Significance
In plants, several DNA methylation pathways help maintain DNA methylation patterns to ensure that transposons remain in a silenced state and cell-specific DNA methylation is preserved after cell division. Here, we demonstrate that loss of function of the chromatin remodeler DDM1 induces a pathway conversion from CMT2- to RdDM-dependency to ensure the maintenance of CHH methylation. In plants defective in both DDM1 and RdDM, there is strong reactivation of TEs and a burst of TE transposition. Thus, our work not only identifies a variant of the RdDM pathway, but also shows that DNA methylation maintenance at a given locus is an adaptable process, revealing the existence of a double-insurance mechanism for the regulation of DNA methylation to guarantee genome integrity.
Keywords: epigenetic regulation, DNA methylation, transposon, genomic stability
Abstract
The CMT2 and RNA-directed DNA methylation (RdDM) pathways have been proposed to separately maintain CHH methylation in specific regions of the Arabidopsis thaliana genome. Here, we show that dysfunction of the chromatin remodeler DDM1 causes hundreds of genomic regions to switch from CMT2 dependency to RdDM dependency in DNA methylation. These converted loci are enriched at the edge regions of long transposable elements (TEs). Furthermore, we found that dysfunction in both DDM1 and RdDM causes strong reactivation of TEs and a burst of TE transposition in the first generation of mutant plants, indicating that the DDM1 and RdDM pathways together are critical to maintaining TE repression and protecting genomic stability. Our findings reveal the existence of a pathway conversion–based backup mechanism to guarantee the maintenance of DNA methylation and genome integrity.
DNA methylation, which refers to the addition of a methyl group to the cytosine bases of DNA to form 5-methylcytosine, is a conserved epigenetic mark important for gene regulation and the silencing of transposable elements (TEs) and other repeats (1–4). In plants, DNA methylation occurs in three sequence contexts: CG, CHG, and CHH, where H is any nucleotide except G. DNA methylation in all sequence contexts is established de novo by the RNA-directed DNA methylation (RdDM) pathway (1, 4–6). Once established, DNA methylation patterns are stably maintained following DNA replication by different mechanisms depending on the sequence context. CG methylation is maintained by MET1, the plant homolog of DNMT1, which recognizes hemimethylated CG dinucleotides following DNA replication and methylates the unmodified cytosine on the daughter strand (7, 8). Maintenance of CHG methylation is catalyzed by CMT3 and is strongly associated with dimethylation of lysine 9 on histone 3 (H3K9me2) (9, 10). The histone methyltransferases KYP, SUVH5, and SUVH6 bind to methylated CHG sites and catalyze H3K9me2 deposition (11, 12); in turn, CMT3 binds to H3K9me2, catalyzing the methylation of CHG sites. This interdependence forms a self-reinforcing loop to maintain repressive CHG methylation and H3K9me2 marks (13).
Two different pathways, RdDM and CMT2, maintain CHH methylation of specific loci, depending on the genomic context (14, 15). The RdDM pathway relies on two plant-specific multisubunit DNA-dependent RNA polymerases, Pol IV and Pol V. At RdDM target loci, Pol IV generates short P4-RNAs (26 to 50 nt) that are converted into double-stranded RNA (dsRNA) by RDR2 and subsequently processed into 24-nt siRNAs by DCL3 (16–18); other DICER-LIKE proteins (DCL1, DCL2, and DCL4) can process the P4-RNAs into 21- or 22-nt siRNA in the absence of DCL3 (16–18). Then, the 24-nt siRNAs are loaded into AGO4 or AGO6 and pair with complementary scaffold RNAs, nascent transcripts produced by Pol V. The resulting complex recruits the DNA methyltransferase DRM2 to catalyze CHH methylation (19). P4-RNAs can also mediate CHH methylation in the absence of the DCLs, a process that is referred to as the Dicer-independent RdDM pathway (17, 18). RdDM maintains CHH methylation at euchromatin regions (short TEs and other repeats in chromosome arms) and at the edge of long TEs, which are usually located in heterochromatin (15). By contrast, CMT2 maintains CHH methylation at heterochromatin regions and in the body of long TEs (15).
Maintenance of DNA methylation in heterochromatin in all sequence contexts also requires the nucleosome remodeling protein DDM1 (15, 20–23). DDM1 can act as a “wrench” to open H1-containing heterochromatin, allowing DNA methyltransferases to access the DNA (15, 24). Without DDM1, DNA methyltransferases cannot efficiently methylate the inaccessible heterochromatic regions, leading to hypomethylation in all sequence contexts. Recent studies found that DDM1 can deposit the histone variant H2A.W to prevent transposon mobility (25). Repeated self-pollination of loss-of-function ddm1 mutants results in ectopic non-CG methylation (26). Extensive hypomethylation in heterochromatic regions in ddm1 causes genome-wide TE transcriptional activation (27) and triggers the onset of RNA interference (RNAi) mechanisms to cleave the TE mRNA into a 21- to 22-nt siRNA (28, 29). Studies on ddm1 mutants have led to the discovery of two distinct branches of RdDM, RDR6-RdDM and DCL3-RdDM, implying that the diversity of RdDM mechanisms is masked by DDM1 (28, 30, 31). Although the molecular function of DDM1 is well described, its interaction with RdDM in regulating DNA methylation, and why its mutation causes TE transposition only after several generations of inbreeding (32–34), remain open questions.
Here, we analyzed the DNA methylomes, transcriptomes, and TE movement in Arabidopsis thaliana plants lacking DDM1 and RdDM. We discovered that CHH methylation in many genomic regions in the ddm1 mutant is maintained through pathway conversion from CMT2 to RdDM. Analyses of Pol IV occupancy and 24-nt siRNA levels confirmed the pathway conversion in these specific regions. Loci showing a change from CMT2- to RdDM-dependent CHH methylation are enriched at the edges of long TEs. RNA-sequencing (RNA-seq) results demonstrate that this pathway conversion is essential to preventing TE reactivation; further, blocking this switch leads to a burst of TE transposition in the first generation of the corresponding mutants, indicating an essential role of the pathway conversion in the protection of genomic stability. Our results reveal a phenomenon of DNA methylation pathway conversion that is critical for the maintenance of genomic stability.
Results
Mutation in DDM1 Induces a Conversion from CMT2-Methylated Loci to RdDM-Methylated Loci.
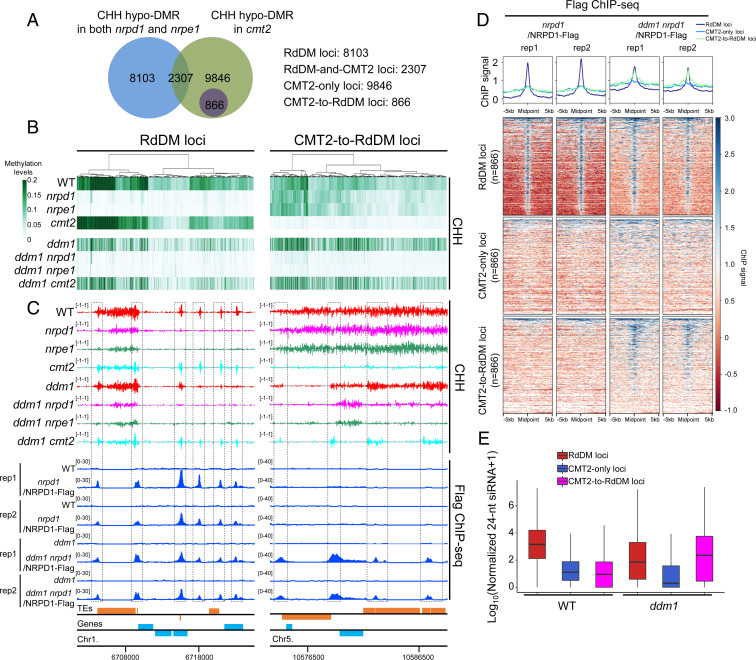
To investigate potential genetic interactions between the CMT2 and RdDM pathways, we generated single-base resolution maps of the DNA methylomes of the nrpd1, nrpe1, cmt2, ddm1, ddm1 nrpd1, ddm1 nrpe1, and ddm1 cmt2 mutants (SI Appendix, Table S1). Because the major function of both the canonical RdDM pathway and the CMT2 pathway is the maintenance of methylation in the CHH context, we compared the CHH methylation levels between each of these mutants and the wild type (WT). Based on these comparisons, we identified 10,410 differentially methylated regions (DMRs) that were CHH hypomethylated (CHH hypo-DMRs) in both nrpd1 and nrpe1 mutants compared with the WT, hence RdDM dependent (Fig. 1A). Similarly, 13,019 CHH hypo-DMRs were uncovered in the cmt2 mutant, therefore considered CMT2 dependent (Fig. 1A). Only limited overlap (2,307) was found between CHH hypo-DMRs from nrpd1 or nrpe1 and cmt2, suggesting that the RdDM and CMT2 pathways function mainly to maintain CHH methylation at separate loci in the WT background (Fig. 1A). Hereafter, the CHH hypo-DMRs found in nrpd1 and nrpe1 only and those found in cmt2 only are referred to as RdDM-methylated loci (RdDM loci) and CMT2-methylated loci (CMT2 loci), respectively (Fig. 1 A and B). Accordingly, the CHH hypo-DMRs found in the overlap between these mutants are referred to as RdDM and CMT2 co-methylated loci (RdDM-and-CMT2 loci) (Fig. 1A and SI Appendix, Fig. S1B). Surprisingly, we found that CHH methylation at 866 CMT2-methylated loci is lost in ddm1 nrpd1 and ddm1 nrpe1 double mutants, but is not changed in ddm1 cmt2 relative to ddm1 (Fig. 1 A and B). These results indicate that the ddm1 mutation causes CHH methylation at these 866 CMT2 loci to be dependent on the RdDM pathway. Therefore, we classified the CMT2 loci into two categories 1): CMT2-only loci, at which CHH methylation is maintained by CMT2 in both WT and ddm1 backgrounds; and 2) CMT2-to-RdDM loci, at which CHH methylation is methylated by CMT2 in the WT background but by RdDM in ddm1 (Fig. 1A). To confirm that the CMT2-to-RdDM loci and RdDM loci are bona fide RdDM targets, we determined the genome-wide profile of Pol IV occupancy in WT and ddm1 mutant backgrounds via chromatin immunoprecipitation followed by sequencing (ChIP-seq) using a previously characterized NRPD1-tagged line (35) and a line where the pNRPD1::NRPD1-3XFlag was introgressed into the ddm1 background (SI Appendix, Table S2). Consistent with our DNA methylation data, Pol IV was enriched at the defined RdDM loci in both the WT and ddm1 mutant backgrounds (Fig. 1 C and D and SI Appendix, Fig. S1C). As expected, enrichment of Pol IV occupancy was not found at CMT2-only loci in either the WT or ddm1 background (Fig. 1D). However, enrichment of Pol IV was detected at CMT2-to-RdDM loci in the ddm1 background but not in the WT (Fig. 1 C and D).
Fig. 1.
Mutation in DDM1 induces a conversion of CMT2-methylated loci to RdDM-methylated loci. (A) Overlap between CHH hypo-DMRs from mutants in the RdDM pathway and the cmt2 mutant. (B) Heatmap of CHH methylation levels in RdDM loci and CMT2-to-RdDM loci. Rows represent data for each indicated genotype; columns represent genomic loci. Columns were sorted by complete linkage hierarchical clustering with Euclidean distance as a distance measure. The 866 CMT2-to-RdDM loci were used for the heatmap on the Right; for easy comparison we randomly selected 866 loci from the 8,103 RdDM loci to make the heatmap on the Left. (C) Screenshots of CHH methylation levels and NRPD1 ChIP-seq signals over representative RdDM (Left) and CMT2-to-RdDM (Right) loci. “rep1” and “rep2” indicate different biological replicates. Black dotted boxes indicate RdDM loci or CMT2-to-RdDM loci. Genes and TEs oriented 5′ to 3′ and 3′ to 5′ are shown above and below the line, respectively. The numbers in parentheses indicate y axis scales. (D) Metaplot and heatmap of NRPD1 ChIP-seq signals in RdDM loci, CMT2-only loci, and CMT2-to-RdDM loci. As in B, we randomly selected 866 loci from the 8,103 RdDM loci and 9,846 CMT2-only loci to make the corresponding metaplots and heatmaps. (E) Levels of 24-nt siRNA at RdDM, CMT2-only, and CMT2-to-RdDM loci in WT and ddm1. The horizontal line within the box represents the median; the whiskers extend to 1.5 times the interquartile range; and the lower and upper boundaries of the box represent the first and third quartiles, respectively.
We downloaded available siRNA sequencing (siRNA-seq) data (29) and analyzed the 24-nt siRNA levels, a hallmark of the RdDM pathway. Consistent with Pol IV occupancy, the 24-nt siRNA levels from RdDM loci were substantially higher than those from CMT2-only loci and CMT2-to-RdDM loci in the WT background (Fig. 1E). However, in ddm1 mutant plants, the 24-nt siRNAs at CMT2-to-RdDM loci increased to levels comparable to those of the RdDM loci, whereas the 24-nt siRNAs at CMT2-only loci remained low (Fig. 1E). These results demonstrate that the loss of function of DDM1 induces a pathway conversion from CMT2 to RdDM in hundreds of genomic regions, suggesting that the pathway determining DNA methylation maintenance at certain genomic loci can switch depending on DDM1 function. We propose that the CMT2-to-RdDM loci represent a variant of the RdDM pathway that is induced by ddm1 loss of function.
Characterization of the CMT2-to-RdDM Loci.
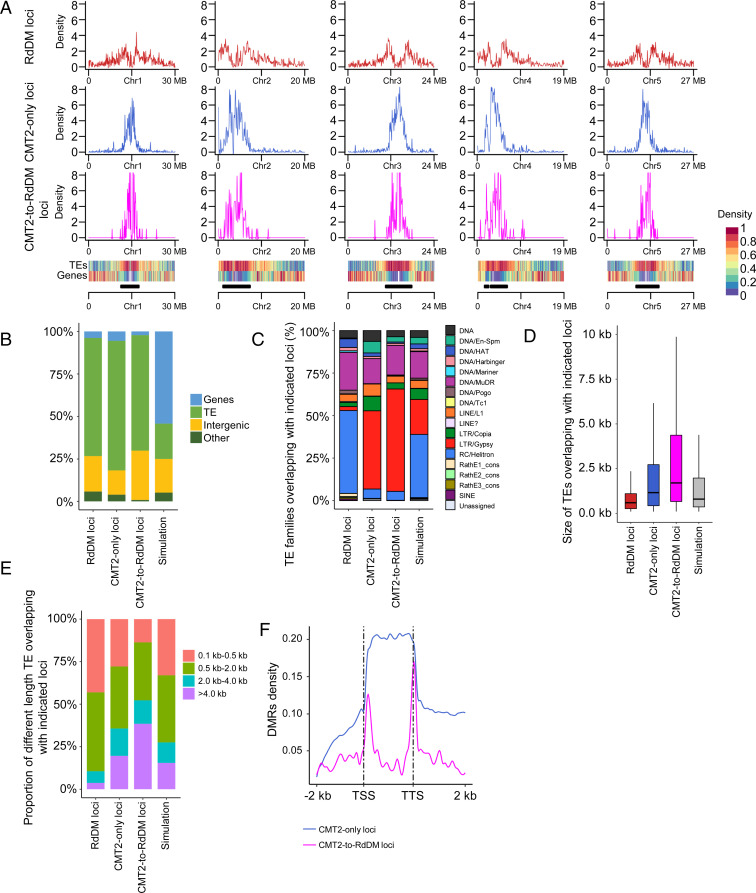
We characterized and compared the genetic features of RdDM, CMT2-only, and CMT2-to-RdDM loci. Both CMT2-only and CMT2-to-RdDM loci are highly enriched in pericentromeric regions, whereas RdDM loci are predominantly distributed along the chromosome arms (Fig. 2A). As expected, TEs are overrepresented in the RdDM, CMT2-only, and CMT2-to-RdDM loci (Fig. 2B). TEs overlapping with CMT2-only and CMT2-to-RdDM loci are enriched in retrotransposons, particularly LTR/Gypsy retrotransposons (Fig. 2C). Consistent with a previous observation that CHH methylation of Athila6A transcription start site is maintained by CMT2 and DRM2 in the WT and ddm1 backgrounds, respectively (31), we found that 40 CMT2-to-RdDM loci belong to Athila6A TEs (SI Appendix, Table S3). On average, the size of TEs overlapping with CMT2-only loci and CMT2-to-RdDM loci is significantly longer than that of TEs overlapping with RdDM loci, with the average size of TEs in CMT2-to-RdDM loci being the longest (Fig. 2D). TEs overlapping with RdDM loci were mostly short (<2 kb), whereas the TEs overlapping with CMT2-only loci and CMT2-to-RdDM loci were enriched in long TEs (>2 kb) (Fig. 2E). We compared CMT2-only loci and CMT2-to-RdDM loci with regard to their loci density along >4-kb TEs and found that the CMT2-to-RdDM loci are concentrated at the edges, whereas the CMT2-only loci are in the bodies of TEs, as expected (Fig. 2F).
Fig. 2.
Characterization of RdDM, CMT2-only, and CMT2-to-RdDM loci. (A) Distribution of RdDM loci, CMT2-only loci, and CMT2-to-RdDM loci across chromosomes. Heatmaps at the Bottom show the TE and gene densities, respectively. Black bars indicate pericentromeric regions. (B) Composition of the genomic location of the RdDM loci, CMT2-only loci, and CMT2-to-RdDM loci. TE, transposable element. Simulation regions served as control loci (the same in C–E). (C) Distribution of TE families overlapping with RdDM loci, CMT2-only loci, and CMT2-to-RdDM loci. (D) Boxplots of sizes of TEs overlapping with RdDM loci, CMT2-only loci, and CMT2-to-RdDM loci. The horizontal line within the box represents the median; the whiskers extend to 1.5 times the interquartile range; and the lower and upper boundaries of the box represent the first and third quartiles, respectively. (E) Composition of sizes of TEs overlapping with RdDM loci, CMT2-only loci, and CMT2-to-RdDM loci. (F) Distribution of CMT2-only loci and CMT2-to-RdDM loci over TEs of >4 kb.
The CMT2-to-RdDM Pathway Is Required for the Repression of TE Transcription and Transposition.
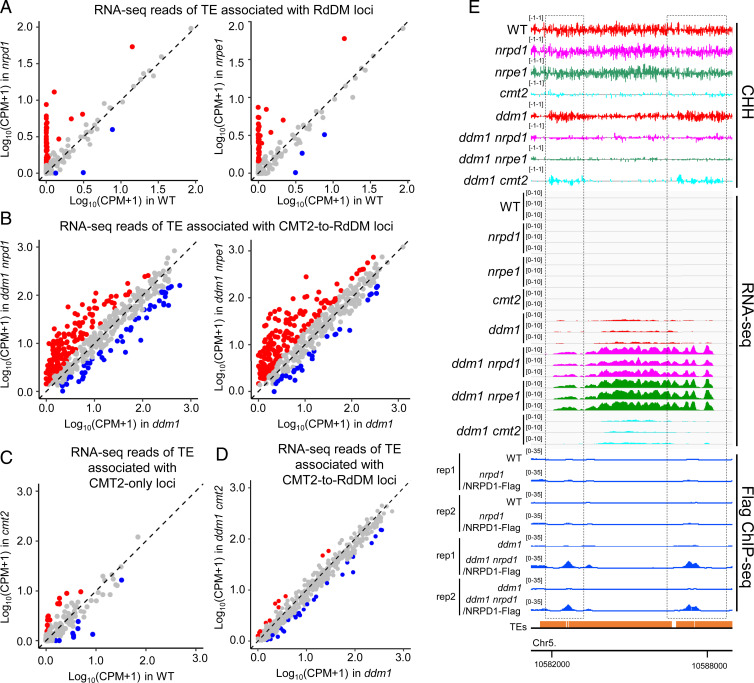
To shed light on the significance of the pathway conversion from CMT2 to RdDM in plants lacking DDM1, we performed RNA-seq in ddm1, ddm1 nrpd1, ddm1 nrpe1, and ddm1 cmt2 mutants (SI Appendix, Table S4 and Fig. S2) and also analyzed publicly available RNA-seq data of WT, nrpd1, nrpe1, and cmt2 (14, 36). The RNA-seq analysis revealed that a few TEs associated with RdDM loci are derepressed in nrpd1 or nrpe1 compared with WT (Fig. 3A). TEs associated with CMT2-only loci are rarely derepressed in cmt2 relative to WT (Fig. 3C). In sharp contrast, a large proportion of TEs associated with CMT2-to-RdDM loci are reactivated in ddm1 nrpd1 or ddm1 nrpe1 relative to ddm1 (Fig. 3 B and E). We compared the RNA-seq reads of TEs associated with CMT2-to-RdDM loci between ddm1 cmt2 and ddm1 and found that very few TEs were reactivated in ddm1 cmt2 relative to ddm1 (Fig. 3D).
Fig. 3.
The CMT2-to-RdDM pathway suppresses the expression of TEs in ddm1. (A–D) Scatterplot showing RNA-seq reads of the indicated TEs in the indicated genotypes. CPMs were averaged from three biological replicates. Each dot represents a single TE. Red dots represent TEs up-regulated in the genotype in the y axis compared with genotype in the x axis (fold change >2, FDR <0.05), whereas blue dots represent TEs down-regulated (fold change >2, FDR <0.05). TEs are considered as associated with the indicated loci when their body or flanking 2-kb regions overlap with the indicated loci. RNA-seq of cmt2 and WT are from ref. 14. (E) CHH methylation levels, TE expression levels, and NRPD1 ChIP-seq signals at a representative region from CMT2-to-RdDM repressed TEs. Black dotted boxes indicate the CMT2-to-RdDM loci. The numbers in parentheses indicate y axis scales.
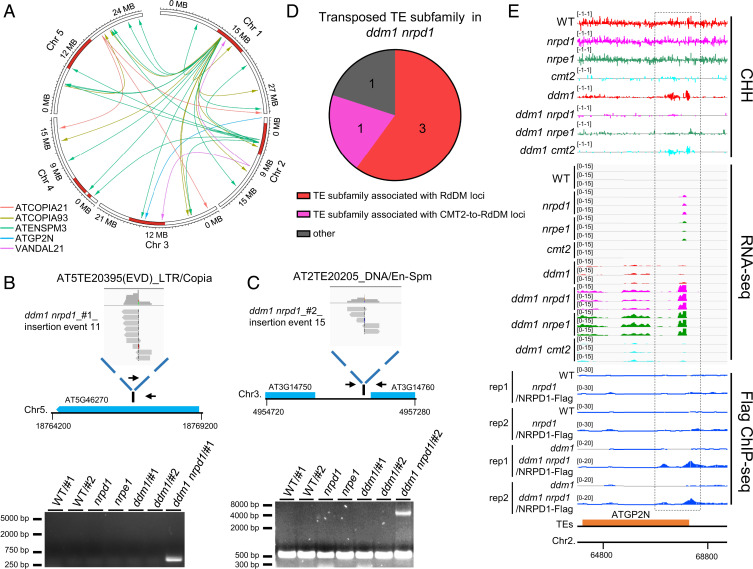
To examine whether reactivation of TEs in plants lacking DDM1 and deficient in RdDM may result in TE transposition, we generated whole-genome DNA sequencing (DNA-seq) data from WT, nrpd1, ddm1, nrpd1 ddm1, and ddm1 cmt2 plants (SI Appendix, Table S5). To avoid the inbreeding effect on TE transposition in ddm1, we only used seeds from the first generation of homozygous plants that were obtained from heterozygous parents. A schematic representation of the strategy used for the generation of materials for DNA-seq is shown in SI Appendix, Fig. S3. By analyzing the DNA-seq data, 11 and 22 putative TE transposition events were identified in ddm1 nrpd1 individuals #1 and #2, respectively (Fig. 4A and SI Appendix, Table S6). We tested 8 of these putative events by PCR and all 8 events were confirmed (Fig. 4 B and C and SI Appendix, Fig. S4). Although 2 putative TE transposition events were identified in the DNA-seq data of ddm1 cmt2 plants (SI Appendix, Table S6), these events could not be confirmed by PCR (SI Appendix, Fig. S5). The 33 insertion events could be classified into five categories of TE subfamilies, encompassing both retrotransposons (ATCOPIA21, ATCOPIA93, and ATGP2N) and DNA transposons (ATENSPM3 and VANDAL21) (Fig. 4A). On average, the expression levels of transposed TE subfamilies in ddm1 nrpd1 and ddm1 nrpe1 are higher compared with the other genotypes (SI Appendix, Fig. S6), suggesting that the TE transposition is correlated with its expression level. AT1TE41580 and AT1TE42210 were found to have transposed within the first homozygous generation in ddm1 nrpd1 plants, while the remaining TEs may have transposed either within the first homozygous generation or after only one generation of selfing (Fig. 4 B and C and SI Appendix, Fig. S4); some TEs transposed more than once (Fig. 4A). Among the five TE subfamilies showing transposition in ddm1 nrpd1 mutant plants, one subfamily corresponds to the CMT2-to-RdDM type of loci, while three subfamilies correspond to RdDM loci (Fig. 4 D and E). These results suggest that the RdDM pathway becomes critical to preventing TE transposition and protecting genomic stability in plants lacking DDM1.
Fig. 4.
TE transposition in ddm1 nrpd1 mutant plants. (A) Circle plots showing TE transpositions. Different colors indicate the subfamily of transposed TEs. Head and bottom of arrow lines represent the insertion and original sites of transposed TEs, respectively. (B and C) Products of PCR amplifications with paired primers flanking the new insertion site or a transposon-specific primer and a primer flanking the new insertion site in the indicated genotypes. Primer positions are indicated (black arrows). Upper panel, Integrative Genomic Viewer (IGV) screenshots showing split-reads at the TE insertion sites. (D) Composition of the subfamily of transposed TEs in ddm1 nrpd1. The TE subfamilies are considered as associated with the indicated loci when they contain at least one TE whose body or flanking 2-kb regions overlap with the indicated loci. (E) CHH methylation levels, TE expression levels, and NRPD1 ChIP-seq signals at a transposed TE associated with a CMT2-to-RdDM locus. Black dotted box indicates the CMT2-to-RdDM locus. The numbers in parentheses indicate y axis scales.
Discussion
Multiple DNA methylation pathways exist to maintain DNA methylation throughout the genome to ensure that transposons remain in a silenced state, hence protecting genome integrity, as well as preserving cell-specific DNA methylation identity after cell division and across generations (1, 2, 4, 37–39). To date, a “static” model is accepted, according to which DNA methylation at a given site in the genome is maintained by a specific pathway. For example, CHH methylation at the body of long TEs or heterochromatic regions is maintained by CMT2, while at the regions located at the edges of some long TEs or euchromatic regions, it is maintained by the RdDM pathway. Here, a careful examination of RdDM- and CMT2-methylated loci in WT and ddm1 mutant plants led us to identify a relationship between the CMT2 and the RdDM pathways in plants lacking DDM1. Maintenance of CHH methylation in hundreds of regions was subjected to a pathway conversion from CMT2 to RdDM in the ddm1 mutant background (Fig. 1 A and B). We analyzed genomic regions in four categories: RdDM loci, RdDM-and-CMT2 loci, CMT2-only loci, and CMT2-to-RdDM loci (Fig. 1A). By analyzing the Pol IV occupancy and 24-nt siRNA levels, we confirmed that CMT2-to-RdDM loci are bona fide RdDM targets in the ddm1 mutant background (Fig. 1 C–E). Our results suggest that the pathway maintaining DNA methylation at a given site can switch depending on the activity of the chromatin remodeling factor DDM1.
Previous studies demonstrated that repeated self-pollination of ddm1 induces ectopic non-CG methylation at a few unmethylated loci; recent studies in other species demonstrated that loss of DDM1 function leads to the global production of 24-nt siRNA and CHH methylation, implying that other types of DNA methylation pathway conversion may exist (26, 40–45). The ddm1-induced pathway conversion is reminiscent of the compensatory effect observed in DNA-deficient mutants, where suppression of the expression of the genes encoding the DNA demethylase ROS1 and/or the H3K9 demethylase IBM1 cause ectopic DNA hypermethylation, compensating for the loss of DNA methylation (46–49). The pathway conversion from CMT2 to RdDM can also to some extent compensate for the decreased DNA methylation in the ddm1 background. Thus, we propose that this switch could be an additional homeostatic mechanism to buffer the loss of repressive chromatin marks.
We found that simultaneous loss of DDM1 and RdDM functions led to a burst of transposition of both retrotransposons and DNA transposons in the first homozygous generation of the corresponding mutants (Fig. 4 and SI Appendix, Fig. S4 and Table S6). Considering that the expression of several core components of the RdDM pathway, including NRPD1, NRPE1, and DRM2, is repressed during male gametogenesis (50, 51), we propose that mutation in DDM1 can generate a small window where the plant lacks both DDM1 and RdDM during sexual reproduction. In one generation, the probability of TE transposition in the small window of male germ cells in the ddm1 mutant is low, and thus it is rarely detected. However, continuous inbreeding of ddm1 would lead to increases in the chances of TE transposition. This model may explain the long-standing observation that ddm1 causes TE transposition only after several generations of inbreeding.
Taken together, our results suggest that plants have evolved a “double insurance” mechanism to protect genomic stability: in the absence of RdDM competency, DDM1-dependent DNA methylation maintenance pathways keep TEs silenced in heterochromatin; in the absence of DDM1 function, RdDM pathways including the CMT2-to-RdDM pathway can repress TE transposition. Interestingly, DDM1 expression is drastically reduced under certain stresses (e.g., osmotic stress or infection by virulent bacteria) (51), suggesting that the pathway conversion described here may become important in specific environmental conditions. Transposon 24-nt siRNAs and CHH methylation in heterochromatin regions, which are controlled by the CMT2 pathway, are dynamically regulated during Arabidopsis embryogenesis (52); it is hence also possible that the CMT2-to-RdDM pathway conversion described here may play a role in suppressing transposons at this stage of the plant life cycle.
Methods
Plant Materials and Growth Conditions.
All plants were grown under long-day conditions (16 h light/8 h dark). For seedling growth, Arabidopsis seeds were plated on 1/2 Murashige and Skoog (MS) medium with 0.6% agar and 1.5% sucrose and stratified for 7 d at 4 °C in darkness before being transferred to the growth chamber (16 h light/8 h dark, 22 °C). For experiments with adult plants, 14-d-old seedlings were transplanted to soil in the growth chamber. All mutant lines used in this study are in the Columbia-0 (Col-0) background. The following mutants have been described previously: nrpd1-3 (53), nrpe1-11 (54), ddm1-1 (21, 55), and cmt2-3 (15). The double mutants used in this study were generated by genetic crossing and subsequent PCR-based genotyping in F2 populations.
Published Genomic Data.
siRNA datasets were taken from ref. 29; data for cmt2 and its WT RNA-seq were taken from ref. 14; and data for WT, nrpd1, and nrpe1 RNA-seq were taken from ref. 36.
PCR Assay.
To confirm new transposon insertions, PCR was performed with a transposon-specific primer and a primer flanking the new insertion or with two primers flanking the new insertion. The DNeasy Plant Mini Kit (Qiagen) was used for DNA extraction. All PCR reactions were carried out using Ex-Taq enzyme (Takara). Primer sequences are listed in SI Appendix, Table S7.
Whole-Genome Bisulfite Sequencing.
Genomic DNA was extracted from 2-wk-old seedlings using the DNeasy Plant Maxi Kit (Qiagen). Bisulfite treatment, library construction, and sequencing were performed at the Shanghai Center for Plant Stress Biology (PSC) Genomics Core Facility. For methylation data, low-quality sequences and adaptors were trimmed using Trimmomatic with parameters “LEADING: 3 SLIDINGWINDOW: 4: 30 minLEN: 36,” and clean reads were mapped to the A. thaliana TAIR 10 genome using the bisulfite sequence mapping program (BSMAP) with parameters “-v 2 -S 1,” allowing two mismatches (56–58). The methratio.py script from BSMAP with parameters “-r -z -p -m 1” was used to extract the methylation ratio from mapping results; only mapped reads after deduplication were considered for subsequent analyses.
Identification of Differentially Methylated Regions.
Identification of CHH DMRs was conducted as previously described with some modifications (59). In brief, for each comparison, only CHH cytosine with a depth of more than four in both libraries were retained for further analyses. In every 200-bp window with a step size of 50 bp, the P value was computed and adjusted for multiple testing using the Benjamini and Hochberg method (60) to control for false discovery, and positions with a false discovery rate (FDR) higher than 0.05 were discarded. Windows with a threefold change or greater in DNA methylation level and four more differentially methylated cytosines (DMCs), defined as CHH cytosine with twofold or greater change in DNA methylation level, were further retained and merged to generate DMRs. Finally, the length of DMRs was adjusted to be from the first methylated CHH to the last methylated CHH in WT. Two independent biological replicates were merged for the identification of DMRs. DMR density over chromosomes was calculated by the number of DMRs identified in 100-kb bins across chromosomes divided by the number of DMRs in the corresponding chromosome. A similar calculation was used for counting TE and gene density over chromosomes. To rule out a random distribution of DMRs, control regions with the same numbers of DMRs were selected by the shuffleBed command in BEDTools (61). Enrichment analysis for the type and length of TEs associated with indicated loci was conducted using Fisher’s exact test by comparing them with TEs associated with control regions.
RNA-Seq and Analysis.
Total RNA was extracted from 2-wk-old seedlings using the RNeasy Plant Mini Kit (Takara). RNA library preparation and paired end sequencing were performed by the PSC Genomics Core Facility. For data analysis, low-quality sequences and adaptors were trimmed using Trimmomatic. Clean reads were mapped to the reference genome by TopHat with the parameter “-g 1” (62). The total number of reads mapping to each gene was calculated with the htseq-count script in HTSeq with the parameter “–nonunique all,” to minimize overestimation of TE expression caused by overlapping genes; read counts for each TE were calculated by htseq-count with parameters “–nonunique all -m intersection-strict” based on the TE-only annotation (63). RNA-seq data for WT, nrpd1, nrpe1, and WT, cmt2 were downloaded from GSE98286 (36) and GSE51304 (14), respectively, and analyzed in the same manner. Genes and TEs with a normalized expression level of at least one count per million mapped reads (CPM) in three or more libraries were considered as expressed. Principal component analysis (PCA) based on the transcript levels of the expressed protein-coding genes and TEs was performed using the prcomp function in R software with default settings. Differentially expressed TEs with at least a twofold change in expression and an FDR of <0.05 were identified by the R package edgeR using the trimmed mean of M-values (TMM) method (64).
ChIP-Seq and Data Analysis.
Seedlings were cross-linked with 1% formaldehyde, and ChIP-seq was performed as previously described (65) with anti-Flag antibodies (Sigma, F3165). Library preparation and sequencing were performed by the PSC Genomics Core Facility. For data analysis, around 15.2 million raw paired-end reads were obtained for each sample and subsequently cleaned by Trimmomatic. Clean reads were mapped to the reference genome by Bowtie2 using the parameter “–very-sensitive –no-unal –no-mixed –no-discordant -k 2” (66). Subsequently, uniquely mapped reads were selected and marked as duplicates using the Picard tool followed by using the SAMtools “rmdup” command (67, 68). Coverage of deduplicated reads was normalized to 1× sequencing depth by bamCoverage in deepTools with parameters “–normalizeUsing RPGC –exactScaling -bs 10” (69). Log transformed normalized coverage of treated samples to control sample were obtained by bigwigCompare in deepTools with parameters “–skipZeroOverZero -bs 10.” Visualization of the log2 transformed coverage over the upstream and downstream 5 kb of DMRs was performed by plotHeatmap in deepTools.
DNA-Seq and Identification of Nonreference TE Insertions.
The DNeasy Plant Mini Kit (Qiagen) was used for DNA extraction. Library construction and sequencing were performed at the PSC Genomics Core Facility. Identification of nonreference TE insertions with target site duplications (TSDs) was conducted using SPLITREADER with some modifications (70). In brief, after trimming low-quality sequences and adaptors using Trimmomatic, clean read pairs were mapped to the reference genome using Bowtie2 with the parameter “-very-sensitive.” Subsequently, unmapped reads from both pairs, including discordantly mapped reads, were extracted and merged together. Those unmapped reads were remapped to a collection of 5′ and 3′ TE sequence extremities (300 bp) with parameters “–local –very-sensitive” (TE families like ARNOLDY2, ATCOPIA62, ATCOPIA95, TA12, and TAG1 families were excluded, since they do not contain copies with intact extremities in the Col-0 genome) (70) and reads with soft-clipped mapping (with one end 20 nt or greater mapped to the TE extremity) were selected. Those selected reads were further recursively soft clipped by 1 nt and mapped to the reference genome using Bowtie2 with parameters “–mp 13 –rdg 8,5 –rfg 8,5 –local –very-sensitive” until the soft-clipped read length reached 20 nt. For the clipped reads that are simultaneously mapped to TE reference and reference genome, we further required that the other pair of the clipped read was also mapped and met one of the following criteria: 1) the other pair was properly mapped (insertion size less than 3,000 bp and on the opposite strand) to the reference genome; 2) the other pair was properly mapped to the same TE reference; and 3) the other pair was also clipped mapped for the same TE reference and reference genome on the opposite strand. Around the TSD insertion sites, read clusters composed of four or more reads clipped from the same extremity and overlapping with read clusters composed of reads clipped from the other extremity were taken to indicate the presence of a bona fide TE insertion only if the size of the overlap was more than 3 and less than 20 bp. Putative nonreference TE insertions overlapping with aberrant genomic regions [3 kb away from the centromeric region based on Repbase annotation (71), 3 kb away from the extremities of each chromosome and regions within 500 bp of “NNNN” sequence] or spanning the corresponding donor TE sequence were filtered out. Nine insertion sites were identified in WT, ddm1, and nrpd1; these insertions were also filtered out in ddm1 nrpd1 and ddm1 cmt2.
Small RNA-Seq Data.
Small RNA-seq data for WT and ddm1 were obtained from GSE52952 (29). After eliminating 3′-adapter sequences by the Cutadapt tool (72), the remaining clean sRNA reads of size 18 to 30 nt were aligned to the reference genome using Bowtie with parameters “-m 1 -v 0 –best” requiring nonmismatch unique mapping. The 24-nt small RNA abundance of DMRs was calculated by counting 24-nt small RNA reads normalized to per million mapped reads per 1,000 bp of the DMR.
Supplementary Material
Acknowledgments
We thank the Genomics Core Facility at the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences, for technical support. This work was supported by the Chinese Academy of Sciences (to J.-K.Z.).
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107320118/-/DCSupplemental.
Data Availability
All high-throughput sequencing data generated in this study have been deposited in the Gene Expression Omnibus database with accession code GSE165877. All data supporting the findings of this study are available within the manuscript and its supporting information or are available from the corresponding author upon request.
References
- 1.Law J. A., Jacobsen S. E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He X. J., Chen T., Zhu J. K., Regulation and function of DNA methylation in plants and animals. Cell Res. 21, 442–465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matzke M. A., Kanno T., Matzke A. J., RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66, 243–267 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Zhang H., Lang Z., Zhu J.-K., Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Haag J. R., Pikaard C. S., Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12, 483–492 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Cuerda-Gil D., Slotkin R. K., Non-canonical RNA-directed DNA methylation. Nat. Plants 2, 16163 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Kankel M. W., et al., Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163, 1109–1122 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finnegan E. J., Dennis E. S., Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 21, 2383–2388 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindroth A. M., et al., Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Du J., et al., Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151, 167–180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du J., et al., Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell 55, 495–504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson L. M., et al., The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17, 379–384 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du J., Johnson L. M., Jacobsen S. E., Patel D. J., DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stroud H., et al., Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21, 64–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zemach A., et al., The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai J., et al., A One Precursor One siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163, 445–455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang D.-L., et al., Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 26, 66–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye R., et al., A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol. Cell 61, 222–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong X., et al., Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157, 1050–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gendrel A. V., Lippman Z., Yordan C., Colot V., Martienssen R. A., Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Vongs A., Kakutani T., Martienssen R. A., Richards E. J., Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Kakutani T., Jeddeloh J. A., Richards E. J., Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 23, 130–137 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kakutani T., Jeddeloh J. A., Flowers S. K., Munakata K., Richards E. J., Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. U.S.A. 93, 12406–12411 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons D. B., Zilberman D., DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes. eLife 6, e30674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osakabe A., et al., The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A.W. Nat. Cell Biol. 23, 391–400 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Ito T., et al., Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis. PLoS Genet. 11, e1005154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lippman Z., et al., Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Nuthikattu S., et al., The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creasey K. M., et al., miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508, 411–415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panda K., et al., Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17, 170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCue A. D., et al., ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 34, 20–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura A., et al., Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411, 212–214 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Tsukahara S., et al., Bursts of retrotransposition reproduced in Arabidopsis. Nature 461, 423–426 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Singer T., Yordan C., Martienssen R. A., Robertson’s Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15, 591–602 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., et al., Epigenetic memory marks determine epiallele stability at loci targeted by de novo DNA methylation. Nat. Plants 6, 661–674 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Yang R., et al., The developmental regulator PKL is required to maintain correct DNA methylation patterns at RNA-directed DNA methylation loci. Genome Biol. 18, 103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Springer N. M., Schmitz R. J., Exploiting induced and natural epigenetic variation for crop improvement. Nat. Rev. Genet. 18, 563–575 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Heard E., Martienssen R. A., Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 157, 95–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slotkin R. K., et al., Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teixeira F. K., et al., A role for RNAi in the selective correction of DNA methylation defects. Science 323, 1600–1604 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Fu F. F., Dawe R. K., Gent J. I., Loss of RNA-directed DNA methylation in maize chromomethylase and DDM1-type nucleosome remodeler mutants. Plant Cell 30, 1617–1627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan F., et al., DDM1 represses noncoding RNA expression and RNA-directed DNA methylation in heterochromatin. Plant Physiol. 177, 1187–1197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corem S., et al., Redistribution of CHH methylation and small interfering RNAs across the genome of tomato ddm1 mutants. Plant Cell 30, 1628–1644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long J. C., et al., Decrease in DNA methylation 1 (DDM1) is required for the formation of m CHH islands in maize. J. Integr. Plant Biol. 61, 749–764 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Sasaki T., Kobayashi A., Saze H., Kakutani T., RNAi-independent de novo DNA methylation revealed in Arabidopsis mutants of chromatin remodeling gene DDM1. Plant J 70, 750–758 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Lei M., et al., Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, 3553–3557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams B. P., Pignatta D., Henikoff S., Gehring M., Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 11, e1005142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathieu O., Reinders J., Caikovski M., Smathajitt C., Paszkowski J., Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130, 851–862 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Rigal M., Kevei Z., Pélissier T., Mathieu O., DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 31, 2981–2993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calarco J. P., et al., Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151, 194–205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winter D., et al., An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papareddy R. K., et al., Chromatin regulates expression of small RNAs to help maintain transposon methylome homeostasis in Arabidopsis. Genome Biol. 21, 251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onodera Y., et al., Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120, 613–622 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Pontier D., et al., Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19, 2030–2040 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Q., et al., The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov. 2, 16027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xi Y., Li W., BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berardini T. Z., et al., The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krueger F (2015) Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files 516. 517.
- 59.Ausin I., et al., INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 8374–8381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 61.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim D., et al., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S., Pyl P. T., Huber W., HTSeq – A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao L., et al., Integrative analysis of reference epigenomes in 20 rice varieties. Nat. Commun. 11, 2658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toolkit Picard, Version 2.18.8. http://broadinstitute.github.io/picard/. Accessed 8 August 2018.
- 69.Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quadrana L., et al., The Arabidopsis thaliana mobilome and its impact at the species level. eLife 5, e15716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bao W., Kojima K. K., Kohany O., Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All high-throughput sequencing data generated in this study have been deposited in the Gene Expression Omnibus database with accession code GSE165877. All data supporting the findings of this study are available within the manuscript and its supporting information or are available from the corresponding author upon request.