Abstract
Bud dormancy is an evolved trait that confers adaptation to harsh environments, and affects flower differentiation, crop yield and vegetative growth in perennials. ABA is a stress hormone and a major regulator of dormancy. Although the physiology of bud dormancy is complex, several advancements have been achieved in this field recently by using genetics, omics and bioinformatics methods. Here, we review the current knowledge on the role of ABA and environmental signals, as well as the interplay of other hormones and sucrose, in the regulation of this process. We also discuss emerging potential mechanisms in this physiological process, including epigenetic regulation.
Keywords: ABA, perennials, bud dormancy, hormone, sucrose, epigenetics
1. Introduction
Dormancy is an evolved trait of perennial plants that allows vegetative buds to survive harsh environmental conditions, and it was classified into three categories by Lang (1987): ecodormancy, caused by limitations in environmental factors; endodormancy, where the inhibition resides in the dormant structure itself, and paradormancy, inhibited by distal organs. Abscisic acid (ABA) is recognized as an essential phytohormone in dormancy regulation, especially as a central hub in seed dormancy [1,2,3,4], but its regulatory mechanism in bud dormancy is not well understood.
1.1. ABA Metabolism and Signaling
ABA biosynthetic and catabolic pathways are well understood [5,6]. ABA is de novo synthesized from the precursor isopentenyl diphosphate (IPP), which is further converted into lycopene after four desaturation steps of carotenoid. Lycopene undergoes cyclization and hydroxylation to generate zeaxanthin. The first step of ABA biosynthesis is initiated in plastid with epoxidation of zeaxanthin to all-trans-xanthophylls zeaxanthin and violaxanthin, catalyzed by zeaxanthin epoxidase (ZEP) [7,8]. Violaxanthin is further isomerized into cis-violaxanthin, which is cleaved by 9-cis-epoxycarotenoid dioxygenase (NCED) to yield xanthoxin, the first C15 intermediate of ABA biosynthesis [9]. Xanthoxin is transferred from the plastid to the cytosol, where it is converted to abscisic aldehyde by short-chain alcohol dehydrogenase. Finally, abscisic aldehyde is oxidized by aldehyde oxidase 3 to form ABA [5,9]. The catabolic pathway of ABA is mainly established through hydroxylation reaction. ABA can be hydroxylated at the C-8′ position by ABA 8′-hydroxylase, which is encoded by the CYP707A gene family. 8′-hydroxy-ABA is unstable and enzymatically isomerizes to generate phaseic acid [10,11]. In addition, ABA homeostasis can also be altered by enzymes during intracellular and intertissue-mediated transport, such as cytosolic UDP- glucosyltransferases (GTs) or β-glucosidases (BGs) [12,13,14]. In addition, ABA could be transported among cells and organs in plants. The active transporters are ATP-binding cassette (ABCG) transporters, NRT1/PTR FAMILY (NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY; NPF), multidrug and toxic compound extrusion (DETOXIFICATION 50; DTX50), and AWPM-19 (ABA-INDUCED WHEAT PLASMA MEMBRANE POLYPEPTIDE-19) family proteins [6].
Over the past decades, the core components and the regulation of ABA signaling have been characterized. In 2009, soluble receptor proteins (PYRABACTIN RESISTANCE/PYRABACTIN RESISTANCE 1-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR (PYR/PYL/RCAR)) for ABA in Arabidopsis were identified [15,16]. ABA co-receptor, ABI1 and ABI2, are members of clade A Type 2C PP2Cs protein (PROTEIN PHOSPHATASE 2Cs) and act as negative regulators of ABA signaling. PP2Cs inactivate SnRK2s (SNF1-related protein kinase 2s) by dephosphorylating their kinase activation loop and repress ABA response in the absence of ABA [17,18]. In the presence of ABA, PYR/PYL/RCARs interact with PP2Cs in which the phosphatase activity of PP2Cs is inhibited, facilitating activation of SnRK2s. Subsequently, SnRK2s activate ABA-RESPONSIVE ELEMENT-BINDING FACTORS (ABFs or AREBs), which then initiate transcription at ABA-responsive elements (ABREs) in the promoter regions of their target genes [19].
1.2. Bud Dormancy
Perennial bud dormancy can be classified into two forms based on the position of dormant vegetative organs, i.e., woody bud dormancy (aboveground) and geophytes dormancy (underground; corms, tubers, bulbs and rhizomes, underground adventitious buds on the crown and lateral roots).
With respect to bud dormancy in woody plants, there is a wide range of genetic variation within and between species and responses. Here, we take the woody model plant, Populus, as a main example. In autumn, with short photoperiod and low temperature, growth cessation, bud set, and dormancy of the bud meristem occurs sequentially in buds of populus [20]. Among environmental factors, light plays the dominant role in dormancy of most woody plants [21]. Early woody bud dormancy involves a series of states: cessation of vegetative growth, formation of terminal buds, arrangement of abscission layers in leaves, development of cold resistance, establishment of winter rest (endodormancy), and leaf fall [22]. Initiation of bud dormancy can be stimulated by environmental factors like short photoperiod, low or high temperature, low nutrition and water deficit. During the stage of winter rest, there are many metabolic and developmental activities happening in the dormant buds including respiration, photosynthesis, slow cell division, enzyme synthesis, production of growth stimulators, and dissipation of growth inhibitors. In buds, callose disrupts the symplastic pathway in all vascular cells, including sieve elements, by blocking the plasmodesmata, resulting in decreased flow of water, nutrients and other molecules in buds [23]. Phytohormones including abscisic acid (ABA), gibberellin (GA), ethylene (ET), auxin and cytokinins (CKs) are involved in bud dormancy, of which ABA plays an essential role in this process [24]. DAM (DORMANCY-ASSOCIATED MADS-BOX) is a SHORT VEGETATVIE PHASE (SVP) homolog, which responds to environment factors including temperature and photoperiod, and regulates endogenous ABA levels during bud dormancy [25].
As for geophytes, the dormant organs develop in the soil before going to a dormant state. During bulb formation, endogenous ABA is increased, which stimulates starch accumulation and bulb development. Afterwards, increased ABA inhibits organs development and initiates bulb dormancy. Based on the different dormancy traits, the major geophytes can be classed into three groups: (1) species with a relative long dormant period, during which the differentiation of new organs is inhibited, like Gladiolus; (2) species that differentiate flower buds inside the bulbs before/during bulb dormancy, including Tulipa, Asiatic Lilium Hybrids and Hyacinthus; (3) species with no visible dormancy unless in hash environments, like Hippeastrum. Environmental factors (low temperature and short photoperiod) triggers bulb dormancy. Temperature is the main factor in geophytes dormancy, affecting internal metabolites, phytohormones and signal response [26,27]. During geophytes dormancy release, several metabolites change dramatically, including a gradually decreased ABA, increased GA and glucose/sucrose ratio and enhanced respiratory activity, although there are invisible morphological differences [28,29]. Other special chemicals like glycerol, phenolics and peroxidase in scales could regulate bulb dormancy as well [30,31].
Much progress has been achieved on seed dormancy in model plants like Arabidopsis, Zea mays and rice, although research on dormancy in perennial plant is also expanding. This article attempts to mainly review the existing knowledge of winter bud endodormancy in perennials including trees and geophytes. We restrict the review to broadleaf deciduous angiosperms living in seasonally cold environments and discuss the scientific questions that still need to be addressed in the near future.
2. ABA Integrates Environments Signaling in Regulating Bud Dormancy
2.1. ABA Mediates Photoperiod Response of the Bud Dormancy
In temperate ecosystems, the photoperiod is one of the most reliable indexes of the seasonal growth cycle. Light regulates plant dormancy through the circadian clock and flowering pathways [32,33]. In many tree species, the photoperiod (short days; SDs) is below the critical threshold for growth before the onset of winter-induced bud growth cessation [34]. Different light wavelengths are sensed by different photoreceptors, among which the phytochromes (phys) sense red (R) and far-red (FR) light [35]. phyA is the photoreceptor of FR light, while phyB is the R light photoreceptor as well as a thermosensor [35,36]. phyA and phyB transduce the light signals predominantly through their interactors (PHYTOCHROME-INTERACTING FACTORS; PIFs) [37]. In 2006, CO/FT (CONSTANS/FLOWERING LOCUS T) was first revealed to modulate light-regulated perennial bud dormancy in aspen trees. FT is induced by long days and downregulated by SD, and negatively regulates bud growth cassation by promoting the expression of D-type cyclins [38,39]. Other regulators in biological clock and flowering pathways are involved in bud dormancy as well, like MADS-box transcription factor FLOWERING LOCUS C (FLC) and SVP, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), LEAFY and so on [40,41,42].
ABA is induced by SD and disrupts intercellular communication in buds, promoting bud endodormancy. In this process, SVP-LIKE (SVL), an essential gene in bud dormancy, is induced by SD and positively regulates NCED expression, forming a positive feedback regulation with ABA signaling [33,43]. Moreover, SVL upregulates transcript level of CALS1, which encodes for a callose synthetase. CALS1 promotes callose deposition, leading to the closure of plasmodesmata with callosic plugs (dormancy sphincters) and limiting access to growth-active signals, thus blocking intercellular communication and slowing down cell activity in buds [33,41,44]. SVL also stimulates BRC1 (BRANCHED 1) expression under SD and further inhibits FT expression, thus promoting bud dormancy [45]. BRC1 is negatively regulated by LAP1 (orthologous to the Arabidopsis floral meristem identity gene APETALA1) under SD, forming a negative feedback loop that controls seasonal growth by interacting with and antagonizing FT [45]. In addition, ABA signaling genes such as ABI3, which is induced by SD-mediated increased ABA, is expressed in tissues including the young embryonic leaves, the subapical meristem, and the procambial strands, resulting in bud dormancy with incorrect bud formation [46]. ABI3 could physically interact with FDL1 (FD-Like 1) under SDs and regulate the expression of genes involved in bud maturation and adaptive responses for cold tolerance [47]. Recently, a chromodomain protein PICKLE (PKL) was shown to be inhibited by ABA under SD. In abi1-1 plants, defects in dormancy regulation caused by ABA insensitivity can be suppressed by down-regulating PKL expression [33].
2.2. ABA Mediates Temperature Signals Regulated Bud Dormancy
Temperature is possibly the most important factor in regulating bud dormancy especially for geophytes whose dormant organs are embedded in the soil [48]. For many deciduous plants, e.g., Malus pumila, the growth cassation occurs in summer, with long photoperiod and high temperature, while the endodormancy happens in autumn, during SD and low temperature [49]. In most deciduous angiosperms, bud dormancy is followed by blockage of vessels with accumulated callose, which results in decreased metabolic function and transport between buds and branches [23]. In autumn, short-term cold exposure increases endogenous ABA content in perennial buds by CBF, which binds and activates DAM/SVL transcription, thus increasing ABA levels and inducing bud endodormancy [50,51]. ABA could further activate its downstream signaling factors, i.e., ABF2, ABF3 and HB22, which in turn regulate the expression of DAM/SVP during dormancy induction and release [50,52,53]. ABF2 interacts with TCP20, which directly inhibits DAM genes expression [52]. DAM could also be activated by the ICE (Inducers of CBF Expression)-CBF module under cold stress [24,51]. As mentioned above, phyB is also a thermosensor, which is a positive regulator of thermo-controlled bud break in poplar. phyB interacts with PIFs and inhibits its expression, leading to activation of FT2 and repression of BRC1 and CENL1, which promote bud break and growth [54].
After exposure to chilling temperatures, there is an increase in cell connectivity in buds when endodormancy is releasing [23]. Many studies have shown that long terms of cold treatment decrease ABA content by inhibiting ABA biosynthesis while activating ABA catabolism in dormant organs, including tree’s buds and geophytes, like poplar, pear, Gladiolus hybridus and Leafy spurge [21,29,34,53]. Prolonged chilling activates CYP707A genes expression, thus decreasing ABA content in dormant buds. Furthermore, DAM/SVP would be inhibited and followed by activation of genes related to cell cycle, cell expansion, GA biosynthesis and FT [50]. In poplar, low temperature upregulates EBB1 (EARLY BUD-BREAK1), an APETALA 2 (AP2)-family transcription factor, resulting in suppression of SVL expression. Repressing SVL expression breaks the SVL/ABA feedforward loop, leading to the upregulation of EBB3 and consequently to activation of CYCD3.1, and bud dormancy release [55,56].
In addition to the DAM hub, there are several reports showing that ABA signaling mediates cold storage and controls bud dormancy release (Figure 1) [27,29]. In Gladiolus, the transcription level of SVL does not significantly change during corm dormancy release, but ABA dominantly regulates this process [27]. Cold storage inhibits NAC83 expression in dormant corms and activates NAC83-targeted gene, PP2C1. PP2C1 is the ABA co-receptor that plays roles in DNA duplication in dormant buds and inhibits ABA signal response, promoting corm dormancy release [27]. Cold storage could also activate TCP19, a Class I member of TCP family, which binds to the NCED promoter and represses NCED expression, contributing to reduction in endogenous ABA and corm dormancy release [57]. TCP19 plays a role in cell division in buds as well, by positively regulating cyclin A/B/D genes during corm dormancy release [57].
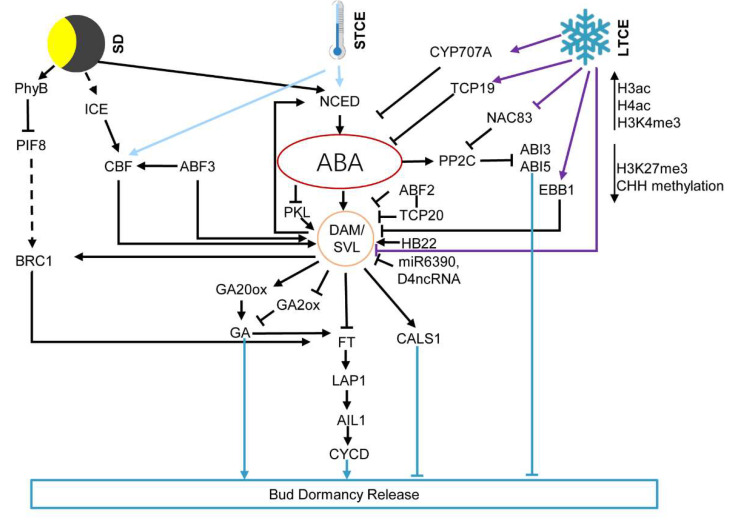
Figure 1.
ABA integrates environments signaling in regulating bud dormancy. During dormancy induction, SD and STCE stimulate ABA biosynthesis and induce bud dormancy via DAM and ABA signaling that blocks the plasmodesmata and slow down the cell cycle. After long term of chilling, endogenous ABA is decreased by cis- and trans- regulation. DAM/SVL is repressed when ABA was decreased, resulting in promoted GA in cells and degradation of callose at the plasmodesmata. Enhanced cell communication leads to active cell division and bud break. Note that this overview summarizes interactions reported in different species, which are not necessarily occurring simultaneously. Full lines and dashed lines specify established and putative/indirect regulation, respectively. LTCE: long term cold exposure; SD: short day; STCE: short term cold exposure.
3. Cross-Talk between ABA and Sugars in Bud Dormancy
Sugars provide energy to cell activity and also act as signaling molecules that regulate plant development and growth, including bud dormancy [58,59,60]. There is crosstalk between sugar signaling and other signaling pathways, including light and plant hormones (e.g., ABA and ethylene) [61,62,63]. Sugar signaling is essential for maintaining paradormacy by affecting cell cycle at the G1/S phase, and the transition from paradormancy to endodormancy in buds [64,65,66,67]. During bud dormancy release, endogenous starch content, which is high in accordance with endogenous ABA level, decreases [65,68]. Meanwhile, soluble sugars (glucose, sucrose, fructose, trehalose, etc.) increase during the transition from endodormancy to ecodormancy in buds of species such as leafy spurge, lily, sweet cherry and tree peony [26,65,69,70,71]. At the bud dormancy induction stage, sucrose in parenchyma cells declines and Tre6P (Trehalose 6-phosphate) decreases, which activates SnRK1 activity and stimulates ABA biosynthesis [72]. Generally, low Tre6P levels and/or high SnRK1 activities are associated with a dormant state/growth cassation, whereas high Tre6P levels and/or low SnRK1 activities are associated with active developmental progression [73]. For example, TPP-overexpressed (Tre6P-phosphatase) potato tubers accumulated higher levels of glucose and sucrose and sprouted prematurely, while tubers of snrk1 mutant displayed strongly delayed sprouting [74].
ABA has been shown to inhibit sucrose transporters in vine and potato dormant buds [72,75]. Moreover, ABA promotes starch accumulation in the dormant phase in grape bud by increasing the expression of starch biosynthesis genes SOLUBLE STARCH SYNTHASE 1 (SS1) and SS3, and inhibiting the expression of starch metabolism genes INVERTASEs (INVs) and sucrose biosynthesis genes SUCROSE PHOSPHATE SYNTHASEs (SUPs) [76].
In plants, the crosstalk between sugars and ABA can be mediated by the Tre6P-SnRK1 and SnRK1-TOR (Target of Rapamycin)-SnRK2 modules (Figure 2). TOR is a Ser/Thr protein kinase that belongs to the phosphatidylinositol 3-kinase-related lipid kinase family and stimulates cell cycle and mRNA translation by phosphorylating of the ribosomal protein S6 kinase S6K1 [77]. As mentioned above, low energy [e.g., low Tre6P, glucose-6-P (G6P), and glucose-1-P (G1P)] activates SnRK1 through ABA signaling [78]. Under stress conditions, SnRK1 and SnRK2 can repress TOR kinase by direct phosphorylation, resulting in TOR complex dissociation [79]. In the absence of ABA (or under low levels of ABA), PP2C phosphatases target SnRK1 and shut down SnRK1 signaling, thus activating TOR and promoting plant growth and development [80]. Moreover, TOR represses the activity of ABA receptors by phosphorylation, preventing SnRK2 activation and ABA downstream response [78]. During the induction of bud endodormancy in grapevine, ABA could induce SnRK1 expression and inhibit cell respiration [81]. However, there is only limited evidence that SnRK1-TOR-SnRK2 cascade may be involved in bud dormancy in grapevine, further genetic evidence is still needed [82].
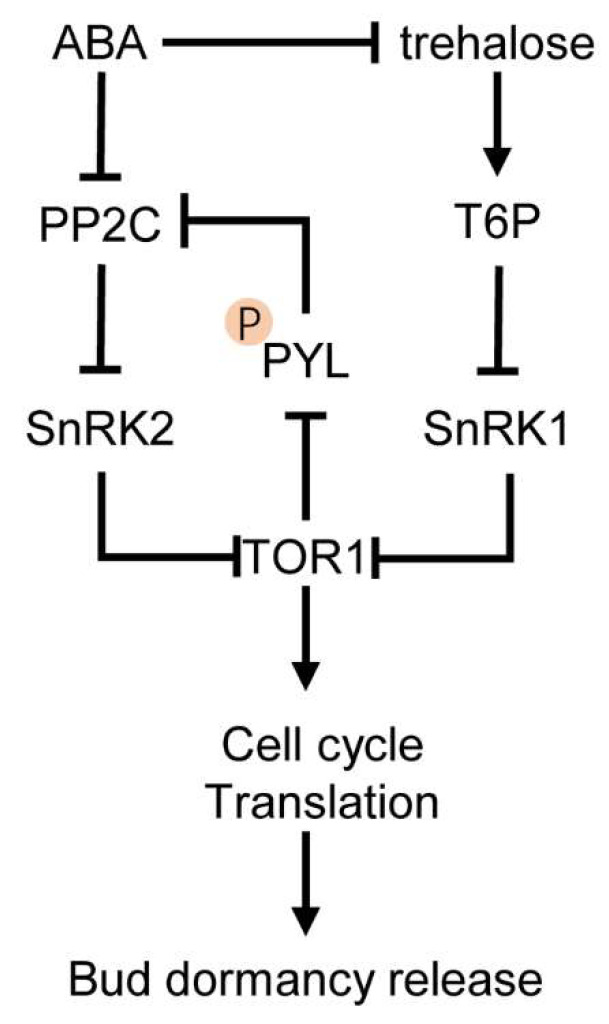
Figure 2.
Schematic overview of the SnRK1-TOR-SnRK2 cascade in mediating the crosstalk between ABA with trehalose during bud dormancy release. SnRK1 signaling keeps and switches energy used for rapid growth and development toward enhanced stress tolerance and survival with low energy. SnRK1 can be functional by repressing plant growth and the activity of TOR kinase. In addition, SnRK1 signaling cross-talks with and actives ABA signaling together with SnRK2. SnRK2 represses TOR signaling by direct phosphorylation, leading to TOR complex dissociation. In favor conditions, TOR kinase backwards represses SnRK2 signaling via phosphorylation of the PYR1-LIKE (PYL) and active PP2C. TOR1 promotes the transcription of genes involved in cell-cycle progression and translation of ribosomal protein mRNAs in plants. SnRK1 is repressed by high energy signals, such as trehalose-6-P (T6P). Note that this overview summarizes interactions reported in different species or environmental condition, which are not necessarily occurring simultaneously. Full lines and dashed lines specify established.
4. Cross-Talks between ABA and Other Hormones in Regulating Bud Dormancy
Hormones, including gibberellin, cytokinins, ethylene, jasmonic acid, are also involved in bud dormancy in perennials, and there is cross-talk between ABA and these hormones in regulating perennial bud dormancy (Figure 3) [24,27,83,84].
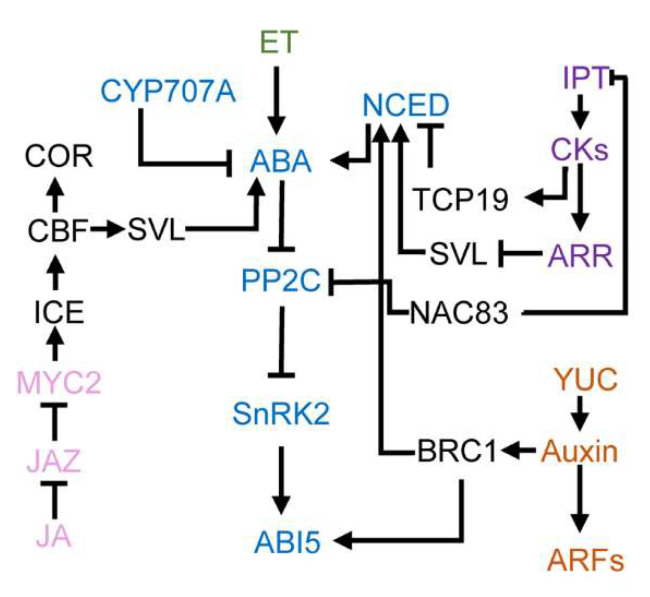
Figure 3.
The crosstalks between ABA and other phytohormones in regulating bud dormancy release. Auxin and ABA crosstalk through BRC1-mediated hormone networks in axillary bud dormancy. BRC1 not only activates ABA signaling factors ABI5 but also up-regulated the expression of the key enzyme gene NCED3 by combining with other proteins. CKs and ABA plays antagonistic role during bud dormancy release. Several transcriptional factors are involved in this process, such as TCP19, SVL and NAC83. In autumn, cold exposure activates JA biosynthesis, both JAZ and MYC2 interact with ICE, forming the ICE-CBF-COR cascade. As the CBF-DAM cascade regulates bud dormancy in popular, it is possible that the crosstalk between JA and ABA can be mediated by MYC2-ICE-CBF-DAM cascade, and induce/maintain bud dormancy. Ethylene has been shown to increase endogenous ABA and maintain bud dormancy in several species, but the mechanism is still need to be investigated. ABA metabolism and signaling is marked in blue; cytokinin metabolism and signaling is marked in purple; auxin metabolism and signaling is marked in orange; ethylene is marked in green; Jasmonic acid metabolism and signaling is marked in pink. Note that this overview summarizes interactions reported in different species or environmental condition, which are not necessarily occurring simultaneously. Full lines and dashed lines specify established.
4.1. ABA and Gibberellins
Gibberellins (GAs) stimulate cell division and elongation at different stages of plant development and in different tissues, including bud dormancy, seed germination, stem elongation, flowering and reproductive organs development [85,86,87]. There are more than 100 types of GAs, but only several forms are bioactive, e.g., GA1, GA3, GA4, and GA7 [85]. In GA metabolism, bioactive GAs are mainly catalyzed by GA 20-oxidases (GA20ox) and GA3ox. The existing bioactive GAs are deactivated to non-bioactive forms, which are catalyzed by GA2ox [88]. In GA signaling, the GA-GID1-DELLA module is considered to be universal in angiosperms. The binding of Gibberellins to the GA receptor GID1 (GIBBERELLIN-INSENSITIVE DWARF1) promotes interaction of GID1 with the DELLA proteins [24]. The GA-GID1-DELLA complex is recognized by the SCFSLY1/GID2 E3 ubiquitin-ligase, which triggers DELLA degradation by the ubiquitin-proteasome pathway [24]. DELLA proteins are classed into RGA (Repressor of Gibberellic Acid), GAI (Gibberellic Acid Insensitive) and RGA-like proteins by protein sequence similarities [89].
The effect of GA on bud dormancy is dependent on spatio-temporal and species’ variety [29,71,90]. In most cases, GA has a positive role in bud endodormancy release. In pear, exogenous GA could positively regulate bud endodormancy release that substitute for partial chilling treatment [91]. However, GA has a limited effect on Gladiolus corm dormancy release when treated at the early stage of endodormancy [92]. In Kiwi, application of GA3 before chilling promoted dormancy, while application after chilling promoted bud break [93]. GA is also involved in paradormancy regulation. The GA deficiency driven by increased expression of GA2ox resulted in increased axillary buds in hybrid aspen [94], whereas GA promotes the outgrowth of branches in Jatropha curcas [95].
Although the role of GA in the process of bud dormancy release and bud outgrowth needs to be further investigated, the antagonistic relationship between GA and ABA has long been shown to regulate key developmental processes, particularly seed dormancy and germination [96,97,98,99,100]. As for bud dormancy release, the antagonism between GA and ABA is also essential for integrating environmental and endogenous signals [44]. For example, in pears, GAST1 (GA-STIMULATED TRANSCRIPTS1) integrates GA biosynthesis and ABA signaling and participates in pear bud dormancy release in winter [91]. GAST1 responds to GA and also plays positive role in GA biosynthesis [91,101]. Indeed, ABA can inhibit GA20ox by downregulation of GAST1, decreasing the level of GA and inhibiting bud dormancy release [91]. For shade avoidance in maize, ABA promotes while GA inhibits axillary bud dormancy (paradormancy) [102]. For bud endodormancy in popular inducted by SD and low temperature, SVL could act as the nexus of GA and ABA metabolism. SVL induces GA2ox8 (a GA catabolic gene) and binds to the promoter of NCED3 (an ABA biosynthesis gene), thereby inhibiting the growth of the buds [41,44]. A study in poplar showed that ectopically expressed RGL from Japanese Apricot delayed the onset of bud dormancy [83]. However, there is still lack of information about how GA signaling factors (e.g., RGL, RGA and GAI) crosstalk with or regulate ABA-related proteins in the process of perennial bud dormancy release.
4.2. ABA and Cytokinins
Cytokinins (CKs) are a group of adenine-derived compounds that are involved in stem and root meristem differentiation, seed germination, leaf senescence delay, stress response and bud dormancy [27,103,104,105]. Adenosine phospho-isopentenyl transferase (IPT) and CYP735As (CYTOCHROME P450, FAMILY 735, SUBFAMILY As) are key rate-limiting enzymes that catalyze CKs biosynthesis, and tans-Zeatin (tZ)-type cytokinins are one of the main types [106,107,108]. Cytokinin oxidase/dehydrogenase (CKX) is the main catalytic enzyme for CK degradation [109]. The core of CK signaling pathway is mainly composed of receptor histidine kinase (AHK), histidine phosphate transfer (HPt) proteins and type-A or type-B response (A-RR or B-RR) regulator, which initiate the expression of CK response genes [110,111,112,113].
In seed and corm dormancy, CKs antagonized ABA to promote seed germination, and a similar regulation has been found in buds [27,114]. First, CKs treatment broke the bud dormancy in strawberry axillary buds and tea buds, which was also shown in potato tubers, while GA3 was found to be insufficient to break dormancy when CKX was overexpressed (CKX-overexpressed tubers showed an extended dormancy period and did not respond to GA3), suggesting that CKs play an important role in terminating tuber dormancy and also indicating that GA is not sufficient to break dormancy in the absence of CK [115,116,117]. By detecting hormone levels before bud break, CKs was found to be the main regulator in the potato tuber buds in the switch from innate dormancy to the non-dormant state [118]. The willow bud dormancy is initiated by high content of ABA and low levels of CKs in the xylem sap [119]. Moreover, the CKs level reached the peak before bud-break, while the ABA content was decreased [120]. It was recently reported that tZ is at low level during bud dormancy and increased during flowering in pears, which supports a positive role for CKs in bud dormancy release [97].
The cross-talk between CKs and ABA in buds of woody plants is mainly through the DAM hub. Overexpression of PmDAM6 increases the ABA content in the late dormancy and bud breaking stages, while decreasing CK level in Prunus [121]. CK-triggered responses down-regulate MdoDAM1 via MdoBRRs, which further reduces ABA content and promotes the release of bud dormancy [122]. In addition to DAM hub, the enhancement of light-mediated CKs signals further negatively regulates ABA content in the dormant bud, indicating that light signal is also involved in the cross-talk between CKs and ABA [123,124,125,126]. It has also been reported that AP2 inhibits meristem activity by negatively regulating CKs signaling, resulting in low mitotic activity and high expression of ABA-responsive genes [127]. Moreover, NAC83-PP2C module and TCP-NCED module indirectly affect the level of CKs by mediating ABA signaling and synthesis pathways, respectively, and jointly regulate corm dormancy in Gladiolus (Figure 3) [27,57].
4.3. ABA and Auxin
Auxin is mainly synthesized in shoot tips and young leaves and is involved in apical dominance/paradormancy, senescence, flowering and other developmental processes [128,129,130]. As one of the most abundant auxin, IAA synthesis starts from tryptophan, which is eventually oxidized to IAA by amino transferase and flavin monooxygenase (YUC), with YUC being an important rate-limiting enzyme in this process [130]. Auxin induces ARF-binding promoter expression by triggering Aux/IAA degradation [131].
Treatments with exogenous auxin, NAA, relieve the phloem dormancy by removing callose from the sieve tubes, resulting in bud dormancy release [132]. Low levels of free IAA were detected in tea buds at deep endodormancy stage, while high IAA levels were found during dormancy release, similar to what was found in Chinese fir, Prunus mume and grapevine buds [68,133,134,135]. In the underground vegetative buds of Canada thistle, auxin and ABA signals act as central regulators of developmental networks as well as paradormancy [136].
The formation of lateral branches can be divided into two steps: initiation of axillary meristems and outgrowth of axillary buds [137,138]. Auxin has long been considered a major signal of apical dominance, primarily inhibiting axillary bud growth [138,139]. The hormone network regulating axillary bud outgrowth mainly includes auxin, strigolactone (SL) and CKs [140,141,142]. Auxin indirectly inhibits the expression of BRC1, a promoter of axillary bud outgrowth, by inhibiting CKs biosynthesis genes while activating SL biosynthesis genes [143]. Auxin and ABA crosstalk through BRC1-mediated hormone networks in axillary bud dormancy (Figure 3). Exogenous ABA treatment inhibited branch development, while fluridone, an ABA biosynthesis inhibitor, promoted the development of branches of Rosa hybrida [142,144,145]. At the same time, plants with low ABA sensitivity produced more branches [146]. Therefore, ABA generally plays an inhibitory role in breaking dormancy in axillary buds [147].
ABA acts downstream of BRC1 [147]. BRC1 not only activates two ABA signaling factors ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABF3 [148,149,150], but also up-regulates the expression of the key enzyme gene, NCED3, by interacting with other proteins [150,151]. In addition, endogenous auxin is inhibited in ABA-treated axillary buds, of which cell cycle-related genes including CYCA2;1 and PCNA1 (POLIFERATING CELL NUCLEAR ANTIGEN1) are repressed [147].
4.4. ABA and Ethylene
Ethylene (ET) is a simple gaseous hormone that regulates a wide range of plant developmental processes, including fruit ripening, seed germination, flowering, abscission, as well as bud dormancy [152,153]. ET biosynthesis starts from the amino acid methionine, which is first converted to SAM (S-adenoysl-methionine) by SAM synthase. SAM is then converted to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (ACS), and finally ET is produced by the conversion of ACC oxidase (ACO) [154,155,156,157]. In the absence of ET, the receptors (i.e., ETR1, ETR2, ERS1, ERS2 and EIN4) activate Constitutive Triple Response 1 (CTR1) and inhibit ET signaling pathway by transmembrane protein EIN2 in Arabidopsis [156,158,159]. However, in the presence of ET, CTR1 is inactivated, and the positive regulatory function of ETHYLENE INSENSITIVE 2 (EIN2) is released [160,161]. EIN2 positively regulates the EIN3 transcription factor members in the nucleus [162,163]. The ETHYLENE RESPONSE FACTOR (ERF) genes, belonging to the AP2/ERF superfamily, are direct targets of EIN3 and activate downstream ET responses [157,164,165].
Many studies have demonstrated that ET biosynthesis and signaling pathways regulate bud dormancy. ET signaling genes EIN3, EIL1, and ERF are abundant during the endodormancy and are decreased during the transition from endodormancy to ecodormancy in populus, suggesting that ET may play roles in dormancy maintenance and release [82,166]. The application of competitive ET antagonist 2,5-Norbornadiene (NBD) leads to premature sprouting in potato tubers [167]. In onion, exogenous ET not only affects the expression of ACO and receptors (EIN4 and EIL3), but also increases the accumulation of ABA by up-regulating the expression of NCED [168]. The etr1-4 mutant in birch had relative low ABA content and fast growth under SD. Moreover, etr1-4 mutant is less sensitive to ABA when sprouting and has weaker paradormancy compared to wild-type plants, suggesting that ET signaling interacts with ABA signaling pathways in dormancy regulation [169]. In addition, ET interacts with GA to participate in bud dormancy by increasing DELLA accumulation in response to photochrome signals and upregulating ERF6 expression [170,171]. However, exogenous ET was also reported to promote bud breaking in some species like grapevine, poplar, while NBD inhibited bud dormancy release and increased ABA level [56,172,173,174]. In pear buds, ET precursor ACC was gradually increased during bud break [175]. ERFs were significantly up-regulated in HC-treated buds or before bud-break [56,174], suggesting that ET may act antagonistically to ABA during bud dormancy release [24].
4.5. ABA and Jasmonic Acid
Jasmonic acid (JA) is a class of plant hormones that regulate plant development and defense processes [176]. The core part of its signaling pathway consists of JA ZIM-DOMAIN (JAZ) protein, F-box protein CORONATINE INSENSITIVE1 (COI1) and several groups of suppressed transcription factors (MYC2, MYC3, MYC4 etc.) [177,178,179]. JAs act synergistically with ABA by inhibiting DNA replication and activating the expression of anti-stress genes during stress tolerance, and exogenous JAs and ABA inhibit seed germination in several species [100,180,181,182,183].
JAs related genes were inhibited in bud dormancy and significantly upregulated in bud dormancy release, and the content of JAs was also significantly increased in bud dormancy release [184,185,186]. Whereas, during bud dormancy release in pear or lily bulbs, the JA content is gradually reduced with the decreases of ABA [175,187]. JA and ABA participate in the process of cold domestication together. In autumn, cold exposure activates JA biosynthesis, and both JAZ and MYC2 interact with ICE, forming the ICE-CBF-COR cascade discussed above. It is possible that the crosstalk between JA and ABA can be mediated by MYC2-ICE-CBF-DAM cascade, to induce/maintain bud dormancy (Figure 3) [188,189].
5. Epigenetic Regulation of Bud Dormancy Mediated by ABA
Epigenetics is the study of molecules and mechanisms that can perpetuate alternative gene activity states in the context of the same DNA sequence, including noncoding RNAs, DNA methylation, histone modification, heterochromatin, and 3D genome architecture [190]. As currently there is little information about heterochromatin and 3D genome architecture on bud dormancy, here we mainly summarize the knowledge of the former three types on perennial bud dormancy.
In plants, DNA methylation occurs in three different cytosine contexts: CpG, CHG and CHH, in which H can be either cytosine, thymine or adenine [191]. De novo methylation can occur by RNA-directed DNA methylation (RdDM), where small interference RNAs (siRNAs) guide DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to homologous sequences in the genome [192]. Total levels of DNA methylation increased in dormant chestnut’s buds compared to non-dormant buds [193]. During bud dormancy release in sweet cherry, changes in DNA methylation precedes transcript changes, and responded to low temperatures including cold signaling, oxidation-reduction process, metabolism of phenylpropanoids, lipids and a DAM gene (i.e., MADS1) [194]. Long-term cold treatment stimulates DNA methylation in the promoter of MADS1 in sweet cherry and increases siRNA that match this region, in accordance with the up-regulation of FT transcripts [191]. In Arabidopsis, miR402 could be induced by ABA and reduces the transcript levels of DML3 (DEMETER-LIKE PROTEIN 3), which results in increased methylation levels at certain loci [195]. Currently, genes regulated by DNA methylation still need to be explored in the process of perennial bud dormancy.
Histone modification is a covalent post-translational modification that fine-tunes gene expression (activation or silencing) by altering chromatin structure, and includes methylation, acetylation, phosphorylation, ubiquitylation and sumoylation [196]. During the transition from dormancy to dormancy release in potato, muti-acetylation of H4 and H3.1/3.2 was increased [197]. Increased acetylation of H4 was also detected in the chestnut [193]. In peach, global H3K27me3 was higher in dormancy-released bud, compared with dormant buds. But, the modification is variable for specific gene [198]. Several studies showed histone modification is involved in bud dormancy by shifting on/off DAM/SVL in this duration [50,198,199]. Coinciding with cold accumulation in peach, several chromatin regions including the region around the translation start of DAM6 in peach are marked by enriched H3K27me3, removal of H3K4me3 and acetylated H3 (H3ac) (Figure 1) [199]. Similar findings were found in Leafy spurge, kiwifruit and sweet cherry [200,201,202]. Other dormancy-related genes like EARLY BUD BREAK3 (EBB3), a target of ABA signaling downstream of the SVL/ABA feedforward loop during dormancy release, is modified by H3K27me3 [55]. In addition, ABA promotes bud dormancy by inhibiting the expression of a chromodomain protein PICKLE (PKL), which inhibits SVL and promotes bud dormancy release in popular [33,41].
ncRNAs are functional RNAs that regulate gene expression at both the transcriptional and post-transcriptional levels, with many ncRNAs involved in histone modification, chromatin remodeling and DNA methylation [203]. In peach, several microRNAs, siRNAs and lncRNAs have been found to be correlated to bud dormancy release for instance miR319, miR6285, miR2275 and D4ncRNA (intronic ncRNA in DAM4) (Figure 1) [204]. Several long ncRNAs (>200 nt), isolated from popular buds at different dormant stage, were reported to be involved in regulating endodormancy release, for example, two lncRNAs acting as endogenous target mimics for gma-miRNA396h, which itself targets CYP707As [205].
6. Conclusions and Outlook
Dormancy is a complex phenomenon in plants and is also a ceasing trait. During bud dormancy, internal changes occur including deposition of plasmodesmatal callose, slow cell division, limiting import of sucrose, and changes of hormones. A large body of studies has shown that ABA is the hub that integrates environmental signals and endogenous chemicals to regulate bud dormancy in perennials (Figure 1). Recent research shows that SL and karrikin can be recognized by the receptor DWARF14 (D14) and KARRIKIN INSENSITIVE2 (KAI2), respectively, and these two receptors are homologs [206,207]. The fact that karrikin has been shown to be involved in bud and seed dormancy, as well as SL control axillary bud development, also suggests that these two hormones play a role in perennial bud dormancy [206,208]. It will be interesting to investigate the cross-talk between ABA and karrikin/SL in endodormancy of perennials. Moreover, it is also worthwhile to compare their role in paradormancy and endodormancy.
Although there are a few studies about epigenetics in regulating bud dormancy, several questions still need to be addressed. For example, how does endogenous ABA affect epigenetics in dormant buds during bud dormancy release with respect to heterochromatin and 3D genome architecture? How do ncRNA or histone modifications regulate ABA metabolism in perennial dormant bud?
When buds are in a dormant state, the plasmodesmata is blocked by callose. Is there any change among the organelles, like mitochondrial, amyloplast and golgi apparatus, during this biological process? How does ABA regulate the activity or development of these organelles?
It will be especially exciting to integrate genetics, omics (e.g., genomics, proteomics and metabolomics) and computational analyses to identify a broad and complex network of perennial bud dormancy, which helps elucidate the molecular components and mechanisms underlying multiple cellular and biological processes. By using omics techniques, it will be much easier to identify domestication loci associated with bud dormancy, which contributes to plant breeding via crossing or molecular methods.
Acknowledgments
Not applicable.
Author Contributions
Conceptualization, writing original draft preparation—J.W., W.P., J.L. (Jiahui Liang); literature research—J.S., J.L. (Jingru Li), C.L., Y.X., Y.Z. (Yanmin Zhang), S.W., Y.Z. (Yajie Zhao), J.Z., review and editing—M.Y., S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by National Natural Science Foundation projects (grants 3217180532; 31701952 to J.W. and 31902047 to J.J.S.), Beijing Natural Science Foundation (6212012 to J.W.), Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032), Natural Science Foundation of Anhui Province (2008085MC79 to J.J.S.), 111 Project of the Ministry of Education (B17043) and Natural Sciences and Engineering Research Council of Canada (NSERC to S.G.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kermode A.R. Role of Abscisic Acid in Seed Dormancy. J. Plant Growth Regul. 2005;24:319–344. doi: 10.1007/s00344-005-0110-2. [DOI] [Google Scholar]
- 2.Zheng C., Halaly T., Acheampong A.K., Takebayashi Y., Jikumaru Y., Kamiya Y., Or E. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 2015;66:1527–1542. doi: 10.1093/jxb/eru519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker M., Pérez M., Steinbrecher T., Gawthrop F., Pavlović I., Novák O., Tarkowská D., Strnad M., Marone F., Nakabayashi K., et al. Molecular mechanisms and hormonal regulation underpinning morphological dormancy: A case study using Apium graveolens (Apiaceae) Plant J. 2021 doi: 10.1111/tpj.15489. [DOI] [PubMed] [Google Scholar]
- 4.Graeber K., Nakabayashi K., Miatton E., Leubner G., Soppe W.J.J. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012;35:1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x. [DOI] [PubMed] [Google Scholar]
- 5.Nambara E., Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 6.Chen K., Li G.J., Bressan R.A., Song C.P., Zhu J.K., Zhao Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2000;62:25–54. doi: 10.1111/jipb.12899. [DOI] [PubMed] [Google Scholar]
- 7.Koornneef M., Jorna M.L., Der Swan D.L.C.B.-V., Karssen C.M. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- 8.Marin E., Nussaume L., Quesada A., Gonneau M., Sotta B., Hugueney P., Frey A., Marion-Poll A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996;15:2331–2342. doi: 10.1002/j.1460-2075.1996.tb00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz S.H., Qin X., Zeevaart J.A. Elucidation of the Indirect Pathway of Abscisic Acid Biosynthesis by Mutants, Genes, and Enzymes. Plant Physiol. 2003;131:1591–1601. doi: 10.1104/pp.102.017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., Hirai N., Koshiba T., Kamiya Y., Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito S., Hirai N., Matsumoto C., Ohigashi H., Ohta D., Sakata K., Mizutani M. Arabidopsis CYP707As Encode (+)-Abscisic Acid 8′-Hydroxylase, a Key Enzyme in the Oxidative Catabolism of Abscisic Acid. Plant Physiol. 2004;134:1439–1449. doi: 10.1104/pp.103.037614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee K.H., Piao H.L., Kim H.Y., Choi S.M., Jiang F., Hartung W., Hwang I., Kwak J.M., Lee I.J., Hwang I. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell. 2006;126:1109–1120. doi: 10.1016/j.cell.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 13.Xu Z.-Y., Lee K.H., Dong T., Jeong J.C., Jin J.B., Kanno Y., Kim D.H., Kim S.Y., Seo M., Bressan R.A., et al. A Vacuolar β-Glucosidase Homolog That Possesses Glucose-Conjugated Abscisic Acid Hydrolyzing Activity Plays an Important Role in Osmotic Stress Responses in Arabidopsis. Plant Cell. 2012;24:2184–2199. doi: 10.1105/tpc.112.095935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietz K.J., Sauter A., Wichert K., Messdaghi D., Hartung W. Extracellular β-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J. Exp. Bot. 2000;51:937–944. doi: 10.1093/jexbot/51.346.937. [DOI] [PubMed] [Google Scholar]
- 15.Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 16.Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F.F., et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melcher K., Ng L.-M., Zhou X.E., Soon F.-F., Xu Y., Suino-Powell K.M., Park S.-Y., Weiner J.J., Fujii H., Chinnusamy V., et al. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazono K.-I., Miyakawa T., Sawano Y., Kubota K., Kang H.-J., Asano A., Miyauchi Y., Takahashi M., Zhi Y., Fujita Y., et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard K., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinne P.L., Welling A., Van Der Schoot C. Genetics and Genomics of Populus. Springer; New York, NY, USA: 2009. Perennial Life Style of Populus: Dormancy Cycling and Overwintering; pp. 171–200. [DOI] [Google Scholar]
- 21.Horvath D.P., Anderson J.V., Chao W.S., Foley M.E. Knowing when to grow: Signals regulating bud dormancy. Trends Plant Sci. 2003;8:534–540. doi: 10.1016/j.tplants.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Perry T.O. Dormancy of Trees in Winter. Science. 1971;171:29–36. doi: 10.1126/science.171.3966.29. [DOI] [PubMed] [Google Scholar]
- 23.Savage J.A., Chuine I. Coordination of spring vascular and organ phenology in deciduous angiosperms growing in seasonally cold climates. New Phytol. 2021;230:1700–1715. doi: 10.1111/nph.17289. [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Sherif S.M. Hormonal Orchestration of Bud Dormancy Cycle in Deciduous Woody Perennials. Front. Plant Sci. 2019;10:1136. doi: 10.3389/fpls.2019.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falavigna V.D.S., Guitton B., Costes E., Andrés F. I Want to (Bud) Break Free: The Potential Role of DAM and SVP-Like Genes in Regulating Dormancy Cycle in Temperate Fruit Trees. Front. Plant Sci. 2019;9:1990. doi: 10.3389/fpls.2018.01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu R.-Y., Niimi Y., Han D.-S. Changes in endogenous abscisic acid and soluble sugars levels during dormancy-release in bulbs of Lilium rubellum. Sci. Hortic. 2006;111:68–72. doi: 10.1016/j.scienta.2006.08.004. [DOI] [Google Scholar]
- 27.Wu J., Jin Y., Liu C., Vonapartis E., Liang J., Wu W., Gazzarrini S., He J., Yi M. GhNAC83 inhibits corm dormancy release by regulating ABA signaling and cytokinin biosynthesis in Gladiolus hybridus. J. Exp. Bot. 2018;70:1221–1237. doi: 10.1093/jxb/ery428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasin H.J., Bufler G. Dormancy and sprouting in onion (Allium cepa L.) bulbs. I. Changes in carbohydrate metabolism. J. Hortic. Sci. Biotechnol. 2007;82:89–96. doi: 10.1080/14620316.2007.11512203. [DOI] [Google Scholar]
- 29.Wu J., Seng S.S., Sui J.J., Vonapartis E., Luo X., Gong B.H., Liu C., Wu C.Y., Liu C., Zhang F.Q., et al. Gladiolus hybridus ABSCISIC ACID INSENSITIVE 5 (GhABI5) is an important transcription factor in ABA signaling that can enhance Gladiolus corm dormancy and Arabidopsis seed dormancy. Front. Plant Sci. 2015;6:960. doi: 10.3389/fpls.2015.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazare S., Bechar D., Fernie A.R., Brotman Y., Zaccai M. The proof is in the bulb: Glycerol influences key stages of lily development. Plant J. 2019;97:321–340. doi: 10.1111/tpj.14122. [DOI] [PubMed] [Google Scholar]
- 31.Benkeblia N., Selselet-Attou G. Effects of low temperatures on changes in oligosaccharides, phenolics and peroxidase in inner bud of onion Allium cepa L. during break of dormancy. Acta Agric. Scand. B Plant Soil Sci. 1999;49:98–102. doi: 10.1080/09064719950135605. [DOI] [Google Scholar]
- 32.Campoy J.A., Ruiz D., Egea J. Dormancy in temperate fruit trees in a global warming context: A review. Sci. Hortic. 2011;130:357–372. doi: 10.1016/j.scienta.2011.07.011. [DOI] [Google Scholar]
- 33.Tylewicz S., Petterle A., Marttila S., Miskolczi P., Azeez A., Singh R.K., Immanen J., Mähler N., Hvidsten T.R., Eklund D.M., et al. Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science. 2018;360:212–215. doi: 10.1126/science.aan8576. [DOI] [PubMed] [Google Scholar]
- 34.Rohde A., Bhalerao R.P. Plant dormancy in the perennial context. Trends Plant Sci. 2007;12:217–223. doi: 10.1016/j.tplants.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Xu D., Deng X.W. CBF-phyB-PIF Module Links Light and Low Temperature Signaling. Trends Plant Sci. 2020;25:952–954. doi: 10.1016/j.tplants.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Legris M., Klose C., Burgie E.S., Rojas C.C., Neme M., Hiltbrunner A., Wigge P.A., Schafer E., Vierstra R.D., Casal J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science. 2016;354:897–900. doi: 10.1126/science.aaf5656. [DOI] [PubMed] [Google Scholar]
- 37.Paik I., Huq E. Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 2019;92:114–121. doi: 10.1016/j.semcdb.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohlenius H., Huang T., Charbonnel-Campaa L., Brunner A.M., Jansson S., Strauss S.H., Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 39.Karlberg A., Bakó L., Bhalerao R.P. Short Day–Mediated Cessation of Growth Requires the Downregulation of AINTEGUMENTALIKE1 Transcription Factor in Hybrid Aspen. PLoS Genet. 2011;7:e1002361. doi: 10.1371/journal.pgen.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar G., Arya P., Gupta K., Randhawa V., Acharya V., Singh A.K. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica) Sci. Rep. 2016;6:20695. doi: 10.1038/srep20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh R.K., Miskolczi P., Maurya J.P., Bhalerao R.P. A Tree Ortholog of SHORT VEGETATIVE PHASE Floral Repressor Mediates Photoperiodic Control of Bud Dormancy. Curr. Biol. 2019;29:128–133.e2. doi: 10.1016/j.cub.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Soto D., Ramos-Sanchez J.M., Alique D., Conde D., Triozzi P.M., Perales M., Allona I. Overexpression of a SOC1-Related Gene Promotes Bud Break in Ecodormant Poplars. Front. Plant Sci. 2021;12:670497. doi: 10.3389/fpls.2021.670497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh R.K., Maurya J.P., Azeez A., Miskolczi P., Tylewicz S., Stojkovic K., Delhomme N., Busov V., Bhalerao R.P. A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busov V.B. Plant Development: Dual Roles of Poplar SVL in Vegetative Bud Dormancy. Curr. Biol. 2019;29:R68–R70. doi: 10.1016/j.cub.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 45.Maurya J.P., Singh R., Miskolczi P.C., Prasad A.N., Jonsson K., Wu F., Bhalerao R.P. Branching Regulator BRC1 Mediates Photoperiodic Control of Seasonal Growth in Hybrid Aspen. Curr. Biol. 2020;30:122–126.e2. doi: 10.1016/j.cub.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Rohde A., Prinsen E., De Rycke R., Engler G., Van Montagu M., Boerjan W. PtABI3 Impinges on the Growth and Differentiation of Embryonic Leaves during Bud Set in Poplar. Plant Cell. 2002;14:1885–1901. doi: 10.1105/tpc.003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maurya J.P., Bhalerao R.P. Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: A molecular perspective. Ann. Bot. 2017;120:351–360. doi: 10.1093/aob/mcx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamenetsky R., Okubo H. Ornamental Geophytes: From Basic Science to Sustainable Production. CRC Press; Boca Raton, FL, USA: 2012. [Google Scholar]
- 49.Heide O.M., Prestrud A.K. Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 2005;25:109–114. doi: 10.1093/treephys/25.1.109. [DOI] [PubMed] [Google Scholar]
- 50.Yang Q., Gao Y., Wu X., Moriguchi T., Bai S., Teng Y. Bud endodormancy in deciduous fruit trees: Advances and prospects. Hortic. Res. 2021;8:139. doi: 10.1038/s41438-021-00575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., Yan X., Yang Q., Ma Y., Yang B., Tian J., Teng Y., Bai S. PpCBFs selectively regulate PpDAMs and contribute to the pear bud endodormancy process. Plant Mol. Biol. 2019;99:575–586. doi: 10.1007/s11103-019-00837-7. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q., Xu G., Zhao X., Zhang Z., Wang X., Liu X., Xiao W., Fu X., Chen X., Gao D., et al. Transcription factor TCP20 regulates peach bud endodormancy by inhibiting DAM5/DAM6 and interacting with ABF2. J. Exp. Bot. 2020;71:1585–1597. doi: 10.1093/jxb/erz516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Q., Yang B., Li J., Wang Y., Tao R., Yang F., Wu X., Yan X., Ahmad M., Shen J., et al. ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ. 2020;43:1360–1375. doi: 10.1111/pce.13744. [DOI] [PubMed] [Google Scholar]
- 54.Ding J., Zhang B., Li Y., Andre D., Nilsson O. Phytochrome B and PHYTOCHROME INTERACTING FACTOR8 modulate seasonal growth in trees. New Phytol. 2021 doi: 10.1111/nph.17350. [DOI] [PubMed] [Google Scholar]
- 55.Azeez A., Zhao Y.C., Singh R.K., Yordanov Y.S., Dash M., Miskolczi P., Stojkovic K., Strauss S.H., Bhalerao R.P., Busov V.B. EARLY BUD-BREAK 1 and EARLY BUD-BREAK 3 control resumption of poplar growth after winter dormancy. Nat. Commun. 2021;12:1123. doi: 10.1038/s41467-021-21449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yordanov Y., Ma C., Strauss S.H., Busov V.B. EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. USA. 2014;111:10001–10006. doi: 10.1073/pnas.1405621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu J., Wu W., Liang J., Jin Y., Gazzarrini S., He J., Yi M. GhTCP19 Transcription Factor Regulates Corm Dormancy Release by Repressing GHNCED Expression in Gladiolus. Plant Cell Physiol. 2018;60:52–62. doi: 10.1093/pcp/pcy186. [DOI] [PubMed] [Google Scholar]
- 58.Lastdrager J., Hanson J., Smeekens S. Sugar signals and the control of plant growth and development. J. Exp. Bot. 2014;65:799–807. doi: 10.1093/jxb/ert474. [DOI] [PubMed] [Google Scholar]
- 59.Wang M., Pérez-Garcia M.-D., Davière J.-M., Barbier F., Ogé L., Gentilhomme J., Voisine L., Péron T., Launay-Avon A., Clément G., et al. Outgrowth of the axillary bud in rose is controlled by sugar metabolism and signalling. J. Exp. Bot. 2021;72:3044–3060. doi: 10.1093/jxb/erab046. [DOI] [PubMed] [Google Scholar]
- 60.Tsai A.Y.-L., Gazzarrini S. Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: The emerging picture. Front. Plant Sci. 2014;5:119. doi: 10.3389/fpls.2014.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibson S.I. Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 2005;8:93–102. doi: 10.1016/j.pbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 62.León P. Sugar and hormone connections. Trends Plant Sci. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 63.Gazzarrini S., McCourt P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 2001;4:387–391. doi: 10.1016/S1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- 64.Arora R., Rowland L.J., Tanino K. Induction and release of bud dormancy in woody perennials: A science comes of age. Hortscience. 2003;38:911–921. doi: 10.21273/HORTSCI.38.5.911. [DOI] [Google Scholar]
- 65.Anderson J.V., Gesch R.W., Jia Y., Chao W.S., Horvath D.P. Seasonal shifts in dormancy status, carbohydrate metabolism, and related gene expression in crown buds of leafy spurge. Plant Cell Environ. 2005;28:1567–1578. doi: 10.1111/j.1365-3040.2005.01393.x. [DOI] [Google Scholar]
- 66.Chao W.S., Serpe M.D., Anderson J.V., Gesch R.W., Horvath D.P. Sugars, hormones, and environment affect the dormancy status in underground adventitious buds of leafy spurge (Euphorbia esula) Weed Sci. 2006;54:59–68. doi: 10.1614/WS-05-088R.1. [DOI] [Google Scholar]
- 67.Chao W.S., Foley M.E., Horvath D.P., Anderson J.V. Signals regulating dormancy in vegetative buds. Int. J. Plant Dev. Biol. 2007;1:49–56. [Google Scholar]
- 68.Zhang Z., Zhuo X., Zhao K., Zheng T., Han Y., Yuan C., Zhang Q. Transcriptome Profiles Reveal the Crucial Roles of Hormone and Sugar in the Bud Dormancy of Prunus mume. Sci. Rep. 2018;8:5090. doi: 10.1038/s41598-018-23108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang C.-Q., Li Q.-H., Jiang X.-Q., Fan Y.-W., Gao J.-P., Zhang C.-Q. Dynamic changes in α-and β-amylase activities and gene expression in bulbs of the Oriental hybrid lily ‘Siberia’during dormancy release. J. Hortic. Sci. Biotechnol. 2015;90:753–759. doi: 10.1080/14620316.2015.11668742. [DOI] [Google Scholar]
- 70.Chmielewski F.M., Gotz K., Homann T., Huschek G., Rawel H. Identification of endodormancy release for cherries (Prunus avium, L.) by abscisic acid and sugars. J. Hortic. 2017;4:210. doi: 10.4172/2376-0354.1000210. [DOI] [Google Scholar]
- 71.Zhang Y., Yuan Y., Liu Z., Zhang T., Li F., Liu C., Gai S. GA3 is superior to GA4 in promoting bud endodormancy release in tree peony (Paeonia suffruticosa) and their potential working mechanism. BMC Plant Biol. 2021;21:323. doi: 10.1186/s12870-021-03106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sonnewald S., Sonnewald U. Regulation of potato tuber sprouting. Planta. 2014;239:27–38. doi: 10.1007/s00425-013-1968-z. [DOI] [PubMed] [Google Scholar]
- 73.Baena-Gonzalez E., Lunn J.E. SnRK1 and trehalose 6-phosphate—Two ancient pathways converge to regulate plant metabolism and growth. Curr. Opin. Plant Biol. 2020;55:52–59. doi: 10.1016/j.pbi.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 74.Debast S., Nunes-Nesi A., Hajirezaei M.R., Hofmann J., Sonnewald U., Fernie A.R., Bornke F. Altering Trehalose-6-Phosphate Content in Transgenic Potato Tubers Affects Tuber Growth and Alters Responsiveness to Hormones during Sprouting. Plant Physiol. 2011;156:1754–1771. doi: 10.1104/pp.111.179903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murcia G., Pontin M., Reinoso H., Baraldi R., Bertazza G., Gomez-Talquenca S., Bottini R., Piccoli P.N. ABA and GA 3 increase carbon allocation in different organs of grapevine plants by inducing accumulation of non-structural carbohydrates in leaves, enhancement of phloem area and expression of sugar transporters. Physiol. Plant. 2016;156:323–337. doi: 10.1111/ppl.12390. [DOI] [PubMed] [Google Scholar]
- 76.Rubio S., Noriega X., Pérez F.J. ABA promotes starch synthesis and storage metabolism in dormant grapevine buds. J. Plant Physiol. 2019;234–235:1–8. doi: 10.1016/j.jplph.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 77.Shi L., Wu Y., Sheen J. TOR signaling in plants: Conservation and innovation. Development. 2018;145:dev160887. doi: 10.1242/dev.160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crepin N., Rolland F. SnRK1 activation, signaling, and networking for energy homeostasis. Curr. Opin. Plant Biol. 2019;51:29–36. doi: 10.1016/j.pbi.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 79.Wang P.C., Zhao Y., Li Z.P., Hsu C.C., Liu X., Fu L.W., Hou Y.J., Du Y., Xie S.J., Zhang C.G., et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell. 2018;69:100–112. doi: 10.1016/j.molcel.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rodrigues A., Adamo M., Crozet P., Margalha L., Confraria A., Martinho C., Elias A., Rabissi A., Lumbreras V., Gonzalez-Guzman M., et al. ABI1 and PP2CA Phosphatases Are Negative Regulators of Snf1-Related Protein Kinase1 Signaling in Arabidopsis. Plant Cell. 2013;25:3871–3884. doi: 10.1105/tpc.113.114066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parada F., Noriega X., Dantas D., Bressan-Smith R., Pérez F.J. Differences in respiration between dormant and non-dormant buds suggest the involvement of ABA in the development of endodormancy in grapevines. J. Plant Physiol. 2016;201:71–78. doi: 10.1016/j.jplph.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 82.Shi Z., Halaly-Basha T., Zheng C., Schwager M.S., Wang C., Galbraith D.W., Ophir R., Pang X., Or E. Identification of potential post-ethylene events in the signaling cascade induced by stimuli of bud dormancy release in grapevine. Plant J. 2020;104:1251–1268. doi: 10.1111/tpj.14997. [DOI] [PubMed] [Google Scholar]
- 83.Lv L., Huo X., Wen L., Gao Z., Khalil-Ur-Rehman M. Isolation and Role of PmRGL2 in GA-mediated Floral Bud Dormancy Release in Japanese Apricot (Prunus mume Siebold et Zucc.) Front. Plant Sci. 2018;9:27. doi: 10.3389/fpls.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tosetti R., Waters A., Chope G., Cools K., Alamar M., McWilliam S., Thompson A., Terry L. New insights into the effects of ethylene on ABA catabolism, sweetening and dormancy in stored potato tubers. Postharvest Biol. Technol. 2020;173:111420. doi: 10.1016/j.postharvbio.2020.111420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhattacharya A. Chapter 6—Effect of High-Temperature Stress on the Metabolism of Plant Growth Regulators. In: Bhattacharya A., editor. Effect of High Temperature on Crop Productivity and Metabolism of Macro Molecules. Academic Press; Cambridge, MA, USA: 2019. pp. 485–591. [DOI] [Google Scholar]
- 86.Sun T.-P. The Arabidopsis Book/American Society of Plant Biologists. Volume 6. ASPB Press; Rockville, MD, USA: 2008. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng C., Acheampong A.K., Shi Z., Halaly T., Kamiya Y., Ophir R., Galbraith D.W., Or E. Distinct gibberellin functions during and after grapevine bud dormancy release. J. Exp. Bot. 2018;69:1635–1648. doi: 10.1093/jxb/ery022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X., Hou X. Antagonistic Regulation of ABA and GA in Metabolism and Signaling Pathways. Front. Plant Sci. 2018;9:251. doi: 10.3389/fpls.2018.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fuentes S., Ljung K., Sorefan K., Alvey E., Harberd N.P., Østergaard L. Fruit Growth in Arabidopsis Occurs via DELLA-Dependent and DELLA-Independent Gibberellin Responses. Plant Cell. 2012;24:3982–3996. doi: 10.1105/tpc.112.103192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamazaki H., Nishijima T., Koshioka M., Miura H. Gibberellins do not act against abscisic acid in the regulation of bulb dormancy of Allium wakegi Araki. Plant Growth Regul. 2002;36:223–229. doi: 10.1023/A:1016577529378. [DOI] [Google Scholar]
- 91.Yang Q., Niu Q., Tang Y., Ma Y., Yan X., Li J., Tian J., Bai S., Teng Y. PpyGAST1 is potentially involved in bud dormancy release by integrating the GA biosynthesis and ABA signaling in ‘Suli’ pear (Pyrus pyrifolia White Pear Group) Environ. Exp. Bot. 2019;162:302–312. doi: 10.1016/j.envexpbot.2019.03.008. [DOI] [Google Scholar]
- 92.Ram R., Mukherjee D., Manuja S. Plant Growth Regulators Affect the Development of Both Corms and Cormels in Gladiolus. HortScience. 2002;37:343–344. doi: 10.21273/HORTSCI.37.2.343. [DOI] [Google Scholar]
- 93.Lionakis S.M., Schwabe W.W. Bud Dormancy in the Kiwi Fruit, Actinidia chinensis Planch. Ann. Bot. 1984;54:467–484. doi: 10.1093/oxfordjournals.aob.a086818. [DOI] [Google Scholar]
- 94.Mauriat M., Sandberg L.G., Moritz T. Proper gibberellin localization in vascular tissue is required to control auxin-dependent leaf development and bud outgrowth in hybrid aspen. Plant J. 2011;67:805–816. doi: 10.1111/j.1365-313X.2011.04635.x. [DOI] [PubMed] [Google Scholar]
- 95.Ni J., Gao C., Chen M.-S., Pan B.-Z., Ye K., Xu Z.-F. Gibberellin Promotes Shoot Branching in the Perennial Woody PlantJatropha curcas. Plant Cell Physiol. 2015;56:1655–1666. doi: 10.1093/pcp/pcv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. Gibberellin Biosynthesis and Response during Arabidopsis Seed Germination[W] Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ito A., Tuan P.A., Saito T., Bai S., Kita M., Moriguchi T. Changes in phytohormone content and associated gene expression throughout the stages of pear (Pyrus pyrifolia Nakai) dormancy. Tree Physiol. 2021;41:529–543. doi: 10.1093/treephys/tpz101. [DOI] [PubMed] [Google Scholar]
- 98.Footitt S., Soler I.D., Clay H., Finch-Savage W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA. 2011;108:20236–20241. doi: 10.1073/pnas.1116325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gazzarrini S., Tsai A.Y.-L. Hormone cross-talk during seed germination. Essays Biochem. 2015;58:151–164. doi: 10.1042/bse0580151. [DOI] [PubMed] [Google Scholar]
- 100.Nambara E., Okamoto M., Tatematsu K., Yano R., Seo M., Kamiya Y. Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 2010;20:55–67. doi: 10.1017/S0960258510000012. [DOI] [Google Scholar]
- 101.Shi L., Gast R.T., Gopalraj M., E Olszewski N. Characterization of a shoot-specific, GA3- and ABA-Regulated gene from tomato. Plant J. 1992;2:153–159. doi: 10.1111/j.1365-313x.1992.00153.x. [DOI] [PubMed] [Google Scholar]
- 102.Dong Z., Xiao Y., Govindarajulu R., Feil R., Siddoway M.L., Nielsen T., Lunn J.E., Hawkins J., Whipple C., Chuck G. The regulatory landscape of a core maize domestication module controlling bud dormancy and growth repression. Nat. Commun. 2019;10:3810. doi: 10.1038/s41467-019-11774-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Argueso C.T., Ferreira F.J., Kieber J.J. Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009;32:1147–1160. doi: 10.1111/j.1365-3040.2009.01940.x. [DOI] [PubMed] [Google Scholar]
- 104.Kieber J.J., Schaller G.E. The Arabidopsis Book/American Society of Plant Biologists. Volume 12. ASPB Press; Rockville, MD, USA: 2014. Cytokinins; p. 0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Werner T., Motyka V., Laucou V., Smets R., Van O.H., Schmülling T. Cytokinin-Deficient Transgenic Arabidopsis Plants Show Multiple Developmental Alterations Indicating Opposite Functions of Cytokinins in the Regulation of Shoot and Root Meristem Activity. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Krall L., Raschke M., Zenk M.H., Baron C. The Tzs protein from Agrobacterium tumefaciens C58 produces zeatin riboside 5′-phosphate from 4-hydroxy-3-methyl-2-(E)-butenyl diphosphate and AMP. Sci. Direct. 2002;527:315–318. doi: 10.1016/s0014-5793(02)03258-1. [DOI] [PubMed] [Google Scholar]
- 107.Sakakibara H. CYTOKININS: Activity, Biosynthesis, and Translocation. Annu. Rev. Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- 108.Li S.-M., Zheng H.-X., Zhang X.-S., Sui N. Cytokinins as central regulators during plant growth and stress response. Plant Cell Rep. 2021;40:271–282. doi: 10.1007/s00299-020-02612-1. [DOI] [PubMed] [Google Scholar]
- 109.Werner T., Schmülling T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009;12:527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 110.Hwang I., Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 111.Kim D.-J., Forst S. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology. 2001;147:1197–1212. doi: 10.1099/00221287-147-5-1197. [DOI] [PubMed] [Google Scholar]
- 112.To J.P., Kieber J.J. Cytokinin signaling: Two-components and more. Trends Plant Sci. 2008;13:85–92. doi: 10.1016/j.tplants.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 113.Arkhipov D.V., Lomin S.N., Myakushina Y.A., Savelieva E.M., Osolodkin D.I., Romanov G.A. Modeling of Protein–Protein Interactions in Cytokinin Signal Transduction. Int. J. Mol. Sci. 2019;20:2096. doi: 10.3390/ijms20092096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y., Li L., Ye T., Zhao S., Liu Z., Feng Y.-Q., Wu Y. Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J. 2011;68:249–261. doi: 10.1111/j.1365-313X.2011.04683.x. [DOI] [PubMed] [Google Scholar]
- 115.Thirugnanasambantham K., Prabu G., Mandal A.K.A. Synergistic effect of cytokinin and gibberellins stimulates release of dormancy in tea (Camellia sinensis (L.) O. Kuntze) bud. Physiol. Mol. Biol. Plants. 2020;26:1035–1045. doi: 10.1007/s12298-020-00786-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Qiu Y., Guan S.C., Wen C., Li P., Gao Z., Chen X. Auxin and cytokinin coordinate the dormancy and outgrowth of axillary bud in strawberry runner. BMC Plant Biol. 2019;19:528. doi: 10.1186/s12870-019-2151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hartmann A., Senning M., Hedden P., Sonnewald U., Sonnewald S. Reactivation of Meristem Activity and Sprout Growth in Potato Tubers Require Both Cytokinin and Gibberellin. Plant Physiol. 2010;155:776–796. doi: 10.1104/pp.110.168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Turnbull C.G.N., Hanke D.E. The control of bud dormancy in potato tubers. Planta. 1985;165:359–365. doi: 10.1007/BF00392233. [DOI] [PubMed] [Google Scholar]
- 119.Alvim R., Hewett E.W., Saunders P.F. Seasonal variation in the hormone content of willow: I. Changes in abscisic Acid content and cytokinin activity in the xylem sap. Plant Physiol. 1976;57:474–476. doi: 10.1104/pp.57.4.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cutting J., Strydom D.K., Jacobs G., Bellstedt D.U., Weiler E.W. Changes in Xylem Constituents in Response to Rest-breaking Agents Applied to Apple before Budbreak. J. Am. Soc. Hortic. Sci. 1991;116:680–683. doi: 10.21273/JASHS.116.4.680. [DOI] [Google Scholar]
- 121.Yamane H., Wada M., Honda C., Matsuura T., Ikeda Y., Hirayama T., Osako Y., Gao-Takai M., Kojima M., Sakakibara H., et al. Overexpression of Prunus DAM6 inhibits growth, represses bud break competency of dormant buds and delays bud outgrowth in apple plants. PLoS ONE. 2019;14:e0214788. doi: 10.1371/journal.pone.0214788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cattani A.M., da Silveira Falavigna V., Silveira C.P., Buffon V., Dos Santos Maraschin F., Pasquali G., Revers L.F. Type-B cytokinin response regulators link hormonal stimuli and molecular responses during the transition from endo- to ecodormancy in apple buds. Plant Cell Rep. 2020;39:1687–1703. doi: 10.1007/s00299-020-02595-z. [DOI] [PubMed] [Google Scholar]
- 123.Tran L.-S.P., Shinozaki K., Yamaguchi-Shinozaki K. Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signal. Behav. 2010;5:148–150. doi: 10.4161/psb.5.2.10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dobisova T., Hrdinova V., Cuesta C., Michlickova S., Urbankova I., Hejatkova R., Zadnikova P., Pernisova M., Benkova E., Hejatko J. Light Controls Cytokinin Signaling via Transcriptional Regulation of Constitutively Active Sensor Histidine Kinase CKI1. Plant Physiol. 2017;174:387–404. doi: 10.1104/pp.16.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roman H., Girault T., Barbier F., Péron T., Brouard N., Pěnčík A., Novák O., Vian A., Sakr S., Lothier J., et al. Cytokinins Are Initial Targets of Light in the Control of Bud Outgrowth. Plant Physiol. 2016;172:489–509. doi: 10.1104/pp.16.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Corot A., Roman H., Douillet O., Autret H., Perez-Garcia M.D., Citerne S., Bertheloot J., Sakr S., Leduc N., Demotes-Mainard S. Cytokinins and Abscisic Acid Act Antagonistically in the Regulation of the Bud Outgrowth Pattern by Light Intensity. Front. Plant Sci. 2017;8:1724. doi: 10.3389/fpls.2017.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martínez-Fernández I., de Moura S.M., Alves-Ferreira M., Ferrándiz C., Balanzà V. Identification of Players Controlling Meristem Arrest Downstream of the FRUITFULL-APETALA2 Pathway. Plant Physiol. 2020;184:945–959. doi: 10.1104/pp.20.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Di D.W., Zhang C., Luo P. An, C.W.; Guo, G.Q. The biosynthesis of auxin: How many paths truly lead to IAA? Plant Growth Regul. 2016;78:275–285. doi: 10.1007/s10725-015-0103-5. [DOI] [Google Scholar]
- 129.Fendrych M., Leung J., Friml J. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. Elife. 2016;5:19048. doi: 10.7554/eLife.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao Y. Auxin Biosynthesis: A Simple Two-Step Pathway Converts Tryptophan to Indole-3-Acetic Acid in Plants. Mol. Plant. 2012;5:334–338. doi: 10.1093/mp/ssr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Leyser O. Auxin Signaling. Plant Physiol. 2017;176:465–479. doi: 10.1104/pp.17.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aloni R., Peterson C.A. Auxin promotes dormancy callose removal from the phloem of Magnolia kobus and callose accumulation and earlywood vessel differentiation in Quercus Robur. J. Plant Res. 1997;110:37. doi: 10.1007/BF02506841. [DOI] [PubMed] [Google Scholar]
- 133.Qiu Z., Wan L., Chen T., Wan Y., He X., Lu S., Wang Y., Lin J. The regulation of cambial activity in Chinese fir (Cunninghamia lanceolata) involves extensive transcriptome remodeling. New Phytol. 2013;199:708–719. doi: 10.1111/nph.12301. [DOI] [PubMed] [Google Scholar]
- 134.Nagar P.K., Sood S. Changes in endogenous auxins during winter dormancy in tea (Camellia sinensis L.) O. Kuntze. Acta Physiol. Plant. 2006;28:165–169. doi: 10.1007/s11738-006-0043-9. [DOI] [Google Scholar]
- 135.Noriega X., Pérez F.J. ABA Biosynthesis Genes are Down-regulated While Auxin and Cytokinin Biosynthesis Genes are Up-regulated During the Release of Grapevine Buds from Endodormancy. J. Plant Growth Regul. 2017;36:814–823. doi: 10.1007/s00344-017-9685-7. [DOI] [Google Scholar]
- 136.Anderson J.V., Dogramaci M., Horvath D.P., Foley M.E., Chao W.S., Suttle J.C., Thimmapuram J., Hernandez A.G., Ali S., Mikel M.A. Auxin and ABA act as central regulators of developmental networks associated with paradormancy in Canada thistle (Cirsium arvense) Funct. Integr. Genom. 2012;12:515–531. doi: 10.1007/s10142-012-0280-5. [DOI] [PubMed] [Google Scholar]
- 137.Yang M., Jiao Y. Regulation of Axillary Meristem Initiation by Transcription Factors and Plant Hormones. Front. Plant Sci. 2016;7:183. doi: 10.3389/fpls.2016.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tantikanjana T. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 2011;15:1577–1588. doi: 10.1101/gad.887301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Müller D., Leyser O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011;107:1203–1212. doi: 10.1093/aob/mcr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Young N.F., Ferguson B., Antoniadi I., Bennett M.H., Beveridge C., Turnbull C.G. Conditional Auxin Response and Differential Cytokinin Profiles in Shoot Branching Mutants. Plant Physiol. 2014;165:1723–1736. doi: 10.1104/pp.114.239996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brewer P.B., Dun E.A., Ferguson B.J., Rameau C., Beveridge C.A. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 2009;150:482–493. doi: 10.1104/pp.108.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chatfield S.P., Stirnberg P., Forde B.G., Leyser O. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 2000;24:159–169. doi: 10.1046/j.1365-313x.2000.00862.x. [DOI] [PubMed] [Google Scholar]
- 143.Rameau C., Bertheloot J., LeDuc N., Andrieu B., Foucher F., Sakr S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015;5:741. doi: 10.3389/fpls.2014.00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Le Bris M., Michaux-Ferrière N., Jacob Y., Poupet A., Barthe P., Guigonis J.M., Le Page-Degivry M.T. Regulation of bud dormancy by manipulation of ABA in isolated buds of Rosa hybrida cultured in vitro. Funct. Plant Biol. 1999;26:273–281. doi: 10.1071/PP98133. [DOI] [Google Scholar]
- 145.Arney S.E., Mitchell D.L. The Effect of Abscisic Acid on Stem Elongation and Correlative Inhibition. New Phytol. 1969;68:1001–1015. doi: 10.1111/j.1469-8137.1969.tb06500.x. [DOI] [Google Scholar]
- 146.Arend M., Schnitzler J.P., Ehlting B., Hansch R., Lange T., Rennenberg H., Himmelbach A., Grill E., Fromm J. Expression of the Arabidopsis mutant ABI1 gene alters abscisic acid sensitivity, stomatal development, and growth morphology in gray poplars. Plant Physiol. 2009;151:2110–2119. doi: 10.1104/pp.109.144956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yao C., Finlayson S.A. Abscisic Acid Is a General Negative Regulator of Arabidopsis Axillary Bud Growth. Plant Physiol. 2015;169:611–626. doi: 10.1104/pp.15.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Grandío E.G., Poza-Carrión C., Sorzano C.O.S., Cubas P. BRANCHED1 Promotes Axillary Bud Dormancy in Response to Shade in Arabidopsis. Plant Cell. 2013;25:834–850. doi: 10.1105/tpc.112.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grandío E.G., Cubas P. Identification of gene functions associated to active and dormant buds in Arabidopsis. Plant Signal. Behav. 2014;9:e27994. doi: 10.4161/psb.27994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.González-Grandío E., Pajoro A., Franco-Zorrilla J.M., Tarancón C., Cubas P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA. 2016;114:E245. doi: 10.1073/pnas.1613199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aguilar-Martinez J.A., Poza-Carrion C., Cubas P. Arabidopsis BRANCHED1 Acts as an Integrator of Branching Signals within Axillary Buds. J. Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Johnson P.R., Ecker J. The ethylene gas signal transduction pathway: A Molecular Perspective. Annu. Rev. Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- 153.Bleecker A.B., Kende H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 154.Yang S.F. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984;35:155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- 155.Kende H. Ethylene Biosynthesis. Annu. Rev. Plant Biol. 1993;44:283–307. doi: 10.1146/annurev.pp.44.060193.001435. [DOI] [Google Scholar]
- 156.Wang K.L.-C., Li H., Ecker J.R. Ethylene Biosynthesis and Signaling Networks. Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Iqbal N., Trivellini A., Masood A., Ferrante A., Khan N.A. Current understanding on ethylene signaling in plants: The influence of nutrient availability. Plant Physiol. Biochem. 2013;73:128–138. doi: 10.1016/j.plaphy.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 158.Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J. CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-B. [DOI] [PubMed] [Google Scholar]
- 159.Huang Y., Hui L., Hutchison C.E., Laskey J., Kieber J.J. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 2010;33:221–233. doi: 10.1046/j.1365-313X.2003.01620.x. [DOI] [PubMed] [Google Scholar]
- 160.Guo H., Ecker J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 161.Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 162.An F., Zhao Q., Ji Y., Li W., Guo H. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in arabidopsis. Plant Cell. 2010;22:2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kim H.J., Hong S.H., Kim Y.W., Lee I.H., Jun J.H., Phee B.-K., Rupak T., Jeong H., Lee Y., Hong B.S., et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 2014;65:4023–4036. doi: 10.1093/jxb/eru112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Solano R., Stepanova A., Chao Q., Ecker J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. et Biophys. Acta (BBA) Bioenerg. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 166.Howe G.T., Horvath D.P., Dharmawardhana P., Priest H.D., Mockler T.C., Strauss S.H. Extensive Transcriptome Changes During Natural Onset and Release of Vegetative Bud Dormancy in Populus. Front. Plant Sci. 2015;6:989. doi: 10.3389/fpls.2015.00989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Suttle J.C. Involvement of ethylene in potato microtuber dormancy. Plant Physiol. 1998;118:843–848. doi: 10.1104/pp.118.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Alamar M.C., Anastasiadi M., Lopez-Cobollo R., Bennett M.H., Thompson A.J., Turnbull C.G.N., Mohareb F., Terry L.A. Transcriptome and phytohormone changes associated with ethylene-induced onion bulb dormancy. Postharvest Biol. Technol. 2020;168:111267. doi: 10.1016/j.postharvbio.2020.111267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ruonala R., Rinne P.L., Baghour M., Moritz T., Tuominen H., Kangasjärvi J. Transitions in the functioning of the shoot apical meristem in birch (Betula pendula) involve ethylene. Plant J. 2006;46:628–640. doi: 10.1111/j.1365-313X.2006.02722.x. [DOI] [PubMed] [Google Scholar]
- 170.Dubois M., Broeck L.V.D., Claeys H., Van Vlierberghe K., Matsui M., Inzé D. The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 Antagonistically Regulate Mannitol-Induced Growth Inhibition in Arabidopsis. Plant Physiol. 2015;169:166–179. doi: 10.1104/pp.15.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pierik R., Cuppens M.L.C., Voesenek L.A.C.J., Visser E.J.W. Interactions between Ethylene and Gibberellins in Phytochrome-Mediated Shade Avoidance Responses in Tobacco. J. Plant Physiol. 2004;136:2928–2936. doi: 10.1104/pp.104.045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ophir R., Pang X., Halaly T., Venkateswari J., Lavee S., Galbraith D., Or E. Gene-expression profiling of grape bud response to two alternative dormancy-release stimuli expose possible links between impaired mitochondrial activity, hypoxia, ethylene-ABA interplay and cell enlargement. Plant Mol. Biol. 2009;71:403–423. doi: 10.1007/s11103-009-9531-9. [DOI] [PubMed] [Google Scholar]
- 173.Mathiason K., He D., Grimplet J., Venkateswari J., Galbraith D.W., Or E., Fennell A. Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Funct. Integr. Genom. 2009;9:81–96. doi: 10.1007/s10142-008-0090-y. [DOI] [PubMed] [Google Scholar]
- 174.Sudawan B., Chang C.S., Chao H.F., Ku M., Yen Y.F. Hydrogen cyanamide breaks grapevine bud dormancy in the summer through transient activation of gene expression and accumulation of reactive oxygen and nitrogen species. BMC Plant Biol. 2016;16:202. doi: 10.1186/s12870-016-0889-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gao Y., Yang Q., Yan X., Wu X., Yang F., Li J., Wei J., Ni J., Ahmad M., Bai S., et al. High-quality genome assembly of ‘Cuiguan’ pear (Pyrus pyrifolia) as a reference genome for identifying regulatory genes and epigenetic modifications responsible for bud dormancy. Hortic. Res. 2021;8:197. doi: 10.1038/s41438-021-00632-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Browse J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 177.Sherif S., El-Sharkawy I., Mathur J., Ravindran P., Kumar P., Paliyath G., Jayasankar S. A stable JAZ protein from peach mediates the transition from outcrossing to self-pollination. BMC Biol. 2015;13:11. doi: 10.1186/s12915-015-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Goossens J., Fernández-Calvo P., Schweizer F., Goossens A. Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 2016;91:673–689. doi: 10.1007/s11103-016-0480-9. [DOI] [PubMed] [Google Scholar]
- 179.Hu Y., Jiang Y., Han X., Wang H., Pan J., Yu D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017;68:1361–1369. doi: 10.1093/jxb/erx004. [DOI] [PubMed] [Google Scholar]
- 180.Swiatek A., Lenjou M., Van Bockstaele D., Inze D., Van Onckelen H. Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol. 2002;128:201–211. doi: 10.1104/pp.010592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang Y., Turner J.G. Wound-Induced Endogenous Jasmonates Stunt Plant Growth by Inhibiting Mitosis. PLoS ONE. 2008;3:e3699. doi: 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wilen R.W., Van Rooijen G.J.H., Pearce D.W., Pharis R.P., Holbrook L.A., Moloney M.M. Effects of Jasmonic Acid on Embryo-Specific Processes in Brassica and Linum Oilseeds. Plant Physiol. 1991;95:399–405. doi: 10.1104/pp.95.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ellis C., Turner J.G. A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta. 2002;215:549–556. doi: 10.1007/s00425-002-0787-4. [DOI] [PubMed] [Google Scholar]
- 184.Juvany M., Müller M., Munné-Bosch S. Bud vigor, budburst lipid peroxidation, and hormonal changes during bud development in healthy and moribund beech (Fagus sylvatica L.) trees. Trees. 2015;29:1781–1790. doi: 10.1007/s00468-015-1259-3. [DOI] [Google Scholar]
- 185.Hao X., Yang Y., Yue C., Wang L., Horvath D.P., Wang X.C. Comprehensive Transcriptome Analyses Reveal Differential Gene Expression Profiles of Camellia sinensis Axillary Buds at Para-, Endo-, Ecodormancy, and Bud Flush Stages. Front. Plant Sci. 2017;8:553. doi: 10.3389/fpls.2017.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Suttle J.C., Huckle L.L., Lulai E.C. The effects of dormancy status on the endogenous contents and biological activities of jasmonic acid, n-(jasmonoyl)-isoleucine, and tuberonic acid in potato tubers. Am. J. Potato Res. 2011;88:283–293. doi: 10.1007/s12230-011-9192-5. [DOI] [Google Scholar]
- 187.Fan X.Y., Yang Y., Li M., Fu L.L., Zang Y.Q., Wang C.X., Hao T.Y., Sun H.M. Transcriptomics and targeted metabolomics reveal the regulatory network of Lilium davidii var. unicolor during bulb dormancy release. Planta. 2021;254:59. doi: 10.1007/s00425-021-03672-7. [DOI] [PubMed] [Google Scholar]
- 188.Zhao M.L., Wang J.N., Shan W., Fan J.G., Kuang J.F., Wu K.Q., Li X.P., Chen W.X., He F.Y., Chen J.Y., et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2012;36:30–51. doi: 10.1111/j.1365-3040.2012.02551.x. [DOI] [PubMed] [Google Scholar]
- 189.Hu Y., Jiang L., Wang F., Yu D. Jasmonate Regulates the INDUCER OF CBF EXPRESSION–C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 Cascade and Freezing Tolerance in Arabidopsis. Plant Cell. 2013;25:2907–2924. doi: 10.1105/tpc.113.112631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cavalli G., Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 191.Rothkegel K., Sánchez E., Montes C., Greve M., Tapia S., Bravo S., Prieto H., Almeida A.M. DNA methylation and small interference RNAs participate in the regulation of MADS-box genes involved in dormancy in sweet cherry (Prunus avium L.) Tree Physiol. 2017;37:1739–1751. doi: 10.1093/treephys/tpx055. [DOI] [PubMed] [Google Scholar]
- 192.Matzke M.A., Kanno T., Matzke A.J. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu. Rev. Plant Biol. 2015;66:243–267. doi: 10.1146/annurev-arplant-043014-114633. [DOI] [PubMed] [Google Scholar]
- 193.Santamaría M., Hasbún R., Valera M., Meijón M., Valledor L., Rodríguez J.L., Toorop P.E., Canal-Villanueva M.J.F., Rodríguez R. Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. J. Plant Physiol. 2009;166:1360–1369. doi: 10.1016/j.jplph.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 194.Rothkegel K., Sandoval P., Soto E., Ulloa L., Riveros A., Lillo-Carmona V., Caceres-Molina J., Almeida A.M., Meneses C. Dormant but Active: Chilling Accumulation Modulates the Epigenome and Transcriptome of Prunus avium during Bud Dormancy. Front. Plant Sci. 2020;11:1115. doi: 10.3389/fpls.2020.01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chinnusamy V., Gong Z., Zhu J.K. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 2008;50:1187–1195. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Albini S., Zakharova V., Ait-Si-Ali S. Chapter 3—Histone Modifications. In: Palacios D., editor. Epigenetics and Regeneration. Volume 11. Academic Press; Cambridge, MA, USA: 2019. pp. 47–72. [DOI] [Google Scholar]
- 197.David Law R., Suttle J.C. Changes in histone H3 and H4 multi-acetylation during natural and forced dormancy break in potato tubers. Physiol. Plant. 2004;120:642–649. doi: 10.1111/j.0031-9317.2004.0273.x. [DOI] [PubMed] [Google Scholar]
- 198.de la Fuente L., Conesa A., Lloret A., Badenes M.L., Ríos G. Genome-wide changes in histone H3 lysine 27 trimethylation associated with bud dormancy release in peach. Tree Genet Genomes. 2015;11:45. doi: 10.1007/s11295-015-0869-7. [DOI] [Google Scholar]
- 199.Leida C., Conesa A., Llacer G., Badenes M.L., Rios G. Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 2012;193:67–80. doi: 10.1111/j.1469-8137.2011.03863.x. [DOI] [PubMed] [Google Scholar]
- 200.Horvath D.P., Sung S., Kim D., Chao W., Anderson J. Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 2010;73:169–179. doi: 10.1007/s11103-009-9596-5. [DOI] [PubMed] [Google Scholar]
- 201.Wu R., Wang T., Richardson A.C., Allan A.C., Macknight R.C., Varkonyi-Gasic E. Histone modification and activation by SOC1-like and drought stress-related transcription factors may regulate AcSVP2 expression during kiwifruit winter dormancy. Plant Sci. 2019;281:242–250. doi: 10.1016/j.plantsci.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 202.Vimont N., Quah F.X., Schöepfer D.G., Roudier F., Dirlewanger E., Wigge P.A., Wenden B., Cortijo S. ChIP-seq and RNA-seq for complex and low-abundance tree buds reveal chromatin and expression co-dynamics during sweet cherry bud dormancy. Tree Genet Genomes. 2019;16:9. doi: 10.1007/s11295-019-1395-9. [DOI] [Google Scholar]
- 203.Bertrand-Lehouillier V., Legault L.-M., McGraw S. Endocrine Epigenetics, Epigenetic Profiling and Biomarker Identification. In: Huhtaniemi I., Martini L., editors. Encyclopedia of Endocrine Diseases. 2nd ed. Academic Press; Cambridge, MA, USA: 2019. pp. 31–35. [DOI] [Google Scholar]
- 204.Yu J., Bennett D., Dardick C., Zhebentyayeva T., Abbott A.G., Liu Z., Staton M.E. Genome-Wide Changes of Regulatory Non-Coding RNAs Reveal Pollen Development Initiated at Ecodormancy in Peach. Front. Mol. Biosci. 2021;8:612881. doi: 10.3389/fmolb.2021.612881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Li L., Liu J., Liang Q., Zhang Y., Kang K., Wang W., Feng Y., Wu S., Yang C., Li Y. Genome-wide analysis of long noncoding RNAs affecting floral bud dormancy in pears in response to cold stress. Tree Physiol. 2021;41:771–790. doi: 10.1093/treephys/tpaa147. [DOI] [PubMed] [Google Scholar]
- 206.Wang L., Xu Q., Yu H., Ma H., Li X., Yang J., Chu J., Xie Q., Wang Y., Smith S.M., et al. Strigolactone and Karrikin Signaling Pathways Elicit Ubiquitination and Proteolysis of SMXL2 to Regulate Hypocotyl Elongation in Arabidopsis. Plant Cell. 2020;32:2251–2270. doi: 10.1105/tpc.20.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhao L.-H., Zhou X.E., Wu Z.-S., Yi W., Xu Y., Li S., Xu T., Liu Y., Chen R.-Z., Kovach A., et al. Crystal structures of two phytohormone signal-transducing α/β hydrolases: Karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 2013;23:436–439. doi: 10.1038/cr.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Imanishi H., Fortanier E.J. Effects of exposing freesia corms to ethylene or to smoke on dormancy-breaking and flowering. Sci. Hortic. 1983;18:381–389. doi: 10.1016/0304-4238(83)90019-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.