Significance
TEM1 is a flowering repressor that suppresses the expression of FT florigen. However, the specific FT targeting mechanism of TEM1 and the molecular basis for the repression function of TEM1 remain unclear. In this research, we performed structural, biochemical, and genetic studies on TEM1. We revealed the molecular mechanism underlying the combinatorial binding of the FT gene 5′-untranslated region (UTR) by TEM1 AP2 and B3 domains. We found that TEM1 interacts with the PRC2 subunit CLF and is responsible for deposition of the repressive histone mark H3K27me3 at the FT 5′-UTR region. Altogether, we revealed a molecular mechanism underlying the flowering repression function of TEM1.
Keywords: TEMPRANILLO 1, FLOWERING LOCUS T, PRC2, combinatorial binding, DNA binding
Abstract
Arabidopsis TEMPRANILLO 1 (TEM1) is a transcriptional repressor that participates in multiple flowering pathways and negatively regulates the juvenile-to-adult transition and the flowering transition. To understand the molecular basis for the site-specific regulation of FLOWERING LOCUS T (FT) by TEM1, we determined the structures of the two plant-specific DNA-binding domains in TEM1, AP2 and B3, in complex with their target DNA sequences from the FT gene 5′-untranslated region (5′-UTR), revealing the molecular basis for TEM1 specificity for its DNA targets. In vitro binding assays revealed that the combination of the AP2 and B3 binding sites greatly enhanced the overall binding of TEM1 to the FT 5′-UTR, indicating TEM1 combinatorically recognizes the FT gene 5′-UTR. We further showed that TEM1 recruits the Polycomb repressive complex 2 (PRC2) to the FT 5′-UTR. The simultaneous binding of the TEM1 AP2 and B3 domains to FT is necessary for deposition of H3K27me3 at the FT 5′-UTR and for the flowering repressor function of TEM1. Overall, our data suggest that the combinatorial recognition of FT 5′-UTR by TEM1 ensures H3K27me3 deposition to precisely regulate the floral transition.
The floral transition in the life cycle of plants marks a shift from vegetative growth to reproductive growth (1, 2). Environmental cues, such as day length and temperature, and developmental signals, such as circadian rhythms, phytohormones, and aging, regulate the floral transition via a number of different pathways including vernalization, photoperiodic, and gibberellin (GA) pathways (3–6). In Arabidopsis, the outputs of these flowering pathways are integrated by flowering integrator genes, such as FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF), which in turn regulate the downstream floral meristem identity genes that coordinate flower development (7). The flowering pathways can regulate the expression of the flowering-time integrators both genetically (utilizing sequence specific transcription factors and cis-regulating elements) and epigenetically (via various epigenetic factors) (8).
In Arabidopsis, the transcription factor TEMPRANILLO 1 (TEM1) and its homolog TEM2 function as flowering repressors. The TEM genes are involved in regulating multiple flowering pathways, including the photoperiodic pathway, the GA pathway, the ambient temperature pathway, and the age-dependent pathway. In the photoperiodic pathway, TEM1/2 antagonize the transcriptional activator CONSTANS (CO) and bind directly to the 5′-UTR of the FT gene to suppress its expression (9). In the GA pathway, TEM1/2 down-regulate the expression of the GA4 biosynthetic genes GA 3-oxidase 1 (GA3OX1) and GA3OX2 (10). In the ambient temperature pathway, SHORT VEGETATIVE PHASE (SVP) up-regulates TEM2, which represses the expression of FT and TSF under low-temperature conditions (11). In the age-dependent pathway, TEM1/2 regulate the juvenile-to-adult growth transition (12). TEM1/2 also binds directly to and represses the expression of the phase-transition regulator microRNA miR172 (13). Together, TEMs function as transcriptional repressors that regulate multiple flowering pathways, finetuning the floral transition in response to both internal and external cues.
TEM1 possesses two putative plant-specific DNA-binding domains, the AP2 and B3 domains, and a C-terminal RLFGV motif (14, 15). The AP2 domain family of plant-specific DNA-binding domains can be subdivided into four subfamilies: AP2 (APETALA2), ERF (ethylene response factors), DREB (dehydration-responsive element–binding proteins), and RAV (RELATED TO ABI3/VP1, to which TEM1 belongs) subfamilies (16). While the molecular mechanisms of ERF subfamily proteins recognizing GCC-box have been revealed (17), how other AP2 subfamilies specifically recognize their targets is not known. The B3 plant-specific DNA-binding domain can also be divided into four subfamilies: LAV (LEC2–ABI3–VAL), ARF (AUXIN RESPONSE FACTOR), RAV, and REM (REPRODUCTIVE MERISTEM) (18). The LAV, ARF, and RAV subfamilies recognizes specific DNA sequences (9, 19–24), while the REM subfamily has not been fully characterized (25). While the DNA recognition mechanisms of ARF and LAV subfamilies have been characterized in detail (23, 26–28), the molecular basis for sequence-specific recognition by RAV subfamily, to which TEM1 belongs, remains unclear.
Published in vitro experiments suggested that TEM1 may bind to the ATCAACA(N)9CACCTGTT sequence located in the 5′-untranslated region (UTR) of the FT gene (9). How the specific recognition of this cis-element is achieved and the mechanism underlying the floral suppression function of TEM1 remain unclear. Here, we determine how the sequence-specific recognition of FT 5′-UTR by the TEM1 AP2 and B3 domains is mediated. Both structural and biochemical data suggest that the dual binding of the TEM1 domains results in stronger and more-specific recognition to the DNA target. Genetic data reveal that both the AP2 and B3 domains are necessary for the flowering repressor function of TEM1 and that TEM1 recruits the Polycomb repressive complex 2 (PRC2) to deposit repressive H3K27me3 marks that inhibit FT expression. Overall, our studies reveal the molecular basis for the suppression of flowering by the combinatorial readout of the FT 5′-UTR by TEM1.
Results
Combinatorial Binding of TEM1 to FT.
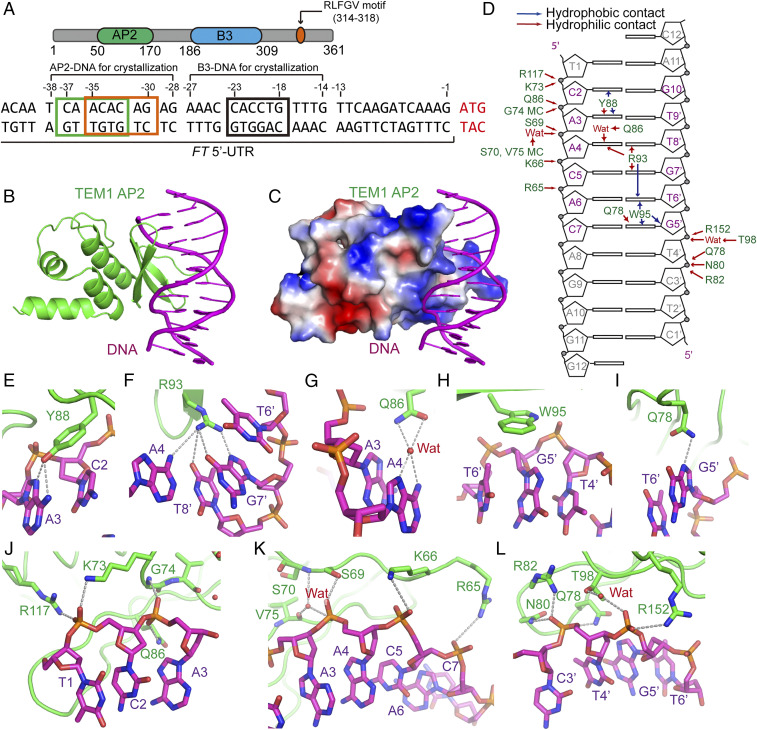
To explore the molecular mechanism of floral repression by Arabidopsis TEM1, we analyzed its primary sequence. TEM1 possess three potential functional domains/motifs: an N-terminal AP2 domain, a central B3 domain, and a C-terminal RLFGV motif (Fig. 1A). Both the AP2 and B3 domains are plant-specific DNA-binding domains. In vitro electrophoretic mobility shift assays have demonstrated that the AP2 and B3 domains bind to CAACA and CACCTG sequences in the 5′-UTR of the FT gene, respectively (9). The FT 5′-UTR CAACA and CACCTG sequences occur within a 20–base pair (bp) region (Fig. 1A), suggesting there could be combinatorial interactions between factors bound at these sites. To investigate the relationship between the AP2 and B3 domains of TEM1, we measure the binding affinity (KD) between different TEM1 fragments and an FT 5′-UTR probe encompassing both the AP2 and B3 binding sites (SI Appendix, Table S1) using a Bio-layer interferometry–based in vitro binding assay. While the B3 and AP2 domains bind to the FT 5′-UTR probe with binding affinities of 102 and 4,610 nM, respectively, the full-length TEM1 binds to this DNA probe with an affinity of 8.85 nM (Table 1). Although the AP2 domain binds to the FT 5′-UTR much more weakly than the B3 domain, the combination of the AP2 and B3 domains substantially increased the overall binding, indicative of a combinatorial effect.
Fig. 1.
Crystal structure of the TEM1 AP2–DNA complex. (A) Schematic representation of the domain architecture of Arabidopsis TEM1 (Upper) and the DNA sequence of FT 5′-UTR. The two AP2 binding sequences and the B3 binding sequence are highlighted by boxes. (B) The overall structure of TEM1 AP2–CAACAC complex with AP2 and DNA colored in green and magenta, respectively. (C) The AP2-CAACAC complex with AP2 in electrostatic surface view and DNA in ribbon view. (D) Schematic representation of the interactions between AP2 and CAACAC DNA. Hydrophilic and hydrophobic contacts are labeled with red and blue arrows, respectively. The core recognition sequence is highlighted in magenta. MC, main chain. (E–L) Details of the interactions between TEM1 AP2 and CAACAC DNA. The interacting residues detailed in stick model and hydrogen bonds by dashed lines. E–I show base-specific interactions. J–L show backbone interactions.
Table 1.
In vitro binding assay of various TEM1 fragments with FT 5′-UTR
| TEM1 protein fragments | DNA probe | KD (nM) |
| TEM1 FL | FT 5′-UTR | 8.85 ± 0.40 |
| TEM1 FL with B3 pentamutant | FT 5′-UTR | 2,660 ± 87 |
| TEM1 FL with AP2 pentamutant | FT 5′-UTR | 90.8 ± 3.8 |
| TEM1 B3 domain WT | FT 5′-UTR | 102 ± 5 |
| TEM1 B3 domain pentamutant | FT 5′-UTR | 3,580 ± 126 |
| TEM1 AP2 domain WT | FT 5′-UTR | 4,610 ± 150 |
| TEM1 AP2 domain pentamutant | FT 5′-UTR | NDB |
| TEM1 FL | FT-AP2 site 1 mutation | 10.6 ± 0.5 |
| TEM1 FL | FT-AP2 site 2 mutation | 11.8 ± 0.4 |
| TEM1 FL | FT-AP2 site 1/2 mutation | 24.4 ± 0.9 |
The in vitro binding assays of various WT and mutant TEM1 fragments to a 29-bp FT 5′UTR segment bearing both the AP2 and B3 recognition sequences or its mutants. The sequences of the DNA oligos can be found in SI Appendix, Table S1. The equilibrium dissociation constant (KD) and SD were calculated using the ForteBio Octet data analysis software; curves of at least five different concentrations were used in KD calculation. FL, full length; NDB, no detectable binding.
Structure of TEM1 AP2 Domain and AP2–DNA Complex.
To understand how TEM1 specifically recognizes the FT 5′-UTR, we carried out structural studies. We first determined the crystal structure of the TEM1 AP2 domain at 1.05-Å resolution (SI Appendix, Fig. S1A and Table S2). The TEM1 AP2 domain adopts a canonical AP2 fold consisting of a three-stranded antiparallel β-sheet and three C-terminal α-helices forming a triangle-like helical subdomain (SI Appendix, Fig. S1A). We also determined the crystal structure of the TEM1 AP2 domain in complex with an FT 5′-UTR–derived DNA at 2.7-Å resolution (Fig. 1B and SI Appendix, Tables S1 and S2). There are four AP2–DNA complexes in the asymmetric unit, with all 11 bp of the DNA used in crystallization clearly interpreted (SI Appendix, Fig. S1B). The superimposition of the free and DNA-bound AP2 structures yielded an RMSD of 0.8 Å, indicating no significant conformational changes (SI Appendix, Fig. S1C). TEM1 AP2 binding does not induce major changes in DNA curvature (SI Appendix, Fig. S1D). Overall, TEM1 AP2 binds to DNA with the antiparallel β-strands inserting into the DNA major groove, while the helical subdomain is opposite against the DNA binding interface and has few interactions with the DNA (Fig.1B). The DNA binding interface of AP2 is highly positively charged, suggesting electrostatic interactions play a key role in binding (Fig. 1C).
Structural Basis for the TEM1 AP2–DNA Interaction.
The TEM1 AP2 domain has been shown to bind the CAACA motif (9). However, in our crystal structure, the four AP2–DNA complexes in the asymmetric unit adopt two different DNA binding patterns. While two of the AP2 domains recognize the CAACAC motif, the other two AP2 molecules recognize the ACACAG motif displaced 2 bp downstream (Fig. 1A and SI Appendix, Fig. S1E). To determine which the canonical TEM1 AP2 domain binding motif is, we maintained the CAACAC or ACACAG motif and mutated the flanking sequences of two sides to TTTT (SI Appendix, Table S1). In vitro binding assays showed that the keeping of a single potential AP2 binding site only slightly decrease the overall binding (Table 1). Mutation of both the CAACAC and ACACAG AP2 binding motifs to poly T significantly weaken the binding (Table 1). The overlay of the two binding modes results in serious steric conflict (SI Appendix, Fig. S1E). Overall, our data suggest that both AP2 binding motifs can be recognized by TEM1 but not at the same time.
A number of hydrophilic and hydrophobic interactions contribute to DNA binding. In the CAACAC binding mode, Tyr88 forms a hydrogen bond with A3 and hydrophobic stacking interactions with A3 and C2 of the DNA forward strand (Fig. 1 D and E). Arg93 forms hydrogen bonds with both bases of the A4-T8’ pair and G7’ of reverse strand and cation–π interaction with T6’ of the reverse strand (Fig. 1 D and F). Gln86 forms a water-mediated hydrogen bond with A4 (Fig. 1 D and G). Trp95 forms hydrophobic stacking interactions with G5′, T6’, and the sugar ring of G5′ of the reverse strand (Fig. 1 D and H). Gln78 forms a hydrogen bond with G5′ (Fig. 1 D and I). Besides base-specific interactions, there are multiple backbone interactions encompassing the recognition sequence (Fig. 1D). Several polar residues including Arg65, Lys66, Ser69, Lys73, Gln78, Asn80, Arg82, Gln86, Thr98, Arg117, and Arg152 and the main chain of Ser70, Gly74, and Val75 form extensive hydrogen bonding and/or electrostatic interactions with the DNA phosphate backbone, further strengthening the overall interaction between TEM1 AP2 and DNA (Fig. 1 D and J–L). Overall, the AP2 has a DNA binding surface area of 791 Å2.
The AP2 domain is shifted 2 bp along and around the DNA to form the ACACAG binding mode, compared to the CAACAC binding mode (SI Appendix, Fig. S1E). Overall, the AP2 ACACAG binding mode is similar to the CAACAC binding mode, with many of the backbone and base interactions largely conserved (SI Appendix, Fig. S1F). For example, recognition of the central ACA (position 3 to 5) consensus of both motifs are the same in the two binding modes (Fig. 1D and SI Appendix, Fig. S1I). Major differences are seen at position 1, 2, and 6. In position 1, the first A of the ACACAG motif, which is equivalent to the first C of the CAACAC motif, is recognized by AP2 via stacking interactions (Fig. 1D and SI Appendix, Fig. S1I). Slight conformational changes in AP2 Tyr88 promote recognition of the A in the CAACAC motif or the C in the ACACAG motif (SI Appendix, Fig. S1G), suggesting a tolerance for different bases. Conformational changes in AP2 Gln78 enable the recognition of G5′, which pairs C7 of the CAACAC motif, or C3′, which pairs with the G9 of the ACACAG motif (SI Appendix, Fig. S1H). Given recognition of position 1 is mediated by stacking interactions, which is not base specific, and positions 2 and 6 can tolerate purine–pyrimidine switches through conformational changes in AP2 residues, we suggest a plausible model that the TEM1 AP2 domain can slide along the DNA. When it meets the central tandem ACA motifs (ACACA) via either of the two binding states, it will steadily bound at only one of the ACA motifs mutually exclusively.
Structural Basis of the TEM1 B3–DNA Interaction.
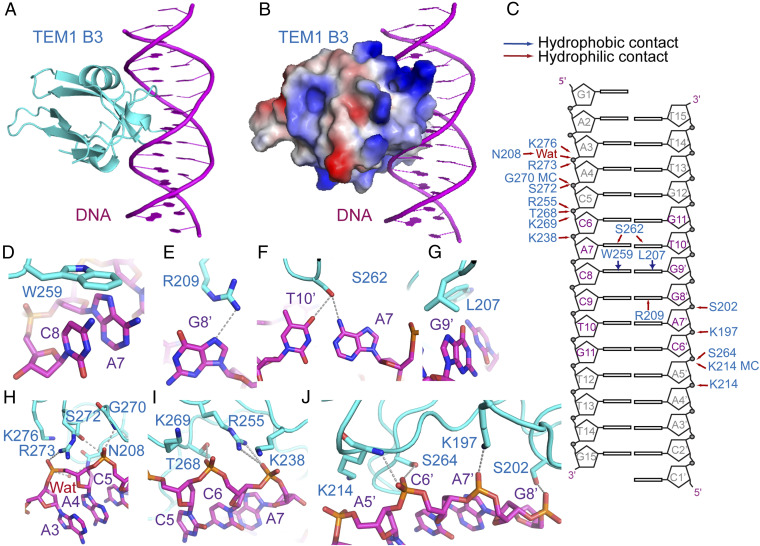
To investigate recognition of the FT gene 5′-UTR by the TEM1 B3 domain, we determined the structure of the TEM1–B3 in complex with an FT 5′-UTR–derived 14-bp DNA at 2.7-Å resolution (Figs. 1A and 2A and SI Appendix, Tables S1 and S2). There are two TEM1–B3 proteins and one DNA segment in an asymmetric unit, with one protein bound via base-specific interactions and the other one bound nonspecifically (SI Appendix, Fig. S2A). The 14-bp DNA segment is clearly traced in the crystal structure and adopts a classical B-form (SI Appendix, Fig. S2 B and C).
Fig. 2.
Crystal structure of the TEM1 B3–DNA complex. (A) The overall structure of TEM1 B3–DNA complex with the TEM1 B3 and DNA colored in cyan and magenta, respectively. (B) Electrostatic surface view of TEM1 B3, showing the positively charged DNA binding interface. (C) Schematic representation of the interactions between TEM1 B3 and DNA. Hydrophilic and hydrophobic contacts are labeled with red and blue arrows, respectively. The core recognition sequence is highlighted in magenta. MC, main chain. (D–J) Details of the interactions between TEM1 B3 and DNA. The interacting residues are highlighted by stick model, and hydrogen bonds are highlighted by dashed silver lines. D–G show base-specific interactions. H–J show backbone interactions.
The TEM1 B3 domain adopts a typical β-barrel–like structure flanked by two short α-helices, resembling other reported B3 domain proteins (23, 25–28) (Fig. 2A). The DNA binding interface of the B3 domain is highly positively charged, similar to the AP2 DNA binding interface (Fig. 2B). There are extensive backbone and base-specific interactions between the B3 domain and DNA (Fig. 2C). In particular, Trp259 forms hydrophobic stacking interactions with C8 of the DNA forward strand (Fig. 2 C and D). Arg209 forms hydrogen bonding and cation–π interactions with G8’ of the DNA reverse strand (Fig. 2 C and E). Ser262 forms hydrogen bonding interactions with A7 of the forward strand and T10’ of the reverse strand (Fig. 2 C and F). Leu207 forms hydrophobic stacking interactions with G9’ (Fig. 2 C and G). Many charged and hydrophilic residues including Lys197, Ser202, Lys214, Lys238, Arg255, Thr268, Lys269, and Lys276 contribute to hydrogen bonding and/or electrostatic interactions with the DNA backbone (Fig. 2 C and H–J). The TEM1 B3 domain has a DNA binding surface area of 917 Å2, which is larger than the AP2 domain. The larger DNA binding surface of B3 than AP2 domain results in more interactions, plausibly explaining our biochemical data that B3–DNA interaction is stronger than AP2–DNA interaction (Table 1).
TEM1 Binding Motifs Are Essential for FT Repression.
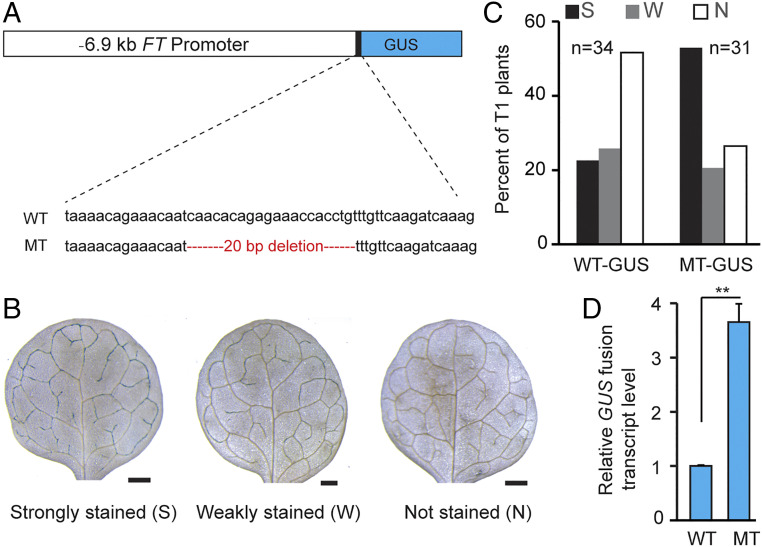
To determine whether the TEM1-binding cis-elements in the FT 5′-UTR are required for to TEM1-mediated transcriptional repression of FT, we constructed a transgene in which the coding region for the β-GLUCURONIDASE (GUS) reporter gene was driven by an FT promoter region. It has been previously shown that a genomic FT fragment containing a 5.7-kb FT promoter region is sufficient to rescue the late flowering of an ft null mutant (29). In this study, we fused a 6.9-kb FT promoter segment containing 5′-UTR with the reporter gene GUS (Fig. 3A), and the FT-GUS expression patterns are similar to what have been described in previous studies (29, 30). The FT promoter segment repressed GUS expression in the first true leaf vasculature, whereas removal of the TEM1 binding fragment (CAACACAGAGAAACCACCTG) resulted in GUS expression (Fig. 3 B–D), suggesting that the motifs function as FT-silencing elements.
Fig. 3.
A cis-regulatory region in FT 5′ UTR mediates FT repression in long days. (A) Illustration of the FT promoter (containing 5′ UTR) in fusion with the GUS reporter gene. The mutant (MT) fragment lacks a 20-bp sequence recognized by TEM1, consisting of putative AP2 and B3 binding motifs. WT, GUS driven by a WT promoter region. (B) Types of GUS staining of transgenic seedlings (T1, first generation) grown under long days. Shown are the seedlings expressing GUS driven by WT FT promoter (harvested at ZT12). (Scale bars, 0.1 mm.) (C) Analysis of GUS staining of transgenic seedlings. Total number (n) of T1 plants scored for each line is indicated. (D) Relative GUS messenger RNA (mRNA) levels in the indicated T1 transgenic plants grown in long days. A total of 25 to 30 independent T1 seedlings from each line were harvested at ZT12 and pooled for RNA extraction. A two-tailed Student's t test was conducted (**P < 0.01). Data are mean ± SD, n = 3 biological replicates.
TEM1 Recruits PRC2 to Deposit H3K27me3 on FT Chromatin.
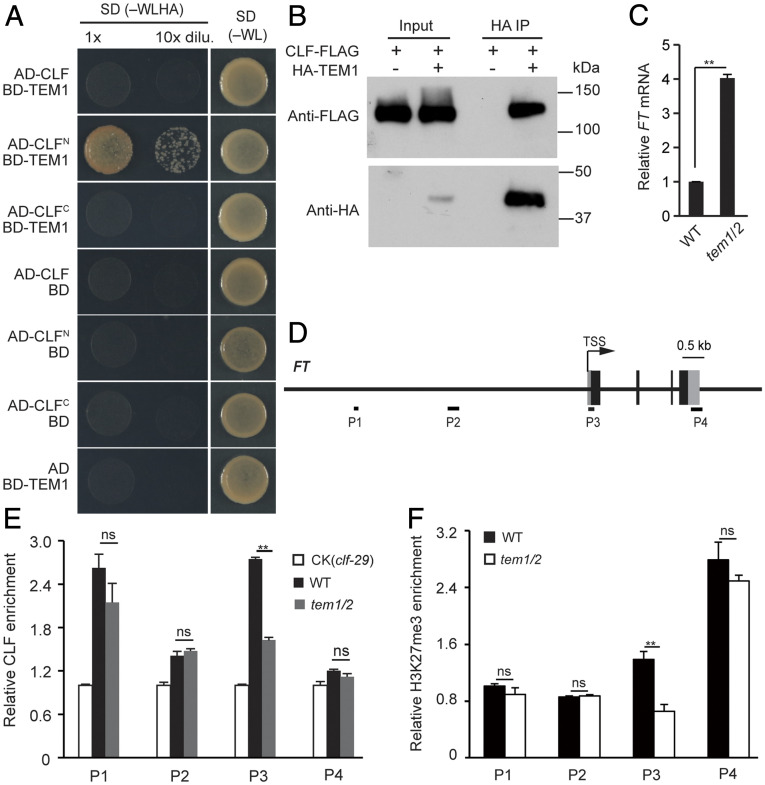
To explore the role of TEM proteins in FT repression, we used yeast two-hybrid assays to identify potential TEM-interacting partners and observed that TEM1 interacted with an N-terminal fragment of CURLY LEAF (CLF) (Fig. 4A). We further found that the AP2 domain mediated the interaction of TEM1 with CLF in yeast cells (SI Appendix, Fig. S3). CLF is the catalytic subunit of the PRC2, which deposits repressive H3K27me3 histone marks (31). PRC2 is known to suppress the FT gene expression (32–36). We confirmed the interaction by coimmunoprecipitation (CoIP) using proteins extracted from Nicotiana benthamiana leaves transiently coexpressing 35S:HA-TEM1 and 35S: CLF-FLAG plasmids. HA-tagged TEM1 was able to pull down the FLAG-tagged CLF, verifying that TEM1 associates with CLF to form a complex in vivo (Fig. 4B).
Fig. 4.
TEM1 recruits CLF-PRC2 to deposit H3K27me3 and repress FT expression. (A) TEM1 interacts with an N-terminal fragment of CLF (residues 1 to 556) in yeast cells. CLF and the full-length TEM1 were fused to the GAL4 AD and BD, respectively. Yeast cells were spotted onto a stringent selection medium. The synthetic dropout medium lacks Trp, Leu, His, and adenine (−WLHA) or a nonselective medium lacking Trp and Leu (−WL; control). (B) CoIP assay of TEM1 with CLF. HA-TEM1 was immunoprecipitated by anti-FLAG that recognizes CLF-FLAG. Nicotiana benthamiana leaves were coinfiltrated with mixed Agrobacteria cells containing 35S-HA-TEM1 or 35S-CLF-FLAG plasmids. (C) Relative FT expression level in WT and tem1/2 seedlings grown under long days. FT expression was normalized to TUBULIN2 (TUB2). (D) Schematic diagram of the FT locus. Arrow, transcription start site; gray boxes, UTR; filled black boxes, exons. (Scale bar, 0.5 kb.) ChIP-examined regions are indicated with black bars. The P3 region contains TEM1-recognized elements. (E) ChIP analysis of CLF enrichment at FT in wild-type (Col-0) and tem1/2 seedlings at ZT8. Genomic fragments immunoprecipitated by a rabbit polyclonal antibody recognizing the native CLF were quantified by qPCR and normalized to TUB2. The fold enrichments of CLF over CK (clf-29) in each examined region in WT or tem1/2 are shown. (F) ChIP analysis of H3K27me3 on FT chromatin in WT and tem1/2 seedlings at ZT8. Ratios of each examined FT region to the endogenous control TUB2 were shown. In C, E, and F, bars indicate SD of three biological replicates, and two-tailed Student’s t tests (**P < 0.01) were conducted (ns, not significant).
Consistent with TEM1 and TEM2 controlling flowering via transcriptional regulation of FT (10), we confirmed that there was significantly increased expression of FT in tem1/2 at midday (ZT8) under long day conditions compared to wild type (WT) (Fig. 4C). Given that TEM1 specifically binds the FT 5′-UTR and forms a protein complex with CLF, we asked whether CLF enrichment on FT chromatin depends on TEM proteins. Using chromatin immunoprecipitation (ChIP) with an antibody that recognizes the native CLF protein (26, 37), we observed that the TEM-binding region (P3 in Fig. 4D) has reduced levels of CLF in tem1/2 relative to WT (Fig. 4E and SI Appendix, Fig. S4), consistent with TEM1/2 recruiting CLF to the FT 5′-UTR. Furthermore, H3K27me3 levels at the TEM1 binding site were significantly reduced in the tem1/2 mutant relative to WT (Fig. 4F). Thus, the TEM genes are required for CLF-mediated H3K27me3 deposition at the FT 5′-UTR.
AP2 and B3 Domains Are Required for Floral Repression.
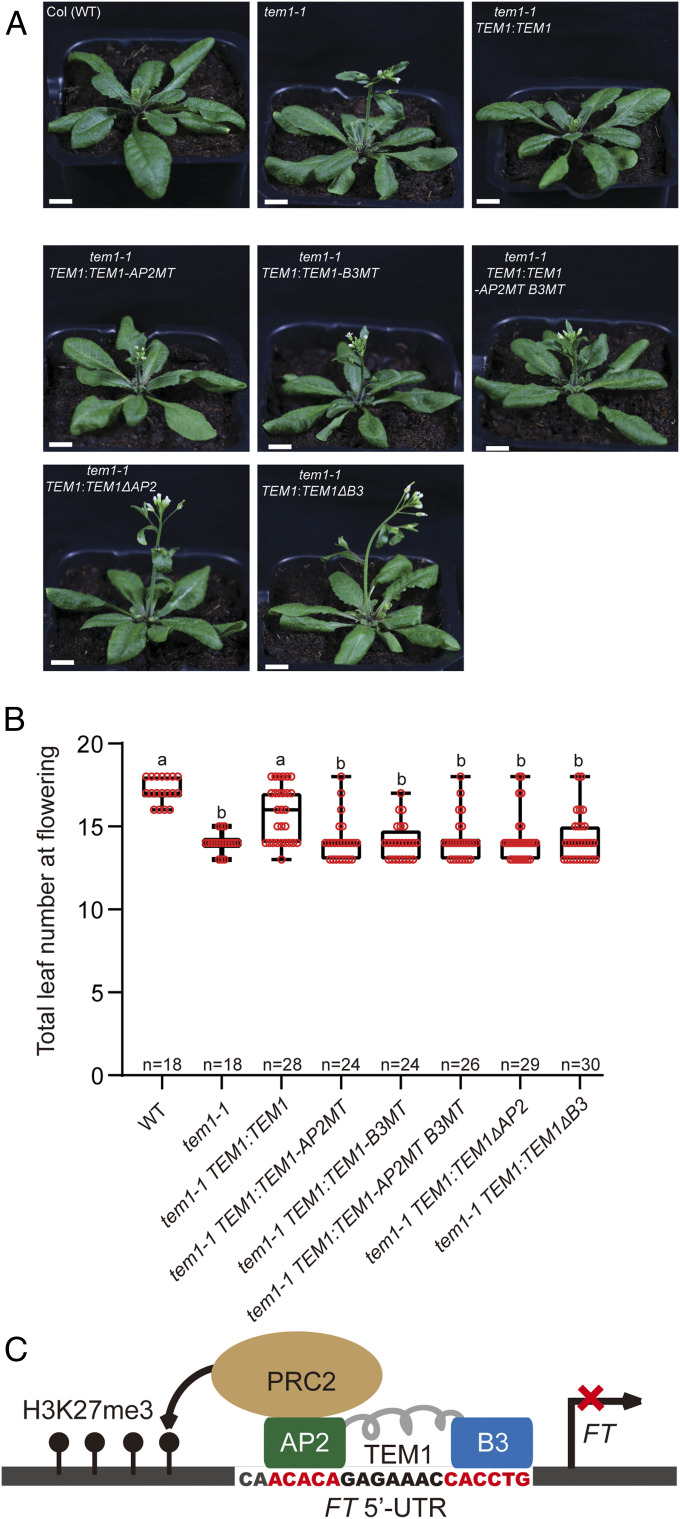
Based on our structures, we selected five key DNA-binding residues in the AP2 or B3 domains and mutated them to negatively charged residues to disrupt the DNA binding. Both the S202E/K214E/K238E/R255E/K269E B3 domain mutant (B3 pentamutation) and K73E/Y88E/R93E/R117E/R152E AP2 domain mutant (AP2 pentamutation) significantly decreased the DNA binding affinity of the corresponding domains (Table 1). In addition, TEM1 protein bearing the B3 pentamutions or AP2 pentamutions had substantially reduced DNA binding affinities (Table 1). TEM1 genes bearing B3 pentamutions (B3MT), AP2 pentamutions (AP2MT), B3/AP2 pentamutions (AP2MTB3MT), B3 deletion (ΔB3), and AP3 deletion (ΔAP2) were engineered into a TEM1 construct (pTEM1:TEM1) and transformed into a tem1-1 mutant background (Fig. 5A). Transgenic plants carrying TEM1 with DNA-binding domain mutations or TEM1 with domain deletion were unable to complement the tem1-induced early flowering phenotype, compared to a full rescue of tem1 by WT TEM1 (Fig. 5 A and B). Thus, the TEM1 AP2 and B3 domains are critical for flowering regulation.
Fig. 5.
AP2 and B3 domains are critical for TEM1-mediated floral repression. (A) Phenotypes of Col, tem1, and the indicated transgenic lines. TEM1:TEM1, WT TEM1; TEM1:TEM1-AP2MT, AP2 domain mutated; TEM1:TEM1-B3MT, B3 domain mutated; TEM1:TEM1-AP2MT B3MT, mutations in both AP2 and B3 domains; TEM1:TEM1∆AP2, AP2 domain deleted; and TEM1:TEM1∆B3, B3 domain deleted. (Scale bars, 1 cm.) (B) Box plots of the flowering times of the indicated lines. Number (n) of T1 plants scored for each line is indicated. Flowering time was measured by the number of total leaves at flowering. Letters mark statistically distinct groups (one-way ANOVA, P < 0.01). (C) A molecular model for FT regulation by TEM1. TEM1, via the AP2 and B3 domains, specifically recognizes FT 5′-UTR and recruits PRC2 to deposit the repressive H3K27me3 mark to suppress FT expression.
Discussion
AP2 domain proteins are a large group of plant-specific transcription factors. In this study, we demonstrate that the TEM1 AP2 domain recognizes a core ACA motif. The presence of tandem ACA motifs in the FT 5′-UTR increases AP2–DNA binding slightly (Table 1). We compared the TEM1 AP2–CAACAC complex with the AtERF1–DNA structure [Protein Data Bank (PDB): 1GCC]. Both AP2 domains adopt a similar topology (SI Appendix, Fig. S5A) (17). In the two structures, most of the key residues participating in base-specific interactions occupy very similar positions (SI Appendix, Fig. S5 A and B). In the antiparallel β-sheets β-1 and β-2, base-interacting residues are highly mutable, resulting in distinct sequence preferences among the different AP2 subfamilies. For example, while ERF1 Trp154 interacts with both the DNA backbone and DNA base, the Asn80 of TEM1 at the equivalent position only recognizes the DNA backbone (SI Appendix, Fig. S5 A and B) (17). However, DNA-contacting residues in β-3 are highly conserved (SI Appendix, Fig. S5 A and B). Thus, selective changes in β-1 and β-2 are responsible for the distinct DNA sequence preference of AP2 domains.
The TEM1 B3 domain belongs to the RAV subfamily of B3 domains that recognize the CACCTG motif. This subfamily includes TEM2, RAV1, RAV1-like, RAV3, RAV3-like, NGA1/2/3/4, NGAL1 (AT2G36080), NGAL2 (AT3G11580), and NGAL3 (AT5G06250) (18). Our TEM1 B3–DNA complex represents a protein–DNA complex structure in this subfamily, enabling us to analyze the basis of their sequence specificity. We compared TEM1 B3 with our recently reported VAL1 B3 structure, which belongs to the TGCATG-specific LAV subfamily (26). TEM1 B3 has an overall similar topology with VAL1 B3, with an RMSD of 1.4 Å. However, the bound DNA is shifted slightly between TEM1 and VAL1 (SI Appendix, Fig. S5C). In both the VAL1 B3–DNA and TEM1 B3–DNA structures, α-1 to β-2 and β-4 and β-5 bind the DNA major groove (SI Appendix, Fig. S5C). However, in the TEM1 B3–DNA structure, α-3 also interacts with the DNA backbone, which results in the shift in DNA orientation. On the other side of the major groove, the antiparallel β-sheets β-4 and β-5 of TEM1 B3 are shorter than β-4 and β-5 in VAL1 B3, resulting in an altered DNA recognition pattern in this region and therefore a different recognition sequence (SI Appendix, Fig. S5 C and D). The structure-based sequence alignment of the RAV subfamily B3 domains indicates that all the base-interacting residues are highly conserved, suggesting the TEM1 B3 domain and other members in this subfamily will have a similar sequence preference (SI Appendix, Fig. S5E).
Unlike prokaryotic transcription factors, which achieve sequence specificity by binding long cis-elements, eukaryotic transcription factors bind short cis-elements (38). Aligning multiple short cis-elements may help eukaryotic transcription factors achieve sufficient specificity to mediate sequence-specific regulation. In TEM1, the presence of both an AP2 and B3 domain overcomes the relatively limited specificity of a single short cis-binding sequence motif. In addition, although AP2 binds to the target DNA with a ∼45-fold lower affinity compared to the B3 domain, AP2 and B3 together bind in a combinatorial manner to the FT 5′-UTR, thereby increasing overall binding affinity ∼11-fold compared to the single B3 domain (Table 1). Recently, we reported that the flowering regulator CO may form a multimeric assembly to bind to multiple cis-elements in the FT promoter (39), suggesting that this type of multimeric binding toward multiple cis-elements may be a common mechanism by which eukaryotic transcription factors are able to achieve high binding affinity and specificity. Epigenetic modifiers often show less-specific locus targeting and more-dynamic activity than eukaryotic transcription factors. Here, we showed that the transcription factor TEM1 bridges these two processes, targeting epigenetic modifiers to regulate limited gene(s) in a specific biological process. Overall, we suggest that TEM1 targets the FT 5′-UTR through its AP2 and B3 domains and recruits PRC2 to deposit repressive H3K27me3 histone marks to suppress FT expression and the floral transition (Fig. 5C).
Materials and Methods
Details are provided in SI Appendix, SI Materials and Methods, including protein expression and purification, crystallization and structure determination, in vitro protein–DNA binding assay, plant materials and growth conditions, yeast two-hybrid analysis, plasmid construction, ChIP, CoIP, histological analysis of GUS staining, and RNA analysis.
Supplementary Material
Acknowledgments
We thank the staffs at beamline BL19U1 at the Shanghai Synchrotron Radiation Facility for assistance during data collection and Dr. Guy Riddihough (Life Science Editors) for text editing. This work was supported by the Shenzhen Science and Technology Program (JCYJ20200109110403829 and KQTD20190929173906742 to J.D.), the National Natural Science Foundation of China (31721001 to Y.H.), and Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes (2019KSYS006 to J.D.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103895118/-/DCSupplemental.
Data Availability
X-ray structure data have been deposited in Research Collaboratory for Structural Bioinformatics PDB (7ET5, 7ET4, and 7ET6).
References
- 1.Turck F., Fornara F., Coupland G., Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594 (2008). [DOI] [PubMed] [Google Scholar]
- 2.McKim S. M., How plants grow up. J. Integr. Plant Biol. 61, 257–277 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Boss P. K., Bastow R. M., Mylne J. S., Dean C., Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 16 (suppl.), S18–S31 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo X., He Y., Experiencing winter for spring flowering: A molecular epigenetic perspective on vernalization. J. Integr. Plant Biol. 62, 104–117 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Bao S., Hua C., Shen L., Yu H., New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 62, 118–131 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Song Y. H., Shim J. S., Kinmonth-Schultz H. A., Imaizumi T., Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D. H., Doyle M. R., Sung S., Amasino R. M., Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25, 277–299 (2009). [DOI] [PubMed] [Google Scholar]
- 8.He Y., Chen T., Zeng X., Genetic and epigenetic understanding of the seasonal timing of flowering. Plant Commun. 1, 100008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillejo C., Pelaz S., The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Osnato M., Castillejo C., Matías-Hernández L., Pelaz S., TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3, 808 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Marín-González E., et al., SHORT VEGETATIVE PHASE up-regulates TEMPRANILLO2 floral repressor at low ambient temperatures. Plant Physiol. 169, 1214–1224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sgamma T., Jackson A., Muleo R., Thomas B., Massiah A., TEMPRANILLO is a regulator of juvenility in plants. Sci. Rep. 4, 3704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilar-Jaramillo A. E., et al., TEMPRANILLO is a direct repressor of the microRNA miR172. Plant J. 100, 522–535 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Ikeda M., Ohme-Takagi M., A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50, 970–975 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Collins J., O’Grady K., Chen S., Gurley W., The C-terminal WD40 repeats on the TOPLESS co-repressor function as a protein-protein interaction surface. Plant Mol. Biol. 100, 47–58 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Yamasaki K., Kigawa T., Seki M., Shinozaki K., Yokoyama S., DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 18, 267–276 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Allen M. D., Yamasaki K., Ohme-Takagi M., Tateno M., Suzuki M., A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swaminathan K., Peterson K., Jack T., The plant B3 superfamily. Trends Plant Sci. 13, 647–655 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Nag R., Maity M. K., Dasgupta M., Dual DNA binding property of ABA insensitive 3 like factors targeted to promoters responsive to ABA and auxin. Plant Mol. Biol. 59, 821–838 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Braybrook S. A., et al., Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 3468–3473 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reidt W., et al., Gene regulation during late embryogenesis: The RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 21, 401–408 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Yuan W., et al., A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 48, 1527–1534 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Boer D. R., et al., Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Kagaya Y., Ohmiya K., Hattori T., RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King G. J., et al., The Arabidopsis B3 domain protein VERNALIZATION1 (VRN1) is involved in processes essential for development, with structural and mutational studies revealing its DNA-binding surface. J. Biol. Chem. 288, 3198–3207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao Z., et al., Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nat. Plants 5, 424–435 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Sasnauskas G., Kauneckaite K., Siksnys V., Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1. Nucleic Acids Res. 46, 4316–4324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu B., et al., Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 501, 415–422 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Adrian J., et al., cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22, 1425–1440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takada S., Goto K., Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu J., Chen C., Li C., Cui Y., The complexity of PRC2 catalysts CLF and SWN in plants. Biochem. Soc. Trans. 48, 2779–2789 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Goodrich J., et al., A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Schubert D., et al., Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25, 4638–4649 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang D., Wang Y., Wang Y., He Y., Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One 3, e3404 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X., et al., Temporal-specific interaction of NF-YC and CURLY LEAF during the floral transition regulates flowering. Plant Physiol. 177, 105–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo X., et al., The NUCLEAR FACTOR-CONSTANS complex antagonizes Polycomb repression to de-repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J. 95, 17–29 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Zeng X., et al., HISTONE DEACETYLASE 9 functions with Polycomb silencing to repress FLOWERING LOCUS C expression. Plant Physiol. 182, 555–565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wunderlich Z., Mirny L. A., Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 25, 434–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv X., et al., Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex. Plant Cell 33, 1182–1195 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
X-ray structure data have been deposited in Research Collaboratory for Structural Bioinformatics PDB (7ET5, 7ET4, and 7ET6).