Transfer machinery, niche-specific, and metabolic genes are predictive of gene transfer events across diverse bacterial species.
Abstract
Phylogenetic distance, shared ecology, and genomic constraints are often cited as key drivers governing horizontal gene transfer (HGT), although their relative contributions are unclear. Here, we apply machine learning algorithms to a curated set of diverse bacterial genomes to tease apart the importance of specific functional traits on recent HGT events. We find that functional content accurately predicts the HGT network [area under the receiver operating characteristic curve (AUROC) = 0.983], and performance improves further (AUROC = 0.990) for transfers involving antibiotic resistance genes (ARGs), highlighting the importance of HGT machinery, niche-specific, and metabolic functions. We find that high-probability not-yet detected ARG transfer events are almost exclusive to human-associated bacteria. Our approach is robust at predicting the HGT networks of pathogens, including Acinetobacter baumannii and Escherichia coli, as well as within localized environments, such as an individual’s gut microbiome.
INTRODUCTION
Horizontal gene transfer (HGT) is a pervasive evolutionary process that results in the distribution of genes between divergent prokaryotic lineages. Although this process has shaped ancient evolution of microorganisms, recent transfer events underlie the spread of antibiotic or metal resistance genes, virulence factors, and other traits that have profound influence on the course of human infection. Widespread sequencing of complete prokaryotic genomes has made it possible to perform systematic, genome-scale comparisons to identify regions of HGT within genomes and to delineate features correlating with HGT rates, although these efforts have fallen short of being able to predict the dispersal of genes across microbes with high precision and accuracy.
Previous efforts to examine the mobility of genes have reported higher rates of HGT in phylogenetically related organisms, compared with those distantly related (1), and between organisms with shared GC content and kmer content (2), or methylation patterns (3). Genome content, such as the presence of specific plasmid replicon and mobilization machinery or shared phage structural proteins, also defines gene flow across microbial species (4–6), whereas restriction-modification genes, the presence of CRISPR-Cas9 adaptive immune systems, and toxin-antitoxin systems serve as barriers to gene flow. Genetic factors intrinsic to bacterial genomes or mobile elements favor transfer between closely related organisms due to their greater compatibility with native molecular machinery or larger stretches of sequence homology.
Alternatively, ecological architecture across various spatial scales also influences HGT rates, enriching HGT among organisms found within the same environment (e.g., marine, host-associated, and soil) or isolated from the same body site across multiple hosts. Many ecological traits are vertically inherited and therefore map onto physiochemical gradients or environmental resource patches (7). Yet, the acquisition and maintenance of mobile functional traits among neighbors increase the potential for ecology-specific adaptation and microbial speciation (8). This environmental selection can be observed in the contents of mobile genetic elements in the gut microbiomes across populations with different diets (9) and in the composition of mobile antibiotic resistance genes (ARGs) within the microbiomes of livestock subject to differing antibiotic burdens (10). This suggests that proximity, in addition to compatibility, is required for HGT.
Despite the appreciation for various factors’ effects on overall HGT rates, it has been difficult to arrive at a holistic understanding of HGT that encompasses and weighs these macro- and microlevel selective pressures. We hypothesized that functional gene content would be a strong determinant of HGT, as gene content reflects phylogenetic, genomic, and ecological factors simultaneously. To test this, we leveraged publicly available genome databases to create a network of HGT events. The network included genome-specific factors, such as functional content (node features), and relative factors, such as phylogenetic distance and cooccurrence (edge features). We implemented several machine learning approaches, namely, logistic regression (LR), random forest (RF), and graphical convolutional neural network (GCN) models, to quantify their effect on HGT, because of their versatility, their demonstrated utility for genomics (11) and bacterial phenotypes (12), and their ability to predict multidimensional links in networks. Furthermore, these methods allow us to use node and edge effects to account for the nonindependence of events and features in the network, which is important when parsing the complex etiologies of HGT events.
RESULTS
The HGT network is highly predictable
We constructed an undirected network of observed gene transfer events (Fig. 1A). After downloading genomes from several public databases, we took special care to remove any potential contaminants (host- or vector-borne) that could be erroneously annotated as HGT among these genomes and any genomes that had low completeness (<90%) or high contamination scores (>5%), as determined by CheckM (13). This resulted in a dataset consisting of 12,518 isolated and sequenced genomes, representing over 10,500 bacterial species, obtained from publicly available datasets (table S1). To reduce sampling bias, we selected a maximum of three isolates per species or 97% 16S ribosomal RNA (rRNA) similarity cluster (fig. S1, A and B). Because of the computational limitations of applying phylogeny-based approaches for HGT detection on a genome set of this size and the challenges of identifying recent transfer between closely related organisms at scale, we used a previously vetted heuristic (1) to identify organisms that have engaged in HGT. HGT-positive edges were defined as those between distantly related organisms (with less than 97% sequence similarity in their 16S rRNA) that harbor near-identical (99% or greater sequence similarity) regions of DNA of at least 500 base pairs (bp) (1). Our final HGT network was sparse, consisting of 147,889 observed HGT events among 6566 genomes or 0.189% of roughly 78.3 million pairwise comparisons (Fig. 1A).
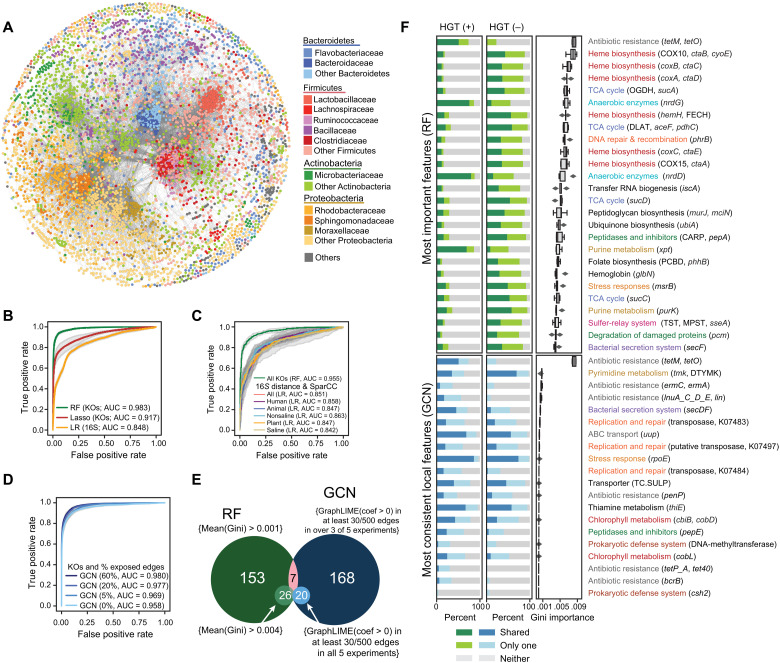
Fig. 1. The functional content of genomes accurately predicts HGT rates.
(A) A network diagram showing organisms (nodes) connected by at least one observed HGT event (edges). Organisms are colored according to taxonomy. (B) Receiver operating characteristic (ROC) curves for an LR using only 16S rRNA sequence similarity (yellow), a Lasso model using functions (KOs) (red), and an RF model using KOs (green). Area under the curve (AUC) values are shown. Details are in the Supplemental Information. (C) ROC curves for LR using full-length 16S rRNA similarity and ecological correlations based on sequences with near-identical similarity with 16S V4 rRNA sequences from the Earth Microbiome Project (EMP) and for an RF model using the KOs of organisms identified in the EMP. Details are in the Supplementary Materials. (D) ROC curves for graphical convolutional neural net (GCN) models, using functions (KOs) for each genome, as well as an uncensored portion of the test set’s adjacency matrix for predictions. AUC values are shown. Details are in the Supplementary Materials. (E) A Venn diagram of the number of KOs deemed important by the RF model and the number of KOs with positive GraphLime coefficients, as stated in the diagram. (F) KOs are listed according to whether they were found important by the RF model [mean(Gini) > 0.004] (top) or consistently had positive GraphLime coefficients in at least 30 of 500 edges in all five experiments (bottom). The mean(Gini) from the RF is shown, in addition to the percentage of HGT-positive and HGT-negative edges for which a feature is shared, present in one or absent from both.
We first tested the extent to which phylogeny alone could be used to predict HGT within the network. Overall, we observed a decay of the HGT rate with 16S rRNA distance, as previously shown (1) (fig. S1, C and D). To evaluate whether we could predict HGT using phylogeny, iteratively, we chose 500 genomes at random for each of five test datasets and chose a balanced set of edges with and without observed HGT (HGT-positive and HGT-negative edges, respectively) for the test set. To avoid overlap between the training and test data, we isolated them from each other by removing from the training set any genome in the same species-level taxon and any genome with greater than 97% 16S rRNA similarity to any genome in the test set. Using LR, we achieved decent HGT predictions using just 16S rRNA distance [mean area under the receiver operating characteristic curve (AUROC) = 0.848] (Fig. 1B and fig. S2).
Functional similarity imperfectly correlates with 16S rRNA distance (Spearman’s rho = 0.679) (fig. S3), owing to both ancient and recent HGT. We hypothesized that functional gene content, which captures traits relevant to survival in a specific niche, may serve as a better predictor of HGT than phylogeny. To test this, we first assigned gene functions to each genome’s open reading frames (ORFs) using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and, for each pair of genomes, recorded the function as shared, in one genome only, or absent in both. As expected, our ability to functionally annotate genomes was limited, with 45.91 ± 9.33% of genes per genome receiving KEGG ortholog (KO) functional assignments (fig. S4). Despite this limitation, the Lasso-regularized model using KO annotations alone (mean AUROC = 0.917) outperformed the model using 16S rRNA distances (Fig. 1B and fig. S2). This was further improved by using an RF-based classifier (mean AUROC = 0.983) with identical input data (Fig. 1B). It is anticipated that RF will outperform Lasso since it effectively uncovers nonlinear signals and predicts high-dimension tasks with a higher degree of efficiency and interpretability.
Shared ecology is often cited as a strong correlate with HGT (1, 14). Documentation of the source for isolates was often nonuniform and incomplete. Instead, to obtain comparable quantitative information about the distribution of each of the organisms in our dataset, we leveraged the Earth Microbiome Project (EMP) dataset (fig. S5, A to C), a single effort to comprehensively sample a range of biomes using standardized procedures. We cross-referenced the 19,724 EMP samples’ 16S rRNA amplicons with our dataset of isolate genomes, obtaining environmental distributions, using SparCC (15), for 9439 genomes (75.4% of our dataset). Our ability to assign functions to genomes according to their distribution was unbiased (fig. S5D). As has been reported, we find elevated HGT rates among organisms with similar ecological distributions (fig. S6A). Since neither phylogeny nor functional capacity correlates strongly with ecological cooccurrence (fig. S6, B and C), we tested them individually for their utility in predicting HGT. Neither ecological cooccurrence nor the combination of ecological cooccurrence with phylogeny predicts HGT as accurately as functional capacity alone (Fig. 1C and fig. S7).
We next hypothesized that there may be signatures of transfer embedded within the HGT network not encoded by gene functions or 16S rRNA similarities. To assess this, we applied a GCN model, a geometric deep learning approach, to the HGT network, to take advantage of GCNs’ ability to deal with hierarchical, as well as localized, patterns in the underlying data (fig. S8). This model considers the traits of each specific genome in the context of its neighbors in the network and, iteratively, the neighbors of their neighbors. We implemented the same conservative evaluation methods and training and test set isolation, as described above, for all models, to avoid label leakage from the test set to the training set and other sources of overestimated performance. With these evaluation measures in place, we titrated the amount of topographical information available to the GCN (16). We determined that the baseline GCN model trained on functional KO-based profiles achieved performance akin to the RF model (mean AUROC = 0.958) and that this was further enhanced as we included increasing amounts of network topographical information as input (Fig. 1D and fig. S9). The increase in the performance strongly suggests that the relationships in the HGT network itself provide added context to new HGT events that extend beyond what can be captured solely by functional annotation.
Niche-specific, metabolic, and mobile traits are important for predicting HGT
Given the performance of the RF and GCN models, we sought to examine which functional traits were exploited to generate predictions of HGT. Whereas feature importance can be readily extracted from RF models, the features used in the GCN’s two hidden layers are less immediately interpretable. We adapted the GraphLIME method (17), which measures the importance of features to particular nodes in the network by examining the features of their local subnetwork, to evaluate edge predictions instead. A subset of features was consistently observed across experiments (Fig. 1E and table S2). Despite large differences in structure and feature selection methods of the RF and GCN models, we found overlap, albeit limited, between the models’ respective important and consistent KOs used to predict HGT (Fig. 1E). Important features in the GCN likely implicate functions that promote/inhibit HGT within closely connected local components, while RF selects features that are broadly important across phylogeny.
Among the top important features used to predict HGT by the RF model (those with Gini impurities >0.001) (Fig. 1F and table S3) were metabolic traits, likely evident of shared environmental niches or cellular physiology, ARGs, and genes involved in the process of HGT. Among the niche-specific traits important for HGT were anaerobic enzymes nrdD and nrdG, which enable organisms to live in strict anaerobic conditions such as the human gut (18). The heme biosynthesis pathway, including ctaA, ctaB, ctaD, and hemH, and iron-containing molecules, such as hemoglobin, both require iron, likely reflecting niche-specific iron availability. Cobalamin (vitamin B12) biosynthesis genes cobD and cobL likely reflect a similar pattern for cobalt (19). ARGs from various classes were identified as important features, although predominantly by the GCN models. In addition, the presence or absence of several transposases, likely involved in HGT, and the CRISPR-associated gene csh2 reveal compatibility factors important for predicting HGT. Emphasized across the results of both models is the complexity of factors affecting the HGT network. Despite this, we find that just 26 KOs were sufficient to accurately predict the HGT network (fig. S10).
HGT network topology improves interphylum HGT predictions
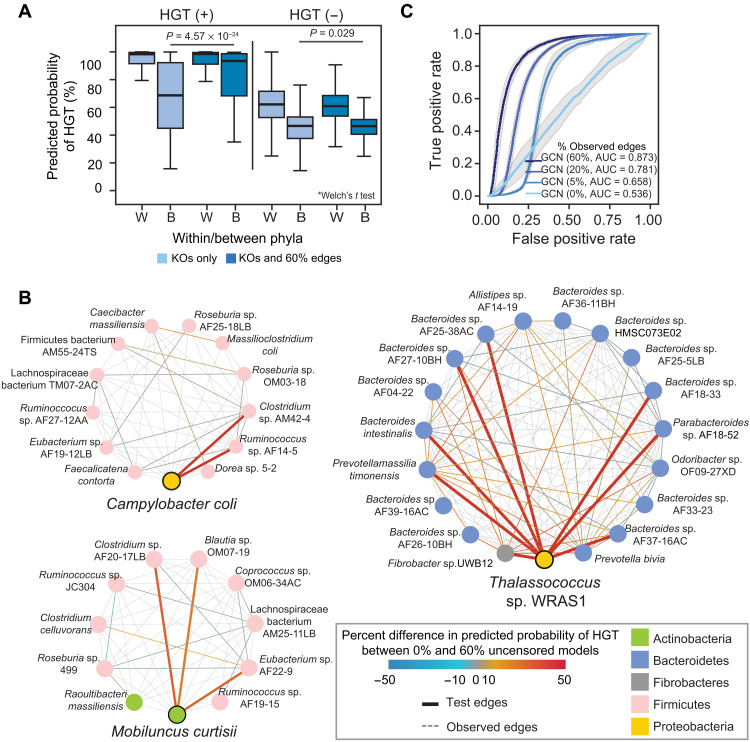
Interphylum HGT events are of particular importance as they likely contribute to the recent emergence of antibiotic resistance in pathogenic organisms after HGT with commensal organisms (20) and prehistorically underlie substantial shifts in speciation (14, 21). “Long-distance” HGT events, between distantly related organisms, are thought to be rare, except between species in certain extreme environments, such as between halophiles, thermophiles, saccharolytic, or fermentative organisms in termite or ruminant guts rich in organic matter (14, 22, 23). Yet, experiments support the feasibility of transfer of ARGs between Actinobacteria and Proteobacteria (24). Recent long-distance HGT events in our dataset represent only 11.87% (17,561 of 147,889) HGT-positive edges and are problematic to predict using 16S rRNA distance alone (mean AUROC = 0.499) (fig. S11A). We were particularly intrigued at how the inclusion of topographical information improved our GCN predictions of interphylum HGT (Fig. 2A). This is further illustrated in the transfer of tetO, a tetracycline resistance gene, between the Proteobacteria Campylobacter coli and the Actinobacteria Mobiluncus curtisii, and members of the Firmicutes, and in the transfer of a peptidoglycan lyase (GH23 family) involved in cell wall recycling between the Proteobacteria Thalassococcus sp. WRAS1 and both Bacteroides and Fibrobacteres genomes (Fig. 2B).
Fig. 2. Network topology is sufficient for predicting HGT.
(A) Predicted values for the GCN used in Fig. 1D of HGT-positive and HGT-negative edges for those involving species from the same phylum (within) or different phylum (between) when predicting on a test set with fully censored or 60% uncensored edges. The middle of the boxplot is the median, and edges are quartiles. P values are provided for the differences between predicted values for interphyla HGT-positive and HGT-negative edges using a Welch’s t test. (B) Three examples of the HGT-positive connections between organisms in the test sets used in the GCNs shown in Fig. 1D. In each, a single organism (Campylobacter coli, Mobiluncus curtisii, or Thalassococcus sp. WRAS1) is depicted with all of HGT-positive connections within the network. Test edges corresponding to interphylum transfers involving these species are thickened. Edges are either colored according to the difference in the HGT prediction in the fully censored and 60% uncensored networks or are dotted, gray edges, representing positive HGT edges that were uncensored in the test data. Genomes (nodes) are colored according to phylum. (C) ROC curves for GCN models using only the adjacency matrix for predictions, with 0, 5, 20, or 60% of the edges uncensored in the test set. These iterations were performed on the same training and test sets as in Fig. 1D. AUC values are shown.
The inclusion of additional network edges in the model resulted in tangible improvements in network predictions. We therefore hypothesized that sufficient information for HGT prediction was embedded in the network structure itself, even in the absence of gene functions or phylogenetic similarities. Inputting a randomized matrix of KO functions and starting with a fully censored network (16, 25), we see increasing accuracy as more observed edges are provided as input, ultimately achieving high performance using network topology alone (mean AUROC = 0.873 with 60% of observed edges) (Fig. 2C and fig. S11B). HGT predictions for observed HGT events were positively associated with a greater number of shared common HGT partners (Spearman’s rho = 0.82), which was not the case for HGT-negative edges (Spearman’s rho = 0.12), and with a higher minimum number of HGT partners between the pair than HGT-negative edges (fig. S12). These network embeddings may compensate for the limited functional annotations available, encounter rates, or other aspects that our model is unable to directly account for.
Transfer involving antibiotic resistance determinants is predictable by orthogonal functions
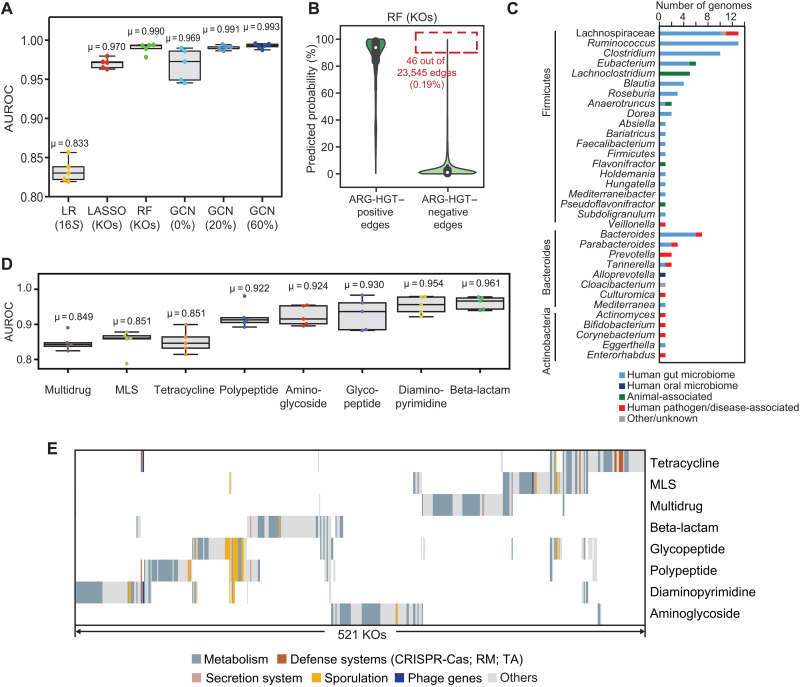
Given the clinical importance of emerging antibiotic resistance, we next evaluated our ability to predict transfers specifically involving one or more ARGs, which comprised 43.63% of observed HGT events and 47.44% of all interphylum transfers. We voided the input KO matrices of 645 KOs that shared even vague similarity to known ARGs. Despite fewer edges, our HGT predictions improved to near-perfect accuracy (RF: mean AUROC = 0.990; GCN with 60% uncensored edges, mean AUROC = 0.993) and the important features remain largely consistent as the model trained on all transfers (Fig. 3A; fig. S13, A and B; and table S4). Given this performance, we chose to examine the rare edges (46 of 23,545 edges, or 0.2%) within our test data that achieved high-prediction probabilities (over 0.9) of ARG-HGT but for which no transfers were detected (Fig. 3B). These edges were nearly exclusive for human-associated gut and oral microbiome members of the Firmicutes, Bacteroides, and Actinobacteria phyla (Fig. 3C and table S5); involved several pathobionts; and were distinct from a randomly permuted sample of HGT-negative edges (fig. S13C). These results highlight not only the promiscuous nature of the human microbiome but also the yet unseen potential for the further spread of ARGs to additional taxa.
Fig. 3. The transfer of ARGs genes is predictable.
(A) Area under the ROC curves for models predicting HGT involving ARGs is plotted for an LR model using only 16S rRNA sequence similarity, a Lasso model using the presence/absence of KOs for each genome, an RF model using KOs, and GCN models using KOs with decreasing censorship of the network (0 to 60% edges). Details are in the Supplementary Materials. Mean AUROC values (μ) are provided. (B) Prediction scores for ARG-HGT–positive and ARG-HGT–negative edges from the RF model using KOs [shown in (A)] are plotted. Of 23,545 total ARG-HGT-negative edges tested over five experiments, 46 HGT-negative edges (0.19%) had prediction scores over 0.9 (red dotted outline). (C) Genomes involved in the 46 ARG-HGT–negative edges that have predictions over 90%, depicted in the outlined box in (B), according to their phylum and origin of isolation, where available. For comparison, 46 HGT-negative edges chosen at random and depicted according to their origin of isolation in fig. S13. (D) Area under the ROC curves for multiclass RF models predicting HGT of genes conferring resistance to each specific class of antibiotics. Mean AUROC values (μ) are provided. (E) The top 1% of important KOs for each ARG class-specific model. Binary matrix was clustered on the basis of the presence and absence of important KOs by Pearson correlation. KOs were colored according to their annotation in the categories listed. KEGG pathway and BRITE functions of important KOs for each antibiotic class are in table S6.
As antibiotic classes have different spectra, mechanisms of action, distributions in nature, and societal uses, we asked, within those transfers involving ARGs, whether it was possible to predict the specific ARG class that was being transferred. Despite the concern over multidrug-resistant plasmids, only 8729 edges in the network involved more than one ARG class (fig. S14A). Overall, we found reliable predictions for all eight well-represented classes of ARGs (mean AUROCs between 0.849 and 0.961), with better performances obtained for classes involving fewer genes, genomes, and transfers (Fig. 3D and fig. S14B).
Models for each antibiotic class resulted in distinct subsets of important gene functions (Fig. 3E and table S6). These include important KOs that reflect each antibiotic’s mechanism of action. For instance, among the important features predicting the transfer of beta-lactamase resistance genes are glmM (26), a cell wall precursor enzyme, and ampG, a membrane permease required for cell wall recycling (27). Whereas the presence of ampG is associated with HGT-positive edges, glmM is absent between beta-lactam resistance HGT partners, aligning with reports showing that mutations in glmM result in increased sensitivity to beta-lactams (28). The muropeptides transported by ampG also act as signals to induce expression of the beta-lactamase, ampC (29). In accordance with the spectrum of glycopeptides and polypeptide antibiotics, we found numerous sporulation genes, largely restricted to Gram-positive Clostridia and Bacilli, as important features in the prediction of the transfer of these ARG classes.
Most of the genes observed across all classes are genes central to cellular metabolism, including amino acid, nucleotide, lipid, and carbohydrate metabolism (Fig. 3E and table S6). These features likely reflect protective metabolic adaptations to antibiotics (12, 30), such as through changes in oxidative phosphorylation (31), and to the acquisition of exogenous DNA (32), including genes that may alter transfer RNA pools (33), or purine and pyrimidine synthesis (12). Similar coping strategies involve altering transcriptional control of core or mobile genes, potentially explaining the large number of transcriptional two-component response systems identified as important features in our models. We also identify members of the Raetz pathway, lpxB, lpxC, lpxD, lpxH, and lpxL, all involved in the biosynthesis of the lipopolysaccharide component lipid A, as important features in predicting beta-lactam and polypeptide ARG transfer, whose members have also been identified as potential antibiotic targets (34). As for aminoglycosides, genes for all members of the sodium-translocating NADH (reduced form of nicotinamide adenine dinucleotide):quinone oxidoreductase complex were found to be important, supporting recent evidence for their role in promoting aminoglycoside resistance through modulating alanine metabolism (35). Metabolic functions are increasingly recognized for their role in promoting antibiotic resistance through experiments performed on Escherichia coli (36). Our approach confirms the importance of many of these functions in selection on mobile elements containing ARGs yet at a much broader taxonomic scale. It also exemplifies how this approach could be used to identify novel antibiotic targets.
Across many ARG classes, we identified prokaryotic defense genes as important features in our predictions, including toxin-antitoxin systems and related components (e.g., parC and mazF), enzymes involved in type I and type II restriction-modification systems (e.g., dam, hsdR, mcrA, and yhdJ, among others), and CRISPR-Cas proteins (e.g., cas1 and cas2). In addition, genes involved in the machinery and process of HGT were also identified: plasmid-segregation systems (e.g., parM), components of type IV coupling systems demarcating plasmid lineages (virD4), phage proteins (xtmB and rstA1), and proteins involved in recombination (xerC, rmuC, ruvC, and recU). We suspect that these barriers to HGT delimit subnetworks where genes are closely associated with specific transposable elements, phage, or plasmid lineages (37–39), as is the case of more recently mobilized colistin-resistance genes (40).
Predictions of ARG transfer involving pathogenic strains
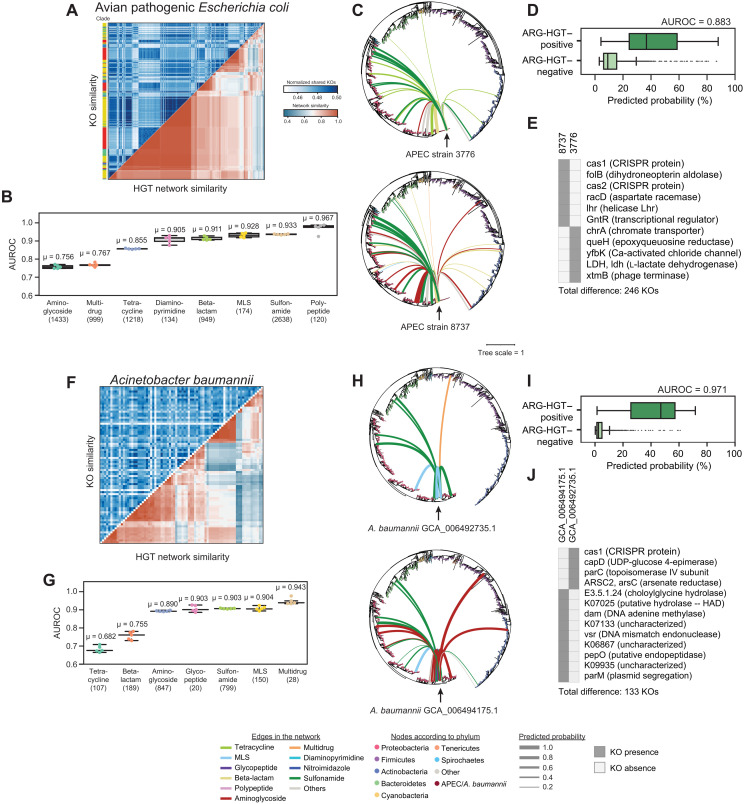
The promise of this work is its ability to predict the potential spread to and from pathogens. As a proof of concept, we retrospectively analyzed the HGT networks of collections of pathogenic isolates with our original dataset. First, we analyzed a collection of 433 diverse avian pathogenic E. coli strains collected over a period of nearly four decades (41) (table S7). Isolates within the same phylogenetic clade varied with respect to their observed HGT networks (Fig. 4A). We achieved reliable predictions in all ARG classes (mean AUROCs ranging from 0.756 to 0.967) (Fig. 4B). The predicted networks of two strains are illustrated (Fig. 4C), along with the presence and absence in important features distinguishing these two promiscuous genomes (Fig. 4, C to E), revealing the effects of relatively few differences in functional content on their respective networks. This mirrors observations that single mutations of metabolic genes lead to large changes in the transcriptome and antibiotic sensitivities (36).
Fig. 4. HGT is predictable across pathogenic strains of the same species.
(A) A heatmap showing normalized shared KOs for 445 avian pathogenic E. coli (APEC) isolates (top left) and their ARG-specific HGT network Jaccard similarity. Isolates are clustered according to HGT network similarity. The phylogenetic clade for each isolate is plotted adjacent to the heatmap. (B) Area under the ROC curves are plotted for the ARG classes for the APEC isolates for five RF models using the same dataset as Fig. 1, excluding E. coli and/or any organisms with 97% similarity to the 16S rRNA of any genome in the test set. Mean AUCs are provided above the boxplots. Boxplots represent median and quartile values. The number of edges containing at least one class-specific ARG is noted below the ARG class names. (C) The ARG-specific HGT network of two APEC isolates. The phylogenetic tree of all isolates includes 12,518 genomes from the original network, and 445 APEC isolates is shown with edges corresponding to predicted HGT events involving ARGs. The color of each edge corresponds to the ARG class, whereas the thickness of each edge is relative to the probability of ARG-HGT. (D) The distribution of ARG-HGT probabilities from the ARG-HGT RF model for ARG-HGT–positive (top) and ARG-HGT–negative (bottom) edges is shown. The AUROC is provided. (E) Important KOs that distinguish the two APEC strains in (C) are shown. (F to J) Same as (A) to (E) but for 96 clinically relevant Acinetobacter baumannii isolates.
We next examined a testing panel of 96 clinically relevant, diverse strains of Acinetobacter baumannii, a World Health Organization Priority 1 pathogen, chosen to represent a large breadth of virulence traits, antibiotic resistance determinants and phylogenetic diversity (42) (table S7). Despite high genetic diversity in this dataset, subsets of strains shared similar HGT networks (Fig. 4F). Class-specific ARG-HGT was generally predictable with mean AUROCs over 0.8 for aminoglycosides, glycopeptides, sulfonamides, MLS (macrolide, lincosamide and streptogramin), and multidrug resistance genes (Fig. 4G and fig. S15B). Features including HGT machinery genes and genes involved in barriers to gene flow captured differences in interphylum transfers between the networks of two A. baumannii clinical isolates (Fig. 4, H and J). Last, we tested our methods on randomly chosen genomes from a database of 4852 Neisseria gonorrhoeae isolates, collected across 15 studies and spanning 65 countries and 38 years (43) (table S7). Although these isolates carry numerous ARGs, mutation and recombination have obscured our ability to detect recent ARG-HGT events with members of the original network using our conservative heuristic. We only observed recent HGT of tetracycline- and beta-lactam–resistant genes, for which we obtain near-perfect classification (mean AUROCs = 1.000 and 0.994, respectively) (fig. S16).
Predictions of HGT are robust across datasets
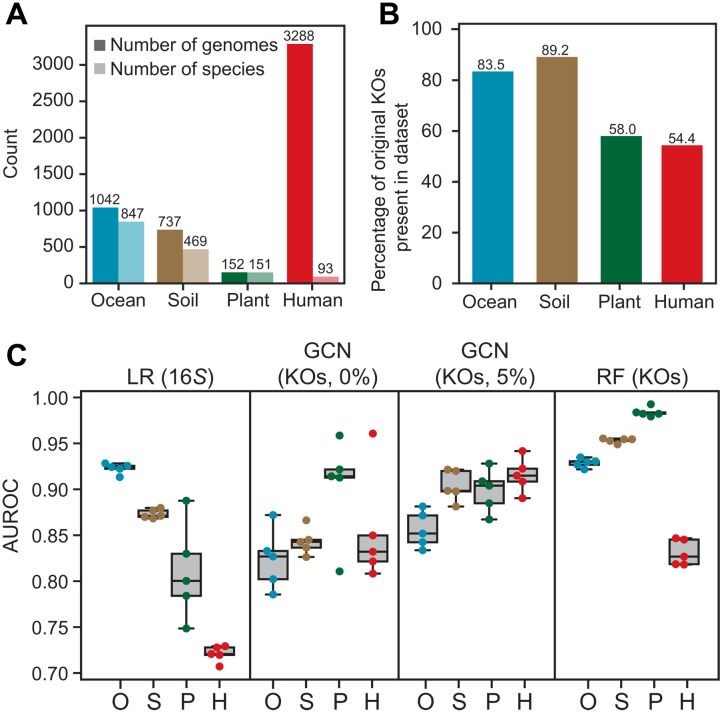
Given that selection may act at differing spatial scales (1), including at the level of individual hosts (9), we sought to determine whether our approach would be robust for HGT prediction within relatively small datasets, sourced from a single environment, or produced by a single laboratory or consortium, where dispersal of mobile genetic elements, rather than selection, may dominate the signal (44). We identified four orthogonal isolate datasets from various environments [ocean (45), soil (46), plant root (47), and human gut (48)] (table S8) and applied our original model built on >12,000 isolates to predict HGT. The ocean dataset comprised 847 species, whereas the human gut dataset contained 3288 high-quality genomes, albeit representing only 93 species (Fig. 5A). The rate of HGT varied significantly between datasets: The highest was within the human gut (17.92%), as has been observed previously (9, 20, 49), the marine and soil datasets were over 60-fold lower (at 0.279 and 0.237%, respectively), and only 26 HGT events were observed in the plant root dataset. Since only a subset of the KOs in the original dataset were represented, as little as 54.4% in the human gut datasets (Fig. 5B), we questioned whether our models would be sufficient at capturing HGT networks within these datasets.
Fig. 5. Predictions of HGT are accurate for small ecology–specific datasets.
(A) The number of genomes and species in each of four datasets: ocean surface sampling, soil microbial communities, rhizomes from three plant species, and human gut microbiomes from 11 individuals. (B) The number of KOs overlapping with the HGT model (shown in Fig. 1B) used for predictions. (C) Model performance, defined by AUROC, for HGT predictions within the ocean, soil, plant and human gut microbiome datasets, using either the LR model based on 16S rRNA distances, the GCN model using gene functions (KOs) with either none or 5% of the network exposed, or the RF model using gene functions (KOs).
Across all datasets, HGT was predictable with high accuracy (Fig. 5C and fig. S17). Within the ocean and soil datasets, HGT rates correlated strongly with phylogeny (fig. S13), reflected in the performance of the LR using 16S rRNA distance. Apart from the ocean dataset, gene functions improved overall predictions of HGT, most notably in the plant root dataset (Fig. 5C). Next, we were curious whether our model would still retain predictive ability for HGT occurring within an individual’s microbiome, where selective pressures may be highly personal (9, 50). Testing on a dataset of 11 individuals, with over 130 high-quality genomes per individual, we observed that the rates of intrapersonal HGT were higher than interpersonal HGT (21.879% versus 17.053%, respectively), but the HGT networks within individuals were predictable (fig. S18), suggesting that our model is capturing generalizable patterns governing HGT, within and across ecosystems.
DISCUSSION
Patterns of HGT can be extracted from large-scale HGT networks. Using machine learning, we identify specific prokaryotic functions that define recent HGT events, gaining insight into the relative contributions of various drivers of HGT, including niche-specific attributes, mechanistic barriers to HGT, and features relevant to subsets of transferred genes. Our analysis reveals a set of likely compensatory and adaptive functions that may enable organisms to accommodate the cellular stresses associated with gene acquisition (32, 33), may be genetically linked at hotspots for mobile genetic element integration (51), or relate to the function of ARGs per antibiotic classes or other functions encoded on mobile elements (52). Machine learning algorithms are sensitive to biases in the training data. Despite our best efforts to select a representative dataset, our approach is sensitive to the quality of input data (i.e., metagenomic-assembled genomes were excessively noisy and were therefore excluded), and there are bound to be blind spots. Further improvements in enhanced culturomics and continued improvements in metagenomic and single-cell assembly would enable microbiome-wide HGT network prediction. Similarly, phylogenetic reconstruction applicable to recent time scales (53) or among closely related strains that can be applied at scale may provide greater resolution and directionality of HGT events.
To our surprise, the predictability of recent HGT events not only was evident at the broadest scale, across phylum and biomes, but also extended to specific environments, such as an individuals’ gut or plant’s rhizome, and even within single clades of pathogenic species, suggesting that signatures of selection dominate over stochastic sampling. The predictability of recent HGT events provides us with a better understanding of bacterial adaptation to rapidly shifting conditions, such as those brought about by the anthropogenic dissemination of antibiotics. This opens the possibility of quantifying the risk of HGT between pathogens and microbiome constituents that lead to the emergence of novel antibiotic resistance strains and the expansion of ARG reservoirs within a localized context. This framework may be leveraged to improve the design of mobile genetic elements intended for engineering the microbiome (44) or inform strategies to reduce ARG burden by curing or eliminating plasmids or inhibiting conjugation.
METHODS
Genome collection
The sequences of 47,373 bacterial and archaeal genomes were downloaded from the National Center for Biotechnology Information (NCBI) and Pathosystems Resource Integration Center (PATRIC) in October 2020, including 1520 recently published nonredundant genomes from cultivated human gut bacteria (54) and 31,911 species-level representatives from the Genome Taxonomy Database (GTDB) (55) release 95 (from NCBI Assembly/Refseq). We excluded metagenomic assembled genomes (MAGs); genomes labeled as coming from an “environmental source,” as it was difficult to consistently determine which of these were MAGs; or single-cell genomes based on NCBI assembly metadata (downloaded on 6 November 2020). To avoid contaminating sequences from appearing as regions of HGT, we carefully screened genomes for host DNA and vector sequence contamination. We used conterminator (56), with default settings, to screen host DNA from their database of hosts, which includes Saccharomyces cerevisiae, Danio rerio, Mus musculus, Drosophila melanogaster, Arabidopsis thaliana, Caenorhabditis elegans, and Homo sapiens. We additionally mapped selected human repeats not present in the current human reference genome (57), GRCh38 (LINE family: L1HS_3end, L1HS_5end, L1MC4_3end, L1P1_orf2, and L1PBa_5end, L2; Alu family: AluJo, AluSg, AluSx, AluSz, AluY, BC200, and FRAM; satellites: ACRO1, ALR, BSR, and HSATII; LTR EVRs: ERVL, MER5A, MIR, MIRb, MST-int, MSTB, and THE1-int; Tigger1) against all prokaryotic genomes, removing genomes that were mapped with HMMER software (58) v3.3 at e values below 10−10. Contigs that were predicted as cross-kingdom contaminations were removed from the genomes. To remove vector contamination, we searched genomes against the UniVec database v10.0 using BLASTn (with Vecscreen parameter -reward 1 -penalty -5 -gapopen 3 -gapextend 3 -dust yes -soft_masking true –evalue 700 -searchsp 1750000000000 -outfmt 6). Contigs with vector contamination less than 100 kb were removed from the genome, or if a genome contained a contaminated contig greater than 100 kb, the entire genome was removed from our analysis.
Genome quality statistics were measured using CheckM (13) v1.1.2 (with parameters lineage_wf --tab_table -x fna), and high-quality genomes (more than 90% completeness and less than 5% contamination) were retained for constructing HGT network in the downstream analysis. 16S rRNA genes from the high-quality genomes were identified with the RNAmmer (59) v1.2. We removed genomes for which we could not identify near–full-length 16S sequences (>1000 bp). Overrepresented species (with over three representative strains) were subsampled down to three genomes, resulting in a total of 12,518 high-quality genomes representing more than 10,500 unique bacterial species included in our dataset (table S1).
Construction of the HGT network
Near full-length 16S rRNA sequences were aligned using MAFFT (60) v7.453 (auto), and a sequence similarity matrix was calculated using Clustal Omega (61) v1.2.4 (with parameter: --full --percent-id). We defined recent HGT between two distantly related genomes (less than 97% 16S rRNA sequence similarity) by the identification of at least one shared region of DNA of at least 500 bp with 99% or greater similarity by BLASTn, as originally described by Smillie et al. (1) and used elsewhere (9). A total of ~78 million pairwise comparisons were performed. A total of 6566 genomes showed evidence of at least one HGT event, which were used to build the binary HGT network using Networkx v2.4.
Functional annotation of high-quality genomes
ORFs were predicted in all high-quality genomes using Prodigal (62) v2.6.3. The resulting coding sequences were annotated by aligning to the KEGG functional database (KEGG database; Release 79.0) using Diamond (63) (blastp --id 50 --max-target-seqs 1). Within a single genome, genes assigned to the same KO were aggregated so that only the binary status (presence/absence) of KOs was considered. We constructed a KO-genome matrix based on KO presence/absence to use as a feature map for downstream modeling. The number of shared KO Nshared between two genomes X and Y was normalized using the following formula
where the number of annotated KOs in genome Y is larger than in X: Ny ≥ Nx.
Functional pathways were annotated using BRITE pathways and grouped into larger categories manually. Some KOs were associated with multiple pathways.
Identification of transferred ARGs
We extracted all transferred DNA sequences from each genome using extractseq from EMBOSS (64) v6.6.0 and predicted ORFs using Prodigal (62) v2.6.3. We excluded partial ORFs in our analysis. Using Diamond (63) v0.9.34 (with parameter: e value <10–5, >40% identity at the protein level, and >80% query sequence coverage), gene sequences were searched against the database ARG-miner (65) v1.1.1.A. Antibiotic resistance classes, as defined by ARG-miner, that were involved in fewer than 1000 edges on the HGT network were regrouped into “other” category (including “nitroimidazole,” “aminocoumarin,” “fosfomycin,” “phenicol,” “fluoroquinolone,” “sulfonamide,” “pleuromutilin,” “nucleoside,” “mupirocin,” “unclassified,” “fosmidomycin,” “rifamycin,” “elfamycin,” “oxazolidinone,” “tetracenomycin,” “triclosan,” “bicyclomycin,” “qa_compound,” and “acridine dye”).
Species-level cooccurrence estimation
All 16S rRNA genes from high-quality genomes were aligned to all V4 operational taxonomic units (OTUs) identified in the EMP (66) (~8 million OTU single-end representative sequence reads of 90 to 151 bp), which include amplicon-sequenced samples from various habitats, i.e., human, animal, plant, and saline/nonsaline environments. 16S rRNA sequences that share over 99% sequence similarity (fig. S5) over a span of at least 80 bp with EMP OTUs were assigned the same species name. The microbial abundance table used in the present study was the open_ref BIOM table from the EMP database (66). The OTU count table was based on the sequence data from the EMP database, which used open-reference OTU picking in QIIME (67). For samples from the same individual’s body site or the same animal, we randomly chose one to include in our analysis. EMP samples in different environments were categorized into five main groups (human, nonhuman animals, saline, nonsaline, and plant), which were used to calculate cooccurrence correlation coefficients using FastSpar (68) (−-iterations 50), a C++ implementation of the SparCC (15) algorithm.
Lasso, LR, and RF models
To predict HGT events using KEGG orthologs, we denote y as the response vector of the HGT state between genomes, y = (y1, y2…, yN) with binary values: 1 (at least one HGT event) or 0 (no detected HGT), and X as the matrix that contains the shared/nonshared status of KOs for each genome pair, X = ([X1, X2…, XN]) with categorical values: 2 (shared), 1 (only present in one), or 0 (in neither). Taking the advantage of scikit-learn (69) v0.22.2 in Python 3.5.5, we implement three other different machine learning models: LR, regularized LR (Lasso), and RF. To predict HGT using 16S rRNA similarity or cooccurrence correlation coefficients, in the case of LR, we used “L2” regularization to avoid overfitting. Best hyperparameters were determined by grid search method (“GridSearchCV”: cv = 5) (69) using a validation set, comprising 500 randomly chosen genomes from the training set. Models were evaluated on five randomly selected training and test sets. Special care was taken to avoid overlap between the test and training sets. Test datasets consist of 500 randomly selected nodes, unless otherwise stated. The corresponding training set for each experiment includes all remaining genomes, excluding those that are of the same species as any genome in the test set and/or those with ≥97% 16S rRNA sequence similarity. After training using the training dataset, the model is then applied to the test dataset, and the predicted results are compared with their original labels. For predicting HGT using KEGG orthologs, we fit a Lasso model (alpha = 0.01, fit_intercept = True, normalize = False, max_iter = 1000, tol = 0.0001) and an RF model [n_estimators = 5000, min_samples_split = 2, min_samples_leaf = 1, min_weight_fraction_leaf = 0.0, max_features = ‘auto’ (same as “sqrt”)] to our labeled training data. For consistency, direct model comparisons were all performed using the same training and test sets.
Prediction of HGT using graph convolutional network
We developed a GCN (70) using the TensorFlow (v1.13.1) and Pytorch (v1.6.0) framework, by first constructing an adjacency matrix, A ϵ RK×K (we assume that diagonal elements are set to 1, i.e., every node is connected to itself), encoding HGT events between its K genomes, and an ortholog feature matrix, X ϵ RK×C. GCN can be represented as a series of neighborhood aggregation layers: , where X(l) is a matrix of node embeddings at the lth layer, X(0) are input node attributes, W is a trainable parameter matrix, σ is a nonlinear activation function, and is the Laplacian-normalized adjacency matrix, defined as . Our model consists of two layers (32 × 16) to learn the HGT network, with a rectified linear unit (ReLU) activation function in the first layer
As the latent embedding vectors cover both functions and network information, the inner product decoder is used to predict potential HGT links. Let H denote the latent vector of the second layer. We calculate embeddings H = GCN(X, A) and the reconstructed adjacency matrix as follows: The score of potential HGT edges is calculated as , where σ is a sigmoid function for binary HGT predictions.
During model training, we minimize the following cost function with L1 regularization
where N denotes the size of adjacency matrix, Wp denotes the ratio of negative and positive edges , and .
To examine the predictive power of the network topology structure, censored networks were generated to mask a proportion of HGT edges and predict these missing edges (fig. S8), with and without including KO features. In the latter, the KO feature matrix X(0) was replaced with binary random matrices generated by “numpy.random.randint(2)” with the same shape of X for both training and test sets.
For consistency, direct model comparisons were all performed using the same training and test sets, as stated above: Five test sets consisting of 500 randomly sampled genomes were selected, and the corresponding training set was then defined by all remaining genomes, excluding any genome with the same species name and/or ≥97% rRNA similarity with any genome in the test set. All hyperparameters are determined through a grid search based on the model’s performance on the validation set, comprising 500 randomly chosen genomes from the training set. We used the ADAM optimizer with learning rate lr = 0.00005 and L1 loss scale (λ = 0.001). The minimum number of epochs for training is 100. To avoid overfitting, the ROC score of the validation set was examined after each epoch.
Model evaluation
The ROC curves were calculated on the relationship between the false positive and true positive rates using the sklearn.metrics.roc_curve function. The precision-recall curves were calculated by the sklearn.metrics.precision_recall_curve function [sklearn (69) v0.22.2]. The area under the curve, calculated using the sklearn.metrics.auc function, was used as a metric to benchmark the accuracy of a prediction model. Test data for evaluations were balanced by decreasing the number of samples of majority class.
Extracting KO importance
For RF, we obtained the “Gini importance” for each KO feature using the “.feature_importances” parameter from the scikit-learn (69) v0.22.2 implementation, which is defined as the total decrease in node impurity (weighted by the probability of reaching the node) averaged over all trees of the ensemble. For graph neural networks, GraphLIME (17) is an algorithm that measures the explanatory value of specific nodes or edge features within small subgraphs by individually considering the union of N-hop neighboring nodes, and then a nonlinear surrogate model, Hilbert-Schmidt Independence Criterion (HSIC) Lasso, is used to fit the local dataset. The subset of important features that explain the HSIC Lasso predictions are considered as the explanations of the original GCN prediction. We slightly modified the output of GraphLIME to perform the same task, but for edge predictions: Instead of extracting the neighborhood of a node, we extracted the union of the neighborhood of two nodes linked by an edge. For each experiment, 500 randomly selected HGT edges were used to learn important features, thereby determining a subset of node features that are most influential for the pretrained GCN prediction. For the GCNs, we focused on the subset of KOs that were consistently important across five separate GCN experiments. For each feature, we calculated the percentage of 10,000 randomly sampled HGT-positive and HGT-negative edges where the KO is present in both, one, or neither genome.
Antibiotic resistance transfer (HGT-ARG) prediction
HGT edges with putative ARGs were used to construct an ARG-specific HGT network. To avoid overfitting, we removed 645 KOs that shared 50% of more sequence similarity to any gene in the ARG-miner database. We then used the RF, Lasso, and GCN frameworks to predict a binary ARG-HGT network using the same hyperparameters as in the binary HGT predictors. The sources of isolates described in Fig. 3C and fig. S13, and listed in table S5, were manually curated according to their records in NCBI or PATRIC. We trained a set of RF (one-versus-rest binary classifiers for multiclass classification) to distinguish the transfer of different ARG classes using parameters (max_features = “auto,” min_samples_leaf = 3, min_samples_split = 4, n_estimators = 5000,class_weight = “balanced”) after tuning by grid search method (“GridSearchCV”: cv = 5) (69). Importance features of each RF predictor were extracted by “.feature_importances.” For the sole purpose of examining the network properties of the ARGs (fig. S14), we clustered putative ARGs using CD-HIT v4.6.8 (-c 0.5 -s 0.8). Longest shortest paths of each cluster were computed using the output of “.shortest_path_length” function from NetworkX.
We further apply these RF predictors to predict ARG transfers between our collected genomes and three sets of single-species isolates from E. coli (41), N. gonorrhoeae (43), and A. baumannii (42) (table S7). HGT links between the original dataset of genomes and these genomes were computed using the methods stated above. Shared regions of DNA were extracted, and putative ARGs in the shared regions were annotated. Each single-species isolate dataset was used as the test data. Genomes with the same species name and/or those with over 97% sequence similarity in 16S rRNA were removed from the training set. For each ARG class, a balanced, random set of negative ARG-specific HGT edges were chosen for evaluation purposes.
Predictions of HGTs in other datasets
Newly isolated genomes from multiple environments [ocean (MARMICRODB) (45), soil (46), plant root (47) and human gut (48)] were downloaded. Genomes were screened for human genome repeats, vector sequence contamination, and contamination from S. cerevisiae, D. rerio, M. musculus, D. melanogaster, A. thaliana, C. elegans, and H. sapiens using the same methods mentioned above. The final list of genomes used in our analysis can be found in table S8. HGT links within each dataset and between the dataset and our original genome dataset were computed. We used similar data splitting strategies as stated above: The training set for each experiment includes all preprocessed genomes, excluding those with the same taxonomy species name as any genome in the test set and those with ≥97% rRNA sequence similarity. LR, RF, and GCN were trained using only the genomes in our dataset and tested using the full dataset from each environment, each time choosing a balanced set of HGT-negative edges for evaluation.
Phylogenetic tree
To construct the phylogenetic tree, MAFFT (60) v7.453 (auto) was used to align 16S sequences. A neighbor-joining tree with nearest-neighbor interchange was estimated by FastTree v2.1.10 using default settings. The phylogenic tree was annotated and plotted using the Interactive Tree of Life (https://itol.embl.de/).
Acknowledgments
We wish to acknowledge members of the Brito Lab, T. Doerr, J. Helmann, and K. Weinberger for comments on the manuscript.
Funding: This work was supported by the NSF (#1661338) and the U.S. Department of Agriculture (2017-03796). I.L.B. is funded by the National Heart, Lung, and Blood Institute (1DP2HL141007-01) and is a Sloan Foundation Research Fellow, a Packard Fellowship in Science and Engineering, and a Pew Foundation Biomedical Scholar.
Author contributions: Conceptualization: H.Z., J.F.B., and I.L.B. Methodology: H.Z., J.F.B., and I.L.B. Investigation: H.Z., J.F.B., and I.L.B. Software: H.Z. and J.F.B. Visualization: H.Z. and I.L.B. Funding acquisition: I.L.B. Project administration: I.L.B. Supervision: I.L.B. Writing—original draft: H.Z. and I.L.B. Writing—review and editing: H.Z., J.F.B., and I.L.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All genome sequences are publicly available on NCBI and PATRIC. Accession numbers are listed for all genomes used in this study in tables S1, S7, and S8. For our analysis, we mainly relied on existing packages using default parameters, unless noted above. The code for the selection of nodes for the training/validation/test split was custom written and is available at https://doi.org/10.5281/zenodo.5337019 and https://github.com/britolab/GCN-HGT.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs S1 to S18
Tables S1 to S8
REFERENCES AND NOTES
- 1.Smillie C. S., Smith M. B., Friedman J., Cordero O. X., David L. A., Alm E. J., Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Azad R. K., Lawrence J. G., Towards more robust methods of alien gene detection. Nucleic Acids Res. 39, e56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaulaurier J., Zhu S., Deikus G., Mogno I., Zhang X.-S., Davis-Richardson A., Canepa R., Triplett E. W., Faith J. J., Sebra R., Schadt E. E., Fang G., Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat. Biotechnol. 36, 61–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acman M., van Dorp L., Santini J. M., Balloux F., Large-scale network analysis captures biological features of bacterial plasmids. Nat. Commun. 11, 2452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redondo-Salvo S., Fernández-López R., Ruiz R., Vielva L., de Toro M., Rocha E. P. C., Garcillán-Barcia M. P., de la Cruz F., Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11, 3602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popa O., Landan G., Dagan T., Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J. 11, 543–554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martiny J. B. H., Jones S. E., Lennon J. T., Martiny A. C., Microbiomes in light of traits: A phylogenetic perspective. Science 350, aac9323 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Soucy S. M., Huang J., Gogarten J. P., Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 16, 472–482 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Brito I. L., Yilmaz S., Huang K., Xu L., Jupiter S. D., Jenkins A. P., Naisilisili W., Tamminen M., Smillie C. S., Wortman J. R., Birren B. W., Xavier R. J., Blainey P. C., Singh A. K., Gevers D., Alm E. J., Mobile genes in the human microbiome are structured from global to individual scales. Nature 535, 435–439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munk P., Knudsen B. E., Lukjancenko O., Duarte A. S. R., Van Gompel L., Luiken R. E. C., Smit L. A. M., Schmitt H., Garcia A. D., Hansen R. B., Petersen T. N., Bossers A., Ruppé E.; EFFORT Group, Lund O., Hald T., Pamp S. J., Vigre H., Heederik D., Wagenaar J. A., Mevius D., Aarestrup F. M., Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 3, 898–908 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Eraslan G., Avsec Ž., Gagneur J., Theis F. J., Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 20, 389–403 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Yang J. H., Wright S. N., Hamblin M., McCloskey D., Alcantar M. A., Schrübbers L., Lopatkin A. J., Satish S., Nili A., Palsson B. O., Walker G. C., Collins J. J., A white-box machine learning approach for revealing antibiotic mechanisms of action. Cell 177, 1649–1661.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caro-Quintero A., Konstantinidis K. T., Inter-phylum HGT has shaped the metabolism of many mesophilic and anaerobic bacteria. ISME J. 9, 958–967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman J., Alm E. J., Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8, e1002687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.L. Franceschi, M. Niepert, M. Pontil, X. He, Learning discrete structures for graph neural networks. arXiv:1903.11960 [cs.LG] (29 March 2019).
- 17.Q. Huang, M. Yamada, Y. Tian, D. Singh, D. Yin, Y. Chang, GraphLIME: Local interpretable model explanations for graph neural networks. arXiv:2001.06216v2 [cs.LG] (27 September 2020).
- 18.Garriga X., Eliasson R., Torrents E., Jordan A., Barbé J., Gibert I., Reichard P., nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem. Biophys. Res. Commun. 229, 189–192 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Waldron K. J., Robinson N. J., How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7, 25–35 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Kent A. G., Vill A. C., Shi Q., Satlin M. J., Brito I. L., Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 11, 4379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loy A., Pfann C., Steinberger M., Hanson B., Herp S., Brugiroux S., Neto J. C. G., Boekschoten M. V., Schwab C., Urich T., Ramer-Tait A. E., Rattei T., Stecher B., Berry D., Lifestyle and horizontal gene transfer-mediated evolution of Mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems 2, e00171-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes M. E., Spear J. R., Oren A., House C. H., Differences in lateral gene transfer in hypersaline versus thermal environments. BMC Evol. Biol. 11, 199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchsman C. A., Collins R. E., Rocap G., Brazelton W. J., Effect of the environment on horizontal gene transfer between bacteria and archaea. PeerJ. 5, e3865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang X., Ellabaan M. M. H., Charusanti P., Munck C., Blin K., Tong Y., Weber T., Sommer M. O. A., Lee S. Y., Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 8, 15784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.P. V. Tran, Multi-task graph autoencoders. arXiv:1811.02798 [cs.LG] (7 November 2018).
- 26.Patel V., Wu Q., Chandrangsu P., Helmann J. D., A metabolic checkpoint protein GlmR is important for diverting carbon into peptidoglycan biosynthesis in Bacillus subtilis. PLOS Genet. 14, e1007689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dik D. A., Fisher J. F., Mobashery S., Cell-wall recycling of the gram-negative bacteria and the nexus to antibiotic resistance. Chem. Rev. 118, 5952–5984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimazu K., Takahashi Y., Uchikawa Y., Shimazu Y., Yajima A., Takashima E., Aoba T., Konishi K., Identification of the Streptococcus gordonii glmM gene encoding phosphoglucosamine mutase and its role in bacterial cell morphology, biofilm formation, and sensitivity to antibiotics. FEMS Immunol. Med. Microbiol. 53, 166–177 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Monteiro C., Fang X., Ahmad I., Gomelsky M., Römling U., Regulation of biofilm components in Salmonella enterica serovar Typhimurium by lytic transglycosylases involved in cell wall turnover. J. Bacteriol. 193, 6443–6451 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zampieri M., Enke T., Chubukov V., Ricci V., Piddock L., Sauer U., Metabolic constraints on the evolution of antibiotic resistance. Mol. Syst. Biol. 13, 917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dwyer D. J., Belenky P. A., Yang J. H., MacDonald I. C., Martell J. D., Takahashi N., Chan C. T. Y., Lobritz M. A., Braff D., Schwarz E. G., Ye J. D., Pati M., Vercruysse M., Ralifo P. S., Allison K. R., Khalil A. S., Ting A. Y., Walker G. C., Collins J. J., Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A. 111, E2100–E2109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiedenbeck J., Cohan F. M., Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Tuller T., Girshovich Y., Sella Y., Kreimer A., Freilich S., Kupiec M., Gophna U., Ruppin E., Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res. 39, 4743–4755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joo S. H., Lipid A as a drug target and therapeutic molecule. Biomol. Ther. 23, 510–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang M., Kuang S., Lai S., Zhang S., Yang J., Peng B., Peng X., Chen Z., Li H., Na+-NQR confers aminoglycoside resistance via the regulation of l-alanine metabolism. MBio 11, 6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopatkin A. J., Bening S. C., Manson A. L., Stokes J. M., Kohanski M. A., Badran A. H., Earl A. M., Cheney N. J., Yang J. H., Collins J. J., Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371, eaba0862 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira P. H., Touchon M., Rocha E. P. C., The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 42, 10618–10631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin J.-E., Lin C., Lim H. N., Horizontal transfer of DNA methylation patterns into bacterial chromosomes. Nucleic Acids Res. 44, 4460–4471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Che Y., Yang Y., Xu X., Břinda K., Polz M. F., Hanage W. P., Zhang T., Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl. Acad. Sci. U.S.A. 118, e2008731118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Dai X., Zeng J., Gao Y., Zhang Z., Zhang L., Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 10, 8113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mageiros L., Méric G., Bayliss S. C., Pensar J., Pascoe B., Mourkas E., Calland J. K., Yahara K., Murray S., Wilkinson T. S., Williams L. K., Hitchings M. D., Porter J., Kemmett K., Feil E. J., Jolley K. A., Williams N. J., Corander J., Sheppard S. K., Genome evolution and the emergence of pathogenicity in avian Escherichia coli. Nat. Commun. 12, 765 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galac M. R., Snesrud E., Lebreton F., Stam J., Julius M., Ong A. C., Maybank R., Jones A. R., Kwak Y. I., Hinkle K., Waterman P. E., Lesho E. P., Bennett J. W., Mc Gann P., A diverse panel of clinical Acinetobacter baumannii for research and development. Antimicrob. Agents Chemother. 64, 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma K. C., Mortimer T. D., Duckett M. A., Hicks A. L., Wheeler N. E., Sánchez-Busó L., Grad Y. H., Increased power from conditional bacterial genome-wide association identifies macrolide resistance mutations in Neisseria gonorrhoeae. Nat. Commun. 11, 5374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronda C., Chen S. P., Cabral V., Yaung S. J., Wang H. H., Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 16, 167–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker J. W., Hogle S. L., Rosendo K., Chisholm S. W., Co-culture and biogeography of Prochlorococcus and SAR11. ISME J. 13, 1506–1519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi J., Yang F., Stepanauskas R., Cardenas E., Garoutte A., Williams R., Flater J., Tiedje J. M., Hofmockel K. S., Gelder B., Howe A., Strategies to improve reference databases for soil microbiomes. ISME J. 11, 829–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levy A., Salas Gonzalez I., Mittelviefhaus M., Clingenpeel S., Herrera Paredes S., Miao J., Wang K., Devescovi G., Stillman K., Monteiro F., Rangel Alvarez B., Lundberg D. S., Lu T.-Y., Lebeis S., Jin Z., McDonald M., Klein A. P., Feltcher M. E., Rio T. G., Grant S. R., Doty S. L., Ley R. E., Zhao B., Venturi V., Pelletier D. A., Vorholt J. A., Tringe S. G., Woyke T., Dangl J. L., Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poyet M., Groussin M., Gibbons S. M., Avila-Pacheco J., Jiang X., Kearney S. M., Perrotta A. R., Berdy B., Zhao S., Lieberman T. D., Swanson P. K., Smith M., Roesemann S., Alexander J. E., Rich S. A., Livny J., Vlamakis H., Clish C., Bullock K., Deik A., Scott J., Pierce K. A., Xavier R. J., Alm E. J., A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 25, 1442–1452 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Groussin M., Poyet M., Sistiaga A., Kearney S. M., Moniz K., Noel M., Hooker J., Gibbons S. M., Segurel L., Froment A., Mohamed R. S., Fezeu A., Juimo V. A., Lafosse S., Tabe F. E., Girard C., Iqaluk D., Nguyen L. T. T., Shapiro B. J., Lehtimäki J., Ruokolainen L., Kettunen P. P., Vatanen T., Sigwazi S., Mabulla A., Domínguez-Rodrigo M., Nartey Y. A., Agyei-Nkansah A., Duah A., Awuku Y. A., Valles K. A., Asibey S. O., Afihene M. Y., Roberts L. R., Plymoth A., Onyekwere C. A., Summons R. E., Xavier R. J., Alm E. J., Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184, 2053–2067.e18 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Zhao S., Lieberman T. D., Poyet M., Kauffman K. M., Gibbons S. M., Groussin M., Xavier R. J., Alm E. J., Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe 25, 656–667.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira P. H., Touchon M., Cury J., Rocha E. P. C., The chromosomal organization of horizontal gene transfer in bacteria. Nat. Commun. 8, 841 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bethke J. H., Davidovich A., Cheng L., Lopatkin A. J., Song W., Thaden J. T., Fowler V. G., Xiao M., You L., Environmental and genetic determinants of plasmid mobility in pathogenic Escherichia coli. Sci. Adv. 6, eaax3173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Press M. O., Queitsch C., Borenstein E., Evolutionary assembly patterns of prokaryotic genomes. Genome Res. 26, 826–833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou Y., Xue W., Luo G., Deng Z., Qin P., Guo R., Sun H., Xia Y., Liang S., Dai Y., Wan D., Jiang R., Su L., Feng Q., Jie Z., Guo T., Xia Z., Liu C., Yu J., Lin Y., Tang S., Huo G., Xu X., Hou Y., Liu X., Wang J., Yang H., Kristiansen K., Li J., Jia H., Xiao L., 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 37, 179–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parks D. H., Chuvochina M., Waite D. W., Rinke C., Skarshewski A., Chaumeil P.-A., Hugenholtz P., A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Steinegger M., Salzberg S. L., Terminating contamination: Large-scale search identifies more than 2,000,000 contaminated entries in GenBank. Genome Biol. 21, 115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breitwieser F. P., Pertea M., Zimin A. V., Salzberg S. L., Human contamination in bacterial genomes has created thousands of spurious proteins. Genome Res. 29, 954–960 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn R. D., Clements J., Eddy S. R., HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lagesen K., Hallin P., Rødland E. A., Stærfeldt H.-H., Rognes T., Ussery D. W., RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K., Kuma K., Toh H., Miyata T., MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sievers F., Higgins D. G., Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hyatt D., Chen G.-L., LoCascio P. F., Land M. L., Larimer F. W., Hauser L. J., Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Rice P., Longden I., Bleasby A., EMBOSS: The European molecular biology open software suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Arango-Argoty G. A., Guron G. K. P., Garner E., Riquelme M. V., Heath L. S., Pruden A., Vikesland P. J., Zhang L., ARGminer: A web platform for the crowdsourcing-based curation of antibiotic resistance genes. Bioinformatics 36, 2966–2973 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Thompson L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., Prill R. J., Tripathi A., Gibbons S. M., Ackermann G., Navas-Molina J. A., Janssen S., Kopylova E., Vázquez-Baeza Y., González A., Morton J. T., Mirarab S., Xu Z. Z., Jiang L., Haroon M. F., Kanbar J., Zhu Q., Song S. J., Kosciolek T., Bokulich N. A., Lefler J., Brislawn C. J., Humphrey G., Owens S. M., Hampton-Marcell J., Berg-Lyons D., McKenzie V., Fierer N., Fuhrman J. A., Clauset A., Stevens R. L., Shade A., Pollard K. S., Goodwin K. D., Jansson J. K., Gilbert J. A., Knight R., A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., Alexander H., Alm E. J., Arumugam M., Asnicar F., Bai Y., Bisanz J. E., Bittinger K., Brejnrod A., Brislawn C. J., Brown C. T., Callahan B. J., Caraballo-Rodríguez A. M., Chase J., Cope E. K., Da Silva R., Diener C., Dorrestein P. C., Douglas G. M., Durall D. M., Duvallet C., Edwardson C. F., Ernst M., Estaki M., Fouquier J., Gauglitz J. M., Gibbons S. M., Gibson D. L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G. A., Janssen S., Jarmusch A. K., Jiang L., Kaehler B. D., Kang K. B., Keefe C. R., Keim P., Kelley S. T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M. G. I., Lee J., Ley R., Liu Y.-X., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B. D., McDonald D., McIver L. J., Melnik A. V., Metcalf J. L., Morgan S. C., Morton J. T., Naimey A. T., Navas-Molina J. A., Nothias L. F., Orchanian S. B., Pearson T., Peoples S. L., Petras D., Preuss M. L., Pruesse E., Rasmussen L. B., Rivers A., Robeson M. S., Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S. J., Spear J. R., Swafford A. D., Thompson L. R., Torres P. J., Trinh P., Tripathi A., Turnbaugh P. J., Ul-Hasan S., van der Hooft J. J. J., Vargas F., Vázquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y., Wang M., Warren J., Weber K. C., Williamson C. H. D., Willis A. D., Xu Z. Z., Zaneveld J. R., Zhang Y., Zhu Q., Knight R., Caporaso J. G., Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watts S. C., Ritchie S. C., Inouye M., Holt K. E., FastSpar: Rapid and scalable correlation estimation for compositional data. Bioinformatics 35, 1064–1066 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay É., Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 70.T. N. Kipf, M. Welling, Variational graph auto-encoders. arXiv:1611.07308 [stat.ML] (21 November 2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs S1 to S18
Tables S1 to S8