Abstract
The fatty acid (FA) compositions of ten seaweeds representative of Chlorophyta, Rhodophyta, and Ochrophyta from Kuwait in the Arabian Gulf region were determined and are discussed in the context of their potential nutritional perspectives for seaweed valorization. All the seaweeds had higher saturated fatty acid (SFA) and lower monounsaturated (MUFA) and polyunsaturated fatty acid (PUFA) contents than those typical of tropical environments. Palmitic, myristic, stearic, oleic, linoleic, α-linolenic, and stearidonic acids were the major FAs detected. Arachidonic, eicosapentaenoic, and docosahexaenoic acids were detected in minor amounts. Conserved fatty acid patterns revealed phylogenetic relationships among phyla, classes, and orders matching the molecular phylogenies at higher taxonomic ranks. Hierarchical clustering analyses clearly segregated different seaweeds (except Codium papillatum and Iyengaria stellata) into distinct groups based on their FA signatures. All but one species (Chondria sp.) had health-beneficial n6/n3 PUFAs (0.33:1–2.94:1) and atherogenic (0.80–2.52) and thrombogenic indices (0.61–5.17). However, low PUFA/SFA contents in most of the species (except Ulva spp.) may limit their utilization in the formulation of PUFA-rich functional foods. Ulva spp. had substantially high PUFAs with PUFA/SFA > 0.4, n6/n3 (0.33–0.66) and atherogenic (0.80–1.15) and thrombogenic indices (0.49–0.72), providing substantial potential for their utilization in food and feed applications.
Keywords: Arabian Gulf, fatty acids, gas chromatography, n6/n3 ratio, PUFA, seaweed
1. Introduction
Seaweeds are photosynthetic, multicellular marine macroalgae that have been utilized for food, animal feed, phycocolloids, and bioactive compounds of pharmacological importance for centuries. In fact, they are considered one of the most important food sources for the coastal communities especially in Asian countries such as Japan, China, and Korea [1]. Today the global seaweed industry is worth USD 6 billion per annum, 85% of which comprise food products for human consumption [1]. These seaweeds are rich sources of essential nutrients and health-promoting compounds including proteins, carbohydrates, polyunsaturated fatty acids (PUFAs), antioxidants, minerals, dietary fibers, and vitamins [2,3,4,5]. As a matter of fact, it is often pointed out that the Japanese, who have eaten seaweeds regularly in their daily cuisines for centuries, have one of the highest life expectancies in the world [6]. Seaweed-digesting enzymes such as porphyranases and agarases were discovered in the Japanese gut bacteria a decade ago, but were absent from American populations [7]. Moreover, increasing awareness of beneficial impacts of seaweed-based food products for health and their popularization in Western and European markets have recently opened a debate on categorizing seaweeds as a ‘healthy superfood’ [8,9,10].
Nevertheless, seaweeds feature low lipid contents (<5% d.w.) [2,4,11,12,13]. Seaweed lipids have received considerable interest due to their high contents of nutritionally essential n3 and n6 PUFAs [3,11,14,15] that cannot be synthesized by humans and are thus obtained only through dietary sources. Moreover, seaweeds also synthesize long-chain PUFAs (LC-PUFAs) such as arachidonic acid (ARA), eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA), which are not present in land plants [2,4,11,14,15,16,17]. These essential LC-PUFAs are physiologically important and are involved in the regulation of membrane structure and function, transcription regulation, cell signaling, and generation of bioactive lipid mediators such as prostaglandins [18]. Among LC-PUFAs, n3 PUFAs are of immense clinical and nutritional importance since a balance of n6 and n3 PUFAs is critical for the prevention of chronic diseases including cardiovascular diseases, colon and breast cancers, neurodegenerative and inflammatory diseases, and is also crucial for infant brain development [18,19,20,21,22,23]. Numerous seaweeds have been recognized to have health-promoting effects reflected in their nutritional indices such as n6/n3 PUFA ratio (0.1:1–3:1), as well as an atherogenic index (AI) and thromogenic index (TI) of both less than one [2,4,11,14,24,25], underlining their potential utilization in nutritional and functional food formulations. AI signifies the relationship between the sum of pro-atherogenic saturated fatty acids (SFAs) such as 12:0, 14:0, and 16:0 and anti-atherogenic unsaturated fatty acids (UFAs) [23,26]. The pro-atherogenic FAs favor the adhesion of lipids to cells of the immunological and circulatory system while anti-atherogenic FAs inhibit the aggregation of plaques and reduce the levels of esterified fatty acid, cholesterol, and phospholipids, thereby preventing the appearance of micro- and macro- coronary diseases [23,27]. TI signifies the relationship between the pro-thrombogenic FAs (12:0, 14:0, and 16:0) and anti-thrombogenic monounsaturated fatty acids (MUFAs), n3 and n6 PUFAs [26]. The dietary intake of food with low AI and TI reduces the threat of plaque formation and of atrial fibrillation respectively, thereby improving cardiovascular health [23].
Furthermore, seaweed FA profiles represent distinct conserved chemotaxonomic traits that have been extensively used to classify seaweeds to different taxonomic levels of genus, family, order, and phylum [4,11,14,15,17]. The taxon-specific understanding of FA profiles of seaweeds is necessary for selecting appropriate seaweed taxa for their valorization as human food, animal feed, or development of other nutraceutical, pharmaceutical, and cosmeceutical products [4,14]. Therefore, it is not surprising that numerous seaweeds have been studied from different parts of the world for their fatty acids and this dataset is continuously growing [15,25,28,29,30]; still, the absolute number of seaweeds studied is low considering the overall seaweed diversity, with approximately 10,000 species been reported worldwide [31].
The Arabian Gulf is a shallow basin located in one of the most arid regions of the world. The coastal environment of Kuwait (approximately 500 km coastline) can be divided into the Northern Region, Kuwait Bay, and the Southern Region. Marine organisms of this region experience the greatest seasonal temperate range in the world as well as the highest annual sea temperature [32] along with high levels of salinity (40–41 g/kg). The unique extreme environmental conditions may well be associated with unusual fatty acid profiles, but surprisingly, only a few seaweeds from the Arabian Gulf coast of Qatar [33], Saudi Arabia [34], and Iran [35,36,37,38] have been studied in this context to date and, in particular, information on FA profiles of seaweeds from hypersaline and warm coastal waters of Kuwait is mostly lacking so far. A notable exception dating back almost two decades is the pioneering study by Al-Hasan et al. [39]. In the present study, we analyzed FA composition by gas chromatography with flame ionization detection (GC-FID) of ten seaweeds exhibiting widespread distribution and high abundance in Kuwait coastal waters, belonging to the genus Ulva, Codium, Chondria, Iyengaria, Feldmannia, Padina, and Sargassum. Our aim was to identify seaweed species containing high levels of nutritionally essential PUFAs that can potentially be valorized for human consumption or other functional food applications. In this pursuit, we also studied FA-based conserved taxonomic differences among different species using hierarchical clustering.
2. Materials and Methods
2.1. Seaweed Sample Collection
A total of 10 different seaweed species, Ulva sp., Ulva chaugulii M. G. Kavale and M. A. Kazi, Ulva ohnoi M. Hiraoka and S. Shimada, Ulva tepida Y. Masakiyo and S. Shimada, Codium papillatum C. K. Tseng and W. J. Gilbert (Chlorophyta), Chondria sp. C. Agardh (Rhodophyta), Iyengaria stellata (Børgesen) Børgesen, Feldmannia indica (Sonder) Womersley and A. Bailey, Padina boergesenii Allender and Kraft, and Sargassum aquifolium (Turner) C. Agardh (Ochrophyta) were collected during May to June 2018 and in February 2021 from different sampling sites of Kuwait’s coastal waters in the Arabian Gulf (Table 1). Seaweed samples were rinsed thoroughly with seawater on-site and placed in plastic bags. Date of collection and location were noted. Samples were transferred to the laboratory in cool packs and washed thrice with sea water. Fresh samples were frozen at −20 °C for 24 h followed by freeze-drying in a freeze dryer (Labconco, Kansas City, MO, USA) at −45 °C for 48 h and stored at −20 °C until analysis.
Table 1.
Locations of macroalgal sampling sites along the Kuwait coastline.
| S. No. | Species | Abbreviations (*) |
Phylogenetic Affinity | Herbarium Code | Date | Location | Coordinates | Offshore Seawater Surface Temperature (°C) |
|---|---|---|---|---|---|---|---|---|
| Chlorophyta | ||||||||
| 1 | Ulva sp. | US | Ulvaceae, Ulvales, Ulvophyceae |
Doh010221-1 | 01/02/2021 | Ras Ushairij | 29°23′00.7″ N 47°49′50.9″ E |
14.8–15.0 |
| 2 | Ulva chaugulii M.G.Kavale and M.A.Kazi | UC | Ulvaceae, Ulvales, Ulvophyceae |
Doh010221-2 | 01/02/2021 | Ras Ushairij | 29°23′00.7″ N 47°49′50.9″ E |
14.8–15.0 |
| 3 | Ulva tepida Masakiyo and S.Shimada | UT | Ulvaceae, Ulvales, Ulvophyceae |
Doh010221-5 | 01/02/2021 | Ras Ushairij | 29°23′00.7″ N 47°49′50.9″ E |
14.8–15.0 |
| 4 | Ulva ohnoi M.Hiraoka and S.Shimada | UO | Ulvaceae, Ulvales, Ulvophyceae |
Doh010221-3 | 01/02/2021 | Ras Ushairij | 29°23′00.7″ N 47°49′50.9″ E |
14.8–15.0 |
| 5 | Codium papillatum C.K.Tseng and W.J. Gilbert | CP | Codiaceae, Bryopsidales, Ulvophyceae |
ABUH030618-2 | 3/06/2018 | Abu Al Hasaniya | 29°12′19.4″ N 48°06′41.5″ E | 30.2–30.3 |
| Rhodophyta | ||||||||
| 6 | Chondria sp. C. Agardh | CS | Rhodomelaceae Ceramiales, Florideophyceae | BNA260518-1 | 26/05/2018 | Bnaider Beach | 28°47′01.5″ N 48°17′50.6″ E | 27.8–27.9 |
| Ochrophyta | ||||||||
| 7 | Iyengaria stellata (Børgesen) Børgesen | IS | Scytosiphonaceae, Ectocarpales, Phaeophyceae |
ABUH060618-1 | 27/05/2018 | Abu Al Hasaniya | 29°12′19.4″ N 48°06′41.5″ E | 30.2–30.3 |
| 8 | Feldmannia indica (Sonder) Womersley and A. Bailey | FI | Acinetosporaceae Ectocarpales, Phaeophyceae | ABUH030618-1 | 3/06/2018 | Abu Al Hasaniya | 29°12′19.4″ N 48°06′41.5″ E | 30.2–30.3 |
| 9 | Padina boergesenii Allender and Kraft | PT | Dictyotaceae, Dictyotales, Phaeophyceae |
ABUH270518-1 | 27/05/2018 | Abu Al Hasaniya | 29°12′19.4″ N 48°06′41.5″ E | 30.2–30.3 |
| 10 | Sargassum aquifolium (Turner) C. Agardh | SA | Sargassaceae, Fucales, Phaeophyceae | ABUH270518-2 | 27/05/2018 | Abu Al Hasaniya | 29°12′19.4″ N 48°06′41.5″ E | 30.2–30.3 |
2.2. Fatty Acid Extraction and Methyl Ester Preparation
Fatty acids were extracted and converted into the respective methyl esters from freeze-dried samples by the base-catalyzed direct transmethylation method modified after Christie and Han [40]. Briefly, 0.3 g of freeze-dried seaweed samples (in triplicates) were homogenized in a mortar and pestle and transferred to Oakridge™ centrifuge tubes (15 mL), to which 3 mL of KOH-MeOH solution (0.2 M) was added. The mixture was heated at 75 °C for 1 h. After cooling to room temperature, 3 mL of n-hexane was added and mixed thoroughly using a vortex. The organic layers containing fatty acid methyl esters (FAMEs) were collected in GC vials and stored at −20 °C until analysis.
2.3. Gas Chromatographic (GC) Analysis
For analysis of FAMEs, 1 µL of esterified sample was injected into a gas chromatograph (Shimadzu GC 2010, Tokyo, Japan) coupled with a flame ionization detector (FID). A cyano-polysiloxane (CP-Sil 88 for FAME, part number 839171) capillary column (100 m × 0.25 mm, 0.20 µm (J&W, Varian, Chrompack, São Paulo, Brazil) was used for the FAMEs separation under the following instrumental conditions: injector and FID detector temperatures were 250 and 270 °C, respectively, with an injector split ratio of 1:50, and carrier gas helium with a constant flow rate of 1.0 mL/min. The initial oven temperature was 80 °C, at which it was held for 5 min, followed by an increase to 220 °C at a rate of 4 °C/min; then, it was held for 5 min, and finally the temperature was increased to 240 °C at a rate of 1 °C/min and was held for an additional 10 min. FAME peaks were identified by comparison of their retention times with those of external standard (FAME Mix C4-C24; Sigma-Aldrich, Laramie, WY, USA) and quantified by area normalization using postrun analysis, GC LabStationsTM software v. 5.96 (Shimadzu, Tokyo, Japan). The content of individual fatty acid was finally reported as relative percentage of the total fatty acid methyl esters (TFAs).
2.4. Nutritional Indices
The unsaturation index (U.I.) was calculated by multiplying the percentage of each fatty acid by the number of double bonds followed by summing up their contributions [41]. Atherogenic and thrombogenic indices (AI and TI) were calculated according to Ulbright and Southgate [26], where:
AI = (12:0 + 4 × 14:0 + 16:0)/(n − 3 PUFAs + n − 6 PUFAs + MUFAs), and
TI = (14:0 + 16:0 + 18:0)/(0.5n − 6 PUFAs + 3n − 3PUFAs + n3/n − 6 PUFAs)
2.5. Statistical Analysis
All analytical determinations were performed in triplicate (n = 3) and the mean values were recorded. The fatty acid contents of different seaweed species were compared by analysis of variance (ANOVA) followed by Tukey’s HSD post-hoc test with differences considered significant at p < 0.01 using SPSS v 22. All multivariate analyses were performed after log-transformation and pareto scaling (mean-centered and divided by the square root of standard deviation of each value) of FA and nutritional data matrices (Supplementary datasheet S1–S2) using the web-based software MetaboAnalyst v 5.0 (https://www.metaboanalyst.ca accessed on 26 September 2021). This data pre-processing was carried out to give equal weight to all variables, regardless of their absolute value as the detected fatty acid levels were of different orders of magnitude. The principal component analysis (PCA) was performed on data matrices without rotation and the principal components were extracted based on scree plot. The dendrogram was obtained by hierarchical clustering based on Ward linkage with Euclidean distance [42]. Additionally, the normalized data matrices obtained in MetaboAnalyst were exported to SPSS v 22 for Kaiser-Meyer-Olkin (KMO) test for measuring sampling adequacy for PCA analysis and Bartlet’s test of sphericity to assess the equality of variance in the data matrices.
3. Results and Discussion
3.1. Fatty Acid Composition
A total of 31 fatty acids were detected in seaweeds using GC-FID listed in Table 2. SFAs constituted of decanoic acid (10:0), dodecanoic acid (12:0), tridecanoic acid (13:0), tetradecanoic acid (14:0; myristic acid), pentadecanoic acid (15:0), hexadecanoic acid (16:0; palmitic acid), heptadecanoic acid (17:0), octadecanoic acid (18:0; stearic acid), icosanoic acid (20:0; arachidic acid), docosanoic acid (22:0; behenic acid), and tetracosanoic acid (24:0; lignoceric acid). MUFAs constituted (9Z)-tetradec-9-enoic acid (9c-14:1; myristoleic acid), (10Z)-pentadec-10-enoic acid (10c-15:1), (9Z)-hexadec-9-enoic acid (9c-16:1; palmitoleic acid), (9Z)-octadec-9-enoic acid (9c-18:1; oleic acid), (9E)-octadec-9-enoic acid (9t-18:1; elaidic acid), (11Z)-icos-11-enoic acid (11c-20:1; gondoic acid), (13Z)-docos-13-enoic acid (13c-22:1; erucic acid), and (15Z)-tetracos-15-enoic acid (15c-24:1; nervonic acid). PUFAs constituted (9Z,12Z)-octadeca-9,12-dienoic acid (9c12c-18:2; linoleic acid, LA), (9E,12E)-octadeca-9,12-dienoic acid (9t12t-18:2; linolelaidic acid), (6Z,9Z,12Z)-octadeca-6,9,12-trienoic acid (6c9c12c-18:3; γ-linolenic acid, GLA), (9Z,12Z,15Z)-octadeca-9,12,15-trienoic acid (9c12c15c-18:3; ɑ-linolenic acid, ALA), (6Z,9Z,12Z,15Z)-octadeca-6,9,12,15-tetraenoic acid (6c9c12c15c-18:4; stearidonic acid, STA), (8Z,11Z,14Z)-icosa-8,11,14-trienoic acid (8c11c14c-20:3; dihomo-γ-linolenic acid, DGLA), (11Z,14Z,17Z)-icosa-11,14,17-trienoic acid (11c14c17c-20:3; dihomolinolenic acid), (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoic acid (5c8c11c14c-20:4; arachidonic acid, ARA), (5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoic acid (5c8c11c14c17c-20:5; eicosapentaenoic acid, EPA), (13Z,16Z)-docosa-13,16-dienoic acid (13c16c-22:2), and (4Z,7Z,10Z,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoic acid (4c7c10c13c16c19c-22:6; docosahexaenoic acid, DHA). The fatty acid contents of seaweeds stated here refer to relative contribution to total fatty acids (% TFA) throughout the manuscript.
Table 2.
Fatty acid composition (% of total fatty acid methyl esters) of different macroalgal species, expressed as means ± SD (n = 3).
| Fatty Acids | Ulva sp. | Ulva chaugulii | Ulva ohnoi | Ulva tepida |
Codium
papillatum |
Chondria sp. |
Iyengaria
stellata |
Feldmannia
indica |
Padina
boergesenii |
Sargassum
aquifolium |
|---|---|---|---|---|---|---|---|---|---|---|
| 10:0 | nd | nd | nd | nd | 0.3 ± 0.01 c | 0.5 ± 0.1 a | 0.3 ± 0.01 c | 0.4 ± 0.003 b | 0.3 ± 0.03 c | 0.3 ± 0.02 c |
| 12:0 | 0.2 ± 0.1 f | 0.4 ± 0.02 de | 1.6 ± 0.2 b | 1.2 ± 0.1 c | 1.9 ± 0.01 a | 0.5 ± 0.01 d | 1.9 ± 0.04 a | 0.2 ± 0.02 f | 1.3 ± 0.1 c | 0.4 ± 0.1 de |
| 13:0 | 0.1 ± 0.05 b | nd | nd | nd | 0.01 ± 0.001 c | 0.2 ± 0.02 a | 0.01 ± 0.001 c | 0.01 ± 0.001 c | 0.04 ± 0.04 c | nd |
| 14:0 | 1.3 ± 0.11 g | 1.7 ± 0.02 fg | 2.5 ± 0.04 e | 2.1 ± 0.05 ef | 3.3 ± 0.07 d | 11.7 ± 0.42 a | 3.2 ± 0.06 d | 7.6 ± 0.04 b | 7.5 ± 0.05 b | 5.6 ± 0.13 c |
| 15:0 | 0.2 ± 0.04 e | 0.2 ± 0.01 e | 0.3 ± 0.001 d | 0.7 ± 0.02 ab | 0.1 ± 0.01 f | 0.8 ± 0.04 a | 0.1 ± 0.02 f | 0.5 ± 0.001 c | 0.7 ± 0.01 ab | 0.4 ± 0.03 d |
| 16:0 | 43.5 ± 5.0 de | 44.6 ± 0.2 cde | 41.3 ± 0.4 e | 35.7 ± 0.6 f | 53.9 ± 0.6 b | 63.9 ± 0.5 a | 53.7 ± 0.8 b | 49.0 ± 0.2 bcd | 49.2 ± 0.3 bc | 49.1 ± 0.5 bc |
| 17:0 | 0.1 ± 0.003 c | 0.2 ± 0.05 bc | 0.2 ± 0.1 bc | 0.1 ± 0.003 c | 0.2 ± 0.02 bc | 0.4 ± 0.02 b | 0.2 ± 0.01 bc | 0.3 ± 0.002 bc | 5.9 ± 0.2 a | 0.1 ± 0.05 bc |
| 18:0 | 3.4 ± 2.0 ab | 1.1 ± 0.05 c | 1.7 ± 0.17 bc | 1.3 ± 0.13 bc | 3.2 ± 0.19 abc | 4.5 ± 0.37 a | 3.0 ± 0.2 abc | 1.9 ± 0.03 bc | 2.3 ± 0.1 abc | 3.2 ± 0.01 abc |
| 20:0 | 1.2 ± 0.3 ab | 1.2 ± 0.01 abc | 0.9 ± 0.01 abcd | 1.4 ± 0.03 a | 0.7 ± 0.5 bcd | 0.5 ± 0.01 d | 0.9 ± 0.1 abcd | 0.5 ± 0.001 d | 0.6 ± 0.02 cd | 0.9 ± 0.02 abcd |
| 22:0 | 1.6 ± 0.01 c | 1.7 ± 0.01 c | 4.5 ± 0.1 b | 2.0 ± 0.04 c | 5.6 ± 0.4 a | 0.2 ± 0.04 de | 5.6 ± 0.31 a | 0.5 ± 0.01 de | 0.1 ± 0.0 e | 0.8 ± 0.02 d |
| 24:0 | nd | 0.5 ± 0.004 c | 1.1 ± 0.02 b | 0.02 ± 0.001 de | 2.7 ± 0.4 a | 0.3 ± 0.03 c | 2.4 ± 0.3 a | 0.3 ± 0.02 c | 0.3 ± 0.03 c | 0.4 ± 0.1 c |
| ΣSFAs | 51.6 ± 3.0 e | 51.4 ± 0.11 e | 54.1 ± 0.6 e | 44.5 ± 0.8 f | 72.0 ± 0.9 b | 83.3 ± 0.3 a | 71.3 ± 0.5 b | 61.3 ± 0.1 d | 68.0 ± 0.4 c | 61.2 ± 0.5 d |
| 9c-14:1 | 0.2 ± 0.04 a | nd | nd | nd | 0.02 ± 0.003 c | nd | 0.02 ± 0.003 c | 0.03 ± 0.004 bc | nd | 0.1 ± 0.01 b |
| 10c-15:1 | nd | nd | 0.04 ± 0.003 a | 0.02 ± 0.001 b | nd | nd | nd | nd | nd | nd |
| 9c-16:1 | 7.3 ± 2.2 a | 6.4 ± 0.03 ab | 3.9 ± 0.04 cd | 1.4 ± 0.02 d | 1.6 ± 0.04 d | 5.4 ± 0.1 abc | 1.5 ± 0.1 d | 4.4 ± 0.1 bc | 3.2 ± 0.3 cd | 4.6 ± 0.1 bc |
| 10c-17:1 | 0.3 ± 0.2 bc | 0.7 ± 0.001 a | 0.1 ± 0.003 c | nd | 0.04 ± 0.002 c | nd | 0.04 ± 0.002 c | 0.5 ± 0.003 ab | 0.1 ± 0.02 c | 0.1 ± 0.02 c |
| 9t-18:1 | 0.1 ± 0.06 b | 3.0 ± 0.03 a | 0.7 ± 0.1 b | 3.2 ± 0.1 a | 0.1 ± 0.05 b | nd | 0.1 ± 0.04 b | 0.04 ± 0.0004 b | 0.1 ± 0.002 b | 0.1 ± 0.01 b |
| 9c-18:1 | 5.9 ± 3.0 de | 8.9 ± 0.03 d | 6.1 ± 0.01 de | 15.7 ± 0.1 c | 18.1 ± 0.7 ab | 4.8 ± 0.1 e | 19.4 ± 0.5 ab | 16.0 ± 0.04 bc | 16.8 ± 0.8 abc | 19.8 ± 0.2 a |
| 11c-20:1 | 1.5 ± 1.2 a | 0.1 ± 0.1 b | 0.2 ± 0.002 ab | 0.2 ± 0.1 ab | 0.1 ± 0.03 b | 0.4 ± 0.01 ab | 0.1 ± 0.03 b | 0.03 ± 0.0003 b | 0.1 ± 0.01 b | 0.9 ± 0.03 ab |
| 13c-22:1 | 0.2 ± 0.01 cd | 0.2 ± 0.05 cd | 0.6 ± 0.1 a | 0.2 ± 0.06 cd | 0.02 ± 0.001 e | 0.20 ± 0.01 cd | 0.02 ± 0.001 e | 0.1 ± 0.01 de | 0.2 ± 0.1 cd | 0.4 ± 0.02 b |
| 15c-24:1 | 0.5 ± 0.1 | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| ΣMUFAs | 15.8 ± 7.0 bc | 19.4 ± 0.1 ab | 11.5 ± 0.9 b | 20.7 ± 0.2 ab | 20.0 ± 0.7 ab | 10.8 ± 0.2 c | 21.2 ± 0.5 ab | 21.1 ± 0.04 ab | 20.5 ± 0.8 ab | 26.0 ± 0.3 a |
| 9t12t-18:2 | nd | nd | nd | nd | 0.3 ± 0.1 ab | 0.5 ± 0.1 a | 0.3 ± 0.11 b | 0.003 ± 0.001 c | 0.3 ± 0.1 ab | nd |
| 9c12c-18:2 | 5.2 ± 0.5 d | 7.0 ± 0.1 c | 11.6 ± 0.1 a | 8.6 ± 0.1 b | 2.3 ± 0.3 f | 1.6 ± 0.1 f | 2.0 ± 0.3 f | 3.7 ± 0.01 e | 3.3 ± 0.1 e | 3.3 ± 0.1 e |
| 6c9c12c-18:3 | 1.5 ± 0.22 b | 1.5 ± 0.03 b | 0.9 ± 0.06 c | 1.8 ± 0.05 a | 0.4 ± 0.03 d | 0.2 ± 0.01 d | 0.4 ± 0.03 d | 0.4 ± 0.01 d | 0.4 ± 0.02 d | 0.2 ± 0.01 d |
| 9c12c15c-18:3 | 13.5 ± 1.6 a | 11.0 ± 0.04 b | 11.4 ± 0.1 b | 13.4 ± 0.2 a | 2.3 ± 0.2 d | 0.5 ± 0.1 d | 2.3 ± 0.1 d | 5.1 ± 0.02 c | 2.2 ± 0.1 d | 1.7 ± 0.04 d |
| 6c9c12c15c-18:4 | 7.4 ± 1.1 ab | 6.4 ± 0.02 bc | 5.6 ± 0.04 c | 7.9 ± 0.1 a | 0.1 ± 0.02 e | 0.3 ± 0.03 e | 0.1 ± 0.02 e | 2.0 ± 0.004 d | 2.2 ± 0.1 d | 0.6 ± 0.01 e |
| 8c11c14c-20:3 | 0.2 ± 0.1 de | 0.3 ± 0.1 bcd | 0.4 ± 0.01 bc | 0.2 ± 0.06 de | 0.1 ± 0.01 e | 0.2 ± 0.01 de | 0.1 ± 0.01 e | 0.4 ± 0.01 bc | 0.5 ± 0.02 b | 1.4 ± 0.05 a |
| 11c14c17c-20:3 | 0.1 ± 0.04 b | nd | nd | 0.1 ± 0.02 b | 0.3 ± 0.03 a | nd | 0.3 ± 0.02 a | 0.1 ± 0.004 b | 0.1 ± 0.01 b | 0.1 ± 0.003 b |
| 5c8c11c14c-20:4 | 0.7 ± 0.03 cde | 0.7 ± 0.01 cde | 0.7 ± 0.02 cde | 0.4 ± 0.1 e | 1.1 ± 0.1 cd | 1.3 ± 0.06 cd | 1.1 ± 0.08 cd | 4.0 ± 0.02 a | 2.0 ± 0.1 b | 4.3 ± 0.1 a |
| 13c16c-22:2 | 0.6 ± 0.1 ab | nd | nd | 0.4 ± 0.01 bc | 0.7 ± 0.1 a | 0.2 ± 0.01 de | 0.6 ± 0.1 a | 0.3 ± 0.01 cd | 0.1 ± 0.01 e | 0.3 ± 0.02 cd |
| 5c8c11c14c17c-20:5 | 1.4 ± 0.0 ab | 1.2 ± 0.01 bc | 1.5 ± 0.04 a | 0.8 ± 0.1 d | 0.4 ± 0.1 e | 0.8 ± 0.1 d | 0.3 ± 0.1 e | 1.5 ± 0.02 a | 0.3 ± 0.02 e | 0.9 ± 0.1 cd |
| 4c7c10c13c16c19c-22:6 | 1.9 ± 0.2 b | 1.1 ± 0.1 c | 2.3 ± 0.1 a | 1.8 ± 0.1 b | nd | nd | 0.2 ± 0.11 d | 0.1 ± 0.003 d | nd | nd |
| ΣPUFAs | 32.4 ± 34.0 ab | 29.2 ± 0.1 b | 34.4 ± 0.2 a | 35.4 ± 0.5 a | 7.9 ± 0.5 ef | 5.6 ± 0.1 f | 7.4 ± 0.4 f | 17.5 ± 0.1 c | 11.4 ± 0.5 de | 12.9 ± 0.3 d |
| ΣC18 PUFAs | 27.6 ± 3.3 a | 25.8 ± 0.1 a | 29.4 ± 0.2 a | 31.7 ± 0.4 a | 5.4 ± 0.3 cd | 3.2 ± 0.2 e | 5.0 ± 0.3 cd | 11.2 ± 0.04 b | 8.4 ± 0.3 bc | 5.8 ± 0.1 cd |
| ΣC20 PUFAs | 2.3 ± 0.3 bc | 2.2 ± 0.1 bc | 2.6 ± 0.03 b | 1.5 ± 0.5 e | 1.8 ± 0.1 cd | 2.3 ± 0.2 bc | 1.7 ± 0.04 cd | 6.0 ± 0.1 a | 2.9 ± 0.2 b | 6.7 ± 0.2 a |
| n6/n3 PUFA | 0.3 ± 0.02 e | 0.5 ± 0.002 e | 0.7 ± 0.002 de | 0.5 ± 0.002 e | 1.7 ± 0.2 b | 2.5 ± 0.3 a | 1.6 ± 0.2 b | 1.0 ± 0.003 cd | 1.4 ± 0.03 bc | 2.9 ± 0.03 a |
| PUFA/SFA | 0.6 ± 0.04 b | 0.6 ± 0.003 b | 0.6 ± 0.003 b | 0.8 ± 0.02 a | 0.1 ± 0.01 e | 0.1 ± 0.002 e | 0.1 ± 0.01 e | 0.3 ± 0.002 c | 0.2 ± 0.01 d | 0.2 ± 0.01 d |
| UI | 123.9 ± 6.3 b | 112.8 ± 0.5 c | 119.2 ± 0.5 bc | 133.3 ± 1.5 a | 42.3 ± 1.3 g | 28.6 ± 0.8 h | 42.2 ± 0.8 g | 78.9 ± 0.4 d | 55.8 ± 0.8 f | 67.7 ± 1.3 e |
| AI | 1.0 ± 0.2 de | 1.1 ± 0.01 de | 1.2 ± 0.03 d | 0.8 ± 0.01 e | 2.5 ± 0.1 b | 6.7 ± 0.7 a | 2.4 ± 0.06 b | 2.1 ± 0.01 bc | 2.5 ± 0.04 b | 1.6 ± 0.04 c |
| TI | 0.6 ± 0.04 e | 0.7 ± 0.003 e | 0.6 ± 0.002 e | 0.5 ± 0.01 e | 5.0 ± 0.2 b | 11.1 ± 0.8 a | 5.2 ± 0.2 b | 2.0 ± 0.01 d | 3.2 ± 0.1 c | 4.0 ± 0.2 c |
nd—not detected; a–g Values in a row for each fatty acids without a common superscript are significantly different between different seaweeds at p < 0.01.
3.1.1. Chlorophyta
The major FAs detected in Chlorophyta species were 16:0, 18:0, 9c-16:1, 9c-18:1, 9c12c-18:2, 9c12c15c-18:3, and 6c9c12c15c-18:4, which together accounted for 81.5% to 86.2% of TFA. The contents of SFAs were high (approximately 44.5% to 51.6% in Ulva spp. to 72.0% in C. papillatum). The contents of MUFAs were low (11.5% in U. ohnoi to 20.7% in U. tepida), while PUFAs ranged from 29.2% in U. chaugulii to 35.4% in U. tepida, except for C. papillatum, which had exceptionally low PUFAs (7.9%) (Table 2).
They had a characteristic FA profile of higher C18 PUFAs than C20 PUFAs, 11.2 to 21.8-fold higher in Ulva spp. and 3.0-fold higher in C. papillatum in congruence with previous studies [4,11,14,15,43,44,45,46,47,48]. The contents of long-chain PUFAs 5c8c11c14c-20:4 and 5c8c11c14c17c-20:5 were considerably lower as compared with red and brown seaweeds (1.2–2.2%) (Table 2). The FA profiles of green seaweeds from Arabian Gulf in the present study were very similar to those reported from tropical or sub-tropical regions of warm climate, exhibiting high SFA and low MUFA and PUFAs [11,35,43,44,45,46].
Further, the members of the same genus exhibited similar FA patterns but differed significantly in their individual FA contents (p < 0.01) as reported in previous studies [4,11,14]. Ulva spp. displayed characteristic FA profiles of high 16:0, 9c-18:1, C18 PUFAs with higher 9c12c15c-18:3 content than 9c12c-18:2 (1.6- to 2.6- times, except in U. ohnoi), and low C20 PUFAs, as reported previously for the same or related Ulva spp. [4,14,15,35,39,46,49]. However, exceptionally higher contents of MUFAs (29.1–32.37%), especially of 9c-18:1 (22.3–26.8%), which had approximately 1.4 to 4.6-fold higher values than the present study, have been reported previously for Ulva species from Iran [35,43].
6c9c12c15c-18:4 is another characteristic FA reported for Ulva spp. [4,14,25,46,49]. Ulva spp. displayed the highest content of 6c9c12c15c-18:4 among all the seaweeds in our study, approximately 5.6% to 7.9%. Ulva spp. also contained 4c7c10c13c16c19c-22:6 in the present study in low amounts (1.1–2.3%) in congruence with previous reports for Ulva spp. (0.1–1.44%) from Yellow Sea [50], Black Sea and Dardenelles [51], Iranian coast [43], southern Australian coast of Tasmania [4], Brazilian coast [46], and Chilean sub-Antarctic region [15]. However, Kumari and co-workers reported relatively higher amounts of 4c7c10c13c16c19c-22:6, approximately 0.7% to 3.4%, from twelve species of fresh Ulva thalli collected during March–October 2011 [14] and 2.15% to 6.05% from shade-dried Ulva species [11] collected during January–April 2008 from the Indian coast. Such large variations in 4c7c10c13c16c19c-22:6 content in Ulva spp. can be due to inter-specific variation, different sampling sites, season, and other environmental factors [14,28,52].
C. papillatum belonging to the Bryopsidales displayed a distinctly different fatty acid profile from those observed for Ulva spp. (Table 2). Specifically, the SFAs content was 1.3 to 1.6- fold higher, mainly due to higher levels in 16:0 (1.2 to 1.5-fold) and 9c-18:1 (n9) (1.2 to 3.1-fold), while the content of PUFAs was significantly lower, especially 9c12c-18:2 (2.2 to 5-fold) and 9c12c15c-18:3 (4.8 to 6.3-fold) as compared with Ulva spp. These differences in FA profiles between Ulva spp. and C. papillatum may be due to genotypic differences as well as different time of sampling. Similar FA profiles with high SFA, 16:0 and low 9c12c-18:2, 9c12c15c-18:3, and minor amounts of C20 PUFAs have been reported for C. dwarkense from Gujarat coast, India [14] and C. bursa from Adriatic Sea, Croatia [29]. However, other studies have detected appreciable amounts of 16:0 (20.9–38.7%), low MUFAs and high PUFAs with C16 PUFAs (16.8–18.9%), C18 PUFAs (25.4–29.5%), and C20 PUFAs (6.4–14.6%) in different Codium species including C. fragile, C. geppi, C. papillatum, C. tomentosum, and Codium sp. across different regions across the world [4,39,48,49,53]. Previously, Dembitsky et al. [54] demonstrated large variations in the FA content of the genus Codium, depending on the species, the season, and the geographic origin of the sample.
3.1.2. Rhodophyta
We investigated only one red seaweed, Chondria sp., belonging to the order Ceramiales (Table 2). Chondria sp. had the highest SFA and the lowest MUFA and PUFA contents among all the seaweeds investigated in the present study. The highest content of SFA was mainly due to high contents of 16:0 (63.9%) and 14:0 (11.7%), in line with the previous report of Govenkar and Wahidullah [55]. These authors reported high SFAs (80.73%), of which 16:0 accounted for 74.3% and low MUFAs (19.27%), while no PUFA was detected in Chondria armata [55]. High 14:0 contents (6.8–13.4%) are characteristic of the seaweeds of the order Ceramiales [11,14]. In contrast to our findings, 5c8c11c14c-20:4 and 5c8c11c14c17c-20:5 have been reported in high amounts together contributing to 43.3% in Chondria dasyphylla and 23.8% in Chondria decipiens from the Sea of Japan, respectively [56]. Vaskovsky et al. [50] reported low 16:0 (27.7%), 9c-18:1 (6.4%), C18 PUFAs <1%, and high C20 PUFAs (45.7%) in Chondria capillaris from the Yellow Sea. Stefanov et al. [57] showed large variations in the FA contents of C. capillaris collected from the Black Sea and Lake Pomorie (Bulgaria), especially in SFAs (42.4–60.2%) and PUFA contents (11.7–27.2%).
3.1.3. Ochrophyta
The FA compositions of four brown seaweeds belonging to the orders Dictyotales, Ectocarpales, and Fucales are given in Table 2. While the contents of individual FAs differed significantly between different individual species, a common trait of different brown seaweed FA profiles was high content of 14:0, 16:0, and 9c-18:1 together accounting for 72.6% to 76.3%. In general, for brown seaweeds investigated in the present study, we found higher SFA contents (61.3–71.3%), similar MUFAs (20.5–26.0%), but lower PUFAs (7.4–17.6%) compared with the same or related brown seaweed species reported from different parts of the world [4,11,14,23,24,37,39,45,46,47,48,58]. Interestingly, Rohani-Gadhikolaei et al. [35] reported comparable FA profiles with high SFAs (51.9–55.2%), MUFAs (27.5–32.9%), and low PUFAs (15.3–17.4%), especially low 5c8c11c14c-20:4 and 5c8c11c14c17c-20:5 (together accounting for only 2.9–5.8%), for brown seaweeds including Sargassum spp. and Colpomenia sinuosa from the Iranian coast. It is worth noting that the two Ectocarpales species investigated by us, I. stellata and F. indica, had distinct FA compositions, which is in line with the large variations in FA profiles reported for different species of Ectocarpales [4,14,39]. I. stellata had higher contents of SFAs (1.2-fold), 12:0 (9.6-fold), 16:0 (1.1-fold), 18:0 (1.5-fold), 22:0 (10.5-fold), but low 14:0 (2.4-fold), 9c-16:1 (3.0-fold) and PUFAs (2.4-fold) as compared with F. indica (Table 2). S. aquifolium (Fucales) had lower 14:0 (1.4-fold) and 6c9c12c15c-18:4 contents (3.8-fold) as compared with P. boergesenii (Dictyotales) as reported previously for Fucales members [4,11,14,24,45]. Furthermore, brown seaweeds differ from red and green seaweeds based on their predominantly higher amounts of both C18 and C20 PUFAs (both DW and FW basis) [4,11,14,24,47,48,58]. Kumari et al. [14] reported FA profiles of 24 brown seaweeds exhibiting higher PUFA contents (23.7–58.0%) and further differentiated them into three groups based on the relative contents of C18 and C20 PUFAs. The first group included brown seaweeds containing higher C18 PUFAs (1.1 to 1.2-fold) such as Padina spp., Sirophysalis trinodis, and Feldmannia mitchelliae. The second group included species containing higher C20 PUFAs such as Sargasuum spp., Dictyota spp. (D. pinnatifida, D. bartayresiana, D. dichotoma), and Hormophysa cuneiformis, while the third group consisted of Dictyopteris delicatula, Canistrocarpus cervicornis, and Dictyota ciliolata containing equal amounts of C18 and C20 PUFAs. Despite low PUFA contents determined in brown seaweeds in our study, P. boergesenii, I. stellata and F. indica had 1.8 to 2.9-fold higher C18 PUFAs than C20 PUFAs, while S. aquifolium had 1.2-fold higher C20 PUFA levels than C18 PUFAs in congruence with the previous reports about Padina spp., Sargassum spp. and related brown seaweeds [11,14]. Similar higher contents of C20 PUFAs than those of C18 PUFAs (1.02 to 3.8-fold; due to higher 5c8c11c14c-20:4 and 5c8c11c14c17c-20:5 contents) were reported for different Sargassum spp. collected from different regions of the world [24,39,48,59]. On the contrary, Verma et al. [45] reported higher C18 PUFAs in all the brown seaweeds studied including Padina tetrastomatica, Spatoglossum asperum, Feldmannia marginatum, I. stellata, and Sargassum linearifolium due to higher 9c12c-18:2contents (17.16–23.15% TFA) while 5c8c11c14c-20:4 and 5c8c11c14c17c-20:5 were detected in minor amounts.
Overall, a substantial variation was observed in the individual FA contents of the same and related species of the same genus among all green, red, and brown seaweeds in our study, which is also reflected in the literature from different regions of the world [4,24,39,45,47,48,52]. These variations are due to species-specific variations, different geographical locations, and environmental factors (temperature, light, salinity, nutrients) [25,28,30,46,59]. Thus, it becomes necessary to screen different seaweeds (both wild and cultivated) from different regions for their FA contents and to monitor them across different seasons to determine the suitable period of harvest for seaweed valorization. The effect of different seasons or other environmental factors on FA composition of seaweeds were not studied in the present study but will be an objective of our future research. Additionally, there can be variations in FA contents of same or related species in literature due to different extraction and derivatization methods employed by researchers [25,60], but it is beyond the scope of this study to compare such FA variations.
3.2. Fatty Acid Chemotaxonomy
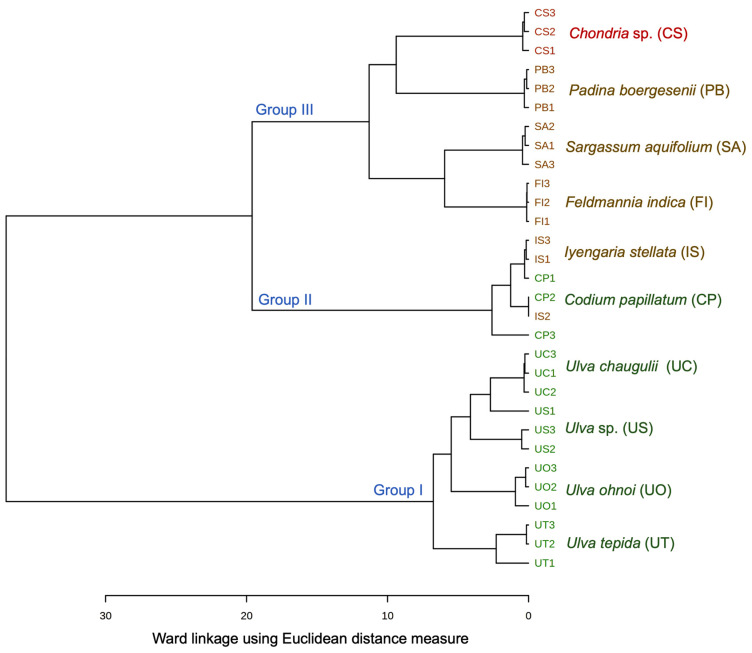
Hierarchical clustering was performed on the FA data matrix (Supplementary datasheet S1) to evaluate the chemotaxonomic relationships between different species at different taxonomic levels. A few FAs, namely, 10:0, 13:0, 24:0, 9c-14:1, 10c-15:1, 10c-17:1, 9t-18:1, 11c-20:1, 13c-22:1, 15c-24:1, 9t12t-18:2, 13c16c-22:2, and 11c14c17c-20:3 were excluded from this FA data matrix due to their insignificant amounts and lack of correlation with the data matrix since such variables often lead to misclassification of species. The dendrogram obtained from Ward hierarchical clustering grouped the seaweed samples into three demarcated clusters (Figure 1).
Figure 1.
Dendrogram obtained from hierarchical clustering of seaweeds samples using Ward linkage with Euclidean distance.
Ward linkage is an agglomerative clustering algorithm which starts with n singleton clusters (each consisting of one element of the data set) and merges two clusters based on similarity measure. All of the Ulva species (Ulvales) were grouped together in group I while C. papillatum (Bryopsidales) was grouped with I. stellata (Ectocarpales) in group II. The single red alga investigated in this study, Chondria sp., belonging to Ceramiales, was grouped together with brown seaweeds P. boergesenii (Dictyotales), S. aquifolium (Fucales), and F. indica (Ectocarpales) in group III. Kumari et al. [14] also showed that Bryopsidales are grouped separately from Ulvales and the latter generally aligns with Ulotrichales, forming the Ulvales-Ulotrichales clade [14]. However, a greater number of replicates as well as species belonging to the genus Codium and Iyengaria are required to resolve their misclassification based on FAs, as observed in our study. Further, group I can be sub-divided into two sub-groups, consisting of U. tepida in one, and U. ohnoi, Ulva sp. and U. chaugulii in another sub-group. Similarly, group III can be further sub-divided into three sub-groups, the first comprising of F. indica and S. aquifolium, the second of P. boergesenii, which was closely related to Chondria sp., forming the third sub-group. Similarly, the species belonging to the genera Padina and Sargassum were grouped in different sub-groups based on their FA profiles [11,14,45] as well as different clades based on their molecular data [61]. However, for adequate comparison of inter-relationships between different groups deduced from FA composition with the clades inferred from genomic data, extensive sampling effort with samples belonging to the same genus as well as same class or orders are imperative.
Thus, our study displayed that FA traits are conserved in seaweeds at higher ordinal levels of families, orders, and phyla, in line with the previous findings [4,11,14,16,17,45]. FA signatures could be potential tools for understanding the chemotaxonomic relationships among different seaweed species, but require proper sampling. Otherwise, higher variations in FA contents at the levels of genus or species may pose difficulty in discriminating species in the absence of adequate taxon sampling and replicates, as observed in our study for C. papillatum and I. stellata.
3.3. Nutritional Assessment for Seaweed Valorization
Our study revealed that Ulva species are rich sources of nutritionally important PUFAs with their unsaturation indices (UI) varying from 119.21 ± 0.45 (U. ohnoi) to 133.28 ± 1.65 (U. tepida) (Table 2) in congruence with the UI values reported in the literature for different species of the genus Ulva [11,14,62]. The UI values for all other species in our study were low, varying from 28.63 ± 0.84 (Chondria sp.) to 78.91 ± 0.35 (F. indica) in agreement with lower PUFA contents in these species.
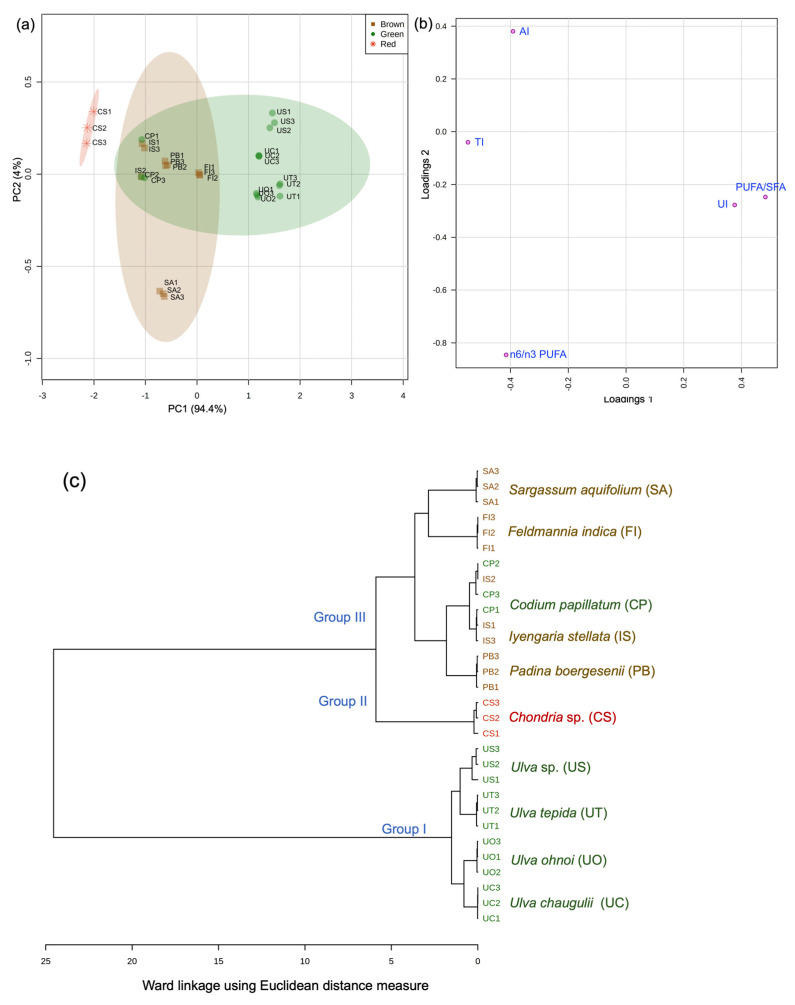
Further, we conducted a principal component analysis (PCA) (without rotation) and hierarchical clustering based on and the nutritional indices data matrix (Supplementary datasheet S2) to identify potential seaweeds that can be valorized for nutritional and functional food applications. We obtained a KMO value of 0.726 and a significant level for the Bartlett’s test (Supplementary Table S1) for the nutritional indices data matrix, suggesting that nutritional indices variables were highly correlated.
The principal components were extracted based on scree plot (Supplementary Figure S1) and the first two principal components, which also presented the maximum explained variance, were used for generating scores and loading plot. PCA of nutritional indices data matrix explained 98.4% of variations (PC1-94.4% and PC2-4%) (Figure 2a). The discriminant variables along PC1 were PUFA/SFA, UI, and TI, and along PC2 were AI and n6/n3 PUFA (Figure 2b). The loadings plot displayed that PUFA/SFA and UI were highly positively correlated, while both these were negatively correlated with TI. Similarly, AI was negatively correlated with n6/n3 PUFAs. Further, PC2 (Y-axis) separated all the Ulva species from brown and red seaweeds owing to their higher loadings of PUFA/SFA and UI, while C. papillatum was positioned along with the brown seaweeds due to its lower contents of UI and PUFA/SFA. S. aquifolium was found to be the outlier, separated from rest of the brown seaweeds by X-axis due to its high loadings of n6/n3 PUFAs. Chondria sp. was separated from the rest of green and brown seaweeds due to its higher loadings of TI and AI in line with its exceptionally high TI and AI contents (Table 2).
Figure 2.
Principal component analysis (PCA) and hierarchical clustering of seaweeds based on nutritional indices. (a) PCA scores plot and (b) PCA loadings plot and (c) dendrogram obtained from hierarchical clustering using Ward linkage and Euclidean distance measures. Seaweed species are labeled as mentioned in Table 1. Abbreviations: AIatherogenic index, PUFApolyunsaturated fatty acids, SFAsaturated fatty acid, STAstearidonic acid, TIThrombogenic index, UIunsaturation index.
The dendrogram obtained from hierarchical clustering of nutritional indices data revealed three demarcated clusters (Figure 2c). All of Ulva spp. (containing high UI and PUFA/SFA) were clustered together in Group I, like the Ulvales clade deduced from the FA data matrix (Figure 1). Contrary to our previous results, where Chondria sp. was grouped with other brown seaweeds in group III (Figure 1), here, Chondria sp. formed a separate clade, group II. C. papillatum was grouped with brown seaweeds in group III, sharing the sub-clade with I. stellata and P. boergesonii.
PUFAs are essential biomolecules to human health since their consumption is associated with decreased risk of cardiovascular and inflammatory diseases as well as cancer [18,19,20,63,64]. 9c12c15c-18:3 is a precursor of 5c8c11c14c17c-20:5 as well as 4c7c10c13c16c19c-22:6 and has anticancer, antiosteoporotic, antioxidant, anti-inflammatory, as well as coronary and neuronal protective effects [65]. 4c7c10c13c16c19c-22:6 is essential for visual and neurological development in infants while 5c8c11c14c-20:4 and 5c8c11c14c17c-20:5 are precursors of prostaglandins, thromboxanes, and other eicosanoids that influence inflammation processes and immune reactions [18,63]. Free PUFAs also have biological effects including induction of an oxidative burst, oxylipin biosynthesis, and induction of resistance against pathogens in seaweeds such as the brown algal kelps Laminaria digitata and Macrocystis pyrifera [66]. The PUFA/SFA ratio, which is an important parameter to assess the nutritional quality of the lipid fraction of food, should be ≥0.4 [67]. In this study, the PUFA/SFA values were in accordance with the nutritional guidelines only for Ulva species (0.57–0.80). However, much higher PUFA/SFA values (≥0.4) have been reported for species of the genera Ulva, Codium, Sargassum, and Padina in previous reports [4,14,15,24,25,34,43,46,49]. The low PUFA/SFA ratio in our study may be due the warm environment of the Arabian Gulf, in agreement with the reports that seaweeds of temperate regions tend to feature a higher degree of unsaturation in their fatty acid composition [46,59,62,68]. High SFA content in tropical seaweeds may be related to their physiological adaptation to warm temperatures, while high PUFA content in cold water may facilitate thermo-adaptive regulation of membrane lipid fluidity [59,69]. All the species investigated in the present study had health-promoting n6/n3 ratios ranging from 0.33 ± 0.02:1 (Ulva sp.) to 2.94 ± 0.03:1 (S. aquifolium) (Table 2) in line with the World Health Organization (WHO) recommendations of an n6/n3 ratio of 5:1 [19,20,70]. The atherogenic indices (AI) varied from 0.8 ± 0.01 (U. tepida) to 2.52 ± 0.04 (P. boergesenii), while thrombogenic indices (TI) varied from 0.49 ± 0.01 (U. tepida) to 5.17 ± 0.15 (I. stellata), except for Chondria sp., which had higher AI and TI values (Table 2). Low AI and TI < 3 have been reported for different green, red, and brown seaweeds in the literature [2,14,24,25,49]. Recently, Chen and Liu [23] compared the nutritional indies of numerous seaweeds reported in the literature with those of plant oils, fish, and dairy products. Accordingly, AI and TI values obtained in our study for all seaweeds except Chondria sp. (Table 2) were comparable to those of fish (AI—0.37–1.22, TI—0.14–0.87), shrimps (AI—0.71–0.82, TI—0.21–0.30), and dairy products (AI—1.42–5.13, TI—0.39–5.04) [23]. There is no recommended level of AI and TI in food products, but the consumption of foods with low AI and TI indices is helpful in reducing the risk of coronary heart diseases [23].
Overall, most of the seaweeds investigated in our study had health-beneficial n6/n3, AI, and TI values, but only Ulva spp. had higher UI and the recommended PUFA/SFA ratio. The multivariate analysis of nutritional indices clearly supported our findings and helped in assessing the nutritional potential of seaweeds from Arabian Gulf. Nevertheless, Ulva spp. are not only rich in essential PUFAs, but also contain high amounts of macro- and micronutrients as reported previously [71]. In addition, the nutritional value of Ulva species in terms of carbohydrates, protein, and fatty acids (especially PUFA content) has been reported to be comparable to some vegetables, nuts, and grains [2,23,72] and it has been consumed traditionally for centuries in many Asian countries [1,2,4,25,72].
4. Final Conclusions
Our study revealed that seaweeds from the Arabian Gulf exhibit typical FA profiles of warm waters with relatively high SFA and low PUFA contents. The green, red, and brown seaweeds exhibit species-specific significant differences in FA contents, but trends of FA profiles were conserved at different taxonomic ranks of genus, class, and order within each phyla. Among all the species investigated, Ulva spp. are the most suitable candidates for developing low-fat foods with PUFA-rich nutraceuticals or utilization in functional food for human consumption and animal feed due to their health beneficial PUFA/SFA, n6/n3, AI, and TI values. However, proper valorization of Ulva species for commercial utilization will require a temporal, spatial, and seasonal consistency in FA contents. Future studies for understanding the environmental and seasonal impacts on FA profiles of Ulva spp. from the Kuwait region will facilitate selecting the correct harvest time for obtaining high PUFA yields.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/foods10102442/s1, Supplementary Figure S1: Scree plot generated from principal component analysis of nutritional indices data matrix. Supplementary Table S1: Kaiser-Meyer-Olkin and Bartlett’s test values obtained from factor analysis of nutritional indices data matrix. Supplementary datasheet S1: Fatty acid data matrix used for hierarchical clustering. Supplementary datasheet S2: Nutritional indices data matrix used for principal component and hierarchical clustering analysis for nutritional assessment.
Author Contributions
Conceptualization, R.E. and F.C.K.; data curation, H.A.-A. and P.K.; formal analysis, H.A.-A. and T.K.A.-S.; funding acquisition, H.A.-A. and F.C.K.; investigation, H.A.-A.; methodology, H.A.-A., T.K.A.-S. and P.K.; project administration, D.A.-B., R.E. and F.C.K.; resources, D.A.-B. and F.C.K.; software, H.A.-A. and P.K.; supervision, D.A.-B., R.E., F.C.K. and P.K.; validation, T.K.A.-S. and P.K.; visualization, H.A.-A. and P.K.; writing—original draft, H.A.-A. and P.K.; writing—review and editing, H. A-A., T.K.A.-S., D.A.-B., R.E., F.C.K. and P.K. All authors have read and agreed to the published version of the manuscript.
Funding
We are grateful to the Kuwait Institute for Scientific Research (KISR) for PhD funding for H.A.-A. We are thankful to the National Unit for Environmental Research and Services (NUERS) at Kuwait University, Project # SRUIL01/13 and the Department of Marine Sciences for providing their facilities and labs. We equally thank the UK Natural Environment Research Council for their support to F.C.K. (program Oceans 2025–WP 4.5 and grants NE/D521522/1 and NE/J023094/1). This work also received support from the Marine Alliance for Science and Technology for Scotland pooling initiative. MASTS is funded by the Scottish Funding Council (grant reference HR09011) and contributing institutions. PK would like to acknowledge European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 839151 for funding.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferdouse F., Holdt S.L., Smith R., Murúa P., Yang Z. The global status of seaweed production, trade and utilization. [(accessed on 12 April 2021)];FAO Globefish. 2018 124:1. Available online: http://www.fao.org/publications/card/en/c/CA1121EN. [Google Scholar]
- 2.Kumar M., Kumari P., Trivedi N., Shukla M.K., Gupta V., Reddy C., Jha B. Minerals, PUFAs and antioxidant properties of some tropical seaweeds from Saurashtra coast of India. J. Appl. Phycol. 2011;23:797–810. doi: 10.1007/s10811-010-9578-7. [DOI] [Google Scholar]
- 3.Wells M.L., Potin P., Craigie J.S., Raven J.A., Merchant S.S., Helliwell K.E., Smith A.G., Camire M.E., Brawley S.H. Algae as nutritional and functional food sources: Revisiting our understanding. J. Appl. Phycol. 2017;29:949–982. doi: 10.1007/s10811-016-0974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid M., Kraft L.G., van der Loos L.M., Kraft G.T., Virtue P., Nichols P.D., Hurd C.L. Southern Australian seaweeds: A promising resource for omega-3 fatty acids. Food Chem. 2018;265:70–77. doi: 10.1016/j.foodchem.2018.05.060. [DOI] [PubMed] [Google Scholar]
- 5.Nunes N., Valente S., Ferraz S., Barreto M.C., de Carvalho M.A.P. Biochemical study of attached macroalgae from the Madeira Archipelago and beach-cast macroalgae from the Canary Islands: Multivariate analysis to determine bioresource potential. Bot. Mar. 2020;63:283–298. doi: 10.1515/bot-2019-0022. [DOI] [Google Scholar]
- 6.Zava T.T., Zava D.T. Assessment of Japanese iodine intake based on seaweed consumption in Japan: A literature-based analysis. Thyroid Res. 2011;4:14. doi: 10.1186/1756-6614-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hehemann J.H., Correc G., Barbeyron T., Helbert W., Czjzek M., Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 8.Goodyear D. A New Leaf: Seaweed Could Be a Miracle Food–If We Can Figure out How to Make it Taste Good. [(accessed on 6 May 2021)]. Available online: http://www.newyorker.com/magazine/2015/11/02/a-new-leaf.
- 9.Mac Monagail M., Cornish L., Morrison L., Araújo R., Critchley A.T. Sustainable harvesting of wild seaweed resources. Eur. J. Phycol. 2017;52:371–390. doi: 10.1080/09670262.2017.1365273. [DOI] [Google Scholar]
- 10.Kemmler W., Von Stengel S., Bebenek M., Engelke K., Hentschke C., Kalender W. Exercise and fractures in postmenopausal women: 12-year results of the Erlangen Fitness and Osteoporosis Prevention Study (EFOPS) Osteoporosis Int. 2012;23:1276. doi: 10.1007/s00198-011-1663-5. [DOI] [PubMed] [Google Scholar]
- 11.Kumari P., Kumar M., Gupta V., Reddy C., Jha B. Tropical marine macroalgae as potential sources of nutritionally important PUFAs. Food Chem. 2010;120:749–757. doi: 10.1016/j.foodchem.2009.11.006. [DOI] [Google Scholar]
- 12.Stengel D.B., Connan S., Popper Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011;29:483–501. doi: 10.1016/j.biotechadv.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Dellatorre F.G., Avaro M.G., Commendatore M.G., Arce L., de Vivar M.E.D. The macroalgal ensemble of Golfo Nuevo (Patagonia, Argentina) as a potential source of valuable fatty acids for nutritional and nutraceutical purposes. Algal Res. 2020;45:101726. doi: 10.1016/j.algal.2019.101726. [DOI] [Google Scholar]
- 14.Kumari P., Bijo A., Mantri V.A., Reddy C., Jha B. Fatty acid profiling of tropical marine macroalgae: An analysis from chemotaxonomic and nutritional perspectives. Phytochemistry. 2013;86:44–56. doi: 10.1016/j.phytochem.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 15.Dos Santos M.A.Z., Berneira L.M., Goulart N.L., Mansilla A., Astorga-España M.S., de Pereira C.M.P. Rhodophyta, Ochrophyta and Chlorophyta macroalgae from different sub-Antarctic regions (Chile) and their potential for polyunsaturated fatty acids. Braz. J. Bot. 2021;44:429–438. doi: 10.1007/s40415-021-00712-0. [DOI] [Google Scholar]
- 16.Gosch B.J., Magnusson M., Paul N.A., de Nys R. Total lipid and fatty acid composition of seaweeds for the selection of species for oil-based biofuel and bioproducts. Glob. Chang. Biol. Bioenergy. 2012;4:919–930. doi: 10.1111/j.1757-1707.2012.01175.x. [DOI] [Google Scholar]
- 17.Galloway A.W., Britton-Simmons K.H., Duggins D.O., Gabrielson P.W., Brett M.T. Fatty acid signatures differentiate marine macrophytes at ordinal and family ranks. J. Phycol. 2012;48:956–965. doi: 10.1111/j.1529-8817.2012.01173.x. [DOI] [PubMed] [Google Scholar]
- 18.Zárate R., El Jaber-Vazdekis N., Tejera N., Pérez J.A., Rodríguez C. Significance of long chain polyunsaturated fatty acids in human health. Clin. Transl. Med. 2017;6:25. doi: 10.1186/s40169-017-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simopoulos A.P. An increase in the omega-6/omega-3 fatty acid ratio increases the risk for obesity. Nutrients. 2016;8:128. doi: 10.3390/nu8030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simopoulos A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008;233:674–688. doi: 10.3181/0711-MR-311. [DOI] [PubMed] [Google Scholar]
- 21.Cornish M.L., Critchley A.T., Mouritsen O.G. Consumption of seaweeds and the human brain. J. Appl. Phycol. 2017;29:2377–2398. doi: 10.1007/s10811-016-1049-3. [DOI] [Google Scholar]
- 22.Lee J.C., Hou M.F., Huang H.W., Chang F.R., Yeh C.C., Tang J.Y., Chang H.W. Marine algal natural products with anti-oxidative, anti-inflammatory, and anti-cancer properties. Cancer Cell Int. 2013;13:55. doi: 10.1186/1475-2867-13-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Liu H. Nutritional indices for assessing fatty acids: A mini-review. Int. J. Mol. Sci. 2020;21:5695. doi: 10.3390/ijms21165695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z., Xu Y., Liu T., Zhang L., Liu H., Guan H. Comparative studies on the characteristic fatty acid profiles of four different Chinese medicinal Sargassum seaweeds by GC-MS and chemometrics. Mar. Drugs. 2016;14:68. doi: 10.3390/md14040068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreira A.S., da Costa E., Melo T., Lopes D., Pais A., Santos S.A., Pitarma B., Mendes M., Abreu M.H., Collén P.N. Polar lipids of commercial Ulva spp. of different origins: Profiling and relevance for seaweed valorization. Foods. 2021;10:914. doi: 10.3390/foods10050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulbricht T., Southgate D. Coronary heart disease: Seven dietary factors. Lancet. 1991;338:985–992. doi: 10.1016/0140-6736(91)91846-M. [DOI] [PubMed] [Google Scholar]
- 27.Yurchenko S., Sats A., Tatar V., Kaart T., Mootse H., Jõudu I. Fatty acid profile of milk from Saanen and Swedish Landrace goats. Food Chem. 2018;254:326–332. doi: 10.1016/j.foodchem.2018.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Britton D., Schmid M., Revill A.T., Virtue P., Nichols P.D., Hurd C.L., Mundy C.N. Seasonal and site-specific variation in the nutritional quality of temperate seaweed assemblages: Implications for grazing invertebrates and the commercial exploitation of seaweeds. J. Appl. Phycol. 2021;33:603–616. doi: 10.1007/s10811-020-02302-1. [DOI] [Google Scholar]
- 29.Cvitković D., Dragović-Uzelac V., Dobrinčić A., Čož-Rakovac R., Balbino S. The effect of solvent and extraction method on the recovery of lipid fraction from Adriatic Sea macroalgae. Algal Res. 2021;56:102291. doi: 10.1016/j.algal.2021.102291. [DOI] [Google Scholar]
- 30.Garcia-Vaquero M., Rajauria G., Miranda M., Sweeney T., Lopez-Alonso M., O′Doherty J. Seasonal variation of the proximate composition, mineral content, fatty acid profiles and other phytochemical constituents of selected brown macroalgae. Mar. Drugs. 2021;19:204. doi: 10.3390/md19040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guiry M.D. How many species of algae are there? J. Phycol. 2012;48:1057–1063. doi: 10.1111/j.1529-8817.2012.01222.x. [DOI] [PubMed] [Google Scholar]
- 32.Sheppard C., Al-Husiani M., Al-Jamali F., Al-Yamani F., Baldwin R., Bishop J., Benzoni F., Dutrieux E., Dulvy N.K., Durvasula S.R.V., et al. Environmental concerns for the future of gulf coral reefs. In: Riegl B.M., Purkis S.J., editors. Coral Reefs of the Gulf: Adaptation to Climatic Extremes. Springer Science and Business Media; New York, NY, USA: 2012. [Google Scholar]
- 33.Heiba H.I., Al-Easa H.S., Rizk A.F.M. Fatty acid composition of twelve algae from the coastal zones of Qatar. Plant Foods Hum. Nutr. 1997;51:27–34. doi: 10.1023/A:1007980227542. [DOI] [PubMed] [Google Scholar]
- 34.Abomohra A.E., El-Naggar A.H., Baeshen A.A. Potential of macroalgae for biodiesel production: Screening and evaluation studies. J. Biosci. Bioeng. 2018;125:231–237. doi: 10.1016/j.jbiosc.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Rohani-Ghadikolaei K., Abdulalian E., Ng W.K. Evaluation of the proximate, fatty acid and mineral composition of representative green, brown and red seaweeds from the Persian Gulf of Iran as potential food and feed resources. J. Food Sci. Tecnol. 2012;49:774–780. doi: 10.1007/s13197-010-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabarsa M., Rezaei M., Ramezanpour Z., Robert Waaland J., Rabiei R. Fatty acids, amino acids, mineral contents, and proximate composition of some brown seaweeds. J. Phycol. 2012;48:285–292. doi: 10.1111/j.1529-8817.2012.01122.x. [DOI] [PubMed] [Google Scholar]
- 37.Akbary P., Liao L.M., Aminikhoei Z., Tavabe K.R., Hobbi M., Erfanifar E. Sterol and fatty acid profiles of three macroalgal species collected from the Chabahar coasts, southeastern Iran. Aquacult. Int. 2021;29:155–165. doi: 10.1007/s10499-020-00616-y. [DOI] [Google Scholar]
- 38.Fariman G.A., Shastan S.J., Zahedi M.M. Seasonal variation of total lipid, fatty acids, fucoxanthin content, and antioxidant properties of two tropical brown algae (Nizamuddinia zanardinii and Cystoseira indica) from Iran. J. Appl. Phycol. 2016;28:1323–1331. doi: 10.1007/s10811-015-0645-y. [DOI] [Google Scholar]
- 39.Al-Hasan R.H., Hantash F.M., Radwan S.S. Enriching marine macroalgae with eicosatetraenoic (arachidonic) and eicosapentaenoic acids by chilling. Appl. Microbiol. Biotechnol. 1991;35:530–535. doi: 10.1007/BF00169763. [DOI] [Google Scholar]
- 40.Christie W.W., Han X. Preparation of derivatives of fatty acids. In: Christie W.W., Han X., editors. Oily Press Lipid Library Series, Lipid Analysis. 4th ed. Woodhead Publishing; Sawston, UK: 2012. pp. 145–158. [Google Scholar]
- 41.Poerschmann J., Spijkerman E., Langer U. Fatty acid patterns in Chlamydomonas sp. as a marker for nutritional regimes and temperature under extremely acidic conditions. Microb. Ecol. 2004;48:78–89. doi: 10.1007/s00248-003-0144-6. [DOI] [PubMed] [Google Scholar]
- 42.Ward J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 43.Pirian K., Piri K., Sohrabipour J., Blomster J. Three species of Ulva (Ulvophyceae) from the Persian Gulf as potential sources of protein, essential amino acids and fatty acids. Phycol. Res. 2018;66:149–154. doi: 10.1111/pre.12212. [DOI] [Google Scholar]
- 44.Pirian K., Jeliani Z.Z., Arman M., Sohrabipour J., Yousefzadi M. Proximate analysis of selected macroalgal species from the Persian Gulf as a nutritional resource. Trop. Life Sci. Res. 2020;31:1–17. doi: 10.21315/tlsr2020.31.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma P., Kumar M., Mishra G., Sahoo D. Multivariate analysis of fatty acid and biochemical constitutes of seaweeds to characterize their potential as bioresource for biofuel and fine chemicals. Bioresour. Technol. 2017;226:132–144. doi: 10.1016/j.biortech.2016.11.044. [DOI] [PubMed] [Google Scholar]
- 46.Santos J., Guihéneuf F., Fleming G., Chow F., Stengel D. Temporal stability in lipid classes and fatty acid profiles of three seaweed species from the north-eastern coast of Brazil. Algal Res. 2019;41:101572. doi: 10.1016/j.algal.2019.101572. [DOI] [Google Scholar]
- 47.Li X., Fan X., Han L., Lou Q. Fatty acids of some algae from the Bohai Sea. Phytochemistry. 2002;59:157–161. doi: 10.1016/S0031-9422(01)00437-X. [DOI] [PubMed] [Google Scholar]
- 48.Pereira H., Barreira L., Figueiredo F., Custódio L., Vizetto-Duarte C., Polo C., Rešek E., Engelen A., Varela J. Polyunsaturated fatty acids of marine macroalgae: Potential for nutritional and pharmaceutical applications. Mar. Drugs. 2012;10:1920–1935. doi: 10.3390/md10091920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopes D., Melo T., Rey F., Meneses J., Monteiro F.L., Helguero L.A., Abreu M.H., Lillebø A.I., Calado R., Domingues M.R. Valuing bioactive lipids from green, red and brown macroalgae from aquaculture, to foster functionality and biotechnological applications. Molecules. 2020;25:3883. doi: 10.3390/molecules25173883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaskovsky V.E., Khotimchenko S.V., Xia B., Hefang L. Polar lipids and fatty acids of some marine macrophytes from the Yellow Sea. Phytochemistry. 1996;42:1347–1356. doi: 10.1016/0031-9422(96)00117-3. [DOI] [Google Scholar]
- 51.Yazici Z., Aysel V., Öksüz E., Köse A., Cumali S., Güven K. Fatty acid composition of marine macroalgae from the Black Sea and Dardanelles. Toxicol. Environ. Chem. 2007;89:371–379. doi: 10.1080/02772240601012366. [DOI] [Google Scholar]
- 52.McCauley J.I., Meyer B.J., Winberg P.C., Skropeta D. Parameters affecting the analytical profile of fatty acids in the macroalgal genus Ulva. Food Chem. 2016;15:332–340. doi: 10.1016/j.foodchem.2016.04.039. [DOI] [PubMed] [Google Scholar]
- 53.Khotimchenko S.V. Fatty acids of species in the genus Codium. Bot. Mar. 2003;46:456–460. doi: 10.1515/BOT.2003.046. [DOI] [Google Scholar]
- 54.Dembitsky V., Řezanková H., Řezanka T., Hanuš L. Variability of the fatty acids of the marine green algae belonging to the genus Codium. Biochem. Syst. Ecol. 2003;31:1125–1145. doi: 10.1016/S0305-1978(03)00043-7. [DOI] [Google Scholar]
- 55.Govenkar M., Wahidulla S. Studies on the fatty acids of the red alga Chondria armata (Kütz.) Okamura. Bot. Mar. 1999;42:3–5. doi: 10.1515/BOT.1999.001. [DOI] [Google Scholar]
- 56.Khotimchenko S., Gusarova I. Red algae of Peter the Great Bay as a source of arachidonic and eicosapentaenoic acids. Russ. J. Mar. Biol. 2004;30:183–187. doi: 10.1023/B:RUMB.0000033953.67105.6b. [DOI] [Google Scholar]
- 57.Stefanov K., Seizova K., Elenkov I., Kuleva L., Popov S., Dimitrova-Konaklieva S. Lipid composition of the red alga Chondria tenuissima (Good et Wood.) Ag., inhabiting waters with different salinities. Bot. Mar. 1994;37:445–448. doi: 10.1515/botm.1994.37.5.445. [DOI] [Google Scholar]
- 58.Khotimchenko S. Fatty acid composition of seven Sargassum species. Phytochemistry. 1991;30:2639–2641. doi: 10.1016/0031-9422(91)85113-E. [DOI] [Google Scholar]
- 59.Narayan B., Miyashita K., Hosakawa M. Comparative evaluation of fatty acid composition of different Sargassum (Fucales, Phaeophyta) species harvested from temperate and tropical waters. J. Aquat. Food Prod. Technol. 2005;13:53–70. doi: 10.1300/J030v13n04_05. [DOI] [Google Scholar]
- 60.Kumari P., Reddy C.R.K., Jha B. Comparative evaluation and selection of a method for lipid and fatty acid extraction from macroalgae. Anal. Biochem. 2011;415:134–144. doi: 10.1016/j.ab.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 61.Silberfeld T., Leigh J.W., Verbruggen H., Cruaud C., De Reviers B., Rousseau F. A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): Investigating the evolutionary nature of the “brown algal crown radiation”. Mol. Phylogenet. Evol. 2010;56:659–674. doi: 10.1016/j.ympev.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 62.Colombo M.L., Rise P., Giavarini F., De Angelis L., Galli C., Bolis C. Marine macroalgae as sources of polyunsaturated fatty acids. Plant Foods Hum. Nutr. 2006;61:64–69. doi: 10.1007/s11130-006-0015-7. [DOI] [PubMed] [Google Scholar]
- 63.Calder P.C., Grimble R.F. Polyunsaturated fatty acids, inflammation and immunity. Eur. J. Clin. Nutr. 2002;56:S14–S19. doi: 10.1038/sj.ejcn.1601478. [DOI] [PubMed] [Google Scholar]
- 64.Elagizi A., Lavie C.J., Marshall K., DiNicolantonio J.J., O′Keefe J.H., Milani R.V. Omega-3 polyunsaturated fatty acids and cardiovascular health: A comprehensive review. Prog. Cardiovasc. Dis. 2018;61:76–85. doi: 10.1016/j.pcad.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 65.Martins R.M., Nedel F., Guimaraes V., Da Silva A.F., Colepicolo P., De Pereira C.M., Lund R.G. Macroalgae extracts from Antarctica have antimicrobial and anticancer potential. Front. Microbiol. 2018;9:412. doi: 10.3389/fmicb.2018.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Küpper F.C., Gaquerel E., Cosse A., Adas F., Peters A.F., Müller D.G., Kloareg B., Salaün J.P., Potin P. Free fatty acids and methyl jasmonate trigger defense reactions in Laminaria digitata. Plant Cell Physiol. 2009;50:789–800. doi: 10.1093/pcp/pcp023. [DOI] [PubMed] [Google Scholar]
- 67.Department of Health London (UK) Nutritional Aspects of Cardiovascular Disease Report on Health and Social. Her Majesty′s Stationery Office; London, UK: 1994. p. 202. Subject No. 46. [Google Scholar]
- 68.Susanto E., Fahmi A.S., Hosokawa M., Miyashita K. Variation in lipid components from 15 species of tropical and temperate seaweeds. Mar. Drugs. 2019;17:630. doi: 10.3390/md17110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson G.A., Jr. Lipids and membrane function in green algae. Biochim. Biophys. Acta Lipids Lipid Metab. 1996;1302:17–45. doi: 10.1016/0005-2760(96)00045-8. [DOI] [PubMed] [Google Scholar]
- 70.WHO. FAO Joint Consultation. Fats and oils in human nutrition. Nutr. Rev. 1995;53:202–205. doi: 10.1111/j.1753-4887.1995.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 71.Al-Adilah H., Al-Bader D.A., Elktob M., Kosma I., Kumari P., Küpper F.C. Trace element concentrations in seaweeds of the Arabian Gulf identified by morphology and DNA barcodes. Bot. Mar. 2021;64:327–338. doi: 10.1515/bot-2021-0027. [DOI] [Google Scholar]
- 72.Roleda M.Y., Lage S., Aluwini D.F., Rebours C., Brurberg M.B., Nitschke U., Gentili F.G. Chemical profiling of the Arctic Sea lettuce Ulva lactuca (Chlorophyta) mass-cultivated on land under controlled conditions for food applications. Food Chem. 2021;341:127999. doi: 10.1016/j.foodchem.2020.127999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article.