Significance
Malaria parasites invade and replicate within human red blood cells, which lack nuclei and have minimal metabolic activity. To survive, the parasites create new pathways that alter the permeability of the red blood cell membrane, allowing them to import nutrients and export waste. Here, we present the native structure of the three-membered RhopH protein complex, which plays a key role in this process. We determined the structure of this essential complex from a heterogeneous mixture of proteins enriched directly from parasite cell lysate, using a cryo-electron microscopy–enabled endogenous structural proteomics approach. The native structure of the RhopH complex in a soluble, trafficking state helps elucidate the long-standing question of how parasite transmembrane proteins are trafficked to the erythrocyte membrane.
Keywords: cryo-EM, malaria, endogenous structural proteomics, trafficking
Abstract
The RhopH complex is implicated in malaria parasites’ ability to invade and create new permeability pathways in host erythrocytes, but its mechanisms remain poorly understood. Here, we enrich the endogenous RhopH complex in a native soluble form, comprising RhopH2, CLAG3.1, and RhopH3, directly from parasite cell lysates and determine its atomic structure using cryo–electron microscopy (cryo-EM), mass spectrometry, and the cryoID program. CLAG3.1 is positioned between RhopH2 and RhopH3, which both share substantial binding interfaces with CLAG3.1 but make minimal contacts with each other. The forces stabilizing individual subunits include 13 intramolecular disulfide bonds. Notably, CLAG3.1 residues 1210 to 1223, previously predicted to constitute a transmembrane helix, are embedded within a helical bundle formed by residues 979 to 1289 near the C terminus of CLAG3.1. Buried in the core of the RhopH complex and largely shielded from solvent, insertion of this putative transmembrane helix into the erythrocyte membrane would likely require a large conformational rearrangement. Given the unusually high disulfide content of the complex, it is possible that such a rearrangement could be initiated by the breakage of allosteric disulfide bonds, potentially triggered by interactions at the erythrocyte membrane. This first direct observation of an exported Plasmodium falciparum transmembrane protein—in a soluble, trafficking state and with atomic details of buried putative membrane-insertion helices—offers insights into the assembly and trafficking of RhopH and other parasite-derived complexes to the erythrocyte membrane. Our study demonstrates the potential the endogenous structural proteomics approach holds for elucidating the molecular mechanisms of hard-to-isolate complexes in their native, functional forms.
Nearly half the world’s population is at risk for contracting malaria. To keep the disease at bay, we rely heavily on artemisinin-based therapies, limiting malaria’s impact to 200 million cases and half a million deaths each year (1). The recently discovered de novo emergence of artemisinin-resistant malaria parasites in Africa heightens the urgent need to define, and thus target, molecular machineries essential for parasites’ survival in human erythrocytes, thereby facilitating the development of new therapeutics with novel modes of action (1–3). To survive inside human red blood cells, the malaria parasite Plasmodium falciparum deploys hundreds of effector proteins that extensively remodel the normally quiescent host erythrocyte, building new infrastructure for nutrient uptake, waste efflux, and immune evasion to support the active growth and replication of the parasite (4, 5). Members of the high-molecular mass rhoptry protein complex known as the RhopH complex—comprising CLAG3, RhopH2, and RhopH3—have been shown to play a key role in this process by directly or indirectly contributing to the activity of the Plasmodium surface anion channel (PSAC), a novel ion channel that appears at the cell surface in malaria parasite–infected erythrocytes and has been shown to mediate increased permeability of the erythrocyte plasma membrane to various solutes (6–18).
The mechanism underlying the role of the RhopH complex in PSAC activity remains poorly understood. How the components of the RhopH complex are trafficked from the parasite out to the erythrocyte membrane is also not known, although it has been hypothesized that they may assume a soluble form or associate with additional proteins that protect them as they traverse the host-cell cytosol (9). Another theory suggests that they may cross the cytosol via a series of membranous compartments created by the parasite, known as the exomembrane system (19, 20). High-resolution structures of the RhopH complex in either membrane-bound or soluble forms would help to answer these open questions and provide a much-needed structural framework to rationalize the large body of seemingly contradictory phenotypic and biochemical data that surround this intriguing complex.
To address the question of how members of the RhopH complex are delivered to the erythrocyte membrane, we sought to investigate whether a soluble complex containing one or more components of the RhopH complex exists in soluble parasite lysate and determine its structure if it did. Unfortunately, the extreme difficulty of producing properly folded and assembled P. falciparum protein complexes via reconstitution or coexpression in heterologous systems has thus far stymied efforts to obtain high-resolution structures of the RhopH complex. Furthermore, the uncertainty about the exact composition of a putative soluble RhopH complex precluded the use of a heterologous system to recapitulate the complex. To address challenges like this, we recently established an endogenous structural proteomics approach that combines the use of cryo–electron microscopy (cryo-EM), mass spectrometry, and cryoID, a program we developed to identify proteins in sub-4.0Å cryo-EM density maps, to identify and determine the structures of unknown protein complexes (Fig. 1) (21). Here, we have used this approach to obtain a near-atomic resolution structure of the unmodified RhopH complex in a soluble state, enriched directly from P. falciparum parasite lysate. The structure explains the body of existing biochemical data, provides insights into the dual roles of the RhopH complex in both invasion and creation of new permeability pathways in host erythrocytes, and sheds light on the long-standing question of how parasite transmembrane proteins are trafficked to the erythrocyte membrane. The study also demonstrates the power of the endogenous structural proteomics approach to provide exciting insights into the molecular mechanisms of hard-to-isolate complexes from challenging native sources where conventional structural biology approaches have failed.
Fig. 1.
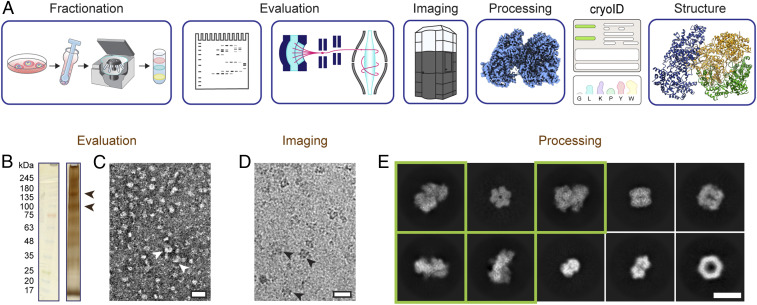
Endogenous structural proteomics workflow using cryoID. (A) Depiction of the workflow. P. falciparum parasites and parasitophorous vacuoles were saponin-released from P. falciparum–infected red blood cells, and the resulting parasite and vacuole pellets were lysed and fractionated using a sucrose gradient. The fractions were evaluated by SDS-PAGE, mass spectrometry, and negative stain EM. Cryo-EM imaging and analysis of the selected fraction yielded a 3.7-Å resolution cryo-EM density map. The proteins in the cryo-EM map were identified using cryoID and then modeled de novo, yielding the final atomic resolution structure of the RhopH complex. (B) Silver-stained SDS-PAGE of the selected fraction of the lysate. Arrowheads denote bands consistent with the molecular weights of the components of the RhopH complex. (C and D) Representative negative stain (C) and cryo-EM (D) micrographs of the selected fraction. (Scale bars, 20 nm.) Arrowheads denote particles consistent with 2D class averages of the RhopH complex. (E) Representative 2D class averages corresponding to multiple protein complexes present in the single dataset of cryo-EM micrographs. Class averages that ultimately gave rise to the structure of the RhopH complex presented here are boxed in green. (Scale bar, 10 nm.)
Results
Identification and Structure Determination of Soluble RhopH Complex.
Following an endogenous structural proteomics workflow (21) (Fig. 1A), we used mass spectrometry to identify fractions of P. falciparum parasite lysate that both looked promising by negative stain EM and contained one or more components of the RhopH complex (Fig. 1 B and C and Dataset S1). Analysis of a cryo-EM dataset collected from one of these fractions in RELION (22) and cryoSPARC (23) yielded numerous promising two-dimensional (2D) class averages (Fig. 1 D and E and SI Appendix, Fig. 1). We identified a subset of these class averages that gave rise to a near-atomic resolution cryo-EM density map of a novel, asymmetric protein complex at an overall resolution of 3.7 Å after ab initio reconstruction and nonuniform refinement in cryoSPARC (Fig. 2 and SI Appendix, Fig. 1).
Fig. 2.
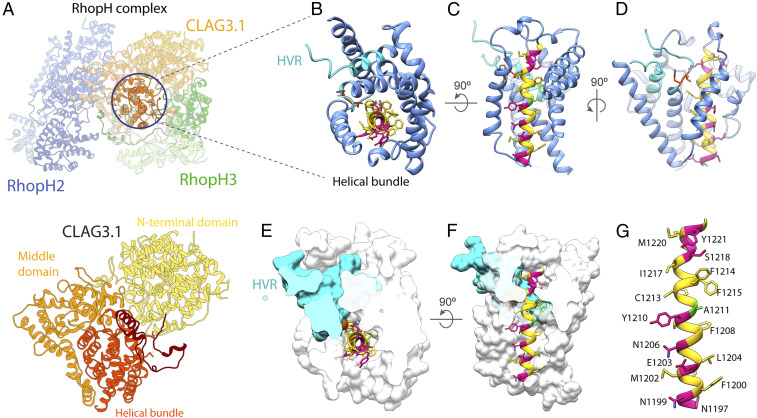
Cryo-EM density map of the soluble RhopH complex. (A) Cryo-EM density map of the RhopH complex viewed from multiple angles, with RhopH2, CLAG3.1, and RhopH3 colored in navy, gold, and green, respectively. Unmodeled regions are shown in gray. (B) Atomic model of the soluble RhopH complex, colored as in A, with the query segments used for protein identification by cryoID colored in pink. (C and D) Local resolution evaluation of the soluble RhopH complex map, calculated using Resmap (57), colored according to resolution, and displayed as a full surface (C) and central slice (D). (E–G) Detailed view of each set of query sequences used for cryoID identification for RhopH2 (E), CLAG3.1 (F), and RhopH3 (G), shown with corresponding cryo-EM density (mesh).
Following further focused classification and refinement in RELION to improve local resolutions throughout the map, we were able to use cryoID to successfully identify all three components of the complex as RhopH2, CLAG3.1, and RhopH3, the three members of the RhopH complex (Fig. 2 B and E–G and SI Appendix, Fig. 1 and Movies 1–3). Clear side-chain densities throughout most of the map enabled us to build de novo atomic models of the three components (Fig. 2B and SI Appendix, Fig. 2), revealing a tripartite protein complex containing a single copy of each of the three proteins, with a total calculated mass of 435 kDa, assuming no posttranslational truncations.
Although the CLAG3.1 and CLAG3.2 isoforms share 96% protein sequence identity, there is a known hypervariable region (10) (HVR) (F1094 to G1142), where CLAG3.1 in NF54 parasites includes an extra two amino acids (a.a.) (T1140 to H1141) that are not found in CLAG3.2 in NF54 parasites. Fortunately, we had strong density in this region that enabled us to definitively resolve that the isoform in our map is CLAG3.1 (SI Appendix, Fig. 3), consistent with previous findings that suggest CLAG3.1 is preferentially expressed in culture (24). The overall map quality was also such that we were able to model the bulk of the complex’s visible density, the main exception being a small auxiliary domain near the C terminus of RhopH2, which at a lower density threshold value (σ = 0.0355) accounts for ∼6.9% of our complex’s total volume (SI Appendix, Fig. 4). Though the density here was of insufficient quality to register residues, we were able to discern that the region mainly comprises several twisted β-strands constituting a β-sheet–rich core.
CLAG3.1 forms the core of the complex and is situated between RhopH2 and RhopH3 (Fig. 2 A and B and Movie 4). RhopH2 and RhopH3 both share substantial binding interfaces with CLAG3.1 but make minimal contacts with each other (Movie 4). Buried and exposed surface areas were calculated using PDBePISA (25). Of the two, the CLAG3.1–RhopH3 interface is more extensive, accounting for a buried surface area of 5,416 Å2 (∼11 to 15% of the total exposed surface area of either CLAG3.1 or RhopH3), compared with a buried surface area of 3,410 Å2 for the CLAG3.1–RhopH2 interface (∼7 to 7.5% of the total exposed surface area of either CLAG3.1 or RhopH2). All three protein components contain an unusually high abundance of cysteines in their primary sequence (13, 10.5, and 15.6% in RhopH2, CLAG3.1, and RhopH3, respectively), which is particularly striking given the relatively low presence of cysteines in the P. falciparum proteome (∼1.7%). Our CLAG3.1 structure contains three sets of disulfide bonds, while RhopH2 and RhopH3 contain five sets of disulfide bonds each (SI Appendix, Fig. 5).
RhopH3 Architecture Explains RhopH3 Truncation Phenotypes.
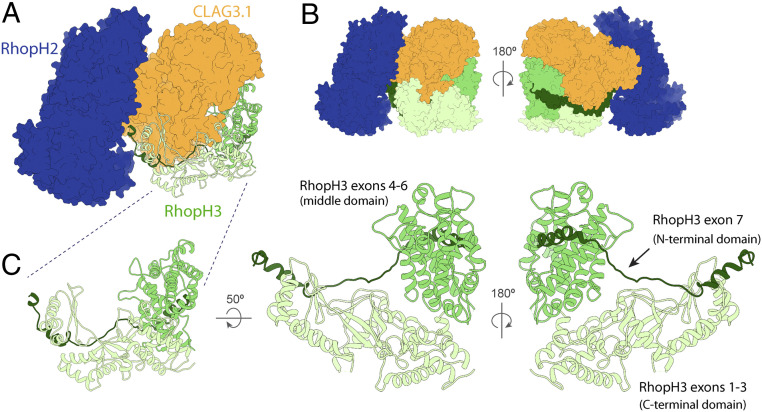
RhopH3 is an 897 a.a. protein, whose visible structure can be divided into three domains (Fig. 3): an elongated N-terminal domain arising from RhopH3 exons 1 to 3 (corresponding to a.a. 1 to 378), composed of a set of interleaved β-sheets flanked by α-helices; a globular α-helix–rich “middle domain” from RhopH3 exons 4 to 6 (a.a. 379 to 675); and an extended C-terminal domain arising from RhopH3 exon 7, characterized by a long loop-rich segment that extends toward RhopH2 (Fig. 3C). The first N-terminal 25 residues were not modeled due to disordered density, nor were residues after a.a. 738, although scattered weak “pigtail” density can be observed after a.a. 738 further extending into a cleft in the RhopH complex between the major bodies of RhopH2 and RhopH3 (SI Appendix, Fig. 4E). The N and C termini of the N-terminal and C-terminal domains, respectively, are positioned adjacent to the RhopH2–CLAG3.1 interface, unlike the middle domain, which is located on the far side of CLAG3.1 (Fig. 3 A and C). This explains previous studies suggesting that exons 1 to 3 and exon 7 help to form the interaction between CLAG3 and RhopH2, while the RhopH3 exons 4 to 6 do not appear to play a role in CLAG3–RhopH2 binding but rather are required for trafficking to the rhoptries (9, 12).
Fig. 3.
Structural details of RhopH3. (A) RhopH3 (ribbon diagram) and RhopH2 and CLAG3.1 (space-filling surface diagram in different blue and orange, respectively) showing the interfaces between RhopH3 and the other components of the RhopH complex. (B) Front and back views showing the arrangement of RhopH3 domains. (C) Multiple views of the RhopH3 N-terminal, middle, and C-terminal domains, corresponding to RhopH3 exons 1 to 3, 4 to 6, and 7, and colored mint, green, and forest, respectively.
The aforementioned abundance of cysteines in the RhopH complex plays an especially important role in the structural organization of RhopH3. Notwithstanding the unmodeled auxiliary domain of RhopH2, RhopH3 is unique among the constituents of the RhopH complex in possessing the only β-sheets within the main complex; in contrast, RhopH2 and CLAG3.1 are dominated by α-helices. Three sets of β-sheets present in RhopH3’s N-terminal domain give rise to an interleaved β-sheet motif, largely facilitated by a 20 residue–long continuous loop (a.a. 225 to 245) that contributes a single β-strand to all three sets of β-sheets, effectively chaining the three β-sheets together (SI Appendix, Fig. 6). Notably, this 20 residue continuous loop contains two cysteine residues, Cys231 and Cys244, each of which participates in a disulfide bond (with Cys157 [from an adjacent loop that also contributes β-strands to the interleaved β-sheets] and Cys253 of RhopH3, respectively) in a manner that could fix this structurally critical β-strand–containing loop in place (SI Appendix, Figs. 5 and 6). We therefore speculate that these cysteine residues are of vital importance to the proper folding and assembly of RhopH3.
Structural Details of RhopH2.
Of the three components of the RhopH complex, RhopH2 is the least studied. We modeled 972 residues of the 1378 a.a. in RhopH2 and five disulfide bonds. Our structure reveals that RhopH2 is an elongated globular protein comprised primarily of α-helices and loops, although a small unmodeled domain putatively extending from RhopH2's C terminus is of sufficient resolution to see that it is rich in β-sheets. Within the tripartite RhopH complex, RhopH2 forms the smaller of two lobes and interfaces with CLAG3.1 solely through residues 385 to 830.
Putative Transmembrane and Extracellular Elements of CLAG3.1 Are Buried.
The 1,209 residues of CLAG3.1 in our structure can also be divided into three domains (Fig. 4A). Residues 52 to 675 form a globular N-terminal domain that shares an interface with the RhopH3 “middle domain” (Fig. 4). Residues 688 to 978 form a squid-shaped “middle” domain, with the head sharing an interface with RhopH2, while the legs wrap around the third CLAG3.1 domain—a helical bundle comprising residues 979 to 1289 (Fig. 4A).
Fig. 4.
Structural details of CLAG3.1. (A) Atomic model of CLAG3.1 shown as a ribbon. (B and C) Detailed views of the CLAG3.1 helical bundle and HVR from the top (B) and side (C). One helix in front is removed for clarity in C. The helical bundle is shown in cornflower blue and the HVR in cyan. For CLAG3.1 residues N1197 to A1222, hydrophobic residues, charged and polar residues, and A1211 are shown in yellow, pink, and green, respectively. The CLAG3.1-specific T1140 to H1141 residues are shown in orange. (D) View rotated 90° from C, better showing the HVR. (E and F) Cutaway space filling top (E) and side (F) views of the helical bundle and HVR, except residues N1197 to A1222, which are displayed in ribbon diagram. (G) Detailed view of CLAG3.1 N1197 to A1222.
CLAG3 has been demonstrated to modulate PSAC activity (10, 26), and it has been postulated that CLAG3 may be a major or sole component of the PSAC ion-conducting pore (10). Based on membrane protein prediction software and helical wheel analysis, CLAG3.2 residues 1203 to 1223 were hypothesized to constitute a transmembrane domain, with F1200 to S1217 predicted to form an amphipathic helix that oligomerizes with the same helix from multiple CLAG3.2 monomers to form the transmembrane pore of PSAC, similar to oligomeric pore-forming bacterial toxins (11, 27). However, recent CLAG3.1/3.2 knockdown and complete knockout studies suggest that CLAG3 isoforms cannot be the sole component of the PSAC transmembrane pore (8, 28).
In our structure, CLAG3.1 residues 1210 to 1223, previously predicted to constitute a transmembrane helix, are embedded in the middle of the helical bundle formed by residues 979 to 1289, near the C terminus of CLAG3.1 (Fig. 4 B–G). As such, they are buried in the core of the RhopH complex and mostly shielded from the solvent by the helical bundle (Fig. 4 B–G). Inspection of the residues corresponding to the putative transmembrane helix (a.a. 1200 to 1223) in the structure reveals that F1200 to S1217 does form an α-helix, but it is not amphipathic across the entire length of that sequence as was previously predicted (Fig. 4G) (11). The amphipathic portion of the helix (N1199 to Y1210) spans three turns (11 residues) and measures ∼15 Å in length (Fig. 4G), 4 to 7 residues shorter than the typical minimum length of a transmembrane helix. As typical biological membranes range from 20 to 40 Å in thickness, it is possible that the predicted transmembrane helix either fails to reach all the way across the membrane or may have to form a coiled-coil interaction with another helix in order to do so. The polar face of the N-terminal 3 turns of this helix, comprising residues N1199, E1203, N1206, and possibly Y1210, are solvent exposed and contribute to the outward, solvent-facing surface of the helical bundle (Movies 5 and 6). All other residues in the N1199 to A1222 helix are buried.
Residues Y1093 to G1142 constitute the HVR previously shown to be exposed on the surface of infected red blood cells in membrane-associated CLAG3.1/3.2 (10) (SI Appendix, Fig. 3). In our structure, this region forms the ends of two helices in the helical bundle, as well as the long loop between them, which extends up from the middle of the CLAG3.1 C-terminal helical bundle through the CLAG3.1–RhopH3 N-terminal domain interface (Fig. 4 and Movie 7). There are 20 residues in the middle of the loop that are unstructured in our density map. Intriguingly, most of the HVR that we can see in our structure is buried deep in the middle of the helical bundle, shielded from solvent by both the CLAG3.1 helical bundle and the RhopH3 N-terminal domain (Fig. 4 B–G and Movies 5–7).
Discussion
High-resolution structures provide invaluable frameworks for interpreting the wealth of biochemical and genetic data surrounding protein complexes implicated in the pathogenesis of malaria parasites (29). Unfortunately, structural studies in malaria have been notoriously difficult (30). Many P. falciparum proteins are extremely challenging to overexpress recombinantly, with multiprotein complexes presenting a particular challenge due to the difficulty of recapitulating proper folding and assembly via reconstitution or coexpression in heterologous systems. Using our endogenous structural proteomics approach (21), we circumvented these obstacles to determine a near-atomic structure of the native RhopH complex, enriched directly from parasite-infected red blood cells. A recent paper (31) reported an atomic structure of the RhopH complex. Their structure was obtained from proteins purified using engineered tags. The two structures are largely consistent with each other, but our native complex contains a different CLAG isoform (CLAG3.1 versus CLAG 3.2), and we were able to model an additional 459 a.a. residues in the RhopH2 subunit of the native complex. This discovery of a hypothesized—but hitherto unobserved—soluble state of the native RhopH complex is a direct observation of an exported P. falciparum transmembrane protein in its soluble, trafficking state. This represents a major step forward in addressing the long-standing question of how parasite effectors, many of which are integral membrane proteins (4, 5), are trafficked to their final site of action in the erythrocyte membrane.
The structure provides exciting insights into how trafficking works, revealing that previously predicted transmembrane and extracellular elements are buried in the middle of the complex, likely to protect these elements from the aqueous cytosolic environment during transit. Taken into account with all the data we have thus far (Movie 8) (8–19, 24, 26, 28, 32, 33), this discovery seems to point toward a model in which CLAG3, RhopH2, and RhopH3 associate early in the secretory pathway (8–10, 12), dissociating only for translocation across the parasitophorous vacuole membrane through PTEX (9, 34) and reforming the soluble complex state in the erythrocyte cytosol to complete the journey out to the erythrocyte surface (9). This suggests that the RhopH complex shares the ability of pathogenic pore-forming proteins to transition from a soluble form for traversing aqueous cytosolic environments to an integral transmembrane protein form upon arriving at its target membrane (27, 35, 36) and that such transitions may represent a general strategy employed by the parasite to deliver protein complexes to the erythrocyte membrane.
The ability to transition from a soluble form for traversing aqueous cytosolic or extracellular environments to an integral transmembrane protein form upon arriving at the target membrane is the unifying key feature shared by pore-forming proteins in biological systems ranging from bacteria to vertebrates (36). As such, there is an argument for considering the RhopH complex to be a pore-forming protein complex. While the use of pore-forming proteins to unrestrictively permeabilize and lyse host-cell membranes during invasion and egress is a well-studied strategy commonly used by a wide variety of pathogens, including P. falciparum itself (27, 36–38), the use of pore-forming proteins to selectively alter host-cell permeability to particular ions or solutes without destroying the membrane is less common and less well understood. This strategy is rare in bacteria but essential for the function of several enveloped viruses, including influenza and HIV (36). In this respect, P. falciparum’s use of the RhopH/PSAC complex to modulate host erythrocyte membrane permeability for nutrient acquisition is more similar to how viruses use viroporins to establish ion-specific permeability across host-cell membranes, making the RhopH complex distinct from the more common use of pore-forming proteins by malaria parasites to perforate host-cell membranes during invasion and egress (38). Similar to most viroporins (35), the vast majority of the RhopH complex is located intracellularly, with only the HVR exposed on the erythrocyte membrane surface (10). The mechanisms underlying viroporins’ transitions from soluble to membrane bound, often triggered by a change in pH or interaction with a lipid or protein in the target membrane, may hold hints about the possible mechanism by which elements of CLAG3.1 are inserted into the erythrocyte membrane. Given the unusual abundance of both cysteines and disulfide bonds in the RhopH complex, we speculated that the large conformational change the RhopH complex would need to undergo at the membrane to expose and insert the putative transmembrane and extracellular elements into the erythrocyte membrane might be triggered or regulated by allosteric disulfide bonds.
First discovered more than a decade ago, the formation and breaking of allosteric disulfide bonds have been firmly established as a strategy used in a wide range of biological systems for regulating and modulating protein structure and function (39, 40). Indeed, when we examined the 13 disulfide bonds in our structure, we found three that exhibited the well-defined strained geometry that has been found to be the hallmark of all allosteric disulfide bonds characterized thus far (SI Appendix, Table S1) (39)—one in RhopH2, and two in RhopH3 (SI Appendix, Fig. 5). As such, it is possible that these bonds may break or be cleaved upon interaction with the erythrocyte membrane, triggering a conformation change that extrudes the putative transmembrane and extracellular elements from the middle of the complex and drives them through membrane. While no parasite-derived protein disulfide isomerases (PDIs) have been identified in the P. falciparum exportome, human erythrocytes do carry active PDIs in the plasma membrane (41, 42), which could cleave the allosteric disulfides and trigger the conformational change upon coming in contact with the RhopH complex at the membrane. Indeed, previous studies have shown that several human PDI inhibitors appear to exhibit anti–P. falciparum activity, although their IC50 values vary (43). Alternatively, changes in redox potential in the cytosol could trigger the breakage of the disulfide bonds to effect the massive conformational changes needed to expose and bring the helical bundle of CLAG3 to interact with red blood cell membrane (Fig. 5 B and C).
Fig. 5.
Model of CLAG3.1 putative transmembrane domain and HVR insertion into the erythrocyte membrane. (A–C) Schematic illustrating a model for how a conformational change in the RhopH complex may drive insertion of the extracellular and putative transmembrane elements into the red blood cell plasma membrane.
Although surface-exposed membrane proteins with essential functions in pathogenesis are often popular targets for drug development, these proteins have thus far proven to be difficult targets for drug and vaccine development in P. falciparum, due to various mechanisms the parasite employs to evade immune detection. If the use of a soluble form is a general strategy employed by the parasite to deliver proteins to the erythrocyte membrane, high-resolution structures of the soluble trafficking forms of these essential membrane protein complexes present an opportunity to approach development of therapies from a new angle, focused on blocking assembly or trafficking of complexes before they have a chance to insert into the erythrocyte membrane. As such, the approach presented here for overcoming the barriers to high-resolution structural studies in malaria parasites opens the door for developing new anti-malarial therapies with novel modes of action and can also be applied to hard-to-isolate complexes in other biological systems to answer long-standing questions.
Materials and Methods
Parasite Culture.
P. falciparum cultures were prepared as described previously (21, 29). Erythrocytes were then lysed with 0.0125% saponin (Sigma, sapogenin content ≥ 10%) in cold phosphate-buffered saline (PBS) with an EDTA-free protease inhibitor mixture (Roche or Pierce). The released P. falciparum parasites were then washed in cold PBS with EDTA-free protease inhibitor mixture. The washed cell pellets were flash frozen in liquid nitrogen and stored at −80 °C.
Sucrose Gradient Fractionation P. falciparum Parasite Lysate and Subsequent Evaluation of Fractions Using Negative Stain EM.
Frozen parasite pellets were resuspended in lysis buffer (25mM Hepes pH7.4, 150mM KCl, 10mM MgCl2, 10% glycerol) and lysed using a glass Dounce tissue homogenizer. The soluble lysates were separated from the membrane fraction by centrifuging at 100,000 g for 1 h and then fractionated on a sucrose gradient as previously described (21). The resulting fractions were evaluated for the presence and relative abundance of proteins of interest as described previously (21). Briefly, fractions were assessed for the abundance of potential particles of interest based on silver-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), tryptic digest liquid chromatography–mass spectrometry, and negative stain EM. The conventional approach of using gel filtration chromatography to assess sample quality was eschewed in favor of using negative stain EM to directly visualize and evaluate the abundance of intact particles giving rise to promising 2D class averages with distinct features. As such, small datasets of ∼100,000 particles were collected for each fraction, and 2D class averages generated in RELION (22, 44) were used to identify fractions containing promising particles (Fig. 1E).
Mass Spectrometry.
Mass spectrometry was performed as previously described (21). The proteomics data will be deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the MassIVE partner repository.
Cryo-EM.
Cryo-EM grids of the selected fraction of parasite lysate were prepared and imaged as described previously (21), yielding a final dataset consisting of a total of 19,449 movies.
Image Processing and Three-Dimensional Reconstruction.
Frames in each movie were aligned, gain reference corrected, and dose weighted to generate a micrograph using MotionCor2 (45) (SI Appendix, Fig. 1). Micrographs that were aligned without dose weighting were also generated for contrast transfer function estimation using CTFFIND4 (46) and automated particle picking in Gautomatch (47).
A total of 2,900,167 particles were extracted from 19,449 micrographs. Two to three rounds of reference-free 2D classification in RELION were used to exclude junk particles. In total, 858,049 particles belonging to 2D class averages exhibiting clear secondary structure features were further classified in CryoSPARC (23). A subset of the 2D class averages, corresponding to 108,872 particles, were identified as likely to have originated from the same asymmetric volume. These 108,872 particles were subjected to an unsupervised single-class ab initio three-dimensional reconstruction followed by a single-class homogenous refinement using C1 symmetry in CryoSPARC. The resulting reconstruction was then subjected to nonuniform refinement using C1 symmetry in CryoSPARC, yielding a density map with a final overall resolution of 3.72 Å (Fig. 2 C and D and SI Appendix, Fig. 7). The 108,872 particles were then subjected to focused classification and refinement in RELION to reach sufficient local resolutions throughout the map to allow successful identification of all three proteins in the map using cryoID.
Identifying Proteins in the Cryo-EM Density Map Using CryoID.
We ran the cryoID Get_queries subprogram (21) on the initial 3.7-Å resolution density map obtained from refinement and postprocessing in RELION, using default symmetry (Any) and a high-resolution limit of 3.6 Å as the input parameters. We then manually inspected the resulting query models, correcting residues incorrectly assigned by Get_queries and extending the queries on both ends as the density permitted (Fig. 2E). This yielded the following degenerate sequences, which were then used for searching:
Query Set 1:
-
1)
GLGYGYYGGLGYYGGGLYYYLLXYYG;
-
2)
LLGGPPGGXGYLYLLGLGLGYYYXLYLL-GXGGYYYXXY.
Using this set of query sequences, cryoID identified two candidates for the protein in this region of the density map from a candidate pool consisting of the 1,170 proteins identified in this sucrose gradient fraction by mass spectrometry (SI Appendix, Table S2). The two candidates, CLAG3.1 and CLAG3.2 from P. falciparum (O77309/ O77310), share 96% protein sequence identity. We confirmed the identification by manually building a de novo atomic model into the rest of the map (Fig. 2B). Focused classification and refinement were used to improve the local resolution around the region corresponding to residues 1110 to 1150, one of the few segments in which CLAG3.1 and CLAG3.2 differ in sequence. The improved resolution was sufficient for us to confidently determine that the sequence in this region matches CLAG3.1 rather than CLAG3.2 based on the presence of two residues, T1140 and H1141, which are found in CLAG3.1 but not CLAG3.2, confirming that the protein in the complex is CLAG3.1 (SI Appendix, Fig. 3).
We completed a backbone trace of the remaining regions of the density map that did not correspond to CLAG3.1, which allowed us to determine that there were likely two additional, distinct polypeptide chains represented in the density map. We then used Get_queries to generate queries from these two remaining regions of the density map, yielding two additional sets of queries (Fig. 2 F and G). We manually inspected the query models, correcting residues incorrectly assigned by Get_queries and extending the queries on both ends as the density permitted. This yielded the following degenerate sequences, which were then used for searching:
Query Set 2:
-
1)
GLYLKKGGGGWGYLLKGLGLYGYLLGLLG;
-
2)
GLGLGLGYGYGLGGKL;
-
3)
GYGLGYLLYLGLGGLL.
Query Set 3:
-
1)
GYYGGGLLLGGYGLGYYLGKYLL;
-
2)
YGKGGGGGGLYGGGGLY.
Using these two sets of query sequences, cryoID identified the proteins in the two regions of the density map, from a candidate pool consisting of the 1,170 proteins identified in this sucrose gradient fraction by mass spectrometry, to be RhopH2 (Query Set 2) and RhopH3 (Query Set 3) from P. falciparum (C0H571 and Q8I395) (SI Appendix, Tables S3 and S4). We subsequently confirmed the identification by manually building de novo atomic models into the corresponding two regions of the map (Fig. 2B).
Manual Model Building and Refinement.
Map interpretation was performed with UCSF Chimera (48) and COOT (49). P. falciparum protein sequences were obtained from the National Center for Biotechnology Information (50) and the PlasmoDB (51) protein databases. Initial sequence registrations during model building for all three proteins in the map were determined using cryoID (21). PHYRE2 (52) secondary structure predictions were also used as a guide during subsequent model building. Each residue in the three proteins was manually traced and built de novo in COOT, using the best focused classification map for each local area. Rotamers for residues were manually selected as assessed by residue-to-map fit using the select_rotamer option in COOT.
Map resolution outside of core regions in the complex were not sufficient to permit the use of real_space_refine_zone found in COOT. As such, manual refinement targeting both protein geometry and fit with the density map was carried out using COOT’s regularize_zone and fixed_atoms_for_refinement functions. Finally, resulting models for the complexes were subjected to iterative cycles of automated refinement using the phenix.real_space_refine program in PHENIX (53) followed by further manual refinement, performed against a map visually determined to possess the best mix of overall features for each local region, to achieve the final structure.
All figures and movies were prepared with UCSF Chimera, Pymol (54), and Resmap (55). Molprobity (56) was used to validate the stereochemistry of the final models.
Supplementary Material
Acknowledgments
This research was supported in part by grants from NIH (R01GM071940, AI094386, and DE025567 to Z.H.Z., 1DP5OD029613 to C.-M.H., and K99/R00 HL133453 to J.R.B.). C.-M.H. acknowledges funding from the Ruth L. Kirschstein National Research Service Award (AI007323). We thank the University of California, Los Angeles (UCLA) Proteome Research Center for assistance in mass spectrometry and acknowledge the use of resources in the Electron Imaging Center for Nanomachines supported by UCLA and grants from the NIH (S10RR23057, S10OD018111, and U24GM116792) and NSF (DBI-1338135 and DMR-1548924).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100514118/-/DCSupplemental.
Data Availability
The atomic models and the cryo-EM density maps have been deposited in the Protein Data Bank and the Electron Microscopy Data Bank under the accession nos. 7MRW and EMD-23959, respectively.
References
- 1.2019 GWHO, World malaria report 2019. (World Health Organization, Geneva, 2019).
- 2.Uwimana A., et al., Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 26, 1602–1608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui F. A., et al., Role of Plasmodium falciparum Kelch 13 protein Mutations in P. falciparum populations from Northeastern Myanmar in mediating artemisinin resistance. MBio 11, e01134-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddey J. A., Cowman A. F., Plasmodium nesting: Remaking the erythrocyte from the inside out. Annu. Rev. Microbiol. 67, 243–269 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Spillman N. J., Beck J. R., Goldberg D. E., Protein export into malaria parasite-infected erythrocytes: Mechanisms and functional consequences. Annu. Rev. Biochem. 84, 813–841 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Alkhalil A., et al., Plasmodium falciparum likely encodes the principal anion channel on infected human erythrocytes. Blood 104, 4279–4286 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Desai S. A., Bezrukov S. M., Zimmerberg J., A voltage-dependent channel involved in nutrient uptake by red blood cells infected with the malaria parasite. Nature 406, 1001–1005 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Gupta A., et al., Complex nutrient channel phenotypes despite Mendelian inheritance in a Plasmodium falciparum genetic cross. PLoS Pathog. 16, e1008363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito D., Schureck M. A., Desai S. A., An essential dual-function complex mediates erythrocyte invasion and channel-mediated nutrient uptake in malaria parasites. eLife 6, e23485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguitragool W., et al., Malaria parasite CLAG3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 145, 665–677 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma P., Rayavara K., Ito D., Basore K., Desai S. A. A., A CLAG3 mutation in an amphipathic transmembrane domain alters malaria parasite nutrient channels and confers leupeptin resistance. Infect. Immun. 83, 2566–2574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherling E. S., et al., The Plasmodium falciparum rhoptry protein RhopH3 plays essential roles in host cell invasion and nutrient uptake. eLife 6, e23239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper J. A., et al., The 140/130/105 kilodalton protein complex in the rhoptries of Plasmodium falciparum consists of discrete polypeptides. Mol. Biochem. Parasitol. 29, 251–260 (1988). [DOI] [PubMed] [Google Scholar]
- 14.Brown H. J., Coppel R. L., Primary structure of a Plasmodium falciparum rhoptry antigen. Mol. Biochem. Parasitol. 49, 99–110 (1991). [DOI] [PubMed] [Google Scholar]
- 15.Kaneko O., et al., The high molecular mass rhoptry protein, RhopH1, is encoded by members of the clag multigene family in Plasmodium falciparum and Plasmodium yoelii. Mol. Biochem. Parasitol. 118, 223–231 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Sam-Yellowe T. Y., Shio H., Perkins M. E., Secretion of Plasmodium falciparum rhoptry protein into the plasma membrane of host erythrocytes. J. Cell Biol. 106, 1507–1513 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling I. T., et al., Characterisation of the rhoph2 gene of Plasmodium falciparum and Plasmodium yoelii. Mol. Biochem. Parasitol. 127, 47–57 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Counihan N. A., et al., Plasmodium falciparum parasites deploy RhopH2 into the host erythrocyte to obtain nutrients, grow and replicate. eLife 6, e23217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vincensini L., Fall G., Berry L., Blisnick T., Braun Breton C., The RhopH complex is transferred to the host cell cytoplasm following red blood cell invasion by Plasmodium falciparum. Mol. Biochem. Parasitol. 160, 81–89 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Sherling E. S., van Ooij C., Host cell remodeling by pathogens: The exomembrane system in Plasmodium-infected erythrocytes. FEMS Microbiol. Rev. 40, 701–721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho C. M., et al., Bottom-up structural proteomics: CryoEM of protein complexes enriched from the cellular milieu. Nat. Methods 17, 79–85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Mira-Martínez S., et al., Expression of the Plasmodium falciparum clonally variant clag3 genes in human infections. J. Infect. Dis. 215, 938–945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Gupta A., et al., CLAG3 self-associates in malaria parasites and quantitatively determines nutrient uptake channels at the host membrane. MBio 9, e02293-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dal Peraro M., van der Goot F. G., Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 14, 77–92 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Mira-Martínez S., et al., Identification of antimalarial compounds that require CLAG3 for their uptake by Plasmodium falciparum-infected erythrocytes. Antimicrob. Agents Chemother. 63, e00052-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho C. M., et al. Malaria parasite translocon structure and mechanism of effector export. Nature 561, 70–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berman H. M., et al., The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schureck M. A., et al., Malaria parasites use a soluble RhopH complex for erythrocyte invasion and an integral form for nutrient uptake. eLife 10, e65282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell G. H., Miller L. H., Hudson D., Franco E. L., Andrysiak P. M., Monoclonal antibody characterization of Plasmodium falciparum antigens. Am. J. Trop. Med. Hyg. 33, 1051–1054 (1984). [DOI] [PubMed] [Google Scholar]
- 33.Holder A. A., Freeman R. R., Uni S., Aikawa M., Isolation of a Plasmodium falciparum rhoptry protein. Mol. Biochem. Parasitol. 14, 293–303 (1985). [DOI] [PubMed] [Google Scholar]
- 34.Beck J. R., Muralidharan V., Oksman A., Goldberg D. E., PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature 511, 592–595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez M. E., Carrasco L., Viroporins. FEBS Lett. 552, 28–34 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Martinac B., Saimi Y., Kung C., Ion channels in microbes. Physiol. Rev. 88, 1449–1490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardenbrook N. J., et al., Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors. Nat. Commun. 11, 840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horta M. F., Pore-forming proteins in pathogenic protozoan parasites. Trends Microbiol. 5, 363–366 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Chiu J., Hogg P. J., Allosteric disulfides: Sophisticated molecular structures enabling flexible protein regulation. J. Biol. Chem. 294, 2949–2960 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt B., Ho L., Hogg P. J., Allosteric disulfide bonds. Biochemistry 45, 7429–7433 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Kallakunta V. M., Slama-Schwok A., Mutus B., Protein disulfide isomerase may facilitate the efflux of nitrite derived S-nitrosothiols from red blood cells. Redox Biol. 1, 373–380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prado G. N., Romero J. R., Rivera A., Endothelin-1 receptor antagonists regulate cell surface-associated protein disulfide isomerase in sickle cell disease. FASEB J. 27, 4619–4629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cobb D. W., et al., A redox-active crosslinker reveals an essential and inhibitable oxidative folding network in the endoplasmic reticulum of malaria parasites. PLoS Pathog. 17, e1009293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheres S. H. W., A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang K., Gautomatch: A GPU-accelerated program for accurate, fast, flexible and fully automatic particle picking from cryo-EM micrographs with or without templates. https://www2.mrc-lmb.cam.ac.uk/download/gautomatch-056/. Accessed 23 August 2021.
- 48.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.NCBI Resource Coordinators , Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44 (D1), D7–D19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aurrecoechea C., et al., PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schrodinger, LLC , The PyMOL Molecular Graphics System, Version 1.8. (2015).
- 55.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EMEM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen V. B., et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swint-Kruse L., Brown C. S., Resmap: Automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic models and the cryo-EM density maps have been deposited in the Protein Data Bank and the Electron Microscopy Data Bank under the accession nos. 7MRW and EMD-23959, respectively.