Abstract
A novel DNA helicase, a homolog of several prokaryotic helicases, including Escherichia coli Rep and UvrD proteins, is encoded by the Saccharomyces cerevisiae nuclear genome open reading frame YOL095c on the chromosome XV. Our data demonstrate that the helicase is localized in the yeast mitochondria and is loosely associated with the mitochondrial inner membrane during biochemical fractionation. The sequence of the C-terminal end of the 80-kDa helicase protein is similar to a typical N-terminal mitochondrial targeting signal; deletions and point mutations in this region abolish transport of the protein into mitochondria. The C-terminal signal sequence of the helicase targets a heterologous carrier protein into mitochondria in vivo. The purified recombinant protein can unwind duplex DNA molecules in an ATP-dependent manner. The helicase is required for the maintenance of the functional ([rho+]) mitochondrial genome on both fermentable and nonfermentable carbon sources. However, the helicase is not essential for the maintenance of several defective ([rho−]) mitochondrial genomes. We also demonstrate that the helicase is not required for transcription in mitochondria.
Helicases are enzymes that can unwind duplex DNA or RNA molecules by using nucleoside triphosphate hydrolysis as the source of energy. DNA helicases play essential roles in DNA replication, repair, recombination, and transcription (20, 22). RNA helicases are involved in transcription, RNA processing, regulation of RNA stability, ribosome assembly, and translation (21). The genes encoding proteins with helicase activity possess seven conserved sequence elements or helicase motifs (11). The data made available by systematic genome sequencing projects predict about 40 helicases in Saccharomyces cerevisiae, and most of these helicases have been characterized to some extent by using genetic or biochemical tools.
Only one thoroughly studied mitochondrial DNA helicase, the product of the nuclear PIF1 gene, has been isolated in yeast S. cerevisiae (9, 17). PIF1 helicase is a 5′-3′ helicase required for a specific type of recombination and DNA repair in mitochondria, and it is essential for mitochondrial DNA (mtDNA) maintenance at higher temperatures (7, 8). PIF1 helicase is not the major replicative helicase in yeast mitochondria since the PIF1 gene is nonessential for mtDNA maintenance under physiological growth conditions. The isolation of two other mitochondrial DNA helicases, from sea urchin and bovine mitochondria, has been reported (12, 27). These enzymes appear to move on DNA with 3′-5′ polarity. Compared to the yeast PIF1 helicase, the corresponding genes have not been isolated, and no extensive purification has been reported.
In this study we characterize a putative yeast helicase encoded by the open reading frame (ORF) YOL095c (the Hmi1p). This helicase belongs to the superfamily I, the best-studied members of which include the Escherichia coli Rep protein, the helicase required for bacteriophage φX174 replication, and the repair helicase UvrD. The YOL095c protein is located in yeast mitochondria, and is required for wild-type (wt) [rho+] mitochondrial genome maintenance. However, the helicase is dispensable for defective [rho−] mitochondrial genome replication. The structure of the C-terminal segment of the protein resembles that of a typical N-terminal mitochondrial targeting signal and is required for proper transport of the protein to the mitochondria.
MATERIALS AND METHODS
Yeast methods.
The parental strains used in this study were W303-1 MATa/α ade2-1/ade2-1 ura3-1/ura3-1his3-11,15/his3-11,15 trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100, W303-1A MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100, and W303-1B MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 (34).
TS103 is W303-1, yol095c::TRP1/YOL095C. SK041, SK061, SK035, and SK048 are haploid [rho−] strains, and SK086 and SK031 are [rho0] haploid strains. These strains were generated via sporulation of the TS103 strain. They originate from different respiratory deficient spore progenies and carry the disrupted allele of the YOL095c ORF. The original haploid colonies formed in tetrad dissection were restreaked, and [rho0] or [rho−] colonies were identified by DAPI (4′,6′-diamidino-2-phenylindole) staining. TS501 and TS502 are URA3+ derivates of W303-1A and W303-1B, respectively. Standard yeast media and procedures for mating, sporulation, and dissection were used (28). The synthetic defined medium was synthetic complete medium complemented with 2% glucose (SC). The medium used for mitochondria preparation was SC supplemented with 0.5% glucose. SCG was complete synthetic medium supplemented with 3% glycerol.
Suppressivity tests of mitochondrial [rho−] genome isolates were performed according to the method of Rickwood et al. (26).
Isolation of the YOL095C genomic clone.
High-molecular-weight genomic DNA was isolated from S. cerevisiae W303-1, partially digested with MboI, and ligated into BamHI-digested pBluescript KS(+). The clone pREP22 was isolated by screening the library by using a PCR probe covering the YOL095c ORF. The 3.3-kb clone REP22 starts at the MboI site 762 nucleotides upstream of the initiator ATG and ends at the MboI site 354 nucleotides downstream of the stop codon of the YOL095c ORF. The complementing yeast shuffle constructs were constructed removing the 3.3-kb SacI-SalI fragment from pREP22 and inserting it into appropriate pRS series vectors (31) to generate pRS315-REP22 and pRS316-REP22.
Disruption of the YOL095c and the PIF1 ORFs.
A 0.57-kb BglII-EcoRI fragment (nucleotides 593 to 1162 of the coding region) was removed from the YOL095c ORF and replaced by a 1.9-kb fragment containing the TRP1 gene (Fig. 1A). The resulting disrupted YOL095c ORF was linearized and used to transform the diploid yeast strain W303-1 and the haploid W303-1A strain. The PIF1 ORF was disrupted with a 1.2-kb fragment containing the URA3 gene, using HindIII sites at positions 1336 and 1985 in the PIF1 coding region. Gene disruptions were verified by PCR with primers that amplify the coding region of the YOL095c gene or the PIF1 gene and by Southern blotting with the corresponding probes.
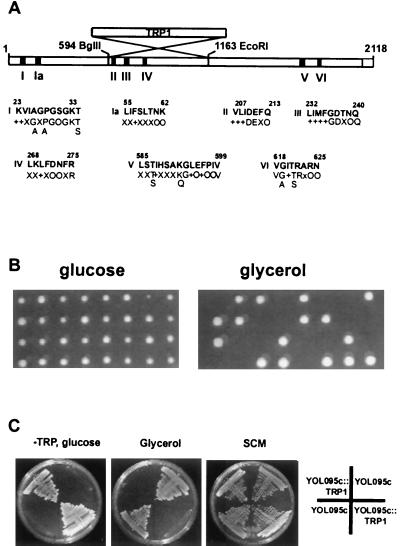
FIG. 1.
Schematic representation of the S. cerevisiae YOL095c gene and loss of mitochondrial respiratory functions in ΔYOL095c strains. (A) YOL095c ORF. The black boxes indicate the positions of the seven helicase motifs. The strategy used to construct disruption strain via replacement of the internal BglII-EcoRI fragment by the TRP1 marker is illustrated. The amino acid sequences of the seven helicase motifs are compared to the corresponding consensus sequences. +, hydrophobic residue; O, polar residue; X, any residue in the consensus structure. (B) Analysis of the phenotypic effect of the YOL095c ORF disruption. The heterozygous strain TS103 was sporulated, and tetrads were analyzed on glucose-containing SCM plates (left panel) or glycerol-containing SCG plates (right panel). (C) Growth of four haploid colonies originating from one tetrad, analyzed on different media. All four haploids are viable on glucose-containing SCM plates (right); two haploids have the YOL095c ORF disrupted with the TRP1 gene, as indicated by growth on the −TRP plate (left). The haploids with disrupted YOL095c ORF fail to grow on the glycerol plate (middle).
Epitope tagging of the YOL095C helicase.
The coding region of the YOL095C helicase was amplified by PCR to generate XbaI site at the 5′ end and BamHI site at the 3′ end of the fragment. This XbaI-BamHI fragment was cloned into pYCAH expression vector (1) to generate pYCAH-HMI1. This construct encodes a fusion protein with the following N-terminal sequence MSSYPYDVPDYASLGGPSRMDKLT... . The amino acid residues of the influenza hemagglutinin (HA) epitope are underlined, and the original initiator methionine in the YOL095c coding region corresponds to the amino acid residue 20 in the fusion protein.
C-terminal deletion mutants of the YOL095C helicase.
Deletion mutants ΔC15Ala and ΔC33Gly were made by replacing the original C terminus of the YOL095c helicase gene in the pYCAH-HMI1 with a PCR fragment carrying the corresponding deletion and a stop codon after the indicated amino acid residue. The last C-terminal amino acid residues in the deletion mutants are ...FGFYRA692-Stop in ΔC15Ala and ...VKVTHG674-Stop in ΔC33Gly.
DHFR-based reporter proteins.
Mouse dihydrofolate reductase (DHFR) was tagged at the N terminus with the HA epitope and expressed in the yeast cells using the YCAH plasmid as the HMI1 helicase. In the DHFR-Sign construct, the region encoding for the C-terminal amino acid residues 616 to 706 of the Hmi1 helicase was fused to the C terminus of the DHFR ORF via the BamH site encoding for an extra Gly-Phe dipeptide.
mtDNA preparation and analysis.
mtDNA was prepared from whole cellular DNA by using CsCl gradient centrifugation in the presence of bisbenzimide (Hoechst 33258) according the the method of Fox et al. (10). To estimate the repeat size, the isolated [rho−] mtDNA was subjected to restriction analysis with Eco47I, SspI, and VspI. The presence of ori sequence in the [rho−] mtDNA clones was checked by using PCR with the oligonucleotides complementary to the A and the C boxes of the mitochondrial ori/rep sequence GGGGGTCCCAATTATTATTTTC (ORI5in) and TAGGGGGAGGGGGTGGGT (ORI3in) and also GAAAATAATAATTGGGACCCCC (ORI5out) and ACCCACCCCCTCCCCCTA (ORI3out). The first pair of oligonucleotides (ORI5in and ORI3in) amplifies the ori sequence, and the second pair of oligonucleotides (ORI5out and ORI3out) amplifies the fragment separating the two ori sequences in the tandem repeats, respectively. The sequence of the hypersuppressive [rho−] repeats was first determined using the amplified material. The neutral [rho−] mtDNA repeats were first cloned as SspI fragments and sequenced. The sequence of the repeats was verified by using isolated mtDNA as the template.
Western blotting.
The whole-cell extracts were prepared by disruption of yeast by vortexing the cell suspension in 50 mM Tris (pH 8.0)–1 mM EDTA– mM phenylmethylsulfonyl fluoride (PMSF) with glass beads for 4 × 30 s at 4°C. The extracts, mitochondria, or mitochondrial fractions were diluted with 2× sodium dodecyl sulfate (SDS) loading buffer and boiled for 5 min. Proteins were fractionated by SDS–10 or –12.5% polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membrane by electroblotting via the semidry transfer protocol. The membranes were blocked with 0.5% nonfat milk, 0.02% Tween 20 in Tris-buffered saline and probed with the 12CA5 mouse monoclonal anti-HA antibody (1:8,000) or with control polyclonal rabbit antibodies (1:1,000) and then incubated with anti-mouse or anti-rabbit secondary antibody conjugated to horseradish peroxidase (1:5,000) and detected with the chemiluminescent substrate (Pierce).
In situ DAPI staining and immunofluorescence analysis.
Logarithmic-phase cells were collected and fixed as described by Pringle et al. (25). Primary anti-HA antibody 12CA5 was incubated overnight at a 1:250 dilution in phosphate-buffered saline–1% bovine serum albumin (BSA). This was followed by a 1-h incubation with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G secondary antibody at a 1:100 dilution and staining with 4′,6′-diamidino-2-phenylindole (DAPI; 0.2 μg/ml). The cells were then observed under a fluorescence microscope Olympus Vanox-S with filters B (FITC) and U (DAPI) and photographed with Fuji 800 film.
Mitochondrial transcript analysis.
Logarithmic-phase cells were collected, and total cellular RNA was isolated using the acid phenol method as described by Köhrer and Domdey (15). Yeast total RNA (15 μg) was treated with 10 U of RNase-free DNase I for 1 h at 37°C and then dot blotted onto Hybond+ nylon membrane. Mitochondrial transcripts were probed with 32P-labeled ori2-specific PCR probe covering the region from the mitochondrial ori box A to box C. The probe was generated by PCR using oligonucleotides ORI5in and ORI3in by using isolated SK041 mtDNA as the template. Hybridization of the filters was carried out at 65°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–2× Denhardt's reagent–0.1% SDS for 14 h; the filters were then washed two times for 5 min in 2× SSC–0.5% SDS and two times for 30 min in 0.2× SSC–0.5% SDS at 65°C. The filters were exposed to X-ray film and quantified with the phosphorimager. The mitochondrial RNA-specific signal was stripped from the filter by heating in 0.5% SDS, and the filters were rehybridized with S. cerevisiae cytoplasmic RNA probe covering nucleotides 2468 to 3116 of the 25S rRNA. The oligonucleotides used to amplify the corresponding region in the 25S rRNA gene were GCGAAACCACAGCCAAGGG and TTGCTGGTAACATTCATCAGTAGG. The filters were hybridized, washed, and exposed as with the mtRNA probes.
Preparation of mitochondria and mitochondrial subfractions.
First, 0.5- to 4-liter yeast cultures at an optical density at 600 nm of 0.8 to 1.0 were used to purify mitochondria and mitochondrial subfractions according to the method of Daum et al. (3). Analysis of solubility of the Hmi1 helicase in 0.1 M sodium carbonate (pH 11.5) was performed using isolated mitochondria frozen in 0.6 M mannitol. The mitochondrial suspension was diluted with 10 mM Tris (pH 8.0)–1 mM EDTA to 0.1 M mannitol, sonicated two times for 5 s, and extracted with the indicated reagent for 30 min on ice, followed by pelleting of the samples for 1.5 h at 35,000 rpm in a Beckman Ti70 rotor.
Rabbit polyclonal antisera to the following marker proteins were used as fractionation controls: Tom40 (outer membrane), Ccpo (intermembrane space), Yta10 (inner membrane), and Mge1 (carbonate soluble matrix protein) (a gift from R. Stuart and W. Neupert). The cytoplasmic marker protein was 3-phosphoglycerate kinase, which was probed with monoclonal antibody 22C5-D8 from Molecular Probes.
Proteinase K treatment of the isolated mitochondrial fraction was performed on ice for 10 min with 50 μg of proteinase K per ml.
Purification of the recombinant Hmi1 protein.
The HMI1 ORF lacking the last 15 codons was cloned into the BamHI site of the pGEX41 vector, and the Hmi1 protein was expressed as a glutathione S-transferase (GST) fusion protein in E. coli BL21. The bacterial culture was propagated at 23°C and induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 6 h. The pelleted bacteria were resuspended in the lysis buffer (50 mM Tris, pH 8.0; 300 mM NaCl; 1 mM EDTA; 1 mM dithiothreitol [DTT], 1 mM PMSF; 10% sucrose) and treated with 1 mg of lysozyme per ml for 1 h on ice, followed by one cycle of freeze-thawing. The GST-Hmi1 fusion protein was bound to glutathione-Sepharose 4B beads and eluted in 10 mM glutathione–20 mM Tris (pH 8.0)–300 mM NaCl–1 mM DTT–20% glycerol. The GST-Hmi1 fusion protein was cleaved with thrombin (10 U/0.4 mg of protein) for 12 h on ice. The sample was diluted to 100 mM NaCl and loaded onto a Q-Sepharose column. The Q-Sepharose flowthrough, containing the Hmi1 protein, was loaded directly onto an S-Sepharose column. The S-Sepharose column was washed with 20 mM morpholine ethane sulfonic acid (pH 6.5)–150 mM NaCl–0.1 mM EDTA–1 mM DTT–20% glycerol, and the Hmi1 protein was eluted in the same buffer containing 400 mM NaCl.
Helicase assay.
A 28-nucleotide oligonucleotide was labeled with polynucleotide kinase and [γ-32P]ATP (specific activity, 3,000 Ci/mmol) and annealed with the single-stranded phagemid pUC119. The annealed substrate was purified on a Sepharose 6B column. DNA unwinding assays (20 μl) were performed in buffer containing 20 mM Tris (pH 8.0)–10 mM MgCl2–1 mM DTT–4 mM ATP–0.1 mg of BSA per ml–substrate DNA (2,000 to 5,000 cpm). The reactions were incubated for 10 min at 30°C and were stopped with 5 μl of 0.5% SDS–50% glycerol–50 mM EDTA–0.1% bromophenol blue; they were then analyzed on a 10% nondenaturing polyacrylamide gel. The gels were dried and exposed to X-ray film.
RESULTS
The helicase encoded by the YOL095c ORF is essential for mitochondrial respiratory activity.
The ORF YOL095c of the S. cerevisiae nuclear genome was identified by the systematic yeast genome sequencing project and encodes a 80-kDa protein with seven conserved helicase motifs (Fig. 1A) (11). The gene has been named HMI1 for “helicase in mitochondria” (SGD). The protein belongs to the helicase superfamily I according to the classification by Gorbalenya and Koonin (11). In order to understand the functions of the helicase, we first analyzed the phenotypic effects of the HMI1 gene disruption. The internal BglII-EcoRI fragment in the HMI1 ORF was replaced with the TRP1 gene (Fig. 1A). The disrupted HMI1 gene version lacks the conserved sequence elements II, III, and IV of the helicase consensus structure, and this deletion will likely destroy the functional helicase gene. The HMI1 gene was disrupted in both haploid W303-1A and diploid W303-1 yeast strains by using the strategy of one-step gene replacement (29). The derivates of W303-1A, carrying the disrupted copy of the HMI1 ORF, were viable on glucose-containing SCM media. However, they failed to grow on nonfermentable carbon sources such as glycerol or a mixture of glycerol and ethanol. This indicated that the mutant strains had a mitochondrial respiratory defect. The diploid strain TS103 with one disrupted allele and the wt W303-1 grew with equal efficiency on both glucose- and glycerol-containing media, confirming that the observed defect was not dominant. Some nuclear gene products, such as the Abf2 protein, are not required for the maintenance of the [rho+] genome when strains are propagated on glycerol (4). In order to check if the Hmi1 helicase is required under selective conditions, we sporulated the diploid HMI1/hmi1::TRP1 heterozygote TS103 and analyzed the tetrads on SCM (glucose-containing medium) and SCG (glycerol-containing medium) (Fig. 1B and C). As expected, we got four viable spores per tetrad on SCM plates (Fig. 1B, left panel); two spores were respiratory defective, and two were respiratory competent (Fig. 1C). The respiratory defective spores grew on −TRP plates, indicating that they carry the disrupted copy of the HMI1 ORF. Direct dissection of the tetrads on YPG plates gave two viable spores out of four (Fig. 1B, right panel). This indicated that the loss of mitochondrial respiratory activity cannot be avoided on selective medium. We also tested the requirement of the helicase at temperatures (24°C) lower than regular growth conditions and, again, only two spores out of four were found to be viable when the tetrads were directly dissected on YPG. Finally, we could complement the respiratory defect caused by disruption of the chromosomal copy of the HMI1 gene with a centromeric plasmid pRS315-REP22. This plasmid contains a 3.2-kb fragment of yeast genomic DNA with the HMI1 gene. We dissected the tetrads, obtained from heterozygous TS103 transformed with pRS315-REP22, and found that out of 18 cases analyzed 3 tetrads gave rise to four viable colonies on YPG.
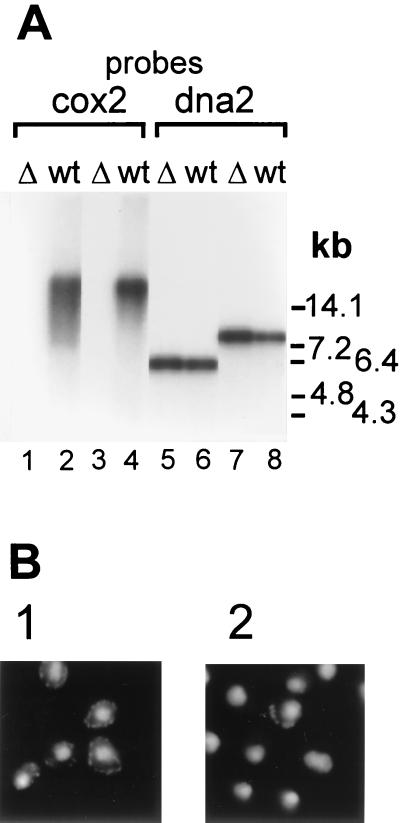
Since the YOL095c helicase was expected to be involved in DNA metabolism and since disruption of the gene caused a mitochondrial defect, we analyzed the effect of HMI1 gene disruption on the integrity of mtDNA. First, we extracted total DNA from four disruptant haploid clones and the W303-1A strain. We digested the isolated DNA with EcoRI or BamHI and probed it with a 32P-labeled fragment from the mitochondrial COX2 gene region covering the first 360 nucleotides of the coding region (Fig. 2A). We also probed the same samples with a fragment from the nuclear DNA2 gene. As expected, the nuclear probe hybridized with the predicted 6-kb EcoRI and an 8-kb BamHI fragments in all analyzed samples (Fig. 2A, lanes 5 to 8). mtDNA from the W303-1A cells hybridized as a high-molecular-weight band (Fig. 2A, lanes 2 and 4). However, we failed to detect a positive hybridization signal that would have corresponded to mitochondrial COX2 DNA in the cells that carried the disrupted copy of the YOL095c ORF (Fig. 2A, lanes 1 and 3). This observation indicated that the mutant disruptant strains lack functional [rho+] mtDNA. Next, we analyzed the wt W303-1A and the mutant cells using in situ staining with DAPI. DAPI staining revealed characteristic extranuclear spots in the wt W303-1A cells that correspond to mtDNA (Fig. 2B, panel 1). However, the mtDNA staining was lost in the majority of the cells that carried a disrupted copy of the YOL095c gene (Fig. 2B, panel 2). The fraction of DAPI-positive cells obtained via sporulation of a heterozygous diploid strain was regularly 1 to 5%. Direct disruption in the haploid strain usually produced slightly more cells that contained mtDNA, and in some colonies up to 20% of the cells retained some mtDNA. However, these cells always failed to grow on YPG, and their respiratory activity was not restored by transformation with pRS315-REP. From these experiments we concluded that the YOL095c gene product is essential for the stability of mtDNA.
FIG. 2.
Loss of functional mtDNA in Δhmi1 strains. (A) Southern blot of total cellular DNA. DNA, isolated from the wt strain W303-1A (wt) and from the haploid Δhmi1 colonies (Δ), was cut with EcoRI (lanes 1, 2, 5, and 6) or BamHI (lanes 3, 4, 7, and 8) and probed with mitochondrial COX2 (lanes 1 to 4) or nuclear DNA2 (lanes 5 to 8) probes. (B) DNA staining in nuclei and mitochondria of the wt W303-1A strain (panel 1) and of a haploid cell population from a colony where the HMI1 ORF has been disrupted (panel 2).
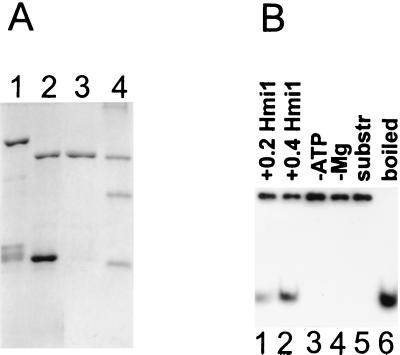
The seven conserved motifs found in the sequence of the HMI1 ORF predict that the Hmi1 protein is a helicase. We overexpressed and purified the Hmi1 protein in E. coli by using the pGEX41-based expression system, as described in Materials and Methods (Fig. 3A). The recombinant Hmi1 protein is an ATPase that is stimulated by single-stranded DNA (data not shown). The helicase activity of the Hmi1 protein was analyzed by using a partially double-stranded DNA substrate (Fig. 3B). The Hmi1 protein displaced the 28-mer oligonucleotide from the DNA duplex when incubated at 30°C (Fig. 3B, lanes 1 and 2). The unwinding reaction was dependent on the presence of ATP and Mg2+ (Fig. 2B, lanes 3 and 4).
FIG. 3.
The recombinant Hmi1 protein is a DNA helicase. (A) Purification of the Hmi1 protein. Lane 1, glutathione agarose eluate; lane 2, thrombin-cleaved material; lane 3, 400 mM S-Sepharose fraction; lane 4, protein size markers BSA (68 kDa), ovalbumin (45 kDa), and trypsinogen (24 kDa). (B) DNA unwinding assay with the recombinant Hmi1 protein. Lane 1, assay with 0.2 μg of Hmi1p; lanes 2 to 4, assays with 0.4 μg of Hmi1; lane 3, omitted ATP; lane 4, omitted MgCl2; lane 5, substrate DNA; lane 6, heat-denatured substrate.
Taken together, the analysis performed with YOL095c ORF disruption strains suggested that the gene product is required for the respiratory activity of mitochondria, and the observed respiration defect is caused by mtDNA instability. Thus, the Hmi1 helicase is the first reported helicase that is required for the maintenance of [rho+] mitochondrial genome at a normal growth temperature.
Hmi1 helicase is not required for [rho−] mtDNA maintenance.
Disruption of the HMI1 ORF in the haploid wt yeast W303-1A strain caused loss of mtDNA in majority of the cells. When propagated in culture, the fraction of mtDNA-containing cells remained relatively constant over 2 weeks, indicating that the defective mtDNA was maintained in a stable manner and that the presence or absence of this DNA did not give the cells an obvious growth advantage. We subcloned some of these mtDNA-containing cells and found that they maintained the mtDNA in almost 100% of cells, as judged by DAPI staining. Four such isolates (SK041, SK061, SK035, and SK048) were analyzed further.
First, we sought to determine whether the mitochondrial genomes maintained in the absence of functional HMI1 ORF are preferentially hypersuppressive or neutral in a genetic cross with a wt strain. The strains SK041, SK061, SK035, and SK048 were crossed to [rho+] strains TS501 and TS502, and the diploid colonies were selected. Replicas of 250 to 300 colonies were made onto glycerol-containing plates in order to estimate the fraction of [rho−] colonies in the cross progeny. A total of 99 and 98% of the diploid clones resulting from crosses to SK041 and SK061 were [rho−], indicating that the mitochondrial genome of the strains SK041 and SK061 was hypersuppressive. In contrast, strains SK048 and SK035 were neutral, since their diploid cross-progeny had almost exclusively [rho+] wt mtDNA. We also determined the sequence of the isolated [rho−] mitochondrial genomes. All four isolates contained small head-to-tail tandem repeats that are typical of mitochondrial [rho−] genomes (Table 1). Three repeats could be identified as fragments of mtDNA, based on sequence homology with the published yeast mtDNA sequences. As expected, the 0.5-kb and 0.8-kb mtDNA repeats of the strains SK041 and SK061 contained mitochondrial rep/ori sequences. In both strains, SK041 and SK061, the ori2 containing fragment was retained. Strains SK048 and SK035 had small repeats that did not contain mitochondrial rep/ori sequences. Clone SK035 had a 0.5-kb fragment from the 21S rRNA gene. The fourth strain SKO48 had a repeat of 100 bp that almost completely consisted of AT base pairs. We could not identify the origin of this repeat in the published yeast S. cerevisiae mtDNA sequence.
TABLE 1.
[rho−] strains used in this studya
| Strain | Suppressivity | Description and/or sequence of the mtDNA repeat |
|---|---|---|
| SK035 | Neutral | 469-bp repeat; nt 234 to 702 of the mitochondrial 21S rRNA |
| SK041 | HS | 536-bp repeat; nt 20301 to 20766 in the GenBank sequence L36897; ori2 region |
| SK048 | Neutral | 100-bp repeat; ATTAATAATA TATTATTAAA AAATAATTAA AATAATATAA TATAAATATA ATAATAATAT AAATATAAAT ATAATATATT AAATATATAT AAGGTAAAAT |
| SK061 | HS | 829-bp repeat; nt 19951 to 20826 in the GenBank sequence L36897; ori2 region |
HS, hypersuppressive; nt, nucleotide.
In conclusion, the analysis of the mtDNAs in strains SK041, SK061, SK035, and SK048 indicated that the Hmi1 helicase, required for wt mtDNA [rho+] maintenance, is dispensable for [rho−] mtDNA replication. Both, hypersuppressive and nonsuppressive, mitochondrial genome isolates were found among the analyzed clones and could be stably maintained without the Hmi1 helicase.
Another DNA helicase, the Pif1 protein, has been detected in yeast mitochondria. Since the Hmi1 helicase is apparently not required for the maintenance of the isolated [rho−] genomes, we next asked whetehr the Pif1 helicase is essential in an Δhmi1 background for the maintenance of these defective [rho−] genomes. We disrupted the PIF1 ORF in the described haploid strains, SK041, SK061, SK048 and SK035, and analyzed the mtDNA using in situ staining with DAPI. The attempts to disrupt the PIF1 ORF in the SK048 that carried a small 100-bp AT-rich repeat were unsuccessful since we did not obtain viable colonies. The neutral SK035 genome was lost in all of the cells analyzed. In contrast, the hypersuppressive [rho−] genomes in SK041 and SK061 were still maintained. The loss of mtDNA in the SK035 cells and the loss of viability of the SK048 cells were not observed in control experiments, where the Δhmi1 cells were transformed with the linearized URA3 gene and selected for the URA+ phenotype.
The Hmi1 helicase is not essential for transcription in mitochondria.
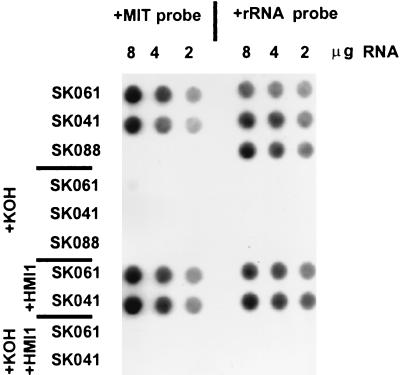
Mutations that block mitochondrial protein synthesis cause instability of the [rho+] mitochondrial genome (24). Therefore, the observed instability of mtDNA in S. cerevisiae strains, which lack the functional Hmi1 helicase, could also be the result of defective transcription, splicing, or translation in mitochondria. Based on its structural homology to well-characterized prokaryotic helicases such as E. coli Rep and UvrD and S. aureus PcrA, as indicated in the Yeast Protein Database, the HMI1 gene product belongs to the class of DNA helicases. Therefore, we think that the involvement of the Hmi1 helicase in mitochondrial RNA processing or translation, which require RNA helicase activity, is unlikely. However, helicase activity is an integral part of the nuclear transcription machinery (6), and we wanted to check if the Hmi1 helicase plays a role in mitochondrial transcription. To examine that possibility, we analyzed mitochondrial RNA transcription in the strains which carried the disrupted version of the HMI1 ORF. The strains SK041 and SK061, which have hypersuppressive mitochondrial genomes, were chosen for analysis. The mtDNA repeats in SK041 and SK061 contain ori2 sequence, which has a promoter close to the conserved box C (2). Strains SK041 and SK061 were transformed with a plasmid that contains an intact copy of the disrupted HMI1 ORF (pRS315-REP22) or with pRS315. As another control, we also included in our analysis the [rho0] strain SK088. Since the strain SK088 does not have mtDNA, there are obviously no mitochondrial transcripts. Total RNA was prepared from mid-log-phase yeast cultures, and mitochondrial RNA was analyzed by dot blot hybridization. Serial dilutions of total cellular RNA were probed with 32P-labeled ori2 PCR fragment of mtDNA. The filters were then stripped of the mtDNA probe and rehybridized with a nuclear rRNA probe. Autoradiographs of the filters are shown in Fig. 4. A strong hybridization signal with ori2 probe was observed in both [rho−] hypersuppressive strains SK041 and SK061, which do not have a functional HMI1 gene. This indicated active transcription from the ori2 promoter in these strains. The signal from [rho0] strain SK088 samples showed only low background level hybridization. This confirms that the detected transcripts in SK041 and SK061 samples originate from mtDNA transcription. These signals originated mostly from hybridizing RNA and not from contaminating DNA, since the signal in alkali-treated samples is reduced approximately 50-fold (compare lines marked KOH-SK041 and KOH-SK061 with lanes marked SKO41 and SKO61 in Fig. 4). Reprobing the filters with a cytoplasmic 25S rRNA probe revealed a hybridization signal in all analyzed strains, including SK088. We could not detect a reproducible difference between samples originating either from pRS315 or pRS315-HMI1 transformed cells. Consequently, the mitochondrial transcription machinery in the analyzed strains SK041 and SK061 seems to be active, and apparently the Hmi1 helicase is not essential for mitochondrial RNA transcription.
FIG. 4.
Active transcription in mitochondria of Δhmi1 strains. Total cellular RNAs from [rho0] strain SK088 and [rho−] hypersuppressive strains SK061 and SK041 were analyzed by dot blot hybridization. The amount of total RNA loaded per dot is indicated. Lanes marked “+KOH” represent alkali-treated samples. Lanes marked “+HMI1” are samples from strains that were transformed with the HMI1 complementing plasmid pRS315-HMI1. The blot was probed with a mitochondrial ori2 probe (left panel) and then stripped and probed with a cytoplasmic 25S rRNA probe (right panel).
The Hmi1 protein is localized in the mitochondria.
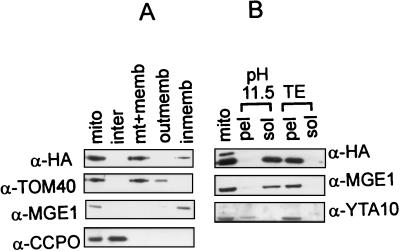
The observed loss of mtDNA phenotype in the Δhmi1 cells prompted us to determine whether the Hmi1 protein is transported into mitochondria. The Hmi protein was tagged with the HA tag at the N terminus of the protein (HA-Hmi1) and expressed using the yeast expression vector pYCAH (1). The pYCAH-HMI1 plasmid expressing the hybrid protein complemented the deletion of the chromosomal YOL095c gene. Four viable spores from one tetrad could be obtained on glycerol plates, when pYCAH-HMI1 plasmid was introduced into HMI1/hmi11::TRP1 heterozygous diploid strain TS103 prior to sporulation (data not shown). Since the tagged HA-Hmi1 protein appeared to be functional, we used it to study intracellular localization of the Hmi1 helicase. W303-1A cells were transformed the YCAH-HMI1 plasmid and grown in selective medium. Mitochondria were isolated and further fractionated into outer membrane, perimembrane space, inner membrane, and matrix fractions as described by Daum et al. (3). Fractions were analyzed by Western blotting with the anti-HA antibody (Fig. 5A). The majority of the total HA-Hmi1 protein in the cell was regularly recovered in the mitochondrial fraction. As revealed by subfractionation of mitochondria, the protein mostly copurifies with the inner membrane (Fig. 5A, lanes 3 and 5). To analyze the nature of the association of the Hmi1 protein with the inner membrane, we extracted the mitochondria with 0.1 M sodium carbonate (pH 11.5) (Fig. 5B). The Hmi1 protein was recovered in the 0.1 M sodium carbonate supernatant (Fig. 5B, lanes 2 and 3). Extraction with Tris (pH 8.0) alone did not solubilize the Hmi1 protein (Fig. 5B, lanes 4 and 5). This indicated that the Hmi1 protein is associated with the inner membrane but is not an integral membrane protein.
FIG. 5.
Biochemical fractionation localizes the Hmi1 protein in mitochondria. (A) Analysis of intracellular localization of the Hmi1 protein. Yeast cells (strain W303-1A), transformed with the expression construct YCAH-HMI1, and mitochondria were isolated by using fractionation by differential centrifugation. Mitochondria were subfractionated further, and the fractions were analyzed by Western blotting. The blots were probed with monoclonal anti-HA antibody 12CA5 (HA) or rabbit polyclonal antibodies against TOM40 (outer membrane protein), CCPO (intermembrane space protein), or MGE1 (peripheral inner membrane protein). Lane 1, mitochondria; lane 2, intermembrane space; lane 3, matrix plus membranes; lane 4, outer membrane; lane 5, inner membrane. (B) The Hmi1 protein is not an integral inner membrane component. Mitochondria were diluted, sonicated briefly, and extracted on ice with 0.1 M sodium carbonate (pH 11.5) (lanes 2 and 3) or with 10 mM Tris (pH 8.0)–1 mM EDTA (lanes 4 and 5). Soluble and membrane-bound fractions were separated by centrifugation for 1.5 h at 35,000 rpm in the Beckman Ti70 rotor. Pellet (pel) and supernatant (sol) fractions were analyzed by Western blotting with anti-HA monoclonal antibody 12CA5 and polyclonal antibodies against YTA10p (integral inner membrane protein) or Mge1p (peripheral inner membrane protein).
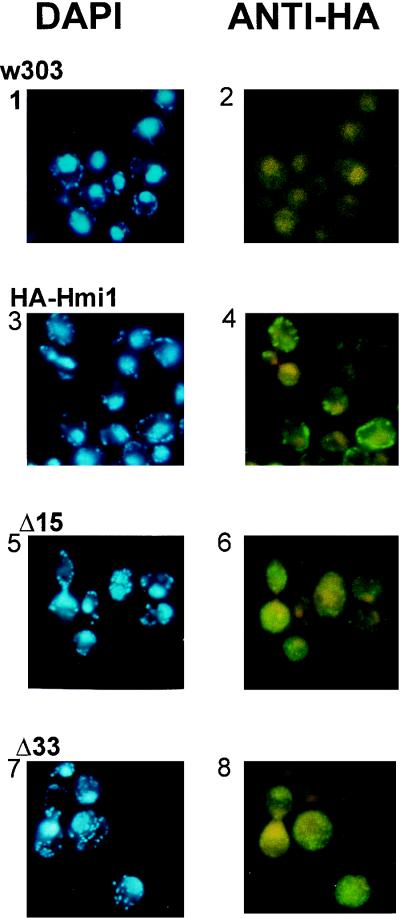
The biochemical fractionation was complemented with the in situ analysis by using indirect immunofluorescence staining with anti-HA antibodies. Distribution of mtDNA was analyzed in the same samples by DAPI staining. The data are presented in Fig. 6. The first two panels (Fig. 6, panels 1 and 2) show the control samples of the strain W303-1A. Panels 3 and 4 are samples of the same strain transformed with YCAH-HMI1. In these cells specific FITC staining was observed that colocalized with DAPI-stained mtDNA, being only slightly more diffuse. Similar in situ staining experiments and biochemical fractionations were performed with cells where no endogenous Hmi1p was present, and identical results were obtained.
FIG. 6.
Colocalization of mtDNA and the Hmi1 protein or the Hmi1 mutants in situ. Yeast cells expressing the tagged Hmi1 protein or the C-terminal deletion mutants were fixed, and indirect immunostaining was used to analyze the intracellular localization of the Hmi1 protein or its mutants (right panels). mtDNA was stained in the same samples by using DAPI (left panels). Panels 1 and 2, W303-1A; panels 3 and 4, W303-1A transformed with YCAH-HMI1; panels 5 and 6, W303-1A transformed with YCAH-ΔC15Ala; panels 7 and 8, W303-1A transformed with YCAH-ΔC33Gly.
The C-terminal segment of the Hmi1 helicase is required for mitochondrial targeting of the protein in vivo.
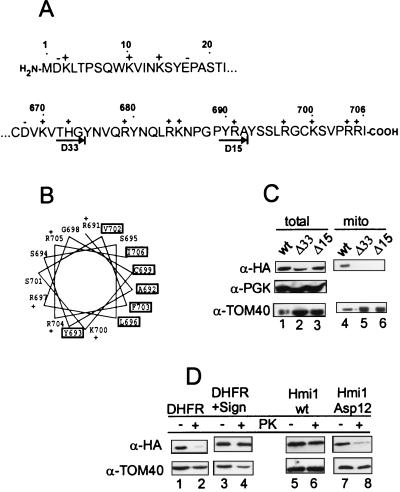
Localization of the YOL095c helicase in the mitochondria raised a question about the structural determinants required for proper targeting of the protein into the mitochondria. The typical N-terminal mitochondrial import signal is an amphiphilic α-helix with one nonpolar face. The other side of the helix has many positively charged and hydroxylated amino acid residues (35). However, several acidic residues are found at the N terminus of the Hmi1 protein (Fig. 7A). Furthermore, the N-terminal fusion of the helicase protein with the HA tag (19 amino acid residues) provided by the YCAH-HMI1 construct was functional in in vivo complementation assays. This indicated that the HA epitope at the N terminus does not interfere with the protein transport into mitochondria as would have been expected if the mitochondrial import signal was N terminal. The C terminus of the Hmi1 protein does not contain any negatively charged residues and can be predicted to form an amphiphilic helix (Fig. 7A and B). In order to investigate whether the C terminus of the Hmi1 protein is required for targeting to the mitochondria in vivo, we made deletions in the HMI1 gene removing 32 (construct YCAH-ΔC33Gly) or 14 (construct YCAH-ΔC15Ala) amino acid residues from the C terminus, respectively (Fig. 7A). The cells, transformed with YCAH-ΔC15Ala and YCAH-ΔC33Gly, expressed proteins of the expected size, and the expression level of the deleted proteins appears to be similar to the expression level of the full-length protein (Fig. 7C, lanes 1 to 3). Localization of the tagged proteins was analyzed as described before, using staining of the fixed cells with DAPI and simultaneous immunofluorescence detection of the HA tag (Fig. 6, panels 5 to 8). The DAPI-stained mtDNA in the cells transformed with YCAH-ΔC15Ala and YCAH-ΔC33Gly had a characteristic spotted pattern (Fig. 6, panels 5 and 7). However, the protein staining pattern of the deletion mutants ΔC15Ala and ΔC32Gly had a nonlocalized staining pattern, indicating that their transport into mitochondria is severely impaired (Fig. 6, panels 6 and 8). Biochemical fractionation of the cell extracts and Western blotting of the fractions with anti-HA antibody confirms the in situ analysis data (Fig. 7C). Only the full-length protein was detected in the mitochondrial fraction (Fig. 7C, lane 4); ΔC32Gly and ΔC15Ala (Fig. 7C, lanes 5 and 6) were not.
FIG. 7.
The C-terminal segment of the Hmi1 helicase is required for correct targeting of the protein into mitochondria. (A) Sequences of the N- and C-terminal segments of the Hmi1 protein. The acidic (−) and basic (+) amino acid residues are indicated above the sequence. The endpoints of the two C-terminal deletion mutants Mrh-ΔC15Ala and Mrh-ΔC33Gly are indicated with arrows below the C-terminal sequence segment. (B) Helical wheel presentation of the 18 C-terminal amino acid residues starting from Arg-691. Hydrophobic residues are indicated with boxes, and the positively charged residues are indicated with a “+.” (C) Western blot analysis of whole-cell extracts and mitochondrial fractions from the W303-1A strain transformants expressing the Hmi1 protein or the corresponding deletion mutant. Lanes 1 and 4, W303-1A transformed with YCAH-HMI1; lanes 2 and 5, W303-1A transformed with YCAH-ΔC33Gly; lanes 3 and 6, W303-1A transformed with YCAH-ΔC15Ala. The blots were probed with the anti-HA antibody 12CA5, anti-TOM40 (mitochondrial marker), and anti-PGK (cytoplasmic marker). (D) The C-terminal segment of the Hmi1 protein targets the DHFR carrier protein into mitochondria, and introduction of negatively charged residues in the C-terminus of the Hmi1 protein abolishes the mitochondrial import. The following proteins were expressed in W303-1A strain cells: lanes 1 and 2, HA-tagged DHFR; lanes 3 and 4, the fusion protein of HA-DHFR and the C-terminal segment (residues 616 to 706); lanes 5 and 6, HA-tagged Hmi1 protein; and lanes 7 and 8, mutant HA-Hmi1-Asp12. A Western blot shows analysis of total cellular protein (lanes 1, 3, 5, and 7) and the mitochondrial fraction treated with proteinase K for 10 min on ice (lanes 2, 4, 6, and 8). The blots were probed with anti-HA antibody 12CA5 and anti-TOM40.
The observed localization defect of the Hmi1 protein deletion mutants could be an indirect phenomena caused, for example, by general misfolding of the protein. To further confirm the role of the C-terminal part of the Hmi1 protein in mitochondrial import, we fused amino acid residues 616 to 706 to DHFR carrier protein and analyzed the intracellular localization of the fusion protein in vivo (Fig. 7D). Both DHFR and the hybrid protein DHFR(616–706) were expressed at similar levels (Fig. 7D, lanes 1 and 3). Both proteins could be detected in the crude mitochondrial fraction (data not shown). However, when isolated crude mitochondria were treated with proteinase K, the DHFR(616–706) fusion protein was protected from the cleavage but not the DHFR protein (Fig. 7D, lanes 2 and 4). We conclude that only the hybrid protein is imported into mitochondria in vivo.
There are no negatively charged amino acid residues at the C terminus of the Hmi1 protein. We next introduced two negatively charged Asp residues in the putative C-terminal targeting signal by mutating the residues Arg705 and Arg704. Intracellular localization of the mutant proteins was analyzed by using biochemical fractionation (Fig. 7D, lanes 5 to 8). Both, the wt Hmi1 and the mutant Hmi1 (Asp12) were expressed at similar levels in the yeast cells (Fig. 7D, lanes 5 and 7). Also, both proteins were detected in the mitochondrial crude fraction. However, only the wt Hmi1 and not the mutant protein was protected against proteinase K treatment, indicating that import of the mutant protein was impaired (Fig. 7D, lanes 6 and 8).
DISCUSSION
In the present study we have characterized a novel DNA helicase, the Hmi1 protein, in S. cerevisiae. The protein is encoded by the YOL095c ORF on chromosome XV and contains seven conserved structural motifs of helicase proteins (11) (Fig. 1). The recombinant Hmi1 protein has single-stranded DNA-stimulated ATPase and DNA helicase activities (Fig. 2). Disruption of the HMI1 ORF led to the loss of functional mtDNA (Fig. 2). Our preliminary analysis of several mutants with point mutations in the conserved helicase motifs indicates that helicase activity is necessary for the functional protein since these mutants do not complement disruption of the HMI1 gene (unpublished data). Biochemical fractionation and in situ immunolocalization data indicated that the protein is localized in the mitochondria (Fig. 4). We can conclude that the Hmi1 is a novel mitochondrial helicase.
The Pif1 helicase has previously been isolated in yeast mitochondria (16, 17). Pif1p is involved in mtDNA repair and recombination, but it is not required for the maintenance of mtDNA at normal growth temperatures (7, 8). The Hmi1 protein, in contrast, was required for the maintenance of the functional mitochondrial genome at regular growth temperatures, since the strains with the disrupted HMI1 gene lost [rho+] mtDNA at 30°C. We also found that, unlike strains with defects in some nuclear genes that will maintain functional mtDNA when propagated on glycerol (4), the strains with the disrupted HMI1 gene could not maintain their functional mtDNA on a selective carbon source.
Homology searches revealed a similarity of the Hmi1 protein to several prokaryotic proteins, mostly ones involved in replication (Yeast Protein Database). The prokaryotic homologs of the Hmi1 protein, the E. coli Rep protein, and Staphylococcus aureus pcrA protein are replicative helicases. The E. coli Rep protein is required for bacteriophage φX174 and M13 replication (32). The PcrA protein of Staphylococcus aureus is involved in pT181 plasmid replication (13, 14). The third well-characterized prokaryotic homolog, the UvrD helicase, is mainly a repair enzyme. However, there are indications that it has some overlapping essential functions with the Rep protein and that these functions include some aspect of chromosomal DNA replication in E. coli (23, 30, 33). It is therefore tempting to speculate that the putative helicase Hmi1 characterized in this study is the major replicative helicase in yeast mitochondria. However, we found that the Hmi1 protein was not required for [rho−] mitochondrial genome replication. The isolated strains SK041, SK061, SK035, and SK048 maintained different mitochondrial [rho−] genomes for weeks in culture despite the lack of the functional HMI1 gene. Several explanations can be proposed for clarification of this phenomena.
First, the Hmi1 protein might only be involved in replication of the [rho+] genome. One can propose that alternative helicases are used to replicate the [rho−] genomes. If this is the case, distinctive differences must exist between the replication machineries that are used for replication of the [rho+] and [rho−] genomes. An obvious candidate of the alternative helicase is the Pif1 protein. Therefore, we tested whether the [rho−] mitochondrial genomes are maintained when the PIF1 gene is disrupted in the Δhmi1 background. mtDNA was lost in one of the strains, SK035. It is remarkable that the SK035 genome contain a region of the mitochondrial 21S rRNA (Table 1), and this region has been suggested to contain a PIF-dependent recombinogenic signal. Therefore, it is likely that PIF1-dependent recombination plays a role in the maintenance of this particular [rho−] mitochondrial genome. Our data indicate that the hypersuppressive isolates SK041 and SK061 can maintain the mtDNA without the Pif1 protein. Therefore, either there is a third mtDNA helicase in yeast or these defective mitochondrial genomes do not require a separate helicase protein for DNA metabolism. Currently, we do not have a good explanation for the observed lethal effect of PIF1 disruption in SK048.
The stability of the wt [rho+] mtDNA depends on additional nuclear genes that are not required for [rho−] mtDNA replication. This includes genes encoding for proteins that are involved in mitochondrial gene expression (5, 24). Involvement of the Hmi1 protein at some stage of gene expression in mitochondria would also lead to the instability of the wt mtDNA. Transcription is the most likely step of gene expression that would require the DNA helicase activity. For this reason we analyzed the effect of the HMI1 gene disruption on transcription in mitochondria (Fig. 3). We found that the mitochondrial hypersuppressive genomes that contained the ori2 promoter were actively transcribed in strains which have only a disrupted copy of the HMI1 ORF. Also, we could not detect any significant change in the transcription level following introduction of the plasmid-encoded HMI1 gene. Thus, the involvement of the Hmi1 helicase in mitochondrial transcription seems unlikely. However, we cannot rule out the possibility that the disruption of the HMI1 gene has a different effect on transcription other than that of the ori2 promoter analyzed here.
Another interesting possibility is that the Hmi1 protein is a helicase involved in recombination. The yeast mitochondrial genome is active in recombination, and several lines of evidence suggest that recombinational processes are important for faithful mitochondrial genome transmission. MGT1-dependent cleavage of recombination intermediates affects transmission of mtDNA during mitotic divisions and has a dramatic effect on the suppressivity of [rho−] genomes (19, 37). Disruption of the ABF2 gene affects mtDNA stability and also significantly reduces recombination (4, 36). It has been discussed that yeast mtDNA replication may be a recombination-dependent process and, in that case, the Hmi1 helicase could affect mtDNA stability through its involvement in recombination-dependent priming of mtDNA replication.
The Hmi1 helicase seems to have a unique C-terminal mitochondrial import signal. The N-terminal segment of the protein contains only a few positively charged amino acid residues, and there are several acidic residues (Fig. 6). This indicates that the N terminus probably does not contain a mitochondrial transport signal. The sequence of the C-terminal part of the Hmi1 protein is predicted to form an amphipathic α-helix (Fig. 6). One side of the C-terminal helix is rich in basic and hydroxylated residues, and there are no negatively charged residues. The other side of the C-terminal helix is hydrophobic. These structural features are characteristic for the classical N-terminal mitochondrial transport signal (35).
A recent in vitro study demonstrates that the C-terminal segment of the Hmi1 protein can target the DHFR protein into mitochondria. In vitro import of the Hmi1 helicase proceeds in a C- to N-terminal direction and requires membrane potential, the TIM17-23 translocase, and the matrix Hsp70 protein (18). The experiments described here demonstrate that the C terminus of the Hmi1 protein is important for mitochondrial targeting in vivo (Fig. 6 and 7). Mutant Hmi1 protein, where the last 14 or 32 amino acids were removed, did not localize in mitochondria (Fig. 6, panels 5 to 8, and Fig. 7C). Acidic residues introduced instead of the C-terminal arginine residues at positions 704 and 705 result in a mutant protein that is not imported into mitochondria (Fig. 7D). Finally, as in the in vitro mitochondrial import system, the C-terminal segment of the Hmi1 protein targets the DHFR carrier protein into the yeast mitochondria in vivo (Fig. 7D).
ACKNOWLEDGMENTS
We thank R. Stuart and W. Neupert for the antibodies against mitochondrial marker proteins and helpful discussions. We are grateful to M. Makarowa and H. Holkeri for their help with spore dissection analysis. We thank J. Remme, A. Stenlund, and our colleagues in the department for the comments on the manuscript.
This work has been supported by Estonian Science Foundation grant 2888 to J.S.
REFERENCES
- 1.Alexandre C, Grueneberg D A, Gilman M Z. Studying heterologous transcription factors in yeast. Methods Companion Methods Enzymol. 1993;5:147–155. [Google Scholar]
- 2.Baldacci G, Bernardi G. Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. EMBO J. 1982;1:987–994. doi: 10.1002/j.1460-2075.1982.tb01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daum G, Böhni P C, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 4.Diffley J F X, Stillman B. A close relative of the nuclear chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Acad Natl Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fangman W L, Henly J W, Brewer B J. RPO41-independent maintenance of [rho−] mitochondrial DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:10–15. doi: 10.1128/mcb.10.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feaver W J, Bardwell S J, Bardwell A J, Buratowski S, Gulyas K D, Donahue T F, Friedberg E C, Kornberg R D. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- 7.Foury F, van Dyck E. A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. EMBO J. 1985;4:3225–3530. doi: 10.1002/j.1460-2075.1985.tb04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foury F, Lahaye A. Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J. 1987;6:1441–1449. doi: 10.1002/j.1460-2075.1987.tb02385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox T D, Folley L S, Mulero J J, McMullin T W, Thorsness P E, Hedin L O, Costanzo M C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 11.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 12.Hehman G L, Hauswirth W W. DNA helicase from mammalian mitochondria. Proc Natl Acad Sci USA. 1992;89:8562–8566. doi: 10.1073/pnas.89.18.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iordanescu S. Characterization of the Staphylococcus aureus chromosomal gene pcrA identified by mutations affecting plasmid pT181 replication. Mol Gen Genet. 1993;241:185–192. doi: 10.1007/BF00280216. [DOI] [PubMed] [Google Scholar]
- 14.Iordanescu S, Bargonetti J. Staphylococcus aureus chromosomal mutations that decrease efficiency of Rep utilization in replication of pT181 and related plasmids. J Bacteriol. 1989;171:4501–4503. doi: 10.1128/jb.171.8.4501-4503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 16.Lahaye A, Leterme S, Foury F. PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J Biol Chem. 1993;268:26155–26161. [PubMed] [Google Scholar]
- 17.Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee C M, Sedman J, Neupert W, Stuart R A. The DNA helicase, Hmip, is transported into mitochondria by a C-terminal cleavable targeting signal. J Biol Chem. 1999;274:20937–20942. doi: 10.1074/jbc.274.30.20937. [DOI] [PubMed] [Google Scholar]
- 19.Lockshon D, Zweifel S G, Freeman-Cook L L, Lorimer H E, Brewer B J, Fangman W L. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell. 1995;81:947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 20.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 21.Luking A, Stahl U, Schmidt U. The protein family of RNA helicases. Crit Rev Biochem Mol Biol. 1998;33:259–296. doi: 10.1080/10409239891204233. [DOI] [PubMed] [Google Scholar]
- 22.Matson S W, Bean D W, George J W. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 23.Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1989;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 24.Myers A M, Pape L K, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–601. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 26.Rickwood D, Dujon B, Darley-Usmar V M. Yeast mitochondria. In: Campell I, Duffus J H, editors. Yeast, a practical approach. Oxford, England: IRL Press; 1988. pp. 185–254. [Google Scholar]
- 27.Roberti M, Musicco C, Polosa P L, Gadaleta M N, Cantatore P. DNA-helicase activity from sea urchin mitochondria. Biochem Biophys Res Commun. 1996;219:134–139. doi: 10.1006/bbrc.1996.0194. [DOI] [PubMed] [Google Scholar]
- 28.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 29.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 30.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 31.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi S, Hours C, Iwaya M, Lane H E D, Denhardt D T. The Escherichia coli rep gene. In: Denhardt D T, Dressler D H, Ray D S, editors. The single-stranded DNA phages. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1978. pp. 393–400. [Google Scholar]
- 33.Taucher-Scholtz G, Abdel-Monem M, Hoffman-Berling H. Functions of DNA helicases in Escherichia coli. In: Cozzarelli N R, editor. Mechanism of DNA replication and recombination. New York, N.Y: Liss; 1983. pp. 65–76. [Google Scholar]
- 34.Thomas B J, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of RAD52 and RAD1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Heijne G. Mitochondrial targeting sequences may form amphipathic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zelenaya-Troitskaya O, Newman S M, Okamoto K, Perlman P S, Butow R A. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zweifel S G, Fangman W L. A nuclear mutation reversing a biased transmission of yeast mitochondrial DNA. Genetics. 1991;128:241–249. doi: 10.1093/genetics/128.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]