Significance
To address the unmet clinical need for effectively suppressing estrogen receptor (ER) activity with both de novo resistance and in advanced ER-positive breast cancers that are resistant to standard-of-care antiestrogens, we have developed dual-mechanism ER inhibitors (DMERIs) that employ two distinct ER-targeting moieties. These DMERI elicited noncanonical structural perturbations of the receptor ligand-binding domain and stabilized multiple antagonist substates within the dimer to generate highly efficacious antagonism of proliferation in ER-positive breast cancer cells and in allele-specific resistance models. This work reveals conformational modes by which the activity of ER can be effectively suppressed to block breast cancer proliferation.
Keywords: breast cancer, estrogen receptor, cancer therapy, SERM, X-ray crystallography
Abstract
Efforts to improve estrogen receptor-α (ER)–targeted therapies in breast cancer have relied upon a single mechanism, with ligands having a single side chain on the ligand core that extends outward to determine antagonism of breast cancer growth. Here, we describe inhibitors with two ER-targeting moieties, one of which uses an alternate structural mechanism to generate full antagonism, freeing the side chain to independently determine other critical properties of the ligands. By combining two molecular targeting approaches into a single ER ligand, we have generated antiestrogens that function through new mechanisms and structural paradigms to achieve antagonism. These dual-mechanism ER inhibitors (DMERIs) cause alternate, noncanonical structural perturbations of the receptor ligand-binding domain (LBD) to antagonize proliferation in ER-positive breast cancer cells and in allele-specific resistance models. Our structural analyses with DMERIs highlight marked differences from current standard-of-care, single-mechanism antiestrogens. These findings uncover an enhanced flexibility of the ER LBD through which it can access nonconsensus conformational modes in response to DMERI binding, broadly and effectively suppressing ER activity.
The estrogen receptor-α (ER) plays a critical role in breast cancer in which it functions as a major driver of tumor growth in ∼70% of breast cancers. The suppression of ER function with endocrine therapy is initially quite effective, either by inhibiting estrogen production with aromatase inhibitors (AIs) or by blocking ER activity with antiestrogens (1, 2). However, many ER-positive breast cancers recur in forms that have become resistant to standard-of-care AIs and/or antiestrogens, while de novo resistance also occurs. In these resistant cases, it is possible that antiestrogens of novel design might still prove effective because most of the tumor cells continue to express ER (3).
Currently, there are two types of approved antiestrogens. Tamoxifen (4) and its newer generation analogs, raloxifene, bazedoxifene, and lasofoxifene, are called selective ER modulators (SERMs) (5) due to their estrogenic activity in some tissues. SERMs all contain an additional aromatic ring that we call the E-ring (named with respect to the four rings of steroids that are lettered A–D; SI Appendix, Fig. S1 A and B). The E-ring is attached to an aminoalkyl side chain via a two-carbon ether that exits from the ligand-binding pocket and interacts on the receptor surface with a single H-bond at Asp351 (SI Appendix, Fig. S1A); in this stabilized position, the side chain controls antagonism of breast cancer growth as well as selective modulation of ER activity in other target tissues (6–10), all within a very tightly defined structural space on the receptor.
Fulvestrant, the only approved ER antagonist for treatment of tamoxifen- or AI-resistant breast cancer (11), and other full antagonists such as RU 58668 are termed selective ER downregulators (SERDs), because they also reduce ER protein levels, although this effect may not be required for their antagonist activity (12–14). Both of these SERDs contain an extended, terminal, fluorine-substituted alkyl sulfinyl or sulfonyl side chain (SI Appendix, Fig. S1B) but suffer from poor pharmaceutical properties. Newer, orally active SERDs under clinical development have acrylate side chains (SI Appendix, Fig. S1C), again having a two-carbon linker but now to a carboxyl group (12, 15–20). Thus, the US Food and Drug Administration–approved SERMs and potential oral SERD replacements for fulvestrant contain a single, carefully positioned aminoalkyl or acrylate side chain that through direct interaction with helix 12 (h12) in the ER ligand-binding domain (LBD) moves it to disrupt the surface-binding site for transcriptional coactivators that drive proliferative gene expression, thus operating by a direct antagonism mechanism of action.
Here, we take a different approach to block the activity of ER by combining two distinct molecular elements that disrupt the conformation of the LBD within one ligand. In addition to side chains typically used to effect direct antagonism, we add bulky chemical groups that cause indirect antagonism by distorting structural epitopes inside the receptor ligand-binding pocket. We show here that indirect antagonism independently drives full antagonism, enabling the direct antagonist side chain to adopt new structural and functional roles that are associated with identified noncanonical conformations of the receptor. Coregulator peptide binding and structural studies highlight marked differences from current standard-of-care, single-mechanism antiestrogens. Thus, these dual-mechanism ER inhibitors (DMERIs) represent a class of antiestrogens that generate full antagonism of proliferation in wild-type (WT) ER-positive breast cancer cells and in a number of allele-specific resistance models. Our findings uncover an enhanced flexibility of the ER LBD through which it can access nonconsensus conformation modes in response to DMERI binding that very effectively suppress ER activity.
Results
Formulation of DMERIs.
Indirect antagonists produce a range of activity profiles by interfering in new ways with the docking of h12 across h3 and h11 of the ER, which is required for formation of the surface-binding site for transcriptional coactivator complexes, called Activation Function-2 (AF-2). Starting from a bulky oxabicyclic scaffold that contain aromatic rings corresponding to the A and E rings of other ER ligands (Fig. 1A and SI Appendix, Fig. S1 A–C), we appended a sulfonamide linker to prepare a 7-oxabicyclo[2.2.1]heptene sulfonamide (OBHS-N) core scaffold, as illustrated by the parental OBHS-N compound 13 (Fig. 1A). Despite lacking the canonical side chains required for direct antagonism by standard-of-care antiestrogens, we previously showed that 13 was a full antagonist SERD, equivalent to fulvestrant in inhibiting proliferation of breast cancer cells with WT ER (21). The sizable –SO2-N(CH2CF3)(p-Cl-phenyl) group in 13 shifted the position of h11 by 2.4 Å; this blocked the interaction between the N terminus of h3 and the C terminus of h11 (SI Appendix, Fig. S1D) and disrupted the agonist binding site for h12 against h11 (SI Appendix, Fig. S1E), resulting in antagonist activity indirectly, that is, without direct interaction with h12. Compounds in this parental series, however, were not effective against the constitutively active mutants of ER that drive treatment-resistant disease (SI Appendix, Fig. S1E) (21, 22).
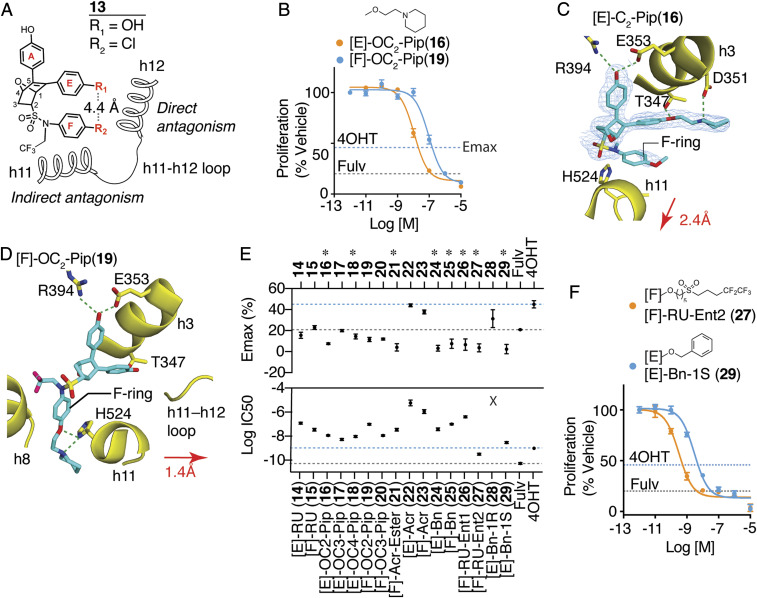
Fig. 1.
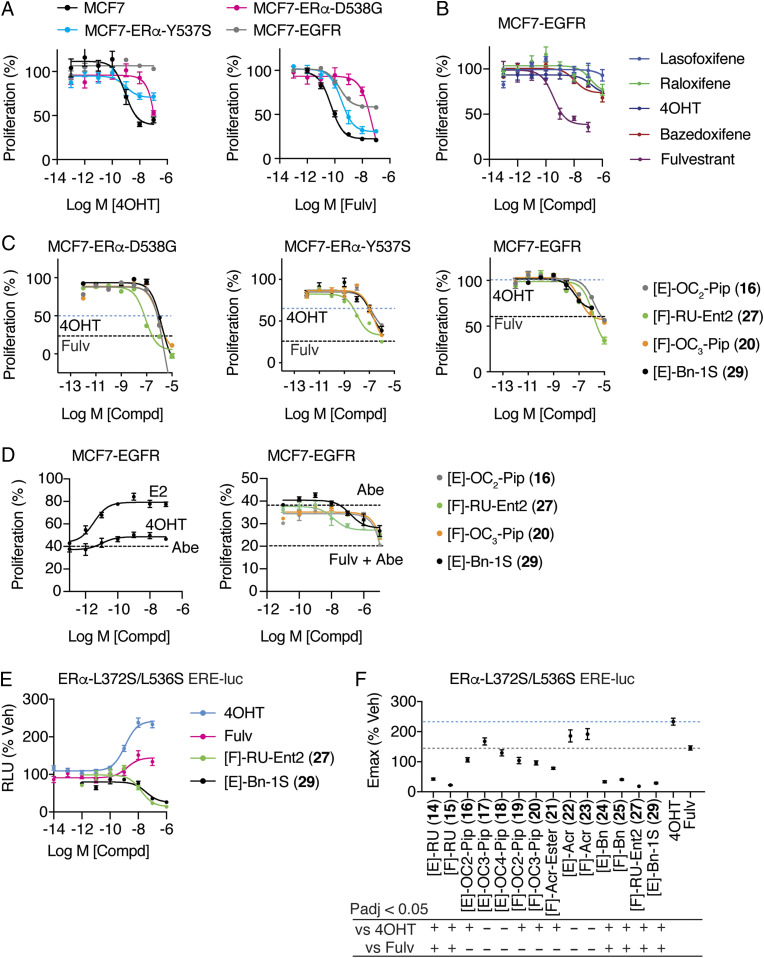
Dual-mechanism ER inhibitors fully suppress breast cancer cell proliferation. (A) Chemical structure of the OBHS-N scaffold and the orientation of substituents R1 and R2, with respect to h11 and h12 in the ER LBD (when R1 has a substituent, R2 is −OCH3 group; when R2 has a substituent, R1 is −OH for compounds 14–29; SI Appendix, Fig. S2 A–H). (B) Proliferation of MCF-7 cells treated for 5 d with 4OHT, fulvestrant (Fulv), or the indicated compounds. Datapoints are mean ± SEM, N = 6. M, Molarity. The horizontal lines indicate the Emax for 4OHT and fulvestrant (Fulv). (C) The structure of [E]-OC2-Pip (16)-bound ER LBD showed that the E-ring substituted piperidine H-bonding to Asp351 in helix 3 (h3), while the F-ring shifts helix 11 (h11) by 2.4 Å compared to an agonist bound structure. 2Fo-Fc electron density map contoured to 0.9 σ within 2 Å of the ligand. (D) The structure of [F]-OC2-Pip (19)–bound ER LBD shows that its [F]-OC2-Pip side chain exits the ligand-binding pocket between h8 and h11, H-bonds to His524, and shifts h11 toward h12. (E) Summary of dose–response curves for compound inhibition of proliferation of MCF-7 cells, shown in SI Appendix, Fig. S2 A–H. Datapoints are mean ± SEM, n = 6. * indicates pAdj < 0.05 (one-way ANOVA) for compounds with Emax > fulvestrant. (F) Selected dose curves from E. The lines indicate the Emax for 4OHT and Fulv.
The position of the E-ring inherent in the OBHS core closely matches the site from which the direct-acting antagonist side chains of standard-of-care SERMs and SERDs emanate. The sulfonamide substitutions that drive indirect antagonism of the OBHS-N compounds also contain another aromatic ring that we call the F-ring, and the flexibility of its linkage to the bicyclic ligand core enables the F-ring to extend in generally the same direction as the E-ring, in both cases, toward h12 and the h11-h12 loop, typical targets of direct antagonism (Fig. 1A). Thus, the OBHS-N compounds provide two sites from which to launch a set of direct-acting antagonist side chains, the canonical E-ring site and the noncanonical F-ring site; the latter, being 4.4 Å away, directs substituents into more unexplored structural space for ER antagonism (Fig. 1A). Our OBHS-N system thus allowed us to evaluate and compare two structure-based design approaches for ER antagonism: 1) combining indirect antagonism with the canonical direct antagonism emanating from substituents on the E-ring and 2) combining direct and indirect antagonism from substituents emanating from the noncanonical F-ring location. To facilitate these comparisons, we chose in both cases to use the same set of substituents typically found in standard-of-care SERMs and SERDs, aminoalkoxy ethers, acrylates, and a Roussel58668-like extended alkyl sulfonyl group with a perfluorinated terminus, similar to the side chain on fulvestrant. We also included some length variations in the aminoalkoxy ether groups as well as ester precursors of the acrylate carboxylates and some benzyl ethers. None of these latter compounds were expected to have antagonist activity.
Comparison of E- versus F-Ring Raloxifene Side-Chain OC2-Piperidine (Pip) Attachments.
We first compared OBHS-N compounds having the most traditional two-carbon aminoalkyl ether SERM side chain attached to the E-ring ([E]-OC2-Pip (16)) or the F-ring ([F]-OC2-Pip (19)). Crystallographic statistics are described in Dataset S1. Both of these compounds inhibited the proliferation of MCF-7 breast cancer cells with greater efficacy (i.e., greater extent of proliferation suppression) than 4OHT (the active 4-hydroxy metabolite of tamoxifen) and equivalent efficacy to fulvestrant (Fig. 1B), but 16 was more potent than 19 (with 11 nM and 97 nM IC50s, respectively). We obtained X-ray crystal structures of these compounds in complexes with the ER LBD in the antagonist conformer, in which h12 was displaced from the agonist position and flipped onto the AF-2 surface to block coactivator binding (SI Appendix, Fig. S1F).
As expected, the [E]-OC2-Pip side chain of 16 directly displaced h12 and formed a tight H-bond with h3 Asp351, whereas the unsubstituted F-ring in this ligand shifted h11 2.4 Å away from the position required for optimal agonist activity (Fig. 1C and SI Appendix, Fig. S1F). To our surprise, the side chain of [F]-OC2-Pip (19) did not point toward h12 but instead exited between h8 and h11, where it is stabilized by H-bonds with h11 His524 (Fig. 1D and SI Appendix, Fig. S1G). In this orientation, the F-ring attached antagonist side chain in 19 pushes h11 toward the agonist position of h12 by 1.4 Å, from which it is also expected to prevent formation of the agonist conformer. This noncanonical orientation of an ER side chain also results in antagonism, which, though less potent than that of [E]-OC2-Pip (16), still fully inhibited breast cancer cell proliferation (Fig. 1B). Thus, the ER LBD complex with the F-ring-substituted OBHS-N compound 19 represents another form of indirect antagonism, which was possible due to the presence of two rotatable bonds in the sulfonamide that allowed the F-ring to adopt multiple positions relative to the core (SI Appendix, Fig. S1H). Examples of this were observed again in our series of crystal structures described below and likely contribute to the unusual activity profiles of these ligands.
Dual-Mechanism Inhibitors Fully Antagonize Breast Cancer Cell Proliferation Irrespective of Their Side Chain Structure and Site of Substitution.
To find other structural conformations that might arise from combining direct and indirect antagonism, we explored a wider range of side chains on both the E- and F-rings (SI Appendix, Fig. S2 A–H), including the one found in RU 58,668 ([E]-RU (14); [F]-RU (15)) and the acrylate found in orally active SERDs ([E]-Acr (22); [F]-Acr (23)). We also explored different side chains, including piperidines with longer three- or four-carbon linkers ([E]-OC3-Pip (17)), ([E]-OC4-Pip (18)), and ([F]-OC3-Pip (20)) as well as an acrylate ester ([F]-AcrEster (21)) and simple benzyl substitution ([E]-Bn (24) or [F]-Bn (25)), these latter ones being available from synthetic intermediates.
In terms of MCF-7 antiproliferative activity, aside from a few low-potency compounds (acrylates 22 and 23 and the disfavored E-ring benzyl 28), all the E- and F-ring–substituted compounds profiled as full antagonists (Fig. 1E and SI Appendix, Fig. S2 A–H). We consider this to be representative of the broad structural tolerance that results in full antagonism for ligands of the DMERI class through an expanded array of noncanonical conformations of their ER complexes. Notably, eight of the compounds showed maximal efficacy (Emax) significantly greater than fulvestrant (Fig. 1E, asterisks at top).
While most of the compounds induced full antagonism, they differed in potency by almost four logs of the half-maximal inhibitory concentration (IC50) values, demonstrating that with DMERI, unlike traditional agonists, the side chain can be used to optimize potency separately from effects on Emax. Because all of the OBHS-N compounds are racemates, we used chiral high-performance liquid chromatography to resolve two of the compounds 15 and 24, and we found that one enantiomer of each (27 from 15, and 29 from 24) accounted for essentially all of their affinity and cellular activity. These preferred enantiomers (27 and 29) had very good antiproliferative IC50s of 0.3 and 3 nM, respectively (Fig. 1 E and F and SI Appendix, Fig. S2 G and H), and they were the focus of subsequent studies.
To verify on-target mechanism of action, we inhibited cell growth with a subset of the compounds and showed full pharmacological reversal with increasing doses of estradiol (SI Appendix, Fig. S2I). We also showed that the compounds completely antagonized E2-induced expression of the ERα-target gene, GREB1 (SI Appendix, Fig. S2J), and notably, the compounds also had no effect on proliferation of MDA-MB-231 cells, a triple-negative breast cancer cell line that lacks ER (SI Appendix, Fig. S2K), again supporting ER specificity.
Atypical Side Chains Perturb the ER LBD Helix 12 Conformation.
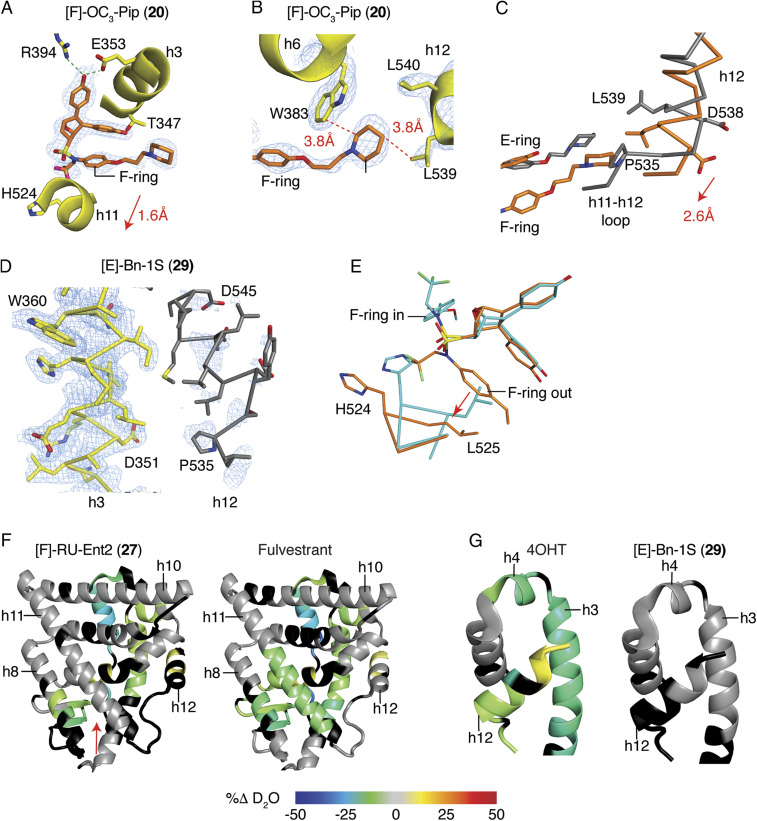
To further understand ligand-dependent effects on receptor structure, we compared X-ray crystal structures of ERα LBD complexes with dual-mechanism inhibitors and other antagonists. In the LBD, the [F]-OC3-Pip (20) longer side chain exited toward h12 and, in doing so, also shifted h11 by 1.6 Å to induce indirect antagonism (Fig. 2A). Here, the piperidine head group made van der Waals contacts with Trp383 in the same location where Pro535 in the h11-h12 loop typically resides in contact with Trp383 (Fig. 2B). This 20-bound ER structure differed from those stabilized by SERMs such as raloxifene, as h12 was shifted 2.6 Å toward the C terminus of h11, allowing Leu539 to directly contact the piperidine group of 20 (Fig. 2 B and C). Unlike traditional SERM side chains that are stabilized by H-bonding or the rigid acrylates of SERDs, the longer side chain of 20 is flexible with many degrees of freedom, suggesting that the shift in h12 is driven by both the shift in h11, which pulls on the h11-h12 loop and additionally by the position of the atypical F-ring side chain.
Fig. 2.
Dual-mechanism inhibitors destabilize helix 12 of ERα. (A) Structure of the ER LBD bound to [F]-OC3-Pip (20). 2Fo-Fc electron density map contoured at 1.0 σ shows the 20 F-ring facing outward between h3 and 11 toward h12, shifting h11 1.6 Å compared to an agonist bound structure. The 2Fo-Fc electron density map is contoured at 0.9 σ within 2 Å of the ligand. (B) The structure of ER with 20 shows that 20 is stabilized by contacts with Trp383, which stabilizes the altered conformer of h12 by contacting L539. (C) The structures of the ER LBD bound to 20 (coral) or raloxifene (gray) were superimposed, showing the 2.6-Å shift of h12 to contact the piperidine ring of 20. (D) Structure of the ER LBD with [E]-Bn-1S (29) shows that h12 could not be modeled in two of four subunits due to poor electron density. The A chain of h3 (yellow) is shown with the B chain superimposed (gray) to show the expected location of h12, which was not modeled. The 2Fo-Fc electron density map is contoured at 1.0 σ. (E) Structure of [F]-AcrEster (21)-bound ER showing different ligand-binding positions in the dimeric subunits. The A and B chains were superimposed and colored blue or coral. (F and G) Changes in ER-Y537S H/D exchange compared to the apo receptor. ER-Y537S LBD was incubated with the indicated ligands and then assayed for exchange of amide hydrogens with deuterium over time, as measured by mass spectrometry. Regions colored black were not detected (SI Appendix, Fig. S4).
The structure of the [E]-Bn (29)-bound LBD showed even more dramatic effects on h12, with very weak electron density in which h12 was expected to be positioned, demonstrating that h12 was disordered (Fig. 2D). This is important, as the original two-position model of h12 (7) (SI Appendix, Fig. S1 D versus F) does not account for how certain antagonists recruit transcriptional corepressors. Structural and biochemical data indicate that the disordering or displacement of h12 renders a more open or accessible AF-2 surface, which is required for binding a longer helical peptide motif found in corepressors (8–10, 23) to support a more complete antagonism of proliferation.
In our structural studies, we noted that several DMERI-bound LBD complexes were ligand-induced conformational heterodimers (Fig. 2E), in which genetically identical monomers bind the same ligand but adopt different conformations in the context of the dimer (SI Appendix, Fig. S3 A–E). This suggests that the ligand-induced shift in h11 of the dimer interface alters the conformer of the other monomer to favorably bind the second ligand differently. Many of the structures reveal the F-ring facing outward in one monomer and facing inward in the other monomer, associated with a smaller shift in h11 (SI Appendix, Fig. S3 C–E). The [F]-acrylate (23) and [F]-acrylate ester (21) side chains differentially H-bonded to the N terminus of h3 (SI Appendix, Fig. S3 A and B), which likely explained their widely different potencies. These effects were ligand selective (SI Appendix, Fig. S3E) and thus not driven by crystal packing. In addition to enforcing noncanonical receptor conformations, these ligands thus also stabilize an ensemble of both direct and indirect antagonist conformers, often within the same dimer.
Hydrogen–Deuterium Exchange Mass Spectrometry Reveals Alternate ER Conformers in Solution.
To validate the destabilizing effects of the ligands on the ER LBD in solution, we examined the dynamics of secondary structural elements through analysis of the exchange of amide hydrogens for deuterium using mass spectrometry (HDX-MS) (24). These included 30, the high-affinity enantiomer of the parental compound 13 (Fig. 1A), the F-ring–substituted Roussel side-chain compound 27, and the E-ring–substituted benzyl compound 29. While 4OHT and fulvestrant stabilized the C-terminal half of h11 proximal to the ligands, the parental OBHS-N 30 and the dual-mechanism inhibitors 27 and 29 did not, consistent with indirect antagonism directed at h11 (Fig. 2F and SI Appendix, Fig. S4A). All of the compounds stabilized h3 and h4 in the AF-2 surface, except for 29, the compound that destabilized h12 in the crystal structure (Fig. 2G and SI Appendix, Fig. S4B). With 27, the extended hydrophobic RU side chain may directly contact the AF-2 surface to stabilize its secondary structural elements, as was seen with the fulvestrant analog ICI 164,384 in ERβ, which is the only available crystal structure of members of this class of SERDs (25) (SI Appendix, Fig. S4C). With the other compounds, the stabilization of the AF-2 surface is likely through h12 binding to the AF-2 surface in the inactive conformer (SI Appendix, Fig. S1F) (6, 7), highlighting the ability of 29 to destabilize h12 in solution and in the crystal structure. These studies demonstrate that with indirect antagonism, the shifts in h11 destabilize or reposition h12 of the ER LBD, allowing the side chains to have distinct roles in stabilizing alternate conformers of h12.
DMERIs Show Ligand Selective Activity Profiles in Other Cellular Contexts.
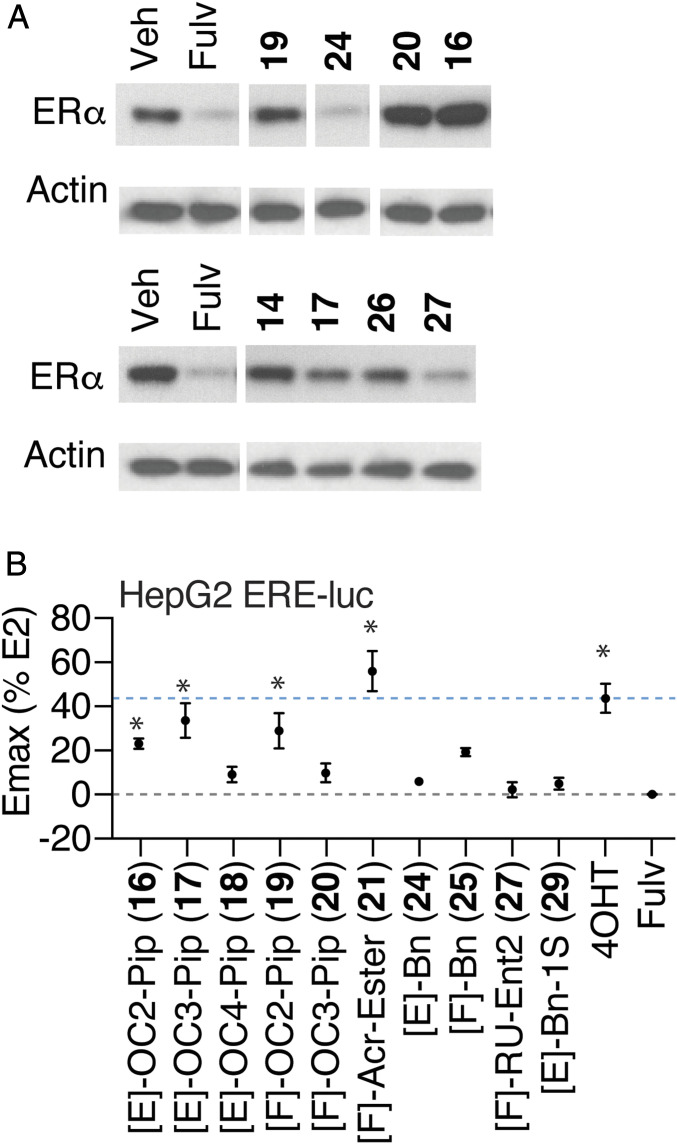
It was striking that the DMERI as a class showed similarly robust Emax for inhibition of growth of WT ERα-driven breast cancer (Fig. 1 B–F and SI Appendix, Fig. S2 A–H) with diverse structural effects on the receptor (Fig. 2 and SI Appendix, Fig. S3). To probe whether the side chains supported different effects on SERM- or SERD-like activities of the ligands, we tested several compounds for effects on degradation of ERα and found that while the parental indirect antagonists are SERDs (21), direct antagonist side chains determined whether compounds displayed SERM- or SERD-like properties (Fig. 3A). [E]-Bn (24) and the higher-affinity enantiomer of [F]-RU, [F]-RU-Ent2 (27), were efficient ER degraders (Fig. 3A). These effects were reversed by 4OHT, demonstrating on-target mechanism of action (SI Appendix, Fig. S5A). In contrast, the compounds with piperidine side chains were more SERM like, with either minimal effects on receptor stability (14, 17, and 19) or showing some stabilization of the receptor (16 and 20; Fig. 3A). [E]-Bn (24) required different ER domains for ligand-dependent degradation than seen with fulvestrant (SI Appendix, Fig. S5B), demonstrating that they have different effects on receptor structure.
Fig. 3.
SERM and SERD properties of DMERI ligands. (A) ER and β-actin levels in MCF-7 cells treated with the indicated compounds for 24 h. Whole-cell lysates were analyzed by Western blot. See also SI Appendix, Fig. S5 A and B. (B) Summary of dose–response curves of HepG2 cells transfected with 3xERE-luciferase reporter and treated with the indicated ligands. Datapoints are mean ± SEM, N= 3. *Significantly different from fulvestrant by one-way ANOVA, Sidak’s test adjusted P value (padj) < 0.05. n = 3 to 6. Dose curves are shown in SI Appendix, Fig. S5C.
To further probe for the cell-type selective activity, we tested the compounds in HepG2 liver cells, because estrogenic effects on liver metabolism are an important contribution to protection from metabolic diseases (26). In these cells, 4OHT displays significant SERM agonist activity through the amino-terminal AF-1 domain of ER (27). We found that some of the compounds with piperidine-containing SERM side chains (16, 17, and 19) showed cell-type–specific agonist activity, as did [F]-acrylate ester (21), while the degraders 24 and 27 were full antagonists (Fig. 3B and SI Appendix, Fig. S5C). Thus, the dual-mechanism inhibitor approach can produce compounds with SERM or SERD properties that are highly efficacious and contain noncanonical side chains.
Dual-Mechanism Inhibitors Induce Unique ER Solution Structures by Peptide Interaction Profiling.
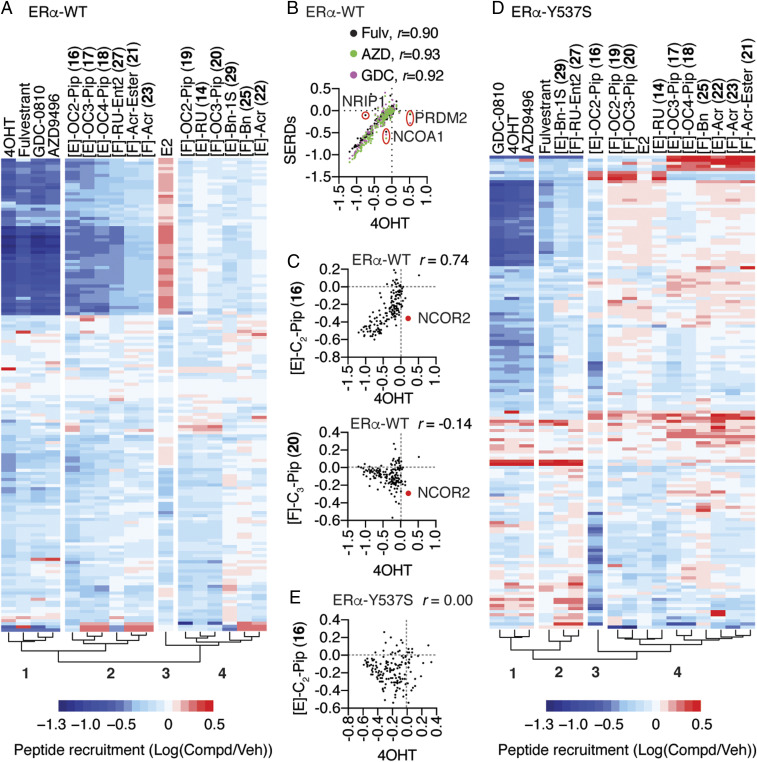
To probe more deeply for molecular insights into the noncanonical activity of DMERIs, we examined the interaction of full-length ER-WT or ER-Y537S complexes of DMERIs versus reference agonist and SERM and SERD compounds with a library of 154 peptides using the Microarray Assay for Real-time Coregulator-Nuclear receptor Interaction (MARCoNI) as a probe for solution structure, which has been widely used for pharmacological profiling (12, 28). This is similar to phage display approaches with both random peptides and peptides constrained to have canonical coactivator or corepressor motifs to characterize solution structures for different ligands (29). Hierarchical clustering of the fluorescence resonance energy transfer (FRET) data describing interaction of ER bound to 19 compounds × 154 peptide interaction profiles is shown in Fig. 4A and Dataset S1, demonstrating a clustering of E2-induced peptide interactions (Cluster 3) that were strongly dismissed by 4OHT, fulvestrant, and the full antagonist SERDs, GDC-0810 and AZD9496 (Cluster 1 versus 3). We found individual peptides that showed some specificity, including PRDM2 (amino acids 948 through 970), which was recruited by 4OHT, NCOA1 (amino acids 737 through 759), which was selectively dismissed by the three SERDs compared to 4OHT, and NRIP1 (amino acids 805 through 831), which was not dismissed by fulvestrant (Fig. 4B). While it is possible to identify individual peptides that are selective for ER bound to these compounds (16, 30), most of the peptide interactions showed identical responses to the ligands in Cluster 1, with Pearson correlations (r) ≥ 0.90 between ligand-dependent peptide interaction profiles (Fig. 4B), highlighting the structural similarities of single-mechanism inhibitors including SERMs and SERDs.
Fig. 4.
Dual-mechanism inhibitors promote conformations of ER that are distinct from traditional single mechanism inhibitors. (A) Hierarchical clustering of MARCoNI FRET assay for interaction of full-length and WT ER with 154 peptides derived from nuclear receptor-interacting proteins and the indicated ligands. (B and C) MARCoNI Pearson correlations for 4OHT versus the indicated ligands. Fulvestrant (Fulv), GDC-0810 (GDC), or AZD9496 (AZD). r = Pearson correlation ligand versus 4OHT. (D) Hierarchical clustering of MARCONI data with ER-Y537S and the indicated ligands. (E) MARCoNI Pearson correlations for 4OHT versus [E]-C2-Pip (16) with the ER-Y537S. r = Pearson correlation (SI Appendix, Fig. S7).
Cluster 2 was in the same clade as Cluster 1 and contained the E-ring piperidine-substituted compounds, as well as the compounds with F-ring Roussel and acrylate side chains, all of which showed dismissal of the E2-induced peptide interactions (Fig. 4A). However, Cluster 2 also has many more unique peptide interactions than Cluster 1, reflected in lower Pearson correlations (r = 0.65 through 0.74 versus 4OHT). These included NCOR2 peptide (amino acids 649 through 671), which is derived from a protein with context-selective coactivator or corepressor activity (31, 32), and was dismissed by 16 (and 20 in Cluster 4) but not 4OHT (Fig. 4C). Cluster 4 displayed peptide interaction patterns most different from the traditional antagonists and included the compounds with F-ring piperidines, E-ring RU, or acrylate side chains, and both the E- and F-ring–substituted benzyl compounds. For example, 20 showed very little overlap in peptide interaction patterns with 4OHT and showed many peptides that were selective for 20 compared to 4OHT (Fig. 4C, Bottom). Thus, the dual-mechanism inhibitors displayed a variety of different solution structural features that differentiate them from the traditional antagonists, all of which displayed highly similar interaction profiles.
The ER-Y537S mutation changed the clustering pattern of ligand-dependent peptide interactions. 4OHT, AZD9496, and GDC-0810 still clustered together and dismissed many of the same E2-induced peptide interactions (Cluster 1, Fig. 4D), while fulvestrant now clustered with [F]-RU-Ent2 (27) and [E]-Bn-1S (29) in Cluster 2 from the same clade. Despite this clustering pattern, fulvestrant still displayed a higher Pearson correlation with 4OHT (SI Appendix, Fig. S6A), as they strongly dismissed many of the peptides (SI Appendix, Fig. S6 B and C). The other major clade includes Cluster 3, which contains only [E]-OC2-piperidine (16), and Cluster 4, which contained all the remaining piperidine-containing compounds, the acrylates, as well as the [E]-RU (14) and [F]-Bn (25) compounds. The dramatic shift in peptide interaction patterns is underscored by 16, which with the ER-Y537S displayed no overlapping effect on peptide interaction patterns with 4OHT (Fig. 4 E versus C). These observations highlight the similarities in solution structures of ER-WT or ER-Y537S bound to the traditional direct antagonists but point to a range of distinct solution structures for many of the dual-mechanism inhibitors, some of which are unique to the mutant ER-Y537S. While these peptides may not reflect in vivo coregulator binding, they identify unique solution conformations of ER that likely contribute to the distinct activity profiles of the dual-mechanism inhibitors.
Activity of Dual-Mechanism Inhibitors in Allele-Driven Models of Anti-Estrogen Resistance.
Approximately one-third of patients with recurrent ER+ breast cancers present with constitutively active ER mutations, including Y537S and D538G (28, 33–35), while de novo EGFR overexpression drives a worse outcome and tamoxifen resistance in a significant subset of newly presenting breast cancer patients (36, 37). To explore these two modes of endocrine therapy resistance, we overexpressed EGFR in MCF-7 cells (SI Appendix, Fig. S7), which we compared to parental MCF-7 cells as well as those engineered to express ER-Y537S or ER-D538G (34). With these models, we observed the expected loss of both potency and efficacy in response to 4OHT or fulvestrant, with the EGFR model showing a complete loss of response to 4OHT and other SERMs in clinical use (Fig. 5 A and B). We tested all the compounds in luciferase assays (SI Appendix, Fig. S8). For proliferation, we tested the two high-potency enantiomers, 27 and 29, and two ligands with SERM properties (16 and 20) that had shown unusual peptide binding (Fig. 4) and structural features (Fig. 1 A–C versus SI Appendix, Fig. S3C and Fig. 2 A–C). All the ligands showed reduced proliferation efficacy, but 27 showed better potency in the mutant ER models, while 20 and 29 showed slightly better potency in the EGFR resistance model (Fig. 5C). To test for effects in a cotreatment setting, the MCF-7 EGFR cells were treated with the CDK4/6 inhibitor abemaciclib and the ERα ligands, which improved the treatment response to fulvestrant and the DMERI, while E2 partially reversed the effects of abemaciclib (Fig. 5D). Despite their diverse side chains for direct antagonism, all these compounds suppressed proliferation across resistance models, highlighting the important role of the dual mechanism for antagonizing ER actions.
Fig. 5.
Activity of ligands in allele-specific models of tamoxifen resistance. (A–C) WT MCF7 cells and MCF7 cells engineered to express the mutant ERα-Y537S or ERα-D538G or overexpress EGFR were treated with the indicated SERM (16, 20) or SERD (27, 29) for 5 d and analyzed for inhibition of cell proliferation. Fulvestrant (Fulv). n = 3. Dashed lines indicate Emax values for 4OHT or fulvestrant (SI Appendix, Fig. S8). (D) MCF7-EGFR cells were treated with the indicated 1μM abemaciclib (Abe) and the indicated ligands for 5 d and analyzed for inhibition of cell proliferation. Fulvestrant (Fulv). n = 3. Dashed lines indicate Emax values for abemaciclib alone or fulvestrant (+Abe). (E and F) Structure-based model of tamoxifen and fulvestrant resistance. HepG2 liver cells were treated for 24 h with the indicated ligands. n = 6, except for 4OHT and Fulvestrant where n = 18. Data were analyzed by one-way ANOVA. Data are mean ± SEM.
To further explore endocrine-resistance modes in breast cancer, we used a model that we previously developed as a structure-based design model of resistance for tamoxifen (27). Using the tamoxifen-bound ER structure to design L372S/L536S as a set of mutations to stabilize h12 as seen in that structure docked into the AF-2 surface, we found that these two mutations blocked binding of the NCOR1 corepressor to ER and enforced AF1-dependent SERM agonist activity for tamoxifen (21, 38). Mutations of L536 have since been identified in metastatic patient samples, highlighting its important role in regulating h12 dynamics (35, 39). Here, we demonstrate that this model also renders fulvestrant an agonist, in essence inverting the activity profile for these two standard-of-care antiestrogens from antagonists to agonists. In using this model for compound profiling (Fig. 5 C and D), we found most of the compounds to be more efficacious than fulvestrant, including those with SERM-like side chains, while the RU (14, 15, and 27) and benzyl (24, 25, and 29) side-chain compounds were significantly more efficacious, almost completely blocking the AF1-driven activity that was enhanced by tamoxifen or fulvestrant (Fig. 5 C and D). In this context, nearly all the dual-mechanism ligands were significantly more efficacious than 4OHT (Fig. 5D). These mutations discriminate DMERI from traditional agonists and reveal that the DMERI show superior ability to displace helix 12.
Discussion
In this work, we show DMERIs to function as a flexible chemical platform for the generation of ligands with tailored SERM- or SERD-like properties that are broadly efficacious across different breast cancer anti-estrogen resistance models, including a structure-based design model of tamoxifen and fulvestrant agonist activity. Probing of ER with a library of interacting peptides revealed that the DMERIs imposed unique solution structures that have important and favorable functional characteristics, an insight that was supported by HDX studies, whereas traditional single-mechanism inhibitors—whether SERM or SERD—overall stabilized very similar structures. Our crystallographic analyses then demonstrated that these ligands induced unique perturbations to h11 and h12 to support their strong antagonism, including the formation of conformational heterodimers, in which each monomer component of the receptor is in effect “reading” the same ligand in two different ways, an interaction that seems to be associated with the most efficacious DMERIs. Overall, the best DMERI SERMs and SERDs showed similar efficacy profiles in most contexts. SERM DMERIs represent a breakthrough in enabling the targeting of early stage breast cancer with de novo treatment resistance in the EGFR overexpression setting, which drives tamoxifen resistance and worse outcome in patients (36, 37) and was broadly resistant to clinically available SERMs here.
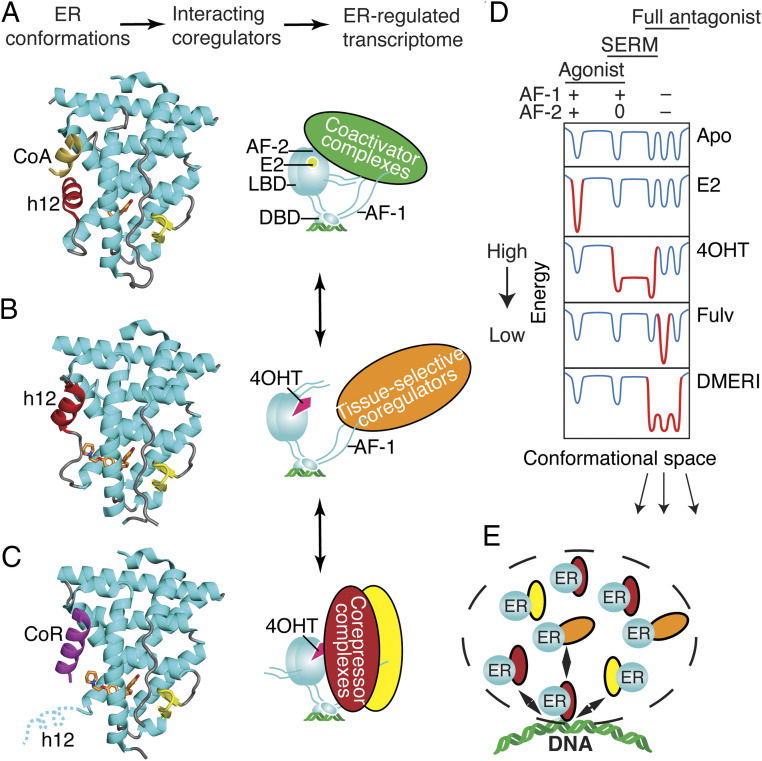
Our findings provide insights beyond the traditional view of nuclear receptor allostery, which is based on a single, direct-acting mechanism in which the ligand adopts a single pose to control the conformation of the protein (Fig. 6 A–C) (40). The single-mechanism ligands select or induce lowest-energy conformations of the receptor associated with specific activity profiles that can be active, inactive, or tissue selective (Fig. 6 A–D). The targeting of multiple antagonist substates with DMERIs (Fig. 6E) may provide a therapeutic targeting advantage similar to the effects of targeting multiple growth pathways with combination therapies or the combined use of bazedoxifene and conjugated estrogens to achieve unique ER-mediated signaling characteristics (41).
Fig. 6.
Ligand-dependent control of ERα-LBD conformation and of ER coregulator recruitment and selection of activity states. (A) The active LBD conformation (7). (Left) Ribbon diagram of the ER LBD bound to estradiol. Helix 12 (h12, colored red) forms one side of the coactivator binding site, shown here binding to a peptide from Steroid Receptor Coactivator-2 (CoA colored yellow) from PDB entry 3UUD. (Right) Schematic of ERα bound to estradiol (E2), DNA, and a coactivator complex. With full agonists, the coactivator recruitment to the LBD surface, AF-2 nucleates binding of multiprotein coactivator complexes to other domains including AF-1 (Activation Function-1). DBD, DNA binding domain. Steroid Receptor Coactivators (SRCs) 1–3 bind to both AF-1 and AF-2 through separate interactions. (B) The inactive LBD conformer (7, 27). Left, Ribbon diagram of the ER LBD bound to an antagonist. Antagonists can flip h12 (colored red) into the coactivator/corepressor binding site, rendering the LBD inactive by blocking both coactivator and corepressor binding to AF-2, from PDB entry 2QXS. (Right) When h12 blocks both coactivators and corepressors from binding the LBD, the activity of AF-1 is cell-type specific. (C) The transcriptionally repressive LBD conformation (9, 12, 25, 53). (Left) Ribbon diagram of the ER LBD bound to a corepressor peptide, colored violet. When h12 is disordered by an antagonist, the LBD can bind an extended peptide motif found in transcriptional corepressors (8) from PDB entry 2JFA. (Right) Cartoon of ERα bound to 4OHT and a corepressor complex, repressing both AF-1 and AF-2 activity and mediating mediate chromatin compaction and inhibition of proliferative gene expression. (D) Energy diagram illustrating how ER ligands differ in stabilizing, specific, low-energy receptor conformations associated with transcriptional activity (+), inactivity (0), or repression (−) that are being driven by the activity state of AF-2 or AF-1. The dips in the curves represent different LBD conformations associated with the three AF-1/AF-2 activity states shown at the top, leftmost being the active state (a), the rightmost representing substates of the repressive state (b), and the middle the inactive state (c). When a state is stabilized by a particular type of ligand, the curves become deeper, with gray changed to red; the barrier heights between states indicate the ease of dynamic interchange among the states or substates. The DMERI showed multiple mechanisms of antagonism, represented by the multiple-favored repressor substates with reduced exchange barriers. (E) Dashed line indicates a transcriptional phase condensate with multiple receptor–coregulator complexes exchanging at an ER binding site, enabling multiple mechanisms of antagonism.
A key feature of the OBHS-N scaffold used here is that the indirect antagonism drives full suppression of ER activity, which we showed with the parental compounds lacking a side chain (21). This then enabled the added side chains to take on different functional roles. Since the development of tamoxifen in the 1970s and fulvestrant in the 1990s, the next-generation SERMs and SERDs have directed either an aminoalkyl group to push on the h11-h12 loop or the acrylate unit to pull on it, leading to compounds with side chains that were localized around a very tight structural interface with ER. With DMERI, the h11-h12 loop was pulled indirectly via h11 to destabilize h12. This enabled a diversity of side-chain activities from either the E-ring or F-ring in the DMERIs to dial back in SERM activity (Fig. 3B), bind directly to h12 to produce altered antagonist conformers (Fig. 2 B and C), or produce efficacy even greater than fulvestrant by fully destabilizing h12 with full antagonists (Figs. 1B, 2 D and G, and 5 B–D).
This advance greatly expands the potential design principles for the ligand side chain that is not being used as the primary driver of antagonism and explains why we observed strong antagonism even when the side chain did not engage in the known modes of antagonism or was completely disordered. Thus, combining two chemical targeting approaches—direct and indirect antagonism—into a single ligand provides a flexible platform for ER-directed therapies with different targeted signaling outcomes and broad efficacy across different treatment resistance models.
Materials and Methods
Cell Culture.
MCF7, MCF7-ERα-Y537S, MCF7-ERα-D538G, MCF7-EGFR, HepG2, and MDA-MB231 cells were maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum. MCF7-ERα-Y537S and MCF7-ERα-D538G were a gift from Steffi Oesterreich, University of Pittsburgh Medical Center. The cell lines above were cultured with 1% penicillin/ streptomycin/ neomycin antibiotics, 1% MEM nonessential amino acids, and 1% GlutaMAX (all from Gibco by Thermo Fisher Scientific), maintained at 37 °C in a 5% CO2 incubator. Cells were tested regularly for mycoplasma contamination.
Luciferase cotransfection assay and cell proliferation assay are as previously described (21, 42) and detailed in SI Appendix, Supplemental Methods.
qPCR.
Total RNA was isolated using the RNeasy kit with on-column DNase I digest (QIAGEN). Four-microgram total RNA samples were reverse-transcribed in 40-µL reactions using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, catalog 4387406). Complementary DNA samples were analyzed by real-time PCR in triplicate 10-μL reactions using the 2X TaqMan gene expression master mix (Applied Biosystems by Thermo Fisher Scientific, catalog 4369016) with human GREB1 (Hs00536409_m1) and GAPDH (Hs02758991_g1) expression assays. Relative messenger RNA levels were compared using the ΔΔCt method.
Western Blot.
Cells were lysed in ice-cold radioimmunoprecipitation assay buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM ethylenediaminetetraacetic acid, and 0.1% sodium dodecyl sulfate). Protein samples were loaded on Any kDa Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, Hercules) and transferred onto polyvinylidene difluoride membranes (Thermo Fisher Scientific, Rockford). The membranes were blocked with 1× phosphate-buffered saline + 0.1% Tween-20 + 5% nonfat dry milk and probed with primary antibodies overnight. The next day, the membranes were washed with 1× Tris-buffered saline + 0.1% Tween-20 and incubated with horseradish peroxidase (HRP)–conjugated probes (Santa Cruz Biotechnology) and developed using an enhanced chemiluminescence detection system (GE Healthcare Bio-Sciences, Pittsburgh).
Antibodies and Probes.
ERα (F-10) mouse mAb (1:1,000, catalog sc-8002), ERα (H222) rat mAb (1:1,000 dilution, catalog sc-53492), β-Actin (C4) mouse mAb (1:10,000 dilution, catalog sc-47778), EGFR (528) mouse mAb (1:50 dilution, catalog sc-120), HRP-conjugated mouse IgG kappa binding protein (catalog sc-516102), and HRP-conjugated goat anti-rat IgG antibody (catalog sc-2006) were purchased from Santa Cruz Biotechnology, Inc.
Macromolecular X-ray Crystallography.
The ERα-L372S/L536S double-mutant LBD (amino acid residues 298 through 554) was expressed in BL21 (DE3) Escherichia coli cells, purified by immobilized metal affinity chromatography using a Ni2+ column, dialysis, Tobacco Etch Virus protease digest, ion exchange, and size exclusion chromatography to remove the HA tag, as previously described (22). The purified LBD was cocrystallized with various ligands through sitting drop vapor diffusion method using trial gradients of 20 to 25% (weight/volume) PEG 3350, 200 mM MgCl2, and pH 6.5 through 8.0, as previously described (38, 43). Data were collected at the Stanford Synchrotron Radiation Lightsource (Beamline: 12–2) and Advanced Photon Source (Beamlines: SER-CAT BM22, ID-22), both at a temperature of 100 K and wavelength of 1.0 Å and scaled using AutoPROC (44) with the application of STARANISO (Globalphasing) to accommodate anisotropic diffraction. The structures were solved by molecular replacement of the starting model, Protein Data Bank (PDB) entry 2QXS, and then rebuilt and refined using the PHENIX software suite version 1.16 (45, 46). Ligand restraints were built on the PHENIX electronic Ligand Builder and Optimization Workbench (47). Ligand docking was automated with LigandFit in PHENIX and visually inspected using Crystallographic Object-Oriented Toolkit (COOT) version 0.8.9.2, as previously described (48, 49). New structures were further refined on the PDB-REDO server (50), before final refinement and validation in the PHENIX environment. Structures were analyzed using COOT and imaged using PyMOL (Schrodinger).
MARCoNI Coregulator Interaction Profiling.
MARCoNI was performed as previously described (51). Human embryonic kidney–293T cells were transfected with full-length, HA-tagged WT ERα or Y537S-ERα. A PamChip peptide micro array with 154 unique coregulator-derived nuclear receptor interaction motifs (#88101, PamGene International) was incubated with extracts from the 293T transfected cells in the presence of 10 µM compound or solvent only (2% dimethyl sulfoxide, apo). Receptor binding to each peptide on the array was detected using fluorescently labeled HA-antibody, recorded by charge-coupled device, and quantified. Per compound, three technical replicates (arrays) were analyzed to calculate the log-fold change (modulation index, MI) of each receptor–peptide interaction versus apo. Significance of this modulation was assessed by Student’s t test.
HDX Detected by MS.
Differential HDX-MS experiments were conducted as previously described with a few modifications (52), described in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
Funding comes from National Key Research and Development Program of China, NIH Grant No. R01 CA220284 (K.W.N., T.I., B.S.K., and J.A.K.), Breast Cancer Research Foundation Grant Nos. BCRF-083 to B.S.K. and BCRF-084 to J.A.K. and B.S.K., NIH Training Grant No. GM 070421 (V.S.G.), 345 Talent Project from Shengjing Hospital of China Medical University China (S.Y.). K.W.N. is supported by the Frenchman’s Creek Women for Cancer Research.
Footnotes
Competing interest statement: J.A.K. is a founder and stockholder of Radius Health Inc. and a consultant of Celcuity Inc. B.S.K. is a consultant of Celcuity Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101657118/-/DCSupplemental.
Data Availability
Proliferation source data are in Dataset S1. X-ray crystallography coordinates will be released upon acceptance. X-ray crystal structure data have been deposited in Research Collaboratory for Structural Bioinformatics Protein Data Bank (https://www.rcsb.org) and are available with the following accession numbers: 7RS4, 7RS9, 7RRZ, 7RS0, 7RRX, 7RRY, 7RS1, 7RS2, 7RS9, 7RS3, and 7RS7. All other study data are included in the article and/or supporting information.
References
- 1.Rugo H. S., et al., Endocrine therapy for hormone receptor-positive metastatic breast cancer: American Society of Clinical Oncology guideline. J. Clin. Oncol. 34, 3069–3103 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Tryfonidis K., Zardavas D., Katzenellenbogen B. S., Piccart M., Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer Treat. Rev. 50, 68–81 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Clarke R., Tyson J. J., Dixon J. M., Endocrine resistance in breast cancer–An overview and update. Mol. Cell. Endocrinol. 418, 220–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerner H. J., Band P. R., Israel L., Leung B. S., Phase II study of tamoxifen: Report of 74 patients with stage IV breast cancer. Cancer Treat. Rep. 60, 1431–1435 (1976). [PubMed] [Google Scholar]
- 5.Jordan V. C., Phelps E., Lindgren J. U., Effects of anti-estrogens on bone in castrated and intact female rats. Breast Cancer Res. Treat. 10, 31–35 (1987). [DOI] [PubMed] [Google Scholar]
- 6.Brzozowski A. M., et al., Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389, 753–758 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Shiau A. K., et al., The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Huang H. J., Norris J. D., McDonnell D. P., Identification of a negative regulatory surface within estrogen receptor alpha provides evidence in support of a role for corepressors in regulating cellular responses to agonists and antagonists. Mol. Endocrinol. 16, 1778–1792 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Webb P., Nguyen P., Kushner P. J., Differential SERM effects on corepressor binding dictate ERalpha activity in vivo. J. Biol. Chem. 278, 6912–6920 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Heldring N., et al., Structural insights into corepressor recognition by antagonist-bound estrogen receptors. J. Biol. Chem. 282, 10449–10455 (2007). [DOI] [PubMed] [Google Scholar]
- 11.DeFriend D. J., et al., Investigation of a new pure antiestrogen (ICI 182780) in women with primary breast cancer. Cancer Res. 54, 408–414 (1994). [PubMed] [Google Scholar]
- 12.Guan J., et al., Therapeutic ligands antagonize estrogen receptor function by impairing its mobility. Cell 178, 949–963.e18 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Wardell S. E., Marks J. R., McDonnell D. P., The turnover of estrogen receptor α by the selective estrogen receptor degrader (SERD) fulvestrant is a saturable process that is not required for antagonist efficacy. Biochem. Pharmacol. 82, 122–130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wardell S. E., et al., Pharmacokinetic and pharmacodynamic analysis of fulvestrant in preclinical models of breast cancer to assess the importance of its estrogen receptor-α degrader activity in antitumor efficacy. Breast Cancer Res. Treat. 179, 67–77 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Savi C., et al., Optimization of a novel binding motif to (E)-3-(3,5-difluoro-4-((1R,3R)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indol-1-yl)phenyl)acrylic acid (AZD9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist. J. Med. Chem. 58, 8128–8140 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Joseph J. D., et al., The selective estrogen receptor downregulator GDC-0810 is efficacious in diverse models of ER+ breast cancer. eLife 5, 1–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B., et al., Unexpected equivalent potency of a constrained chromene enantiomeric pair rationalized by co-crystal structures in complex with estrogen receptor alpha. Bioorg. Med. Chem. Lett. 29, 905–911 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Tria G. S., et al., Discovery of LSZ102, a potent, orally bioavailable selective estrogen receptor degrader (SERD) for the treatment of estrogen receptor positive breast cancer. J. Med. Chem. 61, 2837–2864 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Fanning S. W., et al., Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity. Nat. Commun. 9, 2368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanning S. W., Greene G. L., Next-generation ERα inhibitors for endocrine-resistant ER+ breast cancer. Endocrinology 160, 759–769 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Srinivasan S., et al., Full antagonism of the estrogen receptor without a prototypical ligand side chain. Nat. Chem. Biol. 13, 111–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nettles K. W., et al., NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat. Chem. Biol. 4, 241–247 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H. E., et al., Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature 415, 813–817 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Dai S. Y., et al., Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method. Proc. Natl. Acad. Sci. U.S.A. 105, 7171–7176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pike A. C., et al., Structural insights into the mode of action of a pure antiestrogen. Structure 9, 145–153 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Della Torre S., Non-alcoholic fatty liver disease as a canonical example of metabolic inflammatory-based liver disease showing a sex-specific prevalence: relevance of estrogen signaling. Front. Endocrinol. (Lausanne) 11, 572490 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stender J. D., et al., Structural and molecular mechanisms of cytokine-mediated endocrine resistance in human breast cancer cells. Mol. Cell 65, 1122–1135.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanning S. W., et al., The SERM/SERD bazedoxifene disrupts ESR1 helix 12 to overcome acquired hormone resistance in breast cancer cells. eLife 7, e37161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norris J. D., et al., Peptide antagonists of the human estrogen receptor. Science 285, 744–746 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Connor C. E., et al., Circumventing tamoxifen resistance in breast cancers using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer Res. 61, 2917–2922 (2001). [PubMed] [Google Scholar]
- 31.Blackmore J. K., et al., The SMRT coregulator enhances growth of estrogen receptor-α-positive breast cancer cells by promotion of cell cycle progression and inhibition of apoptosis. Endocrinology 155, 3251–3261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith C. L., et al., Elevated nuclear expression of the SMRT corepressor in breast cancer is associated with earlier tumor recurrence. Breast Cancer Res. Treat. 136, 253–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fanning S. W., et al., Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. eLife 5, e12792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahreini A., et al., Mutation site and context dependent effects of ESR1 mutation in genome-edited breast cancer cell models. Breast Cancer Res. 19, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toy W., et al., Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 7, 277–287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutsui S., Ohno S., Murakami S., Hachitanda Y., Oda S., Prognostic value of epidermal growth factor receptor (EGFR) and its relationship to the estrogen receptor status in 1029 patients with breast cancer. Breast Cancer Res. Treat. 71, 67–75 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Nicholson S., et al., Expression of epidermal growth factor receptors associated with lack of response to endocrine therapy in recurrent breast cancer. Lancet 1, 182–185 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Bruning J. B., et al., Coupling of receptor conformation and ligand orientation determine graded activity. Nat. Chem. Biol. 6, 837–843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katzenellenbogen J. A., Mayne C. G., Katzenellenbogen B. S., Greene G. L., Chandarlapaty S., Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat. Rev. Cancer 18, 377–388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nettles K. W., Greene G. L., Ligand control of coregulator recruitment to nuclear receptors. Annu. Rev. Physiol. 67, 309–333 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Pickar J. H., Boucher M., Morgenstern D., Tissue selective estrogen complex (TSEC): A review. Menopause 25, 1033–1045 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nwachukwu J. C., et al., Predictive features of ligand-specific signaling through the estrogen receptor. Mol. Syst. Biol. 12, 864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma N., et al., Exploring the structural compliancy versus specificity of the estrogen receptor using isomeric three-dimensional ligands. ACS Chem. Biol. 12, 494–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vonrhein C., et al., Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams P. D., et al., The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nwachukwu J. C., et al., Improved crystallographic structures using extensive combinatorial refinement. Structure 21, 1923–1930 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D., electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Debreczeni J. E., Emsley P., Handling ligands with Coot. Acta Crystallogr. D Biol. Crystallogr. 68, 425–430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joosten R. P., Long F., Murshudov G. N., Perrakis A., The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aarts J. M., et al., Robust array-based coregulator binding assay predicting ERα-agonist potency and generating binding profiles reflecting ligand structure. Chem. Res. Toxicol. 26, 336–346 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Chalmers M. J., et al., Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Traboulsi T., El Ezzy M., Dumeaux V., Audemard E., Mader S., Role of SUMOylation in differential ERα transcriptional repression by tamoxifen and fulvestrant in breast cancer cells. Oncogene 38, 1019–1037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proliferation source data are in Dataset S1. X-ray crystallography coordinates will be released upon acceptance. X-ray crystal structure data have been deposited in Research Collaboratory for Structural Bioinformatics Protein Data Bank (https://www.rcsb.org) and are available with the following accession numbers: 7RS4, 7RS9, 7RRZ, 7RS0, 7RRX, 7RRY, 7RS1, 7RS2, 7RS9, 7RS3, and 7RS7. All other study data are included in the article and/or supporting information.