Following exit from mitosis, RNA Pol II is required for reestablishing 3D genome folding by loading cohesin complexes onto DNA.
Abstract
Mammalian chromosomes are three-dimensional entities shaped by converging and opposing forces. Mitotic cell division induces marked chromosome condensation, but following reentry into the G1 phase of the cell cycle, chromosomes reestablish their interphase organization. Here, we tested the role of RNA polymerase II (RNAPII) in this transition using a cell line that allows its auxin-mediated degradation. In situ Hi-C showed that RNAPII is required for both compartment and loop establishment following mitosis. RNAPs often counteract loop extrusion, and in their absence, longer and more prominent loops arose. Evidence from chromatin binding, super-resolution imaging, and in silico modeling allude to these effects being a result of RNAPII-mediated cohesin loading upon G1 reentry. Our findings reconcile the role of RNAPII in gene expression with that in chromatin architecture.
INTRODUCTION
The evolution and expansion of chromosome conformation capture (3C) technologies (1) has profoundly renewed our understanding of the spatial organization of eukaryotic chromosomes and of how it underlies their function and maintenance (2, 3). It is now well accepted that chromosomes are dynamic entities (4), and that their dynamics result from converging and opposing forces acting on chromatin (5). These forces include tethering to nuclear landmarks like lamina or the nucleolus (6), the interplay between transcription factor (TF)–bound cis-elements (7), and the dynamic extrusion of loops via cohesin complexes (5, 8).
Cohesin-extruded loops, almost invariably anchored at convergent CTCF-bound sites, are found along mammalian chromosomes representing a prominent feature of three-dimensional (3D) genome organization. The combination of high-resolution Hi-C with acute and reversible degradation (9) of chromatin-organizing factors has shed light on loop emergence. CTCF degradation causes loss of insulation at thousands of topologically associated domain (TAD) boundaries (10). Cohesin depletion leads to the elimination of essentially all CTCF-anchored loops (11, 12). Depletion of the cohesin-release factor WAPL promotes loop enlargement and aberrant looping by also engaging nonconvergent CTCF-bound anchors (13). These observations, together with the recently documented ability of cohesin to extrude loops in vitro (14, 15) and the finding that CTCF-STAG interactions protect cohesin from chromatin release (16, 17), have crystallized a model for how architectural loops form and dissolve.
In addition to cohesin, another molecular motor known for its ability to translocate DNA is the RNA polymerase (RNAP) (18). However, its contribution to chromatin folding is still debated. Different lines of evidence point to a connection between RNAPII binding to chromatin and the differential formation of spatial interactions. To cite some recent examples, allele-specific Hi-C showed that the mouse inactive X chromosome lacks active/inactive compartments and TADs, which, however, form around “escapee” genes and in the active allele (19); the transcriptional state of variably sized domains across eukaryotes, from Caenorhabditis elegans and Drosophila melanogaster to Arabidopsis thaliana and mammals, is a robust predictor of interactions mapped via Hi-C and explains chromatin partitioning to a great extent (20, 21); and TAD emergence coincides with transcriptional activation in zygotes (22). Pharmacological abrogation of transcription compacts chromatin (23) and weakens, but does not alleviate, TAD boundaries (22, 24), and treating native (25) or fixed nuclei (24) with RNase A does not affect TADs but eliminates specific contacts. Single-nucleosome imaging upon acute RNAPII depletion showed that polymerases act to constrain and direct chromatin movement in 3D space (26), compatible with the idea of transcription-based chromatin organization.
In contrast, RNAPII and Mediator-complex components were found to be dispensable for bringing cis-elements into spatial proximity (27) and inhibition of transcription in parallel with RAD21 reintroduction in cohesin-depleted cells did not affect loop reestablishment (11). Nevertheless, CTCF or cohesin depletion from mammalian cells had rather limited impact on gene expression (10, 11), and upon cohesin elimination, a comparable number of loops formed on the basis of chromatin identity (11) or did not dissolve at all (28). Most recently, Micro-C, a sub-kbp (kilobase pair) Hi-C variant, unveiled thousands of fine-scale loops connecting transcriptionally active loci in mouse and human cells, often without association to CTCF/cohesin (29). Thus, the direct effects of active RNAPs on chromatin folding remain unclear.
On top of its potentially direct effects, RNAPs and the act of transcription may remodel genome folding via interplay with cohesin-CTCF complexes. For example, transcription can relocate cohesin by many kilobases (30). This transcription-mediated displacement can even disrupt prominent CTCF loops and rewire spatial interactions (31, 32). In addition, RNAPs are essential for domain formation and often counteracted by condensin complexes (33, 34). This and other data highlight the need to dissect and reconcile the contribution of RNAPII to chromatin organization. To this end, and as pharmacological inhibition of RNAPs is inefficient, we exploited a human cell line that allows rapid and reversible RNAPII depletion (26). We combined in situ Hi-C and super-resolution imaging of RNAPII with in silico models to disentangle the role of RNAPII in gene expression from that in genome architecture to reconcile the aforementioned observations.
RESULTS
Acute RNAPII depletion affects loop-level interphase chromatin folding
RNAPII is essential for cell viability, so its depletion may only be transient. Thus, we exploited a human DLD-1 colorectal cancer line, in which the largest RNAPII subunit, RPB1, is N-terminally tagged with a mini-AID (mAID) domain. This allows for its acute and reversible degradation upon addition of auxin [and of doxycycline to activate the plant ubiquitin ligase TIR1 recognizing this mAID domain; see (9, 26)]. In our hands, 2 hours of dox/auxin treatment reduce RNAPII protein levels by >60%, while 14 hours of treatment result in >80% degradation as assessed by Western blotting—without affecting RNAPI or RNAPIII levels (Fig. 1A and fig. S1A). Degradation is less impactful on chromatin-embedded RNAPs (e.g., those marked by phosphorylated Ser5 residues in their C-terminal domain). Washing out auxin in the presence of its competitive inhibitor, auxinole, largely restores RNAPII-Ser5 levels (fig. S1A), suggesting that soluble (nonphosphorylated) RNAPII is more susceptible to degradation and that any residual polymerases will be chromatin bound [as was the case for CTCF-mAID; see (10)]. Thus, quantitative RNAPII removal can be achieved via this system to assess its contribution to genome folding.
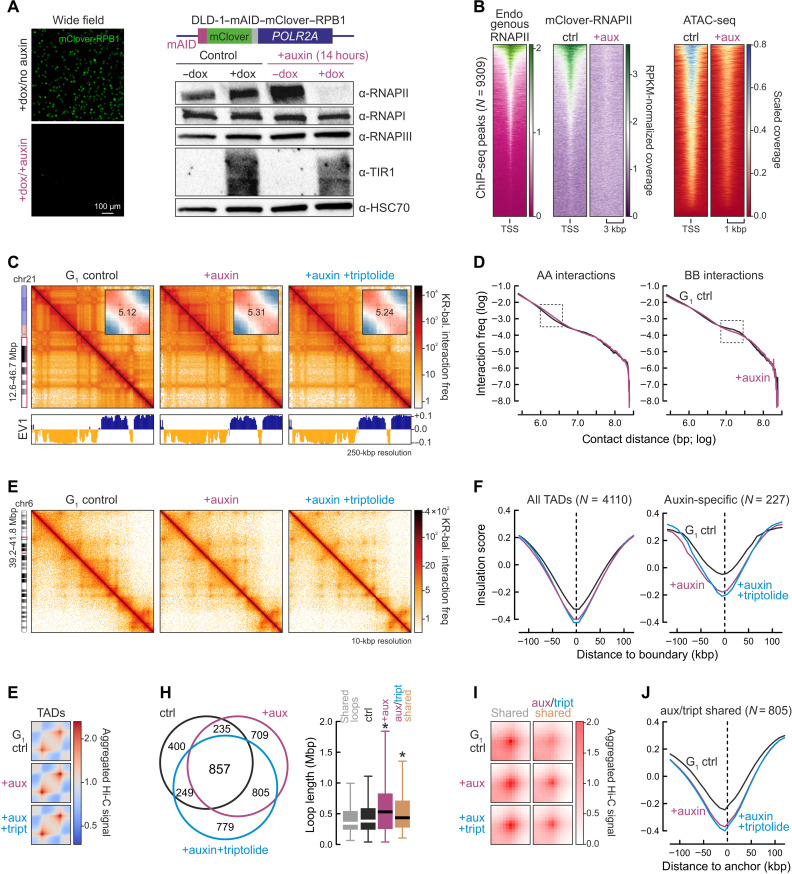
Fig. 1. Effects of acute RNAPII degradation on interphase chromatin folding.
(A) Left: DLD-1–mAID–RPB1 cells (schematic) degrade RNAPII upon 14-hour doxycycline (dox)/auxin treatment. Right: TIR1-mediated RNAPII degradation confirmed by Western blotting; HSC70 levels provide a control. (B) Heatmaps showing mClover-RNAPII ChIP-seq signal loss upon auxin treatment (middle) overlapping RNAP-bound positions in parental DLD-1 (left) concomitant with decreasing accessibility (right). (C) The 250-kbp resolution Hi-C maps from G1-sorted control (left), auxin-treated (middle), and auxin + triptolide–treated cells (right) aligned to first eigenvector values (EV1). Insets: Saddle plots showing compartment insulation. (D) Plots showing Hi-C interaction frequency decay in A- or B-compartments as a function of genomic distance in control (black) or auxin-treated cells (purple). Rectangles indicate distances where values deviate most. (E) The 10-kbp resolution Hi-C maps from control (left), auxin-treated (middle), and auxin + triptolide–treated cells (right). (F) Line plots showing mean insulation from control (black), auxin-treated (purple), and auxin + triptolide–treated cells (blue) around all or degron-specific TAD boundaries. N, number of TADs queried. (G) Heatmaps showing aggregated TAD-level interactions in control (top), auxin-treated (middle), and auxin + triptolide–treated cells (bottom). (H) Left: Venn diagram showing shared and unique loops for control (black), auxin-treated (purple), and auxin + triptolide–treated Hi-C (blue). Right: Loop lengths displayed as boxplots (right). *P < 0.01, Wilcoxon-Mann-Whitney test. (I) Plots showing aggregated Hi-C signal for the loops in (I). (J) As in (F), but for loop anchors shared by auxin- and auxin + triptolide–treated cells. The Hi-C data presented in (C) to (J) are from individual replicates.
To further characterize this line, we performed chromatin immunoprecipitation sequencing (ChIP-seq) using an antibody targeting the mClover tag in RPB1. Compared to public RNAPII ChIP-seq data from DLD-1 (Gene Expression Omnibus: GSM2769059), mClover-tagged polymerases occupied the same positions and could be depleted from chromatin genome-wide upon auxin treatment (Fig. 1B). Polymerase degradation was accompanied by a strong decrease in chromatin accessibility at Transcription start sites (TSSs) (Fig. 1B), similar to that recently seen using mouse embryonic stem cells (mESCs) (35). We also queried the H3K27ac (marking active chromatin) and H3K27me3 histone modifications (marking facultative heterochromatin). Upon RNAPII depletion, significant H3K27ac reduction was observed concomitant with increased H3K27me3 levels (fig. S1, B and C). Last, we monitored changes in nascent RNA levels using “factory” RNA sequencing (RNA-seq) (36). Control and auxin-treated samples separated well in principal components analysis (PCA) plots, with ~1500 genes changing their nascent transcription levels significantly (fig. S1, D and E). Of these, >90% were down-regulated and mainly involved in chromatin assembly and gene expression regulation (fig. S1F).
We next asked whether the spatial organization of interphase chromatin is also altered upon RNAPII depletion. We applied in situ Hi-C to G1-sorted DLD-1–RPB1–mAID cells treated or not with auxin for 14 hours or to cells in which auxin was complemented by triptolide treatment [an inhibitor abrogating transcriptional initiation to further enhance RNAPII degradation; see (37)]. The use of G1 cells removes heterogeneity arising from S-/G2-phase cells to generate Hi-C maps of greater detail (38). Following data analysis, we saw only marginal differences in A-/B-compartments (Fig. 1, C and D). TADs also showed only mild disruptions (Fig. 1, E and F), with <20% of the 4110 identified in control Hi-C data changing in cells lacking RNAPII. Two hundred twenty-seven new TADs could be detected in auxin-/triptolide-treated cells and displayed reinforced insulation at their boundaries (Fig. 1F). Average contact profiles in/around TADs revealed stronger definition of their borders at the expense of intra-TAD interactions (Fig. 1G).
Our data so far agreed well with recent observations in mESCs (35). However, we also detected >1500 loops exclusively in auxin-treated cells. These were significantly larger than those in control cells or than those shared between conditions (Fig. 1, H and I). Of these, 805 loops shared by auxin- and auxin-/triptolide-treated cells were also larger and displayed increased insulation at their anchors (Fig. 1, H to J). To understand the emergence of these stronger loops, we calculated the cumulative nascent RNA expression levels within the loop domains they form. Compared to all or shared loops from control cells, these 805 were significantly more associated with top-quantile loop domains (fig. S1, G and H), suggesting that highly active RNAPII can counteract loop extrusion and its removal leads to enlarged loops. However, although more “stripes” were detected in RNAPII-depleted Hi-C [indicative of loop extrusion; see (39)], these were, on average, shorter (fig. S1I). In addition, almost 10% of these 805 loops also associated with zero-expression domains but were still enhanced upon RNAPII depletion (fig. S1, G and H). We attribute this to Polycomb-mediated interactions that become accentuated upon RNAPII depletion. This is in agreement with the elevated H3K27me3 levels (fig. S1C), with the enrichment of H3K27me3 ChIP-seq signal at loop anchors (fig. S1, J and K), and with recent literature (40).
Last, note that analysis of Hi-C data from mAID-RPB1 cells treated with auxin for 2 hours (where 60 to 70% RPB1 is degraded; fig. S1A) did not reveal changes at any level of spatial genome organization (fig. S2, A to J). Together, our data reveal subtle yet discernible effects at the levels of TAD and loop organization occurring upon acute RNAPII depletion in interphase.
Reestablishment of spatial chromatin organization after mitosis requires RNAPII
Because RNAPII depletion did not markedly affect interphase chromatin organization in asynchronized cell populations, we hypothesized that it may be implicated in reestablishing chromatin folding upon exit from mitosis. This was based on two observations: first, on the detailed description of chromatin refolding dynamics in the mitosis-to-G1 transition, where contacts among cis-elements form early and rapidly, often not related to CTCF/cohesin (41, 42), and second, on the fact that, early in this transition, >50% of all active enhancers and genes exhibit a strong spike in transcription (43).
To study chromatin refolding following mitotic exit, we synchronized mAID-RPB1 cells at the G2-M checkpoint using the CDK1 inhibitor RO3306 (blocking ~90% cells in G2; Fig. 2, A and B), before releasing them via mitosis into G1. Six hours after washing out the inhibitor, >70% cells reentered G1 and were collected by fluorescence-activated cell sorting (FACS) (Fig. 2, A and B). RNAPII degradation, initiated by adding auxin to cells arrested in G2, was maintained throughout mitosis and G1 reentry without compromising progression past early G1 (as also exemplified by cell cycle markers; fig. S3A). RNAPII degradation was confirmed by fractionation and whole-cell blots (Fig. 2C and fig. S3B), immunofluorescence of Ser5-phosphorylated RNAPs, and 5-ethynyl-UTP (EUTP) labeling of nascent RNA (fig. S3, C and E). Like in asynchronous cells, the decrease in H3K27ac levels was accompanied by increased H3K27me3 levels in G1-reentry cells depleted of RNAPII (fig. S3, C and D). At the same time, the levels of abundant SWI/SNF chromatin remodeler subunits in chromatin were altered, but CTCF incorporation remained largely unchanged (Fig. 2C).
Fig. 2. RNAPII affects genome refolding following mitosis.
(A) Top: DLD-1–mAID–RPB1 synchronization. Bottom: Propidium iodine FACS profiles. (B) Bar plots showing cell percentage in each phase from (A). *P < 0.01, Fisher’s exact test. (C) Fractionation blots showing RNAPII, chromatin remodeler, and histone mark levels; HSC70 provides a control. (D) The 250-kbp resolution Hi-C maps from control (left) and auxin-treated cells (right) aligned to first eigenvector values (EV1). Insets: Saddle plots showing compartment insulation. (E) Plots showing interaction frequency decay as a function of genomic distance in control (black) and auxin-treated cells (purple). Rectangles indicate where values deviate most. (F) The 10-kbp resolution Hi-C maps from control and auxin-treated cells. (G) Heatmaps showing aggregated TAD interactions in control and auxin-treated cells. (H) Line plots showing mean insulation from control (black) and auxin-treated cells (purple) around all or control-specific TAD boundaries. N, number of TAD boundaries queried. (I) Boxplots of TAD sizes in control (black) and auxin-treated cells (purple). *P < 0.01, Wilcoxon-Mann-Whitney test. (J) Line plot showing RNAPII (magenta), H3K4me3 (blue), and H3K27ac ChIP-seq signal (orange) around control TAD boundaries. (K) Left: Venn diagram showing shared and unique loops between control (black) and auxin-treated cells (purple). Right: Boxplots of loop lengths. *P < 0.01, Wilcoxon-Mann-Whitney test. (L) Plots of aggregated Hi-C signal for shared loops from (J). (M) As in (H), but for shared loop anchors. (N) Boxplots of loop anchor insulation in control (black) and auxin-treated cells (purple) harboring zero or top-quantile expression levels. *P < 0.01, Wilcoxon-Mann-Whitney test. The Hi-C data presented in (D) to (N) come from two merged replicates (see table S1).
We next performed Hi-C on G1-reentry cells treated or not with auxin. We obtained >740 million and >1 billion Hi-C contacts from control and auxin-treated samples, respectively (table S1). Our first observation was that RNAPII-depleted cells showed increased interchromosomal contacts at the expense of intrachromosomal ones (fig. S3, F and G), also confirmed by high-throughput 3D-DNA fluorescence in situ hybridization (FISH; fig. S3H). At the same time, compartment boundaries were markedly blurred (Fig. 2D and fig. S3I), with interactions at distances of >1 Mbp (million base pairs) between A- and B-compartment segments becoming stronger (Fig. 2E).
Loss of interactions at the TAD scale (<1 Mbp) in auxin-treated cells (fig. S3J) led us to analyze 10-kbp resolution Hi-C maps. There, we observed strong and widespread erosion of domain structure, local insulation, and loop formation (Fig. 2, F and G, and fig. S3K). Insulation was weakened across all ~4500 TADs identified in control cells, and even more in the ~10% of TAD boundaries that were not reestablished in the absence of RNAPII (Fig. 2H). RNAP-depleted reentry cells also had fewer and larger TADs than control cells (Fig. 2I), indicative of boundary collapse and TAD merging. These effects are in line with RNAPII and active histone mark enrichment at TAD boundaries (Fig. 2J).
At the loop level, ~1900 were essentially lost from RNAPII-depleted cells concomitantly with the emergence >1150 new and significantly longer loops (Fig. 2K). Curiously, these longer loops emerged at sites of existing insulation, whereas loops weakened in RNAP-depleted cells showed reduced insulation (fig. S3, L and M). The 2443 loops detected in both control and auxin-treated reentry cells were weakened in the absence of RNAPII while also displaying reduced insulation at their anchors (Fig. 2, K to M). Again, we compared loop domains of no gene expression to those harboring top-quantile nascent RNA levels. We found that insulation was significantly weakened in both cases (Fig. 2N). Looking at stripe formation, we found both accentuated and dissolved ones in RNAP-depleted cells (fig. S3, N and O).
In summary, these data suggest that RNAPII is implicated in reestablishing both higher-order and fine-scale chromatin folding following exit from mitosis, and its depletion compromises loop formation. Critically, the folding changes that follow polymerase depletion do not simply reflect structures of an earlier G1 time point [e.g., compared to data from (42)] but rather compromised refolding.
Topoisomerase II depletion does not affect G1-reentry chromatin folding
Transcription enforces supercoiling onto DNA, and topoisomerase I (TOP1) is stimulated by RNAPII to resolve supercoils during elongation. However, TOP1 binding alongside initiating polymerases at TSSs was not matched by high TOP1 activity (44). In contrast, TOP2 has been linked to chromatin organization along the cell cycle and to transcription, with TOP2A affecting RNAPII kinetics (45) and marking its pausing sites (46). Moreover, TOP2B flanks TAD boundaries in human cells alongside CTCF/cohesin complexes to confine RNAPII (47) and preserves domain boundaries in yeast (48).
Given that no elongating RNAPs remain in auxin-treated cells, and that TOP2A-mAID cells prolong, but do conclude, mitosis (49), we asked whether TOP2 depletion from G1-reentry cells explains the effects we observe in RNAPII-depleted cells. To this end, we exploited another colorectal cancer line, HCT116, carrying or not a full knockout of the TOP2B gene and homozygously expressing mAID-tagged TOP2A. We verified >70% auxin-induced depletion of TOP2A in TOP2B-knockout cells and applied the same synchronization and FACS sorting scheme as before to obtain G1-reentry cells (fig. S4, A and B). Using factory RNA-seq to compare wild-type cells with those lacking both TOP2A and TOP2B, <400 genes, mostly linked to cell cycle control and cell morphogenesis, were affected by TOP2 elimination (fig. S4C).
Hi-C performed on G1-reentry cells carrying or not TOP2A/B activity revealed marginal changes across all scales of chromatin organization. Compartments were not affected, interactions remained unchanged irrespective of distance, and no increase in trans contacts was seen (fig. S4, D to F). Negligible changes to TAD boundary insulation were observed, and the mean size of TOP2A/B-depleted TADs did not differ from that in control cells (fig. S4, G to J). Although ~600 loops were lost or gained upon TOP2 depletion and condition-specific loops were again larger (fig. S3K), the increase/reduction in Hi-C signal at these loops was significantly less than that recorded upon RNAPII depletion and not followed by changes in insulation at their anchors (fig. S4, L and M). Together, these data suggest that the effects inflicted on chromatin refolding by RNAPII degradation cannot be recapitulated by TOP2A/B depletion, so they must rather be polymerase-centric.
RNAPII removal compromises cohesin chromatin reloading and loop formation
Hi-C data from G1-reentry cells depleted of RNAPII demonstrate A/B-compartment mixing, TAD erosion, and differential loss/gain of loops. Given that loop formation relies on the loading and DNA extrusion by cohesin complexes ending up at CTCF-marked anchors (10–13, 16), we examined how the levels of CTCF/cohesin subunits change in reentry cells following auxin treatment. Fractionation Western blots showed little fluctuation in CTCF, SMC1A, or Rad21 levels on chromatin, which was confirmed by quantification of RAD21 levels in individual cells using immunofluorescence (Fig. 3, A and B). However, the chromatin-bound levels of the two cohesin loaders, NIPBL and MAU2, were markedly reduced, as were the levels of the factor responsible for cohesin unloading, WAPL (concomitant with an increase in its soluble pool titers; Fig. 3A). Critically, this is not due to general down-regulation of these proteins, because Western blots showed that TIR1 activation to degrade RNAPII does not change their abundance in G1-reentry or G2-M–arrested cells (fig. S5, A and B).
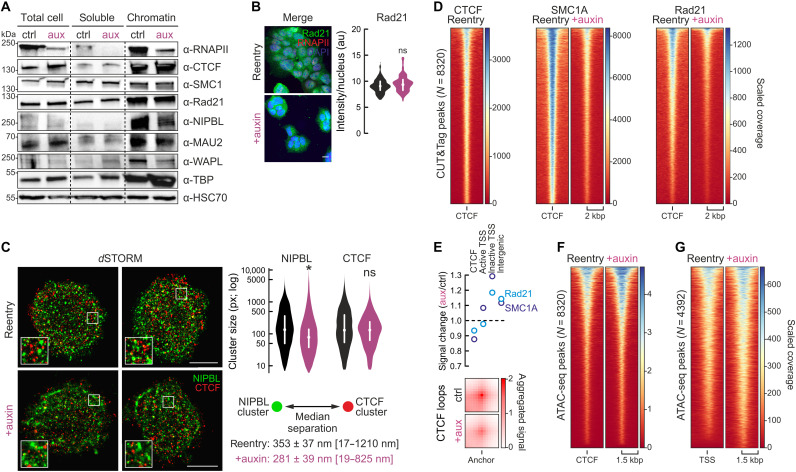
Fig. 3. RNAPII degradation affects CTCF/NIPBL distribution and cohesin loading following mitosis.
(A) Fractionation blots showing changes in chromatin-bound RNAPII, cohesin loaders NIPBL and MAU2, the WAPL unloader, and TBP; HSC70 provides a loading control. (B) RAD21 and RNAPII immunofluorescence in untreated (top) or auxin-treated reentry cells (bottom) and signal quantification (bean plots). Scale bar, 5 μm. au, arbitrary units. (C) Left: Rendering of 3D-STORM localizations for NIPBL and CTCF from control (top row) and auxin-treated reentry cells (bottom row). Scale bars, 5 μm. Right: Bean plots showing changes in NIPBL and CTCF cluster sizes. Bottom right: Changes in separation between the nearest NIPBL/CTCF clusters (smallest and largest distances shown in square brackets). *P < 0.01, significantly different, Wilcoxon-Mann-Whitney test. (D) Heatmaps showing SMC1A (middle) and Rad21 CUT&Tag signal (right) in control and auxin-treated G1-reentry cells at CTCF positions (left). (E) Top: Plot showing changes in SMC1A (dark blue circles) and Rad21 CUT&Tag signal (light blue circles) assigned to CTCF-bound, active/inactive TSSs or intergenic regions in auxin-treated compared to control cells. Bottom: Plots showing aggregate Hi-C signal for loop categories anchored at the CTCF sites in (D). (F) As in (D), but using ATAC-seq around CTCF-proximal SMC1A/Rad21-bound positions. (G) As in (F), but for active TSSs.
To understand whether our findings are due to changes in NIPBL and CTCF nuclear distribution, we performed super-resolution localizations of these factors. Dual-color dSTORM in control and auxin-treated G1-reentry cells led to the following observations. First, NIPBL localizes in clusters of smaller average size upon RNAPII depletion. At the same time, we also observed more localizations in extended and deformed clusters (Fig. 3C), exemplified by the shift in “eccentricity” of NIPBL clusters from 0.54 in control to 0.69 in auxin-treated cells (eccentricity of 0 refers to a perfect circle, while eccentricity of 1 refers to a line). Second, CTCF clusters do not change as regards their mean size, but 50% of all CTCF clusters in control cells lie <129 px2, while in auxin-treated cells 50% lie <82 px2 (Fig. 3C). Such an increased population of smaller CTCF clusters was also observed via dSTORM upon cohesin removal in Rad21-mAID cells (50) occurring as a result of loop collapse. Last, NIPBL distribution relative to CTCF also changed significantly in the absence of RNAPII. The median separation between NIPBL and its nearest CTCF cluster was reduced from 353 to 281 nm, with the largest recorded distance dropping from >1200 to 825 nm (Fig. 3C).
These results, and the changed NIPBL/MAU2/WAPL levels on chromatin, hint to aberrant cohesin loading to (and most likely unloading from) chromatin in the absence of RNAPII and predict that less cohesin will end up at CTCF-bound sites. To test this prediction, we generated SMC1A and Rad21 CUT&Tag (cleavage under targets and tagmentation) data in control and auxin-treated reentry cells and found cohesin signal significantly reduced at CTCF sites genome-wide (Fig. 3D). We quantified the fraction of scaled CUT&Tag signal falling into CTCF-bound regions, active or inactive TSSs, and non–RNAPII-associated intergenic space. We found that the reduced fraction of reads at CTCF sites was accompanied by a 20 to 30% increase in signal mapping to inactive TSSs in auxin-treated reentry cells and by a >11% increase in widespread intergenic signal (Fig. 3E). It follows that this general reduction of cohesin occupancy at CTCF sites impairs loop formation genome-wide (Fig. 3E). Notably, these changes occurred despite no reduction in CTCF-proximal accessibility as judged by ATAC-seq (assay for transposase-accessible chromatin using sequencing) (Fig. 3F).
One could assume that cohesin loading at TSSs (30, 51) simply results from them being rendered accessible by active RNAPs. Hence, reduced accessibility would readily explain compromised loading. To our surprise, ATAC-seq signal in RNAP-depleted TSSs rather increased (Fig. 3G), as did TBP (TATA box–binding protein) levels on chromatin (Fig. 3A). Looking at CUT&Tag signal at >4000 RNAPII/SMC1A co-occupied TSS, it is markedly reduced upon auxin treatment of reentry cells (fig. S5, C and D). This argues in favor of RNAPs recruiting cohesin loaders and unloaders to these sites, while setting up TSS architecture in G1-reentry cells likely relies on “pioneer” factors (perhaps like TBP) preceding the polymerase. Last, we used coimmunoprecipitations to show that RNAPII directly interacts with WAPL (fig. S5E) and that NIPBL copurifies with RNAPII in G1-reentry cells (fig. S5F).
Modeling dissects RNAP contribution to loop extrusion
To dissect the connection between RNAPII and cohesin reloading onto chromatin, we turned to in silico modeling of chromatin folding. This allowed us to test scenarios that would be challenging to address experimentally. First, we performed 3D chromatin folding simulations using the established HiP-HoP model (52) that accounts for the heteromorphic nature of chromatin and incorporates TF binding and loop extrusion. We modeled a 10-Mbp region from human umbilical cord endothelial cell (HUVEC) chr14 for which gene expression, histone mark, and CTCF positioning ENCODE data are available (www.encodeproject.org).
Control chromatin folding was simulated by assuming that most cohesin loading (90%) occurs at RNAPII-occupied TSSs (with 10% loading randomly). Experimentally defined cohesin residence times on DNA [~20 min; see (53)] were incorporated into the model. Following multiple iterations, our model produced a mean contact map resembling Hi-C data (Fig. 4, A and B). To simulate chromatin folding following RNAPII degradation, we eliminated loading at promoters and only allowed random loading (consistent with low-efficacy NIPBL-independent cohesin loading in vitro) (54). As a result, four major effects were observed: first, a general weakening of interactions and domain insulation across the 10 Mbp modeled (Fig. 4, A and B), similar to what we saw using Hi-C (Fig. 2F); second, individual models of the fiber displayed obvious unfolding (Fig. 4B), likely consistent with the increase in trans interactions in our data (fig. S3, F to H); third, reduced cohesin occupancy at CTCF sites (Fig. 4A), consistent with our CUT&Tag data (Fig. 3D and fig. S5, C and D); fourth, markedly weakened loop formation, but with larger loop sizes (Fig. 4, C and D) matching our experimental results (Figs. 2, K and L, and 3E). Thus, our modeling suggests that inability to load cohesin via RNAPII-bound sites suffices for explaining the major chromatin folding differences observed experimentally.
Fig. 4. Computational modeling of RNAPII depletion effects on loop extrusion.
(A) Top: Heatmaps rendered from simulations of wild-type (left) or RNAPII-depleted models (right) of HUVEC chr14:50-60 Mbp. Bottom: The 10 kbp-resolution heatmaps in the chr14:53-56 Mbp region. Cohesin positioning tracks are aligned below each heatmap. (B) Exemplary 3D chromatin folding models of the chr14:54.5-55.5 Mbp subregion shown in comparison to Hi-C data (from two merged replicates; see table S1). (C) APA plots showing weakened loops in RNAPII-depleted models. (D) Histogram showing looping frequency in the absence of RNAPII (turquoise) compared to wild-type models (black). Inset: Boxplots showing larger loops in RNAPII-depleted models (blue). *P < 0.01, significantly different, Wilcoxon-Mann-Whitney test. (E) Heatmaps rendered from 1D simulations representing wild-type (top) or RNAPII-depleted models (bottom) of the chr14 segment from (A) at 3-kbp resolution. Cohesin positioning (overlaid tracks) and TSS orientation (arrows) are aligned below. (F) As in (D), but using data from the 1D simulations in (E).
To interrogate the interplay between RNAPII and loop-extruding cohesin directly, we performed 1D simulations of minimal composition. We modeled a 3-Mbp region of HUVEC chr14 (chr14:53-56 Mbp, hg19) as a coarse-grained fiber carrying CTCF at the appropriate positions, as well as RNAPs transcribing genes in the correct orientation. As before, cohesin was predominantly loaded at RNAPII-bound TSSs in the control scenario but only randomly in the “degron” model. First, we observed formation of loops and intricate domain compartmentalization under control settings, despite having only two activities operating on the fiber (Fig. 4E). Notably, transcription affects cohesin deposition and loop formation in our model, as exemplified by simulations in which all genes in this 3-Mbp segment were modeled as tandemly transcribed (fig. S6). RNAP depletion eliminated compartmentalization, and the frequency of looping was again markedly reduced. Cohesin occupancy was decreased at most loop anchors, with loops again becoming larger (Fig. 4, E and F). This parsimonious model allows us to deduce that the effects observed in experiments can be explained by a simple relationship between cohesin loading at RNAP-occupied sites and the interplay between active transcription and loop extrusion.
DISCUSSION
Following cell division, chromosomes refold to establish interphase architecture. At this point, cohesin also needs to be reloaded. Its reloading coincides with the extrusion of CTCF-anchored loops and TAD reestablishment (41, 42). However, A/B-compartments, driven by homotypic chromatin interactions, reemerge more rapidly, as do contacts among cis-regulatory elements. The latter display rates that exceed those of extruded loops (42). In parallel, the general TF TBP bookmarks mitotic chromatin to facilitate gene reactivation (55), and as transcription reinitiates in late telophase, a strong activity spike occurs at most genes and enhancers (43). These observations suggest that RNAP activity may play a central role in reestablishing interphase chromatin organization following mitosis.
Here, using an RPB1 degron line (26), we show that the presence of RNAPII on chromatin is necessary for both the establishment of compartments and the formation of loops early in G1. The former is intuitively justified by the homotypic interactions that build the “active” A-compartment and the recent finding that chromatin acetylation can drive compartment formation (56). In our RNAPII-depleted G1-reentry cells, H3K27ac levels are reduced, while H3K27me3 levels increase, and this imbalance most probably underlies many of the compartment-level changes, although alleviating H3K27ac bookmarking had little effect on 3D refolding in mESCs (51).
The latter effect is more perplexing but agrees with NIPBL binding at active gene promoters (30, 57, 58) and, thus, raises two key questions. First, how does RNAP depletion promote formation of hundreds of de novo loops that are also longer? According to our simulations, and depending on the direction of elongation, RNAPs can reel DNA such that it counters extrusion while also acting as physical blockades to it. This is reminiscent of the condensin-polymerase antagonism reported for bacteria (34) and flies (33) and inferred by super-resolution imaging in mESCs (59). Some of these newly emerging loops form on the basis of strengthened Polycomb interactions, justified by the increase in H3K27me3 levels upon RNAPII degradation. Recent work showing that cohesin removal also resulted in enhancer looping of Polycomb-bound regions (39) suggests that RNAPII depletion cross-talks with cohesin loading. As regards increased loop lengths, our simulations argue that this is a consequence of reduced cohesin loading rates to chromatin, as well as of the different loading patterns in control versus RNAP-depleted cells. These translate into fewer cohesin rings acting locally at any given time, and this reduced “crowding” leads to fewer extrusion conflicts allowing longer loops to form. However, when compared to data from Wapl-knockout cells showing loop enlargement (14), our Hi-C data differ (fig. S7A). But when compared to Hi-C data from cells undergoing hyperosmotic shock, which affects RNAP and cohesin binding to chromatin (60), the resulting interaction patterns are much more similar (fig. S7B). These comparisons suggest a distinct and more generalized disruption of the cohesin loading-unloading cycle following RNAPII depletion.
Second, why are the effects of RNAPII depletion more obvious upon G1 reentry? We believe that this is due to a combination of effects. On the one hand, cell synchronization counters the inherent heterogeneity of contacts in individual cells. On the other, early chromatin refolding and transcription bursts in the mitosis-to-G1 transition suggest that RNAPs preempt a central role in establishing a loop-based chromosomal architecture by instructing cohesin loading and compartmentalization. However, we were surprised to find that, in the absence of RNAPII, chromatin accessibility increases following mitotic exit. Although this is counterintuitive, it suggests that the formation of active transcriptional complexes can somehow restrain aberrant accessibility at gene promoters. We speculate that the variable levels of chromatin-bound remodelers (as assessed by Western blots in RNAPII-depleted G1-reentry cells) may partly justify this observation. At the same time, this also suggests that pioneer “bookmarking” factors (like TBP, which we find markedly enriched in chromatin in the absence of RNAPII) may have as their default function the “opening up” of chromatin, which is then constrained by preinitiation RNAPs in conjunction with chromatin remodelers recruited at precise stoichiometries.
In summary, we uncovered a dependency of loop extrusion on RNAPII that predominates genome reorganization following exit from mitosis. This dependency may speak of the significance of mitotic bookmarking, as TF association with mitotic chromatin could dictate RNAP positioning and, in a next step, cohesin loading and loop extrusion. Nonetheless, the precise interplay between polymerases, TFs, and cohesin subunits during this transition remains to be further elucidated.
MATERIALS AND METHODS
Cell synchronization and sorting
mAID-POLR2A(RPB1)-mClover DLD-1 (26) and TO2B−/−-TOP2A-mAID HCT116 cells (V.R. laboratory) were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum under 5% CO2. Inducible depletion of RPB1 or TOP2A initiated via treatment with doxycycline for 24 hours to induce TIR1 expression, before addition of 500 μM indole-3-acetic acid solution (“auxin,” Sigma-Aldrich) for different times to induce RPB1 degradation. For cell synchronization, G2-M arrest was achieved by the addition of 10 μM RO-3306 inhibitor for 21 hours. Following this incubation time, cells were washed with phosphate-buffered saline (PBS), and auxin-supplemented medium was added for up to 6 hours to allow cells to quantitatively enter G1. At this point, synchronized or asynchronous cells treated with auxin for up to 14 hours were harvested, where applicable resuspended in propidium iodide (1 μg/ml) to counterstain DNA, and sorted to isolate G1 cells using a FACSCanto II flow cytometer (Becton Dickinson).
In situ Hi-C and data analysis
All in situ Hi-C was performed using the Hi-C+ Kit (Arima Genomics) as per the manufacturer’s instructions. The resulting Hi-C libraries were paired-end sequenced on a NovaSeq6000 platform (Illumina) to >450 million read pairs per replicate (table S1). Reads were separately aligned to the reference build of the human genome (hg38) using BWA and Juicer (v. 1.11.09) to generate .hic files (61). Only reads with MAPQ >30 were considered for further analysis, and bin-to-bin interactions were extracted from KR-balanced matrices in .hic files using the Juicer “dump” utility at different resolutions. A-/B-compartment stratification was performed using the “eigenvector” Juicer utility on 250-kbp resolution matrices, with both gene and H3K27ac ChIP-seq signal density used to deduce A-compartments. Saddle plots were generated as described previously (42). For TADs, KR-balanced matrices we processed via a combination of “directionality index” plus HMM tools at 10-kbp resolution and in 500-kbp windows. TADs smaller than 150 kbp or found in centromeric regions were filtered out. For a TAD to be considered “shared” between two datasets, boundary positions should not shift by >60 kbp, and their coordinates should overlap at least 90%. Insulation scores at TAD boundaries were calculated using a sliding 120 kbp × 120 kbp window along the matrix diagonal at 10-kbp resolution as previously described (62); squares with a sum of interactions <12 were filtered out. Aggregate TAD plots were generated using Coolpup.py (63). For loop detection, we used SIP (64) and standard parameters: -res 10000 -mat 2000 -g 2 -d 3 -fdr 0.01 -nbZero 4 -cpu 1 -factor 1 -max 2 -min 2 -sat 0.01 -t 2800 -norm KR -del true, and a false discovery rate (FDR) of <0.01 to filter the resulting loop lists. Loops specific to a given condition were determined using pgltools (with -d 29999 due to the smallest allowable loop size of 30 kbp; table S2) (https://github.com/billgreenwald/pgltools). Aggregate peak plots were generated via the Aggregate peak analysis (APA) utility in Juicer using standard parameters (-r 10000 -k KR -q 3 -w 6 -n 15 -u), before scaling between 0 and 2 to facilitate comparison. Last, architectural stripes in Hi-C data were detected using stripenn (https://github.com/ysora/stripenn). Visualizations and plots were performed using data from merged Hi-C replicates (except for TOP2A-mAID HCT1166 and 2-hour RPB1-mAID data); exemplary data from individual Hi-C replicates are shown in fig. S8 to highlight reproducibility. All code used is available at https://github.com/shuzhangcourage/HiC-data-analysis.
High-throughput 3D-DNA FISH
Dual-color DNA FISH was performed using the BAC probes targeting different chromosomes (table S3) and labeled with Alexa 488–deoxyuridine triphosphate (dUTP), Alexa 568–dUTP, or Alexa 647–dUTP by nick translation on G1-sorted control and auxin-treated DLD-1 reentry cells seeded on glass slides. Images were acquired using the Opera Phenix High Content Screening System (PerkinElmer), equipped with four laser lines (405, 488, 568, and 640 nm) and two 16-bit complementary metal-oxide semiconductor (CMOS) cameras. Images for 3D and radial distances were acquired in confocal mode using a 40× water objective [numerical aperture (NA), 1.1] and analyzed as described previously to also quantify DNA content and infer cell cycle phase stratification (65).
ChIP coupled to sequencing
DLD-1 cells cultured to 80% confluence in 15-cm dishes were cross-linked in 1% paraformaldehyde (PFA)/PBS for 10 min at room temperature. Cells were processed using the NEXSON ChIP protocol (66). In brief, nuclei were isolated via sonication using Bioruptor Pico (Diagenode; nine cycles of 10 s on and 30 s off). Chromatin was then sheared in the recommended shearing buffer (27 to 30 cycles, 30 s on and 30 s off) to a range of 200- to 500-bp-long fragments, and immunoprecipitation was performed using 4 μg of the appropriate antibody [anti-CTCF: 61311, Active Motif; anti-RAD21: ab88572, Abcam; anti–green fluorescent protein (GFP): ab290, Abcam]. Paired-end sequencing was performed on a NovaSeq6000 platform (Illumina) yielding >25 million reads per sample. Raw reads were processed for mapping and peak calling using the ENCODE Data Coordinating Center pipeline (DCC v1.5.0; https://github.com/ENCODE-DCC). Coverage plots and heatmaps were generated via Deeptools (67).
Assay for transposase-accessible chromatin using sequencing
Tn5 transposed chromatin was isolated from human DLD-1–mAID–RPB1 cells according to the standard ATAC-seq protocol with a modification (68) aiming at quantitative scaling of the resulting data. In brief, 105 DLD-1 cells per replicate we “spiked” with 200 D. melanogaster S2 cells, washed in 1× PBS, and added to lysis buffer [10 mM tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40, 0.1% Tween 20, and 0.01% digitonin] for 3 min to isolate nuclei. Nuclei were next washed in washing buffer [10 mM tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.1% Tween 20] and pelleted by centrifugation. Isolated nuclei were resuspended in transposase reaction mix (25 μl of 2× TD buffer, 16.5 μl of 1× PBS, 0.5 μl of 10% Tween 20, 0.5 μl of 1% digitonin, 2.5 μl of Tn5, and 5 μl of nuclease-free H2O) and incubated at 37°C for 30 min under constant shaking at 1000 rpm. The transposition reaction was terminated by the addition of stop buffer [50 mM tris-HCl (pH 8), 10 mM EDTA, and 1% SDS] and purified using the DNA Clean & Concentrator Kit (Zymo Research). Following standard library generation, samples were sequenced to >40 million reads on a NovaSeq6000 platform (Illumina). Read pairs were mapped to the hg38 and dm6 reference genome builds for human and Drosophila, respectively, using Bowtie 2 (69). Unmapped, duplicate, and mitochondrial reads were removed before merging replicates. ChIPseqSpike was used for calculating scaling factors (https://bioconductor.org/packages/release/bioc/html/ChIPSeqSpike.html) to produce reads per kilobase per million (RPKM)–normalized and scaled coverage.
Cleavage under targets and tagmentation
Following lifting from plates using accutase and FACS sorting, 0.5 million G1-phase DLD-1 cells were processed according to the manufacturer’s instructions (Active Motif). Samples were paired-end sequenced to obtain at least 107 reads. Reads were processed according to the standard CUT&Tag pipeline (https://yezhengstat.github.io/CUTTag_tutorial/). Briefly, paired-end reads were trimmed for adapter removal and mapped to the human (hg38) and Escherichia coli (ASM584v2) reference genomes using Bowtie 2 (69). E. coli mapped reads were quantified and used for calibrating human-mapped reads. Peak calling was performed with SEACR (70) and using immunoglobulin G controls for thresholding peak calling. For stringency, cohesin-bound sites were considered those shared by both the SMC1A and Rad21 control datasets, while CTCF-bound sites were considered those shared by the CUT&Tag and publicly available CTCF ChIP-seq data (http://chip-atlas.org/view?id=DRX013180). All heatmaps were generated using Deeptools (67).
Immunofluorescence and image quantification
DLD-1 cells grown on coverslips were fixed in 4% PFA/PBS for 10 min at room temperature. After washing once in PBS, cells were permeabilized with 0.5% Triton-X/PBS for 5 min at room temperature, washed three times in PBS, blocked using 1% bovine serum albumin for 1 hour, and incubated with the appropriate primary antibody for 1 hour at room temperature (anti-RNAPII: 1:500, 61086, Active Motif; anti-H3K27ac: 1:500, 39133, Active Motif; anti-H3K27me3: 1:500, 39155, Active Motif; anti-Rad21: 1:800, ab992, Abcam; anti-fibrillin: 1:100, sc-393968, Santa Cruz Biotechnology). For visualizing nascent transcripts, cells were preincubated with 3 mM 5-ethynyl uridine for 30 min at 37°C in their growth medium, fixed, and processed with the Click-iT EdU Chemistry Kit (Invitrogen). Images were acquired on a Leica dmi8 microscope using the LASX software. Quantification of nuclear fluorescence was performed by drawing a mask based on 4′,6-diamidino-2-phenylindole (DAPI) staining and then calculating the mean intensity per area falling under this mask. Colocalization was assessed using the ImageJ plugin JACoP (https://imagej.nih.gov/ij/plugins/track/jacop.html).
Chromatin fractionation and Western blotting
For assessing protein abundance in different sample preparations, approximately 106 cells were gently scraped off 15-cm dishes and pelleted for 5 min at 600g at room temperature, supernatants were discarded, and pellets were resuspended in 100 μl of ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1 mM EGTA (pH 8.0), 1% NP-40, and 1% sodium deoxycholate] containing 1× protease inhibitor cocktail (Roche). Next, lysates were incubated for 20 min on ice and centrifuged for 15 min at >20,000g to pellet cell debris to collect the supernatants. The concentration of each protein extract was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). For fractionation, the protocol previously described was used (71). Following protein separation on precast SDS–polyacrylamide gel electrophoresis gels (Bio-Rad), proteins were detected using different primary antibodies (table S4) and visualized using the Pierce SuperSignal West Pico ECL Kit (Thermo Fisher Scientific).
Factory RNA-seq and data analysis
Nascent RNA from ~10 million mAID-RPB1-mCLover DLD-1 or TO2B−/−-TOP2A-mAID HCT116 cells was isolated according to the “factory-seq” protocol (36). Briefly, cells were gently scraped and lysed in isotonic “physiological buffer” supplemented with 0.5% NP-40 buffer. After assessing lysis and nuclei integrity on a hemocytometer microscopy, nuclei were treated with deoxyribonuclease I (Worthington) for 30 min at 33°C, washed, lysed in “native lysis buffer” and treated with caspase group III enzyme mix (PromoKine), pelleted by centrifugation, before the supernatant holding nascent RNA was collected in TRIzol (Invitrogen), and purified using the Direct-Zol RNA Purification Kit (Zymo). Following standard strand-specific complementary DNA (cDNA) library preparation using the TruSeq Kit (Illumina), sequencing was performed on a NovaSeq6000 platform (Illumina) to >40 million paired-end reads. Raw reads were mapped to human genome (build hg38) using STAR (72) and quantified using iRNAseq (73) and the -gene option, before RUVseq normalization (74) and differential gene expression analysis. For gene set enrichment, GSEA (www.gsea-msigdb.org/gsea/index.jsp) was run on the significantly changing genes (Padj ≤ 0.05; listed in table S5).
Dual-color super-resolution dSTORM imaging and analysis
DLD-1 control and auxin-treated reentry cells were seeded onto coverslips, fixed, stained, and imaged as described previously (50). In brief, fixed and immunostained cells for NIPBL and CTCF (as described above) were mounted to an Attofluor cell chamber (Thermo Fisher Scientific) and in 1 ml of dSTORM buffer [25 mM Cysteamine (MEA), glucose oxidase, 50 mM NaCl, and 10% glucose in 10 mM tris-HCl (pH 8.0)]. The chamber is then sealed with a coverslip and left on the microscope at room temperature for 30 min before imaging to minimize drift. Imaging was performed on a Zeiss Elyra PS1 system fitted with an Andor iXon DU 897, 512 × 512 EMCCD camera. Images were made using a 100× 1.49 NA total internal reflection fluorescence (TIRF) objective in HiLo mode. Movies of 12,000 frames were recorded with an exposure time of 33 ms. Multichannel images were acquired sequentially from high wavelength to lower wavelengths. dSTORM movies for each protein target were analyzed via the Zeiss ZEN 2012 software, and any localizations with a precision of >50 nm were discarded. All remaining localizations were drift-corrected using a model-based approach. All additional analysis was done in R (www.R-project.org/), and localizations from individual nuclei were clustered based on their density using a kernel density estimation (KDE)–based clustering algorithm with the threshold set to 0.05 for all channels. The areas of CTCF or NIPBL clusters were measured using the KDE binary image, and distances between closest neighbors were calculated.
Computational modeling
For the 3D simulations, we used our previously described HiP-HoP model (52), extended to account for interactions in inactive regions. This model combines our initial “transcription factory” model (75) with loop extrusion (76) while also accounting for the heteromorphic nature of the chromatin fiber, which means that the local compaction (in DNA base pairs per nanometer) varies along the fiber. Here, we modeled a 10-Mbp region of HUVEC chr14 as a bead-spring polymer containing N beads of diameter σ, each representing 1 kbp of chromatin. We allowed beads in the polymer to interact via three potentials: (i) a Weeks-Chandler-Andersen (WCA) potential, which provides excluded volume interactions; (ii) a finitely-extensible-nonlinear-elastic (FENE) potential accounting for chain connectivity; and (iii) a Kratky-Porod potential describing the flexibility of the chain with parameters set to give a persistence length of 4 to 5 kbp (in line with that of chromatin in vivo). To model the heteromorphic nature of the chromatin fiber in a simple way, we included additional springs (with constants of 200 kBT/σ2) between next-to-neighbor chromatin beads along the chain (i.e., beads i and i + 2) that are not associated with H3K27ac marks. As H3K27ac marks correlate with active euchromatin regions, these springs cause a local crumpling of the polymer in inactive chromatin fragments, or equivalently a swelling in active regions accounting for their generally more open conformation. TFs were simulated as diffusing beads interacting with each other via steric repulsion, again modeled via WCA potentials. We considered three types of TFs: (i) Generic active TFs bind strongly (potential depth, 7.9 kBT) to chromatin beads associated with accessible chromatin [defined using ENCODE Dnase I hypersensitivity sites sequencing (DHS-seq) data], and weakly to beads associated with H3K27ac (potential depth, 3.4 kBT); (ii) HP1-like inactive TFs bind to beads enriched in H3K9me3 marks (potential depth, 3.4 kBT); and (iii) Polycomb-like TFs bind to beads enriched in H3K27me3 marks (potential depth, 7.9 kBT). All of these interactions were modeled via a truncated-and-shifted Lennard-Jones (LJ) potential. To account for posttranslational modifications, we allowed each TF type to switch between a binding and nonbinding state at a defined rate (ksw = 10−3 τ−1, where τ is the simulation time unit). The binding state was characterized by the aforementioned interaction strengths, whereas the nonbinding state was characterized only by steric interactions with chromatin beads (via WCA potentials). We considered 250 active TFs, 625 HP1-like TFs, and 125 Polycomb-like TFs in the wild-type simulations, whereas the RNAPII-degron simulations were run without active TFs. We also implemented nonspecific interactions between inactive chromatin beads (via a truncated-and-shifted LJ potential with depth 0.45 kBT) to account for the generic “phase separation” between eu- and hetero-chromatin. Loop extrusion was modeled by representing cohesin dimers as further additional springs. Loop extrusion dynamics were determined by the number density of cohesins (nc = 0.01/kbp) on the chromatin fiber and two rates: the unbinding rate (koff = 2.5 × 10−5 τ−1) and the extrusion rate (v = 4 × 10−3 kbp/τ). Upon binding of cohesin, we introduced an additional spring between two nearby beads along the fiber (i and i + 3, because crumpling springs already link i and i + 2); the equilibrium length and the spring constant of cohesin bonds were set to 1.5 σ and 40 kBT/σ2, respectively. When cohesins were removed from chromatin, they were instantly repositioned along the fiber. Wild-type conditions were simulated by background random loading (with 10% probability) but with predominant loading at DHS beads (with 90% probability); the RNAPII-degron was simulated by only considering random loading. A cohesin halted either upon colliding with another extruding complex or upon reaching a CTCF site whose direction was against the direction of extrusion [as shown experimentally; (11)]. Note that CTCF sites and orientation were obtained by ENCODE tracks, taking care to include in our simulations only sequences overlapping cohesin (RAD21) peaks—this procedure singles those CTCF binding sites that are relevant to looping. All constituents of the system (chromatin beads and TFs) were allowed to diffuse, and their dynamics were governed by a Langevin equation as described before (52) and implemented using Python and the LAMMPS molecular dynamics software package (77) as a library.
For the 1D simulations, we considered a 3-Mbp-long chromatin fiber coarse-grained into segments of 1 kbp. Again, we modeled data from a specific subregion of HUVEC chr14. We simulated the dynamics of cohesin rings [total number Nrings = 30, comparable to what was used in (76)], each of which could be in one of two states: either bound (i.e., on the fiber) or unbound (i.e., in the diffuse pool). Binding and unbinding were modeled as stochastic processes with rates kon and koff, respectively. When on the fiber, a cohesin molecule was modeled as a dimer, with each monomer undergoing active extrusion at speed v. Each monomer could proceed until it hit a CTCF site with orientation conflicting with its direction of travel, at which point it became immobile (76). If a cohesin complex was halted on one side, its other side could continue to move independently. When both monomers in a cohesin dimer became stuck at convergent CTCF sites, the unbinding rate of the dimer was decreased by a factor of 10 to model CTCF-mediated stabilization of extruded chromatin loops, in line with results in (76). We let monomers in a cohesin dimer interact with each other via steric exclusion so that extrusion would be halted temporarily if another monomer was in their way. Wild-type conditions were simulated by assuming that cohesin was loaded as described above, with 10% background random loading and 90% at DHS sites, whereas RNAPII-degron conditions were simulated by only retaining the random background loading. To simulate feedback of transcription on extrusion, we assumed that the speed of extrusion was reduced by a factor f when the direction of extrusion and that of transcriptional elongation of an active gene were conflicting. Parameters in the simulations were set to kon = 2 × 10−2 τ−1, koff = 10−3 τ−1, v = 0.16 kBT/τ for the wild type, and to kon = 2 × 10−3 τ−1, koff = 10−3 τ−1, v = 0.16 kBT/τ for the RNAPII-degron. These values can be mapped to kon = 1 min−1, koff = 0.05 min−1, v = 0.133 kbp/s (wild type, corresponding to a residence time on chromatin ~20 min) or kon = 0.067 min−1, koff = 0.033 min−1, v = 0.089 kbp/s (RNAPII-degron, corresponding to a residence time on chromatin ~30 min). The koff value in simulation units (koff = 10−3 τ−1) was chosen to be sufficiently small to allow study of extrusion effects. To align simulation times to real times, we consider a residence time on chromatin of ~20 and 30 min for the wild-type and degron simulations, respectively. The wild-type value is comparable to that reported previously (53). We also set kon = 1 min−1 (wild type; while there is no accurate measurements of this rate, it needs to be significantly larger than koff) and reduced its value by a factor of 10 in RNAPII-degron simulations to have the same rate of random loading both scenarios. Extrusion speed v was chosen such that the chromatin length explored during an extrusion event, λ = v/koff, was the same for wild type and degron, and comparable to that used previously (76). For simulations including the feedback of transcription on extrusion, we varied f (which is unknown experimentally) between 0.1 and 0.9 to simulate a variety of scenarios and presented the case with f = 0.1, which led to the most pronounced effects.
Statistical analyses
P values associated with Student’s t tests and Fisher’s exact tests were calculated using GraphPad (http://graphpad.com/), and those associated with the Wilcoxon-Mann-Whitney test were calculated using R. Unless otherwise stated, P values of <0.01 were deemed significant.
Acknowledgments
We thank P. Cook, N. Gilbert, and J. Vaquerizas for critical reading of the manuscript; the Maeshima laboratory (NIG, Japan) for gifting us the DLD-1–mAID–RPB1 cells; Marteen Fornerod for the anti-TIR1 monoclonal; and the NGS Integrative Genomics UMG unit for ATAC library sequencing.
Funding: Work in the laboratory of A.P. is funded by the Deutsche Forschungsgemeinschaft (DFG) via a Basic module grant (Project Nr.: 290613333) and the SPP2202 (422389065) and TRR 81/3 programs (109546710). S.Z. is supported by a CSC fellowship. S.Z., N.Ü., and N.J. are further supported by the International Max Planck Research School for Genome Science. Work in the laboratory of D.M. is supported by an ERC Consolidator grant (THREEDCELLPHYSICS), work in the laboratory of K.S.W. was supported by the project GENOMETRACK (737.016.014) NWO-BBOL research programme financed by the Dutch Research Council (NWO), and work in the laboratory of V.R. was supported by the DFG via the SFB1361 (393547839) and SPP2202 programs (402733153).
Author contributions: S.Z., N.Ü., and N.J. performed experiments. S.Z., N.J., and E.G.G. performed bioinformatics analyses. G.F., M.C., and D.M. performed computational modeling. H.J.G. and V.R. performed 3D DNA FISH. J.A.S., A.B.H., and K.S.W. performed super-resolution STORM imaging. C.B. and J.A. performed all next-generation sequencing. A.P. conceived and supervised the study, and compiled the manuscript with input from all coauthors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: NGS data generated in this study are available via the NCBI Gene Expression Omnibus (GEO) repository under accession number GSE160321. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1, S3 and S4
Legends for tables S2 and S5
Other Supplementary Material for this manuscript includes the following:
Tables S2 and S5
REFERENCES AND NOTES
- 1.Tjalsma S. J., de Laat W., Novel orthogonal methods to uncover the complexity and diversity of nuclear architecture. Curr. Opin. Genet. Dev. 67, 10–17 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Marchal C., Sima J., Gilbert D. M., Control of DNA replication timing in the 3D genome. Nat. Rev. Mol. Cell Biol. 20, 721–737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo Q., Bantignies F., Cavalli G., Principles of genome folding into topologically associating domains. Sci. Adv. 5, eaaw1668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen A. S., Cattoglio C., Darzacq X., Tjian R., Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus 9, 20–32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rada-Iglesias A., Grosveld F. G., Papantonis A., Forces driving the three-dimensional folding of eukaryotic genomes. Mol. Syst. Biol. 14, e8214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canat A., Veillet A., Bonnet A., Therizols P., Genome anchoring to nuclear landmarks drives functional compartmentalization of the nuclear space. Brief. Funct. Genomics 19, 101–110 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Kim S., Shendure J., Mechanisms of interplay between transcription factors and the 3D genome. Mol. Cell 76, 306–319 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Nuebler J., Fudenberg G., Imakaev M., Abdennur N., Mirny L. A., Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. U.S.A. 115, E6697–E6706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yesbolatova A., Natsume T., Hayashi K. I., Kanemaki M. T., Generation of conditional auxin-inducible degron (AID) cells and tight control of degron-fused proteins using the degradation inhibitor auxinole. Methods 164-165, 73–80 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Nora E. P., Goloborodko A., Valton A.-L., Gibcus J. H., Uebersohn A., Abdennur N., Dekker J., Mirny L. A., Bruneau B. G., Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao S. S. P., Huang S.-C., Glenn St Hilaire B., Engreitz J. M., Perez E. M., Kieffer-Kwon K. R., Sanborn A. L., Johnstone S. E., Bascom G. D., Bochkov I. D., Huang X., Shamim M. S., Shin J., Turner D., Ye Z., Omer A. D., Robinson J. T., Schlick T., Bernstein B. E., Casellas R., Lander E. S., Lieberman-Aiden E., Cohesin loss eliminates all loop domains. Cell 171, 305–320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., Fonseca N. A., Huber W., Haering C. H., Mirny L., Spitz F., Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haarhuis J. H. I., van der Weide R. H., Blomen V. A., Yáñez-Cuna J. O., Amendola M., van Ruiten M. S., Krijger P. H. L., Teunissen H., Medema R. H., van Steensel B., Brummelkamp T. R., de Wit E., Rowland B. D., The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidson I. F., Bauer B., Goetz D., Tang W., Wutz G., Peters J. M., DNA loop extrusion by human cohesin. Science 366, 1338–1345 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Kim Y., Shi Z., Zhang H., Finkelstein I. J., Yu H., Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Haarhuis J. H. I., Cacciatore Á. S., Oldenkamp R., van Ruiten M. S., Willems L., Teunissen H., Muir K. W., de Wit E., Rowland B. D., Panne D., The structural basis for cohesin-CTCF-anchored loops. Nature 578, 472–476 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wutz G., Ladurner R., Glenn St Hilaire B., Stocsits R. R., Nagasaka K., Pignard B., Sanborn A., Tang W., Varnai C., Ivanov M. P., Schoenfelder S., van der Lelij P., Huang X., Durnberger G., Roitinger E., Mechtler K., Finley Davidson I., Fraser P., Lieberman-Aiden E., Peters J.-M., ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinSTAG1 from WAPL. eLife 9, e52091 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papantonis A., Cook P. R., Fixing the model for transcription: The DNA moves, not the polymerase. Transcription 2, 41–44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giorgetti L., Lajoie B. R., Carter A. C., Attia M., Zhan Y., Xu J., Chen C. J., Kaplan N., Chang H. Y., Heard E., Dekker J., Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulianov S. V., Khrameeva E. E., Gavrilov A. A., Flyamer I. M., Kos P., Mikhaleva E. A., Penin A. A., Logacheva M. D., Imakaev M. V., Chertovich A., Gelfand M. S., Shevelyov Y. Y., Razin S. V., Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 26, 70–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowley M. J., Nichols M. H., Lyu X., Ando-Kuri M., Rivera I. S. M., Hermetz K., Wang P., Ruan Y., Corces V. G., Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hug C. B., Grimaldi A. G., Kruse K., Vaquerizas J. M., Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169, 216–228 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Naughton C., Avlonitis N., Corless S., Prendergast J. G., Mati I. K., Eijk P. P., Cockroft S. L., Bradley M., Ylstra B., Gilbert N., Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 20, 387–395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barutcu A. R., Blencowe B. J., Rinn J. L., Differential contribution of steady-state RNA and active transcription in chromatin organization. EMBO Rep. 20, e48068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brant L., Georgomanolis T., Nikolic M., Brackley C. A., Kolovos P., van Ijcken W., Grosveld F. G., Marenduzzo D., Papantonis A., Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol. Syst. Biol. 12, 891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagashima R., Hibino K., Ashwin S. S., Babokhov M., Fujishiro S., Imai R., Nozaki T., Tamura S., Tani T., Kimura H., Shribak M., Kanemaki M. T., Sasai M., Maeshima K., Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J. Cell Biol. 218, 1511–1530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Khattabi L., Zhao H., Kalchschmidt J., Young N., Jung S., Van Blerkom P., Kieffer-Kwon P., Kieffer-Kwon K.-R., Park S., Wang X., Krebs J., Tripathi S., Sakabe N., Sobreira D. R., Huang S.-C., Rao S. S. P., Pruett N., Chauss D., Sadler E., Lopez A., Nóbrega M. A., Lieberman-Aiden E., Asturias F. J., Casellas R., A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178, 1145–1158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiecke M. J., Wutz G., Muhar M., Tang W., Bevan S., Malysheva V., Stocsits R., Neumann T., Zuber J., Fraser P., Schoenfelder S., Peters J. M., Spivakov M., Cohesin-dependent and -independent mechanisms mediate chromosomal contacts between promoters and enhancers. Cell Rep. 32, 107929 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh T. H. S., Cattoglio C., Slobodyanyuk E., Hansen A. S., Rando O. J., Tjian R., Darzacq X., Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78, 539–553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busslinger G. A., Stocsits R. R., van der Lelij P., Axelsson E., Tedeschi A., Galjart N., Peters J.-M., Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz S., Texari L., Hayes M. G. B., Urbanowski M., Chang M. W., Givarkes N., Rialdi A., White K. M., Albrecht R. A., Pache L., Marazzi I., García-Sastre A., Shaw M. L., Benner C., Transcription elongation can affect genome 3D structure. Cell 174, 1522–1536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olan I., Parry A. J., Schoenfelder S., Narita M., Ito Y., Chan A. S. L., St C. Slater G., Bihary D., Bando M., Shirahige K., Kimura H., Samarajiwa S. A., Fraser P., Narita M., Transcription-dependent cohesin repositioning rewires chromatin loops in cellular senescence. Nat. Commun. 11, 6049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowley M. J., Lyu X., Rana V., Ando-Kuri M., Karns R., Bosco G., Corces V. G., Condensin II counteracts cohesin and RNA polymerase II in the establishment of 3D chromatin organization. Cell Rep. 26, 2890–2903 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brandão H. B., Paul P., van den Berg A. A., Rudner D. Z., Wang X., MirnY L. A., RNA polymerases as moving barriers to condensin loop extrusion. Proc. Natl. Acad. Sci. U.S.A. 116, 20489–20499 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y., Huang J., Lun K., Li B., Zheng H., Li Y., Zhou R., Duan W., Wang C., Feng Y., Yao H., Li C., Ji X., Genome-wide analyses of chromatin interactions after the loss of Pol I, Pol II, and Pol III. Genome Biol. 21, 158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caudron-Herger M., Cook P. R., Rippe K., Papantonis A., Dissecting the nascent human transcriptome by analysing the RNA content of transcription factories. Nucleic Acids Res. 43, e95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Lu J. J., He L., Yu Q., Triptolide (TPL) inhibits global transcription by inducing proteasome-dependent degradation of RNA polymerase II (Pol II). PLOS ONE 6, e23993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sati S., Bonev B., Szabo Q., Jost D., Bensadoun P., Serra F., Loubiere V., Papadopoulos G. L., Rivera-Mulia J. C., Fritsch L., Bouret P., Castillo D., Gelpi J. L., Orozco M., Vaillant C., Pellestor F., Bantignies F., Marti-Renom M. A., Gilbert D. M., Lemaitre J. M., Cavalli G., 4D genome rewiring during oncogene-induced and replicative senescence. Mol. Cell 78, 522–538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vian L., Pekowska A., Rao S. S. P., Kieffer-Kwon K.-R., Jung S., Baranello L., Huang S.-C., El Khattabi L., Dose M., Pruett N., Sanborn A. L., Canela A., Maman Y., Oksanen A., Resch W., Li X., Lee B., Kovalchuk A. L., Tang Z., Nelson S., Di Pierro M., Cheng R. R., Machol I., Glenn St Hilaire B., Durand N. C., Shamim M. S., Stamenova E. K., Onuchic J. N., Ruan Y., Nussenzweig A., Levens D., Lieberman-Aiden E., Casellas R., The energetics and physiological impact of cohesin extrusion. Cell 173, 1165–1178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodes J. D. P., Feldmann A., Hernandez-Rodriguez B., Diaz N., Brown J. M., Fursova N. A., Blackledge N. P., Prathapan P., Dobrinic P., Huseyin M. K., Szczurek A., Kruse K., Nasmyth K. A., Buckle V. J., Vaquerizas J. M., Klose R. J., Cohesin disrupts polycomb-dependent chromosome interactions in embryonic stem cells. Cell Rep. 30, 820–835 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abramo K., Valton A. L., Venev S. V., Ozadam H., Fox A. N., Dekker J., A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat. Cell Biol. 21, 1393–1402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H., Emerson D. J., Gilgenast T. G., Titus K. R., Lan Y., Huang P., Zhang D., Wang H., Keller C. A., Giardine B., Hardison R. C., Phillips-Cremins J. E., Blobel G. A., Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576, 158–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsiung C. C. S., Bartman C. R., Huang P., Ginart P., Stonestrom A. J., Keller C. A., Face C., Jahn K. S., Evans P., Sankaranarayanan L., Giardine B., Hardison R. C., Raj A., Blobel G. A., A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes Dev. 30, 1423–1439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baranello L., Wojtowicz D., Cui K., Devaiah B. N., Chung H. J., Chan-Salis K. Y., Guha R., Wilson K., Zhang X., Zhang H., Piotrowski J., Thomas C. J., Singer D. S., Pugh B. F., Pommier Y., Przytycka T. M., Kouzine F., Lewis B. A., Zhao K., Levens D., RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165, 357–371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thakurela S., Garding A., Jung J., Schübeler D., Burger L., Tiwari V. K., Gene regulation and priming by topoisomerase IIα in embryonic stem cells. Nat. Commun. 4, 2478 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Singh S., Szlachta K., Manukyan A., Raimer H. M., Dinda M., Bekiranov S., Wang Y. H., Pausing sites of RNA polymerase II on actively transcribed genes are enriched in DNA double-stranded breaks. J. Biol. Chem. 295, 3990–4000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uusküla-Reimand L., Hou H., Samavarchi-Tehrani P., Rudan M. V., Liang M., Medina-Rivera A., Mohammed H., Schmidt D., Schwalie P., Young E. J., Reimand J., Hadjur S., Gingras A. C., Wilson M. D., Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 17, 182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Achar Y. J., Adhil M., Choudhary R., Gilbert N., Foiani M., Negative supercoil at gene boundaries modulates gene topology. Nature 577, 701–705 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Nielsen C. F., Zhang T., Barisic M., Kalitsis P., Hudson D. F., Topoisomerase IIα is essential for maintenance of mitotic chromosome structure. Proc. Natl. Acad. Sci. U.S.A. 117, 12131–12142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casa V., Gines M. M., Gusmao E. G., Slotman J. A., Zirkel A., Josipovic N., Oole E., Van Ijcken W. F. J., Houtsmuller A. B., Papantonis A., Wendt K. S., Redundant and specific roles of cohesin STAG subunits in chromatin looping and transcriptional control. Genome Res. 30, 515–527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelham-Webb B., Polyzos A., Wojenski L., Kloetgen A., Li J., Di Giammartino D. C., Sakellaropoulos T., Tsirigos A., Core L., Apostolou E., H3K27ac bookmarking promotes rapid post-mitotic activation of the pluripotent stem cell program without impacting 3D chromatin reorganization. Mol. Cell 81, 1732–1748 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buckle A., Brackley C. A., Boyle S., Marenduzzo D., Gilbert N., Polymer simulations of heteromorphic chromatin predict the 3D folding of complex genomic loci. Mol. Cell 72, 786–797.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen A. S., Pustova I., Cattoglio C., Tjian R., Darzacq X., CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6, e25776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davidson I. F., Goetz D., Zaczek M. P., Molodtsov M. I., Huis In ‘t Veld P. J., Weissmann F., Litos G., Cisneros D. A., Ocampo-Hafalla M., Ladurner R., Uhlmann F., Vaziri A., Peters J.-M., Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 35, 2671–2685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teves S. S., An L., Bhargava-Shah A., Xie L., Darzacq X., Tjian R., A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7, e35621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosencrance C. D., Ammouri H. N., Yu Q., Ge T., Rendleman E. J., Marshall S. A., Eagen K. P., Chromatin hyperacetylation impacts chromosome folding by forming a nuclear subcompartment. Mol. Cell 78, 112–126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuin J., Franke V., van IJcken W. F. J., van der Sloot A., Krantz I. D., van der Reijden M. I. J. A., Nakato R., Lenhard B., Wendt K. S., A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLOS Genet. 10, e1004153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y., Denholtz M., Lu H., Murre C., Calcium signaling instructs NIPBL recruitment at active enhancers and promoters via distinct mechanisms to reconstruct genome compartmentalization. Genes Dev. 35, 65–81 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu B., Comerci C. J., McCarthy D. G., Saurabh S., Moerner W. E., Wysocka J., Opposing effects of cohesin and transcription on CTCF organization revealed by super-resolution imaging. Mol. Cell 80, 699–711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amat R., Böttcher R., Le Dily F., Vidal E., Quilez J., Cuartero Y., Beato M., de Nadal E., Posas F., Rapid reversible changes in compartments and local chromatin organization revealed by hyperosmotic shock. Genome Res. 29, 18–28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durand N. C., Shamim M. S., Machol I., Rao S. S. P., Huntley M. H., Lander E. S., Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crane E., Bian Q., McCord R. P., Lajoie B. R., Wheeler B. S., Ralston E. J., Uzawa S., Dekker J., Meyer B. J., Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flyamer I. M., Illingworth R. S., Bickmore W. A., Coolpup.py: Versatile pile-up analysis of Hi-C data. Bioinformatics 36, 2980–2985 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rowley M. J., Poulet A., Nichols M. H., Bixler B. J., Sanborn A. L., Brouhard E. A., Hermetz K., Linsenbaum H., Csankovszki G., Lieberman-Aiden E., Corces G., Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. elegans to mammals. Genome Res. 30, 447–458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roukos V., Pegoraro G., Voss T. C., Misteli T., Cell cycle staging of individual cells by fluorescence microscopy. Nat. Protoc. 10, 334–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arrigoni L., Richter A. S., Betancourt E., Bruder K., Diehl S., Manke T., Bonisch U., Standardizing chromatin research: A simple and universal method for ChIP-seq. Nucleic Acids Res. 44, e67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramírez F., Dündar F., Diehl S., Grüning B. A., Manke T., DeepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meers M. P., Tenenbaum D., Henikoff S., Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watrin E., Schleiffer A., Tanaka K., Eisenhaber F., Nasmyth K., Peters J. M., Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 16, 863–874 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madsen J. G. S., Schmidt S. F., Larsen B. D., Loft A., Nielsen R., Mandrup S., iRNA-seq: Computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res. 43, e40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Risso D., Ngai J., Speed T. P., Dudoit S., Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brackley C. A., Johnson J., Kelly S., Cook P. R., Marenduzzo D., Simulated binding of transcription factors to active and inactive regions folds human chromosomes into loops, rosettes and topological domains. Nucleic Acids Res. 44, 3503–3512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L. A., Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plimpton S., Fast parallel algorithms for short-range molecular dynamics. J. Comp. Phys. 1, 1–19 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1, S3 and S4
Legends for tables S2 and S5
Tables S2 and S5