Significance
Aminoacyl-transfer RNA (tRNA) synthetases (aaRSs) are essential enzymes that mediate accurate expression of the genetic code. More than 96% of analyzed species possess duplicated aaRSs genes, which are presumed to encode canonical aaRSs. However, in this study, we show that some of these seemingly canonical aaRSs are in fact repurposed for deliberate mistranslation of the genetic code, enabling organisms to encode two amino acids using a single sense codon. This finding, along with the abundance of duplicated aaRSs genes, suggests that the deliberate mistranslation of sense codons may be more common than previously thought. More broadly, our work illustrates that extant genomes have many exciting functionalities that are currently hidden under a disguise of canonical tRNA synthetases and tRNAs.
Keywords: genetic code, tRNA, aminoacyl-tRNA synthetase, mistranslation, Streptomyces
Abstract
Inaccurate expression of the genetic code, also known as mistranslation, is an emerging paradigm in microbial studies. Growing evidence suggests that many microbial pathogens can deliberately mistranslate their genetic code to help invade a host or evade host immune responses. However, discovering different capacities for deliberate mistranslation remains a challenge because each group of pathogens typically employs a unique mistranslation mechanism. In this study, we address this problem by studying duplicated genes of aminoacyl-transfer RNA (tRNA) synthetases. Using bacterial prolyl-tRNA synthetase (ProRS) genes as an example, we identify an anomalous ProRS isoform, ProRSx, and a corresponding tRNA, tRNAProA, that are predominately found in plant pathogens from Streptomyces species. We then show that tRNAProA has an unusual hybrid structure that allows this tRNA to mistranslate alanine codons as proline. Finally, we provide biochemical, genetic, and mass spectrometric evidence that cells which express ProRSx and tRNAProA can translate GCU alanine codons as both alanine and proline. This dual use of alanine codons creates a hidden proteome diversity due to stochastic Ala→Pro mutations in protein sequences. Thus, we show that important plant pathogens are equipped with a tool to alter the identity of their sense codons. This finding reveals the initial example of a natural tRNA synthetase/tRNA pair for dedicated mistranslation of sense codons.
Ambiguous translation of the genetic code, known as mistranslation, is traditionally considered unfavorable for cell growth and sustainability (1–3). However, the concept of mistranslation is now becoming a paradigm in microbial studies because organisms from all domains of life have been shown to deliberately mistranslate the genetic code to respond to stress or augment pathogenicity (4–8). A prominent example is the pathogenic yeast Candida albicans, which is known to ambiguously translate CUG codons as either leucine or serine. This dual use of CUG codons leads to Leu→Ser mutations in protein sequences, enabling C. albicans to potentially generate thousands of protein variants from a single gene (9, 10). Some of these variants can promote pathogenicity by improving C. albicans adhesion to its host or by helping evade the host immune response (11). In addition to C. albicans, mistranslation has been identified in a growing number of sequenced microbial pathogens, including Mycoplasma (12), Mycobacteria (13), Acinetobacter (14), Microsporidia (15, 16), and possibly many intracellular parasites and endosymbionts (17), suggesting that mistranslation may be a general adaptive strategy that assists parasite survival.
Mistranslation can be divided into two types: deliberate and stochastic. Stochastic mistranslation occurs because of the inherently promiscuous nature of some translation factors. It can also stem from spontaneous mutations or chemical modifications that affect the accuracy of translation machinery. For example, in the absence of any mutations or modifications, rapidly growing Escherichia coli cells mistranslate ∼1 in ∼3,000 to 10,000 messenger RNA codons, causing rare and random variations in protein sequences (3). More frequent, stochastic mistranslation can be caused by mutations in transfer RNA (tRNA) anticodon or identity elements (18), mutations in editing sites of aminoacyl-tRNA synthetases (aaRSs) (12, 19), and mutations or chemical modifications of aaRSs, amidotransferases, and ribosomes in response to stress or exposure to antibiotics (13, 15, 20, 21). Under these conditions, the rates of stochastic mistranslation can increase by up to two orders of magnitude (5).
In contrast, deliberate mistranslation occurs when, aside from canonical translation factors, organisms possess an additional factor(s) for which mistranslation is the primary biological function. For example, C. albicans encodes an unusual tRNA variant, tRNACAGSer, that recognizes two aaRSs, SerRS and LeuRS. This dual specificity causes a scenario in which the same CUG codons can be translated either as leucine (which happens ∼0.5 to 6% of the time, depending on growth conditions) or as serine (which happens ∼94 to 99.5% of the time) (22). Thus, unlike stochastic mistranslation, deliberate mistranslation is typically limited to one specific type of translation error that occurs because of the presence of an unusual, dedicated protein synthesis factor of endowing the organism with a capacity to deliberately alter the rules of the genetic code.
Current progress in discovering the capacity for deliberate mistranslation in pathogenic organisms is hampered by the lack of generalizable strategies for identifying unusual translation factors in nature. In this study, we address this problem by showing that a capacity for deliberate mistranslation can be discovered by studying duplications of genes encoding aaRSs. By studying duplicated prolyl-tRNA synthetase (ProRS) genes found in plant pathogens from the Streptomyces genus, we determined that one of these genes encodes a translation factor for deliberate mistranslation, unearthing a previously unknown potential of these plant pathogens for deliberate mistranslation of the genetic code.
Results
Plant-pathogenic Streptomyces Encode an Unusual ProRS Isoform.
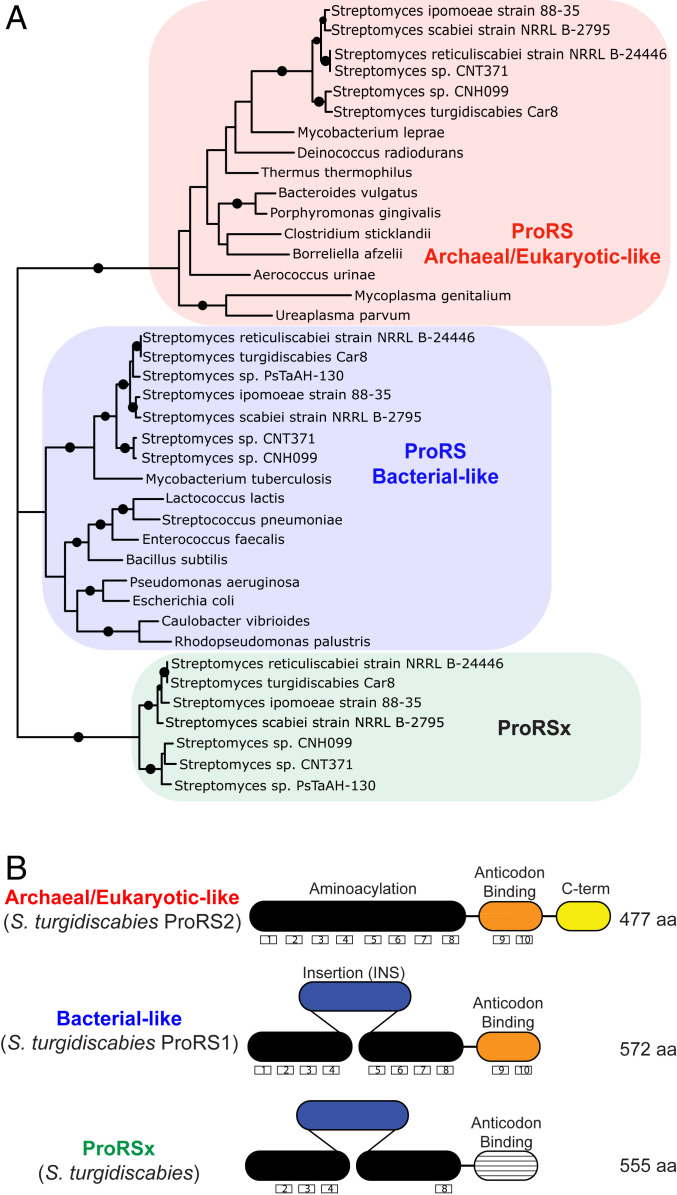
In bacteria, ProRS exists in two isoforms that perform the same essential function but have distinct structures and domain arrangements. These ProRS isoforms are known as the bacteria type and the archaea/eukaryote type (Fig. 1) (23). While most bacteria have a single gene copy of ProRS corresponding to one or other of the isoforms, many bacteria have two ProRS gene copies (24–26). Interestingly, when two ProRS genes are encoded in a single bacterium, one gene encodes a bacteria type and the other gene an archaea/eukaryote type of ProRS (26). Recently, a few Streptomyces species were shown to possess an anomalously high number of three ProRS gene copies (SI Appendix, Table S1) (25), which prompted us to analyze these genes.
Fig. 1.
The plant-pathogenic bacteria Streptomyces encode an atypical isoform of ProRS, ProRSx. (A) Phylogenetic analysis of bacterial ProRS shows that the plant pathogens S. turgidiscabies and S. scabiei encode three isoforms of ProRS, including an archaea/eukaryote-type ProRS, a bacteria-type ProRS, and the previously unknown ProRSx isoform. (B) Domain organization of the ProRS isoforms shows that ProRSx has the same domain structure as bacteria-type ProRS but an altered C-terminal domain that is responsible for tRNAPro binding. Boxed numbers indicate the number of known conserved motifs in ProRS.
We first performed a phylogenetic analysis to determine the origin of each of these three ProRS genes. Consistent with our previous study (26), a phylogenetic tree showed that two of the ProRS genes in Streptomyces correspond to the two canonical ProRS isoforms (i.e., a bacteria-type and a eukaryote-type ProRS) (Fig. 1A). However, the third ProRS gene formed a cluster that is phylogenetically distant from the bacteria- and eukaryote-type ProRSs (Fig. 1A). This result suggests that this ProRS gene evolved divergently from the two canonical genes and thus gave rise to a previously unknown ProRS isoform, which we named ProRSx.
Surprisingly, our basic local alignment search tool (BLAST) searches showed that ProRSx exclusively emerged in a small group of Streptomyces species (Fig. 1A and SI Appendix, Table S1). Among more than 800 Streptomyces species that have been described to date (27), four species encode ProRSx. These species are plant pathogens Streptomyces turgidiscabies, Streptomyces scabiei, Streptomyces reticuliscabiei, and Streptomyces ipomoeae, all of which are a threat to several agricultural crops, primarily potatoes; there they cause noticeable losses of the industry value because of the formation of potato scab (28).
Our primary structure analysis revealed that ProRSx has the same domain organization as bacteria-type ProRS, indicating that ProRSx may have evolved through a duplication of the bacteria-type ProRS gene in Streptomyces. However, the ProRSx sequence has significantly diverged from the bacteria-type ProRS, as it contains just 4 of the 10 motifs that characterize bacteria-type ProRS (Fig. 1B) (24). Most intriguingly, the ProRSx C-terminal domain, which is responsible for the recognition of the tRNAPro anticodon, is the most diverged ProRSx segment and lacks the two bacteria-type ProRS signature motifs that are critical for the recognition of tRNAPro (Fig. 1B). Based on these alterations in the C-terminal domain, we therefore considered that, unlike canonical ProRS variants, ProRSx cannot recognize tRNAPro and possibly has a different biological function.
ProRSx Is Flanked by an Unusual Proline tRNA Gene with Hybrid Identity Elements.
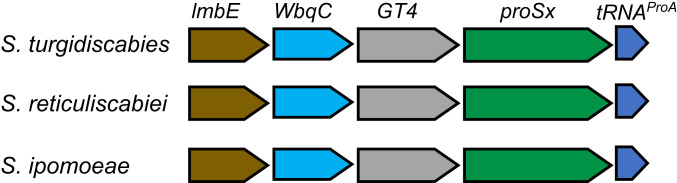
Seeking to gain insights about the possible function of ProRSx, we analyzed the genomic context of ProRSx genes in Streptomyces species. Intriguingly, we found that ProRSx is encoded in an operon that is conserved across multiple Streptomyces species and which includes a tRNA gene adjacent to ProRSx (Fig. 2). Our Northern blot analysis of RNA extracts from laboratory cultures of S. turgidiscabies showed that this tRNA is actively expressed, indicating that this tRNA is encoded by a functional gene (SI Appendix, Fig. S1).
Fig. 2.
The ProRSx gene is colocalized with an unknown tRNA gene. Schematic diagrams show the genomic regions of three Streptomyces species, where the ProRSx gene is located adjacent to a gene encoding an atypical variant of tRNAPro, which we named tRNAProA. The possible operon also includes lmbE, WbqC, and GTA; putative genes encoding a N-acetylglucosaminyl deacetylase; an O antigen; and a glycosyltransferase.
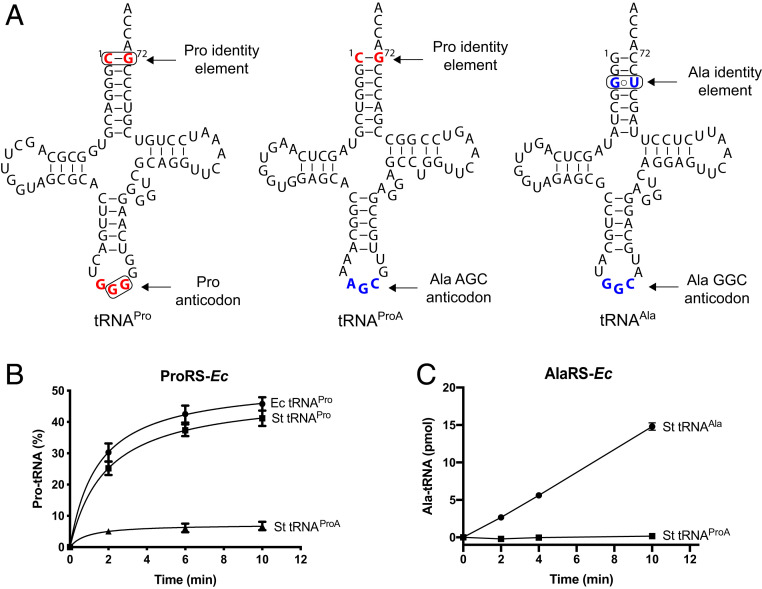
Our analysis (Fig. 3A) of the predicted secondary structures of this tRNA gene revealed that the encoded tRNA has a highly atypical structure. It has an A34G35C36 anticodon, which is a unique alanine anticodon in bacteria because alanine anticodons in bacterial tRNAAla typically avoid using adenine at position 34 (Fig. 3A and SI Appendix, Fig. S2). At the same time, this tRNA lacks a G3:U70 base pair, which is universally conserved in alanine tRNAs and is required for tRNA aminoacylation by alanyl-tRNA synthetase (AlaRS) (Fig. 3A) (29, 30). Instead, this tRNA possesses a C1:G72 base pair, which is a major identity element of bacterial proline tRNAs and is required for tRNA aminoacylation by ProRS. Given these hybrid characteristics, we named this tRNA species tRNAProA to denote that this tRNA resembles tRNAPro but has an alanine anticodon. In addition to those variations, the tRNAProA displays striking sequence elements: 1) it has A33 instead of the highly conserved U33 (only exceptions are in initiator tRNAs with C33 (31) and tRNALeuGAG (32, 33) and G33 in tRNASerCAG); 2) instead of the usual R15oY48 pair, it has a A15oA48 opposition [observed in a subset of AGC isodecoders tRNAAla in eukaryotes (34)]; and 3) it has a G21 instead of the very common A21 (usually A21 forms a base triple with the conserved U8oA14). In multicellular eukaryotes, the AGC tRNAs, normally modified to IGC, are often the most numerous isodecoders.
Fig. 3.
S. turgidiscabies tRNAPro, tRNAAla, and tRNAProA species. (A) The predicted secondary structures of the three tRNAs. The known identity nucleotides critical for aminoacylation of bacterial tRNAAla and tRNAPro are encircled and shown in red and blue, respectively. The sequences reveal tRNAProA to be a hybrid between tRNAAla and tRNAPro, as indicated by the presence of the C1-G72 base pair (a tRNAPro identity element) and the AGC anticodon (a tRNAAla identity element). tRNAProA looks like an evolutionally altered variant of tRNAPro, in which the anticodon progressed from GGG to AGC, repurposing tRNAProA for the recognition of alanine codons. (B and C) In vitro assays show that S. turgidiscabies tRNAProA cannot be effectively aminoacylated by either E. coli ProRS (B) or AlaRS (C) with their cognate amino acids.
Because tRNAProA has a hybrid identity, we next tested whether tRNAProA can cross-react with canonical ProRS or AlaRS. Using in vitro aminoacylation assays, we found that while ProRS or AlaRS from E. coli aminoacylated canonical S. turgidiscabies tRNAPro and tRNAAla, none of these enzymes could react with S. turgidiscabies tRNAProA (Fig. 3 B and C). These data, along with the colocalization of tRNAProA and ProRSx genes, suggested that tRNAProA does not cross-react with canonical aaRSs and instead is a ProRSx substrate.
Coexpression of S. turgidiscabies ProRSx and tRNAProA in E. coli Causes Mistranslation of Alanine Codons with Proline.
We next established a temperature-sensitive assay to investigate the functional activity of S. turgidiscabies ProRSs. For this purpose, we used the temperature-sensitive E. coli strain UQ27, which chromosomally encodes a ProRS variant that is defective at 42 °C (35). At this nonpermissive temperature, E. coli UQ27 cells are only viable when ProRS function is complemented with a functional, plasmid-borne ProRS (e.g., the E. coli wild-type [WT] ProRS) (SI Appendix, Fig. S3A). Using this strain, we showed that S. turgidiscabies ProRS1 (bacteria type) could support E. coli growth at 42 °C but ProRSx could not. This suggests that E. coli tRNAPro can be aminoacylated by S. turgidiscabies ProRS1 but not by ProRSx.
Consistent with these in vivo data, our in vitro enzymatic assays confirmed that S. turgidiscabies ProRS1 can aminoacylate E. coli and S. turgidiscabies tRNAPro with similar efficiency but cannot aminoacylate tRNAProA (SI Appendix, Fig. S3B). We next sought to directly test whether tRNAProA can be aminoacylated by ProRSx. However, we could not perform in vitro assays because ProRSx aggregated into inclusion bodies upon its overexpression in E. coli.
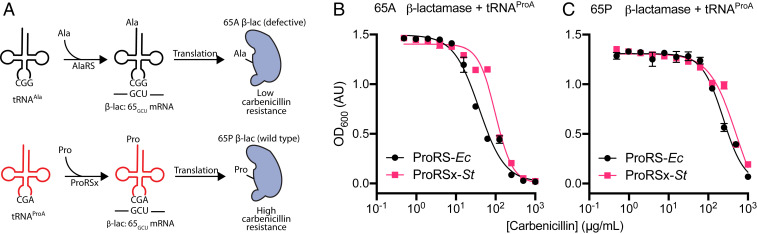
To bypass this problem, we developed an alternative and highly sensitive strategy to measure ProRSx and tRNAProA activity in E. coli cells. The assay was based on a β-lactamase reporter gene that can monitor mistranslation of GCU alanine codons with proline (Ala→Pro mistranslation) (Fig. 4A). The assay arose from our recent finding that the substitution of the conserved Pro residue at position 65 to Ala drastically impairs the activity of β-lactamase (SI Appendix, Fig. S4). Therefore, the β-lactamase Pro65Ala mutant provides E. coli cells with less protection against the antibiotic carbenicillin, compared with the protection conferred by WT β-lactamase (Fig. 4A). However, mistranslation of the Ala codon at position 65 with proline should restore β-lactamase activity, increasing carbenicillin resistance (Fig. 4A).
Fig. 4.
The S. turgidiscabies ProRSx/tRNAProA pair appears to cause mistranslation of alanine codons when expressed in E. coli. (A) A β-lactamase–based assay was used to monitor the mistranslation of alanine codons with proline in E. coli. The assay is based on the fact that the β-lactamase mutant Pro65Ala (β-lac 65A) has poor activity and does not protect E. coli cells against the antibiotic carbenicillin. However, when cells mistranslate alanine codons as proline, this should restore β-lactamase activity and increase the resistance of E. coli to carbenicillin. (B and C) Measurements of E. coli growth in the presence of carbenicillin. (B) E. coli cells harboring β-lactamase mutant (β-lac 65A) and expressing ProRSx-St and tRNAProA tolerate higher doses of carbenicillin, relative to cells expressing ProRS-Ec and tRNAProA. The IC50 values of both datasets are different as estimated by an extra sum-of-squares F test with a P value < 0.0001. (C) Expression of ProRSx-St or ProRS-Ec did not increase carbenicillin resistance in E. coli cells expressing the WT β-lactamase (β-lac 65P) and tRNAProA. Each point represents the average of four biological replicates with the SD indicated.
We anticipated that aminoacylation of tRNAProA by ProRSx would produce prolyl-tRNAProA and cause the mistranslation of alanine codons with proline, which would revert the mutation in the β-lactamase P65A variant and increase carbenicillin resistance. Indeed, we observed a threefold increase in the half-maximal inhibitory concentration (IC50) value in E. coli expressing tRNAProA and ProRSx (Fig. 4B and Table 1). In contrast, coexpression of tRNAProA and ProRSx did not change carbenicillin resistance in our control experiment, in which we used the WT β-lactamase instead of the Pro65Ala mutant (Fig. 4C). These data supported our hypothesis that ProRSx can aminoacylate tRNAProA with proline, causing the decoding of GCU alanine codons with proline.
Table 1.
IC50 values for mistranslation as measured by β-lactamase assay
| IC50 | WT ProRS-Ec | ProRSx-St | ProRS-Ec-5ABD |
| A65 GCU | 38 ± 1 | 99 ± 2 | 99 ± 4 |
Errors represent the SD of four biological replicates.
ProRSx Activity Can Be Engineered in E. coli ProRS.
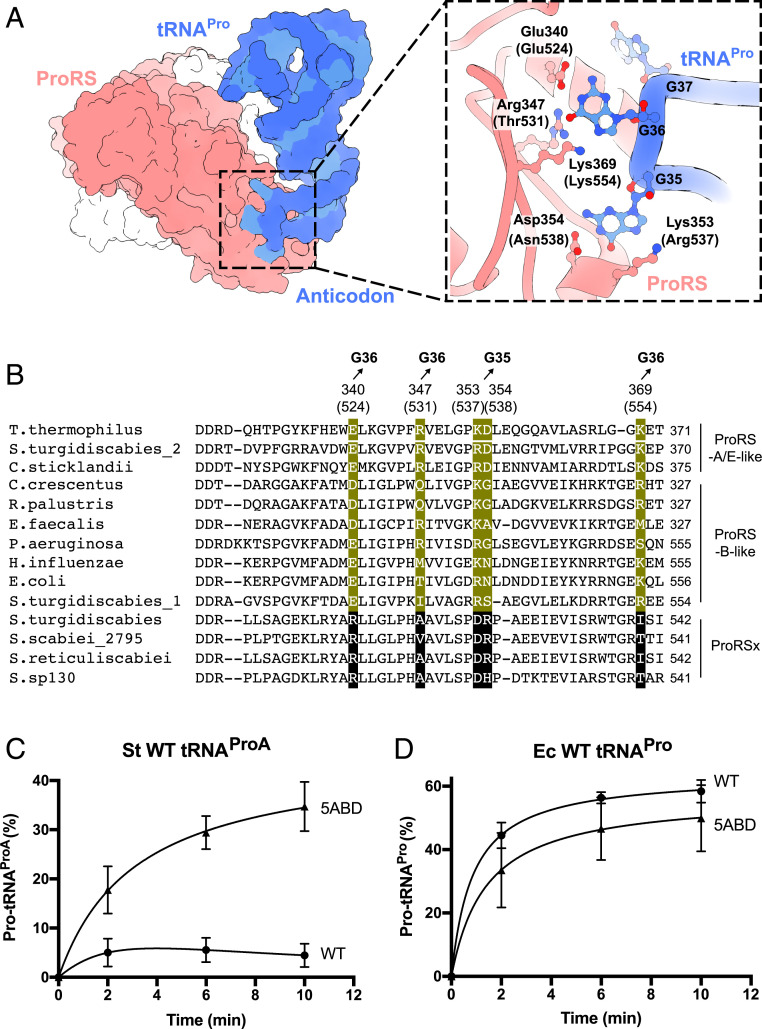
We next sought to understand how tRNAProA can avoid cross-reacting with canonical ProRS but remain specific to ProRSx. Previously, the mechanism of tRNAPro recognition by ProRS was determined from the X-ray structure of tRNAPro–ProRS complex from the bacterium Thermus thermophilus (36). We used this structure, along with multiple sequence alignments, to compare the anticodon-binding residues in ProRS and ProRSx (Fig. 5 A and B).
Fig. 5.
E. coli ProRS can be converted into a ProRSx-like enzyme by “transplanting” anticodon-binding residues from S. turgidiscabies ProRSx. (A) The crystal structure of T. thermophilus ProRS in complex with tRNAPro shows how ProRS recognizes the tRNAPro anticodon. A close-up view illustrates that the anticodon is recognized by five ProRS residues that bind the tRNAPro bases G36 and G35. Residue numbers in parentheses correspond to E. coli ProRS. (B) Multiple sequence alignment-comparing, anticodon-binding residues in canonical ProRS variants (B-type: bacterial-type and A/E-type: archaeal/eukaryotic-type) and the ProRSx variant. Residues that interact with the anticodon bases G35 and G36 of tRNAPro are highlighted in green. The corresponding residues in ProRSx are highlighted in black. Aminoacylation assays of S. turgidiscabies tRNAProA (C) and E. coli tRNAPro (D) with proline by either the WT E. coli ProRS (ProRS-Ec-WT) or its 5ABD variant (ProRS-Ec-5ABD). The results represent the average of three independent trials with the SD indicated.
In the T. thermophilus structure, tRNAPro recognition by ProRS is mediated by five residues that form sequence-specific interactions with the anticodon bases G35 and G36 in tRNAPro. These residues comprise K353, D354, E340, R347, and K369 (T. thermophilus numbering) (Fig. 5A). Strikingly, while these residues are conserved in canonical ProRS, each of them is mutated in ProRSx (R511, A518, D524, R525, and I540 in S. turgidiscabies) (Fig. 5B). This finding, along with the unusual AGC sequence of the tRNAProA anticodon, suggested that ProRSx and tRNAProA may have coevolved to become a tRNA/synthetase pair that mistranslates alanine codons.
To test this hypothesis, we first produced a mutant of E. coli ProRS (ProRS-Ec-5ABD), in which each of the five anticodon-binding domain residues was mutated according to the sequence of ProRSx from S. turgidiscabies. These mutations comprised E524R, T531A, R537D, N538R, and K554I (E. coli numbering; Fig. 5B). We then used the ProRS-Ec-5ABD variant for biochemical assays and found that, although it retained the ability to aminoacylate canonical tRNAPro from E. coli, it also gained the ability to robustly charge tRNAProA with proline (Fig. 5 C and D). Thus, through just five amino acid substitutions, we could engineer a ProRSx-like activity in E. coli ProRS. Overall, this result supported the idea that ProRSx and tRNAProA have evolved a unique codon–anticodon interface that allows these molecules to remain specific to each other and avoid cross-reactions with other translation factors.
In Vivo Aminoacylation of tRNAProA by ProRS-Ec-5ABD Causes Mistranslation of Alanine Codons.
Our engineered, ProRSx-type enzyme expressed well in E. coli and allowed us to measure the impact of ProRSx-type activity on cell fitness and protein synthesis accuracy.
We first tested whether ProRS-Ec-5ABD can aminoacylate tRNAProA in vivo. We expressed ProRS-Ec-5ABD and tRNAProA in cells carrying the P65A β-lactamase reporter and observed a threefold increase in carbenicillin tolerance relative to E. coli that expressed ProRS-Ec-WT and tRNAProA (Table 1). Thus, ProRS-Ec-5ABD appeared to mistranslate alanine codons in vivo.
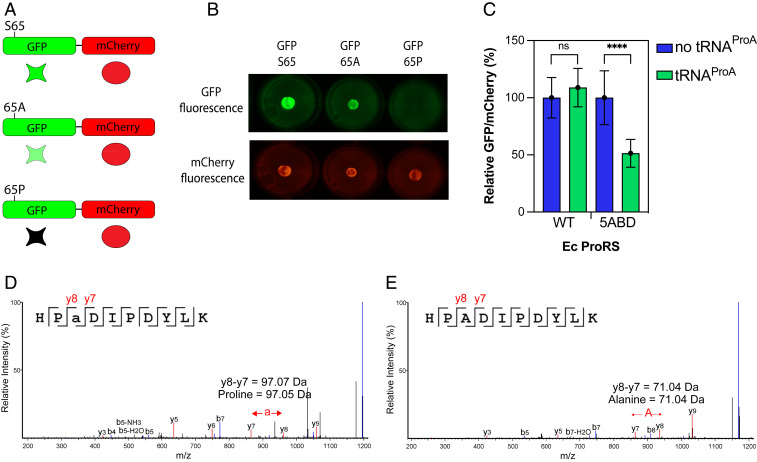
To make our experimental system more quantitative, we established an additional assay that was based on a chimeric protein fusion between green fluorescent protein, GFP, and the red fluorescent protein, mCherry (Fig. 6A). Previously, the substitution of serine 65 with proline was shown to impair GFP fluorescence, but the substitution of serine 65 with alanine resulted in a fluorescence-competent GFP (37). We therefore expected that cells expressing GFP S65A/mCherry fusion would show a high level of green and red fluorescence, but cells expressing the GFP-S65P/mCherry fusion would show red fluorescence only.
Fig. 6.
Coexpression of S. turgidiscabies tRNAProA with ProRS-Ec-5ADB causes mistranslation of alanine codons in E. coli. (A) The scheme shows a dual-fluorescence reporter system to monitor the mistranslation of Ala codons in vivo. This reporter is based on a fusion protein comprising GFP and mCherry. The GFP gene was modified to introduce a Ser65Ala mutation, which produced a GFP-Ser65Ala mutant that was able to fluoresce. Mistranslation of the alanine codon at position 65 with proline decreases the fluorescence emission of GFP, which allows the mistranslation of alanine codons to be monitored. (B) Expression of the GFP-mCherry reporter in E. coli demonstrates that a Ser65Ala GFP variant retains its ability to fluoresce while the GFP Ser65Pro does not. mCherry fluorescence remains unaffected. (C) The ratio of relative GFP/mCherry fluorescence activity was measured for E. coli QU27 cells carrying either E. coli WT or 5ABD ProRS, in the absence or presence of S. turgidiscabies tRNAProA. Bars represent the average of six biological replicates with the SD indicated. ****P < 0.0001; ns, not significant by t test. (D and E) Liquid chromatography with tandem mass spectrometry spectra from the GFP-mCherry peptides HP(a)DIPDYLK (D) and HPADIPDYLK (E). The mistranslated position with proline replacing alanine is indicated as (a). Characteristic b- and y-ion peaks are highlighted in blue and red, respectively. Data are shown as relative peak intensities plotted against mass/charge ratio (m/z) on the x-axis.
In our control experiments, we showed that both GFP S65A and mCherry did indeed emit fluorescence when expressed as a fusion in E. coli cells, but only mCherry fluoresced in cells that expressed the GFP S65P/mCherry reporter (Fig. 6B). We then assessed the fluorescence of the GFP S65A/mCherry reporter in the E. coli UQ27 strain at 42 °C, when the endogenous ProRS is inactive. We found that the GFP/mCherry emission ratio was twofold lower in cells that simultaneously expressed ProRS-Ec-5ABD and tRNAProA compared with cells that expressed ProRS-Ec-5ABD alone (Fig. 6C). From this, the mistranslation rate was estimated (38) to be ∼2%. In contrast, the expression of ProRS-Ec-WT together with tRNAProA or alone did not lead to significant changes in the normalized fluorescence. These data indicated that ProRS-Ec-5ABD can cause significant levels of mistranslation in E. coli. Thus, we decided to directly observe Ala→Pro mistranslation by tandem mass spectrometry (Fig. 6 D and E). This revealed that cells expressing ProRS-Ec-5ABD and tRNAProA indeed contained Ala→Pro mutations (at the GFP position 65) in the GFP/mCherry reporter protein. In contrast, cells expressing ProRS-Ec-5ABD in the absence of tRNAProA contained only WT peptides (Fig. 6D and SI Appendix, Fig. S5). Collectively, these data showed that ProRSx-type activity causes Ala→Pro mistranslation that can be readily observed through changes in reporter gene fluorescence or through mass spectrometry.
Mistranslation by ProRS-Ec-5ABD and tRNAProA Impairs Cell Growth.
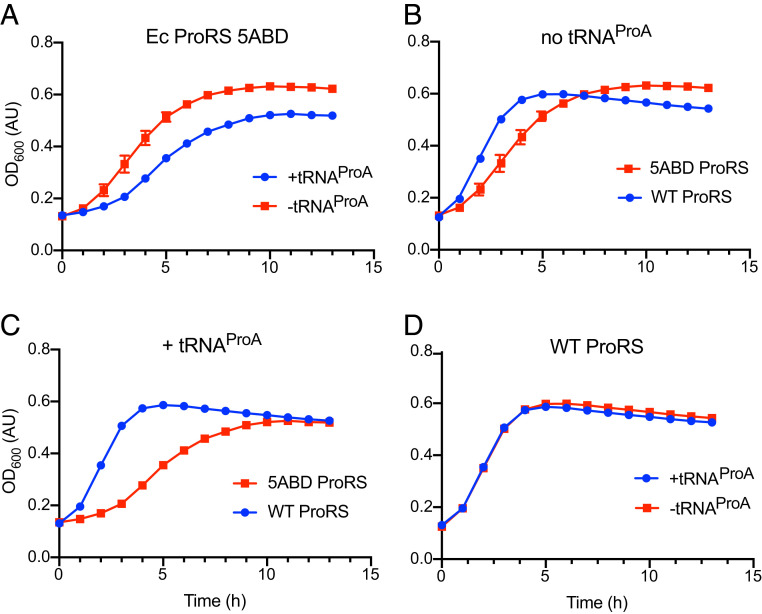
Lastly, we used ProRS-Ec-5ABD to assess how proteome-wide mistranslation of alanine codons affects bacterial fitness. For this, we expressed S. turgidiscabies tRNAProA with E. coli ProRS variants (either WT or 5ABD) in E. coli UQ27 cells at 42 °C (Fig. 7). We observed that, when expressed alone, neither tRNAProA nor ProRS-Ec-5ADB were toxic for E. coli growth (Fig. 7 A and B). However, when tRNAProA and ProRS-Ec-5ADB were coexpressed, E. coli growth was notably inhibited (Fig. 7 C and D). Consistent with previous studies (3), these data showed that mistranslation results impaired growth.
Fig. 7.
Coexpression of S. turgidiscabies tRNAProA with ProRS-Ec-5ADB inhibits E. coli growth. (A–D) The curves compare the growth rates of E. coli strain QU27 at 42 °C, when this strain expressed different variants of ProRS and tRNAPro. (A) tRNAProA expression is not toxic for E. coli growth. (B) Both ProRS-Ec-WT and ProRS-Ec-5ADB can sustain rapid growth of E. coli cells, although ProRS-Ec-5ADB supports a marginally slower growth. (C) The coexpression of S. turgidiscabies tRNAProA and ProRS-Ec-5ADB slows down E. coli growth (compared with the strain that expresses ProRS-Ec-5ADB alone). (D) The coexpression of tRNAProA with the ProRS-Ec-5ADB mutant is highly toxic for E. coli cells (compared with E. coli cells that express tRNAProA with the WT ProRS-Ec).
Discussion
An Aminoacyl-tRNA Synthetase/tRNA Pair for Deliberate Mistranslation.
In this study, we found that an important group of plant pathogens encodes machinery for the deliberate mistranslation of the genetic code. This machinery involves an atypical aaRS, ProRSx, and a dedicated tRNA partner, tRNAProA (Figs. 1 and 3). Together, ProRSx and tRNAProA translate GCU alanine codons as proline, thus altering the identity of alanine codons in cells expressing this unusual aaRS/tRNA pair (Fig. 4).
This finding is particularly intriguing because translation scenarios in which a single codon encodes two different amino acids are currently viewed as exotic. Aside from this study, the dual translation of sense codons has been observed only in the pathogenic yeast C. albicans, in which the CUG codon can encode both serine and leucine (9, 39). Otherwise, the dual use of codons only widely occurs for the UGA and UAG codons. These codons serve as translation termination signals but can additionally encode pyrrolysine, selenocysteine, cysteine, and possibly other amino acids in some species (40). Our discovery of ProRSx/tRNAProA suggests that deliberate mistranslation of sense codons is not exclusive to C. albicans. However, in contrast to C. albicans, which relies on the ambiguous recognition of tRNACAGSer by housekeeping aaRSs, Streptomyces have evolved a dedicated aaRSs/tRNA pair that does not cross-react with other aaRSs and tRNAs and is solely dedicated to deliberate mistranslation of alanine codons. Therefore, ProRSx/tRNAProA represents the first known example of a natural aaRS/tRNA pair that is devoted to mistranslation of sense codons.
Potential Implications for Streptomyces Biology.
ProRSx and tRNAProA are primarily encoded by the plant pathogens S. turgidiscabies, S. scabiei, S. reticuliscabiei, and S. ipomoeae (Fig. 1A and SI Appendix, Table S1). These Streptomyces species are parasites of many economically important agricultural plants, primarily potatoes (41). In potatoes, they are the major causative agents of potato scab, a disease that reduces crop value and tuber marketability (28). In developed countries, these pathogenic Streptomyces cause a moderate market value loss of potato crops (42). The pathogenicity of some of these organisms is directly associated with the production of thaxtomins, a family of toxins that inhibit cellulose biosynthesis and cause necrosis (43). However, additional factors can contribute to the infectivity, virulence, and the severity of the disease. Given its complexity, effective common scab management strategies are not currently available, in part, because of knowledge gaps in the mechanisms of Streptomyces infection of plants (42). Our finding that heterologous S. turgidiscabies ProRSx/tRNAProA expression in E. coli provokes Ala→Pro mutations (Fig. 4) suggests that the same phenomenon may occur in Streptomyces, pointing to a previously unknown ability for proteome diversification in these plant pathogens. It is appealing to consider that by inducing the mistranslation of alanine codons these pathogens can create more infectious or more immune-evasive variants of Streptomyces proteins, as was observed in C. albicans (44).
How ProRSx and tRNAProA gene expression is controlled and whether ProRSx/tRNAProA can contribute to Streptomyces pathogenicity remains unknown. However, based on previous studies, we know that organisms tend to maintain mistranslation at low levels because of its detrimental effects on cell viability (40). It is therefore possible that ProRSx/tRNAProA expression is tightly regulated. Surprisingly, we found that tRNAProA is actively expressed in S. turgidiscabies cell cultures grown in rich or minimal media (SI Appendix, Fig. S1). However, it is unknown whether ProRSx is coexpressed with tRNAProA under these laboratory conditions. Nonetheless, it also is possible that ProRSx aminoacylation activity is inferior, relative to the two canonical ProRS isoforms; consequently, the levels of Pro-tRNAProA may remain low even if ProRSx and tRNAProA are actively expressed. Low activities are observed for the aminoacylation of pyrrolysine and selenocysteine tRNAs by pyrrolysyl- and seryl-tRNA synthetase, respectively (45, 46), which enables the translation of termination codons as pyrrolysine and selenocysteine. This low-aminoacylation activity appears to help balance the negative impact of genome-wide mistranslation of termination codons with pyrrolysine (45). Further studies to establish how ProRSx activity is regulated, and the impact of mistranslation on Streptomyces biology are needed.
Deliberate Mistranslation Might Be More Common Than We Currently Think.
More broadly, our work underlines the current tendency to underestimate the actual content of sequenced genomes. In the era of automated annotation of genes in newly sequenced genomes, there is a tendency to forcefully fit many unusual isoforms of translation factors into previously established protein families (i.e., 22 families of canonical aaRSs). Hence, we tend to underestimate the actual complexity of the protein synthesis machinery by overlooking atypical variants of aaRSs and other translation factors that play a role beyond canonical protein synthesis.
This tendency is even more prevalent for tRNA molecules. Currently, tRNAs are annotated based on their predicted length, cloverleaf secondary structure, and their anticodons (47). The commonly used automatic search program for tRNAs, tRNAscan-SE (48), yields a score aggregating the compliance of the extracted sequence with the covariance model trained on identified and characterized tRNA sequences. Low scores indicate pseudogenes or unusual tRNAs with mispairs. Indeed, our recent studies showed that by relaxing the constraints on stem lengths we were able to reveal thousands of previously unknown tRNAs and tRNA-like molecules that were either overlooked or misannotated because of unusual cloverleaf structures or an unusual combination of identity elements (49).
Similarly, tRNAProA is currently annotated as tRNAAla in several genomic databases because its A34G35C36 anticodon corresponds to alanine codons (47, 48, 50). However, the AGC anticodon is atypical, even for canonical tRNAAla. Out of 4,293 bacterial tRNAAla sequences that are deposited in the tRNA database, only two have the AGC anticodon (51). Thus, even though tRNAProA lacks tRNAAla identity elements and has an atypical alanine anticodon, it is still currently annotated as tRNAAla, highlighting the accepted postulate to systematically annotate the identity of tRNA molecules solely on the basis of the anticodon triplet in sequenced genomes. As shown here, however, any deviation from standard conservations in tRNA sequences should constitute a warning signal for alternative underlying translation processes.
In this regard, it is curious to note that tRNAProA displays some elements typical of eubacteria (the C1:G72 typical of bacterial tRNAPro does not occur in eukaryotes) but also several striking characteristics typical of eukaryotes [the Ala AGC anticodon is almost absent in eubacteria but prevalent in eukaryotes with two isodecoder families characterized by different sequence signatures (34)]. One can therefore wonder whether such tRNAs with hybrid sequence signatures could have been captured by the bacteria from the plants through some horizontal gene transfer process. Slight variations in the genetic code constitute a powerful way to fight pathogens (52). Here, the use of tRNAs of mixed discriminants offers a wider range of variations to the proteins of the pathogen.
To this end, our finding of ProRSx/tRNAProA is particularly intriguing because duplications of aaRS genes are extremely common in nature. To date, more than 25,000 aaRS gene duplications have been observed in ∼96% of organisms with sequenced genomes (25). While most of these duplicated genes are currently annotated as canonical aaRS genes, a growing number of aaRS gene duplicates are being described as noncanonical translation factors. For example, the duplication of the lysyl-tRNA synthetase gene enabled the evolution of the PoxA protein in many bacteria, which is required for the maturation of the translation factor EF-P (53). Similarly, a duplication of the phenylalanyl-tRNA synthetase gene enabled the evolution of pyrrolysyl-tRNA synthetase specific for the aminoacylation of the rare, proteinogenic amino acid, pyrrolysine (54). The duplication of leucyl-tRNA synthetase in the plant pathogen Agrobacterium radiobacter led to the evolution of an additional protein, AsgB, which protects these bacteria from agrocin 84, an inhibitor of leucyl-tRNA synthetase (55). These findings, along with our discovery of ProRSx in pathogenic Streptomyces, provide compelling evidence that some duplicated genes can encode functionally repurposed proteins that are no longer involved in canonical protein synthesis and instead play new roles in cell biology, including mistranslation. Thus, present genomes may hold many more exciting functionalities that are currently hiding under the disguise of canonical tRNA synthetases and tRNAs.
Materials and Methods
Plasmid Construction.
Unless stated otherwise, cloning was performed using the Gibson assembly kit or NEB builder kit (New England Biolabs) and E. coli Stellar cells (Clontech) or NEB Turbo cells (New England Biolabs) following the manufacturers’ protocols. DNA sequencing was performed by Keck Biotechnology Resource Laboratory at Yale University. The S. turgidiscabies ProRS1 and ProRSx gene sequences were codon optimized for expression in E. coli using the codon optimization tool from Integrated DNA Technologies (www.idtdna.com), and the corresponding DNA was purchased from QuintaraBio. The S. turgidiscabies ProRS1 and ProRSx genes were cloned into the pRSF vector. E. coli ProRS DNA was amplified from the EcProRS.pCA24N plasmid (56) and cloned into the pRSF vector. The E. coli ProRS 5ABD mutant (E524R/T531A/R537D/N538R/K554I) was generated by replacing a 133 bp E. coli ProRS in EcProRS-pET15b with a DNA fragment containing the desired mutations. S. turgidiscabies ProRSx DNA was cloned into the pET15b vector. The tRNAProA gene was cloned into the pEVOL vector (57) under the proK promoter. E. coli ProRS (WT and 5ABD variant) were then cloned into the tRNAProA-pEVOL plasmid. The tRNAProA, tRNAPro, and tRNAAla genes from S. turgidiscabies were cloned into the pUC19 vector. The sfGFP-mCherry reporter was constructed by inserting the mCherry gene downstream of the sfGFP gene within a previously described pET plasmid (58). The bla gene, coding for β-lactamase, was amplified from the pUC19 vector and cloned under a pro1 promoter in a plasmid with the pSC101 origin of replication and a kanamycin resistance marker using USER cloning.
Bioinformatics Analysis of ProRSx and tRNAProA Genes.
The amino acid sequences of ProRS1, ProRS2, and ProRSx (formerly ProRS3) from S. turgidiscabies Car8, Streptomyces species (sp.) CNT371, and Streptomyces sp. CNH099 (25) were analyzed using Clustal Omega (59) and MEGAX (60) together with representative ProRS sequences from the organisms listed on Fig. 1A. Sequences of representative ProRSs and S. turgidiscabies, Streptomyces sp. CNT371, Streptomyces sp. CNH099 ProRS1, and ProRS2 were classified into one of the two previously known bacterial ProRS isoforms, bacteria type or archaeal/eukaryote type (23). S. turgidiscabies Car8, Streptomyces sp. CNT371, and Streptomyces sp. CNH099 ProRSx sequences were used as query to search for homologous genes in bacteria using National Center for Biotechnology Information protein–protein BLAST. The final ProRS phylogenetic tree was generated using the maximum likelihood method, and the consensus tree was inferred from 100 replicates in MEGAX (60). The tree was displayed using Interactive Tree of Life (61). tRNAProA genes from the organisms that are listed in SI Appendix, Table S1 were found downstream of the putative ProRSx operon using the genomic feature in Pathosystems Resource Integration Center (50). tRNAProA genes were analyzed and their secondary structures were predicted using tRNAscan-SE (48).
Protein Purification.
E. coli cells from the ASKA library (56) harboring the EcProRS.pCA24N plasmid were used for the overexpression of E. coli ProRS. E. coli ProRS was then purified as described (62) using TALON Metal Affinity Resin (TAKARA). The ProRS-Ec-5ABD and S. turgidiscabies ProRS1 (in pET15b) were expressed in E. coli BL21 (DE3) cells. The expression was induced with 0.05 mM IPTG once the cells reached an optical density of a wavelength of 600 nm (OD600) of 0.6. After the induction, cells were grown overnight at 24 °C. Then, the cells were harvested and lysed with lysozyme (0.6 mg/mL), followed by sonication in a buffer containing 50 mM Tris (pH 8.0), 300 mM NaCl, and cOmplete EDTA-free protease inhibitor mixture tablets (Roche). Lysed cells were centrifuged at 18,000× g for 40 min at 4 °C. The lysate was run through the TALON Metal Affinity Resin, and the protein was eluted with an imidazole gradient. Protein concentrations were calculated using the Bradford assay (Bio-Rad). His-tagged bacteriophage T7 RNA polymerase was overexpressed from the pAR1219 (63) in E. coli BL21 (DE3) Codon+. Cells were grown to an OD600 of 0.6, followed by addition of 1 mM IPTG. Induction was carried out for 5 h at 37 °C. Cells were harvested and lysed with lysozyme and by sonication in buffer containing 50 mM Tris (pH8), 300 mM NaCl, 0.5 mM DTT, and cOmplete EDTA-free protease inhibitor mixture tablets (Roche). The His-tagged protein was purified using TALON Metal Affinity Resin. The enzyme was stored in 20 mM Tris⋅HCl (pH8), 100 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10% glycerol. His-tagged E. coli AlaRS and CCA-adding enzyme were purified from E. coli cells using the ASKA library, as described above for E. coli ProRS.
Preparation of tRNA Transcripts.
DNA templates for in vitro transcription were PCR-amplified from pUC19 plasmids encoding the tRNA genes using an M13F primer together with reverse primers that were complementary to the 3′-end of each tRNA. Transcription reactions were carried out with ∼10 μg DNA; 10 mM Tris⋅HCl (pH 8); 20 mM MgCl2; 2 μg/mL yeast pyrophosphatase; 1 mM spermidine; 0.01% triton X-100; 5 μg/mL bovine serum albumin (BSA); 5 mM DTT; 4 mM ATP, GTP, UTP, and CTP together with T7 RNA polymerase for 7 h at 37 °C. tRNAs were separated on a 12% urea–polyacrylamide gel electrophoresis and extracted with 500 mM ammonium acetate and 1 mM EDTA (pH 8). tRNA transcripts were labeled at the 3′-end adenosine using [32P]-ATP (PerkinElmer) and the E. coli CCA-adding enzyme following a previously described protocol (64).
Aminoacylation Assays.
Aminoacylation of E. coli tRNAPro, S. turgidiscabies tRNAPro, and S. turgidiscabies tRNAProA by E. coli WT or 5ABD ProRS was carried out with 2 mM Pro, 0.5 μM ProRS, and 4 μM tRNA (with trace amounts of [32P]-labeled tRNA) in buffer containing 25 mM Hepes (pH7.3), 4 mM ATP, 25 mM MgCl2, 0.1 mg/mL BSA, 20 mM KCl, and 20 mM two-mercaptoethanol. The time-course of the reactions was monitored by quenching 2 μL of the reaction mixture with 5 μL of a solution containing 0.1 U/μL P1 nuclease (Millipore-Sigma) and 200 mM sodium acetate, pH 5. Aminoacylation of S. turgidiscabies tRNAPro, tRNAProA, and E. coli tRNAPro by S. turgidiscabies ProRS1 were performed using [3H]-Pro (PerkinElmer) using the same reaction conditions as for E. coli ProRS. Aminoacylation of S. turgidiscabies tRNAAla and tRNAProA by E. coli AlaRS was carried out using 150 μM [14C]-Ala (American Radiolabeled Chemicals), 50 mM Hepes (pH 7.3), 4 mM ATP, 10 mM MgCl2, 0.1 mg/mL BSA, 1mM dithiothreitol, 0.5 μM AlaRS, and 4 μM tRNA. The reaction course of aminoacylation assays with radiolabeled amino acids was monitored, as previously described (52). tRNAs were refolded prior to the reaction by heating to 80 °C for 2 min immediately followed by incubation at 60 °C for 2 min and the addition of 10 mM MgCl2. tRNA was cooled down for 5 min at room temperature. All reactions were performed at 37 °C. All assays were repeated three times, and the SD is shown.
Complementation Assays in E. coli.
E. coli UQ27 cells carrying E. coli WT ProRS, S. turgidiscabies ProRSx, or S. turgidiscabies ProRS1 in the pRSF vector were grown overnight at 30 °C in Luria broth (LB) media containing kanamycin (50 μg/mL). The overnight cultures were grown on two LB-agar plates containing kanamycin. Complementation assays with E. coli WT and 5ABD ProRS or S. turgidiscabies ProRSx (in pEVOL) in the absence or presence of S. turgidiscabies tRNAProA were performed in LB-agar plates containing chloramphenicol and 0.1% arabinose. The cells were serially diluted from an overnight culture (at 30 °C), and 4 μL cell culture was spotted. The plates were incubated separately at 30 °C and 42 °C overnight.
β-Lactamase–based Assays.
S2060 chemically competent cells were cotransformed with β-lactamase reporter (WT or P65A) alongside the pEVOL vector encoding S. turgidiscabies tRNAProA and either E. coli ProRS (WT or 5ABD) or S. turgidiscabies ProRSx. Transformations were recovered for 45 min with shaking at 37 °C incubator using 2× YT liquid medium. Following recovery, transformants were streaked on 1.8% agar–2× YT plates supplemented with kanamycin (50 µg/mL) and chloramphenicol (35 µg/mL). Plates were grown in a 37 °C incubator for 16 h. Colonies were picked into DRM (United States Biological), supplemented with kanamycin (50 µg/mL) and chloramphenicol (35 µg/mL), and grown overnight at 37 °C at 900 rpm. Following overnight growth, cultures were back diluted 1,000-fold into a 96-well deep well plate with fresh DRM supplemented with kanamycin, chloramphenicol, and a gradient of twofold, increasing concentrations of carbenicillin (1,000 µg/mL to 0.5 µg/mL). Following an additional 20 h of growth at 37 °C at 900 rpm, 150 µL culture was aliquoted into a 96-well black wall, clear bottom plate (Costar). OD600 was measured using a Spark (Tecan) plate reader. IC50 curves were fit using GraphPad Prism version 9.
Fluorescence-based Assays.
The Ser65 of GFP was mutated to Ala (GCT) or Pro (CCT) in the plasmid containing the GFP-mCherry fusion reporter. The effect of each mutation on GFP was assessed in E. coli TOP10 cells. A total of 4 µL overnight culture grown overnight in LB media with ampicillin at 37 °C was spotted on LB-agar plates containing ampicillin and 0.1 mM IPTG. The plate was incubated at 37 °C overnight. GFP and mCherry fluorescence was detected using a Bio-Rad ChemiDoc system. To examine mistranslation, E. coli UQ27 cells harboring the E. coli ProRS WT or 5ABD ProRS (in pEVOL) in the absence or presence of S. turgidiscabies tRNAProA together with the GFP_S65A-mCherry reporter plasmid were grown in LB media with chloramphenicol and ampicillin at 30 °C overnight. A total of 99 µL fresh LB media (with antibiotics, 0.2 mM IPTG, and 0.05% arabinose) were inoculated with 1 µL of overnight culture and transferred into a 96-well black plate with a clear flat bottom (Corning). Cells were grown at 42 °C in a BioTek synergy plate reader, and GFP (485 nm/528 nm) and mCherry (590 nm/645 nm) fluorescence and cell density were measured after 5 h. The GFP and mCherry fluorescence signals were corrected by subtracting the background fluorescence from cells in which GFP and mCherry expression is repressed with 1% glucose. The mistranslation rate was estimated, as described (38). To confirm the incorporation of Pro in Ala codons using mass spectrometry, E. coli UQ27 cells harboring the E. coli ProRS in pEVOL and the GFP-mCherry reporter were grown in LB media with the appropriate antibiotics at 42 °C to an OD600 of 0.6. Expression of GFP-mCherry was induced with 0.2 mM IPTG for 5 h at 42 °C. Cells were harvested by centrifugation and lysed with 1× Bugbuster, 50 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole, and cOmplete EDTA-free protease inhibitor mixture tablets. The resuspended cells were mixed with Benzonase (Sigma) and rotated at room temperature for 20 min. The lysate was cleared by centrifugation and ran through TALON resin. The immobilized His-tagged GFP-mCherry was washed twice with 10 mM imidazole, 50 mM Tris⋅HCl (pH 8.0), and 150 mM NaCl and eluted with 300 mM imidazole. The protein buffer was exchanged to 50 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl and stored at −20 °C. The samples were analyzed by liquid chromatography–tandem mass spectrometry (details in SI Appendix).
Growth Curve Assays.
E. coli UQ27 cells with E. coli WT or 5ABD ProRS-tRNAProA-pEVOL were grown overnight in LB media with chloramphenicol at 30 °C. Cells were diluted to an OD600 of ∼0.02 in 125 μL LB media with chloramphenicol. Growth was monitored in 96-well black plates with clear flat bottoms (Corning) for 13 h at 42 °C using a BioTek synergy plate reader. The results represent the average of six biological replicates, and the SD is indicated as error bars in each time point.
Northern Blot Analysis.
S. turgidiscabies Car8 cells were grown in 5 mL tryptic soy broth (1.7% tryptone, 0.5% NaCl, 0.3% soy flour, 0.25% glucose, and 0.25% K2HPO4) or minimal medium (0.05% l-asparagine, 0.05% K2HPO4, 0.02% MgSO4, and 0.001% FeSO4) and 1% glucose for 24 h. A total of 600 mL fresh media was inoculated with the small culture, and cells were grown for 4 d at 29 °C. Cells were lysed by sonication in buffer containing 200 mM sodium acetate, 1 mM EDTA, and 4 M guanidinium hydrochloride (pH 5.2). Total RNA was isolated by mixing equal volumes of cell lysate and cold phenol (pH 4.5) followed by incubation for 15 min on ice. The RNA was precipitated using ethanol and stored −20 °C. Total RNA was also isolated from E. coli Stellar cell carrying either the EcProRS.tRNAProA.pEVOL or EcProRS.pEVOL plasmid. A total of 4 μg RNA was electrophoresed, transferred, and cross-linked to a Nylon membrane (GE Healthcare), as described previously (53). A 32P-labeled, single-stranded DNA (5′-CAGGCTCCTCGGCAACG-3′) probe was used for the detection of tRNAProA.
Statistical Analysis.
All experiments were repeated at least three times. The averages of the results are shown in figures with the corresponding SDs. Unpaired t tests and SDs were calculated using GraphPad Prism version 9.
Supplementary Material
Acknowledgments
We thank Christina Chung, Natalie Krahn, Jonathan Fischer, Jeffrey Tharp, and Kazuaki Amikura for critical comments on the manuscript and Dr. Dawn Bignell and Gustavo Diaz-Cruz for inspired discussions. Research was supported by National Institute of General Medical Sciences Grants R35GM122560 (to D.S.) and R35GM128742 (to Y.D.) and Royal Society Research Grant RGS\R2\202003 (to S.M.). A.H.B. was supported by the NIH Director’s Early Independence Award (DP5-OD-024590), the NASA Exobiology Program Award (NNH17ZDA001N-EXO), and the Broad Institute of Massachusetts Institute of Technology and Harvard.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110797118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (65) partner repository with the dataset identifier PXD026636.
References
- 1.Schimmel P., Guo M., A tipping point for mistranslation and disease. Nat. Struct. Mol. Biol. 16, 348–349 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Schimmel P., Mistranslation and its control by tRNA synthetases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2965–2971 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drummond D. A., Wilke C. O., The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 10, 715–724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribas de Pouplana L., Santos M. A. S., Zhu J.-H., Farabaugh P. J., Javid B., Protein mistranslation: Friend or foe? Trends Biochem. Sci. 39, 355–362 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Mohler K., Ibba M., Translational fidelity and mistranslation in the cellular response to stress. Nat. Microbiol. 2, 17117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samhita L., Raval P. K., Agashe D., Global mistranslation increases cell survival under stress in Escherichia coli. PLoS Genet. 16, e1008654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan T., Adaptive translation as a mechanism of stress response and adaptation. Annu. Rev. Genet. 47, 121–137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchner S., Ignatova Z., Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 16, 98–112 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Rocha R., Pereira P. J. B., Santos M. A. S., Macedo-Ribeiro S., Unveiling the structural basis for translational ambiguity tolerance in a human fungal pathogen. Proc. Natl. Acad. Sci. U.S.A. 108, 14091–14096 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki T., Ueda T., Watanabe K., The ‘polysemous’ codon—A codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 16, 1122–1134 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miranda I., et al., A genetic code alteration is a phenotype diversity generator in the human pathogen Candida albicans. PLoS One 2, e996 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L., et al., Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl. Acad. Sci. U.S.A. 108, 9378–9383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su H.-W., et al., The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat. Microbiol. 1, 16147 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Bacher J. M., Waas W. F., Metzgar D., de Crécy-Lagard V., Schimmel P., Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi. J. Bacteriol. 189, 6494–6496 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling J., Söll D., Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. U.S.A. 107, 4028–4033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans C. R., Fan Y., Ling J., Increased mistranslation protects E. coli from protein misfolding stress due to activation of a RpoS-dependent heat shock response. FEBS Lett. 593, 3220–3227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melnikov S. V., van den Elzen A., Stevens D. L., Thoreen C. C., Söll D., Loss of protein synthesis quality control in host-restricted organisms. Proc. Natl. Acad. Sci. U.S.A. 115, E11505–E11512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giegé R., Sissler M., Florentz C., Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26, 5017–5035 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melnikov S. V., et al., Error-prone protein synthesis in parasites with the smallest eukaryotic genome. Proc. Natl. Acad. Sci. U.S.A. 115, E6245–E6253 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loenarz C., et al., Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl. Acad. Sci. U.S.A. 111, 4019–4024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witzky A., Tollerson R. II, Ibba M., Translational control of antibiotic resistance. Open Biol. 9, 190051 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sárkány Z., Silva A., Pereira P. J. B., Macedo-Ribeiro S., Ser or Leu: Structural snapshots of mistranslation in Candida albicans. Front. Mol. Biosci. 1, 27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crepin T., Yaremchuk A., Tukalo M., Cusack S., Structures of two bacterial prolyl-tRNA synthetases with and without a cis-editing domain. Structure 14, 1511–1525 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Chaliotis A., et al., The complex evolutionary history of aminoacyl-tRNA synthetases. Nucleic Acids Res. 45, 1059–1068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubio M. A., et al., Trans-oligomerization of duplicated aminoacyl-tRNA synthetases maintains genetic code fidelity under stress. Nucleic Acids Res. 43, 9905–9917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vargas-Rodriguez O., Musier-Forsyth K., Exclusive use of trans-editing domains prevents proline mistranslation. J. Biol. Chem. 288, 14391–14399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisberg A. J., et al., A novel species-level group of Streptomyces exhibits variation in phytopathogenicity despite conservation of virulence loci. Mol. Plant Microbe Interact. 34, 39–48 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Loria R., Kers J., Joshi M., Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol. 44, 469–487 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Hou Y. M., Schimmel P., A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333, 140–145 (1988). [DOI] [PubMed] [Google Scholar]
- 30.McClain W. H., Foss K., Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 240, 793–796 (1988). [DOI] [PubMed] [Google Scholar]
- 31.Drabkin H. J., Helk B., RajBhandary U. L., The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J. Biol. Chem. 268, 25221–25228 (1993). [PubMed] [Google Scholar]
- 32.Perreau V. M., et al., The Candida albicans CUG-decoding ser-tRNA has an atypical anticodon stem-loop structure. J. Mol. Biol. 293, 1039–1053 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Marck C., et al., The RNA polymerase III-dependent family of genes in hemiascomycetes: Comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 34, 1816–1835 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westhof E., et al., Unusual tertiary pairs in eukaryotic tRNAAla. RNA 26, 1519–1529 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohman K., Isaksson L. A., A temperature-sensitive mutant in prolinyl-tRNA ligase of Escherichia coli K-12. Mol. Gen. Genet. 177, 603–605 (1980). [DOI] [PubMed] [Google Scholar]
- 36.Yaremchuk A., Cusack S., Tukalo M., Crystal structure of a eukaryote/archaeon-like prolyl-tRNA synthetase and its complex with tRNAPro(CGG). EMBO J. 19, 4745–4758 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakano H., Okumura R., Goto C., Yamane T., In vitro combinatorial mutagenesis of the 65th and 222nd positions of the green fluorescent protein of Aequarea victoria. Biotechnol. Bioprocess Eng. 7, 311–315 (2002). [Google Scholar]
- 38.Chen Y.-X., Pan M., Chen Y.-M., Javid B., Measurement of specific mycobacterial mistranslation rates with gain-of-function reporter systems. J. Vis. Exp. 146, e59453 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Bezerra A. R., et al., The role of non-standard translation in Candida albicans pathogenesis. FEMS Yeast Res. 21, foab032 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling J., O’Donoghue P., Söll D., Genetic code flexibility in microorganisms: Novel mechanisms and impact on physiology. Nat. Rev. Microbiol. 13, 707–721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bignell D. R., Huguet-Tapia J. C., Joshi M. V., Pettis G. S., Loria R., What does it take to be a plant pathogen: Genomic insights from Streptomyces species. Antonie Van Leeuwenhoek 98, 179–194 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Wanner L. A., Kirk W. W., Streptomyces—From basic microbiology to role as a plant pathogen. Am. J. Potato Res. 92, 236–242 (2015). [Google Scholar]
- 43.Li Y., Liu J., Díaz-Cruz G., Cheng Z., Bignell D. R. D., Virulence mechanisms of plant-pathogenic Streptomyces species: An updated review. Microbiology (Reading) 165, 1025–1040 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Miranda I., et al., Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio 4, e00285-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herring S., Ambrogelly A., Polycarpo C. R., Söll D., Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 35, 1270–1278 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baron C., Böck A., The length of the aminoacyl-acceptor stem of the selenocysteine-specific tRNA(Sec) of Escherichia coli is the determinant for binding to elongation factors SELB or Tu. J. Biol. Chem. 266, 20375–20379 (1991). [PubMed] [Google Scholar]
- 47.Chan P. P., Lowe T. M., GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowe T. M., Chan P. P., tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukai T., et al., Transfer RNAs with novel cloverleaf structures. Nucleic Acids Res. 45, 2776–2785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis J. J., et al., The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 48, D606–D612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin B. Y., Chan P. P., Lowe T. M., tRNAviz: Explore and visualize tRNA sequence features. Nucleic Acids Res. 47, W542–W547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shackelton L. A., Holmes E. C., The role of alternative genetic codes in viral evolution and emergence. J. Theor. Biol. 254, 128–134 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Yanagisawa T., Sumida T., Ishii R., Takemoto C., Yokoyama S., A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 17, 1136–1143 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Kavran J. M., et al., Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. U.S.A. 104, 11268–11273 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chopra S., et al., Structural characterization of antibiotic self-immunity tRNA synthetase in plant tumour biocontrol agent. Nat. Commun. 7, 12928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitagawa M., et al., Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 12, 291–299 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Young T. S., Ahmad I., Yin J. A., Schultz P. G., An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 395, 361–374 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Ko J.-h., et al., Pyrrolysyl-tRNA synthetase variants reveal ancestral aminoacylation function. FEBS Lett. 587, 3243–3248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sievers F., Higgins D. G., Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong F.-C., Beuning P. J., Nagan M., Shiba K., Musier-Forsyth K., Functional role of the prokaryotic proline-tRNA synthetase insertion domain in amino acid editing. Biochemistry 41, 7108–7115 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W., Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 81, 2035–2039 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ledoux S., Uhlenbeck O. C., [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44, 74–80 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perez-Riverol Y.et al., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (65) partner repository with the dataset identifier PXD026636.