Abstract
PML fuses with retinoic acid receptor α (RARα) in the t(15;17) translocation that causes acute promyelocytic leukemia (APL). In addition to localizing diffusely throughout the nucleoplasm, PML mainly resides in discrete nuclear structures known as PML oncogenic domains (PODs), which are disrupted in APL and spinocellular ataxia cells. We isolated the Fas-binding protein Daxx as a PML-interacting protein in a yeast two-hybrid screen. Biochemical and immunofluorescence analyses reveal that Daxx is a nuclear protein that interacts and colocalizes with PML in the PODs. Reporter gene assay shows that Daxx drastically represses basal transcription, likely by recruiting histone deacetylases. PML, but not its oncogenic fusion PML-RARα, inhibits the repressor function of Daxx. In addition, SUMO-1 modification of PML is required for sequestration of Daxx to the PODs and for efficient inhibition of Daxx-mediated transcriptional repression. Consistently, Daxx is found at condensed chromatin in cells that lack PML. These data suggest that Daxx is a novel nuclear protein bearing transcriptional repressor activity that may be regulated by interaction with PML.
Acute promyelocytic leukemia (APL) arises as a result of chromosomal translocation involving the retinoic acid (RA) receptor α (RARα) gene on chromosome 17 fused with either the promyelocytic leukemia gene (PML) on chromosome 15, the promyelocytic leukemia zinc finger gene (PLZF) on chromosome 11, the nucleophosmin/B23 (NPM) gene on chromosome 5, or the nuclear mitotic apparatus gene (NuMA) on chromosome 11 (30, 39). The t(15;17) translocation between PML and RARα accounts for nearly all APL cases. This translocation creates an oncogenic fusion protein, PML-RARα, which contains both the DNA-binding domain (DBD) and ligand-binding domains of RARα and the N terminus of PML. Transgenic mice that overexpress PML-RARα or PLZF-RARα developed an APL-like phenotype (9, 21, 26), suggesting that these fusion proteins are directly involved in APL pathogenesis. Recent studies have focused on analyzing the functional properties of PML-RARα and PLZF-RARα (20, 22, 25, 40) in order to understand the molecular basis of leukemogenesis. Both fusion proteins form homodimers that bind to RA response elements and interact with the nuclear receptor corepressors SMRT (silencing mediator for retinoid and thyroid hormone action) and N-CoR (nuclear receptor corepressor), which in turn recruit a histone deacetylase complex (1, 27, 40, 46). Pharmacological concentrations of all-trans-RA (atRA) induce dissociation of the corepressors from PML-RARα, but not PLZF-RARα, due to the presence of an additional, RA-insensitive corepressor-interacting surface on PLZF. This differential degree of dissociation of corepressors induced by atRA correlates with the ability of histone deacetylase inhibitors and atRA to induce terminal differentiation of these two subtypes of APL cells. These findings indicate that abnormalities in transcriptional repression by the oncogenic fusion proteins may be involved in leukemogenesis.
PML belongs to a family of proteins characterized by the presence of a RING finger domain (8). RING finger proteins are implicated in transcriptional regulation, and some members of the RING family are associated directly with chromatin (53). Ablation and overexpression experiments suggest an important role of PML in the regulation of cell growth, hematopoietic cell differentiation, tumorigenesis, apoptosis, and RA signaling (44, 63). In normal cells, PML is concentrated within 10 to 20 nuclear structures known as nuclear domains 10 (ND10), Krüppel bodies, nuclear bodies, or PML-oncogenic domains (PODs) (2, 17, 33, 59, 65). The POD structure is disrupted in the t(15;17) translocated APL cells (17, 33, 65), presumably through interaction of wild-type PML with PML-RARα. Interestingly, the POD structure reorganizes upon treatment with atRA or arsenic trioxide (As2O3), a process that correlates with differentiation of APL cells, indicating that the POD structure might affect promyelocyte differentiation.
In addition to PML, the POD contains several other proteins, including the 100-kDa nuclear protein antigen (Sp100) (2), the small ubiquitin-related modifier (SUMO-1 [41], also known as PML-interacting clone 1 [PIC1] [7], ubiquitin-like 1 [UBL1] [57], or sentrin [48]), and the 140-kDa protein (Sp140) (6). Sp100 is a nuclear antigen recognized by autoantibodies from patients with primary biliary cirrhosis (62). Expression of both PML and Sp100 are upregulated by interferon (23). SUMO-1 was recently identified as a ubiquitin-like protein that forms covalent conjugates with PML and Sp100 (7, 58). In addition, the CREB-binding protein (CBP) and the retinoblastoma tumor suppressor (pRB) have been found in the PODs (35, 61). Also, the PODs are targets of several viral proteins, which alter POD structure (11, 14, 18). Although there is evidence for POD's role in transcriptional activation (15, 35), DNA replication (19), apoptosis (51, 64), and viral infection (14, 42), the precise function of PODs in these processes remains unclear.
We have sought to understand the function of PODs through identification of PML-interacting proteins that also localize in the PODs. By using the yeast two-hybrid system, we identified SUMO-1 and the Fas-binding protein Daxx (68) (J. D. Chen and R. M. Evans, unpublished data). Daxx has been shown to promote Fas-mediated apoptosis through activation of the Jun NH2-terminal kinase (JNK) and JNK kinase kinase ASK1 (apoptosis signal-regulating kinase 1) (12). Recent data suggest that Daxx is not sufficient for Fas-mediated apoptosis, since a Fas mutant that selectively binds to Daxx but not the Fas-adaptor death domain-containing protein (FADD/MORT1) failed to induce apoptosis (13). Other evidence suggests that Daxx may interact with the centromeric protein-c (CENP-C) and may bind to a steroidogenic factor 1 (SF-1)-like DNA element (32, 50). Therefore, the exact mechanism by which Daxx regulates Fas-mediated apoptosis may involve nuclear processes.
In the present study, we have characterized both biochemical and functional interactions between Daxx and PML. Daxx resides primarily in the cell nucleus, where it forms a complex with PML. Confocal immunofluorescence data demonstrate that Daxx colocalizes with PML in the PODs, and such colocalization persists in NB4 APL cells (36) before and after treatment with atRA and As2O3. Daxx possesses strong transcriptional repressor activity and appears to interact directly with histone deacetylases. Intriguingly, overexpression of PML inhibits Daxx-mediated transcriptional repression and, in cells that lack PML, Daxx is preferentially associated with condensed chromatin. Our data reveal a new role for Daxx in transcriptional repression and suggest a novel function of PML and the POD structure in the suppression of transcriptional repression.
MATERIALS AND METHODS
Yeast two-hybrid system.
The screening of PML-interacting proteins was conducted by the yeast two-hybrid system by using the Y190 strain as previously described (16). The Gal4 DBD (amino acids 1 to 147) fusion of full-length PML (29) was constructed in the yeast vector pAS1 (16). The resulting Gal4 DBD-PML fusion protein was used as bait to screen a Gal4 activation domain (AD)-fused human B-lymphocyte cDNA library in the pACT expression vector (16). About 106 yeast transformants were screened on selection plates containing 50 mM 3-aminotriazole (Sigma). For ligand treatment, the culture was incubated in the presence of ligand or solvent (control) for 24 h before measuring the β-galactosidase (β-Gal) activity.
Biochemical cell fractionation.
HeLa cells (2 × 106) were harvested into 500 μl of CLB buffer (10 mM HEPES, 10 mM NaCl, 1 mM KH2PO4, 5 mM NaHCO3, 1 mM CaCl2, 0.5 mM MgCl2)–5 mM EDTA–1 mM phenylmethylsulfonylfluoride–proteinase inhibitors. Cells were allowed to swell for 5 min on ice, Dounce homogenized 35 times, and centrifuged at 7,500 rpm for 5 min to pellet nuclei and debris. The supernatant (cytosol plus plasma membrane) was then spun at 25,000 rpm for 30 min to pellet the membrane. The nucleus-debris pellet was resuspended in 1 ml of TSE buffer (10 mM Tris, pH 7.5; 300 mM sucrose; 1 mM EDTA) and Dounce homogenized 30 times, followed by centrifugation at 5,000 rpm for 5 min. The pellet was resuspended and washed twice to obtain the final nucleus pellet. Equal amounts of protein in each fraction were analyzed Western blotting.
Western blotting.
Western blotting was conducted by using the enhanced chemiluminescence reagents according the manufacturers' recommendation (Amersham). The affinity purified anti-Daxx polyclonal antibodies were raised against glutathione S-transferase (GST)-Daxx (amino acids 556 to 740) fusion protein and subsequently purified with the GST-Daxx protein column as described earlier (24). Anti-Gal4-DBD antibody was purchased from Santa Cruz, and anti-HDAC1 antibody was from Upstate Biotechnology.
Co-IP.
Coimmunoprecipitation (Co-IP) was conducted according to a standard procedure by using the protein A-agarose beads (Santa Cruz) (24). Nuclear extracts were prepared as described earlier (3). HeLa and NB4 cells were lysed in cell lysis buffer (0.4 M NaCl, 0.2 mM EGTA, 10% glycerol, 1% NP-40), and cell extracts were precleared by incubating them with protein-A agarose beads for 1 h at room temperature. The affinity-purified IP antibodies were conjugated with protein A-agarose beads in cell lysis buffer for 2 h at room temperature. The antibody-protein A-agarose was collected by brief centrifugation and incubated with cell extracts (100 μg) overnight at 4°C. The precipitates were collected by centrifugation and washed five times with excess phosphate-buffered saline containing 0.1% NP-40. The final precipitate was dissolved in sodium dodecyl sulfate A (SDS) sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Immunofluorescence and confocal microscopy.
Cells were grown on cover glasses (VWR Scientific), fixed in a methanol-acetic acid (1:1) mixture on dry ice for 2 min and processed for immunofluorescence staining as described elsewhere (17). For NB4 cells, the cover glasses were coated with poly-l-lysine before seeding the cells. After immunostaining, cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride hydrate) (Sigma). Confocal microscopy was conducted with a Leica TCS SP spectral laser scanning confocal microscope. Channel cross-talk was avoided by reducing the intensity of the excitation laser beam in the absence of the other excitation laser. Standard epifluorescence microscopy was performed on an Olympus IX-70 microscope equipped with a back-illuminated cool charge-coupled device (CCD) camera (Princeton Instruments), and the image was processed by using the MetaMorph software (Universal Imaging Corp.).
Transient-transfection assay.
Transient transfection was conducted using a standard calcium phosphate precipitate method as described earlier (3). Cultured cells were maintained in Dulbecco modified Eagle medium or RPMI medium (for NB4 cells) supplemented with 10% fetal bovine serum (Gibco). Twelve hours prior to transfection, 2 × 104 cells were plated in each well of 12-well plates. Transfected cells were refed with fresh media and harvested 36 to 48 h after transfection. Transfected cells in each well were lysed and processed for luciferase and β-Gal assay as described elsewhere (38). The luciferase activity was determined with an MLX plate luminometer (Dynex) and normalized with the cotransfected β-Gal.
Far-Western blot.
GST fusion proteins were expressed in DH5α cells and purified by standard glutathione agarose beads according to manufacturer's recommendation (Pharmacia). The purified proteins were separated by SDS-PAGE and electroblotted onto a nitrocellulose filter in transfer buffer (25 mM Tris-HCl, pH 8.3; 192 mM glycine; 0.01% SDS). Proteins were denatured with 6 M guanidine hydrochloride (GnHCl) and renatured by stepwise dilution of GnHCl. Filters were blocked and hybridized overnight with 35S-labeled protein as described elsewhere (38). The membrane was then washed three times with hybridization buffer, and the bound probe was detected by autoradiography.
GST pull-down assay.
The GST pull-down assay was conducted according to a protocol as described earlier (24). Briefly, 5 μg of glutathione agarose-protein beads was incubated with 5 μl of in vitro-translated 35S-labeled protein with moderate shaking at 4°C overnight in binding buffer (20 mM HEPES, pH 7.7; 75 mM KCl; 0.1 mM EDTA; 2.5 mM MgCl2; 0.05% NP-40; 1 mM dithiothreitol; 1 mg of bovine serum albumin per ml). The bound protein was washed three times with the binding buffer, and the beads were collected by centrifugation. The bound protein was eluted in SDS sample buffer and analyzed by SDS-PAGE and autoradiography.
Site-directed mutagenesis.
Site-directed mutagenesis was conducted by using the Quick-Change site-directed mutagenesis kit according to manufacturer's instruction (Stratagene). A mammalian hemagglutinin (HA)-PML vector was used as a template, and mutagenesis was conducted in three rounds consecutively on the same template. The mutated construct was confirmed by DNA sequencing by using dideoxynucleotide chain-termination reactions and Sequenase (U.S. Biochemicals).
RESULTS
Identification of Daxx as a PML-interacting protein.
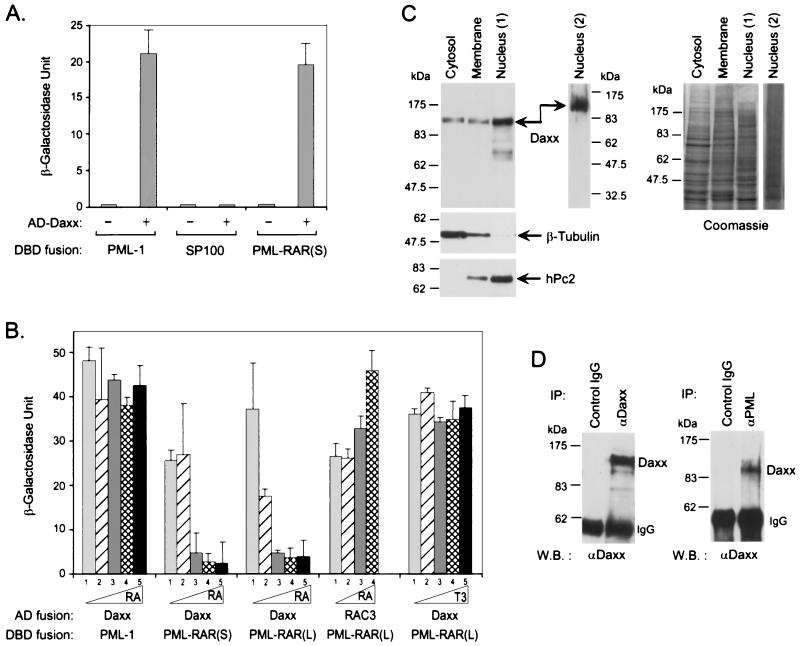
In the yeast two-hybrid screen, we identified a PML-interacting clone that encodes the C-terminal 184 amino acids of Daxx (32, 50). Yeast two-hybrid assay shows that this Daxx clone interacts with Gal4 DBD fusions of both PML and PML-RARα but not SP100 (Fig. 1A), suggesting that Daxx may be a PML-interacting protein. Since atRA binds to PML-RARα in a way similar to that of wild-type RARα (4), we determined the effect of atRA on interaction between Daxx and PML-RARα (Fig. 1B). atRA inhibits the two-hybrid interaction between Daxx and PML-RARα efficiently and in a dose-dependent manner. The inhibition of binding is slightly more sensitive with the long form of PML-RARα than with the short form, a finding consistent with the higher affinity of the long form of PML-RARα for atRA (4). This atRA-dependent inhibition of binding is specific, for atRA has no effect on the interaction between PML and Daxx while it enhances the interaction between PML-RARα and the coactivator RAC3 (38). Also, the thyroid hormone triiodothyronine that does not bind PML-RARα also has no effect on the interaction between Daxx and PML-RARα. These data suggest that Daxx is a PML-interacting protein that may also associate with the oncoprotein PML-RARα in the absence of atRA.
FIG. 1.
Interaction between Daxx and PML in vivo. (A) Interaction of Daxx with PML in yeast two-hybrid system. The average β-Gal activities of three transformants expressing the indicated combinations of Gal4 AD and DBD fusion proteins were determined as described in Materials and Methods. The AD-Daxx fusion protein contains amino acids 556 to 740 of human Daxx. The DBD fusion proteins contain full-length PML-1, SP100, and PML-RARα short form, respectively. The minus sign indicates empty vector alone. (B) atRA disrupts the interaction between Daxx and PML-RARα. The effect of atRA on Daxx-PML-RAR interaction was determined after a 24-h incubation of the culture in the presence of the indicated concentrations of respective ligands. Columns: 1, solvent only; 2, 1 nM; 3, 10 nM; 4, 100 nM; and 5, 1,000 nM. T3, 3,3′,5-triiodo-l-thyronine. (C) Subcellular fractionation of Daxx. HeLa cells were fractionated into cytosolic, membrane, and nuclear fractions, and an equal amount of protein was analyzed by Western blotting for Daxx (left panel). The distribution of the cytoplasmic protein β-tubulin and the nuclear protein hPc2 in each fraction was also determined by immunoblotting to validate the fractionation. Two independent preparations of HeLa nuclear extracts are shown. The right panel is a Coomassie blue-stained gel that shows the relative amount of proteins in each fraction used in the Western blot. (D) Co-IP of Daxx with PML. NB4-cell extracts were immunoprecipitated with affinity-purified anti-Daxx and anti-PML antibodies, and the presence of Daxx in the immunoprecipitates was determined by immunoblotting with anti-Daxx antibodies. The antibodies used for the IP and the Western blot (W.B.) are indicated.
Daxx forms a complex with PML in vivo.
In addition to being diffusely distributed in the cytoplasm, PML is mainly a nuclear protein, while Fas is a transmembrane cell surface receptor. Since Daxx interacts with both PML and Fas, it is important to determine whether Daxx is a nuclear or cytoplasmic protein. We analyzed the subcellular distribution of Daxx by using biochemical fractionation followed by Daxx immunoblotting. In this assay, Daxx cofractionates primarily with nuclear fraction, with a minority also present in the cytosolic and membrane fractions (Fig. 1C). Control antibodies against the cytoplasmic protein β-tubulin and the nuclear protein polycomb hPc2 (55) show no cross-contamination between the cytoplasmic and nuclear fractions. All of these proteins were detected in the membrane fraction, presumably because this fraction also contains insoluble organelles involved in protein synthesis and transportation. These results demonstrate that Daxx resides mainly in the cell nucleus, suggesting that Daxx may interact with PML in the nucleus.
To confirm that the interaction between Daxx and PML also occurs in mammalian cells, we performed Co-IP assays from HeLa and NB4 cell extracts (Fig. 1D). Both anti-Daxx and anti-PML antibodies, but not preimmune serum, efficiently coimmunoprecipitate endogenous Daxx. These data suggest that Daxx may form a stable complex with PML in vivo. In the immunoprecipitates of Daxx and PML antibodies, we also detected weak signals of the 90-kDa PML and two SUMO-1-conjugated forms of PML (data not shown), confirming the presence of PML in the IP. We also attempted to demonstrate an interaction between Daxx and PML in vitro in GST pull-down and far-Western assays, but all experiments failed to show a convincing interaction. We reasoned that this might be due to the fact that PML is extensively modified by SUMO-1 in vivo (44, 45, 58) or that an additional factor may bridge the interaction between PML and Daxx.
Daxx colocalizes with PML in the PODs.
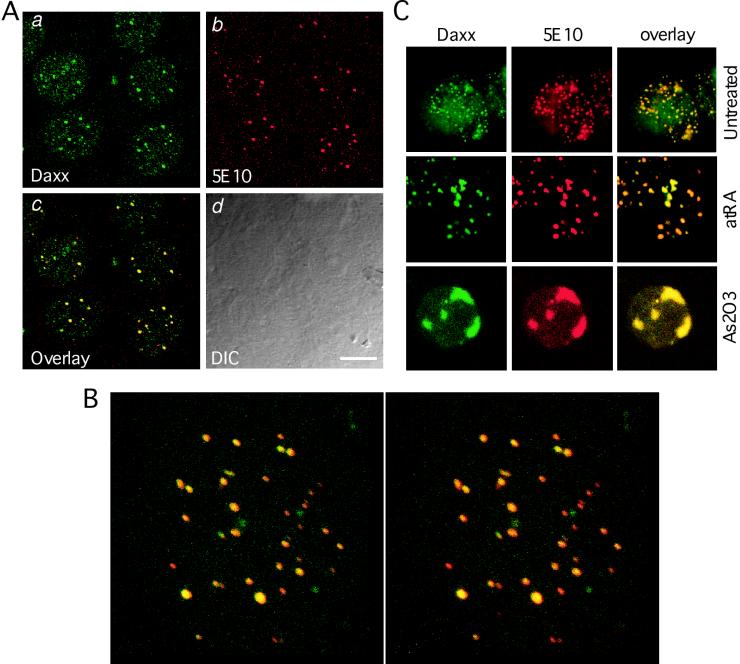
We then wished to determine if Daxx colocalizes with PML in the PODs in order to provide further evidence for a physiological interaction between Daxx and PML. Confocal immunofluorescence microscopy using affinity-purified anti-Daxx antibodies reveals discrete nuclear structures in interphase HEp2 cells, in addition to an evenly distributed nucleoplasmic staining (Fig. 2Aa). Double immunostaining, together with use of anti-PML antibodies, demonstrates that the Daxx foci colocalize perfectly with the PODs in cell nuclei (Fig. 2Aa to d). Such colocalization occurs in many different cell types, including HeLa, HEK293, and A549 cells and normal human fibroblasts, suggesting that colocalization between Daxx and PML may be a common phenomenon in different cell types. The colocalization has been confirmed by using antibodies against different POD antigens, including SP100 and SUMO-1, as well as under conditions that modify the POD structure, such as with interferon, As2O3 treatments, and viral infections (unpublished data). A three-dimensional topographic analysis of the colocalization between Daxx and PML demonstrates an extensive colocalization between Daxx and PML in the PODs (Fig. 2B).
FIG. 2.
Daxx colocalizes with PML at the PODs. (A) Confocal immunofluorescence analysis of endogenous Daxx and PML. HEp2 cells were fixed and immunostained with affinity-purified rabbit anti-Daxx polyclonal antibodies and mouse anti-PML 5E10 monoclonal antibodies as described in Materials and Methods. The sample was analyzed by use of a confocal microscope. Panels a and b show the signals of Daxx (green) and PML (red) on a single confocal section. Panel c shows colocalization (yellow signals) of Daxx and PML in the merged image. Panel d is a differential interference contrast image showing the surfaces of the cells and nuclei. Bar, 10 μm. (B) Three-dimensional presentation of the colocalization between Daxx and PML. Total of 32 consecutive z-sections at increments of 0.08 μm were reconstructed into a three-dimensional image by using the Leica confocal software. The right and left projected images were rotated 4.5° at opposite directions along the x (horizontal) axis. Yellow represents the colocalization between Daxx (green) and PML (red). (C) Colocalization of Daxx and PML in APL cells. NB4 cells were plated on cover glasses coated with poly-l-lysine. The control (untreated), atRA-treated (1 μM for 72 h), and As2O3-treated (1 μM for 72 h) cells were fixed and immunostained with anti-Daxx polyclonal and anti-PML monoclonal antibodies. Colocalization of Daxx and PML was revealed by confocal laser microscopy (except for the untreated cells).
Colocalization of Daxx and PML in NB4 APL cells.
We next analyzed the distribution of Daxx in the NB4 APL cells (Fig. 2C), in which the PODs are disrupted into “microparticulate” structures. Similar to PML, Daxx is also disrupted in the NB4 cells, in which it remains colocalized with PML. The presence of PML-RARα fusion protein in the microparticulate structures (17) supports the observed interaction between Daxx and PML-RARα (Fig. 1). Upon atRA treatment, PML-RARα is degraded in NB4 cells (47), and these microparticulate structures reorganize into normal size of the PODs (17, 65), where Daxx and PML remain colocalized. The colocalization between Daxx and PML is more evident in NB4 cells treated with As2O3, in which larger and fewer PODs are observed. These results suggest that Daxx and PML colocalize in APL NB4 cells, and such colocalization persists after reorganization of the PODs induced by atRA or As2O3.
Daxx represses basal transcription.
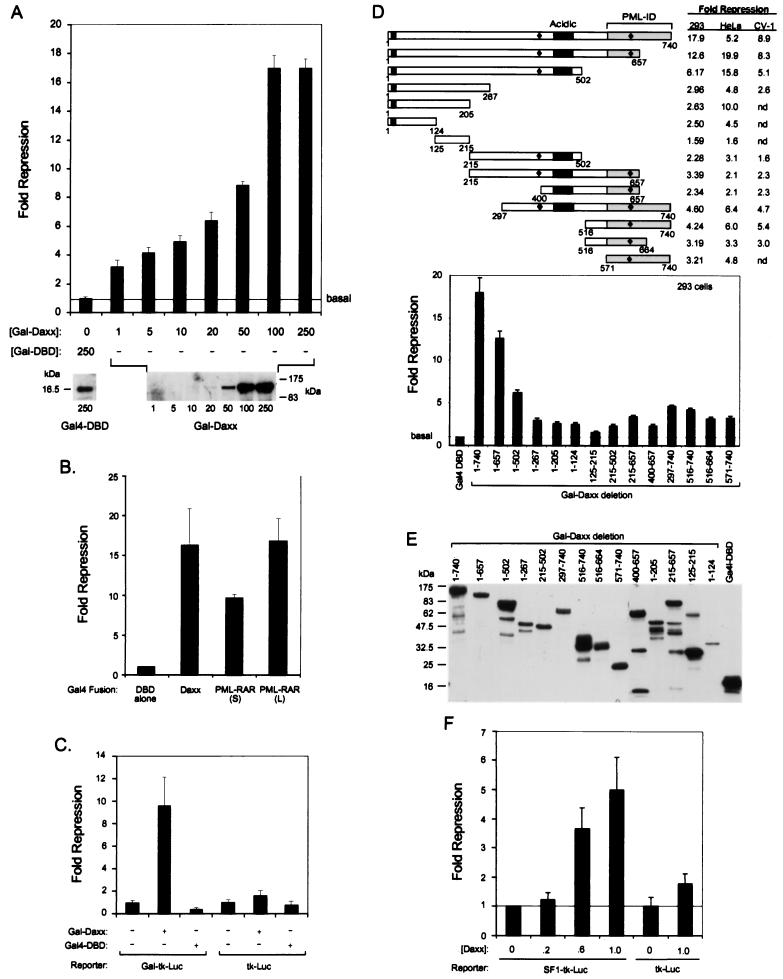
Several POD-associated proteins, including PML, are implicated in transcriptional regulation (for reviews see references 34 and 39). Since Daxx interacts with PML and localizes at the PODs, we decided to test whether Daxx might regulate transcription. Transfection of the Gal4-DBD full-length Daxx fusion protein (Gal-Daxx) in HEK293 cells strongly inhibits basal transcription of the Gal4-tk-luciferase reporter in a dose-dependent manner (Fig. 3A, top). Western blotting using anti-Gal4 DBD antibodies confirms increased expression of Gal-Daxx in transfected cells in the presence of higher concentrations of DNA (Fig. 3A, bottom). Comparison of Daxx-mediated transcriptional repression with that of PML-RARα fusion proteins indicates that Daxx represses as strongly as the PML-RARα oncoprotein (Fig. 3B). Moreover, repression by Gal-Daxx requires Gal4-binding sites (Fig. 3C) and occurs in multiple cell types (Fig. 3D), demonstrating the specificity of the observed Daxx-mediated transcriptional repression.
FIG. 3.
Modulation of promoter activity by Daxx. (A) Transcriptional repression by Gal-Daxx. Recruitment of Daxx to a promoter via Gal4-DBD results in inhibition of basal transcription in a dose-dependent manner. Transient transfection was conducted in HEK293 cells with increasing concentrations (nanograms) of Gal-Daxx as indicated. The relative fold repression of the basal promoter activity in the presence of Gal-Daxx was compared to that of Gal4-DBD alone. The bottom panels show immunoblots with anti-Gal4-DBD antibodies of the transfected fusion protein at indicated concentrations of expression vector. (B) Daxx represses basal transcription as strong as PML-RARα. HEK293 cells were transfected with equal amounts (250 ng) of each expression vector, and the relative repression was determined as described in Materials and Methods. The results show that Gal-Daxx represses basal transcription as strongly as Gal-PML-RARα. (C) Requirement of binding sites for transcriptional repression by Gal-Daxx. HEK293 cells were transfected with 250 ng of Gal-Daxx or Gal4-DBD alone, and the effects on the promoter activities of Gal-tk-luciferase (luc) and tk-luc reporters were determined. The Gal-tk-luc reporter contains four copies of Gal4-binding sites in front of the minimal tk promoter, while the tk-luc lacks the binding sites. (D) Mapping of the Daxx sequences required for repression. Schematic presentation of Gal-Daxx deletion mutants and their effects on promoter activity in HEK293, HeLa, and CV-1 cells are summarized. The two acidic regions are indicated by black bars, and the two potential nuclear localization signals are marked with diamonds. The bottom graph shows a column presentation of the repression activity of various Gal-Daxx deletion mutants in HEK293 cells. (E) Expression of Gal-Daxx mutants in transfected cells. The transfected lysates were analyzed by immunoblotting by using mouse anti-Gal4-DBD monoclonal antibodies. The top band in each lane represents the expected molecular weights of the Gal-Daxx mutants, except for Gal-Daxx (125-215), where the lower band is the expected product. The deviation in protein expression level was compensated by normalization of luciferase activity with the coexpressed β-Gal activity. (F) Inhibition of basal transcription from a natural promoter by Daxx. Wild-type Daxx was transfected into HEK293 cells together with either the SF1-tk-luciferase or tk-luciferase reporter. The fold repression of the luciferase activity at increasing concentrations (micrograms) of Daxx is presented.
We attempted to determine the sequences in Daxx that are responsible for the repression activity by standard deletion analysis (Fig. 3D). Progressive deletion from the C terminus to residue 124 gradually reduces the repression activity of Daxx in a cell-type-dependent manner. Deletions of the N terminus and several other mutants also show a significant decrease in repression. Equal expression of these Gal-Daxx deletion proteins in transfected cells is confirmed by Western blotting using the anti-Gal4 DBD antibodies (Fig. 3E). These data suggest that multiple regions of Daxx may be important for transcriptional repression in a cell-type-dependent manner.
Daxx was previously isolated in a yeast one-hybrid screen using a reporter containing a SF-1-like element (32). We decided to investigate whether Daxx can repress transcription from a promoter containing the SF-1-like element in a transient-transfection assay (Fig. 3F). As expected, overexpression of wild-type Daxx represses basal transcription from the SF1-tk-luciferase reporter that contains four copies of the SF1-like element, while it has little effect on the tk-luciferase reporter lacking the SF-1 sites. These data indicate that Daxx may repress the basal transcription of natural promoters containing SF1-like elements, a result consistent with the strong repressor activity observed with the Gal-Daxx fusion protein.
Daxx interacts with HDACs.
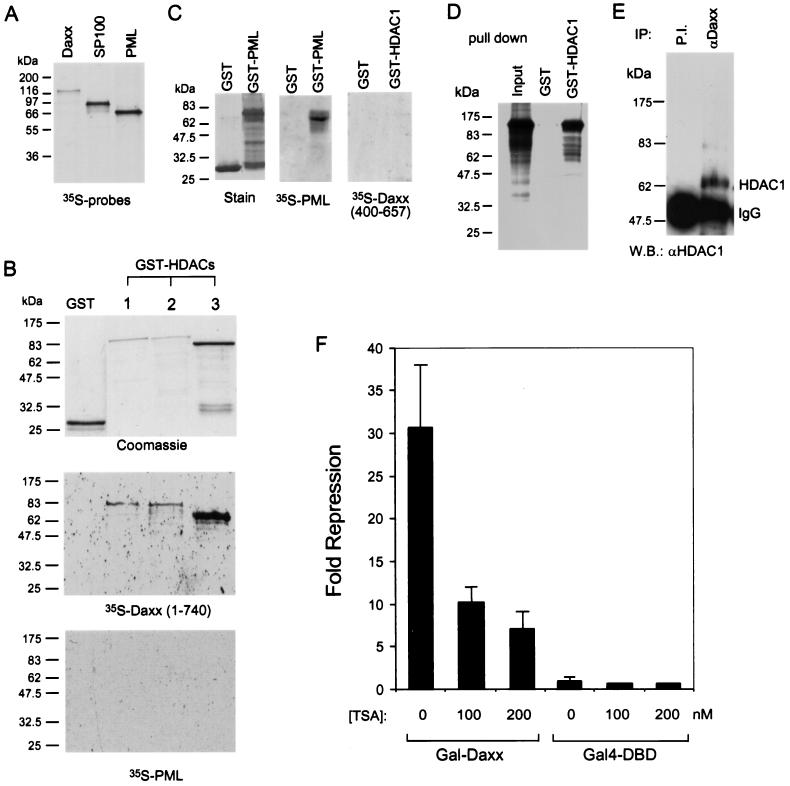
Histone deacetylation has been demonstrated to play a central role in transcriptional repression by inducing chromatin assembly and condensation (49, 66). To determine whether histone deacetylation is required for Daxx-mediated transcriptional repression, we analyzed the interaction between Daxx and the three available human histone deacetylases (HDACs) (67). The three human HDACs are highly conserved in structure and function. All of them repress basal transcription in the Gal4-DBD fusion assay, and all display histone deacetylase activity (67). Far-Western analyses demonstrate interactions between Daxx and all three GST-HDAC fusion proteins, but not GST alone, while PML and SP100 show no interaction with any of these GST-HDACs under the assay conditions (Fig. 4A and B and data not shown). A positive control shows that PML interacts efficiently with GST-PML under identical conditions (Fig. 4C). Furthermore, a Daxx mutant (amino acids 400 to 657) that possesses weak repression activity also does not interact with HDAC1 (Fig. 4C). These data support a role for HDAC interaction in Daxx-mediated transcriptional repression. The interaction between Daxx and HDAC1 is further confirmed in a GST pull-down assay (Fig. 4D), in which GST-HDAC1, but not GST alone, precipitates about 20% of input 35S-labeled Daxx. Moreover, an interaction between Daxx and HDAC1 in vivo is also observed by Co-IP of HeLa nuclear extracts (Fig. 4E), in which HDAC1 coimmunoprecipitates with Daxx antibodies but not with the preimmune serum. Together, these experiments provide strong evidence that Daxx and HDACs interact in vitro and in vivo.
FIG. 4.
Interaction of Daxx with HDACs. (A) 35S-labeled protein probes used in the far-Western assays. In vitro-translated [35S]methionine-labeled Daxx, PML, and SP100 were analyzed by SDS-PAGE and detected by autoradiography. (B) Far-Western analysis of interaction between Daxx and HDACs. The top panel shows the Coomassie blue-stained proteins used in the far-Western assay. The middle panel shows the far-Western blot of GST-HDACs with the 35S-labeled Daxx probe. The bottom panel shows the far-Western blot with the 35S-labeled PML probe. (C) PML interacts with GST-PML in the far-Western assay. A positive control showing that PML interacts with GST-PML in the far-Western assay was conducted under conditions identical to those for panel B. A far-Western blot showing that the Daxx mutant (400-657) fails to interact with GST-HDAC1 fusion protein. (D) GST pull-down assay showing interaction between GST-HDAC1 and Daxx. The input 35S-labeled Daxx contains one-third of the lysate used in the pull-down reaction, which was conducted as described in the Materials and Methods. (E) Co-IP of HDAC1 and Daxx. HeLa nuclear extracts were incubated with affinity-purified anti-Daxx antibody or an equal concentration of the preimmune serum. The immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blot by using anti-HDAC1 polyclonal antibodies. (F) TSA reverses transcriptional repression by Gal-Daxx. HEK293 cells were transfected with 250 ng of Gal4-DBD or Gal-Daxx mammalian expression vector together with a Gal4-dependent luciferase reporter. The fold repression by Gal-Daxx at different concentrations of TSA was determined relative to that for the Gal4-DBD alone.
HDAC inhibitor reverses Daxx-mediated repression.
The physical interaction observed between Daxx and HDAC suggests that Daxx may recruit a HDAC corepressor complex to repress basal transcription via histone deacetylation and chromatin condensation. To provide more evidence for this possibility, we assayed the effect of a histone deacetylase inhibitor, trichostatin A (TSA), on the repressor activity of Gal-Daxx in a transient-transfection assay (Fig. 4F). As expected, TSA reverses transcriptional repression by Gal-Daxx in a dose-dependent manner, while it has little effect on Gal4-DBD alone under identical conditions. These data indicate that histone deacetylation is involved in transcriptional repression by Daxx.
Inhibition of Daxx-mediated transcriptional repression by PML.
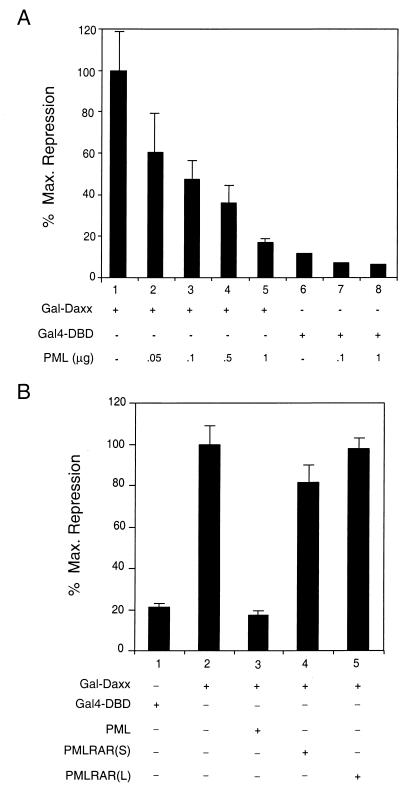
Since Daxx was identified as a PML-interacting protein and subsequently demonstrated to possess strong transcriptional repression activity, we decided to investigate the role of PML in the regulation of transcriptional repression by Daxx. To do this, Gal-Daxx was cotransfected with increasing amounts of full-length PML into HEK293 cells and the activity of the luciferase reporter was measured (Fig. 5A). As observed above, Gal-Daxx represses reporter expression strongly when compared to the Gal4-DBD alone (Fig. 5A, compare lanes 1 and 6). Interestingly, coexpression of increasing amounts of PML inhibits this repression in a dose-dependent manner, abolishing nearly all of the repressor function of Gal-Daxx (lanes 2 to 5). This effect is specific to Gal-Daxx, for cotransfection of PML with the Gal4-DBD alone has little effect on reporter activity (lanes 7 and 8). These data suggest that PML may inhibit Daxx-mediated transcriptional repression.
FIG. 5.
Inhibition of Daxx-mediated transcriptional repression by PML. (A) PML inhibits Daxx-mediated transcriptional repression. HEK293 cells were transiently transfected with 100 ng of the Gal4-DBD or Gal-Daxx mammalian expression vectors in the absence or presence of the indicated amounts of PML expression vector together with a Gal4-dependent luciferase reporter. Data are presented as the percentage of maximum repression, where Gal-Daxx activity is represented as 100% repression. (B) PML-RARα has no effect on Daxx-mediated transcriptional repression. HEK293 cells were transiently transfected with 100 ng of the Gal4-DBD or Gal-Daxx mammalian expression vectors in the absence or presence of expression vectors for PML, PML-RARα (short form), or PML-RARα (long form), together with a Gal4-dependent luciferase reporter. Data are presented as the percentage of maximum repression, where Gal-Daxx activity is represented as 100% repression.
Similar experiments were then performed to determine if PML-RARα might also regulate the function of Gal-Daxx. When either the short or long forms of PML-RARα were cotransfected with Gal-Daxx, the repression activity of Gal-Daxx was unchanged (Fig. 5B). Thus, despite the observation that both PML and PML-RARα interact with Daxx, only PML can inhibit the ability of Daxx to repress transcription, suggesting a differential role of PML and its oncogenic fusion protein in regulation of Daxx function.
PML recruits Daxx to the POD.
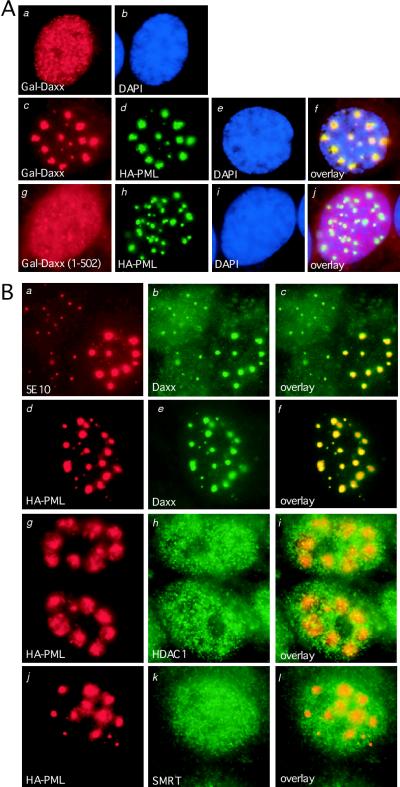
To elucidate the mechanism by which PML blocks Daxx-mediated transcriptional repression, immunofluorescence microscopy was used to investigate the subcellular localization of Gal-Daxx upon coexpression of PML. In these experiments, HEp2 cells were transiently transfected with Gal-Daxx in the absence or presence of HA-PML and subsequently stained with the mouse anti-Gal4-DBD and rabbit anti-HA antibodies (Fig. 6A). When Gal-Daxx was overexpressed alone in HEp2 cells, a fairly diffuse, evenly distributed staining pattern is observed in the nucleus (Fig. 6A, panels a and b). Cotransfection of PML drastically alters the distribution of Gal-Daxx, for nearly all of the Gal-Daxx protein is recruited to the PODs, even at very high levels of Gal-Daxx expression (Fig. 6A, panels c to f). Examination of the localization of these enlarged PODs indicates that they occupy the loose chromatin regions (Fig. 6Ae and f), similar to the localization of PODs in the absence of PML overexpression. On the contrary, cotransfection of PML does not recruit a Daxx mutant (Gal-Daxx 1-502) lacking the PML-interacting domain to the PODs (Fig. 6A, panels g to j), suggesting the specificity of the assay. The abilities of PML to reverse Daxx-mediated repression and to recruit Daxx to the PODs support the hypothesis that PML may inhibit Daxx repressor function by sequestration of Daxx to the PODs.
FIG. 6.
Recruitment of Daxx to POD domains by overexpression of PML. (A) Overexpression of PML recruits transfected Gal-Daxx into the PODs. Gal-Daxx or Gal-Daxx (1-502) were transiently transfected into HEp2 cells in the absence or presence of HA-tagged PML and subsequently stained with the mouse anti-Gal4-DBD and rabbit anti-HA antibodies. Primary antibodies were detected with rhodamine-conjugated anti-mouse immunoglobulin G and fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G secondary antibodies and analyzed by immunofluorescence microscopy. Panels a and b show diffuse nuclear staining of Gal-Daxx in the absence of PML. Panels c to f show Gal-Daxx and HA-PML colocalization at the PODs. Panels g to j show that HA-PML cannot recruit a Gal-Daxx (1-502) mutant lacking the PML-interacting domain to the PODs. (B) Recruitment of endogenous Daxx but not HDAC1 and SMRT to the PODs. HA-PML was transfected into HEp2 cells, and the localization of endogenous Daxx, HDAC1, and SMRT was analyzed by immunofluorescence microscopy. Panels a to c show colocalization of transfected and endogenous PML with endogenous Daxx by using anti-PML monoclonal 5E10 and anti-Daxx rabbit polyclonal antibodies. Panels d to f show HA-PML and Daxx colocalization by using the anti-HA monoclonal and anti-Daxx polyclonal antibodies. Panels g to l demonstrate that HA-PML does not recruit HDAC1 or SMRT to the PODs by using anti-HA, anti-HDAC1, or anti-SMRT antibodies. Yellow signals in the overlay images indicate colocalization.
PML recruits endogenous Daxx to the PODs.
To address whether recruitment of Daxx to the PODs also occurs at the endogenous levels of Daxx, HA-PML was transfected into HEp2 cells alone, and the localization of endogenous Daxx was analyzed by immunofluorescence staining by using anti-Daxx antibodies (Fig. 6B). Double immunostaining with anti-PML antibodies reveals that overexpression of PML leads to accumulation of endogenous Daxx to the PODs, resulting in reduced nucleoplasmic staining (Fig. 6B, panels a to c). Recruitment of endogenous Daxx to the PODs is confirmed with anti-HA antibodies that detect only the transfected HA-PML (Fig. 6B, panels d to f). These data indicate that PML is able to recruit endogenous nucleoplasmic Daxx to the PODs.
PML does not recruit HDAC1 to the PODs.
So far we have shown that Daxx interacts with HDACs (Fig. 4) and that PML recruits Daxx to the PODs (Fig. 6). Accordingly, we wished to determine the localization of HDAC and other corepressors, such as SMRT, after PML overexpression. We find that overexpression of PML does not alter the distribution of HDAC1 or SMRT (Fig. 6B, panels g to l), suggesting that PML may segregate Daxx away from the corepressor complex. These observations are consistent with a speculative mechanism by which PML may inhibit transcriptional repression of Daxx via sequestrating Daxx to the PODs.
SUMO-1 modification of PML is required for recruitment of Daxx to the PODs.
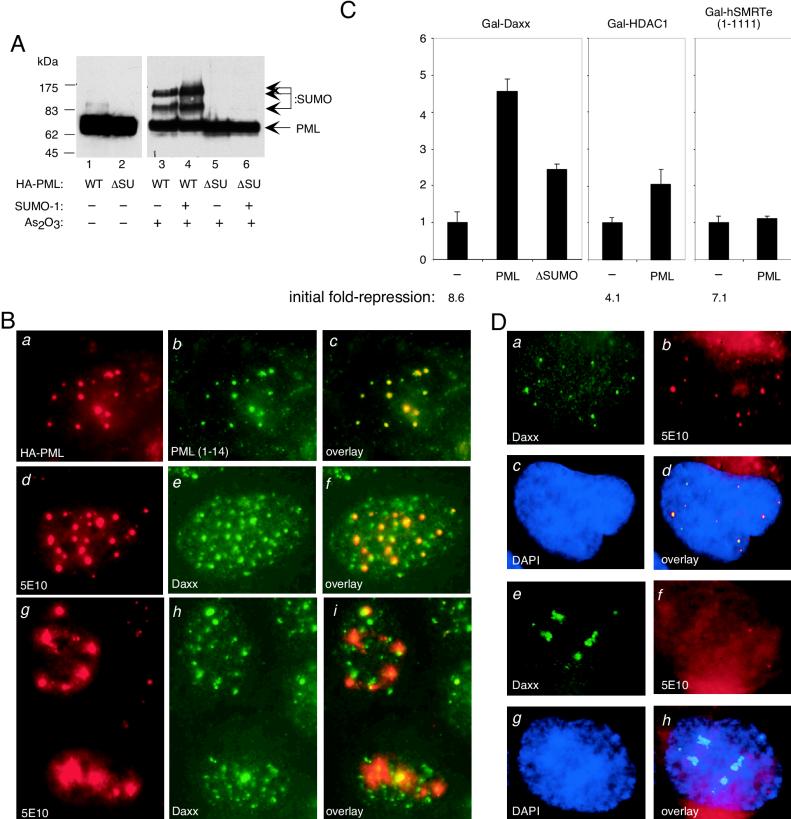
To determine if SUMO-1 modification of PML may play a role in Daxx interaction, we generated a PML mutant with all three SUMO-1 modification lysine residues replaced with arginines by site-directed mutagenesis, based on a prior study that mapped the modification sites (31). Upon mutation of the three lysine residues of PML, we no longer observe SUMO-1-conjugated forms of PML, even after treatment of the transfected cells with As2O3 and coexpression with SUMO-1 (Fig. 7A). This PML ΔSUMO mutant behaves similarly to the wild-type protein in localizing to the PODs and in enlarging the POD structure (Fig. 7B, panels a, d, and g). Interestingly, while this SUMO-1-deficient mutant is capable of localizing to PODs (panels b and c and panels d and e), overaccumulation of the mutant protein in the PODs fails to recruit nucleoplasmic Daxx (Fig. 7B, panels g to i). In contrast, many of the enlarged PODs show reduced staining of Daxx (Fig. 7B, panels g to i), suggesting that accumulation of the unmodified form of PML in the PODs may lead to the disappearance of Daxx in PODs. These data suggest SUMO-1 modification as being the underlying mechanism for the observed interaction and colocalization between Daxx and PML in vivo.
FIG. 7.
SUMO-1 modification of PML is required for sequestration of Daxx to the POD and inhibition of Daxx-mediated transcriptional repression. (A) The PML ΔSUMO (ΔSU) mutant lacks SUMO-1 modification. This mutant was created by replacing all three lysines at residues 65, 160, and 490 with arginines. The wild-type (WT) PML and the ΔSUMO mutant were transfected into HEK293 cells alone or in combination with a SUMO-1 expression vector. Cells were treated with 1 μM arsenic trioxide (As2O3) for 6 h where indicated. The total cell lysates were analyzed by Western blotting by using anti-HA monoclonal antibodies. The upper bands in the wild-type proteins represent SUMO-1 conjugated forms of PML. (B) The PML ΔSUMO mutant localizes to the PODs but fails to recruit Daxx. The HA-PML ΔSUMO mutant was transfected into HEp2 cells and analyzed by immunofluorescence microscopy to detect localization of the transfected mutant protein and the distribution of endogenous Daxx. Panels a to c show localization of the transfected HA-PML ΔSUMO mutant protein (detected by a HA antibody) in the PODs that were revealed by a PML polyclonal antibody (1-14). Panels d to f show that the enlarged PODs in the HA-PML ΔSUMO mutant transfected cells do not result in prominent recruitment of Daxx to the PODs. Panels g to i show that many enlarged PODs containing PML-ΔSUMO mutant have little or no Daxx protein. The 5E10 antibodies also detect PODs in untransfected cells that show smaller structures colocalized with Daxx foci. (C) The PML ΔSUMO mutant is deficient in reversing transcriptional repression by Daxx. The transfection was conducted in HEK293 cells, and the initial fold repression mediated by the Gal4-DBD fusion proteins is as indicated at the bottom. The y axis indicates the fold reversal of repression. The wild-type PML does not reverse transcriptional repression mediated by HDAC1 or SMRTe. (D) Association of Daxx with condensed chromatin in cells that lack PODs. The human neuronal NT2 stem cells were analyzed by double immunofluorescence staining with anti-Daxx polyclonal and anti-PML 5E10 monoclonal antibodies. The NT2 cells display heterogeneous staining for PML. In cells that contain normal PML nuclear bodies (panels a to d), Daxx appears normal and shows complete colocalization with PML. In contrast, cells that contain only two or fewer PML nuclear dots show aggregated Daxx surrounding the condensed chromatin areas stained with DAPI (panels e to h).
SUMO-1 modification of PML is required for efficient inhibition of Daxx-mediated repression.
If our hypothesis that recruitment of Daxx to the POD is inhibitory to its transcriptional repression activity, one would predict that the PML ΔSUMO mutant that fails to recruit Daxx to the PODs will be defective in reversing transcriptional repression by Daxx. As expected, we found that the PML ΔSUMO mutant is less effective in reversing transcriptional repression by Daxx in transient transfection (Fig. 7C). Furthermore, we found that the wild-type PML is incapable of reversing transcriptional repression by Gal-HDAC1 and Gal-SMRTe (Fig. 7C). These data correlate with immunofluorescence studies demonstrating PML recruitment of Daxx, but not HDAC or SMRT, to the POD, where it presumably is unable to repress transcription.
Daxx is associated with condensed chromatin in the absence of PML.
To provide further evidence that the demonstrated functional interactions between Daxx and PML may be physiologically relevant, we screened several cell lines to find a cell type that may display abnormal localization of Daxx and/or PML. We identified the embryonic carcinoma NT2 cell line; upon staining with the anti-PML antibody, it is evident that only a subset of these cells express PML and thus contain PODs (Fig. 7D). In these cells, PML and Daxx colocalize in the PODs (panels a to d). However, in cells lacking detectable PODs, Daxx forms aggregates around the condensed chromatin (Fig. 7D, panels e to h). Therefore, the localization and thus the function of Daxx may depend on the level of PML in the cell. At low PML levels, Daxx is concentrated at condensed chromatin, where it may repress transcription. When PML levels are higher, it is able to recruit Daxx away from condensed chromatin to the PODs, where Daxx no longer represses basal transcription.
DISCUSSION
In the present study, we have identified Daxx as a PML-interacting protein and characterized the functional interaction between Daxx and PML. We find a majority of Daxx in the nucleus of HeLa and HEp2 cells where it colocalizes with PML in the PODs. In the NB4 APL cell line, Daxx is distributed in the microparticulate structures that contain the PML-RARα oncoprotein (17). The repressor function of Daxx is observed upon tethering it to a reporter gene via a heterologous DNA binding domain, as well as from a reporter containing a natural SF1-like promoter element. The mechanism by which Daxx represses basal transcription is found as involving histone deacetylation, for Daxx interacts with HDACs in vitro and in vivo and the histone deacetylase inhibitor, TSA, blocks the repressor activity. Coexpression of PML reverses the transcriptional repression by Daxx, which, in turn, correlates with the recruitment of Daxx to the PODs. In addition, we show that SUMO-1 modification of PML is required for both recruitment of Daxx to the PODs and efficient inhibition of Daxx-mediated repression. The physiological role of Daxx in transcriptional repression is further supported by the observation that Daxx associates with condensed chromatin in cells that lack PML. Together, these data establish novel roles for Daxx, as a transcriptional repressor, and for PML, as a protein that can potentially regulate the repressor function of Daxx.
Consistent with our findings, Daxx has recently been identified as an inhibitor of transcriptional activation by Pax3, a member of the homeodomain family of transcription factors (28). Thus, Daxx not only is able to repress basal transcription, as suggested from our data, but can also inhibit transcriptional activation via interactions with DNA-binding transcription factors. While the exact mechanism of the inhibition of Pax3 transactivation by Daxx is unclear, our data elucidate the mechanism of Daxx-mediated repression of basal transcription as involving histone deacetylation. We observe Daxx localization to condensed chromatin in NT2 cells that lack detectable PML. Condensed chromatin is considered as a site of transcriptional repression that also includes transcriptionally silent centromeric heterochromatin. Other POD-associated proteins, such as SP100, have been demonstrated to interact with heterochromatin protein 1 (HP1) and also colocalize with centromeric chromatin (10, 54). Consistent with this idea, Daxx has been shown to interact with CENP-C in a yeast two-hybrid assay and partially to colocalize with interphase centromeres (50). Also, Daxx has been shown to interact with DNA methyltransferase 1, which plays a role in gene silencing (43).
Previous studies have implicated the PODs as sites of transcriptional activation. For example, PML has been demonstrated to interact with the transcription coactivator CBP and recruit CBP to the PODs (15, 35). Furthermore, PML can enhance the transactivation functions of both CBP and members of the nuclear receptor superfamily (15). PML also induces genes of the major histocompatibility complex, while PML−/− mice display reduced transactivation responses to atRA (64, 69). Finally, the transcriptional activator Sp140 (5, 6) and nascent RNA (35) have been found in at least a subset of PODs. Our findings that Daxx represses basal transcription and PML inhibits such repressor activity suggest a new role for the POD structure in gene regulation. The POD may enhance transcription of target genes not only through recruitment of activators but also through the inactivation of repressors such as Daxx via recruitment by PML. Because other transcriptional repressors, such as PLZF, pRB, and Sp100, have also been found in the PODs, it will be interesting to determine if PML can regulate the repressor activities of these proteins as well.
Our observations that PML-RARα can interact with Daxx but not inhibit transcriptional repression by Daxx suggest a potential role for Daxx in acute promyelocytic leukemia. Support for this hypothesis is evident in our finding that Daxx, PML-RARα, and PML colocalize at diffusely distributed microparticulate structures in nucleus of the APL NB4 cells. The PML-RARα fusion protein disrupts the POD structure in these cells and, through its interaction with Daxx, may direct Daxx to the microparticulate structures, where it is capable of repressing gene expression. PML-RARα itself is a potent transcriptional repressor, which acts via the recruitment of the corepressors SMRT, N-CoR, and HDAC1 (40). The POD structure is reorganized upon treatment of these cells with atRA or arsenic trioxide, leading to the degradation of the PML-RARα fusion protein and colocalization of Daxx and PML in the PODs (47). Therefore, Daxx inactivation through localization to the PODs may be critical to the differentiation of normal hematopoietic cells. Since expression of the PML-RARα fusion protein disrupts the integrity of the PODs, Daxx may act as a constitutive repressor in the APL cells, which along with the repressor function of PML-RARα, may block expression of specific genes that are critical for cell differentiation and culminate in the subsequent APL pathology.
Daxx was initially identified as a Fas-binding protein that promoted Fas-mediated apoptosis via activation of the JNK kinase cascade pathway (12, 68). Interestingly, PML has also been found to be involved in apoptosis triggered by Fas, tumor necrosis factor alpha, and type I and II interferons, possibly by recruitment of the death effector Bax and cdk inhibitor p21 (37, 51, 64). In contrast, expression of PML-RARα prevents apoptosis in response to these signals (51). Our findings, together with these reports, suggest that the regulation of Daxx repressor function by PML may also be important in programmed cell death. Consistent with this possibility, several transcriptional repressors are known to play a role in apoptosis. For example, the adenovirus E1B and the cellular Bcl-2 oncoprotein block p53-mediated apoptosis by inhibiting transcriptional repression by p53, suggesting that p53 induces apoptosis via transcriptional repression (52, 56). In the case of Daxx, PML may recruit it to the PODs, where it is inactivated, thus allowing the expression of certain genes required for apoptosis. Conversely, PML-RARα might inhibit apoptosis in APL cells through disruption of the PODs, thereby promoting enhanced or constitutive repression of these target genes by Daxx and the PML-RARα fusion protein itself, which leads to the APL phenotype. Retinoic acid treatment would stimulate degradation of PML-RARα and restoration of the POD structure (17, 47, 65). This would allow Daxx to be inactivated through sequestration to the PODs and allow apoptosis to proceed and would eventually lead to remission of the APL phenotype. Because PML can shuttle between the nucleus and cytoplasm (59, 60), it is possible that Daxx may be brought along with PML to regulate cytoplasmic events relevant to Fas-mediated apoptosis. However, a recent study reports that the loss of Daxx leads to extensive apoptosis in early mouse development (43), a result seemingly opposite to other findings concerning the function of Daxx in apoptosis (12, 13, 68). Therefore, the precise role of Daxx in apoptosis remains to be further elucidated.
Our data provide strong evidence for the roles of PML and the PODs in regulating the function of Daxx as a transcriptional repressor. Daxx and PML interact in vivo and colocalize in the PODs. Overexpression of PML recruits Daxx to the PODs, which correlates with a complete inhibition of transcriptional repression by Daxx. Although the detailed mechanism of this inhibition of Daxx by PML remains to be determined, our data provide several possibilities. First, PML might inactivate Daxx by transporting it to the PODs and separating it from HDAC and putative target genes. In response to certain stimuli such as interferon, PML levels increase in the PODs, which, via competition for Daxx binding or conformational change of Daxx upon PML binding, might result in the dissociation of Daxx from HDAC and recruitment of Daxx, but not HDAC, to the PODs. Confinement of Daxx in the PODs would thus block access to target genes, whose expression level would then increase to at least the basal level in the absence of Daxx repression. Our findings that PML overexpression results in increased Daxx levels in the PODs, while having no effect on HDAC1 distribution or repression by HDAC1, support this possibility. Alternatively, the increased PML levels may dissociate HDAC from Daxx and recruit both Daxx and its putative target genes, but not HDAC, to the PODs. Because Daxx requires HDAC and histone deacetylation for its repressor activity, the target genes may be expressed in the absence of HDAC. The presence of transcriptional activators in the PODs would facilitate transcription of target genes. With either possibility, it is evident that the POD is involved in maintaining the balance of Daxx function, depending on the PML level. At normal, physiological levels of PML, Daxx might repress transcription at areas of condensed chromatin. However, with increased PML expression, more Daxx is recruited to the PODs, thus reducing its overall repression activity. Although the precise mechanism of the inhibition of Daxx repression by the PODs awaits further investigation, our data clearly reveal a novel connection between Daxx and PML in regulating transcriptional repression that may play a critical role in acute promyelocytic leukemia and apoptosis.
ACKNOWLEDGMENTS
We are grateful to W. M. Yang for the GST-HDAC constructs and to J. F. Strauss III for the SF1-tk-luc construct. We thank M. Lanotte for the NB4 cell line, N. Stuurman for 5E10 monoclonal antibodies, and A. P. Otte for hPc2 antibodies, as well as R. M. Evans for PML-1 and PML-RAR vectors and K. S. Chang for PML vector and antibodies. We also thank colleagues, including W. F. Greenlee, J. Lawrence, D. Ludlum, A. Ross, G. Stein, J. Stein, C. Sagerström, and D. Schroen, for reading and comments on the manuscript. We thank M. Bhaumik for technical assistance, J. Nickerson for confocal microscopy and M. Nadler for epifluorescence microscopy.
C.L. is a predoctoral fellow of the Army Breast Cancer Program. J.D.C. is a junior scholar of the American Society of Hematology. This work was made possible by grant PRG-98-085-01-LBC from American Cancer Society.
H.L. and C.L. contributed equally to this work.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Benedetti L, Levin A A, Scicchitano B M, Grignani F, Allenby G, Diverio D, Lo Coco F, Avvisati G, Ruthardt M, Adamo S, Pelicci P G, Nervi C. Characterization of the retinoid binding properties of the major fusion products present in acute promyelocytic leukemia cells. Blood. 1997;90:1175–1185. [PubMed] [Google Scholar]
- 5.Bloch D B, Chiche J D, Orth D, de la Monte S M, Rosenzweig A, Bloch K D. Structural and functional heterogeneity of nuclear bodies. Mol Cell Biol. 1999;19:4423–4430. doi: 10.1128/mcb.19.6.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloch D B, de la Monte S M, Guigaouri P, Filippov A, Bloch K D. Identification and characterization of a leukocyte-specific component of the nuclear body. J Biol Chem. 1996;271:29198–29204. doi: 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- 7.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 8.Borden K L, Boddy M N, Lally J, O'Reilly N J, Martin S, Howe K, Solomon E, Freemont P S. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci P G, Atwater S, Bishop J M. A PMLRARalpha transgene initiates murine acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown K E, Guest S S, Smale S T, Hahm K, Merkenschlager M, Fisher A G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 11.Carvalho T, Seeler J S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang H Y, Nishitoh H, Yang X, Ichijo H, Baltimore D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 13.Chang H Y, Yang X, Baltimore D. Dissecting Fas signaling with an altered-specificity death-domain mutant: requirement of FADD binding for apoptosis but not Jun N-terminal kinase activation. Proc Natl Acad Sci USA. 1999;96:1252–1256. doi: 10.1073/pnas.96.4.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 15.Doucas V, Tini M, Egan D A, Evans R M. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc Natl Acad Sci USA. 1999;96:2627–2632. doi: 10.1073/pnas.96.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 17.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 18.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grande M A, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort H T, de Jong L, van Driel R. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J Cell Biochem. 1996;63:280–291. doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 20.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 21.Grisolano J L, Wesselschmidt R L, Pelicci P G, Ley T J. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RAR alpha under control of cathepsin G regulatory sequences. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 22.Guidez F, Ivins S, Zhu J, Soderstrom M, Waxman S, Zelent A. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood. 1998;91:2634–2642. [PubMed] [Google Scholar]
- 23.Guldner H H, Szostecki C, Grotzinger T, Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1998. [Google Scholar]
- 25.He L Z, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi P P. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 26.He L Z, Tribioli C, Rivi R, Peruzzi D, Pelicci P G, Soares V, Cattoretti G, Pandolfi P P. Acute leukemia with promyelocytic features in PML/RARalpha transgenic mice. Proc Natl Acad Sci USA. 1997;94:5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 28.Hollenbach A D, Sublett J E, McPherson C J, Grosveld G. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 1999;18:3702–3711. doi: 10.1093/emboj/18.13.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakizuka A, Miller W, Jr, Umesono K, Warrell R, Jr, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 30.Kalantry S, Delva L, Gaboli M, Gandini D, Giorgio M, Hawe N, He L Z, Peruzzi D, Rivi R, Tribioli C, Wang Z G, Zhang H, Pandolfi P P. Gene rearrangements in the molecular pathogenesis of acute promyelocytic leukemia. J Cell Physiol. 1997;173:288–296. doi: 10.1002/(SICI)1097-4652(199711)173:2<288::AID-JCP38>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Kamitani T, Kito K, Nguyen H P, Wada H, Fukuda-Kamitani T, Yeh E T. Identification of three major sentrinization sites in PML. J Biol Chem. 1998;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- 32.Kiriakidou M, Driscoll D A, Lopez-Guisa J M, Strauss J F., III Cloning and expression of primate Daxx cDNAs and mapping of the human gene to chromosome 6p21.3 in the MHC region. DNA Cell Biol. 1997;16:1289–1298. doi: 10.1089/dna.1997.16.1289. [DOI] [PubMed] [Google Scholar]
- 33.Koken M H, Puvion-Dutilleul F, Guillemin M C, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 35.LaMorte V J, Dyck J A, Ochs R L, Evans R M. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3) Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- 37.Le X F, Vallian S, Mu Z M, Hung M C, Chang K S. Recombinant PML adenovirus suppresses growth and tumorigenicity of human breast cancer cells by inducing G1 cell cycle arrest and apoptosis. Oncogene. 1998;16:1839–1849. doi: 10.1038/sj.onc.1201705. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin R J, Egan D A, Evans R M. Molecular genetics of acute promyelocytic leukemia. Trends Genet. 1999;15:179–184. doi: 10.1016/s0168-9525(99)01710-2. [DOI] [PubMed] [Google Scholar]
- 40.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 41.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 42.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 43.Michaelson J S, Bader D, Kuo F, Kozak C, Leder P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 1999;13:1918–1923. doi: 10.1101/gad.13.15.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mu Z M, Le X F, Vallian S, Glassman A B, Chang K S. Stable overexpression of PML alters regulation of cell cycle progression in HeLa cells. Carcinogenesis. 1997;18:2063–2069. doi: 10.1093/carcin/18.11.2063. [DOI] [PubMed] [Google Scholar]
- 45.Muller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 47.Nervi C, Ferrara F F, Fanelli M, Rippo M R, Tomassini B, Ferrucci P F, Ruthardt M, Gelmetti V, Gambacorti-Passerini C, Diverio D, Grignani F, Pelicci P G, Testi R. Caspases mediate retinoic acid-induced degradation of the acute promyelocytic leukemia PML/RARalpha fusion protein. Blood. 1998;92:2244–2251. [PubMed] [Google Scholar]
- 48.Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei C F, Chang H M, Yeh E T. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- 49.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 50.Pluta A F, Earnshaw W C, Goldberg I G. Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J Cell Sci. 1998;111:2029–2041. doi: 10.1242/jcs.111.14.2029. [DOI] [PubMed] [Google Scholar]
- 51.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J C, de The H. PML induces a novel caspase-independent death process. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 52.Sabbatini P, Chiou S K, Rao L, White E. Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol Cell Biol. 1995;15:1060–1070. doi: 10.1128/mcb.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satijn D P, Gunster M J, van der Vlag J, Hamer K M, Schul W, Alkema M J, Saurin A J, Freemont P S, van Driel R, Otte A P. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sewalt R G, van der Vlag J, Gunster M J, Hamer K M, den Blaauwen J L, Satijn D P, Hendrix T, van Driel R, Otte A P. Characterization of interactions between the mammalian polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen Y, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular Bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 58.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 60.Stuurman N, Floore A, Middelkoop E, van Driel R, de Jong L. PML shuttles between nuclear bodies and the cytoplasm. Cell Mol Biol Lett. 1997;2:137–150. [Google Scholar]
- 61.Szekely L, Pokrovskaja K, Jiang W Q, de The H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szostecki C, Guldner H H, Netter H J, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 63.Wang Z G, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi P P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi P P. Pml is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 65.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 66.Wolffe A P. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 67.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 68.Yang X, Khosravi-Far R, Chang H Y, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng P, Guo Y, Niu Q, Levy D E, Dyck J A, Lu S, Sheiman L A, Liu Y. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 1998;396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]