Abstract
Porcine epidemic diarrhea virus (PEDV) is an enteric coronavirus that causes acute watery diarrhea and vomiting in unweaned piglets, and is associated with high mortality, thus causing severe economic losses in the pig industry. Currently, although attenuated vaccines are commonly used in commercial pig farms in China, they do not completely protect against all mutated wild-type strains. Existing nucleic acid assays have high sensitivity and specificity, but the complexity of the assay process and expensive instrumentation hinder disease detection. Here, reverse transcription–enzymatic recombinase amplification (RT-ERA) was combined with the CRISPR-Cas12a system to develop a rapid diagnostic method to distinguish PEDV wild-type strains from attenuated vaccine strains. The protocol used crRNA and RT-ERA amplification primers against open reading frame 3 (ORF3), followed by Cas12a/crRNA complex detection of predefined target sequences at 37 °C for 30 min, thus producing results visible to the naked eye under LED blue light. The assay is highly sensitive and specific, detecting as few as two copies of the target gene per test and showing no cross-reactivity with other porcine pathogens. Overall, this integrated RT-ERA pre-amplification and Cas12a/crRNA cleavage assay is a practical tool for reliable and rapid detection of PEDV for diagnostic differentiation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00216-021-03716-7.
Keywords: RT-ERA, CRISPR-Cas12a, PEDV wild-type strains, Attenuated vaccine strains, Differentiation
Introduction
Porcine epidemic diarrhea virus (PEDV) was first identified in the UK in the early 1970s as a pathogen severely affecting the intestinal tract in feeder and fattening pigs [1, 2]. This virus is the causative agent of porcine epidemic diarrhea (PED), which is characterized clinically by severe watery diarrhea, vomiting, and dehydration, thus leading to high mortality in unweaned piglets [3]. In China, PED outbreaks have been reported since 1973, but can be well controlled with the CV777-derived vaccine. However, since the late 2010s, PEDV outbreaks have suddenly re-emerged in most pig farms, and the vaccine does not provide effective protection. Analysis of the genome-wide characterization of PEDV revealed that the virus has evolved, and the continued spread of the virus has caused severe economic losses in Asian countries over the past 10 years [4, 5]. To combat this variant of PEDV, two new commercial vaccines based on the ZJ08 (classic) and AJ1102 (variant) strains have been developed in China in recent years, thus allowing for relative control and mitigation of disease progression [6, 7].
PEDV is a genus of alphacoronavirus belonging to the Coronaviridae family, with a genome length of approximately 28 kb; it is an enveloped, single-stranded positive RNA virus [8, 9]. The genome of PEDV contains seven open reading frames (ORFs) encoding four structural proteins and two nonstructural polyproteins—the spike glycoprotein, ORF3 (a hypothetical protein gene), envelope, membrane, and nucleocapsid—as well as 16 nonstructural proteins (nsp1 to nsp16). Spike proteins are trimeric glycoproteins that play key roles in viral attachment, receptor binding, and virus-cell membrane fusion when viruses enter cells. ORF3 is a key factor determining the production and virulence of the virus [2, 10, 11]. In PEDV wild-type strains, the 672–675 nt ORF3 gene encodes 223–224 aa, whereas attenuated vaccine strains contain a 51 nt nucleotide deletion. The differences in ORF3 may serve as markers of adaptation to cell culture in attenuated vaccine and wild-type PEDV strains, and may be a valuable tool for PEDV diagnosis [12–14].
Rapid and sensitive nucleic acid testing is essential for the effective prevention and control of infectious diseases. Currently, the approaches to detect PEDV include viral isolation, TaqMan real-time PCR, reverse-transcription loop-mediated isothermal amplification, reverse-transcription recombinase polymerase amplification, and enzyme-linked immunosorbent assay [15–19]. However, these methods have disadvantages, such as being time-consuming, yielding false positives at high rates, and requiring expensive instruments or well-trained personnel [20, 21]. More sensitive, less time-consuming, more specific, and less instrument-dependent assays are also needed to achieve immediate detection of pathogens and to differentiate between wild-type and attenuated vaccine strains of PEDV.
The crRNA-guided CRISPR/Cas nuclease combined with isothermal amplification technology has shown great promise in recent years, because of its high sensitivity, specificity, and reliability in the development of next-generation POC molecular diagnostic technologies for viral infectious diseases. The enzymes Cas12a, Cas12b, and Cas13a have strong collateral cleavage activity, wherein Cas nucleases activated by crRNA-target duplexes indiscriminately cleave surrounding non-target single-stranded nucleic acids [22, 23]. Isothermal nucleic acid amplification method–RT-ERA was developed by Suzhou GenDx Biotech Co., Ltd. and is capable of reversing template RNA to cDNA [24]. Here, we report the first combination of RT-ERA with CRISPR/Cas12a to develop a visual assay that can instantly detect and distinguish PEDV attenuated vaccine strains from wild-type virus strains. The optimal conditions include RT-ERA reactions at 37 °C for 20 min and a Cas12a/crRNA cleavage assay at 37 °C for 5 min. The developed assay has the advantages of being less time-consuming and highly specific, thus providing results comparable to those of conventional RT-PCR, and suitability for clinical applications to reduce livestock losses by enabling early control of the spread of infectious diseases.
Materials and methods
Design, in vitro transcription, and purification of crRNAs
A target sequence of 20–23 bp after the protospacer adjacent motif sequence was designed according to the website CRISPR-DT (http://bioinfolab.miamioh.edu) for a 51-nt length nucleotide region of the ORF3 gene of the attenuated vaccine strain compared with a deleted segment of the wild-type strain. The T7 promoter (TAATACGACTCACTATAGG) and the scaffold sequence of LbCas12a (AATTTCTACTAAGTGTAGAT) were added to the 3′ end of the target sequence to form crRNA-F, and the reverse complement was crRNA-R. The detailed sequences are listed in Table 1. The FAM-N6-BHQ1 probe used in fluorescent reporter assays was synthesized by General Biological System (Anhui) Co. The two oligonucleotide chains crRNA-F/R were sent to TsingKe Co., Ltd, in China for synthesis. Two oligonucleotides were annealed to a double‐stranded DNA by using Annealing Buffer for DNA Oligos (Beyotime). Subsequently, DNA fragments were purified by gel extraction and transcribed into crRNA with a T7 High Yield RNA Transcription Kit (Vazyme, Nanjing). Finally, crRNA was purified using NucAway™ Spin columns according to the manufacturer’s instructions (Invitrogen) and stored at − 80 ℃ until use.
Table 1.
The crRNA, primers, and probes used in this study
| Primers | Sequences (5′–3′) | Size/bp |
|---|---|---|
| ORF3-F | CTTGGACTTTTTCAATACACGATTG | 619 |
| ORF3-R | TCATTCACTAATTGTAGCATACTCGT | |
| RT-qPCR-F | ATTGCCCACTTTTATATTATTGTGG | 286 |
| RT-qPCR-R | TGCCGCCCACGTATAGCTAGATACA | |
| RT-ERA-F | TTCCAATTAGACAAGCTTCAAATGTGACGGG | 238 |
| RT-ERA-R | CGCCAGGAGTAAAAGCAGACTAAACAAAGCC | |
| crRNA-1-F | GAAATTAATACGACTCACTATAGGGTAATTTCTACTAAGTGTAGATTATTATTGTGGTGCATTTTT | 66 |
| crRNA-1-R | AAAAATGCACCACAATAATAATCTACACTTAGTAGAAATTACCCTATAGTGAGTCGTATTAATTTC | |
| crRNA-2-F | GAAATTAATACGACTCACTATAGGGTAATTTCTACTAAGTGTAGATGATGCAACTATTATTTGTTG | 66 |
| crRNA-2-R | CAACAAATAATAGTTGCATCATCTACACTTAGTAGAAATTACCCTATAGTGAGTCGTATTA ATTTC | |
| Probe | FAM-N6-BHQ1 |
Extraction of viral genomic nucleic acids and reverse transcription–enzymatic recombinase amplification reaction
The viral genomic nucleic acids of the PEDV wild-type strain and attenuated vaccine strain CV777 or ZJ08, Pseudorabies virus (PRV), Porcine circovirus type 2 (PCV2), Porcine reproductive and respiratory syndrome virus (PRRSV), and Classical swine fever virus (CSFV) were extracted with a TIANamp Virus DNA/RNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions and stored at – 80 °C until use. The RT-ERA upstream and downstream primers were designed to target the ORF3 gene, and the sequences of the amplified products were sequenced to contain the 51-nt sequence of the wild-type strain in the absence of the ORF3 gene of the PEDV vaccine strain CV777. The RT-ERA primers were synthesized by TsingKe Co., Ltd. (Table 1). The standard RT-ERA reaction was performed with a basic RT-ERA nucleic acid amplification kit from Suzhou GenDx Biotech Co., Ltd. A total of 48 μL of premix was prepared for each sample, including 20 μL lysate, 2.5 μL forward primer (10 μM), 2.5 μL reverse primer (10 μM), 6 μL template RNA, and 17 μL ddH2O. Then, 48 μL of premix was transferred to each tube of RT-ERA amplification reagent and shaken and mixed. Afterward, 2 μL of ERA activator was added to the lid of the reaction tube, and the tube was tightly capped and centrifuged. The tubes were incubated at 37 ~ 42 °C for 20 min. Finally, gel electrophoresis experiments or purification of the RT-ERA amplification product was performed for subsequent applications.
Cas12a/crRNA nucleic acid detection and real-time qPCR
EnGenVR Lba Cas12a (Cpf1) (M0653T) and NEBuffer 2.1 (B7203S) were purchased from New England Biolabs (NEB). Murine RNase inhibitor was purchased from Vazyme Biotech Co., Ltd (R301-01). The Cas12a reaction system consisted of 20 μL containing 500 nM crRNA, 250 nM Cas12a, 200 nM single-stranded DNA fluorescence quantifier-labeled probe sensor, 10 U of RNase inhibitor, and 2 μL NEBuffer 2.1 with 3 μL RT-ERA product. Reactions were performed in an ABI StepOnePlus™ (Applied Biosystems) instrument at 37 °C for 30 min, and fluorescence intensity kinetics was recorded every 5 min. Meanwhile, the brightness and color changes in the reaction tubes under LED blue light, UV light, and no excitation light were observed with the naked eye. The results were recorded with a smartphone camera or a Bio-Rad Molecular Imager Gel Doc TMXR+imaging system with an exposure time of 15 ms. For real-time RT-qPCR assays, primers were designed for the deleted site sequence of the PEDV CV777 vaccine strain, as shown in Table 1. The reaction system was performed in a 20.0 μL total volume containing 10 μL 2 × One Step SYBR Green Mix (Vazyme, Nanjing, China), 1 μL One Step SYBR Green Enzyme Mix, 0.4 μL 50 × ROX Reference Dye 1, 0.4 μL (10 μM) forward and reverse primers for PEDV, 1.2 μL template, and 6.6 μL RNase-free ddH2O. The thermal cycling conditions were as follows: 50 °C for 3 min, 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s.
Evaluation of the sensitivity and specificity of Cas12a-based nucleic acid detection
The ORF3 fragments of PEDV wild-type strain were cloned into the pMD-19 T vector. The copy number of the pMD19T-ORF3 plasmid was calculated with the following formula: number of copies = (amount × 6.022 × 1023)/(length × 1 × 109 × 650). Tenfold serial dilutions of pMD19T-ORF3 (2 × 107 to 2 × 100 copies/μL) were prepared as a template for Cas12a/crRNA assays, then visually observed. The fluorescence intensity was read to determine the limit of detection to evaluate the sensitivity. The assay specificity was evaluated by testing nucleic acids from the PEDV wild-type strain, attenuated vaccine strain CV777 and ZJ08, PCV2, PRRSV, PRV, and CSFV. Complementary DNA (cDNA) was synthesized from an RNA template through reverse transcription with the FastQuant RT Kit (Tiangen, Beijing), according to the manufacturer’s instructions.
Performance of Cas12a/crRNA in PEDV detection of clinical samples
A total of 15 fecal and intestinal tissue samples were collected from 2020 to 2021 from farms located in Anhui Province which had commercial piglets with diarrhea. All samples were diluted threefold with sterile phosphate-buffered saline, ground into a homogenate, then vortexed and centrifuged at 12,000 × g for 5 min at 4 °C. Genomic RNA was extracted from the supernatant with a TIANamp Virus DNA/RNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions and stored at − 80 °C. The assay was performed with the RT-qPCR and Cas12a/crRNA method developed in this study.
Results
The Cas12a/crRNA reaction can directly detect PEDV RNA
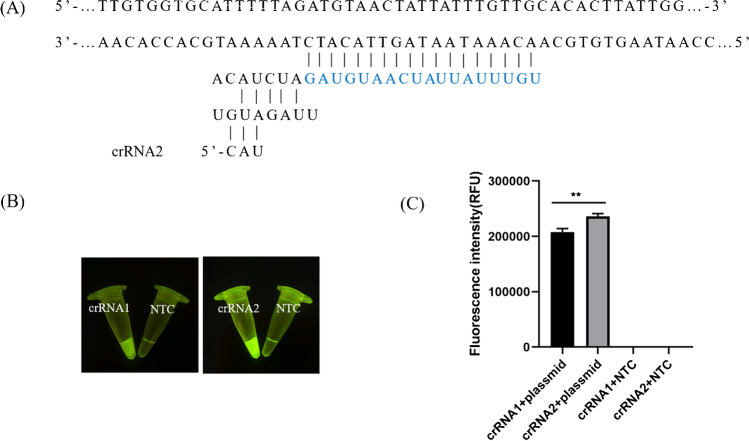
We performed RT-ERA combined with CRISPR/Cas12a to detect PEDV RNA according to the schematic diagram in Fig. 1. First, as shown in Fig. 2, we compared the ORF3 genes of different PEDV wild-type and attenuated vaccine strains, and designed RT-ERA amplification primers based on the two ends of the deletion region of the ORF3 gene. Using PEDV genomic RNA as a template, we performed exponential amplification at a constant temperature. The results of agarose gel electrophoresis showed that the primers effectively amplified the target sequences within 20 min at 37 °C. Second, we searched for the protospacer adjacent motif site (TTT) recognized by Cas12a in the deletion region (ATTGCCCACTTTTATATTACTGTGGTGCATTTTTAGATGCAACTATTAT) of the ORF3 gene, and designed the two crRNAs. To verify the effectiveness of the designed primers, we performed Cas12a experiments using the RT-ERA amplified products as templates. The experimental group that performed the reaction using both crRNAs showed a naked eye orange color (Fig. 3B), while the negative control group showed no change. Subsequently, endpoint fluorescence intensity assays were performed and revealed that the positive group using crRNA2 had higher fluorescence values than crRNA1, while the negative group had negligibly low values (Fig. 3A, C). Therefore, crRNA2 was chosen as the primer for downstream experiments. This experiment showed that the combination of RT-ERA with CRISPR-Cas12a is reliable and implementable.
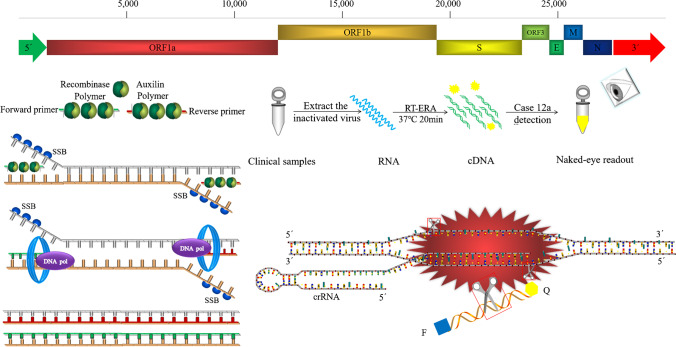
Fig. 1.
Outline of the Cas12a/crRNA assay. (A) Schematic diagram of different gene locations in the whole genome of PEDV. (B) Flow chart of the amplification principle of RT-ERA. (C) Schematic illustration of the workflow for Cas12a/crRNA PEDV testing
Fig. 2.
Comparison of partial sequences of ORF3 genes of PEDV wild-type and attenuated vaccine strains CV777
Fig. 3.
Establishment of the validity of the Cas12a/crRNA reaction. A Sequences of crRNA2 for the target sites. B Visual detection under LED blue light. C Quantification of the maximum fluorescence signal of the Cas12a/crRNA reaction collected using the qPCR instrument (n = 3, error bars showed mean ± SEM). Statistical analyses were performed using the Student’s t test in GraphPad Prism
Optimization of the Cas12a/crRNA assay for PEDV detection
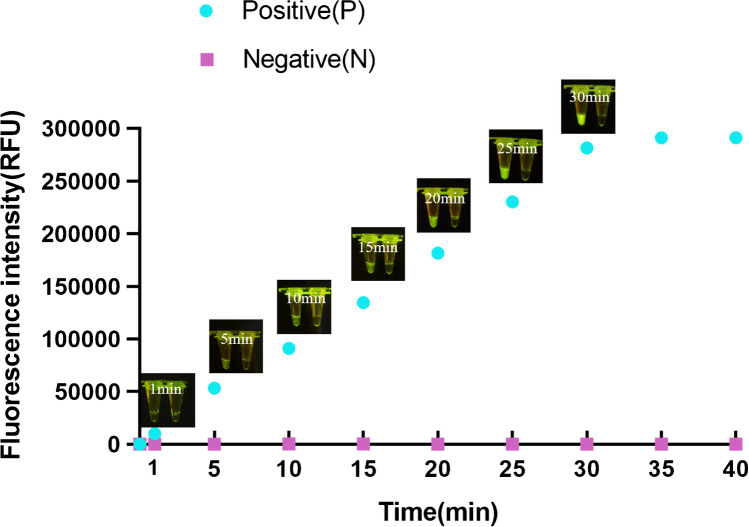
Thereafter, we optimized the reaction time in the experiment. The RNase-free ddH2O was used as a negative control, and the template containing 350 μg/mL PEDV RNA was used as a positive sample. The RT-ERA amplified product was mixed with Cas12a and incubated at 37 °C for different times, and fluorescence values were observed and collected at each time point visually. The fluorescence intensity and signal were observable after 5 min of incubation, and the fluorescence intensity continued to increase with time (Fig. 4). No fluorescence was seen in the negative control. The real-time fluorescence signal was collected with a ABI StepOnePlus™ (Applied Biosystems) instrument, and the fluorescence reached a plateau after 30 min. Therefore, 30 min was determined to be the optimal reaction time in Cas12a/crRNA assays.
Fig. 4.
Real-time fluorescence and LED blue light recorded for Cas12a/crRNA experiments at different time points. P, positive; N, negative control
Sensitivity and specificity of the Cas12a/crRNA-enhanced fluorescence assay
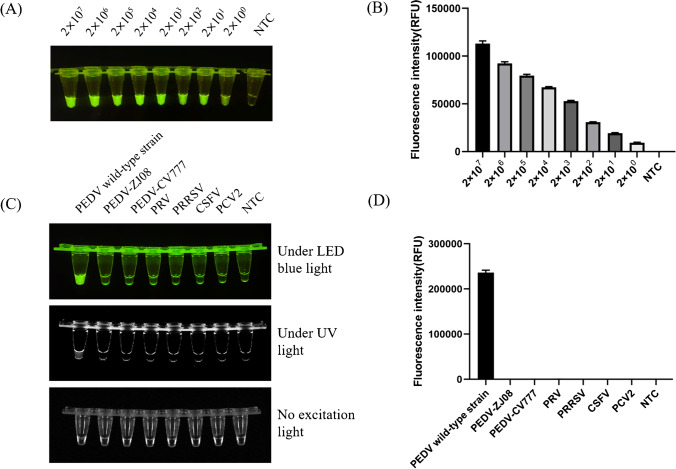
The pMD-19 T-ORF3 plasmid at 2 × 107 to 2 × 100 copies/μL was used to evaluate the sensitivity of Cas12a/crRNA detection by both the naked eye and fluorescence assays. Using different plasmid concentrations as templates for the Cas12a/crRNA-enhanced fluorescence assay, we found that the limit of detection by Cas12a/crRNA was 2 × 100 copies/μL, and the process required half an hour. Subsequently, the fluorescence value was detected at the endpoint of a traditional qPCR instrument, and the sensitivity was consistent with the results of naked eye observation; the fluorescence value showed a gradient decline (Fig. 5A, B). In addition, we added 2 copies of the target sequences in complex samples and detected them using the Cas12a/crRNA method and qPCR. The results showed that Cas12/crRNA was able to detect a weak signal, while qPCR was not (Figure S1). The Cas12a/crRNA-enhanced fluorescence assay was used to assess the specificity detection of the PEDV wild-type and attenuated vaccine strain cDNAs, as well as other porcine viruses, including PCV2, PRRSV, PRV, and CSFV at concentrations of 746, 478, 596, and 224 μg/mL, respectively. No fluorescence was seen by visual observation after the reaction with other viral DNA or cDNAs as templates, and no fluorescence intensity was detected with the qPCR instrument. The results indicated that the Cas12a/crRNA-enhanced fluorescence assay can be used for specific detection of PEDV wild-type strains (Fig. 5C, D).
Fig. 5.
Sensitivity and specificity analysis. A and B The sensitivity of Cas12a/crRNA for the detection of a tenfold serial dilution of plasmid pMD-19 T-ORF3. C and D Assessment of the specificity of Cas12a/crRNA for the detection of PEDV wild-type strains, attenuated vaccine strains, PRV, PRRSV, CSFV, and PCV2
Comparative evaluation of clinical sample detection by using Cas12a/crRNA-enhanced fluorescence assay and RT-qPCR
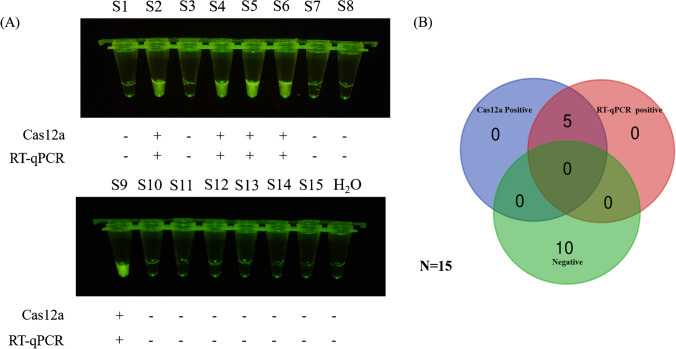
To rapidly detect and distinguish PEDV wild-type strains from attenuated vaccine strains in clinical samples, we further optimized the Cas12a/crRNA assay. With this optimization, the entire assay process was able to be completed in approximately 1 h, including RNA extraction (30 min), RT-ERA (20 min), Cas12a/crRNA detection (10 min), and result reporting (within 2 min). Subsequently, intestinal and fecal samples collected from farms with pigs with diarrhea were used to perform detection and comparison of Cas12a/crRNA-enhanced fluorescence and RT-qPCR assays. As shown in Fig. 6A, high agreement was observed between the Cas12a/crRNA and RT-qPCR methods after detection, with a positive detection rate of 33.3% for both (Fig. 6B, Fig.S2). This finding indicates that our Cas12a/crRNA assay has high specificity and sensitivity in diagnostic performance for PEDV, and can be used for rapid detection of clinical samples.
Fig. 6.
Use of Cas12a/crRNA and RT-qPCR assays to validate and test clinical samples (N = 15) for comparison
Discussion
PEDV is an infectious porcine enteric pathogen that causes severe economic losses in the pig industry and is highly contagious. To date, PEDV outbreaks have been reported in major pig production areas in China. Although PEDV vaccines have been widely used in pig farms, the classical vaccine CV777 strain does not provide complete protection against PEDV infection, and multiple variant strains of vaccines have evolved [25]. However, outbreaks of PEDV still occur in many areas, and we hypothesize that the nucleotide and amino acid site variation in PEDV has persisted. Timely knowledge and monitoring of PEDV through diagnosis are crucial for early control of infectious disease epidemics. The ideal diagnostic method should be inexpensive, accurate, and fast, and should also be easy to operate (not requiring professional expertise), and should not require specialized equipment and infrastructure (such as electricity). At present, most available methods for PEDV monitoring have numerous disadvantages, such as unsuitability for field detection and low sensitivity. Therefore, there is an urgent need to develop a specific and sensitive in situ PEDV detection method that does not require the use of expensive equipment.
The development of Cas12a and Cas13a systems has enabled the production of novel, fast, and accurate portable diagnostic equipment. Chen et al. have combined the RPA and Cas12a-crRNA complex to establish a novel method for the rapid identification of HPV16 and 18, and named it the DETECTR (DNA endonuclease-targeted CRISPR trans reporter) detection system [23]. Ren et al. have developed an RPA-CRISPR-lwCas13a-based nucleic acid detection method to diagnose ASF, which can detect as little as a single copy of ASFV plasmid and genomic DNA with sensitivity comparable to or even higher than those of qPCR methods [26]. Broughton et al. have reported a CRISPR-Cas12a-based diagnostic tool for rapid detection of SARS-CoV-2. This diagnostic tool produces results in approximately 45 min and is comparable in accuracy to conventional RT-PCR assays [27]. CRISPR-Cas12a-based nucleic acid detection methods have significantly higher specificity than PCR, RT-qPCR, RPA, or LAMP alone.
In the present study, the entire genomes of PEDV wild-type strains and attenuated vaccine strains were compared and analyzed. On the basis of the comparison results, crRNAs were designed for the ORF3 deletion region to establish a clinical rapid RT-ERA-CRISPR/Cas12a detection method. The lower limit of detection was two copies. Liu et al. have developed a TaqMan probe–based real-time PCR technique targeting the ORF3 deletion region to detect and quantify the viral load of virulent PEDV in diarrhea samples from infected piglets; this method is able to detect as few as 37 DNA copies and PEDV with a TCID50 of 0.995 [15]. A real-time reverse-transcription recombinase polymerase amplification assay for PEDV developed by Wang et al. has a PEDV RNA detection limit of 300 copies in 20 min at 39 °C [19]. The sensitivity of the Cas12a/crRNA method developed in this study is similar to or higher than those of the above methods. In addition, we observed no cross-reactivity with DNA/RNA of other swine disease viruses, and the wild-type strain of PEDV was able to be specifically distinguished from the vaccine strain within 1 h. For clinical applications, we validated both Cas12a/crRNA and RT-qPCR assays on clinically collected diarrhea samples and intestinal tissues, and we found that the test results for each sample were corresponding and identical, with a positive rate of 33.3%, thus demonstrating the reliability and usability of our developed method.
In summary, a visual, simple, and reliable nucleic acid detection method based on Cas12a/crRNA was established to identify PEDV classic attenuated vaccine strains and wild-type strains in clinical and resource-limited environments. This method has the advantages of high specificity and sensitivity. This is the first report using RT-ERA and Cas12a/crRNA methods to detect PEDV. The concept of this method might also be applied to the development of SARS-CoV-2 detection methods.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank International Science Editing (http://www.internationalscienceediting.com ) for editing this manuscript.
Funding
This work was supported financially by the National Natural Science Foundation of China (no. 31972642).
Declarations
Ethics approval
This article does not contain studies conducted on human participants or animals by any of the authors.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yueqiao Liang, Yanan Li, and Kankan Yang contributed equally to this work.
Contributor Information
Jian Tu, Email: tujian1980@126.com.
Kezong Qi, Email: qkz@ahau.edu.cn.
References
- 1.Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44(2):167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Si F, Chen B, Hu X, Yu R, Dong S, Wang R, et al. Porcine epidemic diarrhea virus ORF3 protein is transported through the exocytic pathway. Journal of virology. 2020;94(17) [DOI] [PMC free article] [PubMed]
- 3.Tian Y, Yang X, Li H, Ma B, Guan R, Yang J, et al. Molecular characterization of porcine epidemic diarrhea virus associated with outbreaks in southwest China during 2014–2018. Transbound Emerg Dis. 2020 [DOI] [PubMed]
- 4.Lu Y, Su X, Du C, Mo L, Ke P, Wang R, et al. Genetic diversity of porcine epidemic diarrhea virus with a naturally occurring truncated ORF3 gene found in Guangxi, China. Fron Vet Sci. 2020;7:435. doi: 10.3389/fvets.2020.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang W, Zhou D, Geng C, Yang K, Duan Z, Guo R, et al. Isolation and evolutionary analyses of porcine epidemic diarrhea virus in Asia. PeerJ. 2020;8:e10114. doi: 10.7717/peerj.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Wang G, Wang J, Man K, Yang Q. Cell attenuated porcine epidemic diarrhea virus strain Zhejiang08 provides effective immune protection attributed to dendritic cell stimulation. Vaccine. 2017;35(50):7033–7041. doi: 10.1016/j.vaccine.2017.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Fang L, Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Li Y, Liu Y, Chen Y, Jiao W, Feng H, et al. Isolation and identification of a recombinant porcine epidemic diarrhea virus with a novel insertion in S1 domain. Front Microbiol. 2021;12:667084. doi: 10.3389/fmicb.2021.667084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren J, Ding Z, Fang P, Xiao S, Fang L. ATPase and helicase activities of porcine epidemic diarrhea virus nsp13. Vet Microbiol. 2021;257:109074. doi: 10.1016/j.vetmic.2021.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turlewicz-Podbielska H, Pomorska-Mol M. Porcine coronaviruses: overview of the state of the art. Virol Sin. 2021 [DOI] [PMC free article] [PubMed]
- 11.Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Y, Liu Y, Chen Y, Zhao B, Ji P, Xing G, et al. Detection and phylogenetic analysis of porcine epidemic diarrhea virus in central China based on the ORF3 gene and the S1 gene. Virology journal. 2016;13(1):192. doi: 10.1186/s12985-016-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun M, Ma J, Wang Y, Wang M, Song W, Zhang W, et al. Genomic and epidemiological characteristics provide new insights into the phylogeographical and spatiotemporal spread of porcine epidemic diarrhea virus in Asia. J Clin Microbiol. 2015;53(5):1484–1492. doi: 10.1128/JCM.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Li LM, Han JQ, Sun TR, Zhao X, Xu RT, et al. A TaqMan probe-based real-time PCR to differentiate porcine epidemic diarrhea virus virulent strains from attenuated vaccine strains. Mol Cell Probes. 2019;45:37–42. doi: 10.1016/j.mcp.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Valko A, Albert E, Csagola A, Varga T, Kiss K, Farkas R, et al. Isolation and characterisation of porcine epidemic diarrhoea virus in Hungary - short communication. Acta Vet Hung. 2019;67(2):307–313. doi: 10.1556/004.2019.031. [DOI] [PubMed] [Google Scholar]
- 16.Pan Z, Lu J, Wang N, He WT, Zhang L, Zhao W, et al. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence. 2020;11(1):707–718. doi: 10.1080/21505594.2020.1771980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X, Shi L, Lv X, Yao W, Cao M, Yu H, et al. Development of a real-time reverse transcription loop-mediated isothermal amplification method for the rapid detection of porcine epidemic diarrhea virus. Virology journal. 2015;12:76. doi: 10.1186/s12985-015-0297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Li X, Shang Y, Wu J, Dong Z, Cao X, et al. Rapid differentiation of PEDV wild-type strains and classical attenuated vaccine strains by fluorescent probe-based reverse transcription recombinase polymerase amplification assay. BMC Vet Res. 2020;16(1):208. doi: 10.1186/s12917-020-02424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan B, Sun J, Zhu L, Zhou J, Zhao Y, Yu Z, et al. Development of a novel double antibody sandwich quantitative enzyme-linked immunosorbent assay for detection of porcine epidemic diarrhea virus antigen. Frontiers in veterinary science. 2020;7:540248. doi: 10.3389/fvets.2020.540248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X, Wang X, Li S, Luo W, Zhang X, Wang C, et al. Rapid, ultrasensitive, and highly specific diagnosis of COVID-19 by CRISPR-based detection. ACS Sens. 2021;6(3):881–888. doi: 10.1021/acssensors.0c01984. [DOI] [PubMed] [Google Scholar]
- 21.Jiao J, Kong K, Han J, Song S, Bai T, Song C, et al. Field detection of multiple RNA viruses/viroids in apple using a CRISPR/Cas12a-based visual assay. Plant Biotechnol J. 2021;19(2):394–405. doi: 10.1111/pbi.13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding X, Yin K, Li Z, Lalla RV, Ballesteros E, Sfeir MM, et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11(1):4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia S, Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell discovery. 2020;6:37. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Han F, Yan X, Liu L, Shu X, Hu H. Prevalence and phylogenetic analysis of spike gene of porcine epidemic diarrhea virus in Henan province, China in 2015–2019. Infect Genet Evol. 2021;88:104709. doi: 10.1016/j.meegid.2021.104709. [DOI] [PubMed] [Google Scholar]
- 26.Ren M, Mei H, Zhou M, Fu ZF, Han H, Bi D, et al. Development of a super-sensitive diagnostic method for African swine fever using CRISPR techniques. Virol Sin. 2021;36(2):220–230. doi: 10.1007/s12250-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.