Summary
Activation of nucleic acid sensing Toll-like receptors (TLRs) in B cells is involved in antiviral responses by promoting B cell activation and germinal center responses. In order to take advantage of this natural pathway for vaccine development, synthetic pathogen-like antigens (PLAs) constructed of multivalent antigens with encapsulated TLR ligands can be used to activate B cell antigen receptors and TLRs in a synergistic manner. Here we report a PLA-based coronavirus disease 2019 (COVID-19) vaccine candidate designed by combining a phage-derived virus-like particle carrying bacterial RNA as TLR ligands with the receptor-binding domain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) S protein as the target antigen. This PLA-based vaccine candidate induces robust neutralizing antibodies in both mice and non-human primates (NHPs). Using a NHP infection model, we demonstrate that the viral clearance is accelerated in vaccinated animals. In addition, the PLA-based vaccine induces a T helper 1 (Th1)-oriented response and a durable memory, supporting its potential for further clinical development.
Keywords: COVID-19, SARS-CoV-2, RBD, vaccine, B cells, Toll-like receptor, Th1, preclinical study, virus-like particle, pathogen-like antigen
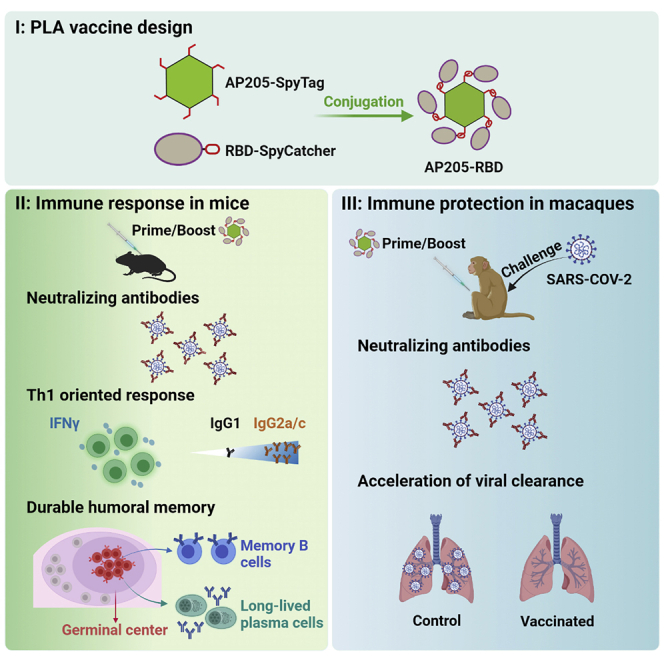
Graphical abstract
Highlights
-
•
AP205-RBD elicits neutralizing antibodies against SARS-CoV-2 in mice and macaques
-
•
AP205-RBD induces Th1-oriented immune response and durable memory
-
•
Vaccination of AP205-RBD accelerates viral clearance in infected macaques
Guo et al. constructed a COVID-19 vaccine candidate that mimics SARS-CoV-2 virus structurally. They tested this vaccine in animals and found that it could induce robust neutralizing antibodies that last for more than a year. Most importantly, the vaccine provided immune protection when the animals were challenged by viral infections.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a new virus that causes coronavirus disease 2019 (COVID-19), has caused more than 4 million deaths at 18 months after its emergence (World Health Organization [WHO]). The pandemic has imposed enormous burdens on medical care, economies, and social lives. Several types of vaccine have been approved for clinical use worldwide, including inactivated virus, non-replicating viral vector, and mRNA-based vaccines.1 Although most countries are actively promoting the vaccination process, new waves of infections with different viral variants continue to be the major concern for the public health.2 Reluctance to vaccination is one of the major problems for achieving the herd immunity, and concerns for the safety and side effects of vaccination have always been an issue. In addition, the long-term efficacy and potential serious side effects for the current approved COVID-19 vaccines are still under examination. It is therefore worthy to continue developing other types of COVID-19 vaccine with less concern of safety and a more durable effect. Indeed, in addition to the above-mentioned three types of vaccine, DNA, protein subunit, and virus-like particle (VLP)-based vaccine candidates are also under clinical or pre-clinical development worldwide (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines).
Although there have been many successful vaccines in human history, such as those for smallpox, polio, and measles, most of these prophylactic vaccines were developed by a trial-and-error approach. Not all infectious diseases can be prevented by vaccines despite the advances in both basic research and biopharmaceutical technology. An incomplete understanding of how our immune system responds to different types of infection, as well as to those successful vaccines, has hindered our progress in vaccine development. It is generally accepted that antigen-presenting cells (APCs), especially dendritic cells (DCs), are important in the initial activation of the adaptive immune responses.3 However, we recently found that B cells instead of DCs could serve as the dominant APCs to activate CD4+ T cells upon immunization with phage Qβ-derived VLPs (Qβ-VLPs) or inactivated influenza virus,4 suggesting that alternative APCs could be used to activate the immune response. The nucleic acid sensing Toll-like receptors (TLRs) in B cells are essential for their antigen-presenting function because they activate B cells to secrete factors that can promote CD4+ T cells differentiating toward T follicular helper (Tfh) and T helper 1 (Th1) cells.4 Moreover, B cell TLR signaling has been shown to be involved in anti-viral responses in multiple cases through promoting B cell proliferation and differentiation, including germinal center (GC) response.5, 6, 7, 8, 9 Dependence on B cell TLR signaling for anti-viral responses is likely an evolutionarily conserved mechanism in both mice and humans. Human B cells express similar endosomal nucleic acid-sensing TLRs as mice do.10 In addition, the pathological role of TLRs in systemic lupus erythematosus, an autoimmune disease characteristic with anti-nuclear antibody, seems to be conserved between mice and humans, suggesting that the same pathway might be reserved for the immune responses to infections.11,12 Interestingly, a recent study identified loss-of-function TLR7 variants being associated with severe COVID-19 in young male patients,13 supporting further that TLRs in humans are involved in anti-viral immunity.
To take the advantage of this natural anti-viral mechanism in B cells for vaccine development, antigens need to be presented in a multivalent form to maximize B cell antigen receptor (BCR) activation, as well as to carry the TLR ligands inside, so that the uptake of the TLR ligands could be coupled to the BCR-mediated endocytosis to achieve a synergistic signaling effect in B cells.5,14, 15, 16 To distinguish this type of VLP from the other VLPs or nanoparticles that do not encapsulate TLR ligands, we named them pathogen-like antigens (PLAs). In this study, we conjugated the receptor-binding domain (RBD) of the SARS-CoV-2 S protein to a PLA platform to build a COVID-19 vaccine candidate. We found that this PLA-based COVID-19 vaccine candidate induced robust neutralizing antibodies, a Th1-oriented immune response, a long-lasting GC response, and the production of long-lived PCs and memory B cells in mice, all of which fit well with our prediction for B cell TLR activation. We further tested this vaccine candidate in non-human primates (NHPs) and found that it could also induce neutralizing antibodies and, most importantly, accelerate the viral clearance upon SARS-CoV-2 challenge in macaques. These results supported that a PLA-based COVID-19 vaccine candidate could potentially serve as an alternative option for coping with the SARS-CoV-2 pandemic.
Results
A PLA-based COVID-19 vaccine candidate elicited the antigen-specific antibody response
To generate a PLA platform that can activate the B cell TLR signaling pathway, we chose bacterial phage-derived VLPs as the building block because they contain host cell-derived single-stranded RNA that can stimulate TLR7 and downstream MyD88 in murine B cells.5,9,17 Acinetobacter phage AP205-derived VLPs were chosen because they could accommodate fusion proteins better than other phages from the same family.18 We chose the SpyTag:SpyCatcher system to conjugate foreign antigens to the PLA platform. SpyTag:SpyCatcher is a peptide and protein domain pair designed by Zakeri et al.,19 which can form a covalent bond under diverse conditions of pH, temperature, and buffer and has been widely used in protein engineering. Indeed, the strategy using SpyTag:SpyCatcher in combination with VLPs has been tested for generating other candidate vaccines previously.20 We chose the RBD part instead of the full-length S protein from SARS-CoV-2 as the target antigen because the antibodies targeting RBD are more likely to be able to neutralize the virus directly.21, 22, 23
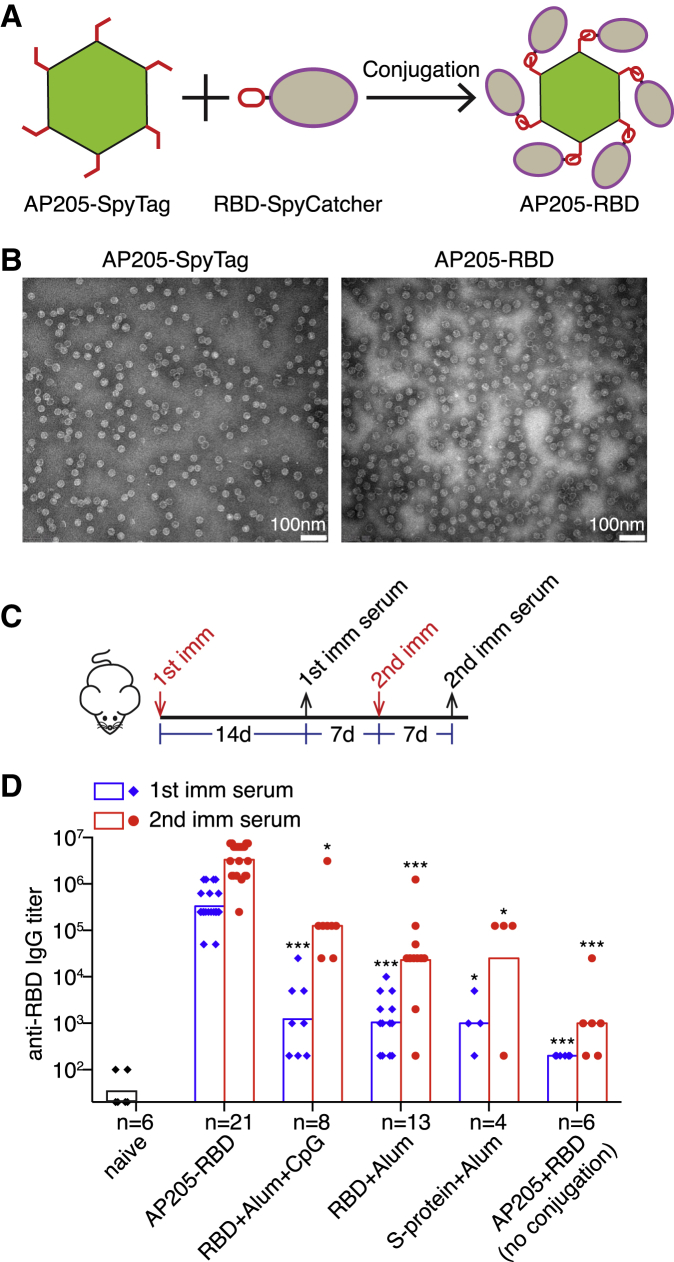
To construct a PLA-based COVID-19 vaccine candidate, we conjugated the fusion protein of RBD and SpyCatcher, RBD-SpyCatcher, to AP205-SpyTag, a particle composed from the fusion protein of AP205 capsid protein and SpyTag (Figure 1A). RBD-SpyCatcher was added at approximately 20 molecules per AP205-SpyTag particle and was almost completely conjugated to the AP205-SpyTag (Figure S1A). The resulting AP205-RBD retained the spherical structure because the intact AP205-SpyTag (Figure 1B) and the nucleic acids inside the particle were preserved after conjugation (Figure S1A).
Figure 1.
AP205-RBD elicited high titers of anti-RBD antibody in mice
(A) Construction strategy of AP205-RBD. Purified AP205-SpyTag and RBD-SpyCatcher are mixed in vitro to form the SpyTag:SpyCatcher covalent bond.
(B) Transmission EM images of intact AP205-SpyTag and AP205-RBD.
(C) Time points for the first (1st) and second (2nd) immunization (imm) and serum collection in mice.
(D) Serum anti-RBD IgG from mice immunized with the indicated antigens was measured by ELISA. Endpoint titers were presented. Symbols indicate data collected from individual mice. Bars indicate the geometric mean of each group. The number (n) of mice examined in each group was shown. Kruskal-Wallis test was used to compare the five immunized groups within either the “1st imm serum” or the “2nd imm serum.” There are significant differences among the five groups for both the “1st” and the “2nd imm serum.” Dunn’s multiple comparisons test was then used to compare between the AP205-RBD-immunized group and one of the other immunized groups, and the adjusted p value was used to determine the statistical significance and indicated in the graph (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
See also Figure S1.
To test the efficacy of AP205-RBD as a vaccine candidate, we immunized mice intraperitoneally (i.p.) with AP205-RBD twice 3 weeks apart (Figure 1C). Soluble RBD protein adjuvanted with alum or with additional CpG oligodeoxynucleotides (ODN) or full-length S protein with alum was also used to immunize mice for comparison. Following the first immunization, both anti-RBD IgM and IgG antibodies were elicited (Figures 1D and S1B). The anti-RBD IgG was further increased by about 10-fold after the second immunization (Figure 1D). Soluble RBD protein and S protein also elicited anti-RBD IgM and IgG, but to a significantly lesser degree than AP205-RBD (Figures 1D and S1B). It should be noted that the soluble RBD and S protein used in this study are not engineered for trimer stabilization and thus likely exhibit much lower immunogenicity than their engineered dimer or trimer counterparts, which were indeed more commonly applied as vaccine candidates.24, 25, 26, 27 The potency of AP205-RBD in inducing anti-RBD antibodies depended on the conjugation of the soluble RBD to AP205-SpyTag VLP, because the mixture of the two parts without conjugation elicited a very low level of anti-RBD IgG (Figure 1D), consistent with our previous finding that the B cell antigen and the TLR ligands need to be physically associated to engage B cell TLR signaling for enhancing the antibody response.5 Because the AP205 capsid protein is also part of the antigen, we also examined the anti-AP205 antibody. Indeed, both anti-AP205 IgM and IgG antibodies were elicited (Figure S1C), suggesting that the antigenic epitopes on the AP205 were not completely covered by the RBD protein.
The PLA-based COVID-19 vaccine candidate elicited neutralizing antibodies against SARS-CoV-2
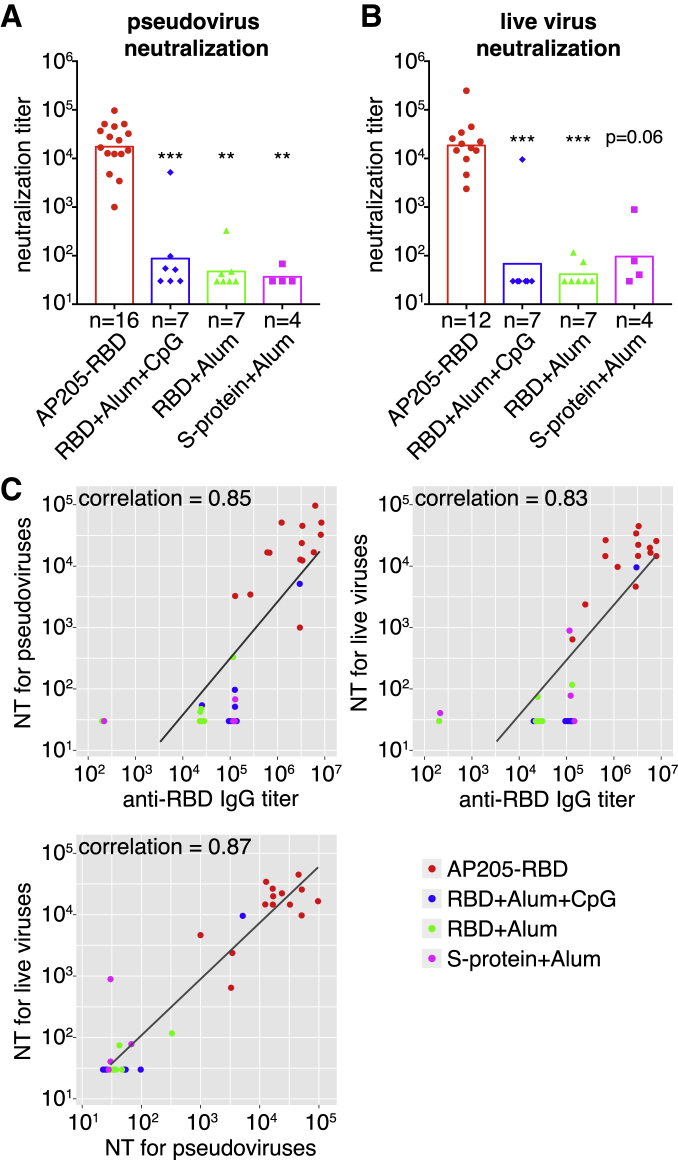
To test whether AP205-RBD-induced anti-RBD antibodies are neutralizing antibodies, we used a pseudovirus generated from the human immunodeficiency virus backbone expressing the SARS-CoV-2 S protein as the envelope protein for neutralization assay first.22 Sera from mice immunized with AP205-RBD exhibited robust neutralization activity against the pseudovirus, whereas the sera from mice immunized with the soluble RBD or full-length S protein exhibited much lower neutralization activity (Figure 2A). To confirm further, we used live SARS-CoV-2 virus to test the neutralizing activity in the above sera.28 Again, the sera from mice immunized with AP205-RBD exhibited the highest neutralization activity compared with the other immunization groups (Figure 2B). Overall, the anti-RBD IgG and the neutralization activity against either the pseudovirus or the live SARS-CoV-2 virus correlated with each other strongly (Figure 2C), suggesting that RBD is a valid target antigen, and the anti-RBD antibody level could be used to indicate the neutralization activity under this condition.
Figure 2.
AP205-RBD elicited neutralizing antibodies in mice
(A and B) Neutralization titers (defined as half-maximal inhibitory concentrations) against pseudovirus (A) or live SARS-CoV-2 virus (B) in sera collected after the second immunization were presented for the indicated immunization groups. Symbols indicate data collected from individual mice. Bars indicate the geometric mean of each group. The number (n) of mice examined in each group was shown. A Kruskal-Wallis test was used to compare the four groups. There are significant differences among the four groups for both (A) and (B). Dunn’s multiple comparisons test was then used to compare between the AP205-RBD-immunized group and one of the other immunized groups, and the adjusted p value was used to indicate the statistical significance (∗∗p < 0.01, ∗∗∗p < 0.001).
(C) Pairwise correlations between anti-RBD IgG and neutralization titers (NTs) against the pseudovirus or NTs against live SARS-CoV-2 virus measured from the same set of serum samples are shown. Dots represent the data from individual mice, and colors indicate different immunization groups. The same raw data were used to generate Figures 1D, 2A, and 2B. Spearman’s correlation coefficient is shown.
See also Figure S2.
To check whether immunization routes could affect the potency of AP205-RBD, we immunized mice subcutaneously (s.c.) or intramuscularly (i.m.) with the same dose of AP205-RBD as previous i.p. immunization. In fact, there was no significant difference among the groups immunized by different methods in either anti-RBD IgG levels or the neutralization activities (Figures S2A–S2C), suggesting that the immunogenicity of AP205-RBD was not affected by the immunization routes tested here.
The PLA-based COVID-19 vaccine candidate elicited a Th1-oriented immune response
One concern in the development of vaccines against respiratory viruses is the potential risk for enhanced respiratory disease (ERD), which was originally reported for an inactivated vaccine against the respiratory syncytial virus and is thought to be caused partly by the improper Th2-biased response.29 For intracellular pathogens such as viruses, Th1-oriented immune response is usually required to effectively clear the pathogens. Interferon γ (IFNγ) is the characteristic cytokine produced by Th1 and is also produced by cytotoxic CD8+ T cells. Both types of T cells are involved in the anti-viral response. From the aspect of B cells, cytokines associated with Th1 response tend to promote Ig isotype switch to IgG2a/c in mice, which is equivalent to IgG1 in humans, whereas Th2-associated cytokines promote Ig isotype switch to IgG1 in mice, which is equivalent to IgG4 in humans. Th1-associated IgG subclass is more potent in mediating antibody-dependent cellular cytotoxicity and phagocytosis compared with the Th2-associated IgG subclass and thus could contribute to the anti-viral response more efficiently.30
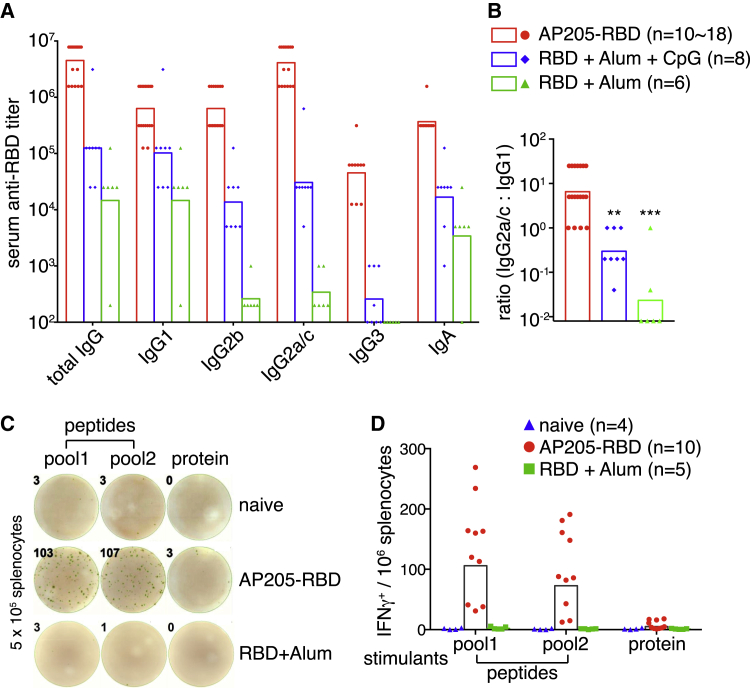
We previously found that activation of TLR/MyD88 signaling in B cells by Qβ-VLP strongly promoted the immune response toward Th1 direction, with the induction of T-bet in cognate CD4+ T cells and Ig class switch to IgG2a/c.4,9 To determine whether the AP205-RBD-induced response is Th1 oriented, we first examined the Ig isotypes of anti-RBD antibodies. As expected, AP205-RBD-induced high titers of IgG2a/c (Figure 3A) and the ratio of IgG2a/c to IgG1 was significantly higher in the AP205-RBD-immunized group compared with the soluble RBD-immunized groups (Figure 3B). Addition of CpG on top of alum could indeed increase the IgG2a/c-to-IgG1 ratio in soluble RBD-immunized groups as expected but could not bring it to the same level as that of the AP205-RBD-immunized group (Figure 3B).
Figure 3.
AP205-RBD induced the Th1-oriented immune response
(A) The indicated Ig isotypes of anti-RBD were examined in sera collected after the second immunization. Colors indicate different immunization groups.
(B) The ratio of RBD-specific IgG2a/c titer to IgG1 titer. One-way ANOVA test was used to determine the significant difference among the three groups. Tukey’s multiple comparisons test was then used to compare between the AP205-RBD-immunized group and one of the RBD protein-immunized groups. The adjusted p value was used to indicate the statistical significance (∗∗p < 0.01, ∗∗∗p < 0.001).
(C) Representative data of IFNγ ELISpot assay. Splenocytes from naive mice or mice immunized with the second dose of AP205-RBD or RBD+Alum 5 d previously were examined for IFNγ+ cells. 15-mer peptide pools derived from the RBD sequence were used to stimulate the cells in vitro. Pool1 and pool2 contain peptides derived from 420–459 and 511–549 aa of SARS-CoV-2 S protein, respectively. Intact RBD protein was also used for stimulation.
(D) Quantification of the IFNγ+ cells as in (C). Symbols indicate data collected from individual mice. Bars indicate the geometric mean of each group. The numbers (n) of mice examined in each group are shown. See also Figure S3.
Because the absolute quantity of different Ig isotypes cannot be compared directly by ELISA, we further quantified RBD-specific plasma cells (PCs) by enzyme-linked immune absorbent spot (ELISpot) assay. There were significantly more IgG2a/c+ PCs than IgG1+ or IgG2b+ PCs in both splenocytes and bone marrow (BM) cells (Figures S3A and S3B), consistent with the ratio of anti-RBD IgG2a/c to IgG1 determined by ELISA (Figure 3B). The potential effect of LPS on AP205-RBD-induced antibody response was also examined. Using AP205-RBD with the LPS level differing by ∼100,000 fold, we found no significant difference in either the total anti-RBD IgG titer or the IgG subclass distribution from the immunized mice (Figure S3C), suggesting that LPS did not contribute to the Th1-oriented response induced by AP205-RBD.
To further evaluate whether AP205-RBD induced a Th1 response, we examined IFNγ-secreting cells in splenocytes from naive or immunized mice. The 15-mer peptides derived from the RBD sequence31 or the intact RBD proteins were used for in vitro stimulation. There were a number of cells secreting IFNγ upon peptide stimulation in AP205-RBD-immunized mice, whereas very few of these cells were found in naive or mice immunized with soluble RBD plus alum (Figures 3C and 3D), suggesting that AP205-RBD indeed induced a Th1-oriented response. We cannot distinguish whether these IFNγ-secreting cells were CD4+ or CD8+ T cells, although we found previously that a similar kind of VLP induced robust T-bet expression in CD4+ T cells4 and a rather moderate CD8+ T cell response (unpublished data). Overall, the IgG2a/c dominated antibody response, and the generation of abundant IFNγ-secreting cells supported that AP205-RBD induced a Th1-oriented response.
The PLA-based COVID-19 vaccine candidate elicited an antigen-specific GC response
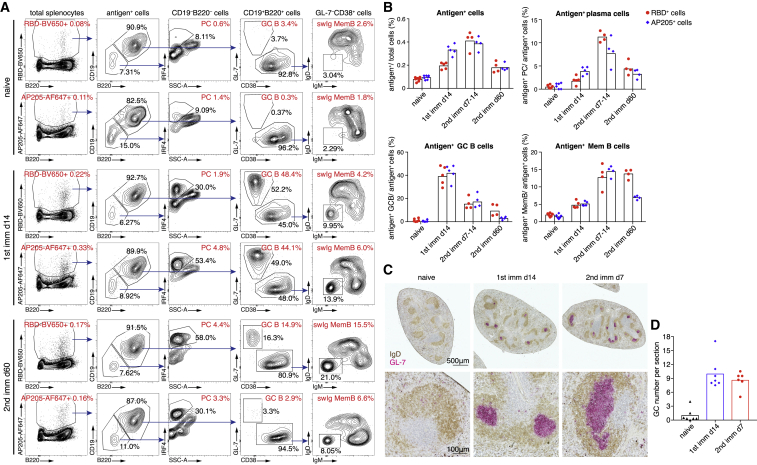
GC response is a specialized form of T-dependent antibody response, in which GC B cells with help from Tfh cells undergo multiple rounds of cell divisions and go through somatic hypermutation for affinity maturation.32 GC response is also involved in the generation of long-lived PCs and memory B cells. We previously found that Qβ-VLP could induce not only a robust but also a prolonged GC response,33 which depends on B cell TLR/MyD88 signaling.9 To test whether AP205-RBD could induce a GC response in mice, we examined antigen-specific B cells labeled with fluorophore-conjugated antigens by flow cytometry. Upon AP205-RBD immunization, both RBD-specific and AP205-specific PCs, GC B cells, and memory B cells with switched isotype (swIg MemB) were detected, whereas naive mice contained only IgD+/IgM+ naive antigen+ B cells, supporting that AP205-RBD induced an antigen-specific GC response (Figure 4A). Fluorophore-binding B cells maintained as IgD+/IgM+ naive B cells upon AP205-RBD immunization (Figures S4A–S4C), supporting our staining strategy. The antigen+ GC B cells reached the peak level at about 2 weeks after the first immunization (Figure 4B), which is consistent with what we have observed previously for the other PLAs.33 Interestingly, a second immunization did not further increase the GC B cell number (Figure 4B), suggesting that one dose of AP205-RBD is enough to launch the full-scale GC response. Nevertheless, a second immunization led to a boost of antigen+ PCs (Figure 4B), which is consistent with the increased antibody titers upon the second immunization and presumably would benefit the vaccinees upon infection. Moreover, the AP205-RBD-induced GC response lasted quite long, with RBD-specific GC B cells being detectable even 2 months after the second immunization (Figures 4A and 4B), which is also consistent with the duration of the GC response elicited by a similar kind of PLA.33 Anatomical GC structures in spleens were also identified by immunohistochemistry in the immunized mice (Figures 4C and 4D).
Figure 4.
AP205-RBD induced a prolonged GC response
(A and B) Splenocytes from naive mice or mice immunized with AP205-RBD previously were examined for antigen-specific cells by RBD-BV650 or AP205-AF647 labeling. RBD-BV650+ and AP205-AF647+ cells are defined as antigen+ cells for simplicity, although they may consist of a fraction of fluorophore-specific cells, which are IgD+IgM+ (Figures S4A and S4B). Antigen-specific plasma cells (PCs) (CD19–B220–IRF4+), germinal center (GC) B cells (CD19+B220+GL7+CD38–), and Ig-switched memory B cells (swIg MemB) (GL7–CD38+IgD–IgM–) can be found in AP205-RBD-immunized animals. The percentages of the antigen+ cells in the total splenocytes and the percentages of the indicated subsets within the total antigen+ cells are labeled in red. The percentages of the gated cells within the parental group of cells are labeled in black. Representative data are shown in (A), and the summary data are shown in (B). Symbols indicate data of individual mice. Bars indicate the geometric mean of each group.
(C and D) Spleen sections were stained for GC structures with IgD (brown) and GL-7 (pink) by immunohistochemistry. Representative data are shown in (C), and quantification data are shown in (D). Symbols indicate data collected from individual mice, and bars indicate the mean of each group.
See also Figure S4.
The PLA-based COVID-19 vaccine candidate elicited durable humoral memory
Humoral memory, the immunological memory formed by B cells, consists of long-lived PCs and memory B cells. The long-lived PCs are responsible for maintaining antigen-specific antibody level and are generated through the GC response. Memory B cells could be highly heterogeneous, with IgM+ memory B cells usually formed during the early response but with low affinity to the antigen, and the swIg memory B cells formed through the GC response with increased affinity and other gene expression changes.34,35 This latter group of memory B cells is usually more responsive and tends to differentiate into PCs more readily upon the reencountering of antigen.
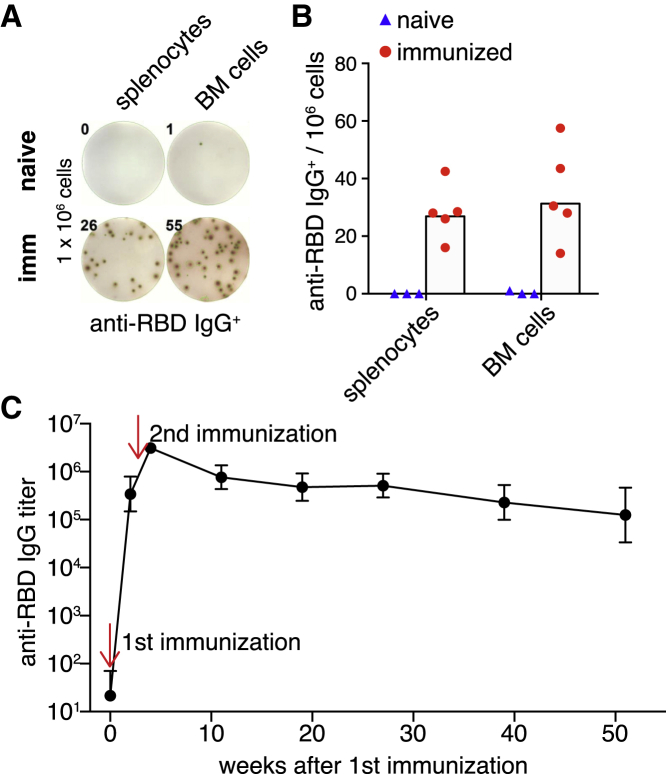
To evaluate AP205-RBD-induced humoral memory, we examined RBD-specific long-lived PCs in mice immunized 3–4 months previously. Because long-lived PCs reside in BM36 and can also be found in spleen as we previously showed,33 both splenocytes and BM cells were examined by ELISpot for RBD-specific IgG-secreting cells. Indeed, RBD-specific PCs were abundant in both spleen and BM even 4 months after immunization (Figures 5A and 5B). Consistently, we found that the anti-RBD IgG was relatively stable from 2 months after the second immunization up until a year as we have followed (Figure 5C). Besides long-lived PCs, we also found that a large proportion of RBD-specific swIg memory B cells were IgG2a/c+ (Figure S5A). We previously showed that IgG2a/c+ memory B cells induced by a similar PLA could last for more than a year in mice.33 Therefore, AP205-RBD induced not only a robust antibody response but also a durable humoral memory.
Figure 5.
AP205-RBD induced durable humoral memory
(A and B) Splenocytes or bone marrow (BM) cells from mice immunized with AP205-RBD twice 3–4 months previously were examined for anti-RBD IgG-secreting cells by ELISpot assay. Representative data are shown in (A), and quantification data are shown in (B). Symbols indicate data collected from individual mice, and bars indicate the geometric mean of each group.
(C) Anti-RBD IgG in mice immunized with AP205-RBD was examined for up to 1 year after immunization. The geometric mean and standard deviation for each time point are shown. There are six to eight individuals for each time point, except for the last time point there are four individuals.
See also Figure S5.
Because AP205-RBD induced both anti-AP205 and anti-RBD antibodies (Figure S1C), one concern using AP205-RBD as a vaccine is whether the induced anti-AP205 antibody could reduce the effect of other vaccines using the same AP205 as a carrier in the future. To address this question, we immunized with AP205-RBD a group of mice that had received other AP205-based vaccine candidates 4–6 months ago. We found that there was no significant difference of the AP205-RBD-induced anti-RBD antibody levels between the mice with and those without a prior immunization history (Figure S5B). In contrast, the anti-AP205 antibody titers were significantly higher in mice that have been immunized with another AP205-based antigen previously, as one might expect (Figure S5B). This result suggested that the pre-existing antibodies against the PLA carrier part did not affect significantly the antibody response toward a new antigen target.
The PLA-based COVID-19 vaccine candidate elicited neutralizing antibodies in NHPs
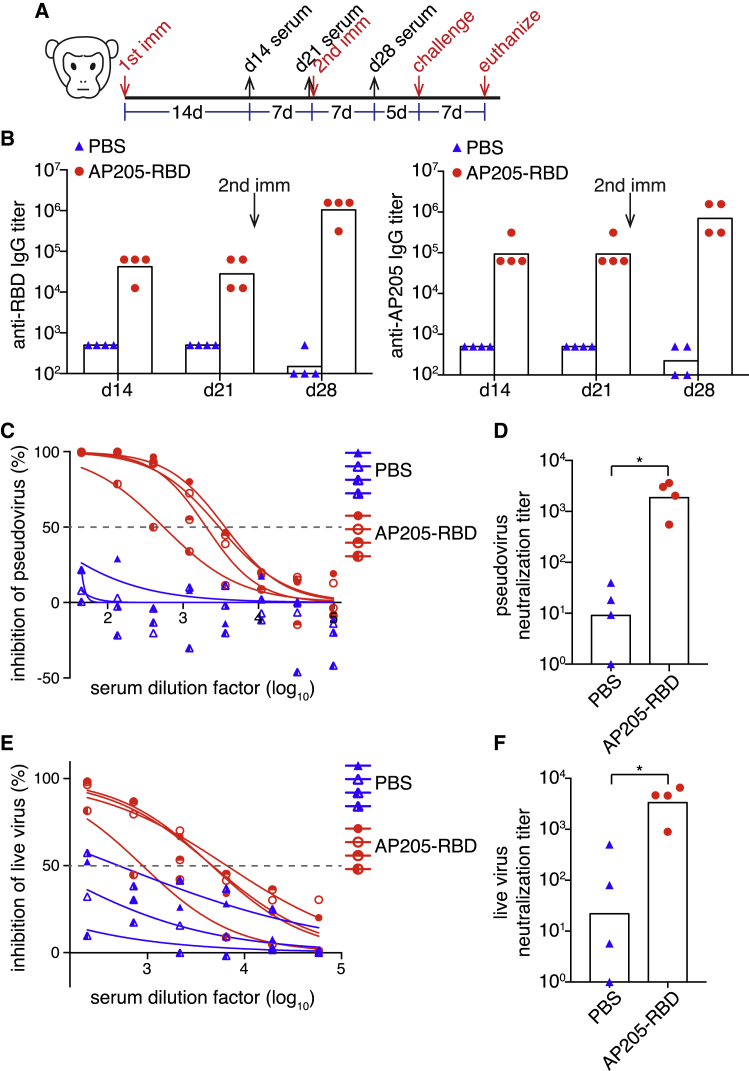
To further evaluate the potential of AP205-RBD as a COVID-19 vaccine candidate for humans, we immunized rhesus macaques i.m. with AP205-RBD twice 3 weeks apart (Figure 6A). Both anti-RBD and anti-AP205 IgG were elicited after the first immunization and further increased upon the second immunization (Figure 6B). In addition, sera collected from the immunized macaques exhibited neutralizing activity against both the pseudovirus and the live SARS-CoV-2 virus (Figures 6C–6F). Curiously, it seems that sera from some unimmunized macaques already exhibited a low level of neutralizing activity (Figure 6F). It is unclear whether these macaques have been exposed to other coronaviruses previously because they were not raised in a strictly pathogen-free environment. Nevertheless, the background neutralizing activity in the unimmunized animals varied a lot, whereas immunization with AP205-RBD significantly increased the neutralizing activity against live SARS-CoV-2 virus, supporting the efficacy of AP205-RBD as a candidate vaccine.
Figure 6.
AP205-RBD elicited neutralizing antibodies in non-human primates
(A) Time points for immunizations, serum collections, and viral challenge in macaques.
(B) Serum anti-RBD and anti-AP205 IgG from macaques immunized with PBS or AP205-RBD were examined at the indicated time points.
(C–F) Neutralization activity against the pseudovirus (C and D) or the live SARS-CoV-2 virus (E and F) in the sera collected after the second immunization was examined. Curve fitting was used to determine the half-maximal inhibitory concentrations (IC50) (C and E), which were presented as the neutralization titers in (D) and (F). Symbols indicate data of individual animals, and bars indicate the geometric mean of each group. Mann-Whitney test was used for statistical analysis (∗p < 0.05).
Immunization with PLA-based COVID-19 vaccine candidate accelerated the viral clearance in the infected animals
To test whether AP205-RBD could indeed provide any protection for the immunized animals, we challenged rhesus macaques with SARS-CoV-2 virus 12 days after the second immunization. Although rhesus macaques can be infected by SARS-CoV-2, they generally develop only mild symptoms and recover spontaneously.37 To ensure that the viral infection can be uniformly established in all experimental animals, a relatively high dose, 1 × 107 median tissue culture infectious dose (TCID50), of SARS-CoV-2 virus in total was inoculated to the animals through a combination of intranasal and intratracheal instillation. Both the control group and the immunized group of animals exhibited no significant changes of body temperatures, weights, and peripheral blood cell counts after the infection (Figure S6A). All the animals exhibited similar levels of the viral loads in the nasal swabs and the rectal swabs on the first day after viral challenge (Figures 7A–7C and S6B–S6D), suggesting that the infection procedure was consistent across all the animals. Notably, the establishment of infection in the immunized animals suggested that the neutralizing antibodies cannot prevent the infection, at least when the inoculation dose is high. Preclinical studies on a few other vaccine candidates have found similar results.38, 39, 40
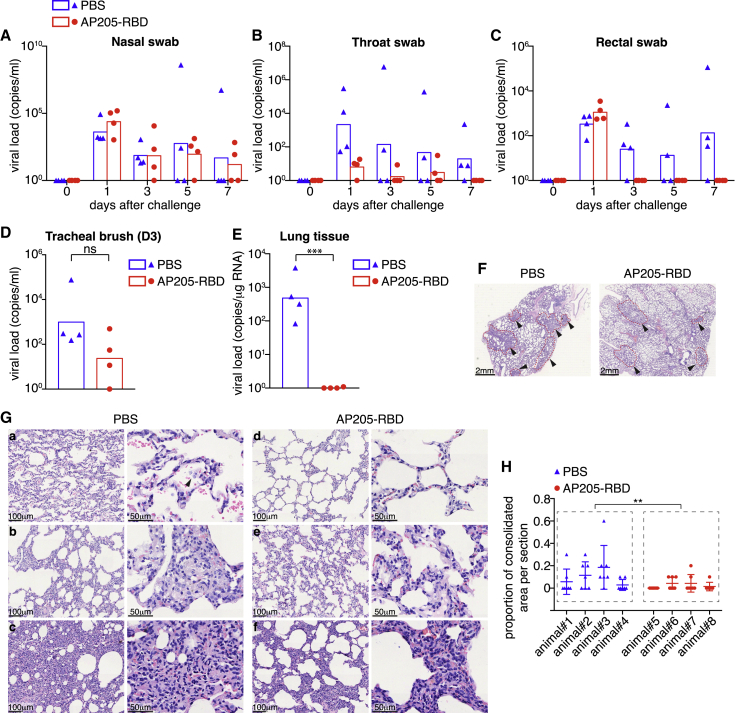
Figure 7.
Vaccination with AP205-RBD accelerated the viral clearance in the infected animals
(A–C) Swab samples were taken from the nasal cavity, throat, and rectum at the indicated time points after SARS-CoV-2 challenge and examined for viral loads.
(D) Tracheal brush samples taken at day 3 after viral challenge were examined for viral loads.
(E) Lung tissue samples taken from the euthanized animals at day 7 after viral challenge were examined for viral loads. Mann-Whitney test was used for statistical analysis in (D) and (E) (∗∗∗p < 0.001). (A–E) Symbols indicate data collected from individual animals, and bars indicate the geometric mean of each group.
(F–H) Histology examination of lung tissues. (F) Representative images of H&E staining at low magnification. Arrowheads point to lesion regions, which were surrounded with dashed lines. (G) Representative images at higher magnifications. Images were chosen from different individuals in both groups. The right panels are enlarged regions from the left panels. The arrowhead in (a, right panel) points to detached cells in the airspace. The severely infiltrated area, which is quantified as the consolidated area in (H), is illustrated in (c). (H) Summary of the proportion of the consolidated area in each tissue section. One section was taken from every lung lobe, with seven sections taken in total for each animal. Symbols indicate data of individual sections. Mean and standard deviation for each animal are shown. Nested ANOVA was used to compare the PBS- and AP205-RBD-immunized groups (∗∗p < 0.01).
ns, non-significant. See also Figure S6.
However, the viral clearance seemed to be accelerated upon immunization with AP205-RBD. The viral loads in the rectal swabs became undetectable since day 3 after viral challenge for all the immunized animals, whereas in three of four control animals the viral RNA persisted in rectal swabs until day 7 after challenge (Figures 7C and S6D). The viral loads in the trachea at day 3 postinfection exhibited a tendency of decrease in the immunized group, although without a statistical significance (Figure 7D). Most strikingly, although all four animals in the control group carried a large amount of virus in at least several, if not all, of the seven lung lobes at day 7 postinfection, all the immunized animals almost completely cleared the virus by this time (Figures 7E and S6E). The clearance of the virus in the upper respiratory tract seemed to take longer, especially in the nasal cavity, with two of four immunized animals continuing to carry the viruses for up to 7 days after challenge (Figures 7A, 7B, S6B, and S6C).
Lung tissues taken at day 7 postchallenge were further processed for histochemistry examinations. Although the virus was cleared much more quickly in the immunized animals, the pathological changes characteristic of interstitial pneumonia were still evident in these animals, which was likely caused by the high dose of the viral infections initially. At low magnification, lesions with a patchy or diffuse pattern were seen in both control and immunized animals (Figure 7F). At higher magnifications, the pathological changes mainly manifested as thickened alveolar septa and infiltration of lymphocytes (Figure 7G). Occasionally, cell debris could be found in the air spaces (Figure 7Ga), which might reflect the infection-related cell damage. There was no infiltration of eosinophils or other granulocytes in all the tissue sections examined from both the control and the immunized animals. In regions where cell infiltration was severe, the air spaces almost completely disappeared (Figure 7Gc). We quantified the proportion of such kinds of consolidated areas in each tissue section and found that the immunized animals tended to have a lower proportion of the consolidated areas than the control animals (Figure 7H). We found no other significant differences of the pathological changes between the control and the immunized groups. Overall, it seemed that immunization with AP205-RBD could accelerate the viral clearance, especially in lungs and guts, but may not prevent the viral infection completely.
Discussion
Here we presented evidence supporting targeting B cell TLR signaling, a physiological anti-viral mechanism, as a valid strategy to develop a COVID-19 vaccine. The PLA structure enables the coupling of the BCR signaling with the TLR signaling in B cells, which serves as a message to B cells to initiate the anti-viral response. The activated B cells could then go through proliferation and differentiation and could also promote Tfh development, which in turn fuels back the GC response, all of which contribute to the anti-viral humoral response.16 In this study, we found that AP205-RBD induced (1) high titers of anti-RBD antibodies, which could neutralize SARS-CoV-2 virus; (2) a Th1-oriented response; (3) a robust GC response with long-lived PCs and memory B cells; and (4) the protection against the viral infection in animals. These results are consistent with what we previously observed using other VLPs containing TLR ligands,9,33 suggesting that the constructed AP205-RBD could indeed elicit the B cell response in the proposed manner.
We chose RBD as the targeted antigen because it mediates the binding of the virus to the host cells.22,41,42 The strong correlation between the anti-RBD antibody level and the viral neutralization activity in the immunized sera confirmed that the RBD constructed in this study adopted the proper conformation. The full-length S protein presumably consisting of more B cell and T cell epitopes has been used in the nucleic acid and the viral-vector vaccines, as well as in some recombinant protein vaccines.43 However, it seems that regardless of the RBD or the full-length S protein used and regardless of the vaccine platforms chosen, the induced neutralizing activity is most strongly correlated with the anti-RBD antibody level,40,44,45 supporting that RBD itself could serve as the targeted antigen. Multiple T cell epitopes have also been identified within RBD, as well as the other parts of the S protein in both immunized animals and convalescent patients, although the T cell epitopes alone may not be sufficient for the protective response.31,46, 47, 48, 49
Constructing antigen into a multivalent form has been of interest to the vaccine field for a long time. Multivalent antigens may enhance the initial B cell activation by simultaneously engaging multiple BCRs, which subsequently may promote many aspects of the B cell response, including the PC formation, the GC formation, and the recruitment of low-affinity antigen-specific cells into the GC response.50, 51, 52 Protein carriers that can assemble into polymers are often used as platforms to build such multivalent antigens.53 Although many carrier platforms could be used to display antigens, only some of them could carry nucleic acids inside, which could activate B cell TLR signaling.18 For multivalent antigens carrying no TLR ligands inside, such as those built from ferritin, lumazine synthase, and some computationally designed carrier proteins, additional adjuvant is required to achieve the proper antibody response,52,54, 55, 56, 57, 58, 59 presumably via activating the innate signaling in DCs. In contrast, multivalent antigens built on capsid proteins such as bacterial phage-derived VLPs or HBcAg-VLPs could carry natural nucleic acids or artificial CpG-ODN, which could activate B cell TLRs and do not require additional adjuvant for immunization.18 Although both types of multivalent antigen have been demonstrated to induce robust antibody responses, the underlying cellular mechanisms presumably are different and need further study. Nevertheless, both types of platform have been used to develop the COVID-19 vaccine candidates, some of which have entered the clinical development stages. Examples of such vaccines include VLPs composed of SARS-CoV2 S protein produced from Nicotiana benthamiana60 and artificially designed VLPs assembled from two components in vitro.54 Preclinical studies of two other AP205 phage-based vaccine candidates, which were very similar to the one described in this study, indeed showed similar antibody responses in mice as in this study, although they have not tested the vaccine efficacy in viral challenge conditions in NHPs.61,62
Advances in the adenovirus vector and mRNA-based vaccine platforms over the last several decades have greatly accelerated the development of the COVID-19 vaccines.63 These vaccine platforms facilitated a much-shortened preclinical development process compared with the traditional vaccine technology or the recombinant protein-based methods. Both vaccine platforms seem to generate relatively strong immunogenicity, especially they likely induce a more robust CD8+ T cell response because the host cells are transfected to express the antigen.64 The relatively pronounced reactogenicity elicited by the adenoviral vector and mRNA vaccines compared with the traditional flu vaccines is consistent with their unique modes of activation of the immune system.65 Although the current clinically approved COVID-19 vaccines have greatly reduced the morbidity and mortality rate of the disease and relieved some public health burdens, the long-term application of these new technologies as a routine vaccination regimen needs further assessment. Some very rare but severe side effects related to these new vaccine platforms need to be addressed. Cases of thrombosis with thrombocytopenia syndrome (TTS) were related with the adenoviral vector vaccines.66 Cases of myocarditis and pericarditis were related with mRNA vaccines, especially after the second dose,67,68 implying that the disease might not be caused simply by the non-specific inflammatory response, raising concerns about whether a third dose of mRNA vaccine would increase the probability of the side effects. The adenoviral vector vaccines face a different problem in the situation when additional vaccination is needed. It is known that the preexisting antibodies against adenoviruses could reduce the vaccine efficacy.69 Indeed, the Gamaleya vaccine used two different adenoviral vectors for the two doses of the vaccinations to overcome this problem.70 The recombinant protein-based vaccines including the different nanoparticle platforms, although taking a few months longer in the preclinical development stage, might provide a choice with less safety concern and suit for long-term clinical application.
In this study, we chose rhesus macaques as the animal model of SARS-CoV2 infection because they probably represent most closely to the human infection.71 All the rhesus macaques in this study exhibited no apparent severe symptoms, such as fever or dyspnea, even after a relatively high dose of viral challenge (Figure S6), which is consistent with that the majority of human COVID-19 patients were not in severe situations.72 The characteristic interstitial pneumonia histological change in the macaques (Figure 7) is also consistent with the chest radiology changes in many human patients.73 We found in this study that although our COVID-19 vaccine could undoubtedly accelerate the viral clearance, it did not prevent the initial viral entry completely (Figure 7). This is also consistent with the current clinical data for other vaccines that the vaccine protected against the severe disease more effectively than preventing the infection.74,75 The viral load in the nasal cavity continued to be detectable at day 7 after infection even in the immunized macaques (Figure 7A), raising the possibility that the vaccinated individual could still transmit the disease. Other preclinical studies have not observed such phenomena either because the nasal swab examination was not performed,38,76,77 or the inoculation doses were much lower and the initial infection was already reduced significantly.40,44 Our study suggested that the vaccinees could potentially carry the virus for a period of time once being infected even though they themselves might have only mild or no symptoms. Indeed, a recent study from the UK that examined the secondary cases from household contacts found that vaccinations with an adenoviral vector (ChAdOx1 nCoV-19) or a mRNA vaccine (BNT162b2) could reduce the transmission by 40%–50%, which is lower than the protection efficacy for these vaccines.78 Thus, quarantine procedures may remain important before a large proportion of populations are immunized.
Limitations of the study
In this study, the AP205-RBD-induced immune response was not compared with those induced by other vaccine platforms, such as mRNA-based vaccines or other nanoparticle vaccines; thus, it is unclear how “potent” AP205-RBD is as a vaccine candidate in comparison with the currently available vaccines. Although AP205-RBD induced durable memory in mice, whether it could induce analogously long-term memory in primates needs further assessment. AP205-RBD lacks the antigenic epitopes in the other parts of the SARS-CoV-2 S protein and potentially induces a “narrower” response.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-mouse-CD45R/B220 PE-CF594(clone RA3-6B2) | BD Biosciences | Cat#562290; RRID: AB_11151901 |
| Rat monoclonal anti-mouse-CD19 APC-Cy7(clone 1D3) | BD Biosciences | Cat#557655; RRID: AB_396770 |
| Rat monoclonal anti-mouse-IgM PE-Cy7(clone II/41) | Invitrogen | Cat#25-5790-82; RRID: AB_469655 |
| Fluorescein (FITC)-AffiniPure Goat Anti-Mouse IgM, μ Chain Specific | Jackson Immunoresearch | Cat#115-095-075; RRID: AB_2338598 |
| Rat monoclonal anti-mouse-IgD BV711 (clone 11-26c.2a) | BioLegend | Cat#405731; RRID: AB_2563342 |
| Rat monoclonal anti-mouse-GL-7 PE (clone GL7) | BD Biosciences | Cat#561530; RRID: AB_10715834 |
| Rat monoclonal anti-mouse-CD38 Alexa Fluor 700 (clone 90) | Invitrogen | Cat#56-0381-82; RRID: AB_657740 |
| Rat monoclonal anti-mouse-CD8a PerCP-Cy5.5 (clone 53-6.7) | Invitrogen | Cat#45-0081; RRID: AB_1107004 |
| Rat monoclonal anti-mouse-GL-7 FITC (clone GL7) | BioLegend | Cat#144603; RRID: AB_2561696 |
| Rat monoclonal anti-mouse/human-IRF4 EF450 (clone 3E4) | Invitrogen | Cat#48-9858-82; RRID: AB_2574135 |
| Biotin Rat Anti-Mouse IgG1(clone A85-1) | BD Biosciences | Cat#553441; RRID: AB_394861 |
| Biotin Mouse Anti-Mouse IgG2a [b] (clone 5.7) | BD Biosciences | Cat#553504; RRID: AB_394889 |
| Goat polyclonal Secondary Antibody to Monkey IgG - H&L (HRP) | Abcam | Cat#ab112767 |
| Goat Anti-Mouse IgM-HRP | SouthernBiotech | Cat#1021-05; RRID: AB_2794240 |
| Goat Anti-Mouse IgG2c, Human ads-HRP | SouthernBiotech | Cat#1079-05; RRID: AB_2794466 |
| Goat Anti-Mouse IgG1, Human ads-HRP | SouthernBiotech | Cat#1070-05; RRID: AB_2650509 |
| Goat Anti-Mouse IgG2b, Human ads-HRP | SouthernBiotech | Cat#1090-05; RRID: AB_2794521 |
| Goat Anti-Mouse IgG3, Human ads-HRP | SouthernBiotech | Cat#1100-05; RRID: AB_2794573 |
| Goat anti-Mouse IgA Heavy Chain Antibody HRP Conjugated | Bethyl Laboratories | Cat# A90-103P |
| Goat anti-Mouse IgG-Fc Fragment Antibody HRP Conjugated | Bethyl Laboratories | Cat# A90-131P |
| Goat anti-Mouse IgG-Fc Fragment Antibody Affinity Purified | Bethyl Laboratories | Cat# A90-131A |
| IFN gamma Monoclonal Antibody (clone AN-18), Functional Grade | Invitrogen | Cat#16-7313-85; RRID: AB_469247 |
| IFN gamma Monoclonal Antibody (clone R4-6A2), Biotin | Invitrogen | Cat#13-7312-85; RRID: AB_466939 |
| IgD Monoclonal Antibody (clone 11-26c), Biotin | Invitrogen | Cat#13-5993-82; RRID: AB_466860 |
| BV650 streptavidin | BioLegend | Cat#405231 |
| PE streptavidin | BioLegend | Cat#405204 |
| HRP-conjugated streptavidin | Jackson Immunoresearch | Cat#016-030-084; RRID: AB_2337238 |
| Bacterial and virus strains | ||
| E. coli BL21(DE3) | TransGen Biotech | Cat# CD601 |
| SARS-CoV-2 (nCoV-2019BetaCoV/Wuhan/WIV04/2019) | The National Virus Resource, China | N/A |
| SARS-CoV-2 strain 107 | The Guangdong Provincial Center for Disease Control and Prevention, Guangdong, China | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| CpG-ODN | Shanghai Generay Biotech | Sequence:TCCATGACGTTCCTGACGTT |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Yeasen biotech, China | Cat#367-93-1 |
| Polyethylenimine (PEI) | Polyscience | Cat#23966-1 |
| 3,3′-Diaminobenzidine tetrahydrochloride hydrate | Sigma-Aldrich | Cat# D5637-1G |
| 3,3′,5,5′-Tetramethylbenzidine | Sigma-Aldrich | Cat#860336-1G |
| Fast Red | Sigma-Aldrich | Cat# F8764-1G |
| D-biotin | BBI Life Sciences | Cat# A100340-0500 |
| Imject Alum | Thermo | Cat# 77161 |
| RBD peptides pool1 | Smith, 2020 #31 | N/A |
| RBD peptides pool2 | Smith, 2020 #31 | N/A |
| Recombinant AP205 protein | This paper | N/A |
| Recombinant AP205-SpyTag protein | This paper | N/A |
| Recombinant RBD protein | This paper | N/A |
| Recombinant RBD-SpyCatcher protein | This paper | N/A |
| Recombinant RBD-AviTag protein | This paper | N/A |
| Recombinant GST-BirA protein | This paper | N/A |
| Recombinant S protein | This paper | N/A |
| Critical commercial assays | ||
| Toxin Sensor Chromogenic LAL Endotoxin Assay Kit | GenScript | Cat# L00350C |
| Alexa Fluor 647 Protein Labeling Kit | Invitrogen | Cat# A20173 |
| THUNDERBIRD Probe One-Step qRT-PCR kit | TOYOBO | Cat# QRZ-101 |
| High Pure Viral RNA Kit | Roche | Cat#11858882001 |
| TRIzol Plus RNA Purification Kit | Thermo Fisher | Cat# A33254 |
| MiniBEST Viral RNA/DNA Extraction Kit | TaKaRa | Cat#9766 |
| PrimeScript RT reagent Kit with gDNA Eraser | Takara | Cat# RR047A |
| Ni-NAT 5mL (Pre-Packed Gravity Column) | BBI Life Sciences | Cat# C600793-0010 |
| GST Fusion Protein Purification Kit | GenScript | Cat# L00207 |
| Bright-Glo Luciferase Assay Vector System | Promega | Cat# E2650 |
| Experimental models: Cell lines | ||
| Expi293F | GIBCO | Cat# A14635 |
| Vero-E6 | ATCC | CRL-1586; RRID: CVCL_0574 |
| Human: HEK293T | ATCC | Cat# CRL-11268 |
| Human: Huh7 | NICR | Cat#1101HUM-PUMC000679 |
| Experimental models: Organisms/strains | ||
| C57BL/6 | The Institute of Biophysics, Chinese Academy of Sciences | N/A |
| Rhesus macaques | Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS) | N/A |
| Oligonucleotides | ||
| SARS-CoV-2- forward primer: 5′-GGGGAACTTCTCCTGCTAGAAT-3′ | Song, 2020 #37 | N/A |
| SARS-CoV-2- reverse primer: 5′-CAGACATTTTGCTCTCAAGCTG-3′ | Song, 2020 #37 | N/A |
| SARS-CoV-2-probe FAM-TTGCTGCTGCTTGACA GATT-TAMRA-3′ |
Song, 2020 #37 | N/A |
| SARS-CoV-2-S- forward primer: 5′-CAATGGTTTAACAGGCACAGG-3′ | This paper | N/A |
| SARS-CoV-2-S-reverse primer: 5′-CTCAAGTGTCTGTGGATCACG-3′ | This paper | N/A |
| Recombinant DNA | ||
| pET21-AP205 | This paper | N/A |
| pET21-AP205-SpyTag | This paper | N/A |
| pCEP4-RBD | This paper | N/A |
| pCEP4-RBD-SpyCatcher | This paper | N/A |
| pCEP4-RBD-AviTag | This paper | N/A |
| pGEX-BirA | This paper | N/A |
| pcDNA3.1-SARS-Cov-2 S | Ju, 2020 #22 | N/A |
| Software and algorithms | ||
| FlowJo software Version 10.7.1 | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism 8.0 | GraphPad Software | https://www.graphpad.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Baidong Hou (baidong_hou@ibp.ac.cn).
Materials availability
Requests for plasmids of PLA vaccine components and recombinant protein should be directed to and will be fulfilled by the Lead Contact, Dr. Baidong Hou. All reagents will be made available on request following completion of a Material Transfer Agreement.
Experimental model and subject details
Experimental Animals
All mice used in this study were housed under specific pathogen-free conditions, and their use was approved by the Animal Care and Use Committee of the Institute of Biophysics, Chinese Academy of Sciences. C57BL/6 mice were bred in-house or purchased from SPF Biotechnology Co., Ltd (China). Roughly equal number of male and female mice of 8-16 weeks were used for immunization.
The experiments with non-human primates (NHPs) were performed at Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS) (Yunnan Province, China). All NHP experiments were approved by the animal ethics committee of KIZ, CAS (certificate number: IACUC20016). The selection of experimental animals and experimental operations follow the guidelines and principles of the experimental animal ethics committee to ensure the welfare of experimental animals. Eight rhesus macaques (males, age 3-6 years) used in this study were sourced from the Kunming Primate Center of CAS. They were kept in similar and independent primate cages, which facilitated communication between animals. The viral challenge experiment was performed in the Animal Biosafety Level 3 Laboratory at the National Kunming High-Level Biosafety Primate Laboratory Center, which is accredited by the China National Accreditation Service for Conformity Assessment and performs related operations in accordance with the standard operating procedures approved by the Biosafety Committee of the KIZ, CAS.
Cell lines
Expi293F cells (GIBCO, USA) were cultured in SMM 293-TII medium (Sino BiologicalInc, China) in suspension at 37°C, 5% CO2, with shaking at 125 rpm/min. HEK293T(ATCC), Huh7(NICR) and Vero-E6(ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS at 37°C with 5% CO2.
SARS-CoV-2 virus
The SARS-CoV-2 strain 10737 was obtained from the Guangdong Provincial CDC, Guangdong, China. The SARS-CoV-2 virus (nCoV-2019BetaCoV/Wuhan/WIV04/2019)28 was obtained from the National Virus Resource. SARS-CoV-2 virus was passaged in Vero-E6 cells.
Method details
Expression and purification of AP205 derived virus-like particles (VLPs)
The DNA sequence encoding the major AP205 coat protein (Gene ID: 956335) was synthesized and cloned into the pET21 vector. The sequence encoding SpyTag was fused at the C terminus of AP205 gene with a flexible linker to generate AP205-SpyTag. AP205-SpyTag encoded VLPs were expressed and purified in a similar way as Qβ-VLP as described before.33 Basically, E. coli BL21 (DE3) transformed with pET21-AP205 or pET21-AP205-Spytag was grown in LB medium and protein expression was induced with 0.1 mM IPTG (Yeasen biotech, China) at 37°C for 4-5 h. VLPs were purified by CsCl density gradient centrifugation at 200,000 g for 22 h. LPS was removed by repetitive extraction with Triton X-114. Endotoxin detection kits (ToxinSensor Chromogenic LAL Endotoxin Assay Kit, GenScript, L00350C) was used to determine the LPS level in purified AP205-Spytag VLPs according to the manufacturer’s instructions. Purified VLPs were examined by SDS-PAGE with Commassie Blue staining, or electrophoresis in 1% agarose gel followed by ethidium bromide staining. The assembly of VLPs was confirmed by transmission electron microscopy. The protein concentration of the VLPs was determined using Bradford assay.
Expression and purification of RBD of SARS-CoV-2 S protein
The coding sequences for the receptor binding domain (RBD) corresponding to the 319aa – 541aa of SARS-CoV-2 (NCBI Reference Sequence: YP_009724390.1) S protein and the full-length S protein were synthesized and cloned into pCEP4 vector (Addgene). The sequence encoding the signal peptide from the S protein and SpyCatcher was synthesized and fused at the N terminus of the RBD, and a 12-mer of histidine tag was fused at the C terminus of the RBD to generate RBD-SpyCatcher. The sequence encoding AviTag and a 12-mer of histidine tag was fused at the C terminus of RBD to generate RBD-AviTag. pCEP4-RBD, pCEP4-S, pCEP4-RBD-SpyCatcher or pCEP4-RBD-AviTag was transiently transfected into Expi293F cells using the PEI (Polyscience, 23966-1) following the manufacturer’s recommendations. Recombinant proteins were then collected from the supernatants of the cell culture and purified by affinity chromatography using Ni-NTA agarose (BBI Life Sciences, China). Purified proteins were examined by SDS-PAGE with Commassie Blue staining, and the protein concentration was determined using Bradford assay. To generate AP205-RBD, every 100 μg of RBD-SpyCatcher was added to 400 μg of AP205-SpyTag in PBS buffer and incubated on ice for more than 1 h.
Fluorescent labeling of AP205 and RBD
To detect antigen-specific cells, AP205-AF647 was generated by conjugating AF647 to the purified AP205 VLPs using the AF647 labeling kit from Thermo Fisher Scientific as described before.33 Qb-AF64733 was used to indicate potential AF647+ cells.
The DNA sequence encoding the BirA (Gene ID: 948469) was synthesized and cloned into the pGEX vector (Addgene). E. coli BL21 (DE3) transformed with pGEX-BirA was grown in LB medium and protein expression was induced with 0.1 mM IPTG at 18°C for 18 h. GST-BirA protein was purified by affinity chromatography using GST Fusion Protein Purification Kit (GenScript,Cat.NO L00206) according to the manufacturer’s instructions. The biotinylated RBD was generated by mixing 100 μM RBD-AviTag, 1 μM GST-BirA, 10mM magnesium chloride, 10mM ATP, and 150 μM D-Biotin (BBI Life Sciences, China). The mixture was incubated at 30°C for 1 h as described before.79 Biotinylated RBD was then mixed with fluorochrome-conjugated streptavidin at room temperature for 60 min to generate fluorescently labeled RBD tetramer.
Immunization
10 μg of AP205-RBD (dose indicates the mass of the RBD part) was injected once or twice 3 weeks apart intraperitoneally when not specified. For comparison of immunization routes, the same doses of AP205-RBD were injected subcutaneously or intramuscularly. For comparison of different antigens, 10 μg of RBD or 50 μg of full-length S protein mixed with alum, or with additional 50 μg of CpG-ODN was injected intraperitoneally. For immunization of AP205 and RBD without conjugation, AP205 instead of AP205-SpyTag was mixed with RBD-SpyCatcher.
Enzyme-linked immunosorbent assay (ELISA)
To determine the amount of antigen-specific immunoglobulins in serum, purified RBD without SpyCatcher fusion region, or purified AP205-SpyTag was used to coat the plate. Serial diluted sera were incubated with the plate, and the horseradish peroxidase (HRP)-conjugated anti-mouse IgA, anti-mouse IgG (Bethyl Laboratories, USA), anti-mouse IgM, anti-mouse IgG1, anti-mouse IgG2b, anti-mouse IgG2a/c, anti-mouse IgG3 (Southern Biotech, USA), or anti-monkey IgG (Abcam, UK) was used for detection. The 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich) was used as the HRP substrate, and the optical density at 450nm was measured by a microplate reader (SpectraMax, Molecular Devices, USA). Antibody titers were determined as the reciprocal of the highest dilution that gave an optical density value that was above ten times of the standard deviation value measured from the serum-free wells.
Flow cytometry
Cells were suspended and stained in FACS buffer (2% newborn calf serum, 2mM EDTA, 0.1% NaN3 in PBS). For intracellular staining, cells were treated with Cytofix/Cytoperm solution (BD Biosciences) following the manufacturer’s instruction. Antibodies and reagents used for flow cytometry include PE-CF594 anti-B220 (RA3-6B2), allophycocyanin-Cy7 anti-CD19 (1D3), PE-Cy7 anti-IgM (II/41), FITC anti-IgM (Jackson Immunoresearch, USA), BV711 anti-IgD (11-26c.2a), PE anti-GL-7 (GL7), FITC anti-GL-7 (GL7), AF700 anti-CD38 (90), PerCP-Cy5.5 anti-CD8 (53-6.7), EF450 anti-IRF4 (3E4), BV650 streptavidin (BioLegend, USA), PE streptavidin (BioLegend, USA), biotin anti-IgG1 (A85-1), biotin anti-IgG2a[b] (IgG2c) (5.7). All data were collected on an LSR II cytometer (Becton Dickinson, USA) and analyzed with FlowJo software (TreeStar, USA).
Enzyme-linked immune absorbent spot (ELISpot) assay of IFNγ + cells
Mouse splenocytes were incubated in anti-IFNγ (clone AN-18, Invitrogen) pre-coated plates at 37°C for 20 h with stimulating peptides or protein. Parallel wells were incubated with vehicles as the negative control or PMA/ionomycin as the positive control (data not shown). Pools of 15-mer peptides derived from the RBD sequence or the intact RBD protein were added at 10 μg/ml for stimulation. Pool 1 contains the peptides corresponding to the amino acids 420-434, 426-440 and 445-459 in the full-length SARS-CoV-2 S protein, and Pool 2 contains the peptides corresponding to the amino acids 511-525, 517-531 and 535-549 of S protein.31 Biotin-labeled IFNγ antibody (clone R4-6A2, Invitrogen) and subsequent HRP-conjugated streptavidin (Jackson ImmunoResearch, USA) were used for detection. Spots were scanned and counted by ImmunoSpot analyzer (Cellular Technology Limited, USA).
ELISpot assay for antibody secreting cells
Splenocytes and bone marrow cells from mice were incubated in RBD protein pre-coated plates at 37°C for 5 h. HRP-conjugated anti-mouse IgG (Bethyl Laboratories), anti-mouse IgG1, anti-mouse IgG2b, or anti-mouse IgG2c (Southern Biotech) were used for detection. Spots were scanned and counted by ImmunoSpot analyzer (Cellular Technology Limited, USA).
Immunohistochemistry for mouse spleen sections
Mouse spleen sections at 7 μm thickness were stained with biotinylated anti-IgD (clone 11-26C, Invitrogen) and FITC anti-GL-7 (clone GL7, BioLegend) and then with streptavidin-conjugated peroxidase and alkaline phosphatase–conjugated anti-FITC as previously described.33 The sections were then developed with 3,3′-diaminobenzidine and Fast Red (both from Sigma-Aldrich).
NHP study design
Four rhesus macaques were injected intramuscularly with 20 μg of AP205-RBD twice 3 weeks apart. The other four rhesus macaques were injected with PBS in parallel. Sera were collected at day 14 and 21 after the first immunization and day 7 after the second immunization. 12 days after the second immunization, SARS-CoV-2 virus (strain 107, provided by the Guangdong Provincial Center for Disease Control and Prevention, Guangdong, China)37 cultured in Vero-E6 cells at 5x106 median tissue culture infective dose (TCID50) per milliliter were given to the animals via a combination of intranasal (0.4ml / nostril) and intratracheal (1.2ml, fiberoptic bronchoscopy) instillation, so that a total of 1x107 TCID50 virus particles were used in the viral challenge for each animal. Body temperature, weight and blood cell counts were monitored at day0, 1, 3, 5, 7 after viral challenge and no significant changes were found for both immunized and control groups of animals. Nasal swab, throat swab and rectal swab samples were taken at day0, 1, 3, 5, 7 after viral challenge and examined for viral load. Tracheal brush samples were taken at day 3 after viral challenge and examined for viral load. Euthanasia was performed on the day 7 after the challenge and tissues from each of the seven lung lobes were taken for viral load and histology examination.
Viral load measurement
Viral load was determined by quantitative real-time RT-PCR (qRT-PCR). Briefly, swab and tracheal brush samples were resuspended in 1 mL of PBS before the total RNA was extracted using the High Pure Viral RNA Kit (Roche, Germany) in accordance with the manufacturer’s instruction. Lung tissue samples were homogenized before the total RNA was extracted using TRIzol reagent (Thermo Fisher, USA). A THUNDERBIRD Probe One-Step qRT-PCR kit (TOYOBO, Japan) was used to detect SARS-CoV-2 RNA, with previously reported primers and probe (5′-GGGGAACTTCTCCTGCTAGAAT-3′, 5′-CAGACATTTTGCTCTCAAGCTG-3′, FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3′).37 Viral copy number was calculated based on the standard provided by National Institute of Metrology, China. The viral loads for swab and tracheal brush samples were normalized as viral RNA copies per milliliter, and the viral load for lung tissues was normalized as viral RNA copies per microgram of total extracted RNA.
Lung histopathology
Tissues were fixed in 4% paraformaldehyde for minimum of seven days, then embedded in paraffin, sectioned at 4 μm for HE staining. Estimation of the proportion of the severely infiltrated areas in tissue sections was performed in a blind manner with a scale from 0.0 to 1.0.
Pseudovirus neutralization assay
The 293T cells were transfected with human immunodeficiency virus backbones expressing firefly luciferase (pNL43R-E-luciferase) and pcDNA3.1 (Invitrogen) expression vectors encoding the SARS-Cov-2 (NCBI Reference Sequence: YP_009724390.1) S protein concurrently to acquire the pseudovirus.22 The supernatants containing the pseudovirus were collected 48 h after transfection. Viral titers were measured as luciferase activity in relative light units (Bright-Glo Luciferase Assay Vector System, Promega Biosciences, USA). In the neutralization test, the pseudovirus was incubated with serial dilutions of mouse serum or macaque serum at 37°C for 1 h before added to Huh7 cells in triplicates (approximately 1.5x104 per well). Luciferase activity was measured 48 h after infection. Half-maximal inhibitory concentrations (IC50) of the serum was determined by curve fitting using GraphPad Prism and used to represent the neutralization titer against the pseudovirus.
Live SARS-CoV-2 neutralization assay
Neutralization assay for live SARS-CoV-2 virus was performed at the Biosafety Level 3 Laboratory at Wuhan Institute of Virology, CAS. SARS-CoV-2 virus (nCoV-2019BetaCoV/Wuhan/WIV04/2019)28 with MOI = 0.01 (about 400 PFU) was incubated with serial dilutions of mouse serum or macaque serum at 37°C for 1 h before being added to 4x104 Vero-E6 cells that have been seeded in a 48-well plate the night before. The infection was maintained for 1 h at 37°C, and the supernatants containing the virus were removed and replaced with fresh medium. The supernatants were then collected 24 h after infection and examined for viral loads. Viral RNA was isolated with MiniBEST Viral RNA/DNA Extraction Kit (Takara, Japan) as described in the instruction, and cDNA was transcribed with PrimeScript RT reagent Kit with gDNA Eraser (Takara). Viral copies were quantified by qPCR from viral cDNA with a pair of primers targeting the S gene of SARS-CoV-2. (5′-CAATGGTTTAACAGGCACAGG-3′, 5′-CTCAAGTGTCTGTGGATCACG-3′). Half-maximal inhibitory concentrations (IC50) of the serum was determined by curve fitting using GraphPad Prism and used to represent the neutralization titer.
Quantification and statistical analysis
Statistics
GraphPad Prism was used to generate the data graphs and perform the statistical tests indicated in the manuscript. A p value < 0.05 was considered as statistically significant.
Acknowledgments
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29050600 to B.H.), the National Natural Science Foundation of China (NSFC 31870882, 81991495, and 82041040 to B.H.), the National Key Research and Development Program of China (2020YFC0861100 to B.H.), a grant from the Chinese Academy of Sciences (KJZD-SW-L05 to H.D.), and the National Key Research and Development Program of China (2018YFA0900803 to Z.H.). This work was supported by additional funding from Rongsen Biotechnology (Beijing) Ltd, China. The graphical abstract was created with BioRender (https://biorender.com).
Author contributions
C.G., Y.P., and L.L. performed the experiments at IBP, CAS. X.P. and Leike Zhang designed and performed the SARS-CoV-2 neutralization experiments at WIV, CAS. M.F. and Linqi Zhang designed and performed the pseudovirus neutralization experiments at Tsinghua University. Y. Zhao and H.H. expressed and purified the RBD and related proteins. K.B. and P.Z. designed and performed the EM experiments on AP205-related VLPs. J.H., T.-Z.S., X.-L.F., and Y. Zheng designed and performed the NHP study at KIZ, CAS. R.L., J.C., and Y. Zhou assisted some experiments at IBP. G.Z. provided the peptide pools. B.H. and H.D. initiated the study. B.H. designed and coordinated the whole study. Z.H. and B.H. wrote the manuscript. H.D. provided suggestions for the manuscript.
Declaration of interests
B.H. and C.G. are listed as inventors on a patent application for a PLA-based COVID-19 vaccine.
Published: October 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100448.
Contributor Information
Zhaolin Hua, Email: zlhua@moon.ibp.ac.cn.
Hongyu Deng, Email: hydeng@ibp.ac.cn.
Baidong Hou, Email: baidong_hou@ibp.ac.cn.
Supplemental information
Data and code availability
-
•
All data for this study are available without restriction from the Lead Contact upon request.
-
•
This study did not generate code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- 1.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 2.Krause P.R., Fleming T.R., Longini I.M., Peto R., Briand S., Heymann D.L., Beral V., Snape M.D., Rees H., Ropero A.M., et al. SARS-CoV-2 Variants and Vaccines. N. Engl. J. Med. 2021;385:179–186. doi: 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 4.Hong S., Zhang Z., Liu H., Tian M., Zhu X., Zhang Z., Wang W., Zhou X., Zhang F., Ge Q., et al. B Cells Are the Dominant Antigen-Presenting Cells that Activate Naive CD4+ T Cells upon Immunization with a Virus-Derived Nanoparticle Antigen. Immunity. 2018;49:695–708.e4. doi: 10.1016/j.immuni.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Hou B., Saudan P., Ott G., Wheeler M.L., Ji M., Kuzmich L., Lee L.M., Coffman R.L., Bachmann M.F., DeFranco A.L. Selective utilization of Toll-like receptor and MyD88 signaling in B cells for enhancement of the antiviral germinal center response. Immunity. 2011;34:375–384. doi: 10.1016/j.immuni.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne E.P. Toll-like receptor 7 controls the anti-retroviral germinal center response. PLoS Pathog. 2011;7:e1002293. doi: 10.1371/journal.ppat.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh K.B., Teijaro J.R., Zuniga E.I., Welch M.J., Fremgen D.M., Blackburn S.D., von Tiehl K.F., Wherry E.J., Flavell R.A., Oldstone M.B. Toll-like receptor 7 is required for effective adaptive immune responses that prevent persistent virus infection. Cell Host Microbe. 2012;11:643–653. doi: 10.1016/j.chom.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clingan J.M., Matloubian M. B Cell-intrinsic TLR7 signaling is required for optimal B cell responses during chronic viral infection. J. Immunol. 2013;191:810–818. doi: 10.4049/jimmunol.1300244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian M., Hua Z., Hong S., Zhang Z., Liu C., Lin L., Chen J., Zhang W., Zhou X., Zhang F., et al. B Cell-Intrinsic MyD88 Signaling Promotes Initial Cell Proliferation and Differentiation To Enhance the Germinal Center Response to a Virus-like Particle. J. Immunol. 2018;200:937–948. doi: 10.4049/jimmunol.1701067. [DOI] [PubMed] [Google Scholar]
- 10.Bekeredjian-Ding I., Jego G. Toll-like receptors--sentries in the B-cell response. Immunology. 2009;128:311–323. doi: 10.1111/j.1365-2567.2009.03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawlings D.J., Schwartz M.A., Jackson S.W., Meyer-Bahlburg A. Integration of B cell responses through Toll-like receptors and antigen receptors. Nat. Rev. Immunol. 2012;12:282–294. doi: 10.1038/nri3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFranco A.L., Rookhuizen D.C., Hou B. Contribution of Toll-like receptor signaling to germinal center antibody responses. Immunol. Rev. 2012;247:64–72. doi: 10.1111/j.1600-065X.2012.01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R.C., Steehouwer M., van Reijmersdal S.V., Jaeger M., et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark M.R., Tanaka A., Powers S.E., Veselits M. Receptors, subcellular compartments and the regulation of peripheral B cell responses: the illuminating state of anergy. Mol. Immunol. 2011;48:1281–1286. doi: 10.1016/j.molimm.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leadbetter E.A., Rifkin I.R., Hohlbaum A.M., Beaudette B.C., Shlomchik M.J., Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 16.Hua Z., Hou B. The role of B cell antigen presentation in the initiation of CD4+ T cell response. Immunol. Rev. 2020;296:24–35. doi: 10.1111/imr.12859. [DOI] [PubMed] [Google Scholar]
- 17.Jegerlehner A., Maurer P., Bessa J., Hinton H.J., Kopf M., Bachmann M.F. TLR9 signaling in B cells determines class switch recombination to IgG2a. J. Immunol. 2007;178:2415–2420. doi: 10.4049/jimmunol.178.4.2415. [DOI] [PubMed] [Google Scholar]
- 18.Gomes A.C., Mohsen M., Bachmann M.F. Harnessing Nanoparticles for Immunomodulation and Vaccines. Vaccines (Basel) 2017;5:6. doi: 10.3390/vaccines5010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakeri B., Fierer J.O., Celik E., Chittock E.C., Schwarz-Linek U., Moy V.T., Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thrane S., Janitzek C.M., Matondo S., Resende M., Gustavsson T., de Jongh W.A., Clemmensen S., Roeffen W., van de Vegte-Bolmer M., van Gemert G.J., et al. Bacterial superglue enables easy development of efficient virus-like particle based vaccines. J. Nanobiotechnology. 2016;14:30. doi: 10.1186/s12951-016-0181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 23.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020;10:14991. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richmond P., Hatchuel L., Dong M., Ma B., Hu B., Smolenov I., Li P., Liang P., Han H.H., Liang J., Clemens R. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: a phase 1, randomised, double-blind, placebo-controlled trial. Lancet. 2021;397:682–694. doi: 10.1016/S0140-6736(21)00241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 26.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S., Li Y., Dai L., Wang J., He P., Li C., Fang X., Wang C., Zhao X., Huang E., et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021;21:1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan X., Zhou P., Fan T., Wu Y., Zhang J., Shi X., Shang W., Fang L., Jiang X., Shi J., et al. Immunoglobulin fragment F(ab’)2 against RBD potently neutralizes SARS-CoV-2 in vitro. Antiviral Res. 2020;182:104868. doi: 10.1016/j.antiviral.2020.104868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acosta P.L., Caballero M.T., Polack F.P. Brief History and Characterization of Enhanced Respiratory Syncytial Virus Disease. Clin. Vaccine Immunol. 2015;23:189–195. doi: 10.1128/CVI.00609-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bournazos S., Ravetch J.V. Fcγ Receptor Function and the Design of Vaccination Strategies. Immunity. 2017;47:224–233. doi: 10.1016/j.immuni.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Victora G.D., Nussenzweig M.C. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 33.Liao W., Hua Z., Liu C., Lin L., Chen R., Hou B. Characterization of T-Dependent and T-Independent B Cell Responses to a Virus-like Particle. J. Immunol. 2017;198:3846–3856. doi: 10.4049/jimmunol.1601852. [DOI] [PubMed] [Google Scholar]
- 34.Kurosaki T., Kometani K., Ise W. Memory B cells. Nat. Rev. Immunol. 2015;15:149–159. doi: 10.1038/nri3802. [DOI] [PubMed] [Google Scholar]
- 35.Weisel F., Shlomchik M. Memory B cells of mice and humans. Annu. Rev. Immunol. 2017;35:255–284. doi: 10.1146/annurev-immunol-041015-055531. [DOI] [PubMed] [Google Scholar]
- 36.Oracki S.A., Walker J.A., Hibbs M.L., Corcoran L.M., Tarlinton D.M. Plasma cell development and survival. Immunol. Rev. 2010;237:140–159. doi: 10.1111/j.1600-065X.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- 37.Song T.Z., Zheng H.Y., Han J.B., Jin L., Yang X., Liu F.L., Luo R.H., Tian R.R., Cai H.R., Feng X.L., et al. Delayed severe cytokine storm and immune cell infiltration in SARS-CoV-2-infected aged Chinese rhesus macaques. Zool. Res. 2020;41:503–516. doi: 10.24272/j.issn.2095-8137.2020.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., et al. A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733.e11. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 43.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing Activity of BNT162b2-Elicited Serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quadeer A.A., Ahmed S.F., McKay M.R. Epitopes targeted by T cells in convalescent COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.08.26.267724. [DOI] [Google Scholar]
- 50.Kim Y.M., Pan J.Y., Korbel G.A., Peperzak V., Boes M., Ploegh H.L. Monovalent ligation of the B cell receptor induces receptor activation but fails to promote antigen presentation. Proc. Natl. Acad. Sci. USA. 2006;103:3327–3332. doi: 10.1073/pnas.0511315103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Batista F.D., Neuberger M.S. B cells extract and present immobilized antigen: implications for affinity discrimination. EMBO J. 2000;19:513–520. doi: 10.1093/emboj/19.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato Y., Abbott R.K., Freeman B.L., Haupt S., Groschel B., Silva M., Menis S., Irvine D.J., Schief W.R., Crotty S. Multifaceted Effects of Antigen Valency on B Cell Response Composition and Differentiation In Vivo. Immunity. 2020;53:548–563.e8. doi: 10.1016/j.immuni.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen B., Tolia N.H. Protein-based antigen presentation platforms for nanoparticle vaccines. NPJ Vaccines. 2021;6:70. doi: 10.1038/s41541-021-00330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marcandalli J., Fiala B., Ols S., Perotti M., de van der Schueren W., Snijder J., Hodge E., Benhaim M., Ravichandran R., Carter L., et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell. 2019;176:1420–1431.e17. doi: 10.1016/j.cell.2019.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abbott R.K., Lee J.H., Menis S., Skog P., Rossi M., Ota T., Kulp D.W., Bhullar D., Kalyuzhniy O., Havenar-Daughton C., et al. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity. 2018;48:133–146.e6. doi: 10.1016/j.immuni.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yassine H.M., Boyington J.C., McTamney P.M., Wei C.J., Kanekiyo M., Kong W.P., Gallagher J.R., Wang L., Zhang Y., Joyce M.G., et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 58.Kanekiyo M., Wei C.J., Yassine H.M., McTamney P.M., Boyington J.C., Whittle J.R., Rao S.S., Kong W.P., Wang L., Nabel G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jardine J., Julien J.P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P.S., MacPherson S., Jones M., et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ward B.J., Gobeil P., Séguin A., Atkins J., Boulay I., Charbonneau P.Y., Couture M., D’Aoust M.A., Dhaliwall J., Finkle C., et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021;27:1071–1078. doi: 10.1038/s41591-021-01370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan T.K., Rijal P., Rahikainen R., Keeble A.H., Schimanski L., Hussain S., Harvey R., Hayes J.W.P., Edwards J.C., McLean R.K., et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 2021;12:542. doi: 10.1038/s41467-020-20654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X., Chang X., Rothen D., Derveni M., Krenger P., Roongta S., Wright E., Vogel M., Tars K., Mohsen M.O., Bachmann M.F. AP205 VLPs Based on Dimerized Capsid Proteins Accommodate RBM Domain of SARS-CoV-2 and Serve as an Attractive Vaccine Candidate. Vaccines (Basel) 2021;9:403. doi: 10.3390/vaccines9040403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 64.Theisen D., Murphy K. The role of cDC1s in vivo: CD8 T cell priming through cross-presentation. F1000Res. 2017;6:98. doi: 10.12688/f1000research.9997.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chapin-Bardales J., Gee J., Myers T. Reactogenicity Following Receipt of mRNA-Based COVID-19 Vaccines. JAMA. 2021;325:2201–2202. doi: 10.1001/jama.2021.5374. [DOI] [PubMed] [Google Scholar]
- 66.Franchini M., Liumbruno G.M., Pezzo M. COVID-19 vaccine-associated immune thrombosis and thrombocytopenia (VITT): Diagnostic and therapeutic recommendations for a new syndrome. Eur. J. Haematol. 2021;107:173–180. doi: 10.1111/ejh.13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montgomery J., Ryan M., Engler R., Hoffman D., McClenathan B., Collins L., Loran D., Hrncir D., Herring K., Platzer M., et al. Myocarditis Following Immunization With mRNA COVID-19 Vaccines in Members of the US Military. JAMA Cardiol. 2021;6:1202–1206. doi: 10.1001/jamacardio.2021.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shay D.K., Shimabukuro T.T., DeStefano F. Myocarditis Occurring After Immunization With mRNA-Based COVID-19 Vaccines. JAMA Cardiol. 2021;6:1115–1117. doi: 10.1001/jamacardio.2021.2821. [DOI] [PubMed] [Google Scholar]
- 69.Tatsis N., Ertl H.C. Adenoviruses as vaccine vectors. Mol. Ther. 2004;10:616–629. doi: 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., Tukhvatulin A.I., Zubkova O.V., Dzharullaeva A.S., Kovyrshina A.V., Lubenets N.L., Grousova D.M., Erokhova A.S., et al. Gam-COVID-Vac Vaccine Trial Group Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 73.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang L., Hijano D.R., Gaur A.H., Geiger T.L., Neufeld E.J., Hoffman J.M., Hayden R.T. Asymptomatic and Symptomatic SARS-CoV-2 Infections After BNT162b2 Vaccination in a Routinely Screened Workforce. JAMA. 2021;325:2500–2502. doi: 10.1001/jama.2021.6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Voysey M., Costa Clemens S.A., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Oxford COVID Vaccine Trial Group Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 77.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harris R.J., Hall J.A., Zaidi A., Andrews N.J., Dunbar J.K., Dabrera G. Effect of Vaccination on Household Transmission of SARS-CoV-2 in England. N. Engl. J. Med. 2021;385:759–760. doi: 10.1056/NEJMc2107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fairhead M., Howarth M. Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 2015;1266:171–184. doi: 10.1007/978-1-4939-2272-7_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data for this study are available without restriction from the Lead Contact upon request.
-
•
This study did not generate code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.