SUMMARY
53BP1 influences genome stability via two independent mechanisms: (i) regulating DNA double-strand break (DSB) repair, (ii) enhancing p53 activity. We discovered a protein, Tudor-interacting repair regulator (TIRR), that associates with the 53BP1 Tudor domain and prevents its recruitment to DSBs. Here we elucidate how TIRR impacts 53BP1 function beyond its recruitment to DSBs and biochemically links the two distinct roles of 53BP1. Loss of TIRR causes an aberrant increase in p53’s gene transactivation function, impacting several p53-mediated cell-fate programs. TIRR inhibits the complex formation between the Tudor domain of 53BP1 and a dimethylated form of p53 (K382me2) that is poised for transcriptional activation of its target genes. TIRR mRNA expression levels negatively correlate with expression of key p53 target genes in breast and prostate cancers, and loss of TIRR is selectively not tolerated in p53-proficient tumors. Therefore, we establish that TIRR is an important inhibitor of the 53BP1-p53 complex.
INTRODUCTION
The p53-binding protein 1 (53BP1) is pivotal in maintaining the balance of competing DNA double-strand break (DSB) repair pathways which is crucial for genomic stability (Chapman et al., 2012, Zimmermann and de Lange, 2014). The function of 53BP1 beyond DSB repair has recently emerged in the context of its interaction with p53 (Durocher and Pelletier, 2016). Perturbing ‘normal’ mitosis caused a p53-dependent cell cycle arrest which was shown to be dependent on 53BP1 and its known interactor USP28 (Fong et al., 2016, Lambrus et al., 2016, Meitinger et al., 2016). The proposed mechanism was that 53BP1 influenced the stability and localization of nuclear p53. A parallel study reported that ionizing radiation (IR) or Nutlin-3 treatment of 53BP1-deficient cells did not alter p53 protein levels but significantly diminished the transcriptional activation of several p53 target genes including cell cycle (CDKN1A/p21), auto-regulatory (MDM2), and pro-apoptotic (BAX, PUMA/BBC3) genes (Cuella-Martin et al., 2016a). This p53-mediated transactivation was mediated by the tandem-BRCT domain of 53BP1. However, structural studies have revealed that two independent domains of 53BP1 (tandem Tudor and tandem BRCT, referred to as Tudor and BRCT hereafter) directly bind p53 with comparable affinity (Derbyshire et al., 2002, Joo et al., 2002, Roy et al., 2010, Tong et al., 2015). The role of Tudor, in the context of p53 transactivation, remained unclear. Together these studies emphasized the importance of 53BP1 in p53 function and highlighted the fundamental gap in our knowledge regarding the molecular mechanism and physiological relevance of 53BP1-mediated regulation of p53.
We and others recently identified TIRR (Tudor Interacting Repair Regulator), as a direct interactor and regulator of 53BP1 function in DSB repair (Drane et al., 2017, Zhang et al., 2017). 53BP1 recruitment to DSBs by recognition of histone H4 dimethylated at lysine K20 (H4K20me2) is inhibited by TIRR binding to the Tudor of 53BP1 (Botuyan et al., 2018a, Dai et al., 2018, Wang et al., 2018). An important yet unanswered question is whether TIRR impacts the functions of 53BP1 that are not reliant upon its recruitment to DSBs. Here, we show that the loss of endogenous TIRR causes an aberrant increase in p53-mediated gene transactivation under different conditions of cellular stress. Phenotypically, TIRR deficiency results in decreased cell survival, higher senescence, and an increase in CDKN1A(p21)-mediated checkpoint arrest. From the mechanistic standpoint, TIRR modulates the stress-induced interaction of 53BP1 and p53. Using Nuclear Magnetic Resonance (NMR) spectroscopy, we demonstrate that TIRR competes with p53 dimethylated at residue K382 (p53K382me2), for the Tudor domain of 53BP1. We further show that the impact of TIRR on p53 is specifically dependent on SET8-mediated methylation of p53. Analysis of cancer genomes reveals that high TIRR expression levels negatively correlates with expression of key p53 targets, thereby mimicking diminished p53 activity. Furthermore, amplification of the TIRR genomic locus was mutually exclusive from TP53 mutation/deletions and amplifications of the MDM2/4 loci suggesting that these are distinct mechanisms of suppressing p53 function in human cancers. Thus, we report for the first time, the significance of TIRR as an upstream inhibitor of the 53BP1-p53 complex.
RESULTS
TIRR loss increases 53BP1-dependent gene transactivation property of p53.
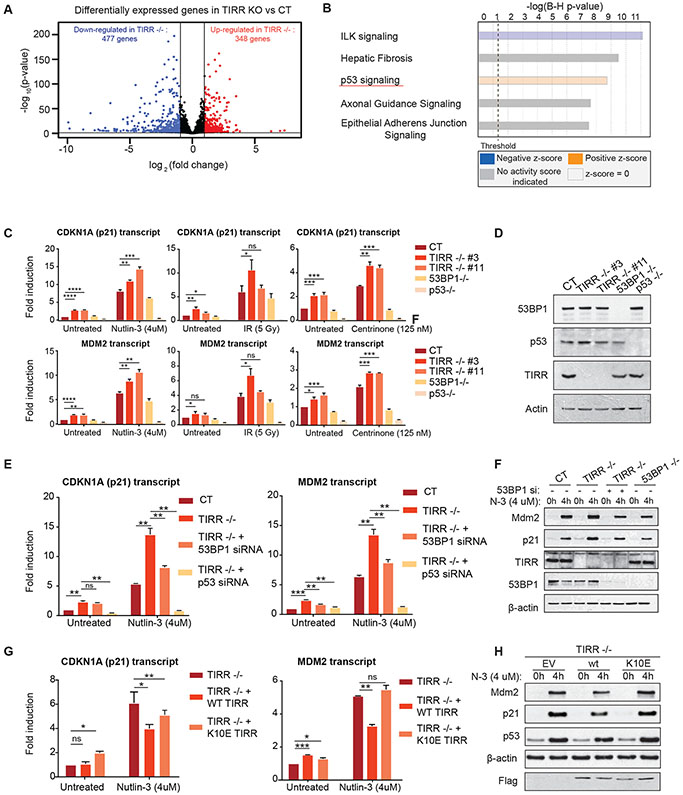
To identify the cellular pathways that are influenced by TIRR, we analyzed gene expression by bulk RNA-sequencing in two clones of U2OS TIRR-knockout (KO) cell line. Relative to control, loss of TIRR resulted in increased expression of 348 genes (log2 (fold change) >1) and decreased expression of 477 genes (log2 (fold change) <−1) (Fig. 1A). Gene annotation and pathway analysis revealed that the p53 pathway was one of the statistically ‘top’ ranked pathways that were upregulated in TIRR-KO cells (p <0.005) (Fig. 1B). To validate this result and further analyze the p53 pathway in the context of exogenous activation (via Nutlin-3 treatment), the expression of ninety-two p53 pathway genes comprising of upstream activators, downstream transcriptional targets, and co-activators was examined by qRT-PCR in control and TIRR-KO cells (S1A). Only p53 transcriptional transcripts were significantly upregulated in both untreated and Nutlin-3 treated TIRR-KO cells. Next, we utilized a subset of p53 target genes, CDKN1A (p21), MDM2, BAX, and BBC3/PUMA, as a read-out of p53 activity, to examine the impact of TIRR in the context of different stress signals [Nutlin-3, IR and Centrinone (PLK-4 inhibitor)]. Loss of TIRR caused a consistent increase in p53-target gene transcripts independent of the mode of p53 activation (Fig. 1C, 1D S1B, S1C). Since loss of TIRR impacts 53BP1-mediated DNA repair functions, we investigated whether TIRR-KO cells harbor increased endogenous DNA damage which could activate p53. However, there was no significant increase in γ-H2AX levels in untreated or Nutlin-3 treated TIRR-KO cells (S1D). To further address whether DNA damage induced signaling is involved in the enhanced p53 response in TIRR-KO cells we used the ATM kinase inhibitor (KU 55933). Inhibition of ATM signaling did not impact p53 activity in TIRR-KO cells (S1E).
Figure 1: TIRR loss increases 53BP1-dependent gene-transactivation property of p53.
A) Volcano plot showing differential gene expression in U2OS TIRR-KO cells. p-values on the y-axis were calculated using the DESeq2 algorithm (see Methods).
B) Gene annotation and pathway analysis (core analysis of IPA, Qiagen) of the bulk RNA sequencing data showing the top ranked pathways that are significantly altered in TIRR-KO cells.
C) qRT-PCR measurements from 3 independent experiments in RPE1 cells of indicated genotypes that were treated either with IR (5Gy) or Nutlin-3 (4 μM) or Centrinone (100nM). Fold induction of CDKN1A(p21) and MDM2 transcripts was normalized to HPRT1 expression. (Mean ± SD) (p-value < 0.0001 (****), p-value = 0.0002-0.0001 (***), p-value = 0.002-0.0002(**), p-value = 0.0332-0.002 (*), p-value= 0.1234 (ns))
D) Western blot in RPE1 cells of indicated genotype to confirm gene knockout.
E) p21 and MDM2 transcript abundance measured by qRT-PCR in RPE1 WT and TIRR-KO cells that were transfected with indicated siRNAs and treated with Nutlin-3 for 4 hours. (Mean ± SD)
F) p21 and MDM2 protein levels measured by immunoblot corresponding to Fig. 1E.
G) p21 and MDM2 transcript abundance measured by qRT-PCR in RPE1 TIRR-KO cells that stably expressed either WT or K10E mutant TIRR or an empty vector control through lentivirus-mediated transduction. (Mean ± SD)
H) p21 and MDM2 protein levels measured by immunoblot corresponding to Fig. 1G.
TIRR binds and regulates 53BP1 activity, therefore the impact of TIRR on p53 might be mediated by 53BP1. Furthermore, a 53BP1-USP28 complex was identified as an activator of p53-mediated transactivation (Cuella-Martin et al., 2016a, Fong et al., 2016, Lambrus et al., 2016, Meitinger et al., 2016). Therefore, we co-depleted either 53BP1 or USP28 in TIRR-KO cells and observed that the increased p21 and MDM2 expression in TIRR-KO cells was significantly suppressed upon depletion of 53BP1 or USP28 protein (Fig. 1E, 1F, S1F). We next used previously established binding-null mutants of TIRR (K10E and R107S mutants) that do not interact with 53BP1 (Botuyan et al., 2018a, Dai et al., 2018, Wang et al., 2018). We stably complemented the TIRR-KO cells with either WT-TIRR, or the binding null mutants of TIRR and examined p53-mediated gene transactivation. Consistent with our prior observations, the increase in p21 and MDM2 expression in TIRR-KO was rescued by WT-TIRR but not by the K10E mutant (Fig. 1G, 1H) or the R107S mutant (S2A, S2B). Together, these results indicate that TIRR loss causes aberrant p53-mediated transactivation in a 53BP1-dependent manner without inducing detectable DNA damage.
TIRR impacts p53’s promoter binding ability independent of its stability or localization
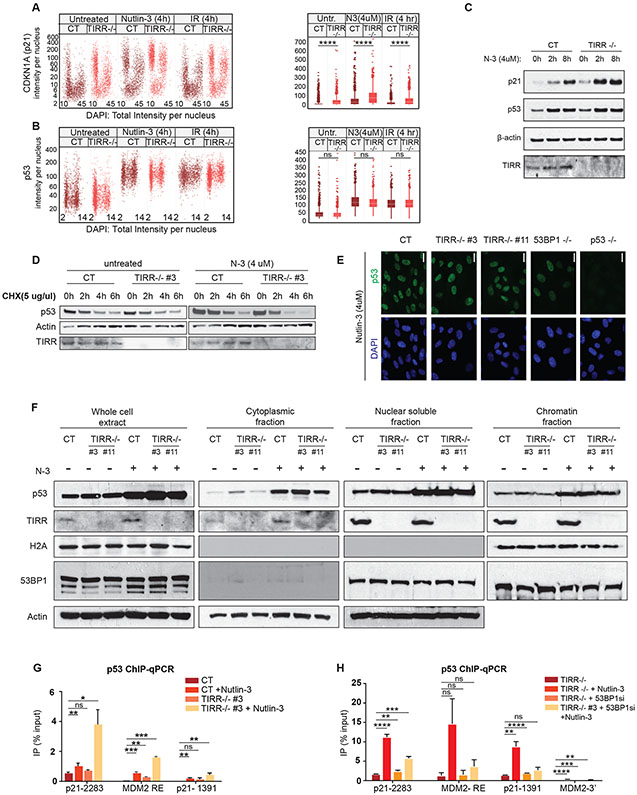
To further investigate p53 activation in TIRR-depleted cells, we conducted quantitative image-based cytometry (QIBC) to quantify nuclear levels of p53 and its transcriptional target, p21, in individual cells. Stress-dependent induction of p21 nuclear intensity was increased primarily in G1- and G2-cells, consistent with previous work (Michelena et al., 2019, Sheng et al., 2019), with at least two-fold increase in TIRR-KO cells in all conditions (Fig. 2A). In contrast to p21, no marked change in p53 nuclear intensity was observed (Fig. 2B). Importantly, TIRR loss also did not significantly change the steady state levels of p53 protein (Fig. 2C). Loss of TIRR did not impact the synthesis or degradation of p53 protein (Fig. 2D, S2C). We also found no significant changes in p53 localization upon TIRR loss by immunofluorescence (Fig. 2E) or biochemically in different sub-cellular fractions (Fig. 2F). p53-mediated transactivation can be modulated by the stability, localization, or promoter binding efficiency of p53 protein (Lavin and Gueven, 2006). Since there were no significant changes in p53 protein stability or localization, we examined p53 occupancy at promoter-responsive elements (RE) by chromatin immunoprecipitation (ChIP) and qPCR in WT and TIRR-KO cells. Loss of TIRR increased p53 occupancy at the promoter RE site in p21 by 4-fold and MDM2 by 1.5-fold after Nutlin-3 treatment (Fig. 2G). Depleting 53BP1 by siRNA suppresses the higher p53 occupancy at promoter sites observed in TIRR-KO cells (Fig. 2H). Together these results suggest that TIRR-deficiency enhances p53 promoter binding efficiency independent of its stability or localization.
Figure 2: TIRR impacts p53’s promoter binding ability but has no effect on p53 protein turnover, stability or localization.
A-B) WT and TIRR-KO U2OS cells were treated with different conditions (IR, Nutlin-3) for indicated time periods, and stained for p21 (Fig. 2A) or p53 (Fig. 2B) and DNA content, for cell cycle resolved quantification of nuclear levels of p21 and p53 by QIBC; Box plots to the right show the quantification of nuclear protein intensities by QIBC analysis (Median ± SD).
C) p21 and p53 protein levels in RPE1 WT and TIRR-KO cells treated with Nutlin-3 at indicated timepoints. Actin was used as a loading control.
D) p53 protein levels at different timepoints after treatment with 5 μg/μl cycloheximide in RPE1 CT and TIRR-KO cells.
E) Immunofluorescence to show p53 localization in RPE1 cells of indicated genotype. (Scale bar = 20 μM)
F) Western blot showing p53 proteins levels in different cellular fractions of RPE1 cells of indicated genotype that were either untreated or treated with Nutlin-3. Cells were first lysed in a hypotonic lysis buffer to extract the cytoplasmic fraction. The nuclear soluble fraction was extracted using a sucrose buffer and 350mM NaCl and the chromatin fraction was extracted using sucrose buffer containing 10% SDS. Actin was used as a loading control for the whole cell extract, cytoplasmic and nuclear soluble fractions and H2A was used as a loading control for the chromatin fraction.
G-H) ChIP-qPCR results measuring p53 occupancy at unique sites in p21 and MDM2 promoter regions. The data shown is representative of two experimental replicates. The bar plots indicate the ratio of immunoprecipitated DNA calculated relative to % total input, error bars indicate Standard Error. (p-value < 0.0001 (****), p-value = 0.0002-0.0001 (***), p-value = 0.002-0.0002(**), p-value = 0.0332-0.002 (*), p-value= 0.1234 (ns))
TIRR alters p53-mediated cell fate programs by inhibiting the 53BP1-p53 interaction.
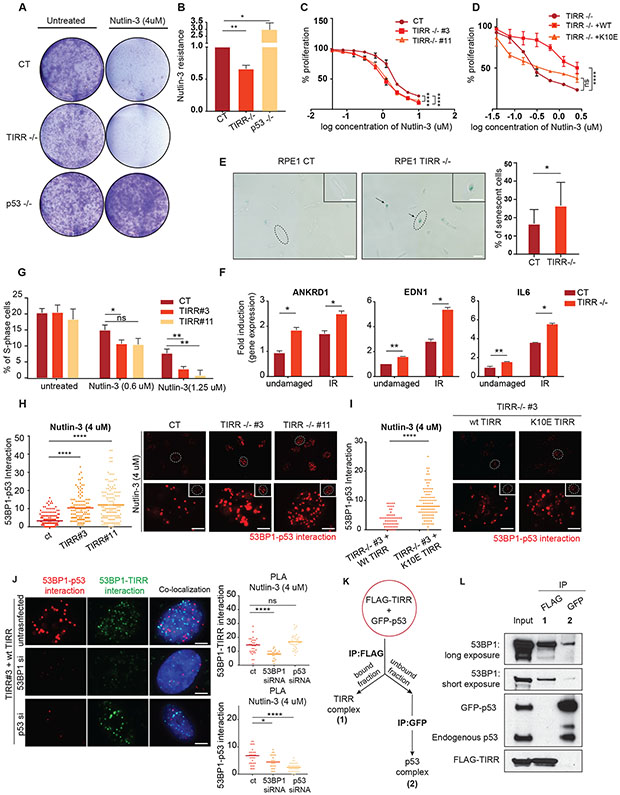
p53 induces CDKN1A(p21) to promote cell-cycle arrest, BAX and BBC3/PUMA to initiate apoptosis, and CDKN2A (p16Ink4a)for cellular senescence (Mello and Attardi, 2018). Since we observed a significant upregulation of these target transcripts in TIRR-KO cells, we asked if this translated to a change in overall cell fate outcomes. To this end, we first tested the effect of p53 activation by Nutlin-3 on survival in TIRR-KO cells. As anticipated, there was a ~ 40% reduction in cell survival due to Nutlin-3 induced growth arrest in TIRR-KO cells relative to control and a 200% increase in p53-KO cells (Fig. 3A, Fig. 3B). We used resazurin cell viability assay as an alternative method to further validate these results and determined that TIRR loss significantly reduced survival in the presence of Nutlin-3 (CT IC50: 1.72 μM, TIRR−/− #3 IC50: 1.034 μM, TIRR−/− #11 IC50: 1.11 μM) (Fig. 3C). The impact of TIRR loss on cell survival was rescued by WT TIRR but not by K10E TIRR, again indicating that the effect of TIRR on p53 function was mediated by its interaction with 53BP1 (Fig. 3D). Next, we measured basal cellular senescence by beta-galactosidase staining and expression of senescence-associated genes (ANKRD1, EDN1 and IL-6) (Noren Hooten and Evans, 2017) after treatment with IR. Consistent with our previous observations, there was a significant increase in the percentage of basal senescent cells in the absence of TIRR (Fig. 3E) and at least 2-fold increase in the expression of the senescence-associated genes in TIRR-KO cells after IR (Fig. 3F). Finally, we tested the impact of TIRR on cell cycle arrest and observed that there was more than 5-fold decrease in the number of TIRR-KO cells in the S-phase relative to WT cells after treatment with low doses of Nutlin-3 (Fig. 3G, S2D). We also confirmed that this increase in p53 activity was not due to accumulated endogenous damage due to the longer treatments with Nutlin-3 (S2E).
Figure 3: TIRR alters p53-mediated cell fate programs by inhibiting the p53-53BP1 interaction.
A-B) Sensitivity to Nutlin-3 measured by crystal violet staining. Representative images (Fig 3A) and quantification of data (Fig 3B) in RPE1 cells of indicated genotype. Experiment was performed in triplicates.
C-D) Resazurin-based cell survival assay of RPE1 cells of indicated genotype treated with indicated concentrations of Nutlin-3 for 4 days. Experiments were performed in three replicates (p-value <0.0001 (****), p-value > 0.05 (ns)).
E) Representative images of senescent cells in RPE1 WT and TIRR-KO genetic backgrounds. Cells were fixed and stained for beta-galactosidase as a senescence marker. Bar plot represents the quantification of senescent cells (beta gal+) from three different replicates. (Scale bar = 100 μM). (Lighter background on one half of the images was an artifact generated due to the positioning of the apotome).
F) RPE1 WT and TIRR-KO cells were treated with IR (5Gy) and recovered for 10 days. mRNA transcript levels of three senescence biomarkers - ANKRD1, EDN1, and IL6 normalized to Actin. (Mean ± SD)
G) RPE1 cells of indicated genotype were treated with Nutlin-3 for 72 hours. Cells were fixed and stained for Propidium Iodide. Data represents the number of cells in S-phase from three different experiments measured by FACS analysis using ModFit software (see Methods).
H-I) Representative images and quantification of endogenous 53BP1-p53 interaction in RPE1 cells of indicated genotype treated with Nutlin-3 using Proximity Ligation Assay (PLA; see Methods). Positive signals are indicated as red dots. (Scale bar = 5 μM)
J) A 3 probe PLA assay assessing co-localization of TIRR-53BP1 and p53-53BP1 complexes in RPE1 TIRR-KO cells stably expressing WT TIRR (FLAG-WT-TIRR). Cells were transfected with indicated siRNAs. TIRR-p53 complex interaction is indicated by the green dots and p53-53BP1 complex interaction is indicated by the red dots. (Scale bar = 5 μM)
K) Schematic of immunoprecipitation shown in Fig 3L. GFP-p53 was transfected in RPE1 TIRR-KO cells that were stably transduced by FLAG-tagged WT TIRR. FLAG-TIRR was first immunoprecipitated to identify TIRR interacting proteins. The fraction that was unbound to FLAG was used to immunoprecipitate GFP-p53 to identify p53 interacting proteins.
L) Immunoblot of indicated proteins in two different interacting complexes (schematic in Fig. 3K). Complex 1 shows interaction of FLAG-TIRR and 53BP1. Complex 2 shows the interaction of GFP-p53 and 53BP1.
Taken together, these results establish that increased p53 activity caused by the loss of TIRR results in a significant change in p53-mediated cell fate programs.
TIRR loss enhances the interaction of 53BP1 with p53
Previously, we had demonstrated that TIRR loss results in an ‘altered’ form of 53BP1, which has increased binding capacity for its DNA repair effectors RIF1 and PTIP (Drane et al., 2017). A mutant 53BP1(F1553R) that does not interact with TIRR also mimics this altered activity (Botuyan et al., 2018a). Based on these findings, we hypothesized that TIRR loss might impact the interaction of 53BP1 and p53. Indeed, in TIRR-KO cells we observed a striking three-fold increase in endogenous 53BP1-p53 interaction by proximity ligation assay (PLA) after Nutlin-3 treatment (Fig. 3H) or after exposure to IR and UV (S2F). The increased 53BP1-p53 interaction in the absence of TIRR can be rescued by WT-TIRR but not by K10E TIRR (Fig. 3I). To further investigate the dynamics of the TIRR/53BP1/p53 associations, we used three probes in the PLA to visualize and quantify the 53BP1/p53 complex and the 53BP1/TIRR complex. We observed that these two complexes are distinct from each other and do not co-localize (Fig. 3J). As a control, we either depleted 53BP1, which prevents the visualization of both complexes, or depleted p53, which specifically prevents tagging the 53BP1/p53 complex. To complement this cytological single-cell based approach, we used a biochemical strategy using TIRR-KO cells. GFP-tagged p53 was transiently transfected in these cells that stably expressed FLAG-TIRR. The first step was FLAG immunoprecipitation to detect TIRR interacting proteins. 53BP1 was found to be in the TIRR-bound fraction but not endogenous or GFP-tagged p53. Next, the unbound fraction was used to immunoprecipitate GFP-p53-bound proteins. Here again, 53BP1 was identified as a p53 interacting partner but not FLAG-TIRR (Fig. 3K, 3L). We further confirmed that TIRR interacted with 53BP1 after p53 activation but there was no detectable interaction between TIRR and p53 (S2G). Together, these results strongly suggest that p53 is excluded from a 53BP1-TIRR complex and TIRR is likely to impede the association of 53BP1 with p53.
TIRR inhibits the interaction of methylated p53 with the Tudor domain of 53BP1.
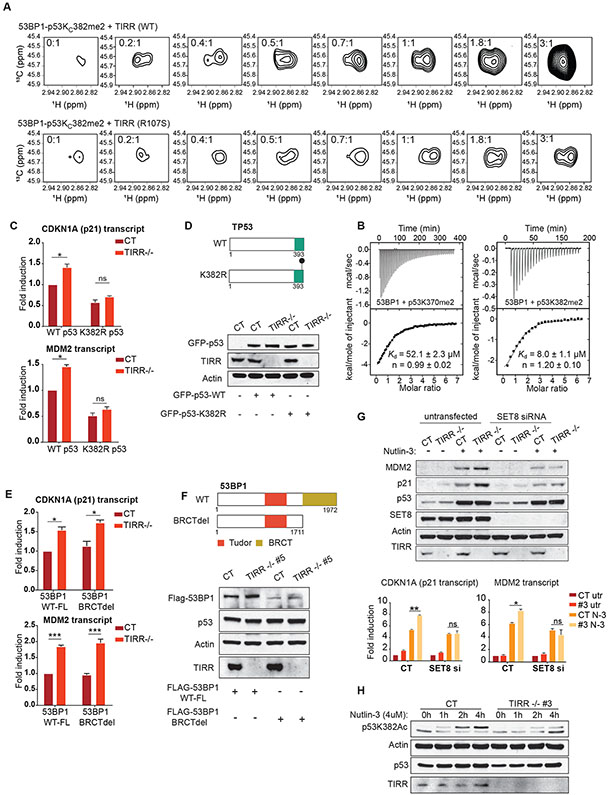
To further understand the mechanism by which TIRR inhibits the 53BP1-p53 interaction, we considered the structural basis of interactions of both TIRR and p53 proteins with 53BP1. Cell-based studies have suggested that the BRCT domain of 53BP1 is indispensable in regulating p53 activity (Cuella-Martin et al., 2016a). However, structural studies suggest that the Tudor domain of 53BP1 interacts with p53 with an affinity comparable to that of the BRCT domain (Roy et al., 2010, Tong et al., 2015, Ekblad et al., 2004). The Tudor domain specifically interacts with dimethylated p53 (p53K370me2, p53K382me2). These modifications are associated with the gene transactivation functions of p53 (Huang et al., 2007, Kachirskaia et al., 2008, Tong et al., 2015). Therefore, we speculated that TIRR directly competes with dimethylated p53 for binding the Tudor domain, thereby negating the positive impact of the Tudor domain on the transactivating function of p53. To test this possibility, we used NMR spectroscopy to probe the interactions of 53BP1 alone and 53BP1-TIRR complex with the CTD region of p53 harboring a 13C-labeled dimethyl lysine mimic at K370 (p53KC370me2) or K382 (p53KC382me2), or at both sites (p53KC370me2KC382me2). By monitoring the decrease in 1H-13C signal intensity associated with complex formation, we could demonstrate that TIRR impairs the interaction of 53BP1-Tudor with p53KC370me2, p53KC382me2 and p53KC370me2KC382me2 (S3A, S3B). Further, we tested whether purified WT TIRR can compete with p53 for the Tudor domain of 53BP1 in a similar NMR-based competition assay. We had previously established that the R107S mutant (S2A, S2B) of TIRR does not bind 53BP1 Tudor domain in vitro and in cells (Botuyan et al., 2018a). Here we observed that WT TIRR, but not the R107S binding-null mutant, can displace p53KC382me2 from the Tudor domain of 53BP1 (Fig. 4A). Ubiquitin was used as a positive control in this assay (S3C). We selected this K382 residue over K370 for our experiments because p53 dimethylated at K382 (p53K382me2) has significantly higher affinity for the Tudor domain than p53 dimethylated at K370 (p53K370me2), with respective Kd of 8 μM and 52 μM determined by isothermal titration calorimetry (ITC) (Fig. 4B).
Figure 4: TIRR inhibits the interaction of methylated p53 with the Tudor domain of 53BP1.
A) Changes in the 1H-13C correlation methyl NMR signals of the preformed 53BP1-Tudor-p53KC382me2 complex harboring 13C-labeled KC382me2 methyl groups upon titration with WT TIRR and binding-null mutant TIRR (R107S). The ratios of WT TIRR and R107S TIRR with respect to 53BP1-Tudor-p53KC382me2 are indicated. Increase in signal intensity with increased concentration of TIRR indicates that, as expected, WT TIRR displaces p53KC382me2 from 53BP1.
B) 53BP1-Tudor was titrated with p53K370me2 (366-375) and p53K382me2 (377-386) peptides. For each titration, the raw data and integrated heat measurements with curve fitting using a one-site binding model, are indicated. The Kds and stoichiometry numbers n are indicated with associated SD determined by nonlinear least squares analysis.
C) p21 and MDM2 transcript levels calculated by qRT-PCR in RPE1 cells of indicated genotype. Cells were treated with p53 siRNA to deplete endogenous p53 and transiently transfected with siRNA- resistant constructs expressing either WT p53 or the K382R mutant.
D) Schematic representation of the p53 constructs used in Fig 4C; Immunoblots comparing expression levels of the two p53 constructs used in Fig 4C in cells of indicated genotype.
E) p21 and MDM2 transcript levels calculated by qRT-PCR in U2OS cells of indicated genotype. Cells were treated with a 3’ UTR siRNA to deplete endogenous 53BP1 and transiently transfected with either a full-length 53BP1 or a truncated mutant lacking the BRCT domain.
F) Schematic representation of 53BP1 constructs used in Fig 4E; Immunoblots comparing expression levels of the two 53BP1 constructs used in Fig 4E in cells of indicated genotype.
G) RPE1 cells of indicated genotype were first transfected with an siRNA against SET8. 48 hours after transfection, the cells were either untreated or treated with Nutlin-3. The western blot indicates p21 and MDM2 proteins levels in the indicated genotypes and treatments. The corresponding p21 and MDM2 transcript abundance calculated by qRT-PCR is represented in the bar graphs.
H) Western blot depicts the levels of indicated proteins. RPE1 CT and TIRR-KO cells, either untreated or treated with Nutlin-3, were extracted at different timepoints to examine the levels of acetylated p53 at residue K382 (p53K382ac).
The transactivation property of p53 is dependent on both the core DNA binding domain (DBD) and the CTD. In a previous cell-based analysis of 600 p53 RE sites, the WT p53 is bound to 355 sites, whereas the CTD mutant is bound to 210 of these sites, suggesting that both the DBD and CTD are important but not sufficient for p53’s TF activity (Laptenko et al., 2015, Sullivan et al., 2018). Therefore, we asked whether TIRR regulates p53 activity by blocking the p53-CTD and 53BP1-Tudor interaction. Endogenous p53 was transiently replaced with a CTD-deletion mutant or with a K382R mutant (S4A). We observed that consistent with our previous results, p21 and MDM2 transcripts were induced in the TIRR-KO cells harboring WT-p53. However, this TIRR loss-mediated induction of p53 targets was completely suppressed by both CTD deletion (S3B) and the K382R mutation of p53 (Fig. 4C, 4D, S4A). We also showed that the increased interaction of p53 and 53BP1 observed in TIRR KO cells is dependent on the CTD of p53 (S4B). Based on these results, we inferred that the impact of TIRR on p53 activity is via its direct competition for the Tudor domain and is independent of the BRCT domain. To further validate this inference, we expressed either the full length 53BP1 or a BRCT-truncated form of 53BP1 in both WT and TIRR-KO cells and assessed the expression of p21 and MDM2 as a read-out of p53 activity. We observed that the enhanced expression of p21 and MDM2 observed in TIRR-KO cells occurs independently of the BRCT domain of 53BP1 (Fig. 4E, 4F).
Based on these findings, we hypothesized that in TIRR-KO cells there is increased availability of an uninhibited Tudor domain of 53BP1 which binds dimethylated p53 (p53K382R). This would suggest that the methylation of K382 is key to the phenotype induced by TIRR loss. To test this idea, we used an siRNA to inhibit SET8, the monomethyl transferase that specifically methylates p53 at residue K382 (Shi et al., 2007). The induction of p53 target transcripts that was observed in TIRR-KO cells, was significantly suppressed by inhibition of SET8 (Fig. 4G). Previously it was shown that methylation of certain key residues in the CTD of p53 (K370, K382) prevents another PTM, acetylation, at the same residues(Shi et al., 2007, Kurash et al., 2008). Consistently, there was decreased acetylated p53 at residue K382 at different time points after Nutlin-3 treatment in TIRR-KO cells (Fig. 4H). Together these results strongly support our model (Fig. 5A) that TIRR inhibits p53-mediated transactivation by directly inhibiting the association of the CTD (primarily dimethylated at K382) of p53 with the 53BP1-Tudor domain.
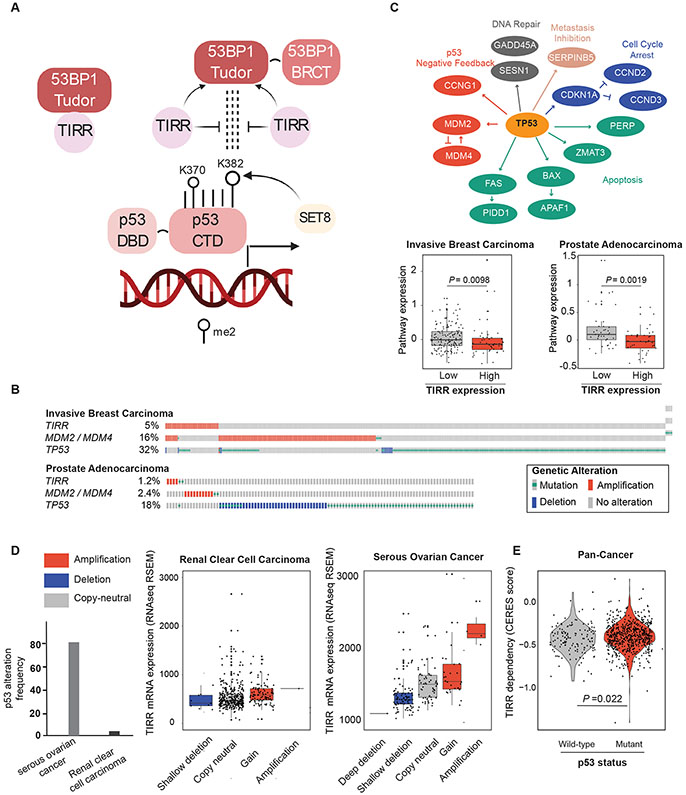
Figure 5: TIRR negatively regulates p53 signaling in cancer.
A) Model depicting TIRR as an upstream regulator of the 53BP1-p53 axis involved in p53 transactivation. The Tudor domain of 53BP1 interacts and with p53K382me2, a PTM of p53 that is associated with higher transactivation of p53. TIRR competes with dimethylated p53 for the Tudor domain of 53BP1.
B) OncoPrint showing the frequency at which TIRR, MDM2 / MDM4 and TP53 are altered in invasive breast carcinoma and prostate adenocarcinoma cohorts from TCGA (Cancer Genome Atlas, 2012, Hoadley et al., 2018). For visualization purposes, only samples with genetic alterations are shown in the plot. Putative driver mutations were chosen based on OncoKB driver annotations and copy number alterations were selected on GISTIC calls. OncoPrints were generated in the cBioPortal for Cancer Genomics [cbioportal.org].
C) Genes belonging to the p53 pathway for which expression data were used to calculate pathway expression estimates. Pathway expression in invasive breast carcinoma (top) and prostate adenocarcinoma (bottom) samples with and without TIRR amplification. Pathway expression scores were calculated as the average mRNA expression of p53 pathway members. RNAseq data for each p53 pathway gene were obtained from TCGA and were evaluated as z-scores relative to diploid samples. P-values, Wilcoxon test.
D) The level at which loss of TIRR is tolerated in human cancers is dependent on p53 status. Cancers with high frequencies of p53 alterations (serous ovarian cancer, right) can tolerate changes in TIRR expression through genomic loss, as a compensating mechanism for p53 activity. On the other hand, cancers with low frequency of p53 alterations (renal cancer) do not tolerate changes in TIRR expression that cause changes in p53 activity. Copy number events have been colored to indicate losses (blue) or gains (red) of genomic material. Gene expression (z-scores from RNAseq V2 RSEM) and copy number data were downloaded from TCGA (Cancer Genome Atlas, 2012).
E) Comparison of TIRR dependency scores (CERES) between 136 p53 WT and 633 p53 mutant cancer cell lines. p53 WT cell lines have a significantly higher dependency on TIRR. Dependency scores were calculated using DepMap’s Achilles Avana 20Q2 public release. [https://portals.broadinstitute.org/ccle]
TIRR negatively regulates p53 signaling in cancer.
Our in vitro and cell line data thus far provide conclusive evidence that TIRR is a negative regulator of p53 trans-activation. The tumor suppressor gene TP53 is one of most commonly altered genes in cancer, with loss-of-function mutations in >40 % of all tumors. In ~6% of remaining cancers, p53 signaling is altered by amplification of the negative regulators MDM2 or MDM4 (Zehir et al., 2017). To assess whether the mutational profile of TIRR resembles that of other p53 negative regulators, we compared the frequency of genetic alterations present in TIRR with MDM2 and MDM4 (Fig. 5B). We focused our analysis on the TCGA invasive breast carcinoma and prostate adenocarcinoma cohorts, as these tumor types exhibit among the highest frequencies of TIRR amplification. Mutation and copy number data revealed that TIRR amplifications are enriched in cancers that are TP53 wild-type and lack MDM2 or MDM4 amplification (P =0.015 one-tailed Fisher’s exact test), suggesting that TIRR amplifications are an alternative mechanism for suppression of p53 activity. Next, we compared expression of p53 pathway members in tumors that were classified as ‘high’ or ‘low TIRR’ according to TIRR mRNA expression levels (using a z-score cut-off of 3). We used the average expression of 15 downstream targets of p53, including negative feedback regulators as well as genes involved in DNA repair, cell cycle arrest, apoptosis, and metastasis inhibition (Fig. 5C). As anticipated, both breast and prostate tumors with high TIRR expression showed decreased levels of p53 pathway expression (Fig. 5C; P = 0.0098 and P = 0.0019, Wilcoxon test). This was comparable to the correlation of MDM2/MDM4 amplification and the expression of the same set of p53 target genes in breast and prostate carcinomas, indicating that TIRR acts as a negative regulator of p53 (S4C). Loss of TIRR amplifies the function of p53, which would be detrimental for tumorigenesis. Therefore, we predicted that p53 proficient tumors are likely to have stable TIRR expression or may enhance TIRR expression to suppress p53. This would be in contrast with p53-deficient tumors which would not have selective pressure to retain TIRR expression. To test this prediction, we analyzed and contrasted TIRR mRNA expression between renal clear cell carcinomas, less than 4% of which exhibit TP53 mutations, and serous ovarian cancers, wherein TP53 mutation frequencies are greater than 80% (Fig. 5D). While ovarian cancers frequently exhibit copy-loss of TIRR, renal cancers do not. Moreover, TIRR expression changes markedly with copy-number in ovarian cancers but not renal cancers. These findings suggest that renal cancers, with intact TP53, do not tolerate changes, especially losses in TIRR expression, whereas ovarian cancers, which have lost TP53 are less reliant on TIRR. This is further supported by TIRR ‘dependency’ analysis of 769 Cancer Cell Line Encyclopedia (CCLE) lines (136 of which are p53 WT, 633 are p53 mutant) which showed that p53-proficient lines were significantly more dependent on TIRR (Fig. 5E). Altogether, these analyses suggested that TIRR amplification may be an important mechanism by which cancers suppress p53 activity.
DISCUSSION
Over 25 years ago, 53BP1 was discovered as a p53 interacting protein (Iwabuchi et al., 1994). A decade later, animal model-based studies showed that the DNA repair function of 53BP1, and the role of p53 in checkpoint control and apoptosis, synergized to prevent tumorigenesis (Morales et al., 2006, Ward et al., 2005). However, 53BP1’s link to p53 faded over the years as 53BP1 emerged as a central player in the repair of DSBs. More recently, 53BP1, USP28, and p53 were identified in three related screens aimed at identifying suppressors of the cell-cycle arrest induced either by reducing centriole number or by prolonged mitotic arrest (Fong et al., 2016, Lambrus et al., 2016, Meitinger et al., 2016). The mechanism proposed was that 53BP1 and USP28 promote p53 protein stability specifically in the context of mitotic stress, but not by DNA damage or by Nutlin-3. In contrast, Chapman and colleagues(Cuella-Martin et al., 2016b) observed that the loss of 53BP1 did not impact p53 levels but significantly diminished p53 transactivation function after IR or Nutlin-3 treatment. Structural studies have shown that both the BRCT and Tudor domains of 53BP1 can directly interact with p53 (Derbyshire et al., 2002, Joo et al., 2002, Kachirskaia et al., 2008, Roy et al., 2010, Tong et al., 2015). More recently, however, the BRCT domain and the oligomerization domain of 53BP1 were implicated in the activation of p53 (Cuella-Martin et al., 2016a, Kilic et al., 2019). Collectively, these studies have highlighted the importance of the 53BP1/p53 complex and raised several unanswered questions, (i) does 53BP1 indeed regulate p53 function beyond mitotic stress? (ii) what is the underlying molecular mechanism of 53BP1-mediated regulation of p53’s function (iii) are there any molecular/biochemical links between 53BP1’s function in DSB repair and p53 activation? Here our results provide insight on these unresolved issues by elucidating the functional and biochemical interplay of TIRR, 53BP1 and p53 in cells and in vitro.
The site-specific DNA binding ability of p53 is mediated by its core DBD and the CTD (Laptenko et al., 2015, Anderson et al., 1997, Laptenko and Prives, 2006, Luo et al., 2004, Bieging et al., 2014) and this in turn influences its transactivation function. p53 dimethylated at residues K370 and K382 in the CTD is poised for higher transactivation (DeHart et al., 2014, Huang et al., 2007, Kachirskaia et al., 2008) and these residues are also involved in the direct interaction with the Tudor domain of 53BP1 (Kachirskaia et al., 2008, Roy et al., 2010, Tong et al., 2015). From our results we have inferred that TIRR down-modulates p53 activity by specifically inhibiting the Tudor-p53K382me2 interaction. TIRR’s inhibition of p53 activity, is dependent on the Tudor domain of 53BP1 and independent of the BRCT domain. Prior work suggested that the role of USP28 in regulating p53 was downstream of 53BP1 and it was via the BRCT domain of 53BP1. However, it was not clear whether USP28 deubiquitinates p53 to stabilize it or regulates its DNA binding function, or whether its impact on p53 is indirect, via other factors. Intriguingly, USP28 is also functionally involved in TIRR-mediated regulation of p53 (S1E). Future work will ascertain the precise role of USP28 in the TIRR/53BP1/p53 signaling network. Analysis of TCGA data revealed the striking similarity between MDM2/4, the established negative regulator of p53, and TIRR. Most intriguingly, we observed cancers which had low p53 alteration frequency maintained stable expression of TIRR with almost no loss or deletion of the TIRR gene. This is consistent with the notion that TIRR loss in these p53-proficient tumors would enhance p53 activity and impair tumor growth. In contrast, p53-deficient tumors have no such incentive to retain TIRR, which is evident by the significant number of tumors with deletions of the TIRR gene locus. Together, our results indicated that TIRR may represent a distinct, and hitherto undiscovered mechanism, for inactivating p53 in primary tumors. Further analysis of cancer databases is necessary to solidify this intriguing concept.
A comprehensive analysis of 44 datasets from human p53 ChIP-seq studies revealed that only 11% of all p53-bound sites near the transcription start site (TSS) of genes result in differential expression (Nguyen et al., 2018). The lack of correspondence between binding and expression suggests that additional context-dependent signals, such as cofactors like 53BP1, are required for an efficient p53 transcriptional response. A limitation of this study is that we have not addressed which p53 target transcripts are regulated directly by 53BP/TIRR. Furthermore, TIRR may impact p53 function beyond its regulation of 53BP1. These two issues should be addressed in future work.
STAR Methods
Resource availability
Lead contact:
Further requests for reagents and resources should be directed to and will be fulfilled by the lead contact, Dr. Dipanjan Chowdhury (dipanjan_chowdhury@dfci.harvard.edu).
Materials availability:
Materials used in this study are available upon reasonable request.
Data and code availability:
The datasets generated in this study are available on NCBI SRA portal under the project id: PRJNA661632.
Experimental Model and Subject Details:
All RPE1, U2OS, and HEK 293T cell lines used in this work were grown in Dulbecco’s Modified Eagle’s medium supplemented with 10% FBS and 1% penicillin-streptomycin. All cell lines were grown in 37°C and 5% CO2. Cell lines tested negative for mycoplasma.
Gene editing:
RPE1 and U2OS TIRR-KO cell lines were generated using the Alt-R CRISPR-Cas9 technology (IDT). CRISPR guides (sgRNAs) were designed using CHOPCHOP and DeskGen CRISPR guide design tools. The RNP complex was generated by first combining 2.4 μl sgRNA (100 μM/L stock) and 2.4 ul tracrRNA (100 μM/L stock) in a PCR tube with 5.2 μl nuclease free water, generating a 240 pmol sgRNA solution. The sgRNA solution was combined by the following PCR reaction: 95°C 5min, 1°C/min decrease to 12°C, then infinite hold at 12°C. Next, 200 pmol Cas9 was combined with 120 pmol sgRNA solution and incubated at room temperature for 10 mins to allow RNP formation. The RNP complexes were introduced into RPE1 cells by electroporation using the Lonza Nucleofector X system. After 72 hours, the knockout cell pools were tested by immunoblotting using relevant antibodies. After confirming a decrease in expression of proteins, single clones were sorted and tested by both immunofluorescence and immunoblotting.
METHOD DETAILS
Retrovirus production:
HEK 293T cells were transfected with TIRR (WT/K10E/R107S) – pOZ-FH-N constructs and viral packaging plasmids. Retroviral particles were collected after 48 hours and used to infect RPE1 TIRR-KO cells. TIRR-KO cells stably complemented with pOZ-TIRR vectors were selected using magnetic CD25-Dyna beads.
RNAi:
p53 ORF siRNA: 5’ ccagtggtaatctactgggacggaa 3’, 53BP1 ORF siRNA: AGAACGAGGAGACGGUAAUAGUGGG, 53BP1 UTR siRNA: AAAUGUGUCUUGUGUGUAA, USP28 siRNA: CUGCAUUCACCUUAUCAUU
10μM siRNA was transfected in cells using Invitrogen Lipofectamine RNAiMAX using the manufacturer’s protocol. The medium was changed 24 hours after the transfection. Cells were harvested for RNA and protein extraction after 48 hours.
RNA extraction and RT-PCR:
Total RNA was extracted from cells using RNeasy mini kit (Qiagen) using the manufacturer’s protocol. 1-5 μg of total RNA was used to generate cDNA using SuperScript III First-strand synthesis system (Invitrogen). qPCR was done using the 2X SybrGreen Mastermix (Applied Biosystems) and the relevant primers. All qPCR primers used in the study are listed in the Key Resources table.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 53BP1 | CST | 4937S |
| 53BP1 | BD Pharmingen | 612522 |
| 53BP1 | NB | NB100-304 |
| p53 | SCBT | DO-1 |
| CDKN1A/p21 | CST | 2947S |
| MDM2 | SCBT | sc-965 |
| β-Actin | SCBT | sc-47778 |
| TIRR | Sigma | HPA044186 |
| FLAG | sigma | M2 clone-F3165 |
| GFP | CST | 2555 |
| p53 | Invitrogen | MA5-12571 |
| p21 | SCBT | sc-756 |
| p53 (mouse) | Thermofisher Scientific | MA5-12571 |
| p21 (rabbit) | SCBT | sc-756 |
| p53K382ac | Abcam | ab75754 |
| SET8 | CST | 2996S |
| USP28 | Abcam | EPR4249 |
| y-H2AX | Millipore | 05-636 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Nutlin-3 | Cayman Chemical | 10004372 |
| Dulbecco’s modified Eagle medium | Gibco | 11995-065 |
| Penicillin-Streptomycin | Gibco | 15140 |
| RNeasy mini extraction kit | Qiagen | 74104 |
| Superscript iii first strand synthesis system | Invitrogen | 18080-051 |
| Power Sybr green master mix | Applied Biosystems | 4367659 |
| Flag-M2 agarose beads | Sigma | A2220 |
| GFP-trap | Chromotrek | GTA-20 |
| DAPI | Sigma | D9542 |
| Cycloheximide | Sigma | C7698 |
| MG132 | Selleckchem | S2619 |
| Triton | Fisher | AC327371000 |
| Tris-HCL | Invitrogen | 15567-027 |
| Glycine | Sigma | G8898 |
| Resazurin | Sigma | R7017 |
| Crystal violet | Millipore | C0775 |
| Dynabeads CD25 | Life Technologies | 11157D |
| Pierce™ ChIP-grade Protein A/G Magnetic Beads | Thermofisher Scientific | 26162 |
| Lipofectamine™ RNAiMAX Transfection Reagent | Invitrogen | 13778150 |
| TrueCut™ Cas9 Protein v2 | Thermofisher Scientific | A36497 |
| Lipofectamine 2000 | Invitrogen | 11668019 |
| MNase enzyme | Thermofisher Scientific | 10107921001 |
| Centrinone | Cayman chemicals | CFI-400945 |
| PI/RNAse staining buffer | BD biosciences | 550825 |
| P53K370me2 | (Cui et al., 2009) | N/A |
| P53k382me2 | (Cui et al., 2009) | N/A |
| 53BP1 TTD | (Botuyan et al., 2018) | N/A |
| Critical Commercial Assays | ||
| Duolink PLA red in situ mouse/rabbit starter kit | Sigma | DUO92101 |
| Duolink PLA multi probe reagent kit | Sigma | DUO96000 |
| TaqMan™ Array Human p53 Signaling | Thermofisher Scientific | 4414168 |
| Q5 site directed mutagenesis kit | NEB | E0554S |
| Beta galactosidase staining kit | CST | 9860S |
| Deposited Data | ||
| RNA sequencing data in CT and TIRR KO | This paper | PRJNA661632 |
| Experimental Models: Cell Lines | ||
| RPE1 | ATCC | ATCC® CRL-4000™ |
| RPE1 CT | This paper | N/A |
| RPE1 TIRR −/− #3 | This paper | N/A |
| RPE1 TIRR −/− #5 | This paper | N/A |
| RPE1 53BP1 −/− | (Drane et al., 2017) | N/A |
| RPE1 p53 −/− | This paper | N/A |
| U2OS | ATCC | ATCC® HTB-96™ |
| U2OS TIRR −/− #5 | This paper | N/A |
| U2OS TIRR −/− #17 | This paper | N/A |
| Oligonucleotides | ||
| HPRT1, Actin, GAPDH MM | IDT | Housekeeping mastermixes |
| TP53 sgRNA #1: 1-F: ACCGGCAGTCACAGCACATGACGG |
IDT | N/A |
| TP53 sgRNA #1: 1-R: AAACCCGTCATGTGCTGTGACTGCC |
IDT | N/A |
| TP53 sgRNA #2: 2F:CACCGGCTTGTAGATGGCCATGGCG |
IDT | N/A |
| TP53 sgRNA #2: 2R: AAACCGCCATGGCCATCTACAAGCC |
IDT | N/A |
| TIRR sgRNA: CAGTGCCAAGATGTCGACGG(CGG) | IDT | N/A |
| 53BP1 sgRNA: AGAACGAGGAGACGGUAAUAGUGGG | IDT | N/A |
| Primers for qRT-PCR, see Table S1 | IDT | N/A |
| Recombinant DNA | ||
| pOZ-FH-N 53BP1 wt (full-length) | (Botuyan et al., 2018) | N/A |
| pOZ-FH-N 53BP1 wt (del BRCT) | (Drane et al., 2017) | N/A |
| pOZ-FH-N TIRR (WT) | (Botuyan et al., 2018, Drane et al., 2017) | N/A |
| pOZ-FH-N TIRR (K10E) | (Botuyan et al., 2018, Drane et al., 2017) | N/A |
| pOZ-FH-N TIRR (R107S) | (Botuyan et al., 2018) | N/A |
| GFP-p53 | Addgene | Addgene plasmid # 12092 |
| GFP-p53 (del CTD) | This paper | N/A |
| GFP-p53 (K382R) | This paper | N/A |
| Software and Algorithms | ||
| FlowJo | FlowJo | https://www.flowjo.com/ |
| DEseq2 | (Love et al., 2014) | https://github.com/mikelove/DESeq2 |
| EdgeR | (Robinson et al., 2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| IPA | Qiagen | https://digitalinsights.qiagen.com/products/ingenuitypathway-analysis;%20release%202019-06-15/ |
| Modfit | Modfit | http://www.vsh.com/products/mflt/index.asp |
| Cell Profiler | Cell Profiler | https://cellprofiler.org/ |
| Biorender | Biorender | https://biorender.com/ |
| Rstudio v1.2.1335 | Rstudio, Inc. | https://rstudio.com/ |
| Graphpad Prism 7 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
Immunoprecipitation and Western Blotting:
Whole cells protein extracts were generated using a lysis buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.65), 0.5% NP-40, 5 mM EDTA, 5% glycerol and protease and phosphatase cocktail inhibitors (Roche). 30 μg of lysates measured by Bradford assay were loaded into precast gels (Nu-PAGE Invitrogen, 4-12%). Immunoprecipitation was carried out by incubating 1 mg of protein lysate with Anti-Flag M2 resin (sigma) or GFP-trap, overnight at 4 °C. The following day, the beads were washed [20 mM Tris-HCl (pH 7.65), 150 mM NaCl, 3 mM MgCl2, 10% Glycerol, 0.01% NP-40] and eluted in either glycine or 4x SDS loading buffer. With GFP-trap, proteins were eluted directly in 4x loading buffer.
Proximity Ligation Assay:
Cells were plated at 25% confluency on coverslips placed in 12 well-plates. After 48 hours, cells were treated with 4 μM Nutlin-3 for 4 hours and fixed with 4% paraformaldehyde (PFA) solution for 15 minutes. Cells were permeabilized with 0.5 % triton for 5 minutes and washed with TBS solution. The steps of blocking, primary antibody incubation, probe incubation, ligation and amplification were followed according to the manufacturer’s protocol. The three-probe PLA assay was done in a similar manner with an additional first step of conjugating primary antibodies to the probes (according to manufacturer’s protocols).
Chromatin immunoprecipitation:
RPE1 cells were grown in 150 mm dishes and were 90% confluent at the time of treatment with Nutlin-3 and extraction. Cells were either untreated or treated with 4 μM Nutlin-3 for 4 hours after which DNA-protein complexes were crosslinked using 1% formaldehyde for 15 minutes. Crosslinking was terminated by adding 125mM Glycine (pH 3.5). Next, the cells were collected after washing twice with 1X PBS and lysed with Lysis Buffer I (50 mmol/L HEPES-KOH, pH7.4; 140 mmol/L NaCl; 10% glycerol; 0.5% NP-40; 0.25% Triton X-100; protease inhibitors) and incubated at 4°C for 15 minutes on a rocker. Upon centrifugation, the nuclei from the cells were collected and lysed with Lysis Buffer II (10 mMol/L Tris-HCl, pH 8.0; 200 mMol/L NaCl; protease inhibitors) for 10 minutes at 4C. The lysates were then washed thrice with MNase buffer and incubated with MNase enzyme (30U/ul). Following this, the nuclei were further lysed with 10 mMol/L Tris-HCl, pH 8.0; 100 mMol/L NaCl; 1 mMol/L EDTA; 0.5 mMol/L EGTA; 0.1% sodium deoxycholate; 0.5% N-lauroylsarcosine; protease inhibitors). The DNA-protein complexes that were extracted were then ruptured using a sonicator (30 sec on 30 sec off cycles were used for 15 minutes). Dynabeads-G coupled with either p53 (DO-1) (IgG mouse antibody was used as a control) were used to pull down p53 protein and its DNA targets. The nucleoprotein complex was washed and eluted, and the crosslinking was reversed at 65°C for 16 hours followed by proteinase K and RNase treatment. qPCR with relevant primers (provided in Key resources) was done.
PI staining and FACS:
RPE1 cells were plated at 25% confluency and treated with (1.25, 2.5 μM) of Nutlin-3 for 48 hours. Cells were trypsinized, collected and resuspended in 100% methanol and fixed overnight. Cells were washed with PBS and incubated with Propidium Iodide solution (Thermofisher Scientific) for 30 minutes to measure the DNA content. The cell profiles were recorded by Flow Cytometry and analyzed by Modfit software.
Senescence Assay:
RPE1 Cells were plated at a low confluency in a 6-well plate and allowed to grow for 6 days. Cells were washed twice with 1X PBS, fixed, and stained using solutions provided with beta-galactosidase staining kit (CST). ANKRD1, EDN1, IL6 RNA transcripts were measured according to the RNA extraction and qPCR protocol provided above.
Cell Survival assays:
In a 96-well plate, 1000 RPE1 cells were plated per well, for treatment with different concentrations of Nutlin-3. The next day, the cells were treated with serially diluted concentrations ranging from 0-10 uM of Nutlin-3. On the fourth day after the addition of Nutlin-3, the medium was replaced with medium containing Resazurin (1X solution; 100X stock: 0.5g Resazurin salt dissolved in 100 ml of 1X PBS) for 3-4 hours at 37°C. Cell viability was calculated by measuring absorbance at 570-600 nm wavelength using a microplate reader.
Bioinformatics:
Copy number, mutation and gene expression data were downloaded from the TCGA invasive breast carcinoma (n = 963) and prostate adenocarcinoma (n = 492) cohorts1,2. Samples were filtered for TP53 mutation status and copy number alterations, to retain wild-type samples. 283 breast cancer samples and 75 prostate cancer samples with wild-type TP53 status and no copy number changes were analyzed. Expression levels for TIRR and selected p53 pathway proteins (indicated in Fig. 5C) were obtained as z-scores relative to diploid samples available from RNAseq V2 RSEM data. TIRR wild-type samples were defined as tumors with a z-score < 0, while TIRR amplified samples contained a z-score > 1. Pathway expression was calculated as the average of expression for each p53 pathway member.
Preparation of proteins and peptides for NMR spectroscopy and ITC studies:
Human 53BP1-Tudor (residues 1484-1603) was expressed in E. coli and purified as previously reported (Botuyan et al., 2006). To prepare the 53BP1-TIRR complex, human 53BP1-Tudor and TIRR were co-expressed in E. coli and purified has previously reported (Botuyan et al., 2018b). The three p53 peptides (residues 363-389), with replacement of Lys370, Lys382 or both by a cysteine, were expressed in E. coli BL21 DE3 cells as a C-terminal fusion to a hexahistidine-GB1 (B1 domain of streptococcal protein G) tag. After purification of the peptides by Nickel-NTA affinity chromatography (Qiagen) and size exclusion chromatography using a Superdex S75 column (GE Healthcare), the dimethyllysine analogs were installed by reductive cysteine alkylation using 13C-enriched dimethylated 2-chloroethylamine. The modified peptides were then purified by size exclusion chromatography using a Superdex S75 column followed by cleavage of the hexahistidine-GB1 tag. Final purification and separation of the p53 peptides from the hexahistidine-GB1 tag was done by reversed-phase chromatography using a preparative C18 column (Phenomenex). The detailed procedure for preparing peptides harboring methyllysine analogs was previously published (Cui et al., 2009). The p53K370me2 (residues 366-375) and p53K382me2 (residues 377-386) peptides used for ITC were purchased from GenScript and purified by reversed-phase chromatography using a preparative C18 column (Phenomenex).
NMR spectroscopy:
The NMR experiments were performed at 25 °C using a 700 MHz Bruker Avance III spectrometer equipped with a cryoprobe. The NMR titration data were processed using NMRViewJ (Johnson, 2018) and analyzed using standard approaches (Benirschke et al., 2010). The samples were in 50 mM sodium phosphate buffer, pH 7.5, 300 mM NaCl, 1.5 mM NaN3, 0.3 mM DSS, 10% D2O and 90% H2O. The p53 peptides at a concentration of 20 μM for p53KC370me2 and p53KC382me2, and 10 μM for p53KC370me2KC382me2, were titrated with a stock solution of 53BP3-Tudor at a concentration of 1 mM. Similar molar ratios were prepared for the three p53 peptides mixed with the co-purified 53BP1-Tudor-TIRR complex. For straightforward comparison, the molar ratios were reported with respect to the concentration in dimethyllysine analog.
Isothermal titration calorimetry:
The ITC measurements were done at 10 °C using a VP-ITC instrument (MicroCal – Malvern Panalytical). 53BP1-Tudor in the calorimeter cell and the p53 peptides in the injection syringe were at concentrations of 20-100 μM and 1-4 mM, respectively in 50 mM Tris/HCl (pH 7.5) and 20 mM NaCl. The titrations were paired with injections of the peptides in buffer solution to determine the heat of dilution. Data were fit with a one-site binding model using Levenberg-Marquardt nonlinear regression in Origin 7.0 (OriginLab).
QIBC:
QIBC was performed as described previously (Kilic et al., 2019). Asynchronously growing cells were seeded on sterile 12 mm glass coverslips and allowed to proliferate until they reached a cell density of 70-90%. They were fixed in 4% formaldehyde for 15 minutes at room temperature, washed once in PBS, permeabilized for 5 minutes at room temperature in 0.2% Triton X-100 (Sigma-Aldrich) in PBS, washed twice in PBS and incubated in blocking solution (filtered DMEM containing 10% FBS and 0.02% Sodium Azide) for 15 minutes at room temperature. Primary antibodies were diluted in blocking solution and incubated for 2h at room temperature. Alexa fluor secondary antibodies (Thermo Fischer Scientific) were diluted 1:500 in blocking solution and incubated at room temperature for 1h. Cells were washed once with PBS and incubated for 10 minutes with DAPI, in PBS at room temperature. Following three washing steps in PBS, coverslips were briefly washed with distilled water and mounted on 5 μl Mowiol-based mounting media (Mowiol 4.88 (Calbiochem) in Glycerol/TRIS). 9-16 images per condition were acquired on an Olympus ScanR high-content screening system (IX83 with a Lumencor SpectraX light engine and a Hamamatsu ORCA-FLASH 4.0 V2 sCMOS camera, 2048 x 2048 pixel of size 6.5 x 6.5 um, 12 bit dynamics) using a UPLSAPO 20x air objective (NA 0.75), and analyzed using the Olympus ScanR Image Analysis software version 3.0.1. A dynamic background correction was applied, and nuclei segmentation was performed using an integrated intensity-based object detection module based on the DAPI signal. Downstream analyses focused on properly detected nuclei containing a 2C-4C DNA content as measured by total and mean DAPI intensities. Nuclear fluorescence intensities were quantified and are depicted as arbitrary units. Color-coded scatter plots of asynchronous cell populations were generated with TIBCO Spotfire data visualization software version 7.0.1. Within one experiment, similar cell numbers were compared for the different conditions.
RNA-seq Differential gene expression analysis and pathway analysis:
RNA-seq data were preprocessed as follows. First, for each sample Kallisto (Gingrich et al., 2016) (version 0.44.0) was used to pseudoalign paired-end sequencing reads to the transcriptome and produce estimated expression abundance for each transcript. GENCODE (Frankish et al., 2019) (release 29) was used as the reference transcriptome annotation. Next, the tximport R package (Soneson et al., 2015) (version 1.10) was used to aggregate the transcript-level abundance results from all samples and produce the gene-level estimated expression counts. These estimated counts were corrected for a potential bias associated with changes in average transcript length across samples. Finally, genes were filtered out if they are not expressed in most samples, using a low threshold of counts per million (CPM).
Differential gene expression analysis was performed using the edgeR package (Robinson et al., 2010), and the DESeq2 package (Love et al., 2014). The estimated log-fold-changes of the two algorithms were confirmed to be highly similar. Multiple testing correction to control the false discovery rate (FDR) was performed using the Benjamini-Hochberg procedure on the p-values.
Pathway analysis was performed using the Core Analysis of IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis; release 2019-06-15).” The input data used for IPA were the differentially expressed genes (DEGs) obtained from the edgeR algorithm with an FDR of 0.1%.
Quantification and statistical analysis:
All statistical analyses were carried out using Graphpad tools. p-values for qRT-PCR, ChIP-qPCR experiments were calculated by unpaired t-tests. p-values for PLA experiments were calculated by Mann-Whitney tests. IC50 and p-value measurements for cell survival curves were assessed by non-regression curve analysis in Graphpad. Significance was described by the following measurement of p-values: p-value < 0.0001 (****), p-value = 0.0002-0.0001 (***), p-value = 0.002-0.0002(**), p-value = 0.0332-0.002 (*), p-value= 0.1234 (ns).
Supplementary Material
ACKNOWLEDGEMENTS
D.C. is supported by R01 CA208244, Gray Foundation Team Science Award, DOD Ovarian Cancer Award W81XWH-15-0564/OC140632, Tina’s Wish Foundation, and the Claudia Adams Barr Program in Innovative Basic Cancer Research. G.M. is supported by NIH grants R01 CA132878 and R35 GM136262. M.V.B. acknowledges support from an OCRA Liz Tilberis Award. M.A. is supported by the Swiss National Science Foundation (PP00P3_179057 & 310030_197003) and by the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation program (ERC-2016-STG 714326). We thank Sharmistha Pal for sharing protocols and reagents, and Shrabasti Roychoudhury for comments on the manuscript.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- ANDERSON ME, WOELKER B, REED M, WANG P & TEGTMEYER P 1997. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol Cell Biol, 17, 6255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BENIRSCHKE RC, THOMPSON JR, NOMINE Y, WASIELEWSKI E, JURANIC N, MACURA S, HATAKEYAMA S, NAKAYAMA KI, BOTUYAN MV & MER G 2010. Molecular basis for the association of human E4B U box ubiquitin ligase with E2-conjugating enzymes UbcH5c and Ubc4. Structure, 18, 955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIEGING KT, MELLO SS & ATTARDI LD 2014. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer, 14, 359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOTUYAN MV, CUI G, DRANE P, OLIVEIRA C, DETAPPE A, BRAULT ME, PARNANDI N, CHAUBEY S, THOMPSON JR, BRAGANTINI B, ZHAO D, CHAPMAN JR, CHOWDHURY D & MER G 2018a. Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein. Nat Struct Mol Biol, 25, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOTUYAN MV, CUI G, DRANÉ P, OLIVEIRA C, DETAPPE A, BRAULT ME, PARNANDI N, CHAUBEY S, THOMPSON JR, BRAGANTINI B, ZHAO D, CHAPMAN JR, CHOWDHURY D & MER G 2018b. Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein. Nat Struct Mol Biol, 25, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOTUYAN MV, LEE J, WARD IM, KIM JE, THOMPSON JR, CHEN J & MER G 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell, 127, 1361–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANCER GENOME ATLAS N 2012. Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAPMAN JR, TAYLOR MR & BOULTON SJ 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell, 47, 497–510. [DOI] [PubMed] [Google Scholar]
- CUELLA-MARTIN R, OLIVEIRA C, LOCKSTONE HE, SNELLENBERG S, GROLMUSOVA N & CHAPMAN JR 2016a. 53BP1 Integrates DNA Repair and p53-Dependent Cell Fate Decisions via Distinct Mechanisms. Mol Cell, 64, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUELLA-MARTIN R, OLIVEIRA C, LOCKSTONE HELENE, SNELLENBERG S, GROLMUSOVA N & CHAPMAN JR 2016b. 53BP1 Integrates DNA Repair and p53-Dependent Cell Fate Decisions via Distinct Mechanisms. Molecular Cell, 64, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUI G, BOTUYAN MV & MER G 2009. Preparation of recombinant peptides with site- and degree-specific lysine 13C-methylation. Biochemistry, 48, 3798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAI Y, ZHANG A, SHAN S, GONG Z & ZHOU Z 2018. Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR. Nat Commun, 9, 2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEHART CJ, CHAHAL JS, FLINT SJ & PERLMAN DH 2014. Extensive post-translational modification of active and inactivated forms of endogenous p53. Mol Cell Proteomics, 13, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DERBYSHIRE DJ, BASU BP, SERPELL LC, JOO WS, DATE T, IWABUCHI K & DOHERTY AJ 2002. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO J, 21, 3863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRANE P, BRAULT ME, CUI G, MEGHANI K, CHAUBEY S, DETAPPE A, PARNANDI N, HE Y, ZHENG XF, BOTUYAN MV, KALOUSI A, YEWDELL WT, MUNCH C, HARPER JW, CHAUDHURI J, SOUTOGLOU E, MER G & CHOWDHURY D 2017. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature, 543, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUROCHER D & PELLETIER L 2016. 53BP1 Goes Back to Its p53 Roots. Mol Cell, 64, 3–4. [DOI] [PubMed] [Google Scholar]
- EKBLAD CM, FRIEDLER A, VEPRINTSEV D, WEINBERG RL & ITZHAKI LS 2004. Comparison of BRCT domains of BRCA1 and 53BP1: a biophysical analysis. Protein Sci, 13, 617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FONG CS, MAZO G, DAS T, GOODMAN J, KIM M, O'ROURKE BP, IZQUIERDO D & TSOU MF 2016. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKISH A, DIEKHANS M, FERREIRA AM, JOHNSON R, JUNGREIS I, LOVELAND J, MUDGE JM, SISU C, WRIGHT J, ARMSTRONG J, BARNES I, BERRY A, BIGNELL A, CARBONELL SALA S, CHRAST J, CUNNINGHAM F, DI DOMENICO T, DONALDSON S, FIDDES IT, GARCIA GIRON C, GONZALEZ JM, GREGO T, HARDY M, HOURLIER T, HUNT T, IZUOGU OG, LAGARDE J, MARTIN FJ, MARTINEZ L, MOHANAN S, MUIR P, NAVARRO FCP, PARKER A, PEI B, POZO F, RUFFIER M, SCHMITT BM, STAPLETON E, SUNER MM, SYCHEVA I, USZCZYNSKA-RATAJCZAK B, XU J, YATES A, ZERBINO D, ZHANG Y, AKEN B, CHOUDHARY JS, GERSTEIN M, GUIGO R, HUBBARD TJP, KELLIS M, PATEN B, REYMOND A, TRESS ML & FLICEK P 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res, 47, D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GINGRICH TR, ROTSKOFF GM, CROOKS GE & GEISSLER PL 2016. Near-optimal protocols in complex nonequilibrium transformations. Proc Natl Acad Sci U S A, 113, 10263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOADLEY KA, YAU C, HINOUE T, WOLF DM, LAZAR AJ, DRILL E, SHEN R, TAYLOR AM, CHERNIACK AD, THORSSON V, AKBANI R, BOWLBY R, WONG CK, WIZNEROWICZ M, SANCHEZ-VEGA F, ROBERTSON AG, SCHNEIDER BG, LAWRENCE MS, NOUSHMEHR H, MALTA TM, CANCER GENOME ATLAS N, STUART JM, BENZ CC & LAIRD PW 2018. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell, 173, 291–304 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG J, SENGUPTA R, ESPEJO AB, LEE MG, DORSEY JA, RICHTER M, OPRAVIL S, SHIEKHATTAR R, BEDFORD MT, JENUWEIN T & BERGER SL 2007. p53 is regulated by the lysine demethylase LSD1. Nature, 449, 105–8. [DOI] [PubMed] [Google Scholar]
- IWABUCHI K, BARTEL PL, LI B, MARRACCINO R & FIELDS S 1994. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci U S A, 91, 6098–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON BA 2018. From Raw Data to Protein Backbone Chemical Shifts Using NMRFx Processing and NMRViewJ Analysis. Methods Mol Biol, 1688, 257–310. [DOI] [PubMed] [Google Scholar]
- JOO WS, JEFFREY PD, CANTOR SB, FINNIN MS, LIVINGSTON DM & PAVLETICH NP 2002. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev, 16, 583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KACHIRSKAIA I, SHI X, YAMAGUCHI H, TANOUE K, WEN H, WANG EW, APPELLA E & GOZANI O 2008. Role for 53BP1 Tudor domain recognition of p53 dimethylated at lysine 382 in DNA damage signaling. J Biol Chem, 283, 34660–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KILIC S, LEZAJA A, GATTI M, BIANCO E, MICHELENA J, IMHOF R & ALTMEYER M 2019. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J, 38, e101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KURASH JK, LEI H, SHEN Q, MARSTON WL, GRANDA BW, FAN H, WALL D, LI E & GAUDET F 2008. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell, 29, 392–400. [DOI] [PubMed] [Google Scholar]
- LAMBRUS BG, DAGGUBATI V, UETAKE Y, SCOTT PM, CLUTARIO KM, SLUDER G & HOLLAND AJ 2016. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J Cell Biol, 214, 143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAPTENKO O & PRIVES C 2006. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ, 13, 951–61. [DOI] [PubMed] [Google Scholar]
- LAPTENKO O, SHIFF I, FREED-PASTOR W, ZUPNICK A, MATTIA M, FREULICH E, SHAMIR I, KADOURI N, KAHAN T, MANFREDI J, SIMON I & PRIVES C 2015. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell, 57, 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAVIN MF & GUEVEN N 2006. The complexity of p53 stabilization and activation. Cell Death Differ, 13, 941–50. [DOI] [PubMed] [Google Scholar]
- LOVE MI, HUBER W & ANDERS S 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUO J, LI M, TANG Y, LASZKOWSKA M, ROEDER RG & GU W 2004. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A, 101, 2259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEITINGER F, ANZOLA JV, KAULICH M, RICHARDSON A, STENDER JD, BENNER C, GLASS CK, DOWDY SF, DESAI A, SHIAU AK & OEGEMA K 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J Cell Biol, 214, 155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MELLO SS & ATTARDI LD 2018. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol, 51, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICHELENA J, GATTI M, TELONI F, IMHOF R & ALTMEYER M 2019. Basal CHK1 activity safeguards its stability to maintain intrinsic S-phase checkpoint functions. J Cell Biol, 218, 2865–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORALES JC, FRANCO S, MURPHY MM, BASSING CH, MILLS KD, ADAMS MM, WALSH NC, MANIS JP, RASSIDAKIS GZ, ALT FW & CARPENTER PB 2006. 53BP1 and p53 synergize to suppress genomic instability and lymphomagenesis. Proc Natl Acad Sci U S A, 103, 3310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NGUYEN TT, GRIMM SA, BUSHEL PR, LI J, LI Y, BENNETT BD, LAVENDER CA, WARD JM, FARGO DC, ANDERSON CW, LI L, RESNICK MA & MENENDEZ D 2018. Revealing a human p53 universe. Nucleic Acids Res, 46, 8153–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOREN HOOTEN N & EVANS MK 2017. Techniques to Induce and Quantify Cellular Senescence. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON MD, MCCARTHY DJ & SMYTH GK 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROY S, MUSSELMAN CA, KACHIRSKAIA I, HAYASHI R, GLASS KC, NIX JC, GOZANI O, APPELLA E & KUTATELADZE TG 2010. Structural insight into p53 recognition by the 53BP1 tandem Tudor domain. J Mol Biol, 398, 489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHENG C, MENDLER IH, RIEKE S, SNYDER P, JENTSCH M, FRIEDRICH D, DROSSEL B & LOEWER A 2019. PCNA-Mediated Degradation of p21 Coordinates the DNA Damage Response and Cell Cycle Regulation in Individual Cells. Cell Rep, 27, 48–58 e7. [DOI] [PubMed] [Google Scholar]
- SHI X, KACHIRSKAIA I, YAMAGUCHI H, WEST LE, WEN H, WANG EW, DUTTA S, APPELLA E & GOZANI O 2007. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell, 27, 636–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SONESON C, LOVE MI & ROBINSON MD 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res, 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SULLIVAN KD, GALBRAITH MD, ANDRYSIK Z & ESPINOSA JM 2018. Mechanisms of transcriptional regulation by p53. Cell Death Differ, 25, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TONG Q, CUI G, BOTUYAN MV, ROTHBART SB, HAYASHI R, MUSSELMAN CA, SINGH N, APPELLA E, STRAHL BD, MER G & KUTATELADZE TG 2015. Structural plasticity of methyllysine recognition by the tandem tudor domain of 53BP1. Structure, 23, 312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG J, YUAN Z, CUI Y, XIE R, YANG G, KASSAB MA, WANG M, MA Y, WU C, YU X & LIU X 2018. Molecular basis for the inhibition of the methyl-lysine binding function of 53BP1 by TIRR. Nat Commun, 9, 2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARD IM, DIFILIPPANTONIO S, MINN K, MUELLER MD, MOLINA JR, YU X, FRISK CS, RIED T, NUSSENZWEIG A & CHEN J 2005. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol Cell Biol, 25, 10079–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZEHIR A, BENAYED R, SHAH RH, SYED A, MIDDHA S, KIM HR, SRINIVASAN P, GAO J, CHAKRAVARTY D, DEVLIN SM, HELLMANN MD, BARRON DA, SCHRAM AM, HAMEED M, DOGAN S, ROSS DS, HECHTMAN JF, DELAIR DF, YAO J, MANDELKER DL, CHENG DT, CHANDRAMOHAN R, MOHANTY AS, PTASHKIN RN, JAYAKUMARAN G, PRASAD M, SYED MH, REMA AB, LIU ZY, NAFA K, BORSU L, SADOWSKA J, CASANOVA J, BACARES R, KIECKA IJ, RAZUMOVA A, SON JB, STEWART L, BALDI T, MULLANEY KA, AL-AHMADIE H, VAKIANI E, ABESHOUSE AA, PENSON AV, JONSSON P, CAMACHO N, CHANG MT, WON HH, GROSS BE, KUNDRA R, HEINS ZJ, CHEN HW, PHILLIPS S, ZHANG H, WANG J, OCHOA A, WILLS J, EUBANK M, THOMAS SB, GARDOS SM, REALES DN, GALLE J, DURANY R, CAMBRIA R, ABIDA W, CERCEK A, FELDMAN DR, GOUNDER MM, HAKIMI AA, HARDING JJ, IYER G, JANJIGIAN YY, JORDAN EJ, KELLY CM, LOWERY MA, MORRIS LGT, OMURO AM, RAJ N, RAZAVI P, SHOUSHTARI AN, SHUKLA N, SOUMERAI TE, VARGHESE AM, YAEGER R, COLEMAN J, BOCHNER B, RIELY GJ, SALTZ LB, SCHER HI, SABBATINI PJ, ROBSON ME, KLIMSTRA DS, TAYLOR BS, BASELGA J, SCHULTZ N, HYMAN DM, ARCILA ME, SOLIT DB, LADANYI M & BERGER MF 2017. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med, 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG A, PENG B, HUANG P, CHEN J & GONG Z 2017. The p53-binding protein 1-Tudor-interacting repair regulator complex participates in the DNA damage response. J Biol Chem, 292, 6461–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIMMERMANN M & DE LANGE T 2014. 53BP1: pro choice in DNA repair. Trends Cell Biol, 24, 108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in this study are available on NCBI SRA portal under the project id: PRJNA661632.