Significance
During the aging process, senescent cells secrete inflammatory factors, causing various age-related pathologies. Thus, controlling the senescence-associated secretory phenotype (SASP) can tremendously benefit human health. Although SASP seems to be induced by the alteration of chromosomal organization, its underlying mechanism remains unclear. Here, it has been revealed that noncoding RNA (ncRNA) transcribed from pericentromeric repetitive elements impairs the DNA binding of CCCTC-binding factor, resulting in the alteration of chromosomal accessibility and the activation of SASP-like inflammatory genes. Notably, the ncRNA was transferred into surrounding cells via small extracellular vesicles, acting as a tumorigenic SASP factor. Our study highlights a novel mechanism regulating chromatin interaction and inflammatory gene expression in senescence and cancer.
Keywords: pericentromeric RNA, senescence-associated secretory phenotype, CTCF, small extracellular vesicles, senescence
Abstract
Cellular senescence causes a dramatic alteration of chromatin organization and changes the gene expression profile of proinflammatory factors, thereby contributing to various age-related pathologies through the senescence-associated secretory phenotype (SASP). Chromatin organization and global gene expression are maintained by the CCCTC-binding factor (CTCF); however, the molecular mechanism underlying CTCF regulation and its association with SASP gene expression remains unclear. We discovered that noncoding RNA (ncRNA) derived from normally silenced pericentromeric repetitive sequences directly impairs the DNA binding of CTCF. This CTCF disturbance increases the accessibility of chromatin and activates the transcription of SASP-like inflammatory genes, promoting malignant transformation. Notably, pericentromeric ncRNA was transferred into surrounding cells via small extracellular vesicles acting as a tumorigenic SASP factor. Because CTCF blocks the expression of pericentromeric ncRNA in young cells, the down-regulation of CTCF during cellular senescence triggers the up-regulation of this ncRNA and SASP-related inflammatory gene expression. In this study, we show that pericentromeric ncRNA provokes chromosomal alteration by inhibiting CTCF, leading to a SASP-like inflammatory response in a cell-autonomous and non–cell-autonomous manner and thus may contribute to the risk of tumorigenesis during aging.
Cellular senescence is a state of essentially irreversible cell cycle arrest induced by several stressors under various physiological and pathological conditions (1). Senescent cells that accumulate in vivo over the course of aging communicate with surrounding tissues through the production of proinflammatory proteins, termed the senescence-associated secretory phenotype (SASP), which are thought to promote multiple age-related diseases, including some cancers, such as breast and colon cancer (2–7). Therefore, elucidating the regulatory mechanism of the SASP is essential for developing new preventive and therapeutic strategies against age-related cancer.
Recent studies have reported that abnormal nuclear morphologies, observed as micronuclei or nuclear buds, induce SASP gene expression via the activation of the DNA-sensing pathway during cellular senescence (8–12). In addition, cellular senescence causes a dramatic alteration of chromatin organization, characterized by an increase in short-range chromatin contacts and genome-wide shrinkage of chromosome arms (13). Chromatin organization and global gene expression are coordinately maintained by the CCCTC-binding factor (CTCF), a zinc-finger (ZF) nucleic acid–binding protein, and the cohesin complex; together, these factors orchestrate higher-order chromatin conformation through the formation of intrachromosomal and interchromosomal loops (14–16). Given that the nuclear localization and RNA-binding capacity of CTCF dynamically change due to cellular stress (17) and the alteration of CTCF distribution and/or followed by chromatin reorganization occur during cellular senescence (13, 18), postulating that CTCF distribution is associated with SASP gene expression in senescent cells is reasonable. However, the molecular mechanism underlying the connection between CTCF regulation and its association with SASP gene expression remains elusive.
In this study, we demonstrate that noncoding RNA (ncRNA) transcribed from pericentromeric repetitive satellite sequences changes the distribution of CTCF binding on the genome, thereby inducing SASP-like inflammatory gene expression via the functional impairment of CTCF in senescent cells. Furthermore, pericentromeric satellite RNA provokes tumorigenesis in a cell-autonomous or non–cell-autonomous manner via a pathway involving exosomes, a type of small extracellular vesicle (EV). Our findings reveal a mechanism of CTCF regulation by pericentromeric satellite RNA during cellular senescence, which may contribute to the risk of tumorigenesis.
Results
Pericentromeric hSATII RNA Promotes SASP-Like Inflammatory Gene Expression in Senescent Cells.
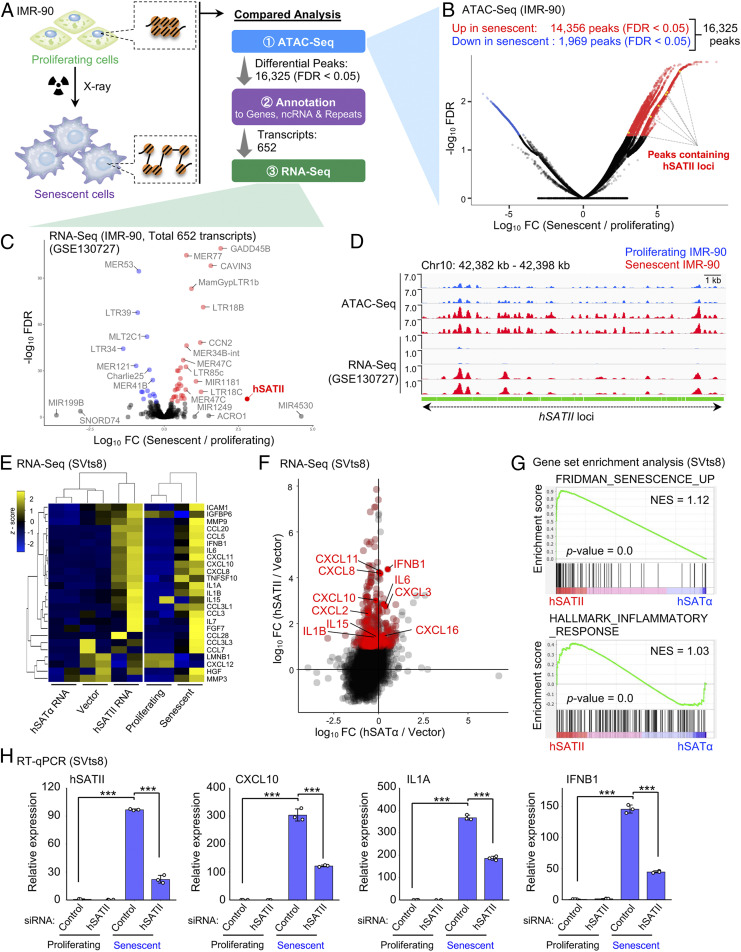
To elucidate the molecular mechanism underlying the alteration of chromatin organization and gene expression during cellular senescence, we first compared genome-wide chromatin accessibility between X-ray–induced senescent and proliferating IMR-90 cells, which are normal human diploid fibroblasts. Assay for transposase-accessible chromatin sequencing (ATAC-seq) analysis revealed that the peak intensities in 16,325 regions were dramatically altered (false discovery rate [FDR] < 0.05) during cellular senescence (Fig. 1 A and B), and a high incidence of distal intergenic regions and introns was identified (SI Appendix, Fig. S1A). From ATAC-seq analysis, 14,356 peaks were identified as higher chromatin accessibility (red) in X-ray–induced senescent cells versus proliferating IMR-90 cells; 1,969 peaks were identified as lower chromatin accessibility (blue; Fig. 1B). The 16,325 ATAC-seq peaks that showed differential chromatin accessibility values in X-ray–induced senescent cells compared to those in proliferating cells (Fig. 1B) were annotated to 652 transcripts using databases, including GRCh37/hg19 (coding genes and some noncoding regions) and RepeatMasker (repetitive elements). Next, we focused on these 652 transcripts and reanalyzed their expression level using published RNA-sequencing (RNA-seq) data of proliferating and X-ray–induced senescent IMR-90 cells (GSE130727; Fig. 1C) (19). Thus, loci containing pericentromeric repetitive sequences called human satellite II (hSATII), which are epigenetically silenced in normal somatic cells, were highly accessible (yellow; Fig. 1B), and hSATII ncRNA expression was markedly up-regulated in X-ray–induced senescent IMR-90 cells compared to proliferating cells (log10 fold change = 2.8) among the transcripts showing an FDR < 1010 (Fig. 1C). When we integrated our ATAC-seq data with published RNA-seq data of proliferating and X-ray–induced senescent IMR-90 cells (GSE130727) (19), a comparative analysis of senescent and proliferating cells represented both higher chromatin accessibility and increased transcription at hSATII loci in senescent cells versus proliferating cells (Fig. 1D). In accordance with previous studies of senescent cells and many types of cancer (20–23), we detected hSATII RNA expression by RT-qPCR and Northern blot analysis in H-RasV12– and serial passage–induced senescent cells (SI Appendix, Fig. S1 B–F). To interpret the biological effects of hSATII RNA expression, we overexpressed hSATII RNA in SVts8 cells, a conditionally immortalized human fibroblast cell line suitable for transfection analysis (24). The ectopic expression of hSATII RNA, but not centromeric human satellite alpha RNA (hSATα), induced SASP-like inflammatory gene expression, which was shown as the enrichment of signatures related to the inflammatory response and SASP by gene set enrichment analysis and altered the chromatin accessibility of the loci of SASP genes (Fig. 1 E–G and SI Appendix, Fig. S2 A–D) (25). Importantly, the knockdown of hSATII RNA diminished the expression of SASP genes in X-ray–induced senescent SVts8 cells (Fig. 1H) or X-ray– and serial passage–induced senescent IMR-90 cells (SI Appendix, Fig. S2 E and F). These data suggest that hSATII RNA regulates SASP-like inflammatory gene expression by altering chromatin accessibility during cellular senescence.
Fig. 1.
Pericentromeric hSATII RNA regulates SASP factor gene expression during cellular senescence. (A–C) Screening of unique transcripts showing increased chromatin accessibility and active transcription during X-ray–induced senescence in IMR-90 cells. (A) A scheme of the screening steps. (B) Volcano plot of ATAC-seq signals showing fold change (FC) (x-axis) and FDR (y-axis) of chromatin accessibility between proliferating and X-ray–induced senescent IMR-90 cells. Red peaks show significantly increased chromatin accessibility in X-ray–induced senescent cells. Blue peaks showing significantly increased chromatin accessibility in proliferating cells. Black peaks show no significant changes. Yellow peaks containing hSATII loci show significantly increased chromatin accessibility. (C) Volcano plot of RNA-seq data (GSE130727) showing FC (x-axis) and FDR (y-axis) concerning 652 transcripts involved in an increased chromatin accessibility region between proliferating and X-ray–induced senescent IMR-90 cells from ATAC-seq analysis in B. The 47 transcripts showing FDR < 1010 are shown as red (up-regulated) or blue (down-regulated) dots. (D) Peaks of uniquely mapped reads by ATAC-seq and RNA-seq (GSE130727) in hSATII loci in proliferating or X-ray–induced senescent IMR-90 cells. Two biological replicates are shown. (E–G) RNA-seq analysis of hSATα– or hSATII-overexpressed and X-ray–induced senescent SVts8 cells. (E) Heatmap regarding SASP-related gene expression in hSATα– or hSATII-overexpressed and X-ray–induced senescent SVts8 cells. (F) Scatterplot showing FC in hSATα (x-axis) or hSATII (y-axis) RNA-overexpressed SVts8 cells compared to empty vector–expressed cells. Red dots indicate genes up-regulated (FC > 10) in vicinity of specific chromatin accessible peaks in hSATII RNA–overexpressed cells. (G) Gene set enrichment analysis of signatures associated with senescence (Upper) and inflammatory response (Lower) in hSATα– or hSATII RNA–overexpressed SVts8 cells. NES, normalized enrichment score. (H) The effect of hSATII RNA knockdown on hSATII RNA and SASP gene expression in proliferating or X-ray–induced senescent SVts8 cells by RT-qPCR. The relative expression is shown as the FC from control small-interfering RNA–treated proliferating cells. Each bar represents mean ± SD of three biological replicates. ***P < 0.001 by one-way ANOVA, followed by the Tukey’s multiple comparisons post hoc test.
Pericentromeric hSATII RNA Regulates SASP-Like Inflammatory Gene Expression by Binding to CTCF.
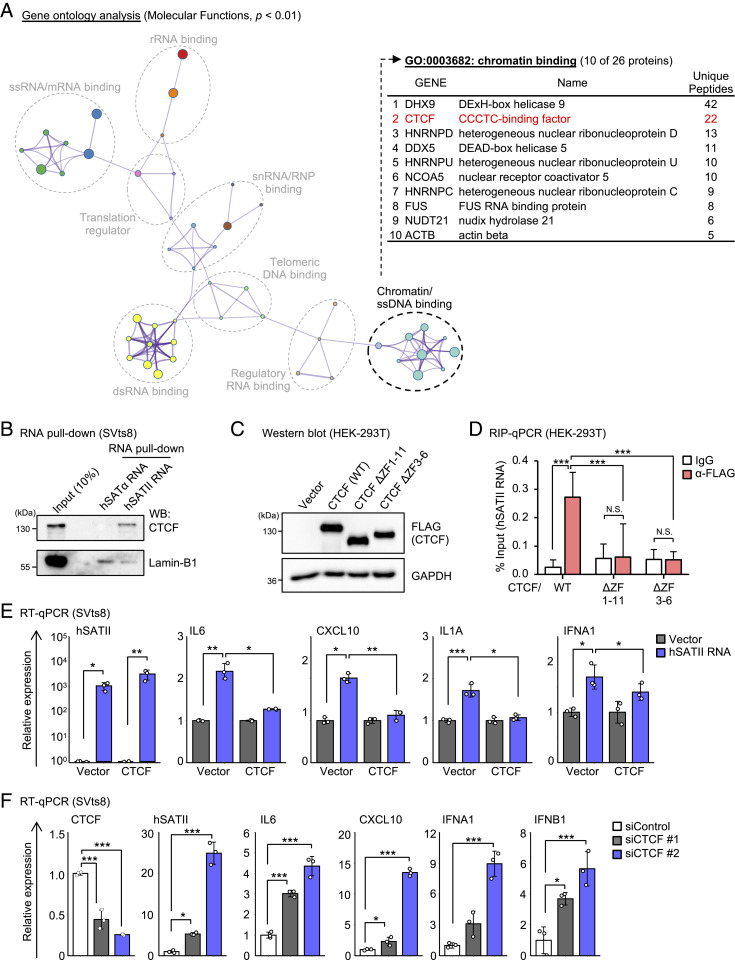
To understand how hSATII RNA promotes SASP-like inflammatory gene expression, we attempted to identify hSATII RNA–binding proteins. Several studies have reported the association of centromeric hSATα RNA with specific proteins (26); however, thus far, none have reported such an association for pericentromeric hSATII RNA. RNA pull-down and mass spectrometry analysis identified 280 hSATII RNA–binding proteins (Fig. 2A and SI Appendix, Fig. S3A and Table S1). Among these proteins, we identified 26 chromatin-binding proteins by Gene Ontology (GO) analysis (GO: 0003682) and focused on CTCF because of both its high intensity score (unique peptides) and its relevance to chromatin organization (15, 16, 27) (Fig. 2A). Unlike hSATα RNA, hSATII RNA bound to CTCF, whereas both ncRNAs bound to lamin B1 (Fig. 2B) (26). Because CTCF binding to genomic DNA is important for the maintenance of genomic integrity and CTCF is an RNA-binding protein (28–31), we performed RNA immunoprecipitation (RIP) analysis, demonstrating that the ZF DNA- and RNA-binding domains of CTCF are important for their binding to not only an exogenous hSATII RNA in human embryonic kidney (HEK)-293T cells (Fig. 2 C and D and SI Appendix, Fig. S3 B and C) but also an endogenous hSATII RNA in X-ray–induced senescent IMR-90 cells (SI Appendix, Fig. S3D). Of the 11 ZF domains of CTCF, the binding of ZF1 or ZF10 of CTCF to RNA is important for CTCF to form chromatin loops and regulate gene expression (31); however, we found that hSATII RNA bound to neither ZF1 nor ZF10 (SI Appendix, Fig. S3 B and C). Further analysis revealed that ZF3-ZF6 of CTCF-, known as DNA-binding domain (32), deficient mutant (CTCF ΔZF3-6) could not bind to hSATII RNA (Fig. 2 C and D), indicating that ZF3-ZF6 of CTCF is important for binding to hSATII RNA. Note that CTCF also binds to RNA through ZF domains; therefore, ΔZF3-6 might be an unfolding protein and not function appropriately. Further analysis will be needed to determine the interaction between CTCF and hSATII RNA. Importantly, the up-regulation of SASP-like inflammatory gene expression caused by hSATII RNA was canceled in the presence of excessive CTCF in SVts8 cells (Fig. 2E and SI Appendix, Fig. S3E). In contrast, CTCF depletion by RNA interference resulted in SASP-like inflammatory gene expression in proliferating cells (Fig. 2F and SI Appendix, Fig. S3F). Based on these results, we considered it likely that SASP-like inflammatory gene expression induced by hSATII RNA depends on the functional impairment of CTCF. Unexpectedly, we found that hSATII RNA expression was also up-regulated by CTCF depletion (Fig. 2F). Moreover, CTCF expression decreased during cellular senescence (SI Appendix, Fig. S3 G and H) (33). Together, these findings imply that CTCF regulates hSATII RNA expression during cellular senescence.
Fig. 2.
Pericentromeric hSATII RNA binds to CTCF. (A) GO analysis of 280 hSATII RNA–binding proteins (Left). Among these proteins, 26 genes were categorized as chromatin-binding (GO: 0003682), and the top 10 ranked genes and unique peptides are listed (Right). (B) RNA pull-down assay using SVts8 cell lysate followed by Western blotting confirmed hSATII RNA but not hSATα RNA bound to CTCF. (C) Western blot analysis of FLAG-tagged CTCF (WT: wild type) or CTCF ΔZF (deletion of ZF domain) in HEK-293T cells. (D) RIP followed by qPCR confirmed the binding of FLAG-tagged CTCF WT, but not CTCF ΔZF1-11 or ΔZF3-6, to hSATII RNA in HEK-293T cells. (E) RT-qPCR analysis of SASP-like inflammatory genes in hSATII RNA–overexpressed SVts8 cells with excess CTCF. The relative expression shows the value normalized from empty vector–expressed cells. (F) RT-qPCR analysis of SASP-like inflammatory genes in CTCF-depleted SVts8 cells. The relative expression shows the value normalized from small-interfering control (siControl)–treated cells. Each bar represents mean ± SD of three technical replicates repeated in two independent experiments (D, E, and F). *P < 0.05, **P < 0.01, ***P < 0.001, or N.S. (not significant) by one-way ANOVA, followed by the Tukey’s (D and E) or Dunnett’s (F) multiple comparisons post hoc test.
Furthermore, we investigated the expression of mouse major satellite (MajSAT) RNA, which is located at the pericentromeric locus of chromosomes, as well as human hSATII RNA. In mouse embryonic fibroblasts (MEFs), DNA damage induced by doxorubicin increased MajSAT RNA expression along with some canonical markers of cellular senescence (SI Appendix, Fig. S3I). As expected, the induction of MajSAT RNA was negatively correlated with the expression of CTCF (SI Appendix, Fig. S3I), and CTCF bound to pericentromeric MajSAT RNA but not mouse centromeric minor satellite (MinSAT) RNA, resulting in the up-regulation of SASP-like inflammatory genes (SI Appendix, Fig. S3 J and K). Taken together, these findings indicate that CTCF is crucial for the regulation of both pericentromeric satellite RNA and the expression of SASP-like inflammatory gene during cellular senescence (SI Appendix, Fig. S3M).
Pericentromeric hSATII RNA Alters CTCF Binding to Genomic DNA.
Because the ZF3-ZF6 DNA binding domain of CTCF was relevant to its binding to hSATII RNA (Fig. 2D), we hypothesized that hSATII RNA changes the DNA-binding capacity of CTCF via direct binding to its ZF domains. As expected, the ectopic expression of hSATII RNA altered the distribution of CTCF at its binding sites (Fig. 3 A and B). Remarkably, both chromatin immunoprecipitation (ChIP)-qPCR and electrophoretic mobility shift assay (EMSA) revealed that hSATII RNA inhibited the DNA-binding capacity of CTCF to an imprinting control region (ICR) positioned between IGF2 and H19, a well-known representative CTCF binding site, in a dose-dependent manner (Fig. 3 C and D) (15). In accordance with this data, pericentromeric MajSAT RNA bound to CTCF and diminished the binding of CTCF to ICR (SI Appendix, Fig. S3L). These notions raised the possibility that pericentromeric satellite RNA could change chromatin interaction as the binding of CTCF to DNA is important to maintain genomic integrity. To validate this assumption, we performed a chromosome conformation capture (3C) assay of the SASP genes in the vicinity of the CXCL10/CXCL11 locus because a robust interaction was noted in proliferating fibroblasts and various cell lines (SI Appendix, Fig. S4 A–F) (34). We discovered that the ectopic expression of hSATII RNA significantly weakened interactions in the T2 and T22 regions, as revealed by 3C-qPCR assay, and increased chromatin accessibility within these loci, as revealed by ATAC-seq analysis, followed by the up-regulation of SASP-like inflammatory gene expression, as also observed in X-ray–induced senescent cells (Fig. 3 E and F). Together, these data indicate that the up-regulation of hSATII RNA in senescent cells causes a conformational change of chromatin structure in some SASP gene loci. Chromatin organization and global gene expression are coordinately regulated by CTCF during a variety of physiological and pathological events, such as embryonic development and carcinogenesis (27). However, the molecular mechanism underlying the connection between CTCF regulation and its association with SASP gene expression during cellular senescence has not been elucidated. Our findings demonstrated that pericentromeric satellite RNA influences chromatin interaction by interfering with CTCF function, resulting in changes in SASP-like inflammatory gene expression (SI Appendix, Fig. S4G).
Fig. 3.
Pericentromeric hSATII RNA changes chromosomal interaction via CTCF disturbance. (A and B) Venn diagram showing overlap of CTCF binding sites from ChIP-seq analysis. (B) Enrichment of peaks from ChIP-seq analysis whose signals on peak summit ± 2 kb region are shown as profile plot (Left) and heatmaps split into two clusters using the k-means algorithm (Right) over sets of genomic regions in SVts8 cells. Wilcoxon rank-sum test P values are shown. (C) ChIP-qPCR for CTCF binding to an ICR positioned between IGF2 and H19. (D) EMSA showing the effect of hSATα or hSATII RNA on CTCF binding to ICR. (E and F) RNA-seq, CTCF ChIP-seq, and ATAC-seq profiles of SVts8 cells in representative loci of the SASP factor genes, CXCL10 and CXCL11 (E), and chromatin conformation by 3C-qPCR assay (F). The interaction of a constant primer (C) with each target primer (T) is shown. Each bar represents mean ± SD of three technical replicates repeated in two independent experiments (C and F). *P < 0.05, ***P < 0.001, or N.S. (not significant) by one-way ANOVA, followed by the Tukey’s multiple comparisons post hoc test (C) or unpaired two-sided t test (F).
Pericentromeric hSATII RNA Promotes Tumor Development in a Cell-Autonomous and Non–Cell-Autonomous Manner.
Human and murine satellite RNAs have the potential to induce chromosomal instability (CIN), leading to tumorigenesis (26, 35, 36). Hence, we explored the possibility that the loss of CTCF could contribute to satellite RNA–induced CIN. In accordance with previous reports, we confirmed that the ectopic expression of hSATII RNA provoked multipolarity and chromosomal bridge formation (SI Appendix, Fig. S5 A–C), which are typical characteristics of CIN (37). Furthermore, hSATII RNA–overexpressing cells exhibited obvious phenotypes of tumor cells, such as an abnormal chromosomal number and anchorage-independent growth (SI Appendix, Fig. S5 D and E). Notably, we found that excessive CTCF expression abolished CIN induced by hSATII RNA (SI Appendix, Fig. S5 F and G), a finding suggesting that CTCF plays a role in hSATII RNA–induced CIN, which is a risk factor for tumor development. Similarly, the ectopic expression of MajSAT RNA in MEFs also provoked multipolarity and chromosomal bridge formation in mitosis (SI Appendix, Fig. S6 A–C), causing the formation of transformed foci and polyploidy transition (SI Appendix, Fig. S6 D and E). Surprisingly, these cells exhibited the ability to form tumors in immunodeficient mice, although control cells did not (SI Appendix, Fig. S6F). Collectively, we concluded that pericentromeric satellite RNA may promote susceptibility to carcinogenesis.
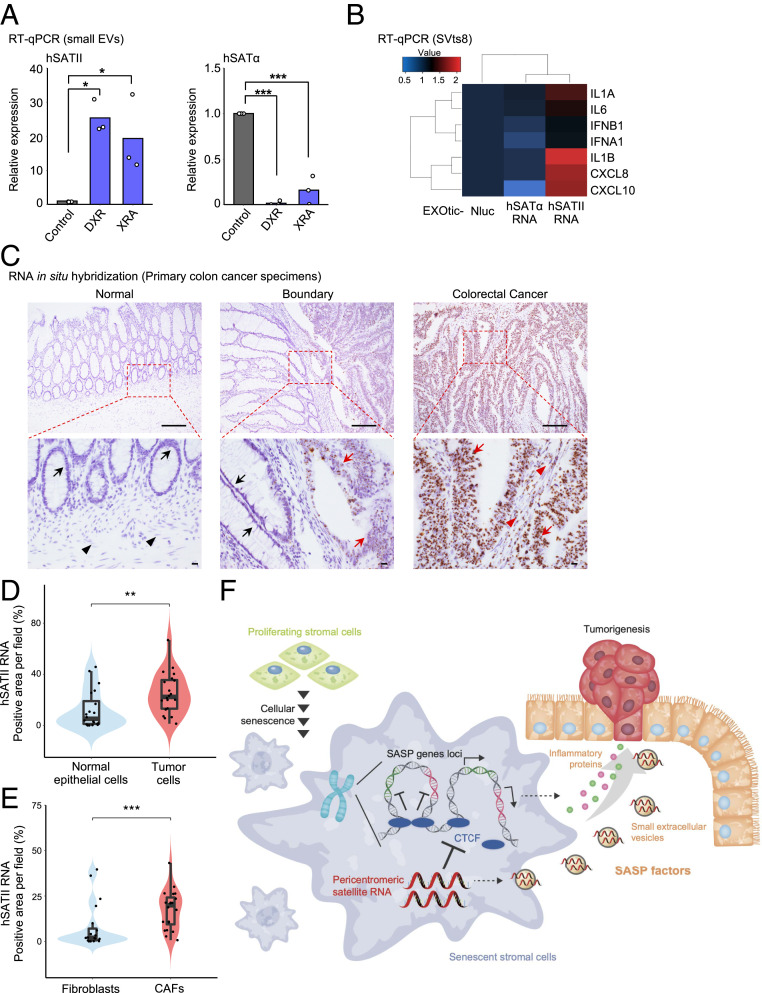
Furthermore, to gain insight into the biological significance of our findings, we focused on the function of hSATII RNA in the tumor microenvironment. We and others recently reported that small EVs secreted from cancer cells and/or senescent stromal cells dynamically contribute to tumor incidence and progression in a non–cell-autonomous manner in the tumor microenvironment (38–41). Intriguingly, the amounts of hSATII RNA, but not those of hSATα, were higher in small EVs derived from senescent cells than in those derived from proliferating cells (Fig. 4A). From an analysis of recently published RNA-seq data (42), we discovered that hSATII RNA could be detected in exosomes, a type of EV, secreted from cells of different human colon cancer cell lines (SI Appendix, Fig. S7A). Based on these observations, we speculated that hSATII RNA derived from senescent stromal cells would be transferred into surrounding cells through small EVs and function as a SASP-like inflammatory factor. Supporting this hypothesis, small EVs derived from senescent cells promoted anchorage-independent growth and CIN in normal cells (SI Appendix, Fig. S7 B and C). To assess the involvement of hSATII RNA in these phenotypes, we used EXOsomal transfer into cells (EXOtic) synthetic biology (43) to establish a designer exosome that contains hSATII RNA and found that these designer exosomes have tumorigenic activity similar to those of small EVs derived from senescent cells (SI Appendix, Fig. S7 D–F). Importantly, these designer exosomes promoted SASP-like inflammatory gene expression (Fig. 4B). Together, these findings show that pericentromeric hSATII RNA in small EVs secreted from senescent cells promote SASP-like inflammatory gene expression and CIN in neighboring cells in the tumor microenvironment.
Fig. 4.
Pericentromeric hSATII RNA promotes tumor development in a cell-autonomous and non–cell-autonomous manner. (A) RT-qPCR analysis of hSATII and hSATα RNA in the same number of small EVs derived from RPE-1/hTERT cells. Each value represents three biological replicates. *P < 0.05 or ***P < 0.001 by one-way ANOVA followed by the Dunnett’s multiple comparisons post hoc test. (B) An effect of the designer exosome produced by the EXOtic devices on SASP-like inflammatory gene expression in SVts8 cells was evaluated by RT-qPCR. Each value was normalized from EXOtic-Nluc-treated cells. (C–E) Representative and magnified (×100) images (C) or quantified data (D and E) of RNA-ISH with hSATII RNA probe in colon cancer specimens. Black and red arrows indicate normal epithelial and tumor cells, respectively. Black and red arrowheads indicate fibroblasts and cancer-associated fibroblasts, respectively. (Scale bar, 200 μm.) In the boxplot, the bottom and top hinges indicate the first and third quartile, respectively. The horizontal lines into the boxes indicate the median. The upper and lower whiskers define the highest and lowest value within 1.5 times of the interquartile range, respectively. n = 20 for each sample. **P < 0.01 or ***P < 0.001 by the Wilcoxon rank-sum test. Statistical analysis was performed using all samples and included outliers. (F) Schematic representation of this study. The up-regulation of pericentromeric satellite RNA during cellular senescence or aging provokes the expression of aberrant SASP-like inflammatory gene by interfering with the function of CTCF. In the tumor microenvironment, inflammatory proteins and hSATII RNA in small EVs are secreted from senescent stromal cells into surrounding tissue, where they act as SASP factors, thereby increasing the risk of carcinogenesis.
Finally, we checked the expression of pericentromeric ncRNA in the tumor microenvironment. Because the expression of murine satellite RNA is higher in malignant organoids derived from colon cancer (ApcΔ716 Trp53R270H/R270H) than in nonmalignant organoids (ApcΔ716) and accompanied by the reduction of CTCF and the up-regulation of SASP-like inflammatory genes (44) (SI Appendix, Fig. S7G), we evaluated the expression of hSATII RNA in surgical resection specimens from patients with primary colon carcinoma. We found that colon cancer cells expressing hSATII RNA were highly abundant in specimens compared with normal epithelial cells by RNA in situ hybridization (RNA-ISH) analysis (Fig. 4 C and D). Strikingly, we also observed that the population of hSATII RNA–positive cells was significantly higher among cancer-associated fibroblasts than among fibroblasts in normal stromal tissues (Fig. 4 C and E). In summary, our findings suggest that senescent stromal cells expressing hSATII RNA support tumor development in a non–cell-autonomous manner in the tumor microenvironment via the secretion of SASP-like inflammatory factors and small EVs containing hSATII RNA (Fig. 4F).
Discussion
Cellular senescence causes a dramatic alteration of chromatin organization (13, 18); however, its effect on gene expression and implication for senescent cells are not fully understood. We identified ncRNA derived from pericentromeric repetitive elements as a novel inducer of SASP-like inflammatory gene expression that acts by altering chromatin interaction. Importantly, pericentromeric satellite RNA is up-regulated during cellular senescence and aging in vivo (Fig. 1 A–D and SI Appendix, Fig. S1 B–F), which decreases CTCF binding to genomic DNA and alters both chromatin interaction and transcription in SASP-like inflammatory gene loci, thereby increasing the risk of tumor development (Fig. 4F). To verify the physiological role of pericentromeric satellite RNA, we confirmed that the expression level of ectopic ncRNA was equivalent to that found in senescent cells. Furthermore, the knockdown of endogenous pericentromeric satellite RNA diminished the expression of SASP-like inflammatory genes in senescent cells despite having no effect on the induction of senescence, clearly indicating that endogenous pericentromeric satellite RNA plays a role in the expression of SASP-like inflammatory genes.
CTCF and the cohesin complex are essential for stabilizing chromatin organization, which has divergent effects on gene regulation, embryonic development, and tumorigenesis (27), through their binding ability to specific sequences in genomic DNA (15, 16, 45). Recently, some groups have reported that CTCF shows a high affinity for specific RNA and depends on binding with RNA to form chromatin loops and genome organization in mouse embryonic stem cells (30, 31). Another group also has shown that the interaction of CTCF with long ncRNA, such as Tsix and Xite, mediates long-range chromosomal interactions, inducing homologous X chromosome pairing in mouse embryonic stem cells (29). In contrast, in senescent cells (in pathological conditions), we have revealed a novel molecular mechanism in which pericentromeric satellite RNA regulates chromatin interaction and gene expression by CTCF disturbance. Because pericentromeric satellite RNA expression is at an extremely low level in normal cells (SI Appendix, Fig. S1 B–F) (20), we considered that it is insufficient for the RNA to disturb CTCF function in physiological conditions but not in senescent and tumor cells that aberrantly express pericentromeric satellite RNA. In the previous study, Zirkel et al. revealed that, upon senescence entry, the high-mobility group B protein (HMGB2) nuclear depletion provokes the alteration of CTCF distribution and CTCF spatial clustering (18). Moreover, Lehman et al. showed that stressors, such as acute oxidative stress, cause CTCF reduction from nuclear speckles and changes in CTCF RNA interaction (17). These reports support our findings that pericentromeric satellite RNA up-regulated during cellular senescence directly binds to CTCF and disturbs CTCF function.
Furthermore, our observations also showed that CTCF maintains pericentromeric satellite RNA expression at extremely low levels in normal cells by directly binding the pericentromeric hSATII locus (46); however, CTCF expression significantly decreased during cellular senescence (SI Appendix, Fig. S3 G and H). Therefore, once pericentromeric satellite RNA is predominantly expressed by CTCF depression, satellite RNA alters chromatin interaction and induces CIN and SASP-like inflammatory gene expression via CTCF disturbance (SI Appendix, Fig. S3M). Previous studies have revealed that CTCF binds to pericentromeric/centromeric regions and recruits the centromeric protein CENP-E to these regions in mitotic chromosomes (46, 47). In addition to these findings, we have identified a regulatory machinery involving CTCF that controls pericentromeric satellite RNA expression.
In patients with gastrointestinal cancer, driver mutations were detected in the ZF domains of CTCF that provoke CIN and aberrant gene expression (48). Although CTCF-knockout mouse embryos die at early implantation stages (49), CTCF haploinsufficient (Ctcf+/−) mice are markedly susceptible to cancer, and transformed foci are observed in Ctcf+/− mouse–derived MEFs (35). These reports strongly support our conclusion that the disturbance of CTCF function caused by pericentromeric satellite RNA results in aberrant gene expression, CIN, and tumorigenesis. Centromeric satellite RNA (hSATα and MinSAT RNA) is associated with CIN in some cell lines (26), but we believe that there must be another mechanism, as these centromeric satellite RNAs neither bind to CTCF nor contribute to SASP-like inflammatory gene expression in normal fibroblasts (Figs. 1 E and F and 2B and SI Appendix, Figs. S2 A and B and S3K).
We conclude that pericentromeric satellite RNA plays a prominent role in tumorigenesis by cell-autonomous and non–cell-autonomous pathways in vivo: 1) DNA damage caused by various oncogenic stresses induces cellular senescence of normal epithelial cells, thereby acting as a tumor-suppressor mechanism. In these senescent cells in benign tumors, pericentromeric satellite RNA is up-regulated and leads to SASP-like inflammatory induction (1, 2, 4, 50). If these senescent cells override their cell cycle arrest, pericentromeric satellite RNA may contribute to transformation from a benign to a malignant tumor through CIN (51). 2) Pericentromeric satellite RNA could be transferred into cancer cells via small EVs from senescent stromal cells, provoking CIN and SASP-like inflammatory gene expression, resulting in tumor progression. Our data suggest that secreted pericentromeric satellite RNA also functions as a tumorigenic SASP factor via small EVs in the tumor microenvironment. Furthermore, the down-regulation of CTCF expression with age may trigger the up-regulation of pericentromeric satellite RNA expression and diminish CTCF function via a positive-feedback loop, subsequently promoting SASP-related inflammation and tumorigenesis during aging (Fig. 4F). Our findings clearly indicate that pericentromeric satellite RNA represents a therapeutic target for age-related pathologies.
Materials and Methods
A full description of the following methods is described in the SI Appendix, Supplementary Information Methods: cell culture, plasmid construction, RNA interference, RT-PCR, RT-qPCR, Northern blot, RNA pull-down assay, Western blotting, mass spectrometric analysis, ChIP followed by ChIP sequencing (ChIP-seq), 3C-qPCR, ATAC-seq, RIP, EMSA, immunofluorescence imaging, karyotype analysis, focus formation assay, anchorage-independent soft agar colony formation assay, RNA-seq, extraction and application of exosome-like EVs, RNA-ISH, organoid culture experiments, in vivo allograft assays, bioinformatical analysis, and statistical analysis.
Cell Culture.
TIG-3 cells (11, 38, 52) and IMR-90 cells were obtained from the Japanese Cancer Research Resources Bank and American Type Culture Collection, respectively. TIG-3 cells, IMR-90, and IMR-90/ER:H-RasV12 cells (52) were cultured in Dulbecco’s Modified Eagle’s (DME) medium (Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (Sigma-Aldrich) at physiological oxygen conditions (92% N2, 5% CO2, and 3% O2) at 37 °C. RPE-1/hTERT cells (39) and HEK-293T cells (52) were cultured in DME medium (Nacalai Tesque) supplemented with 10% FBS and penicillin/streptomycin (Sigma-Aldrich) in a 5% CO2 incubator at 37 °C. SVts8 cells (24) were cultured in DME medium (Nacalai Tesque) supplemented with 10% FBS and penicillin/streptomycin (Sigma-Aldrich) in a 5% CO2 incubator at 34 °C. MEFs were generated from CD-1 mice as previously described (53) and then cultured in DME medium (Nacalai Tesque) supplemented with 10% FBS and penicillin/streptomycin (Sigma-Aldrich) at physiological oxygen conditions (92% N2, 5% CO2, and 3% O2) at 37 °C. All cell lines used were negative for mycoplasma.
Western Blotting.
Cell pellets were lysed in lysis buffer (0.1 M Tris–HCl pH 7.5, 10% glycerol, and 1% sodium dodecyl sulfate [SDS]), boiled for 5 min, and then centrifuged for 10 min at 15,000 rpm. All protein concentrations were determined by BCA Protein Assay Reagent (Pierce). Each cell lysate was electrophoresed by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). After blocking with 5% skim milk (Megumilk) or 5% bovine serum albumin (Sigma-Aldrich) in Tris-buffered saline with 0.1% Tween 20 (TBST), the membrane was treated with primary antibodies to p16 (IBL, #11104, 1:250 dilution), lamin B1 (Abcam, #ab16048, 1:1,000 dilution), GAPDH (Proteintech, #60004-1-lg, 1:10,000 dilution), vinculin (Sigma-Aldrich, #V9131, 1:1,000), CTCF (Cell Signaling Technology, #3418, 1:1,000 dilution), DDDK-tag (MBL, #M185-3L, 1:5,000), and ras (Oncogene, #OP41, 1:1,000 dilution) overnight at 4 °C in blocking buffer. Membranes were then washed three times in TBST and incubated with an enhanced chemiluminescence (ECL) anti-mouse IgG, horseradish peroxidase–linked whole antibody (GE Healthcare, NA931V) or ECL anti-rabbit IgG, horseradish peroxidase–linked whole antibody (GE Healthcare, NA934V) for 1 h at room temperature. After washing the membrane three times in TBST, the signal was resolved with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and imaged on a FUSION imaging system (Vilber Lourmat).
RNA-ISH.
hSATII RNA was detected on formalin-fixed paraffin-embedded (FFPE) sections in primary colon cancer specimens using an Advanced Cell Diagnostics (ACD) RNAscope 2.5 HD Reagent Kit-BROWN (ACD, #322300) and the RNAscope Target Probe Hs-HSATII (ACD, #504071) according to the manufacturer’s instructions. For each sample (n = 10), two images (×100) of normal mucosa, submucosa, and tumor were randomly selected. The areas of hSATII RNA positivity and total cells were analyzed using the ImageJ software (https://imagej.nih.gov/ij/docs/faqs.html). The hSATII RNA–positive area per field (percent) of each type of cell was calculated as the proportion of the total positive area to the total area of cells.
The FFPE sections in primary colon cancer specimens were collected from patients who provided informed consent for genetic and cell biological analyses. All methods were performed in accordance with protocols approved by the Institutional Review Board (approval number: 2013-1090) of the Japanese Foundation for Cancer Research (JFCR).
In Vivo Allograft Assays.
MEF/Vector or MEF/MajSAT RNA (5 × 106 cells) in Hank’s Balanced Salt Solution (Gibco/Thermo Fisher Scientific) were subcutaneously injected with an equal volume of Matrigel (BD Pharmingen) into 4- or 5-wk-old female BALB/c-nu/nu mice (Charles River Laboratories). After 20 or 30 d of cell injection, the mice were euthanized and tumor weight was measured. All animal procedures were performed using protocols approved by the JFCR Animal Care and Use Committee in accordance with the relevant guidelines and regulations (approval number: 1804-05).
Statistical Analysis.
Parametric statistical analyses were performed using the unpaired two-tailed Student’s t test (Fig. 3F and SI Appendix, Figs. S1C, S3 G, I, J, and L, S5 B–E, S6 B–E, and S7 E–G) or one-way ANOVA, followed by the Dunnett’s (Figs. 2F and 4A and SI Appendix, Figs. S2A and S7C) or Tukey’s (Figs. 1H, 2 D and E, and 3C and SI Appendix, Figs. S2 E and F, S3 C and D, and S5G) multiple comparisons post hoc test using the R software for statistical computing (64-bit version 3.6.1). Nonparametric statistical analyses were performed using the Wilcoxon rank-sum test (Figs. 3B and 4 D and E) or the Kruskal–Wallis H test (one-way ANOVA on ranks) followed by the Steel’s multiple comparisons post hoc test (SI Appendix, Fig. S6F) using the R software for statistical computing. P < 0.05 was considered statistically significant. All experiments, except for mass spectrometric analysis, were repeated at least twice.
Supplementary Material
Acknowledgments
We thank K. Nagasaka for valuable suggestions; T. Yamamoto and N. Saitoh for technical assistance for chromatin conformation analysis; K. Matsumoto for technical assistance for northern blot; S. Kuraku, C. Obuse, S. Adachi, and T. Natsume for mass spectrometry; N. Tanaka for bioinformatics analysis; G. Hannon and D. Beach for providing the MaRX vector; R. Asaka, K. Baba, and K. Takeuchi for technical advice for in situ experiments; H. Siomi for technical assistance for the RNA–protein interaction study; and members of the A.T. laboratory for helpful discussions during the preparation of this manuscript. This work was supported in part by the Japan Science and Technology Agency (JST)-Precursory Research for Embryonic Science and Technology (PRESTO) under grant number JPMJPR17H7; JST-Moonshot R and D under grant number JPMJPS2022; the Japan Agency of Medical Research and Development–Advanced Research and Development Programs for Medical Innovation (PRIME) under Grant 19gm6110023h0001; the Japan Society for the Promotion of Science (JSPS) under Grants 19H03507, 18K15254, and 20K16344; the Princess Takamatsu Cancer Research Fund; the Mitsubishi Foundation; and the Takeda Science Foundation. This research was also supported by the Research Fellowships for Young Scientists from JSPS under Grant 19J00796.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025647118/-/DCSupplemental.
Data Availability
The sequence and processing data have been deposited in the DNA Data Bank of Japan with the accession numbers DRA009771 for RNA-seq, DRA010750 for ChIP-seq, and DRA010749 for ATAC-seq. All other data supporting the findings of this study are available within the article and SI Appendix.
References
- 1.Muñoz-Espín D., Serrano M., Cellular senescence: From physiology to pathology. Nat. Rev. Mol. Cell Biol. 15, 482–496 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Wiley C. D., Campisi J., From ancient pathways to aging cells-connecting metabolism and cellular senescence. Cell Metab. 23, 1013–1021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiche A., et al., Injury-induced senescence enables in vivo reprogramming in skeletal muscle. Cell Stem Cell 20, 407–414.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 4.He S., Sharpless N. E., Senescence in health and disease. Cell 169, 1000–1011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faget D. V., Ren Q., Stewart S. A., Unmasking senescence: Context-dependent effects of SASP in cancer. Nat. Rev. Cancer 19, 439–453 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Fane M., Weeraratna A. T., How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 20, 89–106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loo T. M., Miyata K., Tanaka Y., Takahashi A., Cellular senescence and senescence-associated secretory phenotype via the cGAS-STING signaling pathway in cancer. Cancer Sci. 111, 304–311 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H., Wang H., Ren J., Chen Q., Chen Z. J., cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 114, E4612–E4620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dou Z., et al., Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glück S., et al., Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi A., et al., Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat. Commun. 9, 1249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Cecco M., et al., L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Criscione S. W., et al., Reorganization of chromosome architecture in replicative cellular senescence. Sci. Adv. 2, e1500882 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parelho V., et al., Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Wendt K. S., et al., Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Wutz G., et al., Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehman B. J., et al., Dynamic regulation of CTCF stability and sub-nuclear localization in response to stress. PLoS Genet. 17, e1009277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zirkel A., et al., HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell 70, 730–744.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Casella G., et al., Transcriptome signature of cellular senescence. Nucleic Acids Res. 47, 7294–7305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ting D. T., et al., Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Cecco M., et al., Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12, 247–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruickshanks H. A., et al., Senescent cells harbour features of the cancer epigenome. Nat. Cell Biol. 15, 1495–1506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bersani F., et al., Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. U.S.A. 112, 15148–15153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi A., et al., Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat. Cell Biol. 8, 1291–1297 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Coppé J. P., et al., Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Q., et al., Heterochromatin-encoded satellite RNAs induce breast cancer. Mol. Cell 70, 842–853.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spielmann M., Lupiáñez D. G., Mundlos S., Structural variation in the 3D genome. Nat. Rev. Genet. 19, 453–467 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Saldaña-Meyer R., et al., CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 28, 723–734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kung J. T., et al., Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol. Cell 57, 361–375 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen A. S., et al., Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol. Cell 76, 395–411.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saldaña-Meyer R., et al., RNA interactions are essential for CTCF-mediated genome organization. Mol. Cell 76, 412–422.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashimoto H., et al., Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol. Cell 66, 711–720.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah P. P., et al., Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 27, 1787–1799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao S. S., et al., A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kemp C. J., et al., CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 7, 1020–1029 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang F., et al., CTCF prevents genomic instability by promoting homologous recombination-directed DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 114, 10912–10917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakhoum S. F., et al., Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi A., et al., Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 8, 15287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takasugi M., et al., Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 8, 15729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fafián-Labora J. A., O’Loghlen A., Classical and nonclassical intercellular communication in senescence and ageing. Trends Cell Biol. 30, 628–639 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Kalluri R., LeBleu V. S., The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeppesen D. K., et al., Reassessment of exosome composition. Cell 177, 428–445.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kojima R., et al., Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 9, 1305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakayama M., et al., Intestinal cancer progression by mutant p53 through the acquisition of invasiveness associated with complex glandular formation. Oncogene 36, 5885–5896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao S. S. P., et al., Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao T., Wongtrakoongate P., Trainor C., Felsenfeld G., CTCF recruits centromeric protein CENP-E to the pericentromeric/centromeric regions of chromosomes through unusual CTCF-binding sites. Cell Rep. 12, 1704–1714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubio E. D., et al., CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. U.S.A. 105, 8309–8314 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Y. A., et al., Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat. Commun. 9, 1520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore J. M., et al., Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS One 7, e34915 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y. H., et al., Senescent tumor cells lead the collective invasion in thyroid cancer. Nat. Commun. 8, 15208 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vredeveld L. C., et al., Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 26, 1055–1069 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi A., et al., DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol. Cell 45, 123–131 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi S., et al., Intrinsic cooperation between p16INK4a and p21Waf1/Cip1 in the onset of cellular senescence and tumor suppression in vivo. Cancer Res. 70, 9381–9390 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence and processing data have been deposited in the DNA Data Bank of Japan with the accession numbers DRA009771 for RNA-seq, DRA010750 for ChIP-seq, and DRA010749 for ATAC-seq. All other data supporting the findings of this study are available within the article and SI Appendix.