown). In agreement with the high biological activity of the Vav Y174F mutant, the transient expression of this mutant protein was capable of inducing changes in the actin cytoskeleton similar to those elicited by Vav Y3xF both in NIH 3T3 and COS-1 cells (data not shown). Taken together, these results indicate that the optimal oncogenic activation of full-length Vav is primarily determined by the elimination of residue Y174 and, to a lesser extent, of an additional mutation affecting any of the other two tyrosine residues located in the Ac domain.
Relationship between the Y-to-F mutations and the Vav CH and SH3-SH2-SH3 regions.
The generation of deletions affecting the Vav CH and Ac regions leads to different levels of oncogenic transformation of the resulting truncated proteins (3). Thus, Vav proteins with a partial deletion of the CH region [Vav (Δ1-66)] display moderate transforming activity and phosphorylation-dependent exchange activity, whereas Vav proteins with deletions of the entire CH-plus-Ac region [Vav (Δ1-186)] have high oncogenic potential and phosphorylation-independent exchange activity (11, 23, 27). To investigate whether the mutations in the phosphorylation sites could be synergistic with the N-terminal deletions to promote a higher transforming activity of Vav, we measured the oncogenic potential of a new Vav mutant lacking part (residues 1 to 66) of the CH region and containing Y-to-F mutations in residues Y142, Y160, and Y174 of the Ac domain (Fig. 5A). Despite the fact that Vav (Δ1-66) was significantly less transforming than the hyperactive Vav (Δ1-186) protein, we found that the incorporation of the Y3xF mutation in the Vav (Δ1-66) background did not increase the oncogenic potential of this N-terminally truncated protein (Fig. 5B). These results indicate that the biological effect of the Y3xF mutation is restricted to full-length Vav, suggesting that these Y-to-F mutations do not mimic functionally the deletion of the Ac domain that takes place in Vav (Δ1-186).
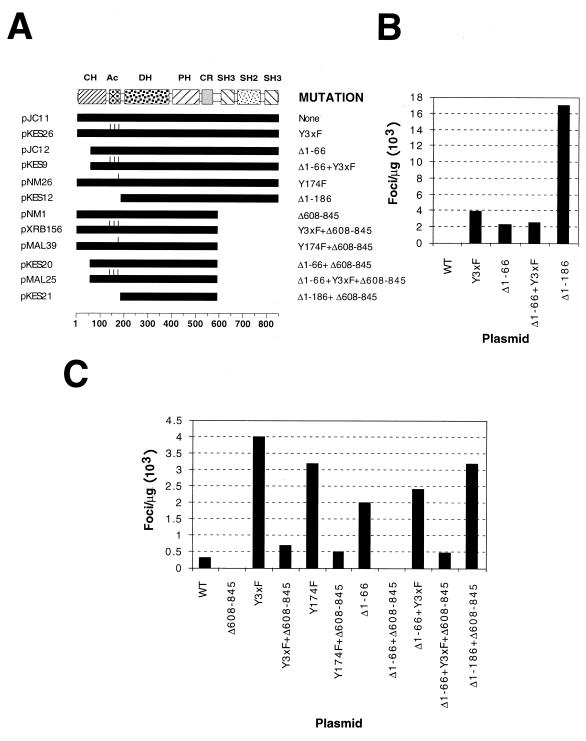
FIG. 5.
(A) Schematic representation of the proteins used in these experiments. The names of plasmids are on the left. The mutation(s) contained in each protein is on the right. Solid bars, lengths of the proteins expressed; tick marks, mutated tyrosine residues in each protein. Amino acid numbers are shown at the bottom. (B) Transforming activity of the indicated Vav mutants in focus formation assays conducted with NIH 3T3 cells. WT, wild type. (C) Transforming activity of the indicated plasmids in focus formation assays conducted with NIH 3T3 cells. To facilitate the study of the effect of ΔSH3-SH2-SH3 deletions in the wild-type version of Vav, the transfections made with pJC11 and pNM1 used ScaI-linearized plasmids. This procedure increases the transforming activity of Vav about 300-fold, probably due to better integration of the transfected DNAs. The other plasmids were used in supercoiled form. Due to the high oncogenic potential of the Vav (Δ1-186) protein, the values obtained with it (>16,000 foci/μg) were not included in the histogram.
Since this lack of synergism between the Y-to-F mutations and the deletion of the CH region could indicate alternatively an identical mode of action, we next tested the biological activity of the same mutants containing an additional deletion that eliminated their entire C-terminal SH3-SH2-SH3 regions (residues 608 to 845) (Fig. 5A). Previous results have demonstrated that such a deletion eliminates the transforming activity of the wild-type Vav and Vav (Δ1-66) but not that of the hyperactive Vav (Δ1-186) (27). In good agreement with those results, these experiments indicated that Vav (Δ608-845) and Vav (Δ1-66+Δ608-845) were totally inactive in cellular transformation (Fig. 5C). In contrast, the deletion of the SH3-SH2-SH3 region in Vav Y3xF, Vav (Δ1-66+Y3xF), and Vav Y174F reduced, but did not abolish, the transforming activity of these proteins (Fig. 5C). This was not due to different transformation levels, because the transforming activities of Vav (Δ1-66) and Vav (Δ1-66+Y3xF) were similar (Fig. 5B and C). The transforming activity of the Vav (Δ1-66+Y3xF+Δ608-845) and Vav (Δ1-66+Δ608-845) mutants was significantly lower than that induced by a similar form of the hyperoncogenic Vav protein, Vav (Δ1-186+Δ608-845), whose activity is independent of tyrosine phosphorylation (Fig. 5A and C). Indeed, the transforming activity of this protein (≈3,500 foci/μg of DNA) was as high as that of the Vav Y3xF and Vav Y174F proteins containing an intact SH3-SH2-SH3 region (Fig. 5C). The sizes of the foci derived from transfections involving Vav (Y3xF+Δ608-845)-, Vav (Y174F+Δ608-845)-, and Vav (Δ1-66+Y3xF+Δ608-845)-encoding vectors were significantly smaller, and the cell densities were less, than those derived from transfections with Vav (Δ1-186+Δ608-845)-encoding plasmids (data not shown). These results indicate that (i) the Vav Y3xF and Y174F mutations do not enhance the transforming activity of the oncogenic Vav (Δ1-66) protein; (ii) these two mutations induce biological properties different from those elicited by the deletion of residues 1 to 66, as they partially eliminate the requirement of the SH3-SH2-SH3 region for the biological activity of Vav proteins; and (iii) this effect is weaker than the one induced by the hyperactive Vav (Δ1-186) deletion mutant, suggesting that the biological activity of Vav Y3xF and Vav Y174F proteins is not totally independent of tyrosine phosphorylation. Collectively, these observations indicate that these point mutations represent a new mechanism of oncogenic activation for the Vav protein.
Expression of Y-to-F Vav mutants leads to enhanced responses in hematopoietic cells.
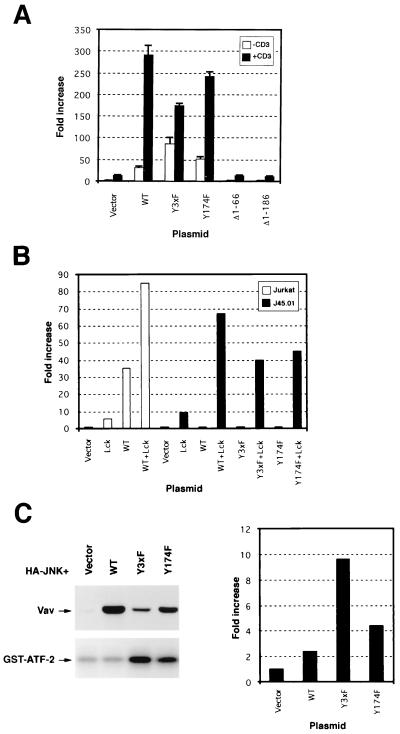
Since Vav is expressed predominantly in hematopoietic cells (5), we decided to test the activity of the Y-to-F Vav mutants in the normal environment of Vav. The best-known activity of Vav in T cells is the stimulation of the transcriptional activity of NF-AT (16, 32). Unlike the proliferative response mediated by Vav in fibroblasts, this response requires an intact CH region. As a consequence, the Vav (Δ1-66) protein cannot elicit NF-AT activation in Jurkat cells (32). The reason for the inactivity of this oncogenic version in T cells remains to be determined. However, Holsinger et al. have shown recently that the cotransfection of the Ca2+-dependent phosphatase calcineurin with either truncated Vav or the constitutively active mutant of Rac1 (Rac-1G12V) rescues the lack of stimulation of NF-AT by these two molecules (15), suggesting that the Vav CH region may be involved in a Ca2+-dependent signaling pathway that is synergistic with the Rac-1 pathway to induce this biological response. To test the activity of our Y-to-F Vav mutants in this alternative signaling system, we electroporated Jurkat cells with a NF-AT-regulated luciferase reporter vector either alone or with pMEX vectors containing wild-type Vav, Vav (Δ1-66), (Δ1-186), Y3xF, or Y174F. After 48 h, cells were left unstimulated or stimulated with anti-CD3 monoclonal antibodies and the activation of NF-AT obtained in each condition was determined using a luciferase assay. In good agreement with previous results (32), we found that the overexpression of the wild-type protein, but not of Vav (Δ1-66), induced a high basal activity of NF-AT in nonstimulated cells (Fig. 6A). Interestingly, Vav (Δ1-186) was also found to be inactive in this biological response despite its high oncogenic potential in NIH 3T3 cells (Fig. 6A). Under the same experimental conditions, the overexpression of Vav Y3xF led to a higher increase in the basal activity of NF-AT in nonstimulated cells than that observed upon wild-type Vav overexpression (Fig. 6A). Vav Y174 induced a smaller, but reproducible, increase in the NF-AT response (Fig. 6A). Stimulation of the T-cell receptor (TCR) via CD3ε cross-linking led to a further increase in NF-AT activity in cells expressing Vav, Vav Y3xF, and Vav Y174F proteins (Fig. 6A). Under these conditions, these Y-to-F Vav mutants did not induce higher responses than wild-type Vav. In fact, Vav Y3xF gave consistently lower levels of NF-AT activation than wild-type Vav and Vav Y174F in all experiments performed (Fig. 6A). This is probably due to the lower expression levels of these two mutants relative to wild-type Vav (Fig. 6C, left). As for nonstimulated cells, both N-terminally truncated versions of Vav failed to induce NF-AT activation after CD3 cross-linking (Fig. 6A).
FIG. 6.
(A) Activation of NF-AT by Vav proteins. Exponentially growing Jurkat cells were electroporated with a luciferase reporter gene with NF-AT sites (pNF-AT/luc; 5 μg) together with empty vector (20 μg) or with plasmids (20 μg) encoding the indicated Vav proteins. After 48 h, cells were either left resting or stimulated with anti-CD3 antibodies for 8 h. After this period, luciferase activity was determined in triplicate. The results shown are the means and standard deviations of three independent transfections, each performed in triplicate. WT, wild type. (B) Exponentially growing wild-type and J45.01 Jurkat cells were electroporated with pNF-AT/luc (5 μg) and with either empty vector or plasmids encoding the indicated Vav proteins (20 μg) and Lck (15 μg). After 48 h, the luciferase activity in each transfected sample was determined as indicated in Materials and Methods. Similar results were obtained in two independent transfections, each performed in duplicate. (C) (Left) Activation of JNK-1 by Y-to-F Vav mutants. Jurkat JMC-T cells were electroporated with a vector encoding HA–JNK-1 (5 μg) together with empty vector or with plasmids encoding the indicated Vav mutants (20 μg each). After 48 h, cells were washed and lysed, and the resulting cellular extracts were immunoprecipitated with anti-HA antibodies. Final immunocomplexes were subjected to in vitro kinase reactions in the presence of [γ-32P]ATP and GST–ATF-2 as the substrate (bottom). Expression of Vav proteins in this transfection was determined by anti-Vav immunoblotting of total cellular lysates (top). The mobilities of Vav proteins and the phosphorylated GST–ATF-2 are indicated by arrows. This is a representative experiment of three independent kinase assays. (Right) Normal Jurkat cells were electroporated with pFR-Luc (5 μg) and pFA2-cJun (2 μg) together with empty pMEX vector (20 μg; Vector) or plasmids (20 μg) encoding wild-type Vav and Vav Y3xF and Y174F. After 48 h, activation of the endogenous JNK was determined in duplicate using a luciferase assay (see Materials and Methods). Similar results were obtained in a second independent electroporation.
To investigate whether the NF-AT response induced by the Y-to-F Vav mutants was still dependent on upstream signals derived from the TCR, we conducted the same type of experiments with J45.01 cells, a Jurkat clone that is deficient in TCR signaling due to lack of expression of CD45. This membrane phosphatase is essential for the dephosphorylation of Lck in the inhibitory residue Y505, an essential step for the activation of Lck and the downstream Syk/Zap-70 kinases during T-cell signaling (8). Vav, Vav Y3xF, and Vav Y174F were incapable of inducing the transactivation of the NF-AT reporter gene in this cellular background (Fig. 6B), indicating that the activity of all these proteins is still dependent on signals emanating from the TCR complex. Lack of activity of wild-type Vav in these cells was previously described (32). Coexpression of these proteins with the constitutively active form of Lck (Y505F mutant) resulted in the rescue of that signaling defect (Fig. 6B), confirming that all the downstream elements of Vav involved in the NF-AT response were intact in J45.01 cells. Interestingly, the transfection of Lck (Y505F) alone in normal Jurkat cells resulted in levels of NF-AT activation lower than those obtained with wild-type Vav (Fig. 6B), suggesting that the limiting step in this biological response is the concentration of Vav.
Since NF-AT activation is not a good biological readout for measuring the catalytic activity of Vav, we decided to check the activation of JNK-1 induced by the Y-to-F Vav mutants in nonstimulated Jurkat cells. To facilitate the detection of the JNK activity in this system, we used JMC-T cells, a Jurkat cell line that allows the episomal amplification of the transfected plasmids. We found that Vav Y3xF and, to a lesser extent, Vav Y174F were fully competent in eliciting this cellular response. However, we could not observe any activation of this kinase by wild-type Vav (Fig. 6C, left). Immunoblot analysis of these cells confirmed that wild-type Vav was expressed at even higher levels than the transforming Vav Y3xF and Y174F proteins (Fig. 6C, left). Similar results were obtained when the activation of the endogenous JNK present in normal Jurkat cells was determined indirectly by testing the transcriptional activation of a fusion protein containing the transactivation domain of c-Jun and the DNA binding domain of Gal-4 (Fig. 6C, right). These results indicate that the Y-to-F Vav mutants increase the basal activity of full-length Vav in both nonhematopoietic and hematopoietic cells, giving a new point of functional divergence from the traditional oncogenic versions of Vav.
Y174F does not affect the phosphorylation-dependent exchange activity of full-length Vav.
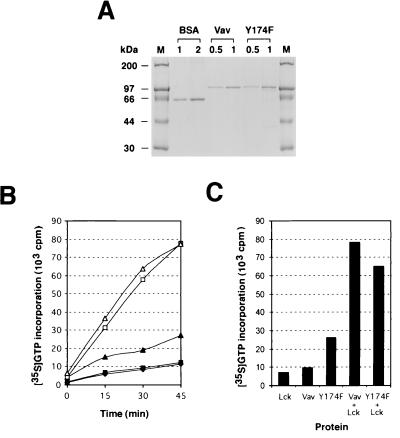
In order to characterize the mechanism of oncogenic activation of the Y-to-F mutations, we determined whether the catalytic activity of the associated proteins was phosphorylation dependent. To this end, we purified to homogeneity polyhistidine-tagged versions of Vav and Vav Y174F from baculovirus-infected Sf9 cells using chromatography onto nickel beads (Fig. 7A). Although Vav Y3xF was also overexpressed efficiently by insect cells, its purification was not possible due to its localization in Triton X-100-insoluble fractions even after prolonged sonication protocols (data not shown). The exchange activities of the nonphosphorylated and phosphorylated versions of Vav and Vav Y174F were then determined by measuring the ability of each protein to enhance the incorporation of γ-35S-GTP into GDP-loaded Rac-1 and RhoG, the two main substrates of Vav (11, 27). Time course experiments indicated that both Vav and Vav Y174F were dependent on tyrosine phosphorylation for the catalysis of the exchange of guanosine nucleotides on Rac-1 (Fig. 7B). However, while the phosphorylated versions of Vav and Vav Y174F showed similar activities, the nonphosphorylated version of Vav Y174F was twofold more active in this response than the wild-type Vav when equivalent molar amounts of each protein were used (Fig. 7B). A similar regulation was observed when these proteins were tested in GDP/GTP exchange assays using RhoG as a substrate (Fig. 7C). These results indicate that the transforming activity induced by the Vav Y174F mutation is not due to the acquisition of a phosphotyrosine-independent exchange activity.
FIG. 7.
(A) Purification of Vav proteins from Sf9 cells. The indicated amounts (micrograms) of aliquots from a representative preparation of six-His-tagged Vav and six-His-tagged Vav Y174F are shown. Bovine serum albumin was used as the standard for concentration. (B) Exchange activity of Vav proteins using γ-35S-GTP incorporation assays. GDP-loaded Rac-1 (15 pmol) was incubated with γ-35S-GTP in the presence of 3.5 pmol of nonphosphorylated Vav (solid squares), phosphorylated Vav (open squares), nonphosphorylated Vav Y174F (solid triangles), or phosphorylated Vav Y174F (open triangles). As a negative control, Rac-1 was incubated with Lck alone (solid diamonds). At each time point, aliquots from each incubation were taken in duplicate and the exchange obtained under each experimental condition was determined using a filter immobilization assay. (C) Exchange activity of Vav proteins towards RhoG. GDP-loaded RhoG was incubated with γ-35S-GTP and the indicated proteins. Exchange rates in each condition were measured as described for panel B after a 45-min incubation. The same results were obtained in three independent experiments, each performed in duplicate.
Increased phosphorylation levels of Vav Y-to-F mutants.
Since activation of Vav is mediated by direct tyrosine phosphorylation by protein tyrosine kinases (11, 13, 27), we next investigated whether the mutations in the tyrosine residues of the Vav Ac domain could result in alterations in the overall phosphorylation of this protein in vivo. To this end, we first made use of established NIH 3T3 cells expressing wild-type (B36-212 clone) and Vav Y3xF proteins (clones X19-61, X19-62, X19-63, and X19-64) to compare their phosphorylation status. Vav proteins were immunoprecipitated from lysates of exponentially growing cells and then subjected to immunoblot analysis using anti-PTyr and anti-Vav antibodies to determine the phosphorylation and protein expression levels of Vav, respectively. After normalization for the different expression levels, the phosphorylation levels of the Vav Y3xF proteins present in four different Vav Y3xF-transformed cell lines were found to be two- to fourfold higher than those found for wild-type Vav (Fig. 8A). To confirm that this effect was independent of the cell type, we evaluated the phosphorylation levels of Vav, Vav Y3xF, and Vav Y174F in transiently transfected COS-1 cells. The two Y-to-F Vav mutants also showed higher levels of tyrosine phosphorylation than Vav in these cells (Fig. 8B).
Since our previous exchange reactions indicated that the Vav Y174F protein shows low levels of exchange activity without prior incubation with Lck, we last compared the phosphorylation levels of purified Vav and Vav Y174F proteins derived from Sf9 cells using immunoblotting with anti-phosphotyrosine (anti-PTyr) antibodies. Vav Y174F displayed higher levels of phosphorylation than wild-type Vav under these conditions (Fig. 8C). Taken together, these results suggest that one of the consequences of the Y3xF and Y174 mutations is the hyperphosphorylation of Vav on tyrosine residues.
Y174 is phosphorylated in vivo in a stimulation-dependent manner in all Vav family members.
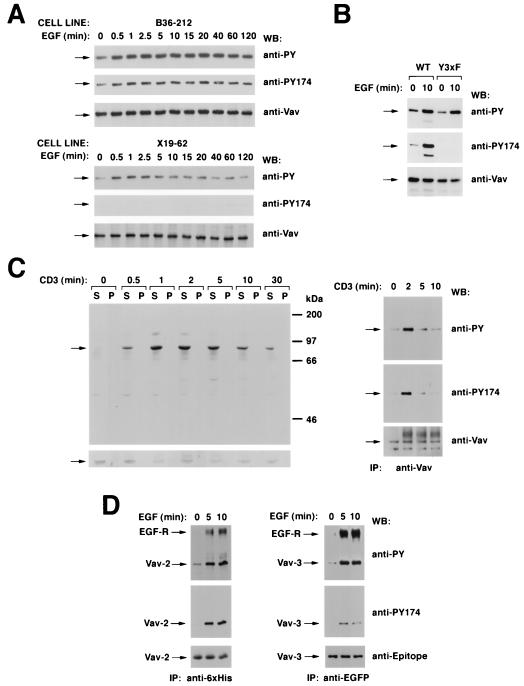
Given the importance of Y174 in Vav regulation, we last investigated its kinetics of phosphorylation in vivo. To this end, we generated a rabbit polyclonal antibody specific for the epitope 168-AEGDEI(p)YEDLMRL-180 of mouse Vav. Enzyme-linked immunosorbent assays indicated that the affinity-purified antibody recognized the phosphorylated antigen at less than 5 ng · ml−1 while the nonphosphorylated antigen required significantly higher levels of antibody (>10 μg · ml−1). The specificity of this antibody was also determined by immunoblot analysis of immunoprecipitated Vav and Vav Y3xF proteins from cell clones stimulated with EGF for several periods of time. As shown in Fig. 9A, the phosphospecific Vav antibody was capable of recognizing the phosphorylated version of wild-type Vav but not the phosphorylated Vav Y3xF mutant protein. Immunoblot analysis using a generic anti-PTyr antibody and anti-Vav antiserum confirmed that Vav and Vav Y3xF were expressed and phosphorylated by EGF in the cell lines used in these experiments (Fig. 9A). Similar results were obtained using Vav proteins obtained from transient transfections in COS-1 cells (Fig. 9B). These experiments demonstrate that this antibody is specific for Vav phosphorylated at position Y174.
FIG. 9.
(A) Phosphorylation of residue Y174 by EGF and specificity of the antibodies to Vav phosphorylated at that position. NIH 3T3 cell clones expressing either wild-type Vav (B36-212 cells; top) or Vav Y3xF (X19-62 cells; bottom) were serum starved for 48 h and then stimulated with EGF for the indicated periods of time. After stimulation, cells were lysed and immunoprecipitated with anti-Vav antibodies and immunocomplexes were subjected sequentially to Western blotting (WB) with antiphosphotyrosine (anti-PY) antibodies, the phosphospecific antibody to residue Y174 of Vav (anti-PY174), and anti-Vav antibodies. Arrows, migration of Vav proteins. (B) Specificity of anti-VavPY174 antibodies in COS-1 cells. Quiescent and EGF-stimulated COS-1 cells expressing the indicated Vav proteins were immunoprecipitated with anti-Vav antibodies and then subjected sequentially to Western blotting with anti-VavPY174, anti-Vav, and antiphosphotyrosine antibodies. WT, wild type. (C) Phosphorylation of Y174 in T cells. (Left) Jurkat cells were stimulated with anti-CD3 antibodies for the indicated periods of time and lysed, and equivalent amounts of total cellular lysates from the Triton X-100-soluble (S) and -insoluble (P) fractions were analyzed by immunoblotting with anti-VavPY174 (top) or anti-Vav (bottom) antibodies. (Right) Vav proteins were immunoprecipitated (IP) from Jurkat cells stimulated for the indicated periods of time with anti-CD3 and then subjected sequentially to Western blot analysis with antibodies as listed for panel A. Arrows, migration of Vav. (D) Phosphorylation of the position equivalent to Y174 in Vav-2 (residue Y172) and Vav-3 (residue Y173). COS-1 cells expressing ectopically six-His-tagged Vav-2 (left) and EGFP-tagged Vav-3 (right) were serum starved overnight and stimulated for the indicated periods of time with EGF. After stimulation, cells were lysed and Vav family proteins were immunoprecipitated with anti-polyhistidine (Vav-2; left) or anti-EGFP (Vav-3; right) antibodies. Washed immunocomplexes were then subjected to sequential Western blot analysis with antiphosphotyrosine, anti-VavY174, and the respective antiepitope antibody. For panel A, signals were developed either using 125I-labeled protein A (anti-Vav immunoblots) or by treatment with an anti-mouse immunoglobulin G antibody followed by incubations with 125I-labeled protein A (antiphosphotyrosine) or by the ECL method (anti-VavPY174). For panels B and C, signals were developed using only the ECL method.
Next, we determined the kinetics of phosphorylation of Y174 in the endogenous Vav protein expressed in Jurkat cells. To this end, we stimulated them for the indicated (Fig. 9C) periods of time with anti-CD3 antibodies and, after lysis, we subjected the cellular lysates derived from the Triton X-100-soluble and -insoluble fractions to immunoblot analysis using the anti-VavPY174 antibody. These experiments indicated that the levels of phosphorylation of Y174 were low in nonstimulated cells (Fig. 9C, left). The engagement of the TCR via CD3 cross-linking led to a rapid (0.5-min) and transient (0.5- to 30-min) phosphorylation of Vav at the Y174 position (Fig. 9C, left). Immunoblot analysis of the same filter with anti-Vav antibodies demonstrated similar levels of expression at all stimulation times (Fig. 9C, left). In spite of the role of endogenous Vav in cytoskeletal organization, no significant relocalization of it to the Triton X-100-insoluble pellet was observed at any stage of the T-cell stimulation cycle (Fig. 9C, left).
To compare the phosphorylation kinetics of Y174 with the overall phosphorylation of Vav occurring during signal transduction, we immunoprecipitated Vav from cellular lysates derived from nonstimulated and CD3-stimulated Jurkat cells and analyzed its phosphorylation levels at each time point using either a generic antiphosphotyrosine monoclonal antibody or the phosphospecific anti-VavPY174 antibody. This analysis indicated that the phosphorylation of position Y174 followed the same kinetics as the phosphorylation of other sites present on the Vav molecule (Fig. 9C, right). The presence of equal amounts of immunoprecipitated Vav protein at all stimulation time points was demonstrated by anti-Vav immunoblots of the same filters (Fig. 9C, right). The same results were obtained when the anti-VavPY174 antibody was used first in the immunoblots, demonstrating that the signals obtained with anti-VavPY174 were not derived from traces of the anti-PTyr antibody remaining bound to Vav after the stripping of the filters (data not shown and Fig. 9B). Taken together, these results confirm that the phosphorylation of Y174 in vivo is by both membrane and cytoplasmic tyrosine kinase receptors and that its phosphorylation is stimulation dependent, indicating that its inhibitory role is circumscribed to poststimulation stages.
Since Y174 is conserved in the two other known members of the Vav family, we last explored the possibility that this phosphospecific antibody could be used to detect the phosphorylation of this residue in Vav-2 and Vav-3. To this end, we used expression vectors encoding a polyhistidine-tagged version of full-length Vav-2 (pAO1) and a EGFP-tagged version of Vav-3 (residues 144 to 847; pNM100) to express these proteins ectopically in COS-1 cells. After transfection, these cells were made quiescent by serum withdrawal and then stimulated for the indicated (Fig. 9D) periods of time with EGF. As shown in Fig. 9D (top), the analysis of the Vav-2 and Vav-3 immunocomplexes by anti-PTyr immunoblot analysis indicated that the treatment with EGF stimulated the phosphorylation of Vav-2 and Vav-3 on tyrosine residues and their physical association with the autophosphorylated EGF receptor. The reblotting of these filters with anti-VavPY174 antibodies confirmed that this position also underwent EGF-dependent phosphorylation both in Vav-2 and Vav-3 (Fig. 9D, middle). Immunoblot analysis with antipolyhistidine and anti-EGFP antibodies confirmed that Vav-2 and Vav-3 were expressed at similar levels at each stimulation time point (Fig. 9D, bottom). These results indicate that the phosphorylation of this negative regulatory site is conserved in all known mammalian members of the Vav family.
DISCUSSION
Tyrosine phosphorylation is one of the most versatile mechanisms to regulate the function of signaling proteins. This posttranslational modification exerts both positive and negative effects on the activity of numerous proteins, either by promoting intramolecular interactions, protein-protein interactions, or allosteric changes in the activity of proteins. One of the most recent examples of the regulation of proteins by such modification is the stimulation of Vav proteins, the only known family of Rho/Rac GEFs whose activity is turned on by direct tyrosine phosphorylation. This activation appears to be due to a conformational change induced by the incorporation of new phosphate groups, because it can be reproduced in vitro with pure preparations of Vav and tyrosine kinases (11, 27). In addition to the regulation of the catalytic activity, tyrosine phosphorylation regulates indirectly the signaling output of the Vav pathway by promoting the interaction of this Rac GEF with other intracellular proteins. Thus, it has been shown that the Vav SH2 region interacts with tyrosine-phosphorylated kinases such as Zap-70, EGF receptor, and platelet-derived growth factor receptor, an interaction that facilitates the subsequent phosphorylation and activation of Vav (4, 18, 21). In addition, the Vav SH2 domain interacts with tyrosine phosphorylated Slp-76 (25, 33), an association that allows the formation of a multiprotein complex among Vav and the Slp-76-associated proteins Nck and PAK. This interaction leads in turn to PAK stimulation due the close proximity of this kinase to the Vav substrate Rac-1 (2). Tyrosine phosphorylation is therefore at the center of the positive regulation of Vav, facilitating its phosphorylation, its activation, and the effective transmission of signals to its downstream elements.
In the present work, we present evidence indicating that tyrosine phosphorylation also plays a negative regulatory role in the function of Vav proteins. We have shown that a single Y-to-F point mutation in one of the phosphorylation sites of wild-type Vav (residue 174) leads to the enhancement of its biological activity in vivo, resulting in high levels of cellular transformation and the constitutive activation of other Rac-1/RhoG-dependent responses. The effect of this Y174F mutation can be further increased by extra mutations affecting either of two neighboring tyrosine residues (Y142 and Y160). This appears to be a synergistic effect, because Vav proteins containing point mutations in either the Y142 or the Y160 residue alone displayed significantly lower biological activities. These results are in agreement with the in vitro kinase assays indicating that Y174 is the main phosphorylation site in the Vav Ac domain.
Using focus formation assays of rodent fibroblasts, we could estimate that the increase in activity of these Y-to-F mutants is about 150- and 2-fold greater than the values promoted by wild-type Vav and the initially described oncogenic version of Vav [Vav (Δ1-66)] (17), respectively. However, these Y-to-F mutants are significantly less active than the hyperoncogenic version of Vav [Vav (Δ1-186)], even when they are compared with Vav (Δ1-186) proteins lacking SH3-SH2-SH3 regions. Since Vav (Δ1-186) has lost completely its regulation by phosphorylation (27), these results suggest that the mechanisms of oncogenic activation of Vav Y174F and Vav (Δ1-186) are different. Several additional observations support this idea. Thus, we have demonstrated biochemically that Vav Y174F, like wild-type Vav, requires tyrosine phosphorylation to stimulate its GDP/GTP exchange activity towards the GTPases Rac-1 and RhoG. Second, we have found that the high basal NF-AT activity induced by Vav Y3xF and Vav Y174F requires upstream signals deriving from a functional TCR complex (Fig. 6A and B), indicating that tyrosine phosphorylation can still enhance the activity of these proteins in T cells. Finally, the lack of activity of the truncated versions of Vav [Vav (Δ1-66) and (Δ1-186)] in the stimulation of NF-AT further highlights the functional disparities between these two mechanisms of Vav oncogenic activation.
Three important clues for the possible regulatory role of Y174 were obtained from our in vivo experiments. Using an antibody specific for Vav phosphorylated at Y174, we have shown that this residue becomes phosphorylated after the stimulation of membrane receptors by mitogens and antigens. In all these cases, the kinetics of phosphorylation of Y174 mimic those of the overall phosphorylation of Vav. These two results indicate that the function of the phosphorylated Y174 residue is to act as a negative-feedback mechanism during cell stimulation rather than to keep Vav inactive in resting cells. Moreover, we have shown that these Y-to-F mutations induce higher levels of Vav tyrosine phosphorylation in exponentially growing cells, indicating that Y174 may be involved in the overall turnover of phosphate groups in Vav. This mechanism appears to be highly conserved, because high levels of phosphorylation are also found in Vav Y174F proteins purified from insect cells. Interestingly, both the Y3xF and the Y174F mutations result in a partial loss of dependency on the SH3-SH2-SH3 domains for the transforming activity of these proteins, a region that is essential for the cellular transformation induced by Vav and Vav (Δ1-66) proteins. Although we have not tested the phosphorylation status of these SH3-SH2-SH3-deficient proteins in transformed cells, preliminary results of experiments conducted with COS-1 cells indicate that the Vav (Y174F+ΔSH3-SH2-SH3) protein still shows low, but detectable, levels of tyrosine phosphorylation in exponentially growing cells (data not shown). Thus, high levels of tyrosine phosphorylation appear to be linked to all oncogenic forms of these Y-to-F Vav mutants. However, it is important to note that while we could observe easily this effect in exponentially growing cells, we could not detect any significant differences in the kinetics of phosphorylation of Vav and Vav Y3xF after the stimulation of quiescent cells with saturating concentrations of EGF (Fig. 9A and data not shown). This suggests that the effect of the Y174F mutation is only relevant when Vav is present under suboptimal phosphorylation conditions. The observation that Vav Y3xF and Vav Y174F promote preferentially higher levels of NF-AT activation than Vav in nonstimulated Jurkat cells is in good agreement with this possibility.
The mechanism by which the Y3xF and Y174F mutations can mediate Vav phosphorylation and activation is still uncertain. One possibility is that, similar to the inhibitory tyrosine residue of Src family members, residue Y174 could bind to other structural domains of Vav, generating an inactive configuration that blocks the access of both tyrosine kinases and GTPases. Several indirect observations strongly argue against this possibility. Using GST pull-down experiments, we have been unable to detect any direct interaction between Vav and GST fusion proteins containing the Vav SH2 domain, the CH region, or the tyrosine-phosphorylated version of the Vav Ac domain (data not shown). Moreover, if that model is correct, it would be expected that mutations in other domains of Vav could induce a Y174F-like activation of full-length Vav, leading to cellular transformation. However, recent experiments indicate that mutations affecting the individual DH, PH, ZF, SH2, and SH3 domains of Vav all result in the inactivation of wild-type Vav, ruling out the possibility that an intramolecular interaction of Y174F with other structural domains is taking place (data not shown). Based on those observations, we currently favor the hypothesis that Y174 may negatively regulate Vav function by mediating the interaction with an inhibitory protein. Consistent with this, residue Y174 is followed by peptide sequences that create an optimal binding site for SH2 domains of class I and III (29), implying that this area of Vav is probably involved in heteromolecular interactions. This functional alternative, although novel for Vav and any other Ras or Rho/Rac GEF, has been described before for other signaling molecules, including the kinase Zap-70 and the adapter molecule CrkL (28, 34). Potential candidates for such an inhibitory molecule include SH2-containing phosphatases similar to PTP-1C or inhibitory proteins with other phosphotyrosine binding domains (e.g., PTB) similar to those of c-Cbl or Cbl-b proteins. We are currently screening expression libraries with this phosphorylated region of Vav to identify this negative regulator.
The observations reported here are also important for the Vav field because they eliminate previous models for the action of Vav. Prior to the discovery that Vav was a Rac GEF, Deckert and coworkers demonstrated that Syk binds to Vav and phosphorylates Y174-containing peptides in vitro, a result that led to the proposal that this residue was an essential step in the activation of the Vav signal transduction pathway, presumably by facilitating the binding of a putative downstream Lck to Vav (12). However, this model was never validated by testing the functional consequences of the Y174F mutation in T cells. Our results demonstrating that Vav Y174F is fully functional strongly argue against any possible effector function for Y174 in T cells. Another study showed that a bacterially expressed Vav Y174F protein could not be phosphorylated properly by Lck in vitro, implying that this site was the main, if not the unique, target for the Lck-mediated stimulation of Vav exchange activity (14). More recently, a different group showed that the Vav Y174F protein acts as a dominant-negative mutant for the Vav-dependent morphological changes induced by G-coupled receptors and activated forms of phosphatidylinositol-3-kinase-γ in COS-7 cells, thereby implying again a major role for Y174 in the phosphorylation-dependent activation of this protein (20). Clearly, these last two observations are not congruent with the in vitro and in vivo observations presented here. Although the reason for this discrepancy is unclear, it is possible that the Y174F Vav mutant used in those studies could contain additional mutations that inactivated this new oncogenic version of Vav.
The results presented here reveal a hitherto-unexpected complexity in the level of regulation of Vav activity by tyrosine phosphorylation. It is now clear that this posttranslational modification will be involved in the activation of Vav, in the regulation of the strength of the signals emanating from this molecule, and also in the negative regulation of its function. The availability of these new gain-of-function mutations of full-length Vav will help us to further dissect these multiple layers of regulation and serve as better experimental tools to study the role of this Rac GEF in cell signaling.
ACKNOWLEDGMENTS
We thank D. Colflesh for his help with microscopy and N. Reich for her comments during the writing of the manuscript. We thank also D. Rothstein and H. Band for the generous gift of T-antigen-expressing Jurkat cells.
This work was supported by an NCI grant (1RO1CA7373501) to X.R.B., who is a Sinsheimer Scholar for Cancer Research. The work of C.C. was supported by a fellowship from the Autonomous Government of Catalonia.
REFERENCES
- 1.Brunati A M, Donella-Deana A, Ruzzene M, Marin O, Pinna L A. Site specificity of p72syk protein tyrosine kinase: efficient phosphorylation of motifs recognized by Src homology 2 domains of the Src family. FEBS Lett. 1995;367:149–152. doi: 10.1016/0014-5793(95)00555-n. [DOI] [PubMed] [Google Scholar]
- 2.Bubeck-Wardenburg J, Pappu R, Bu J Y, Mayer B, Chernoff J, Straus D, Chan A C. Regulation of PAK activation and the T cell cytoskeleton by the linker protein SLP-76. Immunity. 1998;9:607–616. doi: 10.1016/s1074-7613(00)80658-5. [DOI] [PubMed] [Google Scholar]
- 3.Bustelo X R. The VAV family of signal transduction molecules. Crit Rev Oncog. 1996;7:65–88. doi: 10.1615/critrevoncog.v7.i1-2.50. [DOI] [PubMed] [Google Scholar]
- 4.Bustelo X R, Ledbetter J A, Barbacid M. Product of vav proto-oncogene defines a new class of tyrosine protein kinase substrates. Nature. 1992;356:68–71. doi: 10.1038/356068a0. [DOI] [PubMed] [Google Scholar]
- 5.Bustelo X R, Rubin S D, Suen K L, Carrasco D, Barbacid M. Developmental expression of the vav protooncogene. Cell Growth Differ. 1993;4:297–308. [PubMed] [Google Scholar]
- 6.Bustelo X R, Suen K L, Leftheris K, Meyers C A, Barbacid M. Vav cooperates with Ras to transform rodent fibroblasts but is not a Ras GDP/GTP exchange factor. Oncogene. 1994;9:2405–2413. [PubMed] [Google Scholar]
- 7.Bustelo X R, Suen K L, Michael W M, Dreyfuss G, Barbacid M. Association of the vav proto-oncogene product with poly(rC)-specific RNA-binding proteins. Mol Cell Biol. 1995;15:1324–1332. doi: 10.1128/mcb.15.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan A C, Desai D M, Weiss A. The role of protein tyrosine kinases and protein tyrosine phosphatases in T cell antigen receptor signal transduction. Annu Rev Immunol. 1994;12:555–592. doi: 10.1146/annurev.iy.12.040194.003011. [DOI] [PubMed] [Google Scholar]
- 9.Coppola J, Bryant S, Koda T, Conway D, Barbacid M. Mechanism of activation of the vav protooncogene. Cell Growth Differ. 1991;2:95–105. [PubMed] [Google Scholar]
- 10.Crespo P, Bustelo X R, Aaronson D S, Coso O A, Lopez-Barahona M, Barbacid M, Gutkind J S. Rac-1 dependent stimulation of the JNK/SAPK signaling pathway by Vav. Oncogene. 1996;13:455–460. [PubMed] [Google Scholar]
- 11.Crespo P, Schuebel K E, Ostrom A A, Gutkind J S, Bustelo X R. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 12.Deckert M, Tartare-Deckert S, Couture C, Mustelin T, Altman A. Functional and physical interactions of Syk family kinases with the Vav proto-oncogene product. Immunity. 1996;5:591–604. doi: 10.1016/s1074-7613(00)80273-3. [DOI] [PubMed] [Google Scholar]
- 13.Han J, Das B, Wei W, Van Aelst L, Mosteller R D, Khosravi-Far R, Westwick J K, Der C J, Broek D. Lck regulates Vav activation of members of the Rho family of GTPases. Mol Cell Biol. 1997;17:1346–1353. doi: 10.1128/mcb.17.3.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller R D, Krishna U M, Falck J R, White M A, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 15.Holsinger L J, Graef I A, Swat W, Chi T, Bautista D M, Davidson L, Lewis R S, Alt F W, Crabtree G R. Defects in actin-cap formation in Vav-deficient mice implicate an actin requirement for lymphocyte signal transduction. Curr Biol. 1998;8:563–572. doi: 10.1016/s0960-9822(98)70225-8. [DOI] [PubMed] [Google Scholar]
- 16.Holsinger L J, Spencer D M, Austin D J, Schreiber S L, Crabtree G R. Signal transduction in T lymphocytes using a conditional allele of Sos. Proc Natl Acad Sci USA. 1995;92:9810–9814. doi: 10.1073/pnas.92.21.9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzav S, Martin-Zanca D, Barbacid M. vav, a novel human oncogene derived from a locus ubiquitously expressed in hematopoietic cells. EMBO J. 1989;8:2283–2290. doi: 10.1002/j.1460-2075.1989.tb08354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katzav S, Sutherland M, Packham G, Yi T, Weiss A. The protein tyrosine kinase ZAP-70 can associate with the SH2 domain of proto-Vav. J Biol Chem. 1994;269:32579–32585. [PubMed] [Google Scholar]
- 19.Kranewitter W J, Gimona M. N-terminally truncated Vav induces the formation of depolymerization-resistant actin filaments in NIH 3T3 cells. FEBS Lett. 1999;455:123–129. doi: 10.1016/s0014-5793(99)00857-1. [DOI] [PubMed] [Google Scholar]
- 20.Ma A D, Metjian A, Bagrodia S, Taylor S, Abrams C S. Cytoskeletal reorganization by G protein-coupled receptors is dependent on phosphoinositide 3-kinase γ, a Rac guanosine exchange factor, and Rac. Mol Cell Biol. 1998;18:4744–4751. doi: 10.1128/mcb.18.8.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margolis B, Hu P, Katzav S, Li W, Oliver J M, Ullrich A, Weiss A, Schlessinger J. Tyrosine phosphorylation of vav proto-oncogene product containing SH2 domain and transcription factor motifs. Nature. 1992;356:71–74. doi: 10.1038/356071a0. [DOI] [PubMed] [Google Scholar]
- 22.Montaner S, Perona R, Saniger L, Lacal J C. Multiple signalling pathways lead to the activation of the nuclear factor κB by the Rho family of GTPases. J Biol Chem. 1998;273:12779–12785. doi: 10.1074/jbc.273.21.12779. [DOI] [PubMed] [Google Scholar]
- 23.Movilla N, Bustelo X R. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol Cell Biol. 1999;19:7870–7875. doi: 10.1128/mcb.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson M F, Pasteris N G, Gorski J L, Hall A. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- 25.Onodera H, Motto D G, Koretzky G A, Rothstein D M. Differential regulation of activation-induced tyrosine phosphorylation and recruitment of SLP-76 to Vav by distinct isoforms of the CD45 protein-tyrosine phosphatase. J Biol Chem. 1996;271:22225–22230. doi: 10.1074/jbc.271.36.22225. [DOI] [PubMed] [Google Scholar]
- 26.Schuebel K E, Bustelo X R, Nielsen D A, Song B J, Barbacid M, Goldman D, Lee I J. Isolation and characterization of murine vav2, a member of the vav family of proto-oncogenes. Oncogene. 1996;13:363–371. [PubMed] [Google Scholar]
- 27.Schuebel K E, Movilla N, Rosa J L, Bustelo X R. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 1998;17:6608–6621. doi: 10.1093/emboj/17.22.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senechal K, Heaney C, Druker B, Sawyers C L. Structural requirements for function of the Crk1 adapter protein in fibroblasts and hematopoietic cells. Mol Cell Biol. 1998;18:5082–5090. doi: 10.1128/mcb.18.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Songyang Z, Shoelson S E, McGlade J, Olivier P, Pawson T, Bustelo X R, Barbacid M, Sabe H, Hanafusa H, Yi T, Ren R, Baltimore D, Ratnofsky S, Feldman R A, Cantley L C. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Eb A J, Graham F L. Assay of transforming activity of tumor virus DNA. Methods Enzymol. 1980;65:826–839. doi: 10.1016/s0076-6879(80)65077-0. [DOI] [PubMed] [Google Scholar]
- 31.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Katzav S, Weiss A. A functional T-cell receptor signaling pathway is required for p95vav activity. Mol Cell Biol. 1995;15:4337–4346. doi: 10.1128/mcb.15.8.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Motto D G, Koretzky G A, Weiss A. Vav and SLP-76 interact and functionally cooperate in IL-2 gene activation. Immunity. 1996;4:593–602. doi: 10.1016/s1074-7613(00)80485-9. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Q, Weiss A. Enhancement of lymphocyte responsiveness by a gain-of-function mutation of ZAP-70. Mol Cell Biol. 1996;16:6765–6774. doi: 10.1128/mcb.16.12.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]