Abstract
Background & Aims
Gut microbiota have been reported to be sensitive to circadian rhythms and host lipometabolism, respectively. Although melatonin-mediated beneficial efforts on many physiological sites have been revealed, the regulatory actions of oral melatonin on the communication between gut microbiota and host are still not clear. Angiopoietin-like 4 (ANGPTL4) has been shown to be strongly responsible for the regulation of systemic lipid metabolism. Herein, we identified that oral melatonin improved lipid dysmetabolism in ileum and epididymal white adipose tissue (eWAT) via gut microbiota and ileac ANGPTL4.
Methods
Analyses of jet-lag (JL) mice, JL mice with oral melatonin administration (JL+MT), and the control for mRNA and protein expression regarding lipid uptake and accumulation in ileum and eWAT were made. Gut microbiome sequencing and experimental validation of target strains were included. Functional analysis of key factors/pathways in the various rodent models, including the depletion of gut microbiota, mono-colonization of Escherichia coli, and other genetic intervention was made. Analyses of transcriptional regulation and effects of melatonin on E coli-derived lipopolysaccharide (LPS) in vitro were made.
Results
JL mice have a higher level of ileal lipid uptake, fat accumulation in eWAT, and lower level of circulating ANGPTL4 in comparison with the control mice. JL mice also showed a significantly higher abundance of E coli and LPS than the control mice. Conversely, oral melatonin supplementation remarkably reversed these phenotypes. The test of depletion of gut microbiota further demonstrated that oral melatonin-mediated improvements on lipometabolism in JL mice were dependent on the presence of gut microbiota. By mono-colonization of E coli, LPS has been determined to trigger these changes similar to JL. Furthermore, we found that LPS served as a pivotal link that contributed to activating toll-like receptor 4 (TLR4)/signal transducer and activator of transcription 3 (STAT3_/REV-ERBα) signaling to up-regulate nuclear factor interleukin-3-regulated protein (NFIL3) expression, resulting in increased lipid uptake in ileum. In MODE-K cells, the activation of NFIL3 has further been shown to inhibit ANGPTL4 transcription, which is closely associated with lipid uptake and transport in peripheral tissues. Finally, we confirmed that melatonin inhibited LPS via repressing the expression of LpxC in E coli.
Conclusions
Overall, oral melatonin decreased the quantity of E coli-generated LPS, which alleviated NFIL3-induced transcriptional inhibition of ANGPTL4 through TLR4/IL-22/STAT3 signaling in ileum, thereby resulting in the amelioration of ileal lipid intake and lower fat accumulation in eWAT. These results address a novel regulation of oral melatonin originating from gut microbiota to host distal tissues, suggesting that microbe-generated metabolites are potential therapies for melatonin-mediated improvement of circadian rhythm disruption and related metabolic syndrome.
Keywords: Melatonin, Gut Microbiota, Circadian Rhythm Disruption, Lipid Metabolism, ANGPTL
Abbreviations used in this paper: ADFI, average daily food intake; ANGPTL4, angiopoietin-like 4; CHOL, cholesterol; eWAT, epididymal white adipose tissue; HDL-C, high-density lipoprotein-cholesterol; IL-22, interleukin 22; JL, jet-lag; LDL, low-density lipoprotein; LPS, lipopolysaccharide; MT, melatonin; NFIL3, nuclear factor interleukin-3-regulated protein; qPCR, quantitative polymerase chain reaction; SEM, standard error of the mean; STAT3, signal transducer and activator of transcription 3; TG, triglyceride; TLR, toll-like receptor; VLDL-C, very low density lipoprotein-cholesterol; ZT, Zeitgeber time
Graphical abstract
Summary.
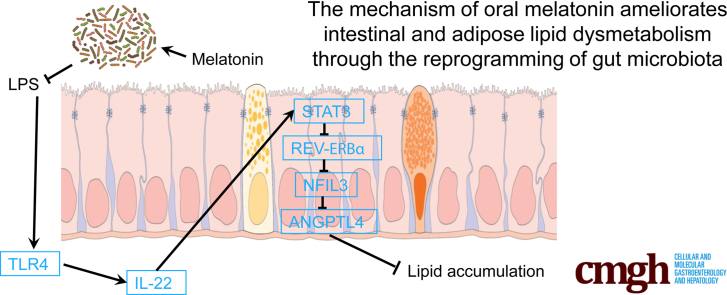
We show that oral melatonin ameliorates lipid dysmetabolism in ileum and epididymal white adipose tissue via decreasing Escherichia coli-generated lipopolysaccharide, suggesting microbial metabolites may be potential therapies target for alleviating and improving circadian rhythm disruption and related lipid dysmetabolism. The mechanism of oral melatonin ameliorates intestinal and adipose lipid dysmetabolism through reducing Escherichia coli-derived lipopolysaccharide.
Many reports highlight the important role of lipid dysmetabolism as a threat to public health; lipid dysmetabolism–induced obesity is a physiological disorder associated with a high incidence of metabolic diseases in multiple organs/tissues.1, 2, 3 Evidence shows that excessive fat uptake is in part responsible for obesity.2 Numerous organs/tissues are involved in lipometabolism; in particular, the gut and adipose tissue are representative of physiological locations that strongly participate in lipid transport and storage.4,5 Previous studies have determined that lipid homeostasis profoundly contributes to protecting individuals from lipid dysmetabolism and obesity.4,5 Notably, in the gut of obese individuals not only do alterations exist in intestinal intracellular environment (ie, the increased permeability of intestinal barrier caused by lower expression of tight junction proteins), but also the pattern of gut microbiota is changed.6
In recent years, the gut microbiota have been revealed to have a close association with host energy metabolism and the development of obesity, which is considered as a significantly potential manipulator of these processes.7 Although gut microbiota-host interactions occur in host intestines, numerous signaling pathways and microbial metabolites involved in their crosstalk also affect distal organs/tissues such as adipose tissue.8,9 Previous studies indicate the pattern of the microbial community in gut is changed in response to dietary intake, which influences host metabolism, including nutrient uptake and prevention of pathogen colonization.7 On the basis of the intensive development of bacterial genome-sequencing during the last decade, the correlation between host physiological activities and alterative pattern of gut microbiota has been gradually revealed.10, 11, 12 One confounding issue is how to explain the partly contrary observations in similar treatment from various studies. For example, as a common observation in human and rodent models, the most abundant microbial phyla in gut are Bacteroidetes and Firmicutes.13 Both these major phyla, Bacteroidetes and Firmicutes, are significantly affected by intermittent fasting.14, 15, 16, 17 But contradictory findings exist regarding how these 2 phyla sometimes produce opposite effects. Therefore, the analysis of gut microbiome has gradually focused on the influence of minor constituents or a rare strain.
In addition to the gut microbiome, circadian rhythm disruptions have been identified as another key factor for triggering obesity and the related metabolic syndrome.18,19 In mammals the circadian clock contributes to synchronizing many metabolic activities with day-night cycle, which ensures that organs/tissues optimally balance anabolic vs catabolic functions.20 In contrast to the master biological clock located in the suprachiasmatic nucleus of the hypothalamus, the circadian clock in many peripheral organs/tissues, including gut and adipose tissue, serves a network of transcription factors that drive rhythmic oscillators of gene expression.20, 21, 22 Considering the strong association between the symbionts and host, the hypothesis has been advanced that significant metabolic changes may be triggered by the overlap or interactive effects. Thus, an interaction of the gut microbiota with peripheral circadian clocks may be important in modulating host obesity.
Angiopoietin-like 4 (ANGPTL4) is highly expressed in liver and adipose tissue. It is also detected in intestines.23,24 Originally, ANGPTL4 was seen as a fasting-induced adipocytokine, whereas recent studies show it to be strongly responsible for the regulation of systemic lipid metabolism via mediating lipoprotein lipase and high-density lipoprotein activity.25,26 In the emerging field of the prevention of obesity, ANGPTL4 has been recognized as a novel target for its involvement under a cold exposure and exercise apart from during fasting condition.24,27 Janssen et al28 first reported the participation of gut microbiota in energy metabolism (including lipometabolism and glucose tolerance) in obese mice with Angptl4-knockout. This suggests that ANGPTL4 may be a key link for delivering the microbial singling to remote organs/tissues. However, this study did not definitively identify which special strains of microbes contributed to shaping the phenotype of mice, and the regulatory pathways still remain unidentified.
Melatonin (MT) is a well-known pineal secretory product that regulates circadian rhythms. The pineal gland was initially identified as the major site of MT secretion.29,30 However, numerous recent studies suggest multiple peripheral organs/tissues likely produce MT, which contributes to maintaining circadian rhythms and protecting against oxidative stress in animals.31, 32, 33, 34 In recent years, recognition of MT-mediated rhythmic control of obesity and subsequent developmental processes stimulated further interest in adipocytes as targets of MT as well as the gut, which controls lipid uptake.35, 36, 37, 38 Unlike MT administration through injection, when taken orally, its level in the gut likely is markedly increased, implying that the possibility of MT-mediated improvements in physiology may be secondary to MT actions on microbiota. Overall, these findings hint that a regulatory pathway of MT alleviates the circadian rhythm disruption-triggered lipid dysmetabolism via shaping the pattern of gut microbiota. In this investigation, we used a mouse model with “jet-lag” and studied the effects of MT on the inhibition of microbial LPS production by altering the intestinal microbiome, which further attenuated the suppression of the ANGPTL4 transcription in ileum and lowered the fat accumulation in epididymal white adipose tissue (eWAT).
Results
Oral Melatonin Decreases Circadian Rhythm Disruption-Mediated Lipid Intake in the Ileum and Lipid Accumulation in Adipose Tissue
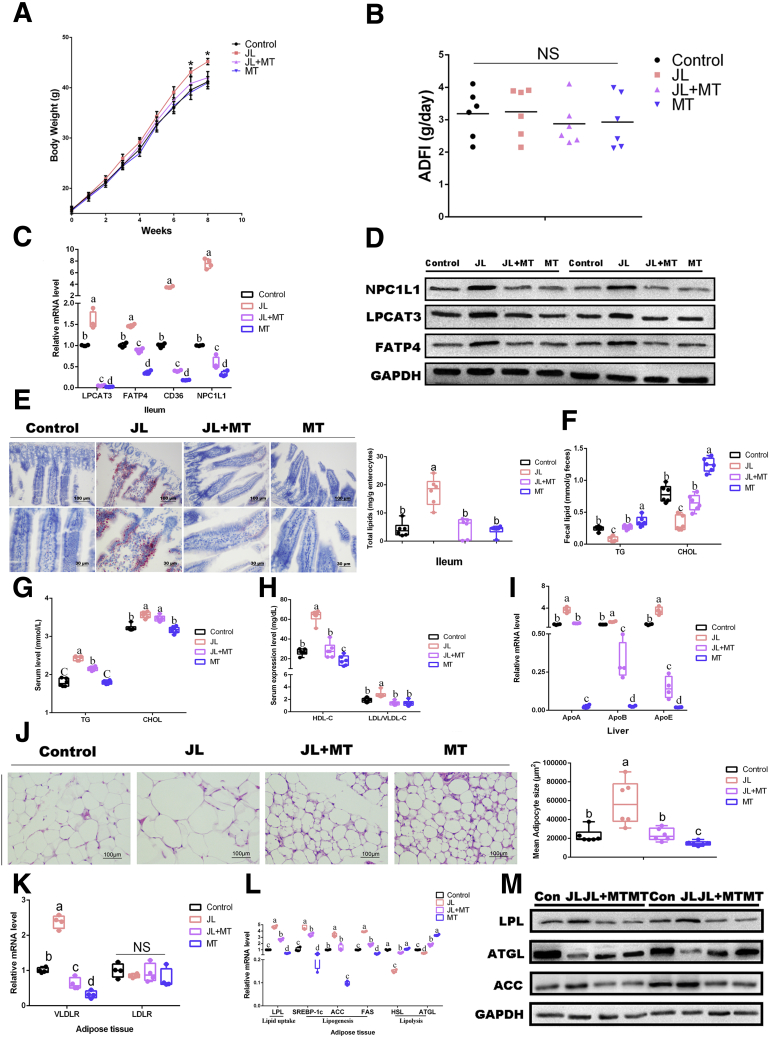
As most visualized influences of oral MT on the jet-lag (JL) mice, we detected the changes of body weight throughout the experiment (Figure 1A). Compared with the other groups, the weight of JL mice exhibited a significant increase (P < .05), whereas average daily feed intake (ADFI) has no statistical difference between the control and JL groups (P > .05) (Figure 1A and B). Meanwhile, JL mice with oral MT treatment (JL+MT group) significantly decreased body weight in comparison with JL treatment (P < .05), without influencing ADFI (P > .05) (Figure 1A and B).
Figure 1.
Oral MT decreases circadian rhythm disruption-mediated lipid intake in the ileum and lipid accumulation in adipose tissue. (A) Body weight (g, n = 6). (B) ADFI (g/day, n = 6). (C) Relative mRNA levels (including mRNAs LPCAT3, FATP4, CD36, and NPC1L1, n = 4). (D) Relative protein levels (including LPCAT3, FATP4, and NPC1L1, n = 5). (E) Representative Oil Red O staining (red dot) of ileal slides from mice with various treatments including the control, JL, JL+MT, and MT; bar: μm (original magnification, ×200 and ×400, n = 6). Right panel: total lipid concentration (mg/g enterocytes) in ileal enterocytes from mice in each group. (F) Fecal levels of TG and CHOL in mice with various treatments including the control, JL, JL+MT, and MT (n = 6). (G) Serum levels of TG and CHOL in mice with various treatments including the control, JL, JL+MT, and MT (n = 6). (H) Serum levels of HDL-C and LDL/VLDL-C in mice with various treatments including the control, JL, JL+MT, and MT (n = 6). (I) Relative mRNA levels (including mRNAs APOA, APOB, APOE, n = 4). (J) Representative H&E staining of adipose tissue from mice with various treatments including the control JL, JL+MT, and MT; bar: μm (original magnification, ×400, n = 6). Right panel: analysis of adipocyte size from mice with various treatments including the control JL, JL+MT, and MT (μm2, n = 6). (K and L) Relative mRNA levels of lipid metabolic indicators in adipose tissue (including VLDLR, LDLR, LPL, SREBP-1c, ACC, FAS, HSL, and ATGL, n = 4). (M) Relative protein levels (including LPL, ACC, and ATGL, n = 5). Values are means ± SEM. Same markup represented no significant difference between 2 groups (P > .05); different markup represented significant difference (P < .05) compared with other groups. ∗P < .05 compared with the other groups.
For the ileum, we further compared the lipid intake/accumulation by means of frozen sections stained with Oil Red O and analyzed the transcriptional level of related genes and proteins expression (Figure 1C–E). The quantitative polymerase chain reaction (qPCR) assays documented that mice under JL had dramatically elevated expressions of mRNA associated with ileal lipid intake, including LPCAT3, FATP4, NPC1L1, and CD36, in comparison with the control (P < .05), whereas oral MT prevented these changes on JL treatment (JL+MT group) (Figure 1C). Notably, oral MT by itself (MT group) has also significantly reduced the transcriptions of these genes compared with the control (P < .05), suggesting that oral MT-mediated inhibition of ileac lipid uptake may be not completely dependent on the improvements of circadian rhythm disruption (Figure 1C). In addition, the expression of relative proteins, including LPCAT3, FATP4, and NPC1L1, showed a similar tendency (Figure 1D). The results of Oil Red O staining showed that lipid levels were elevated in JL mice compared with the control (P < .05), whereas oral MT supplement prevented this alteration in comparison with JL mice (Figure 1E). Overall, these findings suggest that oral MT may contribute to decreasing circadian rhythm disruption-mediated lipid intake in mouse ileum.
To better detect the lipid uptake, the levels of lipid (including triglyceride [TG] and cholesterol [CHOL]) in feces and serum in each group have been measured.39 In feces, the results showed that both TG and CHOL were dramatically decreased under JL treatment compared with the control (P < .05), whereas oral MT supplementation was able to increase the level of fecal lipid in JL mice (JL+MT group). Furthermore, compared with the control, oral MT by itself (MT group) has been detected in a higher level of fecal TG and CHOL (P < .05) (Figure 1F). Meanwhile, JL treatment significantly increased mouse serum concentrations of these molecules compared with the control (P < .05) (Figure 1G). In turn, oral MT did not greatly reduce the concentrations of TG or CHOL in JL mice. Oral MT by itself has similarly shown no significant inhibition on serum lipid in comparison with the control (P > .05) (Figure 1G). Furthermore, we tested the levels of lipoprotein cholesterol (including high-density lipoprotein-cholesterol [HDL-C] and low-density lipoprotein/very low density lipoprotein-cholesterol [LDL/VLDL-C]), and the results showed that the expressions of HDL-C and LDL/VLDL-C were dramatically increased under JL treatment compared with the control (P < .05), whereas MT administration obviously reversed the high level of serum HDL-C and LDL/VLDL-C in JL mice (JL+MT group) (P < .05) (Figure 1H). In addition, compared with the control, the expression of serum HDL-C in MT group was significantly inhibited (P < .05) (Figure 1H). To verify the changes occurred in apolipoproteins (HDL, LDL, and VDLD), we performed qPCR assays to detect the transcriptional levels of hepatic ApoA, ApoB, and ApoE; the results confirmed that their transcriptional levels in liver from JL mice were increased as compared with the control, whereas oral MT markedly inhibited these phenotypes in JL mice (P < .05) (Figure 1I). Oral MT by itself has significantly reduced the hepatic expressions of these genes in naive mice (Figure 1I).
As an important site of lipid accumulation and metabolism, eWAT has finally been tested. Paraffin sections stained with H&E have shown that in comparison with the control mice, JL mice have shown an obviously higher size of adipocytes (P < .05) (Figure 1J), and the qPCR assays similarly showed that JL mice had dramatically elevated expressions of mRNA associated with lipid uptake and lipogenesis (including LPL, SREBP-1c, ACC, and FAS) (P < .05); this administration reduced the transcriptional levels of HSL and ATGL, markers for lipolysis (P < .05) (Figure 1L). The results regarding the level of protein expression, including LPL, ACC, and ATGL, were consistent with their transcriptional levels (Figure 1M). On the contrary, oral MT administration significantly inhibited these changes in JL treatment (JL+MT) (P < .05); oral MT by itself has also exhibited some tendency on naive mice (Figure 1L and M). As shown in Figure 1K, JL mice also presented a higher transcription of VLDL receptor on eWAT, whereas oral MT significantly reduced its high expression in eWAT of JL mice, and simply MT administration has also markedly decreased its expression as compared with the control mice (P < .05) (Figure 1K). Taken together, these findings suggest that oral MT may contribute to improving circadian rhythm disruption-mediated lipid intake in ileum and lipid accumulation in adipose tissue.
Oral Melatonin Reprograms Circadian Rhythm Disruption-Mediated Pattern of Gut Microbiota
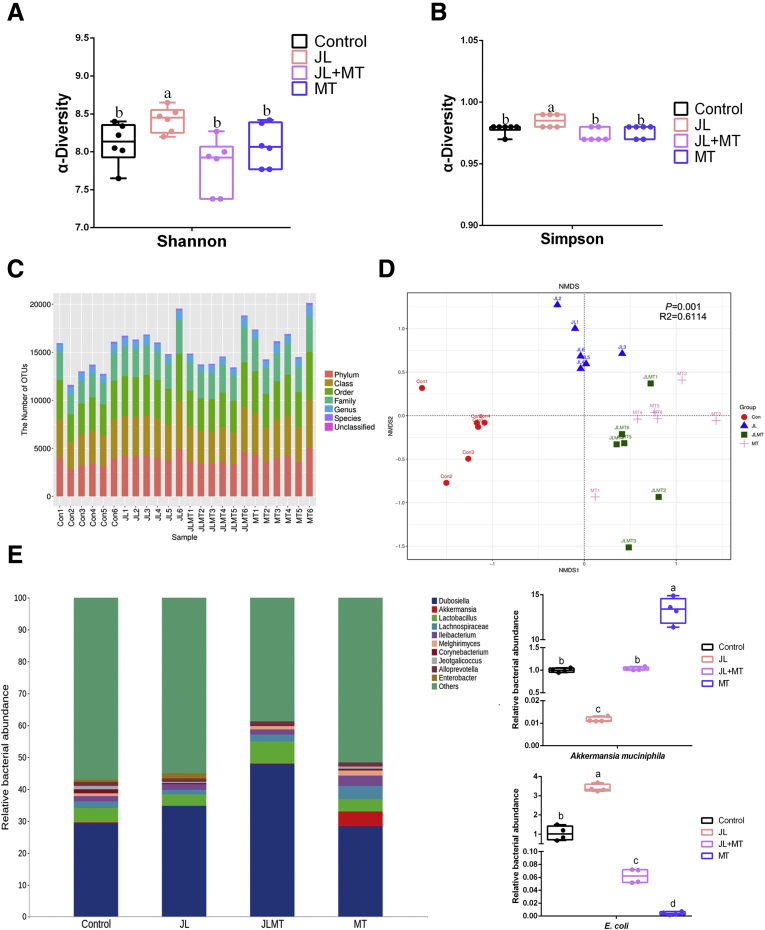
Because previous studies indicated that the pattern of the microbial community in gut is susceptible to drug intake and the possibility of shaped gut microbiota on triggering obesity,7,20,40 we hypothesized that gut microbiota are highly associated with the improvements of oral MT on circadian rhythm disruption-mediated weight gain in mice. Thus, we further investigated the composition of fecal microbiota via 16S rRNA sequencing. The results showed that the Shannon and Simpson index (α-diversity), the parameters of gut microbiota representing bacterial diversity, was markedly elevated in JL mice in comparison with the control, whereas MT supplement prevented this change (P < .05) (Figure 2A and B). Furthermore, the bacterial richness has also shown significant alterations among various groups (Figure 2C). An analysis of nonmetric multidimensional scaling based on unweighted UniFrac documented that the gut microbial community in JL mice was substantially isolated from the other groups, whereas the composition of the gut microbiota in mice with MT treatment was significantly shaped both in JL (JL+MT group) and the naive mice (MT group) (Figure 2D). These findings suggest that oral MT supplement significantly reprograms the pattern of gut microbiota (Figure 2D). As shown in Figure 2E, the relative abundance of Akkermansia muciniphila was lower in JL mice, whereas its relative abundance was higher in the JL+MT and MT groups. Moreover, we identified that the amount of Enterobacteriales was up-regulated in JL mice compared with the control, whereas its relative abundance was lower by MT treatment in the mouse ileum (Figure 2E). To verify the results of bacterial sequencing analysis, we selected Akk muciniphila and E coli (as its high abundance and major contributor of LPS in the intestines) from different groups, which were also closely associated with host obesity, to identify in vitro. The results were similar to sequencing analysis (Figure 2E). Previous studies have demonstrated that Akk muciniphila alleviates multiple lipid dysmetabolism-induced organ/systemic diseases, whereas its lower abundance positively correlates with host hyperlipidemia, type 2 diabetes, and fatty liver.41, 42, 43 Interestingly, this phenotype seems to be conservative among various species, including human, rodent, and even Burmese python.41, 42, 43, 44, 45 Thus, Akk muciniphila has gradually been considered as a key bioindicator for reflecting host lipometabolism and obesity and selected as target strain for representing the alternative pattern of gut microbiota.7 Thus, these species may play an important role in shaping host lipometabolism under circadian rhythm disruption. Overall, these observations suggest host circadian rhythm disruption to be strongly associated with the reprogramming of gut microbiota, whereas oral MT contributes to reversing these alterations.
Figure 2.
Oral MT reprograms circadian rhythm disruption-mediated pattern of gut microbiota. (A and B) Shannon and Simpson index in ɑ-diversity analysis (n = 6). (C) The number of operational taxonomic units from each group at various classified levels (n = 6). (D) Analysis of nonmetric multidimensional scaling (NMDS) based on unweighted UniFrac (n = 6). (E) Principal coordinates analysis plot analysis from each sample (n = 6). (E) Relative bacterial abundance of representative gut microbiota from each group at the level of genus (n = 6). Right panel: relative bacterial abundance of Akk muciniphila and E coli in feces from each group (n = 4). Values are means ± SEM. Same markup represented no significant difference between 2 groups (P > .05); different markup represented significant difference (P < .05) compared with the other groups.
Oral Melatonin Increases Ileal ANGPTL4 Expression and Blocks TLR4/IL-22/STAT3 Signaling in JL Mouse
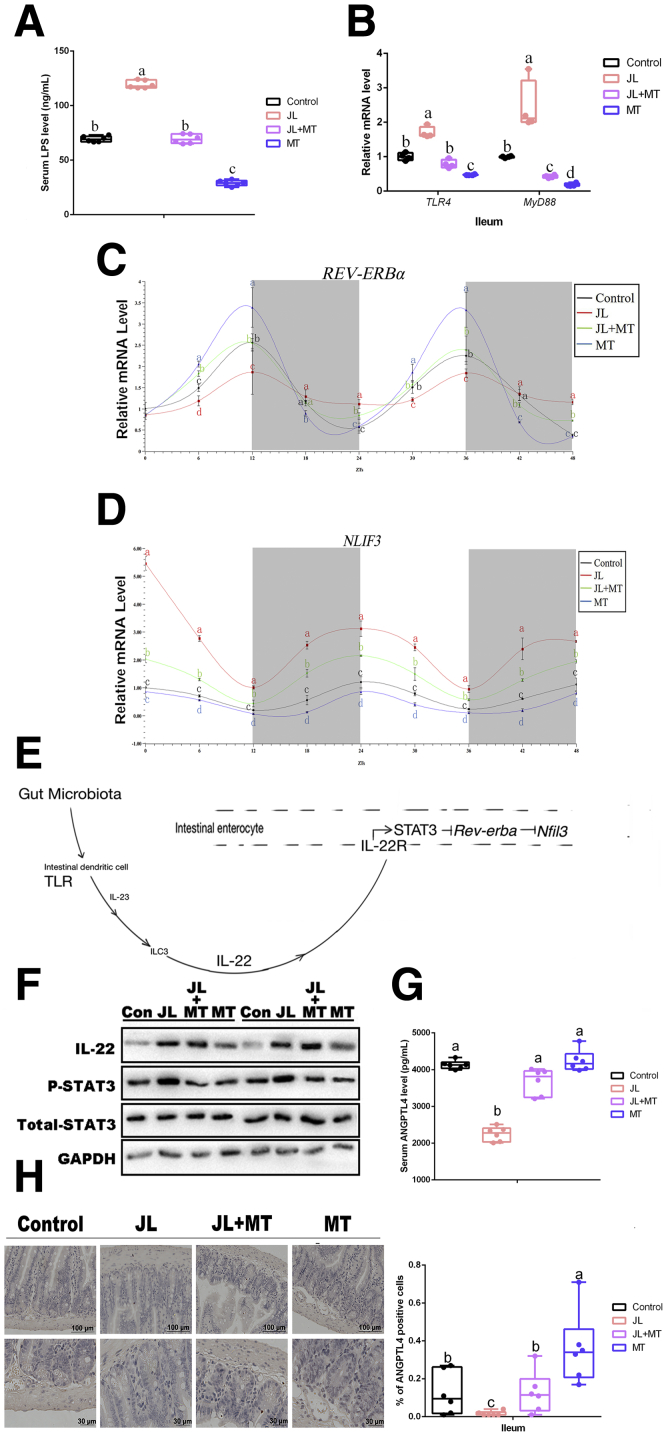
JL treatment contributed to increasing the abundance of E coli. Thus, in serum we assessed the level of lipopolysaccharide (LPS), which is regarded as an important inductor secreted by Gram-negative bacteria to trigger host metabolic disorders and inflammatory responses. The results showed that under the JL condition its concentration was substantially increased in comparison with the control (P < .05), whereas oral MT obviously reduced LPS concentration in the circulation of JL mice (JL+MT group), and there was no statistical difference as compared with the control (P > .05) (Figure 3A). Notably, compared with the control, simply MT administration has also markedly decreased the level of circulating LPS (P < .05) (Figure 3A). Among many toll-like receptors (TLRs), TLR4 is a major receptor for binding to microbial LPS and further induces MyD88 activation to initiate inflammatory activity.46,47 As shown in Figure 3B, the mRNA expressions of TLR4 and its downstream MyD88 were significantly up-regulated in JL mice (P < .05), whereas oral MT markedly inhibited their high expression caused by circadian rhythm disruption in mouse ileum (P < .05). Moreover, MT supplementation by itself also significantly inhibited their transcription in ileum (P < .05) (Figure 3B).
Figure 3.
Oral MT increases ileal ANGPTL4 expression and blocks TLR4/IL-22/STAT3 signaling in JL mouse. (A) Serum levels of LPS in mice (ng/mL, n = 6). (B) Relative mRNA levels of ileal TLR4 and MyD88 (n = 4). (C) REV-ERBα expression (n = 4). (D) NFIL3 expression (n = 4). (E) Gut microbiota-involved regulatory circuit (references 20 and 46). (F) Relative protein levels including total STAT3, phosphorylated STAT3, and IL-22 (n = 5). (G) Serum levels of ANGPTL4 in mice (pg/mL, n = 6). (H) Representative immunohistochemical staining of ileal slides in mice with various treatments including control, JL, JL+MT, and MT; bar: μm (original magnification, ×200 and ×400, n = 6); right panel: percentage of positive cells (n = 6). Values are means ± SEM. Same markup represented no significant difference between 2 groups (P > .05); different markup represented significant difference (P < .05) compared with the other groups.
An important study by Wang et al20 has determined that nuclear factor interleukin-3-regulated protein (NFIL3) is negatively regulated by upstream circadian clock (REV-ERBα) signaling and further shapes intestinal lipometabolism (high-level expression of NFIL3 promoted lipid intake), and the activation of this regulatory pathway is primarily responsible for the existence of gut microbiota. To verify the rhythmic expression of REV-ERBα and NFIL3, we tested their mRNA expression during 48 hours with cosinor analysis and found that compared with the control, the circadian amplitude of REV-ERBα mRNA was significantly inhibited in ileum of JL mice (P < .05), whereas oral MT treatment reversed this suppression in JL mice (JL+MT group, P < .05) (Figure 3C). MT group has shown to have obviously increased circadian amplitude of REV-ERBα mRNA in comparison with the control, suggesting that oral MT contributed to improving circadian rhythm (P < .05) (Figure 3C). Notably, the 48-hour pattern of NFIL3 mRNA was driven in the opposite direction so that the circadian amplitude of NFIL3 mRNA was significantly elevated in ileum of JL mice as compared with the control (P < .05), whereas oral MT treatment reversed this suppression in JL mice (JL+MT group, P < .05) (Figure 3D). The MT group has shown its lowest expression in comparison with other treatment (Figure 3D).
Another investigation has revealed that segmented filamentous bacteria modulate host lipometabolism via intestinal TLRs/interleukin 22 [IL-22]/signal transducer and activator of transcription 3 [STAT3] signaling.48 Thus, we questioned whether LPS/TLR4/IL-22/STAT3 signaling pathway is involved in oral MT-mediated inhibition of lipid uptake in ileum and accumulation in eWAT originating from gut microbiota via the inhibition of NFIL3 expression in mice with circadian rhythm disruption (Figure 3E). Furthermore, we observed the expression of TLR4/IL-22/STAT3 signaling pathway in each group. We further estimated the protein expression of IL-22 and the phosphorylation level of STAT3 by Western blot (Figure 3F). As expected, both of these parameters under JL condition were increased compared with the control (P < .05), whereas oral MT supplementation (JL+MT) attenuated circadian rhythm disruption-activated STAT3 signaling (Figure 3F).
As a vital regulator of systemic lipid metabolism and lipid uptake in distal organ, serum ANGPTL4 in each group has been detected. The results showed that the level of ANGTPL4 was dramatically reduced in the serum from JL mice in comparison with the control (P < .05), whereas oral MT supplementation significantly recovered this change in JL mice (Figure 3G). We further found that these phenotypes in each group existed in the ileum of mice by immunohistochemistry (Figure 3H), suggesting that ileum may be an important site for oral MT-mediated improvement on circadian rhythm disruption-induced inhibition of ANGPTL4 expression. Overall, these results suggest that circadian rhythm disruption-induced high abundance of E coli may increase the level of LPS, which may activate the TLR4/STAT3/REV-ERBα/NFIL3 signaling to inhibit ANGPTL4 expression, whereas oral MT may contribute to blocking this regulatory circuit.
Antibiotics-Mediated Depletion of Gut Microbiota Blocks Oral Melatonin-Induced Improvements on Circadian Rhythm Disruption-Triggered Lipid Intake in Ileum and Lipid Accumulation in eWAT
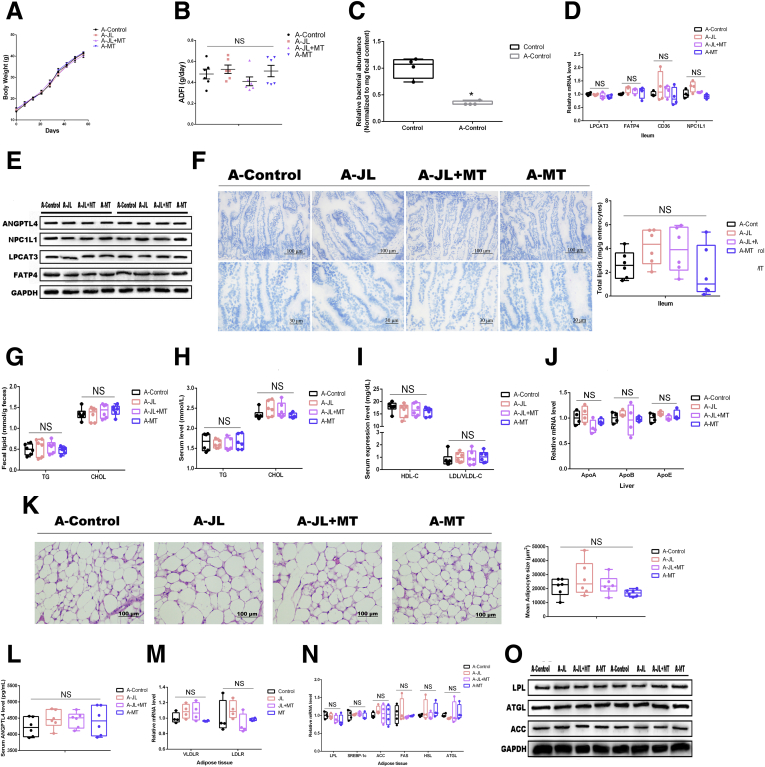
To investigate whether oral MT-induced improvements on circadian rhythm disruption-triggered lipid intake in ileum and lipid accumulation in eWAT were gut microbiota-dependent, we sought to observe these phenotypes in the same administrations under the depletion of gut microbiota by antibiotic mixtures and found that compared with the control (with antibiotics treatment), mice from A-JL, A-JL+MT, and A-MT groups did not exhibit statistical difference on weight gain or ADFI (P > .05) (Figure 4A–C). The qPCR assays and Western blot analysis reflected the same tendency of mRNA associated with ileal lipid intake including LPCAT3, FATP4, and NPC1L1 among various groups (P > .05) (Figure 4D and E). Also, the results of Oil Red O staining on ileum confirmed these observations (Figure 4F). In addition, the protein expression of ANGPTL4 has also shown no significant difference among various groups (P > .05) (Figure 4E).
Figure 4.
Antibiotics-mediated depletion of gut microbiota blocks oral MT-induced improvements on circadian rhythm disruption-triggered lipid intake in ileum and lipid accumulation in eWAT. (A) Body weight (g, n = 6). (B) ADFI (g/day, n = 6). (C) Relative bacterial abundance in feces from mouse with/without antibiotics mixture treatment (n = 4). (D) Relative mRNA levels (including mRNAs LPCAT3, FATP4, CD36, and NPC1L1, n = 4). (E) Relative protein levels (including LPCAT3, FATP4, NPC1L1, and ANGPTL4, n = 5). (F) Representative Oil Red O staining (red dot) of ileal slides from mice with various treatments including A-Control, A-JL, A-JL+MT, and A-MT; bar: μm (original magnification, ×200 and ×400, n = 6). Right panel: total lipid concentration (mg/g enterocytes) in ileal enterocytes from mice in each group. (G) Fecal levels of TG and CHOL in mice with various treatments including A-Control, A-JL, A-JL+MT, and A-MT (n = 6). (H) Serum levels of TG and CHOL in mice with various treatments including A-Control, A-JL, A-JL+MT, and A-MT (n = 6). (I) Serum levels of HDL-C and LDL/VLDL-C in mice with various treatments including A-Control, A-JL, A-JL+MT, and A-MT (n = 6). (J) Relative mRNA levels (including mRNAs APOA, APOB, APOE, n = 4). (K) Representative H&E staining of adipose tissue from mice with various treatments including A-Control, A-JL, A-JL+MT, and A-MT; bar: μm (original magnification, ×400, n = 6). Right panel: analysis of adipocyte size from mice with various treatments including A-Control, A-JL, A-JL+MT, and A-MT (μm2, n = 6). (L) Serum levels of ANGPTL4 in mice (pg/mL, n = 6). (M and N) Relative mRNA levels of lipid metabolic indicators in adipose tissue (including VLDLR, LDLR, LPL, SREBP-1c, ACC, FAS, HSL, and ATGL, n = 4). (O) Relative protein levels (including LPL, ACC, and ATGL, n = 5). Values are means ± SEM. Same markup represented no significant difference between 2 groups (P > .05); different markup represented significant difference (P < .05) compared with the other groups. ∗P < .05 compared with the other groups.
Fecal and serum TG and CHOL in each group with the depletion of gut microbiota have also shown no statistical difference among various treatments (P > .05), although the influenced trend was similar to observations in mice with gut microbiota (Figure 4G and H). Furthermore, the levels of lipoprotein cholesterol (including HDL-C and LDL/VLDL-C) and the transcriptional levels of hepatic ApoA, ApoB, and ApoE have not detected statistical differences among various groups (P > .05) (Figure 4I and J).
In eWAT, compared with the control mice (with antibiotics treatment), the adipocytes from eWAT in JL mice without gut microbiota presented a similar level of lipid accumulation (Figure 4K), and oral MT did not further decrease mice with circadian rhythm disruption under the depletion of gut microbiota. Notably, serum ANGPTL4 in each treatment without gut microbiota has also not detected difference (Figure 4L), which was similar to the results from Western blot analysis in ileum. Moreover, the qPCR assays and Western blot analysis also supported microbiota-deleted mice to be rarely influenced by JL in terms of the expressions of mRNAs and proteins associated with lipid uptake and lipogenesis (P > .05) (Figure 4M–O). Taken together, these results suggest that oral MT-mediated improvements on circadian rhythm disruption-mediated lipid intake in ileum and lipid accumulation in adipose tissue depend on the presence of gut microbiota.
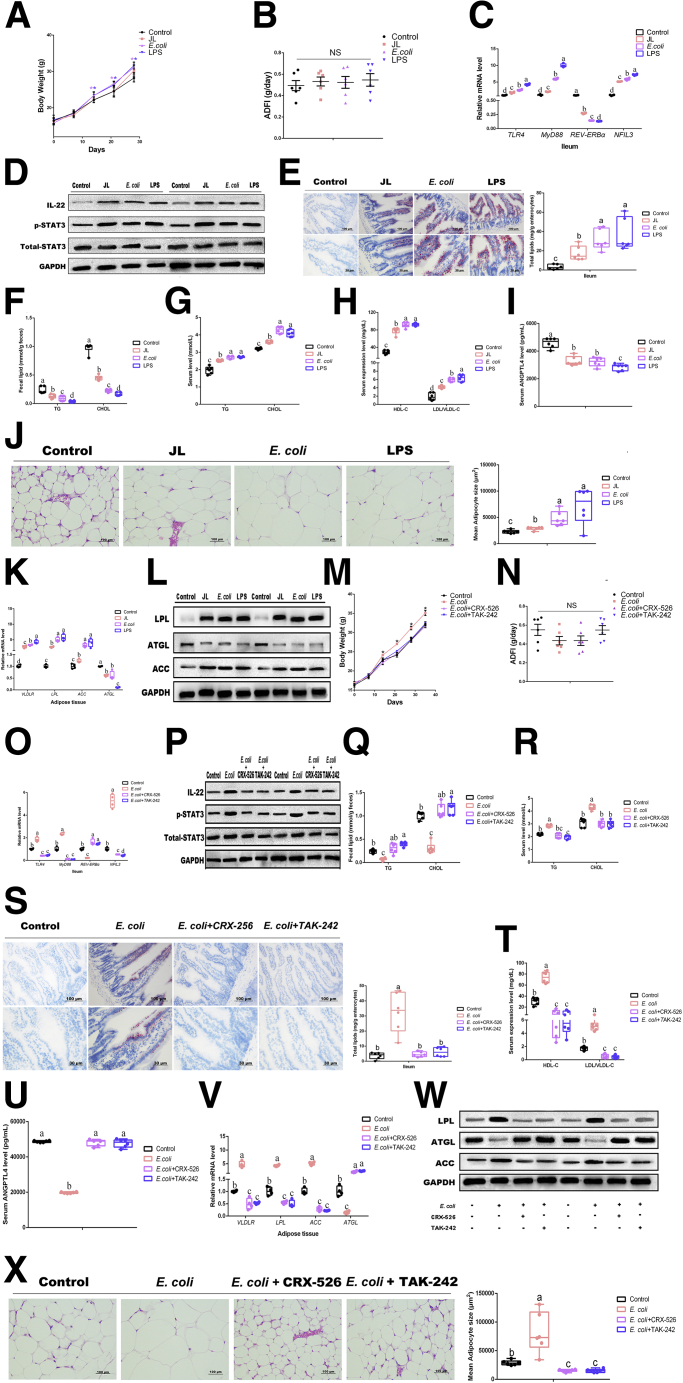
Both E Coli and Microbial LPS Administration Increase Lipid Intake and Inhibit the Expression of ANGPTL4
We found that the abundance of E coli was significantly increased in ileum of JL mice as compared with the control, whereas oral MT administration was responsible for inhibiting its high-level expression under JL treatment (JL+MT group); even oral MT administration further decreased the abundance of E coli in naive mice (MT group). Thus, we hypothesized whether E coli or its LPS served as key targets for MT in the regulatory circuit originating from gut microbiota (microbial metabolites) to host lipometabolism. To explore the effect of E coli and microbial LPS on the lipometabolism of mice, we performed single E coli colonization and LPS supplementation in microbiota-deleted mice and measured the changes of body weight and ADFI (Figure 5A and B). Compared with the control, E coli administration and LPS group were shown to significantly increase body weight at 14 days after treatment (P < .05) (Figure 5A), even higher than JL mice. As shown in Figure 5C, in ileum the mRNA expressions of TLR4 and its downstream MyD88 were significantly up-regulated in E coli and LPS groups as compared with the control and JL treatment (P < .05); the expression of REV-ERBα was significantly inhibited and NFIL3 transcription was obviously increased in both E coli and LPS groups in comparison with the control and JL treatment (P < .05). Notably, LPS administration exhibited more obvious effects in above phenotypes than E coli treatment, suggesting that LPS might be a key molecule for investigating the regulation from E coli to host ileum (P < .05). As expected, the protein expression of IL-22 and the phosphorylation level of STAT3 by Western blot have further shown high-level expression under E coli and LPS treatment as compared with the control (Figure 5D). The Oil Red O staining of ileum for each group has also reflected same observation (Figure 5E).
Figure 5.
(See previous page). Both E coli and microbial LPS administration increase lipid intake and inhibit expression of ANGPTL4. (A) Body weight (g, n = 6). (B) ADFI (g/day, n = 6). (C) Relative mRNA levels (including TLR4, MyD88, REV-ERBα, and NFIL3, n = 4). (D) Relative protein levels (including total STAT3, phosphorylated STAT3, and IL22) (n = 5). (E) Representative Oil Red O staining (red dot) of ileal slides from mice with various treatments including control, JL, E coli, and LPS; bar: μm (original magnification, ×200 and ×400, n = 6). Right panel: total lipid concentration (mg/g enterocytes) in ileal enterocytes from mice in each group. (F) Fecal levels of TG and CHOL in mice with various treatments including control, JL, E coli, and LPS (n = 6). (G) Serum levels of TG and CHOL in mice with various treatments including control, JL, E coli, and LPS (n = 6). (H) Serum levels of HDL-C and LDL/VLDL-C in mice with various treatments including control, JL, E coli, and LPS (n = 6). (I) Serum levels of ANGPTL4 in mice (pg/mL, n = 6). (J) Representative H&E staining of adipose tissue from mice with various treatments including control, JL, E coli, and LPS; bar: μm (original magnification, ×400, n = 6). Right panel: analysis of adipocyte size from mice with various treatments including control, JL, E coli, and LPS (μm2, n = 6). (K) Relative mRNA levels including VLDLR LPL, ACC, ATGL (n = 4). (L) Relative protein levels including total LPL, ACC, and ATGL (n = 5). (M) Body weight (g, n = 6). (N) ADFI (g/day, n = 6). (O) Relative mRNA levels (including TLR4, MyD88, REV-ERBα, and NFIL3, n = 4). (P) Relative protein levels (including total STAT3, phosphorylated STAT3, and IL22) (n = 5). (Q) Fecal levels of TG and CHOL in mice with various treatments including control, E coli, E coli+CRX-526, and E coli+TAk-242 (n = 6). (R) Serum levels of TG and CHOL in mice with various treatments including control, E coli, E coli+CRX-526, and E coli+TAk-242 (n = 6). (S) Representative Oil Red O staining (red dot) of ileal slides from mice with various treatments including control, E coli, E coli+CRX-526, and E coli+TAk-242; bar: μm (original magnification, ×200 and ×400, n = 6). Right panel: total lipid concentration (mg/g enterocytes) in ileal enterocytes from mice in each group. (T) Serum levels of HDL-C and LDL/VLDL-C in mice with various treatments including control, E coli, E coli+CRX-526, and E coli+TAk-242 (n = 6). (U) Serum levels of ANGPTL4 in mice (pg/mL, n = 6). (V) Relative mRNA levels including VLDLR LPL, ACC, and ATGL (n = 4). (W) Relative protein levels including LPL, ACC, and ATGL (n = 5). (X) Representative H&E staining of adipose tissue from mice with various treatments including control, E coli, E coli+CRX-526, and E coli+TAk-242; bar: μm (original magnification,×400, n = 6). Right panel: analysis of adipocyte size from mice with various treatments including control, E coli, E coli+CRX-526, and E coli+TAk-242 (μm2, n = 6). Values are means ± SEM. Same markup represented no significant difference between 2 groups (P > .05); different markup represented significant difference (P < .05) compared with the other groups. ∗P < .05 compared with the other groups.
In feces, under E coli and LPS administration both TG and CHOL were dramatically decreased compared with the control and JL treatment (P < .05), whereas serum lipid (TG and CHOL) and lipoprotein cholesterol (including HDL-C and LDL/VLDL-C) were obviously increased in the E coli and LPS groups (P < .05) (Figure 5F–H). Conversely, we found that the expression of ANGTPL4 was dramatically reduced in the serum of mice after E coli administration and LPS supplementation as compared with the control and JL treatment (P < .05) (Figure 5I).
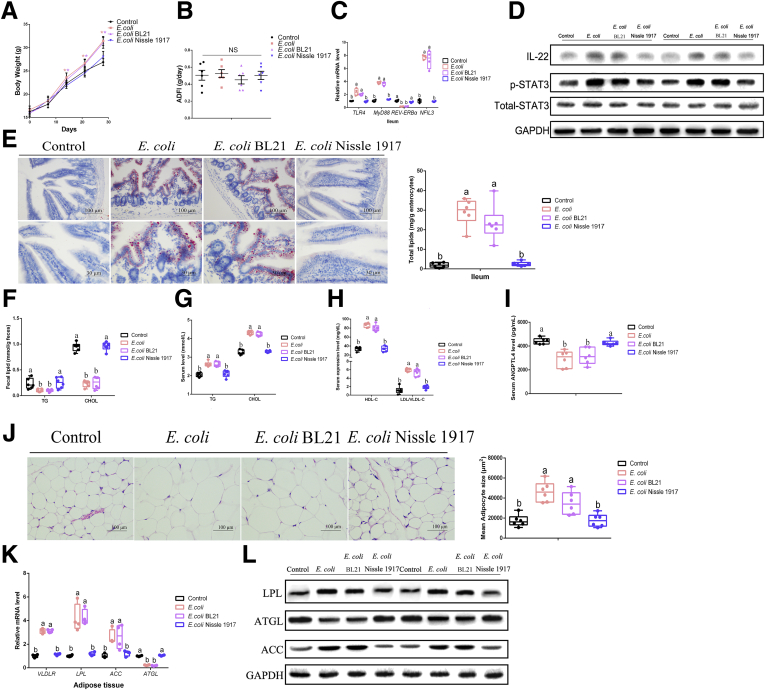
In eWAT, H&E staining has shown that in comparison with the control mice and JL treatment, E coli administration and LPS supplementation induced an obviously higher size of adipocytes in mice (P < .05) (Figure 5J). The qPCR assays and Western blot similarly showed that E coli administration and LPS supplementation had dramatically elevated transcription and translation of the molecule associated with lipid uptake and lipogenesis (including VLDLR, LPL, and ACC), with reducing the transcriptional levels of ATGL (Figure 5K and L). We further performed 2 other strains, E coli Nissle 1917 (a kind of probiotics with LPS-producing defect) and BL21, to verify the conservatism of E coli LPS-triggered effects on host lipometabolism and found similar tendency (Figure6A–L). These findings suggested that E coli might promote host lipid intake and accumulation via microbial LPS.
Figure 6.
E coli-mediated host lipid dysmetabolism depends on microbial LPS. (A) Body weight (g, n = 6). (B) ADFI (g/day, n = 6). (C) Relative mRNA levels (including TLR4, MyD88, REV-ERBα, and NFIL3, n = 4). (D) Relative protein levels (including total STAT3, phosphorylated STAT3, and IL22) (n = 5). (E) Representative Oil Red O staining (red dot) of ileal slides from mice with various treatments including control, E coli, E coli BL21, and E coli Nissle 1917; bar: μm (original magnification, ×200 and ×400, n = 6). Right panel: total lipid concentration (mg/g enterocytes) in ileal enterocytes from mice in each group. (F) Fecal levels of TG and CHOL in mice with various treatments including control, E coli, E coli BL21, and E coli Nissle 1917 (n = 6). (G) Serum levels of TG and CHOL in mice with various treatments including control, E coli, E coli BL21, and E coli Nissle 1917 (n = 6). (H) Serum levels of HDL-C and LDL/VLDL-C in mice with various treatments including control, E coli, E coli BL21, and E coli Nissle 1917 (n = 6). (I) Serum levels of ANGPTL4 in mice (pg/mL, n = 6). (J) Representative H&E staining of adipose tissue from mice with various treatments including control, E coli, E coli BL21, and E coli Nissle 1917; bar: μm (original magnification, ×400, n = 6). Right panel: analysis of adipocyte size from mice with various treatments including control, E coli, E coli BL21, and E coli Nissle 1917 (μm2, n = 6). (K) Relative mRNA levels including VLDLR LPL, ACC, ATGL (n = 4). (L) Relative protein levels including total LPL, ACC, ATGL, and IL22 (n = 5).
Previous research has documented that TLR4 is a major receptor for binding to microbial LPS in multiple tissues and organs including ileum. To verify the effect of E coli-generated LPS on oral MT-mediated improvements on host lipometabolism under circadian rhythm disruption, we used two TLR4 inhibitors, CRX-526 and TAK-242, and further investigated whether E coli administration was able to increase host lipid intake and accumulation under blocking the TLR4 expression. Compared with the control, E coli administration did not induce a higher body weight under CRX-526 supplementation (P > .05); even TAK-242 treatment further decreased the body weight (P < .05) (Figure 5M and N). In addition, the expression of REV-ERBα was significantly increased and NFIL3 transcription was obviously inhibited in both CRX-526 and TAK-242 supplementation in mice with E coli administration (P < .05); E coli-triggered high-level expression of IL-22 and STAT3 signaling were also reduced by CRX-526 and TAK-242 supplementation (Figure 5O and P). The results from fecal and serum lipid (TG and CHOL), lipoprotein cholesterol (including HDL-C and LDL/VLDL-C), and ileal biopsy staining by Oil Red O have similarly indicated that CRX-526 and TAK-242 supplementation reversed E coli-triggered lipid intake (Figure 5Q–T). Moreover, the low level of serum ANGPTL4 in E coli group has obviously recovered (P < .05), with no statistical difference with the control (P > .05) (Figure 5U), suggesting that the lipid accumulation of distal organ (eWAT) might be shaped. As expected, in eWAT E coli administration-induced high size of adipocytes was significantly reduced (P < .05), and the related results from the expression of mRNAs and proteins have similarly supported CRX-526 and TAK-242 supplementation-mediated changes (Figure 5V–X).
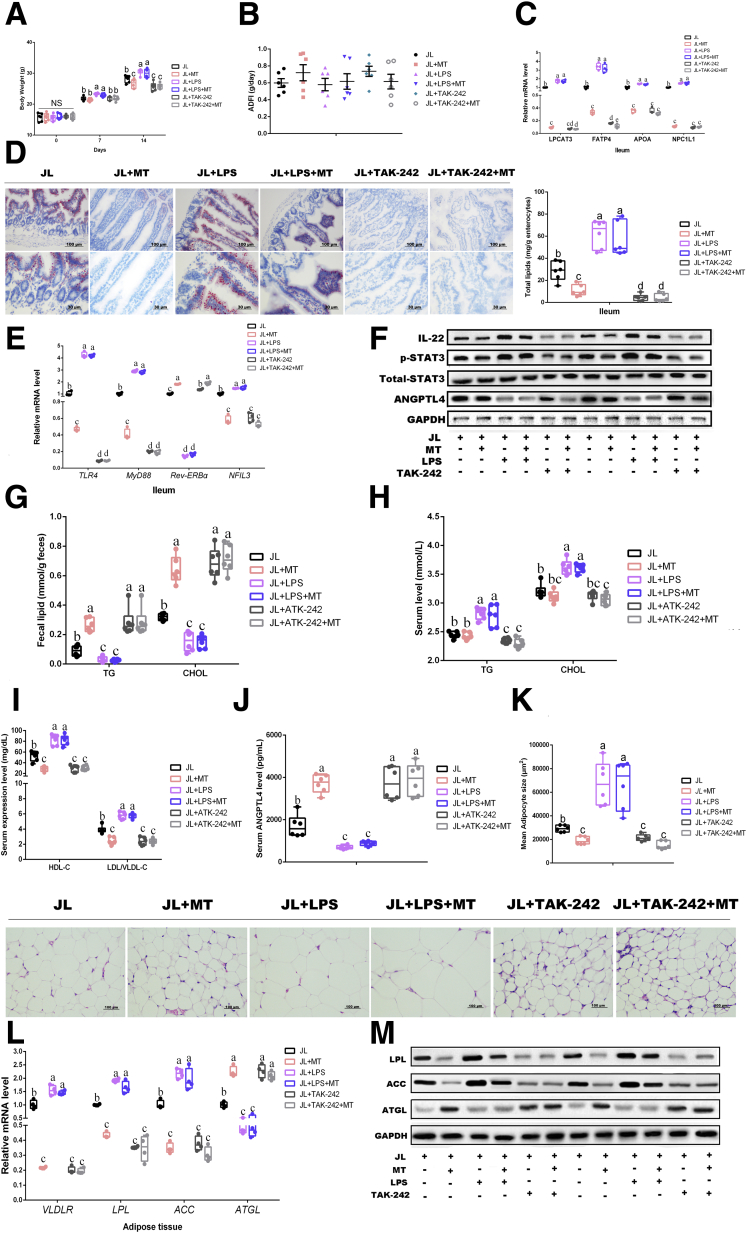
To investigate whether oral MT improves the lipometabolism in mice with circadian rhythm disruption via decreasing the concentration of microbial LPS, we further performed LPS/TAK-242 and MT-cotreated JL mice to detect the body weight gain and lipid accumulation. The results demonstrated that oral MT greatly decreased the body weight as compared with JL mice (P < .05), whereas LPS supplementation further elevated the body weight under JL condition (P < .05) (Figure 7A). However, LPS and MT co-treatment did not reduce the body weight of JL mice in comparison with JL+LPS group with statistical difference (P > .05), implying that oral MT was not able to directly degrade LPS or block TLR4 signaling (Figure 7A). TAK-242 supplementation has obviously decreased the body weight of JL mice (P < .05), whereas TAK-242 and MT co-treatment did not further drive the reduction of body weight as compared with JL+TAK-242 group (P > .05) (Figure 7A). Meanwhile, qPCR and ileal slide with Oil Red O staining reflected same tendency among each group (Figure 7C and D). Notably, compared with JL mice, the transcription of TLR4 and MyD88 was further inhibited in JL+LPS group, and MT supplementation did not obviously reverse this reduction in JL+LPS mice (P > .05) (Figure 7E). Compared with JL+TAK-242 group, the inhibition of TRL4 expression was not promoted by MT supplementation (P > .05) (Figure 7E), supporting that oral MT was not able to directly block TLR4 signaling. At the transcriptional and protein level, LPS supplementation contributed to further inhibiting REV-ERBα expression and increasing NFIL3 transcription as well as the protein expression of IL-22 and the phosphorylation level of STAT3 in ileum of JL mice (P > .05), whereas MT and LPS co-treatment did not reverse these changes (Figure 7E and F). Similarly, compared with inhibition of TLR4 signaling by TAK-242 in JL mice, MT supplementation has not shown obviously opposite effects (Figure 7E and F). As shown in Figure 7G–M, the results regarding fecal and serum lipid, lipoprotein cholesterols, and lipid accumulation in eWAT have also presented same effects among various treatments. Overall, these results suggest that E coli may promote host lipid intake and accumulation via microbial LPS under JL condition, and oral MT-mediated improvements on host lipometabolism do not directly exert on microbial LPS.
Figure 7.
(See previous page). TLR4 plays a vital role in microbial LPS-triggered lipid dysmetabolism. (A) Body weight (g, n = 6). (B) ADFI (g/day, n = 6). (C) Relative mRNA levels (including LPCAT3, FATP4, APOA, and NPC1L1, n = 4). (D) Representative Oil Red O staining (red dot) of ileal slides from mice with various treatments including JL, JL+MT, JL+LPS, JL+LPS+MT, JL+TAK-242, JL+TAK-242+MT; bar: μm (original magnification, ×200 and ×400, n = 6). Right panel: total lipid concentration (mg/g enterocytes) in ileal enterocytes from mice in each group. (E) Relative mRNA levels (including TLR4, MyD88, REV-ERBα, and NFIL3, n = 4). (F) Relative protein levels (including ANGPTL4, total STAT3, phosphorylated STAT3, and IL22) (n = 5). (G) Fecal levels of TG and CHOL in mice with various treatments including JL, JL+MT, JL+LPS, JL+LPS+MT, JL+TAK-242, JL+TAK-242+MT (n = 6). (H) Serum levels of TG and CHOL in mice with various treatments including JL, JL+MT, JL+LPS, JL+LPS+MT, JL+TAK-242, JL+TAK-242+MT (n = 6). (I) Serum levels of HDL-C and LDL/VLDL-C in mice with various treatments including JL, JL+MT, JL+LPS, JL+LPS+MT, JL+TAK-242, JL+TAK-242+MT (n = 6). (J) Serum levels of ANGPTL4 in mice (pg/mL, n = 6). (K) Representative H&E staining of adipose tissue from mice with various treatments including JL, JL+MT, JL+LPS, JL+LPS+MT, JL+TAK-242, JL+TAK-242+MT; bar: μm (original magnification, ×400, n = 6). Right panel: analysis of adipocyte size from mice with various treatments including JL, JL+MT, JL+LPS, JL+LPS+MT, JL+TAK-242, JL+TAK-242+MT (μm2, n = 6). (L) Relative mRNA levels including VLDLR LPL, ACC, ATGL (n = 4). (M) Relative protein levels including total LPL, ACC, and ATGL, n = 5). Values are means ± SEM. Same markup represented no significant difference between 2 groups (P > .05); different markup represented significant difference (P < .05) compared with the other groups. ∗P < .05 compared with the other groups.
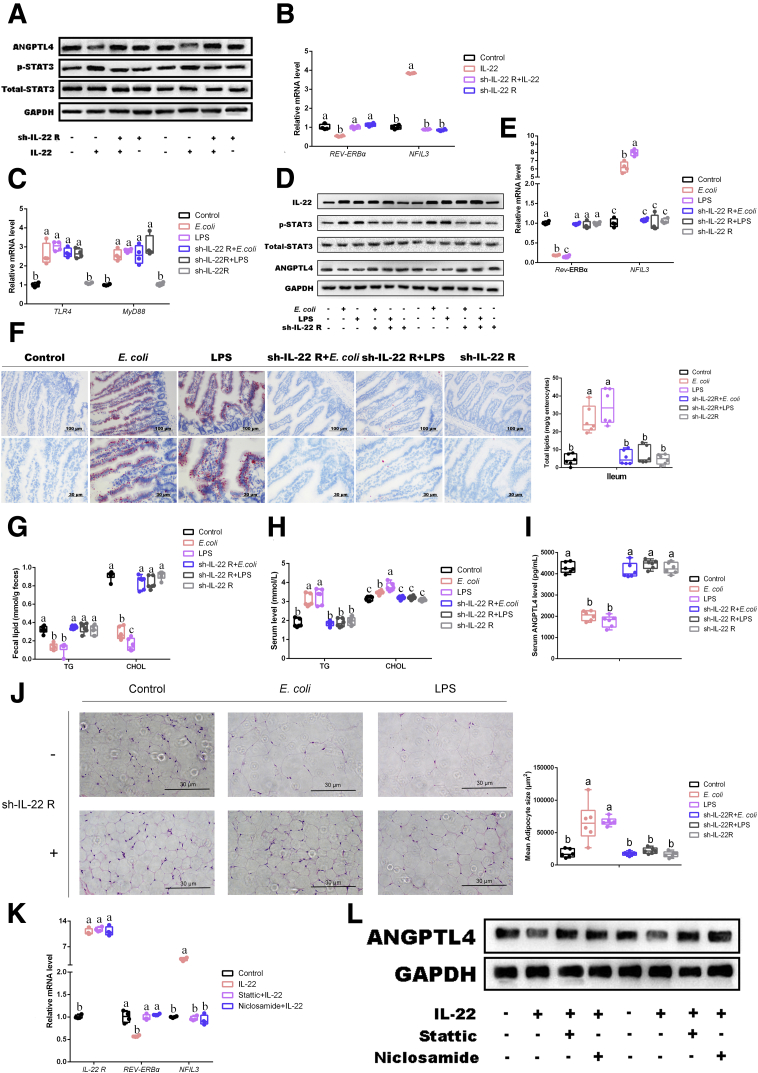
IL-22/STAT3 Signaling Is Involved in the Regulation of Microbial LPS Modulating REV-ERBα/NFIL3 Expression in Mouse Ileum
The results have demonstrated that a common phenotype in JL, E coli and LPS treatment was a high level of IL-22 expression in ileum as compared with control. Because microbial LPS and the activation of ileal TLR4 have been identified as important inducer for triggering host lipid intake in ileum, we then studied the role of IL-22 and STAT3 signaling in the inhibition of REV-ERBα expression, resulting in promoting NFIL3 expression. Wang et al20 have already reported REV-ERBα to negatively modulate NFIL3 expression, which was also negatively regulated by upstream STAT3 signaling and contributed to increasing ileal lipid intake. Moreover, the high level of IL-22 has been considered an important consequence of TLR4 activation in ileum44; we hypothesized that microbial LPS might inhibit REV-ERBα to increase NFIL3 expression via IL-22/STAT3 pathway. Immunoblotting analysis identified that under the interference of IL-22 receptor by sh-IL-22 R, IL-22 administration did not activate the STAT3 signaling (increased the expression of the phosphorylation of STAT3) in primary enterocytes (Figure 8A). Notably, the expression of ANGPTL4 protein was inhibited by IL-22 supplementation, whereas the interference of IL-22 receptor reversed it expression. Moreover, IL-22 supplementation significantly inhibited the transcription of REV-ERBα and elevated NFIL3 mRNA expression (Figure 8B). In vivo, although the interference of IL-22 receptor has not reduced E coli or LPS-induced high transcriptional expression of TLR4 and MyD88 and the protein level of IL-22 in comparison with E coli or LPS groups, this treatment has significantly increased the phosphorylation of STAT3 and NFIL3 in ileum, whereas it inhibited the expression of REV-ERBα mRNA (P < .05) (Figure 8C–E). These results suggested that STAT3 signaling was located in the downstream of IL-22, which is dependent in the activation of IL-22 receptor. Oil Red O staining and TG and CHOL in serum and feces also showed similar observations to above data among various treatments, supporting IL-22 as a key regulated molecule within microbial LPS triggered ileal lipid intake (Figure 8F–H). Notably, the expression of serum ANGPTL4 was obviously reduced by E coli or LPS administrations, whereas its expression in the mice with the blocking of IL-22 receptor was not affected by E coli or LPS administrations (Figure 8I). As expected, the sizes of eWAT in E coli and LPS groups were obviously increased, whereas interference of IL-22 receptor contributed to block this change (Figure 8J).
Figure 8.
IL-22/STAT3 signaling is involved in regulation of microbial LPS modulating REV-ERBα/NFIL3 expression in mouse ileum. (A) Relative protein levels including ANGPTL4, total STAT3, and phosphorylated STAT3 (n = 5). (B and C) Relative mRNA levels (including TLR4, MyD88, REV-ERBα, and NFIL3, n = 4). (D) Relative protein levels including IL-22, ANGPTL4, total STAT3, and phosphorylated STAT3 (n = 5). (E) Relative mRNA levels (including REV-ERBα and NFIL3, n = 4). (F) Representative Oil Red O staining (red dot) of ileal slides from mice with various treatments including control, E coli, LPS, sh-IL-22 R+E coli, sh-IL-22 R+LPS, and sh-IL-22 R; bar: μm (original magnification, ×200 and ×400, n = 6). Right panel: total lipid concentration (mg/g enterocytes) in ileal enterocytes from mice in each group. (G) Fecal levels of TG and CHOL in mice with various treatments including control, E coli, LPS, sh-IL-22 R+E coli, sh-IL-22 R+LPS, and sh-IL-22 R (n = 6). (H) Serum levels of TG and CHOL in mice with various treatments including control, E coli, LPS, sh-IL-22 R+E coli, sh-IL-22 R+LPS, and sh-IL-22 R (n = 6). (I) Serum levels of ANGPTL4 in mice (pg/mL, n = 6). (J) Representative H&E staining of adipose tissue from mice with various treatments including control, E coli, LPS, sh-IL-22 R+E coli, sh-IL-22 R+LPS, and sh-IL-22 R; bar: μm (original magnification, ×400, n = 6). Following panel: analysis of adipocyte size from mice with various treatments including control, E coli, LPS, sh-IL-22 R+E coli, sh-IL-22 R+LPS, and sh-IL-22 R; bar: μm (μm2, n = 6). (K) Relative mRNA levels (including IL-22 R, REV-ERBα, and NFIL3, n = 4). (L) Relative protein levels including ANGPTL4 (n = 5). Values are means ± SEM. Same markup represented no significant difference between 2 groups (P > .05); different markup represented significant difference (P < .05) compared with the other groups.
Finally, qPCR and Western blot further confirmed that STAT3 signaling was closely involved in the regulation from microbial LPS and located in the upstream of REV-ERBα/NFIL3 via inhibitor of the phosphorylation of STAT3 (stattic and niclosamide) (Figure 8K and L). Taken together, these data suggest that E coli-derived LPS may be involved in shaping host lipometabolism in ileum via activating TLR4/IL-22/STAT3 signaling, which inhibits REV-ERBα expression to promote NFIL3 activation, resulting in lipid uptake.
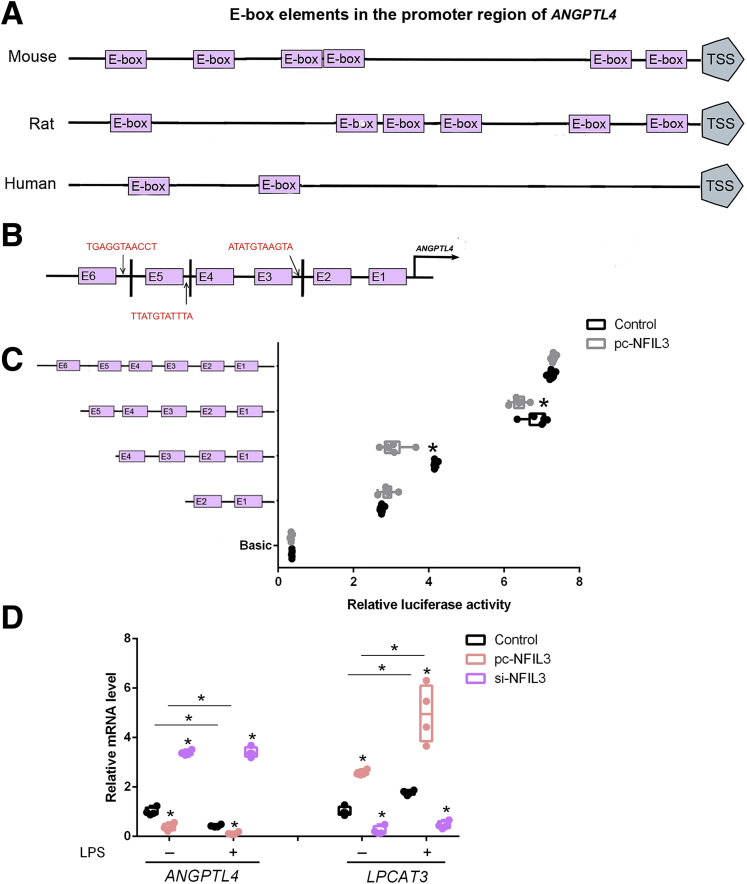
NFIL3 Blocks ANGPTL4 Expression at the Transcription Level
Because we found that ANGPTL4 is obviously inhibited in mice ileum from JL, E coli, and LPS treatments accompanied by highly expressed of NFIL3 transcription, we further tested whether ANGPTL4 mRNA was negatively modulated by the transcription of NFIL3 in enterocytes. On the ANGPTL4 promoter of mouse, NFIL3 has been predicted to own 3 potential binding domains (Figure 9A and B). Using the luciferase assay, 2 binding sites, 795 bp–729 bp and 946 bp–795 bp, were identified as the inhibitor positions of ANGPTL4 (Figure 9C). Moreover, combined with LPS supplementation, we further verified activated NFIL3 to markedly suppress the transcription of ANGPTL4 and LPCAT3 in enterocytes (Figure 9D). Overall, these results indicate that NFIL3 blocks ANGPTL4 expression at the transcription level. Moreover, LPS elevates the expression of NFIL3 mRNA to reduce the transcriptional level of ANGPTL4 in enterocytes, suggesting that NFIL3 may contribute to promoting ileal intake and fat accumulation in distal tissues and organs via increasing the LPCAT3 mRNA expression and inhibiting ANGPTL4 transcription.
Figure 9.
NFIL3 blocks ANGPTL4 expression at transcription level. (A) Schematic of E-box elements in ANGPTL4 promoter. (B) Binding sites of NFIL3 are located in 3 sequences that are defined by E-boxes in ANGPTL4 promoter region. (C) Luciferase activity was corrected for Renilla luciferase activity and normalized to the control activity (n = 6). (D) Relative mRNA level of LPCAT3 and ANGPTL4 of intestinal epithelial cells pretreated with pc-NFIL3 or si-NFIL3 and incubated with LPS or not (n = 4). Luciferase reporter assay of ANGPTL4 and NFIL3. Fragments of ANGPTL4 promoter fused to a luciferase reporter gene were co-transfected into MODE-K cells together with basic (control) or pc-NFIL3. pc-NFIL3: overexpression adenovirus vector of NFIL3; si-NFIL3: interference lentiviral recombinant vectors of NFIL3. Relative mRNA levels were detected by real-time qPCR. Values are means ± SEM. ∗P < .05 compared with other groups.
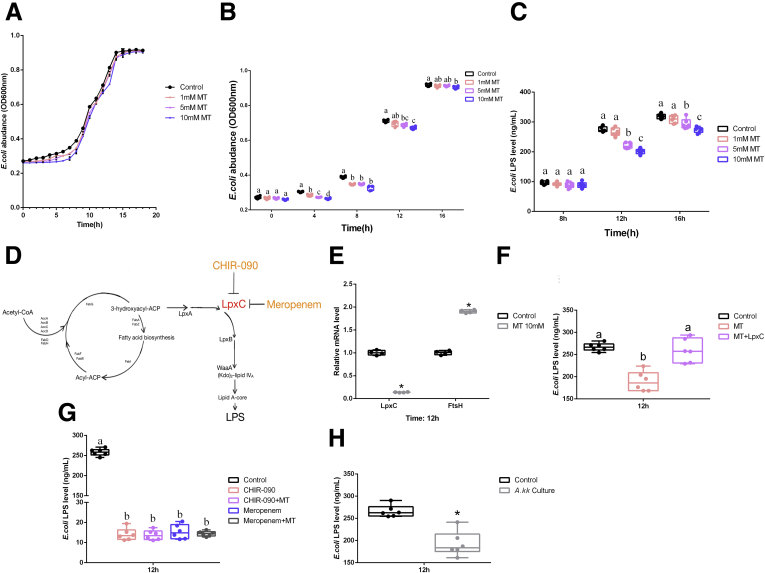
Melatonin Reduces the Production of Microbial LPS via Inhibiting LpxC Expression in E coli
Oral MT did not directly exert on microbial LPS to improve host lipometabolism under JL+LPS treatment. However, the observations from the comparison between JL and JL+MT definitely showed a significant improvement on host lipometabolism. Thus, we hypothesized that oral MT might contribute to inhibit LPS secretion from E coli rather than directly degrading LPS. To explore the effect of MT on E coli-derived LPS in vitro, we measured the growth curve of E coli with/without different concentrations of MT (Figure 10A and B). Compared with the control, MT supplement inhibited bacterial proliferation of E coli in a dose-dependent manner after 8 hours of culture (Figure 10B). E coli-derived LPS was also depressed by MT treatment (5 and 10 mmol/L) at 12 hours (P < .05) (Figure 10C). As shown in Figure 10D, LpxC is a vital regulatory molecule for LPS synthesis in E coli. Thus, we then tested the transcriptional level of LpxC in E coli under MT treatment. The results showed that MT (10 mmol/L) significantly inhibited its mRNA expression (P < .05) (Figure 10E), whereas exogenous UDP-3-O-acyl-N-acetylglucosamine deacetylase recombinant protein (LpxC) reversed this improvement (Figure 10F). By using other known LpxC inhibitors, CHIR-090 and meropenem, we further confirmed that MT decreased LPS production by inhibiting LpxC expression, because MT supplement did not promote lower transcription of LpxC expression in E coli (P > .05) (Figure 10G). Finally, in co-culture of Akk muciniphila and E coli we also observed the inhibition of LPS (Figure 10H). Briefly, oral MT decreases the production of E coli-derived LPS, which further elevates ANGPTL4 expression and triggers ileal lipid intake via attenuating TLR4/STAT3/ REV-ERBα/NFIL3 pathway. The increased level of serum ANGPTL4 may further contribute to inhibiting lipid accumulation in mice eWAT.
Figure 10.
MT reduces production of microbial LPS via inhibiting LpxC expression in E coli. (A) Growth curves of E coli under MT treatment with various dosages in vitro (1, 5, 10 mmol/L) (optical density 600 nm, n = 6). (B) Amount of E coli due to MT treatment with various dosages in vitro (1, 5, 10 mmol/L) (optical density 600 nm, n = 6). (C) Level of E coli LPS under MT treatment with various dosages in culture (1, 5, 10 mmol/L) (ng/mL, n = 6). (D) Regulatory circuit of E coli-derived LPS (references 64 and 66). (E) Relative mRNA levels of LpxC and FtsH in E coli (n = 4). (F) Level of E coli LPS under MT treatment (10 mmol/L) with/without LpxC recombinant protein (0.5 mmol/L) after 12 hours (ng/mL, n = 6) (G) Level of E coli LPS under various treatments including CHIR-090, CHIR-090+MT (10 mmol/L), meropenem, meropenem+MT (10 mmol/L) (ng/mL, n = 6). (H) Level of E coli LPS under co-culture with Akk muciniphila (ng/mL, n = 6). Values are means ± SEM. Same markup represented no significant difference between 2 groups (P > .05); different markup represented significant difference (P < .05) compared with the other groups. ∗P < .05 compared with the other groups.
Discussion
Gut microbiota have a close association with host health and are involved in major energy metabolic processes.7 Enteric flora disturbance is a well-established biomark for many energy dysmetabolism-induced chronic diseases, including excess adiposity, type 2 diabetes, cardiovascular disease, and other metabolic syndromes.47,49 Also, the alterative patterns of microbial metabolites preferentially modulate physiological activities of host digestive tract.40,50 Traditionally, microbial LPS is regarded as an important therapeutic target to prevent host from metabolic disorders and inflammatory responses.51,52 Herein, we hypothesized that microbial LPS may specially serve as an inductor on circadian rhythm disruption-triggered weight gain and lipometabolism. Thus, we performed a comparison of the composition of gut microbiota isolated from various treatments (including the control, JL, and oral MT supplement). With oral administration of MT, we found that circadian rhythm disruption significantly shaped the pattern of microbial community, with higher level of E coli, an LPS-producing species in mouse ileum. In addition, it is noteworthy that antibiotics-mediated microbiota depletion caused an obvious absence of these phenotypes, suggesting that the altered pattern of gut microbiota is responsible for oral MT-mediated anti-obesity actions.
Originally, ANGPTL4 has been considered as a kind of fasting-triggered adipocytokine, referring to a glycosylated protein.53,54 Previous research has already definitely indicated a strong association among circulatory ANGPTL4, serum lipid levels, and the lipometabolism in distal organs.55,56 ANGPTL4 is a secreted protein and an important metabolic regulator produced in the gut in response to changes of nutritional status.55 Endogenous ANGPTL4 can act as an inhibitor for lipoprotein lipase activity in peripheral tissue (adipose tissue) and thereby contributes to the regulation of postprandial plasma TG clearance.57 ANGPTL4 post-translationally inhibits LPL activity under various physiological conditions including fasting and cold exposure, thereby raising plasma triacylglycerol levels.24,27,57 The inactivation of ANGPTL4 has repeatedly been reported to be responsible for decreased plasma TG and high level of HDL cholesterol.25,58 In view of modern medicine, ANGPTL4 plays a major regulator of plasma TG in multiple physiological stresses,54,59, 60, 61 indicating that ANGPTL4 is an essential component of a regulatory circuit to link lipid uptake from dietary and delivery into target tissue.54,61,62 Thus, in this investigation, we detected the expression of serum apolipoproteins (including HDL and VLDL/LDL) and circulating ANGPTL4 to link the crosstalk between gut and distal organs. We supplied the data regarding mRNA expression of VLDL receptor in adipose from each group, which plays an important role in the delivery of lipid (mainly TG) from serum VLDL.59 Our results determined that low level of serum ANGPTL4 and high level expression of LPS were markers in JL mice. On the contrary, ANGPTL4 expression was obviously recovered through oral MT; TLR4 inhibitor supplement and microbiota depletion reversed JL-triggered weight gain, suggesting that MT may shape the LPS-producing strain to improve the level of ANGPTL4 in the ileum, which further suppresses lipid uptake by adipocytes. ANGPTL4 served as a vital mediator/messenger in the regulation of lipometabolism between gut and adipose tissue, which is consistent with other findings.54,61,63,64
In ileum, excessive lipid intake is considered as a basic factor and key link in the development of alimentary obesity.20 Among multiple transcriptional factors, NFIL3 has been identified as a vital link to mammalian rhythms and lipometabolism in intestines.20 Herein, the results documented elevated NFIL3 to contribute to inhibiting ANGPTL4 transcription, noting that NFIL3 played an essential role in triggering lower expression of ANGPTL4, which may further impact its circulatory level. TLR4 is a major receptor for microbial LPS and has been reported to initiate the STAT3/REV-ERBα signaling pathway, which increases NFIL3 expression in intestinal epithelial cells.7,20,44 Our results and previous studies identified that activation of TLR4/STAT3/REV-ERBα is an important signaling pathway in ileal lipometabolism.20 In this research, we reported that microbial LPS plays a crucial role in JL-mediated weight gain, and MT serves as an important inhibitor for LPS production in E coli.
In addition to reducing LPS production from E coli via inhibiting Lpx C expression,65,66 oral MT also promoted the amount of Akk muciniphila, which has been shown to reduce the expression of LPS from E coli. The discovery of discrepant abundance of Akk muciniphila, a mucin-degrading Gram-negative bacteria, between obese and healthy individuals is undoubtedly a significant academic achievement.67, 68, 69 Subsequent studies have demonstrated that this microbe alleviates multiple energy dysmetabolism-induced organ/systemic diseases, whereas the lower abundance of Akk muciniphila positively correlates with host hyperlipidemia, type 2 diabetes, and fatty liver.67, 68, 69, 70, 71 Moreover, the research regarding Akk muciniphila is not limited to this field, but it also extends to other disease preventions such as progeria and multiple sclerosis.67 Notably, the quantity of Akk muciniphila has been reported to inversely correlate with multiple systemic dysmetabolism through its improvements on the physiology of adipose tissue.72, 73, 74, 75 Because this microbe promotes mucus turnover in mucus layer where its colonization improves host intestinal barrier integrity, the amount of Akk muciniphila has become a crucial parameter for intestinal health.7
However, some limitations also remain in this study. First, E coli-derived LPS has been only focused under MT treatment, without considering other microbes in the ileum, and further analysis of microbial composition may identify other species that contribute to MT-mediated positive mechanisms as well as the effects of MT on other microbial metabolites. Second, considering the site-specific differences in various WATs, we preferentially study eWAT as a target for this regulatory circuit originating from gut microbiota, because its initiation of development is earlier than other WAT depots (adipogenesis: eWAT, 4 weeks; subcutaneous WAT, 12 weeks).76 In addition, more mature vessels wrap eWAT, which may elevate the possibility of the effect of circulating ANGPTL4.76 On the basis of some reports, eWAT has been considered as a target site for this regulatory cricuit.77, 78, 79, 80, 81 Finally, the whole circadian variation of gut microbiota has not been focused on. Considering the strong associations between these symbionts and host, the hypothesis has been proposed whether significant metabolic changes would be triggered by the overlap or stagger of both, namely whether rhythmic disorder-induced alternation of gut microbiota contributed to shaping host physiology. By using integrated multiomics analysis, Thaiss et al82 demonstrated the gut microbial diurnal oscillation and these feature-induced metabolic patterns on host. These diurnal oscillations including biogeographic migration and the oscillation of metabolome patterns drive the circadian transcriptional reprogramming at the level of multi-tissues/organs in mice, implying that circadian rhythm disruption-triggered programming of gut microbiota may play a vital role in modulating host metabolism.82 Therefore, in this study we thought that stagger time lag may be a key period for shaping the pattern of gut microbiota, not in overlap time bucket (daytime). However, further study on the whole circadian variation of gut microbiota will be necessary to investigate oral MT-mediated regulatory mechanism as well as the effects of oral MT on the host gut microbial structure.
Taken together, the present results suggest that (1) gut microbiota are responsible to oral MT-mediated weight control in JL mice; (2) ANGPTL4 may play an essential role in delivering message from gut microbiota to host adipose tissue; (3) microbial LPS leads to the inhibition of intestinal ANGPTL4 transcription via activating on the TLR4/STAT3 pathway, which eventually contributes to the high level of NFIL3 expression; and (4) oral MT reduces the level of microbiota-derived LPS via inhibiting LpxC expression. These data are consistent with oral MT-mediated alterations of gut microbiota being a potential therapy for circadian rhythm disruption-induced lipid dysmetabolism via recovering the intestinal ANGPTL4, which may deliver regulatory messages to peripheral target organs/tissues.
Materials and Methods
Animal Experiments
All animal experiments were performed in accordance with the guidelines and regulations and approved by the Animal Ethics Committee (Northwest A&F University, Yangling, Shaanxi). Male C57BL/6J wild-type mice (8-week-old) were purchased from the Laboratory Animal Center of the Fourth Military Medical University (Xi’an, China). In an environmentally controlled room (temperature: 25°C ± 1°C; humidity: 55 % ± 5 %; 12-hour light/dark cycle), all animals had ad libitum access to water and food.
Mice were randomly divided into 4 groups (n = 6) and housed as 1 per cage (30 × 20 × 13) and placed on a layer of bedding (corn cob for animal bedding; Beijing Keao Xieli Feed Co, Ltd, Beijing), which were fed a standard diet (fat provided 30%–31 % of total energy; Boaigang Biotechnology Co Ltd, Beijing). The groups included control, JL, JL+MT, control+melatonin (MT). Melatonin treatment was performed by supplying it in drinking fluid (0.4 mg/mL; Sigma-Aldrich, St Louis, MO). The bottled melatonin water was covered by a layer of aluminum foil to prevent light.83,84 To avoid MT degradation by bacterial activities in water, the water by high temperature sterilization was used to prepare MT solution, and the bottles were daily sterilized by 121°C. The LPS treatment was performed by oral administration in drinking water (10 μg/μL; Sigma-Aldrich), and CRX-526 (2 mg/kg; Invitrogen, Shanghai, China) and TAK-242 (4 mg/kg; MCE, Shanghai, China) were treated by intraperitoneal injection as previously described.85, 86, 87, 88 The JL mice were produced as previously described.31 Zeitgeber time (ZT) 0 corresponds to the time of light onset. In brief, mice were transferred between 2 rooms (Room 1: 6:00 am to 6:00 pm light/6:00 pm to 6:00 am dark, and Room 2: 10:00 am to 10:00 pm dark/10:00 pm to 10:00 am light). Mice were transferred between 2 rooms (Room 1: 6:00 am to 6:00 pm light/6:00 pm to 6:00 am dark, and Room 2: 10:00 am to 10:00 pm dark/10:00 pm to 10:00 am light). Mice were transferred once from room 1 to 2 and then returned to room 1. The time of mice transfer occurs between 9:30 am to 10:00 am, leading to an 8-hour phase advance from room 1 to 2 (light off at ZT4 instead of ZT12 for JL mice on the day of transfer) and an 8-hour phase delay from room 2 to 1 (light off at ZT20 instead of ZT12 for JL mice on the day of transfer) every 2 days. All animals’ body weights were measured weekly, including original weight at the beginning of the experiment (0 day) and affected weight during the period of experiment. The ADFI was calculated through detected daily food consumption by counting the changes of weights.
Mice were killed using an overdose of ethyl ether 2 hours after the last JL treatment. Ileum, eWAT, and liver were immediately collected. Portions of these tissues were fixed in 4 % paraformaldehyde in phosphate buffer and prepared for paraffin or frozen sectioning. Paraffin sections were stained using H&E, and frozen sections were stained using Oil Red O. The sizes of cells and stained area were analyzed by using cell profiler software (cellSens; Olympus, Hamburg, Germany). The immumohistochemical staining of ileal ANGPTL4 was performed in accordance with commercial kit based on paraffin sections (SABC-POD immunohistochemistry kit; Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Images were captured using a Cytation 3 Cell Imaging Multimode Reader (BioTek, Winooski, VT). The immunohistochemistry of ileal ANGPTL4 was performed by paraffin sections in accordance with the guidelines of commercial ELISA kits (Thermo Fisher, Shanghai, China). All immunohistochemistry images were analyzed using Image J (National Institutes of Health, Bethesda, MD) by counting the average gray value of positive cells (staining intensity).
Gut Microbiota Analysis
Total genomic DNA of gut microbiota was extracted from fecal samples and further amplified using primer (16S V3 and V4). Sequencing libraries were built by TruSeq Nano DNA LT Library Prep Kit and MiSeq platform (Illumina, San Diego, CA). Operational taxonomic unit table was obtained using QIIME software and further combined with clustering analysis to form a heatmap. On the basis of the abundance matrix of operational taxonomic units, species diversity was evaluated by observed species, Chao1, Shannon, and Simpson. To directly reflect the differentiation among various groups, microbial hierarchical tree was drawn by using MEGAN software. On the basis of Unifrac, principal coordinate analysis was performed to directly visualize multidimensional data.
E coli and Akk muciniphila Isolation, Identification, and Culture
Stool samples were collected from mice in each group and placed in 1 mL of phosphate-buffered saline. The isolation of E coli was performed on the basis of previous reports by Schmidt et al.89 Briefly, equal volumes of feces (5 g) and brain heart infusion broth (Solarbio, Beijing, China) with 5 % glycerol (5 mL) were homogenized before streaking onto plain eosin methylene blue agar (EMBA; Solarbio, Beijing, China), which was impregnated with third-generation cephalosporins (1 μg/mL ceftazidime and 1 μg/mL cefotaxime, Solarbio, Beijing, China). After overnight aerobic incubation at 37°C, 10 random colonies, morphologically resembling E coli, were selected from plain EMBA, and 1 colony from each (ceftazidime/cefotaxime) impregnated EMBA plate was selected for further investigation. Selected colonies were subcultured onto nutrient agar (Solarbio) for pure growth and incubated aerobically overnight at 37°C before Gram-staining, biochemical analysis (catalase production, absence of oxidase, lactose fermentation, indole production, and inability to use citrate as a carbon source). PCR assays for the 16S rRNA confirmed isolates as E coli. The abundance of E coli was determined through using optical density at 600 nm, and the levels of E coli-derived LPS were measured using optical density at 450 nm in various treatments, including LpxC recombinant protein (MyBioSource, San Diego, CA), CHIR-090 (MedChemExpress, Shanghai, China), meropenem and butyrate (Sigma-Aldrich).
The isolation of Akk muciniphila was performed on the basis of previous reports by Derrien et al.45 In brief, equal volumes of feces (0.5 g) were diluted into sterile anaerobic Ringer’s solution (9 mL, containing 0.5 g cysteine). After thorough mixing, the suspensions were further diluted (10-fold) in Ringer’s solution and then inoculated in triplicate into 9 mL bicarbonate-buffered medium (mucin medium) (inoculums size: 1 mL) at 37°C under anaerobic conditions. Single colonies were selected, grown in mucin medium, and reinoculated in soft agar mucin medium (Sigma-Aldrich). This step was repeated until purity. Selected colonies were defined by Gram-staining and PCR assays for the 16S rRNA of Akk muciniphila.
Mono-colonization
To deplete original gut microbiota, recipient mice were pretreated with multiple antibiotics (1 g/L streptomycin, 0.5 g/L ampicillin, 1 g/L gentamicin, and 0.5 g/L vancomycin) according to Yin et al.84 Mice received antibiotics via the drinking water for 10 days and then were tested for mono-colonization of isolated E coli and 2 commercial strains (E coli BJ21 and E coli Nissle 1917; TSTO, Ningbo, China). Cultures were grown in LB broth (Solarbio) overnight, and 1 × 1010 colony-forming units per mL preparations were made. A single dose of 100 μL was delivered via gavage to mice, as previously reported by Velazquez et al.90
Immunoblotting Analyses
On the basis of previously established protocol of Rong et al,36 proteins were extracted from ileum, eWAT, and liver by using lysing buffer (Solarbio). Roughly 30 μg protein was separated by electrophoresis (12 % and 5 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels) and then transferred onto polyvinylidene difluoride nitrocellulose membranes (Millipore, Burlington, MA), which were further blocked by 5 % non-fat milk in Tris-Tween buffered saline buffer for 2 hours. After blocking, these membranes were incubated with various primary antibodies, including anti-Niemann-Pick C1-Like 1 (NPC1L1), anti-lysophosphatidylcholine acyltransferase 3 (LPCAT3), anti-fatty acid transport protein4 (FATP4), anti-LPL, anti-adipose triglyceride lipase (ATGL), anti-acetyl CoA-carboxylase (ACC), anti-IL-22, anti-STAT3, anti-phosphorylated STAT3 (p-STAT3), anti-ANGPTL4, and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam, Cambridge, UK), and horseradish peroxidase-conjugated secondary antibody. Finally, proteins were imaged using chemiluminescent peroxidase substrate (Millipore) and quantified using ChemiDoc XRS system (Bio-Rad, Richmond, CA).
Quantitative Real-time PCR
On the basis of a previously described method,36 the extraction and reverse transcription of total RNA (500 ng) from ileum, eWAT, and liver were performed (TRIpure Reagent kit and M-MLV reverse transcriptase kit; Takara, Dalian, China), respectively. Primers were designed in accordance with mouse sequence and passed to Invitrogen (Shanghai, China) to synthesize. Quantitative PCR was performed in 25 μL reaction system containing specific primers and SYBR Premix (Vazyme Biotech, Nanjing, China). Amplification was performed in the ABI StepOne plus RT-PCR System (Carlsbad, CA). The levels of mRNA were normalized in relevance to Gapdh and Rpo A (Figure 8E). The relative RNA expressions were analyzed using the method of 2−ΔΔCt.
Fecal and Serum Biochemical Parameters
Serum samples were tested to detect multiple biochemical parameters using commercial enzyme-linked immunosorbent assay kits, including CHOL (Abcam, Cambridge, UK), TG (Abcam), LPS (CUSABIO, Wuhan, China), HDL-C and LDL/VLDL-C (Abcam), and ANGPTL4 (Abcam).
Fecal lipid was determined in accordance with previous studies.39 In brief, fecal samples were collected from all groups when the mice were killed. After being dried at 60°C to a constant weight, samples were ground to a fine powder and then extracted 3 times with ethanol 95 % at 60°C. After filtration and evacuation, fecal TG and CHOL were detected by using commercial kits (Abcam).
Plasmid Transfection and Dual-Luciferase Reporter Assay
A 1181 bp fragment of mouse ANGPTL4 promoter was amplified by PCR and fused to the pGL-3 basic vector. MODE-K cells were transfected after 1-day culture in 96-well plate. The built reporter was referred to ANGPTL4 1181-Luc. Deuterogenic ANGPTL4 946-Luc, ANGPTL4 795-Luc, and ANGPTL4 190-Luc reporters were generated from ANGPTL4-Luc via deletion and relegation, which contained 946, 795, and 190 bp of ANGPTL4 promoter, respectively. Using the ANGPTL4 1181-Luc plasmid as a template, mutant ANGPTL4 reporter plasmids were generated. By co-transfection with ANGPTL4 promoter vector, MODE-K cells were treated together with empty vector, pc-NFIL3-encoding vector (overexpression vector), or si-NFIL3-encoding vector (interference vector) using X-tremeGENETM transfection reagent (Roche, Basel, Switzerland). Luciferase activity was determined as previous described after transfection for 48 hours.36
Primary Enterocytes Culture and Vectors Infection
Primary enterocytes were isolated from sacrificed mice and cultivated in accordance with previous studies.91,92 In brief, ileum was harvested from sacrificed mice in each group and then washed with phosphate-buffered saline after opening longitudinally. Primary crypts were dissociated from ileal mucosa through incubating in gradient EDAT (10 mmol/L and 5 mmol/L) for 15 minutes each. Isolated crypts were further incubated in 1× TrypLE express (Gibco, Shanghai, China) supplemented with 0.8 KU/mL DNase1 (Roche) to obtain single primary enterocytes.
For virus vectors study, primary enterocytes were infected with/without recombinant IL-22 receptor adenovirus interference vector (sh-IL-22 R) for 48 hours at the titer of 1× 109 infectious units/mL, and the control vectors were pAd-GFP. All the vectors were constructed by Gene Pharma (Shanghai, China). Also, sh-IL-22 R was intraperitoneally injected into mice for 2 weeks to study the physiological changes.
Statistical Analysis
Experimental data were analyzed by one-way analysis of variance and two-way analysis of variance in SAS v8.0 (SAS Institute, Cary, NC). Individual means were compared by using Fisher least significant difference. Mean ± standard error of the mean (SEM) represented each dataset, and significance in statistical difference was defined as P < .05. The correlation analysis and cosinor analysis were conducted by linear regression model using GraphPad Prism 5.0 software (GraphPad Software, Inc, San Diego, CA).31
Uncited Reference
This section consists of references that are included in the reference list but are not cited in the article text. Please either cite each of these references in the text or, alternatively, delete it from the reference list. If you do not provide further instruction for this reference, we will retain it in its current form and publish it as an “uncited reference” with your article.35
Acknowledgments
CRediT Authorship Contributions
Bohan Rong (Investigation: Lead; Project administration: Lead; Writing – original draft: Lead)
Qiong Wu (Resources: Supporting)
Russel J. Reiter (Writing – review & editing: Equal)
Chao Sun (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Resources: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by grants from the Key Sci-tech innovation team of Shaanxi province (2017KCT-24), the Joint Funds of the National Natural Science Foundation of China of China (U1804106) and Natural Science Foundation of China (81860762) and Qinghai Fundamental Scientific and Technological Research Plan (2018-ZJ-721) and the Scientific Research Guiding Plan Topic of Qinghai Hygiene Department (2018-wjzdx-131).
Contributor Information
Russel J. Reiter, Email: REITER@uthscsa.edu.
Chao Sun, Email: sunchao2775@163.com.
References
- 1.Hemmingsson E. A new model of the role of psychological and emotional distress in promoting obesity: conceptual review with implications for treatment and prevention. Obes Rev. 2014;15:769–779. doi: 10.1111/obr.12197. [DOI] [PubMed] [Google Scholar]
- 2.Deng T., Lyon C.J., Bergin S., Caligiuri M.A., Hsueh W.A. Obesity, inflammation, and cancer. Annu Rev Pathol. 2016;11:421–449. doi: 10.1146/annurev-pathol-012615-044359. [DOI] [PubMed] [Google Scholar]
- 3.Boutens L., Stienstra R. Adipose tissue macrophages: going off track during obesity. Diabetologia. 2016;59:879–894. doi: 10.1007/s00125-016-3904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scopelliti A., Bauer C., Yu Y., Zhang T., Kruspig B., Murphy D.J., Vidal M., Maddocks O.D.K., Cordero J.B. A neuronal relay mediates a nutrient responsive gut/fat body axis regulating energy homeostasis in adult drosophila. Cell Metab. 2019;29:269–284. doi: 10.1016/j.cmet.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gancheva S., Jelenik T., Álvarez-Hernández E., Roden M. Interorgan metabolic crosstalk in human insulin resistance. Physiol Rev. 2018;98:1371–1415. doi: 10.1152/physrev.00015.2017. [DOI] [PubMed] [Google Scholar]
- 6.Wesemann D.R., Nagler C.R. The microbiome, timing, and barrier function in the context of allergic disease. Immunity. 2016;44:728–738. doi: 10.1016/j.immuni.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rong B., Xia T., Zhang T., Feng R., Huang H., Wu Q., Sun C. Gut microbiota: a potential manipulator for host adipose tissue and energy metabolism. J Nutr Biochem. 2019;64:206–217. doi: 10.1016/j.jnutbio.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Miyamoto J., Igarashi M., Watanabe K., Karaki S.-I., Mukouyama H., Kishino S., Li X., Ichimura A., Irie J., Sugimoto Y., Mizutani T., Sugawara T., Miki T., Ogawa J., Drucker D.J., Arita M., Itoh M., Kimura I. Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat Commun. 2019;10:4007. doi: 10.1038/s41467-019-11978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truax A.D., Chen L., Tam J.W., Cheng N., Guo H., Koblansky A.A., Chou W.-C., Wilson J.E., Brickey W.J., Petrucelli A., Liu R., Cooper D.E., Koenigsknecht M.J., Young V.B., Netea M.G., Stienstra R., Sartor R.B., Montgomery S.A., Coleman R.A., Ting J.P.-Y. The inhibitory innate immune sensor NLRP12 maintains a threshold against obesity by regulating gut microbiota homeostasis. Cell Host Microbe. 2018;24:364–378. doi: 10.1016/j.chom.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R., Sinha R., Gilroy E., Gupta K., Baldassano R., Nessel L., Li H., Bushman F.D., Lewis J.D. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reijnders D., Goossens G.H., Hermes G.D., Neis E.P., van der Beek C.M., Most J., Holst J.J., Lenaerts K., Kootte R.S., Nieuwdorp M., Groen A.K., Olde Damink S.W., Boekschoten M.V., Smidt H., Zoetendal E.G., Dejong C.H., Blaak E.E. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab. 2016;24:341. doi: 10.1016/j.cmet.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Beli E., Yan Y., Moldovan L., Vieira C.P., Gao R., Duan Y., Prasad R., Bhatwadekar A., White F.A., Townsend S.D., Chan L., Ryan C.N., Morton D., Moldovan E.G., Chu F.-I., Oudit G.Y., Derendorf H., Adorini L., Wang X.X., Evans-Molina C., Mirmira R.G., Boulton M.E., Yoder M.C., Li Q., Levi M., Busik J.V., Grant M.B. Restructuring of the gut microbiome by intermittent fasting prevents retinopathy and prolongs survival in db/db mice. Diabetes. 2018;67:1867–1879. doi: 10.2337/db18-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G., Xie C., Lu S., Nichols R.G., Tian Y., Li L., Patel D., Ma Y., Brocker C.N., Yan T., Krausz K.W., Xiang R., Gavrilova O., Patterson A.D., Gonzalez F.J. Intermittent fasting promotes white adipose browning and decreases obesity by shaping the gut microbiota. Cell Metab. 2018;26:801. doi: 10.1016/j.cmet.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cignarella F., Cantoni C., Ghezzi L., Salter A., Dorsett Y., Chen L., Phillips D., Weinstock G.M., Fontana L., Cross A.H., Zhou Y., Piccio L. Intermittent fasting confers protection in CNS autoimmunity by altering the gut microbiota. Cell Metab. 2018;27:1222–1235. doi: 10.1016/j.cmet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarrinpar A., Chaix A., Yooseph S., Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z., Dai X., Zhang H., Shi R., Hui Y., Jin X., Zhang W., Wang L., Wang Q., Wang D., Wang J., Tan X., Ren B., Liu X., Zhao T., Wang J., Pan J., Yuan T., Chu C., Lan L., Yin F., Cadenas E., Shi L., Zhao S., Liu X. Gut microbiota mediates intermittent-fasting alleviation of diabetes-induced cognitive impairment. Nat Commun. 2020;11:855. doi: 10.1038/s41467-020-14676-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buxton O.M., Cain S.W., O’Connor S.P., Porter J.H., Duffy J.F., Wang W., Czeisler C.A., Shea S.A. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med. 2012;4:129ra43. doi: 10.1126/scitranslmed.3003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vetter C., Chang S.-C., Devore E.E., Rohrer F., Okereke O.I., Schernhammer E.S. Prospective study of chronotype and incident depression among middle- and older-aged women in the Nurses’ Health Study II. J Psychiatr Res. 2018;103:156–160. doi: 10.1016/j.jpsychires.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Kuang Z., Yu X., Ruhn K.A., Kubo M., Hooper L.V. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357:912–916. doi: 10.1126/science.aan0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z., Gan L., Zhang T., Ren Q., Sun C. Melatonin alleviates adipose inflammation through elevating α-ketoglutarate and diverting adipose-derived exosomes to macrophages in mice. J Pineal Res. 2018:64. doi: 10.1111/jpi.12455. [DOI] [PubMed] [Google Scholar]
- 22.Sato S., Solanas G., Peixoto F.O., Bee L., Symeonidi A., Schmidt M.S., Brenner C., Masri S., Benitah S.A., Sassone-Corsi P. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell. 2017;170:664–677. doi: 10.1016/j.cell.2017.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattijssen F., Alex S., Swarts H.J., Groen A.K., van Schothorst E.M., Kersten S. Angptl4 serves as an endogenous inhibitor of intestinal lipid digestion. Mol Metab. 2013;3:135–144. doi: 10.1016/j.molmet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dijk W., Heine M., Vergnes L., Boon M.R., Schaart G., Hesselink M.K.C., Reue K., van Marken Lichtenbelt W.D., Olivecrona G., Rensen P.C.N., Heeren J., Kersten S. ANGPTL4 mediates shuttling of lipid fuel to brown adipose tissue during sustained cold exposure. Elife. 2015;4 doi: 10.7554/eLife.08428. pii:08428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romeo S., Pennacchio L.A., Fu Y., Boerwinkle E., Tybjaerg-Hansen A., Hobbs H.H., Cohen J.C. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichtenstein L., Mattijssen F., de Wit N.J., Georgiadi A., Hooiveld G.J., van der Meer R., He Y., Qi L., Köster A., Tamsma J.T., Tan N.S., Müller M., Kersten S. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metab. 2010;12:580–592. doi: 10.1016/j.cmet.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catoire M., Alex S., Paraskevopulos N., Mattijssen F., Evers-van Gogh I., Schaart G., Jeppesen J., Kneppers A., Mensink M., Voshol P.J., Olivecrona G., Tan N.S., Hesselink M.K.C., Berbée J.F., Rensen P.C.N., Kalkhoven E., Schrauwen P., Kersten S. Fatty acid-inducible ANGPTL4 governs lipid metabolic response to exercise. Proc Natl Acad Sci U S A. 2014;111:E1043–E1052. doi: 10.1073/pnas.1400889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen A.W.F., Katiraei S., Bartosinska B., Eberhard D., van Dijk K.W., Kersten S. Loss of angiopoietin-like 4 (ANGPTL4) in mice with diet-induced obesity uncouples visceral obesity from glucose intolerance partly via the gut microbiota. Diabetologia. 2018;61:1447–1458. doi: 10.1007/s00125-018-4583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erren T.C., Reiter R.J., Meyer-Rochow V.B. Frog transparency led to discovery of melatonin. Nature. 2008;451:127. doi: 10.1038/451127c. [DOI] [PubMed] [Google Scholar]
- 30.Spiegel K., Leproult R., Van Cauter E. Impact of sleep debt on metabolic and endocrine function. Lancet. 1999;354:1435–1439. doi: 10.1016/S0140-6736(99)01376-8. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z., Gan L., Luo D., Sun C. Melatonin promotes circadian rhythm-induced proliferation through Clock/histone deacetylase 3/c-Myc interaction in mouse adipose tissue. J Pineal Res. 2017;62 doi: 10.1111/jpi.12383. [DOI] [PubMed] [Google Scholar]
- 32.Han L., Wang H., Li L., Li X., Ge J., Reiter R.J., Wang Q. Melatonin protects against maternal obesity-associated oxidative stress and meiotic defects in oocytes via the SIRT3-SOD2-dependent pathway. J Pineal Res. 2017;63 doi: 10.1111/jpi.12431. [DOI] [PubMed] [Google Scholar]
- 33.Lee K., Hwang O.J., Reiter R.J., Back K. Flavonoids inhibit both rice and sheep serotonin N-acetyltransferases and reduce melatonin levels in plants. J Pineal Res. 2018;65 doi: 10.1111/jpi.12512. [DOI] [PubMed] [Google Scholar]
- 34.Reiter R.J., Ma Q., Sharma R. Melatonin in mitochondria: mitigating clear and present dangers. Physiology (Bethesda) 2020;35:86–95. doi: 10.1152/physiol.00034.2019. [DOI] [PubMed] [Google Scholar]
- 35.Garaulet M., Qian J., Florez J.C., Arendt J., Saxena R., Scheer F.A.J.L. Melatonin effects on glucose metabolism: time to unlock the controversy. Trends Endocrinol Metab. 2020 doi: 10.1016/j.tem.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rong B., Feng R., Liu C., Wu Q., Sun C. Reduced delivery of epididymal adipocyte-derived exosomal resistin is essential for melatonin ameliorating hepatic steatosis in mice. J Pineal Res. 2019;66 doi: 10.1111/jpi.12561. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z., Gan L., Xu Y., Luo D., Ren Q., Wu S., Sun C. Melatonin alleviates inflammasome-induced pyroptosis through inhibiting NF-κB/GSDMD signal in mice adipose tissue. J Pineal Res. 2017;63 doi: 10.1111/jpi.12414. [DOI] [PubMed] [Google Scholar]
- 38.Mannino G., Caradonna F., Cruciata I., Lauria A., Perrone A., Gentile C. Melatonin reduces inflammatory response in human intestinal epithelial cells stimulated by interleukin-1β. J Pineal Res. 2019;67 doi: 10.1111/jpi.12598. [DOI] [PubMed] [Google Scholar]
- 39.Koubaa-Ghorbel F., Chaâbane M., Turki M., Makni-Ayadi F., El Feki A. The protective effects of Salvia officinalis essential oil compared to simvastatin against hyperlipidemia, liver, and kidney injuries in mice submitted to a high-fat diet. J Food Biochem. 2020;44 doi: 10.1111/jfbc.13160. [DOI] [PubMed] [Google Scholar]
- 40.Sun L., Xie C., Wang G., Wu Y., Wu Q., Wang X., Liu J., Deng Y., Xia J., Chen B., Zhang S., Yun C., Lian G., Zhang X., Zhang H., Bisson W.H., Shi J., Gao X., Ge P., Liu C., Krausz K.W., Nichols R.G., Cai J., Rimal B., Patterson A.D., Wang X., Gonzalez F.J., Jiang C. Gut microbiota and intestinal mediate the clinical benefits of metformin. Nat Med. 2018;24:1919–1929. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plovier H., Everard A., Druart C., Depommier C., Van Hul M., Geurts L., Chilloux J., Ottman N., Duparc T., Lichtenstein L., Myridakis A., Delzenne N.M., Klievink J., Bhattacharjee A., van der Ark K.C., Aalvink S., Martinez L.O., Dumas M.E., Maiter D., Loumaye A., Hermans M.P., Thissen J.P., Belzer C., de Vos W.M., Cani P.D. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 42.Roopchand D.E., Carmody R.N., Kuhn P., Moskal K., Rojas-Silva P., Turnbaugh P.J., Raskin I. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes. 2015;64:2847–2858. doi: 10.2337/db14-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Everard A., Plovier H., Rastelli M., Van Hul M., de Wouters d’Oplinter A., Geurts L., Druart C., Robine S., Delzenne N.M., Muccioli G.G., de Vos W.M., Luquet S., Flamand N., Di Marzo V., Cani P.D. Intestinal epithelial N-acylphosphatidylethanolamine phospholipase D links dietary fat to metabolic adaptations in obesity and steatosis. Nat Commun. 2019;10:457. doi: 10.1038/s41467-018-08051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costello E.K., Gordon J.I., Secor S.M., Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 2010;4:1375–1385. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Derrien M., Vaughan E.E., Plugge C.M., de Vos W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(Pt 5):1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 46.Warner N., Núñez G. MyD88: a critical adaptor protein in innate immunity signal transduction. J Immunol. 2013;190:3–4. doi: 10.4049/jimmunol.1203103. [DOI] [PubMed] [Google Scholar]
- 47.Tilg H., Zmora N., Adolph T.E., Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol. 2020;20:40–54. doi: 10.1038/s41577-019-0198-4. [DOI] [PubMed] [Google Scholar]
- 48.Mao K., Baptista A.P., Tamoutounour S., Zhuang L., Bouladoux N., Martins A.J., Huang Y., Gerner M.Y., Belkaid Y., Germain R.N. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 2018;554:255–259. doi: 10.1038/nature25437. [DOI] [PubMed] [Google Scholar]
- 49.Chagwedera D.N., Ang Q.Y., Bisanz J.E., Leong Y.A., Ganeshan K., Cai J., Patterson A.D., Turnbaugh P.J., Chawla A. Nutrient sensing in CD11c sells alters the gut microbiota to regulate food intake and body mass. Cell Metab. 2019;30:364–373. doi: 10.1016/j.cmet.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canfora E.E., Meex R.C.R., Venema K., Blaak E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol. 2019;15:261–273. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed] [Google Scholar]
- 51.Fabbiano S., Suárez-Zamorano N., Chevalier C., Lazarević V., Kieser S., Rigo D., Leo S., Veyrat-Durebex C., Gaïa N., Maresca M., Merkler D., de Agüero M.G., Macpherson A., Schrenzel J., Trajkovski M. Functional gut microbiota remodeling contributes to the caloric restriction-induced metabolic improvements. Cell Metab. 2018;28:907–921. doi: 10.1016/j.cmet.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mauer J., Chaurasia B., Goldau J., Vogt M.C., Ruud J., Nguyen K.D., Theurich S., Hausen A.C., Schmitz J., Brönneke H.S., Estevez E., Allen T.L., Mesaros A., Partridge L., Febbraio M.A., Chawla A., Wunderlich F.T., Brüning J.C. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat Immunol. 2014;15:423–430. doi: 10.1038/ni.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koliwad S.K., Kuo T., Shipp L.E., Gray N.E., Backhed F., So A.Y.-L., Farese R.V., Jr., Wang J.-C. Angiopoietin-like 4 (ANGPTL4, fasting-induced adipose factor) is a direct glucocorticoid receptor target and participates in glucocorticoid-regulated triglyceride metabolism. J Biol Chem. 2009;284:25593–25601. doi: 10.1074/jbc.M109.025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh A.K., Aryal B., Chaube B., Rotllan N., Varela L., Horvath T.L., Suárez Y., Fernández-Hernando C. Brown adipose tissue derived ANGPTL4 controls glucose and lipid metabolism and regulates thermogenesis. Mol Metab. 2018;11:59–69. doi: 10.1016/j.molmet.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dijk W., Kersten S. Regulation of lipid metabolism by angiopoietin-like proteins. Curr Opin Lipidol. 2016;27:249–256. doi: 10.1097/MOL.0000000000000290. [DOI] [PubMed] [Google Scholar]
- 56.Kroupa O., Vorrsjö E., Stienstra R., Mattijssen F., Nilsson S.K., Sukonina V., Kersten S., Olivecrona G., Olivecrona T. Linking nutritional regulation of Angptl4, Gpihbp1, and Lmf1 to lipoprotein lipase activity in rodent adipose tissue. BMC Physiol. 2012;12:13. doi: 10.1186/1472-6793-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jahn D., Dorbath D., Schilling A.-K., Gildein L., Meier C., Vuille-Dit-Bille R.N., Schmitt J., Kraus D., Fleet J.C., Hermanns H.M., Geier A. Intestinal vitamin D receptor modulates lipid metabolism, adipose tissue inflammation and liver steatosis in obese mice. Biochim Biophys Acta Mol Basis Dis. 2019;1865:1567–1578. doi: 10.1016/j.bbadis.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 58.Myocardial Infarction Genetics, CARDIoGRAM, Exome Consortia Investigators Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med. 2016;374:1134–1144. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashizume M., Yoshida H., Koike N., Suzuki M., Mihara M. Overproduced interleukin 6 decreases blood lipid levels via upregulation of very-low-density lipoprotein receptor. Ann Rheum Dis. 2010;69:741–746. doi: 10.1136/ard.2008.104844. [DOI] [PubMed] [Google Scholar]
- 60.Gusarova V., O’Dushlaine C., Teslovich T.M., Benotti P.N., Mirshahi T., Gottesman O., Van Hout C.V., Murray M.F., Mahajan A., Nielsen J.B., Fritsche L., Wulff A.B., Gudbjartsson D.F., Sjögren M., Emdin C.A., Scott R.A., Lee W.-J., Small A., Kwee L.C., Dwivedi O.P., Prasad R.B., Bruse S., Lopez A.E., Penn J., Marcketta A., Leader J.B., Still C.D., Kirchner H.L., Mirshahi U.L., Wardeh A.H., Hartle, Habegger L., Fetterolf S.N., Tusie-Luna T., Morris A.P., Holm H., Steinthorsdottir V., Sulem P., Thorsteinsdottir U., Rotter J.I., Chuang L.-M., Damrauer S., Birtwell D., Brummett C.M., Khera A.V., Natarajan P., Orho-Melander M., Flannick J., Lotta L.A., Willer C.J., Holmen O.L., Ritchie M.D., Ledbetter D.H., Murphy A.J., Borecki I.B., Reid J.G., Overton J.D., Hansson O., Groop L., Shah S.H., Kraus W.E., Rader D.J., Chen Y.-D.I., Hveem K., Wareham N.J., Kathiresan S., Melander O., Stefansson K., Nordestgaard B.G., Tybjærg-Hansen A., Abecasis G.R., Altshuler D., Florez J.C., Boehnke M., McCarthy M.I., Yancopoulos G.D., Carey D.J., Shuldiner A.R., Baras A., Dewey F.E., Gromada J. Genetic inactivation of ANGPTL4 improves glucose homeostasis and is associated with reduced risk of diabetes. Nat Commun. 2018;9:2252. doi: 10.1038/s41467-018-04611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingerslev B., Hansen J.S., Hoffmann C., Clemmesen J.O., Secher N.H., Scheler M., de Angelis M.H., Häring H.U., Pedersen B.K., Weigert C., Plomgaard P. Angiopoietin-like protein 4 is an exercise-induced hepatokine in humans, regulated by glucagon and cAMP. Mol Metab. 2017;6:1286–1295. doi: 10.1016/j.molmet.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dewey F.E., Gusarova V., O’Dushlaine C., Gottesman O., Trejos J., Hunt C., Van Hout C.V., Habegger L., Buckler D., Lai K.M., Leader J.B., Murray M.F., Ritchie M.D., Kirchner H.L., Ledbetter D.H., Penn J., Lopez A., Borecki I.B., Overton J.D., Reid J.G., Carey D.J., Murphy A.J., Yancopoulos G.D., Baras A., Gromada J., Shuldiner A.R. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl J Med. 2016;374:1123–1133. doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruscica M., Zimetti F., Adorni M.P., Sirtori C.R., Lupo M.G., Ferri N. Pharmacological aspects of ANGPTL3 and ANGPTL4 inhibitors: new therapeutic approaches for the treatment of atherogenic dyslipidemia. Pharmacol Res. 2020;153:104653. doi: 10.1016/j.phrs.2020.104653. [DOI] [PubMed] [Google Scholar]
- 64.Kristensen K.K., Leth-Espensen K.Z., Mertens H.D.T., Birrane G., Meiyappan M., Olivecrona G., Jørgensen T.J.D., Young S.G., Ploug M. Unfolding of monomeric lipoprotein lipase by ANGPTL4: insight into the regulation of plasma triglyceride metabolism. Proc Natl Acad Sci U S A. 2020;117:4337–4346. doi: 10.1073/pnas.1920202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emiola A., Andrews S.S., Heller C., George J. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc Natl Acad Sci U S A. 2016;113:3108–3113. doi: 10.1073/pnas.1521168113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.May K.L., Silhavy T.J. The escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. mBio. 2018;9 doi: 10.1128/mBio.00379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belzer C., de Vos W.M. Microbes inside: from diversity to function—the case of Akkermansia. ISME J. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emiola A., Andrews S.S., Heller C., George J. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mithieux G. Does Akkermansia muciniphila play a role in type 1 diabetes? Gut. 2018;67:1373–1374. doi: 10.1136/gutjnl-2017-315732. [DOI] [PubMed] [Google Scholar]
- 70.Thingholm L.B., Rühlemann M.C., Koch M., Fuqua B., Laucke G., Boehm R., Bang C., Franzosa E.A., Hübenthal M., Rahnavard A., Frost F., Lloyd-Price J., Schirmer M., Lusis A.J., Vulpe C.D., Lerch M.M., Homuth G., Kacprowski T., Schmidt C.O., Nöthlings U., Karlsen T.H., Lieb W., Laudes M., Franke A., Huttenhower C. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe. 2019;26:252–264. doi: 10.1016/j.chom.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shao T., Zhao C., Li F., Gu Z., Liu L., Zhang L., Wang Y., He L., Liu Y., Liu Q., Chen Y., Donde H., Wang R., Jala V.R., Barve S., Chen S.-Y., Zhang X., Chen Y., McClain C.J., Feng W. Intestinal HIF-1α deletion exacerbates alcoholic liver disease by inducing intestinal dysbiosis and barrier dysfunction. J Hepatol. 2018;69:886–895. doi: 10.1016/j.jhep.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shin N.-R., Lee J.-C., Lee H.-Y., Kim M.-S., Whon T.W., Lee M.-S., Bae J.-W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 73.Shen J., Tong X., Sud N., Khound R., Song Y., Maldonado-Gomez M.X., Walter J., Su Q. Low-density lipoprotein receptor signaling mediates the triglyceride-lowering action of Akkermansia muciniphila in genetic-induced hyperlipidemia. Arterioscler Thromb Vasc Biol. 2016;36:1448–1456. doi: 10.1161/ATVBAHA.116.307597. [DOI] [PubMed] [Google Scholar]
- 74.Giannoudaki E., Hernandez-Santana Y.E., Mulfaul K., Doyle S.L., Hams E., Fallon P.G., Mat A., O’Shea D., Kopf M., Hogan A.E., Walsh P.T. Interleukin-36 cytokines alter the intestinal microbiome and can protect against obesity and metabolic dysfunction. Nat Commun. 2019;10:4003. doi: 10.1038/s41467-019-11944-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Depommier C., Everard A., Druart C., Plovier H., Van Hul M., Vieira-Silva S., Falony G., Raes J., Maiter D., Delzenne N.M., de Barsy M., Loumaye A., Hermans M.P., Thissen J.-P., de Vos W.M., Cani P.D. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q.A., Tao C., Gupta R.K., Scherer P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu E., Pereira M.M.A., Karakasilioti I., Theurich S., Al-Maarri M., Rappl G., Waisman A., Wunderlich F.T., Brüning J.C. Temporal and tissue-specific requirements for T-lymphocyte IL-6 signalling in obesity-associated inflammation and insulin resistance. Nat Commun. 2017;8:14803. doi: 10.1038/ncomms14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sato C., Shikata K., Hirota D., Sasaki M., Nishishita S., Miyamoto S., Kodera R., Ogawa D., Tone A., Kataoka H.U., Wada J., Kajitani N., Makino H. P-selectin glycoprotein ligand-1 deficiency is protective against obesity-related insulin resistance. Diabetes. 2011;60:189–199. doi: 10.2337/db09-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J., Kwon E.-Y., Park S., Kim J.-R., Choi S.-W., Choi M.-S., Kim S.-J. Integrative systems analysis of diet-induced obesity identified a critical transition in the transcriptomes of the murine liver and epididymal white adipose tissue. Int J Obes (Lond) 2016;40:338–345. doi: 10.1038/ijo.2015.147. [DOI] [PubMed] [Google Scholar]
- 80.de Farias TdSM., de Oliveira A.C., Andreotti S., do Amaral F.G., Chimin P., de Proença A.R.A., Leal F.L.T., Sertié R.A.L., Campana A.B., Lopes A.B., de Souza A.H., Cipolla-Neto J., Lima F.B. Pinealectomy interferes with the circadian clock genes expression in white adipose tissue. J Pineal Res. 2015;58:251–261. doi: 10.1111/jpi.12211. [DOI] [PubMed] [Google Scholar]
- 81.Kim D.-H., Sartor M.A., Bain J.R., Sandoval D., Stevens R.D., Medvedovic M., Newgard C.B., Woods S.C., Seeley R.J. Rapid and weight-independent improvement of glucose tolerance induced by a peptide designed to elicit apoptosis in adipose tissue endothelium. Diabetes. 2012;61:2299–2310. doi: 10.2337/db11-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thaiss C.A., Levy M., Korem T., Dohnalová L., Shapiro H., Jaitin D.A., David E., Winter D.R., Gury-BenAri M., Tatirovsky E., Tuganbaev T., Federici S., Zmora N., Zeevi D., Dori-Bachash M., Pevsner-Fischer M., KartvelishvilyE, Brandis A., Harmelin A., Shibolet O., Halpern Z., Honda K., Amit I., Segal E., Elinav E. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 83.Yin J., Li Y., Han H., Ma J., Liu G., Wu X., Huang X., Fang R., Baba K., Bin P., Zhu G., Ren W., Tan B., Tosini G., He X., Li T., Yin Y. Administration of exogenous melatonin improves the diurnal rhythms of the gut microbiota in mice fed a high-fat diet. mSystems. 2020;5:e00002–20. doi: 10.1128/mSystems.00002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yin J., Li Y., Han H., Chen S., Gao J., Liu G., Wu X., Deng J., Yu Q., Huang X., Fang R., Li T., Reiter R.J., Zhang D., Zhu C., Zhu G., Ren W., Yin Y. Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. J Pineal Res. 2018;65:e12524. doi: 10.1111/jpi.12524. [DOI] [PubMed] [Google Scholar]
- 85.Chen L.-W., Chang W.-J., Chen P.-H., Hsu C.-M. Commensal microflora induce host defense and decrease bacterial translocation in burn mice through toll-like receptor 4. J Biomed Sci. 2010;17:48. doi: 10.1186/1423-0127-17-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 87.Lin M., Yiu W.H., Li R.X., Wu H.J., Wong D.W.L., Chan L.Y.Y., Leung J.C.K., Lai K.N., Tang S.C.W. The TLR4 antagonist CRX-526 protects against advanced diabetic nephropathy. Kidney Int. 2013;83:887–900. doi: 10.1038/ki.2013.11. [DOI] [PubMed] [Google Scholar]
- 88.Wu X., Wu J., Chen T., Li Q., Peng W., Li H., Tang X., Fu X. Fusobacterium nucleatum potentiates intestinal tumorigenesis in mice via a toll-like receptor 4/p21-activated kinase 1 cascade. Dig Dis Sci. 2018;63:1210–1218. doi: 10.1007/s10620-018-4999-2. [DOI] [PubMed] [Google Scholar]
- 89.Schmidt V.M., Pinchbeck G., McIntyre K.M., Nuttall T., McEwan N., Dawson S., Williams N.J. Routine antibiotic therapy in dogs increases the detection of antimicrobial-resistant faecal Escherichia coli. J Antimicrob Chemother. 2018;73:3305–3316. doi: 10.1093/jac/dky352. [DOI] [PubMed] [Google Scholar]
- 90.Velazquez E.M., Nguyen H., Heasley K.T., Saechao C.H., Gil L.M., Rogers A.W.L., Miller B.M., Rolston M.R., Lopez C.A., Litvak Y., Liou M.J., Faber F., Bronner D.N., Tiffany C.R., Byndloss M.X., Byndloss A.J., Bäumler A.J. Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat Microbiol. 2019;4:1057–1064. doi: 10.1038/s41564-019-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lindemans C.A., Calafiore M., Mertelsmann A.M., O’Connor M.H., Dudakov J.A., Jenq R.R., Velardi E., Young L.F., Smith O.M., Lawrence G., Ivanov J.A., Fu Y.-Y., Takashima S., Hua G., Martin M.L., O’Rourke K.P., Lo Y.-H., Mokry M., Romera-Hernandez M., Cupedo T., Dow L., Nieuwenhuis E.E., Shroyer N.F., Liu C., Kolesnick R., van den Brink M.R.M., Hanash A.M. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao L., Wu J., Wang J.-Y., Chung H.K., Kalakonda S., Rao J.N., Gorospe M., Wang J.-Y. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology. 2018;154:599–611. doi: 10.1053/j.gastro.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]