Abstract
Myocardial infarction (MI) is defined as cardiomyocyte death in a clinical context consistent with ischemic insult. MI remains one of the leading causes of morbidity and mortality worldwide. Although there are a number of effective clinical methods for the diagnosis and treatment of MI, further investigation of novel biomarkers and molecular therapeutic targets is required. Circular RNAs (circRNAs), novel non-coding RNAs, have been reported to function mainly by acting as microRNA (miRNA) sponges or binding to RNA-binding proteins (RBPs). The circRNA-miRNA-mRNA (protein) regulatory pathway regulates gene expression and affects the pathological mechanisms of various diseases. Undoubtedly, a more comprehensive understanding of the relationship between MI and circRNA will lay the foundation for the development of circRNA-based diagnostic and therapeutic strategies for MI. Therefore, this review summarizes the pathophysiological process of MI and various approaches to measure circRNA levels in MI patients, tissues, and cells; highlights the significance of circRNAs in the regulation MI pathogenesis and development; and provides potential clinical insight for the diagnosis, prognosis, and treatment of MI.
Keywords: circRNA, myocardial infarction, pathological process, measuring approaches, biomarker, therapy
Graphical abstract
Myocardial infarction (MI) is an ischemic and life-threatening disease, and circular RNAs (circRNAs) could play important roles in the processes of MI. A comprehensive understanding of the emerging roles of circRNAs in the pathological process of MI is significant for circRNAs to serve as the diagnosis, prognosis, and therapeutic intervention of MI.
Introduction
Myocardial infarction (MI) is a prevalent and frequently occurring cardiovascular disease (CVD) and includes acute MI (AMI) and chronic MI. AMI refers to sudden myocardial necrosis caused by a sudden loss of oxygen supply due to acute coronary artery occlusion or major epicardial vessel stenosis.1,2 After the acute phase, MI can develop into chronic MI. MI is a severe manifestation of coronary artery disease (CAD) or acute coronary syndrome (ACS) or ischemic heart disease (IHD) with very high morbidity and mortality.3 For example, the 30-day mortality rate of AMI is approximately 10% in Europe and North America.4,5
MI can be recognized by its specific clinical characteristics, including typical ischemia symptoms, increases in the levels of myocardial necrosis biomarkers, pathological Q waves on electrocardiogram (ECG), and imaging evidence of myocardium loss.6,7 Cardiac tissue damage resulting from MI usually triggers a wide range of intense pathological processes, including cardiomyocyte apoptosis,8 autophagy,9 inflammatory response,10 proliferation,11 angiogenesis,12 cardiomyocyte hypertrophy,13 fibrosis,14 cardiac healing or repair,15 and cardiac remodeling,16 possibly culminating in heart failure (HF)17 or sudden cardiac death.
Circular RNAs (circRNAs) are single-stranded transcripts with a covalently closed-loop structure varying in lengths from dozens to thousands of base pairs.18 circRNAs are generated through backsplicing, in which the 5′ and 3′ termini are covalently connected by alternative splicing of exons from a single pre-mRNA.19,20 Various circRNA detection and characterization approaches have gradually revealed the biological features of circRNAs in recent decades: circRNAs are diverse, highly abundant, evolutionarily conserved, dynamically and specifically expressed, and they play crucial roles in different tissues.21, 22, 23 circRNAs are expressed in specific patterns in tissues, saliva, blood, and exosomes, and the methods used to detect circRNAs are simpler than those used to detect proteins.24,25
circRNAs, sometimes referred to as exon-only circRNAs, perform their physiological epigenetic functions and play non-negligible roles in various cellular processes in a variety of ways.26,27 circRNAs can function as microRNA (miRNA) sponges, RNA-binding protein (RBP) sequestering agents, or regulators of transcription and alternative splicing.18,28, 29, 30 Interestingly, some circRNAs can also be translated into functional proteins.31 Notably, a few circRNAs, such as circ_0060745, circ_0062389, circRNA ZNF292, and circ-Foxo3, might directly interact with proteins instead of sponging miRNAs via the circRNA/miRNA/protein axis during MI.32, 33, 34, 35 All of these biological properties and functions of circRNAs indicate that they have potential as sensitive and specific biomarkers and therapeutic targets for various diseases.36, 37, 38 Significantly, increasing evidence has shown that circRNA expression is changed in an MI mouse model and MI patients and that circRNAs may participate in the diverse pathophysiologic processes of MI, providing new diagnostic and therapeutic strategies for MI.28,39, 40, 41
Pathophysiology of MI
Common etiology of MI: Ischemia and I/R injury
Although the management of MI has improved over the past several decades, exploring the pathogenesis of MI is crucial. It is believed that MI damage is attributed to ischemia, reactive oxygen species (ROS), and other factors.42 In the context of ischemia, cardiomyocytes exhibit a significant decrease in ATP production, a reduced intracellular pH, and inhibited activity of sodium pump, but they tend to be overloaded with intracellular sodium ions, inducing the lysis of organelles and plasma membranes, and extensive ischemia causes cardiac tissue damage and infarction.43,44
Early and timely reperfusion, usually achieved by reopening of the occluded coronary artery, has widely been used in the clinic to protect the ischemic myocardium from MI.45, 46, 47 However, in the early period after reperfusion, paradoxical and irreversible injury, such as further cardiomyocyte death due to altered Ca2+ handling and permeability transition pore opening, known as ischemia/reperfusion (I/R) injury, is induced by myocardial reperfusion itself.48,49 Reperfusion-induced myocardial inflammation has been implicated as a secondary injury mechanism after I/R.50 Overall, I/R, which acts as a double-edged sword, not only salvages the ischemic myocardium but also induces further aggravation of myocardial injury,51 depending on the ischemic time and degree of reperfusion.52,53 Generally, if myocardial reperfusion is carried out within 2 to 3 h after the onset of ischemia, the degree of myocardial salvage exceeds the damage from ROS, calcium loading, and inflammatory cells induced by reperfusion.54, 55, 56
I/R injury induces and amplifies some myocardial injuries, such as myocardial apoptosis, autophagy, and inflammation, through multiple pathological mechanisms.9,57 The oxidative stress theory, which posits that myocardial damage results from excessive production and accumulation of ROS and/or decreased antioxidant capacity of the organism during I/R, is a current hot topic.58 Furthermore, cardiac damage due to MI has been widely studied in animal models of MI or I/R injury, generated by permanent ligation of the left anterior descending (LAD) coronary artery or LAD ligation followed by reperfusion (removing the ligation and restoring normal blood flow) after the ischemic period (30 to 60 min of ischemia), respectively.32,59,60
MI at the cellular and molecular level: Apoptosis and autophagy
As mentioned above, MI manifests as ischemia or I/R-induced cardiomyocyte death, mainly apoptosis and autophagy.9,61 The balance between apoptosis and autophagy has a direct link with cell survival and cardiac function in the MI heart and is a pivotal cellular and subcellular factor of MI.62,63
Cardiomyocytes undergo apoptosis during ischemia or I/R injury, as shown by Saraste’s research64 on myocardial samples from deceased AMI patients and Fliss’s research.65 In addition, cardiomyocyte apoptosis has been found to occur in many areas in MI hearts, such as the ischemic region, the area near the ischemic border, and even areas distant from ischemic regions.66,67 Therefore, it is suggested that apoptosis, the major determinant of infarct size, is of great significance in MI progression.9
Autophagy is elevated in ischemia and in the reperfusion phase of I/R injury.57,61 At a moderate level, cardiomyocyte autophagy can decrease the degree of apoptosis and act as an inflammatory suppressor, enhance the resistance of cardiomyocytes, and reduce the infarct size.9,61,68 However, overactivation or dysregulation of autophagy may cause it to play a maladaptive role, increasing the infarct degree and leading to adverse cardiac remodeling.9,69 In addition, mitophagy, a key mechanism of the degradation of mitochondria and the best understood pathways of selective autophagy, exerts a protective effect on cardiac function by coping with mitochondrial toxic conditions such as hypoxia and cytosolic Ca2+ overload.70
MI at the tissue and organ levels: Infarct healing and cardiac remodeling
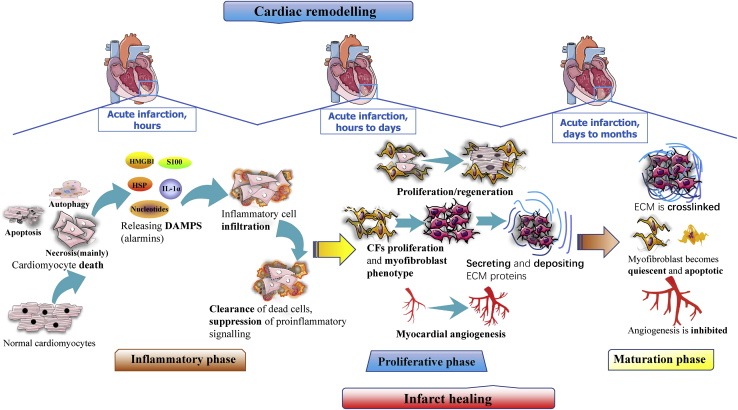
The pathophysiologic progression of MI at the tissue and organ levels basically involves two intertwined processes: infarct healing and cardiac remodeling.15,71 Infarct healing occurs mostly in the infarct zone (IZ) through a series of processes. Cardiac remodeling takes place in the IZ as well as the noninfarct zone (NIZ) of the left ventricular (LV) wall; hence, it is often referred to as LV remodeling. Cardiac remodeling in the IZ is almost in sync and involves many pathological processes occurring simultaneously with infarct healing; thus, some authors consider infarct healing to be part of cardiac remodeling.72 Simultaneous pathological events, such as inflammatory responses and angiogenesis, are reviewed and highlighted in the process of infarct healing.10, 11, 12,73 Following MI, two types of fibrotic responses occur:74 replacement fibrosis, which is involved in the proliferation phase because of its association with the activity of cardiac fibroblasts (CFs),75 and reactive fibrosis, which mostly occurs during adverse remodeling in the NIZ (Figure 1).
Figure 1.
Three phases of post-MI infarct healing and cardiac remodeling
Inflammation phase: necrotic cells release alarmins, mainly damage-associated molecular patterns (DAMPs); activate innate immune pathways; and thereby mediate leukocyte activation and infiltration into inflamed tissues to clear the dead cells and matrix debris. Proliferation phase: CFs proliferate and transform into the synthetic and myofibroblast-like phenotype, damaged cardiomyocytes are repaired and proliferated, extracellular matrix (ECM) proteins are secreted and deposited, and myocardial angiogenesis is activated. Maturation phase: ECM proteins are crosslinked and decreased, myofibroblasts become quiescent and apoptotic, angiogenesis is suppressed, and vast-formed microvessels disappear.
In general, infarct healing refers to the process through with the IZ is replaced with capillary-rich granulation tissue, leaving a firm but noncontractile scar after MI.47,76,77 Accumulating studies have divided this healing process into three distinct but overlapping phases: the inflammatory, proliferation, and maturation phases.43 Timely resolutions and a proper balance among the above three phases are crucial for an appropriate wound-healing response.78 These three phases, which are summarized in Figure 1, are characterized by dynamic and orchestrated variations in the numbers of inflammatory cells, mesenchymal cells such as CFs and myofibroblasts, extracellular matrix levels, and especially the angiogenesis response.
The response to an MI usually involves activation of progressive cardiac remodeling, which is simply defined as the geometric and functional changes in the LV. Cardiac remodeling involves a series of initially adaptive and subsequently maladaptive alterations. Put simply, post-MI hearts have reduced contractility due to expansive IZ or scarring, and this contributes to compensatory hypertrophy of the viable NIZ; however, the hypertrophic myocardium generally decompensates and causes chamber dilation, eliciting cardiac dysfunction and even precipitating HF.79,80
The prevalence of HF caused by MI-induced cardiac remodeling is high, approximately 25%, as reported by epidemiological studies.17 Moreover, the infarct size, infarct healing, and ventricular wall stress are critical factors that determine the degree of cardiac remodeling.81,82 Cardiac remodeling after MI is often caused by the activation of compensatory mechanisms involving persistent pressure and volume overload, in combination with the activation of neurohormones and inflammatory mediators.83,84
In addition, adverse remodeling in the NIZ is closely linked with exaggerated reactive fibrosis, which could cause net accumulation of extracellular matrix proteins in the cardiac interstitium, thereby inducing adverse changes in cardiac geometry and function.14,75,85 Zhang et al.86 speculated that interstitial fibrosis can be enhanced by excessive ROS production and impairment of calcium homeostasis and lead to adverse cardiac remodeling.
Biological functions and clinical significance of circRNAs in MI
As mentioned above, the significant pathological events of MI mainly include cardiomyocyte apoptosis and autophagy, the inflammatory response, angiogenesis, and the proliferation of mesenchymal cells, cardiac remodeling, and myocardial fibrosis. circRNAs are highly stable and widely expressed, and their expression is tissue and time dependent. Many circRNAs that are dynamically expressed in the plasma/serum of MI patients, MI tissues, and cells have been found to significantly participate in the regulation of the above cellular processes and the pathophysiology of MI. Therefore, we summarized the differential expression, molecular mechanism, and important biological functions of circRNAs in MI, paying attention to the clinical significance of circRNAs in MI, such as their ability to serve as sensitive biomarkers and act as new therapeutic targets.
circRNAs and cardiomyocyte apoptosis
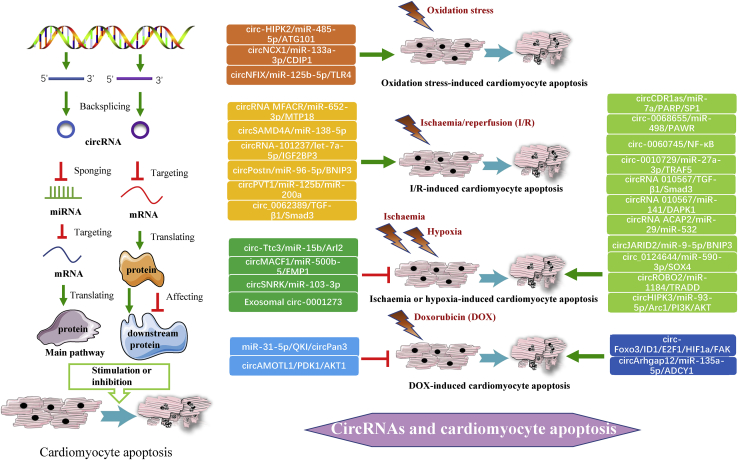
Cardiomyocyte apoptosis can be induced by I/R, hypoxia, or ischemia,9,42 and toxins such as doxorubicin (DOX),87 and these induction factors are used to construct a well-characterized mouse model of myocardial dysfunction, in which many circRNAs are implicated.88,89 Therefore, we divided the circRNAs that regulate cardiomyocyte apoptosis into the following four categories according to the different induction factors that change the expression of circRNAs (Figure 2).
Figure 2.
General pathways and specific regulatory axes of circRNAs in various cardiomyocyte apoptosis
circRNAs stimulate or inhibit MI-associated cardiomyocyte apoptosis via the circRNA/miRNA/protein axis or by directly regulating expression of protein. A wide range of circRNAs regulates four categories of cardiomyocyte apoptosis, and their expressions are changed by related induction factors.
First, three circRNAs were demonstrated to promote oxidation stress-induced cardiomyocyte apoptosis, which is a form of I/R-induced cardiomyocyte apoptosis. It was found that circHIPK2 can promote oxidation-induced cardiomyocyte apoptosis by sponging miRNA (miR)-485-5p and targeting ATG101.90 Li et al.91 revealed that similar to circHIPK2, circNCX1 functions as a powerful miR-133a-3p sponge to regulate cell death-inducing protein 1 (CDIP1) expression in cardiomyocytes, significantly enhancing oxidative stress-induced cardiomyocyte apoptosis. Additionally, circNFIX was observed to act as a pro-apoptosis factor that markedly promotes oxidative stress-induced cell apoptosis.92 The circNFIX/miR-125b-5p/Toll-like receptor 4 (TLR4) axis is suppressed by carvedilol, which thus reduces the progression of AMI and H2O2-induced cell dysfunction in vivo and in vitro.93 Notably, circNCX1 has been demonstrated to improve cardiac function in vivo in an I/R mouse model, and circNFIX has shown to do so in a mouse model of ischemia.
Second, six circRNAs are overexpressed in cardiac tissues and cells subjected to I/R injury and facilitate general I/R-induced cardiomyocyte apoptosis in response to I/R, hypoxia/reoxygenation (H/R), or anoxia/reoxygenation (A/R). Mechanistically, circRNA MFACR positively regulates mitochondrial fission and I/R injury-induced cardiomyocyte apoptosis by directly sequestering miR-652-3p and consequently facilitating the translation and activity of mitochondrial 18 kDa protein (MTP18).94 In addition, circSAMD4A was found to promote H/R-induced apoptosis and cardiac injury by sponging miR-138-5p.95 Next, Gan et al.96 verified that knockdown of circRNA-101237 has an inhibitory effect on A/R-induced cardiomyocyte apoptotic death via modulation of the circRNA-101237/let-7a-5p/insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) axis. circPostn depletion also inhibits H/R-induced myocardial apoptosis and injury in MI mice, as circPostn sponges miR-96-5p and positively regulates miR-96-5p-targeted Bcl2-interacting protein 3 (BNIP3) expression in human cardiomyocytes.97 Moreover, Luo et al.98 found that silencing circPVT1 inhibits H/R-induced cardiomyocyte apoptosis and alleviates MI and H/R injury by sponging miR-125b and miR-200a and suppressing miR-125b- and miR-200a-mediated apoptotic signaling. Zhang and Chen33 indicated that silencing circ_0062389 reduces H9c2 cell apoptosis by regulating the activity of the circ_0062389/transforming growth factor (TGF)-β1/Smad3 signaling pathway. Remarkably, circRNA MFACR and circPVT1 have the capacity to increase the infarct size in an I/R mouse model, whereas circSAMD4A and circPostn exacerbate H/R-induced injury and dysfunction in vivo in an MI mouse model.
Third, abundant circRNAs are emerging as vital regulators of ischemia or hypoxia-induced cardiomyocyte apoptosis. The expression levels of ten circRNAs were shown to be significantly increased in MI mice or rats, and these circRNAs function as positive regulators of hypoxia-induced apoptosis. First, circCDR1as was observed to induce cardiomyocyte apoptosis and aggravate injury.99 Specially, circCDR1as sponges miR-7a and reduces the activity of miR-7a, thus suppressing the expression and function of poly (ADP-ribose) polymerase (PARP) and Sp1 transcription factor (SP1). Moreover, circ-0068655 can contribute to hypoxia-induced apoptotic death and impairment of cell migration via the circ-0068655/miR-498/protein serine/threonine kinase (PRKC) apoptosis WT1 transcription factor (WT1) regulator (PAWR) regulatory axis.100 Additionally, Zhai’s group32 demonstrated that enhanced expression of circ-0060745 aggravates cell apoptosis by decreasing Bcl-2 expression and increasing Bax expression and that knockdown of circ_0060745 suppresses hypoxia-induced cardiomyocyte apoptosis through inhibition of nuclear factor κB (NF-κB) activation. Moreover, researchers found that circ-0010729 sponges miR-27a-3p, evokes tumor necrosis factor receptor (TNFR)-associated factor 5 (TRAF5) expression, and then aggravates hypoxia-induced cardiomyocyte apoptosis and injury.101 In addition, circRNA 010567 was identified to significantly promote cardiomyocyte apoptosis and exacerbate MI via modulation of the circRNA 010567/TGF-β1/Smad3 signaling pathway and circRNA 010567/miR-141/death-associated protein kinase 1 (DAPK1) axis.102,103
circRNA ACAP2 was found to sponge miR-29 and miR-532 to facilitate cardiomyocyte apoptosis after MI.104,105 Moreover, circJARID2 sponges miR-9-5p and indirectly upregulates BNIP3 expression, promoting hypoxia-induced cardiomyocyte apoptosis and cell injury.106 Additionally, circ_0124644 acts as a target gene of miR-590-3p and has a positive effect on SRY-box transcription factor 4 (SOX4). Silencing of circ_0124644 attenuates hypoxia-induced cardiomyocyte injury, such as by suppressing apoptosis and oxidative stress and inhibiting AMI progression through the circ_0124644/miR-590-3p/SOX4 axis.107 In addition, it was demonstrated that knockdown of circROBO2 alleviates hypoxia-induced cardiomyocyte injury and apoptosis and improves cardiac dysfunction by modulating the circROBO2/miR-1184/TNFR1-associated death domain protein (TRADD) axis.108 Finally, Wu et al.109 found the circHIPK3 silencing can upregulate miR-93-5p expression and abrogate the activation of the Rac1/phosphatidylinositol 3-kinase (PI3K)/serine/threonine kinase (Akt) pathway, inhibiting hypoxia-induced cardiomyocyte apoptosis and improving myocardial function after MI. In contrast, Cai et al.110 found that circ-Ttc3 is overexpressed in MI tissues and cells but has an inhibitory effect on hypoxia-induced cardiomyocyte apoptosis. Mechanistically, circ-Ttc3 could serve as an endogenous miR-15b sponge and had a positive influence on ADP ribosylation factor like GTPase 2 (Arl2) targets, suppressing hypoxia-induced ATP depletion and cardiomyocyte apoptosis.
Conversely, the expression of a few circRNAs, such as circMACF1 and circSNRK, is downregulated in mouse models subjected to hypoxia or hypoxia/serum deprivation (H/SD). circMACF1 can downregulate miR-500b-5 expression by sponging it and then increase the expression of epithelial membrane protein 1 (EMP1), potently inhibiting cardiomyocyte apoptosis and ischemic heart injury.111 Overexpression of circSNRK suppresses primary cardiomyocyte apoptosis and increases the resistance of cardiomyocytes to H/SD by sponging and inhibiting miR-103-3p.112 Significantly, in a MI mouse model, CDR1as, circ_0060745, circRNA 010567, circRNA ACAP2, circROBO2, and circHIPK3 can promote MI development and cardiac dysfunction and increase the infarct size, whereas circ-Ttc3, circMACF1, and circSNRK play a protective role during MI and improve myocardial function under hypoxic insult in vivo.
circRNAs have been inserted into exosomes and transplanted into the ischemic hearts of rats. For instance, Li et al.113 performed a contrast experiment—downregulation of the expression of umbilical cord mesenchymal stem cell (UMSC)-derived exosome circRNA by designing small interfering (si)-circ-0001273—and the results revealed that exosomes delivery of circ-0001273 inhibits cardiomyocyte apoptosis and promotes myocardium repair and regeneration in vivo in MI model rats exposed to hypoxia.
Finally, DOX is toxic to cardiomyocytes and can induce cardiomyocyte apoptosis by modulating multiple signaling pathways,87 and the circRNAs mentioned below are pertinent to DOX-induced cardiomyopathy, such as cardiomyocyte apoptosis. Du et al.35,114 discovered that during DOX treatment, ectopic or overexpressed circ-Foxo3 promotes cardiac senescence and apoptosis by binding to ID1, E2F1, HIF1a, and FAK, thus restricting their translocation to the nucleus and inhibiting their functions. They also found that silencing endogenous circ-Foxo3 abrogates the cytotoxic effect of DOX in cardiomyocytes. Similarly, circArhgap12 was found to elevate the apoptotic cell rate and exacerbate oxidative stress injury by sponging miR-135a-5p and potentially targeting adenylate cyclase 1 (ADCY1).115 Conversely, Ji’s group116 demonstrated that circPan3 suppresses DOX-induced apoptotic myocardial death, that circPan3 is regulated negatively by miR-31-5p, and that its maturation is impacted by decreased quaking (QKI) expression. Additionally, circAMOTL1 suppresses cardiomyocyte apoptosis and enhances cardiomyocyte proliferation and survival by interacting with pyruvate dehydrogenase kinase 1 (PDK1) and AKT1, thereby evoking AKT phosphorylation and promoting the nuclear translocation of AKT1 and PDK1.117 Importantly, in a mouse model of DOX-induced cardiomyopathy, circPan3, circAMOTL1, and silencing of circ-Foxo3 were demonstrated to have a powerful, protective impact on cardiac tissues and to alleviate cardiac impairment and DOX-induced cardiomyopathy in vivo.
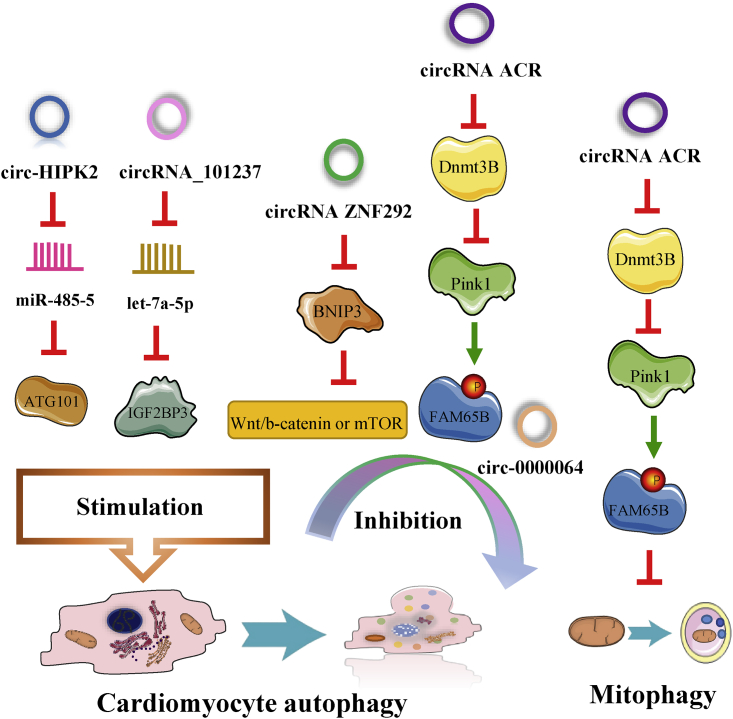
circRNAs and cardiomyocyte autophagy
Recent studies have shown that cardiomyocyte autophagy, which has detrimental impacts on MI,10 is overwhelmingly induced by ischemia and cardiac I/R injury.65,118 The circRNAs that we summarized are capable of regulating MI and influencing cardiac function by modulating cardiomyocyte autophagy (Figure 3).
Figure 3.
circRNAs are capable of regulating MI-associated cardiomyocyte autophagy and mitophagy
Some circRNAs aggravate cardiomyocyte autophagy and MI; for example, circHIPK2 worsens oxidative stress-induced autophagy, and circRNA_101237 facilitates A/R-induced autophagy.90,96 Zhou’s group90 discovered that circHIPK2 and ATG101 expression is upregulated in vitro in mouse cardiomyocytes treated with H2O2 and that circHIPK2 can promote autophagy in myocardial oxidative injury by sponging miR-485-5 and positively regulating the expression of ATG101. circRNA_101237 expression is markedly upregulated in response to A/R, and circRNA_101237 functions as a let-7a-5p sponge and increases the expression of its target protein IGF2BP3.96 Additionally, the researchers verified that circRNA_101237 can excessively activate A/R-induced autophagy, thereby promoting cardiomyocyte apoptosis by modulating the circRNA_101237/let-7a-5p/IGF2BP3 signaling axis.
However, unlike the above circRNAs, the circRNAs discussed below inhibit cardiomyocyte autophagy. Ren et al.34 observed that circRNA ZNF292 is significantly overexpressed in ischemic H9c2 cells subjected to oxygen glucose deprivation (OGD) and can inhibit apoptosis and autophagy by activating Wnt/b-catenin or the mechanistic target of rapamycin kinase (mTOR) signaling pathway through negatively targeting BNIP3. circRNA ZNF292 could ameliorate OGD-stimulated IHD, including MI. Coincidentally, circ-0000064 levels are increased by salidroside (Sal) pretreatment in the hearts of rats subjected to I/R injury, and circ-0000064 can ameliorate I/R-induced MI by inhibiting apoptosis and autophagy.119 Sal treatment attenuates I/R-induced autophagy and protects cardiac function during MI by upregulating circ-0000064 expression. Moreover, circRNA ACR protects cardiac function by inhibiting A/R-induced autophagy and mitophagy. circRNA ACR expression is significantly downregulated in the hearts of mice subjected to I/R injury, and circRNA ACR can reduce the number of autophagic vacuoles and inhibit I/R-induced mitophagy in anoxic cardiomyocytes.120 Mechanistically, circRNA ACR (autophagy-related circRNA) upregulates the expression of its downstream target phosphatase and tensin homolog-induced putative kinase 1 (Pink1) and suppresses the DNA methylation of Pink1 by interacting with DNA methyltransferase 3B (Dnmt3B). Then, Pink1 targets family with sequence similarity 65 member B (FAM65B) and phosphorylates it at S46. Importantly, circ-0000064 and circRNA ACR alleviate I/R-induced autophagy, decrease the infarct size in the context of MI, and protect cardiac tissue in an I/R injury model in vivo.
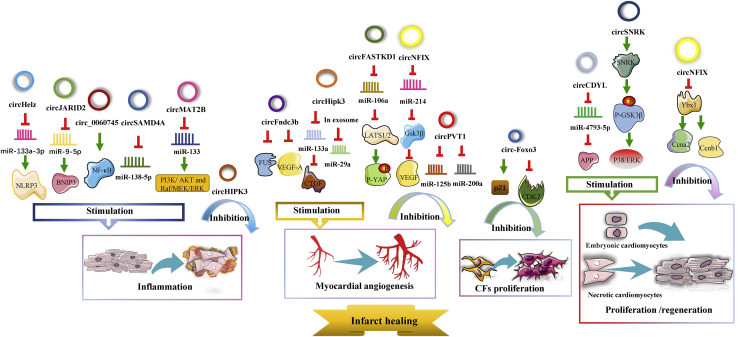
circRNAs and infarct healing after MI
The post-MI infarct healing process involves three superbly orchestrated phases as shown in the above figure: the inflammatory phase, the proliferative phase, and the maturation phase.43 We found that numerous circRNAs are prominently involved in all three phases of infarct healing and thereby influence the progression of MI. Details are provided in the following paragraphs and Figure 4.
Figure 4.
The regulatory roles of circRNAs in post-MI infarct healing
Many circRNAs are prominently involved in various events during infarct healing, such as inflammation, myocardial angiogenesis, CF proliferation, and regeneration of cardiomyocytes.
The following three circRNAs are positively associated with the inflammatory response during infarct healing. circSAMD4A not only dramatically enhances the H/R-induced inflammatory response but also stimulates H/R-induced apoptosis.95 Likewise, circ_0060745 participates in an intense inflammatory response and promotes hypoxia-induced cardiomyocyte apoptosis.32 Additionally, circJARID2 contributes to the inflammatory response and promotes apoptosis.106 In addition, Zhu et al.121 noticed that circMAT2B is highly expressed in ischemic H9c2 cells subjected to OGD. Knockdown of circMAT2B decreases ROS production and the expression of inflammatory factors, exerting a remarkable anti-inflammatory effect by upregulating miR-133 expression and thereby activating the PI3K/AKT and Raf/mitogen-activated protein (MAP) kinse-extracellular regulated MAP kinase (ERK) kinase (MEK)/ERK pathways.121 Furthermore, circHelz expression is significantly upregulated in both MI mouse models and ischemic cardiomyocytes exposed to hypoxia, and when overexpressed, it sponges and inhibits miR-133a-3p and activates the NLRP3 inflammasome, thereby inducing a proinflammatory response and pyroptosis and exacerbating hypoxia-induced cardiomyocyte injury and cardiac dysfunction in vivo and in vitro.122
Many circRNAs are relevant to proliferation and intense angiogenic responses in MI tissues or cells. Four circRNAs serve as proliferative repair promoters. Zhang’s group123 verified that circCDYL expression was downregulated during MI and that circCDYL overexpression enhances myocardial regeneration and proliferation in vitro. Further investigations have revealed that circCDYL can sponge miR-4793-5p and increase APP protein levels. circSNRK has been found to promote post-MI cardiac regeneration in adults by positively modulating phosphorylation of glycogen synthase kinase 3 β (GSK3β) and intracellular accumulation of β-catenin with the help of enhancive SNRK.112 Similarly, circFndc3b expression continuously decreases 6 weeks after MI. Moreover, overexpression of circFndc3b inhibits cardiomyocyte apoptosis and enhances the function of endothelial cells, the angiogenesis response, and cardiac repair via the circFndc3b/FUS RNA binding protein (FUS)/vascular endothelial growth factor (VEGF)-A signaling axis.124 In addition, the expression of circHIPK3 is increased in fetal and neonatal hearts compared with adult hearts, and this increase facilitates coronary artery endothelial cell proliferation, migration, and tube formation in angiogenesis by modulating the circHIPK3/miR-133a/connective tissue growth factor (CTGF) axis and promoting cardiomyogenesis by regulating the stability of the Notch1 intracellular domain (N1ICD).125 Interestingly, Wang et al.126 found that exosomal circHIPK3 in hypoxia-exposed cardiomyocytes can accelerate the cell cycle and migration of cardiac endothelial cells and promote the angiogenesis response by sponging miR-29a.
However, four other circRNAs inhibit proliferation and angiogenesis during MI. The circPVT1/miR-125b/miR-200a axis was found to suppress cell viability and proliferation in MI tissues and H/R-exposed cardiomyocytes.98 Du et al.35 demonstrated that circ-Foxo3 expression is elevated in mouse CFs and that ectopic expression of circ-Foxo3 arrests the function of CDK2 and enhances p21 activity to suppress cell-cycle progression as well as CF proliferation by forming the circ-Foxo3-p21-CDK2 ternary complex. Additionally, when ectopically or excessively expressed, circFASTKD1 can directly sponge miR-106a and then increase the expression of large tumor suppressor kinases 1 and 2 (LATS1/2), suppressing the angiogenesis of vascular endothelial cells.127 circFASTKD1 was also demonstrated to have the highest expression in human cardiac microvascular endothelial cells (HCMEC). Furthermore, loss of circNFIX enhances cardiomyocyte proliferation and angiogenesis. The regulatory mechanism involves circNFIX interacting with Y-box binding protein 1 (Ybx1) and promoting its degradation, and the circNFIX/miR-214/Gsk3β pathway regulates the release of VEGF.128 Notably, in vivo in an adult MI mouse model, circSNRK, circFndc3b, and silencing circPVT1 were found to contribute to cardioprotection and inhibit MI progression such as by decreasing the infarct size, and circHIPK3 and downregulation of circFASTKD1 expression promote angiogenesis and cardiac regeneration, thus alleviating hypoxia-induced cardiac dysfunction.
circRNAs and cardiac remodeling after MI
Cardiac remodeling, which involves extensive alterations to ventricular geometry caused by a series of profound cellular and molecular changes in both the IZ and NIZ, is relevant because it is related to a higher incidence of arrhythmias, an adverse prognosis, and increased mortality in AMI patients.129
Several research groups have investigated the role of circRNAs in cardiac remodeling. In rat models of MI, circ-Ttc3 expression in the ventricular myocardium is notably increased at the 5th week post-MI, and circ-Ttc3 prevents cardiomyocytes from ischemia-related apoptosis and adverse remodeling by directly sponging miR-15b and regulating Arl2.110 Similarly, overexpression of circCDR1as can also protect the heart from infarct-related LV remodeling and improve ventricular function, and two natural compounds, bufalin and lycorine, exert their effects by increasing circCDR1as expression.130 Conversely, upregulation of circPostn expression was found to facilitate MI-induced cardiac remodeling and dysfunction via modulation of circPostn/miR-96-5p/BNIP3.97 Furthermore, the above two circRNAs have cardioprotective effects and can prevent adverse remodeling, whereas circPostn negatively affects post-MI cardiac remodeling and function in vivo.
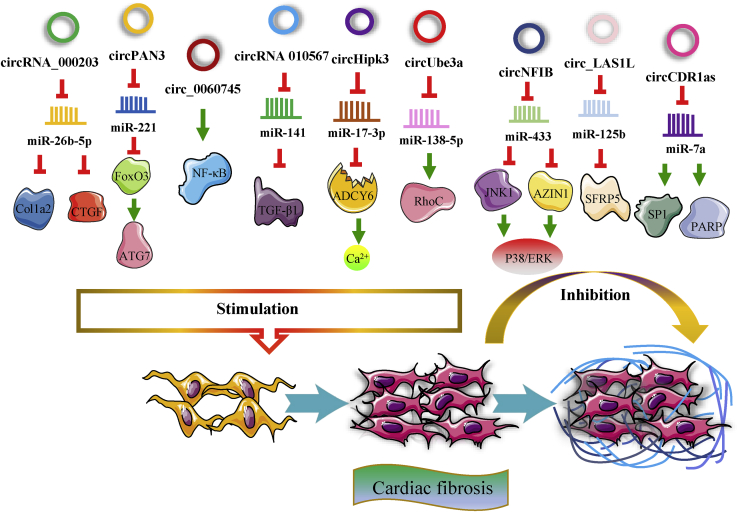
Cardiac fibrosis, divided into replacement and reactive fibrosis, is a common pathological manifestation of most post-MI cardiac remodeling.15,74 Some circRNAs have shown a remarkable capacity to regulate cardiac fibrosis (Figure 5). For instance, when its expression is elevated, circRNA_000203 can promote cardiac fibrosis by sponging miR-26b-5p and abolishing its effect, thus upregulating the expression of collagen type I alpha 2 chain (Col1a2) and CTGF in Ang-II-treated CFs.131 Additionally, circPAN3 expression is markedly increased in the IZ of MI rat hearts, and circPAN3 promotes autophagy-activated fibrosis.132 circPAN3 can facilitate CF proliferation and migration and induce excessive autophagy by modulating the circPAN3/miR-221/FoxO3/ATG7 axis in CFs stimulated by TGF-β1. Moreover, circUbe3a, which is transported into CFs from M2 macrophages (M2Ms) through small extracellular vesicles (SEVs), was also verified to sponge miR-138-5p and indirectly upregulate the expression of the target protein RhoC to accelerate proliferation, migration, and myofibroblastic transformation of CFs.133 In contrast, circNFIB alleviates fibroblast proliferation, and its expression is significantly decreased in post-MI mouse hearts and TGF-β-treated primary adult CFs.134 Mechanistically, circNFIB can directly sponge miR-433 and consequently promote the expression of target genes such as AZIN1 and JNK1 and inhibit their downstream signaling pathways, including the p38, ERK, and Smad3 pathways. Similarly, circ_LAS1L expression is downregulated in AMI patients and in activated CFs.135 circ_LAS1L directly binds to and sponges miR-125b, increases the expression of secreted frizzled-related protein 5 (SFRP5), and inhibits the expression of alpha-smooth muscle actin (SMA) and collagen I and III. Ultimately, the above mentioned circ_LAS1L/miR-125b/SFRP5 pathway leads to the inhibition of CF activation, growth, proliferation, and migration.
Figure 5.
Many circRNAs have a remarkable capacity to regulate cardiac fibrosis
In addition to acting as regulators of cardiac regeneration and remodeling, some of the circRNAs mentioned above also have a significant influence on cardiac fibrosis; for example, circ_0060745, circRNA 010567, and circHIPK3 aggravate cardiac fibrosis,32,102,126,136 whereas circCDR1as has a moderate beneficial effect in terms of reducing fibrosis.99,130 It was shown that in an MI model in vivo, circPAN3, circUbe3a, circ_0060745, and circHIPK3 facilitate cardiac fibrosis, whereas circNFIB, circRNA 010567, and circCDR1as inhibit cardiac fibrosis.
circRNAs as diagnostic and prognostic biomarkers for MI
Thousands of circRNAs have been reproducibly detected in human peripheral whole blood, and the activity of hundreds of coding genes can be revealed and quantified by evaluating circRNA expression in human blood.137 circRNAs exist extensively in cardiac tissue; sequencing data have revealed the presence of more than 15,000 circRNAs in the human heart, and an analysis by Tan et al.138 identified 1664 circRNAs specifically expressed in cardiac tissue. Moreover, it has been demonstrated that the expression of some circRNAs is specifically changed in AMI patients. Yin et al.139 identified 650 circRNAs, including 535 upregulated circRNAs and 115 downregulated circRNAs, that were differentially expressed in AMI patients compared with control subjects. Myocardial expression of circRNAs is very high in AMI patients, and due to the stability and conserved nature of circRNAs, they are likely to be robust and sensitive biomarkers for MI.140 The circRNAs that are discussed below are listed in Table 1.
Table 1.
circRNAs as diagnostic and prognostic biomarkers for MI
| Related process or special significance | Name | Dysregulation | Function | Ref. |
|---|---|---|---|---|
| Apoptosis | circNFIX | upregulated (early) downregulated (subsequently) |
auxiliary diagnostic biomarker for AMI | 92,93,141 |
| circRNA MFACR | upregulated | biomarker for MI | 94 | |
| circROBO2 | upregulated | evidence for the clinical prognosis of MI | 108 | |
| circSNRK | Downregulated (continuously; follow-up of 7 days post-MI) | biomarker for clinical prognosis after MI | 112 | |
| circSLC8A1 | upregulated | auxiliary diagnostic biomarker for IHD-like AMI | 141 | |
| Angiogenesis | circFASTKD1 | unclear | prognostic biomarker in MI patients | 127 |
| Porcine post-MI heart tissue | circCDR1as | upregulated | prognostic biomarker for the AMI | 99,130 |
| circ-RCAN2 | downregulated | prognostic biomarker for the AMI | 143 | |
| Plasma samples | circPostn | upregulated | precision diagnostic biomarker in MI patients | 97 |
| circRNA_081881 | downregulated | precision diagnostic biomarker in AMI patients | 142 | |
| hsa_circ_0124644 (introducing hsa_circ_009896) | upregulated | diagnostic biomarker of CAD patients, including AMI | 107,146 | |
| Post-MI risk classification | circRNA MICRA | downregulated | prognostic biomarker for post-MI patients | 144,145 |
| Cardiac fibrosis | circPAN3 | upregulated | potential prognostic biomarker for cardiac fibrosis during MI | 132 |
| circ_LAS1L | downregulated | diagnostic biomarker in AMI patients and cardiac fibrosis | 135 | |
| circRNA_000203, circRNA_010567 |
upregulated | biomarkers for cardiac fibrosis | 131,147 | |
| DOX-induced cardiomyopathy | circ-Foxo3 | upregulated | biomarker for DOX-induced MI | 35 |
| circRNAs derived from Ttn, Strn3, and Fhod3 | downregulated | possible biomarkers of DOX-induced cardiotoxicity | 89 | |
Cui et al.92 speculated that circNFIX is a promising biomarker for MI because it acts as a proapoptotic factor for cardiomyocyte apoptosis and because its expression is downregulated in H9c2 cells under oxidative stress and in ischemic heart tissues. Similarly, the apoptosis-related circRNA MFACR could be a sensitive biomarker for MI, as it regulates miR-652-3p and then facilitates the activity of MTP18.94 circROBO2, which increases its expression in a mouse MI model and may be a potential pathogenic factor, provides evidence for the clinical prognosis of AMI.108 Additionally, the expression of circSNRK was found to continuously downregulate throughout the follow-up of 7 days after MI and locate mainly in the cytoplasm of cardiomyocytes in rat MI hearts, suggesting circSNRK has diagnostic value in the context of MI.112 Tian et al.141 demonstrated that circSLC8A1 and circNFIX can also serve as auxiliary diagnostic markers for sudden cardiac death caused by sudden IHD, such as AMI.
In contrast to the above circRNAs, circFASTKD1 was suggested to exert a suppressive influence on angiogenesis during MI and had the potential to be a novel prognostic biomarker in MI patients.127 In addition, Cheng et al.97 observed that the expression of circPostn is markedly upregulated in plasma samples of MI patients, the MI mouse model, and the I/R-treated cell model, and they speculated that circPostn expression is closely associated with MI. Conversely, circRNA_081881 expression is dramatically decreased in plasma samples from AMI patients, suggesting that circRNA_081881 levels in the plasma can be measured to precisely diagnose AMI.142 Downregulation of circRNA_081881 expression can reduce PPARc levels because circRNA_081881 acts as a competing endogenous RNA of miR-548, abrogating the protective effect of PPARc in AMI.142 Moreover, Mester-Tonczar et al.130 revealed that circCDR1as is expressed in pig hearts and that its value as a biomarker is obvious and could be further validated in vivo in the hearts of several porcine MI models. They also demonstrated that the expression of circ-RCAN2 is significantly downregulated in vivo in reperfused infarcted porcine hearts, whereas circRNA expression exhibits an obvious ischemia time-dependent increase in vitro in hypoxic porcine cardiac progenitor cells. The different expression patterns of circ-RCAN2 between MI models and hypoxia-exposed cardiomyocytes imply that it has diagnostic significance for MI.143 Additionally, the expression of circRNA MICRA was reported to be lower in MI patients than in healthy people, and circRNA MICRA could serve as a novel prognostic biomarker for post-MI patients, predicting LV outcome and improving risk classification. In addition, researchers suggested that the biomarker circRNA MICRA may assist in fine tuning risk stratification after MI.144,145
circ_0124644 was demonstrated to be a diagnostic biomarker for CAD, including MI, because it is easily detected in peripheral blood with high specificity and sensitivity and because its expression is upregulated in serum samples from AMI patients, suggesting that its diagnostic value would be greater when combined with circ_0098964.107,146 Li et al.132 proposed that the circPAN3/miR-221/Foxo3/ATG7 axis, a strong predictor of cardiac fibrosis during MI, may serve as a sensitive biomarker for cardiac fibrosis during MI. circ_LAS1L expression is downregulated in AMI patients and activated CFs, suggesting its diagnostic value.135 Similarly, the Tang group131 and Zhou and Yu147 investigated whether circRNA_000203 and circRNA_010567 are implicated in cardiac fibrosis and lead to increased levels of profibrotic proteins by sponging their corresponding miRNAs, which may also be possible biomarkers for cardiac fibrosis.
In a previous study, circ-Foxo3 was found to be related to DOX-induced senescence and to promote cardiac senescence by modulating cytoprotective proteins in different cellular compartments, suggesting it could be a good biomarker for DOX-induced MI.35 It was observed that circRNAs derived from Ttn, Strn3, and Fhod3 can be regulated by DOX treatment;89 of course, whether these circRNAs can serve as biomarkers for DOX-induced MI remains to be elucidated. Finally, it is worth noting that circRNA_081881, circRNA MICRA, and circ_LAS1L expression levels are decreased in the blood samples of patients with AMI and that hsa_circ_0124644 expression is upregulated in the peripheral blood of CAD patients.107,135,142,145,146
Unfortunately, the sensitivity and reliability of most circRNAs as biomarkers for MI are currently less than satisfactory. Schulte et al.’s comparative analysis148 revealed that the detectability of circRNAs in plasma is insufficient, although some circRNAs are abundant and readily detectable in cardiac tissue. Moreover, they concluded that cardiac circRNAs are less sensitive cardiac biomarkers than some miRNAs or protein biomarkers, such as cMyC and cardiac troponin I (cTnI).
Moreover, detection of circRNAs in blood or exosomes is more expensive, laborious, and time consuming than existing protein analysis methods.149 However, there is great interest in using circRNAs as diagnostic and prognostic biomarkers for MI, and Lin’s group150 demonstrated that the regulation of circRNA-miRNA networks is closely related to the pathological process of MI. In addition, Schulte et al.148 revealed that combined protein/non-coding RNA (ncRNA) biomarker approaches can integrate the different characteristics of various biomarkers and be more comprehensive and valid, suggesting that circRNAs may be useful in multibiomarker combinations.
circRNA-based approaches as potential therapeutic strategies for MI
Various pieces of evidence have indicated that numerous endogenous ncRNAs, which are being or will be assessed as new therapeutic tools and in clinical trials, have great potential for the treatment of CVD, including MI.151, 152, 153, 154 RNA interference (RNAi) approaches, which include the use of siRNAs,155,156 organ-targeted RNAi using on viral vectors,157,158 and modulation of long ncRNAs (lncRNAs)159,160 and miRNAs,161,162 are particularly advanced. Innovatively, circRNAs, a newly identified and studied ncRNA that has high cytoplasmic stability, can be knocked down by RNAi (such as with siRNAs), and functions mainly by sponging miRNAs and indirectly upregulating the expression of miRNA-targeted proteins, have been employed to specifically target and regulate the various aforementioned pathophysiological processes of MI.145,163,164 Therefore, blocking or mimicking specific circRNA/miRNA/protein axes through regulation of circRNAs might result in effective inhibition of myocardial I/R injury, myocardial cell death, and adverse cardiac remodeling. Thus, when properly packaged and delivered, circRNAs represent novel long-acting therapeutic targets and attractive tools for improving MI-induced cardiac dysfunction and preventing MI development.63,125,165, 166, 167 In the following paragraphs and Tables 2 and 3, we review published studies connecting circRNAs with MI and summarize the wide range of circRNAs that may be potential therapeutic targets for MI pathogenesis (Tables 2 and 3), emphasizing the more promising circRNAs that exert multiple therapeutic effects via various regulatory mechanisms in multiple processes of MI (Table 3).
Table 2.
circRNAs exert therapeutic effects by affecting only one process involved in MI
| Effects | Name | Expression in MI | Regulatory pathway | Function | Ref. |
|---|---|---|---|---|---|
| Anti-apoptotic effects | circRNA MFACR | upregulated (A/R treatment) | circRNA MFACR/miR-652-3p/MTP18 | inhibition of circRNA MFACR alleviates mitochondrial fission and cardiomyocyte apoptosis and improves I/R injury-induced MI in vivo and in vitro | 94 |
| circ_0062389 | upregulated (H/R treatment) | circ_0062389/TGF-β1/Smad3 (upregulates both TGF-β1 and Smad3) | inhibition of circ_0062389 alleviates H/R-induced cardiomyocyte apoptosis in vitro | 33 | |
| circ-0068655 | upregulated (hypoxia treatment) | circ-0068655/miR-498/PAWR | inhibition of circ-0068655 alleviates hypoxia-induced cardiomyocyte apoptosis and impaired cell migration in vitro | 100 | |
| circ_0010729 | upregulated (hypoxia treatment) | circ_0010729/miR-27a-3p/TRAF5 | inhibition of circ_0010729 alleviates hypoxia-induced myocardial apoptosis and injury in vitro |
101 | |
| circRNA ACAP2 | unregulated (hypoxia treatment) | sponge miR-29 and miR-532 | inhibition of circRNA ACAP2 alleviates hypoxia-induced cardiomyocyte apoptosis and MI in vivo and in vitro | 104,105 | |
| circ_0124644 | unregulated (hypoxia treatment) | circ_0124644/miR-590-3p/SOX4 | inhibition of circ_0124644 alleviates hypoxia-induced cardiomyocyte injury in vitro | 107 | |
| circROBO2 | unregulated (hypoxia treatment) | circROBO2/miR-1184/TRADD | inhibition of circROBO2 alleviates hypoxia-induced cardiomyocyte injury and apoptosis in vivo and in vitro | 108 | |
| circMACF1 | downregulated (hypoxia treatment) | circMACF1/miR-500b-5p/EMP1 | circMACF1 suppresses hypoxia-induced cardiomyocyte apoptosis and injury and improves the progression of AMI in vivo and in vitro | 111 | |
| circArhgap12 | upregulated (DOX treatment) | circArhgap12/miR-135a-5p/ADCY1 | inhibition of circArhgap12 alleviates cardiomyocyte apoptosis and oxidative stress injury in vitro | 115 | |
| Anti-autophagic effects | circRNA ACR | downregulated (A/R treatment) | circRNA ACR/Pink1/FAM65B (binds to Dnmt3B, relieves DNA methylation of Pink1, and phosphorylates FAM65B) | circRNA ACR suppresses I/R-induced autophagy and mitophagy and reduces MI sizes in vivo and in vitro | 120 |
| Anti-inflammatory effects | circHelz | upregulated (hypoxia treatment) | circHelz/miR-133a-3p/NLRP3 | circHelz promotes hypoxia-induced cardiac inflammatory response and cardiomyocyte pyroptosis in vivo and in vitro | 122 |
| Proliferative and proangiogenic effects | circCDYL | downregulated (hypoxia treatment) | circCDYL/miR-4793-5p/APP | circCDYL promotes the proliferation of cardiomyocytes and angiogenesis after MI in vitro | 123 |
| circFASTKD1 | highest (HCMECs) | circFASTKD1/miR-106a/LATS1/2/YAP (increases LATS1/2 and then suppresses YAP signaling pathway) | inhibition of circFASTKD1 promotes angiogenesis and ameliorates MI in vivo and in vitro | 127 | |
| Anti-fibrotic effects | circRNA_000203 | upregulated (CF with fibrotic phenotype) | circRNA_000203/miR-26b-5p/Col1a2/CTGF (increases both Col1a2 and CTGF) | inhibition of circRNA_000203 promotes cardiac fibrosis in vitro | 131 |
| circUbe3a | derived from M2M-SEVs | circUbe3a/miR-138-5p/RhoC | circUbe3a promotes CF proliferation, migration, myofibroblastic transformation, and cardiac fibrosis in vivo and in vitro | 133 | |
| circNFIB | downregulated (CFs with TGF-β treatment) | circNFIB/miR-433 (promotes AZIN1 and JNK1 but inhibits p38 and ERK enzyme and Smad3 signaling pathway) | circNFIB alleviates CF proliferation and cardiac fibrosis in vivo and in vitro | 134 | |
| circ_LAS1L | downregulated (AMI patients, CFs with TGF-β1 treatment) | circ_LAS1L/miR-125b/SFRP5 and then inhibits alpha-SMA and collagen I and III | circ_LAS1L suppresses activation, growth, and migration of CFs in vitro | 135 |
Table 3.
circRNAs exert therapeutic effects through various regulatory mechanisms in multiple processes of MI
| Effects | Name | Expression in MI | Regulatory pathways | Function | Ref. |
|---|---|---|---|---|---|
| Anti-apoptotic and anti-autophagic effects | circRNA ZNF292 | upregulated (OGD treatment) | circRNA ZNF292/BNIP3 (inhibits BNIP3 and activates Wnt/β-catenin and mTOR signaling pathways) | circRNA ZNF292 suppresses ischemic H9c2 cell apoptosis and autophagy and alleviates OGD-induced injury in vitro | 34 |
| circ-0000064 | upregulated (Sal pretreatment) | unclear | circ-0000064 suppresses cardiomyocyte autophagy and apoptosis and improves myocardial I/R injury in vivo and in vitro | 119 | |
| circHIPK2 | upregulated (H2O2 treatment) | circHIPK2/miR-485-5p/ATG101 | inhibition of circHIPK2 suppresses cardiomyocyte autophagy to alleviate myocardial apoptosis and oxidative-induced injury in vitro | 90 | |
| circRNA_101237 | upregulated (A/R treatment) | circRNA_101237/let-7a-5p/IGF2BP3 axis (sponge let-7a-5p and upregulates IGF2BP3) | inhibition of circRNA_101237 suppresses cardiomyocyte autophagy and apoptosis and improves A/R injury in vitro | 96 | |
| Anti-apoptotic and anti-inflammatory effects | circSAMD4A | upregulated (H/R treatment) | circSAMD4A/miR-138-5p (then possibly through the SIRT1-PGC-1α pathway) | inhibition of circSAMD4A alleviates H/R-induced cardiomyocyte apoptosis and inflammatory response in vivo and in vitro | 95 |
| circJARID2 | upregulated (hypoxia treatment) | circJARID2/miR-9-5p/BNIP3 | inhibition of circJARID2 alleviates hypoxia-induced cardiomyocyte apoptosis and inflammatory response in vitro | 106 | |
| circMAT2B | upregulate (OGD treatment) | circMAT2B/miR-133/PI3K/AKT/Raf/MEK/ERK | circMAT2B knockdown reduces ROS production, hypoxia-induced apoptosis, and release of inflammatory factors in vitro | 121 | |
| Anti-apoptotic and regenerative effects | circ-0001273 | downregulated (post-MI hearts) | circ-0001273 in exosomes | circ-0001273 suppresses ischemia-induced cardiomyocyte apoptosis and promotes myocardial repair and regeneration after MI in vitro | 113 |
| Anti-apoptotic and proliferative and proangiogenic effects | circSNRK | downregulated (post-MI hearts) | sponge miR-103-3p (anti-apoptotic) and circSNRK/GSK3β/β-catenin (proliferative) | circSNRK suppresses H/SD-induced cardiomyocyte apoptosis and promotes myocardial repair and proliferation after MI in vivo and in vitro | 112 |
| circNFIX | downregulated (H2O2 treatment) and upregulated (adult hearts) | unclear (anti-apoptotic) and circNFIX/miR-214/Gsk3β and interacts with Ybx1(proliferative and proangiogenic) | inhibition of circNFIX suppresses oxidative stress-induced apoptosis in vitro and enhances cardiomyocyte proliferation and angiogenesis in vivo and in vitro | 92,93,128 | |
| circFndc3b | downregulated (post-MI hearts) | circFndc3b/FUS/VEGF (downregulates FUS but upregulates VEGF) | circFndc3b suppresses cardiomyocyte and endothelial cell apoptosis and promotes angiogenesis response and cardiac repair in vivo and in vitro | 124 | |
| circPVT1 | at high levels (MI tissue and H/R model) | circPVT1/miR-125b/miR-200a | inhibition of circPVT1 promotes cell viability and proliferation and suppresses H/R-induced cardiomyocyte apoptosis and enlargement of IZ in vivo and in vitro | 98 | |
| circ-Foxo3 | upregulated (cell cycle was arrested; older hearts) | circ-Foxo3-p21-CDK2 ternary complex (proliferative) and binding to ID1, E2F1, HIF1a, and FAK (anti-apoptotic) | inhibition of circ-Foxo3 promotes cell proliferation in vitro, and circ-Foxo3 suppresses cardiomyocyte apoptosis and exerts the anti-senescent and anti-stress roles in vivo and in vitro | 35,114 | |
| circAMOTL1 | downregulated (adult hearts) | binds to PDK1 and AKT1 and leads to phosphorylation and nuclear translocation of AKT1 | circAMOTL1 suppresses cardiomyocyte apoptosis, protects DOX-induced cardiomyopathy, and enhances cardiomyocyte proliferation and survival in vivo and in vitro | 117 | |
| Anti-apoptotic and anti-remodeling effects | circPostn | upregulated (H/R model) | circPostn/miR-96-5p/BNIP3 | inhibition of circPostn suppresses H/R-induced cardiomyocyte apoptosis and injury and post-MI remodeling in vivo and in vitro |
97 |
| circ-Ttc3 | upregulated (hypoxia model) | circ-Ttc3/miR-15b-Arl2 | suppress hypoxia-induced ATP depletion and cardiomyocyte apoptosis and enlargement of IZ and cardiac dysfunction in vivo and in vitro | 110 | |
| Anti-apoptotic and anti-fibrotic effects | circNCX1 | abundant | circNCX1/miR-133a-3p/CDIP1 | inhibition of circNCfX1 suppresses cardiomyocyte apoptosis and improves myocardial fibrosis and I/R injury in vivo and in vitro |
91 |
| circPAN3 | downregulated (DOX treatment) and upregulated (MI hearts) | miR-31-5p directly inhibits QKI and downregulates circPAN3 (anti-apoptotic); circPAN3/miR-221/FoxO3/ATG7 (anti-fibrotic) |
circPAN3 suppresses DOX-induced cardiomyocyte apoptosis, and inhibition of circPAN3 alleviates autophagy-activated fibrosis after MI in vivo and in vitro | 116,132 | |
| circRNA 010567 | downregulated (hypoxia treatment) and upregulated (CF with fibrotic phenotype) | upregulates both TGF-β1 and Smad3; circRNA 010567/miR-141/DAPK1 (anti-apoptotic); circRNA 010567/miR-141/TGF-β1 (anti-fibrotic) | inhibition of circRNA 010567 suppresses cardiomyocyte apoptosis, alleviates cardiac fibrosis, and improves MI degree in vivo and in vitro | 102,103,147 | |
| Anti-apoptotic, anti-fibrotic, and anti-inflammatory effects | circ_0060745 | upregulated (hypoxia treatment) | activates NF-κB pathway | inhibition of circ_0060745 suppresses hypoxia-induced cardiomyocyte apoptosis, alleviates myocardial fibrosis and inflammation, and limits MI size in vivo and in vitro | 32 |
| Anti-apoptotic, anti-fibrotic, and anti-remodeling effects | circCDR1as | upregulated (MI mice hearts) and exists (IZ of porcine hearts) | circCDR1as/miR-7a/PARP/SP1 (anti-apoptotic) and sponge miR-7 (anti-remodeling) | inhibition of circCDR1as suppresses hypoxia-induced cardiomyocyte apoptosis and MI injury in vivo and in vitro, and circCDR1as alleviates myocardial fibrosis, protects MI hearts from LV remodeling, and improves post-MI cardiac function in vivo and in vitro | 99,130 |
| Anti-apoptotic, anti-inflammatory, proangiogenic, and anti-fibrotic effects | circHIPK3 | upregulated (fetal, neonatal, and post-MI hearts) in exosomes |
circHPK3/miR-93-5p/Rac1/PI3K/AKT (anti-apoptotic); circHIPK3/miR-133a/CTGFs, circHIPK3/Notch1, and circHIPK3/miR-29a/VEGFA (proangiogenic); and circHIPK3/miR-17-3p/ADCY6 (anti-fibrotic) | circHIPK3 promotes angiogenesis and cardiac-regenerative repair and alleviates hypoxia-induced cardiac dysfunction in vivo and in vitro, and inhibition of circHIPK3 suppresses cardiomyocyte apoptosis, alleviates inflammation and cardiac fibrosis, and improves post-MI cardiac function in vivo and in vitro | 109,125,126,136,168 |
There are a large number of circRNAs that may serve as targets for the treatment of MI. For example, circRNA MFACR, circ_0062389, circ-0068655, circ_0010729, circRNA ACAP2, circ_0124644, circROBO2, circMACF1, and circArhgap12 may exert potential anti-apoptotic effects to inhibit AMI progression.33,94,100,101,104,105,107,108,111,116 In addition, circRNA ACR may have an effective anti-autophagic effect.120 During infarct healing, circHelz can exert anti-inflammatory effects,122 circCDYL and circFASTKD1 are likely to contribute to proliferation and proangiogensis,123,127 and circUbe3a can exert anti-fibrotic effects.133 Moreover, circRNA_000203, circNFIB, and circ_LAS1L can have anti-fibrotic effects.131,134,135
Unlike the abovementioned circRNAs, which exert therapeutic effects by affecting only one process involved in MI (Table 2), many circRNAs affect multiple processes involved in MI and may exert therapeutic effects through various regulatory mechanisms simultaneously (Table 3). For instance, circRNA ZNF292, circ-0000064, circHIPK2, and circRNA_101237 could be employed for their anti-apoptotic and anti-autophagic effects,37,90,96,119 and circSAMD4A, circJARID2, and circMAT2B may have important anti-apoptotic and anti-inflammatory roles.95,106,121 In addition, circ-0001273 probably exerts anti-apoptotic and reparative effects,113 and circSNRK, circNFIX, circFndc3b, circPVT1, circ-Foxo3, and circAMOTL1 can have vital anti-apoptotic, proliferative, and proangiogenic effects.35,92,98,112,114,117,124,128 The details are as follows: circSNRK was found to suppress cardiomyocyte apoptosis by sponging miR-103-3p and to promote cardiac regeneration and angiogenesis by regulating the circSNRK/GSK3β/β-catenin axis when it is overexpressed.112 Knocking down circNFIX inhibits oxidative stress-induced H9c2 cell apoptosis, but the signaling pathway involved is unclear.92 Carvedilol diminishes H2O2-induced damage to CFs via the circNFIX/miR-125b-5p/TLR4 axis,93 and circNFIX alleviates cardiomyocyte proliferation and angiogenesis by regulating the circNFIX/miR-214/Gsk3β axis and binding with Ybx1.128 Silencing of circ-Foxo3 has a protective effect against cardiac senescence and DOX-induced cardiomyopathy, as circ-Foxo3 binds to ID1, E2F1, HIF1a, and FAK,114 and disruption of the circ-Foxo3-p21-CDK2 ternary complex can promote cell-cycle progression as well as CF proliferation.35
Moreover, circPostn and circ-Ttc3 may be applied for anti-apoptotic and anti-remodeling effects.97,110 circNCX1, circPAN3, and circRNA 010567 are capable of exerting anti-apoptotic and anti-fibrotic effects.91,102,103,116,132,147 circPAN3 suppresses cardiomyocyte apoptosis, but the regulatory pathway involved is unclear, miR-31-5p downregulates circPAN3 expression, and QKI expression has a positive impact on the maturation of circPan3.116 Furthermore, knockdown of circPAN3 can promote CF proliferation and migration and autophagy-activated myocardial fibrosis by regulating the circPAN3/miR-221/FoxO3/ATG7 axis.132 circRNA 010567 siRNA can reduce cardiomyocyte apoptosis via the circRNA 010567/TGF-β1/Smad3 signaling pathway102 and the circRNA 010567/miR-141/DAPK1 axis,103 whereas silencing of circRNA 010567 can contribute to alleviation of myocardial fibrosis through the circRNA 010567/miR-141/TGF-β1 axis.147 Additionally, circ_0060745 exerts anti-apoptotic, anti-fibrotic, and anti-inflammatory effects via the modulation of the circ_0060745/NF-κB pathway.32 Furthermore, circCDR1as is a potential therapeutic target due to its anti-apoptotic, anti-fibrotic, and anti-remodeling effects, as inhibition of circCDR1as suppresses cardiomyocyte apoptosis via modulation of the circCDR1as/miR-7a/PARP/SP1 axis,99 and circCDR1as, being upregulated by bufalin and lycorine, alleviates LV remodeling and cardiac fibrosis by sponging miR-7.130 circHIPK3 may be an ideal anti-apoptotic, anti-inflammatory, proangiogenic, and anti-fibrotic candidate, as silencing of circHIPK3 suppresses cardiomyocyte apoptosis via the circHPK3/miR-93-5p/Rac1/PI3K/AKT axis.109 circHIPK3 promotes myocardial angiogenesis by modulating the circHIPK3/miR-133a/CTGF axis,125 the stability of N1ICD, and the circHIPK3/miR-29a/VEGFA axis,126 and circHIPK3, the expression of which is upregulated by adrenaline, is beneficial to the heart in the short term, but a reduction in its expression can maintain post-MI cardiac function in the long term through regulation of myocardial fibrosis via the circHIPK3/miR-17-3p/ADCY6 axis.136,168
In addition, Gallet et al.169 proposed that cardiosphere-derived, cell-secreted exosomes are potential cell-free agents for the treatment for MI, and some circRNAs, such as circ-0001273 and circHIPK3, are derived from exosomes and have the potential to be applied for exosome-associated treatment of MI.113,126 However, although having good potential according to extensive basic research, there have been an insufficient number of clinical trials testing the application of circRNAs for MI treatment, and some issues, such as the use of appropriate preclinical animal models, the identification of drug-delivery systems that specifically target MI tissues and cells, the heterogeneity of different populations, and the in vivo safety and possible long-lasting effects of circRNA mimics or inhibitors, need to be addressed.
Strategies for measuring circRNA levels in the context of MI
A variety of approaches and strategies have been developed to detect, characterize, enrich, and verify the presence of circRNAs and reveal the interaction between circRNAs and miRNAs in MI patients, in vivo in MI animal models, and in vitro in cardiomyocytes exposed to MI-related conditions. In recent years, next-generation RNA sequencing and microarray technology have been widely used for the identification of new circRNA species, profiling of circRNA expression, and analysis of the correlation between epigenetics and genetic phenotype.170, 171, 172 For example, Lin et al.150 detected a total of 266 circRNAs (121 upregulated and 145 downregulated) that were differentially expressed in peripheral blood between patients in the control group and patients with ACS. Wu et al.173 used Arraystar microarray analysis to identify 63 circRNAs with distinct expression patterns (29 upregulated and 34 downregulated) between mice in the MI-induced HF and sham groups.
The accuracy and comprehensiveness of circRNA identification strategies are determined by the rigor and reliability of the algorithm; however, there is little overlap in the results of the identification of various algorithms; thus choosing appropriate algorithms and combination of different bioinformatics tools should be used to address these biases.174,175 A variety of bioinformatics tools and algorithms have been developed for the prediction and identification of circRNAs, such as circRNA finder, find-circ, CIRI, MapSplice, CIRCexplorer, and Acfs.176,177 For instance, by using bioinformatic tools, such as Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses, Yu et al.178 identified dysregulated circRNAs (70 downregulated and 30 upregulated) in coronary heart disease (CHD) patients compared to controls and predicted 12 circRNAs that might act as circRNA-miRNA sponges to regulate their CHD-related parental genes.
RNase R, a member of the E. coli RNR superfamily, can degrade most linear RNA molecules (e.g., miRNAs) in the 3′−5′ direction but has no effect on circRNAs with closed loop structures; therefore, RNase R treatment is used to increase the relative concentration of circRNAs, also known as circRNA enrichment.112,179 For example, RNase R treatment, polyadenylation, and the poly(A)+ RNA depletion (RPAD) method are used to isolate circRNAs, thus enabling systematic identification and characterization of highly enriched, highly pure populations of circRNAs.180 Then, two approaches can be employed to verify and validate circRNAs: reverse transcription-polymerase chain reaction (RT-PCR) and Northern blotting.
cDNA for RT-PCR is gotten from RNase R-digested RNA through reverse transcription and then amplified with divergent primers and convergent primers. Subsequent agarose gel electrophoresis reveals that the divergent primer generates a band, but the convergent primer does not in samples treated with RNase R, whereas both divergent and convergent primers produce bands in samples not treated with RNase R. This proves that circRNAs are present and resistant to digestion by RNase R; therefore, RT-PCR can serve as a feasible quantitative estimation of circRNA abundance.120 In addition, online resources such as CircInteractome can be employed to design divergent primers that span the circRNA junction for quantitative real-time PCR (qRT-PCR) analysis of circRNA.181 Northern blotting can be conducted by using circRNA- and mRNA-specific probes, which specifically hybridize with circRNAs and linear mRNAs. Only a circRNA band is visible in samples treated with RNase R, as mRNAs are digested, whereas both circRNA bands and mRNA bands can be seen in samples not treated with RNase R. This indicates that Northern blotting can be used to convincingly verify the circular configuration of putative circRNAs.182
Furthermore, some approaches, such as dual-luciferase reporter assay and fluorescence in situ hybridization (FISH) coupled with high-resolution microscopy, are widely applied to analyze the abundance, subcellular localization, and binding sites of circRNAs, and circRNA-miRNA interactions in cells and tissues. The circRNA-miRNA interaction can be predicted with the help of Arraystar’s proprietary miRNA target-prediction software Interaction.133,183 In addition, the RNA pull-down assay and RNA immunoprecipitation (RIP) assay followed by circRNA sequencing are practical strategies for predicting circRNA-protein interactions.184
Conclusions
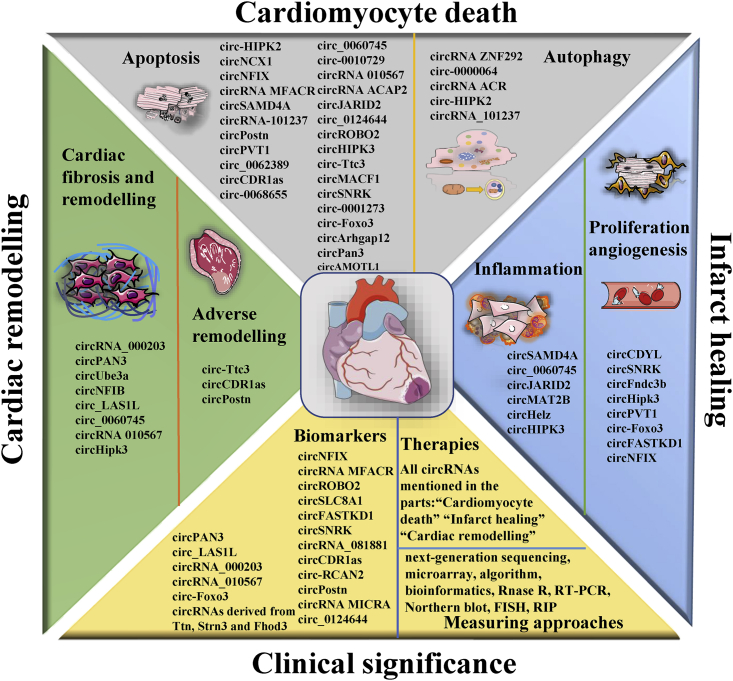
MI is an ischemic and life-threatening emergency endangering human life. There are a series of physiological and pathological processes involved in the progression of MI, such as cardiomyocyte apoptosis and autophagy, myocardial I/R, post-MI infarct healing, and cardiac remodeling. Recently, novel diagnostic and therapeutic approaches for MI have attracted much attention, and numerous candidate targets, especially circRNAs, have been investigated and developed. A growing body of evidence has verified that circRNAs function by binding to miRNAs or RBPs and that circRNAs prominently participate in and regulate the pathogenesis of MI and the pathophysiological processes and events of MI mainly through different circRNA/miRNA/protein axes. In this review, we discussed the pathogenesis and pathophysiology of MI and the biological functions of circRNAs in the process of MI, emphasizing the clinical application of circRNAs in MI, including their diagnostic and therapeutic values and approaches for measuring circRNAs in the context of MI (Figure 6).
Figure 6.
The biological functions and clinical significances of circRNAs for MI
circRNAs, which can be measured by various approaches in MI patients, tissues, and cells, significantly participate in regulation of the cellular processes and the pathophysiology of MI and have excellent biological functions and clinical significances for MI. Considerable circRNAs are involved in cardiomyocyte apoptosis and autophagy, inflammation, proliferation and angiogenesis, cardiac remodeling, and fibrosis, providing novel diagnostic and treatment strategies for MI.
The advancement of novel bioinformatic approaches (such as microarray analysis and algorithms), coupled with biochemical enrichment strategies (such as RNase R treatment and RPAD), deep sequencing, and gene-editing strategies (such as CRISPR-Cas9 technologies), will help allow comprehensive analysis of the involvement of circRNAs in the regulation of MI. In conclusion, the future holds promising application prospects for circRNAs serving as specific biomarkers and therapeutic targets for MI. However, unlike that of coding RNAs, lncRNAs and miRNAs, our current understanding of the biology of circRNAs is far from complete, and application of circRNAs in the clinic is still very difficult and requires much more research. For example, circRNAs have multiple cellular target sites and regulatory mechanisms and may induce unwanted adverse influences and off-target effects in other cells or tissues. Moreover, there are individual differences in the expression and function of circRNAs, which are affected by inter- and intra-individual factors such as ethnicity, gender, diet, and activity. Additionally, the safety and efficacy of circRNA-based therapeutic approaches, including of the vectors used in some gene therapies, are unknown and need to be thoroughly evaluated.
There are some suggestions and directions related to circRNAs and MI in future research and clinical trials. First, MI is known to involve large areas of the myocardium, including cardiomyocytes, CFs, cardiac inflammatory cells, and endothelial cells; hence, the exact mechanism by which circRNAs in various kinds of cells participate in the development of MI deserves additional attention. Second, there is a pressing need to perform a large number of in vivo experiments, such as targeting circRNAs at specific sites and observing their long-term effects, to improve the diagnostic and therapeutic specificity of circRNA blockers and mimics for MI. Third, circRNA detection technology needs to be improved, such as by making high-throughput sequencing and molecular bioinformatics approaches faster and more reliable. Fourth, the safety and effectiveness of multiple circRNAs need to be further monitored and evaluated in numerous preclinical trials. Finally, through advancing circRNA detection technology and attaining a comprehensive understanding of the pathogenesis of MI, we believe that the novel circRNA-based approaches will someday be widely applied for the monitoring, diagnosis, treatment, and prognostication of MI, thereby dramatically alleviating the high morbidity and mortality of MI and benefiting all of mankind.
Acknowledgments
No ethics approval was required for this review that did not involve patients or patient data. All authors consent to publication. This work was supported by the National Natural Science Foundation of China (22006084), Qingdao Applied Basic Research Project (19-6-2-49-cg and 19-6-2-39-cg), and Hubei Key Laboratory of Environmental and Health Effects of Persistent Toxic Substances (PTS2019-05).
Author contributions
Z.-J.W., Y.-C.W., and H.-W.L. searched the literature. Y.-F.Z. and H.X. provided inspiration and guidance for writing. Z.-J.W. wrote the manuscript and prepared all of the figures and tables. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Agewall S., Beltrame J.F., Reynolds H.R., Niessner A., Rosano G., Caforio A.L., De Caterina R., Zimarino M., Roffi M., Kjeldsen K., WG on Cardiovascular Pharmacotherapy ESC working group position paper on myocardial infarction with non-obstructive coronary arteries. Eur. Heart J. 2017;38:143–153. doi: 10.1093/eurheartj/ehw149. [DOI] [PubMed] [Google Scholar]
- 2.Reed G.W., Rossi J.E., Cannon C.P. Acute myocardial infarction. Lancet. 2017;389:197–210. doi: 10.1016/S0140-6736(16)30677-8. [DOI] [PubMed] [Google Scholar]
- 3.Smilowitz N.R., Mahajan A.M., Roe M.T., Hellkamp A.S., Chiswell K., Gulati M., Reynolds H.R. Mortality of Myocardial Infarction by Sex, Age, and Obstructive Coronary Artery Disease Status in the ACTION Registry-GWTG (Acute Coronary Treatment and Intervention Outcomes Network Registry-Get With the Guidelines) Circ. Cardiovasc. Qual. Outcomes. 2017;10:e003443. doi: 10.1161/CIRCOUTCOMES.116.003443. [DOI] [PubMed] [Google Scholar]
- 4.Townsend N., Wilson L., Bhatnagar P., Wickramasinghe K., Rayner M., Nichols M. Cardiovascular disease in Europe: epidemiological update 2016. Eur. Heart J. 2016;37:3232–3245. doi: 10.1093/eurheartj/ehw334. [DOI] [PubMed] [Google Scholar]
- 5.Benjamin E.J., Virani S.S., Callaway C.W., Chamberlain A.M., Chang A.R., Cheng S., Chiuve S.E., Cushman M., Delling F.N., Deo R., American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation. 2018;137:e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 6.Thygesen K., Alpert J.S., Jaffe A.S., Simoons M.L., Chaitman B.R., White H.D., Thygesen K., Alpert J.S., White H.D., Jaffe A.S., Writing Group on the Joint ESC/ACCF/AHA/WHF Task Force for the Universal Definition of Myocardial Infarction. ESC Committee for Practice Guidelines (CPG) Third universal definition of myocardial infarction. Eur. Heart J. 2012;33:2551–2567. [Google Scholar]
- 7.Armstrong P.W. Defining myocardial infarction: a work in progress: Ischaemic heart disease. Heart. 2008;94:1076–1079. doi: 10.1136/hrt.2007.135202. [DOI] [PubMed] [Google Scholar]
- 8.Krijnen P.A., Nijmeijer R., Meijer C.J., Visser C.A., Hack C.E., Niessen H.W. Apoptosis in myocardial ischaemia and infarction. J. Clin. Pathol. 2002;55:801–811. doi: 10.1136/jcp.55.11.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D., Zhang K., Hu P. The Role of Autophagy in Acute Myocardial Infarction. Front. Pharmacol. 2019;10:551. doi: 10.3389/fphar.2019.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frangogiannis N.G. The inflammatory response in myocardial injury, repair, and remodelling. Nat. Rev. Cardiol. 2014;11:255–265. doi: 10.1038/nrcardio.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malliaras K., Zhang Y., Seinfeld J., Galang G., Tseliou E., Cheng K., Sun B., Aminzadeh M., Marbán E. Cardiomyocyte proliferation and progenitor cell recruitment underlie therapeutic regeneration after myocardial infarction in the adult mouse heart. EMBO Mol. Med. 2013;5:191–209. doi: 10.1002/emmm.201201737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badimon L., Borrell M. Microvasculature Recovery by Angiogenesis After Myocardial Infarction. Curr. Pharm. Des. 2018;24:2967–2973. doi: 10.2174/1381612824666180629162726. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura M., Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018;15:387–407. doi: 10.1038/s41569-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 14.Talman V., Ruskoaho H. Cardiac fibrosis in myocardial infarction-from repair and remodeling to regeneration. Cell Tissue Res. 2016;365:563–581. doi: 10.1007/s00441-016-2431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broughton K.M., Wang B.J., Firouzi F., Khalafalla F., Dimmeler S., Fernandez-Aviles F., Sussman M.A. Mechanisms of Cardiac Repair and Regeneration. Circ. Res. 2018;122:1151–1163. doi: 10.1161/CIRCRESAHA.117.312586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schirone L., Forte M., Palmerio S., Yee D., Nocella C., Angelini F., Pagano F., Schiavon S., Bordin A., Carrizzo A. A Review of the Molecular Mechanisms Underlying the Development and Progression of Cardiac Remodeling. Oxid. Med. Cell. Longev. 2017;2017:3920195. doi: 10.1155/2017/3920195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minicucci M.F., Azevedo P.S., Polegato B.F., Paiva S.A., Zornoff L.A. Heart failure after myocardial infarction: clinical implications and treatment. Clin. Cardiol. 2011;34:410–414. doi: 10.1002/clc.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 19.Ebbesen K.K., Hansen T.B., Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14:1035–1045. doi: 10.1080/15476286.2016.1271524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfafenrot C., Preußer C. Establishing essential quality criteria for the validation of circular RNAs as biomarkers. Biomol Detect. Quantif. 2019;17:100085. doi: 10.1016/j.bdq.2019.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu T., Wu J., Han P., Zhao Z., Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patop I.L., Wüst S., Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Mo Y., Gong Z., Yang X., Yang M., Zhang S., Xiong F., Xiang B., Zhou M., Liao Q. Circular RNAs in human cancer. Mol. Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S., Li Y., Chen B., Zhao J., Yu S., Tang Y., Zheng Q., Li Y., Wang P., He X., Huang S. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen I., Chen C.Y., Chuang T.J. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: Identification, biogenesis and function. Biochim. Biophys. Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhai C., Qian G., Wu H., Pan H., Xie S., Sun Z., Shao P., Tang G., Hu H., Zhang S. Knockdown of circ_0060745 alleviates acute myocardial infarction by suppressing NF-κB activation. J. Cell. Mol. Med. 2020;24:12401–12410. doi: 10.1111/jcmm.15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Chen B. Silencing circ_0062389 alleviates cardiomyocyte apoptosis in heart failure rats via modulating TGF-β1/Smad3 signaling pathway. Gene. 2021;766:145154. doi: 10.1016/j.gene.2020.145154. [DOI] [PubMed] [Google Scholar]
- 34.Ren Q., Li H., Wang X. The circular RNA ZNF292 alleviates OGD-induced injury in H9c2 cells via targeting BNIP3. Cell Cycle. 2019;18:3365–3377. doi: 10.1080/15384101.2019.1676585. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayoumi A.S., Aonuma T., Teoh J.P., Tang Y.L., Kim I.M. Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacol. Sin. 2018;39:1100–1109. doi: 10.1038/aps.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol. Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Aufiero S., Reckman Y.J., Pinto Y.M., Creemers E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S., Wang W., Wu X., Zhou X. Regulatory Roles of Circular RNAs in Coronary Artery Disease. Mol. Ther. Nucleic Acids. 2020;21:172–179. doi: 10.1016/j.omtn.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J.Y., Shi Y., Cai X.Y., Liu J. Potential diagnostic and therapeutic value of circular RNAs in cardiovascular diseases. Cell. Signal. 2020;71:109604. doi: 10.1016/j.cellsig.2020.109604. [DOI] [PubMed] [Google Scholar]
- 42.Dorn G.W., 2nd Apoptotic and non-apoptotic programmed cardiomyocyte death in ventricular remodelling. Cardiovasc. Res. 2009;81:465–473. doi: 10.1093/cvr/cvn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frangogiannis N.G. Pathophysiology of Myocardial Infarction. Compr. Physiol. 2015;5:1841–1875. doi: 10.1002/cphy.c150006. [DOI] [PubMed] [Google Scholar]
- 44.Neri M., Riezzo I., Pascale N., Pomara C., Turillazzi E. Ischemia/Reperfusion Injury following Acute Myocardial Infarction: A Critical Issue for Clinicians and Forensic Pathologists. Mediators Inflamm. 2017;2017:7018393. doi: 10.1155/2017/7018393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heusch G., Gersh B.J. The pathophysiology of acute myocardial infarction and strategies of protection beyond reperfusion: a continual challenge. Eur. Heart J. 2017;38:774–784. doi: 10.1093/eurheartj/ehw224. [DOI] [PubMed] [Google Scholar]
- 46.Gerczuk P.Z., Kloner R.A. An update on cardioprotection: a review of the latest adjunctive therapies to limit myocardial infarction size in clinical trials. J. Am. Coll. Cardiol. 2012;59:969–978. doi: 10.1016/j.jacc.2011.07.054. [DOI] [PubMed] [Google Scholar]
- 47.O’Gara P.T., Kushner F.G., Ascheim D.D., Casey D.E., Jr., Chung M.K., de Lemos J.A., Ettinger S.M., Fang J.C., Fesmire F.M., Franklin B.A., CF/AHA Task Force 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2013;127:529–555. doi: 10.1161/CIR.0b013e3182742c84. [DOI] [PubMed] [Google Scholar]
- 48.Hausenloy D.J., Yellon D.M. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J. Clin. Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hausenloy D.J., Barrabes J.A., Bøtker H.E., Davidson S.M., Di Lisa F., Downey J., Engstrom T., Ferdinandy P., Carbrera-Fuentes H.A., Heusch G. Ischaemic conditioning and targeting reperfusion injury: a 30 year voyage of discovery. Basic Res. Cardiol. 2016;111:70. doi: 10.1007/s00395-016-0588-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neri M., Fineschi V., Di Paolo M., Pomara C., Riezzo I., Turillazzi E., Cerretani D. Cardiac oxidative stress and inflammatory cytokines response after myocardial infarction. Curr. Vasc. Pharmacol. 2015;13:26–36. doi: 10.2174/15701611113119990003. [DOI] [PubMed] [Google Scholar]
- 51.Hausenloy D.J., Botker H.E., Engstrom T., Erlinge D., Heusch G., Ibanez B., Kloner R.A., Ovize M., Yellon D.M., Garcia-Dorado D. Targeting reperfusion injury in patients with ST-segment elevation myocardial infarction: trials and tribulations. Eur. Heart J. 2017;38:935–941. doi: 10.1093/eurheartj/ehw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalogeris T., Baines C.P., Krenz M., Korthuis R.J. Ischemia/Reperfusion. Compr. Physiol. 2016;7:113–170. doi: 10.1002/cphy.c160006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heusch G. Myocardial ischaemia-reperfusion injury and cardioprotection in perspective. Nat. Rev. Cardiol. 2020;17:773–789. doi: 10.1038/s41569-020-0403-y. [DOI] [PubMed] [Google Scholar]
- 54.DeVon H.A., Hogan N., Ochs A.L., Shapiro M. Time to treatment for acute coronary syndromes: the cost of indecision. J. Cardiovasc. Nurs. 2010;25:106–114. doi: 10.1097/JCN.0b013e3181bb14a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deleon K.Y., de Castro Brás L.E., Lange R.A., Lindsey M.L. Extracellular matrix proteomics in cardiac ischemia/reperfusion: the search is on. Circulation. 2012;125:746–748. doi: 10.1161/CIRCULATIONAHA.111.086835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burke A.P., Virmani R. Pathophysiology of acute myocardial infarction. Med. Clin. North Am. 2007;91:553–572, ix. doi: 10.1016/j.mcna.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Ma S., Wang Y., Chen Y., Cao F. The role of the autophagy in myocardial ischemia/reperfusion injury. Biochim. Biophys. Acta. 2015;1852:271–276. doi: 10.1016/j.bbadis.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Cadenas S. ROS and redox signaling in myocardial ischemia-reperfusion injury and cardioprotection. Free Radic. Biol. Med. 2018;117:76–89. doi: 10.1016/j.freeradbiomed.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 59.Michael L.H., Entman M.L., Hartley C.J., Youker K.A., Zhu J., Hall S.R., Hawkins H.K., Berens K., Ballantyne C.M. Myocardial ischemia and reperfusion: a murine model. Am. J. Physiol. 1995;269:H2147–H2154. doi: 10.1152/ajpheart.1995.269.6.H2147. [DOI] [PubMed] [Google Scholar]
- 60.Xu Z., McElhanon K.E., Beck E.X., Weisleder N. A Murine Model of Myocardial Ischemia-Reperfusion Injury. Methods Mol. Biol. 2018;1717:145–153. doi: 10.1007/978-1-4939-7526-6_12. [DOI] [PubMed] [Google Scholar]
- 61.Yan L., Vatner D.E., Kim S.J., Ge H., Masurekar M., Massover W.H., Yang G., Matsui Y., Sadoshima J., Vatner S.F. Autophagy in chronically ischemic myocardium. Proc. Natl. Acad. Sci. USA. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X., Guo Z., Ding Z., Mehta J.L. Inflammation, Autophagy, and Apoptosis After Myocardial Infarction. J. Am. Heart Assoc. 2018;7:e008024. doi: 10.1161/JAHA.117.008024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buja L.M., Vela D. Cardiomyocyte death and renewal in the normal and diseased heart. Cardiovasc. Pathol. 2008;17:349–374. doi: 10.1016/j.carpath.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 64.Saraste A., Pulkki K., Kallajoki M., Henriksen K., Parvinen M., Voipio-Pulkki L.M. Apoptosis in human acute myocardial infarction. Circulation. 1997;95:320–323. doi: 10.1161/01.cir.95.2.320. [DOI] [PubMed] [Google Scholar]
- 65.Fliss H., Gattinger D. Apoptosis in ischemic and reperfused rat myocardium. Circ. Res. 1996;79:949–956. doi: 10.1161/01.res.79.5.949. [DOI] [PubMed] [Google Scholar]
- 66.Kajstura J., Cheng W., Reiss K., Clark W.A., Sonnenblick E.H., Krajewski S., Reed J.C., Olivetti G., Anversa P. Apoptotic and necrotic myocyte cell deaths are independent contributing variables of infarct size in rats. Lab. Invest. 1996;74:86–107. [PubMed] [Google Scholar]
- 67.Olivetti G., Quaini F., Sala R., Lagrasta C., Corradi D., Bonacina E., Gambert S.R., Cigola E., Anversa P. Acute myocardial infarction in humans is associated with activation of programmed myocyte cell death in the surviving portion of the heart. J. Mol. Cell. Cardiol. 1996;28:2005–2016. doi: 10.1006/jmcc.1996.0193. [DOI] [PubMed] [Google Scholar]
- 68.Takagi H., Matsui Y., Hirotani S., Sakoda H., Asano T., Sadoshima J. AMPK mediates autophagy during myocardial ischemia in vivo. Autophagy. 2007;3:405–407. doi: 10.4161/auto.4281. [DOI] [PubMed] [Google Scholar]
- 69.Zhu H., Tannous P., Johnstone J.L., Kong Y., Shelton J.M., Richardson J.A., Le V., Levine B., Rothermel B.A., Hill J.A. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bravo-San Pedro J.M., Kroemer G., Galluzzi L. Autophagy and Mitophagy in Cardiovascular Disease. Circ. Res. 2017;120:1812–1824. doi: 10.1161/CIRCRESAHA.117.311082. [DOI] [PubMed] [Google Scholar]
- 71.Jugdutt B.I. Ventricular remodeling after infarction and the extracellular collagen matrix: when is enough enough? Circulation. 2003;108:1395–1403. doi: 10.1161/01.CIR.0000085658.98621.49. [DOI] [PubMed] [Google Scholar]
- 72.Zamilpa R., Zhang J., Chiao Y.A., de Castro Brás L.E., Halade G.V., Ma Y., Hacker S.O., Lindsey M.L. Cardiac wound healing post-myocardial infarction: a novel method to target extracellular matrix remodeling in the left ventricle. Methods Mol. Biol. 2013;1037:313–324. doi: 10.1007/978-1-62703-505-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frantz S., Bauersachs J., Ertl G. Post-infarct remodelling: contribution of wound healing and inflammation. Cardiovasc. Res. 2009;81:474–481. doi: 10.1093/cvr/cvn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van den Borne S.W., Diez J., Blankesteijn W.M., Verjans J., Hofstra L., Narula J. Myocardial remodeling after infarction: the role of myofibroblasts. Nat. Rev. Cardiol. 2010;7:30–37. doi: 10.1038/nrcardio.2009.199. [DOI] [PubMed] [Google Scholar]
- 75.Deb A., Ubil E. Cardiac fibroblast in development and wound healing. J. Mol. Cell. Cardiol. 2014;70:47–55. doi: 10.1016/j.yjmcc.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frangogiannis N.G. The mechanistic basis of infarct healing. Antioxid. Redox Signal. 2006;8:1907–1939. doi: 10.1089/ars.2006.8.1907. [DOI] [PubMed] [Google Scholar]
- 77.Ma Y., Halade G.V., Lindsey M.L. Extracellular matrix and fibroblast communication following myocardial infarction. J. Cardiovasc. Transl. Res. 2012;5:848–857. doi: 10.1007/s12265-012-9398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weil B.R., Neelamegham S. Selectins and Immune Cells in Acute Myocardial Infarction and Post-infarction Ventricular Remodeling: Pathophysiology and Novel Treatments. Front. Immunol. 2019;10:300. doi: 10.3389/fimmu.2019.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhatt A.S., Ambrosy A.P., Velazquez E.J. Adverse Remodeling and Reverse Remodeling After Myocardial Infarction. Curr. Cardiol. Rep. 2017;19:71. doi: 10.1007/s11886-017-0876-4. [DOI] [PubMed] [Google Scholar]
- 80.Sutton M.G., Sharpe N. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation. 2000;101:2981–2988. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 81.Pfeffer M.A., Braunwald E. Ventricular remodeling after myocardial infarction. Experimental observations and clinical implications. Circulation. 1990;81:1161–1172. doi: 10.1161/01.cir.81.4.1161. [DOI] [PubMed] [Google Scholar]
- 82.Nahrendorf M., Pittet M.J., Swirski F.K. Monocytes: protagonists of infarct inflammation and repair after myocardial infarction. Circulation. 2010;121:2437–2445. doi: 10.1161/CIRCULATIONAHA.109.916346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dong H., Mosca H., Gao E., Akins R.E., Gidding S.S., Tsuda T. Integrated wall stress: a new methodological approach to assess ventricular workload and myocardial contractile reserve. J. Transl. Med. 2013;11:183. doi: 10.1186/1479-5876-11-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanai E., Frantz S. Pathophysiology of Heart Failure. Compr. Physiol. 2015;6:187–214. doi: 10.1002/cphy.c140055. [DOI] [PubMed] [Google Scholar]
- 85.Kong P., Christia P., Frangogiannis N.G. The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci. 2014;71:549–574. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang W., Zhang H., Yao W., Li L., Niu P., Huo Y., Tan W. Morphometric, Hemodynamic, and Multi-Omics Analyses in Heart Failure Rats with Preserved Ejection Fraction. Int. J. Mol. Sci. 2020;21:3362. doi: 10.3390/ijms21093362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y.W., Shi J., Li Y.J., Wei L. Cardiomyocyte death in doxorubicin-induced cardiotoxicity. Arch. Immunol. Ther. Exp. (Warsz.) 2009;57:435–445. doi: 10.1007/s00005-009-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boeckel J.N., Jaé N., Heumüller A.W., Chen W., Boon R.A., Stellos K., Zeiher A.M., John D., Uchida S., Dimmeler S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 89.Gupta S.K., Garg A., Bär C., Chatterjee S., Foinquinos A., Milting H., Streckfuß-Bömeke K., Fiedler J., Thum T. Quaking Inhibits Doxorubicin-Mediated Cardiotoxicity Through Regulation of Cardiac Circular RNA Expression. Circ. Res. 2018;122:246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou J., Li L., Hu H., Wu J., Chen H., Feng K., Ma L. Circ-HIPK2 Accelerates Cell Apoptosis and Autophagy in Myocardial Oxidative Injury by Sponging miR-485-5p and Targeting ATG101. J. Cardiovasc. Pharmacol. 2020;76:427–436. doi: 10.1097/FJC.0000000000000879. [DOI] [PubMed] [Google Scholar]
- 91.Li M., Ding W., Tariq M.A., Chang W., Zhang X., Xu W., Hou L., Wang Y., Wang J. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–5869. doi: 10.7150/thno.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cui X., Dong Y., Li M., Wang X., Jiang M., Yang W., Liu G., Sun S., Xu W. A circular RNA from NFIX facilitates oxidative stress-induced H9c2 cells apoptosis. In Vitro Cell. Dev. Biol. Anim. 2020;56:715–722. doi: 10.1007/s11626-020-00476-z. [DOI] [PubMed] [Google Scholar]
- 93.Wang X., Sun Q., Hu W. Carvedilol protects against the H2O2-induced cell damages in rat myoblasts by regulating the circ_NFIX/miR-125b-5p/TLR4 signal axis. J. Cardiovasc. Pharmacol. 2021 doi: 10.1097/FJC.0000000000001095. Published online June 25, 2021. [DOI] [PubMed] [Google Scholar]
- 94.Wang K., Gan T.Y., Li N., Liu C.Y., Zhou L.Y., Gao J.N., Chen C., Yan K.W., Ponnusamy M., Zhang Y.H., Li P.F. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu X., Ma R., Cao J., Du X., Cai X., Fan Y. CircSAMD4A aggravates H/R-induced cardiomyocyte apoptosis and inflammatory response by sponging miR-138-5p. J. Cell. Mol. Med. 2020 doi: 10.1111/jcmm.16093. Published online November 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gan J., Yuan J., Liu Y., Lu Z., Xue Y., Shi L., Zeng H. Circular RNA_101237 mediates anoxia/reoxygenation injury by targeting let-7a-5p/IGF2BP3 in cardiomyocytes. Int. J. Mol. Med. 2020;45:451–460. doi: 10.3892/ijmm.2019.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng N., Wang M.Y., Wu Y.B., Cui H.M., Wei S.X., Liu B., Wang R. Circular RNA POSTN Promotes Myocardial Infarction-Induced Myocardial Injury and Cardiac Remodeling by Regulating miR-96-5p/BNIP3 Axis. Front. Cell Dev. Biol. 2021;8:618574. doi: 10.3389/fcell.2020.618574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Luo C., Ling G.X., Lei B.F., Feng X., Xie X.Y., Fang C., Li Y.G., Cai X.W., Zheng B.S. Circular RNA PVT1 silencing prevents ischemia-reperfusion injury in rat by targeting microRNA-125b and microRNA-200a. J. Mol. Cell. Cardiol. 2021;159:80–90. doi: 10.1016/j.yjmcc.2021.05.019. [DOI] [PubMed] [Google Scholar]
- 99.Geng H.H., Li R., Su Y.M., Xiao J., Pan M., Cai X.X., Ji X.P. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS ONE. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chai Q., Zheng M., Wang L., Wei M., Yin Y., Ma F., Li X., Zhang H., Liu G. Circ_0068655 Promotes Cardiomyocyte Apoptosis via miR-498/PAWR Axis. Tissue Eng. Regen. Med. 2020;17:659–670. doi: 10.1007/s13770-020-00270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lei D., Wang Y., Zhang L., Wang Z. Circ_0010729 regulates hypoxia-induced cardiomyocyte injuries by activating TRAF5 via sponging miR-27a-3p. Life Sci. 2020;262:118511. doi: 10.1016/j.lfs.2020.118511. [DOI] [PubMed] [Google Scholar]
- 102.Bai M., Pan C.L., Jiang G.X., Zhang Y.M. CircRNA 010567 improves myocardial infarction rats through inhibiting TGF-β1. Eur. Rev. Med. Pharmacol. Sci. 2020;24:369–375. doi: 10.26355/eurrev_202001_19935. [DOI] [PubMed] [Google Scholar]
- 103.Zhao Q., Li W., Pan W., Wang Z. CircRNA 010567 plays a significant role in myocardial infarction via the regulation of the miRNA-141/DAPK1 axis. J. Thorac. Dis. 2021;13:2447–2459. doi: 10.21037/jtd-21-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu X., Wang M., Li Q., Liu W., Song Q., Jiang H. CircRNA ACAP2 induces myocardial apoptosis after myocardial infarction by sponging miR-29. Minerva Med. 2020 doi: 10.23736/S0026-4806.20.06600-8. Published online May 13, 2020. [DOI] [PubMed] [Google Scholar]
- 105.Zhang J., Tang Y., Zhang J., Wang J., He J., Zhang Z., Liu F. CircRNA ACAP2 is overexpressed in myocardial infarction and promotes the maturation of miR-532 to induce the apoptosis of cardiomyocyte. J. Cardiovasc. Pharmacol. 2021;78:247–252. doi: 10.1097/FJC.0000000000001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cai X., Li B., Wang Y., Zhu H., Zhang P., Jiang P., Yang X., Sun J., Hong L., Shao L. CircJARID2 Regulates Hypoxia-Induced Injury in H9c2 Cells by Affecting miR-9-5p-Mediated BNIP3. J. Cardiovasc. Pharmacol. 2021;78:e77–e85. doi: 10.1097/FJC.0000000000001033. [DOI] [PubMed] [Google Scholar]
- 107.Tan J., Pan W., Chen H., Du Y., Jiang P., Zeng D., Wu J., Peng K. Circ_0124644 Serves as a ceRNA for miR-590-3p to Promote Hypoxia-Induced Cardiomyocytes Injury via Regulating SOX4. Front. Genet. 2021;12:667724. doi: 10.3389/fgene.2021.667724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen T.P., Zhang N.J., Wang H.J., Hu S.G., Geng X. Knockdown of circROBO2 attenuates acute myocardial infarction through regulating the miR-1184/TRADD axis. Mol. Med. 2021;27:21. doi: 10.1186/s10020-021-00275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu Y., Wu M., Yang J., Li Y., Peng W., Wu M., Yu C., Fang M. Silencing CircHIPK3 Sponges miR-93-5p to Inhibit the Activation of Rac1/PI3K/AKT Pathway and Improves Myocardial Infarction-Induced Cardiac Dysfunction. Front. Cardiovasc. Med. 2021;8:645378. doi: 10.3389/fcvm.2021.645378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Cai L., Qi B., Wu X., Peng S., Zhou G., Wei Y., Xu J., Chen S., Liu S. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J. Mol. Cell. Cardiol. 2019;130:10–22. doi: 10.1016/j.yjmcc.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 111.Zhao B., Li G., Peng J., Ren L., Lei L., Ye H., Wang Z., Zhao S. CircMACF1 Attenuates Acute Myocardial Infarction Through miR-500b-5p-EMP1 Axis. J. Cardiovasc. Transl. Res. 2021;14:161–172. doi: 10.1007/s12265-020-09976-5. [DOI] [PubMed] [Google Scholar]
- 112.Zhu Y., Zhao P., Sun L., Lu Y., Zhu W., Zhang J., Xiang C., Mao Y., Chen Q., Zhang F. Overexpression of circRNA SNRK targets miR-103-3p to reduce apoptosis and promote cardiac repair through GSK3β/β-catenin pathway in rats with myocardial infarction. Cell Death Discov. 2021;7:84. doi: 10.1038/s41420-021-00467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li C.X., Song J., Li X., Zhang T., Li Z.M. Circular RNA 0001273 in exosomes derived from human umbilical cord mesenchymal stem cells (UMSCs) in myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 2020;24:10086–10095. doi: 10.26355/eurrev_202010_23228. [DOI] [PubMed] [Google Scholar]
- 114.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 115.Wang X., Cheng Z., Xu J., Feng M., Zhang H., Zhang L., Qian L. Circular RNA Arhgap12 modulates doxorubicin-induced cardiotoxicity by sponging miR-135a-5p. Life Sci. 2021;265:118788. doi: 10.1016/j.lfs.2020.118788. [DOI] [PubMed] [Google Scholar]
- 116.Ji X., Ding W., Xu T., Zheng X., Zhang J., Liu M., Liu G., Wang J. MicroRNA-31-5p attenuates doxorubicin-induced cardiotoxicity via quaking and circular RNA Pan3. J. Mol. Cell. Cardiol. 2020;140:56–67. doi: 10.1016/j.yjmcc.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 117.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sciarretta S., Yee D., Shenoy V., Nagarajan N., Sadoshima J. The importance of autophagy in cardioprotection. High Blood Press. Cardiovasc. Prev. 2014;21:21–28. doi: 10.1007/s40292-013-0029-9. [DOI] [PubMed] [Google Scholar]
- 119.Jin P., Li L.H., Shi Y., Hu N.B. Salidroside inhibits apoptosis and autophagy of cardiomyocyte by regulation of circular RNA hsa_circ_0000064 in cardiac ischemia-reperfusion injury. Gene. 2021;767:145075. doi: 10.1016/j.gene.2020.145075. [DOI] [PubMed] [Google Scholar]
- 120.Zhou L.Y., Zhai M., Huang Y., Xu S., An T., Wang Y.H., Zhang R.C., Liu C.Y., Dong Y.H., Wang M. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019;26:1299–1315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu Y., Zou C., Jia Y., Zhang H., Ma X., Zhang J. Knockdown of circular RNA circMAT2B reduces oxygen-glucose deprivation-induced inflammatory injury in H9c2 cells through up-regulating miR-133. Cell Cycle. 2020;19:2622–2630. doi: 10.1080/15384101.2020.1814025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bian Y., Pang P., Li X., Yu S., Wang X., Liu K., Ju J., Wu H., Gao Y., Liu Q. CircHelz activates NLRP3 inflammasome to promote myocardial injury by sponging miR-133a-3p in mouse ischemic heart. J. Mol. Cell. Cardiol. 2021;158:128–139. doi: 10.1016/j.yjmcc.2021.05.010. [DOI] [PubMed] [Google Scholar]
- 123.Zhang M., Wang Z., Cheng Q., Wang Z., Lv X., Wang Z., Li N. Circular RNA (circRNA) CDYL Induces Myocardial Regeneration by ceRNA After Myocardial Infarction. Med. Sci. Monit. 2020;26:e923188. doi: 10.12659/MSM.923188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garikipati V.N.S., Verma S.K., Cheng Z., Liang D., Truongcao M.M., Cimini M., Yue Y., Huang G., Wang C., Benedict C. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Si X., Zheng H., Wei G., Li M., Li W., Wang H., Guo H., Sun J., Li C., Zhong S. circRNA Hipk3 Induces Cardiac Regeneration after Myocardial Infarction in Mice by Binding to Notch1 and miR-133a. Mol. Ther. Nucleic Acids. 2020;21:636–655. doi: 10.1016/j.omtn.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Y., Zhao R., Shen C., Liu W., Yuan J., Li C., Deng W., Wang Z., Zhang W., Ge J., Shi B. Exosomal CircHIPK3 Released from Hypoxia-Induced Cardiomyocytes Regulates Cardiac Angiogenesis after Myocardial Infarction. Oxid. Med. Cell. Longev. 2020;2020:8418407. doi: 10.1155/2020/8418407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gao W.Q., Hu X.M., Zhang Q., Yang L., Lv X.Z., Chen S., Wu P., Duan D.W., Lang Y.H., Ning M. Downregulation of circFASTKD1 ameliorates myocardial infarction by promoting angiogenesis. Aging (Albany NY) 2020;13:3588–3604. doi: 10.18632/aging.202305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W. Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.St John Sutton M., Lee D., Rouleau J.L., Goldman S., Plappert T., Braunwald E., Pfeffer M.A. Left ventricular remodeling and ventricular arrhythmias after myocardial infarction. Circulation. 2003;107:2577–2582. doi: 10.1161/01.CIR.0000070420.51787.A8. [DOI] [PubMed] [Google Scholar]
- 130.Mester-Tonczar J., Winkler J., Einzinger P., Hasimbegovic E., Kastner N., Lukovic D., Zlabinger K., Spannbauer A., Traxler D., Batkai S. Association between Circular RNA CDR1as and Post-Infarction Cardiac Function in Pig Ischemic Heart Failure: Influence of the Anti-Fibrotic Natural Compounds Bufalin and Lycorine. Biomolecules. 2020;10:1180. doi: 10.3390/biom10081180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tang C.M., Zhang M., Huang L., Hu Z.Q., Zhu J.N., Xiao Z., Zhang Z., Lin Q.X., Zheng X.L., -Yang M. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li F., Long T.Y., Bi S.S., Sheikh S.A., Zhang C.L. circPAN3 exerts a profibrotic role via sponging miR-221 through FoxO3/ATG7-activated autophagy in a rat model of myocardial infarction. Life Sci. 2020;257:118015. doi: 10.1016/j.lfs.2020.118015. [DOI] [PubMed] [Google Scholar]
- 133.Wang Y., Li C., Zhao R., Qiu Z., Shen C., Wang Z., Liu W., Zhang W., Ge J., Shi B. CircUbe3a from M2 macrophage-derived small extracellular vesicles mediates myocardial fibrosis after acute myocardial infarction. Theranostics. 2021;11:6315–6333. doi: 10.7150/thno.52843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhu Y., Pan W., Yang T., Meng X., Jiang Z., Tao L., Wang L. Upregulation of Circular RNA CircNFIB Attenuates Cardiac Fibrosis by Sponging miR-433. Front. Genet. 2019;10:564. doi: 10.3389/fgene.2019.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun L.Y., Zhao J.C., Ge X.M., Zhang H., Wang C.M., Bie Z.D. Circ_LAS1L regulates cardiac fibroblast activation, growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem. Funct. 2020;38:443–450. doi: 10.1002/cbf.3486. [DOI] [PubMed] [Google Scholar]
- 136.Deng Y., Wang J., Xie G., Zeng X., Li H. Circ-HIPK3 Strengthens the Effects of Adrenaline in Heart Failure by MiR-17-3p - ADCY6 Axis. Int. J. Biol. Sci. 2019;15:2484–2496. doi: 10.7150/ijbs.36149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS ONE. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tan W.L., Lim B.T., Anene-Nzelu C.G., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 139.Yin L., Tang Y., Jiang M. Research on the circular RNA bioinformatics in patients with acute myocardial infarction. J. Clin. Lab. Anal. 2021;35:e23621. doi: 10.1002/jcla.23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tian M., Cao Z., Pang H. Circular RNAs in Sudden Cardiac Death Related Diseases: Novel Biomarker for Clinical and Forensic Diagnosis. Molecules. 2021;26:1155. doi: 10.3390/molecules26041155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tian M., Xue J., Dai C., Jiang E., Zhu B., Pang H. CircSLC8A1 and circNFIX can be used as auxiliary diagnostic markers for sudden cardiac death caused by acute ischemic heart disease. Sci. Rep. 2021;11:4695. doi: 10.1038/s41598-021-84056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deng Y.-Y., Zhang W., She J., Zhang L., Chen T., Zhou J., Yuan Z. GW27-e1167 Circular RNA Related to PPARγ Function as ceRNA of microRNA in Human Acute Myocardial Infarction. J. Am. Coll. Cardiol. 2016;68:C51–C52. [Google Scholar]
- 143.Mester-Tonczar J., Einzinger P., Winkler J., Kastner N., Spannbauer A., Zlabinger K., Traxler D., Lukovic D., Hasimbegovic E., Goliasch G. Novel Identified Circular Transcript of RCAN2, circ-RCAN2, Shows Deviated Expression Pattern in Pig Reperfused Infarcted Myocardium and Hypoxic Porcine Cardiac Progenitor Cells In Vitro. Int. J. Mol. Sci. 2021;22:1390. doi: 10.3390/ijms22031390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Salgado-Somoza A., Zhang L., Vausort M., Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int. J. Cardiol. Heart Vasc. 2017;17:33–36. doi: 10.1016/j.ijcha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vausort M., Salgado-Somoza A., Zhang L., Leszek P., Scholz M., Teren A., Burkhardt R., Thiery J., Wagner D.R., Devaux Y. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J. Am. Coll. Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 146.Zhao Z., Li X., Gao C., Jian D., Hao P., Rao L., Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhou B., Yu J.W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem. Biophys. Res. Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 148.Schulte C., Barwari T., Joshi A., Theofilatos K., Zampetaki A., Barallobre-Barreiro J., Singh B., Sörensen N.A., Neumann J.T., Zeller T. Comparative Analysis of Circulating Noncoding RNAs Versus Protein Biomarkers in the Detection of Myocardial Injury. Circ. Res. 2019;125:328–340. doi: 10.1161/CIRCRESAHA.119.314937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Z., Yang T., Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lin F., Yang Y., Guo Q., Xie M., Sun S., Wang X., Li D., Zhang G., Li M., Wang J., Zhao G. Analysis of the Molecular Mechanism of Acute Coronary Syndrome Based on circRNA-miRNA Network Regulation. Evid. Based Complement. Alternat. Med. 2020;2020:1584052. doi: 10.1155/2020/1584052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Poller W., Dimmeler S., Heymans S., Zeller T., Haas J., Karakas M., Leistner D.M., Jakob P., Nakagawa S., Blankenberg S. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur. Heart J. 2018;39:2704–2716. doi: 10.1093/eurheartj/ehx165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ho P.Y., Yu A.M. Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip. Rev. RNA. 2016;7:186–197. doi: 10.1002/wrna.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lim G.B. MicroRNA-directed cardiac repair after myocardial infarction in pigs. Nat. Rev. Cardiol. 2019;16:454–455. doi: 10.1038/s41569-019-0216-z. [DOI] [PubMed] [Google Scholar]
- 154.Gao J., Chen X., Wei P., Wang Y., Li P., Shao K. Regulation of pyroptosis in cardiovascular pathologies: Role of noncoding RNAs. Mol. Ther. Nucleic Acids. 2021;25:220–236. doi: 10.1016/j.omtn.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Majmudar M.D., Keliher E.J., Heidt T., Leuschner F., Truelove J., Sena B.F., Gorbatov R., Iwamoto Y., Dutta P., Wojtkiewicz G. Monocyte-directed RNAi targeting CCR2 improves infarct healing in atherosclerosis-prone mice. Circulation. 2013;127:2038–2046. doi: 10.1161/CIRCULATIONAHA.112.000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Haussecker D., Kay M.A. RNA interference. Drugging RNAi. Science. 2015;347:1069–1070. doi: 10.1126/science.1252967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hulot J.S., Ishikawa K., Hajjar R.J. Gene therapy for the treatment of heart failure: promise postponed. Eur. Heart J. 2016;37:1651–1658. doi: 10.1093/eurheartj/ehw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Suckau L., Fechner H., Chemaly E., Krohn S., Hadri L., Kockskämper J., Westermann D., Bisping E., Ly H., Wang X. Long-term cardiac-targeted RNA interference for the treatment of heart failure restores cardiac function and reduces pathological hypertrophy. Circulation. 2009;119:1241–1252. doi: 10.1161/CIRCULATIONAHA.108.783852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Boon R.A., Jaé N., Holdt L., Dimmeler S. Long Noncoding RNAs: From Clinical Genetics to Therapeutic Targets? J. Am. Coll. Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 160.Ounzain S., Micheletti R., Beckmann T., Schroen B., Alexanian M., Pezzuto I., Crippa S., Nemir M., Sarre A., Johnson R. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 2015;36:353–368. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Boon R.A., Dimmeler S. MicroRNAs in myocardial infarction. Nat. Rev. Cardiol. 2015;12:135–142. doi: 10.1038/nrcardio.2014.207. [DOI] [PubMed] [Google Scholar]
- 162.Gao J., Chen X., Shan C., Wang Y., Li P., Shao K. Autophagy in cardiovascular diseases: role of noncoding RNAs. Mol. Ther. Nucleic Acids. 2020;23:101–118. doi: 10.1016/j.omtn.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 164.Li M., Duan L., Li Y., Liu B. Long noncoding RNA/circular noncoding RNA-miRNA-mRNA axes in cardiovascular diseases. Life Sci. 2019;233:116440. doi: 10.1016/j.lfs.2019.04.066. [DOI] [PubMed] [Google Scholar]
- 165.Riquelme J.A., Chavez M.N., Mondaca-Ruff D., Bustamante M., Vicencio J.M., Quest A.F., Lavandero S. Therapeutic targeting of autophagy in myocardial infarction and heart failure. Expert Rev. Cardiovasc. Ther. 2016;14:1007–1019. doi: 10.1080/14779072.2016.1202760. [DOI] [PubMed] [Google Scholar]
- 166.Haider H.Kh., Akbar S.A., Ashraf M. Angiomyogenesis for myocardial repair. Antioxid. Redox Signal. 2009;11:1929–1944. doi: 10.1089/ars.2009.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kirkton R.D., Bursac N. Genetic engineering of somatic cells to study and improve cardiac function. Europace. 2012;14(Suppl 5):v40–v49. doi: 10.1093/europace/eus269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fan S., Hu K., Zhang D., Liu F. Interference of circRNA HIPK3 alleviates cardiac dysfunction in lipopolysaccharide-induced mice models and apoptosis in H9C2 cardiomyocytes. Ann. Transl. Med. 2020;8:1147. doi: 10.21037/atm-20-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gallet R., Dawkins J., Valle J., Simsolo E., de Couto G., Middleton R., Tseliou E., Luthringer D., Kreke M., Smith R.R. Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. Eur. Heart J. 2017;38:201–211. doi: 10.1093/eurheartj/ehw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.López-Jiménez E., Rojas A.M., Andrés-León E. RNA sequencing and Prediction Tools for Circular RNAs Analysis. Adv. Exp. Med. Biol. 2018;1087:17–33. doi: 10.1007/978-981-13-1426-1_2. [DOI] [PubMed] [Google Scholar]
- 171.Li S., Teng S., Xu J., Su G., Zhang Y., Zhao J., Zhang S., Wang H., Qin W., Lu Z.J. Microarray is an efficient tool for circRNA profiling. Brief. Bioinform. 2019;20:1420–1433. doi: 10.1093/bib/bby006. [DOI] [PubMed] [Google Scholar]
- 172.Hao X.D., Liu Y., Li B.W., Wu W., Zhao X.W. Exome sequencing analysis identifies novel homozygous mutation in ABCA4 in a Chinese family with Stargardt disease. Int. J. Ophthalmol. 2020;13:671–676. doi: 10.18240/ijo.2020.04.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wu H.J., Zhang C.Y., Zhang S., Chang M., Wang H.Y. Microarray Expression Profile of Circular RNAs in Heart Tissue of Mice with Myocardial Infarction-Induced Heart Failure. Cell. Physiol. Biochem. 2016;39:205–216. doi: 10.1159/000445617. [DOI] [PubMed] [Google Scholar]
- 174.Szabo L., Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 176.Hansen T.B., Venø M.T., Damgaard C.K., Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang J., Chen S., Yang J., Zhao F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat. Commun. 2020;11:90. doi: 10.1038/s41467-019-13840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yu F., Tie Y., Zhang Y., Wang Z., Yu L., Zhong L., Zhang C. Circular RNA expression profiles and bioinformatic analysis in coronary heart disease. Epigenomics. 2020;12:439–454. doi: 10.2217/epi-2019-0369. [DOI] [PubMed] [Google Scholar]
- 179.Cheng Z.F., Deutscher M.P. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 180.Pandey P.R., Rout P.K., Das A., Gorospe M., Panda A.C. RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods. 2019;155:41–48. doi: 10.1016/j.ymeth.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schneider T., Schreiner S., Preußer C., Bindereif A., Rossbach O. Northern Blot Analysis of Circular RNAs. Methods Mol. Biol. 2018;1724:119–133. doi: 10.1007/978-1-4939-7562-4_10. [DOI] [PubMed] [Google Scholar]
- 183.Zirkel A., Papantonis A. Detecting Circular RNAs by RNA Fluorescence In Situ Hybridization. Methods Mol. Biol. 2018;1724:69–75. doi: 10.1007/978-1-4939-7562-4_6. [DOI] [PubMed] [Google Scholar]
- 184.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]