Significance
Resistance to endocrine therapy is a major obstacle to managing breast cancer. Here, we introduce that tripartite motif protein TRIM47 enhances endocrine therapy resistance and facilitates breast cancer tumorigenesis. We showed that TRIM47 immunoreactivity scores in breast cancer specimens were positively associated with shorter disease-free survival of the patients with postoperative endocrine therapy. Overexpression of TRIM47 in breast cancer cells activated NF-κB signaling, which is partly due to the stabilization of PKC-ε and PKD3 by interacting with TRIM47. As an underlying mechanism, we demonstrated TRIM47-dependent modification of PKC-ε by atypical lysine 27–linked polyubiquitination. Our study implies that TRIM47 and its associated kinases can be potential diagnostic and therapeutic targets for patients with breast cancer refractory to endocrine therapy.
Keywords: breast cancer, tripartite motif containing 47, nuclear factor kappa-B signaling, protein kinase C epsilon, protein kinase D3
Abstract
Increasing attention has been paid to roles of tripartite motif–containing (TRIM) family proteins in cancer biology, often functioning as E3 ubiquitin ligases. In the present study, we focus on a contribution of TRIM47 to breast cancer biology, particularly to endocrine therapy resistance, which is a major clinical problem in breast cancer treatment. We performed immunohistochemical analysis of TRIM47 protein expression in 116 clinical samples of breast cancer patients with postoperative endocrine therapy using tamoxifen. Our clinicopathological study showed that higher immunoreactivity scores of TRIM47 were significantly associated with higher relapse rate of breast cancer patients (P = 0.012). As functional analyses, we manipulated TRIM47 expression in estrogen receptor–positive breast cancer cells MCF-7 and its 4-hydroxytamoxifen (OHT)-resistant derivative OHTR, which was established in a long-term culture with OHT. TRIM47 promoted both MCF-7 and OHTR cell proliferation. MCF-7 cells acquired tamoxifen resistance by overexpressing exogenous TRIM47. We found that TRIM47 enhances nuclear factor kappa-B (NF-κB) signaling, which further up-regulates TRIM47. We showed that protein kinase C epsilon (PKC-ε) and protein kinase D3 (PKD3), known as NF-κB–activating protein kinases, are directly associated with TRIM47 and stabilized in the presence of TRIM47. As an underlying mechanism, we showed TRIM47-dependent lysine 27–linked polyubiquitination of PKC-ε. These results indicate that TRIM47 facilitates breast cancer proliferation and endocrine therapy resistance by forming a ternary complex with PKC-ε and PKD3. TRIM47 and its associated kinases can be a potential diagnostic and therapeutic target for breast cancer refractory to endocrine therapy.
Breast cancer is the most commonly diagnosed cancer worldwide and is the leading cause of cancer death among females in the majority of countries (1). More than 70% of breast cancer cases are estimated as hormone-receptor (HR) positive (2). For HR-positive primary breast cancer, endocrine therapy has been recommended as a standard treatment (3). A first-generation selective estrogen receptor modulator, tamoxifen, has been the only drug clinically applied to breast cancer management for years. Aromatase inhibitors (AIs) now have become the first-line therapy for postmenopausal HR-positive breast cancer, while tamoxifen is used as the first-line therapy for premenopausal HR-positive breast cancer and second-line therapy for tumors which acquired resistance to AIs (3). In terms of the mechanisms underlying endocrine therapy resistance, several factors have been proposed: mutation of target genes such as ESR1 (coding estrogen receptor alpha; ER-α) or CYP19A1 (coding aromatase) (4) altered genomic functions of estrogen by transcription factors such as forkhead box A1 (FOXA1) (5, 6), activation of nongenomic signaling of estrogen (7–9), gene polymorphisms of CYP2D6 that catalyze tamoxifen (10), activation of growth factor cascades such as epidermal growth factor (EGF) and insulin-like growth factor 1 (IGF1) signaling (11–13), and activated nuclear factor kappa-B (NF-κB) signaling (14, 15). Efforts have been paid to dissect the mechanism, yet endocrine therapy resistance is a fundamental clinical issue to be overcome to improve the patient prognosis.
We previously showed that some tripartite motif (TRIM) proteins such as TRIM25/Efp (estrogen-responsive finger protein) and TRIM44 play oncogenic roles in breast cancer and predict worse prognosis (16–20). TRIM-family proteins typically have a structure containing RING finger, B-box, and coiled-coil domains (21) and consist of more than 70 members sharing the similar domain structure (22–24). It has been reported that TRIM proteins are involved in various physiological and pathological processes such as antiviral functions, immunity, and carcinogenesis (25–32).
In this study, we focused on the role of TRIM47 in breast cancer biology, as this protein is associated with tumorigenesis in some cancers (33–36). TRIM47 and TRIM25/Efp belong to the same C-IV class of TRIM proteins (28), as they share similar domain structure. Here, we showed that TRIM47 is a potential diagnostic biomarker for tamoxifen resistance, as its protein expression in clinical breast cancer specimens was significantly associated with prognosis of patients who underwent surgical resection of tumor followed by adjuvant tamoxifen therapy. In studies using HR-positive breast cancer cells, we showed that TRIM47 enhances NF-κB signaling through the protein stabilization of its interactors protein kinase C epsilon (PKC-ε) and protein kinase D3 (PKD3), leading to tamoxifen resistance and cell proliferation. As an underlying mechanism, TRIM47-dependent lysine 27–linked polyubiquitination of PKC-ε was demonstrated.
Results
Immunohistochemistry of TRIM47 in HR-Positive Breast Cancer Patients.
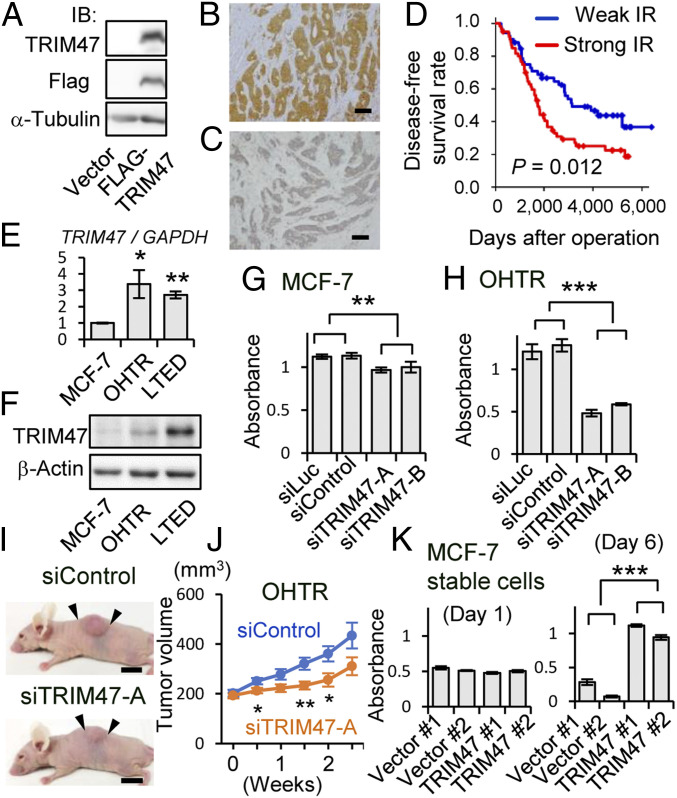
In the present study, we investigated the significance of TRIM47 expression in breast cancer recurrence during endocrine therapy using tamoxifen. Prior to immunohistochemical evaluation of human tissue samples, the reactivity of the antibody against TRIM47 was examined. We showed that the antibody raised against recombinant mouse TRIM47 protein recognized human TRIM47 protein by Western blotting using cell lysate derived from HEK293 cells overexpressing Flag-tagged human TRIM47 (Fig. 1A). This antibody was also used in a previous immunohistochemical analysis of human prostate cancer patients (34). Immunohistochemical analysis was performed with 116 patients who underwent surgical treatment and adjuvant tamoxifen therapy of primary HR-positive breast cancers. Representative images for strong (Fig. 1B) and weak (Fig. 1C) TRIM47 immunoreactivity are shown, respectively. In the cases with the TRIM47 immunoreactivity, it was observed mainly in the cytoplasm of the breast tumor cells. At the time of diagnosis, the TRIM47 immunoreactivity was not significantly related to any of the clinicopathological parameters examined (SI Appendix, Table S1).
Fig. 1.
Overexpression of TRIM47 is associated with endocrine therapy resistance. (A) Western blot analysis showing antibody recognition of human TRIM47 protein. Whole cell lysates from HEK293 cells transfected with a plasmid encoding Flag-TRIM47 or an empty vector were subjected to Western blot analysis with indicated antibodies. (B and C) Representative micrographs of breast cancer tissue with strong (B) and weak (C) TRIM47 immunoreactivity (IR) (Scale bars, 100 μm). (D) Disease-free survival of breast cancer patients with strong or weak TRIM47 IR. Disease-free survival rate was shown by the Kaplan–Meier method. The red line represents cases with strong TRIM47 IR (n = 62), and the blue line represents cases with weak TRIM47 IR (n = 54). (E) Expression of TRIM47 in MCF-7 cells, OHTR MCF-7 cells, and LTED MCF-7 cells analyzed by qRT-PCR. The relative RNA levels were determined by normalizing with GAPDH expression and presenting as mean fold change ± SEM (n = 3). *P < 0.05, **P < 0.01 (Student’s t test compared with MCF-7). (F) Western blot analysis of TRIM47 expression in MCF-7 cells, OHTR cells, and LTED cells. β-actin protein was blotted as an internal control. (G) Cell proliferation of MCF-7 cells transfected with siTRIM47 was measured by MTS assay. Two kinds of siRNAs (10 nM) for TRIM47 (siTRIM47-A and B) and two kinds of siRNAs (10 nM) not targeting human transcripts (siLuc and siControl) were used. Absorbance of 490 nm 6 d after siRNA transfection was evaluated. Results are expressed as mean ± SEM (n = 4). **P < 0.01 (two-way ANOVA). (H) Cell proliferation of OHTR cells transfected with siTRIM47 was measured by MTS assay. The cells were cultured under treatment with OHT (1 μM). ***P < 0.001 (two-way ANOVA). (I) Effect of siTRIM47-A on growth of OHTR-derived xenograft tumors in nude mice. siControl or siTRIM47-A was injected twice a week into the xenograft tumors generated by OHTR cells in the flanks of the mice. Representative photographs of xenografted mice 3 wk after the beginning of siRNA administration are shown. The black arrows indicate margins of tumors (Scale bars, 1 cm). (J) Growth of OHTR-derived xenograft tumors injected with siControl (n = 11) and siTRIM47-A (n = 10). Tumor volumes are presented as mean ± SEM *P < 0.05, **P < 0.01 (Student’s t test). (K) Cell proliferation of MCF-7 cells stably expressing TRIM47 (two independent clones: TRIM47 #1 and #2) and vector clones (two independent clones: Vector #1 and #2) was measured by MTS assay. The cells were cultured under treatment with OHT (10 μM). Absorbance of 490 nm at day 1 and day 5 was evaluated. Results are expressed as mean ± SEM (n = 4). ***P < 0.001 (two-way ANOVA).
Relationship between TRIM47 Immunoreactivity and Patients’ Prognosis.
We next examined the relationship between TRIM47 immunoreactivity and clinical prognosis of breast cancer patients. TRIM47 immunoreactivity was significantly associated with a higher rate of recurrence (Fig. 1D) in breast cancer patients. Univariate analysis of disease-free survival using Cox proportional regression analysis demonstrated that four factors, including strong TRIM47 immunoreactivity, are significant poor prognostic factors for disease-free survival of the patients treated with tamoxifen. Other than strong TRIM47 immunoreactivity, younger age (50 y old and below), larger tumor size, and positive lymph node metastasis were selected (SI Appendix, Table S2). Then, multivariate Cox proportional regression analysis with these four factors was performed. As a result, three factors, strong TRIM47 immunoreactivity, larger tumor size, and positive lymph node metastasis, were identified as independent prognostic factors for predicting tamoxifen resistance (SI Appendix, Table S2).
Our results were supported by the public database on transcriptional analyses. According to Oncomine database (37), expression of TRIM47 is significantly elevated in invasive breast cancer tissues compared with normal breast tissues (SI Appendix, Fig. S1). According to the Kaplan–Meier plotter (38), higher expression of TRIM47 is significantly related with shorter relapse-free survival (RFS) (SI Appendix, Fig. S2 A–D). This relationship is markedly observed when the analyses were confined to the patients who underwent endocrine therapy as well as tamoxifen therapy (SI Appendix, Fig. S2 C and D). We did not observe statistically significant associations between TRIM47 expression and patients’ prognosis in triple-negative breast cancer subtype and human epidermal growth factor receptor type 2-positive subtype (SI Appendix, Fig. S2 E and F). Then, we compared the expression of TRIM47 among the endocrine therapy–resistant clones of MCF-7 cells, an estrogen receptor (ER)-positive breast cancer cell line. Expression of TRIM47 was elevated in a 4-hydroxytamoxifen-resistant (OHTR) derivative of MCF-7, and long-term estrogen-deprived (LTED) cells compared with their parental MCF-7 cells in the messenger RNA (mRNA) (Fig. 1E) and protein (Fig. 1F) levels. These results suggested that high expression of TRIM47 could be related to poor prognosis of breast cancer patients who underwent endocrine therapy.
TRIM47 Knockdown Inhibits the Proliferation of Breast Cancer Cells.
Since our clinical data are based on the patients who underwent endocrine therapy targeting estrogen signaling, we utilized MCF-7 cells and OHTR cells to evaluate biological functions of TRIM47. We transfected specific small interfering RNAs (siRNAs) for TRIM47 (siTRIM47-A/-B) in MCF-7 cells and OHTR cells. Suppression of TRIM47 expression by TRIM47 siRNAs was confirmed by both qRT-PCR (SI Appendix, Fig. S3A) and Western blotting (SI Appendix, Fig. S3B). Then, the effect of TRIM47 knockdown on cell proliferation was analyzed by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. The amounts of both MCF-7 and OHTR cells estimated with the absorbance of 490 nm were suppressed by TRIM47 knockdown (Fig. 1 G and H). Next, we evaluated the effect of TRIM47 knockdown on tumor growth of xenografts derived from OHTR cells using athymic mice. OHTR cells were implanted into the flanks of athymic female mice and then they were injected with siTRIM47-A or control siRNA (siControl). Notably, tumor volume was significantly reduced in mice treated with siTRIM47 compared with mice treated with control siRNA (Fig. 1 I and J). This effect was further analyzed by MCF-7 cell clones stably overexpressing TRIM47. When the two independent clones overexpressing TRIM47 and the two independent vector clones as controls were cultured in the presence of 1 μM OHT, the clones overexpressing TRIM47 kept proliferating, while the vector clones decreased their cell number compared with that of the first day, suggesting that the clones overexpressing TRIM47 acquired OHT-resistant characteristics (Fig. 1K).
Activation of NF-κB Signaling by TRIM47.
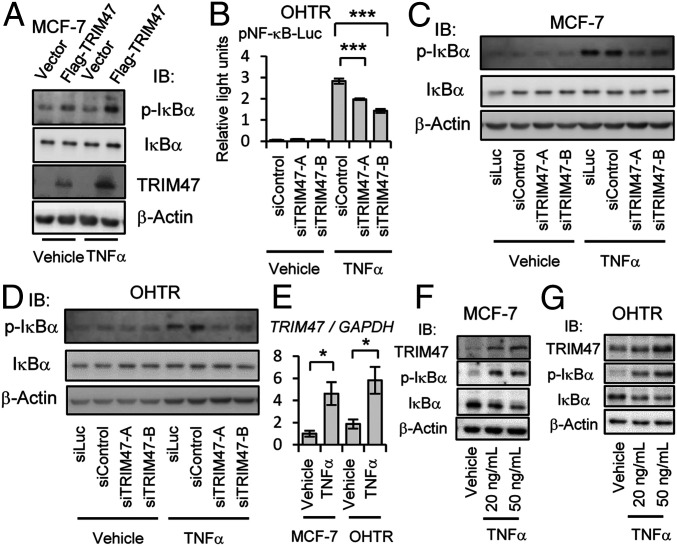
NF-κB signaling has pathological relevance in malignancies, and enhanced NF-κB signaling is reported to have tumor-promoting function in ER-positive breast cancer cells (14, 15). We thus evaluated the effects of TRIM47 on NF-κB signaling pathway by Western blot analysis and luciferase reporter assay. Activation of NF-κB signaling is caused by phosphorylation of NF-κB inhibitor-α (IκBα) and subsequent degradation of IκBα (39). Transient overexpression of TRIM47 in MCF-7 cells resulted in enhanced phosphorylation of IκBα (Fig. 2A). This effect was observed both in the presence and the absence of tumor necrosis factor-α (TNF-α), an activator of NF-κB signaling. NF-κB–mediated transcriptional activity evaluated by the intensity of luminescence when NF-κB luciferase reporter plasmid was transfected (SI Appendix, Fig. S4). This effect was observed both in the presence and the absence of TNF-α in a RING finger domain–dependent manner. In line with the augmented NF-κB signaling by TRIM47 overexpression, TRIM47 knockdown caused impaired NF-κB–mediated transcriptional activity evaluated by NF-κB luciferase reporter assay in OHTR cells (Fig. 2B). Attenuated phosphorylation of IκBα by treatment with TRIM47 siRNAs was observed in both MCF-7 and OHTR cells (Fig. 2 C and D). This effect was remarkable when the cells were treated with TNF-α. These results indicated that TRIM47 activates NF-κB signaling. We analyzed the effect of TRIM47 on the upstream molecule of NF-κB signaling. Knocking down of TRIM47 attenuated phosphorylation of TGF-β–activated kinase 1 (TAK1), which is known to phosphorylate I-κB kinase alpha (IKK-α) and I-κB kinase beta (IKK-β) (40), in MCF-7 cells (SI Appendix, Fig. S5). MAPK pathway, another pathway involved in the promotion of breast cancer, was not affected by knocking down of TRIM47 in MCF-7 and OHTR cells (SI Appendix, Fig. S6 A and B). With genome-wide analysis using microarray, we evaluated the effect of TRIM47 knocking down on the transcriptome. Validation of individual genes revealed that TRIM47 affected downstream genes of NF-κB signaling such as APAF1 and CPEB3 in the selected manner (SI Appendix, Fig. S7 A–D). Both genes are reported to have tumor-suppressive function (41, 42) and are down-regulated by NF-κB signaling (SI Appendix, Fig. S7 E and F).
Fig. 2.
Activation of NF-κB signaling by TRIM47. (A) Flag-TRIM47 expression vector or empty vector were transfected to MCF-7 cells. At 48 h after transfection, cells were treated with TNF-α (50 ng/mL) or vehicle (PBS; phosphate buffered saline) for 5 min before cell lysis. Phosphorylated form–specific IκBα antibody (p-IκBα) and antibody for total IκBα were used for Western blotting. (B) At 24 h after OHTR cells were transfected with a NF-κB reporter plasmid and a plasmid encoding renilla luciferase, the cells were detached from the dish, and indicated siRNAs (10 nM) were transfected by reverse transcription method. siControl was used as a negative control. At 48 h after siRNA transfection, cells were treated with TNF-α (10 ng/mL) or vehicle (PBS) for another 5 h before measuring luciferase activities. Results are expressed as mean ± SEM (n = 6). Luciferase activities of TNF-α–stimulated group analyzed by one-way ANOVA were significantly different. Post hoc Dunnett’s test was performed in comparison with siControl group. ***P < 0.001. (C and D) At 48 h after transfection of indicated siRNAs (10 nM), MCF-7 cells or OHTR cells were treated with TNF-α (20 ng/mL) or vehicle (PBS) for 5 min before cell lysis. Western blotting was performed to detect phosphorylated IκBα (p-IκBα) and total IκBα. (E) MCF-7 cells or OHTR cells were treated with TNF-α (20 ng/mL) for 24 h, and expression of TRIM47 was analyzed by qRT-PCR. The relative RNA levels were shown as mean fold change ± SEM *P < 0.05 (Student’s t test). (F and G) MCF-7 or OHTR cells were treated with TNF-α (20 or 50 ng/mL) or vehicle (PBS) for 24 h before cell lysis. Western blotting was performed to detect phosphorylated IκBα (p-IκBα), total IκBα, and TRIM47.
On the other hand, we also found that transcription of TRIM47 was increased by TNF-α treatment in both MCF-7 and OHTR cells (Fig. 2E). Moreover, the protein levels of TRIM47 were increased in a dose-dependent manner when MCF-7 and OHTR cells were treated with TNF-α for 24 h (Fig. 2 F and G). These results suggest that TRIM47 is one of the responsive genes of NF-κB signaling, and its transcriptional induction results in further activation of NF-κB signaling. Moreover, treatment of MCF-7 cells with estrogen or OHT did not affect transcription of TRIM47 significantly in the condition that GREB1, a typical estrogen-responsive gene was significantly affected (SI Appendix, Fig. S8 A–D).
Stabilization and Polyubiquitination of PKC-ε by TRIM47.
Then, we asked how TRIM47 activates NF-κB signaling. Among several factors reported to modulate NF-κB signaling, we focused on PKC-ε (encoded by PRKCE gene), which was identified as one of the binding partners with TRIM47 by cell-free protein kinase array analysis (SI Appendix, Table S3). PKC-ε is a serine/threonine kinase reported to be an activator of NF-κB signaling in several kinds of cancer including triple negative breast cancer (43–45).
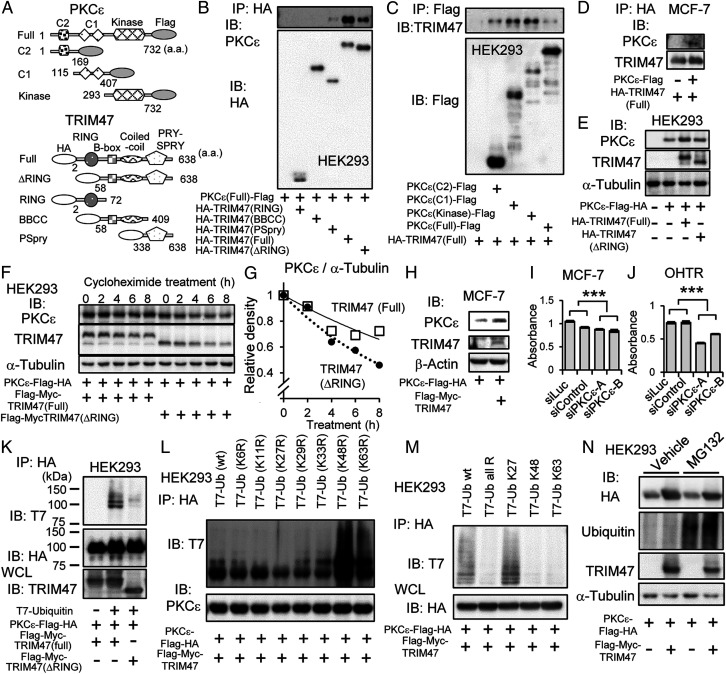
We showed interaction of TRIM47 with PKC-ε by immunoprecipitation using HEK293 cells expressing deletion mutants of TRIM47 or PKC-ε (Fig. 3A). As a result, PRY-SPRY (PSpry) domain of TRIM47 was important for interaction of TRIM47 and PKC-ε (Fig. 3B). On the other hand, each examined domain of PKC-ε was able to interact with TRIM47 (Fig. 3C). The interaction of TRIM47 with PKC-ε was also detected in MCF-7 cells when these proteins were overexpressed (Fig. 3D).
Fig. 3.
Association and stabilization of PKC-ε by TRIM47. (A) Schematic representation of deletion mutants of Flag epitope–tagged PKC-ε and HA epitope–tagged TRIM47. a.a. indicates amino acids. (B) HEK293 cells were cotransfected with plasmid encoding PKCε(Full)-Flag and one of plasmids encoding HA epitope–tagged TRIM47 deletion mutants. After 48 h, cells were lysed, immunoprecipitated with anti-HA agarose, and eluted with HA peptide. Elutants were subjected to Western blot analysis with anti–PKC-ε and anti-HA antibodies. (C) HEK293 cells were cotransfected with HA-TRIM47(Full) plasmid and one of Flag epitope–tagged PKC-ε deletion-mutant plasmids. After 48-h culture, cells were lysed, immunoprecipitated with anti-Flag agarose, and eluted with Flag peptide. Elutants were subjected to Western blot analysis with anti-TRIM47 and anti-Flag antibodies. (D) MCF-7 cells were transfected with plasmids encoding PKCε-Flag and HA-TRIM47. Empty vector (pcDNA3) was used as a negative control. After 48 h, cells were lysed, immunoprecipitated with anti-HA agarose, and eluted with HA peptide. Elutants were subjected to Western blot analysis with anti–PKC-ε and anti-TRIM47 antibodies. (E) HEK293 cells were transfected with plasmids encoding PKCε-Flag-HA and HA-TRIM47(Full) or HA-TRIM47(ΔRING). Empty vector (pcDNA3) was used as a negative control. At 1 d after transfection, the cells were lysed and subjected to Western blot analysis with indicated antibodies. (F) HEK293 cells were transfected with plasmids encoding PKCε-Flag-HA, and Flag-Myc-TRIM47(Full) or Flag-Myc-TRIM47(ΔRING) as indicated. At 1 d after transfection, the cells were treated with 50 μg/mL cycloheximide for indicated duration. Cell lysates were subjected to Western blot analysis with indicated antibodies. (G) PKC-ε and α-tubulin protein levels in F were quantified by densitometry. Densities of PKC-ε blots were normalized to each corresponding density of α-tubulin blot. Then, each value was divided by the value of the sample without cycloheximide treatment (T = 0). The open squares and the solid approximate curve indicate PKC-ε with TRIM47 (full). The black circles and the dotted approximate curve indicate PKC-ε with TRIM47 (ΔRING). (H) MCF-7 cells were transfected with plasmids encoding PKCε-Flag-HA and HA-TRIM47. Empty vector (pcDNA3) was used as a negative control. At 1 d after transfection, the cells were lysed and subjected to Western blot analysis with indicated antibodies. (I and J) Cell proliferation of MCF-7 cells and OHTR cells transfected with siPKC-ε was measured by MTS assay. Two kinds of siRNAs (2 nM) for PKC-ε (siPKC-ε A and B) and two kinds of siRNAs (2 nM) not targeting human transcripts (siLuc and siControl) were transfected by reverse transcription method. Absorbance of 490 nm 6 d after siRNA transfection was evaluated by MTS assay. Results are expressed as mean ± SEM (n = 4). ***P < 0.001 (two-way ANOVA). (K) HEK293 cells were transfected with plasmids encoding PKCε-Flag-HA, T7-Ubiquitin, and Flag-Myc-TRIM47(Full) or TRIM47 lacking Flag-Myc-TRIM47(ΔRING) as indicated. After 48 h, cells were lysed, immunoprecipitated with anti-HA agarose, and eluted with HA peptide. Elutants were subjected to Western blot analysis with indicated antibodies. Whole cell lysates (WCL) were subjected to Western blot analysis with anti-TRIM47 antibody. (L) HEK293 cells were transfected with plasmids encoding PKCε-Flag-HA, Flag-Myc-RIM47, and one of T7 epitope–tagged ubiquitin mutants containing arginine (R) substitution of one of lysine (K) residues at the indicated position. Ub (wt) indicates wild-type ubiquitin without mutation. Ub (K6R) indicates arginine substitution of lysine 6. Substitutions of other lysines are shown in the same way. After 40 h, cells were lysed, immunoprecipitated with anti-HA agarose, and eluted with HA peptide. Elutants were subjected to Western blot analysis with indicated antibodies. (M) HEK293 cells were transfected with plasmids encoding PKCε-Flag-HA, Flag-Myc-RIM47, and one of T7 epitope–tagged ubiquitin mutants containing arginine (R) substitution of all lysine (K) residues except one of the lysine residues at the indicated position. Ub K27 indicates all arginine substitutions except lysine 27. Other ubiquitin mutants are shown in the same way. T7-Ub all R indicates all arginine substitutions. After 40 h, cells were lysed, immunoprecipitated with anti-HA agarose, and eluted with HA peptide. Elutants and WCL were subjected to Western blot analysis with indicated antibodies. (N) HEK293 cells were transfected with plasmids encoding PKCε-Flag-HA, Flag-Myc-TRIM47, or empty vector as indicated. On the next day, cells were treated with MG132 (10 μM) or vehicle (DMSO) for 6 h before they were lysed. Total cell lysates were subjected to Western blot analysis with indicated antibodies.
During the experiments confirming interaction of TRIM47 and PKC-ε, we noticed that the amount of PKC-ε protein is higher when TRIM47 was expressed together, and this effect was RING finger–domain dependent (Fig. 3E). Since the expression of PKC-ε was not significantly regulated at the transcription level (SI Appendix, Fig. S9A), we evaluated the stability of PKC-ε protein by treating cells with cycloheximide for various durations. While the half-life of PKC-ε was less than 6 h in HEK293 cells transfected with exogenous TRIM47 lacking RING finger domain, it was more than 8 h in HEK293 cells overexpressing full-length TRIM47 (Fig. 3 F and G). Stabilization of PKC-ε protein by overexpressing TRIM47 was also observed in MCF-7 cells (Fig. 3H).
When expression of PKC-ε was knocked down by its specific siRNAs, MTS assay showed that proliferation of MCF-7 cells and OHTR cells was suppressed (Fig. 3 I and J), suggesting PKC-ε has growth-promoting effects in breast cancer cells. Knocking down of PKC-ε attenuated phosphorylation of I-κB kinase alpha (IKK-α) and I-κB kinase beta (IKK-β) in MCF-7 cells and OHTR cells (SI Appendix, Fig. S9 A and B), suggesting PKC-ε mediates activation of NF-κB signaling also in breast cancer cells. Therefore, stabilization of PKC-ε by TRIM47 could be one of the mechanisms for TRIM47 to activate NF-κB signaling.
Since PKC-ε stabilizing effect was weak when PKC-ε was cotransfected with plasmid encoding TRIM47 lacking RING finger domain (Fig. 3 E and F), we assumed that this effect is dependent on E3 ubiquitin ligase activity of TRIM47. By cotransfection of plasmids encoding PKC-ε, TRIM47, and ubiquitin, modification of PKC-ε by ubiquitin was detected by immunoprecipitating PKC-ε. This modification was diminished when TRIM47 lacking RING finger domain was expressed (Fig. 3K), suggesting that PKC-ε can be a substrate for TRIM47. Next, we made plasmids encoding a series of ubiquitin mutants in which one of lysine residues was substituted by arginine. By cotransfection of a plasmid encoding a ubiquitin mutant with plasmids encoding TRIM47 and PKC-ε, we observed modification of PKC-ε by ubiquitin mutants in which lysine 48 (K48R) or lysine 63 (K63R) were substituted by arginine. Instead, modification of PKC-ε was not observed when plasmid encoding a ubiquitin mutant in which lysine 27 was substituted by arginine (K27R) (Fig. 3L). In line with this result, modification of PKC-ε by ubiquitin mutants in which all lysines except lysine 27 (K27) are mutated was observed (Fig. 3M).
To evaluate involvement of proteasomal degradation system, we treated HEK293 cells expressing PKC-ε and TRIM47 with MG132, a proteasome inhibitor. Treatment with MG132 increased total amount of ubiquitinated proteins, whereas it did not increase the amount of PKC-ε (Fig. 3N), showing proteasomal degradation system is not relevant in the PKC-ε stability.
Stabilization of PKD3 by TRIM47.
We then focused on another serine/threonine kinase, PKD3, which was also identified as one of the interactive proteins with TRIM47 by cell-free protein kinase array analysis (SI Appendix, Table S3). PKD3 was reported to be an activator of NF-κB signaling in prostate cancer cells (46). Moreover, PKC-ε is reported to phosphorylate and activate PKD3 in prostate cancer cells (47).
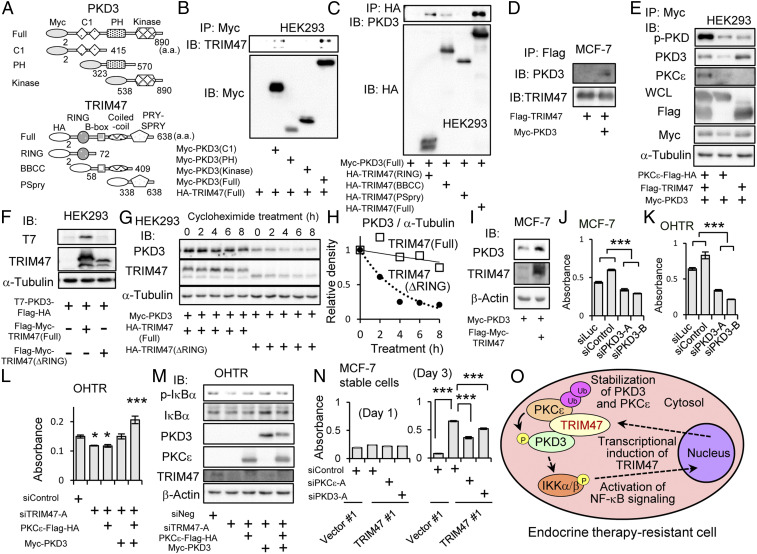
We showed association of TRIM47 with PKD3 by immunoprecipitation using HEK293 cells expressing deletion mutants of TRIM47 or PKD3 (Fig. 4A). As a result, RING finger domain, B-box domain, and coiled-coil domain of TRIM47 were able to interact with PKD3 (Fig. 4B). On the other hand, C1 domain of PKD3 was important for interaction of TRIM47 and PKD3 (Fig. 4C). The interaction of TRIM47 with PKD3 was also detected in MCF-7 cells when these proteins were overexpressed (Fig. 4D).
Fig. 4.
Association and stabilization of PKD3 by TRIM47. (A) Schematic representation of deletion mutants of Myc epitope–tagged PKD3 and HA epitope–tagged TRIM47. a.a. indicates amino acids. (B) HEK293 cells were cotransfected with a plasmid encoding HA-TRIM47 and one of plasmids encoding Myc epitope–tagged PKD3 deletion mutants. After 48 h, cells were lysed, immunoprecipitated with anti-Myc agarose, and eluted with Myc peptide. Elutants were subjected to Western blot analysis with indicated antibodies. (C) HEK293 cells were cotransfected with Myc-PKD3 plasmid and one of HA epitope–tagged TRIM47 deletion-mutant plasmids. After 48-h culture, cells were lysed, immunoprecipitated with anti-HA agarose, and eluted with HA peptide. Elutants were subjected to Western blot analysis with indicated antibodies. (D) MCF-7 cells were transfected with plasmids encoding Myc-PKD3 and Flag-TRIM47. Empty vector (pcDNA3) was used as a negative control. After 48 h, cells were lysed, immunoprecipitated with anti-Flag agarose, and eluted with Flag peptide. Elutants were subjected to Western blot analysis with anti-PKD3 and anti-TRIM47 antibodies. (E) HEK293 cells were cotransfected with plasmids encoding PKCε-Flag-HA, Flag-TRIM47, and Myc-PKD3 as indicated. After 48 h, cells were lysed, immunoprecipitated with anti-Myc agarose, and eluted with Myc peptide. Elutants were subjected to Western blot analysis with indicated antibodies including phosphorylated form–specific PKD antibody (p-PKD). Whole cell lysates (WCL) were subjected to Western blot analysis with indicated antibodies. (F) HEK293 cells were transfected with plasmids encoding T7-PKD3-Flag-HA, and Flag-Myc-TRIM47(Full) or Flag-Myc-TRIM47(ΔRING) as indicated. Two days after transfection, the cells were lysed and subjected to Western blot analysis with indicated antibodies. (G) HEK293 cells were transfected with plasmids encoding Myc-PKD3, and HA-TRIM47(Full) or HA-TRIM47(ΔRING) as indicated. One day after transfection, the cells were treated with 50 μg/mL cycloheximide for indicated duration. Cell lysates were subjected to Western blot analysis with indicated antibodies. (H) PKCε and α-tubulin protein levels in G were quantified. Densities of PKD3 blots were normalized to corresponding densities of α-tubulin blot. Then, each value was divided by the value of the sample without cycloheximide treatment (T = 0). Open squares and the solid approximate curve indicate PKD3 with TRIM47 (full). Black circles and the dotted approximate curve indicate PKD3 with TRIM47 (ΔRING). (I) MCF-7 cells were transfected with plasmids encoding Myc-PKD3, and Flag-Myc-TRIM47. Empty vector (pcDNA3) was used as a negative control. One day after transfection, the cells were lysed and subjected to Western blot analysis with indicated antibodies. (J and K) Cell proliferation of MCF-7 and OHTR cells transfected with siPKD3 were measured by MTS assay. Two kinds of siRNAs (10 nM) for PKD3 (siPKD3-A and B) and two kinds of siRNAs (10 nM) not targeting human transcripts (siLuc and siControl) were transfected. Absorbance of 490 nm 5 d after siRNA transfection was evaluated by MTS assay. Results are expressed as mean ± SEM (n = 4). ***P < 0.001 (two-way ANOVA). (L) OHTR cells transfected with siTRIM47 (10 nM) were transfected with plasmids encoding PKCε-Flag-HA and/or Myc-PKD3 on the next day. Cell proliferation on 4 d after siRNA transfection was measured by MTS assay. Absorbance of 490 nm analyzed by one-way ANOVA were significantly different. Post hoc Dunnett’s test was performed in comparison with siControl transfected cells. ***P < 0.001, *P < 0.05. (M) OHTR cells transfected with siTRIM47 (10 nM) were transfected with plasmids encoding PKCε-Flag-HA and/or Myc-PKD3 on the next day. Forty-eight h after siRNA transfection, cells were lysed and subjected to Western blot analysis. Indicated antibodies including phosphorylated form–specific IκBα antibody (p-IκBα) were used for blotting. (N) Cell proliferation of MCF-7 cells stably expressing TRIM47 (TRIM47 #1 clone) and vector clones (Vector #1 clone) was measured by MTS assay. Transfection of indicated siRNAs (10 nM) was performed by reverse transcription method. The cells were cultured under treatment with OHT (10 μM). Absorbance of 490 nm at day 1 and day 3 was evaluated. Results are expressed as mean ± SEM (n = 4). Absorbance of 490 nm analyzed by one-way ANOVA were significantly different. Post hoc Dunnett’s test was performed in comparison with siControl transfected TRIM47#1 cells. ***P < 0.001. (O) A schematic model of TRIM47 functions contributing to endocrine therapy resistance of breast cancer cells.
The effect of PKC-ε on phosphorylation of PKD3 was evaluated by phosphorylated form–specific PKD antibody, which detects phosphorylated form of PKD1, PKD2, and PKD3. Phosphorylation of immunoprecipitated PKD3 was shown to be increased when a plasmid encoding PKD3 was cotransfected with plasmids encoding PKC-ε and TRIM47 compared with cotransfection of TRIM47 coding plasmid alone (Fig. 4E). This result was in line with the previous report showing that PKC-ε phosphorylates PKD3 (47). In the present experiment, interaction of PKC-ε and PKD3 was detected in the presence of TRIM47, and absence of TRIM47 resulted impaired interaction (Fig. 4E).
The interaction of PKC-ε and PKD3 may be also influenced by the amount of PKD3 protein. Like stabilization of PKC-ε by TRIM47, we noticed that the amount of PKD3 protein is higher when TRIM47 was expressed together, and this effect was also RING finger–domain dependent (Fig. 4F). The expression of PKD3 was not significantly regulated at the transcription level (SI Appendix, Fig. S9B). Then, we evaluated the stability of PKD3 protein by treating cells with cycloheximide for various durations. While the half-life of PKD3 was less than 4 h in HEK293 cells transfected with exogenous TRIM47 lacking RING finger domain, it was more than 8 h in HEK293 cells overexpressing full-length TRIM47 (Fig. 4 G and H). Stabilization of PKD3 protein by overexpressing TRIM47 was also observed in MCF-7 cells (Fig. 3I). The stabilization of PKD3 may be independent of proteasomal degradation system, since treatment with MG132 did not affect the protein amount of PKD3 (SI Appendix, Fig. S11).
When expression of PKD3 was knocked down by its specific siRNAs, proliferation of MCF-7 cells and OHTR cells was suppressed (Fig. 4 J and K), suggesting that PKD3 has growth-promoting effects in breast cancer cells. Knocking down of PKD3 attenuated phosphorylation of IKK-α and IKK-β in MCF-7 and OHTR cells (SI Appendix, Fig. S12 A and B), suggesting that PKD3 mediates activation of NF-κB signaling in breast cancer cells. Therefore, stabilization of PKD3 by TRIM47 can be one of the mechanisms for TRIM47 to activate NF-κB signaling.
To further examine the role of TRIM47/PKC-ε/PKD3 in the breast cancer cell, we overexpressed PKC-ε and/or PKD3 in the OHTR cells where TRIM47 was knocked down. The ectopic expression of PKC-ε and/or PKD3 rescued the suppressed growth (Fig. 4L) and the diminished NF-κB signaling (Fig. 4M). Moreover, knockdown of PKC-ε or PKD3 impaired activation of NF-κB signaling by overexpressing TRIM47 (SI Appendix, Fig. S13 A and B), showing that PKC-ε and PKD3 are downstream effectors of TRIM47 in activating NF-κB signaling. Finally, knockdown of PKC-ε or PKD3 partially reverse the OHT resistance of MCF-7 cells stably expressing TRIM47 (Fig. 4N), showing the involvement of TRIM47/PKC-ε/PKD3 axis in the endocrine therapy resistance of breast cancer cells (Fig. 4O).
Discussion
In the present study, we demonstrated that strong TRIM47 immunoreactivity is a poor prognostic factor that predicts recurrence during or after adjuvant tamoxifen therapy. The high TRIM47 immunoreactivity was an independent predictive factor among other clinicopathological prognostic factors. We observed increased expression of TRIM47 in LTED cells as well as OHTR cells. Considering that LTED cells are a model mimicking hormone deprivation therapy with AIs, our clinical results could be expanded to general endocrine therapies. Indeed, the analysis based on public microarray databases revealed that high expression of TRIM47 mRNA is significantly related with shorter RFS in the patients who underwent any kind of endocrine therapies. Notably, we evaluated TRIM47 immunoreactivity in the clinical samples before adjuvant endocrine therapy and showed their values to predict poor prognosis. Therefore, we speculate elevated expression of TRIM47 in OHTR and LTED cells reflects selection of the MCF-7 cells with high expression of TRIM47, which survived during prolonged exposure with OHT or during prolonged estrogen deprivation.
We showed that TRIM47 activates NF-κB signaling in MCF-7 and OHTR cells. There are several lines of evidence supporting the important roles of NF-κB signaling in developing endocrine therapy resistance of ER-positive breast cancer cells. It was clinically reported that enhanced NF-κB signaling is associated with tamoxifen resistance of primary breast cancer (14). In a genome-wide association study on tamoxifen response and single-nucleotide polymorphisms, NF-κB signaling pathway was extracted as one of the key pathways related to tamoxifen resistance, along with apoptosis, p53, DNA repair, and cell cycle pathways (15). It was shown that inhibiting NF-κB signaling restored tamoxifen sensitivity of tamoxifen-resistant MCF-7 cells (48). In another study, AI-resistant cells derived from MCF-7 and T47D cells exhibited enhanced NF-κB signaling activity, and knocking down NF-κB suppressed proliferation of AI-resistant cells (49). As an underlying mechanism, NF-κB signaling is shown to affect transcription mediated by ER. It was reported that a distinct gene set is shown to be up-regulated by the costimulation of estrogen and TNF-α in MCF-7 cells (50). In another study of MCF-7 cells using chromatin immunoprecipitation sequencing, new ER-binding sites were shown to be created by the binding of NF-κB to FOXA1 in some enhancers (51).
With the validation process of microarray experiments, two endogenous target genes of NF-κB signaling, APAF1 and CPEB3, were identified. APAF1 is a protein mediating apoptosis of several cancers, including breast cancer (41). CPEB3 is an RNA-binding protein and reported to bind to 3′ untranslated region (UTR) of glutamate receptor 2 (GluR2) mRNA and induced translation of GluR2 protein in neural cells (52). In terms of cancer biology, lower expression of CPEB3 was observed in hepatocellular carcinoma (HCC) tissues compared with adjacent tissues. In the same study, high expression of CPEB3 was shown to be related with better prognosis, and overexpression of CPEB3 in HCC cell lines suppressed growth and motility of the cells (42), suggesting tumor-suppressive function of CPEB3. We demonstrated expression of these genes were suppressed TNF-α–dependent manner in MCF-7 and OHTR cells. Together with the results showing knocking down of TRIM47 induced their expression, it can be assumed that NF-κB signaling enhanced by TRIM47 contributes to promotion of breast cancer by suppressing these target genes.
In addition to the activation of NF-κB signaling by TRIM47, we also revealed that expression of TRIM47 is up-regulated by NF-κB activator, TNF-α. Interestingly, the mutual up-regulation of TRIM47 and NF-κB signaling indicates existence of positive feedback activation of NF-κB signaling by TRIM47. This could be an underlying mechanism for sustained activation of NF-κB signaling by TRIM47, which can result in endocrine therapy resistance (Fig. 4O).
In this study, we proposed that protein stabilization of PKC-ε and PKD3 is one of the mechanisms underlying the TRIM47-dependent activation of NF-κB signaling in breast cancer. These are the kinases identified as binding partners of TRIM47 by our cell-free interaction assay. Notably, both kinases were reported to activate NF-κB signaling in prostate cancer cells (43, 46). PKC-ε was reported to activate NF-κB signaling also in glioblastoma (44), breast epithelial (46), and triple negative breast cancer cells (46). PKC-ε belongs to PKC, which is a family of structurally related serine/threonine kinases. Among them, PKC-α is known to be associated with poor response to endocrine therapy of breast cancer (53, 54). In the present study, we focused on the function of PKC-ε. PKC-ε is generally up-regulated in invasive breast cancers (55) and prostate cancers (56) and is characterized as a positive regulator of mitogenic and survival pathways. In prostate cancer cells, PKC-ε overexpression with Pten loss leads to CXCL13 up-regulation through noncanonical NF-κB signaling, leading to the invasive tumor phenotype (43).
We showed that the stabilization of PKC-ε and PKD3 requires RING finger domain of TRIM47, which suggested that E3 ligase activity was important for this effect. We detected RING finger domain–dependent polyubiquitination of PKC-ε in the presence of TRIM47. Of note, we revealed by mutation analysis that PKC-ε was modified by atypical lysine 27–linked polyubiquitination, not by lysine 48– or lysine 63–linked polyubiquitinations, which are commonly observed modes of ubiquitin modification. Although biological significance of lysine 27–linked polyubiquitination is poorly characterized, similar K27-linked polyubiquitination-dependent events have been reported in the innate immune systems. TIR domain–containing adaptor inducing interferon-β (TRIF) and TAK1 are functionally active with K27-linked polyubiquitination, whereas their deconjugated forms lose activities for inflammatory response and TLR3/4-mediated innate immune signaling, respectively (57, 58). Notably, a recent study revealed that deubiquitinating enzyme UCHL3 is functionally inhibited by K27 ubiquitin dimers and K27-ubiquitinated substrates (59), suggesting that K27-ubiquitinated substrates may remain with a relatively longer stability by impairing UCHL3 activity and exert their functions in various mechanisms, including innate immunity. We thus consider that TRIM47 exhibits K27-linked polyubiquitination of oncogenic PKC-ε with increased protein stability, leading to the phosphorylation and activation of PKD3 and the subsequent activation of NF-κB pathway. While the K63-linked polyubiquitination is also known to stabilize substrates and positively regulate substrate functions, the stabilizing mechanism of K63 polyubiquitination may be different from that of K27-linked polyubiquitination. For example, TAK1 is also conjugated with K63-linked polyubiquitination chains by TRIM8 (60) and TRAF6 (61), and K63-ubiquitinated TAK1 plays a role in the complex aggregation of TAK1 and TAK1-binding proteins (TABs) and subsequently recruits IKK complex to activate NF-κB pathway (61). On the contrary, K27-ubiquitinated TAK1 is critical for the interaction between TABs, the early event in the NF-κB activation signal (58). We thus assume that K27- and K63-linked polyubiquitination may exhibit distinct roles in the protein stabilization and substrate activation such as the step-wise NF-κB activation mediated by TAK1 activation. Future studies will reveal whether TRIM47 regulates NF-κB activation step-wisely together with other E3 ligases. We also showed that a proteasome inhibitor did not affect stability of PKC-ε or PKD3, which suggested those proteins are not degraded by proteasome. These proteins may be degraded by another intracellular protein degradation system such as autophagy, which is inferred by several examples that some TRIM family members regulate autophagy by catalyzing polyubiquitination on substrate proteins (62).
Activation of PKD3 with phosphorylation by PKC-ε was reported in prostate cancer cells (47). We showed the interaction of PKC-ε and PKD3 was more evident in the presence of TRIM47. This result could reflect the role of TRIM47 as a scaffold-forming ternary complex of TRIM47, PKC-ε, and PKD3 considering PKC-ε and PKD3 are identified as direct binding partners with TRIM47 with the cell-free system. This scaffolding function of TRIM47 may facilitate phosphorylation of PKD3 by bringing PKD3 in the proximity of PKC-ε (Fig. 4O).
There are several reports on the clinical importance of TRIM47 in other cancer types. We previously reported that TRIM47 is a poor prognostic factor of prostate cancer patients (34). Recently, TRIM47 was shown to be a poor prognostic factor for nonsmall lung cell cancer (35) and colorectal cancer (36). It was shown that in the nonsmall lung cancer cell line A549 and H358, TRIM47 has an inhibitory effect on p53 and facilitating effect on NF-κB signaling (35). Our results are in line with the latter mechanism by showing that TRIM47 can activate NF-κB signaling in breast cancer cell lines, which could explain poor prognosis of HR-positive breast cancer patients. In colorectal cancer, TRIM47 was reported to cause ubiquitination and degradation of SMAD4, which lead enhanced growth and invasion of colorectal cancer cells (35). During the preparation of this manuscript, a study of TRIM47 in breast cancer cells including both ER-positive and -negative cell lines was reported (63). Prognostic value was not analyzed in this study, while involvement of phosphatidylinositol 3-kinase (PI3K)/Akt was shown. This finding supports our results, considering that Akt activates NF-κB signaling by phosphorylating IKK-α (64). In addition, TRIM47 was recently identified as a responsible E3 ubiquitin ligase for cylindromatosis (CYLD), a deubiquitinating enzyme (65). Degradation of CYLD by TRIM47 was shown to be associated with pathogenesis of nonalcoholic steatohepatitis (65). In terms of cancer biology, CYLD is known as a tumor suppressor. In a clinical study using breast cancer tissues, reduced expression of CYLD was associated with estrogen receptor negativity and poor prognosis (66). CYLD is reported to inhibit NF-κB signaling by deubiquitinating tumor necrosis factor receptor-associated factor (TRAF) 2 (67), as well as to suppress cell proliferation by deubiquitinating BCL3 (68). These findings might be involved in another mechanism explaining tumor-promoting function of TRIM47. In the cell-free interaction assay, we identified several kinases as candidates of binding partner with TRIM47. Tousled-like kinase 2 (TLK2), which is related to genome stability, is one of them. In HR-positive breast cancer, TLK2 is often amplified and correlated with worse clinical outcome (69). PKCθ is another kinase identified by the cell-free interaction assay. PKCθ is reported to phosphorylate and stabilize Fra-1, which is related to progression of ER-negative breast cancer cells (70). Another protein identified by the cell-free interaction assay was A6-related protein (A6r), which is also known as protein tyrosine kinase 9 like or twinfilin-2. A6r is reported to be involved in neurite outgrowth (71), although its oncological significance is yet to be clarified.
In conclusion, we demonstrate that TRIM47 is a poor prognostic factor for breast cancer patients who underwent endocrine therapy with tamoxifen. As an underlying mechanism, we revealed that formation of TRIM47/PKC-ε/PKD3 ternary complex, stabilization of PKC-ε and PKD3 proteins by TRIM47, and TRIM47-dependent lysine 27–linked polyubiquitination of PKC-ε. Our study suggests that TRIM47 and its associated kinases can be potential diagnostic and therapeutic targets for patients with breast cancer refractory to endocrine therapy and provides clues to develop effective strategies for breast cancer management.
Materials and Methods
Collection of Human Tissue Samples and Clinical Data.
Tissue samples of invasive breast cancer were obtained from 116 Japanese female breast cancer patients who underwent surgical treatment and adjuvant tamoxifen treatment at three institutions (National Hospital Organization Shikoku Cancer Center, Matsuyama, Japan; National Cancer Center Hospital, Tokyo, Japan; and Tokyo Metropolitan Cancer and Infectious Diseases Center, Komagome Hospital, Tokyo, Japan) from 1989 to 1998. No patients received other hormonal therapies, chemotherapies, or molecular target therapies before and after surgery. This study was approved by institutional review boards of National Hospital Organization Shikoku Cancer Center, and Saitama Medical University. All the patients provided written informed consent to participate in this study. Anonymized data created for this study are available in Dataset S1.
Detailed materials and methods regarding antibodies and reagents, immunohistochemistry, cell culture, plasmid construction and transfection, Western blot analysis, quantitative reverse transcription PCR, small interfering RNA transfection, cell proliferation assay, in vivo tumor growth assay, luciferase assay, microarray analysis, protein kinase array analysis, immunoprecipitation, and statistical analyses are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. S. Saji and C. Shimizu for recruitment of patients and sample collection. We thank Dr. T. Sawasaki for performing protein interaction analysis. We also thank Ms. N. Sasaki for her technical assistance. Discussion with Drs. T. Urano, H. Kawabata, and T. Ogura was helpful. This study was supported by grants from the Cell Innovation Program (S.I.), P-DIRECT (S.I.), and P-CREATE (S.I.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan; by Grants-in-Aid for S.I. (Grant No. 15K15353) and K. Azuma (Grant Nos. 17K10571 and 20K08954) from the Japan Society for the Promotion of Science, Japan by a grant from Uehara Memorial Foundation (S.I.), by a grant from Yamaguchi Endocrine Research Foundation (K. Azuma), and by a grant from Kao Health Science Foundation (K. Azuma).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100784118/-/DCSupplemental.
Data Availability
Microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus Datasets (GSE174612; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174612). All other study data are included in the article and/or supporting information.
References
- 1.Bray F., et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C. E., Ma J., Goding Sauer A., Newman L. A., Jemal A., Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J. Clin. 67, 439–448 (2017). [DOI] [PubMed] [Google Scholar]
- 3.National comprehensive cancer network (NCCN) guideline for treatment of breast cancer. https://www.nccn.org/professionals/physician_gls/default.aspx#site (Accessed 21 September 2020).
- 4.Hanker A. B., Sudhan D. R., Arteaga C. L., Overcoming endocrine resistance in breast cancer. Cancer Cell 37, 496–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurtado A., Holmes K. A., Ross-Innes C. S., Schmidt D., Carroll J. S., FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ijichi N., et al., Association of double-positive FOXA1 and FOXP1 immunoreactivities with favorable prognosis of tamoxifen-treated breast cancer patients. Horm. Cancer 3, 147–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang L., et al., Involvement of estrogen receptor variant ER-alpha36, not GPR30, in nongenomic estrogen signaling. Mol. Endocrinol. 24, 709–721 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ignatov A., Ignatov T., Roessner A., Costa S. D., Kalinski T., Role of GPR30 in the mechanisms of tamoxifen resistance in breast cancer MCF-7 cells. Breast Cancer Res. Treat. 123, 87–96 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Azuma K., et al., Association of estrogen receptor alpha and histone deacetylase 6 causes rapid deacetylation of tubulin in breast cancer cells. Cancer Res. 69, 2935–2940 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Hoskins J. M., Carey L. A., McLeod H. L., CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat. Rev. Cancer 9, 576–586 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Knowlden J. M., Hutcheson I. R., Barrow D., Gee J. M., Nicholson R. I., Insulin-like growth factor-I receptor signaling in tamoxifen-resistant breast cancer: A supporting role to the epidermal growth factor receptor. Endocrinology 146, 4609–4618 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Hurtado A., et al., Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature 456, 663–666 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., et al., IGF1R signaling drives antiestrogen resistance through PAK2/PIX activation in luminal breast cancer. Oncogene 37, 1869–1884 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y., et al., Activation of nuclear factor-kappaB (NFkappaB) identifies a high-risk subset of hormone-dependent breast cancers. Int. J. Biochem. Cell Biol. 37, 1130–1144 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Hicks C., Kumar R., Pannuti A., Miele L., Integrative analysis of response to tamoxifen treatment in ER-positive breast cancer using GWAS information and transcription profiling. Breast Cancer (Auckl.) 6, 47–66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urano T., et al., Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature 417, 871–875 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T., et al., Estrogen-responsive finger protein as a new potential biomarker for breast cancer. Clin. Cancer Res. 11, 6148–6154 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Kawabata H., et al., TRIM44 is a poor prognostic factor for breast cancer patients as a modulator of NF-κB signaling. Int. J. Mol. Sci. 18, E1931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato J., et al., Combined A20 and tripartite motif-containing 44 as poor prognostic factors for breast cancer patients of the Japanese population. Pathol. Int. 71, 60–69 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Sato J., et al., Combined use of immunoreactivities of RIG-I with Efp/TRIM25 for predicting prognosis of patients with estrogen receptor-positive breast cancer. Clin. Breast Cancer, 10.1016/j.clbc.2020.12.001 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Reymond A., et al., The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda K., Inoue S., TRIM proteins as RING finger E3 ubiquitin ligases. Adv. Exp. Med. Biol. 770, 27–37 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Napolitano L. M., Meroni G., TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life 64, 64–71 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Horie-Inoue K., TRIM proteins as trim tabs for the homoeostasis. J. Biochem. 154, 309–312 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Nisole S., Stoye J. P., Saïb A., TRIM family proteins: Retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3, 799–808 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Wolf D., Goff S. P., TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57 (2007). [DOI] [PubMed] [Google Scholar]
- 27.McNab F. W., Rajsbaum R., Stoye J. P., O’Garra A., Tripartite-motif proteins and innate immune regulation. Curr. Opin. Immunol. 23, 46–56 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Hatakeyama S., TRIM proteins and cancer. Nat. Rev. Cancer 11, 792–804 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Cambiaghi V., et al., TRIM proteins in cancer. Adv. Exp. Med. Biol. 770, 77–91 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Fletcher A. J., Towers G. J., Inhibition of retroviral replication by members of the TRIM protein family. Curr. Top. Microbiol. Immunol. 371, 29–66 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Ozato K., Shin D. M., Chang T. H., H. C. Morse, 3rd, TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8, 849–860 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versteeg G. A., et al., The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38, 384–398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandeputte D. A., et al., GOA, a novel gene encoding a ring finger B-box coiled-coil protein, is overexpressed in astrocytoma. Biochem. Biophys. Res. Commun. 286, 574–579 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Fujimura T., et al., Increased expression of tripartite motif (TRIM) 47 is a negative prognostic predictor in human prostate cancer. Clin. Genitourin. Cancer 14, 298–303 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Han Y., Tian H., Chen P., Lin Q., TRIM47 overexpression is a poor prognostic factor and contributes to carcinogenesis in non-small cell lung carcinoma. Oncotarget 8, 22730–22740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang Q., et al., TRIM47 is up-regulated in colorectal cancer, promoting ubiquitination and degradation of SMAD4. J. Exp. Clin. Cancer Res. 38, 159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhodes D. R., et al., ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Györffy B., et al., An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Chen Z., et al., Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev. 9, 1586–1597 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Wang C., et al., TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Fang H., Jiang W., Jing Z., Mu X., Xiong Z., miR-937 regulates the proliferation and apoptosis via targeting APAF1 in breast cancer. OncoTargets Ther. 12, 5687–5699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang H., et al., Mir-452-3p: A potential tumor promoter that targets the CPEB3/EGFR axis in human hepatocellular carcinoma. Technol. Cancer Res. Treat. 16, 1136–1149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garg R., et al., Activation of nuclear factor κB (NF-κB) in prostate cancer is mediated by protein kinase C epsilon (PKCepsilon). J. Biol. Chem. 287, 37570–37582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W., et al., EGFR-induced and PKCε monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell 48, 771–784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Körner C., et al., MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J. Biol. Chem. 288, 8750–8761 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou Z., et al., PKD2 and PKD3 promote prostate cancer cell invasion by modulating NF-κB- and HDAC1-mediated expression and activation of uPA. J. Cell Sci. 125, 4800–4811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J., Deng F., Singh S. V., Wang Q. J., Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCepsilon/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 68, 3844–3853 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Yde C. W., Emdal K. B., Guerra B., Lykkesfeldt A. E., NFκB signaling is important for growth of antiestrogen resistant breast cancer cells. Breast Cancer Res. Treat. 135, 67–78 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Kubo M., et al., Inhibition of the proliferation of acquired aromatase inhibitor-resistant breast cancer cells by histone deacetylase inhibitor LBH589 (panobinostat). Breast Cancer Res. Treat. 137, 93–107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frasor J., et al., Positive cross-talk between estrogen receptor and NF-kappaB in breast cancer. Cancer Res. 69, 8918–8925 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franco H. L., Nagari A., Kraus W. L., TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol. Cell 58, 21–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y. S., Kan M. C., Lin C. L., Richter J. D., CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 25, 4865–4876 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tonetti D. A., Morrow M., Kidwai N., Gupta A., Badve S., Elevated protein kinase C alpha expression may be predictive of tamoxifen treatment failure. Br. J. Cancer 88, 1400–1402 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Assender J. W., et al., Protein kinase C isoform expression as a predictor of disease outcome on endocrine therapy in breast cancer. J. Clin. Pathol. 60, 1216–1221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan Q., et al., Protein kinase C epsilon is a predictive biomarker of aggressive breast cancer and a validated target for RNA interference anticancer therapy. Cancer Res. 65, 8366–8371 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Aziz M. H., et al., Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Cancer Res. 67, 8828–8838 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Wu X., et al., Regulation of TRIF-mediated innate immune response by K27-linked polyubiquitination and deubiquitination. Nat. Commun. 10, 4115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lei C. Q., et al., USP19 Inhibits TNF-α- and IL-1β-Triggered NF-κB Activation by Deubiquitinating TAK1. J. Immunol. 203, 259–268 (2019). [DOI] [PubMed] [Google Scholar]
- 59.van Tilburg G. B. A., et al., K27-linked diubiquitin inhibits UCHL3 via an unusual kinetic trap. Cell Chem. Biol. 28, 191–201.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Li Q., et al., Tripartite motif 8 (TRIM8) modulates TNFα- and IL-1β-triggered NF-κB activation by targeting TAK1 for K63-linked polyubiquitination. Proc. Natl. Acad. Sci. U.S.A. 108, 19341–19346 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan Y., et al., Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor alpha- and interleukin-1beta-induced IKK/NF-kappaB and JNK/AP-1 activation. J. Biol. Chem. 285, 5347–5360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mandell M. A., Saha B., Thompson T. A., The tripartite nexus: Autophagy, cancer, and tripartite motif-containing protein family members. Front. Pharmacol. 11, 308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., Liu C., Xie Z., Lu H., Knockdown of TRIM47 inhibits breast cancer tumorigenesis and progression through the inactivation of PI3K/Akt pathway. Chem. Biol. Interact. 317, 108960 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Ozes O. N., et al., NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature 401, 82–85 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Ji Y. X., et al., The deubiquitinating enzyme cylindromatosis mitigates nonalcoholic steatohepatitis. Nat. Med. 24, 213–223 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Hayashi M., et al., Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res. Treat. 143, 447–457 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Trompouki E., et al., CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424, 793–796 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Massoumi R., Chmielarska K., Hennecke K., Pfeifer A., Fässler R., Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 125, 665–677 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Kim J. A., et al., Comprehensive functional analysis of the tousled-like kinase 2 frequently amplified in aggressive luminal breast cancers. Nat. Commun. 7, 12991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belguise K., et al., The PKCθ pathway participates in the aberrant accumulation of Fra-1 protein in invasive ER-negative breast cancer cells. Oncogene 31, 4889–4897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamada S., et al., Identification of twinfilin-2 as a factor involved in neurite outgrowth by RNAi-based screen. Biochem. Biophys. Res. Commun. 363, 926–930 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus Datasets (GSE174612; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174612). All other study data are included in the article and/or supporting information.