Significance
N6-methyladenosine (m6A) RNA methylation is the most-abundant modification in messenger RNA and noncoding RNA in eukaryotic cells. However, the regulatory and functional role of m6A in response to DNA damage remains poorly understood. Here, using the combination of in vitro models and mice with skin-specific genetic deletion of the key m6A writer factor METTL14, we have demonstrated a crucial role of METTL14 in global genome repair and ultraviolet B (UVB) radiation–induced skin tumorigenesis. Such a role of METTL14 may serve as an emerging epitranscriptomic mechanism to coordinate RNA metabolism and UVB damage response in tumorigenesis. We anticipate that these findings may be translated into potential strategies to improve genome stability and cancer prevention by targeting the METTL14 pathway.
Keywords: METTL14, m6A RNA methylation, nucleotide excision repair, UVB, ultraviolet radiation
Abstract
Global genome repair (GGR), a subpathway of nucleotide excision repair, corrects bulky helix-distorting DNA lesions across the whole genome and is essential for preventing mutagenesis and skin cancer. Here, we show that METTL14 (methyltransferase-like 14), a critical component of the N6-methyladenosine (m6A) RNA methyltransferase complex, promotes GGR through regulating m6A mRNA methylation–mediated DDB2 translation and suppresses ultraviolet B (UVB) radiation-induced skin tumorigenesis. UVB irradiation down-regulates METTL14 protein through NBR1-dependent selective autophagy. METTL14 knockdown decreases GGR and DDB2 abundance. Conversely, overexpression of wild-type METTL14 but not its enzymatically inactive mutant increases GGR and DDB2 abundance. METTL14 knockdown decreases m6A methylation and translation of the DDB2 transcripts. Adding DDB2 reverses the GGR repair defect in METTL14 knockdown cells, indicating that METTL14 facilitates GGR through regulating DDB2 m6A methylation and translation. Similarly, knockdown of YTHDF1, an m6A reader promoting translation of m6A-modified transcripts, decreases DDB2 protein levels. Both METTL14 and YTHDF1 bind to the DDB2 transcript. In mice, skin-specific heterozygous METTL14 deletion increases UVB-induced skin tumorigenesis. Furthermore, METTL14 as well as DDB2 is down-regulated in human and mouse skin tumors and by chronic UVB irradiation in mouse skin, and METTL14 level is associated with the DDB2 level, suggesting a tumor-suppressive role of METTL14 in UVB-associated skin tumorigenesis in association with DDB2 regulation. Taken together, these findings demonstrate that METTL14 is a target for selective autophagy and acts as a critical epitranscriptomic mechanism to regulate GGR and suppress UVB-induced skin tumorigenesis.
Genomic stability is constantly challenged by damage to DNA from endogenous and exogenous genotoxic stresses. To preserve their genome integrity, cells are equipped with multiple DNA repair machineries to remove specific DNA damage lesions. In particular, nucleotide excision repair (NER) is a versatile DNA repair pathway that eliminates a broad range of helix-distorting DNA lesions induced by genotoxic exposures, including solar ultraviolet (UV) radiation and air pollutants (1–7).
In mammalian cells, canonical NER consists of two specialized pathways: global genome repair (GGR) that repairs damaged bases throughout the entire genome and transcription-coupled repair (TCR) that removes lesions on the transcribed DNA strand of active genes (4, 5, 8–10). One major difference between these two repair pathways is in the phenotypes of human patients possessing defects of either repair pathway. Defects in GGR in affected individuals can cause xeroderma pigmentosum (XP), an autosomal recessive disorder predisposing affected individuals to cancer development not only in the skin but also in the brain and lungs, while defects in TCR can cause premature aging and developmental retardation (4, 5, 9, 11). Another major difference is in the repair factors employed in DNA damage recognition. Among all the indispensable GGR factors (XPA-XPG) identified, DDB2 (also known as XPE) and XPC are required for damage recognition in GGR but not in TCR (4, 12). DDB2 is a cofactor of the CUL4A–RING ubiquitin ligase (CRL4) complex, which comprises the DDB1 (DNA damage-binding protein 1), DDB2, CUL4A (Cullin-family E3-ligase adaptor protein), and ROC1 (E3-ligase RING domain) subunits. This complex, with DDB2, induces ubiquitination and activation of XPC following DNA damage (13–15). In addition, DDB2 mediates chromatin decondensation, independent of the CRL4 complex (16). The expression and activity of DDB2 and other NER factors are regulated by transcription, posttranslational protein modifications, and protein degradation (17–23).
One emerging mechanism of gene expression regulation is chemical RNA modifications at the posttranscriptional level. Among those modifications, N6-methyladenosine (m6A) RNA methylation is the most abundant modification in messenger RNA (mRNA) and noncoding RNA in eukaryotic cells (24–26). m6A modification is installed on the RNA by the writer complex composed of factors including METTL3, METTL14, WTAP, and KIAA1429, and removed by the erasers FTO or ALKBH5 (24). Analogous to the reversible epigenetic modifications to DNA and histone, posttranscriptional m6A methylation of adenosines in RNA controls RNA’s fate and its functions, such as mRNA stability, nuclear processing, translation, RNA–protein interactions, and gene transcription (27–33). At the cellular and organism level, it modulates development, stem cell homeostasis, and response to stresses such as heat-shock, genotoxins, and control of the circadian clock (25, 34–38). Recently, Xiang and colleagues showed that m6A RNA methylation is activated at DNA damage sites to promote noncanonical NER through polymerase κ in A375 and U2OS cells specifically in the S/G2/M phase of the cell cycle in response to UVC and UVA radiation (39). However, the role of METTL14 and m6A in canonical NER including GGR and the mechanism for METTL14 regulation under the relevant carcinogenic ultraviolet B (UVB) stress remain unclear.
In this study, we have demonstrated the regulatory and functional role of METTL14 in GGR of UVB-induced DNA damage and skin tumorigenesis and the underlying mechanism. We found that METTL14 regulates GGR, suppresses tumorigenesis, and is degraded through NBR1-dependent selective autophagy. Our results provide a mechanistic link between m6A RNA methylation and GGR in epithelial tumorigenesis.
Results
UVB Down-Regulates METTL14 through Autophagy.
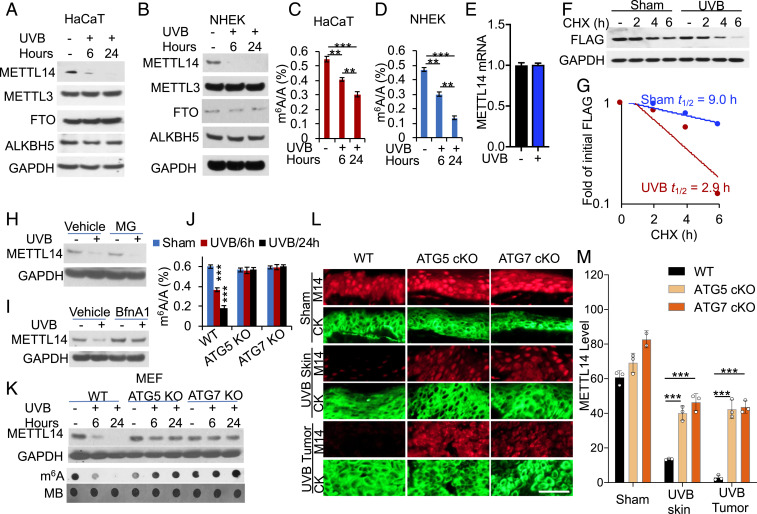
To determine the role of METTL14 in UVB damage response, we assessed whether UVB irradiation regulates METTL14 abundance in keratinocytes. In both HaCaT (human keratinocytes) and normal human epidermal keratinocyte (NHEK) cells, UVB down-regulated METTL14, while it had no effect on other m6A regulators, such as METTL3, FTO, or ALKBH5 (24) (Fig. 1 A and B). Similarly, UVB irradiation decreased the m6A/A ratio by ultra–high-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) assay in the polyadenylated RNAs and the m6A levels in total RNA by dot blot assay in both HaCaT and NHEK cells (Fig. 1 C and D and SI Appendix, Fig. S1 A and B).
Fig. 1.
UVB induces METTL14 down-regulation through autophagy. (A and B) Immunoblot analysis of METTL14, METTL3, FTO, ALKBH5, and GAPDH in sham- or UVB-irradiated HaCaT cells (A) and NHEK cells (B) at 6 and 24 h post-UVB (20 and 30 mJ/cm2 for HaCaT and NHEK, respectively). (C and D) Quantification of the m6A/A ratios in polyadenylated RNA by UHPLC-MS/MS in HaCaT (C) and NHEK cells (D) at 6 or 24 h post–UVB- or sham-irradiation (n = 3). (E) qPCR analysis of METTL14 mRNA levels in sham- or UVB-irradiated HaCaT cells at 24 h post-UVB (mean ± SD, n = 6). (F) Immunoblot analysis of FLAG (METTL14) in HaCaT cells transfected with METTL14-FLAG, followed by UVB irradiation and subsequent treatment with cycloheximide (CHX) over a time course. (G) Quantification of F. The half-life of METTL14-FLAG is indicated. (H and I) Immunoblot analysis of METTL14 and GAPDH in sham- or UVB-irradiated HaCaT cells treated with or without MG132 (MG, 10 μM) (H) or bafilomycin A1 (BfnA1, 50 ng/mL) (I) at 24 h post-UVB. (J) Quantification of the m6A/A ratios in polyadenylated RNA by LC-MS/MS in in WT, ATG5 KO, and ATG7 KO MEF cells at 6 or 24 h post–UVB- or sham-irradiation (n = 3). (K) Immunoblot analysis of METTL14 and GAPDH and dot blot analysis of m6A levels in cells as in K. (L) Immunofluorescence analysis of METTL14 (red) in mouse skin treated with (UVB Skin) or without (Sham) chronic UVB irradiation or UVB-induced skin tumors (UVB Tumor) from mice with WT or skin-specific deletion of ATG5 (ATG5 cKO) or ATG7 (ATG7 cKO). Cytokeratin (CK, green) is used as a keratinocyte marker. (Scale bar, 50 μm.) (M) Quantification of METTL14 protein levels in the epidermis in L. WT: n = 3 for sham and UVB groups. ATG5 cKO: n = 3 for sham and UVB groups). ATG7 cKO: n = 2 for sham group, n = 3 for UVB group. **P < 0.05; ***P < 0.001; Student’s t test (M).
Next we determined the mechanism by which UVB down-regulates METTL14. We found that UVB irradiation has no effect on METTL14 mRNA levels (Fig. 1E). Instead, it decreased the protein stability of METTL14 (Fig. 1 F and G). Then, we assessed the role of two critical protein degradation pathways, the proteasomal and the autophagic–lysosomal pathways (40–42). The proteasome inhibitor MG132 (MG) had no effect on UVB-induced METTL14 down-regulation (Fig. 1H). In contrast, inhibition of the lysosomes by bafilomycin A1 (BfnA1) blocked UVB-induced METTL14 down-regulation (Fig. 1I). Emerging evidence has shown that autophagy can be induced by a variety of physiological and pathological stresses (43), including UV radiation (44, 45); thus, we reckon that UV-induced autophagy may mediate METTL14 down-regulation. Indeed, in mouse embryonic fibroblast (MEF) cells, genetic inhibition of autophagy by deletion of the essential autophagy gene ATG5 or ATG7 inhibited UVB-induced down-regulation of METTL14 and m6A enrichment (Fig. 1 J and K). Genetic inhibition of autophagy by deleting the essential autophagy genes ATG5 or ATG7 in mouse skin inhibited UVB-induced METTL14 down-regulation in nontumor skin and skin tumors (Fig. 1 L and M and SI Appendix, Fig. S1C). These findings demonstrate that UVB down-regulates METTL14 through autophagy.
NBR1 Mediates METTL14 Autophagic Degradation under Homeostatic Condition and UVB Stress.
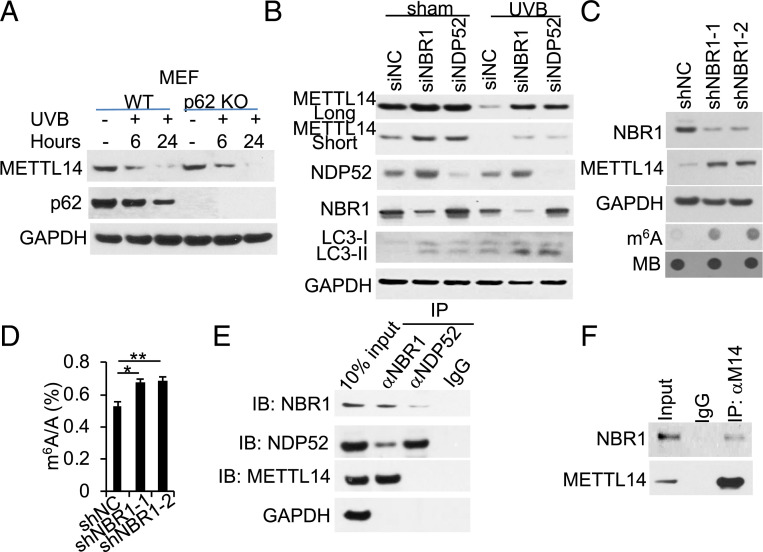
To determine the autophagy receptor for UVB-induced METTL14 down-regulation, we first assessed the role of the autophagy receptor p62 (also known as SQSTM1) (46, 47). In MEF cells, p62 deletion had no effect on UVB-induced METTL14 down-regulation (Fig. 2A), suggesting a role for other selective autophagy receptors. We next focused on NBR1 and NDP52 (12, 48). Indeed, knockdown of either NBR1 or NDP52 increased basal METTL14 levels and inhibited UVB-induced down-regulation of METTL14 (Fig. 2B and SI Appendix, Fig. S2). Knockdown of NBR1 increased the protein levels of METTL14 and m6A enrichment (Fig. 2 C and D). Coimmunoprecipitation analysis showed that METTL14 binds with NBR1 but not NDP52 (Fig. 2E) and that NBR1 binds with METTL14 (Fig. 2F). Future investigation is needed to assess whether NBR1 also interacts with other proteins in the m6A methyltransferase complex. These findings demonstrate that autophagy down-regulates METTL14 abundance and m6A enrichment through NBR1.
Fig. 2.
NBR1 binds to METTL14 and mediates METTL14 autophagic degradation. (A) Immunoblot analysis of METTL14, p62, and GAPDH in WT and p62 KO MEF cells. (B) Immunoblot analysis of METTL14, NDP52, NBR1, LC3-I/II, and GAPDH in HaCaT cells with small interfering RNA (siRNA) targeting negative control (siNC), NBR1 (siNBR1, Santa Cruz), or NDP52 (siNDP52, Santa Cruz). (C) Immunoblot analysis of METTL14, NBR1, and GAPDH and dot blot analysis of m6A levels in HaCaT cells stably infected with short hairpin RNA (shRNA) targeting negative control (shNC) or NBR1 (shNBR1, two independent shRNAs). (D) Quantification of the m6A/A ratios in polyadenylated RNA by LC-MS/MS in cells as in C (n = 3). (E) Immunoblot analysis of NBR1, NDP52, or METTL14, following immunoprecipitation using control species-matched IgG, anti-NBR1, or anti-NDP52 antibody in HaCaT cells. (F) Immunoblot analysis of NBR1 or METTL14, following immunoprecipitation using control species-matched IgG or anti-METTL14 (M14) in HaCaT cells. *P < 0.05; **P < 0.01; Student’s t test, significant differences.
METTL14 Regulates UVB-Induced DNA Damage Repair through m6A.
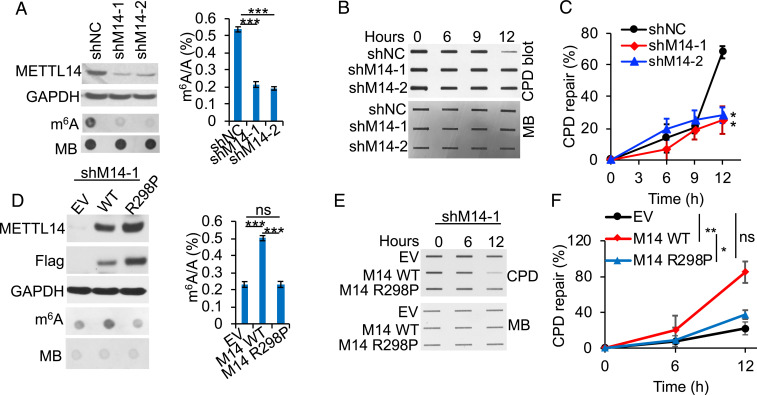
To determine the role of METTL14 in UVB-induced DNA damage repair, we compared the differences in repair of UVB-induced cyclobutene pyrimidine dimers (CPDs), the main tumorigenic DNA damage products induced by UVB radiation (49), in control and METTL14-inhibited HaCaT keratinocytes. METTL14 knockdown decreased the m6A levels and CPD repair (Fig. 3 A–C). Overexpression of wild-type (WT) METTL14, but not mutant METTL14 [R298P, loss of METTL14’s target recognition function for the m6A methyltransferase complex (50)] increased the m6A levels and CPD repair (Fig. 3 D–F). These results demonstrate that METTL14 regulates UVB damage repair through m6A modification.
Fig. 3.
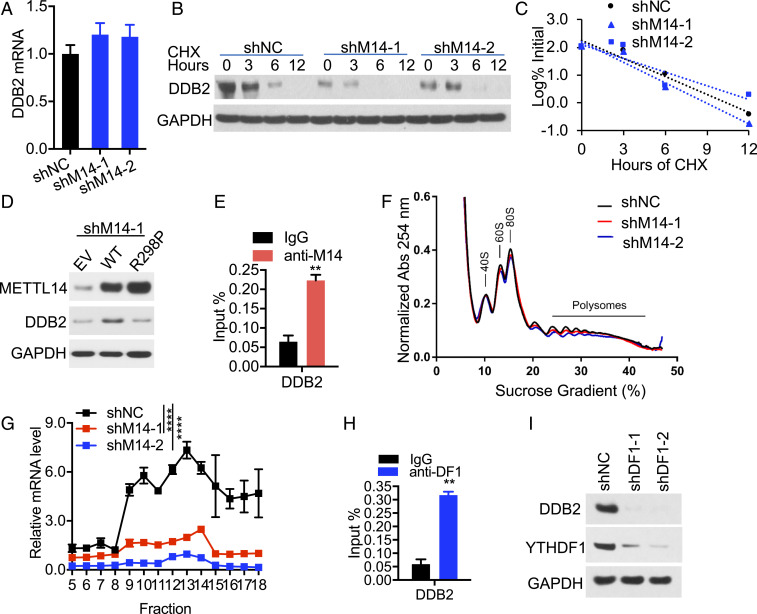
METTL14 inhibition suppresses repair of UVB-induced DNA damage. (A) Immunoblot analysis confirming METTL14 knockdown, dot blot assay of the m6A levels (Left), and quantification of the m6A/A ratios in polyadenylated RNA by UHPLC-MS/MS (Right, n = 3) in HaCaT cells stably infected with shRNA targeting negative control (shNC) or METTL14 (shM14-1, shM14-2). (B) Slot blot analysis of UVB-induced CPDs in control or METTL14-knockdown HaCaT at different time points post-UVB irradiation (20 mJ/cm2). (C) Quantification of B (mean ± SE, n = 3). (D) Immunoblot analysis confirming overexpression of WT METTL14 or R298P mutant METTL14, dot blot assay of the m6A levels (Left), and quantification of the m6A/A ratios in polyadenylated RNA by LC-MS/MS (Right, n = 3) in HaCaT cells transfected with empty vector (EV), WT, or R298P mutant METTL14. (E) Same as B except that cells in D were used. (F) Quantification of E (mean ± SE, n = 3). Methylene blue (MB) was used as an equal loading control for total RNA (A and D) and DNA (B and E). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant; Student’s t test, significant differences from shNC (C) or EV (F) groups.
METTL14 Facilitates UVB Damage Repair through Regulating DDB2 Abundance.
To determine the molecular target of METTL14 in regulating UVB-induced DNA damage repair, we performed immunoprecipitation (IP) of m6A-methylated transcripts followed by sequencing (51) to determine the transcriptome-wide transcript targets for METTL14-mediated m6A RNA methylation. In cells with or without either METTL14 knockdown or UVB irradiation, sequence analysis of m6A peaks showed the previously identified m6A target sites (GGACU) (51) (SI Appendix, Fig. S3A). m6A peaks detected in poly(A)+-enriched RNAs showed reproducible patterns of methylation (SI Appendix, Fig. S3B). Either METTL14 knockdown or UVB irradiation decreased the number of m6A-hypomethylated peaks (SI Appendix, Fig. S4 A and B). Gene ontology analysis of the hypomethylated peaks showed a number of pathways affected by either METTL14 knockdown or UVB irradiation (SI Appendix, Fig. S4 C and D).
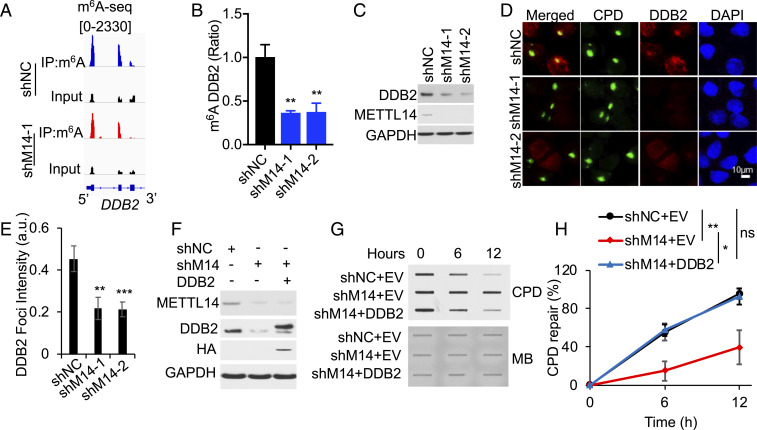
Next, we reckoned that METTL14-mediated m6A methylation may regulate the expression of an essential GGR factor. Indeed, we found that METTL14 knockdown remarkably decreased the m6A enrichment across the transcript of the key GGR factor DDB2, in particular in the first three exons (Fig. 4A, SI Appendix, Fig. S5 A and B). m6A IP qPCR analysis confirmed that METTL14 knockdown significantly reduced the m6A level of the DDB2 transcript (Fig. 4B) and DDB2 protein level (Fig. 4C). A microirradiation assay demonstrated that METTL14 knockdown reduced the recruitment of DDB2 to subnuclear UVB-induced CPD sites (Fig. 4 D and E). In METTL14 knockdown cells, repletion of DDB2 to a level similar to control cells reversed CPD repair defects in METTL14 knockdown cells (Fig. 4 F–H). These findings demonstrate that DDB2 is the critical molecular target responsible for METTL14’s function in GGR.
Fig. 4.
METTL14 facilitates UVB damage repair through regulating the m6A enrichment and availability of DDB2. (A) Distribution of m6A peaks in the first three exons of the DDB2 transcript in HaCaT cells with or without METTL14 knockdown. (B) qPCR analysis of the m6A levels of the DDB2 transcript following m6A IP (mean ± SD, n = 3). (C) Immunoblot analysis of DDB2, METTL14, and GAPDH in shNC or shM14-1/2 HaCaT cells. (D) Immunofluorescence assay of the colocalization of DDB2 with subnuclear CPD in HaCaT cells as in A at 15 min post-UV (10 mJ/cm2) through a 5-mm micropore filter. (E) The relative intensity of DDB2 was calculated by analyzing ≥50 foci and normalized to that of CPD (mean ± SE, n = 3). (F) Immunoblot analysis of DDB2, HA (DDB2), METTL14, and GAPDH in shNC or shM14-1/2 HaCaT cells transfected with empty vector or DDB2-HA. (G) Slot blot analysis of UVB-induced CPDs in cells as in F. (H) Quantification of G (mean ± SE, n = 3). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant; Student’s t test, significant differences from shNC (B, E) or comparison groups (H).
METTL14 Regulates DDB2 Translation through YTHDF1.
Next, we investigated the mechanism by which METTL14 regulates DDB2. We found that METTL14 knockdown had no effect on DDB2 mRNA levels (Fig. 5A) or protein stability (Fig. 5 B and C). Overexpression of WT METTL14 but not R298P mutant increased DDB2 protein level (Fig. 5D), indicating that METTL14 regulates DDB2 via METTL14’s target recognition function of the m6A methyltransferase complex. Using crosslinking and immunoprecipitation (CLIP) assay, we showed that METTL14 binds to the DDB2 transcript (Fig. 5E). To determine whether METTL14 regulates DDB2 translation, we carried out polysome profiling analysis. We found that knockdown of METTL14 decreased the level of DDB2 mRNA in the translating pool (Fig. 5 F and G, fractions 9 through 18), indicating that METTL14 facilitates DDB2 translation.
Fig. 5.
METTL14 regulates DDB2 translation through YTHDF1. (A) qPCR analysis of DDB2 mRNA levels in GAPDH in shNC or shM14-1/2 HaCaT cells (mean ± SD, n = 6). (B) Immunoblot analysis of DDB2 and GAPDH in shNC or shM14-1/2 HaCaT cells treated with or without cycloheximide (CHX, 100 μg/mL) over a time course. (C) Quantification of B. (D) Immunoblot analysis of METTL14, DDB2, and GAPDH in HaCaT cells transfected with empty vector (EV), WT, or R298P mutant METTL14. (E) CLIP-qPCR analysis for binding of METTL14 to the DDB2 transcript (mean ± SD, n = 3). (F and G) Polysome profiling assays in fractionations of HaCaT cell lysates (F) and qPCR analysis of DDB2 mRNA levels in different fractions of ribosomes (G, mean ± SD, n = 3). (H) CLIP-qPCR analysis for binding of YTHDF1 to the DDB2 transcript (mean ± SD, n = 3). (I) Immunoblot analysis of DDB2 and YTHDF1 in shNC or shDF1-1/2 HaCaT cells. **P < 0.01; ****P < 0.0001; Student’s t test, significant differences from IgG (E and H) or shNC groups (G).
Next, we investigated how METTL14 regulates DDB2 translation. Since the effect of m6A on translation is mediated through its readers, we assessed the role of YTHDF1, an m6A reader that promotes translation of m6A-modified transcripts (29). CLIP analysis showed that YTHDF1 binds to the DDB2 transcript (Fig. 5H). Furthermore, knockdown of YTHDF1 decreased protein levels of DDB2 (Fig. 5I). YTHDF1 knockdown had no effect on the other YTHDF proteins including YTHDF2 or YTHDF3 (SI Appendix, Fig. S6A). YTHDF1-3 protein levels were not affected by METTL14 knockdown (SI Appendix, Fig. S6B). These findings reveal that METTL14 regulates DDB2 translation through YTHDF1.
METTL14 Acts as a Tumor Suppressor in UVB-Induced Skin Cancer.
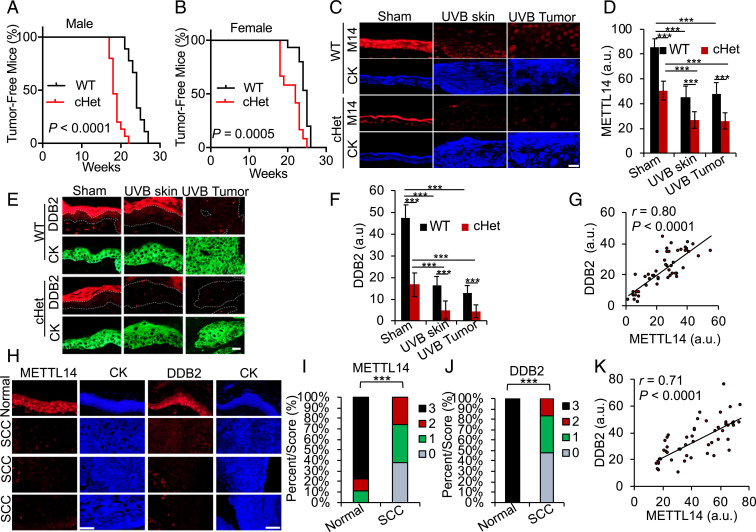
Because of notable involvement of METTL14 in UVB damage repair, we last investigated the potential roles of METTL14 in UVB-induced skin cancer. To determine the specific role of METTL14 in skin tumorigenesis, we assessed the difference in tumor onset between WT mice and mice with epidermis-specific METTL14 deletion. METTL14 global deletion is embryonic lethal (52). Intriguingly, skin-specific homozygous METTL14 deletion in the SKH-1 background was also pre- or postnatally lethal, since we did not obtain mice with homozygous skin-specific METTL14 deletion (conditional knockout; cKO; SI Appendix, Fig. S7), indicating that skin METTL14 is essential for skin development and/or function. After chronic UVB irradiation, skin-specific heterozygous METTL14 deletion (cHet) accelerated skin tumorigenesis in both male (Fig. 6A) and female (Fig. 6B) mice, demonstrating that METTL14 suppresses UVB-induced skin tumorigenesis. To determine whether the levels of METTL14 and DDB2 are altered and whether METTL14 is associated with DDB2 in skin cancers, we next assessed their levels in skin tissue and tumors from sham-irradiated and chronic UVB-irradiated mice and human skin cancer. We found that METTL14 levels are decreased in both UVB-irradiated mouse skin and skin tumors as compared with sham-treated skin (Fig. 6 C and D), suggesting that METTL14 down-regulation is an early event in skin cancer. In parallel, DDB2 levels were also reduced in UVB-irradiated mouse skin and skin tumors (squamous cell carcinoma [SCC]) as compared with sham-treated skin (Fig. 6 E and F), suggesting that DDB2 down-regulation is also an early event in skin cancer. Furthermore, METTL14 heterozygous deletion in skin reduced DDB2 levels (Fig. 6 E and F), supporting the regulation of DDB2 by METTL14 in vivo. In addition, METTL14 levels were positively correlated with DDB2 levels (Fig. 6G). We found that METTL14 and DDB2 are high in normal human skin, while they are decreased in skin SCC (Fig. 6 H–J). Consistent with mouse tumors, METTL14 levels were positively correlated with DDB2 levels in human skin and SCC samples (Fig. 6K). These findings demonstrate that METTL14 regulates DDB2 abundance in vivo and acts as a tumor suppressor in skin cancer.
Fig. 6.
METTL14 is a tumor suppressor in UVB-induced skin cancer. (A and B) Percentage of tumor-free mice in WT and skin-specific METTL14 heterozygous conditional knockout (cHet, K14Cre;Mettl14fl/+) male (A) and female (B) mice following chronic UVB irradiation. n = 9 (WT male), n = 15 (cHet male), n = 15 (WT female), and n = 12 (cHet female). (C and E) Immunofluorescence staining of METTL14 (C) or DDB2 (E) in sham- and chronically UVB-irradiated mouse skin and skin tumors. (Scale bar, 20 μm.) (D and F) Quantification of METTL14 (D) or DDB2 (F) in C and E (mean ± SD, n = 4). (G) Positive correlation of DDB2 levels with METTL14 levels by Spearman test from C and E. (H) Immunofluorescence staining of METTL14 (red) in normal human skin (n = 7) and skin SCC tumors (n = 57). (Scale bar, 50 μm.) Cytokeratin (blue) is used as a keratinocyte marker. (I and J) Percentage of tumors (in stacked column format) for each score of METTL14 (I) or DDB2 (J). (K) Positive correlation of DDB2 levels with METTL14 levels by Spearman test from H. ***P < 0.001; Mann–Whitney U test (I and J), Student’s t test (D and F), and Log-rank test (A and B).
Recently, METTL14 has been shown to promote leukemia and facilitates proliferation of leukemia cells (53). To determine the function of METTL14 in keratinocytes (NHEK and HaCaT) and skin cancer cells (HaCaT-Ras and A431) without UVB irradiation, we assessed the effect of METTL14 knockdown or overexpression on cell proliferation, migration, and invasion in these cells. Knockdown of METTL14 in NHEK and HaCaT cells decreased cell proliferation, migration, and invasion, while overexpression of WT METTL14 but not mutant METTL14 (R298P) increased them (SI Appendix, Figs. S8–S12). These results suggest that under certain conditions or tumor cells, METTL14 may serve as a protumorigenic factor. However, following UVB irradiation, METTL14 promotes GGR and thus serves as a tumor suppressor in UVB-induced skin tumorigenesis. Taken together, our findings together with previous work (53) suggest that the role of METTL14 and m6A RNA methylation in carcinogenesis is context dependent.
Discussion
RNA m6A modification regulates a number of physiological and pathological processes, including maintenance of established cancer. However, the role of the m6A machinery in GGR and epithelial tumor initiation and how m6A effectors are regulated remain poorly understood. Here, we show that METTL14 regulates GGR through facilitating m6A- and YTHDF1-mediated translation of DDB2. In addition, we demonstrate that METTL14 is down-regulated by UVB irradiation through NBR1-mediated selective autophagy. METTL14 is down-regulated in human and UVB-induced mouse skin cancers, in association with DDB2 down-regulation. Furthermore, heterozygous METTL14 deletion in the skin accelerated UVB-induced skin tumorigenesis in mice. Our findings demonstrate that METTL14 positively regulates GGR, is degraded through NBR1-mediated selective autophagy, and acts as a tumor suppressor in skin cancer.
First, we show that m6A RNA methylation positively regulates GGR through promoting translation of DDB2. METTL14 knockdown decreased UVB damage repair and DDB2 availability, while METTL14 overexpression increased them. DDB2 repletion reversed the GGR defects in METTL14 knockdown cells. METTL14 knockdown reduced m6A enrichment in the DDB2 transcript. Furthermore, using CLIP analysis, we found that both METTL14 and YTHDF1 bind to the DDB2 mRNA. These findings support a crucial role of m6A in regulating GGR through regulating DDB2 translation. Earlier studies by Xiang and colleagues reported that in cells in the S/G2/M phase, m6A is activated at DNA damage sites and promotes DNA polymerase κ-mediated noncanonical NER or translesion synthesis following UVC or UVA irradiation (39). A recent report showed that m6A deposition at the DNA damage sites were observed in U2OS and HeLa cells, but not HaCaT keratinocytes (54), supporting a cell type–specific effect. It is possible that keratinocytes, as the skin epithelial barrier cells, utilize canonical NER predominantly for UVB-induced DNA damage repair, including GGR and TCR, since they are the primary cell targets for UV damage. In addition, UVB irradiation may induce molecular and cellular damage that is different from UVC or UVA. Nevertheless, our findings showed that m6A promotes GGR capacity in epidermal keratinocytes under UVB stress.
We observed that METTL14 regulates DDB2 translation but has no effect on DDB2 mRNA abundance. In keratinocytes, YTHDF1 knockdown has an effect similar to METTL14 knockdown, supporting the role of m6A-dependent translation in UVB damage repair. However, it is possible that either m6A did not affect DDB2 mRNA stability and transcription, or it affected them in opposite directions. Recent work has shown that METTL3-mediated m6A modification in chromatin-associated regulatory RNA (carRNA) controls chromatin accessibility and transcription (33). It remains to be determined whether such m6A modification in carRNA plays a role in UVB damage repair.
Although METTL14 has been shown to play critical roles in development and diseases such as cancer, the mechanism by which METTL14 is regulated remain unclear. Previous studies showed that METTL14 expression is inhibited by the transcription regulator SPI1 in hematopoietic stem/progenitor cells and acute myeloid leukemia cells (53). Here, we show that METTL14 protein is degraded by NBR1-mediated selective autophagy. In keratinocytes, the autophagic degradation of METTL14 was detected both under basal conditions and in response to UVB damage. Our earlier work has shown that UVB irradiation induces autophagy in keratinocytes and mouse skin (44, 55), which can lead to accelerated autophagic degradation of METTL14 protein through NBR1. Although we found that METTL14 is down-regulated by UVB irradiation at 6 h, cells can repair CPD at 12 h, suggesting that it may take longer time in affecting DDB2 expression and protein levels following METTL14 down-regulation by UVB irradiation. Further investigations can elucidate the temporal response and interaction of METTL14 down-regulation, DDB2 expression, and GGR process post–UVB irradiation and the functional significance of the autophagy-METTL14 axis in other physiological and stress responses.
Previous studies have demonstrated that the stability of METTL3 and METTL14 is dependent on each other in installing m6A (56, 57). We detected significant loss of METTL14 at 24 h post–UVB irradiation. However, we can still detect m6A modification in NHEK and HaCaT cells. We think this may be due to the following reasons (1). UVB irradiation only down-regulates METTL14, while the remaining METTL14 interacting with METTL3 as well as other proteins in the writer complex is still functional in installing m6A modification (2). UVB irradiation may also affect other m6A enzymes, counteracting the effect of METTL14 down-regulation. Future investigation is needed to elucidate the specific connection between UVB irradiation, METTL14 regulation, and m6A alteration.
METTL14 has been suggested to be either a tumor suppressor or an oncogene in established cancers depending on cell origins and genetic contexts (36, 53). Using mice with skin-specific METTL14 deletion, we demonstrate that in skin cancer, METTL14 is a tumor suppressor. Skin-specific heterozygous METTL14 deletion increased susceptibility of mice to UVB-induced skin tumorigenesis. Such a tumor-suppressing role of METTL14 is consistent with its function in promoting GGR capacity and its down-regulation in human skin cancer as compared with normal skin. It is possible that by maintaining the pool of DDB2 protein, METTL14 enhances GGR capacity and thus inhibits tumorigenic mutagenesis. In addition, other mechanisms may play important roles in METTL14’s function in epithelial cancer, which requires additional investigations. Nevertheless, our data demonstrate that METTL14 is a critical tumor suppressor in the epithelium following genotoxic stress.
In summary, we have demonstrated the crucial role of METTL14 in GGR, the regulation of METTL14 protein stability by autophagic degradation, and the tumor-suppressive role of METTL14 in UVB-induced skin tumorigenesis. The regulatory and functional role of METTL14 may serve as an emerging epitranscriptomic mechanism to coordinate RNA metabolism and UVB damage response in tumorigenesis. We anticipate that these findings may be translated into potential strategies to improve genome stability and cancer prevention by targeting the METTL14 pathway.
Materials and Methods
All human specimens were studied after approval by the University of Chicago Institutional Review Board. All animal procedures have been approved by the University of Chicago Institutional Animal Care and Use Committee. WT, Atg5 KO, Atg7 KO, p62 KO MEF cells, NHEK, A431 (human squamous carcinoma cells), and HaCaT cells were used. Detailed descriptions of all methods are provided in SI Appendix, Methods online.
Supplementary Material
Acknowledgments
We thank Dr. Masaaki Komatsu for providing the Atg7fl/fl mice and ATG7 KO and p62 KO MEF cells, Dr. Noboru Mizushima for providing the Atg5fl/fl mice and ATG5 KO MEF cells, Dr. Norbert Fusenig for providing the HaCaT cells, Terri Li for immunohistochemistry, Drs. Jiping Xie and Xiaoyang Wu (University of Chicago) for providing the Cas9 lentiviral vector, and Dr. Ann Motten for a critical reading of the manuscript. This work was supported in part by NIH Grants ES030576 (Y.-Y.H.), ES024373 (Y.-Y.H.), ES030546 (C.H.), the University of Chicago Comprehensive Cancer Center P30 CA014599, the Clinical and Translational Science Awards (CTSA) (UL1 TR000430), the Chicago Center for Health and Environment (CACHET) (ES027792), and the University of Chicago Friends of Dermatology Endowment Fund. C.H. is an investigator of the HHMI.
Footnotes
Competing interest statement: C.H. is a scientific founder and a member of the scientific advisory board of Accent Therapeutics, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025948118/-/DCSupplemental.
Data Availability
m6A IP sequencing and RNA sequencing data are deposited and accessible at the Gene Expression Omnibus repository under accession no. GSE145924.
References
- 1.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S., Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Niggli H. J., Röthlisberger R., Cyclobutane-type pyrimidine photodimer formation and induction of ornithine decarboxylase in human skin fibroblasts after UV irradiation. J. Invest. Dermatol. 91, 579–584 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Vink A. A., Berg R. J., de Gruijl F. R., Roza L., Baan R. A., Induction, repair and accumulation of thymine dimers in the skin of UV-B-irradiated hairless mice. Carcinogenesis 12, 861–864 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Cleaver J. E., Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer 5, 564–573 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Cleaver J. E., Lam E. T., Revet I., Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 10, 756–768 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Braithwaite E., Wu X., Wang Z., Repair of DNA lesions induced by polycyclic aromatic hydrocarbons in human cell-free extracts: Involvement of two excision repair mechanisms in vitro. Carcinogenesis 19, 1239–1246 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Kad N. M., Wang H., Kennedy G. G., Warshaw D. M., Van Houten B., Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol. Cell 37, 702–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoeijmakers J. H., Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 (2001). [DOI] [PubMed] [Google Scholar]
- 9.DiGiovanna J. J., Kraemer K. H., Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 132, 785–796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugasawa K., Xeroderma pigmentosum genes: Functions inside and outside DNA repair. Carcinogenesis 29, 455–465 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Bradford P. T., et al., Cancer and neurologic degeneration in xeroderma pigmentosum: Long term follow-up characterises the role of DNA repair. J. Med. Genet. 48, 168–176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khaminets A., Behl C., Dikic I., Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6–16 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Sugasawa K., et al., UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121, 387–400 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Sugasawa K., UV-DDB: A molecular machine linking DNA repair with ubiquitination. DNA Repair (Amst.) 8, 969–972 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Huang T. T., D’Andrea A. D., Regulation of DNA repair by ubiquitylation. Nat. Rev. Mol. Cell Biol. 7, 323–334 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Luijsterburg M. S., et al., DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell Biol. 197, 267–281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah P., He Y. Y., Molecular regulation of UV-induced DNA repair. Photochem. Photobiol. 91, 254–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah P., Zhao B., Qiang L., He Y. Y., Phosphorylation of xeroderma pigmentosum group C regulates ultraviolet-induced DNA damage repair. Nucleic Acids Res. 46, 5050–5060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q. E., Zhu Q., Wani G., Chen J., Wani A. A., UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis 25, 1033–1043 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Wang Q. E., et al., Cellular ubiquitination and proteasomal functions positively modulate mammalian nucleotide excision repair. Mol. Carcinog. 42, 53–64 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Jarrett S. G., et al., PKA-mediated phosphorylation of ATR promotes recruitment of XPA to UV-induced DNA damage. Mol. Cell 54, 999–1011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Jarrett S. G., et al., Sirtuin 1-mediated deacetylation of XPA DNA repair protein enhances its interaction with ATR protein and promotes cAMP-induced DNA repair of UV damage. J. Biol. Chem. 293, 19025–19037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Qiang L., Shah P., Barcellos-Hoff M. H., He Y. Y., TGF-β signaling links E-cadherin loss to suppression of nucleotide excision repair. Oncogene 35, 3293–3302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B. S., Roundtree I. A., He C., Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frye M., Harada B. T., Behm M., He C., RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer K. D., Jaffrey S. R., The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., et al., N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X., et al., FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., et al., N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer K. D., et al., 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alarcón C. R., Lee H., Goodarzi H., Halberg N., Tavazoie S. F., N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu N., et al., N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J., et al., N 6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batista P. J., The RNA modification N6-methyladenosine and its implications in human disease. Genomics Proteomics Bioinformatics 15, 154–163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S., et al., Roles of RNA methylation by means of N6-methyladenosine (m6A) in human cancers. Cancer Lett. 408, 112–120 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Deng X., et al., RNA N6-methyladenosine modification in cancers: Current status and perspectives. Cell Res. 28, 507–517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson M., Shah P., Cui Y. H., He Y. Y., The role of dynamic m6 A RNA methylation in photobiology. Photochem. Photobiol. 95, 95–104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H., Weng H., Chen J., m6A modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell 37, 270–288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang Y., et al., RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amm I., Sommer T., Wolf D. H., Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 182–196 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Klionsky D. J., Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J., Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kroemer G., Mariño G., Levine B., Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiang L., Wu C., Ming M., Viollet B., He Y. Y., Autophagy controls p38 activation to promote cell survival under genotoxic stress. J. Biol. Chem. 288, 1603–1611 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L. H., et al., Targeting protective autophagy exacerbates UV-triggered apoptotic cell death. Int. J. Mol. Sci. 13, 1209–1224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bjørkøy G., et al., p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pankiv S., et al., p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Johansen T., Lamark T., Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jans J., et al., Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 15, 105–115 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Wang P., Doxtader K. A., Nam Y., Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dominissini D., et al., Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Meng T. G., et al., Mettl14 is required for mouse postimplantation development by facilitating epiblast maturation. FASEB J. 33, 1179–1187 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Weng H., et al., METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 22, 191–205.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svobodova Kovarikova A., et al., N(6)-Adenosine methylation in RNA and a reduced m3G/TMG level in non-coding RNAs appear at microirradiation-induced DNA lesions. Cells 9, 360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiang L., et al., Autophagy gene ATG7 regulates ultraviolet radiation-induced inflammation and skin tumorigenesis. Autophagy 13, 2086–2103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J., et al., A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., et al., N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
m6A IP sequencing and RNA sequencing data are deposited and accessible at the Gene Expression Omnibus repository under accession no. GSE145924.