Abstract
The neurofibromatosis type 2 (NF2) protein, known as schwannomin or merlin, is a tumor suppressor involved in NF2-associated and sporadic schwannomas and meningiomas. It is closely related to the ezrin-radixin-moesin family members, implicated in linking membrane proteins to the cytoskeleton. The molecular mechanism allowing schwannomin to function as a tumor suppressor is unknown. In attempt to shed light on schwannomin function, we have identified a novel coiled-coil protein, SCHIP-1, that specifically associates with schwannomin in vitro and in vivo. Within its coiled-coil region, this protein is homologous to human FEZ proteins and the related Caenorhabditis elegans gene product UNC-76. Immunofluorescent staining of transiently transfected cells shows a partial colocalization of SCHIP-1 and schwannomin, beneath the cytoplasmic membrane. Surprisingly, immunoprecipitation assays reveal that in a cellular context, association with SCHIP-1 can be observed only with some naturally occurring mutants of schwannomin, or a schwannomin spliced isoform lacking exons 2 and 3, but not with the schwannomin isoform exhibiting growth-suppressive activity. Our observations suggest that SCHIP-1 interaction with schwannomin is regulated by conformational changes in schwannomin, possibly induced by posttranslational modifications, alternative splicing, or mutations.
Neurofibromatosis type 2 (NF2) is an autosomal dominant disease which causes a predisposition to the development of nervous system tumors, mainly vestibular schwannomas involving the eighth pair of cranial nerves and to a lesser extent schwannomas of other locations, meningiomas, and ependymomas (13). In 1993, positional cloning led to identification of the NF2 gene, which was shown to be mutated in one allele in NF2 patients and in both alleles in NF2 patient tumors (41, 58). Thus, the NF2 gene was classified as a typical tumor suppressor gene requiring functional inactivation of both copies to promote NF2-related tumor development. Later, the NF2 gene was also found to be mutated in sporadically occurring schwannomas, meningiomas (3, 5, 35, 42, 44), and primary malignant mesotheliomas (4, 9). The NF2 gene is composed of 17 exons and codes for a protein known as schwannomin or merlin (41, 58). Alternative splicing events lead to the expression of several isoforms of this protein (1, 3, 21, 25, 37, 38). The two most abundant isoforms either lack residues corresponding to exon 16 (isoform 1) or exon 17 (isoform 2), leading to variant C-terminal ends of the proteins. Less frequently encountered isoforms lack residues in the N-terminal domain of the protein encoded by one to three exons.
Schwannomin is strikingly similar to certain members of the membrane-associated cytoplasmic proteins of the band 4.1 superfamily, known as ezrin, radixin, and moesin (the ERMs). These ERM proteins are 45 to 47% identical to schwannomin overall, with homology mainly located in the N-terminal ERM domain (41, 58). The ERM proteins appear to function as molecular linkers between the cytoplasmic membrane and the cytoskeleton. They are found in actin-rich surface protrusions of the cortical plasma membrane such as filopodia, lamellipodia, membrane ruffles, and microvilli (2, 14, 45, 56). In these structures, N-terminal domains of the ERMs are apparently bound to transmembrane proteins whereas the C-terminal ends are anchored to the cortical actin cytoskeleton (7, 33, 59).
The homology between the ERMs and schwannomin suggest that the latter may also act as a linker between membrane and cytoplasmic components. Like the ERMs, schwannomin localizes mainly to the interface between the plasma membrane and the actin cytoskeleton, with an enrichment in ruffling edges (10–12, 17, 43, 46, 48, 50). Besides, schwannomin shares common molecular partners with the ERMs as it colocalizes with the cell surface marker CD44 and a regulatory cofactor of Na+-H+ exchanger, NHE-RF (36, 43). However, several observations suggest that schwannomin and ERMs have somewhat divergent functions. First, schwannomin, unlike the ERMs, is also found in the perinuclear region, sometimes in uncharacterized granules (11, 12, 28, 50, 53). In addition, schwannomin lacks the high-affinity actin-binding domain present in the C-terminal end of ERM proteins (24, 50, 60). Instead, it may contact actin either indirectly via the actin-binding protein βII spectrin/fodrin (49) or directly through a more central domain of the protein (61).
Another important functional difference between ERMs and schwannomin is the ability of schwannomin isoform 1 (SCH-Iso1) to function as a growth suppressor when exogenously expressed in immortalized cells (31, 52). SCH-Iso1 can also revert a ras-induced malignant phenotype in fibroblasts (57). Such activities have never been observed for any of the ERMs. Unlike SCH-Iso1, isoform 2 expressed in rat schwannoma cells does not inhibit cell growth (52). The functional difference between schwannomin isoforms 1 and 2 may rely on conformational properties of the two proteins. Indeed, isoform 1, like the ERM proteins (15, 32, 33), is able to form intramolecular interactions between domains distant on the sequence (19, 24, 33, 52). The domains involved in these interactions include a central region spanning residues 288 to 400 and the C-terminal exon 17 present in SCH-Iso1 but absent in isoform 2. According to this model, only isoform 1 can exist in two conformations (open and closed), possibly affecting its ability to interact with molecular partners. As will be discussed below, other isoforms, as well as mutants expressed in NF2 tumors, may also be affected in their protein folding (61).
How NF2 genetic alterations contribute to tumorigenesis remains obscure. Mutations of the NF2 gene can be classified in two categories. The majority are nonsense, frameshift, or splice site mutations that result in premature termination of translation. The putative truncated proteins encoded by these mutant genes are not accumulated to detectable levels in tumors (20, 30, 44, 54), possibly due to calpain-dependent degradation (27). The second class of mutations, which occur at a low frequency, are missense mutations, in-frame interstitial deletions, or splice site mutations causing exon skipping without frameshift. The proteins encoded by this second class of mutants are apparently stable (19; L. Goutebroze, unpublished results), but in several cases, these mutant proteins were found to have a weakened interaction with the plasma membrane and to be delocalized from the membrane to the cytoplasm (10, 28). Interestingly, some of the mutant proteins show also increased in vitro interaction with polymerized microtubules (61), suggesting that the mutations, like the naturally occurring splicing events, may disrupt intramolecular interactions and favor interactions with different molecular partners by forcing the schwannomin protein into a specific conformation.
In an attempt to understand the molecular mechanism allowing schwannomin to function as a tumor suppressor, we have used the yeast two-hybrid system to identify novel molecular partners of schwannomin. Screening with a bait expressing only the conserved N-terminal ERM domain of the protein, we isolated cDNAs encoding two previously noncharacterized proteins that we called SCHIP-1 and SCHIP-2. Here, we described the cloning and characterization of one of these proteins.
MATERIALS AND METHODS
DNA constructs.
For expression in mammalian cells, cDNAs encoding C-terminally vesicular stomatitis virus (VSV)-tagged versions of the various schwannomin isoforms and mutants were inserted in the pCB6 cytomegalovirus expression vector. SCH-Iso1, SCH-Δ118, SCH-Δ(39-121), and SCH-219 constructs have been described before (10). To obtain the SCH(1-314) construct, the corresponding region was amplified from the schwannomin cDNA by PCR and the product was inserted in pCB6 as a KpnI/SmaI fragment. The pCB6-HA-SCHIP-1 expression vector contains a full-length SCHIP-1 cDNA fused at the level of the initiating ATG to a hemagglutinin epitope (HA) tag-encoding sequence and inserted into pCB6 digested by BglII and EcoRI.
For in vitro expression, SCH-Iso1, SCH-Δ118, SCH-Δ(39-121), SCH-219, SCH(1-288), SCH(1-314), SCH(289-595), SCH(315-595), SCH(19-288), and SCH(19-314) constructs were obtained by introducing corresponding schwannomin cDNA fragments, generated by either restriction digestion or PCR amplification, into pBluescript (pBls; Stratagene). For SCHIP-1 and SCHIP-1-Δ(22-253) in vitro expression, we used the pBls and pGEM-T (Promega) vectors obtained during the isolation of human SCHIP-1 cDNA or the cloning of reverse-transcribed and amplified mRNA of SCHIP-1. In vitro expression of the two truncated forms of SCHIP-1, SCHIP-1(1-305) and SCHIP-1(1-413), was performed with the pBls-SCHIP-1 construct digested by HincII and DraI, respectively. DNA for in vitro expression of the partial mouse SCHIP-1 protein SCHIP-1(120-487) was obtained from a PCR product containing a T3 RNA polymerase recognition site upstream of a ATG initiation codon in frame with the mouse SCHIP-1 sequence.
For expression in Escherichia coli, glutathione S-transferase (GST)–SCHIP-1(306-487) and GST–SCHIP-1(112-305) constructs were obtained by inserting corresponding fragments from the human SCHIP-1 cDNA into pGEX-3X (Pharmacia Biotech). The GST–SCHIP-1(120-487) construct was obtained by inserting a corresponding fragment of mouse SCHIP-1 cDNA into pGEX-2T (Pharmacia Biotech). To express GST–SCH(1-314), GST-SCH(1-27), and GST-SCH(280-323), PCR fragments encoding amino acids 1 to 314, 1 to 27, and 280 to 323, respectively, of schwannomin were inserted in pGEX-2T. For all of the constructs, cloned PCR products were entirely sequenced to verify that no spurious mutation had been introduced during the PCR amplification step.
Yeast two-hybrid screening.
Yeast and two-hybrid procedures were handled according to published methods (26). A cDNA library from mouse fetal brain polyadenylated RNA, constructed in fusion with the GAL4 activating domain in a pGAD vector, was used. It was provided by J. Camonis. The N-terminal part (amino acids 1 to 314) of schwannomin fused in frame with the GAL4 DNA-binding domain [pGBT-SCH(1-314) vector] was defined as the bait. The Saccharomyces cerevisiae reporter strain HF7, containing two reporter genes, HIS3 and lacZ, was sequentially transformed with pGBT-SCH(1-314) and the mouse fetal brain cDNA library by the lithium acetate method. Double transformants were plated on synthetic medium lacking histidine, leucine, and tryptophan but containing 20 mM 3-amino-1,2,4-triazole (Sigma). Plates were incubated at 30°C for 3 days. His+ colonies were assayed for β-galactosidase activity by filter assay. Positive clones were rescued and tested for specificity by retransformation into the HF7 strain either with pGBT-SCH(1-314) or with irrelevant baits (pGBT-snf4 and pGBT-lamin). Specific clones were then tested for the ability to give a positive response in cotransfection with a bait expressing full-length SCH-Iso1 fused in frame with the GAL4 DNA-binding domain.
Human SCHIP-1 cDNA cloning and nucleotide sequence analysis.
To obtain a human SCHIP-1 full-length cDNA, the mouse 32P-labeled (bp 1 to 347) fragment of SCHIP-1 was hybridized to a human fetal brain λgt10 cDNA library (Clontech, Palo Alto, Calif.). The cDNA inserts of positive phages were subcloned into the pBls SK+ vector (Stratagene) and sequenced. The putative SCHIP-1 alternatively spliced isoforms SCHIP-1-Δ(241-253) and SCHIP-1-Δ(22-253) were isolated by reverse transcriptase PCR amplification experiments on human brain poly(A)+ RNA (Clontech), followed by subcloning of the amplified DNA fragments in the pGEM-T vector (Promega).
Sequencing of both mouse and human cDNAs was performed on both strands with an automatic sequencer (Applied Biosystems [Foster City, Calif.] model 373A) using an ABI PRISM DyeDeoxy terminator kit (Applied Biosystems). Sequences were assembled using the Autoassembler software (Applied Biosystems). Sequence comparisons were done with the BLAST program. Protein signatures were sought in PROSITE. Predicted primary and secondary structures were analyzed through the tools of the ExPaSy molecular biology server at the Swiss Institute of Bioinformatics.
Northern blotting.
Hybridizations were performed sequentially with three random-primed 32P-labeled probes corresponding to the KpnI/EcoRI (bp 402 to 2112) and EcoRI/KpnI (bp 1 to 405) fragments of the full-length human cDNA of SCHIP-1 and to β-actin, using human multiple-tissue northern blotting (Clontech) where 2 μg of poly(A)+ mRNAs from heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas were represented. The filter was washed to a final stringency of 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 65°C and then exposed to X-ray film.
In vitro binding assays.
GST fusion proteins were expressed in E. coli and purified according to recommendations of the manufacturer of pGEX vectors (Pharmacia Biotech). [35S]methionine-labeled proteins were prepared by in vitro transcription-translation using the TNT coupled rabbit reticulocyte lysate system as recommended by the supplier (Promega). Four microliters of translated lysates was incubated with 5 μg of either GST fusion protein or GST, bound to glutathione-agarose beads, in 350 μl of TKT40 or TKT150 buffer (40 or 150 mM KCl, 50 mM Tris [pH 7.4], 0.05% Tween 20). After 2 h of incubation at 4°C, the beads were washed twice with the appropriate TKT incubation buffer, and then bound proteins were eluted by competition with an excess of glutathione (10 mM in TKT buffer) and solubilized in sample buffer. The supernatant was subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and 35S-labeled polypeptides were detected by autoradiography.
Antibodies.
The rabbit affinity-purified polyclonal antibody A19, raised against a peptide corresponding to amino acids 2 to 21 of schwannomin, was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). The polyclonal antibody C14, raised against amino acids 469 to 595 of schwannomin, was obtained by injecting rabbits with a GST fusion protein produced in E. coli. HA peptide tags were detected using mouse monoclonal antibody (MAb) 12CA5. MAb P5D4, raised against the 11-amino-acid C terminus of VSV G protein, was purchased from Boehringer Mannheim.
For preparation of anti-SCHIP-1 antibodies, GST fusion proteins with SCHIP-1 were produced in E. coli, purified on glutathione-agarose beads according to the manufacturer's recommendations, and injected into either chickens or rabbits. Antibodies 959 and 964 were obtained by injecting chickens with a purified GST fusion protein with amino acids 306 to 487 of human SCHIP-1 [GST–SCHIP-1(306-487)]. Antibodies 17014 and 17141 were generated by injecting rabbits with a purified GST fusion protein containing amino acids 112 to 305 of human SCHIP-1 [GST–SCHIP-1(112-305)]. These four antibodies were tested for the ability to specifically recognize an HA-tagged human SCHIP-1 protein overexpressed in HeLa cells in Western blots, immunoprecipitations, and indirect immunofluorescence.
Cell culture and transient expression in HeLa cells.
ST88.14 is a schwannoma cell line established from a malignant peripheral nerve sheath tumor from an NF1 patient (8); it was provided by D. Lowy. SF1335 is a cell line established from a benign meningioma (39); it was kindly provided by S. Rempel. BAN and DUN cell lines were established by M.-C. Jaurand from specimens obtained from two patients with confirmed malignant mesothelioma (9). HeLa, SF1335, BAN, and DUN cells were grown in Dulbecco's minimum essential medium (DMEM; Gibco) supplemented with 10% fetal calf serum and antibiotics, at 37°C, under a humidified 5% CO2 atmosphere. The ST88.14 cell line was maintained in DMEM supplemented with 15% fetal calf serum and antibiotics.
HeLa cells, seeded 24 h before transfection (106/10-cm-diameter dish, 8 × 104/3.5-cm-diameter dish), were transfected with plasmids (either pCB6 or pCB6-HA-SCHIP-1 plus either schwannomin or schwannomin variant expression vector pCB6; 8 μg of each plasmid/10-cm-diameter dish, 2.5 μg of each plasmid/3.5-cm-diameter dish), using the calcium phosphate method previously described (10). After transfection, cells were either grown for 24 h and then lysed with the appropriate lysis buffer for further analysis or processed for indirect immunocytochemistry.
Immunoblotting.
Cells, seeded 24 h before (106/10-cm-diameter dish), were washed twice with phosphate-buffered saline (PBS) and harvested by scraping on ice in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris [pH 8], 1% NP-40, 0.1% deoxycholate 0.1% SDS) in the presence of protease inhibitors (Complete; Boehringer Mannheim). After centrifugation at 7,000 rpm for 10 min at 4°C, proteins in the supernatant (50 μg) were denatured in sample buffer, fractionated by SDS-PAGE (10% gel), electrotransferred onto nitrocellulose membrane (Bio-Rad), and incubated with chicken polyclonal antibodies 959 and 964 at 1:500 dilution or rabbit polyclonal antibodies 17014 and 17141 at 1:1,000 dilution. Results were visualized by chemiluminescence with peroxidase-conjugated AffiniPure rabbit anti-chicken immunoglobulin Y* (IgY*; IgG [heavy plus light chain]; Jackson Immunoresearch Laboratories) or a horseradish peroxidase-conjugated sheep anti-rabbit whole antibody (Amersham), using the chemiluminescence Western blotting system (POD; Boehringer Mannheim) according to the manufacturer's protocol.
Immunoprecipitations and coimmunoprecipitations.
For immunoprecipitations, cells were lysed in RIPA buffer in the presence of protease inhibitors (Complete). Extracts were precleared on protein A-Sepharose beads (Sigma) and incubated for 3 h at 4°C in the presence of either rabbit anti-SCHIP-1 antibodies or chicken anti-SCHIP-1 antibodies supplemented with 5 μl of AffiniPure rabbit anti-chicken IgY* (Jackson Immunoresearch Laboratories), previously bound to protein A-Sepharose beads. After washing with lysis buffer, proteins bound to beads were denatured in sample buffer, subjected to SDS-PAGE, electrotransferred, and submitted to immunoblotting using adequate rabbit or chicken anti-SCHIP-1 antibodies as described above. For coimmunoprecipitations, transfected cells were harvested in buffer B (150 mM NaCl, 50 mM Tris [pH 7.5], 1% Tween 20, 0.05% deoxycholate, 0.1% SDS buffer) in the presence of protease inhibitors (Complete). Extracts were precleared on protein G-Sepharose beads (Sigma). Twenty-microliter aliquots of these supernatants were retained for protein expression analysis by immunoblotting with appropriate antibodies. The remaining supernatant was incubated for 3 h at 4°C with the anti-HA MAb 12CA5 previously bound to protein G-Sepharose beads. After washing with buffer A (150 mM NaCl, 20 mM Tris [pH 8], 0.5% Triton X-100), proteins bound to beads were denatured in sample buffer, subjected to SDS-PAGE, electrotransferred, and submitted to two rounds of immunoblotting using antischwannomin (anti-SCH) polyclonal antibody A19 at 1:400 dilution and then anti-SCHIP-1 polyclonal antibody 17141 at 1:500 dilution as described above.
Indirect immunocytochemistry.
After being washed three times with PBS, transfected cells spread on coverslips were fixed for 20 min in 3% paraformaldehyde and permeabilized with 0.02% Triton X-100 for 5 min. Fluorescein isothiocyanate (FITC)-phalloidin, primary antibodies (anti-HA MAb 12CA5 at 1:1,000 dilution, anti-SCH polyclonal antibody C14 at 1:400 dilution), and appropriate tetramethyl rhodamine isothiocyanate-conjugated anti-mouse or FITC-conjugated anti-rabbit secondary antibodies (Amersham) were applied for 30 min each. All the incubations were done in PBS supplemented with 0.1 mM CaCl2 and 0.1 mM MgCl2. Coverslips were mounted in Vectashield medium (Vector Laboratories) in the presence of DAPI (4′,6-diamidino-2-phenylindole) (Sigma). Cells were examined with a Leica epifluorescence microscope or with a confocal Leica microscope.
Nucleotide sequences accession numbers.
The cDNA sequences reported in this paper have been deposited into GenBank under the following accession numbers: human SCHIP-1, AF145713; human SCHIP-1-Δ(22-253), AF145714; human SCHIP-1-Δ(241-253), AF145715; and mouse SCHIP-1, AF145716.
RESULTS
Identification of schwannomin-binding proteins SCHIP-1 and SCHIP-2.
To identify molecular partners of human schwannomin, we performed a yeast two-hybrid screen using the conserved N-terminal ERM domain (amino acids 1 to 314) as bait and an oligo(dT)-primed mouse fetal brain cDNA library. This screen resulted in the isolation of two yeast clones scoring positive for interaction with N-terminal as well as with full-length schwannomin but not with unrelated proteins (lamin and snf4). The plasmids harbored by these clones contained two nonhomologous cDNAs of 1,726 and 627 bp, each with an open reading frame contiguous with the GAL4 activation domain. The putative proteins encoded by these cDNAs have not been previously characterized and were designated SCHIP-1 and SCHIP-2, respectively, for Schwannomin-interacting proteins 1 and 2. In the text that follows, we focus on SCHIP-1.
Isolation of a full-length SCHIP-1 cDNA and nucleotide sequence analysis.
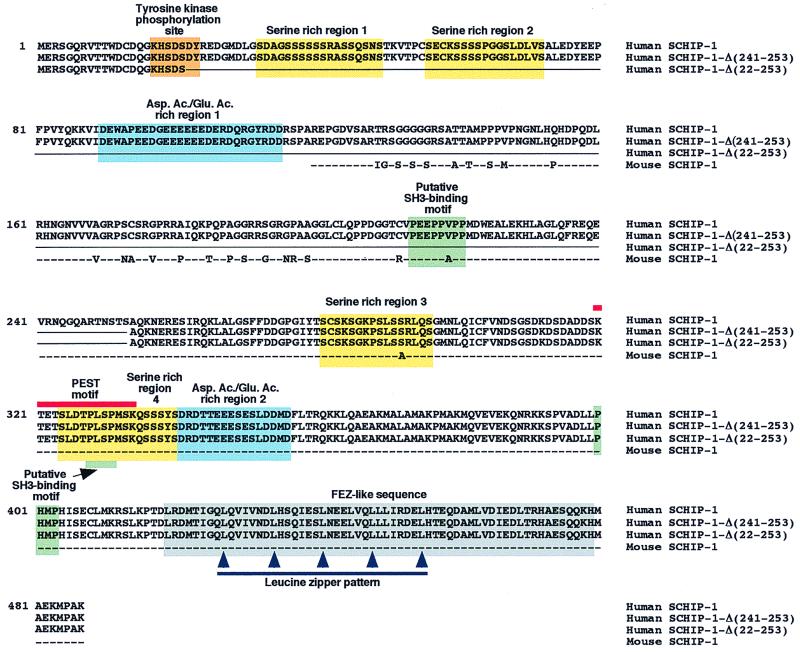
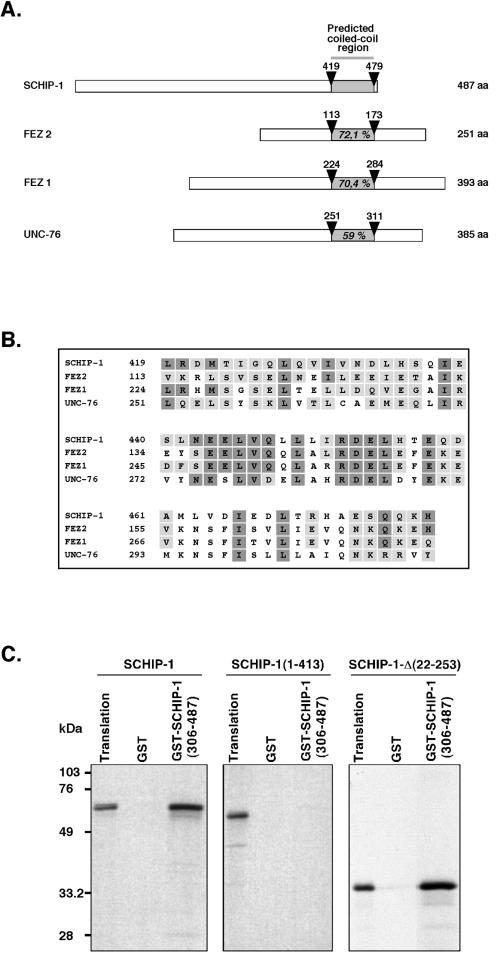
A full-length SCHIP-1 cDNA was isolated from a λgt10 human fetal brain library. This 2,112-bp cDNA encoded a putative protein of 487 amino acids (Fig. 1, line 1), presenting more than 99% conservation in amino acids with its mouse counterpart (Fig. 1, line 4). The amino acid composition of human SCHIP-1 predicts a highly acidic protein with a predicted isoelectric point at 4.98 and a theoretical molecular mass of 53.5 kDa. A computer search for conserved protein motifs revealed the presence of a leucine zipper coiled-coil domain located between amino acids 419 and 479. Within this domain, SCHIP-1 is homologous to two human proteins, FEZ1 and FEZ2 (more than 70% homology), and the related Caenorhabditis elegans gene product UNC-76 (59% homology), involved in axonal outgrowth and fasciculation (Fig. 2A and B) (6). Outside of the coiled-coil domain, SCHIP-1 and the FEZ proteins showed no obvious homology. Further sequence analysis of SCHIP-1 revealed an organization in two serine-rich regions followed by an acidic region. This motif is repeated twice (Fig. 1). The SCHIP-1 amino acid sequence also contains a PEST domain between residues 320 and 334 (PEST-SEARCH [40]), suggesting that the protein could have a short half-life. Three putative SH3-binding motifs and a potential sequence for tyrosine kinase phosphorylation were also detected in the protein sequence (Fig. 1).
FIG. 1.
Protein sequence and putative domain structure of SCHIP-1. Line 1, amino acid sequence of human SCHIP-1, deduced from the nucleotide sequence of a SCHIP-1 cDNA, isolated from a λgt10 human fetal brain library. Lines 2 and 3, amino acid sequences of two putative isoforms of human SCHIP-1, named SCHIP-1-Δ(241-253) and SCHIP-1-Δ(22-253). The corresponding cDNAs were identified by reverse transcriptase PCR amplification experiments on human brain poly(A)+ RNA. These cDNAs may correspond to in-frame alternative splicing events. Amino acids absent in SCHIP-1-Δ(241-253) and SCHIP-1-Δ(22-253) are replaced by a continuous line. Line 4, amino acid sequence encoded by the partial mouse SCHIP-1 cDNA isolated from the two-hybrid screen. Dashes mark identity between human and mouse sequences; nonconserved amino acids between human and mouse proteins are directly indicated. Yellow and blue boxes indicate the four serine-rich regions and the two aspartic acid/glutamic acid-rich regions of the human SCHIP-1 protein, respectively. Green boxes show the localization of three putative SH3-binding domains conserved between the mouse and human SCHIP-1 proteins, and the orange box shows the localization of a putative tyrosine kinase phosphorylation site in human SCHIP-1. The grey box indicates the region of homology between the SCHIP-1 and FEZ proteins, predicted to adopt a coiled-coil conformation. The red line defines a putative PEST motif conserved in mouse and human SCHIP-1 proteins, and the blue line indicates the leucine zipper pattern in the putative coiled-coil region.
FIG. 2.
SCHIP-1 can homodimerize through its coiled-coil domain homologous to the FEZ and UNC-76 proteins. (A) Schematic alignment of human SCHIP-1 with human FEZ1 and FEZ2 and C. elegans UNC-76 proteins. Open boxes represent full-length SCHIP-1, FEZ1, and UNC-76 proteins and the 251 C-terminal amino acids (aa) of the FEZ2 protein. The grey color highlights the localization of the coiled-coil domain in each protein. Arrowheads indicate amino acids delimiting coiled-coil domains. Percentages of homology with SCHIP-1 in the coiled-coil domains are indicated. (B) Amino acid alignment of human SCHIP-1 with human FEZ1 and FEZ2 and C. elegans UNC-76 proteins within the predicted coiled-coil domain. Identical amino acids are presented in dark boxes; conserved residues are displayed in grey boxes. (C) The predicted coiled-coil domain is required for SCHIP-1 homodimerization. SCHIP-1 and SCHIP-1 deletion mutants were expressed in vitro in the presence of [35S]methionine and incubated in TKT150 buffer with either GST or GST–SCHIP-1(306-487) bound to glutathione-agarose beads. After washing, retained SCHIP-1 and SCHIP-1 variant proteins were eluted, resolved by SDS-PAGE, and visualized by autoradiography. Aliquots of the labeled proteins corresponding to 1/40 of the input were loaded on the same gel (Translation). The C-terminal region of SCHIP-1 including the coiled-coil domain [GST–SCHIP-1(306-487)] interacts with full-length SCHIP-1 or SCHIP-1-Δ(22-253) but not with a truncated SCHIP-1 missing the predicted coiled-coil domain [SCHIP-1(1-413)].
Expression of SCHIP-1 mRNA and protein.
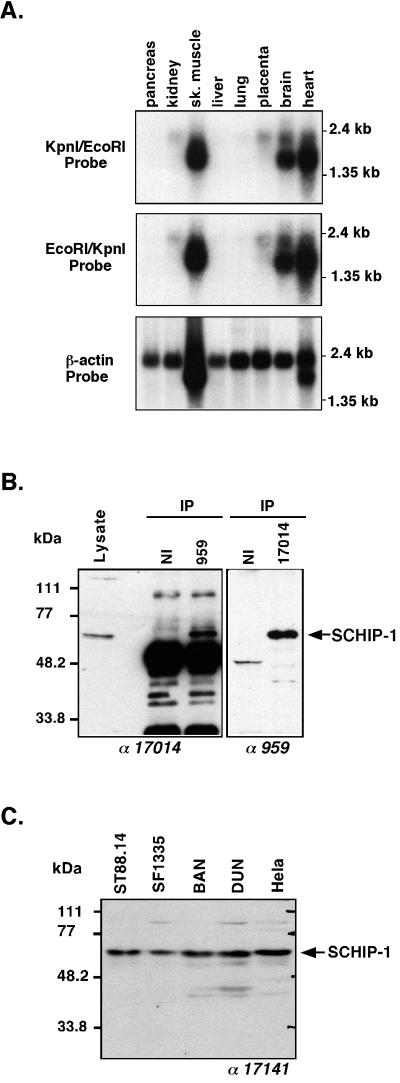
Upon screening of an expressed sequence tag data bank with the SCHIP-1 nucleotide sequence, 65% of the hits were isolated from brain tissues. Other organs expressing SCHIP-1 expressed sequence tags included skeletal muscles, heart, uterus, liver, and lung. To further investigate the tissue distribution of SCHIP-1, a human multiple-tissue RNA blot was probed sequentially with fragments spanning either nucleotides 1 to 405 (EcoRI/KpnI probe) or nucleotides 402 to 2112 (KpnI/EcoRI probe) of the human SCHIP-1 cDNA. Both probes detected multiple transcripts with sizes ranging from 1.5 to 2.1 kb, a size range consistent with the length of the isolated cDNA. Strongest expression was detected in brain, skeletal muscles, and heart (Fig. 3A). Long exposure of the blots showed that low levels of SCHIP-1 message also could be detected in pancreas, kidney, liver, lung, and placenta. Taken together, these data indicate that SCHIP-1 mRNA is present mainly in the brain, heart, and skeletal muscles but that other tissues also express this gene at low levels.
FIG. 3.
Expression pattern of SCHIP-1 mRNAs and proteins. (A) Northern blot analysis of SCHIP-1 expression. Human mRNAs from the indicated organs were analyzed by Northern blotting using either SCHIP-1 cDNA bp 1 to 405 (EcoRI/KpnI fragment), SCHIP-1 cDNA bp 402 to 2112 (KpnI/EcoRI fragment), or β-actin cDNA as the probe. Strong expression of SCHIP-1 is detected in skeletal (sk.) muscles, brain, and heart. (B) Immunoprecipitation of endogenous SCHIP-1 in ST88.14 cells. Endogenous SCHIP-1 from the schwannoma cell line ST88.14 was immunoprecipitated (IP) using either a chicken polyclonal antibody detecting a C-terminal region of SCHIP-1 (lane 959) or a polyclonal rabbit antibody detecting a central region of SCHIP-1 (lane 17014). Precipitates were resolved by SDS-PAGE on a 10% gel and detected with either rabbit antibody 17014 (left) or chicken antibody 959 (right). Twenty micrograms of crude protein extract from the ST88.14 cell line (Lysate) was loaded on the same gel. Antibodies used for Western blotting are indicated below the gels. NI, preimmune IgY or rabbit serum. The two independent antibodies 959 and 17014 both immunoprecipitate and detect in Western blots a protein migrating with an apparent molecular mass of 65 kDa. (C) SCHIP-1 expression in five human cell lines. Proteins (50 μg) from each cell line were resolved by SDS-PAGE on a 10% gel and detected with the rabbit polyclonal antibody 17141. Expression of the 65-kDa SCHIP-1 protein is detected in each cell line.
To visualize the endogenous SCHIP-1 proteins in cells, we produced two sets of polyclonal antibodies raised against two nonoverlapping regions of SCHIP-1, a central region spanning from amino acids 112 to 305 (antibodies 17014 and 17141) and a complete C-terminal region from amino acids 306 to 487 (antibodies 959 and 964). In Western blots on extracts from the human schwannoma cell line ST88.14, the four antibodies detected a species migrating as a 65-kDa protein (Fig. 3B, left, [Lysate] for antibody 17014; other data not shown). To demonstrate that this species corresponded to endogenous SCHIP-1 protein, we performed immunoprecipitation using antibody 959 followed by Western blotting with antibody 17014. Detection of the 65-kDa species indicated that these two independent SCHIP-1 antibodies (959 and 17014) recognize the same protein (Fig. 3B, left). A reciprocal experiment using the 17014 antibody for immunoprecipitation and the 959 antibody for Western blot analysis led to detection of the same 65-kDa species (Fig. 3B, right). The apparent molecular mass (65 kDa) of the SCHIP-1 protein detected was higher than the theoretical molecular mass (53.5 kDa). However, expression of SCHIP-1 in vitro yielded a protein comigrating with endogenous cellular SCHIP-1 (Fig. 2C). The slow migration in SDS-PAGE may be due to specific physical properties of the protein. The two sets of antibodies were then used in Western blots to study the expression of SCHIP-1 in several cell lines, including HeLa cells and cell lines established from tumor types in which the NF2 gene is frequently inactivated. These cell lines included two human mesothelioma cell lines (BAN and DUN), a human meningioma cell line (SF1335), and the human schwannoma cell line ST88.14 as a control. Expression of the 65-kDa SCHIP-1 protein was detected in each cell line with both sets of antibodies (Fig. 3C for the 17141 antibody; data not shown for the other antibodies).
Homodimerization properties of SCHIP-1.
Coiled-coil domains are usually associated with homo- or heteromeric protein-protein interactions. We therefore tested the ability of SCHIP-1 to self-associate. For that purpose, the predicted coiled-coil region (amino acids 306 to 487) of SCHIP-1 was expressed in E. coli as a GST fusion protein [GST–SCHIP-1(306-487)]. This fusion protein, when bound to glutathione-agarose beads, was found to associate specifically with in vitro-translated [35S]methionine-labeled full-length SCHIP-1 or with an in vitro-translated SCHIP-1 construct corresponding to a putative SCHIP-1 isoform lacking amino acids 22 to 253 [SCHIP-1-Δ(22-253) (Fig. 1 and 2C)] but not with an in vitro-translated SCHIP-1 construct missing the predicted coiled-coil region [SCHIP-1(1-413) (Fig. 2C)]. These results demonstrated that SCHIP-1 can form homodimers via its predicted coiled-coil region.
SCHIP-1 associates with schwannomin in vitro.
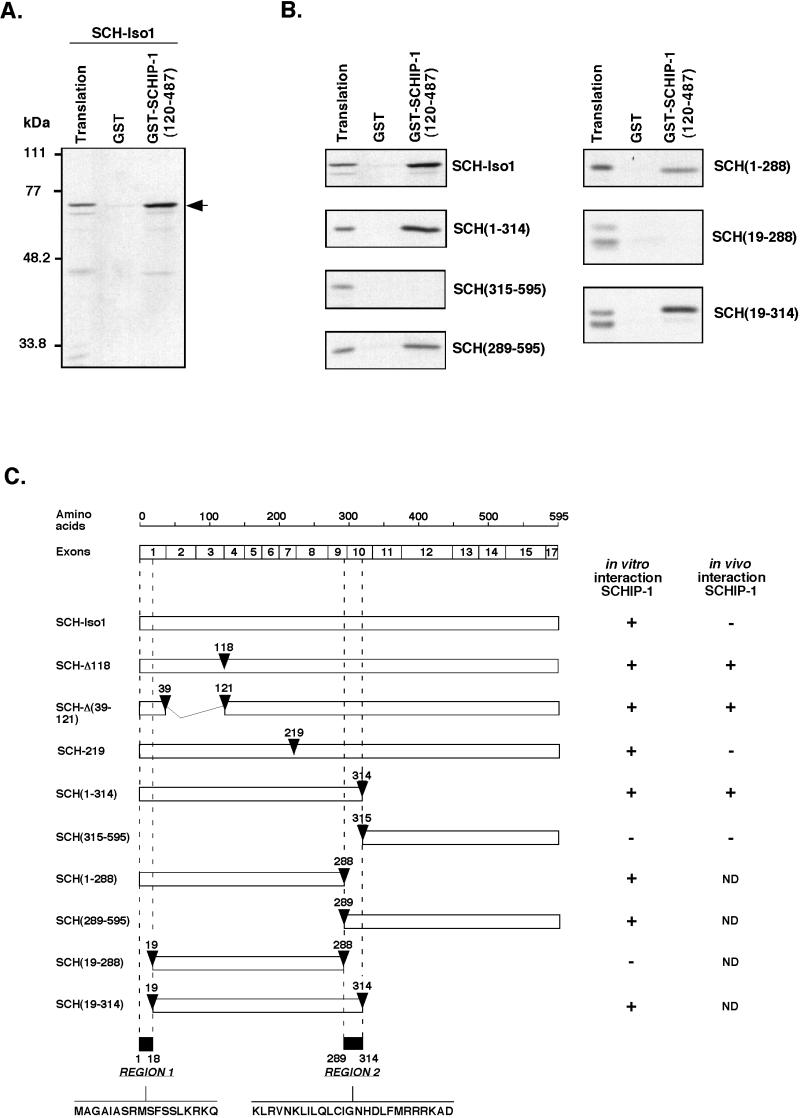
To confirm and extend the observation of schwannomin–SCHIP-1 interaction in the yeast two-hybrid system, we tested the interaction between the two proteins by in vitro GST pull-down experiments. The partial SCHIP-1 cDNA isolated in the initial two-hybrid screen was expressed as a GST fusion protein [GST–SCHIP-1(120-487)]. As shown in Fig. 4A, this fusion protein, when bound to glutathione-agarose beads, was able to associate specifically with in vitro-translated [35S]methionine-labeled full-length SCH-Iso1. The GST–SCHIP-1(120-487) protein also interacted with an in vitro translated schwannomin construct expressing only the N-terminal part of schwannomin from amino acids 1 to 314 [SCH(1-314) (Fig. 4B and C)] but not with an isoform 1 construct missing this N-terminal region [SCH(315-595) (Fig. 4B and C)].
FIG. 4.
SCHIP-1 interacts with two distinct regions of schwannomin in vitro. (A and B) In vitro interaction of schwannomin with SCHIP-1. Either GST or GST–SCHIP-1(120-487) was bound to glutathione-agarose beads and incubated in TKT40 buffer with [35S]methionine-labeled SCH-Iso1 or deletion mutants SCH(1-314), SCH(315-595), SCH(289-595), SCH(1-288), SCH(19-288), and SCH(19-314) as indicated. After washing, retained schwannomin and schwannomin variant proteins were eluted, resolved by SDS-PAGE, and visualized by autoradiography. Aliquots of the labeled proteins corresponding to 1/40 of the input were loaded on the same gel (Translation). SCHIP-1 interacts with SCH-Iso1 in vitro (A). This interaction requires two regions of schwannomin spanning amino acids 1 to 18 and 289 to 314 (B). (C) Map of schwannomin constructs and summary of the properties of the corresponding proteins. (Left) Map of schwannomin constructs. Open boxes indicate the regions of schwannomin that are encoded by the different constructs. Positions of the truncations or amino acids substitutions and deletions are shown by arrowheads. The two regions found to be implicated in the interaction with SCHIP-1 are indicated at the bottom. Relative positions of the NF2 exons and schwannomin amino acid residues are illustrated on the top. (Right) Summary of properties of the various proteins. In the two columns are indicated the ability of the various proteins to interact in GST pull-down experiments with SCHIP-1 (column 1) and the ability of the various proteins to coimmunoprecipitate with SCHIP-1 when the proteins are overexpressed in HeLa cells (column 2). +, interaction in vitro or coimmunoprecipitation detected; −, interaction in vitro or coimmunoprecipitation not detected; ND, ability to coimmunoprecipitate not determined.
To identify more precisely the sites of interaction on schwannomin, we tested a schwannomin-derived construct spanning from amino acids 289 to 595 [SCH(289-595) (Fig. 4C)]. This construct was only 25 amino acids longer than SCH(315-595) and was able to interact with SCHIP-1 (Fig. 4B). This experiment showed that the region of schwannomin located between residues 289 and 314 is required for interaction with SCHIP-1. To determine if other regions of schwannomin were involved in the interaction, we tested association between GST–SCHIP-1(120-487) and a N-terminal schwannomin construct missing residues 289 to 314 [SCH(1-288) (Fig. 4C)]. This construct was still able to associate with SCHIP-1 in GST pull-down experiments, indicating the presence of a second SCHIP-1 interaction domain on schwannomin (Fig. 4B). Further deletion of the first 18 amino acids of schwannomin [construct SCH(19-288) (Fig. 4C)] was required to abolish SCHIP-1 interaction (Fig. 4B). This observation allowed us to map the additional SCHIP-1-binding site to the complete N terminus of schwannomin. The two domains of schwannomin implicated in interaction with SCHIP-1 were called region 1 and region 2 (Fig. 4C). In agreement with the mapping of the interaction to these two regions, we found that two naturally occurring mutants of schwannomin deleted of either Phe118 (SCH-Δ118 [Fig. 4C]) or residues encoded by exons 2 and 3 [SCH-Δ(39-121) (Fig. 4C)], retained the ability to interact with SCHIP-1 (data not shown).
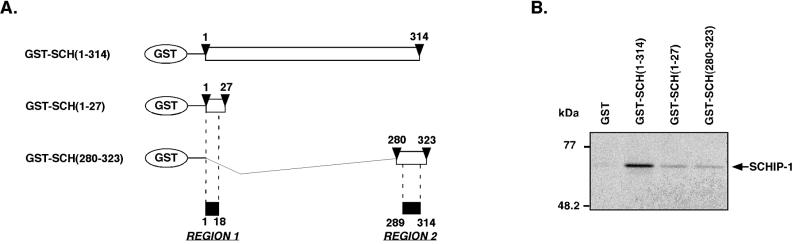
To demonstrate that region 1 and region 2 of schwannomin that we had identified were sufficient to mediate binding to SCHIP-1, we expressed three GST fusion proteins containing either the entire N-terminal region of schwannomin [GST–SCH(1-314)] or short regions of schwannomin centered on region 1 and region 2 [GST–SCH(1-27) and GST-SCH(280-323), respectively (Fig. 5A)]. These proteins were used in pull-down experiments with in vitro-translated [35S]methionine-labeled SCHIP-1. As shown in Fig. 5B, each putative interaction region of schwannomin was able to associate independently with SCHIP-1. However, extracting these regions from the context of the protein appeared to decrease their affinity for SCHIP-1.
FIG. 5.
Schwannomin region 1 and region 2 are sufficient for interaction with SCHIP-1 in vitro. (A) Map of GST-schwannomin constructs. Open boxes indicate the regions of schwannomin that are encoded by the different constructs. Amino acids delimiting these regions are indicated by arrowheads. (B) In vitro interaction of schwannomin region 1 and region 2 with SCHIP-1. Either GST, GST–SCH(1-314), GST–SCH(1-27), or GST–SCH(280-323) was bound to glutathione-agarose beads and incubated in TKT150 buffer with [35S]methionine-labeled SCHIP-1 as indicated. After washing, retained SCHIP-1 protein was eluted, resolved by SDS-PAGE, and visualized by autoradiography. Region 1 [GST–SCH(1-27)] and region 2 [GST-SCH(280-323)] are able to associate independently with SCHIP-1 in vitro.
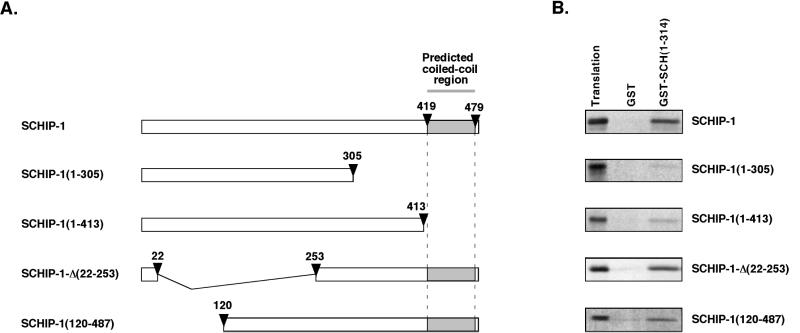
To identify the region of SCHIP-1 required for the interaction with schwannomin, we performed a third set of GST pull-down experiments using GST-SCH(1-314) and several in vitro-translated [35S]methionine-labeled SCHIP-1 mutants (Fig. 6A). Deletion of the 182 most C-terminal amino acids of SCHIP-1 completely abolished binding to schwannomin [SCHIP-1(1-305) (Fig. 6B)]. Interaction was also strongly diminished when we used a truncated SCHIP-1 protein missing the 74 C-terminal amino acids [SCHIP-1(1-413) (Fig. 6B)]. This observation indicated that the predicted coiled-coil region of SCHIP-1 is required for efficient interaction between SCHIP-1 and schwannomin. In agreement with this, two proteins lacking part of the N-terminal domain of SCHIP-1, SCHIP-1-Δ(22-253) and SCHIP-1(120-487) (Fig. 6A), were shown to maintain an interaction with GST–SCH(1-314) (Fig. 6B).
FIG. 6.
The coiled-coil region of SCHIP-1 is required for efficient interaction with schwannomin in vitro. (A) Map of SCHIP-1 constructs. Open boxes indicate the regions of SCHIP-1 encoded by the different constructs. The grey color highlights the localization of the coiled-coil domain. Arrowheads indicate the amino acids delimiting the coiled-coil domain and the positions of truncations and deletions. (B) In vitro interaction of SCHIP-1 with schwannomin. Either GST or GST–SCH(1-314) was bound to glutathione-agarose beads and incubated in TKT150 buffer with [35S]methionine-labeled full-length SCHIP-1, SCHIP-1(1-413), SCHIP-1(1-305), SCHIP-1-Δ(22-253), or SCHIP-1(120-487) as indicated. After washing, retained SCHIP-1 and SCHIP-1 variants were eluted and visualized by autoradiography. Aliquots of the labeled proteins corresponding to 1/40 of the input were loaded on the same gel (Translation). Schwannomin interacts in vitro with full-length SCHIP-1 or SCHIP-1 proteins deleted in the N-terminal domain [SCHIP-1-Δ(22-253) and SCHIP-1(120-487)] but not or poorly with truncated SCHIP-1 proteins missing the coiled-coil region [SCHIP-1(1-413) and SCHIP-1(1-305)].
In vivo interaction with SCHIP-1 is correlated with altered schwannomin protein conformation.
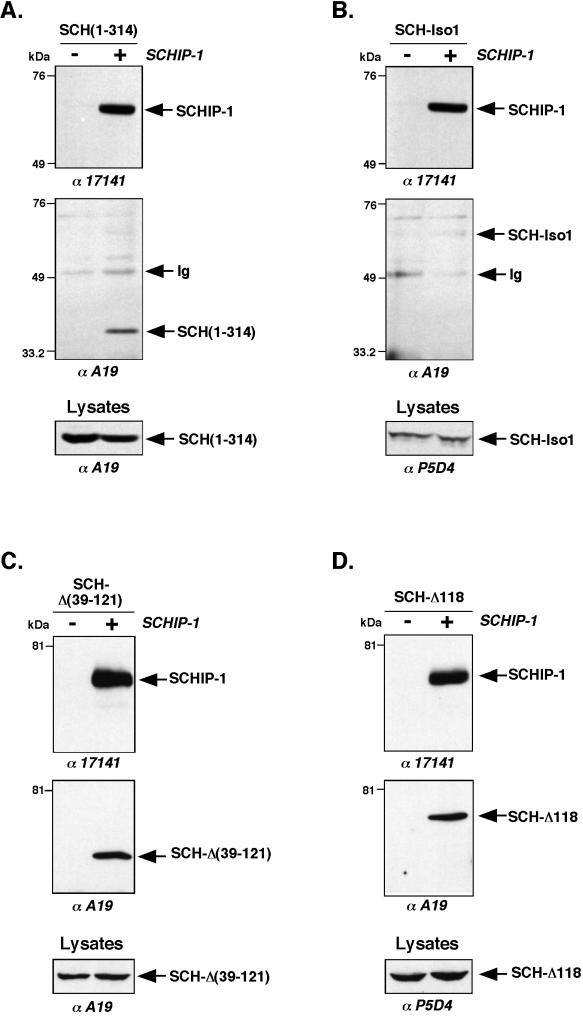
To determine if the SCHIP-1 interaction domains identified in vitro were also functional in vivo, we cotransfected HeLa cells with a vector expressing HA-tagged SCHIP-1 together with constructs expressing either VSV-tagged SCH-Iso1 or the SCH(1-314) deletion mutant. Coimmunoprecipitation experiments using HA-specific MAb 12CA5 demonstrated the ability of SCH(1-314) to associate with SCHIP-1 under these conditions (Fig. 7A). However, no clear interaction was detected between SCHIP-1 and SCH-Iso1 (Fig. 7B), even when buffers with reduced salt and detergent concentrations were used. The reverse experiment using anti-SCH antibodies for immunoprecipitation confirmed the interaction between SCHIP-1 and SCH(1-314) and the apparent lack of stable interaction between SCHIP-1 and SCH-Iso1 (data not shown).
FIG. 7.
In vivo interaction of schwannomin and SCHIP-1. HeLa cells were cotransfected with either pCB6-HA-SCHIP-1 expression vector (+) or empty pCB6 vector (−) and vector expressing schwannomin variants SCH(1-314) (A), SCH-Iso1 (B), SCH-Δ(39-121) (C), and SCH-Δ118 (D). Twenty-four hours after transfection, proteins extracts were prepared as described in Materials and Methods, and coimmunoprecipitations were performed with the anti-HA MAb 12CA5. Precipitates were resolved by SDS-PAGE on an 8% gel, electroblotted, and submitted to two rounds of immunoblotting, first with the anti-SCH polyclonal antibody A19 and then with the anti-SCHIP-1 polyclonal antibody 17141. Twenty microliters of crude protein extract (Lysates) was also subjected to immunoblotting with either the anti-SCH polyclonal antibody A19 or the anti-VSV monoclonal antibody MAb P5D4, to verify expression of schwannomin proteins. Antibodies used for Western blotting are indicated below the gels. SCHIP-1 is able to associate in vivo with the schwannomin variant SCH(1-314) (A) and the two naturally occurring mutants SCH-Δ(39-121) and SCH-Δ118 (C and D) but not with SCH-Iso1 (B).
The absence of conspicuous in vivo interaction between SCHIP-1 and SCH-Iso1 prompted us to test naturally occurring schwannomin variants in the coimmunoprecipitation assay. We first used SCH-Δ(39-121) (Fig. 4C). This protein lacks residues encoded by exons 2 and 3. It is a naturally occurring rare isoform of schwannomin resulting from alternative splicing in normal cells (1, 37, 38) and also a mutant form of schwannomin found in some NF2 tumors. This mutant is believed to have a modified conformation because, in contrast to isoform 1, it is able to interact with polymerized microtubules (61). As described above, SCH-Δ(39-121) was shown to associate with SCHIP-1 in vitro (Fig. 4C). After cotransfection with the HA-tagged SCHIP-1 expression vector, SCH-Δ(39-121) protein was found to coimmunoprecipitate with SCHIP-1 (Fig. 7C). The reverse experiment using anti-SCH antibodies to coimmunoprecipitate SCHIP-1 confirmed the interaction between the two proteins in vivo (data not shown). We next tested another naturally occurring mutant of schwannomin deleted of Phe118 and known as SCH-Δ118 (Fig. 4C). Like SCH-Δ(39-121), SCH-Δ118 was found to interact with SCHIP-1 in vitro (Fig. 4C). Besides, it was shown to associate with polymerized microtubules more efficiently than SCH-Iso1 (data not shown), suggesting that the Phe118 mutation may induce conformational changes similar to those suspected for SCH-Δ(39-121). SCH-Δ118 was found to coimmunoprecipitate with SCHIP-1 when we used an anti-HA antibody (Fig. 7D) and when we used an anti-SCH antibody (data not shown). Another naturally occurring schwannomin mutant carrying a single amino acid substitution at residue 219 (Val to Met) was also tested (SCH-219 [Fig. 4C]). Unlike SCH-Δ(39-121) and SCH-Δ118, this mutant did not interact with microtubules (data not shown), suggesting that the amino acid mutation V219M does not induce conformational changes within the protein. In our assays, SCH-219 was able to interact with SCHIP-1 in vitro but not in vivo (Fig. 4C and data not shown). Two other mutants carrying single amino acid substitutions at residues 220 (Asn to Tyr) and 360 (Leu to Pro) (10) behaved similarly in our assays (data not shown), suggesting that the missense mutations N220Y and L360P also leave the conformation of schwannomin intact.
SCHIP-1 colocalizes partially with schwannomin.
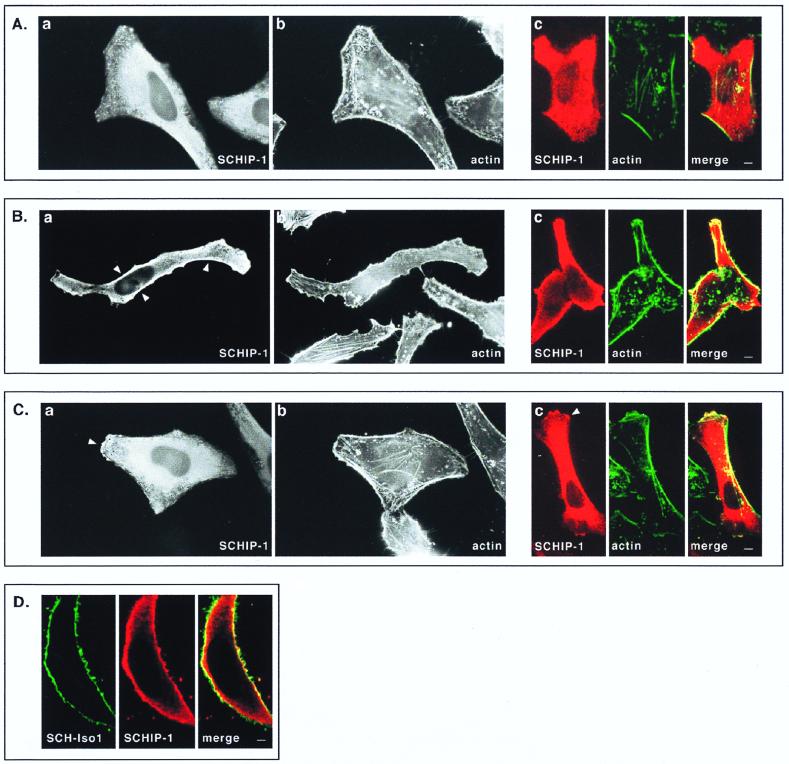
Wild-type schwannomin is primarily concentrated at the cytoplasmic membrane (10–12, 17, 43, 46, 48, 50). To determine whether SCHIP-1 is localized similarly, we transfected HeLa cells with constructs expressing SCHIP-1 either alone or in combination with SCH-Iso1. For cells expressing SCHIP-1 alone, indirect immunofluorescent staining showed that in most cells, SCHIP-1 was distributed throughout the cytoplasm, with higher concentrations in the perinuclear area (Fig. 8A). In some cells, SCHIP-1 was found to be enriched in regions beneath the cytoplasmic membrane (Fig. 8B) or concentrated in actin-rich membrane-associated structures (Fig. 8C). However, actin destabilization experiments with cytochalasin D did not lead to clear redistribution of SCHIP-1, suggesting that SCHIP-1 is not systematically associated with the actin-rich cytoskeleton (data not shown). In addition, we observed that cellular distribution of SCHIP-1 could not be correlated with variable degrees of cell confluency.
FIG. 8.
Colocalization of SCHIP-1 and schwannomin in overexpressing cells. HeLa cells were transfected with expression vector pCB6-HA-SCHIP-1 alone (A to C) or together with an SCH-Iso1 expression vector (D). Twenty-four hours later, cells were processed for indirect immunofluorescence using MAb 12CA5 to detect the HA-tagged SCHIP-1 protein (A to D), FITC-phalloidin to reveal F-actin (A to C), and the C14 rabbit polyclonal antibody to detect schwannomin (D). Coverslips were examined with a Leica epifluorescence microscope (A to C, views a and b) and with a Leica confocal microscope (A to C, views c; D). Red indicates anti-mouse secondary antibody staining (SCHIP-1) (A to D), whereas green represents actin staining (A to C) or anti-rabbit secondary antibody staining (schwannomin) (D). Overlays are shown with yellow indicating colocalization (A to D). (A to C) Representative examples of the three main types of localization that have been observed for SCHIP-1 on a single coverslips. Arrowheads indicate regions where SCHIP-1 localizes below the cytoplasmic membrane (B) or colocalizes with cortical actin (C). (D) Cell in which SCHIP-1 localizes beneath the cytoplasmic membrane and colocalizes partially with SCH-Iso1. Scale bars, 5 μm (A to C) and 2 μm (D).
In cells coexpressing SCH-Iso1 and SCHIP-1, schwannomin did not appear to modify the subcellular localization of SCHIP-1; as when it was transfected alone, SCHIP-1 was found either in the cytoplasm or at the cytoplasmic membrane, depending on the individual cell. In cells where SCHIP-1 was present in regions beneath the cytoplasmic membrane, immunofluorescent staining of schwannomin revealed a partial colocalization of the two proteins (Fig. 8D). This overlapping distribution supports the biological relevance of the SCHIP-1–schwannomin interaction detected in vitro.
DISCUSSION
SCHIP-1 is a novel coiled-coil protein.
Using the amino-terminal half of schwannomin as bait in a yeast two-hybrid screen, we have identified a cDNA encoding a novel human protein that we called SCHIP-1. This protein contains in its C-terminal domain a region predicted to adopt a coiled-coil conformation. It has no overall homology to other known proteins, but the coiled-coil domain shows intriguing sequence similarities with human FEZ proteins and the C. elegans homolog UNC-76. The FEZ1 and UNC-76 proteins are involved in axonal outgrowth and fasciculation (6). The UNC-76 protein may play a structural role in the formation and the maintenance of axonal bundles or alternatively may mediate signal transduction from cell surface molecules to intracellular machineries regulating axonal extension and adhesion. A rat homolog of FEZ1 was also reported to be a cellular substrate of protein kinase C ζ, which has been implicated in a wide range of cellular functions (29). Other rat FEZ1 and FEZ2 homologs, named zyginI and zyginII, were described as binding proteins for synaptotagmins (6). The latter are membrane proteins required for synaptic vesicles exocytosis and are believed to function also in other membrane fusion events in nonneural cells (47). Our in vitro studies showed that SCHIP-1 can self associate via its coiled-coil domain. Further investigations will be necessary to determine if SCHIP-1 can also form heterodimers with FEZ proteins or share with the FEZ proteins some cellular partners interacting via the coiled-coil region.
Restricted SCHIP-1–schwannomin interaction in a cellular context.
GST pull-down experiments demonstrated the ability of all tested schwannomin isoforms or variants to interact physically with SCHIP-1 in vitro. The region of SCHIP-1 required for schwannomin interaction was mapped to a domain overlapping with the coiled-coil structure. In schwannomin, the interaction with SCHIP-1 was mediated by two regions, located in the N-terminal and central parts of the protein (Fig. 4C). In our in vitro experiments, these two regions were able to associate with SCHIP-1 independently of each other. Using transient transfection experiments, we surprisingly found that in a cellular context, interaction between SCHIP-1 and schwannomin was drastically modified. Under these conditions, overexpressed SCH-Iso1 was not found to interact with SCHIP-1. However, interaction was still observed with two naturally occurring schwannomin mutants, SCH-Δ(39-121) and SCH-Δ118.
Schwannomin, like the ERM proteins, is able to form intramolecular associations between domains that are distant on the sequence (15, 19, 24, 32, 33, 52). This protein folding is likely to control the interaction of schwannomin with its molecular partners. A recent study has shown that SCH-Iso1 associates with ezrin in COS-1 and U251 glioma cells. However, full-length SCH-Iso1 is unable to heterodimerize with ezrin in yeast two-hybrid system. To obtain this heterotypic interaction with ezrin in yeast, schwannomin has to be expressed as a truncated protein (18). Similarly, SCH-Iso1 does not interact with polymerized microtubules in vitro but the interaction is recovered with several truncated or deleted schwannomin variants including the SCH-Δ(39-121) (61). This last observation led Xu et al. (61) to propose a model in which the microtubule interaction site would be occluded by protein folding in SCH-Iso1 but exposed in some other naturally occurring mutants or spliced variants. One of the SCHIP-1 interaction sites of schwannomin, located in the central region of the protein (region 2 [Fig. 4C]), overlaps with the above-mentioned microtubule interaction site. Together, these observations lead us to suggest that the interactions of schwannomin with ezrin, polymerized microtubules, and SCHIP-1 are ruled by a common mechanism and that the association between schwannomin and SCHIP-1 could be determined and possibly regulated by schwannomin conformation changes.
The predicted occlusion of the central SCHIP-1 interaction site of schwannomin by intramolecular associations implies that the interaction that we observed in vitro between SCHIP-1 and SCH-Iso1 relies primarily on the second interaction domain of schwannomin located in the most N-terminal part of the protein (region 1 [Fig. 4C]). In a cellular context, this N-terminal interaction site may also be masked, possibly by other molecular partners present in the cytoplasm, thus explaining the lack of interaction between SCH-Iso1 and SCHIP-1 in vivo. That point will be delicate to address since it requires the ability to inactivate the N-terminal site without exposing the central site.
Possible functional significance of interaction between SCHIP-1 and schwannomin.
Schwannomin and SCHIP-1 strongly interact in vitro. Besides, these two proteins have very similar expression patterns, with an enrichment in brain, heart, and skeletal muscles. Finally, the two proteins were found to partially colocalize beneath the cytoplasmic membrane. These observations strongly suggest that SCHIP-1 is involved in schwannomin activity. However, as discussed above, we were unable to detect interaction between SCHIP-1 and SCH-Iso1 in transfected cells, possibly because the schwannomin protein expressed under these conditions assumes a conformation that prevents it from associating with some of its molecular partners. In the case of the ERMs, phosphorylation and acidic phospholipids such as phosphatidylinositol 4,5-bisphosphate are believed to regulate the conformation of the proteins and to promote their interactions with cellular partners (22, 23, 34; for reviews, see references 7 and 33). Endogenous schwannomin may be regulated in a similar way. Several studies have shown that the protein is phosphorylated on both serine and threonine residues (51, 55). Dephosphorylation of schwannomin is associated with loss of cell adhesion, and the ratio of phosphorylated to unphosphorylated schwannomin is regulated by serum growth factor starvation and cell confluency in NIH 3T3 cells (51). Thus, it is possible that cues such as cell-cell contact, growth factor microenvironment, or changes in cell shape could result in modified schwannomin conformation and thereby potentiate its interaction with SCHIP-1.
Alternatively, the association of SCHIP-1 with schwannomin may be restricted to rare natural spliced isoforms of schwannomin. We find that in transfected cells, SCHIP-1 interacts with the naturally occurring spliced variant SCH-Δ(39-121) under conditions where it is apparently not interacting with SCH-Iso1. We have recently observed that overexpression of the SCH-Δ(39-121) mutant in murine Schwann cells promotes the development of schwannomas (16). It is possible that the normal function of the schwannomin protein is the result of an equilibrium between various isoforms and conformational states of the protein. In tumor cells, this equilibrium may be shifted in favor of variant proteins with an altered conformation, resulting in malignancy. Validation of this hypothesis will require evaluation of the relative expression levels of the different schwannomin isoforms in normal tissues and tumors.
ACKNOWLEDGMENTS
We thank R. Hellio for help in performing the confocal microscopy analysis; we thank S. Rempel, D. Lowy, and M.-C. Jaurand for cell lines SF1335, ST88.14, BAN, and DUN. We are grateful to M. Giovannini, B. Deguen, J. Seeler, H. McKhann, and M. Arpin for numerous stimulating discussions and critical reading of the manuscript. We thank M. Yaniv for support during this project.
This work was supported by grants from the European Commission (BMH4-CT95 0090) and the Ligue Française contre le Cancer.
REFERENCES
- 1.Arakawa H, Hayashi N, Nagase H, Ogawa M, Nakamura Y. Alternative splicing of the NF2 gene and its mutation analysis of breast and colorectal cancers. Hum Mol Genet. 1994;3:565–568. doi: 10.1093/hmg/3.4.565. [DOI] [PubMed] [Google Scholar]
- 2.Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105:1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi A B, Hara T, Ramesh V, Gao J, Klein-Szanto A J P, Morin F, Menon A G, Trofatter J A, Gusella J F, Seizinger B R, Kley N. Mutations in transcript isoforms of the neurofibromatosis 2 gene in multiple human tumour types. Nat Genet. 1994;6:185–192. doi: 10.1038/ng0294-185. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi A B, Mitsunaga S-I, Cheng J Q, Klein W M, Jhanwar S C, Seizinger B, Kley N, Klein-Szanto A J P, Testa J R. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc Natl Acad Sci USA. 1995;92:10854–10858. doi: 10.1073/pnas.92.24.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bijlsma E K, Merel P, Bosch D A, Westerveld A, Delattre O, Thomas G, Hulsebos T J M. Analysis of mutations in the SCH gene in schwannomas. Genes Chromosomes Cancer. 1994;11:7–14. doi: 10.1002/gcc.2870110103. [DOI] [PubMed] [Google Scholar]
- 6.Bloom L, Horvitz H R. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc Natl Acad Sci USA. 1997;94:3414–3429. doi: 10.1073/pnas.94.7.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bretscher A. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr Opin Cell Biol. 1999;11:109–116. doi: 10.1016/s0955-0674(99)80013-1. [DOI] [PubMed] [Google Scholar]
- 8.DeClue J E, Papageorge A G, Fletcher J A, Diehl S R, Ratner N, Vass W C, Lowy D R. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- 9.Deguen B, Goutebroze L, Giovannini M, Boisson C, van der Neut R, Jaurand M C, Thomas G. Heterogeneity of mesothelioma cell lines as defined by altered genomic structure and expression of the NF2 gene. Int J Cancer. 1998;77:554–560. doi: 10.1002/(sici)1097-0215(19980812)77:4<554::aid-ijc14>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Deguen B, Merel P, Goutebroze L, Giovannini M, Reggio H, Arpin M, Thomas G. Impaired interaction of naturally occurring mutant NF2 protein with actin-based cytoskeleton and membrane. Hum Mol Genet. 1998;7:217–226. doi: 10.1093/hmg/7.2.217. [DOI] [PubMed] [Google Scholar]
- 11.den Bakker M A, Riegman P H J, Arnold R, Hekman C P, Boersma W, Janssen P J A, van der Kwast T H, Zwathoff E C. The product of the NF2 tumor suppressor gene localizes near the plasma membrane and is highly expressed in muscle cells. Oncogene. 1995;10:757–763. [PubMed] [Google Scholar]
- 12.den Bakker M A, Tascilar M, Riegman P H J, Hekman A C P, Boersma W, Janssen P J A, de Jong T A W, Hendriks W, van der Kwast T H, Zwathoff E C. Neurofibromatosis type 2 protein co-localizes with elements of the cytoskeleton. Am J Pathol. 1995;147:1339–1349. [PMC free article] [PubMed] [Google Scholar]
- 13.Evans D G, Huson S M, Donnai D, Neary W, Blair V, Newton V, Harris R. A clinical study of type 2 neurofibromatosis. Q J Med. 1992;84:603–618. [PubMed] [Google Scholar]
- 14.Franck Z, Gary R, Bretscher A. Moesin, like ezrin, colocalizes with actin in the cortical cytoskeleton in cultured cells, but its expression is more variable. J Cell Sci. 1993;105:219–231. doi: 10.1242/jcs.105.1.219. [DOI] [PubMed] [Google Scholar]
- 15.Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovannini M, Robanus-Maansdag E, Niwa-Kawakita M, van der Valk M, Woodruff J M, Goutebroze L, Merel P, Berns A, Thomas G. Schwann cell hyperplasia and tumors in transgenic mice expressing a naturally occurring mutant NF2 protein. Genes Dev. 1999;13:978–986. doi: 10.1101/gad.13.8.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Agosti C, Xu L, Pinney D, Beauchamp R, Hobbs W, Gusella J, Ramesh V. The merlin tumor suppressor localizes preferentially in membrane ruffles. Oncogene. 1996;13:1239–1247. [PubMed] [Google Scholar]
- 18.Gronholm M, Sainio M, Zhao F, Heiska L, Vaheri A, Carpen O. Homotypic and heterotypic interaction of the neurofibromatosis 2 tumor suppressor protein merlin and the ERM protein ezrin. J Cell Sci. 1999;112:895–904. doi: 10.1242/jcs.112.6.895. [DOI] [PubMed] [Google Scholar]
- 19.Gutmann D H, Geist R T, Xu H-M, Kim J S, Saporito-Irwin S. Defects in neurofibromatosis 2 protein function can arise at multiple levels. Hum Mol Genet. 1998;7:335–345. doi: 10.1093/hmg/7.3.335. [DOI] [PubMed] [Google Scholar]
- 20.Gutmann D H, Giordano M J, Fishback A S, Guha A. Loss of merlin expression in sporadic meningiomas, ependymomas and schwannomas. Neurology. 1997;49:267–270. doi: 10.1212/wnl.49.1.267. [DOI] [PubMed] [Google Scholar]
- 21.Haase V H, Trofatter J A, MacCollin M, Tarttelin E, Gusella J F, Ramesh V. The murine NF2 homologue encodes a highly conserved merlin protein with alternative forms. Hum Mol Genet. 1994;3:407–411. doi: 10.1093/hmg/3.3.407. [DOI] [PubMed] [Google Scholar]
- 22.Heiska L, Alfthan K, Gronholm M, Vilja P, Vaheri A, Carpen O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J Biol Chem. 1998;273:21893–21900. doi: 10.1074/jbc.273.34.21893. [DOI] [PubMed] [Google Scholar]
- 23.Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S, Tsukita S. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Ichimaru E, Pestonjamasp K, Cui X, Nakamura H, Lo G Y, Lin F I, Luna E J, Furthmayr H. Merlin differs from moesin in binding to F-actin and in its intra- and intermolecular interactions. Biochem Biophys Res Commun. 1998;248:548–553. doi: 10.1006/bbrc.1998.9009. [DOI] [PubMed] [Google Scholar]
- 25.Huynh D P, Nechiporuk T, Pulst S-M. Alternative transcripts in the mouse neurofibromatosis type 2 (NF2) gene are conserved and code for schwannomins with distinct C-terminal domains. Hum Mol Genet. 1994;3:1075–1079. doi: 10.1093/hmg/3.7.1075. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- 27.Kimura Y, Koga H, Araki N, Mugita N, Fujita N, Takeshima H, Nishi T, Yamashima T, Saido T C, Yamasaki T, Moritake K, Saya H, Nakao M. The involvement of calpain-dependent proteolysis of the tumor suppressor NF2 (merlin) in schwannomas and meningiomas. Nat Med. 1998;4:915–922. doi: 10.1038/nm0898-915. [DOI] [PubMed] [Google Scholar]
- 28.Koga H, Araki N, Takeshima H, Nishi T, Hirota T, Kimura Y, Nakao M, Saya H. Impairment of cell adhesion by expression of the mutant neurofibromatosis type 2 (NF2) genes which lack exons in the ERM-homology domain. Oncogene. 1998;17:801–810. doi: 10.1038/sj.onc.1202010. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda S, Nakagawa N, Tokunaga C, Tatematsu K, Tanizawa K. Mammalian homologue of the Caenorhabditis elegans UNC-76 protein involved in axonal outgrowth is a protein kinase C zeta-interacting protein. J Cell Biol. 1999;144:403–411. doi: 10.1083/jcb.144.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J H, Sundaram V, Stein D J, Kinney S E, Stacey D W, Golubic M. Reduced expression of schwannomin/merlin in human sporadic meningiomas. Neurosurgery. 1997;40:578–587. doi: 10.1097/00006123-199703000-00031. [DOI] [PubMed] [Google Scholar]
- 31.Lutchman M, Rouleau G A. The neurofibromatosis type 2 gene product, schwannomin, suppresses growth of NIH 3T3 cells. Cancer Res. 1995;55:2270–2274. [PubMed] [Google Scholar]
- 32.Magendantz M, Henry M D, Lander A, Solomon F. Interdomain interactions of radixin in vitro. J Biol Chem. 1995;270:25324–25327. doi: 10.1074/jbc.270.43.25324. [DOI] [PubMed] [Google Scholar]
- 33.Mangeat P, Roy C, Martin M. ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 1999;9:187–192. doi: 10.1016/s0962-8924(99)01544-5. [DOI] [PubMed] [Google Scholar]
- 34.Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merel P, Hoang-Xuan K, Sanson M, Moreau-Aubry A, Bijlsma E K, Lazaro C, Moisan J P, Resche F, Nishisho I, Estivill X, Delattre J Y, Poisson M, Theillet C, Hulsebos T, Delattre O, Thomas G. Predominant occurence of somatic mutations of the NF2 gene in meningiomas and schwannomas. Genes Chromosomes Cancer. 1995;13:211–216. doi: 10.1002/gcc.2870130311. [DOI] [PubMed] [Google Scholar]
- 36.Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, Gusella J, Ramesh V. NHE-RF, a regulatory cofactor for Na+-H+ exchange, is a common interactor for merlin and ERM (MERM) proteins. J Biol Chem. 1998;273:1273–1276. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- 37.Nishi T, Takeshima H, Hamada K, Yoshizato K, Koga H, Sato K, Yamamoto K, Kitamura I, Kochi M, Kuratsu J-I, Saya H, Ushio Y. Neurofibromatosis 2 gene has novel alternative splicings which controls intracellular protein binding. Int J Oncol. 1997;10:1025–1029. doi: 10.3892/ijo.10.5.1025. [DOI] [PubMed] [Google Scholar]
- 38.Pykett M J, Murphy M, Harnish P R, George D L. The neurofibromatosis 2 (NF2) tumor suppressor gene encodes multiple alternatively spliced transcripts. Hum Mol Genet. 1994;3:559–564. doi: 10.1093/hmg/3.4.559. [DOI] [PubMed] [Google Scholar]
- 39.Rempel S A, Schwechheimer K, Davis R L, Cavenee W K, Rosenblum M L. Loss of heterozygosity for loci on chromosome 10 is associated with morphologically malignant meningioma progression. Cancer Res. 1993;53:2386–2392. [PubMed] [Google Scholar]
- 40.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 41.Rouleau G A, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, Pulst S M, Lenoir G, Bijlsma E, Fashold R, Dumanski J, de Jong P, Parry D, Eldrige R, Aurias A, Delattre O, Thomas G. Alteration in a new gene encoding a putative membrane-organizing protein causes neurofibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- 42.Ruttledge M H, Sarrazin J, Rangaratnam S, Phelan C M, Twist E, Merel P, Delattre O, Thomas G, Nordenskjold M, Collins V P, Dumanski J P, Rouleau G A. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat Genet. 1994;6:180–184. doi: 10.1038/ng0294-180. [DOI] [PubMed] [Google Scholar]
- 43.Sainio M, Zhao F, Heiska L, Turunen O, den Bakker M, Zwarthoff E, Lutchman M, Rouleau G A, Jääskeläinen J, Vaheri A, Carpén O. Neurofibromatosis 2 tumor suppressor protein colocalizes with ezrin and CD44 and associates with actin-containing cytoskeleton. J Cell Sci. 1997;110:2249–2260. doi: 10.1242/jcs.110.18.2249. [DOI] [PubMed] [Google Scholar]
- 44.Sainz J, Huynh D P, Figueroa K, Ragge N K, Baser M E, Pulst S M. Mutations of the neurofibromatosis type 2 gene and lack of the gene product in vestibular schwannomas. Hum Mol Genet. 1994;3:885–891. doi: 10.1093/hmg/3.6.885. [DOI] [PubMed] [Google Scholar]
- 45.Sato N, Funayama N, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. A gene family consisting of ezrin, radixin and moesin. Its specific localization at actin filament/plasma membrane association sites. J Cell Sci. 1992;103:131–143. doi: 10.1242/jcs.103.1.131. [DOI] [PubMed] [Google Scholar]
- 46.Scherer S S, Gutmann D H. Expression of the neurofibromatosis 2 tumor suppressor gene product, merlin, in Schwann cells. J Neurosci Res. 1996;46:595–605. doi: 10.1002/(SICI)1097-4547(19961201)46:5<595::AID-JNR8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 47.Schiavo G, Osborne S L, Sgouros J G. Synaptotagmins: more isoforms than functions? Biochem Biophys Res Commun. 1998;248:1–8. doi: 10.1006/bbrc.1998.8527. [DOI] [PubMed] [Google Scholar]
- 48.Schmucker B, Ballhausen W G, Kressel M. Subcellular localization and expression pattern of the neurofibromatosis type 2 protein merlin/schwannomin. Eur J Cell Biol. 1997;72:46–53. [PubMed] [Google Scholar]
- 49.Scoles D R, Huynh D P, Morcos P A, Coulsell E R, Robinson N G G, Tamanoi F, Pulst S M. Neurofibromatosis 2 tumour suppressor schwannomin interacts with βII-spectrin. Nat Genet. 1998;18:354–359. doi: 10.1038/ng0498-354. [DOI] [PubMed] [Google Scholar]
- 50.Shaw R J, McClatchey A I, Jacks T. Localization and functional domains of the neurofibromatosis type II tumor suppressor, merlin. Cell Growth Differ. 1998;9:287–296. [PubMed] [Google Scholar]
- 51.Shaw R J, McClatchey A I, Jacks T. Regulation of the neurofibromatosis type 2 tumor suppressor protein, merlin, by adhesion and growth arrest stimuli. J Biol Chem. 1998;273:7757–7764. doi: 10.1074/jbc.273.13.7757. [DOI] [PubMed] [Google Scholar]
- 52.Sherman L, Xu H-M, Geist R T, Saporo-Irwin S, Howells N, Ponta H, Herrlich P, Gutmann D H. Interdomain binding mediates tumor growth suppression by the NF2 gene product. Oncogene. 1997;15:2505–2509. doi: 10.1038/sj.onc.1201418. [DOI] [PubMed] [Google Scholar]
- 53.Stemmer-Rachamimov A O, Gonzalez-Agosti C, Xu L, Burwick J A, Beauchamp R, Pinney D, Louis D N, Ramesh V. Expression of NF2-encoded merlin and related ERM family proteins in the human central nervous system. J Neuropathol Exp Neurol. 1997;56:735–742. [PubMed] [Google Scholar]
- 54.Stemmer-Rachamimov A O, Xu L, Gonzalez-Agosti C, Burwick J A, Pinney D, Beauchamp R, Jacoby L B, Gusella J F, Ramesh V, Louis D N. Universal absence of merlin, but not other ERM family members, in schwannomas. Am J Pathol. 1997;151:1649–1654. [PMC free article] [PubMed] [Google Scholar]
- 55.Takeshima H, Izawa I, Lee P S, Safdar N, Levin V A, Saya H. Detection of cellular proteins that interact with the NF2 tumor suppressor gene product. Oncogene. 1994;9:2135–2144. [PubMed] [Google Scholar]
- 56.Takeuchi K, Sato N, Kasahara H, Funayama N, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Perturbation of cell adhesion and microvilli formation by antisense oligonucleotides to ERM family members. J Cell Biol. 1994;125:1371–1384. doi: 10.1083/jcb.125.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tikoo A, Varga M, Ramesh V, Gusella J, Maruta H. An anti-ras function of neurofibromatosis type 2 gene product (NF2/merlin) J Biol Chem. 1994;269:23387–23390. [PubMed] [Google Scholar]
- 58.Trofatter J A, MacCollin M M, Rutter J L, Murrell J R, Duyao M P, Parry D M, Eldridge R, Kley N, Menon A G, Pulaski K, Haase V H, Ambrose C M, Munroe D, Bove C, Haines J L, Martuza R L, MacDonald M E, Seizinger B R, Short M P, Buckler A J, Gusella J F. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- 59.Turunen O, Sainio M, Jaaskelainen J, Carpen O, Vaheri A. Structure-function relationships in the ezrin family and the effect of tumor-associated point mutations in neurofibromatosis 2 protein. Biochim Biophys Acta. 1998;1387:1–16. doi: 10.1016/s0167-4838(98)00103-4. [DOI] [PubMed] [Google Scholar]
- 60.Turunen O, Wahlström T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu H M, Gutmann D H. Merlin differentially associates with the microtubule and actin cytoskeleton. J Neurosci Res. 1998;51:403–415. doi: 10.1002/(SICI)1097-4547(19980201)51:3<403::AID-JNR13>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]