Abstract
In our database searches, we have identified mammalian homologues of yeast actin-binding protein, twinfilin. Previous studies suggested that these mammalian proteins were tyrosine kinases, and therefore they were named A6 protein tyrosine kinase. In contrast to these earlier studies, we did not find any tyrosine kinase activity in our recombinant protein. However, biochemical analysis showed that mouse A6/twinfilin forms a complex with actin monomer and prevents actin filament assembly in vitro. A6/twinfilin mRNA is expressed in most adult tissues but not in skeletal muscle and spleen. In mouse cells, A6/twinfilin protein is concentrated to the areas at the cell cortex which overlap with G-actin-rich actin structures. A6/twinfilin also colocalizes with the activated forms of small GTPases Rac1 and Cdc42 to membrane ruffles and to cell-cell contacts, respectively. Furthermore, expression of the activated Rac1(V12) in NIH 3T3 cells leads to an increased A6/twinfilin localization to nucleus and cell cortex, whereas a dominant negative form of Rac1(V12,N17) induces A6/twinfilin localization to cytoplasm. Taken together, these studies show that mouse A6/twinfilin is an actin monomer-binding protein whose localization to cortical G-actin-rich structures may be regulated by the small GTPase Rac1.
Actin is a ubiquitous protein, being found in all eukaryotic species and cell types. In muscle cells, actin filaments are stable and organized into highly ordered structures, sarcomeres. These actin filament structures, together with myosin filaments, form the framework for muscle contraction. In other cell types, actin filaments are often very dynamic and form a number of different structures called the actin cytoskeleton. In nonmuscle cells, the actin cytoskeleton is involved in multiple cell biological processes including cell morphogenesis, motility, endo- and exocytosis, and intracellular signal transduction (30).
To regulate the structure and dynamics of the actin cytoskeleton, a number of actin-binding proteins have evolved in eukaryotic organisms. These proteins can specifically interact with actin monomers or actin filaments to regulate the structural and biochemical properties of actin monomers and/or filaments. From the recent increase in sequence and structural data, it has become evident that a large number of actin-binding proteins are composed of a relatively small number of protein modules that interact with actin in a specific manner (25, 32). These protein modules include motifs that specifically interact with actin monomers (thymosin-β-4 repeat) as well as those that interact only with actin filaments (CH domain) (32).
An important class of actin-binding modules is the ADF (actin-depolymerizing factor) homology (ADF-H) domain, which has been found in three functionally distinct classes of actin-binding proteins so far. The ADF-H domain is a 15- to 20-kDa protein module that is able to interact both with actin monomers and actin filaments (15). This domain was originally found in cofilin/ADF proteins, which are composed of a single ADF-H domain (3). Cofilin/ADF proteins regulate actin dynamics in cells by increasing the rate of actin dissociation from the pointed end of the filament (9), and they also appear to have a weak actin filament-severing activity (18). In addition to their interactions with actin filaments, cofilin/ADF proteins also interact with actin monomers. The physiological roles of these actin monomer interactions of ADF/cofilin are not well understood. The ADF-H domain has more recently also been identified in the drebrin/Abp1 class of actin-binding proteins, which probably serve as a linker between actin filaments and other actin filament-associated proteins (16). Drebrin/Abp1 proteins interact only with actin filaments; they do not appear to have any effects on actin filament structure or dynamics (31). In drebrin/Abp1 proteins, the ADF-H domain is located at the N terminus of the protein and is followed by a variable region and in most cases by a C-terminal SH3 domain (15).
We have recently identified a third class of ADF-H domain proteins, twinfilins, which are composed of two ADF-H domains (15). Our biochemical studies with yeast twinfilin showed that it is an actin monomer-binding protein able to prevent actin filament polymerization in vitro. Despite the sequence homology to ADF/cofilin proteins, twinfilin does not bind or depolymerize actin filaments. Deletion of the twinfilin gene in yeast did not result in significant effects on growth kinetics, but twfΔ mutants appeared to have somewhat enlarged cortical actin patches and defects in bipolar budding pattern. These results, together with observed genetic interactions between twinfilin and the actin filament-depolymerizing protein cofilin, suggest that yeast twinfilin would be involved in actin filament dynamics in vivo (10). However, due to the lack of antibodies against twinfilin, we were unable to determine the localization of twinfilin in yeast cells and study how the activity and localization of this protein are regulated in vivo.
In our database searches, we also identified mammalian proteins with approximately 25% sequence identity to yeast twinfilin at the amino acid level. Interestingly, previous studies with these human and mouse proteins did not reveal any actin-related activities in these proteins but instead suggested that they would constitute a novel class of protein tyrosine kinases. Therefore, these proteins were named A6 tyrosine kinases (4, 5).
To elucidate whether these mammalian proteins are functional homologues of yeast twinfilin or whether they indeed are tyrosine kinases, we have carried out a biochemical and cell biological analysis of mouse A6/twinfilin protein. We found no evidence for the putative tyrosine kinase activity of this protein. However, we showed that mouse A6/twinfilin has an actin monomer binding/sequestering activity in vitro and that it is expressed in those mouse tissues that are characterized by high levels of actin dynamics. Furthermore, in mouse cell lines A6/twinfilin localizes to the highly dynamic areas of the cortical actin cytoskeleton, suggesting its role in the regulation of actin filament dynamics in vivo.
MATERIALS AND METHODS
Plasmid construction.
A DNA fragment corresponding to mouse A6/twinfilin cDNA was amplified by PCR from mouse cDNA which had been transcribed from NIH 3T3 cell mRNA. The oligonucleotides used in this amplification create NcoI and HindIII sites at the 5′ and 3′ ends of the PCR fragment, respectively. This fragment was then digested with NcoI and HindIII and ligated to NcoI-HindIII-digested plasmid pGAT2 (24) to create plasmid pPL64. The construct was sequenced by chain termination method, and clones containing undesired mutations were discarded. To create green fluorescent protein (GFP)-Rac1 fusion constructs, the mouse Rac1(12V) and Rac1(12V,17N) genes (a gift from Alan Hall, Ludwig Institute for Cancer Research, London, United Kingdom) were cloned into the BglII-HindIII sites of a modified pEGFP-C1 vector called pEGFP-C1A (J. Peränen et al., unpublished data). Constructs pEGFP-Cdc42(12V) and pEGFP-Cdc42(17N) were obtained by cloning the canine Cdc42 mutants into the EcoRI-HindIII sites of pEGFP-C1A. The GFP-RhoA fusions were constructed by cloning the human RhoA(14V) and RhoA(19N) genes into the EcoRI-HindIII sites of pEGFP-C1A (Peränen et al., unpublished data). The A6/twinfilin overexpression plasmid was constructed by subcloning the full-length mouse A6/twinfilin cDNA into EcoRV-HindIII sites of Myc-tagged pEGFP-N1 (Clontech)-based plasmid (Peränen, unpublished data). In cells, this plasmid drives expression of Myc-tagged full-length A6/twinfilin under control of the cytomegalovirus promoter.
Protein expression and purification.
Mouse A6/twinfilin was expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli JM109(DE3) cells under control of the T7 lac promoter. Cells were grown in 6,000 ml of Luria broth medium to an optical density of 0.5 at 600 nm, and expression was induced with 0.2 mM isopropyl-β-d-thiogalactosidase (IPTG). Cells were harvested 3 h after induction, washed with 100 ml of 20 mM Tris (pH 7.5), resuspended in 30 ml of phosphate-buffered saline (PBS)–0.15 mM phenylmethylsulfonyl fluoride, lysed by sonication, and centrifuged for 15 min at 15,000 × g. GST fusion protein was enriched from the lysis supernatant by using glutathione-agarose beads (2). A6/twinfilin-GST fusion proteins bound to GST-beads were incubated overnight with thrombin (0.05 mg/ml) at room temperature to cleave A6/twinfilin away from GST. The beads were washed five times with 20 mM Tris (pH 7.5)–150 mM NaCl. The supernatants were concentrated in Centricon 10-kDa-cutoff tubes to 1 ml and loaded onto a Superdex-75 HiLoad gel filtration column (Pharmacia Biotech) which had been equilibrated with 10 mM Tris (pH 7.5)–50 mM NaCl. The peak fractions containing A6/twinfilin eluted from the column at 60 ml were pooled and concentrated in Centricon 10-kDa-cutoff devices to a final concentration of 100 to 200 μM. The concentrated protein was divided into small aliquots, frozen in liquid N2, and stored at −70°C. The mouse A6/twinfilin was >95% pure, based on analysis of a Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gel. Human platelet nonmuscle actin (APH99; Cytoskeleton Inc.) was dissolved in G buffer in a concentration of 50 μM, divided into small aliquots, frozen in liquid N2, and stored at −70°C. Purified human Csk tyrosine kinase was a generous gift from Mathias Bergman (Department of Biochemistry, University of Helsinki, Helsinki, Finland).
In vitro kinase assay.
The kinase assay was carried out as described by Beeler et al. (4). Purified mouse A6/twinfilin (0.4 ng) or Csk tyrosine kinase (0.2 ng) was mixed with 2 μg of poly(Glu-Tyr) (4:1; Sigma Chemical Co.) in 20 μl of kinase buffer (50 mM HEPES [pH 7.5], 20 μM ATP, 1% NP-40, 10 mM MgCl2, 5 mM MnCl2); 10 μCi of γ-[32P]ATP (Amersham Pharmacia Biotech) was added to each reaction. After incubation for 15 min at room temperature, 7 μl of 4× SDS gel sample buffer (12) was added to each reaction and the samples were boiled for 2 min. Proteins were then resolved on an SDS–12% polyacrylamide gel, and the incorporation of 32P into poly(Glu-Tyr) was visualized with a phosphoimager.
Cosedimentation assay.
For actin filament cosedimentation assays, 40-μl aliquots of 1.1, 2.2, 4.4, 6.6, or 8.8 μM (human platelet) actin were prepared in G buffer (20 mM Tris-HCl, 0.2 μM ATP, 0.2 mM dithiothreitol, 0.2 mM CaCl2). Actin was then polymerized for 30 min by turning reaction buffer into F buffer by adding 5 μl of 10× polymerization initiation mix (20 mM MgCl2, 10 mM ATP, 1 M KCl); 5 μl of 5, 10, 20, 40, 60, or 80 μM mouse A6/twinfilin in G buffer (pH 7.5) was applied to premade filaments, and the mixture was incubated for 30 min. Samples were then centrifuged in Beckman Airfuge at 180,000 × g for 30 min in a 30° angle rotor. Equal proportions of supernatants and pellets were loaded on SDS–12% polyacrylamide gels, the gels were Coomassie blue stained, and the intensities of the actin and A6/twinfilin bands were quantified. All steps were carried out at room temperature.
Rate zonal gradient centrifugation.
Rate zonal gradient analysis to determine the association of human platelet actin and mouse A6/twinfilin was performed according to Juuti et al. (11) by using a linear 5 to 20% (wt/vol) sucrose gradient and a 40-h centrifugation for 35,000 rpm at 5°C in a Beckman SW40.1 rotor. Proteins were diluted in G buffer to a concentration of 0.5 mg/ml in a 100-μl reaction volume, which was then applied on top of the sucrose gradient prior to centrifugation. Gradients were fractioned manually using a peristaltic pump starting from the bottom of the tubes. Ovalbumin (43 kDa) and bovine serum albumin (BSA; 67 kDa) from Pharmacia Biotech were used as molecular weight standards. Protein distribution in the fractions was analyzed by using the silver staining protocol (7). A gel silver stain method was chosen over Coomassie blue staining of sucrose gradient proteins because of its inherently higher sensitivity, detection limits being 1 to 10 ng, versus 50 to 100 ng for the latter.
Northern blotting.
Plasmid pPL64 was digested with NcoI and HindIII and loaded on 1.0% agarose gel, and the mouse A6/twinfilin cDNA fragment was then purified with a Qiaex II kit (Qiagen). The purified twinfilin cDNA fragment and mouse β-actin cDNA control probe (Clontech) were labeled with [32P]CTP (Amersham Pharmacia Biotech) by using a random primed DNA labeling kit (Boehringer Mannheim) according to manufacturer's instructions. Labeled DNA fragments were then purified by passage through NICK columns (Amersham Pharmacia Biotech). Mouse A6/twinfilin and β-actin DNA probes were then hybridized to premade mouse multiple-tissue Northern blots (Clontech) for 60 min at 68°C, and each blot was washed according to the manufacturer's instructions. The Northern blot filter was then exposed on a phosphoimager screen for 120 min.
Antibodies.
One New Zealand White rabbit was immunized with purified recombinant mouse A6/twinfilin, and the serum was collected after four immunizations. Purified recombinant mouse A6/twinfilin was immobilized to CNBr-activated Sepharose 4B beads (Pharmacia), and the antibody was affinity purified from 1.5 ml of serum according to manufacturer's instructions. Fluorescein isothiocyanate (FITC)- and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated phalloidin and DNase I were obtained from Molecular Probes. Secondary antibodies (FITC-, TRITC-, and Cy5-conjugated goat anti-rabbit immunoglobulin G (IgG) and peroxidase-conjugated goat anti-rabbit (IgG) were all from Jackson Immunoresearch Inc.
Cell culture and immunofluorescence.
NIH 3T3 and N18 (mouse neuroblastoma) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (GIBCO), 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). For immunofluorescence studies NIH 3T3 cells were grown on glass coverslips coated with human fibronectin (5 μg/ml) to 60 to 80% confluence. N18 cells were grown at a density of 10,000 to 20,000 cells/coverslip on glass coverslips coated with laminin (20 μg/ml). NIH 3T3 cells were transfected with 1 μg of plasmid, using the Boehringer Mannheim FuGENE6 transfection system according to manufacturer's instructions. Cells were fixed with 3% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Primary antibodies were diluted in phosphate buffer containing 0.2% BSA. Dilutions were as follows DNase I, 1/500; FITC-phalloidin, 1/200; TRITC-phalloidin, 1/300; and anti-A6/twinfilin, 1/30. After incubation at room temperature for 30 min, coverslips were washed at least three times for 10 min each and then treated with fluorescent secondary antibodies, which were used at 1/200 dilution. After a further 30-min incubation, the coverslips were again washed, postfixed with 3% paraformaldehyde, and mounted in Mowiol containing 10% DABCO [1,4-diazobicyclo(2.2.2)octane].
Western blotting.
Cells were grown on Falcon 10-cm-diameter dishes to confluence, washed and harvested in cold PBS, and then lysed with 1% Triton X-100 in PBS containing 1/1,000 protease inhibitor cocktail (500 μg each of antipain, leupeptin, pepstatin, chymostatin, and aprotin [all from Sigma] per ml). The proteins in these cell lysates were separated in an SDS–12.5% polyacrylamide gel and electroblotted to a nitrocellulose membrane. The membrane was blocked at room temperature for 1 h using 5% dried milk in PBS containing 0.1% Tween 20 (PBST) and then incubated with anti-A6/twinfilin at a dilution of 1/500 in 5% milk–PBST. After 1 h, the membrane was washed three times with PBST and incubated for another hour with peroxidase-conjugated goat anti-rabbit IgG at 1/10,000 dilution. After washing, bound antibody was detected by an enhanced chemiluminescence method (Amersham).
Miscellaneous.
Polyacrylamide gel electrophoresis in the presence of SDS was carried out by using the buffer system of Laemmli (12). Protein concentrations were determined with a Hewlett-Packard 8452A diode array spectrophotometer and using calculated extinction coefficients for mouse twinfilin at 280 nm (ɛ = 36.4 mM−1 cm−1) and for human platelet nonmuscle actin at 290 to 320 nm (26.6 mM−1 cm−1). Protein distribution in polyacrylamide gels was quantified by using a Hewlett-Packard ScanJet 6200C and TINA software version 2.09c. The TINA program was also exploited to process the signal from a Fujifilm Basreader 1500 phosphorimager.
RESULTS
Mammalian A6 proteins are structural homologues of yeast twinfilin.
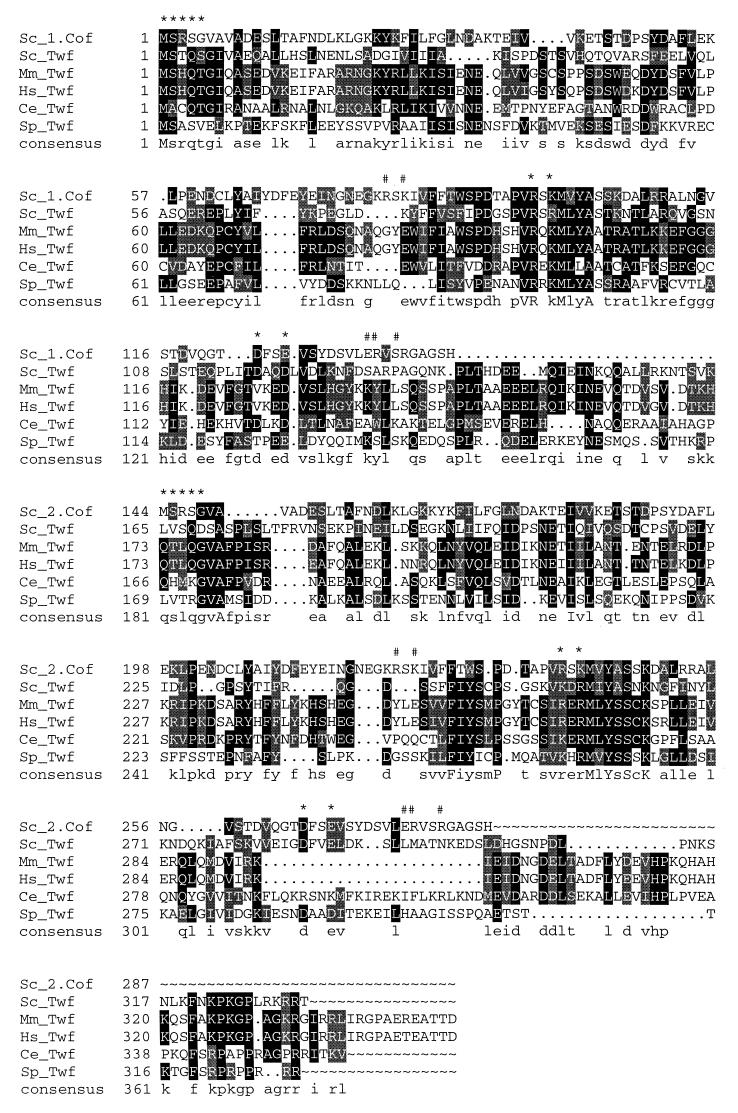
Yeast Saccharomyces cerevisiae twinfilin is a small actin monomer-binding protein that is composed of two cofilin-like repeats, the ADF-H domains (10, 15). In our database searches we have also identified homologues of S. cerevisiae twinfilin in humans, mice, Caenorhabditis elegans and Schizosaccharomyces pombe, demonstrating that twinfilins form an evolutionarily conserved family of proteins (Fig. 1). These proteins have 23 to 30% sequence identity to yeast twinfilin at the amino acid level (Fig. 1). They also appear to be similar in domain structure to yeast twinfilin, with two ADF-H domains (see also reference 15). The positions of secondary structure elements, as well as the residues that have been shown to be essential for actin monomer binding in cofilin, are well conserved in all twinfilin homologues (Fig. 1 and reference 15). This suggests that these proteins may be similar to each other with respect to three-dimensional core structures and biochemical activities. However, previous studies with the human homologue of twinfilin suggested that this protein has a tyrosine kinase activity, and therefore this protein was named A6 tyrosine kinase (4). More recently, Beeler et al. (5) have also identified a mouse homologue of human A6 tyrosine kinase and showed that this protein is widely expressed in various mouse cell lines and tissues.
FIG. 1.
Sequence alignment of S. cerevisiae (Sc), human (Hs), mouse (Mm), C. elegans (Ce), and S. pombe (Sp) A6/twinfilin proteins. Residues either identical or similar to yeast twinfilin (Twf) are highlighted with black or grey, respectively. Yeast cofilin (Sc_1.Cof and Sc_2.Cof) is included into this sequence alignment to bring mutagenesis and structural data of cofilin into alignment. Residues that have been shown to be important for F-actin binding in yeast cofilin (13) are indicated above the sequences (#), as are the residues that have been shown to be important for both G- and F-actin binding in yeast cofilin (∗). A linker region approximately 30 residues in length is clearly observed between the two cofilin-like regions (ADF-H domains) in A6/twinfilins. This linker region contains a conserved proline followed by up to three conserved negatively charged residues, indicating that it may also have more specific biological function(s). Protein names and accession numbers for the sequences are as follows (where no database is stated, the accession number refers to GenBank): S. cerevisiae cofilin, Q03048 (Swissprot); S. cerevisiae twinfilin, YGR080W (SGD); Mus musculus twinfilin, U82324; Homo sapiens twinfilin, A55922 (PIR); C. elegans twinfilin: U46668 (ID, g1166579); S. pombe twinfilin, AL034490 (ID, g4008554). The software used were PileUp and LineUp in the Genetics Computer Group Wisconsin Package together with Boxshade 3.21.
These mammalian A6 tyrosine kinases do not have detectable sequence homology with known protein kinases, nor do they contain sequence motifs typical for the catalytic domains of protein kinases. However, since they show relatively high (25%) sequence identity to yeast twinfilin, we decided to carry out a biochemical and cell biological characterization of mouse A6 tyrosine kinase.
Biochemical activities of mouse A6/twinfilin.
Mouse A6/twinfilin was expressed in E. coli as a GST fusion protein and separated from GST by a thrombin treatment. Recombinant A6/twinfilin was further purified by gel filtration. A6/twinfilin eluted in a Superdex-75 gel filtration column in a sharp peak at 55 to 60 ml, indicating that it was monomeric and, based on SDS-gels, >95% pure.
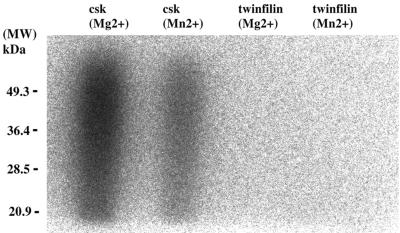
To test the putative tyrosine kinase activity of the purified A6/twinfilin, we carried out an in vitro tyrosine kinase assay by using substrate and reaction conditions identical to those described by Beeler et al. In these studies, Beeler et al., demonstrated that purified recombinant A6 protein can phosphorylate poly(Glu-Tyr) in Mg- and Mn-containing buffers. As a positive control in our kinase assay we used purified Csk kinase, which is the smallest tyrosine kinase found to date (6). As shown in Fig. 2, Csk kinase promotes a strong incorporation of 32P into poly(Glu-Tyr) in both Mg- and Mn-containing buffers (Fig. 2, lanes 1 and 2, respectively). In contrast, we detected no incorporation of 32P into poly(Glu-Tyr) with purified A6/twinfilin under either of these buffer conditions (lanes 3 and 4). These results show that our purified recombinant mouse A6/twinfilin does not exhibit tyrosine kinase activity.
FIG. 2.
In vitro kinase assay was carried out as described in Materials and Methods with purified Csk tyrosine kinase (6) and mouse A6/twinfilin. The reaction buffer contained either 10 mM MgCl2 or 5 mM MnCl2. The incorporation of 32P into the substrate, poly(Glu-Tyr), was visualized by an autoradiogram from an SDS–12% polyacrylamide gel. As expected, the Csk tyrosine kinase shows a strong tyrosine kinase activity under both buffer conditions. In contrast, no tyrosine kinase activity can be detected with purified mouse A6/twinfilin in either Mg- or Mn-containing buffers. MW, molecular weight markers.
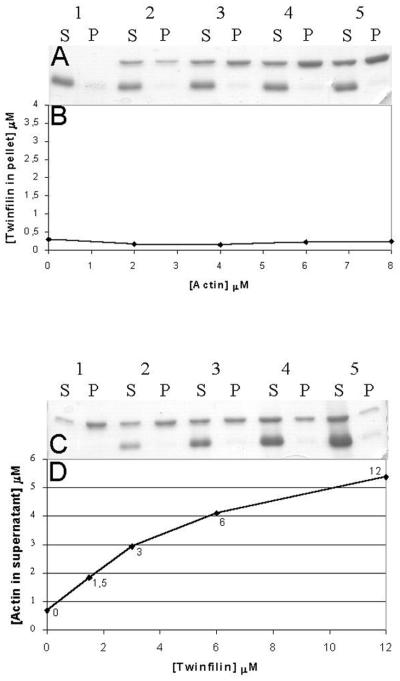
To study the possible actin interactions of A6/twinfilin, we carried out actin filament cosedimentation assays. We first tested whether A6/twinfilin binds to actin filaments. In these experiments, we used a constant A6/twinfilin concentration (2 μM) and a range of different actin concentrations (0 to 8 μM). There is no significant increase in the amount of A6/twinfilin in the pellet fractions with actin concentrations ranging from 0 to 8 μM, suggesting that mouse A6/twinfilin does not bind to actin filaments with detectable affinity (Fig. 3A and B). However, as shown in Fig. 3C and D, A6/twinfilin is able to prevent actin assembly in a concentration-dependent manner. In this assay, we used a constant actin concentration (6 μM) and incubated prepolymerized actin filaments with different concentrations (0 to 12 μM) of A6/twinfilin. A6/twinfilin shifts actin from the pellet fractions (F-actin) to the supernatant fractions (G-actin) in a manner that is dependent on the concentration of A6/twinfilin (Fig. 3C and D). This actin monomer-sequestering activity is similar to the one previously observed for yeast twinfilin (10).
FIG. 3.
A6/twinfilin-actin cosedimentation assays. Mouse A6/twinfilin does not cosediment with actin filaments but increases the amount of actin in the supernatant (S) in a concentration-dependent manner. (A) Mouse A6/twinfilin (2 μM) was mixed with 0 (lane 1), 2 (lane 2), 4 (lane 3), 6 (lane 4), and 8 (lane 5) μM prepolymerized human platelet actin, and the samples were centrifuged for 30 min at 180,000 × g to sediment actin filaments. There is not a significant increase in the amount of A6/twinfilin in the pellet (P) with actin concentration between 0 and 8 μM. (B) Quantification of the amount of A6/twinfilin cosedimented with actin concentrations of 0 to 8 μM. (C) Human platelet actin (6 μM) was polymerized for 30 min and mixed with 0 (lane 1), 1.5 (lane 2), 3 (lane 3), 6 (lane 4), and 12 (lane 5) μM mouse A6/twinfilin. A6/twinfilin increases the amount of actin in the supernatant, and with 12 μM A6/twinfilin there is hardly any visible amount of actin in the pellet fraction. (D) Graphical presentation of the data from panel C.
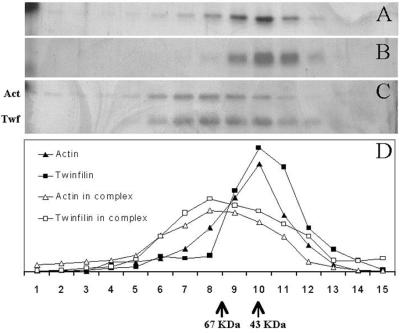
To evaluate whether A6/twinfilin forms a complex with actin monomers and to determine the stoichiometry of this putative protein complex, we carried out a rate zonal centrifugation analysis of purified mouse A6/twinfilin, human platelet actin, and a 1:1 mixture of A6/twinfilin and actin. These experiments were carried out in G buffer (see Materials and Methods) to keep actin in a monomeric form. As shown in Fig. 4, the strongest density of both A6/twinfilin (molecular mass of 40 kDa) and actin (42 kDa) are found in the same fraction as the 43-kDa marker protein, ovalbumin, suggesting that these proteins are monomers in solution. However, when A6/twinfilin and actin are mixed 1:1 prior to ultracentrifugation, there is a clear shift in the sedimentation behavior of twinfilin and actin, and both proteins sediment somewhat faster than the 67-kDa marker, BSA (Fig. 4). This result shows that A6/twinfilin and actin form a rather strong complex with each other in vitro. Furthermore, because this protein complex sediments close to a 67-kDa molecular weight marker protein (Fig. 4C), the stoichiometry of this complex most probably is 1:1 (A6/twinfilin:actin). It is also important to note that in the silver-stained gel, A6/twinfilin and actin in this protein complex appear to have similar densities. The formation of the complex between A6/twinfilin and actin monomers was also confirmed by native gel electrophoresis as described by Safer (29) (data not shown).
FIG. 4.
Rate zonal gradient centrifugation analysis of the stoichiometries of the interaction of human platelet nonmuscle actin and mouse A6/twinfilin. Samples in panels A (actin alone), B (A6/twinfilin alone), and C (actin and A6/twinfilin together) were floated independently but simultaneously in identical reaction conditions. In panel C, actin and A6/twinfilin were incubated together only during the 40-h centrifugation. (D) Graphic representation of distribution of proteins in panels A to C. Fractions from the left correspond to the heaviest or least globular proteins or protein complexes. Actin (42 kDa) and A6/twinfilin (40 kDa) alone peak at the same fraction as the 43-kDa size standard. Actin is distributed more broadly and toward larger molecular masses. This is understandable in the light the autoaffinity of actin and its ability to form filaments. A6/twinfilin does not seem to possess intrinsic affinity to itself under these conditions. In panel C, the distribution of actin and A6/twinfilin is broadened and shifted to the left by a distance that corresponds to a formation of a >67-kDa complex. The peak positions of the sedimentation assay protein standards (ovalbumin [43 kDa] and BSA [67 kDa]) are indicated at the bottom.
Tissue distribution of mouse A6/twinfilin.
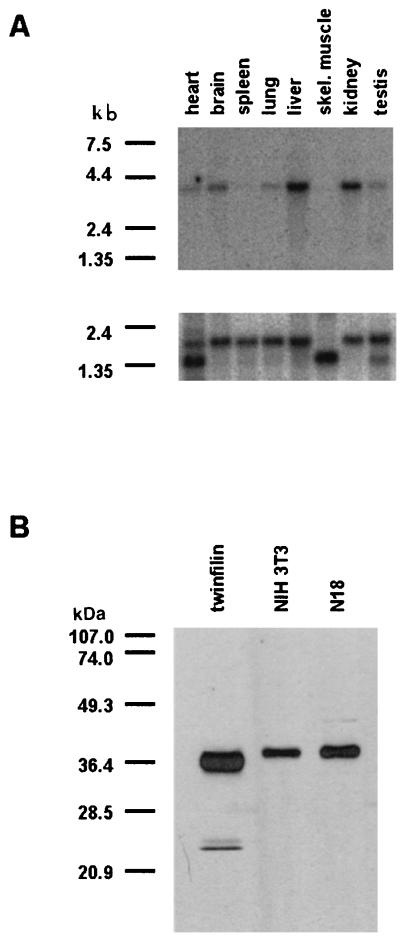
We carried out a multiple-tissue Northern blot analysis to examine the expression of the A6/twinfilin mRNA in various adult mouse tissues. The A6/twinfilin cDNA probe used in this study recognizes only a single ∼4-kb mRNA in these mouse tissues, indicating that the probe was specific and that there are no A6/twinfilin splice variants in the mouse tissues examined in this study (Fig. 5A). A6/twinfilin mRNA appears to be widely expressed in different mouse tissues, with particularly strong expression in the brain, liver, and kidney (Fig. 5A). The ∼4-kb A6/twinfilin mRNA is also expressed in the heart, lung, and testis but is virtually absent from the spleen and from skeletal muscle. To control the amount of mRNAs in different tissue samples of this Northern blot, we also probed the blot with a β-actin control probe (Fig. 5A, lower panel).
FIG. 5.
(A) Northern blot analysis of the expression of A6/twinfilin mRNA in adult mouse tissues, using a mouse A6/twinfilin cDNA probe (top) and mouse β-actin control probe (bottom). The ∼4-kb A6/twinfilin mRNA is expressed strongly in the brain, liver, and kidney and at a lower level in the heart, lung, and testis. There is no detectable expression of A6/twinfilin mRNA in the spleen and in skeletal muscle tissues. The β-actin mRNA is expressed at equal levels in all tissues in this Northern blot, indicating that all lanes contained similar amounts of mRNA. (B) Specificity of anti-A6/twinfilin antibody in NIH 3T3 and N18 mouse cell lines. The anti-A6/twinfilin antibody recognizes only one protein of ∼40 kDa from NIH 3T3 and N18 cell extracts. This protein has also the same molecular weight as the purified recombinant mouse A6/twinfilin.
Localization of A6/twinfilin in NIH 3T3 and N18 cells.
To study the localization of A6/twinfilin in mouse cells, we generated a polyclonal antibody against mouse A6/twinfilin in rabbits. The serum from a rabbit that had been immunized with recombinant mouse A6/twinfilin was affinity purified with mouse A6/twinfilin, and its specificity was tested by Western blotting. The affinity-purified anti-A6/twinfilin antibody recognizes purified recombinant mouse A6/twinfilin in these Western blots (Fig. 5B, lane 1) and also reacts with only a single, approximately 40-kDa protein in NIH 3T3 mouse fibroblast and N18 mouse neuroblastoma cell extracts. Therefore, this antibody appears to be specific for A6/twinfilin and does not cross-react with other ADF-H domain family proteins. The native A6/twinfilin in these cells also seems to be identical in mobility to our purified recombinant A6/twinfilin on SDS-gels.
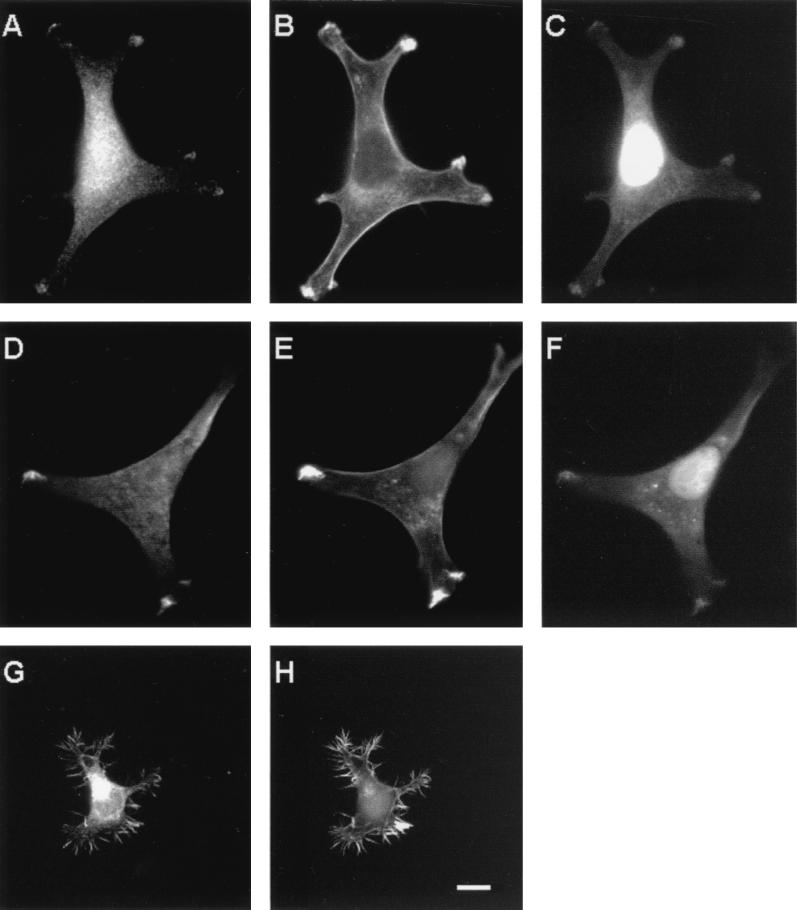
We then examined the subcellular localization of the A6/twinfilin protein in the two mouse cell lines. In NIH 3T3 cells, A6/twinfilin shows a strong punctate cytoplasmic staining which in many cells is most prominent in the perinuclear area. In addition to the strong cytoplasmic staining, a significant portion of A6/twinfilin is also localized to cell processes in NIH 3T3 cells (Fig. 6A and D). These processes are rich in F-actin as visualized by phalloidin staining (Fig. 6B and E). Furthermore, these A6/twinfilin-rich cell processes also colocalize with G-actin as visualized by DNase I staining (Fig. 6C and F). It is important to note that in addition to G-actin, DNase I also stains the DNA, which results in a bright nuclear staining in these cells. Interestingly, the G-actin-rich processes in NIH 3T3 cells appear to be the only actin structures where A6/twinfilin is concentrated, because A6/twinfilin is not found in other F-actin-rich cortical areas (Fig. 6A to E). Identical staining patterns were also obtained with A6/twinfilin antibody and DNase I alone. This shows that bleed-through did not contribute to the observed double staining patterns.
FIG. 6.
Localization of A6/twinfilin in NIH 3T3 (panels A to F) and N18 (G and H) cells. The cells were stained with FITC-conjugated DNase I (C and F) to visualize G-actin, with rhodamine-phalloidin (B, E, and H) to visualize F-actin, and with anti-A6/twinfilin antibody (A, D, and G). DNase I also shows bright nuclear staining and therefore could not be used for visualization of G-actin in N18 cells. A6/twinfilin shows strong punctate cytoplasmic staining in NIH 3T3 and N18 cells, but in NIH 3T3 cells it is also concentrated in those cortical actin structures which overlap with high concentrations of G-actin. In N18 cells, A6/twinfilin is concentrated at the F-actin-rich filopodia. Scale bar, 10 μm.
In N18 neuroblastoma cells, A6/twinfilin also shows bright perinuclear cytoplasmic staining similar to that shown with NIH 3T3 cells above. In these cells, A6/twinfilin also strongly localizes to the F-actin-rich filopodia and cell processes (Fig. 6G and H). However, due to a very bright nuclear DNase I staining in N18 cells, we could not evaluate whether A6/twinfilin in these cells localizes to the G-actin-rich structures similarly to NIH 3T3 cells.
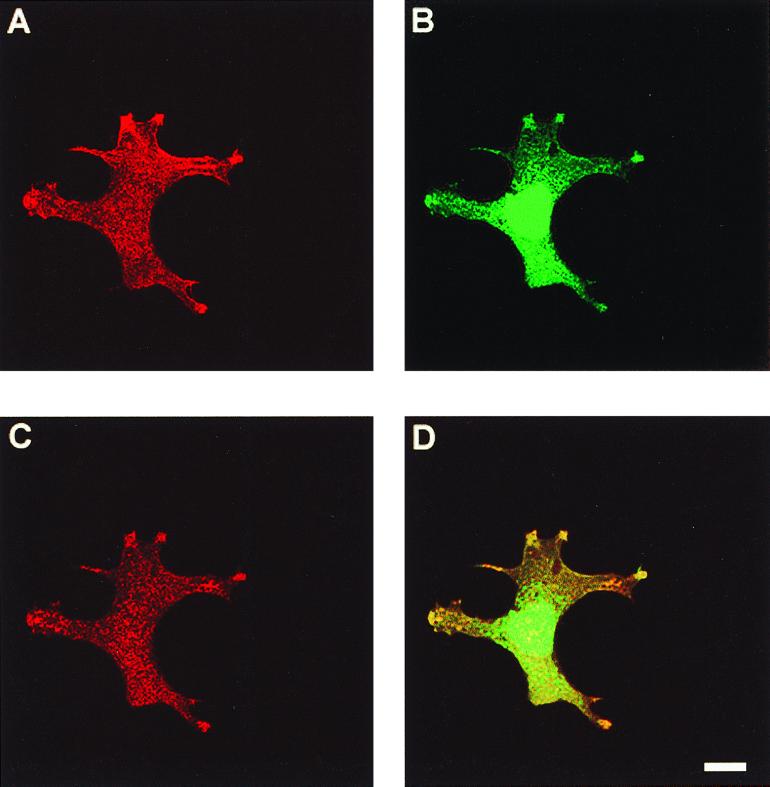
We also used confocal light microscopy to confirm that a fraction of A6/twinfilin indeed colocalizes with G-actin at the cortical actin cytoskeleton and to confirm that the observed localization pattern is not a secondary consequence of a cell thickness in these regions. Figure 7A to C show confocal microscope images of an NIH 3T3 fibroblast stained with phalloidin, DNase I, and A6/twinfilin, respectively. A6/twinfilin is indeed enriched at cortical areas in these cells. The A6/twinfilin localization overlaps with cortical DNase I staining (G-actin) as well as with a population of actin filaments (phalloidin staining). However, it is important to note that A6/twinfilin localizes exclusively to G-actin-rich areas of the actin cytoskeleton and does not show significant colocalization with other F-actin structures.
FIG. 7.
Confocal microscope images of colocalization of A6/twinfilin with F-actin and G-actin in NIH 3T3 cells. The cells were stained with rhodamine-phalloidin to visualize F-actin (A), with DNase I to visualize G-actin (B), and with anti-A6/twinfilin antibody (C). (D) Colocalization of G-actin and twinfilin. Scale bar, 10 μm.
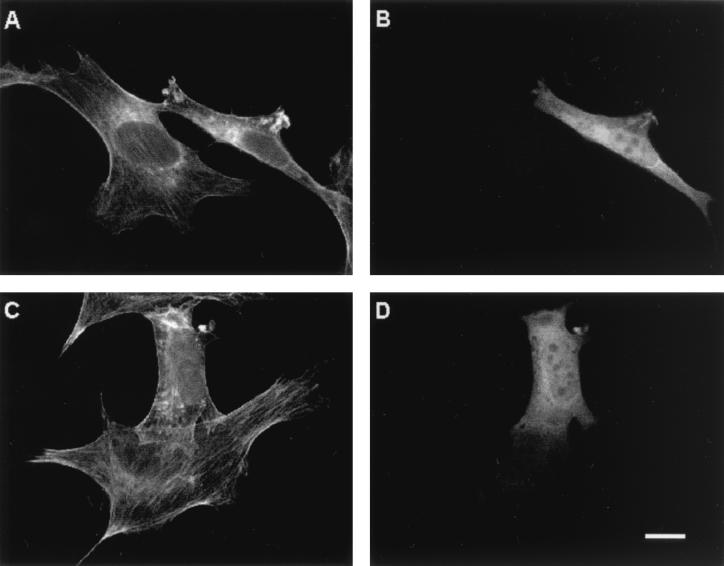
To study whether A6/twinfilin can also regulate the structure or dynamics of the actin cytoskeleton in vivo, we overexpressed full-length A6/twinfilin in NIH 3T3 cells. Overexpression of A6/twinfilin leads to a decrease in the amount of stress fibers in these cells and to the appearance of abnormal worm-like actin filament structures in the cytoplasm (Fig. 8).
FIG. 8.
Overexpression of A6/twinfilin in NIH 3T3 cells. Cells were stained with fluorescein-phalloidin to visualize F-actin (A and C) and with anti-A6/twinfilin antibody (B and D). Both fields contain one cell overexpressing A6/twinfilin. These cells have fewer stress fibers than untransfected cells in the same field. The cells overexpressing A6/twinfilin also show an accumulation of abnormal worm-like actin structures in the cytoplasm. Scale bar, 10 μM.
Effects of Rho family GTPases on A6/twinfilin localization in vivo.
Rho family small GTPases belong to the Ras superfamily of small GTP-binding proteins. The three most-studied members of the Rho family are RhoA, Rac1, and Cdc42. In fibroblasts, RhoA regulates stress fiber and focal adhesion formation (26), Rac1 regulates membrane ruffling and formation of lamellipodia (27), and Cdc42 regulates formation of filopodia (22). It is not well understood how the Rho family proteins mediate the changes in the actin cytoskeleton that lead to these morphological changes in cells. Furthermore, only a small number of downstream effector proteins of these small GTPases have been identified.
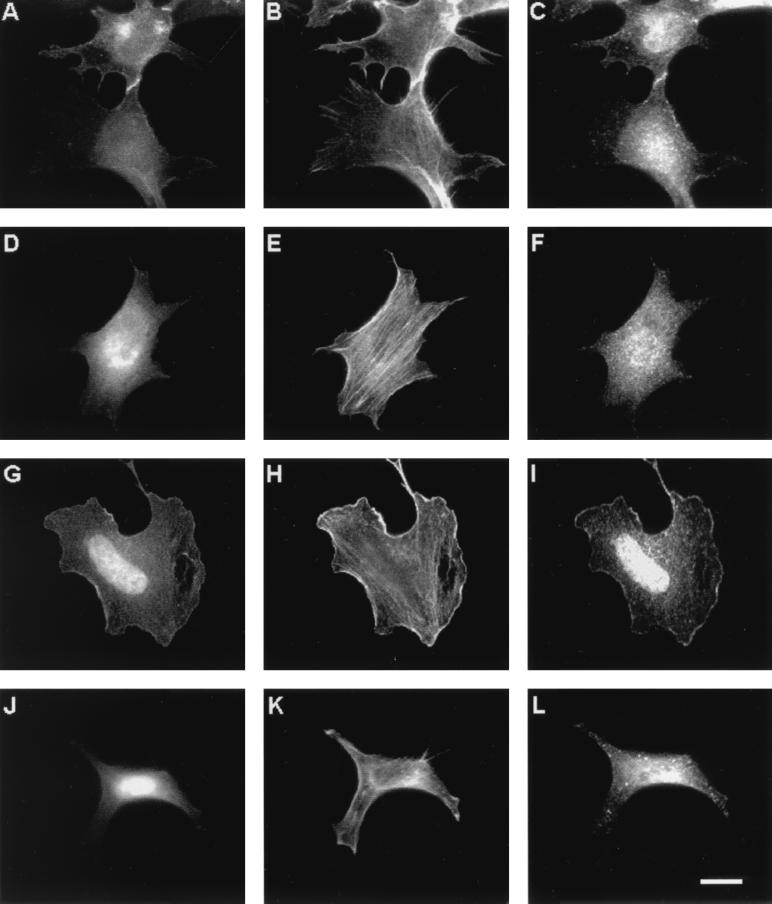
To study if Rho family GTPases would have any effects on A6/twinfilin localization, we transfected NIH 3T3 cells with GFP fusions of the activated (GTP) forms of RhoA, Cdc42, and Rac1. As expected, Cdc42(12V) induces filopodium formation, RhoA(V14) induces stress fiber formation, and Rac1(V12) induces membrane ruffles in NIH 3T3 cells (Fig. 9). In cells expressing activated RhoA(V14), A6/twinfilin localizes mainly to the cytoplasm and shows rather weak localization to certain areas of the cortical actin cytoskeleton but does not colocalize with the stress fibers (Fig. 9D to F). Furthermore, A6/twinfilin does not seem to have any significant colocalization with GFP-RhoA(V14) in these cells. In cells transfected with the activated form of Cdc42(V12), A6/twinfilin shows mainly a punctated cytoplasmic staining pattern but is also found in certain parts of the cortical actin cytoskeleton and in cell-cell contacts. Interestingly, these A6/twinfilin-rich areas also colocalize with GFP-Cdc42(V12) (Fig. 9A to C). This colocalization is particularly obvious in cell-cell contacts. The staining pattern is also similar in a confluent monolayer of cells. Finally, in cells transfected with an activated form of Rac1(V12), A6/twinfilin shows a relatively weak punctate cytoplasmic staining and, in contrast to the wild-type cells, is detected mostly in the nucleus and membrane ruffles (Fig. 9I). The A6/twinfilin staining in these cells also overlaps with the localization of GFP-Rac1(V12) (Fig. 9G and I). In contrast, in cells transfected with a GFP fusion protein of dominant negative form of Rac1(V12,N17), A6/twinfilin localizes strongly to the cytoplasm. In these cells, no significant localization of A6/twinfilin to the cortical actin cytoskeleton could be detected (Fig. 9J to L). Furthermore, these cells show less nuclear staining of A6/twinfilin than cells transfected with Rac(V12). Rac1(V12,N17) itself strongly localizes to the nucleus in these cells. These results suggest that A6/twinfilin in NIH 3T3 fibroblasts colocalizes with the GTP forms of Cdc42 and Rac1 and that the localization of A6/twinfilin in these cells may be regulated by the small GTPase Rac1.
FIG. 9.
Effects of small GTPases on A6/twinfilin localization in NIH 3T3 cells. Cells were transfected with Cdc42(V12) (A to C), RhoA(V14) (D to F), active Rac1(V12) (G to I), and inactive Rac1(V12,N17) (J to L). (A, D, G, and J) Localization of the GTPases as GFP fusion proteins. Cells are stained with rhodamine-phalloidin to visualize F-actin (B, E, H, and K) and with anti-A6/twinfilin antibody (C, F, I, and L). A6/twinfilin colocalizes with the activated forms of Rac1 and Cdc42 to membrane ruffles and to cell-cell contacts, respectively. The activated form of Rac1(V12) induces A6/twinfilin localization to the nucleus and cell cortex. Scale bar, 10 μm.
DISCUSSION
We recently identified a novel actin monomer-binding protein from the budding yeast S. cerevisiae. This protein is composed of two cofilin-like repeats (ADF-H domains) and was therefore named twinfilin (10, 15). In our database searches, we have also identified homologues of yeast twinfilin in mammals, C. elegans, and S. pombe (reference 15 and Fig. 1). However, previous biochemical studies with a human homologue of twinfilin suggested that it was a tyrosine kinase (4).
In this study, we have reexamined the putative tyrosine kinase activity of purified mouse A6/twinfilin protein, which is 95% homologous to the human A6 tyrosine kinase at the amino acid level. We did not find any evidence for the tyrosine kinase activity with this protein (Fig. 2). In this study, we used the same experimental setup (buffers and substrate) as used earlier by Beeler et al. (4). Because Beeler et al. (4) also used a recombinant protein produced in E. coli in their kinase assays, it is unlikely that the difference in source of the A6/twinfilin protein would explain these discrepancies in results. It is also notable that our recombinant protein appeared to be homogenous, monomeric, and active in other biochemical assays (see Materials and Methods and Fig. 3 and 4). Therefore, we feel that it is unlikely that these proteins would be tyrosine kinases. This conclusion is also supported by the fact that A6/twinfilins do not show any sequence homology to known protein kinases or have any sequence motifs typical for kinases.
Various biochemical results in our work with mouse A6/twinfilin show that it has actin-related activities similar to those previously reported for yeast twinfilin (10). Both yeast and mouse proteins appear to form a complex with actin monomers and prevent the actin filament assembly. However, mouse A6/twinfilin appears to be somewhat less efficient in increasing the critical concentration for actin polymerization compared to yeast twinfilin (Fig. 3C and D) (10). Migration of the actin-A6/twinfilin complex in sucrose gradients indicate that this protein forms a 1:1 ratio complex with actin monomer (Fig. 3). This is in agreement with our previous studies with yeast twinfilin, which showed that twinfilin can prevent the assembly of (sequester) an equal molar amount of actin monomers in solution (10). In future studies, it will be important to examine why the twinfilin molecule needs two ADF-H domains in order to interact with one molecule of actin and to determine the molecular mechanism whereby it prevents actin filament assembly.
In our previous studies with yeast twinfilin, it was difficult to judge whether this protein interacts with actin filaments, because a fraction of yeast twinfilin pelleted on its own in actin filament cosedimentation assays (10). However, because the mouse recombinant A6/twinfilin is fully soluble in buffers used in actin cosedimentation assays, we were able to demonstrate that A6/twinfilin does not have a detectable affinity to actin filaments (Fig. 3A and B). This is also in agreement with the fact that only those residues that have previously been shown to be important for both actin monomer and actin filament binding in yeast cofilin (13) are conserved in both ADF-H domains in A6/twinfilin proteins (Fig. 1). In contrast, the yeast cofilin residues that are important specifically for actin filament interactions are not conserved in A6/twinfilin proteins (Fig. 1).
To understand the role of A6/twinfilin in the mouse, we first examined its expression patterns in adult mouse tissues (Fig. 5A). A6/twinfilin mRNA appears to be expressed in most adult mouse tissues examined in this study. However, A6/twinfilin has only very low expression levels in skeletal muscle and spleen. The majority of actin filaments in skeletal muscle are organized into sarcomeres and are capped at both ends of the filaments (33). Therefore, these cells are expected to have less actin filament turnover than other cell types and consequently probably lower requirements for proteins involved in actin filament turnover. We have recently also examined the expression patterns of the three different cofilin/ADF mRNAs in adult mouse tissues and noticed that, similarly to A6/twinfilin, skeletal muscle and spleen are the tissues where expression levels of these cofilin mRNAs are lowest (P. Lappalainen et al. unpublished data). Because the role of cofilin in vivo is to increase actin filament turnover (8, 14, 28), the actin filament dynamics in these tissues probably are slow. Therefore, the expression pattern of A6/twinfilin mRNA in adult mouse tissues is in agreement with its possible role in actin filament turnover in vivo.
We used cultured cells to study the localization of A6/twinfilin at the cellular level. In NIH 3T3 and N18 cells, A6/twinfilin shows a strong cytoplasmic staining but is also concentrated to certain F-actin-rich structures at the cell cortex (Fig. 6 and 7). In previous studies, we localized twinfilin in yeast by using a twinfilin-GFP fusion protein. In these experiments the GFP-twinfilin fusion protein showed a diffuse cytoplasmic staining in yeast (10). However, in the course of our work with mouse A6/twinfilin we also used a GFP-mouse A6/twinfilin construct to localize this protein in various mouse cell lines. As observed with the GFP-twinfilin fusion protein in yeast, the mouse GFP-A6/twinfilin fusion protein showed a diffuse cytoplasmic staining, with no obvious colocalization with the cortical actin cytoskeleton, as can be observed when an antibody is used (data not shown). These results indicate that as a GFP fusion protein, twinfilin does not localize correctly in yeast and mouse cells. We speculate that the GFP at the C terminus of these constructs would interfere twinfilin's interaction with other protein(s) required for its correct localization. Alternatively, the possible elevated expression levels of these GFP constructs could also be responsible for these artifactual localization results. In support of these results, we have recently also examined twinfilin localization in yeast cells by using a polyclonal antibody. These experiments showed that also in yeast the native twinfilin localizes mainly to the cortical actin cytoskeleton (S. Palmgren and P. Lappalainen, unpublished data).
It is important to note that although A6/twinfilin in mouse cells colocalizes with F-actin structures, it does not localize to all actin filament structures. As shown in Fig. 7 and 9, A6/twinfilin does not colocalize with stress fibers or with certain other cortical actin filament structures in RhoA-induced cells. Interestingly, A6/twinfilin staining in NIH 3T3 cells overlaps with G-actin staining as visualized with fluorescent DNase I (Fig. 6 and 7). Because high concentrations of G-actin are believed to localize to the areas of rapid actin filament turnover in cells (9), the A6/twinfilin localization in these cells is in agreement with its possible role in actin filament turnover. The role of A6/twinfilin in actin filament dynamics is also supported by the overexpression data (Fig. 8), which shows that high concentration of A6/twinfilin in NIH 3T3 cells leads to a decrease in the amount of stress fibers and simultaneous appearance of abnormal actin filament structures in these cells.
As shown in Fig. 9, A6/twinfilin also colocalizes with the activated forms of small GTPases Rac1 and Cdc42 to membrane ruffles and to cell-cell contacts, respectively. Therefore, it is possible that the A6/twinfilin localization to these cellular compartments is regulated by these small GTPases. In support of this hypothesis, we also showed that in NIH 3T3 cells expressing an activated form of Rac1(V12), A6/twinfilin localizes strongly to the cortical areas in cells, whereas in cells expressing a dominant negative form of Rac1(V12,N17) A6/twinfilin localizes predominantly to the perinuclear area of the cytoplasm (Fig. 9). Therefore, we speculate that the GTP-bound form of Rac1 could localize A6/twinfilin to membrane ruffles by a direct activation (posttranslational modification) of A6/twinfilin. Alternatively, the Rac1-induced localization of A6/twinfilin could be mediated by an interaction with some other downstream protein(s) of the Rac1 signaling cascade. It has recently been demonstrated that dephosphorylation of another ADF-H domain protein, cofilin, leads to its localization from cytoplasm to nucleus and to the cortical actin cytoskeleton (19, 21, 23). It is therefore interesting that in addition to the cortical localization, the expression of Rac1(V12) in NIH 3T3 cells also appears to lead to an increased nuclear localization of A6/twinfilin (Fig. 9).
Previous reports have shown that Rac1 induces membrane ruffles and lamellipodium formation in fibroblasts (27). Two downstream effectors of Rac1 on the actin cytoskeleton have been identified so far. Rac1 may lead to an activation of an actin filament-nucleating Arp2/3 protein complex through interactions with WASP (Wiscott-Aldrich syndrome protein) family proteins (17). Active Arp2/3 complex promotes actin filament nucleation and assembly in cells (20, 34). Rac1 also regulates cofilin phosphorylation through LIM kinases (1, 35). Because phosphorylation of cofilin leads to its inactivation (19), Rac1 also seems to promote the formation of actin filament structures through a decrease in the rate of actin filament depolymerization. Therefore, the overall effect of Rac1 activation appears to be an increase in actin assembly at the cell cortex. Because Rac1 seems to play a role in A6/twinfilin localization in these cells, we speculate that twinfilin would also be involved in actin assembly. Its role in actin assembly may be to localize actin monomers at the sites of rapid actin filament polymerization. Alternatively, twinfilin might form a complex with some other protein(s) to promote actin filament assembly by a more complex mechanism.
In conclusion, we have shown that mouse A6 protein is a homologue of yeast twinfilin and that it has biochemical activities similar to those of the yeast protein, including a 1:1 actin monomer sequestering activity. In contrast to earlier reports, we did not find any tyrosine kinase activity with this protein. Furthermore, we showed that in mouse cell lines A6/twinfilin localizes to the G-actin-rich cortical actin structures and that its localization to these areas may be regulated by the small GTPase Rac1. These results suggest that A6/twinfilin is involved in the regulation of actin filament dynamics at the cortical actin cytoskeleton.
ACKNOWLEDGMENTS
The purified human Csk tyrosine kinase was kindly provided by Matthias Bergman (Department of Biochemistry, University of Helsinki, Helsinki, Finland). We thank Dennis Bamford for comments on the manuscript. We also acknowledge Alan Hall (Ludwig Institute for Cancer Research, London, United Kingdom) for a generous gift of mouse mutant Rac1 plasmids.
This work was supported by grants from Academy of Finland and Biocentrum Helsinki (to P.L.). P.J.O. is supported by a fellowship from the Viikki Graduate School for Biosciences.
REFERENCES
- 1.Arber S, Barbayannis F A, Hanser H, Schneider C, Stanyon C A, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 3.Bamburg J R, Harris H E, Weeds A. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 1980;121:178–182. doi: 10.1016/0014-5793(80)81292-0. [DOI] [PubMed] [Google Scholar]
- 4.Beeler J F, LaRochelle W J, Chedid M, Tronick S R, Aaronson S A. Procaryotic expression cloning of a novel human tyrosine kinase. Mol Cell Biol. 1994;14:982–988. doi: 10.1128/mcb.14.2.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beeler J F, Patel B K R, Chedid M, LaRochelle W J. Cloning and characterization of the mouse homolog of the human A6 gene. Gene. 1997;193:31–37. doi: 10.1016/s0378-1119(97)00073-5. [DOI] [PubMed] [Google Scholar]
- 6.Bergman M, Mustelin T, Oetken C, Partanen J, Flint N A, Amrein K E, Autero M, Burn P, Alitalo K. The human p50csk tyrosine kinase phosphorylates p56lck at Tyr-505 and down regulates its catalytic activity. EMBO J. 1992;11:2919–2924. doi: 10.1002/j.1460-2075.1992.tb05361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum M, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 8.Carlier M-F, Laurent V, Santolini J, Melki R, Didry D, Xia G-X, Hong Y, Chua N-H, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1323. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlier M-F, Pantaloni D. Control of actin dynamics in cell motility. J Mol Biol. 1997;269:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- 10.Goode B L, Drubin D G, Lappalainen P. Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J Cell Biol. 1998;142:723–733. doi: 10.1083/jcb.142.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juuti J T, Bamford D H, Tuma R, Thomas G J. Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage φ4. J Mol Biol. 1998;279:347–359. doi: 10.1006/jmbi.1998.1772. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Lappalainen P, Fedorov E V, Fedorov A A, Almo S C, Drubin D G. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lappalainen P, Drubin D G. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- 15.Lappalainen P, Kessels M M, Cope M J T V, Drubin D G. The ADF homology (ADF-H) domain, a highly exploited actin-binding module. Mol Biol Cell. 1998;9:1951–1959. doi: 10.1091/mbc.9.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lila T, Drubin D G. Evidence for physical and functional interactions among two Saccharomyces cerevisiaeSH3 domain proteins, and adenyl cyclase-associated protein and the actin cytoskeleton. Mol Biol Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machesky L M, Insall R H. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 18.Maciver S K, Zot H G, Pollard T D. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan T E, Lockerbie R O, Minamide L S, Browning M D, Bamburg J R. Isolation and characterization of a regulated form of actin depolymerizing factor. J Cell Biol. 1993;122:623–633. doi: 10.1083/jcb.122.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullins R D, Heuser J A, Pollard T D. The interaction of Arp2/3 complex with actin: nucleation, high-affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaoka R, Abe H, Obinata T. Site-directed mutagenesis of the phosphorylation site of cofilin: its role in cofilin-actin interaction and cytoplasmic localization. Cell Motil Cytoskel. 1996;35:200–209. doi: 10.1002/(SICI)1097-0169(1996)35:3<200::AID-CM3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 22.Nobes C D, Hall A. Rho, Rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 23.Ohta Y, Nishida E, Sakai H, Miyamoto E. Dephosphorylation of cofilin accompanies heat shock-induced nuclear accumulation of cofilin. J Biol Chem. 1989;264:16143–16148. [PubMed] [Google Scholar]
- 24.Peränen J, Rikkonen M, Hyvönen M, Kääriäinen L. T7 vectors with a modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 25.Puius Y A, Mahoney N M, Almo S C. The modular structure of actin regulatory proteins. Curr Opin Cell Biol. 1998;10:23–34. doi: 10.1016/s0955-0674(98)80083-5. [DOI] [PubMed] [Google Scholar]
- 26.Ridley A J, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 27.Ridley A J, Paterson H F, Johnson C L, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 28.Rosenblatt J, Agnew B J, Abe H, Bamburg J R, Mitchison T J. Xenopus actin depolymerizing factor/cofilin XAC is responsible for the turnover of actin filaments in Listeria monocytogenestails. J Cell Biol. 1997;136:1323–1332. doi: 10.1083/jcb.136.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safer D. An electrophoretic procedure for detecting proteins that bind actin monomers. Anal Biochem. 1989;178:32–37. doi: 10.1016/0003-2697(89)90351-5. [DOI] [PubMed] [Google Scholar]
- 30.Sheterline P, Clayton J, Sparrow J. Actin. Protein Profile. 1995;2:1–103. [PubMed] [Google Scholar]
- 31.Shirao T. The roles of microfilament-associated proteins, drebrins, in brain morphogenesis. J Biochem. 1995;117:231–236. doi: 10.1093/jb/117.2.231. [DOI] [PubMed] [Google Scholar]
- 32.Van Troys M, Vandekerckhove J, Ampe C. Structural modules in actin-binding proteins: towards a new classification. Biochim Biophys Acta. 1999;1448:323–348. doi: 10.1016/s0167-4889(98)00152-9. [DOI] [PubMed] [Google Scholar]
- 33.Weber A, Pennise C R, Babcock B B, Fowler V M. Tropomodulin caps the pointed ends of actin filaments. J Cell Biol. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welch M D, Iwamatsu A, Mitchison T J. Actin polymerization is induced by Arp2/3 complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 35.Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]