Abstract
c-Jun N-terminal kinase (JNK) plays a critical role in coordinating the cellular response to stress and has been implicated in regulating cell growth and transformation. To investigate the growth-regulatory functions of JNK1 and JNK2, we used specific antisense oligonucleotides (AS) to inhibit their expression. A survey of several human tumor cell lines revealed that JNKAS treatment markedly inhibited the growth of cells with mutant p53 status but not that of cells with normal p53 function. To further examine the influence of p53 on cell sensitivity to JNKAS treatment, we compared the responsiveness of RKO, MCF-7, and HCT116 cells with normal p53 function to that of RKO E6, MCF-7 E6, and HCT116 p53−/−, which were rendered p53 deficient by different methods. Inhibition of JNK2 (and to a lesser extent JNK1) expression dramatically reduced the growth of p53-deficient cells but not that of their normal counterparts. JNK2AS-induced growth inhibition was correlated with significant apoptosis. JNK2AS treatment induced the expression of the cyclin-dependent kinase inhibitor p21Cip1/Waf1 in parental MCF-7, RKO, and HCT116 cells but not in the p53-deficient derivatives. That p21Cip1/Waf1 expression contributes to the survival of JNK2AS-treated cells was supported by additional experiments demonstrating that p21Cip1/Waf1 deficiency in HCT116 cells also results in heightened sensitivity to JNKAS treatment. Our results indicate that perturbation of JNK2 expression adversely affects the growth of otherwise nonstressed cells. p53 and its downstream effector p21Cip1/Waf1 are important in counteracting these detrimental effects and promoting cell survival.
The c-Jun N-terminal kinase (JNK) signal transduction pathway, culminating in the phosphorylation of one or more JNK proteins, is known to play an important role in coordinating the cellular response to stress (25, 34). The JNK family includes three genes, JNK1, JNK2, and JNK3, each of which can produce 46- and 54-kDa isoforms (25). JNK1 and JNK2 are ubiquitously expressed, while JNK3 is largely restricted to brain, heart and testis (25, 28, 35). Although some differences in the substrate specificities and activities of various JNK isoforms have been reported (5, 9, 13, 20, 26, 44), their functional distinctions remain unclear. Among the major targets of JNK phosphorylation are the transcription factor c-Jun (12, 22, 29) and the tumor suppressor protein p53 (24, 32). Numerous studies have implicated both JNK activation and c-Jun phosphorylation in the induction of apoptosis following stress (27). However, an opposing function of the JNK pathway has also been suggested by other studies, including two recent reports which demonstrate that JNK-mediated phosphorylation of c-Jun confers protection to cells exposed to UVC irradiation or tumor necrosis factor alpha (3, 42).
In addition to playing a role during stress, there is also evidence to support a role for the JNK pathway in regulating cell growth. While the basal activity of JNK is generally low in cells maintained under normal growth conditions, JNK can be activated by growth factors such as epidermal growth factor and platelet-derived growth factor via mechanisms that appear to rely on phosphoinositide 3-kinase (2, 4, 30, 31, 33). It has long been appreciated that c-Jun promotes proliferation (1), and both cells lacking c-Jun and cells expressing its nonphosphorylatable dominant negative counterpart, c-Jun(Ser63A,Ser73A), display well-defined growth defects (3, 42). Another study has provided evidence suggesting that c-Jun controls cell cycle progression in a p53-dependent manner (41). The JNK pathway has also been implicated in the transformation of pre-B cells by Brc-Abl and in the transformation of fibroblasts by the Met oncogene (36, 37). In addition, both epidermal growth factor-dependent proliferation and anchorage-independent growth of A549 cells can be prevented by either stable expression of c-Jun(Ser63A,Ser73A) or addition of JNK antisense oligonucleotides (JNKAS) (4, 5). While most of the effects of JNK described above have been attributed to its phosphorylated state, it has recently become apparent that nonphosphorylated JNK (which lacks kinase activity) also contributes to the regulation of its substrates. In contrast to activated JNK, which serves to stabilize and enhance the transcriptional activity of its substrates, nonphosphorylated JNK appears to lead to ubiquitination and degradation of its substrates (14–16). Thus, both inactive and active JNK are likely to play a role in regulating a variety of cell processes including growth, apoptosis, and transformation.
To investigate the role of JNK in regulating tumor cell growth, we have used highly specific JNKAS to inhibit the expression of JNK1 and JNK2 (4, 5, 43). A preliminary survey of several different cell lines revealed that JNKAS treatment had little or no effect on the growth of most of the cell lines examined, but three cell lines displayed marked growth inhibition in response to such treatments (Table 1). Interestingly, the growth-inhibitory response to JNKAS treatment appeared to correlate with the p53 status of the cell, in that suppression of growth occurred only in cells harboring mutations in p53 (6; O. Potapova, M. Gorospe, F. Bost, N. M. Dean, R. McKay, S. A. Kim, D. Mercola, and N. J. Holbrook, submitted for publication; our unpublished observations). The present study was designed to further investigate the possibility that p53 directly influences the response of tumor cells to JNKAS treatment. To this end, we have used derivatives of MCF-7 and RKO cells, in which p53 function was abrogated through expression of the viral E6 oncoprotein (38), and HCT116 p53−/− cells, where the p53 gene was disrupted through homologous recombination (8). We demonstrate that loss of p53 function in these cells results in growth inhibition and apoptosis following JNKAS treatment and provide additional evidence indicating that the inability to elevate p21Cip1/Waf1 levels contributes to the enhanced sensitivity of p53-deficient cells to JNKAS. Our findings support an important role for the JNK pathway in maintenance of normal tumor cell growth in the absence of p53 function and suggest the utility of strategies aimed at eliminating JNK for the treatment of tumors harboring mutant p53.
TABLE 1.
Inhibition of growth by JNKAS oligonucleotides in different human tumor cell lines
| Tumor cell line | Growth inhibition by JNKAS treatmenta | p53 status |
|---|---|---|
| PC3 prostate carcinoma | +++ | Null |
| A549 lung carcinoma | ± | Wild type |
| T98G glioblastoma | ++ | Mutant |
| HeLa cervical carcinoma | +++ | Null |
| RKO colorectal carcinoma | − | Wild type |
| MCF-7 breast carcinoma | ± | Wild type |
| HCT116 colorectal carcinoma | ± | Wild type |
Cells were treated with a combination of JNK1AS and JNK2AS (0.2 μM each), and viable cell mass was measured 3 to 5 days after the treatment by using the MTS dye reduction assay as described in Materials and Methods.
MATERIALS AND METHODS
Cells and reagents.
Human breast carcinoma MCF-7 neo and MCF-7 E6 cells, a gift from A. J. Fornace, were cultured in RPMI 1640 (Life Technologies, Bethesda, Md.). Human colorectal carcinoma RKO neo and RKO E6 cells, a gift from M. Kastan, were cultured in minimal essential medium (Life Technologies, Bethesda, Md.). HCT116 and their p21−/− and p53−/− derivatives (8, 41), a gift from B. Vogelstein, were grown in McCoy's 5A medium (Biofluids, Rockville, Md.). The media were supplemented with 10% fetal calf serum (HyClone Laboratories, Logan, Utah), nonessential amino acids, and antibiotics.
Oligonucleotide synthesis, treatment, and confocal microscopy.
The oligonucleotides used in this study were synthesized at Isis Pharmaceuticals, Inc. (Carlsbad, Calif.). The sequences of the oligonucleotides used are as follows: JNK1AS (ISIS12539), 5′-CTC TCT GTA GGC CCG CTT GG-3′; JNK2AS (ISIS 12560), 5′-GTC CGG GCC AGG CCA AAG TC-3′; JNK1Scr (ISIS14321), 5′-CTT TCC GTT GGA CCC CTG GG-3′; and JNK2Scr (ISIS14319), 5′-GTG CGC GCG AGC CCG AAA TC-3′ (4). All oligonucleotides were 2′-O-methoxyethyl chimers containing five 2′-O-methoxyethyl–phosphodiester residues flanking a 2′-deoxynucleotide-phosphorothioate region (43). Cells were treated with 0.3 μM oligonucleotides in the presence of 10 μg of Lipofectin reagent (Life Technologies, Bethesda, Md.) per ml as described previously (11). To determine the efficiency of the oligonucleotide transfection into MCF-7 and RKO cells, a fluorescein isothiocyanate (FITC)-labeled phosphorothioate control oligonucleotide, ISIS13193 (5′-TCC CGC CTG TGA CAT GCA TT-3′), was used. At the indicated times following lipofection with the FITC-labeled oligonucleotide, the cells were fixed in 3.7% formaldehyde and images were taken using a Carl Zeiss LSM 410 confocal microscope (488-nm laser wavelength; ×40/1.3 magnification; pinhole size, 20 nm).
Analysis of mRNA and protein expression.
Total RNA was isolated using Stat-60 solution (Tel-Test, Friendswood, Tex.). RNA samples (20 μg) were denatured, size separated by electrophoresis in 1.2% agarose–formaldehyde gels, and transferred onto GeneScreen Plus membranes (DuPont NEN Research Products, Boston, Mass.). For the detection of JNK1 and JNK2 mRNAs, the respective cDNAs were excised from 3xHA-JNK1-SRα3 and 1xHA-JNK2-SRα3 and labeled with [α-32P]dATP using a random primer labeling kit (Boehringer Mannheim, Indianapolis, Ind.). Expression of p21Cip1/Waf1 was detected by hybridization with an 46-base oligonucleotide specific for human p21Cip1/Waf1 (Integrated DNA Technologies, Coralville, Iowa). Variations in loading and transfer among samples were monitored by hybridizing to a 29-base oligonucleotide complementary to 18S RNA (Clontech, Palo Alto, Calif.). Hybridization and washes were performed by the Church and Gilbert method (10), and radioactive signals were quantified by using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
For Western analysis, total protein was extracted using whole-cell extract buffer (5) and the protein concentration was quantified by the Bradford assay. Protein samples (25 to 50 μg) were separated in by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide) and transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). JNK proteins were identified using C-571 anti-JNK primary antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.), and reactive bands were visualized using the enhanced chemiluminescence detection system (NEN Life Science Products, Boston, Mass.).
In vitro JNK activity assay.
Cells were irradiated with UVC at 40 J/m2 24 h following the lipofection; 30 min later they were washed with cold phosphate-buffered saline and suspended in whole-cell extract (WCE) buffer (5). The JNK assay was performed with a fusion protein containing glutathione S-transferase (GST) linked to the 1–222 fragment of human c-Jun (GST-cJun) as substrate, as described previously (22). Briefly, 50-μg portions of cell lysates were incubated for 3 h at 4°C with 10 μg of GST-cJun bound to glutathione-Sepharose 4B (Pharmacia Biotech, Uppsala, Sweden). After being washed three times in WCE buffer (5) and once in kinase reaction buffer (20 mM HEPES [pH 7.7], 20 mM MgCl2, 20 mM α-glycerophosphate, 20 mM p-nitrophenyl phosphate, 0.1 mM sodium vanadate, 2 mM dithiothreitol), the beads were incubated for 30 min at 30°C with 30 μl of kinase reaction buffer containing 20 μM ATP and 5 μCi of [γ-32P]ATP. The reaction was stopped by the addition of Laemmli sample buffer, and samples were boiled for 5 min and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide). The [32P]GST-cJun was quantified with a PhosphorImager.
Cell viability assays.
Cells were seeded in 96-well cluster plates at a density of 15 × 103 cells/well and transfected with AS oligonucleotides as described above. At the designated times, viable cell mass was measured by detection of 3-(4,5-dimethylthiazol - 2 - yl) - 5 - (3 - carboxymethoxyphenyl) - 2 - (4 - sulfophenyl) - 2H - tetrazolium, inner salt (MTS) dye reduction at 490 nm as described in the manufacturer's protocol (Promega, Madison, Wis.). All viability assays were carried out in triplicate.
Apoptosis detection and flow cytometry analysis.
Apoptosis was assessed using a cell death detection enzyme-linked immunosorbent assay (ELISA) (Boehringer Mannheim, Indianapolis, Ind.) as specified by the manufacturer. Oligonucleosomal fragments, released from the nuclei after internucleosomal degradation of genomic DNA, were detected in cytoplasmic fractions of 2,000 cells. For cell cycle analysis, cells were collected 24 h following treatment, fixed in 70% ethanol, and stained with 1 μg of propidium iodide (Cellular DNA flow cytometric analysis kit; Boehringer Mannheim) per ml. The DNA content was analyzed on FACScalibur flow cytometer (Becton Dickinson, San Jose, Calif.). The percentages of cells in various phases of the cell cycle were determined using Becton Dickinson ModFitLT software.
RESULTS
Specific inhibition of JNK expression by JNKAS.
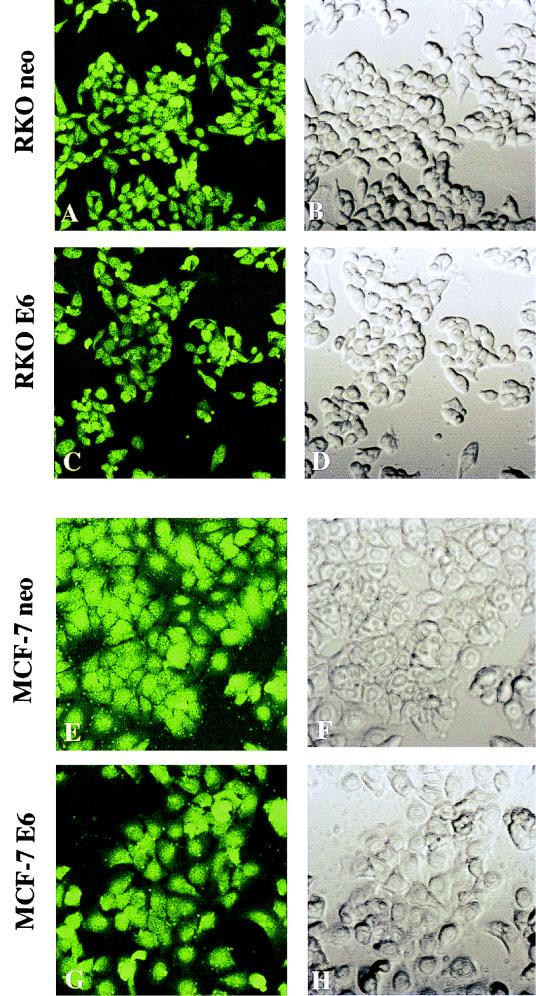
To verify that the lipofection procedure used here resulted in high-efficiency oligonucleotide uptake in MCF-7 and RKO cells, the control FITC-labeled phosphorothioate oligonucleotide ISIS13193 was used. Shown in Fig. 1 is green fluorescence, visualized 24 h after lipofection with 0.4 μM ISIS13193. Virtually 100% of cells showed uptake of the transfected oligonucleotide. Similar results were obtained at the 12- and 48-h time points and over an extended dose-response range (0.1 to 0.5 μM oligonucleotide). Mock-lipofected cells did not excite any fluorescent signal (data not shown). No differences in the efficiency of oligonucleotide uptake were seen between p53-deficient (E6-expressing) and p53-proficient (expressing a neo-containing vector control) cells.
FIG. 1.
MCF-7 and RKO cells display high-efficiency uptake of oligonucleotides delivered via transfection with Lipofectin reagent. Cells were treated with 0.3 μM 3′ FITC-labeled control oligonucleotide as described in Materials and Methods. Twenty-four hours later, they were fixed and examined for fluorescence with a confocal microscope. Images were obtained at a magnification of ×800. (Left) Confocal fluorescent images of transfected cells. (Right) Confocal transparent images of the same fields. Virtually all cells show uptake of the oligonucleotides, irrespective of p53 status.
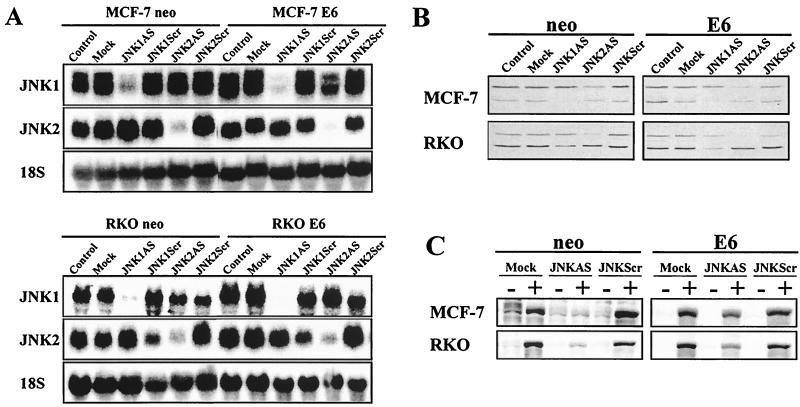
Next, we investigated the effect of JNK1AS and JNK2AS on JNK expression in MCF-7 and RKO cells. Cells were treated either with 0.3 μM JNK1AS or JNK2AS individually or with JNK1AS and JNK2AS in combination (0.15 μM each). Scrambled-sequence oligonucleotides (JNK1Scr and JNK2Scr) were used at the same concentrations and served as controls. Both AS treatments led to >95% reduction in the corresponding mRNA levels (Fig. 2A), while neither mock lipofection nor treatment with scrambled oligonucleotides had any effect on JNK1 or JNK2 mRNA expression.
FIG. 2.
JNKAS treatment effectively inhibits JNK expression in MCF-7 and RKO cells regardless of p53 status. (A) Total RNA was extracted from cells 24 h following treatment with 0.3 μM JNK1AS, JNK2AS, or control oligonucleotides (JNKScr). RNA samples were analyzed by Northern analysis using cDNA probes specific for JNK1 and JNK2 mRNAs. (B) Whole-cell lysates were examined by Western analysis for JNK expression 24 h following treatment with JNKAS oligonucleotides. (C) Total Jun kinase activity was determined 30 min following exposure to UVC (40 J/m2) by an in vitro kinase assay using GST-cJun(1-222) as a substrate. At 24 h prior to UVC treatment, cells were transfected with combinations of JNK1AS plus JNK2AS (0.15 μM each) or JNK1Scr plus JNK2Scr (0.15 μM each) (labeled JNKAS and JNKScr, respectively).
Western analysis of JNK protein expression revealed a corresponding reduction in the level of JNK protein in JNKAS-treated cells (Fig. 2B). Since differential splicing of both JNK1 and JNK2 mRNAs results in the production of 46- and 54-kDa isoforms and since there is significant cross-reactivity between the available JNK1 and JNK2 antibodies, we cannot distinguish with certainty between the JNK1 and JNK2 isoforms. However, based on our observations and those of others, it appears that the slower-migrating proteins (p54JNK) are composed primarily of JNK2 isoforms whereas the faster-migrating forms (p46JNK) contain mostly JNK1 (25). Hence, JNK1AS was more effective in eliminating the p46JNK isoform while JNK2AS was more effective in eliminating the p54JNK form (Fig. 2B).
An immunocomplex kinase assay was used to examine basal and UVC-induced JNK activity in mock-, JNKAS- and JNKScr-treated cells (Fig. 2C). No significant JNK activity was detectable in any of the treatment groups of either wild-type or p53-deficient MCF-7 and RKO cells in the absence of stress. Consistent with the reduction in JNK protein levels, UVC-induced JNK activation was markedly reduced in both wild-type and p53-deficient MCF-7 and RKO cells treated with a combination of JNK1AS and JNK2AS (Fig. 2C, JNKAS). Neither mock lipofection nor treatment with scrambled oligonucleotides affected the JNK kinase activity.
Taken together, the experiments described above demonstrate that, using JNKAS, we can effectively achieve a significant reduction in JNK expression and therefore directly investigate the roles of JNK1 and JNK2 in regulating the growth of p53-proficient and p53-deficient MCF-7 and RKO cells.
Growth inhibition by JNKAS is dependent on p53 status.
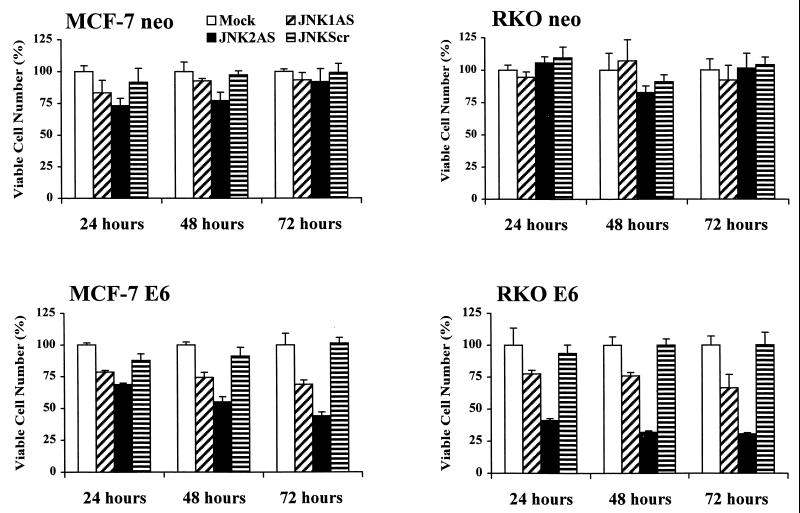
Growth of untreated and JNKAS-treated cells was monitored using an MTS dye reduction assay. JNKAS treatment had little to no effect on the growth of wild-type (neo) MCF-7 and RKO cells (Fig. 3). A small transient reduction in the growth of JNK2AS-treated MCF-7 cells was apparent at 24 to 48 h following lipofection, but the viable cell mass did not differ from that of control cultures by 72 h posttreatment. No significant effect on growth was observed in wild-type RKO cells with any treatment. In contrast, p53-deficient derivatives (E6) of both MCF-7 and RKO cells displayed a marked reduction in cell viability following treatment with JNKAS; the growth-inhibitory effect was greater for JNK2AS than for JNK1AS. Growth suppression by JNK2AS was most pronounced in p53-deficient RKO cells, where viability was reduced >70% by 48 h following treatment with the oligonucleotide.
FIG. 3.
Effect of JNKAS treatment on the viability of p53-proficient and p53-deficient MCF-7 and RKO cells. Cells were seeded in 96-well cluster plates and treated with 0.3 μM oligonucleotides in Lipofectin reagent (JNKScr designates treatment with 0.15 μM JNK1Scr plus 0.15 μM JNK2Scr). Cell viability was assessed at the indicated times by measuring MTS dye in a colorimetric assay as described in Materials and Methods.
Inhibition of JNK2 expression results in apoptosis of p53-deficient cells.
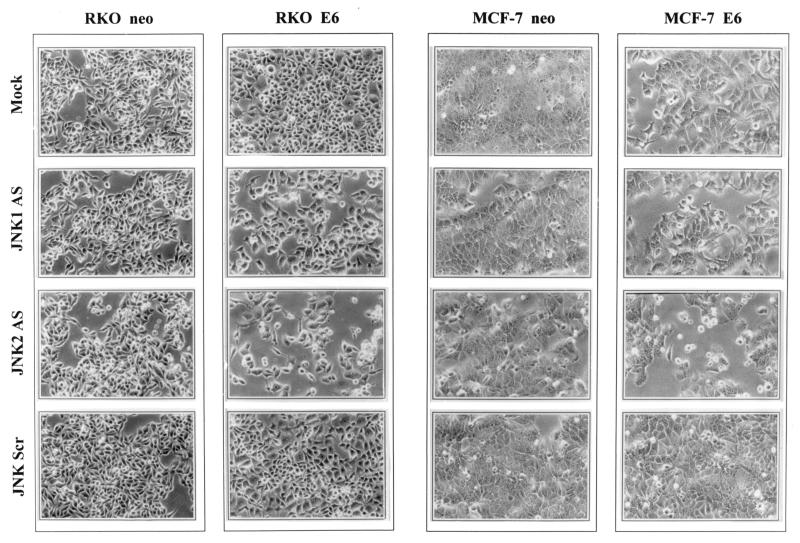
To determine the cause of the decline in growth of JNKAS-treated cell populations, we first examined whether DNA synthesis was altered after application of JNKAS. We observed no differences in bromodeoxyuridine (BrdU) incorporation relative to the number of viable cells among any of the treatment groups (data not shown); therefore, the reduction in growth of p53-deficient cells could not be explained by an inhibition of DNA replication. Next we tested the possibility that the decrease in cell number reflected selective death in JNKAS-treated cultures. Shown in Fig. 4 is the morphology of parental and p53-deficient MCF-7 and RKO cells with or without JNKAS treatment. The p53-deficient (E6) lines treated with JNK2AS revealed features consistent with apoptosis, including cell rounding, membrane blebbing, and detachment from the tissue culture dish (Fig. 4). These effects were also evident to some degree in JNK1AS-treated cultures but were completely absent in p53-deficient cells subjected to either mock treatment or treatment with scrambled oligonucleotides. They were also absent in p53-proficient (neo) cells regardless of treatment.
FIG. 4.
Morphological appearance of JNK2AS-treated MCF-7 and RKO cells with normal (neo) or deficient (E6) p53 function. Cells treated with 0.3 μM of the indicated oligonucleotides were examined by optical microscopy 24 h after lipofection. Cultures that were subjected to either mock lipofection or treatment with scrambled oligonucleotides (the same as described in the legend to Fig. 3) had morphologies similar to those seen in untreated control cultures. The density of JNKAS-treated E6-expressing MCF-7 and RKO cells is lower than that of the neo-expressing counterparts. JNK2AS-treated p53-deficient (E6-expressing) cells exhibit membrane blebbing and an increase in the number of rounded and/or detached cells.
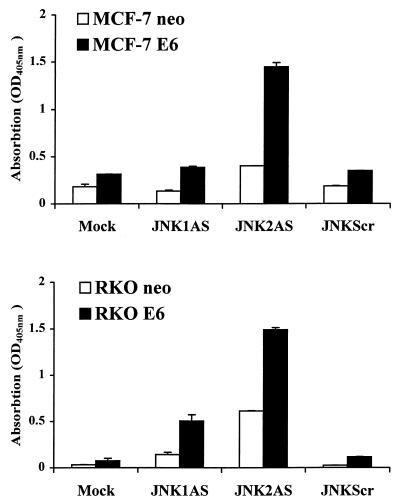
Biochemical evidence of apoptosis in the p53-deficient cells was obtained using an ELISA for detection of cytoplasmic histone-associated DNA fragments (Fig. 5). Both E6-expressing MCF-7 and RKO lines exhibited significant induction of apoptosis 24 h following JNK2AS treatment. In this assay, we also observed some evidence of DNA degradation in JNK2AS-treated RKO neo cells as well as in RKO E6 cells treated with JNK1AS, but quantitatively it was not nearly as great as that seen in JNK2AS-treated RKO E6 cells.
FIG. 5.
Biochemical evidence of DNA degradation in p53-deficient cells treated with JNKAS. Apoptosis of p53-proficient (neo) and p53-deficient (E6) MCF-7 and RKO cells was assessed 24 h after treatment with 0.3 μM JNKAS or control oligonucleotides using an ELISA that measures nucleosomal degradation. JNKScr depicts results for treatments with equal amounts of JNK1Scr plus JNK2Scr (0.15 μM each). Results are expressed as relative absorption (OD) at 405 nm.
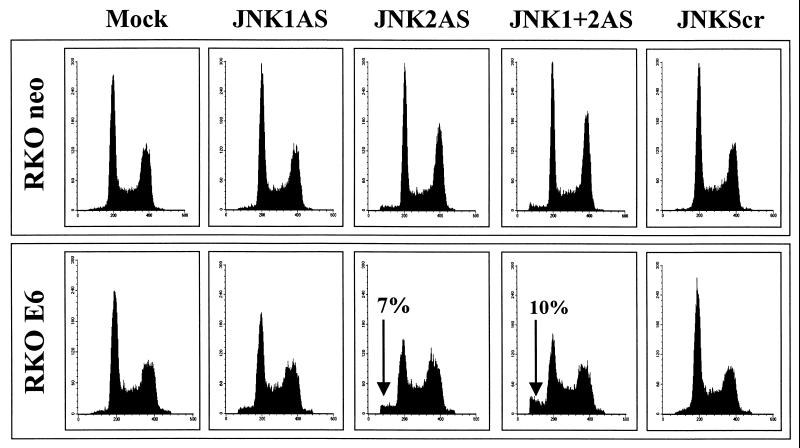
Consistent with the biochemical analysis, fluorescence-activated cell sorter (FACS) analysis of RKO E6 cells as early as 24 h after treatment revealed the presence of a sub-G1 cell population in cultures treated with JNK2AS alone or with JNK1AS and JNK2AS in combination (Fig. 6). No sub-G1 peak was evident in RKO E6 cells subjected to any of the control treatments or in the RKO neo cells regardless of treatment. The number of apoptotic cells in JNK2AS-treated RKO E6 cultures further increased with time, and by 48 h posttreatment had reached 14% (data not shown). Taken together, these experiments indicate that JNK2AS treatment results in apoptosis of p53-deficient MCF-7 and RKO cells, thus largely accounting for the growth-suppressive effects described above (Fig. 3).
FIG. 6.
FACS analysis of mock-transfected and JNKAS-treated RKO cells. RKO neo and RKO E6-expressing cells were treated with either JNK1AS, JNK2AS, or a combination of control scrambled oligonucleotides (JNKScr). They were harvested 24 h following the lipofection, and 2 × 106 cells were subjected to DNA content analysis by FACS. The percentage of total cells contained in the sub-G1 peak of JNK2AS and JNK1AS- plus JNK2AS-treated RKO E6 cell cultures is indicated in the figure (arrow). For all other treatments of E6-expressing RKO and RKO neo cells, the sub-G1 fraction was 1.5% or less.
Effects of JNKAS treatment on HCT116 p53−/− cells.
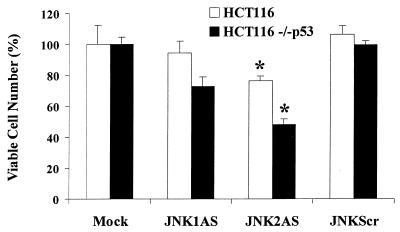
Although expression of the E6 oncoprotein is a well-recognized way to inhibit p53 function, it remained possible that our findings with RKO E6 and MCF-7 E6 cells could reflect other function(s) of E6 protein. To address this possibility, we used another model of p53 deficiency, HCT116 human colorectal carcinoma cells rendered p53-null by a somatic knockout procedure (8). Consistent with our observations with E6-expressing RKO and MCF-7 cells, HCT116 p53−/− cells displayed significantly greater sensitivity (P < 0.003 by Student's t test) to JNK2AS treatment than did parental HCT116 cells with normal p53 status (Fig. 7).
FIG. 7.
Inhibition of JNK expression in HCT116 cells results in growth suppression in a p53-dependent manner. Viable cell mass was measured at 24 h, 48 h (shown here), and 72 h following treatment with JNKAS and control oligonucleotides as described in Materials and Methods. Growth inhibition following JNK2AS treatment is p53 dependent: ∗, statistically significant difference between the number of viable cells in JNK2AS-treated cultures of parental HCT116 cells and HCT116 p53−/− cells (P < 0.003, Student's t test).
Induction of p21Cip1/Waf1 in p53-proficient and p53-deficient cells following JNKAS treatment.
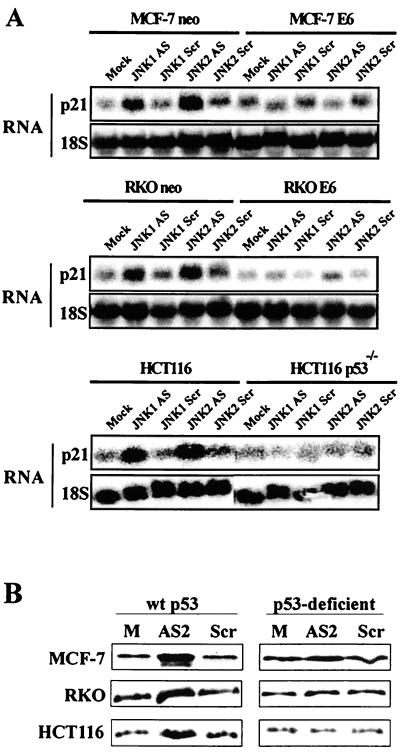
Our finding that inhibition of JNK expression, especially JNK2, resulted in preferential apoptosis of p53-deficient cells suggests that cells with intact p53 signaling possess a protective mechanism that is unavailable to their p53-deficient counterparts. We have previously shown that the ability of cells to elevate expression of the cyclin-dependent kinase inhibitor p21Cip1/Waf1 following exposure to cellular stress leads to enhanced survival (18, 19). Given that p53 is an important regulator of p21Cip1/Waf1 expression during stress and that perturbations in JNK expression could constitute a stress to growing cells, we examined p21Cip1/Waf1 mRNA levels in JNKAS-treated cells. Treatment with JNKAS (especially with JNK2AS) resulted in induction of p21Cip1/Waf1 mRNA in parental MCF-7, RKO, and HCT116 cells. In contrast, no elevation in p21Cip1/Waf1 mRNA was evident in p53-deficient MCF-7 E6, RKO E6, and HCT116 p53−/− cells (Fig. 8A). That the increase in p21Cip1/Waf1 mRNA results in increased p21Cip1/Waf1 protein expression is shown in Fig. 8B, where p21Cip1/Waf1 protein levels were examined 24 h following JNK2AS treatment. All three p53-proficient cell lines showed an increase in p21Cip1/Waf1 protein expression, but this p21Cip1/Waf1 induction was not seen in the p53-deficient derivatives.
FIG. 8.
JNK2AS treatment results in induction of p21Cip1/Waf1 expression in cells with normal p53 function (neo-transfected MCF-7 and RKO, and HCT166 cells) but not in p53-deficient cells (E6-expressing MCF-7 and RKO, and HCT116 p53−/− cells). (A) Total RNA was extracted from cells 24 h following treatment with 0.3 μM antisense (AS) or control (Scr) oligonucleotides. RNA samples were analyzed by Northern analysis using an oligonucleotide probe specific for human p21Cip1/Waf1. Following analysis of p21Cip1/Waf1 expression, Northern blots were stripped and reprobed with an oligonucleotide complementary to 18S rRNA to verify equal loading and transfer of RNA samples. (B) p21Cip1/Waf1 expression was examined by Western blot analyses 24 h following treatment with 0.3 μM JNK2AS or JNKScr oligonucleotides (as described in the legend to Fig. 3).
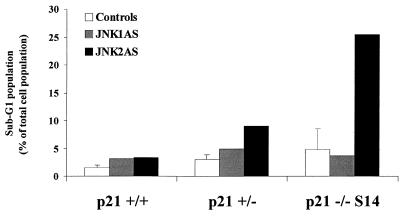
Finally, to more directly examine the influence of p21Cip1/Waf1 on cell survival following JNKAS treatment, we used HCT116 colorectal carcinoma cells in which the p21Cip1/Waf1 genes were disrupted by homologous recombination (41). Wild-type HCT116 cells (+/+) and HCT116 cells in which one (+/−) or both (−/−) p21Cip1/Waf1 alleles were disrupted were examined for apoptosis by flow cytometry following treatment with antisense oligonucleotides (Fig. 9). No significant apoptosis was evident in p21+/+ cells, regardless of treatment (1.5% in control populations versus 3% in JNKAS-treated cultures). p21−/− cells exhibited higher basal apoptosis than did the p21+/+ parental cell lines (4 and 1.5%, respectively), perhaps reflecting the importance of p21Cip1/Waf1 for normal cell homeostasis. Neither mock lipofection nor treatment with JNKScr or JNK1AS altered this level. However, treatment of the p21−/− cells with JNK2AS resulted in a substantial increase in the sub-G1 population within 24 h after treatment (Fig. 9). An intermediate level of apoptosis was observed following JNK2AS treatment of cells containing one functional p21Cip1/Waf1 allele (+/−), suggesting that the relative level of p21 expression might be important in determining the outcome. These results further support the hypothesis that p21 contributes to the survival of cells following JNKAS treatment.
FIG. 9.
HCT116 human colorectal carcinoma cells lacking p21Cip1/Waf1 expression display enhanced sensitivity to JNK2AS treatment. Parental HCT116 cells with normal p21Cip1/Waf1 expression (p21 +/+) and derivative lines in which one (p21 +/−) or both (p21 −/−) p21Cip1/Waf1 alleles have been disrupted through homologous recombination were examined by FACS analysis for apoptotic cells 24 h following treatment with JNK1AS or JNK2AS oligonucleotides.
DISCUSSION
Over the past several years, the functional role of the JNK pathway has been extensively investigated. The vast majority of studies have relied on overexpression of mutant forms (dominant negative or constitutively active) of various upstream signaling components to modulate JNK kinase activity. Although informative, this approach can produce confounding results for several reasons. First, most upstream JNK regulators are also implicated in other signaling pathways (p38, ERK, and others), and thus alterations in these pathways can contribute to the effects observed. Second, considerable redundancy exists in the upstream components (e.g., both MEKK4 and MEKK7 phosphorylate and activate JNK), and therefore interference with the activity of one component often leads to only partial inhibition of JNK activation. Finally, the approaches described above do not alter JNK protein levels and therefore do not allow the investigation of the importance of changes in basal JNK activity or other potential functions of the protein unrelated to kinase activity. This is particularly important given that, in the absence of stress, nonphosphorylated JNK has recently been implicated in down-regulating the expression of its substrates, targeting them to degradation (14–17). While the recent generation of JNK1, JNK2, and JNK3 knockout mice provides the opportunity to address the roles of specific JNK isoforms in normal mouse cells and tissues, such model systems are not currently available for human cells, and they do not address the importance of JNK in regulating tumor cell growth. The AS strategy used here avoids many of the drawbacks noted above and offers several distinct advantages. It produces significant and isoform-specific inhibition of basal JNK mRNA and protein expression in human cells without perturbing other components of the pathway. The availability of AS oligonucleotides highly specific for JNK1 and JNK2 allows investigation into differential roles of these genes. Using this strategy, we have investigated growth-regulatory functions of JNK1 and JNK2 in otherwise unstressed cells and have provided evidence suggesting that JNK2 is required for growth and homeostasis of tumor cells lacking p53 function, since elimination of JNK2 expression in p53-deficient cells results in their apoptosis (Fig. 4 to 6). This role appears not to be shared by JNK1. However, we have shown that the JNK1 protein has a longer half-life than JNK2 protein (Potapova et al., submitted), and therefore it remains possible that the more efficient elimination of JNK2 accounts for its greater inhibitory effects.
The ability of JNK to bind to and phosphorylate p53 was reported several years ago (24, 32). More recently, several reports have appeared indicating that JNK plays opposing roles in regulating the stability of the p53 protein under normal growth conditions and during stress (15, 16). Finally, it was recently shown that p53 transcription is subject to repression by activated c-Jun, a major known substrate of JNK (39). Taken together, these observations suggest important links between the activities of JNK and p53. Our study demonstrating that JNK2AS treatment leads to growth inhibition and apoptosis of RKO, MCF-7, and HCT116 human tumor cells in a p53-dependent manner provides evidence for yet another link between these stress-regulated pathways. The greater sensitivity of p53-deficient tumor cells to JNK2AS treatment is not restricted to these cell types, since we have observed that other human tumor cell lines with null or mutant p53 status display greater growth inhibition and cytotoxicity following JNKAS treatment than do cells with normal p53 function (Table 1) (6; Potapova et al., submitted). Whether reintroduction of wild-type p53 into tumor cells with mutant or null p53 status can alter their response to JNKAS treatment remains to be investigated.
p53 can act in two opposing directions to influence the sensitivity of a cell to stressful conditions: it can promote apoptosis in response to certain cytotoxic agents (e.g., hydrogen peroxide) while conferring protection against the cytotoxic effects of others (e.g., UVC irradiation and tumor necrosis factor alpha) (7, 8, 21, 40). The factors contributing to these seemingly disparate functions of p53 are far from clear, but recent studies have provided strong evidence that the protective influence of p53 is mediated largely through its ability to up-regulate p21Cip1/Waf1 expression (18, 19). Consistent with this view, we observed that MCF-7, RKO, and HCT116 cells with normal p53 function responded to the JNK2AS treatment with induction of p21Cip1/Waf1 while the p53-deficient derivatives did not (Fig. 8). Although JNK1AS treatment resulted in induction of p21Cip1/Waf1 expression (Fig. 8A), this effect was less pronounced than that seen in JNK2AS-treated cells, and p21Cip1/Waf1 protein levels were induced to a much lesser extent (data not shown). These results, taken together with the absence of marked growth suppression following JNK1AS treatment (Fig. 3 and 7), suggest that JNK2 may have a specific function(s) which is not shared by JNK1. That the p21Cip1/Waf1 expression might be important in contributing to the survival of JNK2AS-treated cells was supported by additional studies with HCT116 cells in which targeted disruption of the p21Cip1/Waf1 gene was likewise associated with enhanced apoptosis following JNK2AS treatment (Fig. 9).
Based on these observations, we propose the following model. Tumor cells require JNK2 for normal growth, and the depletion of JNK2 in cells with normal p53 function by the JNK2AS treatment perturbs normal homeostasis, triggering a stress response. This results in the induction of p21Cip1/Waf1, which promotes survival. The lack of p53 in the E6-expressing and p53−/− cells prevents the induction of p21Cip1/Waf1, depriving the cells of this important protective factor and therefore leading to growth suppression and apoptosis. It is not clear, however, how the death program is initiated or which of the mediators of this response are important. It is also important to note that not all p53-deficient cells undergo apoptosis in response to JNK2AS treatment. T98G glioblastoma cells, for example, do not die but, rather, undergo arrest in the S and G2 phases of the cell cycle (Potapova et al., submitted). However, unlike p53-deficient cells used in this study, T98G cells show marked induction of p21Cip1/Waf1 expression following JNK2AS treatment, more so in fact than do parental MCF-7, RKO, and HCT116 cells. It is likely that the high levels of p21Cip1/Waf1 contribute to the cell cycle arrest, since the importance of p21Cip1/Waf1 in mediating G2 arrest has recently been established (8). The mechanisms leading to this p53-independent induction of p21Cip1/Waf1 remain to be clarified.
In summary, there is accumulating evidence suggesting that the JNK pathway plays an important role in regulating the growth of tumor cells and may in fact contribute to cellular transformation (4–6; Potapova et al., submitted). The current findings reported here support this view and further suggest that JNK (JNK2 in particular) is important for growth of human tumor cells lacking functional p53. It may be possible to exploit this preferential sensitivity of p53-deficient cells to JNK2AS treatment for the design of therapeutic approaches for the management of tumors harboring p53 mutations (23).
ACKNOWLEDGMENTS
We are grateful to A. J. Fornace, Jr., M. Kastan, and B. Vogelstein for cells provided for this study; to M. Karin for the gift of plasmid reagents; to R. Roberson for technical assistance; and to J. Chrest and C. Morris for help with flow cytometry analysis.
REFERENCES
- 1.Angel P, Karin M. The role of Jun, Fos, and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 2.Antonyak M A, Moscatello D K, Wong A J. Constitutive activation of c-Jun N-terminal kinase by a mutant epidermal growth factor receptor. J Biol Chem. 1998;273:2817–2822. doi: 10.1074/jbc.273.5.2817. [DOI] [PubMed] [Google Scholar]
- 3.Behrens A, Sibilia M, Wagner E F. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat Genet. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- 4.Bost F, Dean N, McKay R, Mercola D. Activation of the Jun kinase/stress-activated protein kinase pathway is required for EGF-autocrine stimulated growth of human A549 lung carcinoma cells. J Biol Chem. 1997;272:33422–33429. doi: 10.1074/jbc.272.52.33422. [DOI] [PubMed] [Google Scholar]
- 5.Bost F, McKay R, Bost M, Potapova O, Dean N, Mercola D. Jun kinase-2 isoform is preferentially required for epidermal growth factor-induced proliferation of human A549 lung carcinoma. Mol Cell Biol. 1999;19:1938–1949. doi: 10.1128/mcb.19.3.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bost F, Potapova O, Liu C, Yang Y-M, Charbono W, Dean N, McKay R, Mercola D. High frequency regression of established human prostate carcinoma PC3 xenografts by systemic treatment with antisense Jun kinase. Prostate. 1999;38:321. [Google Scholar]
- 7.Brown J M, Wouters B G. Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res. 1999;59:1391–1399. [PubMed] [Google Scholar]
- 8.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 9.Chan E D, Winston B W, Jarpe M B, Wynes M W, Riches D W. Preferential activation of the p46 isoform of JNK/SAPK in mouse macrophages by TNF alpha. Proc Natl Acad Sci USA. 1997;94:13169–13174. doi: 10.1073/pnas.94.24.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean N M, McKay R, Condon T P, Bennett C F. Inhibition of protein kinase C-alpha expression in human A549 cells by antisense oligonucleotides inhibits induction of intercellular adhesion molecule 1 (ICAM-1) mRNA by phorbol esters. J Biol Chem. 1994;269:16146–16424. [PubMed] [Google Scholar]
- 12.Dérijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 13.Dong C, Yang D D, Wysk M, Whitmarsh A J, Davis R J, Flavell R A. Defective T cell differentiation in the absence of Jnk1. Science. 1998;282:2092–2095. doi: 10.1126/science.282.5396.2092. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs S Y, Dolan L R, Davis R J, Ronai Z. JNK targets the ubiquitination of c-Jun in a phosphorylation dependent manner. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- 15.Fuchs S Y, Adler V, Buschmann T, Yin Z, Wu X, Jones S N, Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs S Y, Adler V, Pincus M R, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs S Y, Fried V, Ronai Z. Stress activated kinase regulates protein stability. Oncogene. 1998;12:2558–2563. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- 18.Gorospe M, Wang X, Holbrook N J. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorospe M, Wang X, Holbrook N J. Functional role of p21 during the cellular response to stress. Gene Expression. 1999;7:337–385. [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Dérijard B, Davis R J. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 21.Hawkins D S, Demers G W, Galloway D A. Inactivation of p53 enhances sensitivity to multiple chemotherapeutic agents. Cancer Res. 1996;56:892–898. [PubMed] [Google Scholar]
- 22.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–3148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 23.Hollstein M, Sidransky D, Vogelstein B, Harris C C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 24.Hu M C, Qiu W R, Wang Y P. JNK1, JNK2 and JNK3 are p53 N-terminal serine 34 kinases. Oncogene. 1997;15:2277–2287. doi: 10.1038/sj.onc.1201401. [DOI] [PubMed] [Google Scholar]
- 25.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK) from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 26.Kallunki T, Su B, Tsigelny I, Sluss H K, Dérijard B, Moore G, Davis R, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 27.Karin M, Liu Z G, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 28.Kumagae Y, Zhang Y, Kim O J, Miller C A. Human c-Jun N-terminal kinase expression and activation in the nervous system. Brain Res Mol Brain Res. 1999;67:10–17. doi: 10.1016/s0169-328x(99)00013-3. [DOI] [PubMed] [Google Scholar]
- 29.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 30.Logan S K, Falasca M, Hu P, Schlessinger J. Phosphatidylinositol 3-kinase mediates epidermal growth factor-induced activation of the c-Jun N-terminal kinase signaling pathway. Mol Cell Biol. 1997;7:5784–5790. doi: 10.1128/mcb.17.10.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Ilasaca M, Li W, Uren A, Yu J C, Kazlauskas A, Gutkind J S, Heidaran M A. Requirement of phosphatidylinositol-3 kinase for activation of JNK/SAPKs by PDGF. Biochem Biophys Res Commun. 1997;232:273–277. doi: 10.1006/bbrc.1997.6289. [DOI] [PubMed] [Google Scholar]
- 32.Milne D M, Campbell L E, Campbell D G, Meek D W. p53 is phosphorylated in vitro and in vivo by an ultraviolet radiation-induced protein kinase characteristic of the c-Jun kinase, JNK1. J Biol Chem. 1995;270:5511–5518. doi: 10.1074/jbc.270.10.5511. [DOI] [PubMed] [Google Scholar]
- 33.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 34.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophys Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 35.Mohit A A, Martin J H, Miller C A. p493F12 kinase: a novel MAP kinase expressed in a subset of neurons in the human nervous system. Neuron. 1995;14:67–78. doi: 10.1016/0896-6273(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 36.Raitano A B, Halpern J R, Hambuch T M, Sawyers C L. The Bcr-Abl leukemia oncogene activates Jun kinase and requires Jun for transformation. Proc Natl Acad Sci USA. 1995;92:11746–11750. doi: 10.1073/pnas.92.25.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigues G A, Park M, Schlissinger J. Activation of the JNK pathway is essential for transformation by the Met oncogene. EMBO J. 1997;16:2634–2645. doi: 10.1093/emboj/16.10.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner E F. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsang N M, Nagasawa H, Li C, Little J B. Abrogation of p53 function by transfection of HPV16 E6 gene enhances the resistance of human diploid fibroblasts to ionizing radiation. Oncogene. 1995;10:2403–2408. [PubMed] [Google Scholar]
- 41.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 42.Wisdom R, Johnson R S, Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X S, Vanderziel C, Bennett C F, Monia B P. A role for c-Raf kinase and Ha-Ras in cytokine-mediated induction of cell adhesion molecules. J Biol Chem. 1998;273:33230–33238. doi: 10.1074/jbc.273.50.33230. [DOI] [PubMed] [Google Scholar]
- 44.Yang D D, Conze D, Whitmarsh A J, Barrett T, Davis R J, Rincón M. Differentiation of CD4+ T cell to Th1 cells requires MAP kinase JNK2. Cell. 1998;9:575–585. doi: 10.1016/s1074-7613(00)80640-8. [DOI] [PubMed] [Google Scholar]