Abstract
The p53-inducible gene PC3 (TIS21, BTG2) is endowed with antiproliferative activity. Here we report that expression of PC3 in cycling cells induced accumulation of hypophosphorylated, growth-inhibitory forms of pRb and led to G1 arrest. This latter was not observed in cells with genetic disruption of the Rb gene, indicating that the PC3-mediated G1 arrest was Rb dependent. Furthermore, (i) the arrest of G1-S transition exerted by PC3 was completely rescued by coexpression of cyclin D1 but not by that of cyclin A or E; (ii) expression of PC3 caused a significant down-regulation of cyclin D1 protein levels, also in Rb-defective cells, accompanied by inhibition of CDK4 activity in vivo; and (iii) the removal from the PC3 molecule of residues 50 to 68, a conserved domain of the PC3/BTG/Tob gene family, which we term GR, led to a loss of the inhibition of proliferation as well as of the down-regulation of cyclin D1 levels. These data point to cyclin D1 down-regulation as the main factor responsible for the growth inhibition by PC3. Such an effect was associated with a decrease of cyclin D1 transcript and of cyclin D1 promoter activity, whereas no effect of PC3 was observed on cyclin D1 protein stability. Taken together, these findings indicate that PC3 impairs G1-S transition by inhibiting pRb function in consequence of a reduction of cyclin D1 levels and that PC3 acts, either directly or indirectly, as a transcriptional regulator of cyclin D1.
The control of the cell cycle plays an essential role in cell growth and in the activation of important cellular processes such as differentiation and apoptosis. pRb (retinoblastoma protein) and p53 are two molecules identified as key regulators of the cell cycle.
pRb is a nuclear phosphoprotein whose phosphorylation state oscillates regularly during the cell cycle. Its underphosphorylated forms predominate in G0 and G1, while highly phosphorylated forms exist in S, G2, and M phases (13, 16, 21). The primary biological function of underphosphorylated pRb is to inhibit progression toward S phase by controlling a checkpoint in late G1 (for reviews, see references 8, 22, and 51). In fact, underphosphorylated pRb associates with members of the E2F family of transcription factors, impairing their activity and leading to a cell cycle block in G1. Conversely, the phosphorylation of pRb inactivates its growth suppression activity by freeing E2F molecules, thus enabling them to transactivate genes required for the progression of the cell into S phase and the remainder of the cell cycle (52, 97, 114).
Cyclin-dependent kinases (CDKs) are the molecules responsible for pRb phosphorylation and its consequent inactivation (reviewed in references 70 and 102). Each CDK has its own functional specificity, based on the period of its activity during the cell cycle and on the specific cyclin partner. CDK4, CDK5, and CDK6 form complexes with D-type cyclins during the G1 phase (65, 69, 116). CDK2, when bound to cyclin A or E, is instead essential for G1-to-S transition (28, 78), while the cdc2 kinase, associated with cyclins A and B, determines the G2/M transition (78, 82, 90). Interestingly, the expression of D-type cyclins and also their assembly with their CDK partners are heavily dependent on stimulation by growth factors (101, 102). If stimulation by growth factor(s) ceases, the level of D-type cyclins decreases rapidly, their half-life being short, with a consequent impairment of S-phase entry (7, 87). Since cells lacking a functional Rb gene become independent from D-type cyclins for G1/S progression, this clearly indicates that pRb is the final target (61, 107).
A further level of control in the function of the pRb pathway is exerted by the CDK inhibitors (reviewed in reference 103). These are represented by two families of molecules, the INK4 family (comprising p16INK4a, p15INK4b, p18INK4c, and p19INK4c), which causes G1 arrest by directly binding and inhibiting the activation of CDK4 and CDK6 by D-type cyclins, and the KIP/CIP family, which includes p27Kip1 and p21CIP1/WAF1. This latter was identified as a potent inhibitor of all known cyclin-CDK complexes (39, 42, 115).
Besides regulating cell cycle progression, the G1 checkpoint function of pRb can mediate exit from the cell cycle in response to growth-inhibitory signals or differentiation inducers. These signals in fact activate the pRb growth suppression function by preventing its phosphorylation, thus allowing the cell to attain the postmitotic state, an essential preliminary requirement for terminal differentiation of many cell types (for reviews, see references 44 and 91). A critical role of pRb in the control of differentiation and survival of several cell lineages, such as neurons, lens fiber cells, cells from the cerebellar cortex, and muscle and hematopoietic cells, is clearly indicated by the phenotype of the Rb-deficient mouse (53, 54, 74, 119). Furthermore, pRb enhances the activities of transcription factors such as MyoD and C/EBPs in promoting muscle and adipocyte terminal differentiation, respectively (14, 15, 38, 77).
The G1 checkpoint regulatory pathway also responds to stressful situations and DNA damage. The p53 protein, which is activated by different types of DNA damage, functions by arresting the cell cycle in G1 to allow repair to take place (for reviews, see references 4 and 56). p53 effects G1 arrest mainly by inducing transcription of p21CIP1/WAF1, which inhibits CDK's activity, thus preventing pRb phosphorylation (12, 24, 27, 112, 115). Alternatively, if the growth arrest program fails, p53 can activate an apoptotic program in the cell carrying the DNA damage (4). Recently, the antiproliferative activity of p53 has also been implicated in a G2/M-phase checkpoint that controls the entry into mitosis (3).
In this context, the gene PC3, isolated by us (9) and by others with the alternative names BTG2 (92) and TIS21 (32), plays a role. PC3 is in fact endowed with antiproliferative activity and is induced by p53 (72, 92). We originally isolated PC3 while studying the onset of neuronal differentiation, induced in the rat PC12 cell line by nerve growth factor within its first hour of activity (9, 108). The time window chosen for our analysis of gene induction corresponds to the period of transition between mitosis and growth arrest that serves as a prelude to differentiation (36, 37, 95). The antiproliferative properties displayed by PC3 are consistent with such timing and are peculiar among the immediate-early genes induced by nerve growth factor. Furthermore, PC3 was found to be a marker for neuronal cell birthday (47). In fact, its mRNA expression during embryonic development of the central nervous system is restricted to the neuroblast undergoing the last proliferation before differentiating into postmitotic neuron (47). This led us to hypothesize that PC3 is involved in the growth arrest of the neuronal precursors (47). However, the expression of PC3 during development and in the adult animal is not limited to the nervous system (9, 47). Accordingly, PC3 displayed an antiproliferative effect in different cell types, such as fibroblasts and PC12 cells (72). Such an antiproliferative effect was afterwards confirmed by the work of Rouault et al. (92) for the human counterpart of the PC3 gene, i.e., BTG2. Interestingly, the same group also showed that BTG2/PC3 is induced by p53 and that embryonic stem cells in which BTG2/PC3 had been ablated, underwent apoptosis following DNA damage because of failure in growth arrest (92). These observations raise the question whether PC3 may promote p53-induced cell cycle arrest, similarly to p21CIP1/WAF1, the prototype inhibitor of CDKs. In this regard, it has been recently pointed out that the ability of p53 to arrest the cell cycle in G1 is only partially dependent on the induction of p21CIP1/WAF1 (24).
After the cloning of PC3, other novel related antiproliferative genes were isolated, namely, BTG1 (94), TOB (64), and ANA (118). These genes share 60, 40, and 35% sequence homology with PC3, respectively. Interestingly, the homology of Tob to the entire BTG1 and PC3 molecules is limited to its amino-terminal domain, whereas its carboxyl-terminal domain interacts with the mitogenic receptor p185erbB2 (64). Since no homology to known functional motifs is evident in the cDNA-deduced proteins of these genes, it appears likely that PC3, BTG1, and Tob belong to a novel functional class of cell cycle regulators, but the question about their specific molecular function remains open. In this regard, some suggestions came from a recent report which showed that TIS21/BTG2 interacts with a protein-arginine N-methyltransferase (Prmt1) by positively modulating its activity (58). Prmt1, in turn, has been found to bind the interferon receptors and to be required for interferon-mediated growth inhibition (2). A further interaction was observed between BTG2 and the mCAF1 gene, i.e., the mouse homolog of the yeast CAF/POP2 gene (93). This latter gene is part of the yeast CCR4 multisubunit complex, which is required for the transcriptional regulation of several genes (59, 60).
This report describes our attempts to shed light on the molecular mechanisms by which PC3 impinges on cell cycle activity. We observed that the inhibition of cell cycle progression by PC3 requires functional pRb, and we found the existence of a mutually exclusive interaction between PC3 and cyclin D1. In fact, the latter blocked the PC3 effects on the cell cycle, whereas PC3 inhibited cyclin D1 expression.
MATERIALS AND METHODS
Cell culture, cell lines, and transfections.
NIH 3T3 and Rb−/− 3T3 cells and cyclin D1+/+ and cyclin D1−/− mouse embryo fibroblasts (MEFs) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (HyClone, Logan, Utah) in a humidified atmosphere of 5% CO2 at 37°C.
Transfection of the plasmids was performed by the liposome technique using the Lipofectamine reagent (Life Technologies, Gaithersburg, Md.) as per the manufacturer's instructions. The indicated amount of DNA (see figure legends), diluted in Optimem containing Lipofectamine (5 or 30 μl for 35- or 90-mm-diameter dishes, respectively), was added to the cultures, left to incubate for 5 h, and then replaced with normal DMEM. In the experiments aimed at defining the influence of PC3 on the phosphorylation state of pRb in NIH 3T3 cells and to carry out cell sorting, the calcium phosphate procedure was used (34). To the cell cultures were added the indicated amounts of DNA in calcium phosphate solution (0.5 ml for a 60-mm-diameter dish). Cells were exposed to the DNA precipitates for 20 h, washed twice with phosphate-buffered saline (PBS), and then placed in their medium for an additional 40 h. At the end of this period, cell cultures were used for the procedures indicated.
Plasmids, PC3 expression vectors, and mutants.
The expression vector pSCT was from B. Schäfer (33), who obtained it by adding an artificial polylinker to the vector pSCT GAL 556X (96). pSCT-β-Gal was produced by inserting, in the BamHI site of the pSCT vector, the Escherichia coli β-galactosidase (β-Gal) gene, excised from the vector pCMVβ (Clontech, Palo Alto, Calif.) as a NotI fragment, whose ends were previously ligated to BamHI linkers. Human pRb was expressed by the construct pCMV-pRB1, obtained by D. Livingston (85) by inserting Rb cDNA into the pCMVneoBam vector (6). The different constructs expressing cyclins (pRcCMV-cycA, pRcCMV-cycB1, pRcCMV-cycB2, pRcCMV-cycD1, pRcCMV-cycD3, and pRcCMV-cycE; obtained by R. Weinberg [see reference 45]) and CDKs (CMVcdc2, pRcCMV-CDK2, and pRcCMV-CDK4; obtained by E. Harlow [see reference 110] and by D. Livingston [see reference 29]) were all in cytomegalovirus (CMV) promoter-driven plasmids. The expression vectors for human p16INK4a (pXp16 [99]) and mouse p27Kip1 (pCMX-p27) were gifts of D. Beach and J. Massaguè, respectively.
pSCT-PC3 was constructed by cloning into the pSCT vector the coding region of PC3 cDNA (nucleotides 65 to 541, with the stop codon), amplified by PCR using primers that incorporated 5′ XbaI and 3′ HindIII sites, and confirmed by sequencing. The pSCT-PC3 constructs having a deletion internal to the coding region were generated by cloning in the pSCT vector two fragments amplified by PCR corresponding to the PC3 regions upstream and downstream of the deleted region, joined by a PstI site in frame. The PCR primers used were as follows: (a) mutant PC3Δ50–68; 5′CTCGAGTCTAGAGCACCGGGCCCGCCACCATGAGCCACGGGAAGAGA3′ (PC3-PCR3, upstream sense primer containing a flanking 5′ XbaI site [underlined] and the PC3 initiator codon [underlined]), 5′CGCTGCAGATGATCGGTCAGTGCGTC3′ (downstream antisense primer complementary to PC3 sequence corresponding to amino acids [aa] 44 to 49 [underlined] and flanked by a 3′ PstI site), 5′GGCTGCAGCGCATCAACCACAAGATG3′ (upstream sense primer complementary to PC3 sequence corresponding to aa 69 to 74 [underlined] and flanked by a 3′ PstI site), and 5′GGAAGATCTATCGATAAGCTTGAATTCTCCTCTCTAGCTGGAGAC3′ (PC3-PCR4, downstream antisense primer containing a flanking 5′ HindIII site [underlined] and the PC3 termination codon [CTA in the antisense strand, underlined]); and (b) mutant PC3Δ105–123; PC3-PCR3 primer, 5′CGCTGCAGGACCCACAGGGTCAGCT3′ (downstream antisense primer complementary to PC3 sequence corresponding to aa 99 to 104 [underlined] and flanked by a 3′ PstI site), 5′GGCTGCAGGAGGAGGCGCCGGTGGC3′ (upstream sense primer complementary to PC3 sequence corresponding to aa 124 to 129 [underlined] and flanked by a 3′ PstI site), and a PC3-PCR4 primer. The pSCT-PC3 S147N mutant, bearing a point mutation at nucleotide 504 that mutates serine 147 to asparagine, was produced by cloning in the pSCT vector the insert amplified by PCR with primers that incorporated 5′ XbaI and 3′ SalI sites: 5 ′ GAG TC TAGAGAAT TCGCACCGGGCCCGCCACCATGAGCCACGGGAAGAGA3′ (upstream sense primer containing flanking 5′ XbaI and EcoRI sites [underlined] and the PC3 initiator codon [underlined]) and 5′CGATGTCGACC TAGC TGGAGACAG TCATCACG TAGTTC T TCGATGGAT TGC TCCT3′ (downstream antisense primer containing a flanking 5′ SalI site [underlined], the PC3 termination codon [CTA in the antisense strand, 3′ to SalI site], and the mutated nucleotide [underlined]). The corresponding construct pGEX-PC3 S147N was obtained by restricting pSCT-PC3 S147N in SalI, blunting and adding EcoRI linkers, and then subcloning the insert excised by EcoRI from pSCT-PC3 S147N in frame in the EcoRI site of the pGEX-2T vector. All the constructs obtained were checked by sequencing. The production of a protein was verified by immunoblotting and by immunofluorescence staining with the anti-PC3 A3H polyclonal antibody (72), by which no differences in the efficiency of expression of the different constructs were detected.
The retroviral vector pBABE puro, a Moloney murine leukemia virus-based vector carrying the puromycin resistance gene (75), was used for infection of cell cultures. To obtain the pBABE puro-PC3 construct, the PC3 coding region was subcloned into the BamHI site of pBABE puro, after amplification by PCR using the primers 5′GAGAGATCTGCACCGGGCCCGCCACCATGAGCCACGGGAAGAGA3′ as upstream sense primer (carrying a BglII site) and PC3-PCR4 as downstream antisense primer (see above). The construct was confirmed by sequencing.
Flow cytometry assays and cell sorting.
NIH 3T3 or Rb−/− 3T3 cells cotransfected with pSCT-PC3 or pXp16 and with the CD20 cDNA (pCMVCD20; see reference 122) were washed in PBS-EDTA (5 mM) and then incubated in PBS-EDTA (5 mM) for 10 min at 37°C, harvested, and pelleted. The cell number at the moment of harvesting was about 106 cells in a 90-mm-diameter dish. The cell pellet was then resuspended in DMEM, centrifuged, and resuspended again in 100 μl of DMEM containing fluorescein isothiocyanate (FITC)-conjugated mouse monoclonal anti-CD20 antibody (Caltag Laboratories, San Francisco, Calif.) to a final concentration of 40 μg/ml. Cells were incubated for 1 h at 4°C and then pelleted and washed once in DMEM, to be finally resuspended in PBS.
Then, the cell suspension either was analyzed for cell cycle phase distribution or was sorted for Western blot analysis and for reverse transcriptase PCR (RT-PCR) analysis, using an EPICS 541 flow cytometer (Coulter Electronics, Inc.). For cell cycle analysis, the CD20-stained cells were fixed in 70% ethanol and stained with propidium iodide (50 μg/ml; Sigma Chemical Co.) in PBS containing RNase A (75 kU/ml; Sigma Chemical Co.). Two-color flow cytometry was performed, simultaneously measuring FITC (green channel) and propidium iodide (red channel) fluorescence intensities. The total population was gated on scatter parameters to remove cell debris. The gates to analyze cell cycle distribution of CD20-PC3-expressing cells were established by measuring background levels of FITC fluorescence, by use of vector-transfected cells incubated with a nonspecific immunoglobulin G FITC-conjugated antibody (Caltag Laboratories). DNA histograms were analyzed by a suitable mathematical model (20) to estimate the percentage of cells in the various compartments of the cell cycle. For Western blot analysis or for RT-PCR analysis, transfected cells were sorted on the basis of the FITC-CD20 positivity and, immediately thereafter, lysed in Laemmli buffer with protease inhibitors or homogenized in 4 M guanidine thiocyanate followed by extraction with phenol-chloroform (18), respectively. An aliquot of the Laemmli lysate was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), whereas an aliquot of total RNA was used for RT-PCR analysis.
Immunofluorescence staining and antibodies.
Transfected cells, grown on polylysine-coated coverslips, were washed three times with PBS and fixed for 20 min at room temperature in PBS containing 3.75% paraformaldehyde. The coverslips were then washed three times in PBS and incubated for 2 min in 0.1 M glycine–PBS. Permeabilization was performed with 0.1% Triton X-100 in PBS for 6 min at room temperature. After a PBS wash, the cells were incubated for 60 min at room temperature with one primary antibody, or two where indicated, diluted in PBS. A3H rabbit polyclonal antibody (obtained using the whole PC3 protein as immunogen and affinity purified as described in reference 72) was diluted 1:50, anti-β-Gal rabbit polyclonal antibody (Chemicon International, Inc., Temecula, Calif.) was diluted 1:50, and antibromodeoxyuridine (BrdU) mouse monoclonal antibody (Amersham, Little Chalfont, England) was used undiluted, whereas affinity-purified rabbit polyclonal antibodies anti-cyclin A (C19; Santa Cruz Biotechnology, Heidelberg, Germany) and anti-cyclin E (M-20; Santa Cruz Biotechnology), as well as anti-cyclin D1 mouse monoclonal antibody 72-13G specific for rodent cyclin D1 (Santa Cruz Biotechnology; see reference 66), were used at a final concentration of 2 μg/ml. After three washes in PBS, the cells were incubated for 30 min at room temperature with the secondary antibody(ies), either FITC conjugated (Myles-Yeda, Rehovot, Israel) or TRITC (tetramethylrhodamine isothiocyanate) conjugated (Sigma Chemicals), and then washed three times with PBS. Cells were finally mounted with PBS-glycerol (3:1). The immunofluorescence assay was performed on a Leitz Dialux 22 microscope.
DNA synthesis assays were performed by adding 50 μM BrdU to the culture medium 24 to 18 h before fixation. To detect BrdU, DNA denaturation was obtained by adding 50 mM NaOH for 10 s after permeabilization with 0.1% Triton X-100 and was followed by three PBS washes. BrdU was detected by undiluted anti-BrdU monoclonal antibody (Amersham RPN 202) added together with the other primary antibody as described above, followed by FITC-conjugated goat antibody to mouse immunoglobulin G (Sigma F9006). To detect nuclei, cells were incubated at the end of the immunofluorescence staining procedure for 2 min in Hoechst 33258 dye diluted in PBS at 1 μg/ml (Sigma), washed twice in PBS, and mounted as described above.
Immunoblotting analysis and antibodies.
Transfected cell cultures (after cell sorting where indicated) were lysed into Laemmli buffer (125 mM Tris-HCl [pH 6.8], 10% glycerol, 2.1% SDS) containing 0.5 M β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg of leupeptin per ml, and 10 μg of aprotinin per ml and heated for 5 min at 100°C. An aliquot was analyzed by SDS–10% PAGE. After electrophoresis, proteins were electrophoretically transferred to nitrocellulose (12 to 16 h at 65 mA in 24 mM Tris-HCl [pH 8.3]–166 mM glycine–20% methanol). The filters were then soaked for 2 h in blocking buffer (TBS [10 mM Tris HCl (pH 8), 150 mM NaCl]–0.05% Tween–5% powdered milk) and then incubated in the same buffer for 2 h with the first antibody. This latter was one of the following: the anti-PC3 affinity-purified rabbit polyclonal A3H antibody (diluted 1:1,000); the affinity-purified rabbit polyclonal antibody anti-cyclin A (sc-596), anti-cyclin E (sc-481), anti-cdc2 (sc-53), anti-CDK2 (sc-163), or anti-CDK4 (sc-260); the mouse monoclonal antibody anti-cyclin D1 72-13G specific for rodent cyclin D1 (all from Santa Cruz Biotechnology, diluted 1:200); and the mouse monoclonal antibody anti-β-actin (clone AC-15, diluted 1:5,000; Sigma Chemical). After three washes in TBS with 0.05% Tween, the filter was incubated in blocking buffer containing the second antibody (either goat anti-rabbit or goat anti-mouse horseradish peroxidase-conjugated antibody; Pierce, Rockford, Ill.). After three washes in TBS with 0.05% Tween, detection of the second antibody was performed by chemiluminescent assay. Anti-pRb immunoblotting was performed with the G3-245 monoclonal antibody (Pharmingen, San Diego, Calif.; diluted to a final concentration of 1 μg/ml) used as described above, with the only differences being that SDS-PAGE had 7% polyacrylamide and blocking buffer contained 0.4% gelatin in place of powdered milk. The intensity of the bands of the immunoblots was quantified by an EPA 3000 densitometer (Sanwatsusho, Tokyo, Japan) in the linear range of the film. The intensity values of the sample were normalized to the corresponding values of β-actin.
RNA extraction and RT-PCR assay.
Total cellular RNA was obtained from sorted cells according to the procedure of Chomczynski and Sacchi (18) (see above) and was analyzed by semiquantitative RT-PCR as previously described (72). Four micrograms of total RNA was treated with DNase (RQ1; Promega; 2 U) and then denatured at 75°C for 5 min and added to a total reaction volume of 50 μl containing 1× RT buffer (10 mM Tris-HCl [pH 8.8], 50 mM KCl, 0.1% Triton X-100), 5 mM MgCl2, 0.5 mM (each) deoxynucleoside triphosphate, 1 U of RNasin (Promega), and 600 pmol of random hexamer primers. Moloney murine leukemia virus RT (200 U; Promega) was added to one-half reaction volume (25 μl) and incubated for 2 h at 37°C (the remaining reaction volume without RT was kept to be used as a control in PCR amplifications for possible contamination of the sample with genomic DNA). RT reaction mixtures were stored at −20°C and then used for PCR amplification. Two microliters of each RT reaction mixture was amplified in a 100-μl PCR mixture containing 1× PCR buffer (10 mM Tris-HCl [pH 9] at 25°C, 50 mM KCl, 0.1% Triton X-100), 0.2 mM (each) deoxynucleoside triphosphate, 1.5 mM MgCl2, 20 pmol of each primer (see below), and 2 U of Taq polymerase (Promega). The number of cycles was designed so as to maintain the reactions of amplification in exponential phase (20 cycles for β-actin and 35 cycles for all the other templates). Coamplification of β-actin mRNA gave a measure of the efficiency of the reaction and of the starting RNA amount in each sample, since β-actin is constitutively expressed in the cell lines used. Amplification profiles were the following: denaturation at 95°C for 5 min during the first cycle or at 94°C for 1 min in the remaining cycles, primer annealing at 50°C (for β-actin, cyclin A, and PC3; at 52°C for cyclins D1 and E) for 1 min, and primer extension at 72°C for 1.5 min. About 1/10 of the PCR sample was electrophoresed on a 1.2% agarose gel, blotted onto a nylon filter, and hybridized to 32P-labeled specific oligonucleotides (whose sequence was internal to the region amplified by PCR): (a) cyclin A (5′-CAGAGTGTGAAGATGCCCTGG-3′), (b) cyclin D1 (5′-CCATGCTCAAGACGGAGGAGA-3′), (c) cyclin E (5′-GGCGAGGATGAGAGCAGTTCT-3′), (d) PC3 (5′-CCGTAGGTTTCCTCACCAGTC-3′), and (e) β-actin (5′-CAGCTGAGAGGGAAATCGTGC-3′). The relative product amounts were quantitated by analysis with a Molecular Dynamics 400A PhosphorImager system. The PCR primers used were as follows: (a) cyclin A, 5′ (5′-TGCTCCTCTTAAGGACCTT-3′) and 3′ (5′-TCAGAACCTGCTTCTTGGA-3′); (b) cyclin D1, 5′ (5′-ACACCAATCTCCTCAACGA-3′) and 3′ (5′-TAGCAGGAGAGGAAGTTGT-3′); (c) cyclin E, 5′ (5′-GAAAATCAGACCACCCAGA-3′) and 3′ (5′-ATACAAAGCAGAAGCAGCG-3′); (d) PC3, 5′ (5′-ATGAGCCACGGGAAGAGA-3′) and 3′ (5′-CCTGAAGTTC TCAGCTCT-3′) (this latter primer is reverse complementary to the pSCT polylinker in the region 3′ of the cloning site of PC3, and thus the PC3-amplified product derives only from the PC3 exogenous transcript); and (e) β-actin, 5′ (5′-TTGAGACCTTCAACACCC-3′) and 3′ (5′-GCAGCTCATAGCTCTTCT-3′). All the oligonucleotides except those for PC3 were from mouse cDNA sequences.
Reporter gene assay.
NIH 3T3 cell cultures were transfected with the indicated PC3 or E2F-1 expression constructs. The variations in the amounts of expression vectors were completely compensated for by addition of the corresponding empty DNA plasmid vectors. Transfection of the expression construct cDNAs was performed in parallel with the positive-control simian virus 40 promoter-driven pGL2 control plasmid (Promega). Luciferase activity of each sample (Li) was corrected for differences in transfection by normalization, measuring the amount of plasmid DNA present in each extract of the transfected cells (Di), as determined by dot blot hybridization according to a previously described procedure (1). Plasmid DNA was visualized using as a probe a 3-kb BamHI-SmaI fragment from the noncoding region of vector pGL2, to avoid possible interactions with RNA. The formula used was luciferase activity normalized = Li × Dm/Di, where Dm is the average value for each experiment. The fold activity was then obtained by dividing each normalized value of luciferase activity by the average number of normalized luciferase units of the corresponding control culture.
Metabolic labeling and immunoprecipitation.
NIH 3T3 cultures transfected with Flag-tagged cyclin D1 construct (kindly provided by C. Sherr [26]) were washed twice in DMEM without methionine, preincubated in the same medium for an hour, and then labeled by incubation in methionine-free DMEM containing Pro-mix35S (0.15 mCi of [35S]methionine per ml) for 2 h. Afterwards, cultures were washed twice in DMEM with an excess of cold methionine, incubated in the same medium for the indicated time periods, and lysed by 30 min of incubation at 4°C in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, and 150 mM NaCl, containing 1% Triton X-100, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, and 1 mM PMSF). After clearing by centrifugation at 10,000 × g for 15 min, extracts were assayed for protein concentration (10); 500-μg aliquots were then precleared using a rabbit preimmune serum and protein G-Sepharose (Amersham Pharmacia Biotech) for 1 h at 4°C. After centrifugation at 12,000 × g, the supernatants were incubated with protein G-Sepharose with the anti-Flag M2 mouse monoclonal antibody (Sigma; F3165; 3 μg per sample) for 2 h at 4°C. The immunocomplexes were washed three times in lysis buffer and then resuspended in Laemmli buffer containing protease inhibitors, heat denatured, and run on SDS-polyacrylamide gels.
Production and purification of GST fusion proteins.
The vector pGEX-2T-PC3 was obtained by subcloning in frame into pGEX-2T the coding region of PC3, excised as a 5′ BamHI-3′ EcoRI PCR-amplified fragment from the vector pRSETA-PC3, in which it had been previously cloned. The restriction reaction for the site BamHI was partial, given the existence of a BamHI site internal to PC3. The construct was checked by sequence analysis. Human pGEX-p21 and pGEX-p16 were from Y. Xiong. pGEX-PC3 S147N was obtained as described above. The fusion proteins, after lysis of the bacterial pellet in PBS with 0.5% NP-40, were purified through glutathione S-transferase (GST)–Sepharose beads (Pharmacia) and eluted per the manufacturer's instructions. The proteins were stored at −80°C until use, either bound to GST-Sepharose beads (for in vitro binding assays) or in elution buffer (for in vitro kinase assays; 30 mM reduced glutathione, 60 mM HEPES [pH 7.5], 30 μg of leupeptin per ml, 1 mM PMSF) at a concentration of about 0.3 μg/μl at −80°C until use.
In vitro binding assays.
Expression and purification of GST fusion proteins were performed as described above. Mouse pCMV-cdc2 (110), human pRcCMV-CDK2 and pRcCMV-CDK4 (29), and human pBSK-glob-CDK6 (68) were transcribed and translated in vitro using 35 μl of nuclease-treated rabbit reticulocyte lysate (Promega) as described elsewhere (41). The programmed lysates (1.5 μl) were incubated with GST, GST-PC3, GST-p16, or GST-p21 beads (20 μl, carrying about 15 μg of bound protein) for 2 h at 4°C. The beads were washed five times with 20 volumes of NET-N buffer (20 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% nonfat dry milk containing 10 μg of leupeptin per ml, 1 mM PMSF) and then mixed with 1 volume of 2× SDS loading buffer. Bound proteins were analyzed by SDS-PAGE.
Production of cyclins and CDKs in insect cells.
Baculoviruses expressing His-tagged cyclin A, His-tagged cyclin B1, hemagglutinin-tagged cdc2, and hemagglutinin-tagged CDK2 were provided by D. Morgan (25), while cyclin D1 and CDK4 were provided by C. Sherr (49). In the preparation of insect lysates (essentially as described in reference 115), 2.5 × 106 Sf9 cells were infected with the indicated cyclin and/or CDK viruses at a multiplicity of infection of 10. After 40 h, cells were lysed in 0.4 ml of kinase buffer (see below) by five passages through a 26-gauge needle and used for kinase assays. The cell lysates were then cleared of insoluble material by two centrifugations at 10,000 × g and stored at −80°C or directly used for kinase assays.
Retroviral infections.
High-titered retroviral supernatants (about 1 × 106 to 5 × 106 virus/ml) were generated by transient transfection with calcium phosphate of either pBABE puro vector as a control or pBABE puro-PC3, in the helper-free packaging cell line BOSC23, according to a procedure described elsewhere (80). The supernatants were then used to infect NIH 3T3 cells according to a protocol described elsewhere (111). Briefly, cell cultures (4 × 105 cells for each 90-mm-diameter dish) were infected for 5 h and then replated and exposed for 48 h to puromycin (2 μg/ml). This procedure allowed us to obtain a pure culture of cells expressing the retroviral constructs, given that all noninfected cells detached from the plate. The cultures at the moment of harvesting were subconfluent. Cells were divided into aliquots, either in lysis buffer for kinase assays (used immediately; see below) and Western blotting or in PBS for cell cycle profile analysis (used after fixation).
Kinase assays.
Kinase assays were performed basically as described by Toyoshima and Hunter (109). For the assays of CDK activities in vitro using the baculovirus system, to the lysate of Sf9 cells (in kinase buffer: 50 mM HEPES [pH 7.4], 10 mM MgCl2, 2.5 mM EGTA, 1 mM dithiothreitol [DTT], 10 mM β-glycerophosphate, 0.1 mM Na3VO4, 1 mM NaF, 1 mM PMSF, 10 μg of leupeptin per ml, 5 μg of aprotinin per ml) coinfected with CDKs and cyclins (2 to 16 μl) was added either GST-PC3, GST-p21, or GST-p16 (5 to 1,600 ng, as indicated), and the mixtures were incubated at 30°C for 20 min. Reactions were started by adding 250 ng of GST-Rb (769–921) fusion protein (Santa Cruz Biotechnology) as substrate, 25 μM ATP, and 5 μCi of [γ-32P]ATP (6,000 Ci/mmol; Amersham), and reaction mixtures were incubated for 10 min at 30°C. Differences in the volumes of baculovirus lysates were compensated for by addition of wild-type baculovirus lysate, in order to attain a final reaction volume of 20 μl. Reactions were then terminated by adding 200 μl of stop buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 20 mM EDTA, 1 mM EGTA, 10% glycerol), also containing glutathione-Sepharose beads (Pharmacia) in order to recover GST-pRb, and incubating the mixture for 1 h at 4°C. The GST-Rb protein bound to glutathione-Sepharose beads was then washed twice in stop buffer, eluted by addition of sample buffer, and analyzed by SDS–10% PAGE. 32P-labeled proteins were detected by autoradiography. For analysis of the phosphorylation of PC3 by cyclin A-CDK2, the substrates used were either 800 ng of GST-PC3 S147N, GST-PC3, or GST or 250 ng of GST-Rb.
For the assays of CDK activities in vivo on retrovirus-transduced NIH 3T3 cells, infected cultures were lysed by resuspension in lysis buffer (50 mM HEPES [pH 7.5], 200 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 1 mM DTT, 0.1% Tween 20, 10% glycerol, 0.1 mM sodium orthovanadate, 1 mM NaF, 1 mM PMSF, 10 μg of leupeptin per ml, 5 μg of aprotinin per ml, 10 mM β-glycerophosphate) for 30 min at 4°C, followed by four cycles of 5 s of sonication at low power and clearing by centrifugation at 14,000 rpm for 5 min at 4°C. Supernatants were assayed for protein concentration as described elsewhere (10). Protein samples of 0.5 mg (CDK2 assay) or 2 mg (CDK4 assay) were then precleared with rabbit immunoglobulin G and then immunoprecipitated for 2 to 4 h at 4°C with protein A-Sepharose beads (Amersham Pharmacia Biotech) precoated with saturating amounts of the appropriate antibody (5 μg of either sc-163 or sc-260, respectively; anti-CDK2 or anti-CDK4 from Santa Cruz; 1 h of preincubation at 4°C). Immunoprecipitated proteins on beads were washed twice with 1 ml of lysis buffer and twice with 1 ml of wash buffer (50 mM HEPES [pH 7.5], 1 mM DTT, 10 mM MgCl2, plus the protease inhibitors as described above). The beads were resuspended in 50 μl of kinase buffer (see above) containing 2 μg of GST-pRb (769–921) fusion protein (Santa Cruz Biotechnology, Inc.), 20 μM ATP, and 10 μCi of [γ-32P]ATP (NEN Dupont, Boston, Mass.; 6,000 Ci/mmol). After incubation for 30 min at 30°C, the samples were boiled in 2× Laemmli buffer, separated by SDS-PAGE, and transferred to a nitrocellulose filter. Phosphorylated proteins were visualized and quantitated by densitometry using a Molecular Dynamics 400A PhosphorImager system.
Colony formation assay.
The colony formation assay was used to measure the growth inhibition by the PC3 mutants and was performed on NIH 3T3 cells according to the procedure previously described (72). NIH 3T3 cells (2.3 × 105) were plated into 60-mm-diameter dishes and after 24 h transfected by the Lipofectamine procedure with pSCT-PC3 (3.8 μg), either wild type or mutated, or with the empty vector pSCT (3.8 μg), together with the vector carrying the neomycin resistance gene (pcDNA3; 0.5 μg). After 48 h, two aliquots of each culture were split into 90-mm-diameter dishes (2 × 105 and 1 × 105 cells) and grown in medium containing G418 (0.5 mg/ml), to allow resistant cells to form colonies. A third aliquot (6 × 105 cells) was lysed and used for Western blotting, to measure the expression of the PC3 construct used. The cutoff point for colony size was >20 cells/colony. Percentages of growth inhibition were calculated with the formula pi = ci × 100/vi, where ci is the number of colonies in the dish transfected in experiment (i) with the indicated mutant, and vi is the number of colonies in the dish transfected in the same experiment (i) with the empty vector. Statistical analysis was performed on the original raw number of colonies, comparing the ci with the vi values of all the experiments by Student's t test.
RESULTS
PC3 overexpression leads to pRb hypophosphorylation.
We have previously shown that PC3, when overexpressed, inhibits proliferation, leading to an impairment of the G1/S transition, concomitantly with dephosphorylation of pRb (72). Since the growth-inhibitory activity of pRb is regulated by phosphorylation (13, 16), these findings suggested that PC3 exerts its antiproliferative activity by preventing pRb phosphorylation. Given that our observations were for NIH 3T3 cell clones stably expressing exogenous PC3, in which secondary mutational events might have occurred during the selection procedure, we sought to evaluate the effect of PC3 on pRb phosphorylation in transiently transfected cells. Therefore, asynchronously growing NIH 3T3 mouse fibroblasts, which do not express detectable levels of endogenous PC3 (72), were cotransfected with expression vectors for pRb and PC3. As a positive control, the pRb expression construct was alternatively cotransfected with the dominant interfering Ras mutant RasAsn17 (Fig. 1A). This mutant induces disruption of Ras function, resulting in G1 cell cycle arrest and pRb dephosphorylation (30, 81). Transfected cells were harvested 60 h posttransfection, and cell lysates were analyzed for pRb expression and phosphorylation state by Western blotting. We observed that the ectopic expression of pRb alone was detected as a single band of 115,000 in Mr, corresponding to the hyperphosphorylated (inactive) form (13, 16), while coexpression of pRb with PC3 led to the appearance also of the 105,000-Mr band, corresponding to the hypophosphorylated (active) form of pRb (Fig. 1A). In the presence of RasAsn17, pRb was almost totally detected as a 105,000-Mr singlet.
FIG. 1.
Induction of pRb dephosphorylation by PC3 and its reversal by cyclins. (A) Ectopic expression of PC3 leads to dephosphorylation of pRb. NIH 3T3 cells (1.3 × 105) were seeded onto 60-mm-diameter dishes. After 24 h, cells were transfected with the human Rb expression plasmid pCMVpRb (4.5 μg) together with the pSCT (VEC), pSCT-PC3 (PC3), or pRSVRasAsn17 (Ras N17) expression vector (4.5 μg each), as indicated. Control cells without transfected plasmids were also analyzed (NT). After 60 h, cells were lysed in Laemmli buffer and pRb was detected by Western blotting using the G3-245 monoclonal antibody. (B) Reversal by cyclins of the PC3-mediated pRb dephosphorylation. NIH 3T3 cells (1.3 × 105) were seeded onto 60-mm-diameter dishes. After 24 h, cells were transfected with pCMVpRb (4.5 μg) together with expression vectors for PC3, cyclins, or cyclin-dependent kinases (4.5 μg each), as indicated. In transfections where Rb or PC3 expression constructs were absent, a corresponding amount of the empty vectors (4.5 μg of each) was used. Equal amounts of cell lysates were analyzed for pRb and PC3 expression by immunoblotting. Protein loading was verified by β-actin detection.
The phosphorylation state of pRb depends on the activity of the cyclin-CDK complexes, whose activity, in turn, depends on the cyclin levels (50, 66). Therefore, we sought to assess if the effects of PC3 seen on pRb phosphorylation could be influenced by coexpression of cyclins (Fig. 1B). We observed that all the cyclins tested, namely, cyclins D1, E, and A, led to almost complete disappearance of the hypophosphorylated form of pRb elicited by PC3 (Fig. 1B). In contrast, CDK4 and CDK2 alone did not counteract the effect of PC3 on pRb dephosphorylation (Fig. 1B). These results strongly suggest that the PC3-dependent appearance of the hypophosphorylated, active form of pRb could be responsible for the cell cycle impairment by PC3.
PC3 arrests G1/S progression depending on the presence of Rb.
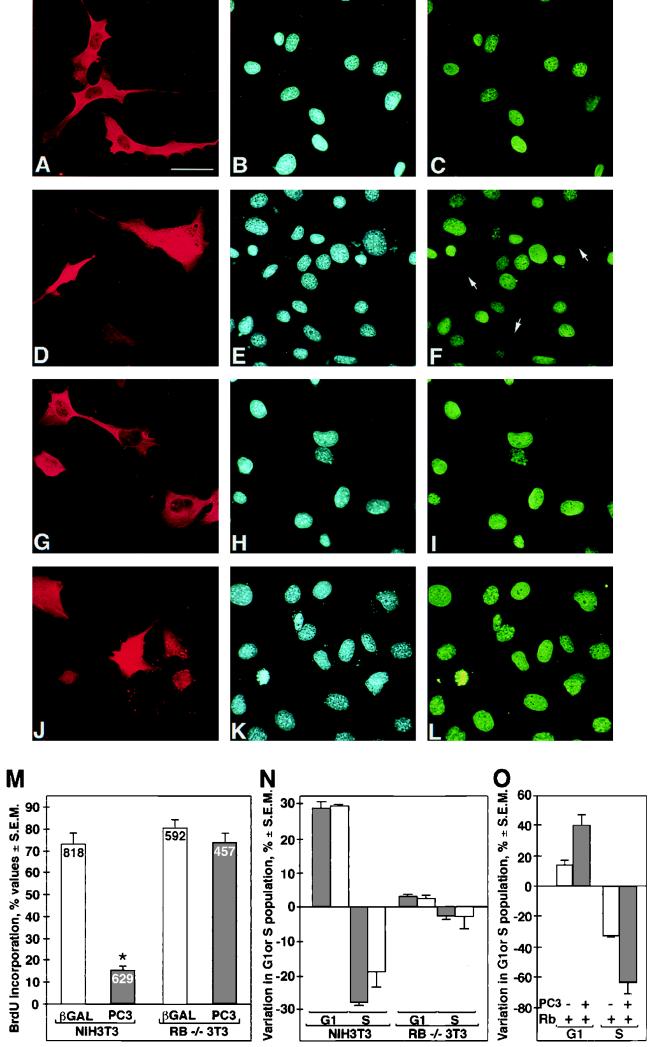
If the mechanism by which PC3 inhibits cell growth is by counteracting pRb phosphorylation, then its ability to induce cell cycle arrest would be lost in cells lacking functional pRb. To ascertain this possibility, we examined the effect of PC3 on cell cycle progression, specifically from the G1 to the S phase, in NIH 3T3 compared to Rb−/− 3T3 cells. These latter cells lack the gene for Rb but remain responsive to signals that restrain proliferation independently from Rb (81). The two cell lines were transiently transfected with PC3 or, alternatively, with a β-Gal expression construct as a negative control, and the DNA synthesis was determined by means of BrdU incorporation. The PC3- or the β-Gal-expressing cells were identified by immunofluorescence staining, either with the anti-PC3 affinity-purified polyclonal antibody A3H (72) or with an anti-β-Gal rabbit polyclonal antibody, while the cells that entered into S phase were identified by staining with an anti-BrdU antibody (Fig. 2A to L, showing a representative experiment). We observed that expression of PC3 led to inhibition of BrdU incorporation in NIH 3T3 cells (Fig. 2D to F), compared to control cultures (Fig. 2A to C). Such inhibition was significant, as clearly indicated by the frequency values for BrdU incorporation (Fig. 2M). In contrast, no significant effect was produced by PC3 on BrdU incorporation in Rb−/− 3T3 cells (Fig. 2J to M). The same result was seen in primary Rb+/+ and Rb−/− MEFs, transiently transfected with PC3 (data not shown). These results indicate that PC3 arrests the progression toward the S phase in an Rb-dependent manner.
FIG. 2.
Rb-dependent inhibition of S-phase entry by PC3. (A to L) Representative immunofluorescence photomicrographs of BrdU incorporation in NIH 3T3 (A to F) and Rb−/− (G to L) cells transfected with PC3. NIH 3T3 and Rb−/− 3T3 cells (0.8 × 105) were seeded onto coverslips in 35-mm-diameter dishes. After 24 h, cells were transfected with the expression vector pSCT-β-Gal or pSCT-PC3 (1.5 μg each). DNA synthesis assays were performed by adding 50 μM BrdU to the culture medium 40 h after transfection. After 18 to 20 h, cells were fixed, permeabilized, and stained. β-Gal and PC3 proteins were revealed using anti-β-Gal (A and G) or anti-PC3 (i.e., A3H [D and J]) polyclonal antibodies followed by incubation with goat anti-rabbit TRITC-conjugated antibody. BrdU was visualized by anti-BrdU monoclonal antibody (corresponding photomicrographs C, F, I, and L) followed by goat anti-mouse FITC-conjugated antibody. Nuclei were detected by Hoechst 33258 dye (corresponding photomicrographs B, E, H, and K). Arrows indicate the positions of nuclei that did not incorporate BrdU. Bar, 30 μm. (M) Percentage of BrdU-incorporating cells (NIH 3T3 or Rb−/− 3T3, as indicated) after transfection with pSCT-PC3 (filled bars) or control pSCT-β-Gal (open bars). Values are calculated as the percentages of cells positive for BrdU, detected between cells positive for β-Gal and those positive for PC3, whose total number within each experiment was assumed to be 100%. Means ± SEM are from three independent experiments (a representative field is shown in A to L). The number of cells counted for each group is indicated at the top of each bar. ∗, P = 0.0000 versus any other group (Student's t test). (N) Flow cytometry analysis of PC3 (filled bars) or p16 (open bars) effects on cell cycle profile in NIH 3T3 and Rb−/− 3T3 cells. NIH 3T3 or Rb−/− 3T3 cells (3 × 105) were seeded onto 90-mm-diameter culture dishes; after 24 h, cells were transfected either with the pSCT empty plasmid (8.5 μg), with pSCT-PC3 (8.5 μg), or with pXp16 plasmid (8.5 μg), together with a plasmid encoding the CD20 cell surface marker (pCMVCD20, 3 μg). After 60 h, transfected cells were identified by staining with an FITC-conjugated anti-CD20 antibody, and their cell cycle distribution was measured by analyzing the DNA content after staining with propidium iodide, using two-color flow cytometry. Data from three independent experiments are shown as means ± SEM of the changes in the percentages of cells in G0/G1 or S cycle phase, compared to the corresponding value of the control transfection with the empty vector pSCT. (O) Effects of PC3 on cell cycle profile of Rb−/− 3T3 cells upon readdition of Rb. Rb−/− 3T3 cells were transfected with pCMVpRb (0.5 μg) or its empty vector. To each of these two treatments was added either pSCT-PC3 (7.5 μg, filled bars) or the pSCT empty plasmid (7.5 μg, open bars). The plasmid encoding the CD20 cell surface marker (pCMVCD20, 3 μg) was used in all transfections. Shown are the changes in the percentages of cells in G0/G1 or S cycle phase induced by addition of pRb (compared for each group to the corresponding value of the control transfection in the absence of pRb), with or without PC3 as indicated. Data are means ± SEM from three independent experiments.
As a further analysis of this point, we sought to measure the cell cycle profile of NIH 3T3 and Rb−/− 3T3 cells expressing ectopic PC3. To this end, we cotransfected the pSCT-PC3 expression construct with the cell surface marker CD20 (105) cloned into an expression vector (122) and analyzed the cell cycle profile of transfected cells by means of two-color flow cytometry (Fig. 2N). The expression of PC3 in NIH 3T3 cells induced a significant percent increase of the cell population in the G1 phase, accompanied by a complementary decrease of the S phase, while no significant effect was seen in the PC3-expressing Rb−/− 3T3 cells (Fig. 2N, filled bars). No evident changes were observed in the G2/M-phase cell population (data not shown). Furthermore, ectopic expression of the pRb-dependent CDK inhibitor p16INK4a gave the same effects on the cell cycle profile as seen with PC3, i.e., a significant increase of G0/G1 and decrease of S phase only in NIH 3T3 cells (Fig. 2N, open bars). Thus, PC3, similar to p16INK4a, caused an Rb-dependent impairment of G1-S transition. Our Rb−/− 3T3 cell clone was, however, fully responsive to the CDK inhibitor p27Kip1, transfected as expression vector pCMX-p27 ([26.2 ± 0.53]% increase of cells in G0/G1 phase and [−16.8 ± 2.97]% decrease of cells in S phase, expressed as changes in percentage ± standard errors of the mean [SEM]). Given that p27Kip1 acts through a pRb-independent pathway (81), this confirmed that the clone did not undergo mutational changes in the course of our experimentation, remaining responsive to Rb-independent stimuli. We also checked whether in Rb−/− 3T3 cells the inhibitory effect of PC3 on G1-S progression could be reinstated after reintroduction of pRb by transfection (Fig. 2O). In fact, the percent changes in the cell populations in G1 or S phases induced by PC3 cotransfected with exogenous pRb (filled bars, Fig. 2O) attained about the same level seen in NIH 3T3 cells transfected with PC3 alone, if the basal effect of exogenous pRb is subtracted (open bars, Fig. 2O).
Cyclin D1 expression reverses the PC3-induced cell cycle block.
As a whole, the above results indicate that PC3 impairs G1-to-S-phase progression by means of an active pRb, also given the ability of cyclin-CDKs to reverse the PC3-induced dephosphorylation of pRb (Fig. 1B).
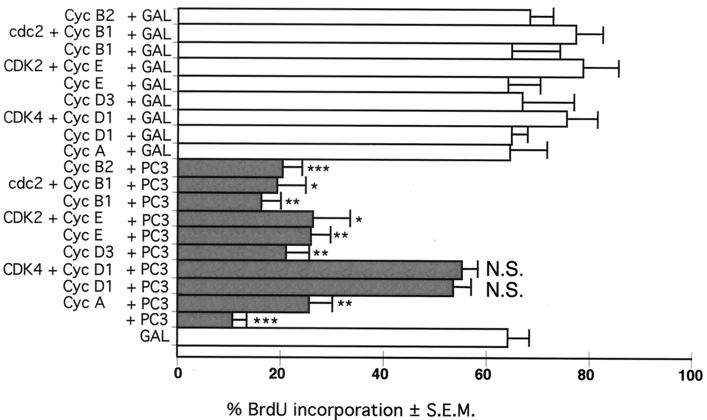
To further elucidate this aspect of the mechanism by which PC3 blocks cell cycle progression, we examined whether overexpression of cyclins could overcome the inhibitory effect of PC3 on G1/S progression. To this aim, NIH 3T3 cells were transiently transfected with PC3 in either the presence or the absence of cyclins and CDKs, and the entry of cells into S phase was monitored by means of BrdU incorporation (Fig. 3). While cyclins A, D3, E, B1, and B2 only partially counteracted the impairment of DNA synthesis elicited by PC3 (with an increase of BrdU incorporation ranging from 16 up to 25 to 40% of the basal level), cyclin D1 led to a significant (80%) recovery of the basal level (86% when cyclin D1 was coexpressed with CDK4). These data point to cyclin D1 as an essential component in the pathway(s) responsible for the PC3 inhibitory activity on the cell cycle. Generally, coexpression of CDKs with cyclins and PC3 led to a recovery of BrdU incorporation that was very similar to that brought about by cyclins alone. This is consistent with the previous observation that CDK2 and CDK4 alone did not modify the PC3-dependent hypophosphorylation of pRb (Fig. 1B).
FIG. 3.
Expression of cyclin D1 rescues the PC3-dependent G1 arrest. NIH 3T3 cells (0.8 × 105) were seeded onto 35-mm-diameter dishes. After 24 h, cells were transfected with the expression vector pSCT-PC3 (PC3, 0.4 μg, filled bars) or pSCT-β-Gal (GAL, 0.4 μg, open bars), together with the indicated cyclins (0.8 μg) and CDKs (0.8 μg). In transfections where the CDK or the cyclin was absent, a corresponding amount (0.8 μg) of the empty CMV vector was cotransfected. Detection of transfected cells expressing PC3 or β-Gal and analysis of their DNA synthesis by measuring BrdU incorporation were performed as described in the Fig. 2A legend. At least 90 cells were scored for each experiment. The results are means ± SEM of at least three independent experiments. ∗∗∗, P = 0.0000 versus GAL; ∗∗, P < 0.0001 versus GAL; ∗, P < 0.001 versus GAL; N.S., P > 0.05 versus GAL (Student's t test).
The ectopic expression of PC3 down-regulates cyclin D1 levels.
The observation that cyclin D1 was able to rescue the cell-growth-inhibitory effect of PC3, taken together with the well-established requirement for cyclin D1 in G1 progression (7), suggested that the cell cycle block imposed by PC3 could be consequent to a reduction of cyclin D1 levels.
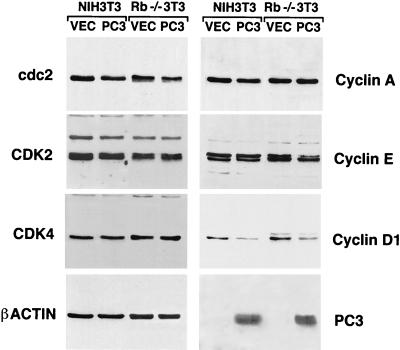
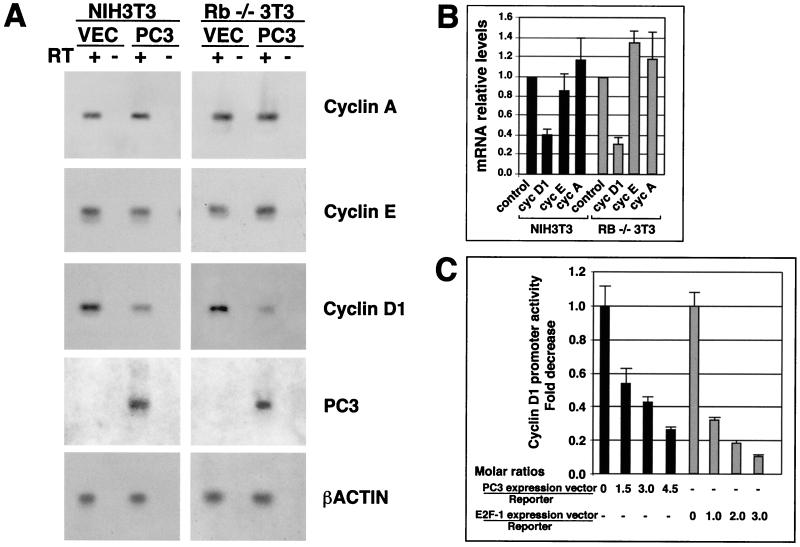
Thus, we wished to evaluate the effects of ectopic PC3 on the endogenous levels of cyclins and CDKs, by immunoblotting. Given that the cells which took up and expressed the transiently transfected PC3 were not more than 10 to 20% in our experimental conditions, to improve the detection of protein levels the population of cells successfully transfected was enriched up to 90% by flow cytometry, using the cotransfected CD20 antigen as a marker protein. It turned out that cyclin D1 indeed was reduced by PC3 expression, in both NIH 3T3 and Rb−/− 3T3 cells (Fig. 4), about threefold, as judged by densitometry scanning. The other cyclins and CDKs analyzed did not show significant changes in their levels, except for a slight reduction of cyclin E in Rb−/− 3T3 cells. As expected, the levels of the different cyclins in control transfections were similar for both NIH 3T3 and Rb−/− 3T3 cells (43, 61).
FIG. 4.
Inhibition of cyclin D1 expression by PC3 in NIH 3T3 and Rb−/− cells. NIH 3T3 and Rb−/− 3T3 cells (3 × 105) were seeded onto 90-mm-diameter culture dishes. After 24 h, cells were transfected with either pSCT empty plasmid (VEC; 21 μg for each dish, total of seven dishes for each cell line) or pSCT-PC3 (21 μg for each dish, total of seven dishes for each cell line), together with a plasmid encoding the CD20 cell surface marker (pCMVCD20, 3 μg). Sixty hours after, extracts from transfected cells isolated by CD20-specific cell sorting were subjected to immunoblotting with antibodies specific for the cyclin and CDK proteins indicated.
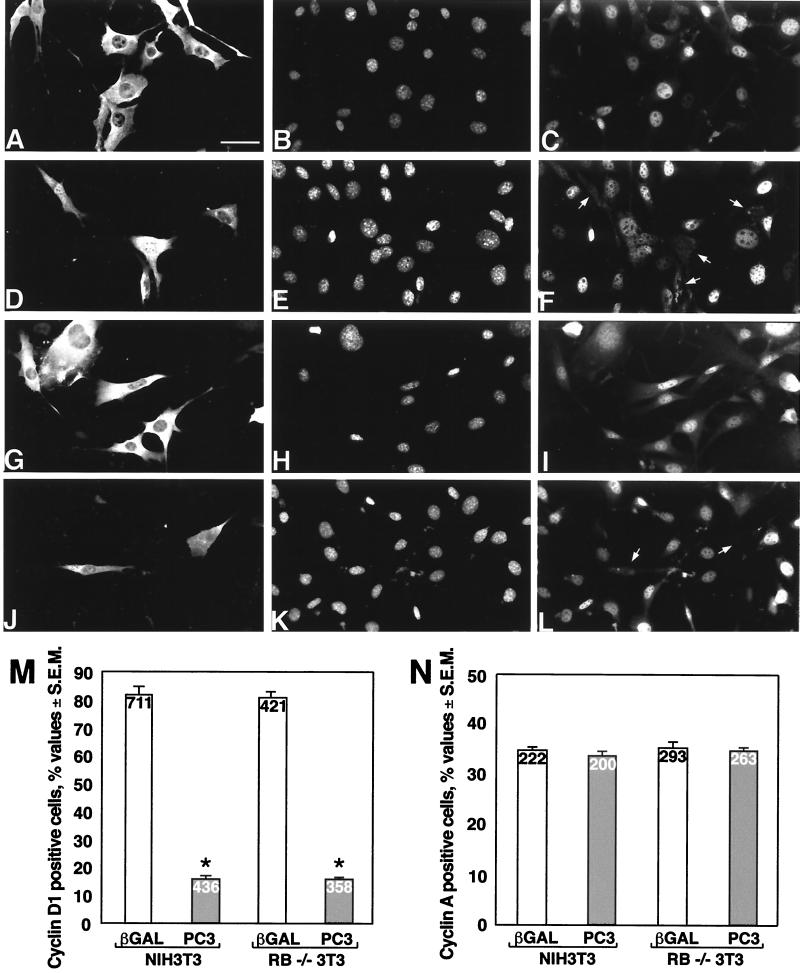
As an independent assessment, we verified by immunofluorescence staining the expression of cyclin D1 and cyclin A in cells transfected with pSCT-PC3 or with the control vector pSCT-β-Gal (Fig. 5A to L). Cells expressing ectopic PC3 were detected by using the anti-PC3 antibody A3H (72). Again, it was observed that in both NIH 3T3 and Rb−/− 3T3 cells expressing ectopic PC3 the cyclin D1 nuclear immunostaining was detectable at a frequency significantly lower (fivefold) than that in cells expressing ectopic β-Gal (Fig. 5A to L and M, showing the frequency values for cyclin D1 nuclear staining). On the other hand, cyclin A expression in both NIH 3T3 and Rb−/− 3T3 cells transfected with PC3 remained the same as in control cultures transfected with β-Gal (Fig. 5N).
FIG. 5.
Inhibition of cyclin D1 nuclear immunofluorescence staining by ectopic PC3 in NIH 3T3 and Rb−/− cells. (A to L) Representative immunofluorescence photomicrographs of cyclin D1 expression in NIH 3T3 (A to F) and Rb−/− 3T3 (G to L) cells transfected with PC3 (or control β-Gal). NIH 3T3 or Rb−/− 3T3 cells (0.8 × 105) were seeded onto coverslips in 35-mm-diameter dishes and transfected with the expression vector pSCT-β-Gal or pSCT-PC3 (1.5 μg each). After 60 h, cells were fixed, permeabilized, and stained. β-Gal and PC3 proteins were detected with anti-β-Gal (A and G) or anti-PC3 (D and J) polyclonal antibodies, followed by goat anti-rabbit TRITC-conjugated antibody. Nuclei were stained by Hoechst 33258 dye (corresponding photomicrographs B, H, E, and K). Cyclin D1 was visualized by anti-cyclin D1 mouse monoclonal antibody, followed by goat anti-mouse FITC-conjugated antibody (corresponding photomicrographs C, I, F, and L). Arrows indicate the positions of nuclei negative for cyclin D1 staining. Bar, 40 μm. (M) Percentage of NIH 3T3 and Rb−/− 3T3 cells positive for cyclin D1 immunofluorescence staining after transfection with pSCT-β-Gal (open bars) or pSCT-PC3 (filled bars). Values are calculated as the percentages of cells positive for cyclin D1 nuclear staining, detected between cells positive for β-Gal and those positive for PC3, whose total number within each experiment was assumed to be 100%. Means ± SEM of three independent experiments, performed as described above for panels A to L, which are a representative field, are shown. ∗, P = 0.0000 versus the corresponding control (Student's t test). The number of cells counted for each group is indicated at the top of each bar. (N) Percentage of cyclin A-positive cells by immunofluorescence staining after transfection with pSCT-β-Gal (open bars) or pSCT-PC3 (filled bars). Transfection and detection of PC3 and β-Gal were performed as described for panels A to L. However, in order to distinguish the reactivity to the rabbit polyclonal anti-cyclin A antibody from that to either anti-PC3 (A3H) or anti-β-Gal (all rabbit polyclonal antibodies), cells were incubated first with anti-cyclin A and then with a mouse anti-rabbit antibody, washed, and fixed. Incubation with A3H (or anti-β-Gal) followed. Anti-cyclin A and anti-PC3 (or anti-β-Gal) antibodies were detected by goat anti-mouse FITC-conjugated and goat anti-rabbit TRITC-conjugated antibodies, respectively. Values are the means ± SEM of three independent experiments. The number of cells counted for each group is indicated at the top of each bar.
PC3 down-regulates the transcription of the cyclin D1 gene.
The above findings raised the question whether the reduction of cyclin D1 protein levels elicited by PC3 was a consequence of down-regulation of cyclin D1 transcription. To this aim, we analyzed the cyclin D1, A, and E mRNA levels in PC3-expressing cells by semiquantitative RT-PCR. Cells expressing exogenous PC3 were enriched as previously indicated by selecting the cell population expressing the CD20 marker cotransfected with PC3. We observed that cyclin D1 mRNA levels were significantly reduced by PC3, about 2.5- and 3-fold, compared to the levels of NIH 3T3 and Rb−/− 3T3 control cells, respectively (Fig. 6A and B). The mRNA levels of cyclin E appeared to be not significantly decreased in NIH 3T3 cells and to be slightly increased by PC3 in Rb−/− 3T3 cells (1.4-fold), whereas those of cyclin A were not significantly increased in both cell types (1.2-fold [Fig. 6A and B]). Thus, to assess the existence of transcriptional regulation by PC3, we analyzed the effect of PC3 on the activity of the cloned cyclin D1 promoter, transiently transfected into NIH 3T3 cells. We used the construct prCD1-1810, which contains 1,810 nucleotides 5′ to the transcription start in front of the luciferase reporter gene in the vector pGL2 (see reference 117). The activity of the cyclin D1 promoter in cells cotransfected with PC3 was compared to that of cells cotransfected with the empty vector. As a control of the efficiency of transfection, we measured the amount of plasmid DNA present in each cell extract by dot blot hybridization, according to a previously described procedure (1). We observed that PC3 reduced the activity of cyclin D1 promoter up to threefold, with a concentration-dependent effect (Fig. 6C). A similar effect on the cyclin D1 promoter was also observed for E2F-1, known to inhibit the cyclin D1 promoter (113), used as an internal experimental control.
FIG. 6.
Inhibition of cyclin D1 transcription by PC3. (A) Inhibition of cyclin D1 mRNA levels in NIH 3T3 and Rb−/− 3T3 cells by PC3. Cells (3 × 105) were seeded onto 90-mm-diameter culture dishes and transfected with either pSCT empty plasmid or pSCT-PC3 (21 μg each) together with a plasmid encoding the CD20 cell surface marker (pCMVCD20, 3 μg), as described for Fig. 4. Sixty hours after, cells were isolated by CD20-specific cell sorting (obtaining 3 × 105 to 5 × 105 cells), and total RNA was extracted. The specific mRNA species indicated were visualized by RT-PCR analysis using specific primers. Equal amounts of RT-PCR products amplified from NIH 3T3 or Rb−/− 3T3 sorted cells, transfected with either pSCT-PC3 or the empty vector, were electrophoresed, blotted on a filter, and hybridized to probes for cyclins A, E, and D1; PC3; and β-actin. RT “+” or “−” indicates the products of amplification performed in parallel on two aliquots of each RNA starting sample preincubated or not with RT, respectively, in order to check the presence of DNA contamination. Control amplifications using as template the cDNA corresponding to each mRNA species gave a signal of the expected size (data not shown). (B) Relative levels of the mRNAs, as means ± SEM of three independent experiments, of which a representative one is shown in panel A. Values were obtained by measuring Southern blot densities of the PCR product of each experiment with a PhosphorImager system and were represented as ratios of the density observed in pC3-transfected cells to the corresponding one in pCST-transfected cells (assumed to be 1; see control bars). Values were then corrected for the corresponding β-actin relative expression, according to the following formula: relative sample density = sample density in PC3-transfected cells × 100/sample density in vector-transfected cells/(β-actin density in PC3-transfected cells × 100/β-actin density in vector-transfected cells). Black bars, NIH 3T3 cells; grey bars, Rb−/− 3T3 cells. (C) Inhibitory effect of PC3 on cyclin D1 promoter activity. NIH 3T3 cells (105) seeded onto 35-mm-diameter culture dishes were transfected after 24 h with either pSCT-PC3 or CMV–E2F-1 or the corresponding empty plasmids. Forty-eight hours after transfections, cell lysates were collected and assayed for luciferase activity. The fold decrease in luciferase activity was calculated relative to the level of control samples (transfected with the empty vectors), which were set to the unit. The bars represent the average fold activities ± SEM of three independent experiments performed in duplicate. The luciferase activities were measured in luciferase units per microgram of protein normalized to the amount of plasmid DNA present in each extract (black or grey bars). The ratio of transfected expression vector to reporter plasmid is shown on the abscissa. The amount of reporter used was 0.5 μg, while the highest amount of pSCT-PC3 and CMV–E2F-1 was 1.5 μg (corresponding to a molar ratio of expression vector to reporter of 4.5 and 3.0, respectively).
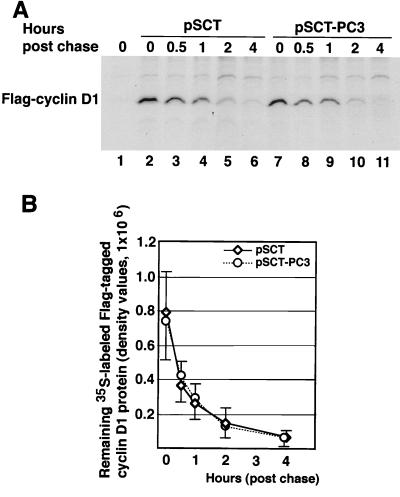
A further analysis was performed to verify whether PC3, in addition to its effects on cyclin D1 transcription, could also affect the stability of cyclin D1 protein. To this aim, we measured the half-life of Flag-tagged cyclin D1, cotransfected in NIH 3T3 cells with either pSCT-PC3 or pSCT control vector. Cells were metabolically labeled with [35S]methionine, and the lysates were immunoprecipitated with an antibody to Flag (Fig. 7A). We observed that PC3 did not produce significant differences in the turnover kinetics of Flag-tagged cyclin D1 protein, given its half-life of 23.3 ± 5 and 21.5 ± 4 min in the absence and the presence of PC3, respectively (average values ± SEM obtained by linear regression analysis of the data of four independent experiments, shown in Fig. 7B).
FIG. 7.
The half-life of Flag-tagged cyclin D1 protein is not changed by PC3. (A) Forty-five hours after cotransfection of NIH 3T3 cultures (2.3 × 105 cells in 60-mm-diameter dishes) with Flag-tagged cyclin D1 (2.15 μg) and either pSCT-β-Gal or pSCT-PC3 (2.15 μg each), cells were metabolically labeled for 2 h with [35S]methionine. Cells were washed with medium containing an excess of unlabeled methionine, collected, and lysed at the indicated times. Cell lysates containing equal amounts of proteins were then immunoprecipitated using the M2 monoclonal antibody against the Flag epitope. A control transfected with pSCT vector without Flag-tagged cyclin D1 is shown in lane 1. Shown are results of a representative experiment. (B) Graphic representation of Flag-tagged cyclin D1 expression. The data at individual time points are the amounts of 35S-labeled Flag-tagged cyclin D1 protein as measured by a PhosphorImager system and are the means ± SEM of four independent experiments. The half-lives of Flag-tagged cyclin D1 protein were calculated for each experiment by linear regression analysis of the density values at the different time points, transformed by common logarithm.
Cell cycle blocking activity of PC3 mutants.
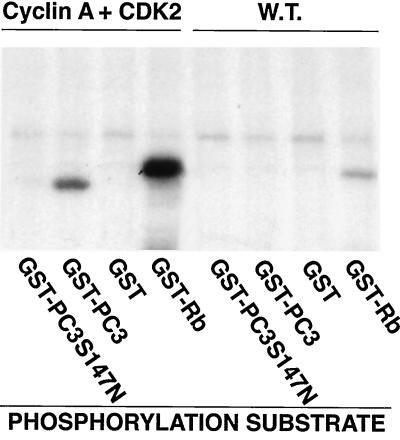
A comparison of the protein sequences of the PC3/BTG/Tob gene family shows the existence of conserved regions with higher homology. By comparing, through the algorithm Align, the protein sequences of rat PC3 (72), human PC3 (whose cDNA, isolated by us with EMBL accession no. Y09943, corresponds to BTG2 [see reference 92]), Tob (64), and BTG1 (94), we identified two conserved regions that correspond, in the PC3 protein, to residues 50 to 68 and 105 to 123 (Fig. 8A). We reasoned that such regions, given the common ability of these genes to inhibit proliferation, might play a role in that effect. To analyze this possibility, we produced two PC3 mutants with an internal deletion, comprising either residues 50 to 68 or 105 to 123 (pSCT-PC3 Δ50–68 and pSCT-PC3 Δ105–123, respectively). Furthermore, the PC3 protein contains a sequence motif known as the consensus site for phosphorylation by cdc2 and/or CDK2 (46). Therefore, we produced a third mutant, pSCT-PC3 S147N, whose serine 147, which belongs to the phosphorylation motif mentioned, was replaced with asparagine (Fig. 8A). We found that cyclin A-CDK2, expressed in baculovirus, was able to phosphorylate the wild-type PC3 molecule but did not phosphorylate the mutant pSCT-PC3 S147N (Fig. 9), indicating that indeed PC3 is phosphorylated by cyclin A-CDK2 at the consensus aa 147 (while PC3 did not appear to be a substrate either of cyclin B1-cdc2 or of cyclin D1-CDK4 [data not shown]).
FIG. 8.
Effects of the ectopic expression of wild-type and mutant PC3 constructs on cell growth arrest and cyclin D1 expression. (A) Schematic representation of PC3 mutants; the hatched boxes inside the PC3 sequence represent the regions conserved among the different members of the PC3 family. (B, C, and D) NIH 3T3 cells were transfected with the indicated pSCT-PC3 construct (either wild type or mutant; 3.8 μg) or with the empty vector pSCT (3.8 μg). The vector carrying the neomycin resistance gene (pcDNA3, 0.5 μg) was included in each transfection. The transfected cultures were then split into three fractions 48 h after transfection, two for the colony formation assay (2 × 105 and 1 × 105 cells [B]) and a second (6 × 105 cells) for protein expression analysis by Western blotting (C and D). (B) For the colony formation assay, the colonies resistant to G418 after 2 weeks of selection, arising from each transfected construct, were counted and expressed as percentages of the number of resistant colonies formed by transfection of the empty vector. Calculations are means ± SEM from four independent experiments. VEC, vector-cotransfected cells, considered 100% of colony formation. ∗, P < 0.05 versus VEC control group (Student's t test); ∗∗, P < 0.01 versus VEC control group (Student's t test). (C and D) Equal amounts of cell lysates were used for Western blot analysis; (C) representative experiment analyzing the PC3 and β-actin protein levels; (D) means ± SEM of protein expression levels as judged by densitometry analysis of the four independent experiments, after normalization to the corresponding β-actin expression level (unity = the expression of PC3 wild-type protein for each experiment). (E) Immunofluorescence photomicrographs showing NIH 3T3 cells expressing either wild-type or mutated PC3, as indicated. Detection was done by the anti-PC3 antibody. The lower panels show nuclear staining, using Hoechst 33258 dye. Bar, 25 μm. (F) Percentage of NIH 3T3 cells positive for cyclin D1 immunofluorescence staining after transfection with pSCT-β-Gal, pSCT-PC3, pSCT-PC3 Δ50–68, pSCT-PC3 Δ105–123, or pSCT-S147N mutants (1.5 μg each). Transfections, as well as detection of proteins (PC3 [wild type and mutated], β-Gal, and cyclin D1), were performed as described for Fig. 5A to L. Values are the means ± SEM of four independent experiments. ∗, P < 0.05 versus β-Gal control group (Student's t test); N.S., P > 0.05 versus β-Gal control group (Student's t test). The number of cells counted for each group is indicated at the top of each bar. w.t. and W.T., wild type.
FIG. 9.
Phosphorylation of PC3 at aa 147 by cyclin A-CDK2. Lysates of Sf9 cells coinfected with either CDK2 and cyclin A or wild-type baculovirus lysates were assayed in 20-μl reaction mixtures for GST-PC3 S147N, GST-PC3, GST, or GST-Rb phosphorylation by measuring incorporation of 32P. Samples were analyzed by SDS-PAGE. Shown is the autoradiograph from the area of the gel containing the substrate proteins. W.T., wild type.
The ability of the PC3 mutants to inhibit cell proliferation was then analyzed by colony formation assay. This revealed that growth inhibition was lost for the Δ50–68 mutant, which even presented a slight paradoxical stimulatory effect, whereas for the Δ105–123 mutant the ability to inhibit growth, although still significant, was severely reduced. The mutation of aa 147 led only to a slight impairment of the growth inhibition of PC3 (Fig. 8B). In parallel with the colony formation assays, the expression of the PC3 mutated proteins was analyzed by Western blotting and was shown to be equivalent to that of the wild-type PC3, indicating that the effects on growth were not due to differences in the expression of the PC3 mutants (Fig. 8C and statistical analysis of the expression in Fig. 8D). The intracellular localization of the mutated PC3 proteins did not differ from that of wild-type PC3, being apparently cytoplasmic, as judged from immunostaining with the antibody A3H (Fig. 8E).
We have previously observed that ectopic expression of PC3 concomitantly induces down-regulation of cyclin D1 levels and G1 arrest, suggesting that these two events are correlated. To verify this possibility, we checked the ability of the PC3 mutants analyzed for their effects on growth to affect cyclin D1 levels. To this aim, the expression of cyclin D1 was assessed by immunofluorescence staining in cells transfected either with pSCT-PC3, pSCT-PC3 Δ50–68, pSCT-PC3 Δ105–123, or pSCT-PC3 S147N or with the control vector pSCT-β-Gal (Fig. 8F). It was shown that the mutants negatively affected the frequency of cyclin D1 expression in the following order of potency: PC3 wild type > PC3 S147N > PC3 Δ105–123 > PC3 Δ50–68 (Fig. 8F).
As a whole, these data suggested the existence of a correlation between the inhibition of proliferation and the reduction of cyclin D1 expression elicited by PC3.
Exclusivity of the cyclin D1 pathway for PC3.
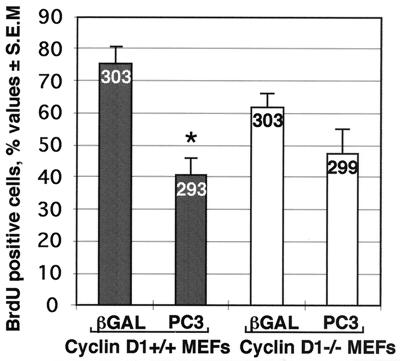
A further question raised by these findings, pointing to a negative regulatory control of cyclin D1 levels exerted by PC3 in correlation with inhibition of G1-S progression, concerned the exclusivity of such control, in regard to the possible involvement of other cell cycle pathways different from cyclin D1. To this aim, we verified the ability of PC3 to inhibit S-phase progression in primary MEF cells explanted from an animal ablated of the cyclin D1 gene, by measuring BrdU incorporation. Cyclin D1+/+ and cyclin D1−/− MEF cells, transiently transfected with the PC3 or the control β-Gal expression constructs, were identified for their expression by immunofluorescence staining with the anti-PC3 or anti-β-Gal polyclonal antibodies and monitored for BrdU incorporation by double labeling with the BrdU monoclonal antibody (Fig. 10). We observed that expression of PC3 led to inhibition of BrdU incorporation in both cell types, although to different extents, i.e., about 45% inhibition in cyclin D1+/+ cells and 23% inhibition in cyclin D1−/− cells, compared to control cultures, being statistically significant only in the former cell type (Fig. 10).
FIG. 10.
Assessment of the inhibition of G1-S progression by PC3 in cyclin D1−/− cells. About 0.8 × 105 cyclin D1+/+ and cyclin D1−/− MEF cells were seeded onto coverslips in 35-mm-diameter dishes and transfected after 24 h with the expression vector pSCT-β-Gal or pSCT-PC3 (1.5 μg each). DNA synthesis assays were performed by adding 50 μM BrdU to the culture medium 36 h after transfection. After 24 h, cells were fixed, permeabilized, and stained. β-Gal and PC3 proteins were revealed with the polyclonal antibodies anti-β-Gal and anti-PC3, respectively, followed by goat anti-rabbit TRITC-conjugated antibody, whereas BrdU was visualized by anti-BrdU monoclonal antibody followed by goat anti-mouse FITC-conjugated antibody, as described for Fig. 2. The percentages of BrdU-incorporating cells shown are means ± SEM of three independent experiments. The number of cells counted for each group is indicated at the top of each bar. ∗, P < 0.001 versus control group (Student's t test).
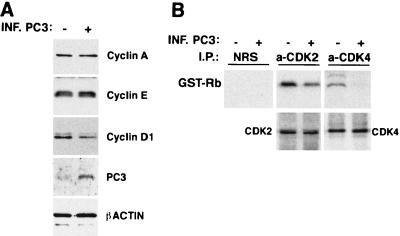
PC3 indirectly inhibits CDK2 and CDK4 activity.
The strong impairment of cyclin D1 levels by PC3 can by itself fully account for the pRb-dependent cell cycle arrest induced by PC3, given that phosphorylation of pRb by cyclin D1-CDK4 complexes is the prerequisite for G1 progression (for reviews, see references 70, 101, and 102). We wished to verify this point by analyzing the effect of PC3 on the activity in vivo of CDK4, and CDK2 as well, in NIH 3T3 cells. The generation of a large population of pure PC3-expressing cells, necessary for the kinase assays, was obtained by expressing PC3 through retroviral infection. The retroviral vector pBABE puro, in which the complete coding region of PC3 cDNA was cloned, was used to generate the high-titered retroviral supernatants employed to infect NIH 3T3 cultures, according to a procedure described elsewhere (80). These cultures offered an additional system in which to verify our previous findings. In fact, in the course of our analyses we observed that cell cultures infected with the PC3 retrovirus presented a high expression of PC3 concomitant with a reduced expression of cyclin D1 protein and mRNA, accompanied by an increase of the cell population in G1 phase and a decrease of cells in S phase (Fig. 11A and data not shown), as already seen in PC3-expressing cells sorted by flow cytometry from transiently transfected cultures. In cultures infected with the PC3 retrovirus, we then observed a decrease, with respect to the control group, of both CDK4- and CDK2-mediated Rb kinase activities, the effect on CDK2 being less evident but well reproducible (Fig. 11B).
FIG. 11.
Effect of PC3 on the protein kinase activities of cyclins-CDKs in vivo. (A) Characterization by Western blotting of NIH 3T3 cultures infected with retrovirus carrying the PC3 coding region or with empty vector (from supernatants of BOSC23 cells transfected with the pBABE puro-PC3 or pBABE puro vector, respectively). Equal amounts of proteins were loaded. (B) In vivo activity of CDK2 and CDK4 in NIH 3T3 cells infected with the PC3 retrovirus or with the empty retrovirus, as indicated. Equal amounts of proteins, from NIH 3T3 lysates of cultures infected with PC3 retrovirus or empty retrovirus, were immunoprecipitated with normal rabbit serum (NRS) or with anti-CDK2 and anti-CDK4 antibodies and then assayed for GST-Rb phosphorylation in 50-μl reaction mixtures by measuring incorporation of 32P. Samples were loaded in SDS-PAGE gels and transferred by electrophoresis to a nitrocellulose filter. This was analyzed for the presence of phosphorylated GST-Rb by a PhosphorImager (upper panels). The immunoprecipitated samples were checked for the presence of equal amounts of CDK2 and CDK4 proteins by Western blot analysis of the nitrocellulose filter (lower panels). INF., infected; I.P., immunoprecipitation.
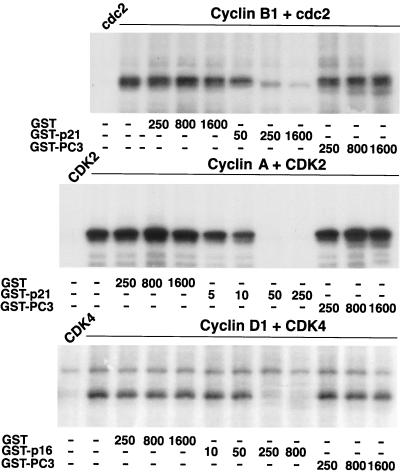
A further analysis that we performed concerned the possibility of a direct inhibition of CDK activities by PC3. This latter, implying a cell cycle arrest by PC3 occurring also independently from its effect on cyclin D1 transcription, was considered on account of the interaction seen between PC3 and CDKs, by means of a GST pull-down analysis using in vitro-translated CDKs. In fact, we found that, while p21CIP1/WAF1 and p16INK4a showed the expected associations with cdc2 and CDK2 (Fig. 12A and B) (see also references 42 and 121) and with CDK4 and CDK6 (Fig. 12C and D) (see also references 40 and 116), respectively, PC3 was shown to be associated with cdc2 to an extent higher than that of p21CIP1/WAF1 (Fig. 12A) and also, although weakly, with CDK4 (Fig. 12C). Thereafter, we tested whether PC3 could inhibit the pRb kinasing activity of different cyclins-CDKs expressed in Sf9 insect cells, using a GST-Rb fusion protein as substrate. Purified GST-PC3 was compared either to purified GST-p21CIP1/WAF1, for the ability to inhibit cyclin B1-cdc2 and cyclin A-CDK2, or to purified GST-p16, for the ability to inhibit cyclin D1-CDK4 (Fig. 13). Purified GST was used as negative control. This choice was in agreement with the observation that p21CIP1/WAF1, although able to inhibit the activity of several CDKs (115), is, however, a more efficient inhibitor of cyclin B-cdc2 and cyclin A- and E-CDK2 activities than of cyclin D-CDK4 (42, 121), while p16INK4a preferentially inhibits CDK4 and CDK6 (86, 99). It turned out that while p21CIP1/WAF1 and p16INK4a dose dependently inhibited the corresponding cyclins-CDKs, PC3 did not show significant effects (Fig. 13). Therefore, under the conditions used, we can rule out a direct inhibitory effect of PC3 on the activity of the CDKs analyzed.
FIG. 12.
In vitro interactions of PC3 with CDKs. Shown is binding of GST, GST-PC3, and GST-p21 or GST-p16 to cdc2 (A), CDK2 (B), CDK4 (C), or CDK6 (D). Equal amounts of [35S]methionine-labeled CDKs (shown in the left lanes of each panel) were incubated with GST-PC3, GST-p21, or GST-p16, as indicated. Bound proteins were eluted and analyzed by SDS-PAGE (6% polyacrylamide for cdc2, 9% polyacrylamide for CDK2 and CDK6, and 12% polyacrylamide for CDK4) and autoradiography. Numbers at left of each panel are molecular masses in kilodaltons.
FIG. 13.
Effect of PC3 on the in vitro protein kinase activities of cyclin B1-cdc2, cyclin A-CDK2, and cyclin D1-CDK4. Lysates of Sf9 cells containing the indicated combination of cyclin-CDK and in the presence of increasing amounts (in nanograms) of GST-p21, GST-p16, GST-PC3, or GST were assayed in 20-μl reaction mixtures for GST-Rb phosphorylation by measuring incorporation of 32P. Samples were analyzed by SDS-PAGE. Shown is the autoradiograph from the area of the gel containing the GST-Rb protein. cdc2, CDK2, and CDK4 denote lysates from Sf9 cells infected with the CDK baculovirus alone as a control.
DISCUSSION
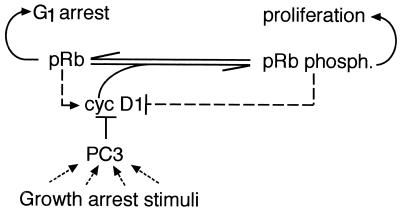
We show in this report that the gene PC3 inhibits S-phase entry in an Rb-dependent manner and that this effect is correlated with its ability to inhibit cyclin D1 expression.
The PC3-mediated arrest of G1/S progression is Rb dependent.
PC3 induces an evident inhibition of cell cycle progression from G1 to S phase, as judged by the severe impairment of DNA synthesis observed in cycling cells expressing ectopic PC3. Accordingly, flow cytometry analysis of cell cultures transfected with PC3 shows that the population of cells in S phase undergoes a significant decrease, while that of cells in G1 shows a parallel increase. The impairment of G1/S transition by PC3 observed in this report confirms and extends our previous observations made with clones stably expressing PC3 (72). Furthermore, our analyses of Rb−/− cells indicate that the inhibition of S-phase entry by PC3 requires the presence of pRb, the key molecule responsible for growth arrest and accumulation of cells in G1 in response to antiproliferative signals (for reviews, see references 44 and 114). In fact, in Rb−/− cells PC3 fails to arrest DNA synthesis, as seen by BrdU incorporation, and to alter the population of cells in S phase, according to the cell cycle profile analysis by flow cytometry. Such findings agree with our observation that the PC3-dependent block of S-phase entry is rescued by coexpression of cyclin D1, whose activity is necessary for G1-to-S progression in an Rb-dependent manner (61, 107).
It is worthwhile to point out that the impairment of G1/S transition is an effect shared by PC3 with another p53-induced gene, p21CIP1/WAF1. Earlier observations indicated that, following DNA damage, MEFs lacking p21CIP1/WAF1 show a partial defect in G1 arrest that is less severe than that of p53-defective fibroblasts, thus making plausible the idea that other p53-dependent G1 arrest pathways exist (24).
Additionally, it has been observed that the G2 arrest that occurs following DNA damage is not detected in embryonic stem cells ablated of PC3/BTG2, thus suggesting that PC3 might also be implicated in the p53-mediated G2 arrest (92). However, it was observed that PC3/BTG2−/− cells presented a marked increase in cell death, and it was not possible to define if the loss of G2/M arrest resulted from the absence of a specific G2 block, ongoing apoptosis, or loss of other factors (92). Nonetheless, a possibility to be considered is that PC3 could inhibit cell cycle progression also at checkpoints different from G1/S, by other, additional Rb-dependent or Rb-independent pathways.
Correlation between down-regulation of cyclin D1 levels and cell cycle impairment by PC3.
The inhibition of the G1-S transition by PC3 is accompanied by down-regulation of cyclin D1 levels. Given the necessity of cyclin D1 for G1 progression (7, 87, 107), the cyclin D1 decrease effected by PC3 might in itself be sufficient to explain the PC3-induced cell cycle arrest. In fact, the formation of an active cyclin D1-CDK4 complex depends on de novo synthesis of D1 protein (66). Furthermore, the impairment in the ability of PC3 to lower cyclin D1 levels, seen in consequence of mutations of the PC3 molecule, correlates with the extent of impairment in growth arrest. This indicates that inhibition of proliferation and down-regulation of cyclin D1 levels by PC3 are events significantly connected. Remarkably, the fact that PC3 significantly inhibits cyclin D1 expression also in cells lacking pRb indicates that such an effect is genuinely dependent on PC3 and is not secondary to other effects consequent to the Rb-dependent cell cycle arrest in G1. In second place, it follows that the signaling pathway between PC3 and cyclin D1 in Rb−/− 3T3 cells is intact.
Conversely, cyclin D1 is able to rescue the PC3-dependent G1 arrest, evidence pointing to cyclin D1 as a target for the PC3 inhibition of the cell cycle. Furthermore, the ability of PC3 to impair cyclin D1 expression, as well as the ability of cyclin D1 to reverse the cell cycle block by PC3, appears to be preferential, since the level of cyclin A is unaffected by PC3, while that of cyclin E is only slightly reduced in 3T3 Rb−/− cells, and the G1 arrest elicited by PC3 is not reversed by these cyclins.
As a whole, this indicates a significant functional correlation between PC3 and cyclin D1, which can be placed upstream of pRb. Given that pRb is the sole critical substrate of CDKs regulated by D-type cyclins, this makes somewhat selective the functional correlation between PC3 and, downstream, pRb.
Nonetheless, it should be considered that the rescue performed by cyclin D1 of the PC3-dependent impairment in S-phase entry did not attain 100% and, more important, that the inhibition by PC3 of G1-S progression is detectable, even though only partially, in cyclin D1−/− MEF cells also. These points, in our view, suggest that the cyclin D1 pathway, although preferential for the PC3-dependent effect on G1-S progression, is not exclusive.
Many pieces of evidence point to the existence of a homeostatic network regulating pRb activity, centered on cyclin D1. Synthesis and accumulation of cyclin D1, triggered by growth factors or by molecules activated along their pathways, e.g., ras (31, 71, 81), lead to inactivation of pRb by CDK4-dependent phosphorylation (for reviews, see references 101 and 114) and thus to the release of molecules that trigger the cell cycle, such as E2F-1 (52, 97). At the end of G1/S transition, a down-regulation of cyclin D1 levels follows, in consequence of at least two events triggered by pRb inactivation, i.e., an increase of free E2F-1 molecules, which leads to repression of cyclin D1 transcription (113), and the increase of p16INK4a, which leads to displacement of cyclin D1 from CDK4 and to its degradation (106). Thus, phosphorylated, inactive pRb exerts negative control on cyclin D1 levels. These remain low until pRb is made active again at the end of mitosis by dephosphorylation. Then, active pRb leads to accumulation of cyclin D1 (76) and, consequently, to CDK4-dependent phosphorylation of pRb, with a negative feedback loop on its activity. Thus, the down-regulation by PC3 of cyclin D1 expression should not only negatively influence cyclin D1-CDK4 activity but also have other effects predictable from the context mentioned above (e.g., on p16INK4a or E2F levels).
Regarding the mechanisms by which PC3 down-regulates cyclin D1 levels, our data clearly show that PC3 negatively influences cyclin D1 transcription and does not affect the stability of cyclin D1 protein. In fact, the decreases of the levels of cyclin D1 transcript and protein caused by PC3 were quantitatively similar, and they were also accompanied by inhibition of cyclin D1 promoter activity, indicating that PC3 can repress cyclin D1 gene transcription, which accounts for the decrease of cyclin D1 protein. Thus, our data suggest that PC3 acts, either directly or indirectly, as a transcriptional regulator. This idea is in agreement with the recent observation that PC3/BTG2 binds the mouse homolog of the yeast protein yCAF1 (93). This gene, together with the yeast cell-cycle-regulated protein kinase DBF2, the NOT proteins, and other yet-unidentified proteins, is part of the yeast transcriptional regulatory complex CCR4, which plays an important general transcriptional role in diverse cellular processes (59, 60). It is interesting that mutations of yeast CCR4, CAF1, and DBF2 lead to a delayed exit from late mitosis (60) and that mutations of yCAF1 are able to suppress defects in DNA repair (98).
The observations of the subcellular localization of PC3 (as determined by immunofluorescence staining of transfected cells, in this report and in reference 72) clearly show its presence in the cytoplasm but also leave open the possibility of a nuclear localization. In the latter case, the participation of PC3 in a transcriptional complex could occur also in the nucleus.
Our data also show that PC3 can associate with CDK4 in vitro. However, assuming that this observation can be reproduced in vivo, it appears unlikely that the binding of PC3 to CDK4 might displace the cyclin D1 protein, thus inducing its degradation (as for instance seen with p16INK4a [106]), since PC3 was unable to directly inhibit pRb phosphorylation by cyclin D1-CDK4 and to influence cyclin D1 protein stability. Rather, we see that PC3 behaves as an indirect CDK inhibitor, as judged from the detection of reduced activity of CDK4 and CDK2 in PC3-expressing cells. While the inhibition of CDK4 may well be consequent to the PC3-dependent reduction of cyclin D1 levels, the effect on CDK2 is less obvious, given that no evident effect of PC3 on the levels of cyclins E and A, cofactors of CDK2, was seen for NIH 3T3 cells. However, it has been shown that the activity of CDK2 can be inhibited by a decrease of cyclin D1-CDK4 complexes. In fact, a reduction of cyclin D1 would lead to a lower fraction of cyclin D1-CDK4 complexes available to bind the CDK inhibitor p27 and consequently to a larger number of active p27 molecules able to inhibit CDK2 activity (89).
Recently, a report appeared showing that exogenous TIS21/PC3, stably expressed in the tumor 293 cell line, induced G1 arrest accompanied by a decrease of cyclin E protein levels and CDK2 activity (57). It is worth noting that these cells lack cyclin D1 and have both nonfunctional pRb and nonfunctional p53, due to the expression of adenovirus type 5 E1A and E1B transforming proteins, respectively (35, 73). In this regard, previous reports suggested that in systems with inactive pRb, such as tumor cells, S-phase progression can be regulated by cyclin E in place of cyclin D (55) and that cyclin E expression can overcome a pRb-dependent block of the cell cycle (5, 62), thus suggesting that cyclin E might induce cell cycle progression through pathways independent from and downstream of pRb. Importantly, we observed that in 3T3 Rb−/− cells PC3 led to a decrease also of cyclin E protein, although considerably less pronounced than that of cyclin D1 (Fig. 4). Given that we found cyclin E to be ineffective in rescuing the inhibition of G1 progression by PC3 in NIH 3T3 cells carrying an active pRb gene and that in our cell system the cycle arrest induced by PC3 depended biunivocally on the presence of pRb, a hypothesis that might account for our data and for those of Lim et al. (57) is that PC3 induces the arrest of the cell cycle through at least two pathways, involving cyclin D1 and cyclin E. The latter could become evident when there is impairment of pRb function and, possibly, of other cell cycle regulators such as, for instance, p53. Further analyses will certainly be necessary for a thorough understanding of this point.
PC3-dependent hypophosphorylation of pRb and its reversal by cyclins.
The PC3-dependent dephosphorylation of pRb in NIH 3T3 cells is a clear indication that PC3 influences the activity of pRb.
However, PC3 does not lead to complete disappearance of the phosphorylated pRb forms. This fact should be the result of concomitant phosphorylation of pRb by CDKs different from CDK4, whose regulatory cyclins are not down-regulated by PC3. This possibility is suggested by the observation that not only cyclin D1 but also cyclins E and A prevent the PC3-dependent impairment of pRb phosphorylation. Noteworthily, this fact is in apparent contrast to the quite exclusive functional correlation seen between PC3 and cyclin D1, i.e., the rescue by cyclin D1 of the G1/S impairment exerted by PC3 and the PC3-dependent down-regulation of cyclin D1 levels. In fact, the position of the pRb residue undergoing phosphorylation is more critical, in terms of impairment of pRb's antiproliferative activity, than the total number of phosphorylated residues. In particular, for a complete inactivation of pRb by CDK2 and cdc2 in S phase, a preliminary phosphorylation in G1/S by CDK4 is required (19, 63, 88). Thus, the complete conversion by cyclins A and E (and their associated CDK2 and cdc2 kinases) of the PC3-dependent dephosphorylated form of pRb into a phosphorylated one, seen in a denaturing gel, should result in a pRb still active in growth arrest. In agreement, cyclins A and E led to a nonsignificant rescue of the PC3-dependent inhibition of proliferation. All together, these data indicate that the final target of PC3 is pRb.
Functional domains of PC3.
Colony formation assay data for PC3 mutants point to a functional role in cell proliferation for the region corresponding to aa 50 to 68 (which we term here GR, for growth regulatory), namely, the more amino terminal of the two conserved regions existing within the PC3/BTG/Tob family. In fact, deletion of GR (mutant PC3Δ50–68) led to a complete impairment of the growth-inhibitory properties of PC3 and to almost complete loss of the ability to inhibit cyclin D1. On the other hand, deletion of the conserved domain spanning aa 105 to 123 partially reduced the inhibitory effect on growth and on cyclin D1 expression by PC3. Therefore, it is possible that the GR domain plays a direct role in conferring on the PC3 molecule its antiproliferative properties. Alternative and not mutually exclusive hypotheses are that the GR motif has a role ancillary to that of other domains and that it requires the cooperation of other regions of the molecule, as would be true if its function were concerned with the conservation of the proper conformation of the molecule, e.g., through intramolecular interactions with the aa 105 to 123 region, given the presence of a cysteine in both domains.
The observation that PC3 is phosphorylated by CDK2 suggests that the effect of growth inhibition exerted by the PC3 molecule might be regulated in a cell-cycle-dependent fashion. However, we have observed that the inhibition of colony formation by the phosphorylation-defective mutant of PC3 is only slightly lower than that of the wild-type molecule. We can also tentatively exclude an effect of the phosphorylation by CDK2 on the stability or the intracellular localization of the PC3 protein, since mutant pSCT-PC3 S147N appears to be expressed to the same extent as the wild-type molecule, with no apparent differences in the intracellular pattern. We cannot exclude the phosphorylation of other sites by other kinases as a requirement to bring an effect into evidence, but presently, the functional role of the CDK2-dependent phosphorylation of PC3 remains to be defined.
Physiological role of the G1 arrest by PC3.
The model for PC3 activity presented here, proposing that PC3 activity induces G1 arrest by targeting pRb function through cyclin D1 down-regulation, raises questions about the functional rationale for such activity. So far, the expression of PC3/TIS21/BTG2 has been shown to increase at the onset of neuronal differentiation in cell lines and in vivo (9, 47), at the onset of and during neuronal apoptosis (67), or following DNA damage through a p53-dependent mechanism (92). Evidence has been presented for a role of PC3/TIS21/BTG2 in the growth arrest of the neuroblast preceding its differentiation into postmitotic neuron (47, 48) and in cell survival after genotoxic damage (92). The cellular condition of DNA damage is associated with a decrease of cyclin D1 levels (79, 83, 100). In fact, it has been shown previously that a decrease of cyclin D1 induced by antisense cDNA accelerates DNA repair (79). In this regard, it is known that cyclin D1 associates directly with the proliferating cell nuclear antigen (PCNA) (116), i.e., the auxiliary factor of DNA polymerases δ and ɛ, required for DNA replication and repair (11, 84, 104), and that a down-regulation of cyclin D1 is necessary for PCNA relocation and DNA repair synthesis (79). It is therefore interesting to hypothesize that PC3 might play a role in DNA damage through an effect on cyclin D1 levels, in an Rb-dependent fashion. Regarding the process of neuronal differentiation, PC3 and pRb are expressed during development mainly in areas where embryonic cells proliferate and eventually differentiate (47, 54). Furthermore, it has been shown that animals ablated of the Rb gene by targeting die in utero, displaying major defects in neuronal differentiation. In fact, neuroblasts continue to proliferate without arrest, until they undergo massive apoptosis (54). Very similar defects were detected in skeletal muscle differentiation (119). It is also very suggestive that PC3 expression occurs in vivo in the period when dephosphorylation of pRb is observed (54), thus mirroring our data in vitro. However, pRb expression continues in the adult neuron, while PC3 is completely shut off. Shown in Fig. 14 is a working model summarizing PC3 action.
FIG. 14.
A working model of PC3 activity. PC3, by modulating cyclin D1 levels, leads to dephosphorylation of pRb and consequent growth arrest. PC3 is normally not expressed in proliferating cells but is induced in response to diverse growth arrest stimuli (e.g., terminal differentiation and DNA damage). The dashed lines indicate pathways not experimentally implicated in this report. phosph., phosphorylation; cyc, cyclin.
We should also consider that the above model for the action of PC3 presents some incongruities. In fact, while PC3 expression is directly induced by p53 (92), it has been observed that exogenous expression of p53 induces cyclin D1 levels (17, 23). Such an increase of cyclin D1 levels by p53 has been shown to be mediated by p21 and to be necessary for G1 arrest, possibly as a consequence of the binding to PCNA (17, 23). Thus, a conflict with our model appears evident in regard to the p53 action, which increases cyclin D1 levels and at the same time induces PC3, which in turn should effect a reduction of cyclin D1 levels. Indeed, we have observed that activation of p53 in the mouse BALB/c cell line Val 5 containing the temperature-sensitive mutant p53Val135, kindly provided by C. Schneider and G. Del Sal (23), induced p21 and cyclin D1 expression as expected but did not induce either PC3 protein or mRNA (data not shown). This suggests that the induction of PC3 by p53 is somewhat dependent on the cell system used. Furthermore, DNA damage induces PC3/BTG2 and p53 levels (92, 120; D. Guardavaccaro and F. Tirone, unpublished data) and reduces cyclin D1 levels (79, 83, 100), but this latter effect has also been shown to occur in the absence of p53 (100). Therefore, this experimental evidence indicates that further studies are necessary to assess the possibility of PC3 being a mediator of G1 inhibition by p53.
ACKNOWLEDGMENTS
We are grateful to J. A. DeCaprio for the gift of Rb−/− 3T3 cells; to R. Weinberg for primary Rb+/+ and Rb−/− MEFs and for the gift of pRcCMV-cycA, pRcCMV-cycD1, pRcCMV-cycD3, and pRcCMV-cycE; to P. Sicinski and R. Weinberg for the gift of primary cyclin D1+/+ and cyclin D1−/− MEFs; to L. Zhu and E. Harlow for the gift of pCMV-CD20 and of CMVcdc2 constructs; to M. Ewen and D. M. Livingston for the gift of pRcCMV-CDK2 and pRcCMV-CDK4; to K. Poliak, D. Morgan, and C. Sherr for the gift of baculoviruses expressing cyclin A, cyclin B1, cdc2, CDK2, cyclin D1, and CDK4; to C. Sherr for the gift of the Flag-cyclin D1 construct; and to D. Beach and J. Massaguè for the gifts of pXp16 and pCMX-p27, respectively. We thank L. Baron for outstanding technical assistance. We are all very grateful to Francesca de Santa for her qualified help with the in vivo kinase experiments, given in a critical moment.
We gratefully acknowledge the support of F.T. by Donazione Bianchi and the help of A. Cesari who made it possible. This work was also carried out under a research contract with N.E.F.A.C., Pomezia, Italy, within the Neurobiological Systems National Research Plan of the Ministero dell' Università e della Ricerca Scientifica e Tecnologica.
REFERENCES
- 1.Abken H, Reifenrath B. A procedure to standardize CAT reporter gene assay. Nucleic Acids Res. 1992;20:3527. doi: 10.1093/nar/20.13.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramovich C, Yakobson B, Chebath J, Revel M. A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 1997;16:260–266. doi: 10.1093/emboj/16.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal M L, Agarwal A, Taylor W R, Stark G R. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal M L, Taylor W R, Chernow M V, Chernova O B, Stark G R. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Alevizopoulos K, Vlach J, Hennecke S, Amati B. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 7.Baldin V, Lukas J, Marcote M J, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 8.Bartek J, Bartkova J, Lukas J. The retinoblastoma protein pathway and the restriction point. Curr Opin Cell Biol. 1996;8:805–814. doi: 10.1016/s0955-0674(96)80081-0. [DOI] [PubMed] [Google Scholar]
- 9.Bradbury A, Possenti R, Shooter E M, Tirone F. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc Natl Acad Sci USA. 1991;88:3353–3357. doi: 10.1073/pnas.88.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Bravo R, Frank R, Blundell P A, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 12.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 13.Buchkovich K, Duffy L A, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 14.Caruso M, Martelli F, Giordano A, Felsani A. Regulation of MyoD gene transcription and protein function by the transforming domains of the adenovirus E1A oncoprotein. Oncogene. 1993;8:267–278. [PubMed] [Google Scholar]
- 15.Chen P-L, Riley D J, Chen Y, Lee W-H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 16.Chen P-L, Scully P, Shew J-Y, Wang J Y J, Lee W-H. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Bargonetti J, Prives C. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 1995;55:4257–4263. [PubMed] [Google Scholar]
- 18.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 19.Connell-Crowley L, Harper J W, Goodrich D W. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean P N, Gray J W, Dolbeare F A. The analysis and interpretation of DNA distributions measured by flow cytometry. Cytometry. 1982;3:188–195. doi: 10.1002/cyto.990030309. [DOI] [PubMed] [Google Scholar]
- 21.DeCaprio J A, Ludlow J W, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang C M, Livingston D M. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 22.Del Sal G, Loda M, Pagano M. Cell cycle and cancer: critical events at the G1 restriction point. Crit Rev Oncog. 1996;7:127–142. doi: 10.1615/critrevoncog.v7.i1-2.80. [DOI] [PubMed] [Google Scholar]
- 23.Del Sal G, Murphy M, Ruaro E, Lazarevic D, Levine A J, Schneider C. Cyclin D1 and p21/waf1 are both involved in p53 growth suppression. Oncogene. 1996;12:177–185. [PubMed] [Google Scholar]
- 24.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 25.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diehl J A, Zindy F, Sherr C J. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 27.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 28.Elledge S J, Richman R, Hall F L, Williams R T, Lodgson N, Harper J W. CDK2 encodes a 33-kDa cyclin A-associated protein kinase and is expressed before CDC2 in the cell cycle. Proc Natl Acad Sci USA. 1992;89:2907–2911. doi: 10.1073/pnas.89.7.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewen M E, Sluss H K, Whitehouse L L, Livingston D M. TGFβ inhibition of Cdk4 synthesis is linked to cell cycle arrest. Cell. 1993;74:1009–1020. doi: 10.1016/0092-8674(93)90723-4. [DOI] [PubMed] [Google Scholar]
- 30.Feig L A, Cooper G M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filmus J, Robles A I, Shi W, Wong M J, Colombo L L, Conti C J. Induction of cyclin D1 overexpression by activated ras. Oncogene. 1994;9:3627–3633. [PubMed] [Google Scholar]
- 32.Fletcher B S, Lim R W, Varnum B C, Kujubu D A, Koski R A, Herschman H R. Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J Biol Chem. 1991;266:14511–14518. [PubMed] [Google Scholar]
- 33.Genini M, Schwalbe P, Scholl F A, Remppis A, Mattei M G, Schäfer B W. Subtractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in rhabdomyosarcoma. DNA Cell Biol. 1997;16:433–442. doi: 10.1089/dna.1997.16.433. [DOI] [PubMed] [Google Scholar]
- 34.Graham F L, Vander Eb A J. A new technique for the assay of infectivity of human adenovirus DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 35.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 36.Greene L A. Nerve growth factor prevents the death and stimulates the neuronal differentiation of clonal PC12 pheochromocytoma cells in serum-free medium. J Cell Biol. 1978;78:747–755. doi: 10.1083/jcb.78.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greene L A, Tischler A S. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 39.Gu Y, Turck C W, Morgan D O. Inhibition of CDK2 activity in vivo by an associated 20K regulatory subunit. Nature. 1993;366:707–710. doi: 10.1038/366707a0. [DOI] [PubMed] [Google Scholar]
- 40.Guan K-L, Jenkins C W, Li Y, Nichols M A, Wu X, O'Keefe C L, Matera A G, Xiong Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 41.Guardavaccaro D, Montagnoli A, Ciotti M T, Lotti L, Di Lazzaro C, Torrisi M-R, Gatti A, Tirone F. Nerve growth factor regulates the sub-cellular localization of the nerve growth factor-inducible protein PC4 in PC12 cells. J Neurosci Res. 1994;37:660–674. doi: 10.1002/jnr.490370514. [DOI] [PubMed] [Google Scholar]
- 42.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 43.Herrera R E, Sah V P, Williams B O, Mäkelä T P, Weinberg R A, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herwig S, Strauss M. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 45.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 46.Holmes J, Solomon M J. A predictive scale for evaluating cyclin-dependent kinase substrates. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 47.Iacopetti P, Barsacchi G, Tirone F, Maffei L, Cremisi F. Developmental expression of PC3 gene is correlated with neuronal cell birthday. Mech Dev. 1994;47:127–137. doi: 10.1016/0925-4773(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 48.Iacopetti P, Michelini M, Stuckmann I, Oback B, Aaku-Saraste E, Huttner W B. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc Natl Acad Sci USA. 1999;96:4639–4644. doi: 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato J, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 50.Koff A, Ohtsuki M, Polyak K, Roberts J M, Massaguè J. Negative regulation of G1 in mammalian cells: inhibition of cyclin E-dependent kinase by TGF-β. Science. 1993;260:536–538. doi: 10.1126/science.8475385. [DOI] [PubMed] [Google Scholar]
- 51.Kouzarides T. Transcriptional control by the retinoblastoma protein. Semin Cancer Biol. 1995;6:91–98. doi: 10.1006/scbi.1995.0012. [DOI] [PubMed] [Google Scholar]
- 52.LaThangue N B. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell cycle control. Trends Biochem Sci. 1994;19:108–114. doi: 10.1016/0968-0004(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 53.Lee E Y-H P, Chang C-Y, Hu N, Wang J, Lai C-C, Herrup K, Lee W-H, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 54.Lee E Y-H P, Hu N, Yuan S-S F, Cox L A, Bradley A, Lee W-H, Herrup K. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 1994;8:2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 55.Leone G, DeGregori J, Sears R, Jakoi L, Nevins J R. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 56.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 57.Lim I K, Lee M S, Ryu M S, Park T J, Fujiki H, Eguchi H, Paik W K. Induction of growth inhibition of 293 cells by downregulation of the cyclin E and cyclin-dependent kinase 4 proteins due to overexpression of TIS21. Mol Carcinog. 1998;23:25–35. doi: 10.1002/(sici)1098-2744(199809)23:1<25::aid-mc4>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 58.Lin W J, Gary J D, Yang M C, Clarke S, Herschman H R. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 59.Liu H Y, Badarinarayana V, Audino D C, Rappsilber J, Mann M, Denis C L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu H Y, Toyn J H, Chiang Y C, Draper M P, Johnston L H, Denis C L. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 1997;16:5289–5298. doi: 10.1093/emboj/16.17.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 63.Lundberg A S, Weinberg R A. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuda S, Kawamura-Tsuzuku J, Oshugi M, Yoshida M, Emi M, Nakamura Y, Onda M, Yoshida Y, Nishiyama A, Yamamoto T. Tob, a novel protein that interacts with p185erbB2, is associated with antiproliferative activity. Oncogene. 1996;12:705–713. [PubMed] [Google Scholar]
- 65.Matsushime H, Ewen M E, Strom D K, Kato J-Y, Hanks S K, Roussel M F, Sherr C J. Identification and properties of an atypical catalytic subunit (p34PKSJ3/CDK4) for mammalian D-type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 66.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J-Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mesner P W, Winters T R, Green S H. NGF withdrawal-induced cell death in neuronal PC12 cells resembles that in sympathetic neurons. J Cell Biol. 1992;119:1669–1680. doi: 10.1083/jcb.119.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meyerson M, Enders G H, Wu C-L, Su L-K, Gorka C, Nelson C, Harlow E, Tsai L-H. A family of human cdc2-related protein kinases. EMBO J. 1991;11:2909–2917. doi: 10.1002/j.1460-2075.1992.tb05360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D1 partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 71.Mittnacht S, Paterson H, Olson M F, Marshall C J. Ras signalling is required for the inactivation of the tumour suppressor pRb cell-cycle control protein. Curr Biol. 1997;7:219–221. doi: 10.1016/s0960-9822(97)70094-0. [DOI] [PubMed] [Google Scholar]
- 72.Montagnoli A, Guardavaccaro D, Starace G, Tirone F. Overexpression of the nerve growth factor-inducible PC3 immediate early gene is associated to inhibition of cell proliferation. Cell Growth Differ. 1996;7:1327–1336. [PubMed] [Google Scholar]
- 73.Moran E. Interaction of adenoviral proteins with pRB and p53. FASEB J. 1993;7:880–885. doi: 10.1096/fasebj.7.10.8344487. [DOI] [PubMed] [Google Scholar]
- 74.Morgenbesser S D, Williams B O, Jacks T, DePinho R A. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- 75.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Müller H, Lukas J, Schneider A, Warthoe P, Bartek J, Eilers M, Strauss M. Cyclin D1 expression is regulated by the retinoblastoma protein. Proc Natl Acad Sci USA. 1994;91:2945–2949. doi: 10.1073/pnas.91.8.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G F. Cyclin A is required at two points in human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pagano M, Theodoras A M, Tam S W, Draetta G F. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 1994;8:1627–1639. doi: 10.1101/gad.8.14.1627. [DOI] [PubMed] [Google Scholar]
- 80.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peeper D S, Upton T M, Ladha M H, Neuman E, Zalvide J, Bernards R, DeCaprio J A, Ewen M. Ras signalling linked to the cell cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 82.Pines J, Hunter T. Isolation of human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 83.Poon R Y, Toyoshima H, Hunter T. Redistribution of the CDK inhibitor p27 between different cyclin-CDK complexes in the mouse fibroblast cell cycle and in cells arrested with lovastatin or ultraviolet irradiation. Mol Biol Cell. 1995;6:1197–1213. doi: 10.1091/mbc.6.9.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prelich G, Tan C-K, Kostura M, Mathews M B, So A G, Downey K M, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 85.Qin X-Q, Chittenden T, Livingston D, Kaelin W G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 86.Quelle D E, Ashmun R A, Hannon G J, Rehberger P A, Trono D, Richter K H, Walker C, Beach D, Sherr C J, Serrano M. Cloning and characterization of murine p16INK4a and p15INK4b genes. Oncogene. 1995;11:635–645. [PubMed] [Google Scholar]
- 87.Quelle D E, Ashmun R A, Shurtleff S A, Kato J-Y, Bar-Sagi D, Roussel M F, Sherr C J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 88.Resnitzky D, Reed S I. Different roles for cyclin D1 and E in regulation of the G1-to-S transition. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reynisdòttir I, Polyak K, Iavarone A, Massaguè J. Kip/Cip and Ink4 Cdk inhibitor cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 90.Riabowol K, Draetta G, Brizuela L, Vandre D, Beach D. The cdc2 kinase is a nuclear protein that is essential for mitosis in mammalian cells. Cell. 1989;57:393–401. doi: 10.1016/0092-8674(89)90914-8. [DOI] [PubMed] [Google Scholar]
- 91.Riley D J, Lee E Y-H P, Lee W-H. The retinoblastoma protein: more than a tumor suppressor. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 92.Rouault J-P, Falette N, Guéhenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, Shaw P, Berger R, Samarut J, Magaud J-P, Ozturk M, Samarut C, Puisieux A. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 93.Rouault J-P, Prevot D, Berthet C, Birot A M, Billaud M, Magaud J-P, Corbo L. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J Biol Chem. 1998;273:22563–22569. doi: 10.1074/jbc.273.35.22563. [DOI] [PubMed] [Google Scholar]
- 94.Rouault J-P, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, Garoccio M, Germain D, Samarut J, Magaud J-P. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rudkin B B, Lazarovici P, Levi B-Z, Abe Y, Fujita K, Guroff G. Cell cycle-specific action of nerve growth factor in PC12 cells: differentiation without proliferation. EMBO J. 1989;8:3319–3325. doi: 10.1002/j.1460-2075.1989.tb08493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rusconi S, Severne Y, Georgiev O, Galli I, Wieland S. A novel expression assay to study transcriptional activators. Gene. 1990;89:211–221. doi: 10.1016/0378-1119(90)90008-f. [DOI] [PubMed] [Google Scholar]
- 97.Sanchez I, Dynlacht B D. Transcriptional control of the cell cycle. Curr Opin Cell Biol. 1996;8:318–324. doi: 10.1016/s0955-0674(96)80004-4. [DOI] [PubMed] [Google Scholar]
- 98.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Serrano M, Hannon G J, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 100.Shapiro G I, Edwards C D, Ewen M E, Rollins B J. p16INK4A participates in a G1 arrest checkpoint in response to DNA damage. Mol Cell Biol. 1998;18:378–387. doi: 10.1128/mcb.18.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sherr C J. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 102.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 103.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 104.Shivji M K K, Kenny M K, Wood R D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 105.Stamenkovic I, Seed B. Analysis of two cDNA clones encoding the B lymphocyte antigen CD20 (B1, Bp35), a type III integral membrane protein. J Exp Med. 1988;167:1975–1980. doi: 10.1084/jem.167.6.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tam S W, Shay J W, Pagano M. Differential expression and cell cycle regulation of the cyclin-dependent kinase 4 inhibitor p16Ink4. Cancer Res. 1994;54:5816–5820. [PubMed] [Google Scholar]
- 107.Tam S W, Theodoras A M, Shay J W, Draetta G F, Pagano M. Differential expression and regulation of cyclin D1 protein in normal and tumor human cells: association with Cdk4 is required for cyclin D1 function in G1 progression. Oncogene. 1994;9:2663–2674. [PubMed] [Google Scholar]
- 108.Tirone F, Shooter E M. Early gene regulation by nerve growth factor: induction of an interferon related gene. Proc Natl Acad Sci USA. 1989;86:2088–2092. doi: 10.1073/pnas.86.6.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-CDK protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 110.Van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 111.Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- 112.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 113.Watanabe G, Albanese C, Lee R J, Reutens A, Vairo G, Henglein B, Pestell R G. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 115.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 116.Xiong Y, Zhang H, Beach D. D-type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 117.Yan G Z, Ziff E B. Nerve growth factor induces transcription of the p21 WAF1/CIP1 and cyclin D1 genes in PC12 cells by activating the SP1 transcription factor. J Neurosci. 1997;17:6122–6132. doi: 10.1523/JNEUROSCI.17-16-06122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yoshida Y, Matsuda S, Ikematsu N, Kawamura-Tsuzuku J, Inazawa J, Umemori H, Yamamoto T. ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene. 1998;16:2687–2693. doi: 10.1038/sj.onc.1201805. [DOI] [PubMed] [Google Scholar]
- 119.Zacksenhaus E, Jiang Z, Chung D, Marth J D, Phillips R A, Gallie B L. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 120.Zhan Q, Carrier F, Fornace A J., Jr Induction of cellular p53 activity by DNA-damaging agents and growth arrest. Mol Cell Biol. 1993;13:4242–4250. doi: 10.1128/mcb.13.7.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang H, Hannon G J, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 122.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]