Abstract
The serine/threonine kinase Cot is a member of the mitogen-activated protein kinase (MAPK) kinase kinase family implicated in cellular transformation. Enhanced expression of this protein has been shown to activate both the MAPK and the c-Jun N-terminal kinase (JNK) pathways and to stimulate the nuclear factor of activated T cells and NF-κB-dependent transcription. However, the nature of the normal functions of the Cot protein and the molecular mechanisms responsible for its oncogenic potential are still largely unknown. Here, we show that overexpression of the cot proto-oncogene is sufficient to stimulate the expression of c-jun and that, in turn, the activity of c-Jun is required for Cot-induced transformation. These observations prompted us to explore the molecular events by which Cot regulates c-jun expression. We found that Cot potently stimulates the activity of the c-jun promoter utilizing JNK-dependent and -independent pathways, the latter involving two novel members of the MAPK family, p38γ (ERK6) and ERK5. Molecularly, this activity was found to be dependent on the ability of Cot to activate, in vivo, members of each class of the MAPK kinase superfamily, including MEK, SEK, MKK6, and MEK5. Furthermore, the use of dominant interfering molecules revealed that Cot requires JNK, p38s, and ERK5 to stimulate the c-jun promoter fully and to induce neoplastic transformation. These findings indicate that Cot represents the first example of a serine/threonine kinase acting simultaneously on all known MAPK cascades. Moreover, these observations strongly suggest that the transforming ability of Cot results from the coordinated activation of these pathways, which ultimately converge on the regulation of the expression and activity of the product of the c-jun proto-oncogene.
The serine/threonine protein kinase Cot was originally identified as a carboxy-terminally truncated protein encoded by the cot oncogene, which was isolated by cellular transformation assays upon transfection of a human thyroid carcinoma cell line DNA onto hamster cells (49). Similarly, the rat cot homolog, designated tpl-2, was identified as a target for provirus insertion in Moloney murine leukemia virus-induced rat T-cell lymphomas, which resulted in the enhanced expression of a carboxy-terminally truncated kinase (11, 41, 56). In addition, the cot proto-oncogene, designated est, was also isolated as a highly transforming gene when using NIH 3T3 cells as the recipient for a human cDNA expression library (12), thus indicating that both gene rearrangement and overexpression may unmask the oncogenic potential of the Cot protein.
Interestingly, the cot gene appears to be highly expressed in a number of tissues, including the spleen, thymus, liver, and lung (56), and is also expressed at lower levels in many other tissues and cell lines (54, 56). Furthermore, mitogenic stimuli such as concanavalin A (56), inflammatory mediators such as interleukin-1 (IL-1) (12), and tumor promoters such as okadaic acid (12) were all shown to potently induce the expression of cot transcripts in a variety of cell types. However, the normal functions of the Cot protein, as well as the molecular mechanisms responsible for its oncogenic potential, are still poorly defined.
In this regard, Cot has been shown to participate in the transcriptional regulation of several important genes, including those for tumor necrosis factor alpha and IL-2 (5, 6, 67). Recently available evidence suggests that Cot is also an integral component of signaling pathways that control the proteolytic processing of the NF-κB1 p105 protein (7) and is able to stimulate NF-κB-dependent transcription through the interaction and activation of the NF-κB-inducing kinase (NIK) (40). In addition, it has been shown that Cot potently stimulates both (i) the mitogen-activated protein kinase (MAPK) pathway by acting downstream from Ras (63) and (ii) the c-Jun N-terminal kinase (JNK) pathway by acting upstream of JNK kinases (30, 63).
Nuclear transcription factors are often the final target of signal-transducing kinase cascades, thereby converting membrane and cytoplasmic signaling events into specific changes in gene expression (66). In this regard, members of the AP-1 (activating protein 1) family of transcription factors are frequently regulated at the transcriptional and posttranscriptional levels by MAPKs. AP-1 complexes have been shown to be necessary for cell cycle progression in several cell systems (39, 51) and for cellular transformation by a variety of oncogenes, including src, ras, and raf (55, 65). Members of the AP-1 family of transcription factors are usually classified into two subfamilies, namely, the Jun (c-Jun, JunB, and JunD) (4, 61, 62) and the Fos (c-Fos, FosB, Fra-1, and Fra-2) (13, 44, 52, 73) families. Homodimerization of Jun proteins or heterodimerization between proteins of the two subfamilies (59) or with other transcription factors, including the ATF2, CREB, NFAT, and SMAD proteins (8, 53, 74), confers on the complexes the ability to recognize specific DNA sequences known as tetradecanoyl phorbol acetate-responsive elements or AP-1 sites (2, 50). Through these elements, the different complexes regulate the expression of many cellular genes, including those for collagenase, metallothionein IIA, stromelysin, IL-2, and transforming growth factor β (45). Interestingly, the c-jun gene itself contains AP-1 sites in its promoter region, thus being the target for a positive autoregulatory loop (1) which is dependent on the ability of JNK to phosphorylate the c-Jun transactivation domain and increase its transcriptional activity (21, 36, 48). However, recent data from our laboratory and others indicate the existence of JNK-independent pathways regulating c-jun expression (16, 34, 42).
In this study, we show that overexpression of the cot proto-oncogene is sufficient to stimulate the expression of c-jun and that the activity of c-Jun, in turn, is required for Cot-induced transformation. In this regard, we found that Cot potently stimulates the activity of the c-jun promoter by enhancing the enzymatic activity of JNK and two novel members of the MAPK family, p38γ (ERK6) and ERK5, through the stimulation of their respective MAPK kinases, SEK, MKK6, and MEK5, respectively. Furthermore, the activity of these MAPK kinases was found to be required for full stimulation of the c-jun promoter by Cot and for Cot-induced neoplastic transformation. Thus, these findings indicate that Cot represents the first example of a serine/threonine kinase acting simultaneously on all known MAPK cascades and that its biological activity results from the coordinated activation of these pathways, which ultimately converge on the regulation of the expression and activity of the product of the c-jun proto-oncogene.
MATERIALS AND METHODS
Cell lines.
NIH 3T3 fibroblasts were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% calf serum.
DNA constructs.
A plasmid encoding a luc gene driven by a wild-type murine c-jun promoter was kindly provided by R. Prywes (33). Plasmids pJC6, pJTX, pJSX, and pJSTX are pBLCAT3-based reporter constructs carrying a chloramphenicol acetyltransferase (CAT)-encoding reporter gene controlled by the wild-type murine c-jun promoter and its mutant forms as previously described (33). The pCEV27Cot (est) expression vector used for the focus-forming assays has already been described (12). The Cot coding sequence was transferred to the pCDNAIIIB expression vector to obtain the pCDNAIIICot plasmid. An expression plasmid for a kinase-deficient mutant form of Cot, Cot KR, was obtained by replacing a lysine residue in position 167 with arginine by PCR-mediated site-directed mutagenesis. ERK5, p38α, p38γ, and p38δ cDNAs were amplified by the PCR technique using human skeletal muscle cDNA (Clontech, Inc.) as the template. The sequences of the oligonucleotides utilized are available upon request. The amplified DNA fragments were subcloned in pCEFL, a modified pcDNAIII expression vector containing the elongation factor 1 promoter driving the expression of an in-frame N-terminal tag of nine amino acids derived from the influenza virus hemagglutinin HA1 protein (HA) (72). The expression vectors containing HA-tagged MAPK and JNK have been previously described (15, 18). MEK, expressed as a glutathione S-transferase (GST) fusion protein, was cloned as a BamHI/NotI fragment in the pEBG vector. SEK-1, expressed as a GST fusion protein from the vector pEBG, was kindly provided by L. I. Zon (64). PCR-amplified MKK6 cDNA was cloned as a BamHI/NotI fragment in a pCEFLGST vector. MEK5 cDNA was obtained from Kevin Walton at Cephalon Inc. and subcloned in pCEFL and in pCEFLGST as a BamHI/NotI fragment. pCEFL MEK5DD and MEK5AA, dominant active and dominant negative forms of MEK5, respectively, were obtained by site-directed mutagenesis (QuickChange Site-Directed Mutagenesis; Stratagene), replacing serine 311 and threonine 315 with aspartate or alanine, respectively. A kinase-deficient mutant form of MKK6, MKK6KR, was obtained using the same method, replacing a lysine residue in position 82 with arginine (58, 64). Raf CAAX-, MEKEE-, and MEKK1-containing expression vectors have already been described (15, 16). The expression vector for Raf CAAX, pEF Raf CAAX, was kindly provided by Andrew Larner. Dominant negative MEK1 was generated by replacing Ser-218 and Ser-222 with alanine; this construct was designated pCDNAIIIMEKAA (17). The pCDNAIII JIP-1-expressing vector was kindly provided by R. Davis (23). The c-Jun dominant negative expression vector pCEFLAU5c-JunTAM67 was obtained by cloning the c-jun gene lacking amino acids 3 to 122 (9) into the pCEFLAU5 vector. The pCEFLAU5 RasV12 expression vector was obtained by cloning murine Ha-RasV12 into the pCEFLAU5 vector. The pAP-1 and pSRE luciferase reporters were obtained from Stratagene. The transactivation domain of the Elk1 (amino acids) 307 to 428) (58) transcription factor was subcloned as a Gal4 fusion protein in a pCDNAIII vector containing the DNA-binding domain of the yeast transcription factor Gal4. TATA-Gal4-driven luciferase reporter plasmid pGal4 Luc was constructed by inserting six copies of a Gal4 responsive element and a TATA oligonucleotide to replace the simian virus 40 minimal promoter in the pGL3 vector (Promega). A GST-MEF2C fusion protein carrying amino acids 174 to 327 of the MEF2C transactivation domain was obtained by PCR using murine MEF2C cDNA as the template and oligonucleotides 5′-GGG GAT CCC CGT CTC TGC AGA GGA AT-3′ and 5′-TAG AAT TCA GCC AGA CAG AGA GGG ACA-3′. The amplified DNA fragment was cloned between the BamHI and EcoRI sites of pGEX4T-3 (Pharmacia) in frame with the GST-encoding gene. The GST-ATF2 fusion protein has been already described (15).
Focus-forming assays.
NIH 3T3 cells were transfected by the calcium phosphate precipitation technique with different expression plasmids together with 1 μg of pCDNAIIIβ-gal, a plasmid expressing the enzyme β-galactosidase, and the total amount of plasmid DNA was adjusted with empty vector. The day after transfection, cells were washed in medium supplemented with 5% calf serum and then maintained in the same medium until foci were scored 2 to 3 weeks later. Duplicate plates were fixed with a phosphate-buffered saline (PBS) solution containing 2% (vol/vol) formaldehyde and 0.2% (vol/vol) glutaraldehyde and stained at 37°C for β-galactosidase activity with a PBS solution containing 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 0.1% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to score transfection efficiency.
Reporter gene assays.
NIH 3T3 cells were transfected by the calcium phosphate precipitation technique with different expression plasmids together with 1 μg of pCDNAIIIβ-gal and 1 μg of each of the reporter plasmids. After 24 h of incubation, the cells were washed and kept for 24 h in serum-free Dulbecco's modified Eagle's medium. Cells were then lysed using reporter lysis buffer (Promega). Luciferase activity present in cellular lysates was assayed using d-luciferin and ATP as substrates, and light emission was quantitated using a Monolight 2010 luminometer as specified by the manufacturer (Analytical Luminescence Laboratory). CAT activity was assayed in the cell extracts by incubation at 37°C for 10 to 16 h in the presence of 0.25 μCi of [14C]chloramphenicol (100 mCi/mmol; ICN) per sample and 200 μg of butyryl coenzyme A (Sigma) per ml in 0.25 M Tris-HCl, pH 7.4. Labeled butyrylated products were extracted using a mixture of xylenes (Aldrich) and counted as previously described (42). β-Galactosidase activity present in each sample was assayed by a colorimetric assay and used to normalize for transfection efficiency.
Kinase assays.
NIH 3T3 cells were transfected by Lipofectamine Plus Reagent in accordance with the manufacturer's (Life Technologies, Inc.) instructions; with different expression plasmids. The phosphorylating activity of epitope-tagged MAPK and JNK was previously described (14). Briefly, cells were seeded at 10% confluence and 2 days later were incubated in serum-free medium overnight for MAPK or for 2 h for JNK, p38α, p38γ, p38δ, and ERK5. After serum starvation, cells were washed with cold PBS and lysed at 4°C in a buffer containing 25 mM HEPES (pH 7.5)–0.3 M NaCl–1.5 mM MgCl2–0.2 mM EDTA–0.5 mM dithiothreitol–20 mM β-glycerophosphate–1 mM vanadate–1% NP-40–1 mM phenylmethylsulfonyl fluoride (PMSF)–20 μg of aprotinin per ml–20 μg of leupeptin per ml. Cleared lysates containing HA-tagged kinases were immunoprecipitated at 4°C for 2 h with anti-HA monoclonal antibody (MAb) HA.11 (Berkeley Antibody Company). Immunocomplexes were recovered with the aid of protein G-Sepharose (Pharmacia Biotech). Beads were washed three times with PBS containing 1% NP-40 and 2 mM vanadate, once with 100 mM Tris (pH 7.5)–0.5 M LiCl, and once with kinase reaction buffer (25 mM HEPES [pH 7.6], 20 mM β-glycerophosphate, 20 mM MgCl2, 0.5 mM sodium fluoride, 0.1 mM vanadate, 2 mM dithiothreitol). Samples were then resuspended in 30 μl of kinase reaction buffer containing 1 μCi of [γ-32P]ATP per reaction and 20 μM of unlabeled ATP. After 20 min at 30°C, the reactions were terminated by addition of 10 μl of 5× Laemmli buffer. In vitro kinase assays were performed using as substrates 1.5 μg of myelin basic protein (MBP; Sigma) per μl for MAPK, p38α, and p38γ; 1 μg of purified, bacterially expressed GST-ATF2 for JNK and p38δ; and GST-MEF2C protein for ERK5. Samples were analyzed by sodium dodecyl sulfate–12% (or 15% for MBP) polyacrylamide gel electrophoresis, and autoradiography was performed with the aid of an intensifying screen.
Bacterial expression of GST fusion proteins.
The BL21Lys strain of Escherichia coli was transformed with the vector pGEX-4T3 encoding the fusion protein GST-ATF2 or GST-MEF2C. The transformed bacteria were grown in 500 ml of Luria-Bertani medium until the optical density was 0.5, at which time isopropyl-β-d-thiogalactopyranoside (1 mM final concentration) was added and the mixture was incubated for 3 h. The cells were collected by centrifugation at 3,000 × g for 30 min and resuspended in 10 ml of PBS–1% Triton X-100–1 mM EDTA–2 μg of aprotinin per ml–2 μg of leupeptin per ml–1 mM PMSF. The cell suspension was sonicated, and cellular debris was removed by centrifugation at 10,000 × g for 15 min. The supernatant was mixed with 300 μl of glutathione-agarose beads (Pharmacia Biotech, Piscataway, N.J.) and centrifuged at 3,000 × g for 5 min. The pellet was washed three times with PBS–1% Triton X-100–1 mM EDTA–2 μg of aprotinin per ml–2 μg of leupeptin per ml–1 mM PMSF and then twice with PBS–2 μg of aprotinin per ml–2 μg of leupeptin per ml–1 mM PMSF. Finally, purified fusion proteins were eluted in 50 mM Tris–10 mM glutathione–2 μg of aprotinin per ml–2 μg of leupeptin per ml–1 mM PMSF.
Western blot analysis and antibodies.
Expression of HA-tagged MAPK, JNK1, ERK5, p38α, p38γ, and p38δ cDNAs in transfected NIH 3T3 cells was analyzed by Western blotting after sodium dodecyl sulfate-polyacryalmide gel electrophoresis using anti-HA MAb HA.11 (Berkeley Antibody Company). Epitope-tagged proteins were visualized by enhanced chemiluminescence detection (Amersham Corp.) using goat anti-mouse immunoglobulin G (IgG) coupled to horseradish peroxidase as the secondary antibody (Cappel). Rabbit polyclonal antisera to the phospho-MKK6 and phospho-SEK1 proteins were purchased from New England Biolabs, Inc. Specific polyclonal antibodies to phosphoserine were purchased from Zymed Laboratories. Rabbit anti-Tpl-2/Cot antibodies were purchased from Santa Cruz Biotechnology, Inc.
Indirect immunofluorescence.
NIH 3T3 cells were transfected by Lipofectamine Plus Reagent in accordance with the manufacturer's (Life Technologies, Inc.) instructions. Cells serum starved for 24 h were washed twice with PBS and then fixed with 4% formaldehyde and 5% sucrose in PBS for 10 min and permeabilized with 0.5% Triton X-100 in PBS for 10 min. The cells were incubated with an anti-c-Jun MAb (Santa Cruz Biotecnology, Inc.) and an anti-Tpl2/Cot (Santa Cruz Biotechnology, Inc.) or anti-MEKK1 antibody for 1 h and washed three times with PBS and then with a 1:100 dilution of fluorescein-conjugated goat anti-mouse F(ab′)2 IgG and tetramethyl rhodamine isocyanate-conjugated anti-rabbit F(ab′)2 IgG antibodies (Jackson ImmunoResearch Laboratories, Inc.). Coverslips were mounted in Gel-Mount (Biomeda Corp., Foster City, Calif.) containing p-phenylenediamine (ICN) at 1 mg/ml to inhibit photobleaching and viewed using a Zeiss Axiophot photomicroscope equipped with an epifluorescence detector. Immunofluorescence was photographed using Kodak TMAX 3200 film.
RESULTS
Cot enhances c-Jun expression.
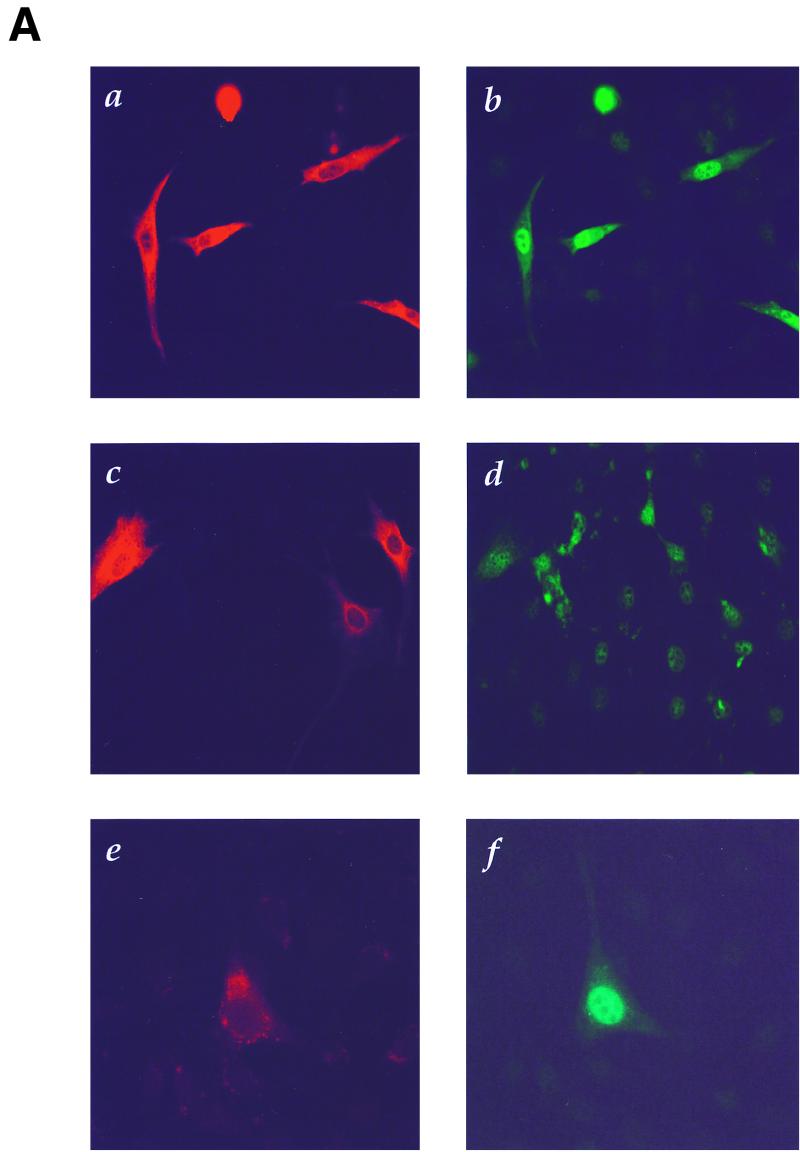
c-Jun is a nuclear proto-oncoprotein whose expression is stimulated by a variety of growth-promoting agents and activated oncogenes (35, 59). Thus, we investigated if overexpression of the Cot proto-oncogene, known to transform NIH 3T3 cells (12), is also able to affect c-Jun protein levels. For these experiments, NIH 3T3 cells were transfected with a Cot expression vector, fixed, and analyzed by double immunofluorescence using specific anti-Cot and anti-c-Jun antibodies. As shown in Fig. 1A, only cells overexpressing Cot (panel a) demonstrated marked nuclear staining with specific anti-c-Jun antibodies (panel b), indicative of an increase in the endogenous c-Jun protein level. This effect was dependent on the Cot kinase activity, as its kinase-deficient mutant, CotKR, was unable to stimulate c-Jun expression (panel d), even if expressed at levels comparable to those of the wild-type protein (panel c). Nearly identical results were obtained when MEKK1, a potent activator of the JNK pathway (47) and of c-jun expression (16), was transfected as a positive control (panels e and f). However, transfection of the vector alone or expression plasmids for the green fluorescent protein or an activated form of Raf did not induce c-Jun expression (data not shown).
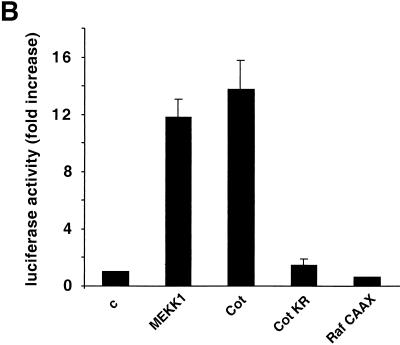
FIG. 1.
Cot kinase enhances c-Jun expression. (A) NIH 3T3 cells were transfected with a wild-type Cot (a and b)-, CotKR (c and d)-, or MEKK1 (e and f)-expressing vector (0.5 μg). At 24 h after transfection, cells were transferred into serum-free medium for an additional 24 h, fixed, and then analyzed by double immunofluorescence with specific anti-c-Jun (b, d, and f) and anti-Cot/Tpl2 (a and c) or anti-MEKK1 (e) antibodies. (B) NIH 3T3 cells were transfected with the reporter plasmid pJLuc, which encodes a luc gene driven by the wild-type c-jun promoter. These cells were also cotransfected with expression vectors (1 μg) encoding the MEKK1 Cot, and Raf proteins, as indicated. At 24 h after transfection, cells were transferred to serum-free medium for an additional 24 h, lysed, and then analyzed for luciferase activity as described in Materials and Methods. Luciferase activity was normalized with respect to that of vector-transfected cells, whose value was taken as 1. β-Galactosidase activity resulting from the cotransfection of a pCDNAIII β-galactosidase expression vector was used to normalize the luciferase values to the corresponding transfection efficiencies. The results shown are averages ± the standard errors of triplicate samples from a typical experiment. Similar results were obtained in three independent experiments. c, control.
To investigate whether the increased level of c-Jun protein upon Cot overexpression results from direct stimulation of the c-jun promoter, we took advantage of the availability of a reporter plasmid carrying the luc gene under the control of the murine c-jun promoter (34). Cotransfection of this reporter with the Cot cDNA in NIH 3T3 cells revealed that Cot can strongly induce the activity of the c-jun promoter (Fig. 1B), to an extent comparable to that induced by MEKK1. However, the kinase-deficient mutant form of Cot, CotKR, failed to elicit a transcriptional response (Fig. 1B). As an additional control, Raf CAAX, a constitutively active form of Raf, a MAPK kinase kinase (MAPKKK) acting on the MAPK pathway (27), failed to stimulate the c-jun promoter (Fig. 1B), further confirming that activation of MAPK is not sufficient to regulate the activity of this promoter (16, 42). These data therefore indicate that Cot can regulate the expression of c-Jun and suggest that c-Jun, in turn, represents a potential candidate to mediate the biological activities elicited by Cot.
A dominant negative mutant form of c-Jun inhibits Cot-induced transformation of NIH 3T3 cells.
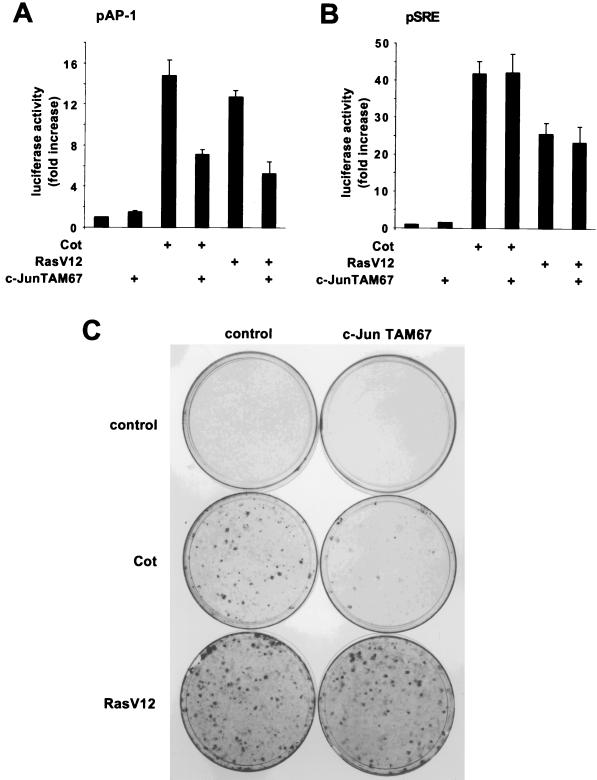
Cellular transformation is the best-characterized biological effect resulting from Cot overexpression and/or carboxy-terminal truncation (3, 11, 12, 26, 56). We thus decided to investigate if the c-Jun/AP-1 transcription factor might participate in Cot-induced cellular transformation. For these experiments, we used a dominant negative c-Jun protein which consisted of a c-Jun molecule with NH2 deleted that still binds DNA but is incapable of transactivation (9, 10). First, we confirmed the specificity of this dominant negative protein. As shown in Fig. 2A, Cot overexpression readily enhanced the transcription of a luc reporter gene under the control of tandemly repeated AP-1 responsive elements, probably through JNK activation (63). Accordingly, coexpression of the c-Jun dominant negative protein strongly reduced Cot-induced transactivation of the AP-1 responsive element (Fig. 2A). As expected, a similar effect of the dominant negative c-Jun protein on RasV12 stimulation of the AP-1 reporter element was observed (Fig. 2A). In contrast, no inhibition by c-Jun TAM67 on Cot or RasV12 stimulation of a serum responsive element-driven reporter was observed (Fig. 2B), confirming the specificity of c-Jun TAM67 as a dominant negative molecule for the c-Jun/AP-1 transcription factor. We next decided to assess the effect of this mutant on Cot-induced cellular transformation. Indeed, whereas transfection of the Cot proto-oncogene into NIH 3T3 cells readily induced the appearance of foci of transformation after 2 to 3 weeks of culture (12), cotransfection with dominant negative c-Jun caused a remarkable inhibition of Cot transforming potential (Fig. 2C). However, in agreement with previous results (37, 43), this mutant c-Jun did not interfere with RasV12-induced transformation of NIH 3T3 cells (Fig. 2C). In each case, a plasmid carrying the β-galactosidase-encoding gene was included in the transfection mixture and parallel plates were stained for β-galactosidase expression, showing comparable transfection efficiencies (data not shown). Taken together, these results suggest that the activity of c-Jun is necessary for the transforming ability of Cot.
FIG. 2.
The c-Jun TAM67 dominant negative protein inhibits Cot-induced transformation of NIH 3T3 cells. (A) NIH 3T3 cells were transfected with the reporter plasmid pAP-1, which encodes a luc gene under the control of six repeated AP-1 responsive elements. The cells were also cotransfected with Cot (1 μg) and RasV12 (1 μg) expression vectors, alone or in combination with a c-Jun TAM67-encoding plasmid (1 μg). Cells were lysed, and luciferase activities were measured as described in Materials and Methods. The data represent luciferase activity normalized to the β-galactosidase activity present in each sample, expressed as fold induction relative to the control, and are averages ± the standard errors of triplicate samples from a typical experiment. Similar results were obtained in three independent experiments. (B) Same as in panel A, using as a reporter plasmid pSRE, which encodes a luc gene under the control of five repeated serum responsive elements. Similar results were obtained in three independent experiments. (C) NIH 3T3 cells were transfected with plasmid pCEV27Cot (0.5 μg) or pCEFLAU5RasV12 (0.5 μg), alone or in combination with pCEFLAU5c-JunTAM67 (0.5 μg). Cells were cultured for 3 weeks in 5% calf serum, fixed, and then stained as described in Materials and Methods. Representative plates for all of the transfections are shown.
JNK-dependent and -independent pathways contribute to the stimulation of the c-jun promoter by Cot.
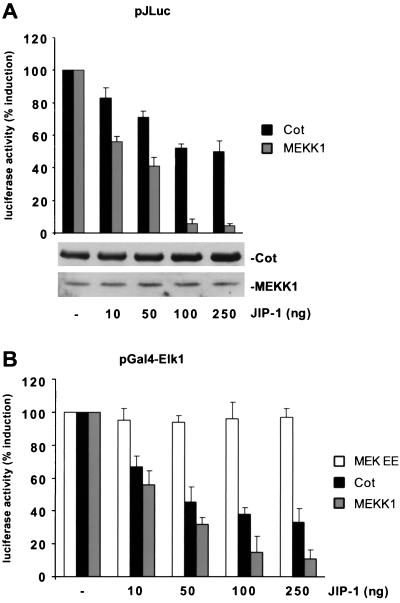
Posttranslational modifications of the c-Jun transactivating domain through JNK phosphorylation have been recognized as a central mechanism of AP-1-mediated regulation of the c-jun promoter itself (1). To test whether the activation of JNK by Cot mediates the stimulation of the c-jun promoter, we took advantage of the observation that overexpression of JNK-interacting protein 1 (JIP-1) blocks the nuclear translocation of JNK, thereby impeding JNK-dependent gene expression (23, 69). For these experiments, we performed parallel cotransfections of Cot and MEKK1 cDNAs together with increasing amounts of the JIP-1 expression plasmid and then evaluated the effect of JIP-1 on expression from the c-jun promoter-driven reporter plasmid. As shown in Fig. 3A, JIP-1 caused strong, dose-dependent inhibition of the MEKK1-induced response. This inhibition was almost complete at the highest doses tested (100 to 250 ng). In contrast, increasing amounts of JIP-1 caused a more modest inhibition of Cot-induced c-jun promoter stimulation (Fig. 3A), suggesting the existence of both JNK-dependent and -independent pathways linking Cot to the c-jun promoter. Equal levels of expression of the Cot and MEKK1 proteins, upon cotransfection with increasing amounts of the JIP-1 expression plasmid, were confirmed by Western blot analysis (Fig. 3A).
FIG. 3.
JNK-dependent and -independent pathways contribute to the stimulation of the c-jun promoter by Cot. (A) NIH 3T3 cells were cotransfected with the pJLuc reporter plasmid, the expression vectors for Cot (1 μg) and MEKK1 (0.5 μg), and increasing amounts of the pCDNAIIIJIP-1 plasmid, as indicated. Cells were then lysed and analyzed as described in Materials and Methods. Resulting luciferase activities were then expressed as percentages of the values from Cot (or MEKK1)-transfected cells. Protein expression in cellular lysates was determined by Western blotting with appropriate antibodies, as indicated. (B) Same as in panel A but using a Gal4-driven luciferase-expressing reporter plasmid to score MEKK1-induced activation of a chimerical Gal4 Elk1 protein. The results shown are averages ± the standard errors of triplicate samples from a typical experiment. Similar results were obtained in three independent experiments.
To further confirm the specificity of the JIP-1 inhibitory activity on the JNK pathway, we performed a similar dose-response analysis testing the effect of this protein on the transactivation of a Gal4-Elk1 fusion protein. The transcription factor Elk1 is a substrate for activating phosphorylation by several members of the MAPK family, including MAPK (28), JNK (71), and p38α (70). The Gal4-Elk1 fusion protein, in which the Elk1 transactivating domain is fused to the DNA-binding domain of the yeast transcription factor Gal4, can respond to the activating effects of each of these kinases when assessed with a luc reporter gene under the control of 6× Gal4-responsive elements and a minimum TATA promoter (pGal4). As shown in Fig. 3B, both a constitutive active form of MEK1, MEKEE (17), and the JNK stimulator MEKK1 (47) activated reporter gene expression. The MEKK1-induced activation of Gal4-Elk1 was readily blocked in a dose-dependent manner by JIP-1 cotransfection, but increasing amounts of the JIP-1 expression plasmid did not affect this response when elicited by MEKEE (Fig. 3B), confirming the specificity of JIP-1 on the JNK pathway in our cellular setting. In addition, JIP-1 cotransfection partially affected the transcriptional activation of Gal4-Elk1 by Cot (Fig. 3B). Taken together, these data support the specificity of the use of JIP-1 overexpression as an approach to evaluate the contribution of JNK to transcriptional regulation. They also further support the existence of both JNK-dependent and -independent pathways participating in the transduction of signals from Cot to the c-jun promoter.
A MEF2 responsive element is critical for induction of the c-jun promoter by Cot.
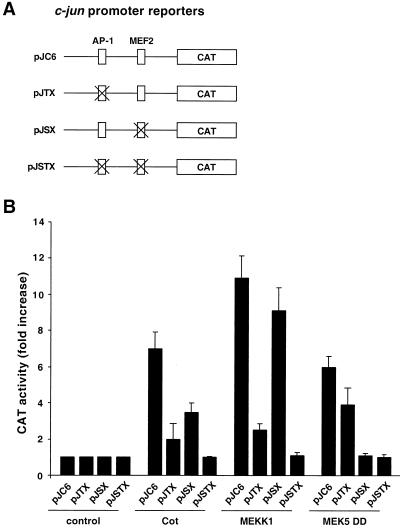
The activity of the c-jun promoter is tightly regulated on key regulatory elements (1, 33, 42). Among them, an AP-1/ATF site at position −71 to −64 and a MEF2 site at position −59 to −50 appear to be critical for the integration of signals transmitted by distinct MAPK signaling pathways (16, 38, 42). More specifically, whereas the JNK pathway regulates the activity of the AP-1/ATF site, the ERK5 pathway acts on the MEF2 responsive element (42). Conversely, different members of the p38 family of MAPK are able to regulate the c-jun promoter by acting on both responsive elements (42). Based on the previous evidence that both JNK-dependent and -independent pathways participate in the induction of the c-jun promoter by Cot, we decided to examine the relative contribution of the AP-1/ATF and MEF2 sites to this transcriptional response. To this end, we used reporter plasmids encoding the CAT-encoding gene under the control of the wild-type c-jun promoter (pJC6) or mutant promoters lacking a functional AP-1/ATF (pJTX) or MEF2 (pJSX) responsive element or both (pJSTX) (33) (Fig. 4A).
FIG. 4.
Distinct regulatory elements on the c-jun promoter mediate its stimulation by Cot. (A) Schematic representation of the reporter for the wild-type murine c-jun promoter (pJC6) and for its derivatives mutated in the AP-1 (pJTX), MEF2 (pJSX), and AP-1/MEF2 (pJSTX) regulatory elements. The crosses represent the sites at which the different reporters were mutated. (B) The pJC6, pJTX, pJSX, and pJSTX reporter plasmids and pCDNAIIIβ-gal were transiently cotransfected into NIH 3T3 cells together with the indicated plasmids (1 μg each); in the case of MEK5DD, an expression vector for ERK5 was also cotransfected (1 μg). At 24 h after transfection, cells were transferred to serum-free medium for an additional 24 h, lysed, and then analyzed for CAT activity as described in Materials and Methods. The data represent CAT activity normalized to the β-galactosidase activity present in each sample, expressed as fold induction relative to that of the corresponding gene fluorescent protein-transfected control, and are averages ± the standard errors of triplicate samples from a typical experiment. Similar results were obtained in three independent experiments.
As shown in Fig. 4B, Cot strongly stimulated the pJC6 reporter plasmid, to an extent similar to that observed with the luc-based reporter construct (see above). When the AP-1/ATF-deficient reporter (pJTX) was tested, the stimulation induced by Cot was strongly affected (Fig. 4B). Surprisingly, however, the mutation in the MEF2 responsive element (pJSX) also had a dramatic effect on the Cot-induced response. As controls, we confirmed, in parallel experiments, that the stimulation of the JNK and ERK5 pathways by overexpression of MEKK1 and an active MEK5 protein, MEK5DD (38), respectively, readily induces CAT expression from the wild-type c-jun promoter (Fig. 4B). Mutations in the AP1/ATF site (pJTX) abolished the response to MEKK1, while these mutations had a much more limited effect on the activation of the c-jun promoter by MEK5DD (Fig. 4B). Work in progress is aimed to address the contribution of transcription factors binding the AP-1/ATF site to the stimulation of the c-jun promoter by the ERK5 pathway. Nonetheless, mutations in the MEF2 site (pJSX) prevented the enhanced expression from the reporter plasmid provoked by MEK5DD but not when induced by MEKK1 (Fig. 4B), supporting a critical role for the MEF2 responsive element in the transcriptional response elicited by ERK5. Furthermore, a double AP-1/MEF2 mutant reporter (pJSTX) was not stimulated by any of these kinases (Fig. 4B). Taken together, these data indicated that signaling pathways acting on MEF2 transcription factors might participate in the activation of the c-jun promoter by Cot, thus raising the possibility that ERK5 or p38 family members act downstream from this oncoprotein.
Novel members of the MAPK family can be activated by Cot.
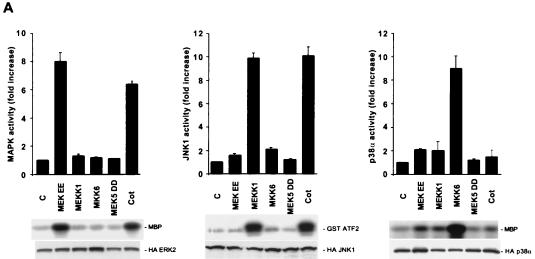
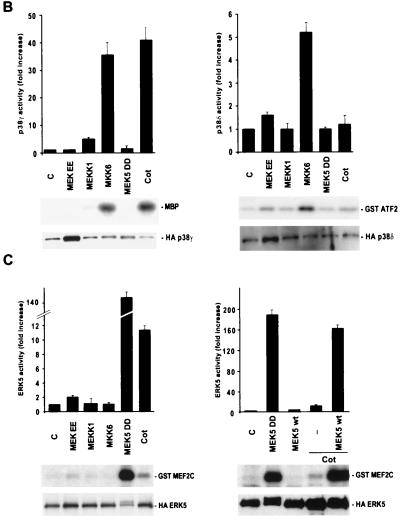
Overexpression of Cot is able to potently activate both the MAPK and JNK pathways in Cos-7 cells, being the activity of p38α only modestly affected (63). However, it has been observed that MAPK does not affect the activity of the c-jun promoter (16) and that Cot can stimulate this promoter utilizing JNK-independent pathways, probably involving kinases such as p38 family members and/or ERK5, involved in the regulation of MEF2 transcription factors (see above). To address this possibility, we transiently cotransfected NIH 3T3 cells with expression vectors containing the Cot cDNA and HA-tagged forms of MAPK, JNK, p38α, and the more recently identified members of the MAPK family p38γ (46), p38δ (29), and ERK5 (75) (Fig. 5). As a positive control, we used MEKEE (15) for MAPK, MEKK1 (47) for JNK, MKK6 for members of the p38 family of kinases (19, 29, 32), and an activated form of MEK5, MEK5DD, for ERK5 (38). After transfection into NIH 3T3 cells, each HA-tagged kinase was immunoprecipitated from cellular lysates with anti-HA antibodies and assayed for kinase activity using MBP as the substrate for MAPK, p38α, and p38γ or bacterially expressed GST-MEF2C for ERK5 and GST-ATF2 for JNK and p38δ.
FIG. 5.
Activation of members of the MAPK family by Cot. (A) NIH 3T3 cells were transfected with 1 μg of pCDNAIIIHAMAPK, pCDNAIIIHAJNK1, or pCEV29HAp38α. For each of these kinases, cells were cotransfected with the vector alone or 1 μg of pCDNAIIIMEKEE, pCEV29MEKK1, pCEFLGSTMKK6, pCEFLAU1MEK5DD, or pCDNAIIICot. Cells were then lysed, and kinase assays were performed as described in Materials and Methods. 32P-labeled substrates are indicated. Phosphorylation of the respective substrates (MBP for MAPK and p38α and GST-ATF2 for JNK1) was quantified by PhosphorImager analysis and is reported in the histograms as kinase activity relative to that of vector-transfected cells, whose value was taken as 1. The data represent the means of four independent experiments. (B) pCEFLHAp38γ (1 μg)- and pCEFLHAp38δ (1 μg)-transfected NIH 3T3 cells were cotransfected and analyzed for kinase activity as described for panel A, using MBP and GST-ATF2 as substrates, respectively. Resulting kinase activities were normalized as described for panel A. (C) HA epitope-tagged ERK5 was cotransfected in NIH 3T3 cells with the indicated expression vectors. Kinase activity was measured as described in Materials and Methods, using GST-MEF2C as the substrate. Normalized results are expressed as described above. C, control.
As shown in Fig. 5A, both MAPK and JNK were potently activated by Cot overexpression in NIH 3T3 cells, whereas no specific effect was observed for p38α or p38δ kinase activity (Fig. 5A and B), both of which were potently stimulated by MKK6 under identical experimental conditions. Interestingly, however, we observed that Cot overexpression was sufficient to stimulate p38γ potently, to an extent even greater than that caused by its upstream activator, MKK6 (Fig. 5B). Cot also activated a novel member of the MAPK superfamily, ERK5, causing a more-than-10-fold increase in its kinase activity (Fig. 5C). In this case, however, the stimulation of ERK5 by Cot was more limited than that caused by the activated form of MEK5, MEK5DD. To explain this difference, we reasoned that the availability of endogenous MEK5 protein might be the limiting step for transducing the Cot signal to the ERK5 kinase. We speculated that if this was the case, then overexpression of wild-type MEK5 protein might overcome this limitation, thus allowing the full activation of ERK5 by Cot. Indeed, as shown in Fig. 5C, overexpression of wild-type MEK5, which itself had no demonstrable effect on ERK5 activity, dramatically enhanced the response to Cot. These data indicated that ERK5 can be regulated by Cot, thus establishing Cot as the first ERK5 kinase kinase. Furthermore, these observations suggested that Cot possesses the unique ability to act simultaneously as an upstream activator of multiple MAPK pathways.
Cot activates in vivo distinct members of the MEK superfamily of MAPK kinases.
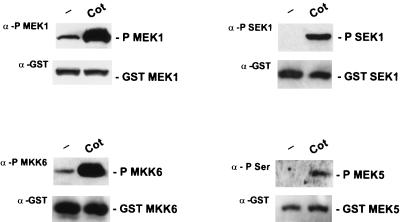
The high degree of homology between the Cot protein and both yeast and mammalian MAPKKKs (63) and its ability to phosphorylate in vitro and activate MEK1 and SEK1 (63) suggest that the Cot protein can itself be considered a MAPKKK. Activation of MEKs by upstream kinases involves the phosphorylation of specific serine and/or threonine residues; in turn, these dual-specificity kinases phosphorylate threonine and tyrosine residues within the TXY motif of the corresponding MAPK (see references 20 and 22 for a review). To investigate whether the activity of the Cot kinase results in the in vivo phosphorylation of different MEKs, we took advantage of the existence of anti-phospho-specific antibodies recognizing the serine and threonine residues responsible for the activation of these proteins (22). As shown in Fig. 6, MEK, SEK1, and MKK6 anti-phospho-specific antibodies revealed that, upon Cot expression, cotransfected MEK, SEK1, and MKK6 were heavily phosphorylated in vivo, at levels comparable to those of the corresponding positive controls (data not shown). MEK5 also presents a conserved S311XXXT315 motif (25, 75), whose phosphorylation has been demonstrated to be necessary for its kinase activity (38). As no anti-phospho-specific antibodies for MEK5 are available, we assessed the status of MEK5 phosphorylation in response to Cot overexpression using antiphosphoserine antibodies upon affinity precipitation of a coexpressed GST-tagged MEK5 protein (Fig. 6). Under these experimental conditions, the serine phosphorylation of MEK5 was greatly enhanced by Cot. These results suggest that, in addition to stimulating the MAPK and JNK pathways, Cot phosphorylates MKK6 and MEK5 in vivo, thus resulting in their activation. Taken together, these observations strongly suggest that the stimulation of several members of the MAPK superfamily by Cot is mediated by its ability to activate their corresponding MAPK kinases, including MEK, SEK1, MKK6, and MEK5.
FIG. 6.
Cot activates several members of the MEK family of kinases in vivo. Plasmids encoding GST-tagged MEK, SEK1, MKK6, and MEK5 (1 μg of each) were transfected by the Lipofectamine Plus method into NIH 3T3 cells without (minus sign) or with the pCDNAIIICot-expressing vector (1 μg). At 24 h after transfection, cells were lysed and subjected to affinity precipitation with glutathione-Sepharose beads. Precipitates were analyzed by Western blotting using specific anti-phospho-MEK1 (New England Biolabs Inc.), anti-phospho-SEK1 (New England Biolabs Inc.), anti-phospho-MKK6 (New England Biolabs Inc.), and antiphosphoserine (Zymed Laboratories) antibodies, as indicated. Specific anti-GST antibodies were used to confirm that there were no significant differences in the relative amounts of the different GST-tagged kinases.
The p38 and ERK5 pathways are involved in the regulation of c-jun promoter activity by Cot.
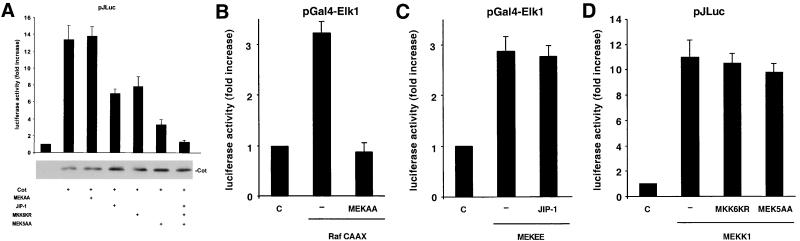
We next decided to explore whether the p38 and ERK5 kinases play a role in the JNK-independent pathway linking Cot to the c-jun promoter, using dominant interfering molecules specific for each MAPK pathway as an experimental approach. As shown in Fig. 7A, both MKK6KR and MEK5AA were able to diminish Cot-induced c-jun promoter activity, to an extent comparable to that observed with JIP-1. None of these molecules affected Cot expression (Fig. 7A). Combination of these dominant negative interfering molecules completely abolished Cot stimulation of the c-jun promoter (Fig. 7A). In contrast, the dominant negative form of MEK1, MEKAA, did not affect this response (Fig. 7A), although it blocked the activation of the Gal4-Elk1 reporter by Raf CAAX (Fig. 7B). As additional confirmation of the specificity of MEKAA, no demonstrable effect of this protein on MEKK1 and MKK6 activation of the Gal4-Elk1 reporter was observed (data not shown). The absence of nonspecific effects of MKK6KR and MEK5AA on pJLuc reporter activity was also confirmed, as they did not exert any demonstrable effect on the activation of this reporter by MEKK1 (Fig. 7D). Similarly, no effect of JIP-1 on MEKEE-induced activation of the Gal4-Elk1 chimera was observed (Fig. 7C and see above). Altogether, these data suggest that at least three MAPK, JNK, p38 (most likely the p38γ isoform), and ERK5, participate in the regulation of the c-jun promoter by Cot.
FIG. 7.
p38γ- and ERK5-dependent pathways participate in the regulation of the c-jun promoter by Cot. (A and D) A pJLuc reporter plasmid and pCDNAIIIβ-gal were transiently cotransfected into NIH 3T3 cells together with the indicated plasmids (1 μg of each; 250 ng of JIP-1). Cells were lysed, and luciferase activities were measured as described in Materials and Methods. In panel A, the level of expression of the Cot protein was also analyzed by Western blotting. (B and C) Same as in panel A but using a Gal4-driven luciferase-expressing reporter plasmid (1 μg) to score Raf CAAX (B)- or MEKEE (C)-induced activation of a chimeric Gal4-Elk1 protein. The data represent luciferase activity normalized to the β-galactosidase activity present in each sample, expressed as fold induction relative to that of the control, and are averages ± the standard errors of triplicate samples from a typical experiment. Similar results were obtained in three independent experiments. C, control; −, empty vector.
The JNK, p38, and ERK5 signaling pathways are integral components of the transforming pathway elicited by Cot in NIH 3T3 cells.
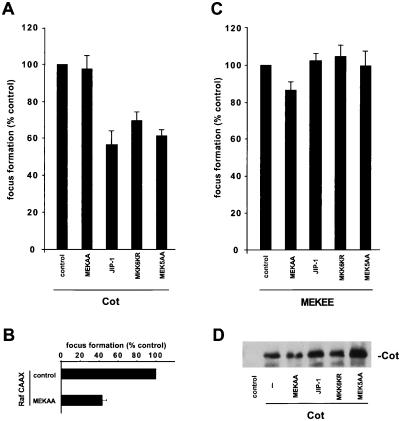
In view of the role of JNK, p38, and ERK5 in the activation of the c-jun promoter by Cot, we next asked whether these kinases also participate in the transforming ability of Cot when expressed in NIH 3T3 cells. For that, we cotransfected the Cot expression plasmid together with dominant negative proteins for each of the known MAPK signaling pathways: MEKAA, JIP-1, MKK6KR, and MEK5AA. As shown in Fig. 8A, molecules interfering with the JNK, p38, and ERK5 signaling pathways reduced the focus-forming activity of Cot in NIH 3T3 cells. In contrast, MEKAA displayed a very limited effect, although it effectively diminished the transforming ability of an activated form of Raf, a MEK kinase (Fig. 8B). As additional controls, none of these dominant interfering molecules affected the focus-forming activity of an activated form of MEK, MEKEE (15) (Fig. 8C) or Cot protein expression (Fig. 8D). In each case, cells were also cotransfected with a plasmid encoding the gene for β-galactosidase and parallel plates were fixed and stained for β-galactosidase activity, showing no substantial differences in transfection efficiency (data not shown). These data indicate that activation of JNK, p38s (probably p38γ), and ERK5 contributes to the transforming potential of the Cot oncoprotein.
FIG. 8.
Involvement of the JNK, p38γ, and ERK5 signaling pathways in Cot-induced transformation. (A) NIH 3T3 cells were transfected with 0.5 μg of the indicated plasmids. Cultures were maintained in 5% calf serum for approximately 3 weeks, fixed, and then stained as described in Materials and Methods. Foci were counted, and results are expressed as percentages of the number of foci observed in plates transfected with Cot alone. (B and C) Same as in panel A but using expression plasmids for Raf CAAX and MEKEE, respectively, as transformating genes. The data shown represent averages ± the standard errors of three independent experiments. (D) NIH 3T3 cells were transfected with 0.5 μg of the Cot expression vector, alone or in combination with plasmids expressing the indicated proteins, and cellular lysates were immunoblotted with specific anti-Cot antibodies. −, empty vector.
DISCUSSION
The complexity of the mechanisms controlling the activity of regulatory elements found in the promoter region of growth-regulating genes, including c-jun and c-fos, has just begun to be appreciated. These elements appear to be under the control of a number of transcription factors, each regulated by one or more members of the MAPK superfamily of proline-directed kinases (66). The transcriptional activation or repression of these genes would then be expected to result from the coordinated activity of multiple kinase cascades, indicating that the transcriptional regulation of these genes represents an important site for signal integration. Specifically for c-jun, recently available evidence suggests that, in addition to the autoregulatory loop by which c-Jun regulates its own expression (1), another transcription factor, MEF2, plays a critical role in the regulation of the activity of the c-jun promoter (16). The transactivating activity of these transcription factors may be tightly regulated by distinct MAPKs: c-Jun is phosphorylated by JNK (21), while MEF2 transcription factors have been demonstrated to be substrates for the p38α, p38γ, and ERK5 MAPKs (31, 38, 42).
Cot has been shown to stimulate both MAPK and JNK, but only the latter is able to regulate the activity of the c-jun promoter (16). However, we observed that inhibition of the JNK pathway exerts only a partial effect on the stimulation of the c-jun promoter by Cot. In addition, mutations in the MEF2 responsive element, whose regulation appears to be independent of the JNK pathway (16), were found to affect deeply the Cot-induced response, thus raising the possibility that Cot activates additional members of the MAPK family, which, in turn, may control the activity of the c-jun promoter. Indeed, both p38γ and ERK5 were potently activated by Cot overexpression, while no activation was observed for p38α and p38δ. Furthermore, the use of dominant negative mutant forms of their respective upstream kinases helped to establish a role for ERK5 and p38s in the regulation of c-jun promoter activity by Cot. From these observations and the remarkable inhibition of Cot-induced focus formation by a dominant negative c-Jun protein, we can conclude that this transcription factor and the multiple MAPK pathways controlling c-jun expression may play a key role as part of the transforming pathway elicited by Cot (Fig. 9).
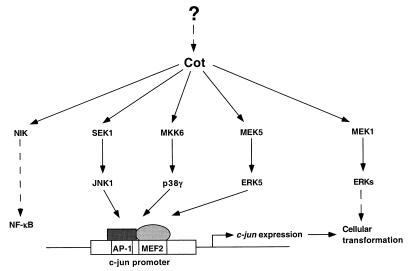
FIG. 9.
Schematic representation of MAPK signaling pathways activated by Cot, leading to c-jun promoter activation. Extracellular stimuli, such as mitogens (56) or inflammatory cytokines (12), may stimulate Cot expression, in turn leading to activation of different MAPK signaling pathways and to cell type-specific cellular responses. Overexpression of Cot may constitutively activate these signaling pathways, most of which converge on the regulation of the c-jun proto-oncogene, thereby increasing c-Jun expression, leading to cellular transformation. NIK, NFκB-inducing kinase.
An intriguing issue is why Cot stimulates MKK6 but this results in the stimulation of p38γ but not of p38α and p38δ. As we have observed that Cot forms stable complexes in vivo with cotransfected GST-MKK6 (our unpublished observations), it is possible that these complexes result in a distinct ability of MKK6 to activate each p38 isoform, p38γ being the best such downstream target. Additionally, it is possible that the Cot-MKK6 complex can also specifically associate with yet to be identified scaffolding proteins, which may discriminate among p38 family members. An example of such a mechanism has already been described in the yeast Saccharomyces cerevisiae, where a MEKK homolog, Ste11, can be used both in the response to high osmolarity and in mating, which results in the activation of HOG1 and FUS3, respectively (24, 57). Two scaffolding proteins, Ste5 and Pbs2, have been shown to be responsible for maintaining the necessary specificity between these two pathways, thus avoiding dangerous cross talks (24, 57). These possibilities, as well as additional possibilities that might help to explain the surprising specificity of the Cot-MKK6 complex for p38γ in vivo, are under investigation.
It is of interest that p38γ has recently been found to activate both the Jun/ATF and the MEF2 regulatory elements in the c-jun promoter (42) and that the MEK5-ERK5 pathway has recently been shown to regulate the activity of members of the MEF2 class of transcription factors (38, 42). Indeed, we have observed that Cot can potently enhance the transcriptional activity of chimeric molecules containing the DNA-binding domain of Gal4 and the transactivating domain of ATF2 through JNK and p38s and that Cot stimulates expression from reporter plasmids under the control of a MEF2 responsive element through ERK5 (data not shown). In line with these observations, mutations in the Jun/ATF and MEF2 regulatory elements of the c-jun promoter strongly affected its ability to be activated by Cot. Thus, we can conclude that the remarkable ability of Cot to stimulate the c-jun promoter and c-Jun expression may result from the concomitant activation of JNK, p38γ, and ERK5 and the consequent stimulation of the activity of transcription factors acting on the distinct regulatory elements within the c-jun promoter.
Typically, molecules controlling the activity of MAPKs are organized in discrete signaling units, constituted by modules of sequentially acting protein kinases: a MAPKKK stimulating a MAPK kinase that, in turn, activates a MAPK (20, 60). Furthermore, a growing number of scaffolding protein might help to ensure the specificity of signal transmission (57). In this regard, our present findings suggest that Cot represents the first example of a MAPKKK which is able to activate several MAPK signaling pathways. At the molecular level, this activity may be dependent on the ability to form complexes in vivo and to phosphorylate and activate different members of the MAPK kinase superfamily, including MEK, SEK, MKK6, and MEK5. This unique biochemical activity might help explain many of the described functions of Cot (5, 6, 30, 67, 68). Furthermore, it may also provide a novel molecular mechanism whereby extracellular stimuli might simultaneously activate more than one MAPK cascade, thereby enhancing the available repertoire of biochemical pathways by which signals are transmitted to the nucleus. In this scenario, stimuli inducing Cot expression (12, 56) would be expected to provoke long-term activation of multiple MAPK signaling pathways. The precise nature of such stimuli, as well as that of the molecules controlling the expression and activity of Cot in physiological, as well as in pathological, situations warrants further investigation.
REFERENCES
- 1.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 2.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 3.Aoki M, Akiyama T, Miyoshi J, Toyoshima K. Identification and characterization of protein products of the cot oncogene with serine kinase activity. Oncogene. 1991;6:1515–1519. [PubMed] [Google Scholar]
- 4.Ball A R, Jr, Bos T J, Loliger C, Nagata L P, Nishimura T, Su H, Tsuchie H, Vogt P K. Jun: oncogene and transcriptional regulator. Cold Spring Harbor Symp Quant Biol. 1988;53:687–693. doi: 10.1101/sqb.1988.053.01.078. [DOI] [PubMed] [Google Scholar]
- 5.Ballester A, Tobena R, Lisbona C, Calvo V, Alemany S. Cot kinase regulation of IL-2 production in Jurkat T cells. J Immunol. 1997;159:1613–1618. [PubMed] [Google Scholar]
- 6.Ballester A, Velasco A, Tobena R, Alemany S. Cot kinase activates tumor necrosis factor-alpha gene expression in a cyclosporin A-resistant manner. J Biol Chem. 1998;273:14099–14106. doi: 10.1074/jbc.273.23.14099. [DOI] [PubMed] [Google Scholar]
- 7.Belich M P, Salmeron A, Johnston L H, Ley S C. TPL-2 kinase regulates the proteolysis of the NF-kappaB-inhibitory protein NF-kappaB1 p105. Nature. 1999;397:363–368. doi: 10.1038/16946. [DOI] [PubMed] [Google Scholar]
- 8.Benbrook D M, Jones N C. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- 9.Brown P H, Alani R, Preis L H, Szabo E, Birrer M J. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene. 1993;8:877–886. [PubMed] [Google Scholar]
- 10.Brown P H, Chen T K, Birrer M J. Mechanism of action of a dominant-negative mutant of c-Jun. Oncogene. 1994;9:791–799. [PubMed] [Google Scholar]
- 11.Ceci J D, Patriotis C P, Tsatsanis C, Makris A M, Kovatch R, Swing D A, Jenkins N A, Tsichlis P N, Copeland N G. Tpl-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Genes Dev. 1997;11:688–700. doi: 10.1101/gad.11.6.688. [DOI] [PubMed] [Google Scholar]
- 12.Chan A M, Chedid M, McGovern E S, Popescu N C, Miki T, Aaronson S A. Expression cDNA cloning of a serine kinase transforming gene. Oncogene. 1993;8:1329–1333. [PubMed] [Google Scholar]
- 13.Cohen D R, Curran T. fra-1: a serum-inducible, cellular immediate-early gene that encodes a Fos-related antigen. Mol Cell Biol. 1988;8:2063–2069. doi: 10.1128/mcb.8.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coso O A, Chiariello M, Kalinec G, Kyriakis J M, Woodgett J, Gutkind J S. Transforming G protein-coupled receptors potently activate JNK (SAPK). Evidence for a divergence from the tyrosine kinase signaling pathway. J Biol Chem. 1995;270:5620–5624. doi: 10.1074/jbc.270.10.5620. [DOI] [PubMed] [Google Scholar]
- 15.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 16.Coso O A, Montaner S, Fromm C, Lacal J C, Prywes R, Teramoto H, Gutkind J S. Signaling from G protein-coupled receptors to the c-jun promoter involves the MEF2 transcription factor. Evidence for a novel c-jun amino-terminal kinase-independent pathway. J Biol Chem. 1997;272:20691–20697. doi: 10.1074/jbc.272.33.20691. [DOI] [PubMed] [Google Scholar]
- 17.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 18.Crespo P, Xu N, Simonds W F, Gutkind J S. Ras-dependent activation of MAP kinase pathway mediated by G-protein beta gamma subunits. Nature. 1994;369:418–420. doi: 10.1038/369418a0. [DOI] [PubMed] [Google Scholar]
- 19.Cuenda A, Cohen P, Buee-Scherrer V, Goedert M. Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6): comparison of the specificities of SAPK3 and SAPK2 (RK/p38) EMBO J. 1997;16:295–305. doi: 10.1093/emboj/16.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis R J. MAPKs: new JNK expands the group. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 21.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 22.Dhanasekaran N, Premkumar Reddy E. Signaling by dual specificity kinases. Oncogene. 1998;17:1447–1455. doi: 10.1038/sj.onc.1202251. [DOI] [PubMed] [Google Scholar]
- 23.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 24.Elion E A. Routing MAP kinase cascades. Science. 1998;281:1625–1626. doi: 10.1126/science.281.5383.1625. [DOI] [PubMed] [Google Scholar]
- 25.English J M, Vanderbilt C A, Xu S, Marcus S, Cobb M H. Isolation of MEK5 and differential expression of alternatively spliced forms. J Biol Chem. 1995;270:28897–28902. doi: 10.1074/jbc.270.48.28897. [DOI] [PubMed] [Google Scholar]
- 26.Erny K M, Peli J, Lambert J F, Muller V, Diggelmann H. Involvement of the Tpl-2/cot oncogene in MMTV tumorigenesis. Oncogene. 1996;13:2015–2020. [PubMed] [Google Scholar]
- 27.Fanger G R, Gerwins P, Widmann C, Jarpe M B, Johnson G L. MEKKs, GCKs, MLKs, PAKs, TAKs, and tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- 28.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb M H, Shaw P E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goedert M, Cuenda A, Craxton M, Jakes R, Cohen P. Activation of the novel stress-activated protein kinase SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6): comparison of its substrate specificity with that of other SAP kinases. EMBO J. 1997;16:3563–3571. doi: 10.1093/emboj/16.12.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagemann D, Troppmair J, Rapp U R. Cot protooncoprotein activates the dual specificity kinases MEK-1 and SEK-1 and induces differentiation of PC12 cells. Oncogene. 1999;18:1391–1400. doi: 10.1038/sj.onc.1202431. [DOI] [PubMed] [Google Scholar]
- 31.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 32.Han J, Lee J D, Jiang Y, Li Z, Feng L, Ulevitch R J. Characterization of the structure and function of a novel MAP kinase kinase (MKK6) J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 33.Han T-H, Lamph W W, Prywes R. Mapping of epidermal growth factor-, serum-, and phorbol ester-responsive sequence elements in the c-jun promoter. Mol Cell Biol. 1992;12:4472–4477. doi: 10.1128/mcb.12.10.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han T-H, Prywes R. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol Cell Biol. 1995;15:2907–2915. doi: 10.1128/mcb.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herschman H R. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 36.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 37.Janulis M, Silberman S, Ambegaokar A, Gutkind J S, Schultz R M. Role of mitogen-activated protein kinases and c-Jun/AP-1 trans-activating activity in the regulation of protease mRNAs and the malignant phenotype in NIH 3T3 fibroblasts. J Biol Chem. 1999;274:801–813. doi: 10.1074/jbc.274.2.801. [DOI] [PubMed] [Google Scholar]
- 38.Kato Y, Kravchenko V V, Tapping R I, Han J, Ulevitch R J, Lee J D. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovary K, Bravo R. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin X, Cunningham J, Mu Y, Geleziunas R, Greene W C. The proto-oncogene Cot kinase participates in CD3/CD28 induction of NF-κB acting through the NF-κB-inducing kinase and IκB kinases. Immunity. 1999;10:271–280. doi: 10.1016/s1074-7613(00)80027-8. [DOI] [PubMed] [Google Scholar]
- 41.Makris A, Patriotis C, Bear S E, Tsichlis P N. Genomic organization and expression of Tpl-2 in normal cells and Moloney murine leukemia virus-induced rat T-cell lymphomas: activation by provirus insertion. J Virol. 1993;67:4283–4289. doi: 10.1128/jvi.67.7.4283-4289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marinissen M J, Chiariello M, Pallante M, Gutkind J S. A network of mitogen-activated protein kinases links G protein-coupled receptors to the c-jun promoter: a role for c-Jun NH2-terminal kinase, p38s, and extracellular signal-regulated kinase 5. Mol Cell Biol. 1999;19:4289–4301. doi: 10.1128/mcb.19.6.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshall-Heyman H, Engel G, Ljungdahl S, Shoshan M C, Svensson C, Wasylyk B, Linder S. Tumorigenic and metastatic properties of two ras-oncogene transfected rat fibrosarcoma cell lines defective in c-jun. Oncogene. 1994;9:3655–3663. [PubMed] [Google Scholar]
- 44.Matsui M, Tokuhara M, Konuma Y, Nomura N, Ishizaki R. Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene. 1990;5:249–255. [PubMed] [Google Scholar]
- 45.Mechta F, Lallemand D, Pfarr C M, Yaniv M. Transformation by ras modifies AP1 composition and activity. Oncogene. 1997;14:837–847. doi: 10.1038/sj.onc.1200900. [DOI] [PubMed] [Google Scholar]
- 46.Mertens S, Craxton M, Goedert M. SAP kinase-3, a new member of the family of mammalian stress-activated protein kinases. FEBS Lett. 1996;383:273–276. doi: 10.1016/0014-5793(96)00255-4. [DOI] [PubMed] [Google Scholar]
- 47.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 48.Minden A, Lin A, Smeal T, Dérijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyoshi J, Higashi T, Mukai H, Ohuchi T, Kakunaga T. Structure and transforming potential of the human cot oncogene encoding a putative protein kinase. Mol Cell Biol. 1991;11:4088–4096. doi: 10.1128/mcb.11.8.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakabeppu Y, Ryder K, Nathans D. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell. 1988;55:907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- 51.Nishikura K, Murray J M. Antisense RNA of proto-oncogene c-fos blocks renewed growth of quiescent 3T3 cells. Mol Cell Biol. 1987;7:639–649. doi: 10.1128/mcb.7.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishina H, Sato H, Suzuki T, Sato M, Iba H. Isolation and characterization of fra-2, an additional member of the fos gene family. Proc Natl Acad Sci USA. 1990;87:3619–3623. doi: 10.1073/pnas.87.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Northrop J P, Ullman K S, Crabtree G R. Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J Biol Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- 54.Ohara R, Miyoshi J, Aoki M, Toyoshima K. The murine cot proto-oncogene: genome structure and tissue-specific expression. Jpn J Cancer Res. 1993;84:518–525. doi: 10.1111/j.1349-7006.1993.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okuno H, Suzuki T, Yoshida T, Hashimoto Y, Curran T, Iba H. Inhibition of jun transformation by a mutated fos gene: design of an anti-oncogene. Oncogene. 1991;6:1491–1497. [PubMed] [Google Scholar]
- 56.Patriotis C, Makris A, Bear S E, Tsichlis P N. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc Natl Acad Sci USA. 1993;90:2251–2255. doi: 10.1073/pnas.90.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 58.Raingeaud J, Whitmarsh A J, Barrett T, Derijard B, Davis R J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ransone L J, Verma I M. Nuclear proto-oncogenes fos and jun. Annu Rev Cell Biol. 1990;6:539–557. doi: 10.1146/annurev.cb.06.110190.002543. [DOI] [PubMed] [Google Scholar]
- 60.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 61.Ryder K, Lanahan A, Perez-Albuerne E, Nathans D. jun-D: a third member of the jun gene family. Proc Natl Acad Sci USA. 1989;86:1500–1503. doi: 10.1073/pnas.86.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryder K, Lau L F, Nathans D. A gene activated by growth factors is related to the oncogene v-jun. Proc Natl Acad Sci USA. 1988;85:1487–1491. doi: 10.1073/pnas.85.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salmeron A, Ahmad T B, Carlile G W, Pappin D, Narsimhan R P, Ley S C. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 1996;15:817–826. [PMC free article] [PubMed] [Google Scholar]
- 64.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki T, Murakami M, Onai N, Fukuda E, Hashimoto Y, Sonobe M H, Kameda T, Ichinose M, Miki K, Iba H. Analysis of AP-1 function in cellular transformation pathways. J Virol. 1994;68:3527–3535. doi: 10.1128/jvi.68.6.3527-3535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 67.Tsatsanis C, Patriotis C, Bear S E, Tsichlis P N. The Tpl-2 protooncoprotein activates the nuclear factor of activated T cells and induces interleukin 2 expression in T cell lines. Proc Natl Acad Sci USA. 1998;95:3827–3832. doi: 10.1073/pnas.95.7.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsatsanis C, Patriotis C, Tsichlis P N. Tpl-2 induces IL-2 expression in T-cell lines by triggering multiple signaling pathways that activate NFAT and NF-kappaB. Oncogene. 1998;17:2609–2618. doi: 10.1038/sj.onc.1202460. [DOI] [PubMed] [Google Scholar]
- 69.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 70.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 71.Whitmarsh A J, Yang S H, Su M S, Sharrocks A D, Davis R J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 73.Zerial M, Toschi L, Ryseck R P, Schuermann M, Muller R, Bravo R. The product of a novel growth factor activated gene, fos B, interacts with JUN proteins enhancing their DNA binding activity. EMBO J. 1989;8:805–813. doi: 10.1002/j.1460-2075.1989.tb03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Feng X H, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 75.Zhou G, Bao Z Q, Dixon J E. Components of a new human protein kinase signal transduction pathway. J Biol Chem. 1995;270:12665–12669. doi: 10.1074/jbc.270.21.12665. [DOI] [PubMed] [Google Scholar]