Abstract
Non-invasive evaluation of renal transplant function is essential to minimize and manage renal rejection. A computer-assisted diagnostic (CAD) system was developed to evaluate kidney function post-transplantation. The developed CAD system utilizes the amount of blood-oxygenation extracted from 3D (2D + time) blood oxygen level-dependent magnetic resonance imaging (BOLD-MRI) to estimate renal function. BOLD-MRI scans were acquired at five different echo-times (2, 7, 12, 17, and 22) ms from 15 transplant patients. The developed CAD system first segments kidneys using the level-sets method followed by estimation of the amount of deoxyhemoglobin, also known as apparent relaxation rate (R2*). These R2* estimates were used as discriminatory features (global features (mean R2*) and local features (pixel-wise R2*)) to train and test state-of-the-art machine learning classifiers to differentiate between non-rejection (NR) and acute renal rejection. Using a leave-one-out cross-validation approach along with an artificial neural network (ANN) classifier, the CAD system demonstrated 93.3% accuracy, 100% sensitivity, and 90% specificity in distinguishing AR from non-rejection . These preliminary results demonstrate the efficacy of the CAD system to detect renal allograft status non-invasively.
Index Terms—: Renal Transplants, BOLD-MRI, mean R2*, pixel-wise R2*, machine learning
1. INTRODUCTION
Over 650,000 patients in the U.S. have end-stage renal disease and renal transplant offers the best outcome for these patients. Over 17,000 kidney transplants are performed annually in the U.S. [1, 2]. However, 15%–27% of renal transplant patients have acute renal rejection (AR) within 5 years, which if not detected and treated promptly, causes renal damage and leads to allograft failure [1–4]. Given the paucity of donor organs, routine post-transplantation clinical evaluation of kidney function is critical to prevent allograft loss [5]. The current diagnostic technique recommended by the national kidney foundation (NKF) is to measure overall kidney function using glomerular filtration rate (GFR). GFR has low sensitivity and is a late marker for renal graft dysfunction (detectable after > 60% loss of renal function) [6]. In addition to nuclear imaging and ultrasonography, conclusive AR diagnosis requires renal biopsy. However, needle biopsy is used as a last resort due to invasiveness, high cost, time, concomitant risk factors (infection, bleeding, etc.), and is prone to over- or under-estimation of inflammation in the entire graft. Thus, there is a critical unmet need for a non-invasive technology that can provide accurate and rapid early diagnosis of renal transplant rejection.
Non-invasive evalutation of renal dysfunction using dynamic contrast-enhanced (DCE)- [7–15], diffusion-weighted (DW)- [16–24], and blood oxygen level-dependent (BOLD)-magnetic resonance imaging (MRI) [16, 25–36] is an ongoing area of research. Using DCE-MRI, kidney kinetic parameters were evaluated by Zikic et al. [9] after correcting kidney motion by applying a template-matching registration, and normalized gradient field as the contrast-invariant similarity measure. However, their study was limited by manual segmentation of kidneys, and visual evaluation of perfusion parameters (plasma volume and tubular flow) by trained physicians. Wentland et al. [12] utilized MRI-based intra-renal perfusion measurement to detect allografts with acute tubular necrosis (ATN) or AR on a cohort of 24 patients with the diagnosis confirmed by biopsy. Cortical and medullary blood flow was demonstrated to be significantly reduced in AR. While DCE-MRI has been used to develop CAD systems to assess the status of renal transplants, DCE-MRIs require contrast agents (CAs) which may induce nephrogenic systemic fibrosis [14], in patients with GFR < 30 ml/min. In contrast, MRI methodologies that do not require CAs (DW- and BOLD-MRIs) are increasingly used to evaluate allograft status.
Liu et al. [16] used DW-MRI along with manually selected cortical and medullary ROIs to early detect renal allograft dysfunction caused by AR and ATN. Their study demonstrated lower values of the measured apparent diffusion coefficients (ADCs) for the AR group compared to the control groups. Kaul et al. [20] evaluated the renal function with cortical and medullary ADC maps and reported a significant change in the medulla and cortex ADCs during AR. Although DW-MRI does not use CAs, it is limited by a low signal to noise ratio (SNR) especially at high gradient field strengths and duration [16, 23], which increases the difficulty of both segmentation and ADC estimation.
BOLD-MRI has the unique advantage of having a higher SNR while avoiding the use of CAs. Therefore, it has been recently used by researchers to study renal rejection [16, 25–27], using the amount of the deoxygenated hemoglobin in the kidney to quantify renal function. The amount of deoxyhemoglobin is quantified by apparent relaxation rate (R2*), which is calculated using the reciprocal of T2*, where T2* is amount of oxygenated hemoglobin [37].
It has been reported that the R2* in medulla is higher in both healthy transplants and native kidneys compared to AR [16, 25–27], while cortical R2* values were reported to be similar [16, 27]. These BOLD-MRI studies have several limitations including (1) manual delineation of the kidney using a 2D ROI, which makes this delineation subjective, (2) only performed statistical analyses to investigate the significant differences between different groups, and (3) non of these studies developed a fully automated CAD system for the early detection of AR renal transplants.
To overcome these limitations, we develop a fully automated CAD system, Fig. 1, to make an early and accurate diagnosis of acute rejection renal transplants, with the ability to: (i) delineate the kidney at different echo-times; (ii) extract global features and local features from the segmented kidney at different echo-times; and (iii) implement a classification model using the global and local features to assess the renal transplant status. To the best of our knowledge, this is the first CAD system of its kind to distinguish AR from non-rejection (NR) renal transplants from BOLD-MRI using both global (mean R2*) and local (pixel-wise R2*) features using state-of-the-art machine learning techniques.
Fig. 1:
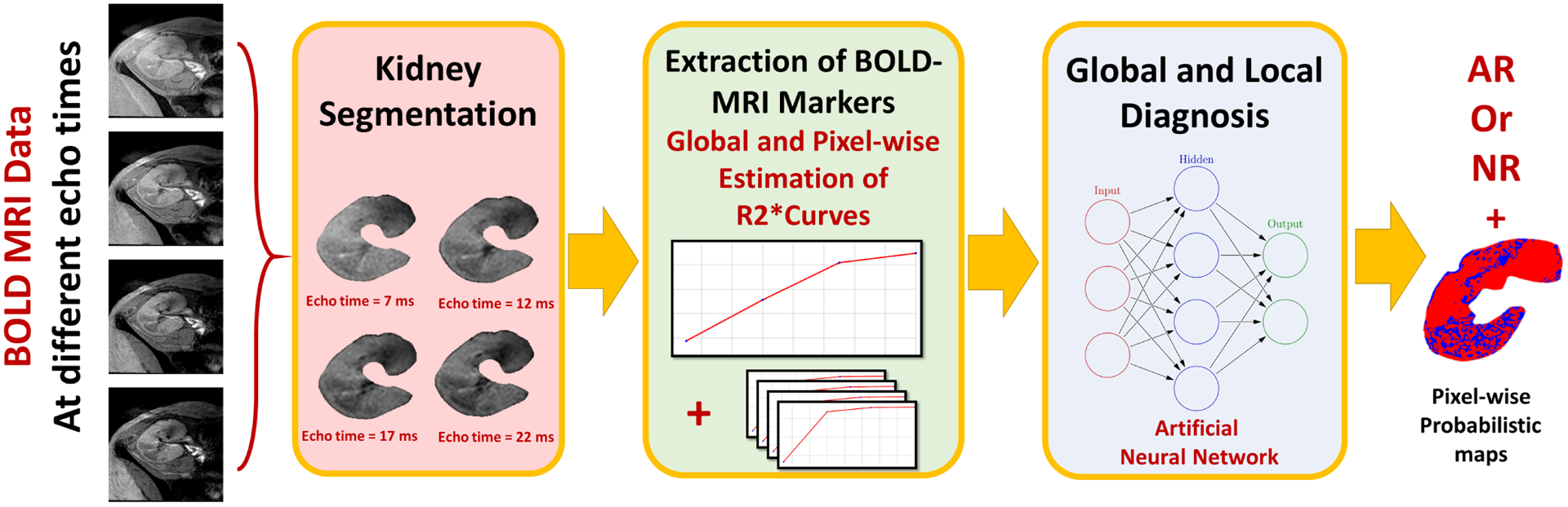
The developed CAD system for early assessment of acute renal rejection post-transplantation. The input blood oxygen level-dependent (BOLD) MRI data acquired at four different echo-times (2, 7, 12, 22) ms is first segmented. Then the global (i.e. mean R2*) and the local (i.e. the pixel-wise R2*) are constructed. Both global and local features are then fed to an artificial neural network to obtain the final global and local diagnosis.
2. METHODS
An accurate and robust CAD system to evaluate renal allograft status was developed. The CAD system consisted of the following key steps: (i) automatic delineation of the kidney from the surrounding abdominal tissues from BOLD-MR images (segmentation); (ii) extraction of both global (i.e. mean R2*) and local features (i.e. the pixel-wise values of R2*) from the segmented kidneys at different echo-times; and (iii) categorization of the renal transplant into one of two categories (i.e. NR vs. AR) by utilizing these global and local features using a state-of-the art machine learning classifier (e.g., artificial neural networks (ANNs)). Details of the proposed CAD system, (see Fig. 1), are discussed below.
2.1. Kidney Segmentation
A nonrigid registration based on using 2D B-splines approach [38] was first applied to handle renal allograft’s motion and to reduce BOLD-MRI inter-patient anatomical variability to improve segmentation accuracy. A 2D BOLD-MRI renal segmentation method based on using level-sets was applied [39]. To enhance segmentation accuracy, a guiding force integrating regional statistics derived from the kidney and background regions was employed. Regional appearance, shape, and spatial BOLD-MRI features were combined using a joint Markov-Gibbs random field (MGRF) image model [40]. Additional details of the segmentation approach can be found in [39, 41].
2.2. BOLD-MRI Markers
Renal function is evaluated by quantifying the amount of deoxygenated hemoglobin in the kidney. BOLD measures T2*, which is the amount of oxygenated hemoglobin [37] in the kidney. R2* (deoxygenated hemoglobin) is the reciprocal of T2* and will be used as our BOLD marker. After segmentation, the global features (i.e. mean R2* over the entire kidney) and the local features (i.e. pixel-wise R2*) are estimated at four different echo-times (7, 12, 17, 22) ms. These R2* values are used as BOLD-MR image-markers for renal transplant status assessment, while the BOLD-MRI data acquired at echo-time = 2 ms was used as the baseline. The pixel-wise T2* maps can be calculated using the following equation [36] as:
| (1) |
while the amount of deoxyhemoglobin (apparent relaxation rate (R2*)) is the reciprocal of T2* and can be calculated using the following equation:
| (2) |
p: a pixel at a location with its 2D coordinates (x, y).
SIt: the signal intensity of the pixel (p) of the segmented BOLD-MR image obtained at the echo-time (t ms).
: the signal intensity of the pixel (p) of the segmented BOLD-MR image obtained at the baseline echo-time (t0 = 2 ms).
2.3. Global and Local Diagnosis of The Kidney Tissue
After obtaining the global (i.e. mean R2*) and local features (i.e. pixel-wise R2*), two stages of classification were employed using ANNs to obtain the final diagnosis. The first stage uses the global features, shown in Fig. 2, extracted from all subjects, along with a leave-one-out cross validation (LOOCV) approach to train and validate an ANN classifier, shown in Fig. 3, with two hidden layers (the first layer with 10 nodes and the second layer with 5 nodes) to obtain a global diagnosis for the entire kidney.
Fig. 2:
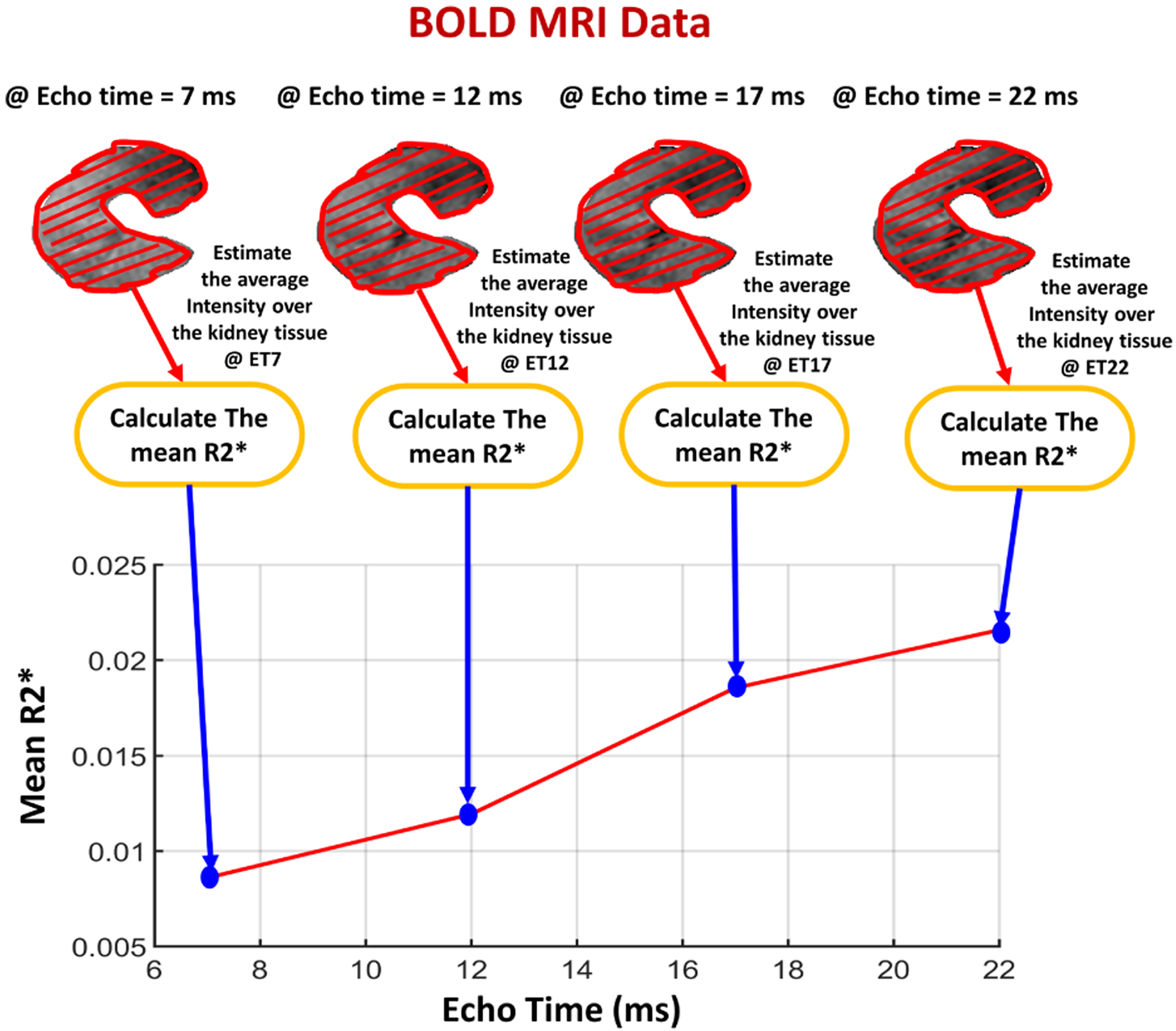
Illustrative figure showing the process of constructing the global features by calculating the mean R2* values from the segmented kidney at four different echo-times (2, 7, 12, 17) ms.
Fig. 3:
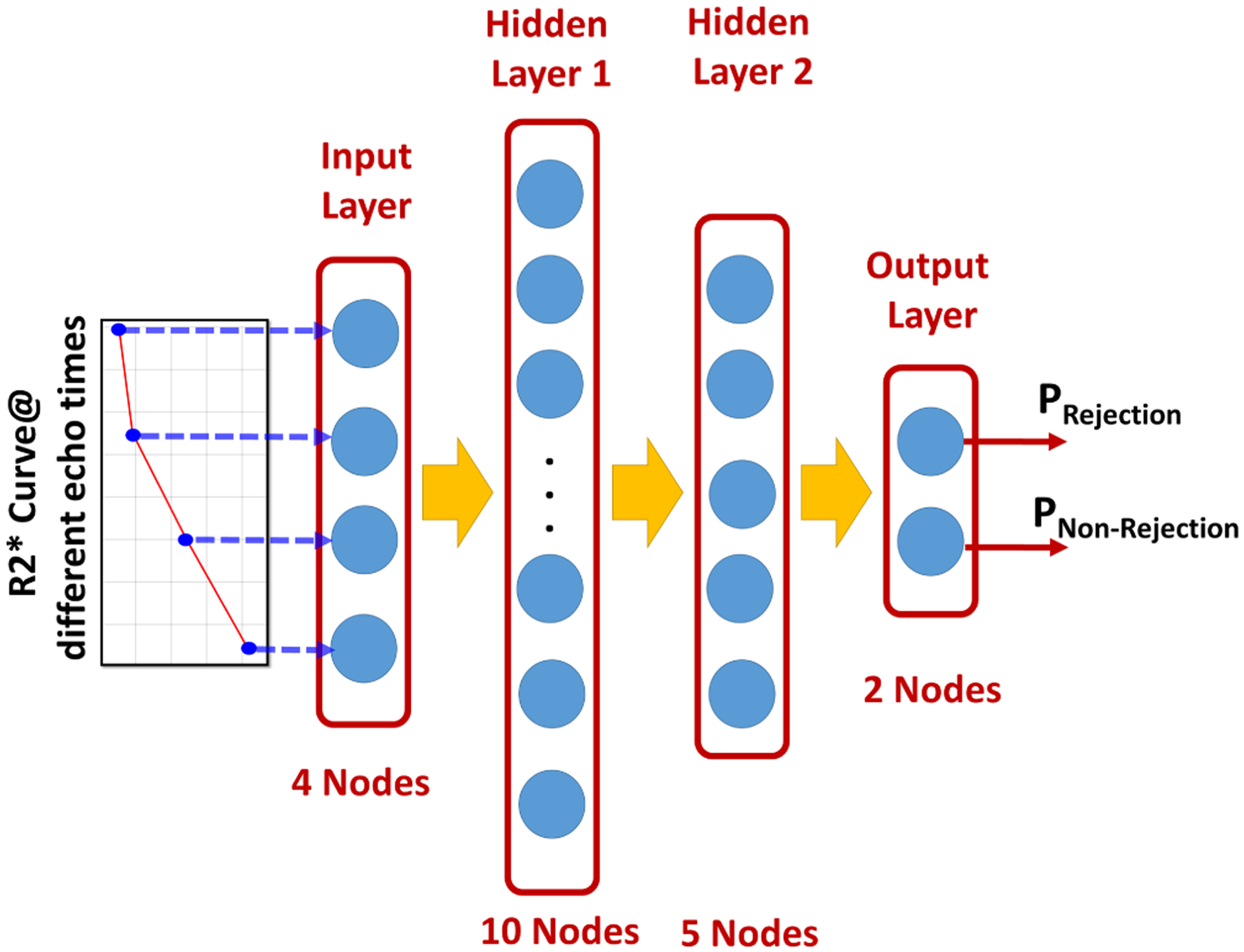
Illustartion of the schematic of the used artificial neural network (ANN) and the classification process starting from feeding the ANN with the global and local features till getting the final output probabilities of being a non-rejection or an acute rejection renal transplant.
The classification model obtained from the first stage was then tested using local features (see Fig. 4) to obtain a pixel-wise probabilistic map representing the probability of each pixel being NR or AR, for a local diagnosis, as shown in Fig. 3.
Fig. 4:
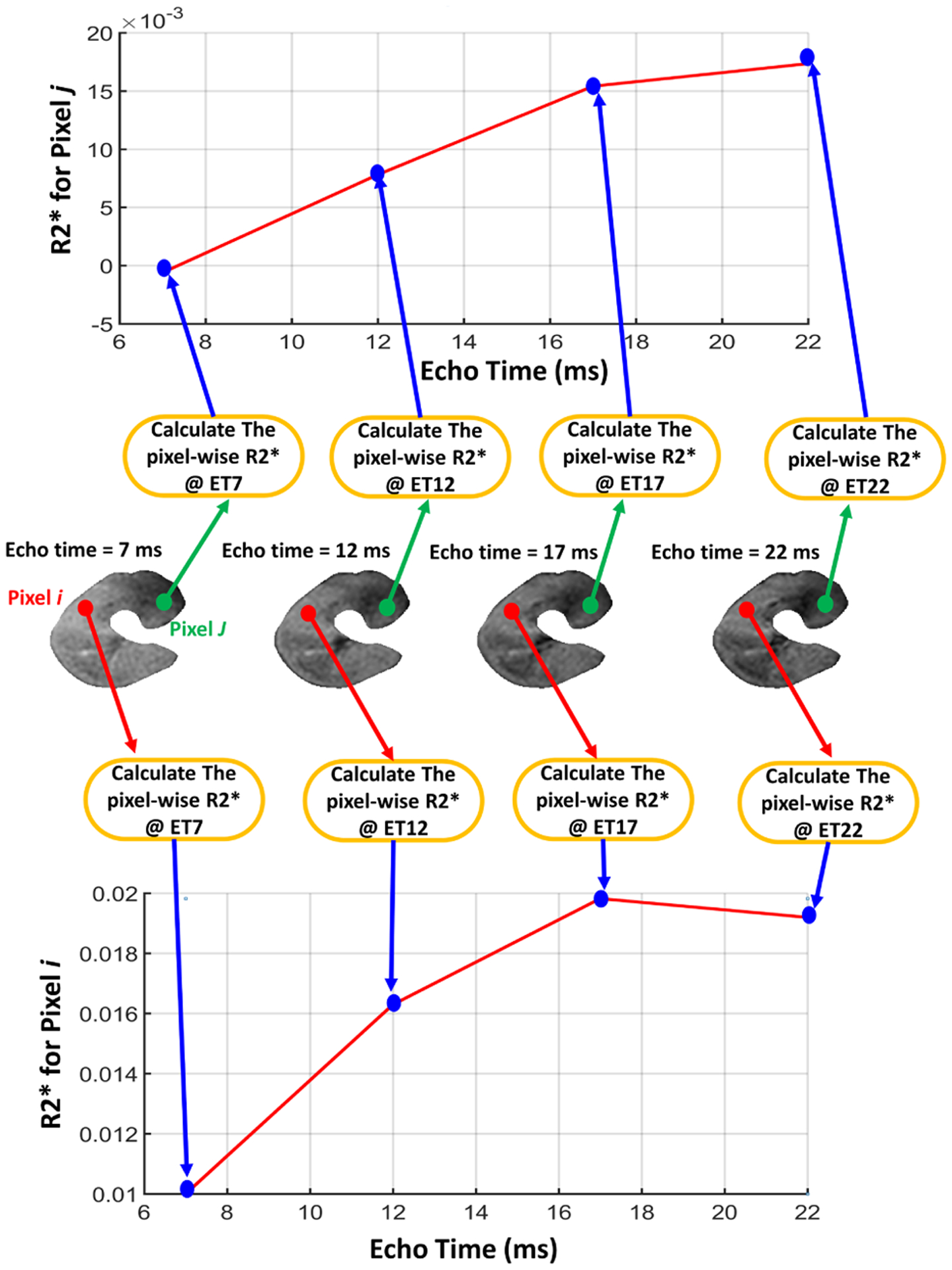
Illustrative figure showing the process of constructing the local features by calculating the pixel-wise R2* values from the segmented kidney at four different echo-times (2, 7, 12, 17) ms.
3. EXPERIMENTAL RESULTS
A total of 24 patients who underwent kidney transplantation from Jan 2018 to Dec 2018, were enrolled in this study after obtaining patient consent and IRB approval. Nine patients were excluded due to incomplete patient participation and/or technical problems, metallic prostheses, artificial valves, or claustrophobia. Scans and biopsies were obtained from the remaining 15 patients (M=10, F=5, age = 27 ± 13.6 years, age range = 12–54 years). Patients were divided into two groups - NR group (10 patients) and AR group (5 patients). Most of the NR patients only underwent BOLD-MRI scans and a clinical biopsy was not indicated. Renal biopsy, histology, and BOLD-MRI was obtained in the AR group. Coronal BOLD-MRIs were acquired before any biopsy procedure. BOLD-MRI scans were obtained using a 3T scanner (Philips Medical System, Netherlands) using a body coil and a gradient single-shot spin-echo echoplanar sequence; repetition time: 140 ms, echo-time: 2 ms, Flip angle: 25 degree, Bandwidth: 150 kHz, slice size: 384×384, number of signals acquired: 1, field of view: 14.4 cm, thickness: 6.0 mm. The largest coronal cross-section was obtained at five different echo-times (2, 7, 12, 17, and 22) ms, so that each subject has five images.
In order to validate the accuracy of the proposed renal classification technique, the developed acute renal rejection CAD system was tested using the 15 BOLD-MRI data sets using different classifiers. The matrix of global features of size 15 × 4 of mean R2* values at 7, 12, 17, and 22 ms were used with a LOOCV approach to train and test 8 different classifiers provided by MATLAB 2017 classification learner Tool Box (random forest (RF), linear discriminant analysis (LDA), logistic regression (logR), quadratic SVM (SVMQuad), cubic SVM (SVMCub), radial basis function SVM ((SVMRBF ), ensemble bagged trees (EBT), and ANNs). The accuracy, sensitivity, and specificity of these classifiers are presented in Table 1. The ANN classifier provided the best diagnostic performance in terms of accuracy, sensitivity, and specificity. Results in Table 1 demonstrate the feasibility of the constructed global features (i.e. mean R2* values) to diagnose AR.
Table 1:
Diagnostic performance evaluation of the proposed CAD system using different machine learning classifiers provided by the MATLAB 2017 Tool Box such that Acc: accuracy, Sens: sensitivity, Spec: specificity, and AUC: area under the curve.
| RF | LDA | logR | SVMQuad | SVMCub | SVMRBF | EBT | ANNs | |
|---|---|---|---|---|---|---|---|---|
| Classification Performance (NR vs. AR) | ||||||||
| Acc % | 86.7 | 66.7 | 73.3 | 66.7 | 80.0 | 80.0 | 73.3 | 93.3 |
| Sens % | 60.0 | 60.0 | 80.0 | 40.0 | 60.0 | 40.0 | 40.0 | 100 |
| Spec % | 100 | 70.0 | 70.0 | 80.0 | 90.0 | 100 | 90.0 | 90.0 |
| AUC | 0.60 | 0.64 | 0.69 | 0.68 | 0.82 | 0.52 | 0.50 | 0.99 |
The local features (i.e. the pixel-wise R2* maps) were used to test the ANN classification model resulting in a pixel-wise probabilistic map for each kidney. The local features analysis outputs the probability of each pixel in the kidney to be AR or NR. These probabilistic maps were then color-coded to assist in the visualization of the local kidney function by the clinicians, Fig. 5. The local features analysis will also enable tracking of the progression of AR or improvement with treatment during follow up. The data in Fig. 5 reveals the expected relation of the the pixel-wise R2* maps for NR and AR status.
Fig. 5:

Pixel-wise color-coded probabilistic maps obtained from the local feature analysis. Where the upper row shows three different examples for non-rejection (NR) renal transplants and the lower row shows three others acute rejection (AR) renal transplants. Note that the red color indicate the probability of being NR, while the blue color indicates the probability of being an AR.
To evaluate the performance of the developed CAD system, we constructed receiver operating characteristics (ROC) [42] for ANN and the compared classifiers. The ANN based-classifier demonstrated the highest area under the curve (AUC) and nearly approached unity, as shown in Table 1. These preliminary results demonstrate the feasibility of the proposed CAD system for early stage, non-invasive AR diagnosis.
4. CONCLUSIONS
A non-invasive CAD system for early diagnosis of AR using BOLD-MRI provided high classification accuracy, sensitivity, and specificity. The CAD system incorporates global and local features to better characterize renal function and evaluate AR. The CAD system will be optimized by training and validating on a larger patient cohort. Furthermore, genomic markers and histopathology image markers will be integrated into the CAD system to further enhance the accuracy of AR classification and to potentially determine the cause of AR.
5. ACKNOWLEDGEMENT
This research is supported by the National Institutes of Health (NIH Grant Number: 1R15AI135924-01A1).
6. REFERENCES
- [1].National Institutes of Health, “Kidney disease statistics for the united states,” Washington: NHI, 2012. [Google Scholar]
- [2].US Department of Health & Human Services, “Kidney disease statistics for the united states,” Bethesda, MD: National Kidney and Urologic Diseases Information Clearinghouse, National Institutes of Health, 2011. [Google Scholar]
- [3].Hollis E et al. , “Towards non-invasive diagnostic techniques for early detection of acute renal transplant rejection: A review,” The Egypt. J. Radiol. Nuc. Med, 2016. [Google Scholar]
- [4].Hollis E et al. , “Statistical analysis of ADCs and clinical biomarkers in detecting acute renal transplant rejection,” Br. J. Radiol, vol. 90, no. 1080, pp. 20170125, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kasiske BL et al. , “Kdigo clinical practice guideline for the care of kidney transplant recipients: A summary,” Kidney international, vol. 77, no. 4, pp. 299–311, 2010. [DOI] [PubMed] [Google Scholar]
- [6].Myers GL et al. , “Recommendations for improving serum creatinine measurement: A report from the laboratory working group of the national kidney disease education program,” Clin. Chem, vol. 52, no. 1, pp. 5–18, 2006. [DOI] [PubMed] [Google Scholar]
- [7].de Priester JA et al. , “Automated quantitative evaluation of diseased and nondiseased renal transplants with MR renography,” Journal of Magnetic Resonance Imaging, vol. 17, no. 1, pp. 95–103, 2003. [DOI] [PubMed] [Google Scholar]
- [8].Rusinek H et al. , “Performance of an automated segmentation algorithm for 3D MR renography,” vol. 57, no. 6, pp. 1159–1167, 2007. [DOI] [PubMed] [Google Scholar]
- [9].Zikic D et al. , “Automatic alignment of renal DCE-MRI image series for improvement of quantitative tracer kinetic studies,” in Proc. of SPIE, Medical Imaging: Image Processing. International Society for Optics and Photonics, 2008, vol. 6914, pp. 1–8. [Google Scholar]
- [10].de Senneville BD et al. , “Improvement of MRI-functional measurement with automatic movement correction in native and transplanted kidneys,” vol. 28, no. 4, pp. 970–978, 2008. [DOI] [PubMed] [Google Scholar]
- [11].Anderlik A et al. , “Quantitative assessment of kidney function using dynamic contrast enhanced MRI-Steps towards an integrated software prototype,” in Proceedings of the 6th International Symposium on Image and Signal Processing and Analysis (ISPA’09), Salzburg, Austria, September 16–18, 2009, pp. 575–581. [Google Scholar]
- [12].Wentland AL et al. , “Quantitative MR measures of intrarenal perfusion in the assessment of transplanted kidneys: initial experience,” Academic radiology, vol. 16, no. 9, pp. 1077–1085, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yamamoto A et al. , “Quantitative evaluation of acute renal transplant dysfunction with low-dose three-dimensional MR renography,” Radiology, vol. 260, no. 3, pp. 781–789, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hodneland E et al. , “In vivo estimation of glomerular filtration in the kidney using DCE-MRI,” in Proc. Int. Sym. Image and Signal Process. Anal, 2011, vol. 1, pp. 755–761. [Google Scholar]
- [15].Khalifa F et al. , “A comprehensive non-invasive framework for automated evaluation of acute renal transplant rejection using DCE-MRI,” NMR in Biomedicine, vol. 26, no. 11, pp. 1460–1470, 2013. [DOI] [PubMed] [Google Scholar]
- [16].Liu G et al. , “Detection of renal allograft rejection using blood oxygen level-dependent and diffusion weighted magnetic resonance imaging: A retrospective study,” BMC Nephrology, vol. 15, no. 1, pp. 158, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Thoeny HC et al. , “Functional evaluation of transplanted kidneys with diffusion-weighted and BOLD MR imaging: Initial experience 1,” Radiology, vol. 241, no. 3, pp. 812–821, 2006. [DOI] [PubMed] [Google Scholar]
- [18].Abou-El-Ghar M et al. , “Role of diffusion-weighted MRI in diagnosis of acute renal allograft dysfunction: A prospective preliminary study,” The British Journal of Radiology, vol. 85, no. 1014, pp. e206e211, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Eisenberger U et al. , “Evaluation of renal allograft function early after transplantation with diffusion-weighted MR imaging,” European Radiology, vol. 20, no. 6, pp. 1374–1383, 2010. [DOI] [PubMed] [Google Scholar]
- [20].Kaul A et al. , “Assessment of allograft function using diffusion-weighted magnetic resonance imaging in kidney transplant patients,” Saudi Journal of Kidney Diseases and Transplantation, vol. 25, no. 6, pp. 1143, 2014. [DOI] [PubMed] [Google Scholar]
- [21].Palmucci S et al. , “Magnetic resonance with diffusion-weighted imaging in the evaluation of transplanted kidneys: Preliminary findings,” in Transplantation Proceedings. Elsevier, 2011, vol. 43, pp. 960–966. [DOI] [PubMed] [Google Scholar]
- [22].Xu J et al. , “[value of diffusion-weighted MR imaging in diagnosis of acute rejection after renal transplantation].,” Journal of Zhejiang University. Medical Sciences, vol. 39, no. 2, pp. 163–167, 2010. [DOI] [PubMed] [Google Scholar]
- [23].Vermathen P et al. , “Three-year follow-up of human transplanted kidneys by diffusion-weighted MRI and blood oxygenation level-dependent imaging,” J. Magn. Reson. Imaging, vol. 35, no. 5, pp. 1133–1138, 2012. [DOI] [PubMed] [Google Scholar]
- [24].Wypych-Klunder K et al. , “Diffusion-weighted MR imaging of transplanted kidneys: Preliminary report,” Polish Journal of Radiology, vol. 79, pp. 94–98, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Han F et al. , “The significance of BOLD MRI in differentiation between renal transplant rejection and acute tubular necrosis,” Nephrology Dialysis Transplantation, vol. 23, no. 8, pp. 2666–2672, 2008. [DOI] [PubMed] [Google Scholar]
- [26].Sadowski EA et al. , “Blood oxygen level-dependent and perfusion magnetic resonance imaging: Detecting differences in oxygen bioavailability and blood flow in transplanted kidneys,” Magnetic Resonance Imaging, vol. 28, no. 1, pp. 56–64, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Djamali A et al. , “Noninvasive assessment of early kidney allograft dysfunction by blood oxygen level-dependent magnetic resonance imaging,” Transplantation, vol. 82, no. 5, pp. 621–628, 2006. [DOI] [PubMed] [Google Scholar]
- [28].Xin-Long P et al. , “A preliminary study of blood-oxygen-level-dependent mri in patients with chronic kidney disease,” Magnetic resonance imaging, vol. 30, no. 3, pp. 330–335, 2012. [DOI] [PubMed] [Google Scholar]
- [29].Pruijm M et al. , “Blood oxygenation level-dependent mri to assess renal oxygenation in renal diseases: progresses and challenges,” Frontiers in physiology, vol. 7, pp. 667, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chrysochou C et al. , “Bold imaging: a potential predictive biomarker of renal functional outcome following revascularization in atheromatous renovascular disease,” Nephrology Dialysis Transplantation, vol. 27, no. 3, pp. 1013–1019, 2011. [DOI] [PubMed] [Google Scholar]
- [31].Hall ME et al. , “Bold magnetic resonance imaging in nephrology,” International journal of nephrology and renovascular disease, vol. 11, pp. 103, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pruijm M et al. , “Determinants of renal tissue oxygenation as measured with bold-mri in chronic kidney disease and hypertension in humans,” PloS one, vol. 9, no. 4, pp. e95895, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wu G.-y. et al. , “The value of blood oxygenation level-dependent (bold) mr imaging in differentiation of renal solid mass and grading of renal cell carcinoma (rcc): analysis based on the largest cross-sectional area versus the entire whole tumour,” PloS one, vol. 10, no. 4, pp. e0123431, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Vivier P-H et al. , “Renal blood oxygenation level–dependent imaging: Contribution of r2 to r2* values,” Investigative radiology, vol. 48, no. 7, pp. 501, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Seif M et al. , “Renal blood oxygenation level–dependent imaging in longitudinal follow-up of donated and remaining kidneys,” Radiology, vol. 279, no. 3, pp. 795–804, 2016. [DOI] [PubMed] [Google Scholar]
- [36].Zhang J et al. , “Blood-oxygenation-level-dependent-(bold-) based r2 mri study in monkey model of reversible middle cerebral artery occlusion,” BioMed Research International, vol. 2011, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Michaely H et al. , “Functional renal imaging: nonvascular renal disease,” Abd. Imaging, vol. 32, no. 1, pp. 1–16, 2007. [DOI] [PubMed] [Google Scholar]
- [38].Glocker B et al. , “Non-rigid registration using discrete MRFs: Application to thoracic CT images,” in MICCAI Workshop Evalu. Methods Pulm. Image Reg, 2010, pp. 147–154. [Google Scholar]
- [39].Shehata M et al. , “A level set-based framework for 3D kidney segmentation from diffusion MR images,” in IEEE Int. Conf. Image Process, 2015, pp. 4441–4445. [Google Scholar]
- [40].El-Baz A et al. , Stochastic modeling for medical image analysis, CRC Press, 2015. [Google Scholar]
- [41].Shehata M et al. , “3D kidney segmentation from abdominal diffusion MRI using an appearance-guided deformable boundary,” PloS one, vol. 13, no. 7, pp. e0200082, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fawcett T, “An introduction to ROC analysis,” Pattern Recog-nit. Lett, vol. 27, no. 8, pp. 861–874, 2006. [Google Scholar]


