Abstract
Although the Drosophila melanogaster erect wing (ewg) gene is broadly transcribed in adults, an unusual posttranscriptional regulation involving alternative and inefficient splicing generates a 116-kDa EWG protein in neurons, while protein expression elsewhere or of other isoforms is below detection at this stage. This posttranscriptional control is important, as broad expression of EWG can be lethal. In this paper, we show that ELAV, a neuron-specific RNA binding protein, is necessary to regulate EWG protein expression in ELAV-null eye imaginal disc clones and that ELAV is sufficient for EWG expression in wing disc imaginal tissue after ectopic expression. Further, analysis of EWG expression elicited from intron-containing genomic transgenes and cDNA minitransgenes in ELAV-deficient eye discs shows that this regulation is dependent on the presence of ewg introns. Analyses of the ewg splicing patterns in wild-type and ELAV-deficient eye imaginal discs and in wild-type and ectopic ELAV-expressing wing imaginal discs, show that certain neuronal splice isoforms correspond to ELAV levels. The data presented in this paper are consistent with a mechanism in which ELAV increases the splicing efficiency of ewg transcripts in alternatively spliced regions rather than with a mechanism in which stability of specific splice forms is enhanced by ELAV. Additionally, we report that ELAV promotes a neuron-enriched splice isoform of Drosophila armadillo transcript. ELAV, however, is not involved in all neuron-enriched splice events.
The Drosophila melanogaster erect wing gene (ewg) provides a function that is vital in the nervous system and essential for the development of specific indirect flight muscles (13, 15, 19). A single 116-kDa protein is sufficient for both neuron- and muscle-associated functions (15, 46). Recent data indicates that, although the functional 116-kDa protein is highly enriched in neurons, the gene is transcribed comparably in both neuron-rich and neuron-poor tissues and that neuron-enriched expression of the functional protein is achieved by posttranscriptional regulation of ewg, which includes both alternative and inefficient splicing (27). Alternative splicing in two regions leads to enrichment in heads of the transcript with an open reading frame (ORF) that encodes the 116-kDa protein containing an unusual DNA binding domain. Further, misregulation is biologically consequential, as global expression of the 116-kDa protein can be lethal (27).
The Drosophila embryonic lethal abnormal visual system (elav) gene encodes a neuron-specific RNA binding protein and is expressed in all neurons (44). Since Drosophila ELAV has been shown to be important for the formation of the neuron-specific protein isoform of Neuroglian (26), we investigated if ELAV also has a role in ewg regulation. ELAV-like proteins are evolutionarily conserved, as several genes encoding proteins with three RNA recognition motifs with high homology to ELAV in the RNA recognition motifs have been identified in both vertebrates and invertebrates (reviewed in reference 2). Data on several proteins of the ELAV family, from mammals and Drosophila, suggest that these proteins bind the 3′ untranslated region (UTR) and regulate mRNA stability (11, 12, 17, 29, 41, 42) and translatability (3, 21). Recently, an autoregulatory function, whereby ELAV regulates its own expression, requiring 3′ UTR sequences has also been demonstrated (47).
In this paper, we report that, indeed, in photoreceptor neurons, the generation of the 116-kDa EWG protein is dependent on ELAV and this dependence is contingent on the presence of ewg introns. Next, using reverse transcription-PCR (RT-PCR), we show that the ewg splicing profile is altered in ELAV-deficient photoreceptors such that transcripts representing splice choices that lead to the 116-kDa ORF are reduced. We also show that ectopic expression of ELAV in nonneural tissue is sufficient both to increase RNAs with neuron-like splicing choices and for the expression of the 116-kDa protein. These data further substantiate an in vivo role of ELAV in promoting neuron-specific splice isoforms. Further, we show that alternative splicing of armadillo (arm), another ubiquitously expressed gene with a neuron-specific isoform (31), is also regulated by ELAV. Finally, we report our initial findings that not all genes with neuron-specific alternatively spliced transcripts are regulated by ELAV.
MATERIALS AND METHODS
Fly stocks and genetic crosses.
Flies were raised at 25°C. Standard genetic marker genes and Balancer chromosomes were as described in reference 30. Canton-S was used as the wild-type strain. The alleles and transgenes used in this study are as follows (abbreviated names are given, followed by full descriptions): ewgl1, an ewg lethal allele which does not allow expression of the 116-kDa EWG protein (27); ewgR19-1 and ewgR19-2, two independent inserts of ewg genomic transgene P{ry+=ewg+} that provide full rescue of ewgl1 (18); elav-EWG, a transgene, P{w+=elav-EWG} (in which ewg cDNA [SC3 ORF] is fused to the elav promoter [53]), which provides full rescue of ewgl1 and is referred to in reference 13 as EWGNS; elav-ewg, a transgene, P{w+=elav-ewg}28 (in which ewg genomic transcribed sequences are fused to the neuron-specific elav promoter [53]), which provides full rescue of ewgl1 (25); elavedr, a specific insert of the transgene P{w+=elav+} which has a wild-type elav genomic rescue fragment (ELAV expression of P{w+=elav+}edr insert is specifically reduced in photoreceptor neurons, but expression in brain neurons is less affected due to the transgene insertion site [25, 26]) and whose expression phenotype is revealed in combination with elave5, an elav null allele; edr, the genotype elave5; elavedr; UAS-ELAV2e2 and UAS-ELAV3e1, two inserts of transgene P{w+=UAS-ELAV} on chromosomes 2 and 3 that express elav cDNA under UAS transcriptional control (26); P{w+=elav)DmORF2, a transgene expressing ELAV under the control of the elav promoter (54); and c309, a P{GAL4}c309 enhancer trap line with the transgene insertion on the second chromosome (34).
EWG expression in photoreceptors under ELAV-deficient conditions or in an ewg-null background was analyzed in eye discs dissected from male larvae carrying an ewg transgene in the edr or the protein-null ewgl1 background. To generate males of the appropriate genotype, elave5; elavedr or ewgl1y w sn/FM6 l females were crossed to males of one of the following genotypes: (i) ewgl1y w sn/Y; elav-EWG/elav-EWG, (ii) ewgl1y w sn/Y; ewgR19-1/ewgR19-1, or (iii) w/Y; elav-ewg/TM6B, Tb. Eye imaginal discs from male larvae were dissected and immunostained.
For ectopic ELAV expression in the wing imaginal discs, UAS-ELAV2e2 or UAS-ELAV2e2; UAS-ELAV3e1 virgin females were crossed to c309 males. For monitoring the effect of ectopically expressed ELAV in wing discs on EWG expression, ewgl1y w sn; ewgR19-1 UAS-ELAV3e1 females were crossed to w/Y; c309 males and wing imaginal discs from third-instar male progeny were double labeled for ELAV and EWG. As controls, ewgl1 y w sn; elav-EWG UAS-ELAV3e1 virgin females were crossed to w/Y; c309 males; in the control male larvae, there is no wing disc transcription from the ewg-ELAV neuron-specific minitransgene.
Somatic clones were generated using the method described in reference 27 by crossing virgin elave5; P{w+=elav)DmORF2/TM3 virgin females to yw/Y; KipP(Δ2-3 ry+) males, a source of transposase (43). Due to transposase activity of Δ2-3, the P element containing the elav rescue construct is excised in cells yielding somatic clones of ELAV-deficient tissue. Clones were detected by double staining eye imaginal discs of third-instar larvae for both ELAV and EWG. Clones were viewed in an MRC-600 confocal microscope.
Immunohistochemistry.
Imaginal discs were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 40 min and washed several times in PBS containing 0.1% bovine serum albumin and 0.3% Triton X-100. Antibody incubations were done overnight at 4°C. Anti-EWG rabbit serum (15) was used at a 1:300 or 1:400 dilution, and anti-ELAV monoclonal antibody 9F (16) was used at a 1:20 dilution. Anti-APPL rabbit serum (33) was used at a 1:200 dilution. Secondary antibodies were from Jackson Immunoresearch Laboratories Inc. (West Grove, Pa.) and were used at a dilution of 1:50 or 1:100. Photography was done using a Zeiss Axiophot fluorescence microscope and Tmax-400 film.
RT-PCR assays.
Total RNA was isolated with Trizol reagent (Life Technologies, Gaithersburg, Md.) in accordance with the manufacturer's instructions. For precipitation of the RNA, 10 to 20 μg of glycogen (Boehringer Mannheim, Indianapolis, Ind.) was used. Sixty eye or 60 wing imaginal discs from wandering third-instar larvae were dissected, collected in Trizol reagent, and used for oligo(dT)-primed RT after treatment with RNase-free DNase I (Life Technologies). RT was done with the Superscript II cDNA synthesis kit (Life Technologies) according to the manufacturer's instructions, except that the RNA was kept at 50°C for 5 min before the RT reaction was started. After RT the cDNA was treated with RNase H. Control experiments were carried out with no reverse transcriptase. PCR was performed using the ewg and rp49 primers described in reference 27. An additional ewg primer was FV, 5′-GCTTGTCCTCATTTTATATTGAG-3′. From 60 discs 1/20 of the disc tissue was used for PCR, yielding very similar amounts of PCR product from different genotypes for the same amount of disc tissue. Semiquantitative PCR was done for 30 cycles, and PCR products were visualized on agarose gels. The sizes of the expected products for each primer pair used are catalogued in Table 1. In addition to the size, the identity of PCR products was further verified by restriction digests or direct sequencing with ABI automated sequencing equipment. Quantitative amplification was done in the presence of 5 μCi of [α-32P]dCTP, and PCR products were separated on nondenaturing 5% polyacrylamide gels. Quantitation was done on a Molecular Dynamics PhosphorImager using Imagequant at cycles 22 and 24, which was in the linear range for all primer pairs. The Appl primers were 5′-CGCCTCGCCGCGATGGGAGC-3′ and 5′-GATCCCTTGCACTTAGCCGCAT-3′. The elav primers were 5′-GTTCGCTTTGTTTGTCCAGCC-3′ and 5′-TGTTGCCGCCACTGCTGCGGC-3′. The arm primers were 5′-GCAGGATTACAAGAAGCGGCT-3′ and 5′-CTCCAGACCCTGCATCGAATC-3′. The nrg primer was 5′-CGGAAAGTACGATGTCCACG-3′, with return primers being 5′-TAAATCAAAGTCCTTTGCGTCC-3′ and 5′-TGATGCGCCGCAGCGGAATTGT-3′. The creb primers were 5′-AAGATCTTCACCGAGATCAGCG-3′ and 5′-GCGCTTACGGGTCTGATCCT-3′, spanning alternatively spliced exon 6 as described in reference 55. The pres primers were as described in reference 35. The fra primers were 5′-TGGTGGATGGTCACAATCTGAA-3′ and 5′-CTTCACTATCTGCGCCACCAGA-3′. The PTP4E primers were 5′-CATCCTGCAGCACATTCACAAG-3′ and 5′-AGATTCCCGTCTCAGCTATGCC-3′. The PTP10D primer was 5′-ATATCTGCATCCACCAGTGCCT-3′, with the return primers being 5′-AGATGGTTGGGTTCTGTTGGGT-3′ and 5′-GCTGGTCATCATGGAGTATCGC-3′. The PTP99A primers were 5′-GAATTCGAGGATGTGACAACGG-3′ and 5′-ACAGGGCAAACGAATTGTTGAA-3′.
TABLE 1.
Summary of RT-PCR products
| PCR primer paira | Product(s)c | Band size (bp)b |
|---|---|---|
| 4F-5R | Ex 4, Ex 5 | 202 |
| 3aF-3aR | In 3a | 449 |
| In 3a, spliced | 150 | |
| 3cF-3cR | In 3a, Ex E, In 3b/c-2 | 961 |
| In 3a, Ex E | 773 | |
| Ex E | 228 | |
| In 3c, spliced | 154 | |
| 3aF-3cR | In 3a, Ex D, Ex E | 1014 |
| In 3a, Ex D | 936 | |
| Ex D, Ex E | 711 | |
| Ex D | 637 | |
| Ex E | 243 | |
| In 3b spliced | 169 | |
| 6F-6R | In 6a, Ex I | 316 |
| In 6, spliced | 278 | |
| 6F-RV | 5′ In 6 | 434 |
| FV-6R | 3′ In 6a, Ex I, In 6b | 356 |
| 2F-2R | Not shown | 177 |
| rp49F-rp49R | Not shown | 318 |
For primer sequences, see reference and Materials and Methods.
Underlining indicates band expected from 116-kDa ORF transcript.
Figure 2B and C shows splicing profiles. In, intron; Ex, exon.
To determine the 3′ end of ewg transcripts, rapid amplification of cDNA 3′ ends (3′-RACE) was carried out with the Life Technologies 3′-RACE kit according to the manufacturer's instructions, with primer 5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′ being used for RT. Subsequent PCR was done with nested primers 5′-ATATCCCGTTTCGGTGAGCAAT-3′ (6F) and 5′-TGGTCCAGGTGGTCAGCTTAAA-3′ (6F2) and return primer 5′-GGCCACGCGTCGACTAGTAC-3′.
Plasmid construction.
The EcoRI-PstI fragment of genomic ewg was cloned into pCaSpeR 4 (14). The elav promoter (53) was digested with NotI and EcoRI and inserted via a NotI-EcoRI adapter oligonucleotide into pBluescript (Stratagene) and subsequently cloned into pCaSpeR 4 containing the genomic ewg fragment as an EcoRI fragment, and Drosophila embryos were transformed by standard techniques as described in reference 54, but Δ2-3 was used as the transposase source (43).
RESULTS
Figures 1A and 2A summarize the aspects of ewg exon and intron structure and splicing patterns relevant to this study and described previously (27). The primary transcript of ewg, which has 10 exons, A to J (exon A is not shown in Fig. 1), is alternatively spliced in two regions (Fig. 2A). We have previously shown that neuron-enriched heads and neuron-poor bodies have different ewg RNA splicing profiles. Heads show enrichment for a transcript encoding a 116-kDa protein, whereas bodies have lower amounts of the transcript that encodes the 116-kDa protein and greater amounts of unprocessed RNA. The head-enriched transcript encoding the 116-kDa protein results from inclusion of exon D and exclusion of exons E and I, which is depicted in Fig. 1 (SC3 ORF). Additionally, splicing of introns 3a, 3c, and 6 is inefficient, as these introns are retained in polyadenylated ewg RNA (27). Despite similar abundance of other splice isoforms, only one protein of 116 kDa could be detected (27).
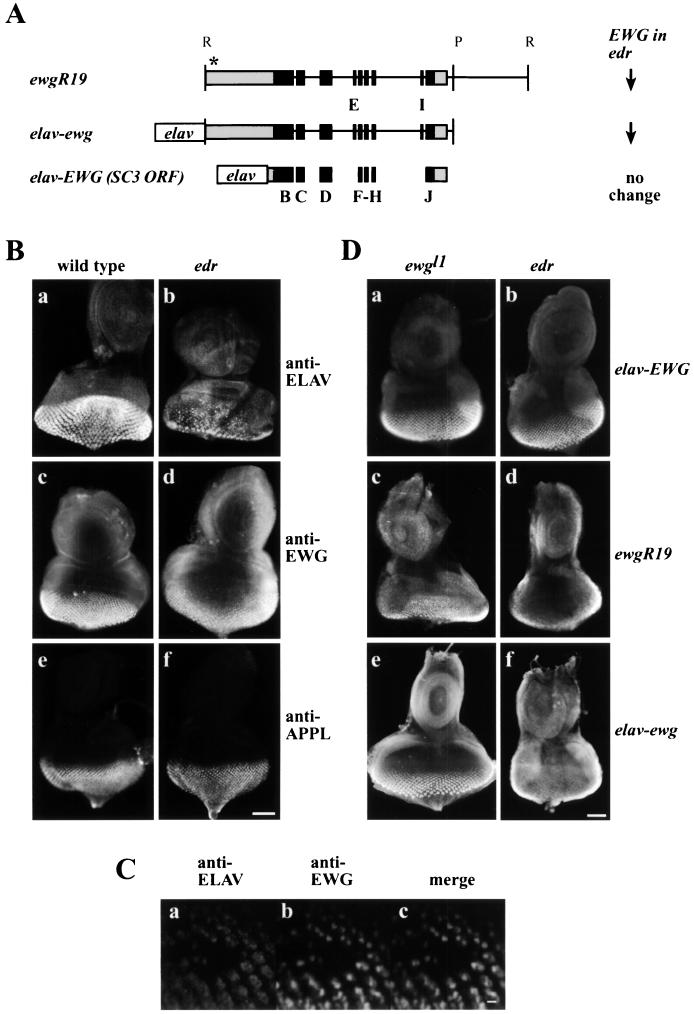
FIG. 1.
(A) Maps of ewg gene and ewg transgenes. ewgR19 is a genomic EcoRI fragment that includes ewg SC3 cDNA. elav-ewg is a transgene in which a genomic EcoRI-PstI fragment is driven by the neuron-specific elav promoter. elav-EWG is a minitransgene in which the elav promoter drives the SC3 cDNA. The black boxes show exons, and the grey boxes indicate 5′ and 3′ UTR sequences present in the SC3 cDNA. All constructs provide full rescue of ewgl1, a protein-null allele. The transcriptional start site of ewgR19 is not known, but its general location is indicated with an asterisk. Exons E and I are not present in the SC3 cDNA. Retention of exons E and I prematurely terminates the 116-kDa ORF. On the right, levels of EWG expression in the third-instar ELAV-depleted edr eye imaginal discs shown in panel D are summarized for the different transgenes. Note that only the genomic fragments show a reduced expression. R and P denote restriction sites EcoRI and PstI. (B) Expression of EWG protein in ELAV-deficient eye discs. Third-instar larval eye discs of the wild type (Ba, c, and e) or an edr mutant (Bb, d, and f) were stained with either anti-ELAV (Ba and b), anti-EWG (Bc and d), or anti-APPL (Be and f) antibodies. Note that both ELAV and EWG levels are reduced compared to that of the control APPL antigen. Bar, 50 μm. (C) ELAV-null clone double labeled for ELAV and EWG. An eye disc of the genotype elave5/Y; elavDMORF2/Ki pp(Δ2-3ry+) depicting an ELAV-null patch simultaneously immunoreacted with anti-ELAV (Ca) and anti-EWG antibodies (Cb) and a merged image (Cc). Note that the ELAV and EWG signals overlap and that the EWG-null patch coincides with the ELAV-null patch. Bar, 5 μm. (D) EWG expression elicited from ewg transgenes in wild-type and ELAV-deficient eye discs. Different ewg transgenes were crossed into the EWG-null (ewgl1) and ELAV-deficient (edr) genetic backgrounds to assess EWG levels by immunohistochemistry. Eye discs of genotype ewgl1/Y; ewg transgene (Da, c, and e) and edr/Y; ewg transgene (Db, d, and f) were immunoprocessed; the transgene designation in each case is noted on the right edge. Note that in ewgl1, expression is only from the transgene, while in edr, EWG levels reflect transgene expression superimposed on the endogenous (reduced) EWG expression. (Da and b) elav-EWG transgene expression. Note the similar level of EWG immunoreactivity in both genotypes. (Dc and d) ewgR19 transgene expression in ewgl1 and edr. Note that the expression of EWG evident in panel Dd is reduced compared to that evident in panels Da, b, and c. (De and f) elav-ewg transgene expression in ewgl1 and elavedr. Note that the expression evident in panel Df is reduced compared to that evident in panels Da, b, c, and e. Bar, 50 μm.
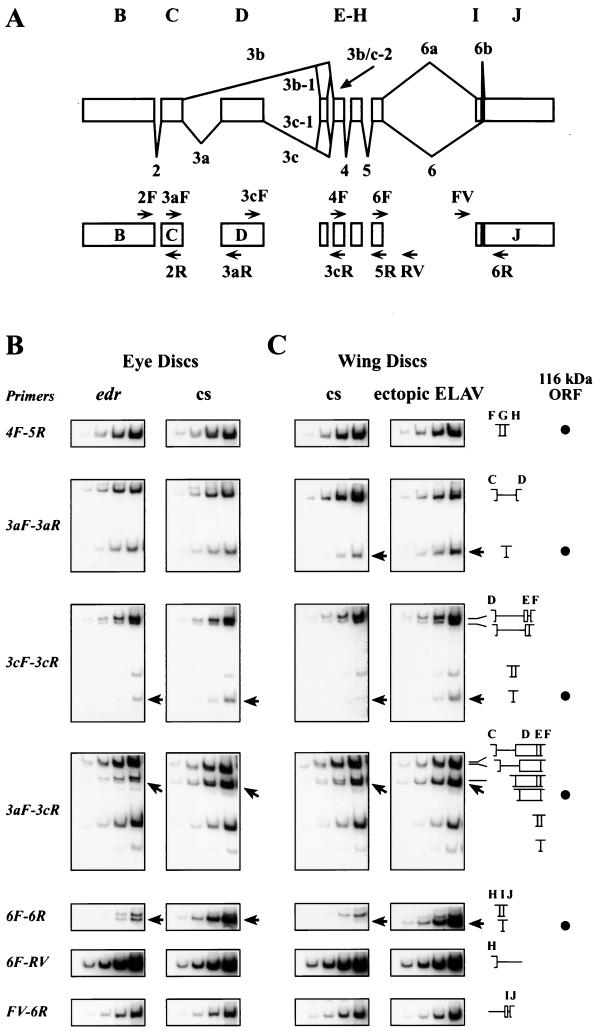
FIG. 2.
(A) Alternative splicing of ewg transcripts and PCR primer map. The top diagram shows ewg introns and exons except exon A. The splicing pattern for the neurally enriched SC3 ORF is drawn below the exons and the predominantly nonneuronal splicing pattern is shown above. The lower diagram depicts the positions of the primers used for RT-PCR. The primer sequences are as described in reference 27. (B and C) Splicing profile of ewg mRNA in wild-type and ELAV-deficient eye discs (B) and wild-type wing discs and wing discs ectopically expressing ELAV (C). Quantitative RT-PCR assays were carried out using DNase I-treated total RNA isolated from eye discs of third-instar wandering larvae. The primer sets used for the four reactions shown in each row of images are listed in the leftmost column. The rightmost columns depicts diagrams of the amplified fragments, and the black dots indicate the fragments expected of 116-kDa ORF-encoding transcripts. Each rectangle shows the results of four successive PCR cycles in the presence of [α-32P]dCTP (cycles 18, 20, 22, and 24 for primer pairs 3cF-3cR and 3aF-3cR and cycles 20, 22, 24, and 26 for primer pairs 4F-5R, 3aF-3aR, 6F-6R, 6F-RV, and FV-6R). Sizes of bands are as listed in Table 1. As shown in panel B, ewg splicing was assessed in wild-type (cs) and edr eye discs. Note the threefold reduction of intron 3c splicing in 3cF-3cR (marked with an arrow) and in 3aF-3cR reactions (marked with an arrow) in edr. Additionally, intron 6 splicing efficiency decreases more than 10-fold in edr as revealed with primer pair 6F-6R (marked with an arrow). As shown in panel C, ewg splicing was assessed in wild-type (cs) and c309/UAS-ELAV2e2; UAS-ELAV3e1 (ectopic ELAV) wing discs (expression pattern shown in Fig. 3). ewg mRNA, but no protein, is expressed in wild-type wing imaginal discs (14). Note the two- to threefold increase of intron 3a splicing in the 3aF-3aR reaction (marked with an arrow) and the 10-fold increase of intron 3c splicing in 3cF-3cR (marked with an arrow) and 3aF-3cR reactions (marked with an arrow) upon ectopic ELAV expression. Additionally, intron 6 splicing efficiency increases by orders of magnitude after ectopic expression of ELAV, as revealed with primer pair 6F-6R (marked with an arrow).
In this analysis, we consider splicing of introns 2 to 6, which encompass the entire coding region, with a special focus on the alternative splicing that leads to the 116-kDa ORF. From previous studies, we know that due to alternative splicing, several conceptual isoforms are encoded by the ewg locus (27). All putative EWG isoforms share exons B and C and thus should be recognized by the anti-EWG antiserum. Inclusion of exon E or I, as well as retention of introns 3a, 3c, and 6, changes the 116-kDa ORF. Since our analysis failed to detect isoforms other than the 116-kDa protein, we concluded that other isoforms are either minor, unstable, or not made (27). These observations together with the ability of the 116-kDa protein to restore all three characterized phenotypes associated with ewg mutations, viability, erect wings, and formation of the flight muscles (15, 27), suggest that the other isoforms are unlikely to be functionally significant. However, since generalized increased expression of the 116-kDa protein is lethal, the splicing regulation of ewg is important.
In these studies, as a source for neuron-enriched RNA we used third-instar larval eye-antenna imaginal discs. In the eye discs, the differentiating field of photoreceptors shows robust ELAV expression (Fig. 1Ba); however, it does contain many cell types in addition to the photoreceptor neurons (51). As a source of nonneural tissue we used wing imaginal discs, which at this stage of development have not yet differentiated neurons and therefore are devoid of any ELAV protein (see below).
Presence of ewg introns confers ELAV regulation of EWG expression.
To analyze whether ELAV has a role in neuron-enriched expression of the 116-kDa EWG protein, we used a synthetic elav mutant genotype, edr, which has dramatically reduced ELAV expression in developing photoreceptors, though expression in central nervous system neurons is only slightly affected (26). Members of our group had previously noted (26) that photoreceptors with elavedr as their exclusive source of ELAV have a low level of EWG compared to wild-type photoreceptors, while expression of many other proteins remains unaffected (Fig. 1B). To further test that ELAV indeed regulates EWG expression, we generated ELAV-null somatic clones in the eye discs as described in Materials and Methods. Figure 1C shows a somatic clone in a disc simultaneously immunoreacted for ELAV and EWG. The merged image reveals that EWG and ELAV immunoreactivity is coincidentally absent from the same cells. Thus, EWG expression in photoreceptors is dependent on ELAV expression and the effect of ELAV is cell autonomous.
The ELAV-dependent regulation of EWG expression in photoreceptors could result from either transcriptional or posttranscriptional mechanisms, for example, regulation of alternative splicing or mRNA stability. To distinguish between these mechanisms, ewg transgenes that contain either the genomic transcribed region or the minigenes that express the SC3 cDNA under the control of a heterologous promoter were used. SC3 cDNA has a 1,962-nucleotide-long (1,962-nt-long) 5′ UTR and a 578-nt-long 3′ UTR (15). 3′-RACE experiments confirmed that the 3′ end of the SC3 cDNA coincides with the 3′ end of the transcript (data not shown). Minitransgene elav-EWG expresses the SC3 cDNA with only 27 bp of the 5′ UTR fused to the neuron-specific elav promoter (Fig. 1A) and terminates 10 nt after a polyadenylation signal (AAUAAA) (13). ewgR19 is a genomic transgene starting 1,852 nt 5′ to exon B, including exons B to J and introns 2 to 6 (Fig. 1A) (18). elav-ewg is a genomic transgene encompassing exons B to J and introns 2 to 6 (EcoRI-PstI fragment from the ewg gene fused to the elav promoter [Fig. 1A]) (53). Also, in constructing elav-EWG and elav-ewg, no additional 3′ UTR sequences were added to avoid masking the regulation of ewg expression. All transgenes provide full rescue of the lethal phenotype of ewgl1, a null allele (13, 27). EWG immunoreactivity for these three transgenes was assessed in ewgl1 and edr male eye discs (Fig. 1D). In edr, EWG signal results from summation of both the endogenous ewg+ gene and the transgene, while in the ewgl1 background only the transgene expression is observed, as ewgl1 is a protein-null allele (27).
In edr, the endogenous EWG signal is reduced compared to wild-type expression (Fig. 1Bc and d), but the signal from elav-EWG is robust and similar to that in the ewgl1 genetic background (Fig. 1Da and b). Therefore, ELAV does not regulate elav-EWG expression. In contrast, the expression of both ewgR19 and elav-ewg depends on ELAV, as indicated by the reduced EWG signal from these transgenes in edr compared to that in the ewgl1 genetic background (Fig. 1Dc and f). These results show that ELAV-dependent EWG expression in photoreceptors is observed when transgenes contain introns 2 to 6, even when the transcription is driven by a heterologous promoter, as in the case of elav-ewg. The requirement for introns makes splicing an attractive step for ELAV control, especially in the light of the known alternative splicing of ewg transcripts (27). Also, since elav-ewg and elav-EWG have the same 3′ UTR, but only elav-ewg responds to reduced ELAV levels, 3′ UTR-mediated regulation by affecting stability is unlikely.
Analysis of splice forms indicative of 116-kDa ORF of ewg.
Generation of the neuron-enriched 116-kDa ORF, which requires inclusion of exon D and exclusion of exons E and I, is achieved by excision of introns 3a, 3c, and 6 (27). Previous analysis had revealed that neuron-rich head RNA has higher concentrations of splice forms indicative of alternatively spliced introns 3a, 3c, and 6, while splicing of introns 2, 3b, 4, and 5 was similar in head and body RNAs. To determine if ELAV affects ewg splicing patterns, the concentration of different spliced isoforms of ewg in wild-type and ELAV-deficient third-instar edr eye discs was assessed by quantitative RT-PCR using nine primer pairs. The number and size of the expected bands for each primer pair are catalogued in Table 1, and the experimental results are shown in Fig. 2B.
Using primers that flank intron 2 and introns 4 and 5, we confirmed that these introns were efficiently spliced in the wild-type and edr eye discs and that the amplified bands indicated equal concentrations of cDNA from the edr and wild-type eye disc samples (Fig. 2B, primers 4F-5R). Further confirmation that the two RNA samples had similar concentrations came from the control rp49 primers, which, as expected, yielded comparable bands (data not shown) (28).
To analyze splicing within the 3b region, three sets of primers were used, as follows: 3aF-3aR (flanking intron 3a), 3aF-3cR (flanking the entire region), and 3cF-3cR (flanking intron 3c). In edr, the PCR products indicative of 3a-spliced and unspliced transcripts remain comparable to wild-type eye discs (Fig. 2B, primers 3aF-3aR). The splicing in the 3c region is complicated by the presence of exon E, which is excluded from the 116-kDa ORF. Primers 3cF-3cR and 3aF-3cR yield four and six products respectively, as described in Table 1 and depicted in Fig. 2B, which shows the band of the product expected from the 116-kDa ORF and another band that includes exon E. A modest effect of ELAV depletion is discerned on intron 3c, as in edr eye disc RNA, transcripts with only exon D included (Fig. 2B, primers 3cF-3cR and 3aF-3cR) show a reduction of about threefold, while those that contain both D and E show no change when compared to the wild-type eye disc RNA, as evidenced in 3cF-3cR and 3aF-3cR reactions. These results show that ELAV depletion results in a reduction of transcripts that splice intron 3c, but splicing of introns 3a, 3c-1, and 3b is relatively unaffected. Based on the spliced product/spliced-plus-unspliced product ratio for introns 3a and 3c (about 0.55 for each) and assuming that the two events are independent, we estimate that the overall splicing efficiency which leads to inclusion of exon D is only about 30%.
In the intron 6 region, a dramatic (greater than 10-fold) reduction of intron 6-spliced transcripts (Fig. 2B, primers GF-GR) is seen in edr. To analyze retention of intron 6 we used primer pairs 6F-RV and FV-6R; in each case, one of the primers is within intron 6a. Marginal differences were observed; they were, however, in the range of variability, suggesting that intron 6 is spliced only from a small amount of all transcripts. Therefore, a likely interpretation of these results is that ELAV's presence enhances splicing of intron 6. In summary, ELAV depletion is accompanied by reduction of intron 6 splicing and a more modest reduction of intron 3c splicing.
Ectopic ELAV induces expression of EWG protein in nonneural tissue.
Next we tested if EWG protein expression can be induced when ELAV is ectopically expressed in nonneural wing imaginal discs. In third-instar wing discs ewg RNA is broadly expressed (14), while EWG protein is not detected at this stage (Fig. 3Ab). ELAV was ectopically expressed in a subset of wing disc cells that do not normally express ELAV using the GAL4/UAS system (8) as described previously (26). Wing discs immunoreacted for EWG show the presence of EWG protein when ELAV is expressed ectopically (Fig. 3Ac). To test if EWG levels correlate with the concentration of ELAV, wing discs with one and two doses of UAS-ELAV transgenes were compared. An increase in the concentration of ELAV clearly shows a dose-dependent enhancement of the EWG signal (Fig. 3Ac and d). Thus, ELAV is sufficient to induce EWG expression, in a dose dependent manner, in nonneural wing disc tissue.
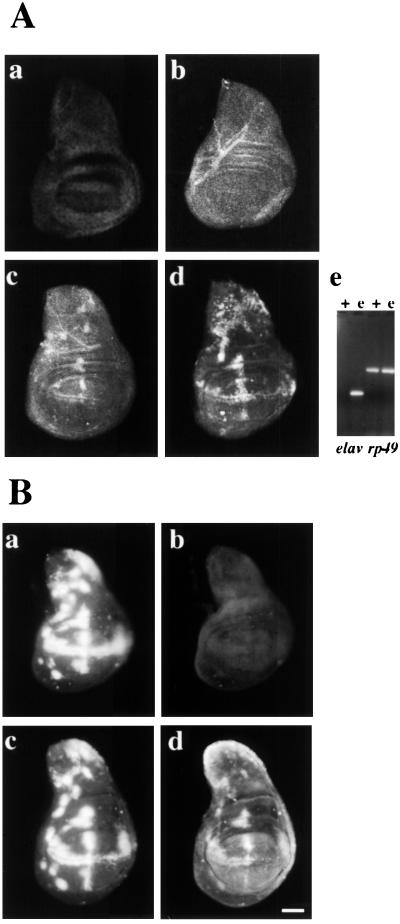
FIG. 3.
(A) Ectopic EWG expression levels correspond to ELAV levels. (Aa to Ad) EWG levels were monitored in wing discs ectopically expressing ELAV. Wing discs were stained with anti-ELAV (Aa) or anti-EWG (Ab to d) antibodies. Note that neither ELAV (Aa) nor EWG (Ab) is expressed at this stage in wild-type wing discs. Wing discs of the genotypes c309/UAS-ELAV2e2 (Ac) and c309/UAS-ELAV2e2; UAS-ELAV3e1 (Ad) were stained with anti-EWG serum. Note that ectopic ELAV expression with two doses of UAS-ELAV further increases EWG levels in nonneural cells. (Ac) Expression of elav transcript in wild-type (+) and ectopically expressing (e) wing discs of the genotype c309/UAS-ELAV2e2; UAS-ELAV3e1 revealed by RT-PCR. Transcripts of ribosomal protein 49 (rp49) were used as a control. (B) Induction of ectopic EWG expression by ectopic ELAV depends on the presence of ewg introns. Wing imaginal discs of the genotypes ewgl1 y w sn/Y; c309; UAS-ELAV3e1 elav-EWG (Ba and b) and ewgl1 y w sn/Y; c309; UAS-ELAV3e1 ewgR19-1 (Bc and d) were double labeled for ELAV (Ba and c) and EWG (Bb and d). Note that ELAV can drive EWG expression from ewg intron-containing transgenes. Bar, 50 μm.
The ability of ectopic ELAV to induce EWG expression was further tested using the genomic transgene ewgR19 (in the ewgl1 background, null for ewg). In wing discs of genotype ewgl1y w sn/Y; c309; ewgR19 UAS-ELAV3e1 ectopic expression of EWG was observed (Fig. 3Bd). No EWG signal was observed in ewgl1y w sn/Y; elav-EWG UAS-ELAV3e1 wing discs (Fig. 3Bb), since elav-EWG is transcribed only in the nervous system (13). These data imply that genomic transgenes spanning ewg exons B to J (Fig. 1B) widely express ewg transcripts similar to endogenous ewg and further, in the presence of ELAV, express EWG protein.
Ectopic ELAV induces expression of 116-kDa ORF-like splice profile in nonneuronal tissue.
To address whether the generation of EWG protein is accompanied by changes in the splicing profiles, polyadenylated RNA from wild-type and ectopic ELAV-expressing (c309/UAS-ELAV2e2; UAS-ELAV3e1) wing discs was analyzed using quantitative RT-PCR. As expected, elav transcript levels in the wild-type wing discs were below the level of detection, but a robust signal was seen in ectopically expressing wing discs (Fig. 3Ae). Wild-type and ELAV-expressing wing disc profiles do not differ in the splicing of ewg introns 2, 3b, 4, and 5 (data not shown for intron 2) (Fig. 2C, primers 3aF-3cR [lowest band] and 4F-5R). However, differences were observed in products of the alternatively spliced ewg introns 3a, 3c, and 6 (see below).
The RT-PCR profiles observed in ectopic ELAV-expressing wing discs are consistent with the expectation that ELAV's presence promotes 116-kDa ORF-like transcripts. In the intron 3c region, a higher concentration (about 10-fold) of transcripts, indicative of a 116-kDa-like ORF, is seen in wing discs ectopically expressing ELAV (Fig. 2C, compare bands in images corresponding to primers 3cF-3cR and 3aF-3cR). Also, a two- to threefold increase in the spliced 3a product, accompanied by a comparable decrease in the unspliced product, is seen in wing discs ectopically expressing ELAV (Fig. 3C, compare bands in images corresponding to primer 3aF-3aR). Overall splicing efficiency which leads to inclusion of exon D is only about 5% but increases to about 25% upon ectopic ELAV expression.
There is a barely discernible intron 6-spliced product in wild-type wing discs, but ectopic ELAV expression results in an increase of orders of magnitude in the intron 6-spliced transcript while transcripts that retain exon I are unchanged (Fig. 2C, primers 6F-6R). Similar to edr and wild-type eye disc RNA, levels of retained intron 6 were unchanged under ectopic ELAV expression (Fig. 2C, primers FV-6R and RV-6F).
In summary, ectopically expressed ELAV in wing discs mimics the splice profile of wild-type eye discs. Wild-type wing discs imitate the ELAV-deficient edr eye discs, except that inclusion of exon D is reduced and splicing of intron 6 is barely detectable (Fig. 2B and C). Thus, ectopic ELAV in nonneural cells leads to the neuronal splicing pattern. Further, the dose-dependence of ectopic EWG expression in the wing disc on ELAV levels implies that ELAV is a rate-limiting factor in this cell type (Fig. 3Ac and d).
ELAV regulates neuronal splicing of arm and nrg.
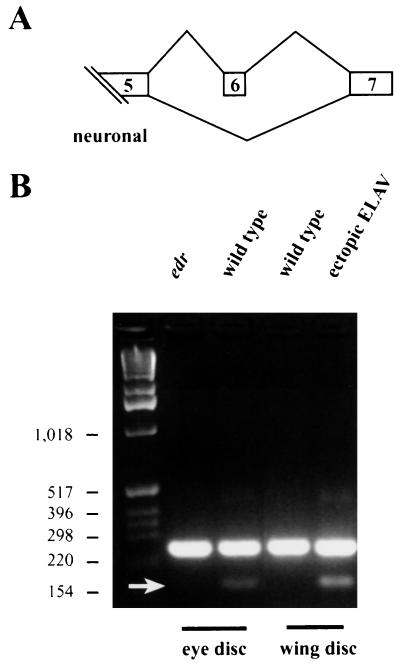
The neuron-specific arm transcript, n-arm, is generated by an alternative splice event that results from the exclusion of exon 6 of ubiquitous-arm (u-arm) (Fig. 4A) (31). The primer pair used amplifies both u-arm and n-arm transcripts; the 147-bp smaller band corresponds to n-arm, while the 244-bp band corresponds to u-arm. To test if ELAV has a role in the formation of n-arm transcripts, RNA from wild-type and edr eye discs, as well as from wild-type eye discs and wing discs ectopically expressing ELAV was analyzed by RT-PCR. The amount of n-arm was reduced in ELAV-deficient eye discs, and in the ectopically expressing wing discs expression of n-arm was clearly induced (Fig. 4B). No change was observed in the band representing u-arm splicing (Fig. 4B). rp49 levels were equal and elav levels were comparable to those shown in Fig. 3Ae. In summary, the presence of n-arm is correlated with the presence of ELAV in both neural and nonneural tissues, implying that arm transcripts are regulated by ELAV. Similar data were obtained for splicing of exons VIIa and VIIb of nrg transcripts (25, 56).
FIG. 4.
Exclusion of exon 6 of arm is dependent on ELAV levels. (A) Schematic representation of arm splicing. The penultimate exon 6 is excluded in neurons (31). (B) arm splicing was monitored in edr and wild-type eye discs and in wild-type and c309/UAS-ELAV2e2; UAS-ELAV3e1 wing discs. c309 is an enhancer trap GAL4 line (expression pattern shown in Fig. 3). Note that exclusion of exon 6 (arrow) correlates with the presence of ELAV.
ELAV does not regulate all neuron-enriched splicing events.
Thus far, we have identified three transcripts, ewg, nrg, and arm, where ELAV affects alternative splicing to produce neuron-enriched or -specific protein isoforms (26). This made us wonder if all genes which are alternatively spliced and which are expressed in neuronal and nonneuronal tissues are regulated by ELAV. Therefore, we analyzed alternative splicing of several ubiquitously expressed genes which are alternatively spliced. While searching for this class of genes, genes which could be involved in the nervous system defects of elav-mutant embryos were favored (22, 45). The DCC homologue frazzled (24), receptor phosphatases PTP4E, PTP10D, and PTP99A (39, 52), CREB (55), and presenilin (35) were tested for ELAV-dependent alternative splicing in edr and wild-type eye discs. None of these genes seem to be ELAV dependent, since their splicing patterns were unaffected, i.e., not reduced in elavedr eye discs (data not shown). Thus, not all genes which are alternatively spliced and expressed in neuronal and nonneuronal tissues are regulated by ELAV.
DISCUSSION
Expression of EWG is regulated posttranscriptionally.
Previous studies demonstrate that, in spite of the broad transcription of the ewg gene, the 116-kDa EWG isoform is almost exclusively generated in neurons and that the other predicted isoforms, if synthesized, are present at very low levels (27). One exception to this is the expression of EWG protein in the developing myoblasts (13) that has been shown to be important in the development of the dorsal longitudinal indirect flight muscles. Whether the myoblast EWG is the same 116-kDa protein is not known; however, a minitransgene expressing SC3 cDNA under the control of heat shock promoter 70 rescues the muscle phenotype (15). In this study, we show that abundance of certain splice isoforms indicative of the 116-kDa EWG ORF are reduced when ELAV is reduced. Additionally, we show that ectopically expressed ELAV can induce EWG expression. How is this positive regulation effected? The broad expression of ewg primary transcript makes transcriptional regulation unlikely to be the primary cause. Nevertheless, we formally excluded that possibility by demonstrating that intron-containing transgenes are ELAV responsive even when driven by a heterologous promoter, while intronless transgenes driven by the same heterologous promoter are ELAV independent.
Mechanism of ELAV-mediated regulation.
Our major findings are that transcripts indicative of the 116-kDa ORF are reduced when ELAV is reduced in the photoreceptors of the eye imaginal discs and are induced when ELAV is ectopically expressed in the imaginal wing discs. We have previously shown that a similar transcript preference is also seen in the neuron-rich adult heads over neuron-poor bodies (27). Together, these two findings suggest that the observations made in the photoreceptors may be extended to adult neurons. Since ELAV is not expressed in the myoblasts, the mechanism of ewg regulation in myoblasts is not addressed here.
Our data was collected by RT-PCR analysis with oligo(dT)-primed cDNA and therefore only relates to that fraction of ewg pre-mRNA that was amplified from the first-strand cDNA synthesis. An additional limitation of these data is that third-instar eye discs, the source for neuron-rich RNA, are composed not only of neurons with high levels of ELAV expression but also of other nonneuronal cell types that have no ELAV. We used eye discs because of the unique genetic advantage offered in having an ELAV-depleted cell type. The wing discs present a more homogeneous population of cells devoid of ELAV.
ELAV could influence splicing efficiency of specific splice sites directly, or affect the stability of specifically spliced transcripts through 3′ UTR interaction, similar to its mammalian family members HuR (17, 42), HuD (11, 12), and Hel-N1 (29). Below we evaluate key data presented in this paper in the context of these two alternatives. Although direct action by ELAV is the simplest alternative, our data do not rule out a scenario in which ELAV orchestrates these events through ELAV-regulated trans-acting protein(s).
First, observations with the ewg transgene expression suggest that the 3′ UTR is neither sufficient nor necessary for ELAV-mediated ewg regulation. Genomic transgenes are ELAV regulated, while minitransgenes without introns which express the 116-kDa isoform and have the same 578-nt-long 3′ UTR as the SC3 cDNA are not ELAV responsive. Furthermore, ewg transgene EWGHS (P{w+=hsp−EWG), in which the SC3 DNA is driven by the hsp70 promoter, also generates a 116-kDa protein in nonneural tissues and is immune to ELAV in the eye disc (25). The requirement for the intron-containing genomic region for ELAV dependence in photoreceptor neurons and in nonneural wing imaginal discs suggests that the 3′ UTR alone is not sufficient. Moreover, if ELAV-mediated regulation stabilizes the 116-kDa ORF encoding transcripts only in neurons, then the 116-kDa ORF transcripts transcribed from a cDNA with the same 3′ UTR under a heterologous promoter should be unstable in ELAV-deficient eye discs, which does not appear to be the case.
Second, although transcripts that include exon J or exons J and I both share the same 3′ ends, only transcripts that include J alone show a correlation with ELAV. For example, the majority of spliced RNA species in the 6F-6R reaction in wing discs (Fig. 2C) include both exons J and I. The spliced RNA species which includes exon J alone is almost not detected. If transcript stabilization were the mechanism, one would expect to see both transcripts increased when ELAV is ectopically expressed.
Third, data presented in this paper show that, in ELAV's presence, intron 6-spliced transcripts increase. However, the transcripts that retain intron 6 do not show a dramatic change, nor is there a major effect on transcripts that retain exon I. This was observed in comparisons of both edr and wild-type eye discs and wild-type wing discs and wing discs ectopically expressing ELAV (Fig. 2B and C). These observations are consistent with the scenario in which ELAV enhances splicing but overall splicing of intron 6 is still very inefficient. Similar arguments also hold for splicing of intron 3c. These data allow a rough estimate of the efficiency of splicing in the wild-type eye disc RNA amplified from a polyadenylated RNA population, resulting in the 116-kDa splice form, to be about 1 to 2%. This calculation assumes that splicing of introns 3a, 3b, and 6 occurs through independent events and that all products in a given reaction are amplified with equal efficiency (intron 3a-spliced product, about 50%; intron 3c-spliced product, about 50%; and intron 6-spliced product, less than 20%).
Fourth, if a specific transcript is more abundant due to stabilization, common regions of transcripts (exons B, C, and F to H) should also show correlation with altered ELAV levels. ELAV levels do not seem to affect these common regions either in edr eye discs or in wing discs ectopically expressing ELAV. However, it is possible that, even in ELAV's presence, the splicing of intron 6 is so inefficient that the exon J-containing transcript constitutes too small a fraction of the commonly spliced transcript.
Taken together, these considerations present a strong case for ELAV-mediated processes influencing specific splice site choices rather than transcript stabilization to regulate EWG protein expression.
ELAV does not affect all splice site choices in ewg.
Many of the splice events, for example, 3b-spliced product or the inclusion of exons E and I, are unaffected by ELAV levels. This could be because these events occur only in nonneuronal cells, e.g., epithelial cells in eye discs. Alternatively, the default splicing could be mediated by another mechanism which does not compete with ELAV. Similar findings were reported in Caenorhabditis elegans for regulated alternative splicing of unc-52 transcripts by mec-8, an RNA-binding protein with two RNA recognition motifs (RRMs) (32). In mec-8 mutants only a subset of alternative splice events of unc-52 transcripts are abolished in a region of several alternative splice events and levels of unaffected alternatively spliced transcripts remain unaltered.
Tissue-specific alternative splicing has been postulated to be controlled by both tissue-specific factors and differential concentrations of general splicing factors (10). Cell culture studies have shown that ubiquitously expressed general splicing factor SR proteins, which, like ELAV, also contain RRMs, influence splice site choice depending on their concentration (9, 48, 49). Also, factors other than SR proteins have been shown to influence alternative splicing (10, 50). The KH-RNA binding motif containing mammalian KSRP is expressed ubiquitously, although at a higher concentration in neurons, where it enhances c-Src neuron-specific splicing in cell culture (38). Drosophila PSI, also a KH-RNA binding motif-containing protein, is expressed predominantly in the soma, where it represses the splicing of the P element third intron (1). Ectopic expression of PSI in the germ line represses splicing of an intron-containing reporter transgene (1).
The role of ELAV in alternative splicing does not preclude it from regulating mRNA function in the cytoplasm, e.g., translatability or stability. Indeed, Drosophila Sex-lethal, an RRM-containing protein with significant homology to ELAV RRMs (7), has been shown to have a role both in splicing (4, 36, 37) and in mRNA translatability (5, 23).
One biological role of ELAV is promoting generation of specific alternatively spliced protein isoforms in neurons.
ELAV ensures that the correct alternatively spliced protein isoforms of certain genes are generated in neurons. Currently we have identified three target genes, ewg, nrg, and arm. Both nrg and arm are vital genes and are broadly transcribed and ubiquitous protein isoforms are broadly expressed, but an additional isoform, encoded by an alternatively spliced transcript, is panneurally expressed (6, 20, 31). The significance of the neural Nrg (n-Nrg) isoform is not known, but the distinct cytoplasmic domain could be important in signaling. The n-Arm isoform differs from the ubiquitous Arm (u-Arm) isoform as it lacks the Wingless interacting domain (40); moreover, it preferentially interacts with DE-cadherins (31). Even with these differences in properties, the current evidence suggest that the u-Arm is sufficient to provide the n-Arm function (31). Perhaps a more detailed phenotypic analysis may reveal a specific role for n-Arm.
ewg, also a vital gene, is broadly transcribed (27), but the protein product, a likely transcriptional regulator, is almost exclusively neural (15). In the case of ewg, it is clear that the expression of the 116-kDa protein isoform is essential for viability in the nervous system and that, when expressed in nonneural tissues it can be lethal (27). These ELAV-regulated genes provide insight into the regulatory role of ELAV in neurons.
Experiments reported here, for the first time, demonstrate that the prevalence of neuron-specific ewg, nrg, and arm transcripts positively correlates with ELAV levels, and these results were achieved through increased use of specific splice sites.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant NS 37169. Confocal microscopy was made possible by NIH shared instrumentation grant RR 05615. M.S. was supported by a fellowship from the Swiss National Science Foundation.
We thank S. Selleck, L. Manseau, and the Bloomington stock center for fly stocks, J. Loureiro and M. Peifer for antibodies, and L. Torroja for discussions. We also thank K.-M. Sun for constructing elav-ewg and E. Dougherty for imaging assistance.
S.P.K. and M.S. contributed equally to this work.
REFERENCES
- 1.Adams M D, Tarng R S, Rio D C. The alternative splicing factor PSI regulates P-element third intron splicing in vivo. Genes Dev. 1997;11:129–138. doi: 10.1101/gad.11.1.129. [DOI] [PubMed] [Google Scholar]
- 2.Antic D, Keene J D. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antic D, Lu N, Keene J D. ELAV tumor antigen, hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker B S. Sex in flies: the splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 5.Bashaw G J, Baker B S. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89:789–798. doi: 10.1016/s0092-8674(00)80262-7. [DOI] [PubMed] [Google Scholar]
- 6.Bieber A J, Snow P M, Hortsch M, Patel N H, Jacobs R J, Traquina Z R, Schilling J, Goodman C S. Drosophila Neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- 7.Birney E, Kumar S, Krainer A. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 9.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 10.Chabot B. Directing alternative splicing: cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 11.Chung S, Eckrich M, Perrone-Bizzozero N, Kohn D, Furneaux H. The Elav-like proteins bind to a conserved regulatory element in the 3′-untranslated region of GAP-43 mRNA. J Biol Chem. 1997;272:6593–6598. doi: 10.1074/jbc.272.10.6593. [DOI] [PubMed] [Google Scholar]
- 12.Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J Biol Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 13.DeSimone S, Coelho C, Roy S, VijayRaghavan K, White K. ERECT WING, the Drosophila member of a family of DNA binding proteins is required in imaginal myoblasts for flight muscle development. Development. 1996;121:31–39. doi: 10.1242/dev.122.1.31. [DOI] [PubMed] [Google Scholar]
- 14.DeSimone S M. Ph.D. thesis. Waltham, Mass: Brandeis University; 1992. [Google Scholar]
- 15.DeSimone S M, White K. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol Cell Biol. 1993;13:3641–3649. doi: 10.1128/mcb.13.6.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis M C, O'Neill E M, Rubin G M. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- 17.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming R J, DeSimone S M, White K. Molecular isolation and analysis of the erect wing locus in Drosophila melanogaster. Mol Cell Biol. 1989;9:719–725. doi: 10.1128/mcb.9.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleming R J, Zusman S, White K. Developmental genetic analysis of lethal alleles at the ewg locus and their effects on muscle development in Drosophila melanogaster. Dev Genet. 1983;3:347–363. [Google Scholar]
- 20.Hortsch M, Bieber A J, Patel N H. Differential splicing generates a nervous system-specific form of Drosophila Neuroglian. Neuron. 1990;4:697–709. doi: 10.1016/0896-6273(90)90196-m. [DOI] [PubMed] [Google Scholar]
- 21.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez F, Campos-Ortega J A. Genes in the subdivision 1B of the Drosophila melanogaster X-chromosome and their influence on neural development. J Neurogenet. 1987;4:179–200. [PubMed] [Google Scholar]
- 23.Kelley R L, Wang J, Bell L, Kuroda M I. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- 24.Kolodziej P A, Timpe L C, Mitchell K J, Fried S R, Goodman C S, Jan L Y, Jan Y N. frazzled encodes a Drosophila member of the DCC immunoglobulin subfamily and is required for CNS and motor axon guidance. Cell. 1996;87:197–204. doi: 10.1016/s0092-8674(00)81338-0. [DOI] [PubMed] [Google Scholar]
- 25.Koushika S P. Ph.D. thesis. Waltham, Mass: Brandeis University; 1998. [Google Scholar]
- 26.Koushika S P, Lisbin M J, White K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol. 1996;6:1634–1641. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- 27.Koushika S P, Soller M, DeSimone S M, Daub D, White K. Differential and inefficient splicing of a broadly expressed Drosophila ewg transcript results in tissue-specific enrichment of the vital EWG protein isoform. Mol Cell Biol. 1999;19:3998–4007. doi: 10.1128/mcb.19.6.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legrain P, Rosbash M. Some cis-acting and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 29.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsley D L, Zimm G G. The genome of Drosophila melanogaster. New York, N.Y: Academic Press, Inc.; 1992. [Google Scholar]
- 31.Louriero J, Peifer M. Roles of Armadillo, a Drosophila catenin, during central nervous system development. Curr Biol. 1998;8:622–632. doi: 10.1016/s0960-9822(98)70249-0. [DOI] [PubMed] [Google Scholar]
- 32.Lundquist E A, Herman R K, Rogalski T M, Mullen G P, Moerman D G, Shaw J E. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development. 1996;122:1601–1610. doi: 10.1242/dev.122.5.1601. [DOI] [PubMed] [Google Scholar]
- 33.Luo L Q, Martin-Morris L E, White K. Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J Neurosci. 1990;10:3849–3861. doi: 10.1523/JNEUROSCI.10-12-03849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philp A V, Yang M, Glover D, Kaiser K, Palter K, Selleck S. GAL4 enhancer traps expressed in embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 35.Marfany G, Del-Favero J, Valero R, De Jonghe C, Woodrow S, Hendriks L, Van Broeckhoven C, Gonzalez-Duarte R. Identification of a Drosophila presenilin homologue: evidence of alternatively spliced forms. J Neurogenet. 1998;12:41–54. doi: 10.3109/01677069809108554. [DOI] [PubMed] [Google Scholar]
- 36.Mattox W, Ryner L, Baker B S. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J Biol Chem. 1992;267:19023–19026. [PubMed] [Google Scholar]
- 37.McKeown M. Sex differentiation: the role of alternative splicing. Curr Opin Genet Dev. 1992;2:299–303. doi: 10.1016/s0959-437x(05)80288-6. [DOI] [PubMed] [Google Scholar]
- 38.Min H, Turck C W, Nikolic J M, Black D L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 39.Oon S H, Hong A, Yang X, Chia W. Alternative splicing in a novel tyrosine phosphatase gene (DPTP4E) of Drosophila melanogaster generates two large receptor-like proteins which differ in their carboxyl termini. J Biol Chem. 1993;268:23964–23971. [PubMed] [Google Scholar]
- 40.Orsulic S, Peifer M. An in vivo structure-function study of armadillo, the beta-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park S J, Yang E S, Kim-Ha J, Kim Y J. Downregulation of extramacrochaete mRNA by a Drosophila neural RNA binding protein Rbp9 which is homologous to human Hu proteins. Nucleic Acids Res. 1998;26:2989–2994. doi: 10.1093/nar/26.12.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng S S Y, Chen C A, Xu N, Shyu A. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D M, Benz W K, Engels W R. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 45.Robinow S N. Ph.D. thesis. Waltham, Mass: Brandeis University; 1989. [Google Scholar]
- 46.Roy S. Ph.D. thesis. Mysore, India: University of Mysore; 1997. [Google Scholar]
- 47.Samson M L. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics. 1998;150:723–733. doi: 10.1093/genetics/150.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valcarcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 49.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Manley J L. Regulation of pre-mRNA splicing in metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 51.Wolff T, Ready D F. Pattern formation in the Drosophila retina. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1277–1325. [Google Scholar]
- 52.Yang X, Seow K T, Bahri S M, Oon S H, Chia W. Two Drosophila receptor-like tyrosine phosphatase genes are expressed in a subset of developing axons and pioneer neurons in the embryonic CNS. Cell. 1991;67:661–673. doi: 10.1016/0092-8674(91)90062-4. [DOI] [PubMed] [Google Scholar]
- 53.Yao K M, White K. Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. J Neurochem. 1994;63:41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
- 54.Yao K M, White K. Organizational analysis of elav gene and functional analysis of ELAV protein of Drosophila melanogaster and Drosophila virilis. Mol Cell Biol. 1991;11:2994–3000. doi: 10.1128/mcb.11.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin J C P, Wallach J S, Wilder E L, Klingensmith J, Dang D, Perrimon N, Zhou H, Tully T, Quinn W G. A Drosophila CREB/CREM homolog encodes multiple isoforms including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Mol Cell Biol. 1995;15:5123–5130. doi: 10.1128/mcb.15.9.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao G, Hortsch M. The analysis of genomic structures in the L1 family of cell adhesion molecules provides no evidence for exon shuffling events after the separation of arthropod and chordate lineages. Gene. 1998;215:47–55. doi: 10.1016/s0378-1119(98)00273-x. [DOI] [PubMed] [Google Scholar]