Significance
Nonreplicating (NR) Mycobacterium tuberculosis (Mtb) was thought to be drug tolerant due to its low metabolic activity. However, drug tolerance often gradually increases by exposure to antibiotics, inferring the role of an adaptive strategy. This study established that drug tolerance of NR Mtb is associated with phosphoenolpyruvate (PEP) anabolic down-regulation. PEP is a substrate of multiple pathways needed for replication; in NR Mtb, their functions were abolished due to PEP depletion. Intriguingly, PEP supplementation partly restored drug sensitivity and prevented the emergence of drug resistance (DR). The results expand our knowledge of how NR Mtb remodels its metabolic networks and propose potential Mtb metabolites as a source of therapeutic adjuvants to synthetically kill NR Mtb and prevent development of DR.
Keywords: tuberculosis, metabolic remodeling, drug tolerance, synthetic lethality, phosphoenolpyruvate
Abstract
Mycobacterium tuberculosis (Mtb) infection is difficult to treat because Mtb spends the majority of its life cycle in a nonreplicating (NR) state. Since NR Mtb is highly tolerant to antibiotic effects and can mutate to become drug resistant (DR), our conventional tuberculosis (TB) treatment is not effective. Thus, a novel strategy to kill NR Mtb is required. Accumulating evidence has shown that repetitive exposure to sublethal doses of antibiotics enhances the level of drug tolerance, implying that NR Mtb is formed by adaptive metabolic remodeling. As such, metabolic modulation strategies to block the metabolic remodeling needed to form NR Mtb have emerged as new therapeutic options. Here, we modeled in vitro NR Mtb using hypoxia, applied isotope metabolomics, and revealed that phosphoenolpyruvate (PEP) is nearly completely depleted in NR Mtb. This near loss of PEP reduces PEP-carbon flux toward multiple pathways essential for replication and drug sensitivity. Inversely, supplementing with PEP restored the carbon flux and the activities of the foregoing pathways, resulting in growth and heightened drug susceptibility of NR Mtb, which ultimately prevented the development of DR. Taken together, PEP depletion in NR Mtb is associated with the acquisition of drug tolerance and subsequent emergence of DR, demonstrating that PEP treatment is a possible metabolic modulation strategy to resensitize NR Mtb to conventional TB treatment and prevent the emergence of DR.
Despite significant efforts to end tuberculosis (TB), TB is still a leading killer among infectious diseases worldwide, claiming 1.5 million lives and making 10.5 million individuals ill each year (1–3). The hallmark of Mycobacterium tuberculosis (Mtb) pathogenesis is its ability to survive under a range of antimicrobial environments. After invading, recruitment of immune cells at the site of infection causes granuloma formation, a multicellular structure that traps uncleared Mtb. The interior of the granuloma is hypoxic, nutritionally starved, acidic, and filled with biochemical antimicrobial effectors such as reactive oxygen species (ROS) and reactive nitrogen intermediates. Mtb is able to survive under these conditions because it enters a nonreplicating (NR) persistent state (4, 5). The reduced efficacy of a conventional TB treatment method is largely attributed to this Mtb phenotypic heterogeneity ranging from replicating to NR states, a bet-hedging strategy that supports Mtb survival in response to antibiotic treatment and the host immune system (6). Although the precise mechanisms of how Mtb enters the NR state are unclear, the proportion of NR Mtb can be gradually enhanced by intermittent exposure to antibiotics, suggesting that NR Mtb is formed by adaptive strategies, to avoid irreversible antimicrobial damage (7). Indeed, our recent reports showed that the tolerance to antibiotics of NR Mtb was largely attributed to metabolic shifts including functional depletion of metabolic processes such as glycolipid biosynthesis, the canonical TCA cycle, and electron transport chain (ETC) activities that are required for Mtb replication (8–11). Conventional TB drugs kill Mtb by inhibiting the very processes that become dispensable for NR Mtb viability, thus rendering NR Mtb resistant to treatment.
During the course of infection, activated macrophages release antimicrobial effectors such as ROS. Recent metabolomics studies using NR Mtb collected under hypoxia or in vitro biofilm culture achieved drug tolerance by remodeling its trehalose metabolism by switching trehalose-carbon flux toward the biosynthesis of central carbon metabolism (CCM) intermediates while decreasing the flux to the biosynthesis of cell wall glycolipids (8, 9). This caused accumulation of intermediates in the early portion of glycolysis (GL) such as glucose 6-phosphate (Glc-P) and fructose 1,6-bisphosphate (FBP), and near complete depletion of phosphoenolpyruvate (PEP), a metabolite in the low portion of GL (8). We separately showed that NR Mtb under hypoxia also altered the TCA cycle by shifting the carbon flux through the glyoxylate shunt to secure succinate. A portion of synthesized succinate was secreted through the membrane to maintain membrane potential (ΔΨm) and ATP levels in the absence of ETC terminal electron acceptors (10, 12). The accompanied bypassing of the oxidative branch of the TCA cycle enabled NR Mtb to avoid overproduction of NADH (the first substrate of the ETC) and ROS.
There is high demand for new TB treatment methods that can quickly eradicate NR Mtb, since the existing arsenal of TB drugs no longer provides effective protection against NR Mtb and drug resistant (DR) TB (13). This ineffectiveness is compounded by antibiotics with reduced penetration efficacy through the NR Mtb cell wall as compared to that of its replicating counterparts (14, 15). Since NR Mtb survival does not rely on the metabolic networks that are targets of conventional TB drugs, novel approaches killing NR Mtb are needed and may include blocking NR Mtb metabolic remodeling and increasing the penetration of TB drugs. Recent reports indicated that agents that restore NR Mtb membrane bioenergetics caused an increase in drug uptake (16). Thus, we hypothesized that metabolic activities altered in NR Mtb play a role in NR Mtb drug tolerance, and reagents that prevent NR Mtb metabolic remodeling can render NR Mtb sensitive to conventional TB drugs.
Our metabolomics profile of NR Mtb collected under hypoxia showed accumulation of intermediates in GL and the reductive branch of the TCA cycle, with reciprocal depletion of PEP and oxidative branch intermediates of the TCA cycle such as α-ketoglutarate (αKG). Accumulating GL intermediates included FBP, which is known to allosterically enhance pyruvate kinase (Pyk), thereby helping deplete PEP in NR Mtb (17, 18). PEP depletion also caused an increase in the pyruvate to PEP ratio, which is known to regulate TCA cycle gene expression (19, 20). Thus, PEP depletion may affect multiple cellular metabolic processes that are involved in NR Mtb metabolic remodeling.
PEP is the ester derived from the enol of pyruvate and phosphate and is an important intermediate in multiple metabolic networks (21). PEP has the high energy phosphate bond that is involved in GL and the TCA cycle in all organisms. In GL, PEP is formed by the action of the enzyme enolase. Subsequent conversion from PEP to pyruvate by Pyk generates ATP via substrate level phosphorylation. PEP is also formed from oxaloacetate (OAA) after decarboxylation and accompanied hydrolysis of GTP. This is catalyzed by PEP carboxykinase (PckA), which is the rate-limiting step in gluconeogenesis (22, 23). PEP is an enol source that bridges the glycan and peptide of nascent peptidoglycan (PG) biosynthesis for bacterial cell wall construction (24, 25). Separately, PEP initiates the shikimate pathway by condensing with erythrose 4-phosphate, thereby biosynthesizing chorismate, dihydrofolate, and aromatic amino acids. Thus, multiple bacterial metabolic processes are presumably affected by intrabacterial PEP availability. Furthermore, PEP can penetrate the cell membrane and transfer its high-energy phosphate group to ADP and replenish ATP levels (26). Taken together, PEP is functionally linked to multiple metabolic processes, all of which play an essential role for Mtb replication.
In the present study, we identified that growth arrest and drug tolerance of NR Mtb are largely attributed to defects in PEP anabolic and catabolic activities. We used isotope metabolomics to show that PEP depletion led to a down-regulation in the activities of the oxidative branch of the TCA cycle and an increase in the carbon flux toward the biosynthesis of triacylglycerol (TAG), which caused a depletion in NADH levels and ETC activities. Down-regulation of ETC activities directly affected membrane bioenergetics and drug uptake, rendering NR Mtb drug tolerant. In parallel, PEP depletion also led to a down-regulation of multiple metabolic pathways required for replication including folate metabolism, aromatic amino acid metabolism, and PG biosynthesis. PEP supplementation was sufficient to partly restore growth and drug susceptibility of NR Mtb even under hypoxia. Finally, we showed that PEP depletion caused emergence of DR mutations. This study demonstrates that the catalytic depletion of PEP is a trigger for the formation of NR Mtb, thereby implying that PEP supplementation can potentially block NR Mtb metabolic remodeling and be a therapeutic option to kill NR Mtb and prevent the emergence of DR.
Results
Growth Arrest and Drug Tolerance of NR Mtb Are Attributed to Metabolic Remodeling.
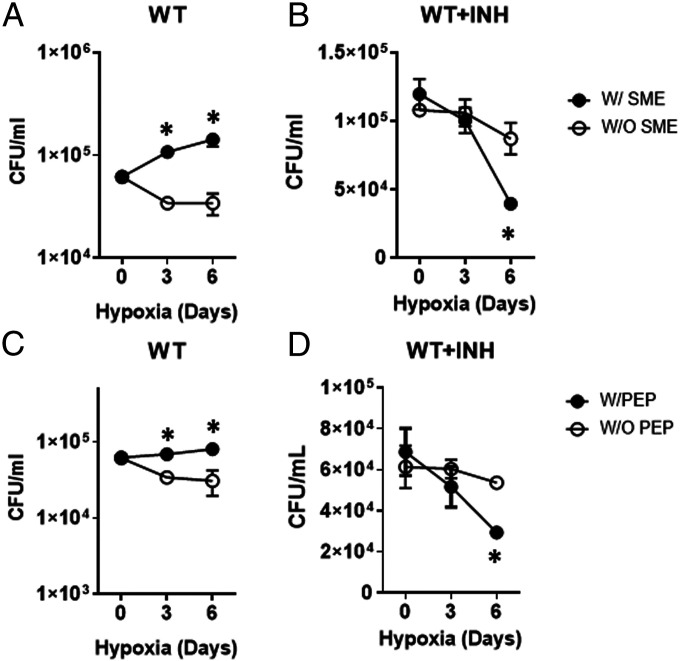
Hypoxia is the most popular environmental stress for modeling NR Mtb in vitro and studying its drug tolerance (27). Our hypoxia chamber achieved 1% O2 and 5% CO2, and a colony forming unit (CFU)-viability test confirmed that NR Mtb under hypoxia (hypoxic Mtb) neither replicated nor died. This growth arrest was accompanied by a high level of drug tolerance against isoniazid (INH) during 6 d of incubation (SI Appendix, Fig. S1A and B), confirming that the Mtb within our hypoxia chamber underwent growth arrest and drug tolerance (10). We monitored the metabolic alterations induced by hypoxia to elucidate NR Mtb metabolic remodeling relevant to growth arrest and drug tolerance. Unsupervised hierarchical clustering of hypoxia-induced changes in the Mtb metabolome identified discrete sets of accumulated or depleted metabolites (SI Appendix, Fig. S2A). A volcano plot identified significantly altered metabolites (SI Appendix, Table S1), and a pathway enrichment analysis indicated that central carbon metabolism, amino acid metabolism, nucleotide metabolism, and pantothenate and CoA pathways belonged to high ranked pathways (SI Appendix, Fig. S2 B and C). The metabolomics analysis implied that growth arrest and drug tolerance of NR Mtb were, directly or indirectly, associated with metabolic remodeling induced by hypoxia. To confirm this, we prepared metabolite extracts of Mtb in a replicating state (small metabolite extract [SME]) by harvesting midlog phase in Middlebrook 7H9 liquid media (m7H9) under 20% O2 (normoxia) and replacing the solvent with 20 mM Tris-Cl (pH 7.4) (28, 29). Then, Mtb-laden filters on Middlebrook 7H10 agar media (m7H10) were inserted to our hypoxia chamber that was preinstalled with two squeezable plastic Pasteur pipettes (10): one for SME and the other for 10× minimal inhibitory concentration (MIC) equivalent INH (SI Appendix, Fig. S3A). After 24 h, Mtb on m7H10 were treated with the SME and then INH after 18 h incubation without breaking hypoxia. CFUs showed that pretreatment with SME was sufficient to protect an initial CFU reduction of hypoxic Mtb and even enhanced its growth over time (Fig. 1A). The enhanced growth was also accompanied by heightened susceptibility against INH by ∼65% compared to that of NR Mtb (Fig. 1B). The improved growth and drug susceptibility of hypoxic Mtb due to a continuous supply of replicating Mtb metabolites suggested that NR Mtb achieves growth arrest and drug tolerance largely by remodeling metabolic networks.
Fig. 1.
Growth arrest and drug tolerance of NR Mtb under hypoxia is associated with metabolic remodeling. The effect of supplementing with SME alone (A) or in the presence of INH (B) on hypoxic Mtb viability. The effect of supplementing with PEP alone (C) or in the prescence of INH (D) on hypoxic Mtb viability. In B and D, day 0 indicated the moment after treatment with INH (at 18 h after treatment with either SME or PEP). Experiments were conducted in m7H9 containing 0.2% acetate. All values were the average of triplicates ± SEM and representative of at least two independent experiments. *P < 0.01 by Student’s t test.
The NR Mtb Phenotype under Hypoxia Is Partly Attributed to PEP Depletion.
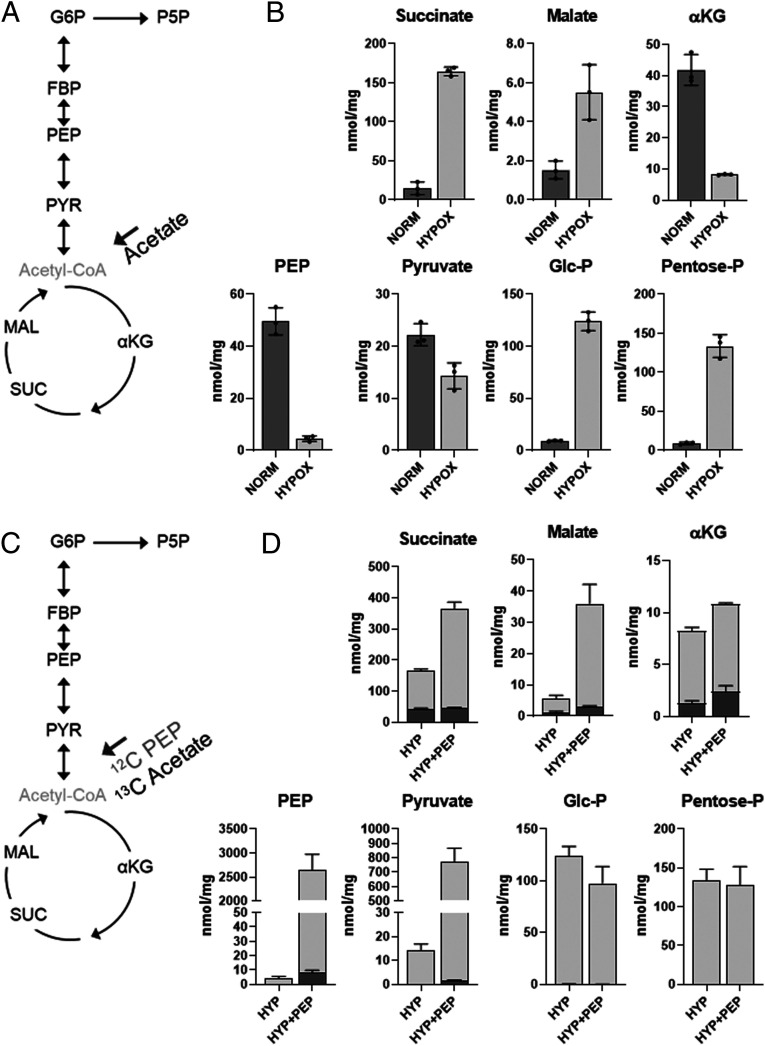
To pinpoint the specific metabolites that contributed to NR Mtb growth arrest and drug tolerance, we analyzed the metabolomics profiles of normoxic and hypoxic Mtb (SI Appendix, Fig. S2A). Our semiuntargeted metabolomics showed that CCM and pyruvate metabolism pathways are high-ranked pathways (SI Appendix, Fig. S2 B and C and Table S1). Targeted metabolomics was conducted by focusing on CCM intermediates because they are abundant and functionally interconnected with multiple pathways, including sucrose metabolism, nucleotide metabolism, and amino acid metabolism. We observed two metabolites that were substantially depleted in hypoxia: PEP, an intermediate bridging GL and the TCA cycle, and αKG, an oxidative branch intermediate of the TCA cycle (Fig. 2 A and B and SI Appendix, Fig. S4). Consistent with recent reports (8, 9), these depletions were catalytically linked to accumulation of intermediates in the early portion of GL (Glc-P and FBP) and reductive branch of the TCA cycle (succinate and malate) (Fig. 2B and SI Appendix, Fig. S4). This implied that the carbon fluxes from both GL and the TCA cycle toward PEP biosynthesis were profoundly decreased. To interrogate which steps in GL contributed to PEP depletion, we monitored mRNA levels of GL genes and identified that expression of aldolase, glyceraldehyde 3-phosphate dehydrogenase (G3PDH), and enolase were lower than that of other GL genes (SI Appendix, Fig. S5), which corroborated our metabolomics profile (Fig. 2B and SI Appendix, Fig. S4). To probe the role of PEP depletion in NR Mtb, we supplemented NR Mtb with PEP. Due to known PEP structural instability, we first monitored the stability of 10 mM PEP after incubating in media and found that its abundance was largely unchanged for at least 3 d with minor production of pyruvate presumably due to the spontaneous hydrolysis of PEP; the concentration of newly produced pyruvate was ∼18 μM at day 3. Intrabacterial PEP abundance in hypoxic Mtb was maintained for 3 d during supplementing with 10 mM PEP, suggesting a continuous PEP supply from the media (SI Appendix, Fig. S6 A and B). In contrast, exogenous pyruvate in media couldn’t support PEP biosynthesis within hypoxic Mtb, indicating that pyruvate is not a substrate for PEP, at least under hypoxia (SI Appendix, Fig. S6C). These findings affirmed that phenotypic restoration of hypoxic Mtb after PEP supplementation was independent of pyruvate arising from PEP hydrolysis in media. This was confirmed by a CFU viability test of hypoxic Mtb after adding pyruvate. Treatment with PEP, but not pyruvate, under continuous hypoxia significantly mitigated the initial CFU reduction of hypoxic Mtb, albeit to a lesser extent than that of SME, and rendered the hypoxic growth by ∼2.5-fold in the final viable biomass compared to controls without PEP (Fig. 1C and SI Appendix, Fig. S3B). Similar to SME, supplementation with PEP, but not pyruvate, partly improved hypoxic Mtb susceptibility to INH (Fig. 1D and SI Appendix, Fig. S3B). Taken together, growth arrest and drug tolerance of NR Mtb are at least in part attributed to down-regulation of PEP biosynthetic activities.
Fig. 2.
The effect of exogenous PEP on hypoxic Mtb CCM intermediates. (A) Schematic depicting Mtb CCM. 12C acetate was used as a single carbon source. G6P, glucose phosphate (Glc-P); P5P, pentose phosphate (Pentose-P); SUC, succinate; MAL, malate. (B) Intrabacterial pool sizes (nmol/mg protein) of select Mtb CCM intermediates following incubation in acetate media for 24 h at either 20% O2 (NORM) or 1% O2 (HYP). Total bar heights indicate intrabacterial concentrations. (C) Schematic depicting Mtb CCM. [U-13C] acetate was used as a carbon source with or without 5 mM 12C PEP. (D) The effect of PEP on abundance and 13C enrichment of hypoxic Mtb CCM metabolites. Total bar heights indicate intrabacterial pool sizes (nmol/mg protein), in which the dark gray of each bar denotes the enrichment of 13C labeling achieved after transfer to [U-13C] acetate under the indicated conditions. All values are the average of three biological replicates ± SEM and representative of two independent experiments.
Multiple Cellular Processes Needed for Replication Were Down-Regulated by PEP Depletion.
The metabolomics profile of hypoxic Mtb showed accumulation of Glc-P and FBP due to a blockage of GL-carbon flux toward PEP biosynthesis. This resulted in an increased pyruvate to PEP ratio, a redox biomarker known to regulate TCA cycle gene expression. Since PEP is a substrate of multiple pathways, including PG biosynthesis, the shikimate pathway, and pyruvate metabolism, we hypothesized that the growth of NR Mtb is slowed or halted by down-regulating the activities of these pathways due to PEP depletion.
-
I.
TCA cycle: We quantified the levels of hypoxic Mtb TCA cycle intermediates and 13C enrichment after supplementing with PEP and fully 13C labeled ([U-13C]) acetate, a potential carbon source encountered during host infection (30). Intrabacterial PEP abundance was maintained following treatment with increasing doses of PEP (1 to 10 mM) (SI Appendix, Fig. S6D). Treatment with PEP increased the 12C fraction of pyruvate and reductive branch intermediates of the TCA cycle (succinate and malate), and both 12C and 13C fractions of αKG, an oxidative branch metabolite of the TCA cycle (Fig. 2D). This implied that PEP was not only a direct substrate for the synthesis of TCA cycle intermediates but also helped increase uptake and consumption of [U-13C] acetate, especially for the biosynthesis of oxidative branch intermediates of the TCA cycle.
-
II.
Pyk activity: PEP is a substrate of Pyk, an enzyme involved in the biosynthesis of pyruvate and ATP. Intriguingly, pyk mRNA was higher in hypoxic Mtb than in normoxic Mtb (SI Appendix, Fig. S5B). Separately, FBP, which accumulated in NR Mtb, is known to enhance Pyk activity. Thus, NR Mtb maintained Pyk activity, thereby facilitating PEP depletion. Inversely, exogenous PEP sustained Pyk activity, resulting in the production of pyruvate and ATP (Fig. 2D), and helped restore the canonical TCA cycle activity even during down-regulated ETC activity.
-
III.
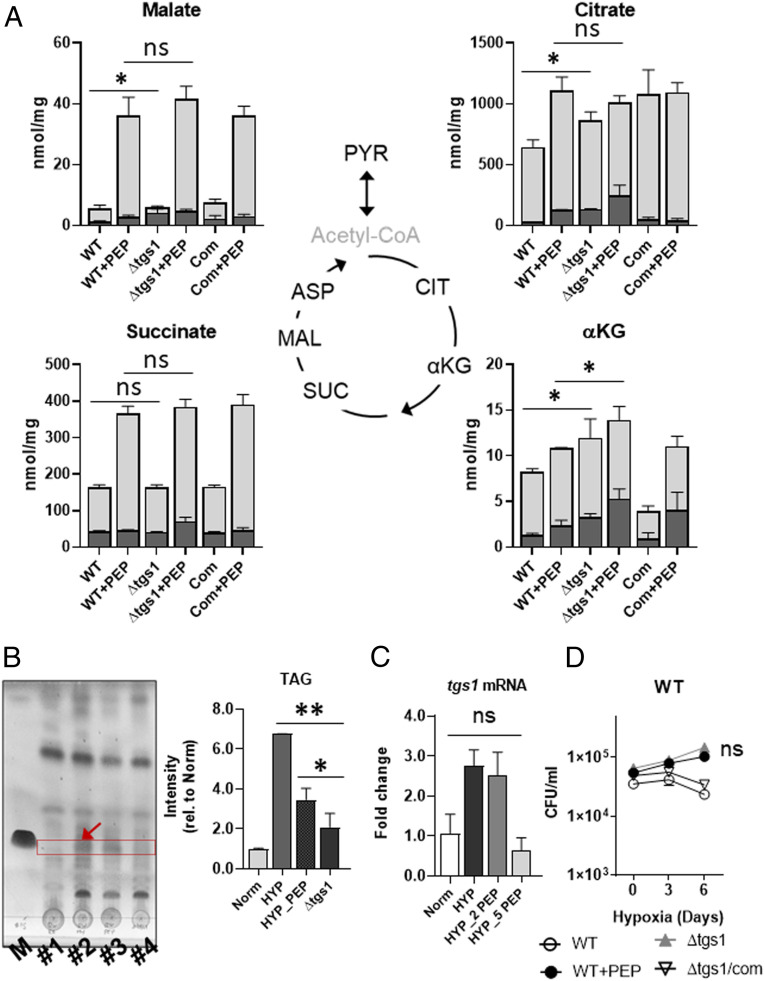
TAG biosynthesis: Baek et al. showed that hypoxic Mtb becomes drug tolerant through a carbon flux shift toward TAG biosynthesis and away from the oxidative branch of the TCA cycle (31, 32). To explore the impact of PEP depletion on this carbon flux shift, we monitored metabolomics profiles and TAG abundance of hypoxic Mtb following PEP supplementation. In response to PEP supplementation, TAG levels of hypoxic Mtb were significantly decreased with an increase in pool size and 13C enrichment of oxidative TCA cycle intermediates such as citrate and αKG (Figs. 2D and 3 A and B). This suggested that PEP depletion helped alter the acetate-carbon flux in NR Mtb toward the biosynthesis of TAG from TCA cycle intermediates. To confirm this, we used tgs1 (encoding a TAG synthase) deficient Mtb (Δtgs1) and conducted isotope metabolomics profiling using [U-13C] acetate under hypoxia. Consistent with previous literature, there was increased abundance and 13C enrichment of oxidative branch intermediates of the TCA cycle in hypoxic Δtgs1 compared to those of wild-type (WT) and Δtgs1/com strains under hypoxia (Fig. 3A). Intriguingly, with the exception of αKG, these levels became similar to those found in hypoxic Mtb supplemented with PEP (Fig. 3A), confirming that the acetate-carbon flux toward TAG biosynthesis in hypoxic Mtb was at least in part attributed to PEP mediated metabolic modulation. This is supported by the findings that adding PEP to hypoxic Mtb restored tgs1 mRNA expression level, hypoxic growth, and TAG abundance to levels that were similar to those of normoxic Mtb and hypoxic Δtgs1 (Fig. 3 B–D).
-
IV.
PG biosynthesis: PEP is a substrate of MurA, the first committed step of PG biosynthesis (SI Appendix, Fig. S7A). The metabolomics profile of hypoxic Mtb showed that hypoxia caused accumulation of Glc-P, GlcNAc-P, and UDP-GlcNAc, the intermediates above the MurA step, and depletion of intermediates lower than the MurA step, such as UDP-MurNAc, UDP-MurNAc-dipeptides, and d-Ala-d-Ala (SI Appendix, Fig. S7 A and B). PEP supplementation restored their abundance to levels similar to those of normoxic Mtb (SI Appendix, Fig. S7B). Similar to the effect of PEP on tgs1 mRNA, exogenous PEP also restored mRNA levels of PG pathway genes murA-G and murX, which were all down-regulated under hypoxia (SI Appendix, Fig. S8 A and B). This rescue caused the biosynthesis of nascent PG at the Mtb cell wall (SI Appendix, Fig. S7C), partly restored hypoxic growth (Fig. 1C), and heightened susceptibility to ampicillin, a covalent PG synthesis inhibitor (SI Appendix, Fig. S7D).
-
V.
Shikimate pathway: PEP is also a substrate for initiation of the shikimate pathway, which is functionally linked to the biosynthesis of chorismate, aromatic amino acids, and folate metabolism intermediates (SI Appendix, Fig. S9A). As expected, we observed significant depletion of chorismate and L-arogenate in hypoxic Mtb, which was kinetically matched to the abundance of phenylalanine, tryptophan, and folate metabolism intermediates such as dihydrofolate and dihydropteroate. Inversely, supplementing with PEP restored their abundance (SI Appendix, Fig. S9B).
Fig. 3.
The effect of exogenous PEP on hypoxic Mtb TAG biosynthesis. (A) The effect of PEP on abundance and 13C enrichment of hypoxic WT, tgs1 deficient Mtb (Δtgs1), and Δtgs1 complemented with tgs1 (COM) TCA cycle intermediates. Total bar heights indicate intrabacterial pool sizes (nmol/mg protein), in which the dark gray of each bar denotes the enrichment of 13C labeling achieved following transfer to [U-13C] acetate for 24 h under 1% O2. (B) TLC (thin layer chromatography) analysis of the extractable TAG from Mtb incubated for 24 h at 20% O2 (#1), 1% O2 (#2), 1% O2 with 10 mM PEP (#3), Δtgs1 for 24 h at 1% O2 (#4), and TAG chemical standard (M). Right panel is the TAG band intensity measured by Adobe Photoshop CS6. (C) mRNA expression levels of Mtb tgs1 for 24 h under 20% O2 (Norm), 1% O2, (HYP), 1% O2 with 2 mM PEP (HYP_2 PEP) or 5 mM PEP (HYP_5 PEP). (D) CFU viability kinetics of Mtb, Δtgs1, and Δtgs1/COM under 1% O2 for 6 d. To compare the effect on hypoxic Mtb growth, 1% O2 with 10 mM PEP was included (WT+PEP). All values were average of triplicates ± SEM; *P < 0.05; **P < 0.001; ns, not significant by Student’s t test.
Taken together, multiple metabolic networks essential for Mtb replication were functionally down-regulated due to PEP depletion, leading to the formation of NR Mtb.
PEP Depletion in NR Mtb Is in Part Attributed to the Down-Regulation of PckA Activity.
The metabolomics profile of hypoxic Mtb in [U-13C] acetate media showed profound depletion of both 12C and 13C fractions of PEP (Fig. 2D), implying that hypoxic Mtb also down-regulates gluconeogenic activities that biosynthesize PEP. To prove this, we measured the specific activity of PckA (SI Appendix, Fig. S10A) by monitoring 13C enrichment patterns and abundance of CCM intermediates of WT, pckA deficient Mtb (ΔpckA) and its complement strain (ΔpckA/COM) following transfer from unlabeled to [U-13C] acetate media at exposure to normoxia or hypoxia. At normoxia, PckA deficiency sequestered the acetate and its intermediates into the reductive branch intermediates of the TCA cycle, resulting in en bloc accumulation of OAA and malate and a complete loss of 13C labeled PEP (SI Appendix, Fig. S10B). Intriguingly, ΔpckA cultured in acetate media under normoxia maintained the PEP level, but there was a complete loss of 13C labeling, revealing the contribution of GL to PEP biosynthesis. Unlike under normoxia, the abundance and 13C enrichment of all GL and TCA cycle intermediates in hypoxic ΔpckA were at levels similar to those of hypoxic WT or ΔpckA/COM (SI Appendix, Fig. S10C). There were no significant changes in the abundance and 13C enrichments of reductive branch intermediates of the TCA cycle such as OAA and malate in hypoxic ΔpckA; this implied that depletion of gluconeogenic carbon flux in hypoxic Mtb is partly attributed to a lack of PckA activity in both directions (SI Appendix, Fig. S10C). Consistent with the metabolomics results, PckA deficiency had little effect on Mtb viability under hypoxia (SI Appendix, Fig. S11 A–D), indicating that PEP depletion of hypoxic Mtb is partly due to the catalytic depletion of PckA and accompanied down-regulation of gluconeogenic carbon flux.
PEP Supplementation Restores NR Mtb Metabolism Similar to that of Replicators.
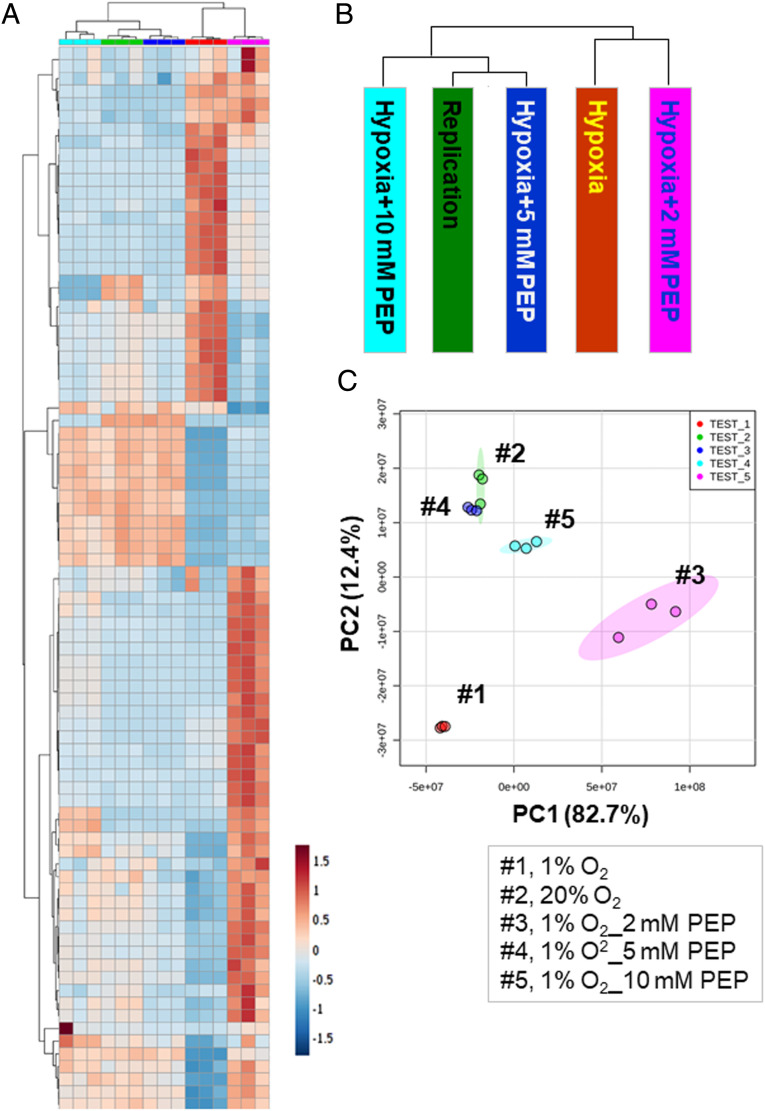
Exogenous PEP partly restored NR Mtb growth and drug sensitivity (Fig. 1 C and D). Our metabolomics analysis showed that the hypoxic growth and improved drug sensitivity of PEP treated hypoxic Mtb were largely attributed to the impaired remodeling of multiple NR Mtb metabolic networks. To interrogate the impact of PEP supplementation on NR Mtb global metabolic networks, we compared abundance of ∼100 annotated Mtb metabolites of hypoxic Mtb with or without 2 to 10 mM PEP. Multivariate hierarchical clustering identified a set of metabolites associated with PEP-mediated metabolic impact. A clustered heatmap and principal component analysis (PCA) indicated that concentrations of PEP of at least 5 mM prevented or significantly restored the metabolic remodeling that occurred in hypoxic Mtb, thereby rendering hypoxic Mtb metabolic networks similar to those of normoxic Mtb (Fig. 4 A–C). Metabolomics analysis confirmed that supplementing with PEP to hypoxic Mtb helped restore metabolic networks, thereby preventing Mtb from entering the NR state. Thus, PEP supplementation can be a metabolic modulation strategy to synthetically heighten hypoxic Mtb drug susceptibility against first line TB drugs.
Fig. 4.
The effect of exogenous PEP on global metabolic networks of hypoxic Mtb. (A) Clustered heatmap depicting levels of ∼100 Mtb metabolites of hypoxic Mtb (1% O2) in the presence of 2, 5, and 10 mM PEP and of replicating Mtb (20% O2); 0.2% acetate was used as a carbon source. Columns depict each condition including 20% O2 (green), 1% O2 (red), 1% O2 with 2 mM PEP (pink), 5 mM PEP (dark blue), and 10 mM PEP (dark blue). Rows indicate individual metabolites. Data were parsed using uncentered Pearson’s correlation with centroid linkage clustering and rendered using MetaboAnalyst (version 4.5). Data are depicted on a log2 scale. (B) Schematic depicting phylogenetic distance of each condition by analyzing the overall metabolome abundance. (C) PCA plot of the Mtb metabolome pattern under all conditions studied. Dot colors match the clustered heatmap condition bar colors.
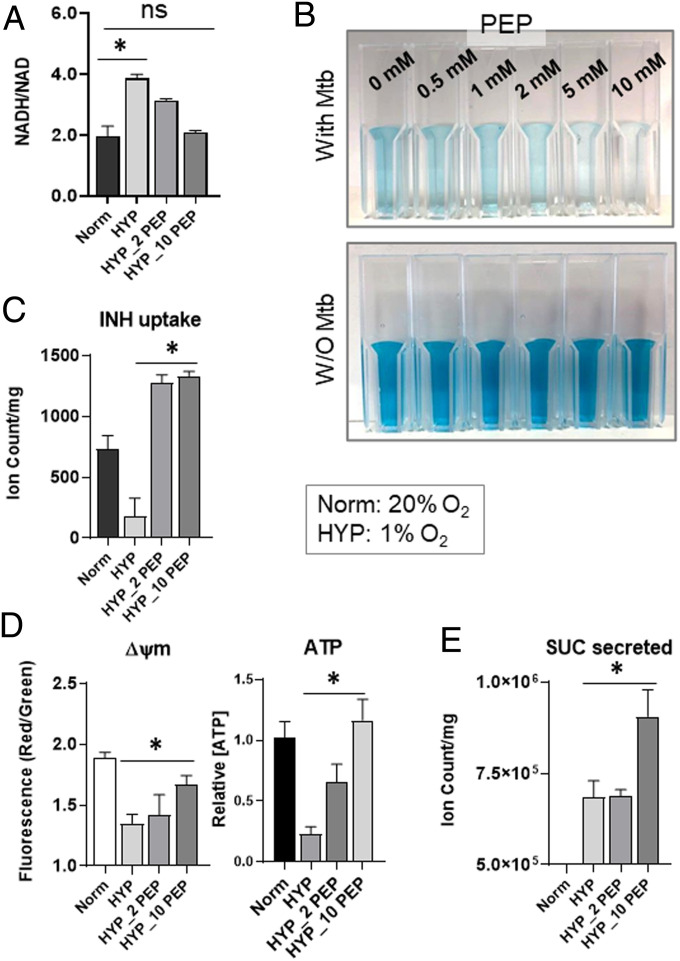
Membrane Bioenergetics of NR Mtb Are Affected by PEP Depletion.
Accumulating evidence has shown that NR Mtb drug tolerance is due to its slowed growth rate, dysregulated proton motive force (ΔΨm), and accompanied restriction of drug uptake (14, 33). Our metabolomics data of hypoxic Mtb revealed that the oxidative branch of the TCA cycle was partly restored by PEP (Fig. 2D and SI Appendix, Fig. S12A). Isotope metabolomics analysis showed that PEP supplementation increased αKG abundance in both 12C and 13C fractions. In the 13C isotopologue pattern of αKG, there was an increased upshift of M+4 from M+2, presumably due to cumulative assimilation of [U-13C] acetate-based C2 units (SI Appendix, Fig. S12B), implying that canonical TCA cycle activity was partly restored even under hypoxia. Separately, we detected a striking induction of succinate and malate 12C fractions but not 13C fractions (SI Appendix, Fig. S12B), implying that PEP was directly consumed by a reversed TCA cycle and/or glyoxylate shunt for the biosynthesis of reductive branch intermediates of the TCA cycle, as previously reported (10, 12). Thus, we hypothesized that PEP helped restore canonical TCA cycle activity, which led to the NADH biosynthesis. In hypoxic Mtb, PEP restored the NADH/NAD ratio so that it was similar to that of normoxic Mtb (Fig. 5A). Rescued NADH was coupled to ETC respiration, whereby PEP supplementation to hypoxic Mtb caused an increase in oxygen consumption (Fig. 5B). These findings confirmed that PEP depletion in NR Mtb was also associated with membrane bioenergetics and respiration, leading us to speculate that drug uptake in hypoxic Mtb may be affected by PEP depletion. To explore this, we modified our filter culture device by replacing the underlying m7H10 with a plastic inset containing chemically identical m7H9 with 10× MIC INH in direct contact with the underside of Mtb-laden filters (10) (SI Appendix, Fig. S13A). This enabled a timed start-stop measurement of INH uptake by sampling Mtb-free liquid medium and quantifying INH remaining in the media using liquid chromatography mass spectrometry (LC-MS). Consistent with previous reports (14, 34), INH uptake by hypoxic Mtb was one-fourth of that of normoxic Mtb but was almost fully recovered by PEP supplementation, likely associated with improved membrane bioenergetics such as ΔΨm and ATP (Fig. 5 C and D). PEP also enhanced succinate biosynthesis and secretion (Figs. 2D and 5E), which are known to maintain ΔΨm and ATP of hypoxic Mtb (10). Intriguingly, ATP restoration in hypoxic Mtb by supplementing with PEP was partly abolished by adding succinate to media because succinate secretion was interfered (10) (SI Appendix, Fig. S13B). Taken together, PEP deficiency in hypoxic Mtb affected TCA cycle remodeling, NADH biosynthesis, and succinate secretion, all of which are associated with hypoxic Mtb membrane bioenergetic remodeling, antibiotic uptake reduction, and drug tolerance.
Fig. 5.
The effect of exogenous PEP on membrane bioenergetics of hypoxic Mtb. Determining the effect of various concentrations of PEP on intrabacterial NADH/NAD ratio of hypoxic Mtb (HYP, 1% O2). 2_PEP, 2 mM PEP; 10_PEP, 10 mM PEP (A), oxygen consumption rate of hypoxic Mtb using the oxygen sensor, methylene blue at 0.001% for 2-h hypoxic incubation (B), INH uptake of hypoxic Mtb (C), ΔΨm (membrane potential) and intrabacterial ATP content of hypoxic Mtb (D), and the level of succinate secreted from hypoxic Mtb (E). Values were compared to those of replicating Mtb (Norm, 20% O2). All values are the average of three biological replicates ± SEM; *P < 0.01; ns, not significant by Student’s t test.
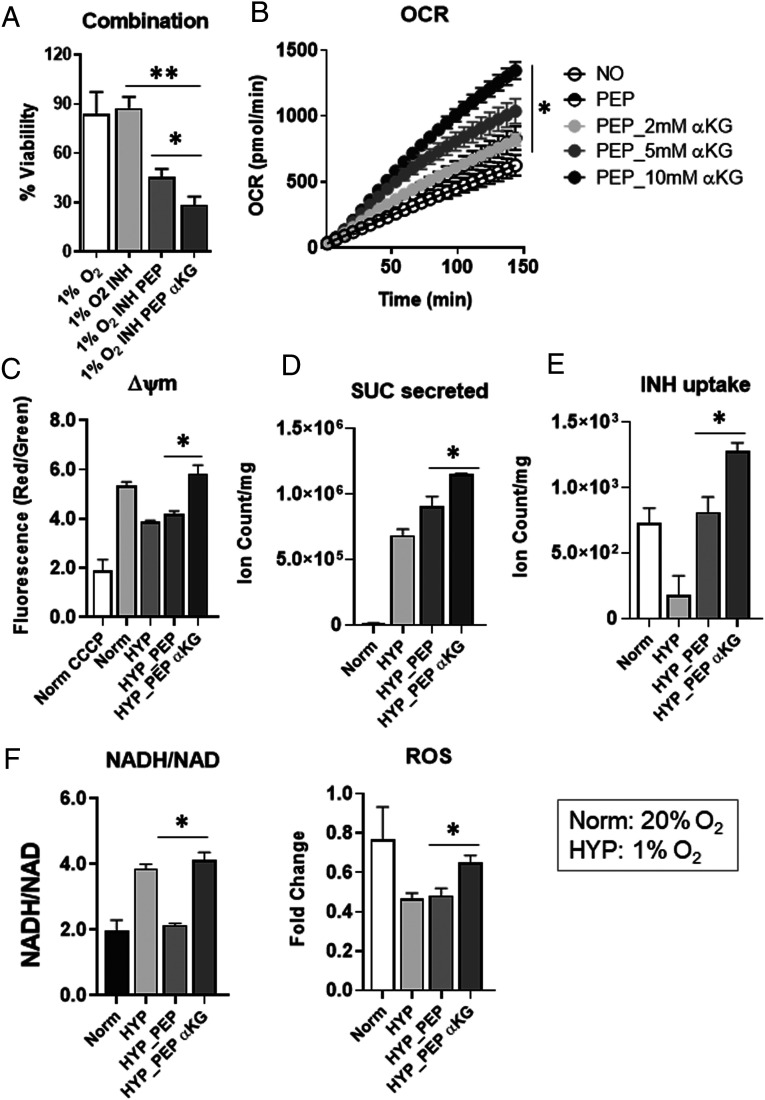
αKG Synergized with PEP on NR Mtb Phenotypic Restoration.
Metabolic networks of hypoxic Mtb were restored by exogenous PEP similar to those of normoxic Mtb (Fig. 4). However, levels of the oxidative branch of the TCA cycle such as αKG only partly recovered, implying that the oxidative TCA cycle branch in hypoxic Mtb was relatively unaffected by PEP depletion. Since αKG participates in many metabolic networks, including glutamate metabolism and the GABA shunt, we hypothesized that PEP-mediated restoration of NR Mtb drug susceptibility can be further improved by additional treatment with αKG. To test this, we treated hypoxic Mtb with cell permeable dimethyl αKG and PEP and observed that hypoxic Mtb became more sensitive to INH compared to treatment with PEP alone in vitro and ex vivo (Fig. 6A and SI Appendix, Fig. S14A). As expected, co-treatment with αKG and PEP improved oxygen consumption rate (OCR), ΔΨm, succinate biosynthesis and secretion, and INH uptake (Fig. 6 B–E). Due to the biosafety issue, we used H37Rv (ΔleuCDΔpanCD) BSL2+ strain in the stationary phase. Surprisingly, the NADH/NAD ratio increased presumably due to vigorous production of NADH and greater levels of ΔΨm and ROS (Fig. 6 C and F). Further, co-treatment of PEP and αKG restored the metabolic profile of hypoxic Mtb so that it more closely resembled that of normoxic Mtb (SI Appendix, Fig. S14B). Taken together, PEP dependent and independent metabolic activities are altered in NR Mtb and a nontoxic Mtb metabolite mixture, including PEP and αKG, can better resensitize NR Mtb to conventional TB drugs.
Fig. 6.
Synergy effects of αKG with exogenous PEP on hypoxic Mtb metabolism. (A) Percent CFU viability of hypoxic Mtb with or without treatment with PEP and/or αKG in the presence of 10× MIC equivalent INH for 6 d incubation compared to individual input. This was conducted under continuous hypoxia using a hypoxia chamber preinstalled with squeezable Pasteur pipettes (SI Appendix, Fig. S3A). Determination of the effect of 10 mM PEP and/or 5 mM αKG on OCR using BSL2+ H37Rv strain (ΔleuCDΔpanCD) in the stationary phase (B), ΔΨm (membrane potential) (C), amount of succinate secreted (D), INH uptake (E), and NADH/NAD ratio and ROS levels (F) of hypoxic Mtb (HYP, 1% O2). Values were compared with those of replicating Mtb (Norm, 20% O2). All values are the average of three biological replicates ± SEM; *P < 0.01; **P < 0.005 by Student’s t test.
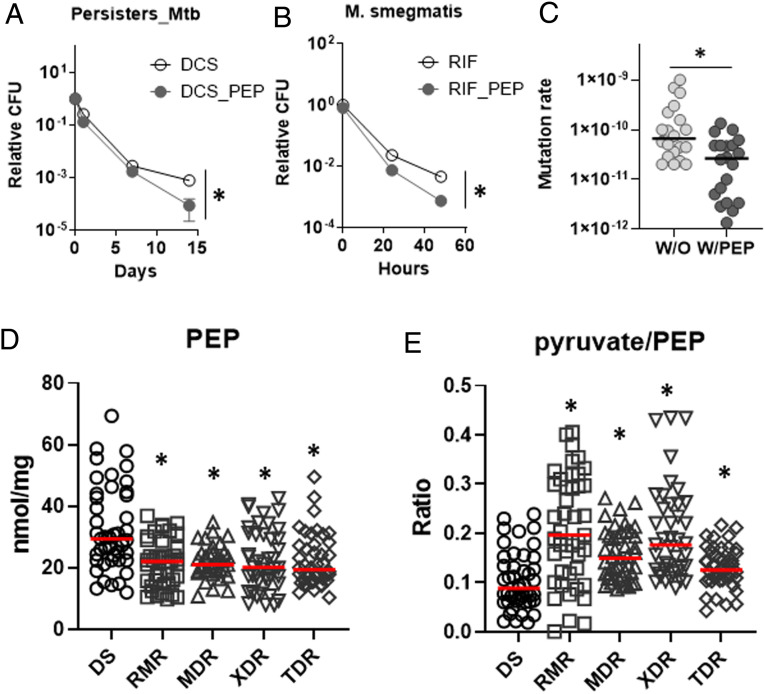
PEP Depletion Helps Form Persisters and Develop DR Mutants.
In 1944, Bigger et al. observed a small fraction of bacterial pathogens that survived after treatment with bactericidal antibiotics and called them persisters (35). Recent metabolomics studies of in vitro persisters using a biofilm culture showed that persister metabolism resembles that of NR Mtb (9). This led us to hypothesize that the metabolic advantages from PEP depletion also play roles in Mtb persister formation. To this end, we treated 100 μg/mL D-cycloserine (DCS) to normoxic Mtb and monitored viable bacilli by CFU to calculate the level of persisters as previously reported (36). PEP supplementation slightly decreased the level of Mtb persisters, especially between days 7 and 14 (Fig. 7A). Notably, Mycobacterium smegmatis, a saprophytic mycobacterial species, significantly down-regulated the PEP levels after treatment with 1× MIC RIF (SI Appendix, Fig. S15), and the bacilli co-treated with PEP remained less viable compared to that without PEP treatment (Fig. 7B). These findings confirmed that the PEP depletion–mediated metabolic remodeling also contributes to antibiotic-induced persister formation. Accumulating evidence showed that bacterial persisters are predisposed to the development of DR mutants. To interrogate the impact of PEP depletion on the emergence of DR, we used a Luria–Delbrück fluctuation assay to determine DR mutation rates by selecting spontaneous RIF resistant mutants (37). A total of 44 cultures of M. smegmatis were prepared in 4 mL with (20 cultures) or without (20 cultures) PEP and individually plated on m7H10 containing 100 μg/mL RIF. After incubating for at least 7 d, RIF-resistant colonies were counted. The remaining 4 cultures were used to determine the total number of inputs. Mutation rates were calculated by the Lea–Coulson method as previously used (38, 39). RIF resistant mutation rates without PEP were around 2 × 10−10 per cell division, but pretreatment with PEP significantly reduced the rates to around 9 × 10−11 per cell division, a roughly 2.2-fold decrease (Fig. 7C). We also collected 15 drug sensitive (DS)-, 15 RIF mono DR (RMR)-, 15 multidrug resistant (MDR)-, 15 extensively DR (XDR)-, and 15 totally DR (TDR)-TB isolates from the International TB Research Center (ITRC, South Korea). All clinical TB isolates had similar growth rates when cultured in sodium butyrate (SB) media (9). Metabolomics analysis of all clinical TB isolates cultured in SB media revealed that the abundance of PEP was significantly greater in DS and lower in RMR, MDR, XDR, and TDR (Fig. 7D). The pyruvate to PEP ratio also positively correlated with the extent of drug resistance (Fig. 7E). These findings suggested that PEP and the pyruvate to PEP ratio can be biomarkers used to diagnose DR-TB infection and help guide the therapeutic direction.
Fig. 7.
The effect of exogenous PEP on Mtb drug tolerance and drug resistance. (A) Killing curves of Mtb with or without PEP following treatment with 100 μg/mL DCS. Viability was monitored by CFU/mL at 1, 7, and 14 d posttreatment. Viable colonies after day 14 are considered Mtb persisters. (B) PEP supplementation sensitizes M. smegmatis to RIF treatment. (C) A fluctuation assay in M. smegmatis was conducted in the presence or absence of 10 mM PEP. RIF resistant rates were calculated using the bz-rate web tool. Black line indicates the median level of mutation rates. (D) Intrabacterial PEP abundance from DS, RMR, MDR, XDR, and TDR clinical isolates. Dots depict intrabacterial PEP concentrations (nmol/mg protein) of 3 replicates from 15 clinical isolates. Red line indicates the median level of PEP in each group. (E) Pyruvate/PEP ratio of DS, RMR, MDR, XDR, and TDR was calculated by nmol/mg concentrations of pyruvate and PEP. All are the average of three biological replicates ± SEM; *P < 0.01 by Student’s t test.
Discussion
NR Mtb has generally been perceived to have minimal metabolic activities (40). However, NR Mtb faces the challenge of surviving in an adverse host environment. Within its etiological niche, Mtb enters a clinically asymptomatic NR state and exhibits nonheritable resistance to nearly all TB drugs. Thus, NR Mtb physiology has emerged as a hallmark of TB pathogenicity. The studies presented herein proved the functional relevance of hypoxic Mtb metabolic remodeling in the acquisition of drug tolerance. Comparative metabolomics profiles between normoxic and hypoxic Mtb led us to pinpoint the specific set of metabolites that are uniquely altered under hypoxia, which included a specific bypass of the biosynthesis of oxidative branch intermediates of the TCA cycle. Meanwhile, levels of intermediates in the reductive branch of the TCA cycle were maintained, presumably due to the reversed reductive branch of the TCA cycle and glyoxylate shunt (10, 12). Hypoxic Mtb also accumulated intermediates in the early portion of GL, which were catalytically linked to near complete depletion of PEP at the lowest portion of GL. Supplementing NR Mtb with SME, a small metabolite extract from normoxic Mtb, was sufficient to improve its growth and drug susceptibility, confirming that metabolic remodeling is a triggering factor that leads to NR Mtb phenotypic shifts (5, 41, 42). Certain SME metabolites continuously activate a specific set of metabolic networks; their inactivation is required for NR Mtb formation and drug tolerance. Targeted metabolomics was used to screen for candidate metabolites, and PEP was identified as a potential metabolic modulator that enables restoration of NR Mtb drug tolerance and growth arrest to levels similar to those of the replicating state. Our LC-MS analysis and generation of a standard curve derived by known amounts of a PEP chemical standard showed that PEP within the SME used was calculated to be ∼145 μM, a reasonable value as it was previously reported to be ∼15 μM in DR Edwardsiella tarda grown in M9 minimal medium (43). The PEP amount in the SME was less than the 1 to 10 mM that was used to conduct PEP functional studies. Nevertheless, the effect on phenotypic restoration of NR Mtb was more prominent when treated with SME than with PEP (Fig. 1), implying a role for other metabolites independent of PEP, such as αKG (Fig. 6). Select Mtb metabolites including PEP and/or αKG can be a potential metabolic modulator that resensitizes NR Mtb to first-line TB drugs, thereby shortening our TB treatment duration and preventing the development of DR-TB strains.
As shown in recent reports (9, 10), hypoxic Mtb shifted the trehalose-carbon flux toward the biosynthesis of GL intermediates such as FBP, and this trehalose catalytic shunting is presumably linked to depletion of trehalose dimycolates (TDM) and sulfolipid-1 (SL-1). As TDM and SL-1 are ligands that induce proinflammatory immune responses and cough-inducing neuronal signals of hosts, respectively, their depletion plays a crucial part in an immune evasion strategy that allows NR Mtb to establish a stealth invasion (44, 45). Separately, FBP allosterically enhances Pyk activity in Escherichia coli and ATP biosynthesis via substrate level phosphorylation. In a functional analogy, NR Mtb Pyk activity may also be activated by accumulated FBP but only until intrabacterial PEP is available, thus temporarily serving as an alternate ATP source while down-regulating ETC-mediated ATP biosynthesis. Thus, Pyk activity may be an alternate ATP source that initiates the metabolic remodeling required for NR Mtb formation. Accumulated FBP may ultimately facilitate PEP depletion, which can inhibit multiple metabolic networks essential for Mtb replication, including PG biosynthesis, folate metabolism, pyruvate metabolism, the TCA cycle, and aromatic amino acid biosynthesis (SI Appendix, Fig. S16A). Notably, many of the foregoing activities have been investigated as potential TB drug targets (46). In E. coli, the pyruvate to PEP ratio is a redox biomarker to regulate expression of genes involved in the TCA cycle (19, 20). We also found that supplementing PEP to hypoxic Mtb altered the abundance of TCA cycle intermediates and restored NADH production, leading to a revival of membrane bioenergetics and improved drug uptake. Intriguingly, PEP-mediated redox balance also restored acetate-carbon flux toward the biosynthesis of oxidative branch intermediates of the TCA cycle and away from TAG biosynthesis. Consequently, the phenotypic restoration of hypoxic Mtb after treatment with PEP resembled that of hypoxic Δtgs1 due to a rescue of canonical TCA cycle activity, corrected NADH/NAD ratio, revived membrane bioenergetics, and enhanced drug uptake. Although it is premature to conclude that PEP depletion is a strategy employed by Mtb to remodel its metabolic networks for NR Mtb survival, it is clear that disproportionate GL activity allosterically regulates Pyk to initially consume PEP as a source of ATP. Consequent metabolic remodeling that arises from limited PEP availability is advantageous to shift the NR Mtb phenotypes.
The signals that lead to NR Mtb metabolic remodeling are unclear. In a recent report, Mtb failed to enter the NR state and became more sensitive to INH under nutrient starvation in the absence of Rel, monofunctional alamone (p)ppGpp synthase (47). Mtb may use Rel to sense environmental threats including antibiotic treatment, as the findings that rel deficient Mtb (Δrel) fails to undergo metabolic remodeling needed to form NR Mtb. Notably, TCA cycle and long and short chain fatty acid metabolism pathways in Δrel under nutrient-starved stress were similar to those of WT in a replicating state. Thus, defects in stress-induced remodeling in the foregoing metabolic networks renders Δrel susceptible to INH under in vitro stress models and in the lungs of chronically infected mice. Indeed, rel transcript levels were increased in hypoxic Mtb but brought back down to normal levels after supplementing with PEP (SI Appendix, Fig. S16B). Thus, Rel may be an Mtb metabolic sensor required to resist environmental stresses and leads to down-regulation of PEP anabolic activities.
Whether drug resistance of bacterial pathogens evolves from drug tolerance is a controversial subject (48, 49). Recent studies of E. coli subjected to intermittent exposure to ampicillin separated by intervals in antibiotic-free medium showed that antibiotic exposure significantly delayed its lag time, a state of drug tolerance, several cycles before it achieved increased MIC of ampicillin, a state of drug resistance (50). This implied that bacterial pathogens may use the same metabolic networks to form both NR Mtb and DR-TB strains. Notably, supplementing hypoxic Mtb with PEP significantly reduced DCS-induced persisters, indicating that metabolic remodeling arising from PEP depletion plays an important role in Mtb persister formation. Furthermore, supplementing with PEP also reduced the DR mutation rates in Mtb, confirming a functional linkage between metabolic remodeling for Mtb persister formation and development of DR-TB strains. Targeted metabolomics profiles of clinical TB isolates supported this notion because PEP and the pyruvate to PEP ratio were significantly altered in MDR-, XDR- and TDR-TB strains compared to those of DS-TB, suggesting that metabolic activities arising from low levels of PEP are not only advantageous to develop DR mutations but also favorable to maintain DR-TB metabolic networks. Recently, two comprehensive genetic studies screened for Mtb genes that are functionally altered to reduce antimicrobial effects using DR-TB clinical isolates (51, 52). Both identified frameshift mutations in a hypervariable homopolymeric region of GlpK (glycerol kinase), an essential gene required for optimal glycerol catabolism. Loss of function of GlpK was shown to be a specific marker of DR-TB. In DR-TB, a catabolic defect of glycerol consumption due to a lack of functional GlpK may cause the alteration of global GL activity, presumably leading to PEP depletion. Thus, investigations on the effects of PEP supplementation on antimicrobial synergy with second-line TB drugs against DR-TB clinical isolates are warranted.
Our metabolomics analysis of hypoxic Mtb showed that gluconeogenic PEP anabolic activities are nearly depleted. A report recently proposed the role of reversing the direction of PckA converting PEP to OAA while adapting to a reducing environment such as hypoxia (53). Our results differ from previous findings because the experiments in the previous study were mostly conducted under an in vitro condition where the ratio of NADH/NAD was artificially altered. Within a biological system, PckA activity is affected by the redox state, its energy state, and substrate availability. We and others showed that hypoxic Mtb maintains ATP levels that are roughly 15 to 20% of that of its replicating counterparts (10, 54). The catalytic reaction of PckA converting OAA to PEP is bioenergetically unfavorable because GTP is required as a cofactor. PckA may serve as an alternate source of ATP while an initial stage of NR Mtb formation, but soon after, PckA may be shut down by PEP depletion. Under normoxia, we also observed that ΔpckA maintained PEP abundance but there was a complete loss of 13C labeling from exogenous acetate, suggesting that glycolytic carbon flux toward PEP biosynthesis is active during replication but abolished during the NR state.
Over the past decades, the TB drug discovery pipeline has focused on the identification of novel drug targets by screening for gene products that are essential for Mtb viability or NR Mtb formation. In the present study, we propose therapeutics to interfere with the metabolic remodeling required for NR Mtb formation and viability, thereby synthetically killing NR Mtb using our conventional TB drugs and preventing the development of DR-TB. This metabolic modulation strategy will simplify the conventional TB drug regimen and shorten treatment duration.
Materials and Methods
Bacterial Strains, Culture Conditions, and Chemicals.
Details are in SI Appendix.
Mtb Metabolite Extraction.
Mtb-laden filters used for metabolomics profiling were generated as previously described (10). Details are in SI Appendix.
In Vitro Hypoxia Model.
In vitro hypoxia was achieved using a type A bio-bag environmental chamber (Becton Dickinson), as specified by the manufacturer’s instructions and validated by a resazurin indicator, which decolorized at oxygen concentrations of 1.0% or less. Mtb-laden filters transferred to new m7H10 were inserted to hypoxic chambers, and then the chambers were immediately sealed. This system rapidly depletes oxygen and generates CO2 via a palladium catalyst; within 4 h, achieving a final atmosphere of 1.0% O2 and 5.0% CO2, levels similar to those encountered in the TB lungs of infected animals. We showed that this system is associated with both reversible biphasic induction of dosR, a validated transcriptional marker of hypoxia (10, 55).
LC-MS Metabolomics Profiling.
LC-MS–based metabolomics was used as previously described (10). Details are in SI Appendix.
Isotopologue Analysis Using [U-13C] Acetate.
Details are in SI Appendix.
Bone Marrow–Derived Macrophages Infection and CFU Enumeration Assay.
Bone marrow was removed from the femur and tibia of 8 to 10 wk old C57BL6 mice after euthanasia as previously described (56). Details are in SI Appendix.
In Vitro CFU Enumeration Assay.
Mtb viability was determined using liquid cultures manipulated under experimentally identical conditions as filter-grown cultures for metabolomics profiling as previously described (10). Details are in SI Appendix.
Membrane Potential, Intracellular ATP, and NADH/NAD Ratios.
Mtb membrane potential, ATP, and NADH/NAD ratios were measured by as previously described and adapted to our in vitro hypoxia apparatus (10). Details are in SI Appendix.
Extraction of RNA and qRT-PCR.
Details are in SI Appendix.
SME Preparation.
SME was prepared as previously described (28). Briefly, water-soluble metabolite fractions were prepared from Mtb in a replicating state. Around 500 mL midlog phase culture was used to gain a sufficient amount of metabolome. After growing in m7H9, cells were resuspended in 10 mL 100% acetonitrile and transferred to 2 mL screw-cap tubes containing 0.1 mm Zirconia beads. Cells were disrupted by 30 s pulses six times (6,800 rpm) twice under continuous cooling at or below 2 °C. Lysates were then centrifuged at 13,000 rpm for 10 min at 4 °C, and supernatant was dried under a continuous nitrogen stream. The pellet was resuspended in 1 mL 20 mM Tris-Cl (pH 7.4). Aliquots were stored at −80 °C until use.
Drug Uptake Measurement.
Our filter culture system was modified by replacing the underlying agar media with a plastic inset containing chemically equivalent m7H9 (with no detergent) in direct contact with the underside of Mtb-laden filters as shown in SI Appendix, Fig. S13A. Details are in SI Appendix.
OCR Measurement.
OCR in Mtb was quantified using the Seahorse XFe96 extracellular flux analyzer (Agilent Technologies) as previously described (57). Details are in SI Appendix.
BODIPY FL Vancomycin-Labeling.
Details are in SI Appendix.
Luria–Delbrück Fluctuation Assay.
The fluctuation assay was performed as previously described (37). Briefly, M. smegmatis, a fast-growing mycobacterial species, was used. Starter cultures were inoculated from freezer stocks for OD595 ∼1.0 culture. The culture at OD595 between 0.7 and 1.1 was diluted in 200 mL complete m7H9 to OD595, 0.0003. This volume was immediately divided to start 44 cultures of 4 mL each in 15-mL round bottom bottles and cultured at 37 °C with shaking for additional 3 d until reaching OD595 of ∼1.0. Twenty cultures were transferred to 15-mL conical tubes and spun at 4,000 rpm for 10 min at 4 °C. Cultures were then resuspended in 300 μL of complete m7H9 and spotted onto m7H10 supplemented with 100 μg/mL RIF. Twenty cultures were plated onto m7H10 supplemented with 100 μg/mL RIF with 10 mM PEP. Input cell counts were determined by serial dilution of the remaining four cultures. The drug resistance rate was calculated using the number of viable cell counts on RIF plates and estimated by the bz-rate web tool (http://www.lcqb.upmc.fr/bzrates) (58).
Triacylglycerol Extraction.
Total lipid extraction was performed as previously described with minor modifications (59). Details are in SI Appendix.
Statistical Analysis.
Analyses were performed using a Student’s t test or ANOVA. A P value of less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Sabine Ehrt for the generous gift of pckA-deficient Mtb and its complement strain, Christopher Sassetti for tgs1-deficient Mtb, and William Jacobs for the H37Rv (ΔleuCDΔpanCD) BSL2+ strains used herein. This work was supported by the Donald E. & Delia Baxter Foundation, the L.K. Whittier Foundation, Wright Foundation, American Lung Association Innovative Award, NIH R21 AI139386 and R56 AI143870 (H.E.), and The Korea Centers for Disease Control and Prevention, Republic of Korea (S.Y.E).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105800118/-/DCSupplemental.
Data Availability
The data that support the findings of this study are available within the article and its supporting information. The metabolomics data underlying figures are provided in Dataset S1.
References
- 1.Nathan C., Taming tuberculosis: A challenge for science and society. Cell Host Microbe 5, 220–224 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Nathan C., Barry C. E. 3rd, TB drug development: Immunology at the table. Immunol. Rev. 264, 308–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization , Global tuberculosis report 2020 (World Health Organization Geneva, Switzerland, 2020). https://www.who.int/publications/i/item/9789240013131. Accessed 11 August 2021.
- 4.Nathan C., Cunningham-Bussel A., Beyond oxidative stress: An immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol. 13, 349–361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrt S., Schnappinger D., Rhee K. Y., Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 496–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gollan B., Grabe G., Michaux C., Helaine S., Bacterial persisters and infection: Past, present, and progressing. Annu. Rev. Microbiol. 73, 359–385 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Yew W. W., Barer M. R., Targeting persisters for tuberculosis control. Antimicrob. Agents Chemother. 56, 2223–2230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eoh H., et al., Metabolic anticipation in Mycobacterium tuberculosis. Nat. Microbiol. 2, 17084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J. J., et al., Transient drug-tolerance and permanent drug-resistance rely on the trehalose-catalytic shift in Mycobacterium tuberculosis. Nat. Commun. 10, 2928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eoh H., Rhee K. Y., Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 6554–6559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nandakumar M., Nathan C., Rhee K. Y., Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat. Commun. 5, 4306 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Watanabe S., et al., Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 7, e1002287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith T., Wolff K. A., Nguyen L., Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr. Top. Microbiol. Immunol. 374, 53–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarathy J., Dartois V., Dick T., Gengenbacher M., Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 1648–1653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raffetseder J., et al., Replication rates of Mycobacterium tuberculosis in human macrophages do not correlate with mycobacterial antibiotic susceptibility. PLoS One 9, e112426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goossens S. N., Sampson S. L., Van Rie A., Mechanisms of drug-induced tolerance in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 34, e00141-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurica M. S., et al., The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure 6, 195–210 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Kapoor R., Venkitasubramanian T. A., Glucose 6-phosphate activation of pyruvate kinase from Mycobacterium smegmatis. Biochem. J. 193, 435–440 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogema B. M., et al., Inducer exclusion in Escherichia coli by non-PTS substrates: The role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol. Microbiol. 30, 487–498 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Sauer U., Eikmanns B. J., The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29, 765–794 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Walsh C. T., Benson T. E., Kim D. H., Lees W. J., The versatility of phosphoenolpyruvate and its vinyl ether products in biosynthesis. Chem. Biol. 3, 83–91 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Liu K., Yu J., Russell D. G., pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiology (Reading) 149, 1829–1835 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S., Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreteau H., et al., Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 168–207 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Crick D. C., Mahapatra S., Brennan P. J., Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11, 107R–118R (2001). [DOI] [PubMed] [Google Scholar]
- 26.Hamasaki N., Hardjono I. S., Minakami S., Transport of phosphoenolpyruvate through the erythrocyte membrane. Biochem. J. 170, 39–46 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wayne L. G., Sohaskey C. D., Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55, 139–163 (2001). [DOI] [PubMed] [Google Scholar]
- 28.de Carvalho L. P., et al., Activity-based metabolomic profiling of enzymatic function: Identification of Rv1248c as a mycobacterial 2-hydroxy-3-oxoadipate synthase. Chem. Biol. 17, 323–332 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nandakumar M., Prosser G. A., de Carvalho L. P., Rhee K., Metabolomics of Mycobacterium tuberculosis. Methods Mol. Biol. 1285, 105–115 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Ehrt S., Schnappinger D., Mycobacterial survival strategies in the phagosome: Defence against host stresses. Cell. Microbiol. 11, 1170–1178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baek S. H., Li A. H., Sassetti C. M., Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 9, e1001065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang X., Wallqvist A., Reifman J., Modeling phenotypic metabolic adaptations of Mycobacterium tuberculosis H37Rv under hypoxia. PLOS Comput. Biol. 8, e1002688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meylan S., et al., Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem. Biol. 24, 195–206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C., et al., Verapamil targets membrane energetics in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 62, e02107-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bigger J. W., Treatment of staphylcoccal infectioins with penicillin by intermittent sterilisation. Lancet 244, 497–500 (1944). [Google Scholar]
- 36.Keren I., Minami S., Rubin E., Lewis K., Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio 2, e00100–e00111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ford C. B., et al., Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45, 784–790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foster P. L., Methods for determining spontaneous mutation rates. Methods Enzymol. 409, 195–213 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huitric E., et al., Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 54, 1022–1028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher R. A., Gollan B., Helaine S., Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Eoh H., Metabolomics: A window into the adaptive physiology of Mycobacterium tuberculosis. Tuberculosis (Edinb.) 94, 538–543 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Galagan J. E., et al., The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su Y. B., et al., Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E1578–E1587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson M. B., Williams S. J., MCL and mincle: C-type lectin receptors that sense damaged self and pathogen-associated molecular patterns. Front. Immunol. 5, 288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruhl C. R., et al., Mycobacterium tuberculosis sulfolipid-1 activates nociceptive neurons and induces cough. Cell 181, 293–305.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mdluli K., Spigelman M., Novel targets for tuberculosis drug discovery. Curr. Opin. Pharmacol. 6, 459–467 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Dutta N. K., et al., Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci. Adv. 5, eaav2104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen N. R., Lobritz M. A., Collins J. J., Microbial persistence and the road to drug resistance. Cell Host Microbe 13, 632–642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levin-Reisman I., et al., Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Kohanski M. A., DePristo M. A., Collins J. J., Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37, 311–320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Safi H., et al., Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc. Natl. Acad. Sci. U.S.A. 116, 19665–19674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellerose M. M., et al., Common variants in the glycerol kinase gene reduce tuberculosis drug efficacy. MBio 10, e00663-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machová I., et al., Mycobacterium tuberculosis phosphoenolpyruvate carboxykinase is regulated by redox mechanisms and interaction with thioredoxin. J. Biol. Chem. 289, 13066–13078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao S. P., Alonso S., Rand L., Dick T., Pethe K., The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 105, 11945–11950 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park H. D., et al., Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J. J., et al., Glutamate mediated metabolic neutralization mitigates propionate toxicity in intracellular Mycobacterium tuberculosis. Sci. Rep. 8, 8506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saini V., et al., Ergothioneine maintains redox and bioenergetic homeostasis essential for drug susceptibility and virulence of Mycobacterium tuberculosis. Cell Rep. 14, 572–585 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gillet-Markowska A., Louvel G., Fischer G., bz-rates: A web tool to estimate mutation rates from fluctuation analysis. G3 (Bethesda) 5, 2323–2327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bligh E. G., Dyer W. J., A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the article and its supporting information. The metabolomics data underlying figures are provided in Dataset S1.