Significance
DEAD-box RNA helicases are essential cellular enzymes that remodel misfolded RNA structures in an adenosine triphosphate (ATP)-dependent process. The DEAD-box helicase DbpA is involved in the complex and highly regulated process of ribosome maturation. To prevent wasteful hydrolysis of ATP by DbpA, the enzyme is only active when bound to maturing ribosomes. Here, we elucidate the structural basis behind this important regulatory mechanism and find that the recruited ribosome substrate is able to stabilize the catalytically important closed state of the helicase. In addition, our data identify the natural site of action for DbpA in the maturing ribosome and provide a molecular explanation for the observed ribosome maturation defects that result from the overexpression of a DbpA mutant form.
Keywords: enzyme regulation, DEAD-box helicase, ribosome assembly, NMR spectroscopy, molecular mechanism
Abstract
The adenosine triphosphate (ATP)-dependent DEAD-box RNA helicase DbpA from Escherichia coli functions in ribosome biogenesis. DbpA is targeted to the nascent 50S subunit by an ancillary, carboxyl-terminal RNA recognition motif (RRM) that specifically binds to hairpin 92 (HP92) of the 23S ribosomal RNA (rRNA). The interaction between HP92 and the RRM is required for the helicase activity of the RecA-like core domains of DbpA. Here, we elucidate the structural basis by which DbpA activity is endorsed when the enzyme interacts with the maturing ribosome. We used nuclear magnetic resonance (NMR) spectroscopy to show that the RRM and the carboxyl-terminal RecA-like domain tightly interact. This orients HP92 such that this RNA hairpin can form electrostatic interactions with a positively charged patch in the N-terminal RecA-like domain. Consequently, the enzyme can stably adopt the catalytically important, closed conformation. The substrate binding mode in this complex reveals that a region 5′ to helix 90 in the maturing ribosome is specifically targeted by DbpA. Finally, our results indicate that the ribosome maturation defects induced by a dominant negative DbpA mutation are caused by a delayed dissociation of DbpA from the nascent ribosome. Taken together, our findings provide unique insights into the important regulatory mechanism that modulates the activity of DbpA.
RNAs are involved in numerous cellular processes, and correct folding of these RNAs is often essential for their function. RNA helicases, which modulate RNA structure, are thus important players in RNA-dependent processes (1). The largest family of RNA helicases is formed by the adenosine triphosphate (ATP)-dependent DEAD-box helicases (2). The double-stranded RNA (dsRNA)-unwinding process that is catalyzed by DEAD-box helicases is nonprocessive and not sequence specific. Only short duplexes up to ∼13 nucleotides (nt) are unwound, and the unwinding rates are inversely correlated with duplex stability (2). DEAD-box helicases consist of a functional core of two RecA-like domains that are connected by a short flexible linker. In the absence of the substrates (ATP and RNA), DEAD-box helicase are in an open conformation, where the two RecA-like domains tumble independently (2). During the catalytic cycle, DEAD-box helicases undergo large conformational changes and transiently adopt a distinct closed conformation in which the bound ATP is sandwiched between both RecA domains. As a result, these domains come in close contact and form a continuous, composite binding site for single-stranded RNA (ssRNA) (3, 4). Structural information on the closed state of the helicase indicates that this state likely represents a postunwinding state, as the bound ssRNA adopts a conformation that is incompatible with the structure of dsRNA. The closed state of the enzyme shows strongly increased ATPase activity. As a consequence, the bound ATP is rapidly hydrolyzed, after which the ssRNA and ADP dissociate, thus preparing the enzyme for another round of unwinding (5). Despite the extensive knowledge about the apo- and ssRNA (product)-bound forms of the enzyme, it is not well understood how the initial interaction between the helicase and an RNA duplex (substrate) takes place. On the one hand, it has been suggested that DEAD-box helicases directly load onto duplex RNA; on the other hand, it has been proposed that the enzyme captures a stretch of free ssRNA during the breathing of the duplex ends (6). In either case, the formation of the closed helicase state is essential, as this destabilizes the RNA duplex, which induces dsRNA unwinding due to spontaneous dissociation of the destabilized duplex (5). RNA substrate specificity of DEAD-box helicases is provided by N- and carboxyl-terminal regions that flank the canonical RecA core domains (2). These regions are in many cases intrinsically disordered, although a number of DEAD-box helicases contain additional folded domains, such as RNA binding or oligomerization domains.
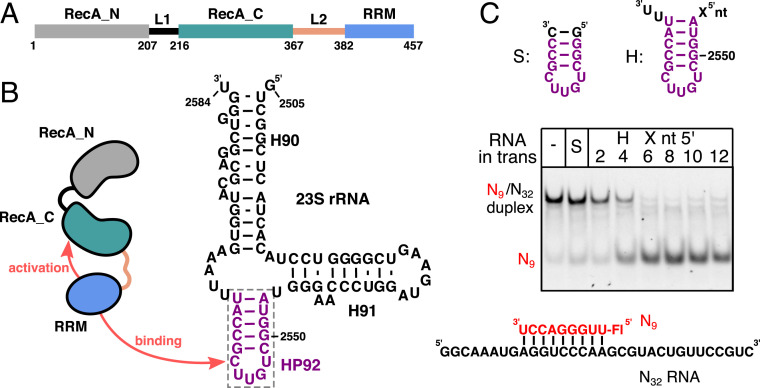
The Escherichia coli DEAD-box helicase DbpA and its homolog YxiN from Bacillus subtilis are important model systems for mechanistic studies of the DEAD-box helicases (4, 7–14). In addition to the two core RecA domains DbpA possesses a carboxyl-terminal RNA recognition motif (RRM), which is connected to the RecA_C domain by a 14–amino acid linker (L2) (Fig. 1A). Neither the linker length (10 to 35 amino acids) nor the linker sequence are well conserved between different DbpA homologs (15). The RRM specifically binds to the loop of hairpin 92 (HP92) of the 23S ribosomal RNA (rRNA) and thereby recruits DbpA to the nascent 50S subunit (16–18) (Fig. 1B). In the mature 50S subunit, HP92 is not accessible and part of the highly conserved peptidyl transferase center (PTC). In vitro, DbpA has been shown to efficiently unwind short RNA oligonucleotides annealed to ssRNA regions that flank HP92 (9, 16). The unwinding activity of DbpA critically depends on the presence of HP92 in the substrate RNA, as the lack of this hairpin dramatically diminishes ATPase activity and abolishes unwinding (16, 19, 20). Biologically, this will prevent the wasteful hydrolysis of ATP when DbpA is not bound to maturing ribosomes. For the DbpA homolog YxiN, it has been shown that addition of HP92 RNA in trans is sufficient to stimulate the unwinding of RNA substrates lacking HP92 (21). The molecular basis behind this important HP92 RNA–mediated activation mechanism of DbpA has, however, not been revealed so far.
Fig. 1.
DbpA domain organization, rRNA binding, and activation of DbpA activity by HP92. (A) Domain organization of E. coli DbpA. The linker L1 and L2 that connect the three domains are shown in black and light brown, respectively. Residue numbers at the domain boundaries are indicated. (B) Secondary structure of the 23S rRNA (nt 2,505 to 2,584) as found in the mature 50S subunit. HP92, which is recognized specifically by the DbpA RRM, is shown in violet. (C) Helicase assays that show that the binding of HP92 to the RRM in trans enables duplex unwinding by DbpA. Unwinding of an RNA duplex consisting of a fluorescently labeled 9mer RNA (red) and the unstructured N32 RNA (Bottom) is followed by native polyacrylamide gel electrophoresis (PAGE) analysis. The nonfluorescently labeled HP92–derived RNAs that were added in trans are indicated on the Top. No unwinding is observed in the absence of HP92 (−) or in the presence of a short, stabilized HP92 fragment without ssRNA overhangs (S). HP92 constructs containing 5′ overhangs of 4 to 12 nt enable duplex unwinding by DbpA.
Interactions between the RRM and the RecA domains likely play a role in the HP92-mediated activation of DbpA. The RecA_N domain tumbles independently of the RecA_C and RRM domains in the apo state (22); however, a fixed-domain RRM-RecA_C orientation is supported by two lines of evidence: First, the isolated RecA_C and RRM domains of DbpA are unstable (22), whereas a construct that contains both domains shows long-term stability. Second, it has been shown that a spacer of several unpaired nt between the stem of HP92 and the duplex is necessary for unwinding activity. The spacer length (3 or 11 nt) depends on whether the duplex is located 5′ or 3′ to HP92, a finding that can be explained by a fixed orientation of the RNA substrate with respect to the helicase core (16). On the other hand, there is also support for the structural independence of core and RRM domain for YxiN and DbpA based on small-angle X-ray scattering data (23) and enzymatic experiments for a DbpA mutant with an elongated L2 linker (24). Single-molecule fluorescence resonance energy transfer (smFRET) experiments indicate a repositioning of the RRM relative to the core domains upon binding of HP92 (21, 25). Structural models that are based on these latter data, however, only display limited contacts between the RRM and the core domains, which led to the suggestion that the L2 linker transmits the allosteric signal between the RRM and the RecA_C domain (21).
In line with its function as a ribosome maturation factor (18, 26, 27), DbpA only interacts with isolated 23S rRNA and 50S assembly intermediates but not with the mature 50S subunit of the ribosome (18, 20), where HP92 is mostly inaccessible (28). The exact ribosomal assembly intermediate on which DbpA acts remains unknown, as HP92 is accessible for interactions with DbpA during the large time window between its transcription and the final stages of 50S subunit maturation (29, 30). In addition, it is not clear whether DbpA acts on a specific rRNA region or if the enzyme stochastically targets rRNA regions that are in the vicinity of HP92 during ribosome assembly. Interestingly, a DbpA knockout mutant shows no ribosome maturation defect or discernible phenotype (31, 32). In contrast, overexpression of a DbpA-R331A mutant leads to a slow-growth phenotype and interferes with ribosome maturation, as evidenced by accumulation of a 45S assembly intermediate of the 50S subunit (18, 26, 27, 32, 33). The highly conserved R331 is located in the RecA_C domain, and its guanidinium group interacts with the ATP γ-phosphate in the closed state. This residue has thus been proposed to act as an arginine finger during ATP hydrolysis, and in line with this, the R331A mutant shows strongly reduced ATP turnover (33). Consequently, the R331A mutation also abolishes the helicase activity of DbpA. Nonetheless, neither the lack of helicase activity nor the reduced ATP turnover explain the effects on ribosome maturation, as no phenotype has been observed for other mutations in DbpA that reduce helicase activity and ATP turnover (33). The molecular basis for the ribosome maturation defect upon overexpression of the R331A mutant thus remains unclear.
Here, we show that a tight interaction between the RRM and the RecA_C domains places HP92 in a position that facilitates the formation of the catalytically important, closed conformation of DbpA. Our results also point toward a region upstream of helix 90 of the 23S rRNA as the physiological target site of DbpA during ribosome assembly and indicate that the phenotype of the DbpA-R331A mutant is caused by delayed dissociation of this mutant from the nascent ribosome. Taken together, these findings enhance our understanding of the mechanism and regulation of DbpA in particular and DEAD-box helicases in general.
Results
HP92 and a 5′ ssRNA Region Are Necessary for the Activation of the Helicase Activity of DbpA.
For the DbpA homolog YxiN, it has been shown that the addition of an RNA composed of HP92 and a 5′ ssRNA overhang in trans is sufficient to stimulate the helicase toward a duplex lacking HP92 (21). This points toward an allosteric activation mechanism, where binding of HP92 to the RRM stimulates the activity. To test whether this mechanism is conserved in E. coli DbpA, we performed unwinding assays using a fluorescently labeled 9mer RNA (N9) that is hybridized to an unstructured 32mer RNA (N32, Fig. 1C). In the absence of HP92 or in the presence of a minimal HP92 construct, the dsRNA is not unwound, whereas addition of HP92 constructs with 5′ overhangs of 2 or 4 nt induces weak unwinding activity. Robust unwinding of the dsRNA substrate takes place in the presence of HP92 RNAs that contain a 6- to 12-nt 5′ ssRNA overhang. These results demonstrate that the HP92-induced allosteric activation mechanism is conserved between E. coli DbpA and the B. subtilis homolog YxiN.
Binding of HP92 to the RRM Stabilizes the Closed Conformation of the Helicase Core.
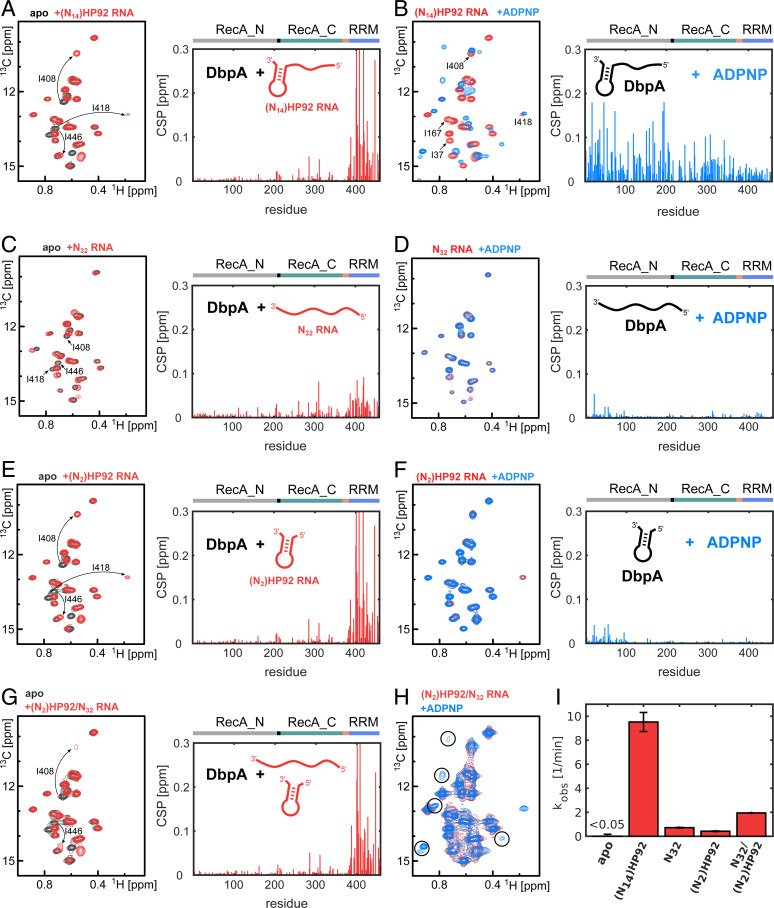
The closed conformation of the helicase core domains has been shown to be the central step in the dsRNA-unwinding cycle. To evaluate how binding of HP92 to the RRM influences the formation of the closed state, we performed nuclear magnetic resonance (NMR) titration experiments with full-length DbpA and saturating amounts of RNA (SI Appendix, Fig. S1) that either include or lack HP92, as well as with the nonhydrolyzable ATP analog ADPNP (Fig. 2 and SI Appendix, Fig. S2). To this end, we recorded methyl transverse relaxation optimized spectroscopy (TROSY) spectra of deuterated DbpA that is 13CH3 labeled in the methyl groups of isoleucine, leucine, methionine, valine, and alanine residues (ILMVA labeling), which yields high-quality NMR spectra. This allowed us to directly observe interactions of DbpA with RNA and ADPNP and to detect the associated structural changes.
Fig. 2.
HP92 is required for the formation of the catalytically important closed conformation of DbpA. (A–F) Titrations of NMR-active (ILMVA-labeled) full-length DbpA with different NMR-inactive RNA constructs and the ATP analog ADPNP: (Left) Isoleucine region of the methyl TROSY spectra of free DbpA (black), after addition of different RNA constructs (red), or after addition of different RNA constructs and ADPNP (blue). (Right) CSPs of the ILMVA methyl groups caused by binding of RNA to DbpA (A, C, and E) or by binding of ADPNP to the DbpA/RNA complex (B, D, and F) are plotted against the residue number. DbpA domains are indicated on top. (A) An RNA that contains a single-stranded region of 14 nt plus HP92 [(N14)HP92 RNA] binds mainly to the RRM. The signals of three Ile Cδ1 methyl groups of the RRM are labeled. (B) Addition of ADPNP to the DbpA/ (N14)HP92 RNA complex induces strong CSPs in both core domains and leads to the formation of the closed helicase conformation. Assignments of several Ile Cδ1 signals are indicated in the spectrum. (C) An unstructured RNA of 32 nt (N32) that lacks HP92 shows weak binding to the RRM. (D) For the unstructured N32 RNA, the helicase does not form the closed conformation upon addition of ADPNP, and only small CSPs in the vicinity of the ATP binding site are observed. (E) The (N2)HP92 RNA that contains HP92 but that lacks the 5′ ss overhang binds to the RRM similar to the (N14)HP92 RNA (compare to A). (F) Addition of ADPNP to the DbpA/(N2)HP92 RNA complex does not result in the formation of the closed helicase conformation and CSPs are only observed in the vicinity of the ATP binding site. (G) Simultaneous addition of the (N2)HP92 and the N32 RNAs leads to similar CSPs in the RRM as are observed for the interaction with the (N2)HP92 RNA (compare to E), although several of the shifted signals are broadened. (H) Addition of ADPPNP to this DbpA/RNA complex causes the partial (∼10 to 15%) formation of the closed conformation. This implies that the helicase can adopt the closed conformation on a ssRNA when HP92 is bound in trans. The characteristic Ile Cδ1 signals of the closed state are indicated by circles (compare to B). (I) Stimulation of the ATPase activity of DbpA by the RNAs that were used in the NMR experiments (A–H). ATP turnover rates were determined using a coupled pyruvate kinase–lactate dehydrogenase assay. The ATPase activity in the absence of RNA is below the detection limit of the assay. The ATPase activity of DbpA thus correlates with the degree to which the closed conformation is adopted.
Binding of an RNA construct comprising HP92 and a 5′ ssRNA overhang of 14 nt ((N14)HP92) to DbpA elicits large chemical shift perturbations (CSPs) on the RRM but only weakly influences either of the core domains (Fig. 2A, black and red spectra). This is in good agreement with previous results for YxiN, which show that HP92 RNA is recruited to the helicase via the RRM (15, 17). Subsequent addition of an ATP analog results in the formation of the ternary DbpA/RNA/ADPNP complex and in a sizable conformational change, as evidenced by large CSPs in both core domains (Fig. 2B; red and blue spectra). From this, we conclude that simultaneous binding of an RNA that includes HP92 and an ATP analog leads to the formation of the closed state of the helicase. The formation of the closed state is confirmed by nuclear Overhauser effect (NOE) measurements [see the model of the complex that we determine below (Fig. 4)] and in agreement with previous results for YxiN (4, 7, 21). Interestingly, the conformational transition from the HP92-bound open state to the HP92-bound closed state affects the RRM markedly less than the core domains (Fig. 2B), indicating that the RRM does not undergo large rearrangements upon the transition from the open to the closed state. Importantly, binding of ADPNP in the absence of RNA does not result in the formation of the closed state, and only small CSPs in the vicinity of the known ATP binding site in the RecA_N domain are observed (SI Appendix, Fig. S3).
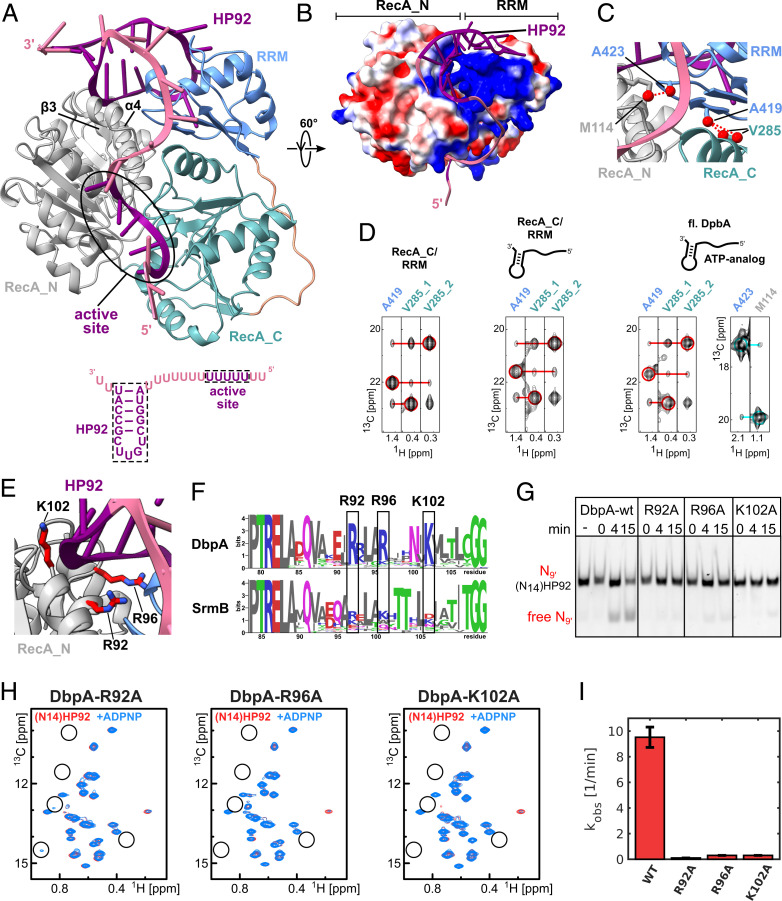
Fig. 4.
A model of the closed state of DbpA reveals stabilizing interactions between the RecA_N domain and HP92 that enable helicase activity. (A) The model of the closed state based on the solution structure of the RecA_C/RRM construct shows that HP92 and the RRM contact the RecA_N domain in the closed state. HP92 and 5 nt of the 5′ ss overhang bound to the ssRNA binding site of the helicase core were modeled based on previously published structures (3, 17). These regions are shown in purple, and the rest of the RNA is shown in pale violet. RRM, RecA_N and RecA_C domains are shown in blue, gray, and green, respectively. β-3 and α-4 of the RecA_N domain that interact with HP92 and the RRM are indicated. The RNA construct that is bound to the helicase in the model is shown on the bottom. (B) Surface representation the modeled DbpA colored according to the electrostatic surface potential (positive, blue; negative, red). The structure is rotated by 60° compared to A. The stem of HP92 packs into a positively charged groove formed between RRM and RecA_N domains. (C) Closeup of the interdomain interface of the closed state. Methyl groups that are predicted to show strong interdomain NOEs (dashed, red lines) are shown in red. The short distance between V285 and A419 (Right) is used to validate the orientation between the RecA_C and RRM domains. The short distance between A423 and M114 reports on the correct orientation between the RecA_N and RRM domains. (D) CCH-NOESY strips along the 13C-NOE dimension that show the interdomain NOEs for the RecA_C/RRM construct in the free form (Left), bound to (N14)HP92 RNA (Middle), or for full length DbpA in the closed state (Right). Interdomain NOEs between the RecA_C (V285) and RRM domains (A419) are indicated by red lines. The interdomain NOE between the RecA_N and RRM domains in the closed state (between A423 and M114) is indicated by a cyan line. Diagonal peaks are indicated by circles. (E) Closeup of the interaction between the RecA_N domain and HP92 in the closed state. The conserved Arg/Lys residues (R92/R96/K102) are shown in stick representation (red). (F) Sequence logos for DbpA (Top) and the unrelated DEAD-box helicase SrmB (Bottom). Residues R92, R96, and K102 are highly conserved across DbpA homologs, whereas the equivalent positions in SrmB are not conserved. The PTRELAXQ (Left) and GG motifs (Right) belong to the characteristic sequence motifs of DEAD-box helicases. (G) Mutation of the conserved Arg/Lys residues abolishes the helicase activity of DbpA. Unwinding of a fluorescently labeled 9mer RNA hybridized to (N14)HP92 RNA is followed by native PAGE after 0-, 4-, and 15-min incubation time. DbpA-wt shows robust unwinding after 15-min incubation time; no unwinding is observed after 15 min for the wt in the absence of ATP (−). Neither of the DbpA-R92A, -R96A, and -K102A mutants shows significant unwinding after 15 min. (H) Ile region of the methyl TROSY spectra of the DbpA-R92A, -R96A, and -K102A mutants bound to (N14)HP92 RNA (red) and after addition of ADPNP (blue). The positions of some of the characteristic signals of the closed state are indicated by circles. The mutations impair the formation of the closed conformation (R96A and K102A) or strongly reduce the population of the closed conformation (R92A) (compare to Fig. 2B). (I) Comparison of the ATPase activity of DbpA-wt with the -R92A, -R96A, and -K102A mutants. All measurements were performed in the presence of (N14)HP92 RNA. The mutations strongly reduce the ATPase activity of DbpA.
To assess whether HP92 is needed for the formation of the closed state, we repeated the NMR titration experiments with an unstructured 32mer RNA lacking HP92 (N32). The N32 RNA interacts only weakly with the RRM, as the CSPs are markedly reduced compared to the (N14)HP92 RNA (Fig. 2C, black and red spectra, compare to Fig. 2A). Addition of ADPNP to the N32/DbpA complex does not result in the formation of the closed conformation, and only very small CSPs are observed (Fig. 2D, red and blue spectra). These CSPs are similar to those elicited by ADPNP binding in the absence of RNA (SI Appendix, Fig. S3). This is in striking contrast to the titration with the (N14)HP92 RNA (Fig. 2B) and shows that HP92 is essential for the formation of the closed state of the helicase.
We next tested whether a construct comprising only HP92 and a 5′ ss overhang of 2 nt ((N2)HP92) is sufficient for the formation of the closed state (Fig. 2E, compare to Fig. 2A). We observe that the (N2)HP92 RNA binds to the RRM in the same manner as the (N14)HP92 RNA does. However, upon addition of ADPNP, the closed state is not formed. Taken together, these results demonstrate that binding of HP92 to the RRM and ssRNA and ATP to the helicase core are both required for the formation of the closed state of the helicase. This is in contrast to most other DEAD-box helicases (e.g., Prp5, Vasa, Dbp5, Mss116, and DDX19), where simultaneous binding of ssRNA and ATP are sufficient for the closure of the core domains (3, 34–37).
Here, we showed that neither the minimal (N2)HP92 RNA hairpin nor the linear N32 RNA are able to induce the closed conformation of the helicase (Fig. 3 D and F). Our initial helicase assays, however, show that addition of HP92 in trans was sufficient for the activation of the helicase activity (Fig. 1C). This prompted us to test whether the simultaneous addition of the minimal hairpin and the linear RNA can induce the formation of the closed helicase state. The methyl TROSY spectrum of DbpA in the presence of both RNAs strongly resembles the spectrum in the presence of (N2)HP92, in agreement with the specific binding of DbpA to HP92 (Fig. 2G). Subsequent addition of ADPNP leads to the formation of a small population (5 to 10%) of the closed conformation (Fig. 2H). Simultaneous binding of the (N2)HP92 and N32 RNAs thus enables the formation of the closed conformation, albeit to a lesser degree than the (N14)HP92 RNA (Fig. 2B), where H92 and a ssRNA region are combined in one construct.
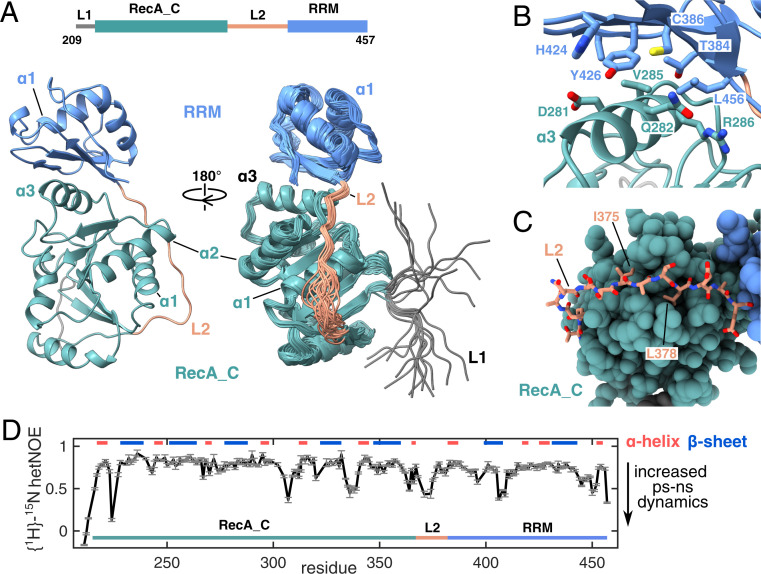
Fig. 3.
Solution structure of the DbpA RecA_C/RRM domains. (A) The construct used for the NMR structure determination is shown on top and includes the RecA_C domain and the RRM of DbpA. A cartoon representation of the structure with the lowest target function is shown on the left. The bundle of 20 NMR structures is shown on the right, rotated by 90°. The structure reveals that the RRM (light blue) tightly interacts with α-3 of the RecA_C domain (green) via its beta sheet surface. Note that the interdomain orientation is well defined. L1 (gray) and the N-terminal half of L2 (light brown) are less well defined due to increased local dynamics (see below). (B) Closeup of the interface between the RecA_C and RRM domains. Central residues are labeled and shown in stick representation. (C) Closeup of the interaction between the carboxyl-terminal part of L2 and the RecA_C domain. I375 and I378 of L2 that form hydrophobic interactions with the RecA_C domain are labeled. (D) {1H}-15N hetNOE values of the RecA_C/RRM construct that report on fast internal protein motions. Decreased values in several loop regions including the N-terminal half of L1 reveal increased dynamics on the ps to ns timescale. Secondary structure elements are indicated on top.
The extent of the closed state formation in the NMR experiments is also in good agreement with the observed stimulation of the ATPase activity of DbpA by the respective RNAs (Fig. 2I and SI Appendix, Fig. S4). In line with previous results (19), the (N14)HP92 RNA increases the ATP turnover of DbpA from <0.05 to 9.5 min−1. In contrast, (N2)HP92 (0.4 min−1) and N32 (0.7 min−1) RNAs show a more-modest ATPase stimulation that is increased to 1.9 min−1 upon addition of both RNAs. In combination with the NMR experiments, these results show that binding of HP92 to the RRM stimulates the helicase activity by stabilizing the closed state of the helicase core, where the composite ATP and ssRNA binding sites are formed.
The RRM Tightly Interacts with the RecA_C Domain.
In the preceding section we showed that HP92 is required for the stabilization of the closed state of DbpA. To reveal the molecular basis of this long-range mechanism, we solved the NMR solution structure of a DbpA construct comprising the RecA_C and RRM domains (residues 209 to 457). The RecA_N domain tumbles independently of the RecA_C/RRM domains (22) and is not influenced by the interaction between HP92 and the DbpA RRM (Fig. 2A). The minimal RecA_C/RRM construct is thus sufficient to recapitulate the interaction of full-length DbpA with HP92.
Due to the size of the RecA_C/RRM domain construct (27 kDa), we used an NMR labeling scheme that combines 2H,15N labeling with 13CH3 labeling of the methyl groups of Ile, Leu, Met, Val, Ala, and Thr and protonation of the Tyr residues (ILMVATY labeling). Based on 3,305 NOE distance restraints and 414 backbone dihedral angle restraints (SI Appendix, Table S1), we obtained a well-defined structure bundle (Fig. 3A) (average RMSD of 0.6 ± 0.1 and 1.1 ± 0.1 Å for the backbone and heavy atoms, respectively, excluding the L1 linker residues). The structures of the RecA_C and RRM domains are very similar to the crystal structures of the isolated RecA_C (38) and RRM (17) domains of YxiN (C-α RMSD 2.5/2.1 Å, SI Appendix, Fig. S5). The important insight that is revealed by our structure is the tight interaction between the two domains: The β-sheet surface of the RRM packs onto helix α-3 of the RecA_C domain. The interactions between the two domains are mainly hydrophobic and formed between the sidechains of Y426, T384, C386, and L456 from the RRM and the sidechains of D281, Q282, V285, and R286 from the RecA_C domain (Fig. 3B). Most of the interaction surface on the side of the RRM is formed by Y426. The selective protonation of the Tyr residues in the NMR sample that was used for the structure determination allowed us to obtain 88 interdomain NOEs in this region, which leads to a well-defined interdomain orientation in the structure bundle. The important structural role of Y426 at the interdomain interface is further supported by our observation that no soluble protein could be obtained for the Y426A or Y426D mutants of DbpA and by the fact that Y426 is highly conserved between DbpA homologs as either a Phe or Tyr.
Based on our structure, the linker L2 (residues 369 to 381) that connects both domains can be divided into two parts. The N-terminal half of the L2 linker (residues 369 to 374) loops over helix α-1 of the RecA_C domain. This region of L2 does not show NOE contacts to the RecA_C domain and is consequently not well defined. By contrast, the carboxyl-terminal half of L2 (residues 375 to 381) interacts tightly with the carboxyl-terminal end of helix α-1 and with a hydrophobic groove that is formed between the loops following helices α-1/α-2 (Fig. 3C); the sidechains of I375 and L378 are deeply inserted into this groove. In addition, we observe hydrogen bonds between the backbone carbonyls of A376 and L378 of L1 and Q266 and S267 of the RecA_C domain in several structures of the NMR bundle. To determine whether the structural heterogeneity observed for the N-terminal half of L2 is caused by inherent flexibility in this region or by insufficient experimental distance restraints, we performed {1H}-15N heteronuclear NOE (hetNOE) experiments (Fig. 3D). Reduced values in these experiments indicate increasing structural fluctuations on the ps-ns timescale, whereas for nonflexible residues, values above ∼0.8 are expected. In our experiments, we observe increased dynamics for residues at the beginning of L2 as well as for three regions in DbpA around residues D306, R334, and T405. The first two regions harbor conserved motifs of the DEAD-box family, and these residues directly interact with the ATP molecule in the closed state, whereas T405 forms part of the HP92 binding site of the RRM. These regions are most likely stabilized upon binding to their respective interaction partners during domain closure.
In summary, the solution structure of the carboxyl-terminal domains of DbpA shows a tight interaction between the beta sheet surface of the RRM and helix α-3 of the RecA_C domain. The observed interdomain orientation leaves the RNA binding site of the RRM exposed. The RNA binding site is mainly formed by helix α-1 (17) and is located opposite to the surface that interacts with the RecA_C domain. The RRM thus anchors HP92 in a well-defined orientation relative to the RecA_C domain.
The RRM and HP92 Directly Interact with the RecA_N Domain in the Closed State.
To understand how the bound HP92 stabilizes the closed conformation of the helicase core, we modeled the interaction between DbpA and the (N14)HP92 RNA in the closed state (Fig. 4A). The model is based on our RecA_C/RRM structure (Fig. 3), the HP92 binding mode observed in the YxiN RRM/rRNA complex (17), and the known domain orientation of the homologous helicase core of the VASA enzyme in the closed conformation (3). As the formation of the closed state requires binding of ssRNA to the helicase core (2) (see also Fig. 2), the ssRNA 5′ overhang (Fig. 4A) was modeled in the composite ssRNA binding site formed by the core domains.
The model of the closed conformation shows a surprisingly compact domain arrangement and places the RRM and HP92 in the direct vicinity of the RecA_N domain. The stem of HP92 and loop l3 (residues 421 to 423) of the RRM contact α-4/β-3 and the connecting loop of the RecA_N domain. This region of the RecA_N domain together with the RRM forms a positively charged groove that accommodates the stem of HP92 (Fig. 4B). A continuous stretch of positive surface charge connects the stem binding groove with the ssRNA binding site of the helicase core. This patch interacts with the nt 5′ of the HP92 stem, which are necessary for the activation of the helicase activity in trans (Fig. 1C).
Our model is predicated on the assumption that the interdomain orientation between the RecA_C domain and the RRM does not change upon HP92 binding or formation of the closed state. To validate this experimentally, we also performed NOE experiments for these two states. We observe virtually identical NOE patterns for the residues at the RecA_C/RRM interface for the three states (free DbpA, (N14)HP92 RNA bound, and closed) (SI Appendix, Figs. S6 and S7). Exemplary NOE strips for the methyl groups of A419 from the RRM and V285 from the RecA_C domain are shown in Fig. 4D. Very similar interdomain NOEs between these methyl groups are observed in all states. These results demonstrate that the interdomain orientation between RecA_C and RRM is not influenced by binding of HP92 to the RRM or by formation of the closed state. We also validated the domain orientation between the RecA_N domain and the RRM in the model. The model predicts a short distance (3.5 Å) between the methyl groups of M114 (RecA_N) and A423 (RRM) (Fig. 4C). In agreement with this, an NOE cross-peak between the two signals is observed for the closed state of full-length DbpA (Fig. 4D). Due to the large structural rearrangements, the resonances of M114 and A423 in the closed conformation were assigned by mutation (SI Appendix, Fig. S8). We also validated the position of the ssRNA overhang by paramagnetic relaxation enhancement experiments with a 5′ spin-labeled RNA (SI Appendix, Fig. S9) and ensured that the stem loop of the (N14)HP92 RNA is still formed based on RNA imino proton spectra (SI Appendix, Fig. S10).
Taken together, these results confirm that our model of full-length DbpA in complex with RNA accurately depicts the domain arrangement and RNA interaction of the closed state. Importantly, the observed domain arrangement reveals a direct interaction between the stem of HP92 and the positively charged patch on the RecA_N domain. This interaction readily rationalizes the stabilization of the active, closed state by HP92 (Fig. 2) and the concomitant activation of the helicase activity (Fig. 1). In addition, in our model, 3′ end of HP92 is further away from the ssRNA binding groove compared to the 5′ end of HP92. This finding explains the observation that a longer linker between the duplex and HP92 is necessary for unwinding when the duplex is located 3′ to HP92 instead of 5′ (16).
A Positively Charged Patch on the RecA_N Domain Is Essential for Helicase Activity.
The positively charged patch in the RecA_N domain that contacts the stem of HP92 is mainly formed by three Arg/Lys residues (R92/R96 and K102, Fig. 4E). These residues are highly conserved between DbpA homologs but not between homologs of the unrelated E. coli DEAD-box helicase SrmB (Fig. 4F). To experimentally verify the importance of this positively charged patch for the helicase activity we performed unwinding assays for wild-type (wt) DbpA and single alanine mutants of these three residues (Fig. 4G). The single mutation of each of these Arg or Lys residues to Ala is sufficient to abolish the helicase activity of DbpA. The mutations also impair (R96A and K102A) or strongly reduce (R92A) the formation of the closed the conformation in the presence of (N14)HP92 RNA and ADPNP (Fig. 4H) and strongly decrease the (N14)HP92 RNA–stimulated ATPase activity of the mutants (Fig. 4I). In summary, these results demonstrate that a positively charged patch that is conserved in the RecA_N domains of the DbpA family of DEAD-box helicases is necessary for the helicase activity, as this stabilizes the active conformation of the enzyme.
Interaction of DbpA with the 23S rRNA.
Previously, the structure of the RRM of the DbpA homolog YxiN in complex with a fragment of the 23S rRNA comprising helices H90, H91, and HP92 has been determined (17) (Fig. 5A and SI Appendix, Fig. S11). The DbpA RRM–bound rRNA fragment shows a similar conformation as in the mature ribosome, suggesting that the RNA structure is not influenced by DbpA binding (SI Appendix, Fig. S11). This notion is supported by RNA probing experiments with a longer rRNA fragment that suggest that the rRNA conformations of H89-H93 are similar in the ribosome, in the DbpA bound state, and after formation of the closed state of DbpA (39). We are therefore able to combine the rRNA-bound RRM structure with the fixed orientation between the RRM and RecA_C domain that we find here (Fig. 3) to visualize how DbpA interacts with the native 23S rRNA substrate in the maturing ribosome (Fig. 5A).
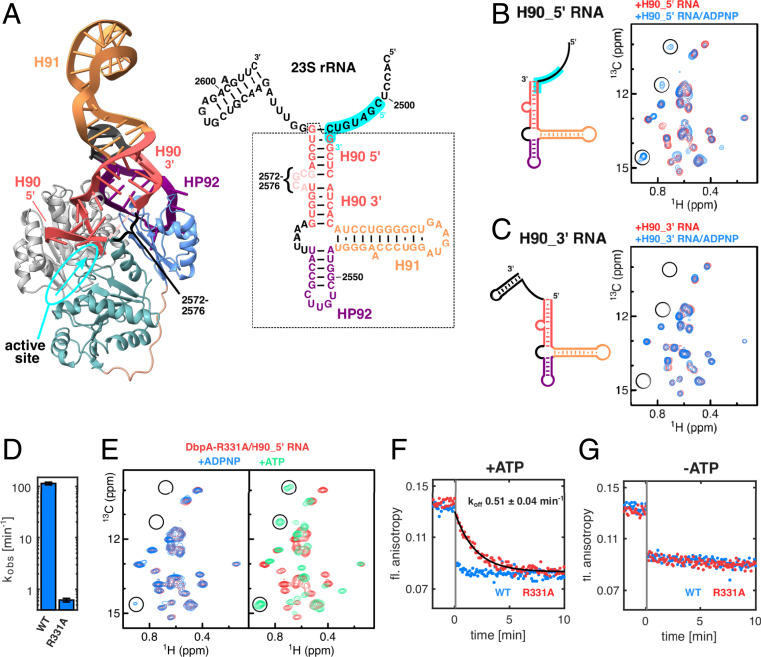
Fig. 5.
Interaction of DbpA with its target sequence in the 23S rRNA and effects of the DbpA-R331A. (A) Model of DbpA in the closed state bound to HP92 in the context of the 23S rRNA [nt 2,508 to 2,581, based on the YxiN RRM/23S rRNA structure (17)] (Left) and secondary structure of the 23S rRNA fragment (nt 2,496 to 2,604) (Right). The part of the rRNA that is observed in the structure is marked by a dashed box, and the secondary structure is depicted according to the crystal structure of the YxiN RRM/23S rRNA complex. The 5′ and 3′ parts of H90 (red) are indicated. The 5′ part of H90 points toward the ssRNA binding site of the helicase core. This places the region 5′ to H90 (nt 2,501 to 2,508, highlighted in cyan) in the correct orientation to interact with the active site (the ssRNA binding site and its 5′ to 3′ directionality are indicated in cyan). Note that nt 2,572 to 2,576 are not resolved in the crystal structure. (B) Secondary structure of the H90_5′ RNA lacking the ss region 3′ to H90 (Left) and Ile region of the methyl TROSY spectra of free DbpA (black), after addition of H90_5′ RNA (red) (Middle) or bound to H90_5′ RNA and ADPNP (blue) (Right). The RNA region that is proposed to interact with the helicase ssRNA binding site is highlighted in cyan. Characteristic signals of the closed state are marked by circles. (C) Same as B but for the H90_3′ RNA, which contains a ssRNA region 3′ to H90. No closed state is formed in the presence of H90_3′ RNA and ADPPNP, indicating that the ssRNA region 3′ to H90 is not a substrate region for DbpA. (D) ATP turnover of DbpA-wt and -R331A mutant in the presence of the H90_5′ RNA. The R331A mutation strongly reduces the ATPase activity of DbpA. (E) Ile region of the methyl TROSY spectra of DbpA-R331A bound to H90_5′ RNA (red) and after addition of ADPNP (Left, blue) or ATP (Right, green). ADPNP induces the formation of a very small population of the closed conformation (note the weak signal at the bottom left), whereas ATP leads to complete conversion to the closed conformation. Characteristic signals of the closed state are marked by circles. (F and G) Fluorescence anisotropy time traces of fluorescently labeled H90_5′ RNA in complex with DbpA in the presence (F) or absence (G) of ATP. At −2 min, the complex between labeled H90_5′ RNA and DbpA was formed (anisotropy ∼0.14). At t = 0, a 100-fold excess of unlabeled H90_5′ RNA was added. Dissociation of the labeled H90_5′ RNA leads to a decrease in fluorescence anisotropy. For DbpA-wt, dissociation is essentially complete after the dead time of the experiment (10 s, indicated by the vertical gray bar). The dissociation rate of the complex is decreased to 0.51 min−1 for the DbpA-R331A mutant.
We observe that the 5′ part of H90 comes in close vicinity to the active site of the helicase core. This finding thus suggests that DbpA preferentially interacts with the 5′ part of H90 and/or the ssRNA region 5′ to it. To experimentally confirm this, we performed NMR titration experiments with a 23S rRNA fragment that comprises H90, H91, HP92, and the ssRNA region 5′ to H90 (H90_5′, Fig. 5B). The CSPs that are induced by this long RNA fragment are highly similar to those that are caused by RNAs that contain HP92 (Fig. 2 A and E), indicating that the interaction between H90_5′ and DbpA is limited to the RRM and HP92. Addition of ADPNP to the H90_5′/DbpA complex leads to the partial formation of the closed state (Fig. 5B), in agreement with a situation where the region 5′ to H90 interacts with the ssRNA binding groove in the expected 5′ to 3′ orientation. Contrarily, an RNA construct that contains the region 3′ to H90 (H90_3′) but that lacks the ssRNA region 5′ to it (Fig. 5C) elicits similar CSPs as H90_5′ but fails to support the formation of the closed state in the presence of ADPNP. Taken together, this demonstrates that the RRM orients the bound 23S rRNA such that the active site of the helicase core is poised to interact with the ssRNA region preceding H90 (nt ∼2,501 to 2,508) (Fig. 5A). In agreement with this, RNA footprinting assays show increased protection for nt 2,502 and 2,505 in the presence of DbpA and ADPNP (15, 39).
The DbpA-R331A Mutation Increases the Lifetime of the DbpA/RNA Complex.
To gain insights into the mechanism by which overexpression of the DbpA-R331A mutant interferes with ribosome maturation, we tested the influence that this mutation has on the interaction between DbpA and the H90_5′ RNA. We find that this mutation does not interfere with H90_5′ RNA binding (SI Appendix, Fig. S12) but results in a 190-fold decrease of the ATPase activity of the DbpA/H90_5′ RNA complex (Fig. 5D). Both results are in good agreement with previous observations for this mutant in combination with a longer RNA construct (33). In addition, NMR titration experiments confirm that the R331A mutant interacts with the H90_5′ RNA in a similar manner as the wt enzyme does (SI Appendix, Fig. S12). However, in contrast to the wt, the mutant protein shows a markedly reduced population of the closed conformation in the presence of H90_5′ RNA and ADPNP (Fig. 5E). The R331A mutation thus destabilizes the closed conformation compared to the wt enzyme (compare to Fig. 5C). We next exploited the strongly reduced ATPase activity of the R331A mutant protein and recorded methyl TROSY spectra in the presence of the natural substrates ATP (as opposed to substrate analog ADPNP) and H90_5′ RNA (Fig. 5E). In that case, we observe a complete conversion into the closed conformation, which shows that ATP stabilizes the closed conformation to a greater extent than the analog ADPNP. More importantly, these results reveal that the R331A mutant enzyme can form a stable and long-lived closed conformation, where the ssRNA binding groove efficiently interacts with the ssRNA region 5′ to H90 in the nascent ribosome. To determine the lifetime of this R331A enzyme–RNA complex, we monitored the dissociation of a fluorescently labeled H90_5′ RNA using time-resolved fluorescence anisotropy measurements (Fig. 5 F and G and SI Appendix, Fig. S13). In this experiment, the dissociation of the RNA from DbpA can be observed by the decrease in fluorescence anisotropy, where rebinding of the fluorescently labeled RNA is prevented by the addition of a 100-fold excess of unlabeled H90_5′ RNA at t = 0. In the presence of ATP, the dissociation rate of the DbpA-R331A/H90_5′ RNA complex is 0.51 ± 0.04 min−1. This dissociation rate is very similar to the ATP turnover rate of the complex (0.61 ± 0.05 min−1). The dissociation of the DbpA-wt/H90_5′ RNA complex is, on the other hand, significantly faster and close to completion within the dead time of the experiment (10 s) (Fig. 5F). This fast dissociation correlates with the high ATPase activity of this complex (114 ± 8 min−1; Fig. 5B). Importantly, the enzyme–RNA dissociation rates in the absence of ATP (Fig. 5G) are high for the wt as well as for the R331A mutant enzymes. Taken together, these findings show that the dissociation rate of the substrate is determined by the nt state of the enzyme and that RNA dissociation only takes place from the open conformation of the enzyme.
Discussion
RNA helicases are ubiquitous enzymes that play an essential role in RNA metabolism. It is thus important to understand how these molecular machines function in detail and how their activity is regulated. To that end, we here elucidated the structural basis for the activation of DbpA by HP92 of the nascent ribosome (Fig. 1 A and B). The key mechanism by which HP92 activates DbpA lies in the direct and stable interaction between the RecA_C domain and the RRM (Fig. 3). This results in the placement of HP92 such that it can interact with a positive surface patch on the RecA_N domain of the helicase core (Fig. 4), which stabilizes the catalytically important closed state of DbpA. The stabilization of the closed state by the correct positioning of the substrate RNA constitutes an effective mechanism for the coupling of helicase activity and substrate recruitment, which prevents wasteful hydrolysis of ATP and ensures that the helicase activity is restricted to its target site.
Similar to DbpA, many DEAD-box RNA helicases contain RNA binding domains that flank the RecA core domains. It seems likely that other helicases also exploit interactions between these flanking regions and the core domains to stabilize catalytically important structural states. Identifying such interactions can be complicated by the fact that the linker regions that connect the ancillary with the core domains are not necessarily conserved in length and sequence, as we also observe here for the functionally important DbpA L2 linker. In the case of DbpA, the positioning of the RRM is achieved by interactions between the RecA_C domain, the carboxyl-terminal half of the L2 linker, and the RRM. Due to this multiplicity in the interactions between the separate regions, the structural importance of the L2 linker had escaped attention. Interestingly, an elongation of the L2 linker by 23 unstructured amino acids only slightly reduced the helicase activity of DbpA (24). This can be explained by our finding that the L2 linker is only partially folded (Fig. 3 A and D), and elongation of this unfolded region is expected to result only in minor distortions of the observed domain orientation. Previous FRET studies have addressed the interdomain orientation between the RRM and the RecA core domains for the DbpA homolog YxiN (21, 25). These low-resolution studies also position the RRM close to the RecA_C domain, although the interaction surfaces differ. In addition, our data shows no indications of the previously observed ∼5-nm positional shift between the RRM and the RecA_C domain upon RNA binding. These discrepancies likely result from the inherent uncertainty of FRET-based distance restraints due the large size and flexibility of the fluorescent labels.
Despite the known interaction between HP92 of the 23S rRNA and the RRM the precise role of DbpA during ribosome maturation remains unclear (18, 24). In that light, our results provide additional insights and identify the ssRNA region preceding H90 (nt ∼2,501 to 2,508) as the binding site for the helicase core. The two interaction sites of DbpA in the 23S rRNA (HP92 and nt 2,501 to 2,508) are part of the PTC, which forms during the later stages of ribosome assembly, and its formation leads to long-range interactions with other rRNA elements (29, 30). Interestingly, nt 2,501 to 2,508 are located deep in the core of the 50S subunit, whereas HP92 is located at the periphery. It is tempting to speculate that the function of DbpA is to block premature formation of the PTC, while protecting nt 2,501 to 2,508 from forming nonnative interactions. ATP hydrolysis would then periodically open short time windows, in which PTC formation could be initiated by folding of nt 2,501 to 2,508 into the 50S core. This ribosome-folding step would then prevent the reformation of the closed conformation of DbpA on the maturing ribosome and consequently facilitate DbpA dissociation followed by the complete folding of the PTC. In addition to a potential role of DbpA in unwinding nonnative rRNA structures, this scenario would also classify DbpA as an RNA chaperone.
Finally, our results also suggest a mechanism for the observed growth phenotype and accumulation of a 45S ribosomal precursor upon overexpression of the DbpA-R331A mutant (18). The lack of helicase activity of the R331A mutant does not explain the observed phenotype, as other mutations that prevent helicase activity do not induce any phenotype (33). Here, we show that DbpA-R331A is still able to form the catalytically important closed conformation (Fig. 5E) and that the mutant enzyme interacts with the RNA substrate in the same manner as the wt protein does (SI Appendix, Fig. S12). The R331A mutation, however, results in a dramatically decreased dissociation rate of the DbpA-R331A/RNA complex, and the lifetime of the DbpA-R331A/H90_5′ RNA complex (1.9 min) is comparable to the total duration of ribosome assembly in vivo (∼2 min) (40). The presence of DbpA in the final steps of ribosome biogenesis is not compatible with the formation of the PTC. The persistent presence of the R331A mutant DbpA is thus expected to lead to the accumulation of preribosomal particles lacking a fully formed PTC. In line with this, the 45S precursor particles that accumulate in the DbpA R331A background lack the ribosomal proteins L16 and L36 (18, 26) that are recruited to the ribosome concomitantly with PTC formation (29). Interestingly, the 45S precursor is converted to the mature 50S subunit much more slowly than regular ribosome intermediates (27), which suggests that the delayed dissociation of DbpA causes structural or compositional abnormalities in the maturing 50S subunit that prevent regular maturation even after dissociation of DbpA and highlights the importance of the precise timing of all steps during ribosome maturation.
Intriguingly, the S. cerevisiae DEAD-box helicase Dbp10, which is also involved in ribosome maturation, interacts with the same regions in the eukaryotic 28S rRNA (HP92 and the ssRNA region preceding H90) (41). This suggests that DbpA and Dbp10 might be functional homologs. In contrast to DbpA, the ∼400 amino acids–long carboxyl-terminal extension of Dbp10 is predicted to be disordered (SI Appendix, Fig. S14). Recruitment of Dbp10 to nascent ribosomes is thus most likely achieved by a different and currently unknown mechanism.
In summary, our results provide unique insights into the activation of DbpA by the nascent ribosome and identify the target site of the helicase core during ribosome assembly. Thereby, our work paves the way for future research into the role of DbpA during ribosome assembly and into the complex process that underlies ribosome biogenesis.
Materials and Methods
Full details relating to protein expression and purification, structure determination of the RecA_C/RRM construct, additional NMR measurements, ATPase assays, RNA production and purification, modeling of the closed state of DbpA, helicase assays, RNA binding measurements by fluorescence anisotropy, and sequence alignments are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Olga Rudi for help with protein preparation. We thank Johanna Stöfl for technical assistance, and Katherine Bohnsack and Markus Bohnsack are acknowledged for helpful advice regarding the ATPase assay. We would also like to thank Sébastien Ferreira-Cerca for stimulating discussions and David Stelzig, Jan Overbeck, and Christina Krempl for careful reading of the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft through Grant No. WU 988/1-1 (to J.P.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105961118/-/DCSupplemental.
Data Availability
The atomic coordinates for the DbpA RecA_C/RRM construct data have been deposited in the Protein Data Bank under accession code 7BBB. All other study data are included in the article and/or SI Appendix.
References
- 1.Linder P., Jankowsky E., From unwinding to clamping - The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Putnam A. A., Jankowsky E., DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim. Biophys. Acta 1829, 884–893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S., Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125, 287–300 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Theissen B., Karow A. R., Köhler J., Gubaev A., Klostermeier D., Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 105, 548–553 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henn A., Bradley M. J., De La Cruz E. M., ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu. Rev. Biophys. 41, 247–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., et al., DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc. Natl. Acad. Sci. U.S.A. 105, 20203–20208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aregger R., Klostermeier D., The DEAD box helicase YxiN maintains a closed conformation during ATP hydrolysis. Biochemistry 48, 10679–10681 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Childs J. J., Gentry R. C., Moore A. F. T., Koculi E., The DbpA catalytic core unwinds double-helix substrates by directly loading on them. RNA 22, 408–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diges C. M., Uhlenbeck O. C., Escherichia coli DbpA is a 3′–> 5′ RNA helicase. Biochemistry 44, 7903–7911 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Henn A., et al., Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc. Natl. Acad. Sci. U.S.A. 107, 4046–4050 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henn A., Cao W., Hackney D. D., De La Cruz E. M., The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J. Mol. Biol. 377, 193–205 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaminker I., et al., Probing conformational variations at the ATPase site of the RNA helicase DbpA by high-field electron-nuclear double resonance spectroscopy. J. Am. Chem. Soc. 133, 15514–15523 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Polach K. J., Uhlenbeck O. C., Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry 41, 3693–3702 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Samatanga B., Klostermeier D., DEAD-box RNA helicase domains exhibit a continuum between complete functional independence and high thermodynamic coupling in nucleotide and RNA duplex recognition. Nucleic Acids Res. 42, 10644–10654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karginov F. V., Caruthers J. M., Hu Y., McKay D. B., Uhlenbeck O. C., YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J. Biol. Chem. 280, 35499–35505 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Diges C. M., Uhlenbeck O. C., Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 20, 5503–5512 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardin J. W., Hu Y. X., McKay D. B., Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J. Mol. Biol. 402, 412–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharpe Elles L. M., Sykes M. T., Williamson J. R., Uhlenbeck O. C., A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 37, 6503–6514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsu C. A., Kossen K., Uhlenbeck O. C., The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA 7, 702–709 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsu C. A., Uhlenbeck O. C., Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry 37, 16989–16996 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Samatanga B., Andreou A. Z., Klostermeier D., Allosteric regulation of helicase core activities of the DEAD-box helicase YxiN by RNA binding to its RNA recognition motif. Nucleic Acids Res. 45, 1994–2006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurm J. P., Assignment of the Ile, Leu, Val, Met and Ala methyl group resonances of the DEAD-box RNA helicase DbpA from E. coli. Biomol. NMR Assign. 15, 121–128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S., Overgaard M. T., Hu Y., McKay D. B., The Bacillus subtilis RNA helicase YxiN is distended in solution. Biophys. J. 94, L01–L03 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore A. F. T., Gentry R. C., Koculi E., DbpA is a region-specific RNA helicase. Biopolymers 107, 10.1002/bip.23001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karow A. R., Klostermeier D., A structural model for the DEAD box helicase YxiN in solution: Localization of the RNA binding domain. J. Mol. Biol. 402, 629–637 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Arai T., et al., Single methylation of 23S rRNA triggers late steps of 50S ribosomal subunit assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E4707–E4716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentry R. C., Childs J. J., Gevorkyan J., Gerasimova Y. V., Koculi E., Time course of large ribosomal subunit assembly in E. coli cells overexpressing a helicase inactive DbpA protein. RNA 22, 1055–1064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noeske J., et al., High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 22, 336–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis J. H., et al., Modular assembly of the bacterial large ribosomal subunit. Cell 167, 1610–1622.e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li N., et al., Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res. 41, 7073–7083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iost I., Dreyfus M., DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 34, 4189–4197 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peil L., Virumäe K., Remme J., Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 275, 3772–3782 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Elles L. M. S., Uhlenbeck O. C., Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucleic Acids Res. 36, 41–50 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beier D. H., et al., Dynamics of the DEAD-box ATPase Prp5 RecA-like domains provide a conformational switch during spliceosome assembly. Nucleic Acids Res. 47, 10842–10851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins R., et al., The DEXD/H-box RNA helicase DDX19 is regulated by an alpha-helical switch. J. Biol. Chem. 284, 10296–10300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Campo M., Lambowitz A. M., Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol. Cell 35, 598–609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montpetit B., et al., A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472, 238–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caruthers J. M., Hu Y., McKay D. B., Structure of the second domain of the Bacillus subtilis DEAD-box RNA helicase YxiN. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 1191–1195 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karginov F. V., Uhlenbeck O. C., Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 32, 3028–3032 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaczanowska M., Rydén-Aulin M., Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71, 477–494 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manikas R.-G., Thomson E., Thoms M., Hurt E., The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res. 44, 1800–1812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates for the DbpA RecA_C/RRM construct data have been deposited in the Protein Data Bank under accession code 7BBB. All other study data are included in the article and/or SI Appendix.