Abstract
Notch receptors participate in a highly conserved signaling pathway that regulates morphogenesis in multicellular animals. Maturation of Notch receptors requires the proteolytic cleavage of a single precursor polypeptide to produce a heterodimer composed of a ligand-binding extracellular domain (NEC) and a single-pass transmembrane signaling domain (NTM). Notch signaling has been correlated with additional ligand-induced proteolytic cleavages, as well as with nuclear translocation of the intracellular portion of NTM (NICD). In the current work, we show that the NEC and NTM subunits of Drosophila Notch and human Notch1 (hN1) interact noncovalently. NEC-NTM interaction was disrupted by 0.1% sodium dodecyl sulfate or divalent cation chelators such as EDTA, and stabilized by millimolar Ca2+. Deletion of the Ca2+-binding Lin12-Notch (LN) repeats from the NEC subunit resulted in spontaneous shedding of NEC into conditioned medium, implying that the LN repeats are important in maintaining the interaction of NEC and NTM. The functional consequences of EDTA-induced NEC dissociation were studied by using hN1-expressing NIH 3T3 cells. Treatment of these cells for 10 to 15 min with 0.5 to 10 mM EDTA resulted in the rapid shedding of NEC, the transient appearance of a polypeptide of the expected size of NICD, increased intranuclear anti-Notch1 staining, and the transient activation of an Notch-sensitive reporter gene. EDTA treatment of HeLa cells expressing endogenous Notch1 also stimulated reporter gene activity to a degree equivalent to that resulting from exposure of the cells to the ligand Delta1. These findings indicate that receptor activation can occur as a consequence of NEC dissociation, which relieves inhibition of the intrinsically active NTM subunit.
Notch receptors are central components of a highly conserved signal transduction pathway that regulates cell fate decisions in multicellular animals (for recent reviews, see references 2 and 17). Numerous genetic and molecular analyses have demonstrated that Notch signaling controls the implementation of differentiative, proliferative, and apoptotic programs of gene expression, which is consistent with it having a broad role in the processes of organ development and morphogenesis (12, 16, 23; S. Kurata, M. Go, S. Artavanis-Tsakonas, and W. Gehring, submitted for publication). Acquired or inherited abnormalities in genes involved in Notch signaling have also been detected in certain human leukemias (13) and Alagille (29, 33) and CADASIL (24) syndromes, indicating that perturbations of Notch signaling underlie several forms of human disease.
Notch receptors are initially synthesized as approximately 300- to 350-kDa single-pass transmembrane proteins, which then undergo proteolytic processing in the trans-Golgi network by a furin-like convertase at a site ∼70 amino acids external to the transmembrane domain (3, 7, 31). This results in the formation of a mature heterodimeric receptor, consisting of N-terminal extracellular (NEC) and C-terminal transmembrane (NTM) subunits, which is subsequently transported to the cell surface (7). The NEC subunits of various Notch receptors consist largely of up to 36 tandemly repeated epidermal growth factor (EGF) modules followed by three iterated Lin12-Notch (LN) modules, a protein motif that is only found in Notch receptors. NTM subunits have a small extracellular domain containing two conserved cysteine residues and intracellular domains that include six to seven ankyrin-CDC10-like repeats that are usually flanked by functional nuclear localization signal sequences and C-terminal PEST sequences. Although NEC and NTM can be coimmunoprecipitated (7), the nature of the interaction between these two subunits has not been defined.
Genetic and molecular analyses have identified several Notch ligands and at least one transcription factor, Suppressor of Hairless [Su(H), which is referred to in vertebrates as CBF1 or RBP-Jκ], that acts downstream of Notch (9, 21, 42). Ligands fall into groups homologous to the Drosophila ligands Delta or Serrate, which are themselves single-pass transmembrane proteins that exhibit partial functional redundancy and interact with EGF repeats 11 and 12 of Drosophila Notch (19, 38). Recent studies have provided the rationale for a seemingly simple and direct activation mechanism whereby the binding of ligand triggers one or more proteolytic cleavages that free the Notch intracellular domain (NICD) from its membrane tether (21, 26, 32). This is thought to permit translocation of NICD to the nucleus, where it associates with CBF1 bound to specific promoter sequences, thereby activating transcription of target genes (11, 21, 26, 28, 44, 45, 47, 48). However, with the exception of a select set of transformed and terminally differentiated cells, extensive immunocytochemical analyses have failed to detect Notch in the nuclei of developing animals (1, 49). In addition, although some studies have shown a positive correlation between the level of nuclear Notch and downstream signaling (41, 44), other studies conducted in flies (15), mice (3, 35), and cultured cells (4) have failed to detect such a relationship. These disparate results suggest that the relationship between nuclear translocation of NICD and signaling events, such as the activation of CBF1, is likely to be complex.
Studies conducted in invertebrates, frogs, and mammals have shown that forms of Notch resembling NTM produce gain-of-function phenotypes when expressed independently of NEC (10, 15, 35, 38), implying that the NEC subunit acts to restrain receptor activation in the absence of ligand. NEC deletions that remove all EGF repeats result in a dysfunctional receptor (21). In contrast, deletions that remove the LN modules and certain LN point mutations have been reported to produce gain-of-function phenotypes (18, 30, 44), suggesting that this domain is a negative regulator of Notch signaling.
Biochemical characterization of prototypical EGF and LN modules of human Notch1 (hN1), has shown that the structural integrity of these modules is dependent on the presence of millimolar Ca2+ (5, 37). Based on the regulatory role of the domains containing these modules, the importance of Ca2+ for heterodimeric stability was investigated. We now demonstrate that the NEC and NTM subunits of both Drosophila Notch and hN1 are noncovalently associated. This association is stabilized by millimolar Ca2+ and disrupted by EDTA, indicating the involvement of Ca2+-binding domains in maintenance of the interaction. Deletion of Ca2+-binding NEC EGF repeats does not disrupt subunit association, whereas deletion of Ca2+-binding LN modules abolishes stable association of NEC and NTM. Brief exposure of NIH 3T3 cells expressing human Notch1 to EDTA leads to the rapid shedding of NEC, which is followed by the appearance of a short-lived polypeptide of the expected size of NICD, increased anti-Notch1 intranuclear staining, and increased CBF1-dependent transcription from a reporter gene. EDTA also stimulates CBF1-dependent reporter gene activity in HeLa cells expressing endogenous Notch1 to a degree approximating that produced by co-cultivation with cells expressing the ligand Delta1. These findings suggest a model for receptor regulation whereby the activity of NTM is restrained, prior to activation, by a Ca2+-dependent interaction with NEC.
MATERIALS AND METHODS
cDNA expression constructs.
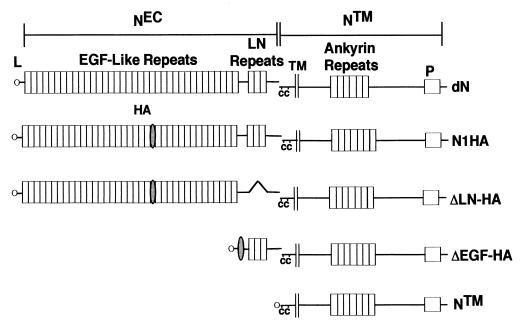
The various forms of Drosophila Notch and hN1 used in this study are summarized in Fig. 1. To permit detection of hN1 NEC, a single copy of an oligonucleotide linker encoding the hemagglutinin (HA) epitope was ligated in the sense orientation into a SalI site at coding sequence position 2602 in the full-length hN1 cDNA. The resultant cDNA encodes a form of hN1 (N1HA) with an HA tag inserted between EGF repeats 22 and 23. Other cDNAs were created using the PCR to introduce silent restriction sites that permitted in-frame deletion and/or insertion of coding sequences of interest. The N1HA cDNA was further modified by introduction of a deletion removing nucleotides 4294 to 4683 (codons 1431 to 1560) to create ΔLIN12-HA, which encodes a form of hN1 lacking the three LN repeats. ΔEGF-HA was created by insertion of the HA-encoding oligonucleotide linker into an engineered SalI site at position 4282 (21 codons 5′ of LN module 1), followed by ligation to a PCR product spanning hN1 bp −15 to +72 encoding the human Notch1 leader peptide. NTM was created by ligation of the same leader peptide-encoding oligonucleotide to nucleotides 4992 to 7665 of the Notch1 open reading frame (codons 1665 to 2555). Expression constructs for Drosophila Notch and Delta (14) and the intracellular domain of human Notch2 (8) have been described previously. An expression construct for the intracellular domain of murine Notch3 (N3IC), cloned into the vector pCMV, was provided by U. Lendahl (6).
FIG. 1.
Predicted structure of engineered Notch polypeptides. cDNAs encoding Drosophila Notch and various forms of human Notch1 were assembled and expressed in Drosophila S2 and mammalian cell lines, respectively, as described in Materials and Methods. dN, full-length Drosophila Notch; NEC, Notch extracellular subunit; NTM, Notch transmembrane subunit; L, leader peptide; HA, hemagglutinin tag; C, conserved cysteine residues; P, PEST sequence; N1HA, HA epitope-tagged human Notch 1; ΔEGF, a form of hN1 lacking all 36 EGF repeats; ΔLN-HA, a form of hN1 with the three LN module repeats deleted; NTM, a form of hN1 with an amino terminus 69 amino acids external to the transmembrane domain.
Expression of Notch proteins in cultured cells.
Schneider S2 cells and Drosophila Kc cells were grown in Sang's M3 medium (JRH Biosciences) with 10% fetal calf serum. Expression of Drosophila Notch and Delta was induced with CuSO4 in stable lines of S2 cells: N-S2 and Dl-S2, respectively (14). Mammalian cell lines were maintained at 37°C in 5% CO2. In transient-expression studies, cDNAs inserted into pcDNA3 (Invitrogen) were transfected into NIH 3T3 or 293A cells using Lipofectamine Plus reagent (Gibco-BRL). In stable expression studies, cDNAs inserted into pBABE were packaged into retroviruses, which were used to infect NIH 3T3 cells as described previously (3, 32). Infected cells were selected by the addition of 2 μg of puromycin per ml to culture media. 293A cells and NIH 3T3 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (D10). Jurkat cells were grown in RPMI 1640 supplemented with 10% fetal calf serum.
Preparation of cell lysates and immunoprecipitates.
Mammalian cells were washed two times with ice-cold Hanks buffered saline (HBS) and collected by centrifugation at 1,000 rpm for 5 min. Cell pellets were lysed in 50 mM Tris (pH 7.5) containing 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, and 1% NP-40 for 15 min on ice. Lysates were cleared by centrifugation at 14,000 × g for 15 min at 4°C.
hN1 polypeptides were immunoprecipitated from whole-cell lysates by incubation for 1 h at 4°C with 3 to 5 μl of polyclonal rabbit serum, termed TC (20), raised against a portion of intracellular hN1 (IC) or by incubation with anti-HA (clone HA.11; BABCO). This was followed by incubation with 10 μl of protein A-Sepharose beads (Pharmacia) for 1 h. Immunoprecipitates were prepared from conditioned cell culture media by prebinding anti-HA to protein A beads for 2 h. Beads were then mixed with conditioned medium for 6 h to overnight with rocking at 4°C.
Gel electrophoresis and Western blotting.
Proteins were solubilized in 50 mM Tris (pH 6.8) containing 3% sodium dodecyl sulfate (SDS) and 10% glycerol with or without 3% β-mercaptoethanol and electrophoresed on discontinuous SDS-polyacrylamide gels (27). Western blots were prepared according to the method of Towbin et al. (48). Drosophila Notch NEC and NTM were detected by staining of blots with monoclonal antibodies F461.3B and C17.9C6, respectively (39). Epitope-tagged human Notch1 NEC and NTM were detected by staining with mouse anti-HA or with a rabbit anti-IC, respectively. Notch2 polypeptides were detected with rat monoclonal antibody bHN6 (7), which was raised against the intracellular domain of Notch2. Staining was developed by using a chemiluminescent detection method (ECL kit; Amersham).
NEC-NTM heterodimer dissociation studies.
Notch expression in N-S2 cells was induced by the addition of 0.7 mM CuSO4 to the medium for 16 to 24 h. To study the release of the NEC subunit, N-S2 cells, or Kc cells were washed one to two times in TBS (20 mM Tris-HCl [pH 7.4] with 150 mM NaCl and 0 to 5 mM CaCl2) and resuspended in TBS containing either 2 to 5 mM CaCl2, 0.5 to 5 mM EDTA, or 5 mM EGTA. The cell suspension was rocked slowly at room temperature for 2 to 60 min, and the supernatant collected after two centrifugations at 2,000 and 16,000 rpm, respectively, and analyzed directly by Western blotting.
In EDTA dissociation experiments, N1HA cells were washed twice with HBS and then incubated at various temperatures in HBS containing 2.5 mM CaCl2 or 0.5 to 10 mM EDTA for 1 to 30 min. Conditioned HBS was then removed, taking care not to disturb weakly adherent cells, and spun at 14,000 × g for 15 min. Free NEC was detected in the cleared HBS by immunoprecipitation with anti-HA followed by Western blot analysis. The cells were gently washed once with HBS and then changed back to D10. Whole-cell extracts were subsequently prepared at various time points as described above.
In experiments studying the dissociation of immunoprecipitated NTM and NEC, immune complexes prepared on protein A beads (see above) were washed three times with ice-cold 50 mM Tris (pH 7.5), 100 mM NaCl, and 1% NP-40. NEC-NTM complexes were then resuspended in the same solution with either no additional ions, 2.5 mM CaCl2, or 10 mM EDTA at 25°C for 30 to 60 min with gentle rocking. After the beads were pelleted, the supernatant was removed and analyzed for release of hN1 subunits by reprecipitation with the appropriate antibody. The remaining beads were washed once with incubation buffer and analyzed by Western blotting.
Aggregation of Notch and Delta cells.
Expression of N and Dl in N-S2 and Dl-S2 cells, respectively, was induced overnight by addition of 0.7 mM CuSO4. After one washing with TBS containing 2 mM CaCl2, N-S2 cells were resuspended in TBS containing 5 mM CaCl2 or 2 mM EDTA and incubated for 30 min at room temperature. The EDTA-treated cells were reconstituted with CaCl2 (5 mM) to saturate the Ca2+-binding sites in the extracellular domain of Notch. After 5 min of further incubation, N-S2 cells were mixed with washed Dl-S2 cells, and aggregation was monitored turbidometrically as previously described (36).
Transcriptional activation assays.
NIH 3T3 cells growing in six-well dishes were cotransfected in triplicate with either 1 μg of the HES-AB-luciferase or 1 μg of the HES-ΔAB-luciferase plasmids containing hairy/enhancer of split 1 (HES1) promoter elements (21) and 20 ng of a Renilla luciferase control plasmid (pRL-TK; Promega) by using Lipofectamine Plus reagent (Gibco-BRL). At 24 h after transfection, the cells were briefly treated with HBS containing various concentrations of EDTA or 2.5 mM CaCl2 as described above and then allowed to recover in D10 for up to 8 h. Firefly and Renilla luciferase activities were determined in whole cell extracts using the Dual Luciferase Assay Kit (Promega) and a Turner Designs TD20 dual luminometer. Luciferase activity assays were also conducted with HeLa cells using a procedure modified from that described by Jarriault et al. (22). Cells were grown in 24-well plates in D10. On day 1, cells were cotransfected with 0.06 μg of CMV-βGal control plasmid (Invitrogen) and 0.6 μg of the HES-AB-luciferase plasmid or the HES-ΔAB-luciferase plasmid (mHES) using Lipofectamine Plus reagent. On day 2, approximately 24 h after transfection, 4 × 105 control quail QT6 cells or Delta-expressing QT6Dl cells (22) were added and cocultured with the transfected HeLa cells. On day 3, individual wells were treated with regular media, phosphate-buffered saline (PBS) or 0.5 mM EDTA in PBS for 15 min at 37°C. Cells were then incubated an additional 6 h in D10 medium prior to analysis of luciferase activities by using the Luciferase Reporter Gene Assay kit (Boehringer Mannheim). Luciferase activities were normalized to levels of β-Gal measured by the β-Gal Elisa kit (Boehringer Mannheim).
Immunostaining.
NIH 3T3 cells growing on eight-well chamber slides (Permanox; Tissue-Tek) were treated with HBS supplemented with CaCl2 or EDTA as described above. After various periods of recovery in D10, the cells were fixed in 3% paraformaldehyde and stained with rabbit anti-IC and goat anti-rabbit-fluorescein isothiocyanate (FITC) (Sigma) as described previously (3).
RESULTS
Noncovalent association of NEC and NTM.
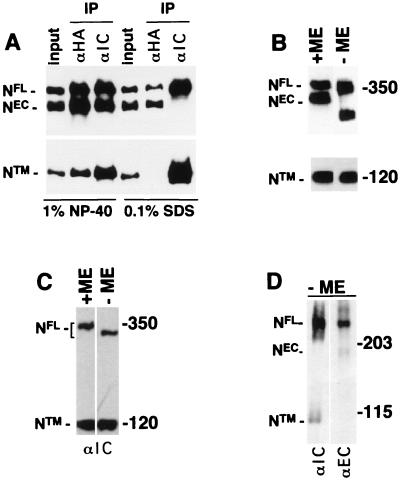
To determine the nature of the interaction between NEC and NTM, a form of hN1 bearing an extracellular HA epitope tag was stably expressed in NIH 3T3 cells (subsequently termed N1HA cells), which do not express endogenous murine Notch1 polypeptides at detectable levels (3). In control experiments with membrane impermeable biotinylation reagents, it was found that NTM and NEC were labeled, whereas full-length unprocessed hN1 (NFL) was not (not shown), indicating that hN1 processing is necessary for surface expression, just as has been previously shown for hN2 (7). The NEC and NTM subunits coimmunoprecipitated in extracts prepared in 1% NP-40 but did not coprecipitate in 0.1% SDS (Fig. 2A), indicating that their association is maintained by noncovalent interactions sensitive to ionic detergents. In addition, the amount of NTM detected was essentially identical when equivalent volumes of a whole-cell extract prepared with NP-40 were subjected to SDS-electrophoresis under reducing and nonreducing conditions (Fig. 2B). It was consistently noted that the mobility of both NFL and NEC were retarded by treatment with β-mercaptoethanol, presumably because reduced extracellular domain EGF and LN modules assume a more extended conformation. Levels of NTM were also roughly equivalent when aliquots of a whole-cell extract prepared from Jurkat cells with NP-40 were analyzed by SDS-electrophoresis under reducing and nonreducing conditions (Fig. 2C), indicating that endogenous NTM is also free of covalently associated polypeptides. Whole-cell extracts from Schneider 2 (S2) cells stably expressing Drosophila Notch (N) prepared in sample buffer containing 2% SDS and no reducing agent again showed that the NEC and NTM subunits were dissociated by nonreducing SDS-electrophoresis (Fig. 2D). These data indicate that noncovalent association is an evolutionarily conserved property of Notch receptor heterodimers.
FIG. 2.
Noncovalent association of Notch receptor subunits. The processed (NEC and NTM) and unprocessed (NFL) forms of Notch expressed in various cell lines were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) in reducing and nonreducing conditions and detected by Western blotting. (A) Proteins in whole-cell extracts (input) prepared from 106 N1HA cells with solutions containing either 1% NP-40 or 0.1% SDS were immunoprecipitated with anti-HA (αHA) or anti-Notch intracellular domain (αIC) antibody as indicated above each lane. After electrophoresis in SDS–6% polyacrylamide gels, Western blotting was done with anti-HA (in the top half of the panel) to detect the NFL and NEC and with anti-IC (in the lower half of the panel) to detect NTM. In the remaining panels, equivalent volumes of whole-cell extract prepared from N1HA (B), Jurkat (C), or Drosophila N-S2 (D) cells were solubilized in SDS loading buffer in the presence or absence of β-mercaptoethanol (ME), resolved by SDS-PAGE, and analyzed by Western blotting. N1HA and Jurkat cell blots were stained with anti-HA or anti-IC, and N-S2 blots were stained with antibodies against Notch intracellular (αIC, clone C17.9C6) or extracellular (αEC, clone F461.3B) domains, as indicated.
Metal ion dependency of NEC-NTM association.
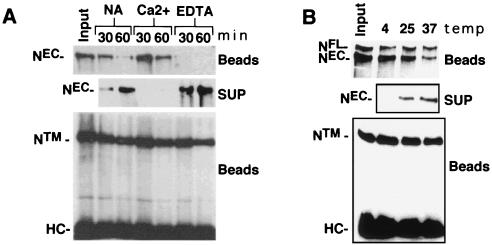
The Ca2+ requirement of multiple EGF (37) and LN (5) modules for maintenance of structural integrity suggested that the noncovalent interaction of NEC and NTM might be Ca2+-sensitive. To investigate this possibility, we studied the dissociation of NEC from immunocomplexes prepared with antibody directed against NTM. Time-dependent release of NEC was accelerated by 10 mM EDTA and prevented by 2.5 mM CaCl2 at 25°C (Fig. 3A), indicating that NEC-NTM heterodimers are stabilized by millimolar Ca2+. We had noted previously that NEC and NTM coprecipitated at 4°C in the presence of 1 mM EDTA (not shown), suggesting that dissociation rates were strongly influenced by temperature. This was confirmed by incubation of NEC-NTM immunocomplexes in 1 mM EDTA at various temperatures (Fig. 3B), which showed that the dissociation of NEC was retarded at 4°C compared to 25 or 37°C (Fig. 3B).
FIG. 3.
EDTA-induced dissociation of NEC from NTM. The effect of EDTA on NEC-NTM stability was examined by using immunoprecipitates prepared from hN1HA cell lysates. (A) NEC-NTM immunocomplexes were prepared from a whole-cell extract of 2 × 106 N1HA cells with anti-IC on protein A-Sepharose beads. The beads were then divided into seven aliquots which were either held on ice (input) or incubated in TBS alone (NA), TBS containing 2 mM CaCl2 (Ca2+), or TBS containing 10 mM EDTA (EDTA) at 25°C for the times indicated. After the beads were pelleted, the resulting supernatants were incubated with anti-HA and protein A-beads to immunoprecipitate any released NEC. Immunoprecipitated proteins were analyzed by SDS-PAGE in 6% gels followed by Western blotting, using anti-HA to detect NEC (top and middle panels) and anti-IC to detect the NTM (bottom panel). HC, immunoglobulin H heavy chain. (B) To investigate the temperature dependence of EDTA-mediated dissociation of NEC, NEC-NTM immunocomplexes were prepared from a whole-cell lysate of 106 N1HA cells with anti-IC and protein A-beads. The beads were then divided into four aliquots that were either held on ice (input) or incubated for 15 min in TBS containing 5 mM EDTA at 4, 25, or 37°C. Beads and supernatants were then processed and analyzed as described for panel A.
Metal ion chelator-induced shedding of NEC from cells.
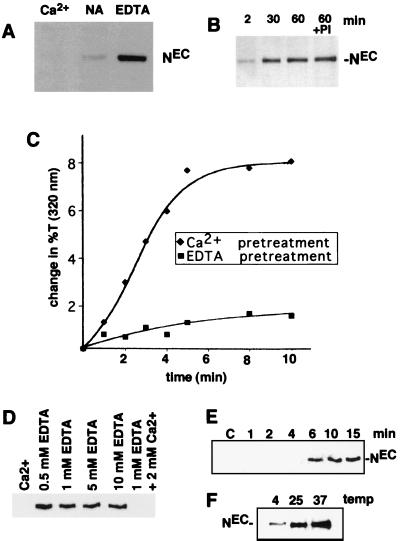
The role of divalent metal ions in stabilization of NEC-NTM heterodimers in cells was first assessed in N-S2 cells (Fig. 4A). Incubation of these cells in TBS resulted in some release of NEC, which was prevented by addition of 5 mM CaCl2 and enhanced by 5 mM EDTA, suggesting that NEC-NTM-interaction in cells is also stabilized by Ca2+. NEC release was also enhanced by 5 mM EGTA (not shown), a chelator that is relatively specific for Ca2+ (42). Additional studies demonstrated that dissociation of endogenous NEC from Drosophila Kc cells was detected as early as 2 min after addition of 0.5 mM EDTA and appeared to be complete by 30 min (Fig. 4B). The EDTA-mediated release of NEC was not affected by addition of a cocktail of protease inhibitors, suggesting that its occurrence is not dependent on proteolysis (Fig. 4B).
FIG. 4.
EDTA-induced dissociation of NEC from cells. (A) A total of 2 × 106 N-S2 cells were incubated for 30 min at 25°C in TBS buffer containing 5 mM CaCl2 (Ca2+), no additional ions (NA), or 5 mM EDTA. Polypeptides within the conditioned media were then resolved by SDS-PAGE on 7% gels and transferred to nitrocellulose. A Western blot is shown that is stained with antibody F461.3B directed against the dN extracellular domain. (B) Kc cells (107) were incubated for various times at 25°C in TBS containing 0.5 mM EDTA. In one experiment, cells were incubated for 60 min in TBS containing 0.5 mM EDTA and a cocktail of protease inhibitors (PI) consisting of 1 mM Preflabloc (Roche), 10 μg of aprotinin per ml, and 10 μg of leupeptin per ml. (C) The effect of divalent metal ion chelation on the depletion of NEC from N-S2 cells was examined by a quantitative aggregation assay (see Materials and Methods). N-S2 cells were pretreated for 30 min at 25°C in 2 mM EDTA (■) or 5 mM CaCl2 (⧫). A molar excess of CaCl2 (5 mM) was then added to the EDTA-treated cells, which were incubated at additional 5 min. Dl-S2 cells were then added to Ca2+- or EDTA pretreated cells at 25°C. Aggregation was monitored by the change in transmitted light at 320 nm. (D) The concentration dependence of EDTA-mediated NEC shedding was investigated by incubating ∼2 × 105 N1HA cells for 15 min at 37°C in HBS containing the indicated concentrations of CaCl2 and/or EDTA. Released NEC was precipitated from the conditioned media with anti-HA on protein A-beads, resolved by SDS-PAGE in a 6% gel, and detected on a Western blot stained with anti-HA. (E) The time course of NEC dissociation from N1HA cells was determined by incubating ∼2 × 105 cells at 37°C in HBS containing either 2 mM CaCl2 (C) for 15 min or 0.5 mM EDTA for the times indicated. (F) The temperature dependence of NEC dissociation from N1HA cells was determined by incubating ∼2 × 105 cells in TBS containing 0.5 mM EDTA for 15 min at the temperatures indicated. In panels E and F, the release of NEC into conditioned media was analyzed as described for panel D. Western blots stained with anti-HA are shown.
The effectiveness of EDTA in depleting NEC from the cell surface was evaluated by using a turbidometric cell aggregation assay that is dependent on Notch-Delta interaction (36). EDTA treatment has been shown to inhibit the aggregation of N-S2 and Dl-S2 cells (13), an effect which could be explained by either quantitative dissociation of NEC subunits or qualitative changes in the structure of Ca2+-binding EGF and LN modules within NEC. If the latter were the prevailing mechanism, it would predict that the effect of EDTA would be rapidly reversed by the addition of Ca2+. However, repletion of EDTA-treated N-S2 cells with Ca2+ was unable to restore aggregation (Fig. 4C), suggesting that the effect of EDTA on aggregation is primarily mediated through the dissociation of NEC from the cell surface.
The characteristics of EDTA-mediated shedding of NEC from cells were defined further in studies conducted with N1HA cells. N1HA cells incubated for 15 min at 37°C in 0.5 to 10 mM EDTA shed roughly equivalent amounts of NEC, an effect that was abolished by addition of a molar excess of Ca2+ (Fig. 4D). A time course study showed that shedding at 37°C in the presence of 0.5 mM EDTA was quite rapid, being detected by 6 min and appearing maximal by 10 to 15 min of treatment (Fig. 4E). The rate of shedding of NEC in the presence of 0.5 mM EDTA was reduced at 4°C compared to 25 or 37°C (Fig. 4F), paralleling the temperature dependency of NEC dissociation from NTM in immunocomplexes (Fig. 3B).
Requirement of LN repeats for NEC-NTM association.
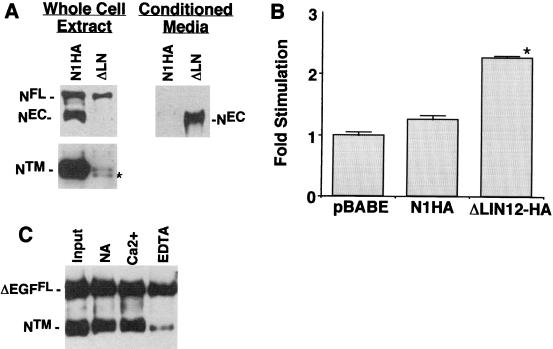
The role of specific Ca2+-binding domains in stabilization of NEC-NTM heterodimers was studied by expressing forms of N1HA bearing deletions in NIH 3T3 cells. To investigate the role of the LN domain, NIH 3T3 cells were transduced with a cDNA, termed ΔLN-HA, encoding an epitope-tagged form of NEC deleted of all three LN modules. Whole-cell extracts of cells expressing ΔLN-HA were found to contain ΔLNFL and NTM but no detectable ΔLNEC (Fig. 5A). These cells also contained a second polypeptide of slightly smaller size than NTM that cross-reacted with anti-IC. The size of this novel polypeptide approximates the size of an intracellular cleavage product implicated in signaling downstream of activated Notch (11, 26, 32, 45, 47, 48). We also noted that the levels of hN1 polypeptides were substantially lower in ΔLN-HA-expressing cells than in NIHA-expressing cells. NTM in N1HA cell extracts was detectable in Western blot exposures of as little as 5 s, whereas exposure times of several minutes were required to detect the processed ΔLN polypeptides. This is consistent with the observation that other forms of constitutively active hN1 are consistently expressed at low levels, relative to forms of hN1 without intrinsic signaling activity, in retrovirus-transduced NIH 3T3 cells (J. C. A., unpublished data).
FIG. 5.
Requirement of LN repeats for NEC-NTM association. (A) Association of NEC and NTM subunits was assessed in NIH 3T3 cells expressing N1HA or a form of N1HA bearing a deletion that removes the coding sequence for all three LN modules (ΔLN; see Fig. 1). Whole-cell extracts (50 μg of total protein) from equivalent numbers of N1HA and ΔLN cells were analyzed on a divided Western blot stained with anti-HA (upper left panel) and anti-IC (lower left panel). The asterisk denotes a polypeptide of slightly smaller size than NTM that was observed only in ΔLN extracts; on shorter exposures, only the upper band was present in N1HA cell extracts. Media (∼8 ml) conditioned by ∼1 × 106 to 2 × 106 N1HA or ΔLN cells for 24 h was analyzed by immunoprecipitation with anti-HA to detect released NEC and ΔLNNEC (right panel). A representative Western blot is shown that is stained with anti-HA. (B) To determine the effect of various Notch1 polypeptides on CBF1-dependent signaling, equivalent numbers of pBABE control, N1HA, and ΔLN cells growing in six-well dishes were cotransfected in four independent experiments with a CBF1-sensitive firefly luciferase plasmid (HES-AB) and a Renilla luciferase control plasmid. Cell lysates prepared 36 h posttransfection were analyzed by dual luciferase assay. The fold stimulation was calculated as the ratio of normalized firefly luciferase activities to the mean activity in pBABE control lysates. Error bars indicate +1 standard error of the mean. The asterisk denotes differences from both pBABE and N1HA at P < 0.05. (C) ΔEGF, a cDNA encoding a form of hN1 bearing an extracellular-domain HA tag and a deletion removing the coding sequence for all 36 EGF repeats (see Fig. 1), was transiently expressed in dishes containing equivalent numbers (∼1 × 105) 293A cells. Processed and unprocessed ΔEGF polypeptides were immunoprecipitated from whole-cell extracts with anti-HA antibody on protein A-beads. Polypeptides retained on protein A-beads after incubation in TBS with either no addition (NA), 2.5 mM CaCl2 (Ca2+), or 10 mM EDTA for 30 min at 20°C were detected on a Western blot stained with anti-IC.
To see if the LN deletion led to the release of ΔLNEC, anti-HA immunoprecipitates were prepared from conditioned media and analyzed by Western blotting. This revealed the presence of substantial amounts of an immunoreactive polypeptide of the expected size of ΔLNEC (∼210 kDa, Fig. 5A) in conditioned medium obtained from ΔLN-HA expressing cells. Of note, spontaneous shedding of NEC was not observed from N1HA cells, despite the presence of higher amounts of processed NTM in these cells relative to ΔLN-HA cells. These findings indicate that the LN modules are essential for the formation of a stable NEC-NTM heterodimer in cultured cells.
To determine whether the expression of ΔLN-HA polypeptides produced increases in Notch signaling, control pBABE cells, N1HA cells, and ΔLN-HA cells were transfected with a CBF1-sensitive reporter gene. Despite relatively low levels of processed ΔLN-HA polypeptides, ΔLN-HA cells demonstrated a small but significant increase in reporter gene activity relative to pBABE and N1HA cells (Fig. 5B). These results are compatible with data showing that expression of LN-deleted forms of Notch in vivo results in phenotypes consistent with gain-of-function (30).
To address the role of the Ca2+-binding EGF repeats in stabilizing the interaction of NEC and NTM, a form of hN1 containing an extracellular epitope tag and all three LN repeats but lacking all 36 EGF repeats, termed ΔEGF-HA (Fig. 1), was transiently expressed in 293A cells. Immunoprecipitates prepared with anti-HA contained large amounts of coprecipitating NTM, indicating that association of NTM and NEC does not require the EGF repeats (Fig. 5C). Further, NTM was released from immune complexes by EDTA, indicating that the interaction of ΔEGFEC and NTM is dependent on divalent metal ions. Together with data obtained through studies of ΔLN-HA, these findings demonstrate that the Ca2+-dependent interaction of NEC and NTM is dependent on Ca2+-binding LN modules.
Activation of hN1 processing and nuclear translocation by EDTA.
The release of NEC from cells treated with EDTA presumably creates a cellular pool of free NTM in a near synchronous fashion (Fig. 4E). Prior work has shown that amino-terminally deleted forms of Notch resembling NTM, commonly termed ΔE, produce gain-of-function phenotypes in several developmental systems (10, 15, 38), suggesting that free NTM might behave like an activated form of Notch. This possibility is also supported by observations showing that a form of ΔE retaining 61 of the predicted 69 extracellular amino acids of human Notch1 NTM induces T-cell leukemia in mice (35) and activates CBF1 in cultured cells (4).
To show formally that NTM has intrinsic signaling activity, a cDNA encoding NTM was transfected into NIH 3T3 cells and scored for its ability to activate a CBF1-sensitive reporter gene (Fig. 6A). NTM was observed to stimulate reporter gene expression to about the same degree as ΔE. Transfected cells also expressed approximately equivalent amounts of NTM or ΔE polypeptides (Fig. 6A, inset), indicating that these two polypeptides are of similar potency. These data suggested that EDTA-mediated release of NEC from NTM should activate Notch signaling, possibly by triggering the processing of NTM to NICD. To investigate the latter possibility, hN1HA cells were treated for 15 min with buffer containing either 10 mM EDTA or 2.5 mM CaCl2 and allowed to recover for various lengths of time in complete medium. Analysis of HBS supernatants showed that release of NEC was observed only in EDTA-treated cells (Fig. 6B). In this and other experiments, shedding of NEC from N1HA cells was incomplete, since substantial amounts of residual NEC were observed in whole-cell extracts prepared from EDTA-treated cells at 0 min. Despite this limitation, extracts prepared from EDTA-treated cells, but not Ca2+-treated control cells, showed the appearance of a short-lived polypeptide of the expected size of NICD that peaked 1 h after EDTA treatment and disappeared by 2 to 4 h after exposure (Fig. 6B). Additional studies showed that exposure to 0.5 mM EDTA for 15 min was sufficient to induce the appearance of this novel polypeptide by 1 h after treatment (Fig. 6C). The appearance of putative NICD correlated temporally with the results of immunostaining with a Notch1 antibody raised against the intracellular domain (Fig. 6D). Treatment for 15 min with 0.5 mM EDTA led to a transient increase in intranuclear staining after 1 to 2 h that was no longer detectable by 4 h after treatment. Taken together, these data suggest that dissociation of NEC induced further processing and nuclear translocation of the NTM subunit.
FIG. 6.
EDTA-induced NTM processing and nuclear translocation. (A) To demonstrate the intrinsic signaling activity of the NTM subunit, NIH 3T3 cells were cotransfected in triplicate with HES-AB luciferase reporter plasmid, Renilla luciferase control plasmid, and the indicated amounts of pcDNA3 plasmids containing either no cDNA insert, a cDNA insert encoding NTM, or a cDNA insert encoding ΔE. Firefly luciferase activities measured in cell lysates prepared 48 h posttransfection were normalized by using the corresponding internal Renilla luciferase control activities. The fold stimulation values were calculated as the ratio of individual normalized mean HES-AB-luciferase activities to the mean activity in control lysates prepared from cells transfected with empty pcDNA3 vector. In parallel experiments, extracts were prepared from NIH 3T3 cells transfected with either pcDNA3, pcDNA3-NTM, or pcDNA3-ΔE and the Renilla control plasmid. A Western blot normalized for differences in transfection efficiency (based on luciferase levels) was prepared and stained with rabbit anti-IC (inset). (B) ∼105 N1HA cells were treated for 15 min with HBS containing 2.5 mM CaCl2 (C) or 10 mM EDTA (E) at 37°C. The HBS-conditioned medium was harvested (SUP) and analyzed for release of NEC by preparation of immunoprecipitates with anti-HA, while cells were changed back to complete medium (D10) and allowed to recover for up to 8 h. Whole-cell lysates (WCE) prepared at various time points and immunoprecipitates prepared from conditioned media were analyzed on Western blots stained with anti-HA (upper and middle panels) or anti-IC (lower panel). An asterisk denotes a novel anti-IC-cross-reactive polypeptide that appeared in lysates prepared from cells 1 h after exposure to EDTA. (C) A total of 2 × 105 N1HA cells were treated for 15 min with HBS containing either no addition (0) or increasing concentrations of EDTA at 37°C and then allowed to recover in D10 for 1 h. Whole-cell lysates were prepared and analyzed on a Western blot stained with anti-IC. An asterisk denotes a novel anti-IC-cross-reactive polypeptide that appeared in lysates prepared from cells exposed to EDTA. (D) N1HA cells growing on slides were treated with HBS containing 0.5 mM EDTA for 15 min at 37°C, allowed to recover in D10 for 1 h (panel 1), 2 h (panel 2), or 4 h (panel 4) and then fixed and stained with anti-IC and goat anti-rabbit secondary antibody linked to FITC. The immunolocalization of Notch1 polypeptides in cells treated with EDTA was compared to control cells treated with HBS containing 2.5 mM CaCl2 for 15 min at 37°C followed by recovery in D10 for 1 h (panel C).
Activation of signaling by NEC shedding.
To correlate EDTA-induced NEC shedding and NTM processing with functional activation, a series of experiments were performed with reporter plasmids. In an initial time course experiment, exposure to 10 mM EDTA, but not 2.5 mM Ca2+, led to a time-dependent increase in the activity of a CBF1-sensitive reporter gene that was apparent by 4 h and which peaked by 6 h after exposure (Fig. 7A). Reporter gene activity slowly declined at time points beyond 6 h (Fig. 7A and data not shown), which is to be expected given the 3- to 5-h half-lives of luciferase mRNA and protein. The effect appeared to be mediated through activation of CBF1, since EDTA treatment did not activate a mutated promoter element that fails to bind CBF1 (Fig. 7B). To further correlate dissociation of NEC and activation of the reporter, a dose-response experiment was carried out. Exposure to 0.5 to 10 mM EDTA for 15 min at 37°C produced roughly equivalent degrees of reporter gene activation, an effect that was completely abolished by the addition of a molar excess of Ca2+ (Fig. 7C).
FIG. 7.
EDTA-induced activation of CBF1-dependent transcription. (A) At 24 h after transfection with the HES-AB luciferase plasmid and a Renilla luciferase control plasmid, ∼2.5 × 104 N1HA cells were treated with HBS containing 2.5 mM CaCl2 (Ca++) or 10 mM EDTA for 15 min at 37°C. Cell lysates were subsequently prepared in triplicate after various periods of additional incubation in complete medium (D10). Firefly luciferase activities in lysates were normalized using the corresponding internal Renilla luciferase control activities, which were not affected by EDTA or CaCl2 treatment (not shown). The fold stimulation values were calculated as the ratio of individual normalized mean HES-AB luciferase activities to the mean activity in control lysates prepared from cells treated with HBS plus CaCl2 for 15 min and then immediately harvested. An asterisk denotes time points at which the mean firefly luciferase fold stimulation differs significantly from the corresponding CaCl2 control (P < 0.05). (B) N1HA cells were transfected as in panel A with the HES-AB-luciferase (HES) plasmid or the HES-ΔAB-luciferase plasmid (mHES) lacking functional CBF1 binding sites, plus a Renilla luciferase control plasmid. At 24 h posttransfection, cells were treated with 10 mM EDTA as in panel A and then allowed to recover for various times in D10. Normalized firefly luciferase activities were determined in cell lysates prepared in triplicate at each time point. The fold stimulation values were calculated as the ratio of individual normalized mean firefly luciferase activities to the mean activities of extracts prepared at 0 min. An asterisk denotes time points at which the fold stimulation of HES-AB luciferase activities differ significantly from the corresponding mHES control (P < 0.05). (C) N1HA cells were transfected as in panel A with the HES-AB-luciferase plasmid and a Renilla luciferase control plasmid. At 24 h posttransfection, cells were treated with HBS containing the concentrations of CaCl2 and/or EDTA indicated for 15 min at 37°C and then incubated an additional 6 h in D10 medium. Normalized mean luciferase activities were determined in cell lysates prepared in triplicate for each treatment. The fold stimulation values were calculated as the ratio of individual normalized mean firefly luciferase activities to the mean activity of lysates prepared after treatment with 2 mM CaCl2. An asterisk denotes the mean firefly luciferase activities that differ significantly from that of the 2 mM CaCl2 control (P < 0.05). (D) N1HA and pBABE control cells were transfected as in panel A with the HES-AB firefly luciferase plasmid and a Renilla luciferase plasmid. At 24 h posttransfection, cells were treated with HBS containing 2.5 mM CaCl2 or 10 mM EDTA for 15 min or 30 min at 37°C. After 6 h of recovery in D10, cell lysates were prepared. The fold stimulation values were calculated as the ratio of normalized mean firefly luciferase activities in EDTA-treated cell lysates to the mean activity in the corresponding CaCl2-treated cell lysates. An asterisk denotes the mean levels of firefly luciferase activity that differ significantly from the corresponding 3T3-pBABE control (P < 0.05). To detect expression of Notch2 NTM, a Western blot (inset) containing extracts from 3T3-pBABE cells (3T3; 50 μg of protein loaded) and was stained with the Notch2-specific rat monoclonal antibody bHN6 (46). An adjacent lane containing an extract of 293A cells transiently transfected with pcDNA3-ICN2 (5 μg of protein loaded) served as a positive control and demonstrated that the cross-reactive polypeptide in 3T3-pBABE cells was slightly larger in size than the intracellular domain of Notch2. (E) 3T3-pBABE cells were transfected as in panel A with the HES-AB firefly luciferase plasmid, a Renilla luciferase plasmid, and either pcDNA3 (2 μg) or pCMV-N3IC (2 μg). At 24 h posttransfection, cells transfected with pcDNA3 (control) or pCMV-N3IC (N3IC) were treated in triplicate with HBS containing 2.5 mM CaCl2 or 0.5 mM EDTA for 15 min at 37°C. After 6 h of recovery in D10, cell lysates were prepared. The fold stimulation values were calculated as the ratio of normalized mean firefly luciferase activities in EDTA-treated cell lysates to the mean activity in the corresponding CaCl2-treated cell lysates. An asterisk denotes the mean levels of firefly luciferase activity that differ significantly from the corresponding pBABE control (P < 0.05). (F) HeLa cells (seeded at 105 cells and grown for 2 days) were cotransfected with 0.06 μg of CMV-βGal plasmid and either 0.6 μg of HES-AB-luciferase or 0.6 μg of mHES-luciferase reporter. At 24 h after transfection, control QT6 and Delta-expressing QT6Dl cells (4 × 105) were added to the HeLa cells and maintained in coculture for 24 h. In separate wells, HeLa cells were also exposed to D10 media (control), PBS, or PBS containing 0.5 mM EDTA for 15 min at 37°C and then incubated an additional 6 h in D10 prior to determination of the luciferase activities. All treatments were performed in duplicate and luciferase activities were normalized by using the β-galactosidase activities. The fold stimulation values represent the ratio between individual mean normalized luciferase activities and the mean normalized activity produced by treatment with complete media alone, except for the QT6Dl-treated samples, which are compared to QT6 samples. The data represent the means of three independent experiments. Notch1 and Notch2 polypeptides in HeLa cell extracts were detected on Western blots (inset) stained with rat monoclonal anti-Notch1 (clone bTan20) or anti-Notch2 (clone bHN6). The error bars in panels A to F correspond to +1 standard error of the mean.
To investigate the dependency of the EDTA effect on hN1 overexpression, the level of activation of the Su(H) reporter was compared in N1HA cells and control 3T3-pBABE cells after exposure to 10 mM EDTA for 15 or 30 min (Fig. 7D). Expression of N1HA produced a modest but significant increase in activation above that seen in 3T3-pBABE cells, indicating that some of the activation can be attributed to N1HA-encoded polypeptides. 3T3-pBABE cells also showed significant transcriptional activation with EDTA treatment compared to cells treated with Ca2+. Although 3T3-pBABE cells contain no detectable Notch1 polypeptides, Western blot analysis showed the presence of Notch2 NTM in 3T3-pBABE cell extracts (Fig. 7D, inset). This observation, which is consistent with the prior Northern blot detection of Notch2 mRNA in 3T3-pBABE cells (32), suggested that the effect of EDTA on 3T3-pBABE control cells stems from activation of Notch2. This possibility was supported by the observation that the EDTA-induced activation of reporter gene activity in 3T3-pBABE cells was antagonized by the intracellular domain of Notch3 (Fig. 7E), which appears to inhibit activating forms of Notch by competing for access to CBF1 (6). We also observed that HES-reporter gene activity in 3T3-pBABE cells increased two- to threefold after treatment with Jagged2-expressing feeder cells (not shown), confirming their ability to respond to a physiologic activator of Notch receptors.
To explore further the activation of endogenous Notch receptors by EDTA, assays were performed with HeLa cells. A 3.9-fold increase in CBF1 reporter activity was observed 6 h after treatment with 0.5 mM EDTA, compared to control cells treated with normal media or buffer without EDTA (Fig. 7F). The activation appeared to be mediated through CBF1 because EDTA had no effect on a mutated promoter element that does not bind CBF1. Although the degree of stimulation by EDTA was modest, it was comparable to that produced by cocultivation of HeLa cells with Delta ligand-expressing cells (Fig. 7F). Western blot analysis confirmed that the HeLa cells used in this experiment express mature Notch1 and Notch2 receptors (Fig. 7F, inset), providing a basis for the response of these cells to EDTA and Delta. Thus, the modest stimulation in CBF1-sensitive gene expression produced by EDTA is comparable in magnitude to that produced by activation of Notch receptors by physiologically relevant ligands.
DISCUSSION
Our data indicate that the NEC subunit of Notch receptors acts to restrain NTM subunit signaling and illustrate two distinct mechanisms through which the effect of NEC can be overcome: (i) divalent metal ion chelator-mediated release of NEC and (ii) internal deletions in NEC that remove Ca2+-binding LN modules.
Disruption of NEC-NTM interaction by EDTA and stabilization by Ca2+ demonstrates a conserved requirement for metal ions in the maintenance of Notch heterodimers. Stabilization by Ca2+ could result from metal ion coordination by amino acid residues in NEC, NTM, or both, the latter possibly stemming from shared metal ion coordination as part of a metal ion bridge. With respect to metal binding by NEC, prior work has shown that the folding and structural integrity of individual EGF and LN modules depends on the presence of millimolar Ca2+ (5, 37). Here we show by deletional analysis that the LN repeat region plays an essential role in stabilization of NEC-NTM heterodimers, whereas the EGF repeats appear not to influence this association. Together, these findings suggest that the release of NEC by chelators may stem from conformational changes in LN modules. Such a critical role for the LN repeats in the restraint of NTM activity is further supported by developmental studies in invertebrates which have shown that disruption of LN modules by deletions or point mutations results in dominant gain-of-function phenotypes (18, 30, 44). It will be of interest to see if activating LN point mutations also disrupt NEC-NTM interaction. The effects attributable to the LN domain could be explained by direct binding of these modules to the extracellular portion of NTM, which might serve to protect NTM from activating proteolytic cleavage(s). The appearance of a novel form of intracellular Notch of the expected size of NICD in hN1 ΔLN-expressing cells is compatible with this possibility. These observations predict that mutations or modifications in NTM that disrupt NEC-binding and/or metal-binding residues should also be activating. Of note, several point mutations in the extracellular portion of NTM have also proven to cause gain-of-function phenotypes (18, 30).
Other lines of evidence support a model in which proteolytic processing of NTM to NICD and its subsequent translocation to the nucleus are critical steps in the activation of downstream genes such as HES1 by Notch (21, 26, 28, 44). Here, after EDTA treatment we have directly observed that transcriptional activation of a CBF1-sensitive reporter gene is preceded by the appearance of an NICD-like cleavage product and increased levels of intranuclear Notch1. To date, nuclear Notch has been very difficult to detect by biochemical or in situ methods in normal cells, possibly because it is present in very low amounts and/or has a very short half-life. The transient appearance of NICD and intranuclear Notch after a brief period of exposure to EDTA would tend to support the latter possibility. The ability to detect NICD and intranuclear Notch after EDTA treatment of NIH 3T3 cells overexpressing hN1 likely stems from the near synchronous release of NEC from NTM. This pool of free NTM presumably serves as a substrate for secondary events, such as proteolytic processing, that permit nuclear translocation.
Certain aspects of our data, however, raise a number of issues that are pertinent to both the specific mechanism of activation by EDTA and, more generally, to the normal mechanism of Notch signaling. Given the apparently large increase in nuclear Notch that is induced by EDTA, it is perhaps surprising that the increases in reporter gene activation are modest (3.5- to 5-fold) compared to those observed with sustained overexpression of NICD from strong promoters (21). Factors that may act to limit the magnitude of the response to EDTA treatment include the transient nature of the stimulus and the resistance of a significant fraction of the cellular NEC to EDTA-induced dissociation. We have observed that a substantial portion of the EDTA-resistant NEC in NIH 3T3 cells is located on the cell surface (J.C.A., unpublished data), suggesting that EDTA sensitivity may be regulated by currently unknown factors that modify NEC-NTM complex stability. Although the magnitude of the signal induced by EDTA is smaller than the “supraphysiologic” signals observed in transiently overexpressing cells, it is comparable to that produced by cocultivation with ligand-expressing cells and therefore within a range that is expected to produce biologically relevant effects.
The apparent delay between the peak appearance of NICD and intranuclear staining (∼1 h after EDTA) and maximal reporter gene activation (∼4 to 6 h after EDTA) was also unanticipated and raises questions about the timing of events that activate downstream target genes. While the temporal relationship between the nuclear translocation of NICD and the activation of target genes has yet to be addressed, a number of other studies have suggested that the level of nuclear Notch and signaling are not strictly correlated. Membrane-tethered forms of Drosophila ΔE resembling NTM that localized largely to extranuclear membranes caused gain-of-function developmental abnormalities in several tissues that were equivalent to, or more severe, than those produced by a form of NICD that localized entirely to the nucleus (2, 15, 38). Similarly, a form of human ΔE that localized largely to extranuclear membranes is as effective as nuclear NICD in activating CBF1 in cultured cells (4) and inducing T-cell leukemia in mice (35). The imperfect relationship between the level of nuclear Notch and phenotype observed in these experiments suggest that additional events besides nuclear translocation may be necessary for signaling. It is possible that a need for other events may underlie the discordance between the timing of peak intranuclear Notch levels and gene activation observed in cells treated with EDTA.
Another observation requiring further investigation is the basis for the relatively small increase in CBF1-dependent reporter gene activation that is observed in cells stably expressing ΔLN-HA. Multiple factors may be acting to limit the degree of sustained CBF1 activation in ΔLN-HA cells. We have observed consistently that the levels of constitutively active Notch1 polypeptides in NIH 3T3 cells fall over a period 3 to 7 days post-retroviral transduction to levels much lower than those observed in transiently expressing cells, in which reporter assays are typically performed (J.C.A., unpublished data). The explanation for this decrease in expression is not clear, but it has also been observed within single clones, suggesting that mechanisms other than negative selection may be responsible. In addition, Notch1 activation may upregulate the expression of several negative regulators, such as Deltex (34; W. Pear et al., in press) or Notch3 (6, 32). Together, these factors may act to limit the activation of CBF1 in cells stably overexpressing constitutively active forms of Notch1 to levels similar to those produced by ligand stimulation.
The observation that EDTA-mediated dissociation of NEC is sufficient to trigger NTM signaling raises the possibility that ligand-induced activation might proceed through an analogous mechanism. The simplest mechanism would be for ligand-binding to induced conformational changes that promote dissociation of NEC. However, the Notch-Delta-dependent aggregation of S2 cells would appear to exclude this possibility as a general mechanism for physiologic activation of Notch signaling. A second possibility is that ligand binding might trigger endocytosis of Notch-ligand complexes, an event clearly documented in Drosophila (14, 25). The importance of endocytosis has been corroborated by genetic analyses demonstrating that dynamin, a protein that participates in endocytosis, is necessary for Notch signaling in vivo (43). Endocytosis could contribute by exposing the ligand-receptor complex to the low pH within endosomes, which might physically destabilize the NEC-NTM interaction by titrating Ca2+-binding acidic residues in the LIN12 domain (5). Alternatively, endocytosis could deliver receptor-ligand complexes to activating protease(s), the subcellular localization of which is currently unknown. A third possibility, which is also compatible with the second, is that ligand binding might facilitate additional extracellular cleavages in NTM or NEC that remove domains required for heterodimer stabilization. We have recently observed that cocultivation of N1HA cells with cells expressing Jagged1 or Jagged2 ligands leads to the appearance of NEC in conditioned media (J.C.A., unpublished data), suggesting that one or more of these mechanisms may be relevant to normal receptor activation. Overexpression of Notch may titrate out factors, such as endocytic trafficking molecules or proteases, that participate in required events prior or subsequent to ligand binding, thus providing an explanation for how ligand might both promote the aggregation of S2 cells and the dissociation of NEC from NIH 3T3 cells. Future investigation of the fate of NTM, NEC, and ligand postinteraction will be needed to sort out these possibilities and uncertainties.
Of more general importance, the presence of 0.5 mM EDTA in trypsin solutions and the expression of one or more Notch receptors by most cultured cell lines implies that routine splitting of adherent cells likely produces transient activation of Notch signaling. In support of this possibility, we have recently noted that standard treatment of certain cell lines with trypsin-EDTA solutions induces the appearance of immunoreactive polypeptides of the expected size of NICD (J.C.A., unpublished data). Given the pleiotropic effects of activated Notch on many cellular processes, the timing of trypsinization may contribute to variability in some cultured cell assays.
ACKNOWLEDGMENTS
This work was supported by NIH grants CA66849 and CA82308 (J.C.A.), CA62450 (J.S.), NS26084 (S.A.T.), NS10735-01 (L.M.G.), HL61001 (S.C.B.), and the Massachusetts General Hospital (S.A.T.). S.C.B. is a Pew Scholar in the Biomedical Sciences.
We thank A. Israel for providing the HES-AB and HES-ΔAB luciferase reporters and U. Lendahl for providing the N3IC expression plasmid.
REFERENCES
- 1.Ahmad I, Zagouras P, Artavanis-Tsakonis S. Involvement of Notch-1 in mammalian retinal neurogenesis: association of Notch-1 activity with both immature and terminally differentiated cells. Mech Dev. 1995;53:73–85. doi: 10.1016/0925-4773(95)00425-4. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Aster J, Pear W, Hasserjian R, Erba H, Davi F, Luo B, Scott M, Baltimore D, Sklar J. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp Quant Biol. 1994;59:125–136. doi: 10.1101/sqb.1994.059.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 5.Aster J C, Simms W B, Zavala-Ruiz Z, Patriub V, North C L, Blacklow S C. The folding and structural integrity of the first LIN-12 module of human Notch1 are calcium-dependent. Biochemistry. 1999;38:4736–4742. doi: 10.1021/bi982713o. [DOI] [PubMed] [Google Scholar]
- 6.Beatus P, Lundkvist J, berg C, Lendahl U. The Notch 3 intracellular domain represses Notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126:3925–3935. doi: 10.1242/dev.126.17.3925. [DOI] [PubMed] [Google Scholar]
- 7.Blaumueller C M, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 8.Capobianco A J, Zagouras P, Blaumueller C M, Artavanis-Tsakonas S, Bishop J M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H) Development. 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 10.Coffman C R, Skoglund P, Harris W A, Kinter C R. Expression of an extracellular deletion of Xotch diverts cell fates in Xenopus embryos. Cell. 1993;73:659–671. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- 11.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 12.Deftos M L, He Y W, Ojala E W, Bevan M J. Correlating notch signaling with thymocyte maturation. Immunity. 1998;9:777–786. doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 14.Fehon R G, Kooh P J, Rebay I, Regan C L, Xu T, Muskavitch M A T, Atravanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 15.Fortini M E, Rebay I, Caron L A, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 16.Go M J, Eastman D S, Artavanis-Tsakonas S. Cell proliferation control by Notch signaling in Drosophila development. Development. 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- 17.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 18.Greenwald I, Seydoux G. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature. 1990;346:197–199. doi: 10.1038/346197a0. [DOI] [PubMed] [Google Scholar]
- 19.Gu Y, Hukriede N A, Fleming R G. Serrate expression can functionally replace Delta activity during neuroblast segregation in the Drosophila embryo. Development. 1995;121:855–865. doi: 10.1242/dev.121.3.855. [DOI] [PubMed] [Google Scholar]
- 20.Hasserjian R H, Aster J C, Davi F, Weinberg D, Sklar J. Modulated expression of NOTCH1 during thymic development. Blood. 1996;88:970–976. [PubMed] [Google Scholar]
- 21.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signaling downstream of activated mNotch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 22.Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong C F, Brou C, Seidah N G, Israel A. Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jehn B M, Bielke W, Pear W S, Osborne B A. Protective effects of notch-1 on TCR-induced apoptosis. J Immunol. 1999;162:635–638. [PubMed] [Google Scholar]
- 24.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis E A, Ruchoux M M, Weissenbach J, Bach J F, Bousser M G, Tournier-Lasserve E. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 25.Klueg K M, Parody T R, Muskavitch M A. Complex proteolytic processing acts on Delta, a transmembrane ligand for Notch, during Drosophila development. Mol Biol Cell. 1998;9:1709–1723. doi: 10.1091/mbc.9.7.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopan R, Schroeter E H, Weintraub H, Nye J S. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lecourtois M, Schweisguth F. Indirect evidence for Delta-dependent intracellular processing of Notch in Drosophila embryos. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Krantz I D, Deng Y, Genin A, Banta A B, Collins C C, Qi M, Trask B J, Kuo W L, Cochran J, Costa T, Pierpont M E, Rand E B, Piccoli D A, Hood L, Spinner N B. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 30.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 31.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah N G, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci USA. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo B, Aster J C, Hasserjian R P, Kuo F, Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997;17:6057–6067. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oda T, Elkahloun A G, Pike B L, Okajima K, Krantz I D, Genin A, Piccoli D A, Meltzer P S, Spinner N B, Collins F S, Chandrasekharappa S C. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 34.Ordentlich P, Lin A, Shen C P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi H, Rand M D, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 37.Rand M D, Lindblom A, Carlson J, Villoutreix B O, Stenflo J. Calcium binding to tandem repeats of EGF-like modules. Expression and characterization of the EGF-like modules of human Notch-1 implicated in receptor-ligand interactions. Protein Sci. 1997;6:1–13. doi: 10.1002/pro.5560061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebay I, Fehon R G, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 39.Rebay I, Fleming R J, Fehon R G, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 40.Schmid R W, Reilly C N. New complexon for the titration of calcium in the presence of magnesium. Anal Chem. 1957;29:264–268. [Google Scholar]
- 41.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 42.Schweisguth F, Posakony J. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- 43.Seugnet L, Simpson P, Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev Biol. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- 44.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 45.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 46.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe M S, Xia W, Ostaszewski B L, Diehl T S, Kimberly W T, Selkoe D J. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 48.Ye Y, Lukinova N, Fortini M E. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 49.Zagouras P, Stifani S, Blaumueller C M, Carcangiu M L, Artavanis Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]