Abstract
Epigenome-wide studies report higher methylation among women than men with decreasing levels with age. Little is known about associations of sex and age with methylation of monoamine oxidase A (MAOA). Methylation of the first exonic and partial first intronic region of MAOA has been shown to strengthen associations of interactions of MAOA-uVNTR genotypes and adversity with aggression and substance misuse. Our study examined associations of sex and age with MAOA first exon and intron methylation levels in 252 women and 157 men aged 14–73 years. Participants included adolescents recruited at a substance misuse clinic, their siblings and parents, and healthy women. Women showed ~ 50% higher levels of exonic, and ~ 15% higher intronic, methylation than men. Methylation levels were similar between younger (M = 22.7 years) and older (M = 46.1 years) participants, and stable across age. Age modified few associations of methylation levels with sex. MAOA genotypes modified few associations of methylation with sex and age. Higher methylation levels among women were not explained by genotype, nor interaction of genotype and sexual abuse. Findings were similar after adjusting for lifetime diagnoses of substance dependence (women = 24.3%; men = 34.2%). Methylation levels were higher among women who experienced sexual abuse than women who did not. Results extend on prior studies by showing that women display higher levels of methylation than men within first intronic/exonic regions of MAOA, which did not decrease with age in either sex. Findings were not conditioned by genotype nor interactions of genotype and trauma, and indicate X-chromosome inactivation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00702-021-02403-2.
Keywords: Epigenetics, Methylation, Age, Sex differences, Child abuse, Gene–environment interaction
Introduction
Methylation of DNA at cytosine–guanine dinucleotides (CpGs) across the genome is a dynamic biological process influenced by environmental and biological factors, that in turn impacts gene activity and the subsequent regulation of biological systems (Xiao et al. 2018; Pagiatakis et al. 2019). Together, these factors contribute to the high degree of inter-individual variation in CpG methylation across the genome associated with mental and physical disorders (Han et al. 2019; Pagiatakis et al. 2019). However, recent reviews of studies of methylation levels characterizing individuals with various disorders report inconsistent findings. These inconsistencies may result from inadequate control for, or examination of sex- and age-related variations in methylation (Barnett Burns et al. 2018; Brown et al. 2019; Fransquet et al. 2019; Gegenhuber and Tollkuhn 2019; Han et al. 2019). Furthermore, little is known about the associations of sex and age with genes on the X-chromosome, for example monoamine oxidase A (MAOA), that has been excluded from some epigenome-wide methylation studies (Chen et al. 2020), even though MAOA plays a major role early in life by setting life-long levels of brain monoamines (Meyer-Lindenberg et al. 2006; Oreland et al. 2007; Booij et al. 2015). Extant evidence also implicates gene-by-environment interactions of a functional variable nucleotide polymorphism in the promoter of MAOA (MAOA-uVNTR) and adversity in modifying risk for negative behavioural outcomes, principally antisocial behaviour, and mental disorders such as depression (Byrd and Manuck 2014; Booij et al. 2015; Nilsson et al. 2018). Polymorphisms of MAOA have also been implicated in treatment response to mirtazapine among individuals with depression (Tzeng et al. 2009), further highlighting the gene’s clinical importance to understanding etiology and treatment.
Sex and DNA methylation
Females typically show higher methylation levels than males in genome-wide studies, although the magnitude and direction of sex differences can vary greatly at specific genomic regions (Nugent and McCarthy 2011). Accordingly, sex differences in methylation have been identified in genes contributing to regulation of biological pathways involved in sex differentiation, endocrine function, and neurodevelopment, in genes that regulate epigenetic processes and mechanisms, and in genes regulating aging (Nugent and McCarthy 2011; Yousefi et al. 2015; Kader and Ghai 2016; Van Dongen et al. 2016; Ratnu et al. 2017; Suderman et al. 2017). Only one study has examined sex differences in methylation of MAOA: The sample was small, 23 of the 66 participants presented with depression, and women showed higher methylation levels than men within the exonic region (Melas and Forsell 2015). Similar levels of MAOA first exon methylation in women, ranging between 40 and 50%, have been reported (Ziegler and Domschke 2018).
Sex differences in methylation have been observed among individuals presenting with mental or physical disorders (Ratnu et al. 2017). Several mental disorders are characterized by alterations of methylation of the MAOA gene including depression, post-traumatic stress disorder, substance abuse disorders, antisocial personality disorder, conduct disorder, borderline personality disorder, and schizophrenia (Ziegler and Domschke 2018), and by different functional variable number tandem repeat (uVNTR) variants of MAOA in males and females (reviewed in Veroude et al. 2016). As the gene sits on the X-chromosome, sex differences in methylation levels may result from X-chromosome inactivation, an epigenetic process whereby one of the two X-chromosomes carried by females is silenced. Inactivation serves as a dosage compensation mechanism in females, relative to males who carry only one X-chromosome (Cotton et al. 2011). X-inactivation is also linked to the exacerbation of numerous sexually dimorphic phenotypes (Carrel and Willard 2005) ranging from cell structure (Garieri et al. 2018) to disease susceptibility (Schurz et al. 2019) and has been hypothesized to play a role in sexually dimorphic behaviors (de Almeida et al. 2015). Notably, women also show greater variability in X-linked gene expression than men (Peeters et al. 2014). This variability may be partly attributable to the approximately 15% of X-linked genes that partially, or fully, escape inactivation (Carrel and Willard 2005) which also varies greatly by cell type (Garieri et al. 2018). As such, sex differences in methylation, particularly among X-linked genes, are key considerations for furthering understanding of links with environmental events, treatment response, fundamental biological processes, and how the epigenome itself functions (Ratnu et al. 2017).
Aging and methylation
Sex differences in methylation levels across the genome are observable in new-borns (Yousefi et al. 2015), remain stable through childhood (Suderman et al. 2017), and are observed in adulthood (Ratnu et al. 2017). There is a global reduction of methylation across the genome with increasing age, such that cumulative inter-individual differences are harmonized in a process called epigenetic assimilation, although methylation levels in some regions do not undergo this reduction (Ciccarone et al. 2018). This global reduction of methylation levels over the lifespan is not strongly associated with changes in gene expression (Ciccarone et al. 2018). It has been hypothesized that regulation of gene expression during this period of lowering methylation may be gradually taken up and maintained by other epigenetic processes over time, although these mechanisms are not currently well understood (Ashapkin et al. 2017; Ciccarone et al. 2018; Pagiatakis et al. 2019; Xiao et al. 2019; Chen et al. 2020). Disturbances in the trajectory of epigenetic aging are associated with health risks across the lifespan, and may be influenced by environmental factors encountered as early as pre- and post-natal life (Ciccarone et al. 2018; Chen et al. 2020). Importantly, despite the pattern of genome-wide hypo-methylation with increasing age, age-related fluctuations in DNA methylation may remain more variable and susceptible to environmental influences within particular genomic loci, including regions that span first exonic/intronic boundaries of genes, and are thus regions of particular interest in health-related outcomes (Ashapkin et al. 2017; Ciccarone et al. 2018).
Disturbances to the trajectory of epigenetic aging processes have been implicated in risk for mental and physical illnesses (Miller et al. 2015; Ciccarone et al. 2018; Vaiserman 2018), influenced by environmental factors (Ciccarone et al. 2018; Vaiserman 2018), and may differ by sex (Van Dongen et al. 2016). One study showed that among children in families with low socioeconomic status, those exposed to positive environmental factors showed greater self-control that in turn predicted less aggressive behavior, fewer depressive symptoms, lower rates of substance use, and fewer externalizing problems, increased academic success, and better psychosocial adjustment in young adulthood (Miller et al. 2015). Exposure to positive factors, however, did not protect against accelerated epigenetic aging, nor poor cardio-metabolic health indicators that predict medical illnesses later in life (Miller et al. 2015). The authors of this study suggested a potential “skin deep” resilience, such that observed protective associations with outcomes that begin in late-adolescence and early adulthood mask an increased risk for other negative outcomes that may emerge over protracted periods of time (Miller et al. 2015). The impact of environmental factors present in prenatal and early life on the re-programming of epigenetic aging processes across the lifespan is further supported by twin and non-human animal studies, and is associated with alterations of metabolic functions that underlie a myriad of health conditions that emerge throughout the lifespan (Vaiserman 2018). A recent study of twins found that genetic factors were strongly associated with stability of methylation levels over time, while novel experiences and exposure to environmental factors accounted for fluctuations in methylation levels (Reynolds et al. 2020). In turn, age is increasingly being recognized as an important consideration in epigenetic studies as mental and physical disorders that emerge over time may be associated with aberrant epigenetic aging processes initiated by environmental factors acting in early life (Miller et al. 2015; Ciccarone et al. 2018; Pagiatakis et al. 2019).
Sex and aging
A recent systematic review and meta-analysis of DNA methylation studies of aging and age-related physical disorders and mortality suggested that the inconsistencies in findings across studies were partly attributable to insufficient statistical control for sex and bias introduced by sampling only males both in human and animal studies (Fransquet et al. 2019). One study found that the interaction of age and sex was associated with methylation levels across the genome, especially in regions associated with key health indicators such as metabolic traits and smoking (Van Dongen et al. 2016). Another recent study found that the majority of sex differences in methylation associated with age were in X-linked genes implicated in sexually dimorphic disorders and traits, including prostate cancer and male pattern baldness (McCartney et al. 2019). Another recent review highlighted that sex differences in age-related methylation may differ widely by genomic region and by tissue type, particularly in the brain (Unnikrishnan et al. 2019). For instance, one study in mice and humans showed that genome-wide hippocampal methylation levels did not differ by sex in early life, but that 95% of age-related changes in methylation levels observed later in life were sexually divergent and particular to specific genomic loci (Masser et al. 2017). Prior methylation studies of persons presenting with mental disorders have often been conducted in tissue samples from post-mortem hippocampal tissues (Barnett Burns et al. 2018). Further understanding of the role of methylation in mental disorders using peripheral tissue samples from afflicted individuals is needed (Szyf 2015; Barnett Burns et al. 2018; Brown et al. 2019). Yet, such studies are presently hindered by the lack of knowledge of sex and age differences across the genome and in candidate genes.
The current understanding of sex, age, and methylation derives primarily from genome-wide studies (Ciccarone et al. 2018; Vaiserman 2018; Chen et al. 2020). Candidate gene studies including regions of interest, such as exonic/intronic junctions, that may be subject to greater fluctuation in methylation across the lifespan have the potential to extend understanding of how environmental and biological factors alter risk for physical and mental health. For example, a recent study found little association of epigenome-wide methylation and aggressive behaviour in general population samples, and none in a sample of adolescents with ADHD nor in a sample of females presenting conduct disorder (van Dongen et al. 2021). By contrast, a recent study from our group found that among men, methylation levels of a region-of-interest (ROI) spanning the MAOA first exon were associated with aggressive behaviour (Checknita et al. 2020).
Furthermore, epigenetic studies of candidate genes have the potential to further specify whether associations with sex and age are modified by genotype, and by interactions of genotype and environmental factors as has been reported in epigenome studies (Van Dongen et al. 2016). Surprisingly, one recent study of a large cohort found no direct association of childhood adversity with genome-wide methylation or methylation of six candidate genes (not including MAOA). The authors hypothesized that other factors, including genotype, may increase vulnerability of genes to epigenetic changes resulting from maltreatment (Marzi et al. 2018). One recent twin study reported that regions across the genome showing enrichments of DNA methylation associated with environmental factors were also strongly influenced by additive genetic factors (Hannon et al. 2018). Furthermore, genome-wide and gene-specific associations between environmental factors, genotypes, and epigenetic processes have been identified in risk for numerous physical illnesses such as cancer, diabetes, and neurodegenerative disorders (Romanowska and Joshi 2019). A recent systematic review of associations of DNA methylation and brain structure and function across the lifespan among individuals presenting with neurodegenerative or mental disorders noted the importance of controlling for, or examining, the impact of genotype (Wheater et al. 2020).
Methylation of MAOA
Two recent studies from our group found that, among men, methylation levels of a ROI spanning a portion of the MAOA core promoter containing the gene’s first exonic and partial first intronic regions further modified the association of the interaction of maltreatment and MAOA-uVNTR genotypes with alcohol consumption (Bendre et al. 2018) and with aggressive behavior (Checknita et al. 2020). Similarly, another recent study found that among women who had experienced adversity in childhood genotype-specific methylation profiles of FKBP5 intron 7 were associated with Post Traumatic Stress Disorder symptom severity (Grasso et al. 2020). Furthermore, recent studies suggest that promoter sequence variants can alter the binding affinity of methyl groups to DNA in upstream regions, which in turn alters the susceptibility of genes to be epigenetically modified by environmental factors, offering one potential mechanism through which genotype and methylation can interact (Ahsan et al. 2017; Ek et al. 2018).
The MAOA gene, located on the X-chromosome (Xp11.3), encodes the monoamine oxidase A (MAO-A) enzyme which metabolizes serotonin (and other aminergic transmitters) following reuptake. Aberrant activity of MAOA has been posited as a critical factor involved in system-wide aminergic dysregulation that, in turn, contributes to the development of multiple negative outcomes including increased aggressive behavior, substance misuse, and mental disorders such as depression (Meyer-Lindenberg et al. 2006; Byrd and Manuck 2014; Booij et al. 2015; Nilsson et al. 2018; Ziegler and Domschke 2018). The promoter region of MAOA contains a functional 30 bp variable tandem number repeats polymorphism (MAOA-uVNTR) that includes short 2–3 repeat low-expressing variants (MAOA-S), and long 3.5–5 repeat high-expressing variants (MAOA-L) (Beach et al. 2010). Direct associations of MAOA genotypes with negative outcomes are not well supported. Rather, the interactions of sex-specific genotypes (in males MAOA-S, in females MAOA-L) with exposure to negative environmental factors in childhood are associated with increased risk of negative outcomes such as Antisocial Personality Disorder and antisocial behavior, criminality, aggressive behavior, and substance misuse (Caspi et al. 2002; Sjöberg et al. 2007; Beach et al. 2010; Wakschlag et al. 2011; Åslund et al. 2011; Byrd and Manuck 2014; Tiihonen et al. 2014; Nilsson et al. 2018). The importance of this robust finding is underlined by the large number of individuals carrying these susceptibility alleles. In Caucasian populations, approximately one-third of males (Caspi et al. 2002) and at least two-thirds of females carry MAOA susceptibility alleles (Åslund et al. 2011). Recent meta-analyses and reviews note, however, that while associations of negative outcomes with interactions of MAOA and childhood trauma are robust, there are notable discrepancies in results. The discrepancies may be accounted for, at least in part, by failure to take account of epigenetic processes that modify gene expression (Nordquist and Oreland 2010; Booij et al. 2015; Ziegler and Domschke 2018).
Altered DNA methylation in an MAOA region of interest spanning the first exonic and partial first intronic regions, among men and women, has been associated with mental disorders including anxiety disorders, depression, substance abuse disorders, post-traumatic stress disorder, antisocial personality disorder, borderline personality disorder, and schizophrenia (Melas and Forsell 2015; Checknita et al. 2015, 2018, 2020; Bendre et al. 2018; Ziegler and Domschke 2018). Notably, the direction of methylation alterations (hyper- or hypo-methylation) associated with these conditions varies and differs by proposed precipitating factors such as exposure to adversity and treatment (Bendre et al. 2018; Checknita et al. 2018, 2020; Ziegler and Domschke 2018). Thus, extant evidence suggests that methylation of the MAOA ROI may vary across different disorders and be highly dynamic in response to environmental factors (Ziegler and Domschke 2018).
The present study
The study aimed to: (1) characterize methylation levels in MAOA ROI CpGs by age, sex, and by the interaction of age and sex, adjusting for genotype and substance dependence; and (2) determine whether MAOA ROI methylation levels were associated with the interaction of MAOA-uVNTR genotype, sex, and sexual abuse among males and/or females. Methylation in the MAOA ROI was assessed in five ways: individual CpGs; mean levels across the ROI; mean levels within intronic and exonic regions, since promotor and first exon methylation associates with suppressed gene expression, while intronic and gene body methylation associates with elevated gene expression (Brenet et al. 2011; Jones 2012; Moore et al. 2013); empirically derived components of CpG methylation levels (since we have previously found that these components vary by sex Bendre et al. 2018; Checknita et al. 2020). Participants were 252 women, mean age 33.2 (SD = 11.98) years, range 15–62 years, and 157 men, mean age 36.3 (SD = 14.48) years, range 14–73 years at the time of saliva collection for DNA extraction. Only 23 participants were less than 18 years old. Most of the sample was recruited at an outpatient clinic for adolescents misusing substances, and included former clients, their siblings, and parents. Given the nature of the sample, analyses were undertaken to determine whether substance dependence affected associations of sex and age with methylation levels of the MAOA ROI. Furthermore, analyses of the associations of age and sex with methylation were adjusted for several factors previously related to MAOA, including depression and anxiety disorders (Booij et al. 2015; Nilsson et al. 2018; Ziegler and Domschke 2018; Checknita et al. 2020), and impulsivity that characterizes individuals with antisocial disorders (Pavlov et al. 2012; Comai et al. 2012; Booij et al. 2015). Among males only, analyses were also adjusted for tobacco use that has been shown to impact methylation levels (Philibert et al. 2010).
Methylation was measured in DNA extracted from saliva. Knowledge of MAOA ROI methylation derives from studies sampling both blood (Philibert et al. 2008; Checknita et al. 2015; Ziegler et al. 2016) and saliva (Melas et al. 2013; Melas and Forsell 2015; Bendre et al. 2018; Checknita et al. 2018, 2020). The concordance of MAOA ROI methylation levels across peripheral tissues and with central tissue has not yet been exhaustively assessed. Methylation of the MAOA ROI in blood has also been reported to be inversely correlated with MAO enzymatic activity in the brain of adult men (Shumay et al. 2012). As such, evidence supports the use of saliva to measure methylation of the MAOA ROI.
We previously found that MAOA-uVNTR and sexual abuse, but not their interaction, were each associated with higher methylation of the MAOA ROI and current diagnosis of depression in a smaller sample of the young women included in the present study (Checknita et al. 2020). The association of sexual abuse with methylation of the MAOA ROI was robust to adjustment for psychoactive medication, alcohol and drug dependence, and current substance use. Given our previous findings (Bendre et al. 2018; Checknita et al. 2020) and current hypotheses that vulnerability to epigenetic changes varies by genotype (Bendre et al. 2018; Checknita et al. 2020), the associations of sex and age with methylation of the MAOA ROI were adjusted for genotype. Genetic polymorphisms may modify the binding affinity of methyl groups to DNA in regions up to 5000 bp downstream (Ahsan et al. 2017; Ek et al. 2018), thus contributing to the facilitation or attenuation of epigenetic modifications to genes. Furthermore, evidence from epigenome studies showed that associations of methylation with sex and age varied by region depending on genotype and environmental factors (Van Dongen et al. 2016).
Method
Participants
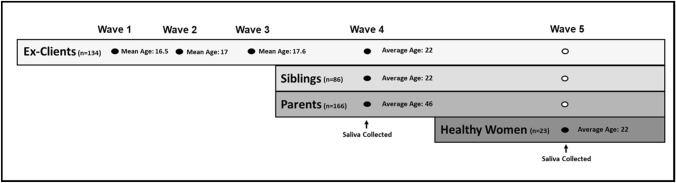
The sample included 409 participants: 134 had been recruited when they were adolescents seeking treatment for substance misuse; 86 of their siblings; 103 of their mothers; 63 of their fathers; and 23 healthy women of similar age and birthplace as the ex-clients. The 134 ex-clients completed structured, validated, diagnostic interviews and questionnaires to report on substance misuse at first contact with the clinic and 6, 12, and 60 months later. At the 60 month follow-up, the siblings and parents completed similar assessments, and all 409 participants provided saliva samples for DNA extraction. At the 75 month follow-up, healthy female participants were recruited, completed assessments similar to those completed by the other participants, and provided saliva for DNA extraction. The study timeline is summarized in Fig. 1. Age was calculated as the chronological age at the time of saliva collection. Sex was coded as male or female. DNA was extracted with a standard in-silica based method from saliva samples collected with the Oragene Self-Collection Kit (DNA Genotek Inc. Ottawa, Ontario, Canada) according to the manufacturer’s guidelines. Additional details about the processing of samples are outlined in Supplementary Material.
Fig. 1.
Study timeline. At waves 1 through 3, male and female ex-clients recruited in adolescence completed interviews and questionnaires. At wave 4, both ex-clients, siblings, and their parents completed interviews and questionnaires and provided saliva samples for DNA extraction. At wave 5, healthy women matched on age to the other women completed interviews and questionnaires and provided saliva samples for DNA extraction. Filled in dots represent time-points at which data were used in the current study and empty dots represent time-points at which data were not used in the current study to ensure consistency between the timing of molecular and clinical observations
Measures
Genotyping and methylation procedures were performed in a blinded manner and included samples from all waves and study subgroups to mitigate potential batch effects.
Methylation of the MAOA ROI
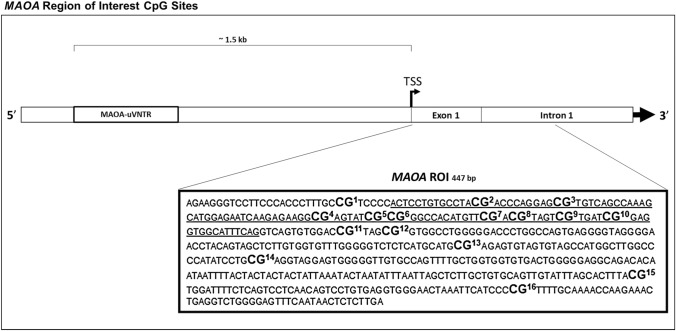
Methylation analysis targeted a previously characterized 448 bp ROI (hg19 chrX: 43,515,544—43,515,991) comprised of 16 CpGs spanning the first exon and part of the first intronic region of MAOA (Shumay et al. 2012; Checknita et al. 2015). Genomic DNA extracted from saliva was first bisulfite-treated using EZ DNA Methylation™ Kit (Zymo Research Corporation, Irvine, California) and then assayed using Sequenom’s EpiTYPER at Karolinska Institutet’s Mutation Analysis Core Facility (MAF). Resulting data represented the percentage of methylation at each CpG to the nearest 0.5%. The 16 CpGs were denoted numerically based on their 5´–3´ position within the ROI based on the forward strand genomic sequence. To optimize technical outcomes, an amplicon designed on the reverse strand covered CpGs 1–13 and another amplicon covering CpGs 13–16 was designed on the forward strand. In addition to the 16 individual CpGs, variables for mean levels of methylation for genomic features (exonic CpGs 2–10, and intronic CpGs 11–16), and overall ROI (all CpGs) were calculated. The MAOA ROI genomic sequence is provided in Fig. 2.
Fig. 2.
Genomic sequence of the MAOA ROI with the 16 CpGs included (numbered) and coding region (underlined) relative to its genomic location relative to the MAOA-uVNTR polymorphism, transcription start site (TSS), and first exonic and intronic regions of the gene. Genomic locations illustrated are scaled for interpretive ease
Genotyping of the MAOA-uVNTR
Genotyping was performed using a standard PCR technique, followed by gel electrophoresis. The target 30-bp repeat target region of MAOA (MAOA-uVNTR) was amplified using forward primer 5' ACA GCC TGA CCG TGG AGA AG 3' and reverse primer 5' GAA CGG ACG CTC CAT TCG GA 3' (Sabol et al. 1998). In accordance with prior in-vitro functional analyses of the MAOA-uVNTR (Beach et al. 2010), the three repeat variants were denoted as the short (MAOA-S) allele, and 3.5, 4, or 5 repeat variants as the long (MAOA-L) allele in men. In women, MAOA genotype was denoted as MAOA-SS or MAOA-LL in homozygous women and MAOA-SL in heterozygous women. Hardy–Weinberg Equilibrium for MAOA-uVNTR genotype was confirmed using an X2 test among women (p = 0.29) but not men since distribution of genotype is the same as the allelic distribution. Furthermore, the allelic frequencies were similar to those reported in other studies including samples of Swedish men and women (Nilsson et al. 2006; Sjoberg et al. 2007; Åslund et al. 2011).
Lifetime history of mental disorders
Clinicians administered the Structured Clinical Interview for DSM-IV (SCID I) (First et al. 2002) to all participants at the time of saliva collection and, to most, also at baseline. Lifetime diagnoses of alcohol and/or drug dependence were acquired by 24.3% (n = 61) women and 34.2% (n = 53) men (X2(2,N = 406) = 4.64 p = 0.040). Lifetime diagnoses of depression disorders (major depression disorder, dysthymia, depression disorder not-otherwise-specified, or substance-induced mood disorder) were observed in 58.2% (n = 146) women and 42.6% (n = 66) men (X2(2,N = 406) = 9.33 p = 0.040). Lifetime diagnoses of anxiety disorders (agoraphobia, generalized anxiety disorder, anxiety disorder not-otherwise-specified, obsessive compulsive disorder, panic disorder, post-traumatic stress disorder, social phobia, specific phobia, or substance-induced anxiety disorder) were observed in 48.2% (n = 121) women and 31.0% (n = 48) men (X2(2,N = 406) = 11.72 p = 0.001). The impulsivity symptom of antisocial personality disorder characterized 2.5% (n = 6) women and 9.9% (n = 15) men (X2(2,N = 396) = 10.24 p = 0.002).
Tobacco use
At the time of DNA collection, male ex-clients and siblings reported on use of cigarettes and snus (a small bag of dried snuff tobacco placed under the lip). Responses were coded as: (0) “No”, and (1) “Yes” if any smoking and/or use of snus were reported. Among these males, 42.0% (n = 66) reported tobacco use.
Sexual abuse
Information on sexual abuse was only available for the ex-clients, siblings, and healthy women (n = 243). These participants reported on sexual abuse at all waves of data collection using items from the Sexual Experience Survey (Koss and Oros 1982), Sexual and Physical Abuse Questionnaire (Kooiman et al. 2002), and McArthur Community Violence Instrument (Steadman et al. 1998). Sexual abuse was coded as absent (0) or present (1) if any of the following were reported as occurring at any time: forced to engage in sexual activity against his or her will by a person in authority, by offering alcohol or drugs, or by physical violence. Among the women, 43.8% (n = 64) reported having experienced sexual abuse as did 14.4% (n = 13) of the men (X2(2,N = 236) = 21.88 p < 0.001).
Statistical analyses
Characterization of MAOA ROI methylation by sex and age
The first set of analyses aimed to provide a cursory descriptive analysis of methylation levels in our study sample. The number of participants included in these models was varied slightly due to missing data as not all participants had useable methylation data for all CpGs, MAOA-uVNTR genotypes were missing for five participants, substance dependence diagnoses were missing for three participants, and reports of sexual abuse were missing for seven participants. Associations of sex and age with methylation of each CpG within the MAOA ROI, across the ROI, of intronic and exonic regions, and of the empirically derived components of CpG methylation were estimated. To identify homogenous groups (components) of methylation within the MAOA ROI, Principal Component Analyses (PCA) of methylation of the 16 MAOA ROI CpGs with Varimax rotation and Kaiser normalization were computed, first among all 409 participants and then for women and men. Pearson correlations were used to verify associations between component methylation and the mean methylation levels of the CpGs included in the component.
Participants’ ages were bimodally distributed. An age-group variable was constructed: young with a mean of 22.7 (SD = 3.61) years; and old with a mean of 46.1 (SD = 7.02) years. Young and old participants were compared on mean methylation levels at each CpG within the MAOA ROI and across the ROI using a two-way mixed-model ANOVA with post hoc Bonferroni corrections for multiple comparisons, on methylation of the intronic and exonic regions, and the homogeneous components of methylation using independent sample t tests.
Women and men were compared on mean methylation levels at each CpG within the MAOA ROI and across the ROI using a two-way mixed-model ANOVA with post hoc Bonferroni corrections for multiple comparisons, and on methylation of the intronic and exonic regions and components of methylation using independent sample t tests.
To determine if age moderated associations of sex with methylation levels of individual CpGs, across the ROI, of intronic and exonic regions, and of homogeneous components, PROCESS 2.16 for SPSS was used with the simple moderation modeling procedure outlined by Hayes (2013). Separate models were computed for levels of individual CpGs, across the ROI, of intronic and exonic regions, and of homogeneous components of methylation as dependent variables (Y). Sex was entered as the independent variable (X), and age (a continuous variable) was entered as the moderating variable (M). Conditional effects (“simple slopes”) of the moderator, age, on associations between sex and methylation levels were used to interpret significant interaction effects. The conditional effects procedure subdivided age using Mean ± 1SD criteria; low (Mean − 1SD, age = 19), moderate (Mean, age = 32), and high (Mean + 1SD, age 45). Significant models were re-run adjusting for MAOA genotype.
Next, we examined associations between age as a continuous variable and methylation separately by sex among the 252 women and 157 men. The sex-specific PCA analyses were performed to identify methylation components. Linear regression models were computed to examine associations of age in years with methylation of individual CpGs, across the ROI, of intronic and exonic regions, and of sex-specific components of methylation. Significant regression models were subsequently adjusted for MAOA-uVNTR genotype.
Because a large proportion of participants had a history of substance misuse, and some substance misuse at the time of DNA extraction, all linear regression analyses that were computed separately by sex to examine the association of age with methylation of individual CpGs, across the ROI, of exonic and intronic regions, and of sex-specific components were re-run adjusting for lifetime diagnoses of substance dependence. Similar adjustments for depression disorders, anxiety disorders, impulsivity, and tobacco use were computed for methylation of the ROI, exonic and intronic regions, and sex-specific components.
Examining the interaction of sex, MAOA-uVNTR genotype, and sexual abuse on methylation
To determine whether sex would continue to be associated with methylation of the intronic and exonic regions and the sex-specific components of methylation within the MAOA ROI when taking account of genotype (females SL and LL versus SS), sexual abuse, and their interactions, four General Linear Models were computed. These analyses were again done separately by sex. Parents were not included in these analyses as they had not reported on their own experiences of sexual abuse. Furthermore, the analyses were also run including ex-clients and healthy participants and excluding siblings.
Results
Characterization of MAOA ROI methylation by sex and age
Analysis in the full sample
Empirically derived components of methylation
As presented in Table 1, among the 409 participants, principal component analysis of methylation across the 16 CpGs in the MAOA ROI identified two components based on Eigen values over Kaiser’s criterion of 1; (1) CpGs 2–14, and (2) CpGs 15 and 16, accounting for 69.96% and 17.22% of the variation in methylation levels, respectively. Pearson’s correlations revealed that component methylation was highly correlated with the mean value of its constituent CpGs (component 1, r = 0.992, p < 0.001; component 2, r = 0.961, p < 0.001).
Table 1.
Correlation coefficients for rotated component matrix from principal component analysis of MAOA ROI CpG methylation in all participants, women only, and men only
| Component | CpG Site | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG 2/3 | CpG 4 | CpG 5/6 | CpG 7/8 | CpG 10 | CpG 11 | CpG 12 | CpG 13 | CpG 14 | CpG 15 | CpG 16 | |||
| All participants | |||||||||||||
| 1 | 0.959 | 0.956 | 0.964 | 0.959 | 0.966 | 0.965 | 0.874 | 0.735 | 0.767 | – | – | ||
| 2 | – | – | – | – | – | – | – | – | – | 0.908 | 0.841 | ||
| Women | |||||||||||||
| 1 | 0.895 | 0.831 | 0.904 | 0.863 | 0.933 | 0.895 | 0.590 | – | – | – | – | ||
| 2 | – | – | – | – | – | – | – | 0.641 | 0.711 | 0.845 | 0.670 | ||
| Men | |||||||||||||
| 1 | 0.872 | 0.795 | 0.882 | 0.861 | 0.848 | 0.788 | 0.482 | – | – | – | – | ||
| 2 | – | – | – | – | – | – | – | 0.847 | 0.607 | 0.895 | 0.927 | ||
Sex
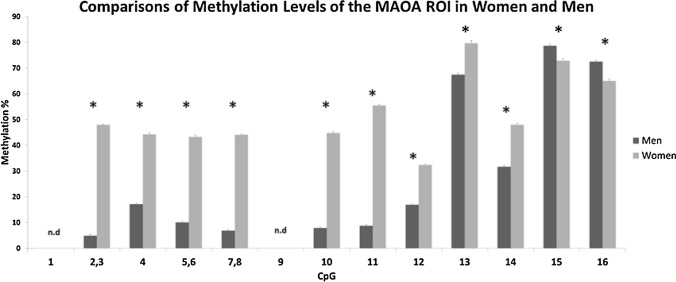
A two-way mixed-model ANOVA examining associations of sex and CpG methylation and overall methylation indicated that women displayed higher levels of ROI methylation than men, with post hoc Bonferroni analyses showing that women had higher methylation levels of CpGs 2–14, while men displayed higher methylation levels of CpGs 15 and 16. Results are illustrated in Fig. 3. Independent samples t tests indicated that women displayed higher exonic (t(409) = 53.21, p < 0.001), and higher intronic (t(409) = 19.95, p < 0.001) methylation levels than men. Independent samples t tests also indicated that women showed higher component 1 methylation levels than men (t(409) = 47.91, p < 0.001), and that men showed higher component 2 methylation levels than women (t(409) = 4.21, p < 0.001). Thus, methylation levels of individual CpGs within the MAOA ROI, in intronic and exonic regions, in components of methylation differed by sex. Mean methylation levels in men and women for all CpGs, the exonic and intronic regions, overall MAOA ROI, and components 1 and 2 are provided in Table S1. Sex-wise comparisons of all CpG, exonic and intronic, overall MAOA ROI, and component 1 and 2 methylation levels by MAOA genotypes are provided in Table S2.
Fig. 3.
Results of comparisons of men and women on MAOA ROI CpG methylation. (*p < 0.05) (n.d: No data)
Age-group
Results of a two-way mixed-model ANOVA detected no differences by age-group of methylation levels of individual CpGs or methylation across the ROI. Independent sample t tests revealed no differences by age-group in exonic and intronic regions or with the empirically derived components. Thus, age-group, older or younger, was not associated with various measures of methylation of the MAOA ROI.
Are the associations of sex with methylation levels modified by age?
Age moderated the association of sex with methylation levels of CpG15 (B = − 0.001, p = 0.024). Follow-up analyses revealed significant conditional effects, such that women at mean (age = 32) and mean + 1SD (age = 45) years exhibited lower methylation levels of CpG15 than men. Only sex was associated with methylation levels of all other CpGs (results are summarized in Table S3). Significant conditional effects are summarized in Table 2. Only sex was significantly associated with methylation levels overall in the ROI methylation and within intronic and exonic regions, such that women showed higher levels than men.
Table 2.
Conditional effects (“Simple Slopes”) of sex and age on MAOA ROI methylation among women compared to men
| Age | Unstandardized simple slope (beta coefficient) | 95% confidence interval | p |
|---|---|---|---|
| Component 2 Methylation (CpGs 15 and 16) | |||
| 1 SD below meana | − 0.2269 | -0.51 to 0.06 | 0.120 |
| Meanb | − 0.4276 | − 0.63 to − 0.22 | <0 .001 |
| 1 SD above meanc | 0.5997 | − 0.91 to − 0.35 | < 0.001 |
| CpG15 methylation | |||
| 1 SD below meana | − 0.0311 | − 0.06 to 0 | 0.051 |
| Meanb | − 0.0556 | − 0.08 to − 0.03 | < 0.001 |
| 1 SD above meanc | − 0.0801 | − 0.11 to − 0.05 | < 0.001 |
a1 SD below mean (age = 19)
bMean (age = 32)
c SD above mean (age = 45)
The interaction of age (a continuous variable) and sex with component 1 methylation levels was not significant (B = − 0.003, p = 0.370). Only sex was significantly associated with component 1 methylation levels, with women displaying higher levels than men (B = 1.98, p < 0.001). The interaction of age and sex with component 2 methylation levels was significant (B = − 0.015, p = 0.04). Follow-up analyses showed a significant conditional effect such that women at mean (age = 32) and mean + 1SD (age = 45) ages exhibited lower methylation levels of component 2 than men. Results are presented in Table 2.
Significant results were adjusted for MAOA genotype. Among women, the interaction of sex and age with CpG15 methylation was robust to adjustment for MAOA genotype (B = − 0.182, p = 0.031). By contrast, among women, the interaction of age and sex with component 2 was not robust to adjustment for genotype (B = − 0.014, p = 0.060). Furthermore, among women, the association of age and component 2 methylation was no longer significant once MAOA genotype was included in the model (B = − 0.009, p = 0.057).
Analyses in women only
Empirically derived components of methylation
Among the 252 women, principal component analysis of methylation across the 16 CpGs in the MAOA ROI identified two components based on Eigen values over Kaiser’s criterion of 1; (1) CpGs 2–12, and (2) CpGs 13–16, accounting for 49.41% and 19.14% of the variation in methylation levels, respectively. Results are presented in Table 1. Pearson correlations revealed that component methylation was highly correlated with the mean values of its constituent CpGs (component 1, r = 0.996, p < 0.001), (component 2, r = 0.991, p < 0.001).
Associations of methylation levels with age
Linear regression models revealed a negative association of age with methylation levels of CpG 13 (F(1,239) = 14.94, p < 0.001, B = − 0.21, r2 = 0.055), no association of age with overall methylation nor with methylation of the exonic and intronic regions, and a negative association of age with component 2 methylation (F(1,239) = 5.13, p = 0.024, B = − 0.01, r2 = 0.017). The association of age with CpG 13 methylation was robust to adjustment for MAOA genotype (B = − 0.208, p < 0.001), while the association of age and component 2 was not (B = − 0.009, p = 0.057).
Analyses in men only
Empirically derived components of methylation
Among the 157 men, principal component analysis of methylation across the 16 CpGs in the MAOA ROI identified two components based on Eigen values over Kaiser’s criterion of 1; (1) CpGs 2–12, and (2) CpGs 13–16, accounting for 44.63% and 23.63% of the variation in methylation levels, respectively (Table 1). Pearson’s correlations revealed that component methylation was highly correlated with the mean value of its constituent CpGs (component 1, r = 0.985, p < 0.001), (component 2, r = 0.994, p < 0.001).
Associations of methylation levels with age
Among the men, linear regressions revealed a positive association of age with CpG 15 methylation (F(1,150) = 4.58, p = 0.034, B = 4.15), r2 = 0.023, and no association with the methylation of the other CpGs or across the ROI. Age was not significantly associated with methylation of intronic and exonic regions, nor with components of methylation. The association of age with CpG 15 methylation was robust to adjustment for MAOA genotype (B = − 0.182, p = 0.031).
A summary of full sample and sex-specific results are presented in Table 3.
Table 3.
Associations of age and sex with methylation levels of the MAOA-ROI: a summary of results
| CpGs | Across ROI | Intronic | Exonic | Component 1 | Component 2 | |
|---|---|---|---|---|---|---|
| All participants | ||||||
| Age-group | x | x | x | x | x | x |
| Sex |
↑women CpG 2–14 ↓women CpGs 15 and 16 |
↑women | ↑women | ↑women | ↑women | ↓women |
| Sex modified by age | In women, at CpG 15B | In womenA | ||||
| Only sex | ↑women | ↑women | ↑women | |||
| Women only | ||||||
| Age | Negative association at CpG 13B | x | x | x | x | Negative associationA |
| Men only | ||||||
| Age | Positive association at CpG 15B | x | x | x | x | x |
A. Not robust to covariation with MAOA-uVNTR genotype
B. Robust to adjustment for MAOA-uVNTR genotype
x. No association
Are associations of sex and age with methylation levels modified by substance dependence, depression disorders, anxiety disorders, impulsivity, or tobacco use?
Given that most of the sample included adolescents presenting with substance misuse, their siblings and parents, analyses were re-run with lifetime substance dependence as a covariate. Results of all linear regression models among women were robust to adjustments for lifetime substance dependence, while, among men, the positive association of age with CpG15 methylation was no longer significant (B = 0.120, p = 0.068). Additionally, among men, the association of age with mean exonic methylation remained non-significant in the adjusted model, though lifetime substance dependence was positively associated with mean exonic methylation (B = 2.876, p = 0.011).
The linear regression models on genomic feature and component methylation were also re-run to determine whether lifetime depression and anxiety disorders, impulsivity, and tobacco use (in men only) modified results. Among women, depression diagnoses were associated with higher levels of exonic (B = 0.136, p = 0.034) and component 1 (B = 0.136, p = 0.035) methylation, though age remained non-significant in these models. The negative association of age with component 2 methylation levels in women was robust to adjustment for depression and anxiety disorders, and impulsivity.
Among the men, most models were robust to adjustments, although a positive association of age with component 2 methylation was observed after adjusting for tobacco use (B = 0.236, p = 0.030). While the association of age with first exon methylation levels remained non-significant in the model, a positive association with anxiety diagnoses (B = 0.189, p = 0.022) was observed.
Is higher methylation among women associated with genotype, sexual abuse, and their interactions?
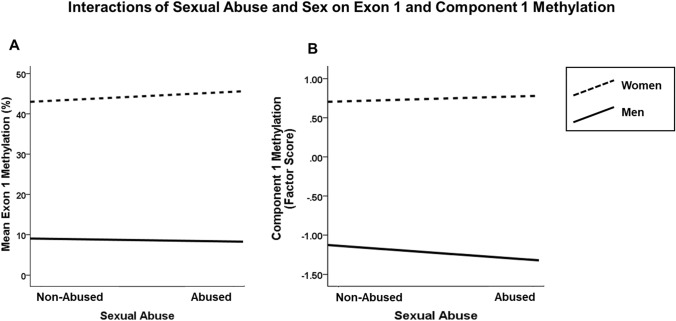
As presented in Table 4, among the younger participants, neither genotype nor sexual abuse was directly associated with methylation in intronic and exonic regions and components. The only significant direct effect was sex on intronic, exonic, and component 1 methylation. Exonic methylation and component 1 methylation were additionally associated with the interaction of sex and sexual abuse. Intronic methylation was only associated with sex. Figure 4 illustrates the two significant interaction terms showing higher methylation levels in women than men regardless of sexual abuse. However, as recommended, we did not interpret direct effects when effects of interaction terms were statistically significant (Nelder 1998). Thus, the results indicate that sexual abuse was associated with higher methylation levels in the MAOA ROI among women. To further explore the association of sexual abuse with methylation levels among women and men, we compared intronic, exonic, and component methylation among abused and non-abused participants within each sex. These exploratory, descriptive analyses (Tables S4 and S5) indicate that among men, methylation levels of those who did and who did not experience sexual abuse did not differ, while among women, those who were sexually abused exhibited higher methylation in the exonic region and in component 1 than the non-abused, and lower methylation in component 2.
Table 4.
General linear models of factors associated with MAOA ROI methylation in exonic and intronic regions and components 1 and 2
| Methylation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exonic | Intronic | Component 1 | Component 2 | |||||||||
| df | F | p | df | F | p | df | F | p | df | F | p | |
| MAOA-uVNTR | 1 | 0.302 | 0.583 | 1 | 0.143 | 0.705 | 1 | 1.501 | 0.221 | 1 | 0.678 | 0.410 |
| Sexual abuse | 1 | 1.510 | 0.219 | 1 | 1.050 | 0.305 | 1 | 3.688 | 0.055 | 1 | 1.363 | 0.243 |
| Sex | 1 | 1747.654 | < 0.001 | 1 | 215.215 | < 0.001 | 1 | 1294.844 | <0 .001 | 1 | 2.414 | 0.120 |
| Sexual abuse × Sex | 4.161 | 0.041 | 1.088 | 0.297 | 4.973 | .026 | 0.931 | 0.335 | ||||
| MAOA-uVNTR × Sexual Abuse | 1.045 | 0.307 | 2.390 | 0.122 | 3.092 | .079 | 1.523 | 0.217 | ||||
| MAOA-uVNTR × Sex | 0.559 | 0.455 | 0.036 | 0.850 | 1.281 | .258 | 0.359 | 0.549 | ||||
| MAOA-uVNTR × Sexual Abuse x Sex | 1.223 | 0.269 | 3.152 | 0.076 | 3.590 | .058 | 2.269 | 0.132 |
Fig. 4.
Interactions of sexual abuse status (no sexual abuse vs. sexual abuse) and sex on exon 1 methylation level (A) and component 1 factor score (B) in the young women and men (ex-clients, siblings, and healthy women)
To mitigate the impact of including related siblings in these analyses, the models were re-run among only the ex-clients and healthy participants (no siblings). Results of the models are summarized in Table S6. Notably, results are very similar to those obtained with the larger sample. Again, the only significant direct effects were the association of sex with intronic, exonic, and component 1 methylation. Only exonic methylation was associated with the interaction of sex and sexual abuse. Unlike results from the larger sample, component 1 methylation was associated with the interaction of MAOA-uVNTR genotype and sex. Further exploration showed that women who had experienced sexual abuse displayed higher exonic methylation than those who did not (p = 0.017). Among men, no difference in exonic methylation was observed between men who did and did not experience sexual abuse (p = 0.978).
Discussion
The first aim of the current study was to characterize associations of sex and age with methylation in a region-of-interest (ROI), spanning the first exonic region and partial first intronic region of MAOA. Methylation levels of individual CpGs, across the entire ROI, in exonic and intronic regions, and in homogeneous empirically derived components of CpG methylation were measured. Women were characterized by higher methylation levels than men and varied little by age. The only previous study to examine sex differences in methylation of MAOA included a small sample and observed higher levels among women than men in the exonic region (Melas and Forsell 2015). The present study confirmed this finding in a larger sample and showed higher levels of methylation within the MAOA ROI among women than men and in the intronic region. These findings are consistent with the results of epigenome-wide methylation studies (Nugent and McCarthy 2011). Additionally, the present study extends knowledge by showing that within the MAOA ROI, methylation levels among women and men generally did not decrease with age. This finding is consistent with epigenome-wide studies, indicating that methylation levels in some regions (Ciccarone et al. 2018), particularly those in first exon/intron junctions (Kochmanski and Bernstein 2020), are more likely than other regions to remain open to fluctuation in spite of global reductions in methylation.
Among women, results of the moderation analyses indicated that the interaction of sex and age with methylation at CpG 15 was robust to adjustment for MAOA-uVNTR genotype, and genotype dependent at component 2. Similarly, among women, the association of age with methylation at CpG 13 was robust to adjustment by genotype, whereas the association of age with component 2 was no longer significant after adjustment for genotype. Among men, the one direct association of age with methylation of CpG 15 was robust to adjustment for genotype. As such, the few associations of age with methylation identified in the women and men were largely independent of MAOA-uVNTR genotype. Following adjustments for lifetime history of substance dependence, depression disorders, anxiety disorders, impulsivity, and tobacco use (only in men), findings were largely unchanged with age showing few associations with methylation levels.
The second aim of the study was to determine whether MAOA ROI methylation levels were associated with the interaction of MAOA-uVNTR genotype, sex, and sexual abuse among males and/or females. Analyses that included only the younger participants revealed that methylation levels were not associated with genotype, nor with the interaction of genotype and sexual abuse, and were slightly increased among survivors of sexual abuse. In fact, the association of methylation levels within the MAOA ROI with a negative environmental factor—sexual abuse—was much weaker than the association with sex. Furthermore, in these models, sex remained the factor most strongly associated with intronic, exonic, and component 1 methylation levels even after taking account of MAOA-uVNTR genotype, sex, and sexual abuse. Exonic and component 1 methylation were associated with the interaction of sex and sexual abuse. To ensure that the inclusion of siblings had not confounded these results, models were re-run with only unrelated ex-clients and healthy participants. Results were similar with those in the larger sample, showing that sex was the factor most strongly associated with exonic, intronic, and component 1 methylation levels, while exonic levels were also associated with the interaction of sex and sexual abuse. Notably, in these models, there were no direct associations of MAOA-uVNTR genotypes with methylation levels. Thus, even when taking account of MAOA-uVNTR genotypes and sexual abuse, and the interactions of these factors, sex remained the principal factor associated with methylation levels observed within the MAOA ROI. Despite the small sample size, and the low number of sexually abused males, results suggested that sex was associated with the magnitude of the difference in methylation levels among participants who had and had not experienced sexual abuse. The finding that these associations were not dependent on genotype is consistent with results from a recent study, showing that genetic factors associate with stability of methylation levels, while environmental factors associate more strongly with alterations to methylation (Reynolds et al. 2020).
Considered together, our findings within the MAOA ROI are consistent with prior evidence of sex differences in methylation across the genome (Yousefi et al. 2015; Ratnu et al. 2017; Suderman et al. 2017), and extend knowledge by showing that within the MAOA ROI women present higher methylation levels than men for approximately 3 decades following puberty. Previous evidence of a decrease in methylation levels across the genome as individuals age (Ciccarone et al. 2018) was not observed in the MAOA ROI, except in intronic CpG13 and component 2 among women. Perhaps, the age range of the participants—approximately 3 decades following puberty—was insufficient to identify age-related changes or did not cover critical periods of change. Alternatively, studies have reported that methylation in first exonic–intronic junctions such as the MAOA ROI, relative to other regions, is more likely to be spared from the global pattern of demethylation as individuals grow older (Ashapkin et al. 2017; Ciccarone et al. 2018; Kochmanski and Bernstein 2020). Perhaps, through adulthood, these regions remain open to fluctuation in response to environmental and biological factors which contribute to health-related outcomes (Ashapkin et al. 2017; Ciccarone et al. 2018). However, as the results examining sexual abuse showed, this may be the case more often among women than men. This interpretation warrants further investigation as the present sample included few males who had experienced sexual abuse.
As EpiTYPER samples methylation randomly from both X-chromosomes in females, the resulting levels represent the averaged level of methylation from both chromosomes (Cotton et al. 2011). As such, the large sex difference, particularly in the exonic region of the MAOA ROI, likely indicates near total methylation from the inactivated chromosome and low methylation from the active chromosome, thus accounting for exonic methylation levels in in the ~ 40–50% range among women (Table S1). Similar levels of MAOA first exon methylation in women have been reported and interpreted similarly by others examining methylation in this region (Melas et al. 2013; Melas and Forsell 2015; Ziegler et al. 2016). As the sex difference in methylation levels in the MAOA first exon remained after taking other factors into account, our findings offer further evidence that the MAOA first exon may be an X-inactivation site in women. The finding that methylation of first exonic regions of genes is associated with total transcriptional silencing of a gene rather than downregulated transcription (Brenet et al. 2011) may indicate that MAOA is not among the ~ 15% of X-linked genes that escape inactivation (Carrel and Willard 2005). MAOA expression may thus be monoallelic rather than biallelic in women. Given the consistency of the sex difference in methylation levels reported in the current and previous studies (Melas et al. 2013; Melas and Forsell 2015), it is highly likely that it is due to X-inactivation.
Other factors could influence sex differences in MAOA methylation on the active X-chromosome in women. Some evidence suggests that MAOA may be a genomic locus where sex hormones influence serotonergic activity and phenotypes that feature prominently in our study sample, such as antisocial behaviour and substance misuse (Nilsson et al. 2018). For instance, one study found that the interaction of MAOA genotype and testosterone concentration in cerebrospinal fluid predicted antisocial behaviour and suggested that this interaction may have been mediated by direct effects on MAOA transcription (Sjöberg et al. 2009). Another study of healthy males showed that those carrying MAOA-S, as compared to those carrying MAOA-L, displayed increased risk-taking behaviours following administration of a topical gel containing testosterone (Wagels et al. 2017). Furthermore, MAOA expression may be modified by interactions between testosterone levels and transcription factor Sp1 at its binding sites throughout the MAOA promoter region (Ou et al. 2006). Given that DNA methylation can modify gene expression by disrupting transcription factor binding (Bird 2011), the impact of sex hormones on MAOA expression may be influenced by epigenetic processes. Although potential influences of female sex hormones on MAOA expression and serotonergic activity in such phenotypes remain unclear (Booij et al. 2015; de Almeida et al. 2015; Raine 2019), investigations of such associations will be important to further elucidate sex differences in MAOA regulation.
Limitations and strengths of the present study
Limitations of the study included the EpiTYPER method that is unable to distinguish between DNA methylation and 5-hydroxymethylation, which is a further modification of a methylated cytosine through enzymatic oxidation and a proposed marker of DNA demethylation processes (Kumar et al. 2018). DNA 5-hydroxymethylation may also have unique functional impacts on gene transcription that are distinct from those conferred by methylation, though these functional impacts are not yet fully understood (Zheleznyakova et al. 2016). In addition, genomic regions spanning exon–intron boundaries (Ciccarone et al. 2018), such as the MAOA ROI, show enrichments of hydroxymethylation (Kochmanski and Bernstein 2020). This could be one reason why no reduction in methylation levels with increasing age was detected. As such, it is vital for future studies of the MAOA ROI to examine the ratio of DNA methylation to 5-hydroxymethylation, their association with age, and their respective contributions to MAOA transcriptional activity.
Our study sample was relatively small, thus providing insufficient power to perform multiple corrections for all statistical models, particularly in the models that included direct effects, and two- and three-way interaction terms to determine associations with exonic, intronic, and component methylation levels. However, based on previous methylation studies (Jones 2012; Moore et al. 2013), these regions were regarded as distinct functional units, greater than their constituent CpG sites, in terms of impact on gene expression, thus making corrections for multiple testing in these models unnecessary. The sample sizes often recommended to examine gene-by-environment interactions stem from earlier critiques, suggesting that the effect sizes of interactions should exceed those for a candidate gene as detected by genome-wide association studies including large sample sizes (Duncan and Keller 2011). However, it has been more recently proposed that there is likely no main effect of MAOA genotypes independent from the influence from environmental factors (Booij et al. 2015; Åslund and Nilsson 2018; Nilsson et al. 2018). If an interaction term is associated with a dependent variable, the main effect becomes difficult to interpret (Nelder 1998; Nilsson et al. 2014), particularly since the main effect will change with any additional variables entered into the interaction (Nilsson et al. 2018). The initial comparisons of methylation levels by age and by sex were made using two-way mixed-model ANOVA which included CpG as a repeated measure. These analyses estimated group differences in overall methylation and differences at each CpG using stringent Bonferroni corrections for multiple comparisons. As such, this approach offered a means to mitigate issues with multiple testing, as has been used in prior epigenetic studies (Labonte et al. 2012; Gross et al. 2013; Cruceanu et al. 2016).
Statistical analyses of X-linked genes in GWAS are typically conducted within sex (Weir et al. 2006; Sillanpää 2011). GWAS typically adjust for relatedness of participants. To address these considerations, we assessed methylation levels both in the full sample and separately by sex and re-ran models that examined interactions of sex, sexual abuse, and genotype including only unrelated participants. Similar exclusions were not used in the first set of analyses aimed at providing a cursory characterization of methylation levels in our study sample.
However, while the sample was small and had limited statistical power, it was well characterized by multiple face-to-face interviews and questionnaires. Some authors have suggested that studies of gene-by-environment interactions in smaller samples may be preferable as dependent variables are more likely to be assessed using in-person interviews with better reliability and validity than the telephone, post, or Internet-based assessments used in studies with large samples (Uher and McGuffin 2008; Caspi et al. 2010; Karg et al. 2011; Moffitt and Caspi 2014; Nilsson et al. 2018). The use of such in-person measures is a key strength of the current study.
Another strength of the study was the use of five different measures of methylation levels, including a novel approach, principal component analyses, to identify components of CpG with similar and distinct methylation levels. The use of component analyses also helped mitigate, to some extent, concerns regarding the low statistical power by reducing the number of variables included in our interaction analyses. Another strength of the study were the analyses conducted to confirm differences in levels of methylation in exonic regions and component 1 between women who had, and who had not, experienced sexual abuse. Our study also benefited from the use of gold-standard genotyping.
As few males in our sample had experienced sexual abuse (n = 13), the lack of association between sexual abuse and methylation of the MAOA ROI observed should be interpreted cautiously, particularly since evidence indicates that males are particularly susceptible to the impacts of adversity in both biological and behavioural consequences (Raine 2019). One previous study by our group in an overlapping sample of these men showed that, while maltreatment defined as having experienced physical and/or sexual abuse was not directly associated with MAOA ROI methylation levels, the interaction of maltreatment, the MAOA-S genotype, and MAOA ROI methylation was associated with increased aggressive behavior (Checknita et al. 2020) and alcohol consumption (Bendre et al. 2018). As such, conclusions regarding associations of maltreatment and MAOA ROI methylation in men may require more nuanced interpretations including other genetic and environmental factors and framed in the context of different outcomes.
We examined methylation of DNA extracted from peripheral cells in saliva, thus limiting the interpretation of our results in relation to brain processes as methylation patterns are often tissue-specific (Deaton and Bird 2011). However, the current evidence shows associations of MAOA ROI methylation in whole blood DNA with MAO-A enzymatic activity in the brain. Furthermore, in-silico cross-tissue examination of two available CpGs within the MAOA ROI using IMAGE-CpG (http://han-lab.org/methylation/ default/imageCpG#) (Braun et al. 2019) showed a fairly strong association between methylation levels from blood and saliva samples at these two CpGs with rho values of 0.64 and 0.79, respectively, and between brain and saliva methylation with rho values of 0.76 and 0.69, respectively. The methylation levels reported for the male and female participants in our study are similar to those reported elsewhere in whole blood samples from men (Checknita et al. 2015; Ziegler et al. 2018) and women (Ziegler et al. 2016; Schiele et al. 2018) and in studies examining salivary methylation of this region in men and women (Melas et al. 2013; Melas and Forsell 2015). Recent reviews in behavioural epigenetics emphasize that the value of investigations of methylation in peripheral tissues is critical for the study of mental and behavioural disorders and responses to trauma (Labonte and Turecki 2012; Lutz and Turecki 2014; Szyf 2015). As such, the extensive characterization of methylation of the MAOA ROI provided in our study contributes to this effort.
The MAOA ROI characterized in the current study was selected based on prior in-vitro analyses, showing that methylation of this region was associated with drastically reduced reporter gene expression (Checknita et al. 2015), and that methylation levels of the ROI were associated with MAO enzymatic activity in the brain (Shumay et al. 2012), highlighting the functional relevance of the MAOA ROI. Furthermore, genome-wide methylation studies have pointed to exonic/intronic junction regions, similar to the MAOA ROI, as being epigenetic “hot-spots” which remain responsive to epigenetic alterations across the lifespan and are associated with health outcomes (Ashapkin et al. 2017; Ciccarone et al. 2018). As such, our findings showing few age-associated methylation differences further support the MAOA ROI as an important target for epigenetic studies, since it may remain responsive to environmental factors as individuals’ age.
The MAOA gene is among the most well-established candidate genes associated with behavioural phenotypes (Comai et al. 2012; Byrd and Manuck 2014; Booij et al. 2015; Nilsson et al. 2018). Converging lines of evidence from gene-by-environment studies and from DNA methylation studies have suggested that MAOA plays a role in antisocial behaviour, substance misuse, depression (Verhoeven et al. 2012; Melas et al. 2013; Byrd and Manuck 2014; Booij et al. 2015; Melas and Forsell 2015; Checknita et al. 2015, 2020; Bendre et al. 2018; Nilsson et al. 2018), and other mental disorders (Reif et al. 2014; Booij et al. 2015; Ziegler et al. 2016, 2018; Checknita et al. 2018). Among women, adjustments for depression disorders revealed a positive association of depression, but not age, with exonic methylation levels. This is an expected finding given previously reported associations of higher exonic methylation and depression diagnoses in a smaller group of the women included in the current study (Checknita et al. 2018), and further highlights the role of MAOA in depression. As such, the comprehensive characterization of MAOA ROI methylation, taking account of MAOA genotypes, that is reported in current study contributes to furthering understanding of MAOA regulation in behavioural phenotypes. It is notable that although our sample was recruited through a substance misuse clinic, almost all the current results were robust to adjustment for substance dependence, depression disorders, anxiety disorders, impulsivity, and tobacco use. This finding suggests that the principal findings may be generalizable to community samples.
Conclusion
Epigenome studies have shown higher levels of methylation among women than men and, decreasing levels with increasing age in both sexes. The present study observed that within the intronic and exonic regions of MAOA, women did present higher levels of methylation but levels did not differ by age, and age did not modify the associations of methylation with sex. This finding is likely attributable to patterns of methylation indicating inactivation of one of the X-chromosomes carried by women. Participants’ ages spanned 3 decades following puberty, and with only a few exceptions, age was not associated with the higher methylation levels among women as compared to men. Older women did display lower levels of CpG13 methylation than younger women, but this finding was not robust to adjustment for MAOA-uVNTR genotype. The stability of methylation in this region across ages needs replication, especially in light of our previous findings, showing that methylation levels in these regions strengthened associations of the interactions of sex-specific genotypes and adversity with aggressive behavior and substance misuse. Few of the observed associations were modified by genotype. None were modified by interactions of genotype and sexual abuse, but sexual abuse of women was associated with slightly increased methylation levels. However, the differences in methylation levels between women who had, and who had not, experienced sexual abuse were small in comparison to the differences in levels between women and men.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The study was funded by grants from the Swedish Council for Working Life and Social Research, Mobilisering mot Narkotika, the county councils of Stockholm and Västmanland, and Karolinska Institutet Strategic Funding.
Funding
Open access funding provided by Uppsala University.
Data availability
Data will be made available on reasonable request.
Declarations
Conflicts of interest
The authors report no biomedical financial interests or potential conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All waves of data collection were approved by the Regional Ethical Review Board in Stockholm. At each wave of data collection, participants signed informed consents, except at baseline when consent for those under age 18 was given by their parents. At collection of saliva samples, all participants were 18 or older, and they specifically consented to providing saliva samples for DNA extraction. Confidentiality of all the information was guaranteed, and at each wave of data collection, participants received gift certificates as compensation for their time.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahsan M, Ek WE, Rask-Andersen M, et al. The relative contribution of DNA methylation and genetic variants on protein biomarkers for human diseases. PLoS Genet. 2017;13:1–24. doi: 10.1371/journal.pgen.1007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin VV, Kutueva LI, Vanyushin BF. Aging as an epigenetic phenomenon. Curr Genomics. 2017;18:385–407. doi: 10.2174/1389202918666170412112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åslund C, Nilsson KW. Individual biological sensitivity to environmental influences: testing the differential susceptibility properties of the 5HTTLPR polymorphism in relation to depressive symptoms and delinquency in two adolescent general samples. J Neural Transm. 2018;125:977–993. doi: 10.1007/s00702-018-1854-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åslund C, Nordquist N, Comasco E, et al. Maltreatment, MAOA, and delinquency: sex differences in gene-environment interaction in a large population-based cohort of adolescents. Behav Genet. 2011;41:262–272. doi: 10.1007/s10519-010-9356-y. [DOI] [PubMed] [Google Scholar]
- Barnett Burns S, Almeida D, Turecki G. The epigenetics of early life adversity: current limitations and possible solutions. Prog Mol Biol Transl Sci. 2018;157:343–425. doi: 10.1016/bs.pmbts.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Beach SRH, Brody GH, Gunter TD, et al. Child maltreatment moderates the association of MAOA with symptoms of depression and antisocial personality disorder. J Fam Psychol. 2010;24:12–20. doi: 10.1037/a0018074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendre M, Comasco E, Checknita D, et al. Associations between MAOA-uVNTR genotype, maltreatment, MAOA methylation, and alcohol consumption in young adult males. Alcohol Clin Exp Res. 2018;42:1–12. doi: 10.1111/acer.13578. [DOI] [PubMed] [Google Scholar]
- Bird A. Putting the DNA back into DNA methylation. Nat Genet. 2011;43:1050–1051. doi: 10.1038/ng.987. [DOI] [PubMed] [Google Scholar]
- Booij L, Tremblay R, Szyf M, Benkelfat C. Genetic and early environmental influences on the serotonin system: consequences for brain development and risk for psychopathology. J Psychiatry Neurosci. 2015;40:5–18. doi: 10.1503/jpn.140099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun PR, Han S, Hing B, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl Psychiatry. 2019;9:1–10. doi: 10.1038/s41398-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, et al. DNA methylation of the first exon Is tightly linked to transcriptional silencing. PLoS ONE. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Fiori LM, Turecki G. Bridging basic and clinical research in early life adversity, DNA methylation, and major depressive disorder. Front Genet. 2019;10:1–10. doi: 10.3389/fgene.2019.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AL, Manuck SB. MAOA, childhood maltreatment, and antisocial behavior: meta-analysis of a gene-environment interaction. Biol Psychiatry. 2014;75:9–17. doi: 10.1016/j.biopsych.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, et al. Genetic sensitivity to the environment: the case of the serotonin transporter. Am J Psychiatry. 2010;167:509–527. doi: 10.1176/appi.ajp.2010.09101452.Genetic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checknita D, Maussion G, Labonté B, et al. Monoamine oxidase A gene promoter methylation and transcriptional downregulation in an offender population with antisocial personality disorder. Br J Psychiatry. 2015;206:216–222. doi: 10.1192/bjp.bp.114.144964. [DOI] [PubMed] [Google Scholar]
- Checknita D, Ekström TJ, Comasco E, et al. Associations of monoamine oxidase A gene first exon methylation with sexual abuse and current depression in women. J Neural Transm. 2018;125:1053–1064. doi: 10.1007/s00702-018-1875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checknita D, Bendre M, Ekstrom TJ, et al. Monoamine oxidase A genotype and methylation moderate the association of maltreatment and aggressive behaviour. Behav Brain Res. 2020;382:1–11. doi: 10.1016/j.bbr.2020.112476. [DOI] [PubMed] [Google Scholar]
- Chen J, Zang Z, Braun U, et al. Association of a reproducible epigenetic risk profile for schizophrenia with brain methylation and function. JAMA Psychiat. 2020;77:628–636. doi: 10.1001/jamapsychiatry.2019.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarone F, Tagliatesta S, Caiafa P, Zampieri M. DNA methylation dynamics in aging: how far are we from understanding the mechanisms? Mech Ageing Dev. 2018;174:3–17. doi: 10.1016/j.mad.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Comai S, Tau M, Pavlovic Z, Gobbi G. The psychopharmacology of aggressive behavior: a translational approach: part 2: clinical studies using atypical antipsychotics, anticonvulsants, and lithium. J Clin Psychopharmacol. 2012;32:237–260. doi: 10.1097/JCP.0b013e31824929d6. [DOI] [PubMed] [Google Scholar]
- Cotton AM, Lam L, Affleck JG, et al. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet. 2011;130:187–201. doi: 10.1007/s00439-011-1007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruceanu C, Kutsarova E, Chen ES, et al. DNA hypomethylation of Synapsin II CpG islands associates with increased gene expression in bipolar disorder and major depression. BMC Psychiatry. 2016;16:1–5. doi: 10.1186/s12888-016-0989-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida RMM, Cabral JCC, Narvaes R. Behavioural, hormonal and neurobiological mechanisms of aggressive behaviour in human and nonhuman primates. Physiol Behav. 2015;143:121–135. doi: 10.1016/j.physbeh.2015.02.053. [DOI] [PubMed] [Google Scholar]
- Deaton M, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek WE, Rask-Andersen M, Karlsson T, et al. Genetic variants influencing phenotypic variance heterogeneity. Hum Mol Genet. 2018;27:799–810. doi: 10.1093/hmg/ddx441. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Miriam G, Williams JBW. Structured clinical interview for DSM-IV-TR axis I disorders, Research Version, Patient Edition (SCID-I/P) New York: Biometrics Research, New York State Psychiatric Institute; 2002. [Google Scholar]
- Fransquet PD, Wrigglesworth J, Woods RL, et al. The epigenetic clock as a predictor of disease and mortality risk : a systematic review and meta-analysis. Clin Epigenet. 2019;11:1–17. doi: 10.1186/s13148-019-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garieri M, Stamoulis G, Blanc X, et al. Extensive cellular heterogeneity of X inactivation revealed by single-cell allele-specific expression in human fibroblasts. Proc Natl Acad Sci USA. 2018;115:13015–13020. doi: 10.1073/pnas.1806811115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenhuber B, Tollkuhn J. Sex differences in the epigenome : a cause or consequence of sexual differentiation of the brain ? Genes (basel) 2019;10:1–17. doi: 10.3390/genes10060432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso DJ, Drury S, Briggs-Gowan M, et al. Adverse childhood experiences, posttraumatic stress, and FKBP5 methylation patterns in postpartum women and their newborn infants. Psychoneuroendocrinology. 2020;114:104604. doi: 10.1016/j.psyneuen.2020.104604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JA, Fiori LM, Labonté B, et al. Effects of promoter methylation on increased expression of polyamine biosynthetic genes in suicide. J Psychiatr Res. 2013;47:513–519. doi: 10.1016/j.jpsychires.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LK, Verhoeven EJ, Tyrka AR, et al. Accelerating research on biological aging and mental health: current challenges and future directions. Psychoneuroendocrinology. 2019;106:293–311. doi: 10.1016/j.psyneuen.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Knox O, Sugden K, et al. Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genet. 2018;14:1–27. doi: 10.1371/journal.pgen.1007544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes AF. Introduction to mediation, moderation, and conditional process analysis: a regression-based approach. New York: Guilford Press; 2013. [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kader F, Ghai M. DNA methylation-based variation between human populations. Mol Genet Genomics. 2016;292:5–35. doi: 10.1007/s00438-016-1264-2. [DOI] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski J, Bernstein AI. The impact of environmental factors on 5-Hydroxymethylcytosine in the brain. Curr Environ Heal Reports. 2020;7:109–120. doi: 10.1007/s40572-020-00268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooiman CG, Ouwehand AW, ter Kuile MM. The sexual and physical abuse questionnaire (SPAQ). A screening instrument for adults to assess past and current experiences of abuse. Child Abuse Negl. 2002;26:939–953. doi: 10.1016/S0145-2134(02)00363-0. [DOI] [PubMed] [Google Scholar]
- Koss MP, Oros CJ. Sexual experiences survey: a research instrument investigating sexual aggression and victimization. J Consult Clin Psychol. 1982;50:455–457. doi: 10.1037/0022-006X.50.3.455. [DOI] [PubMed] [Google Scholar]
- Kumar S, Chinnusamy V, Mohapatra T. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front Genet. 2018;9:1–14. doi: 10.3389/fgene.2018.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Yerko V, Gross J, et al. Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry. 2012;72:41–48. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- Labonte B, Turecki G. Epigenetic effects of childhood adversity in the brain and suicide risk. In: Dwivedi Y, editor. The neurobiologcial basis of suicide. Boca Raton: CRC Press/Taylor & Francis; 2012. [PubMed] [Google Scholar]
- Lutz P, Turecki G. DNA methylation and childhood maltreatment: From animal models to human studies. Neuroscience. 2014;264:142–156. doi: 10.1016/j.neuroscience.2013.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi SJ, Sugden K, Arseneault L, et al. Analysis of DNA methylation in young people: Limited evidence for an association between victimization stress and epigenetic variation in blood. Am J Psychiatry. 2018;175:517–529. doi: 10.1176/appi.ajp.2017.17060693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masser DR, Hadad N, Porter HL, et al. Sexually divergent DNA methylation patterns with hippocampal aging. Aging Cell. 2017;16:1342–1352. doi: 10.1111/acel.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney DL, Zhang F, Hillary RF, et al. An epigenome-wide association study of sex-specific chronological ageing. Genome Med. 2019;12:1–11. doi: 10.1186/s13073-019-0693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas PA, Forsell Y. Hypomethylation of MAOA׳s first exon region in depression: a replication study. Psychiatry Res. 2015;226:2–4. doi: 10.1016/j.psychres.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Melas PA, Wei Y, Wong CCY, et al. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int J Neuropsychopharmacol. 2013;16:1513–1528. doi: 10.1017/S1461145713000102. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Buckholtz JW, Kolachana B, et al. Neural mechanisms of genetic risk for impulsivity and violence in humans. Proc Natl Acad Sci U S A. 2006;103:6269–6274. doi: 10.1073/pnas.0511311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Yu T, Chen E, Brody GH. Self-control forecasts better psychosocial outcomes but faster epigenetic aging in low-SES youth. Proc Natl Acad Sci. 2015;112:10325–10330. doi: 10.1073/pnas.1505063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt TE, Caspi A. Bias in a protocol for a meta-analysis of 5-HTTLPR, stress, and depression. BMC Psychiatry. 2014;14:1–4. doi: 10.1186/1471-244x-14-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelder JA. The selection of terms in response-surface models-how strong is the weak-heredity principle? Am Stat. 1998;52:315–318. doi: 10.2307/2685433. [DOI] [Google Scholar]
- Nilsson KW, Sjöberg RL, Damberg M, et al. Role of monoamine oxidase A genotype and psychosocial factors in male adolescent criminal activity. Biol Psychiatry. 2006;59:121–127. doi: 10.1016/j.biopsych.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Nilsson KW, Comasco E, Hodgins S, et al. Genotypes do not confer risk for delinquency but rather alter susceptibility to positive and negative environmental factors: gene-environment interactions of BDNF Val66Met, 5-HTTLPR, and MAOA-uVNTR. Int J Neuropsychopharmacol. 2014;18:1–10. doi: 10.1093/ijnp/pyu107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson KW, Åslund C, Comasco E, Oreland L. Gene–environment interaction of monoamine oxidase A in relation to antisocial behaviour: current and future directions. J Neural Transm. 2018 doi: 10.1007/s00702-018-1892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordquist N, Oreland L. Serotonin, genetic variability, behaviour, and psychiatric disorders—a review. Ups J Med Sci. 2010;115:2–10. doi: 10.3109/03009730903573246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent BM, McCarthy MM. Epigenetic underpinnings of developmental sex differences in the brain. Neuroendocrinology. 2011;93:150–158. doi: 10.1159/000325264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreland L, Nilsson K, Damberg M, Hallman J. Monoamine oxidases: activities, genotypes and the shaping of behaviour. J Neural Transm. 2007;114:817–822. doi: 10.1007/s00702-007-0694-8. [DOI] [PubMed] [Google Scholar]
- Ou XM, Chen K, Shih JC. Glucocorticoid and androgen activation of monoamine oxidase a is regulated differently by R1 and Sp1. J Biol Chem. 2006;281:21512–21525. doi: 10.1074/jbc.M600250200. [DOI] [PubMed] [Google Scholar]
- Pagiatakis C, Musolino E, Gornati R, et al. Epigenetics of aging and disease: a brief overview. Aging Clin Exp Res. 2019;33:737–745. doi: 10.1007/s40520-019-01430-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov KA, Chistiakov DA, Chekhonin VP. Genetic determinants of aggression and impulsivity in humans. J Appl Genet. 2012;53:61–82. doi: 10.1007/s13353-011-0069-6. [DOI] [PubMed] [Google Scholar]
- Peeters SB, Cotton AM, Brown CJ. Variable escape from X-chromosome inactivation: Identifying factors that tip the scales towards expression. BioEssays. 2014;36:746–756. doi: 10.1002/bies.201400032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Gunter TD, Beach SR, et al. MAOA methylation is associated with nicotine and alcohol dependence in women. Am J Med Genet. 2008 doi: 10.1002/ajmg.b.30778.MAOA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Beach SRH, Gunter TD, et al. The effect of smoking on MAOA promoter methylation in DNA prepared from lymphoblasts and whole blood. Am J Med Genet. 2010 doi: 10.1002/ajmg.b.31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine A. A neurodevelopmental perspective on male violence. Infant Ment Health J. 2019;40:84–97. doi: 10.1002/imhj.21761. [DOI] [PubMed] [Google Scholar]
- Ratnu VS, Emami MR, Bredy TW. Genetic and epigenetic factors underlying sex differences in the regulation of gene expression in the brain. J Neurosci Res. 2017;95:301–310. doi: 10.1002/jnr.23886.Genetic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif A, Richter J, Straube B, et al. MAOA and mechanisms of panic disorder revisited: from bench to molecular psychotherapy. Mol Psychiatry. 2014;19:122–128. doi: 10.1038/mp.2012.172. [DOI] [PubMed] [Google Scholar]
- Reynolds CA, Tan Q, Munoz E, et al. A decade of epigenetic change in aging twins: Genetic and environmental contributions to longitudinal DNA methylation. Aging Cell. 2020;19:1–12. doi: 10.1111/acel.13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowska J, Joshi A. From genotype to phenotype: through chromatin. Genes (basel) 2019;10:1–16. doi: 10.3390/genes10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103:273–279. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
- Schiele MA, Ziegler C, Kollert L, et al. Plasticity of functional MAOA gene methylation in acrophobia. Int J Neuropsychopharmacol. 2018;21:822–827. doi: 10.1093/ijnp/pyy050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurz H, Salie M, Tromp G, et al. The X chromosome and sex-specific effects in infectious disease susceptibility. Hum Genom. 2019;1:1–12. doi: 10.1186/s40246-018-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumay E, Logan J, Volkow ND, Fowler JS. Evidence that the methylation state of the monoamine oxidase A (MAOA) gene predicts brain activity of MAO A enzyme in healthy men. Epigenetics. 2012;7:1151–1160. doi: 10.4161/epi.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää MJ. Overview of techniques to account for confounding due to population stratification and cryptic relatedness in genomic data association analyses. Heredity (edinb) 2011;106:511–519. doi: 10.1038/hdy.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg RL, Nilsson KW, Wargelius H-L, et al. Adolescent girls and criminal activity: role of MAOA-LPR genotype and psychosocial factors. Am J Med Genet. 2007;144B:159–164. doi: 10.1002/ajmg.b.30360. [DOI] [PubMed] [Google Scholar]
- Sjöberg RL, Ducci F, Barr CS, et al. A non-additive interaction of a functional MAO-A VNTR and testosterone predicts antisocial behavior. Neuropsychopharmacology. 2009;33:425–430. doi: 10.1038/sj.npp.1301417.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoberg RL, Nilsson KW, Wargelius H-L, et al. Adolescent girls and criminal activity: role of MAOA-LPR genotype and psychosocial factors. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:159–164. doi: 10.1002/ajmg.b.30360. [DOI] [PubMed] [Google Scholar]
- Steadman HJ, Mulvey EP, Monahan J, et al. Violence by people discharged from acute psychiatric inpatient facilities and by others in the same neighborhoods. Arch Gen Psychiatry. 1998;55:393–401. doi: 10.1001/archpsyc.55.5.393. [DOI] [PubMed] [Google Scholar]
- Suderman M, Simpkin A, Sharp G, et al. Sex-associated autosomal DNA methylation differences are wide-spread and stable throughout childhood. bioRxiv. 2017;44:118265. doi: 10.1101/118265. [DOI] [Google Scholar]
- Szyf M. Prospects for the development of epigenetic drugs for CNS conditions. Nat Rev Drug Discov. 2015;14:461–474. doi: 10.1038/nrd4580. [DOI] [PubMed] [Google Scholar]
- Tiihonen J, Rautiainen M-R, Ollila HM, et al. Genetic background of extreme violent behavior. Mol Psychiatry. 2014;20:786–792. doi: 10.1038/mp.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng D-S, Chien C-C, Lung F-W, Yang C-Y. MAOA gene polymorphisms and response to mirtazapine in major depression. Hum Psychopharmacol. 2009;24:293–300. doi: 10.1002/hup. [DOI] [PubMed] [Google Scholar]
- Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in the aetiology of mental illness: review and methodological analysis. Mol Psychiatry. 2008;13:131–146. doi: 10.1038/sj.mp.4002067. [DOI] [PubMed] [Google Scholar]
- Unnikrishnan A, Freeman WM, Jackson J, et al. The role of DNA methylation in epigenetics of aging. Pharmacol Ther. 2019;195:172–185. doi: 10.1016/j.pharmthera.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiserman A. Developmental tuning of epigenetic clock. Front Genet. 2018;9:1–7. doi: 10.3389/fgene.2018.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen J, Nivard MG, Willemsen G, et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun. 2016;7:1–13. doi: 10.1038/ncomms11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J, Hagenbeek FA, Suderman M, et al. DNA methylation signatures of aggression and closely related constructs: a meta-analysis of epigenome-wide studies across the lifespan. Mol Psychiatry. 2021 doi: 10.1038/s41380-020-00987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven FEA, BooijKruijt LA-W, et al. The effects of MAOA genotype, childhood trauma, and sex on trait and state-dependent aggression. Brain Behav. 2012;2:806–813. doi: 10.1002/brb3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veroude K, Zhang-James Y, Fernandez-Castillo N, et al. Genetics of aggressive behavior: an overview. Am J Med Genet B Neuropsychiatr Genet. 2016;171B:3–43. doi: 10.1002/ajmg.b.32364. [DOI] [PubMed] [Google Scholar]
- Wagels L, Votinov M, Radke S, et al. Blunted insula activation reflects increased risk and reward seeking as an interaction of testosterone administration and the MAOA polymorphism. Hum Brain Mapp. 2017;38:4574–4593. doi: 10.1002/hbm.23685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakschlag LS, Kistner EO, Pine DS, et al. Genotype in pathways to youth antisocial behavior. Mol Psychiatry. 2011;15:928–937. doi: 10.1038/mp.2009.22.Interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Anderson AD, Hepler AB. Genetic relatedness analysis: Modern data and new challenges. Nat Rev Genet. 2006;7:771–780. doi: 10.1038/nrg1960. [DOI] [PubMed] [Google Scholar]
- Wheater ENW, Stoye DQ, Cox SR, et al. DNA methylation and brain structure and function across the life course: A systematic review. Neurosci Biobehav Rev. 2020;113:133–156. doi: 10.1016/j.neubiorev.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao FH, Chen XQ, He YH, Kong QP. Accelerated DNA methylation changes in middle-aged men define sexual dimorphism in human lifespans. Clin Epigenetics. 2018;10:1–6. doi: 10.1186/s13148-018-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Wang H, Kong Q. Dynamic DNA methylation during aging: a “prophet” of age-related outcomes. Front Genet. 2019;10:1–8. doi: 10.3389/fgene.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi P, Huen K, Davé V, et al. Sex differences in DNA methylation assessed by 450K BeadChip in newborns. BMC Genom. 2015;16:1–12. doi: 10.1186/s12864-015-2034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheleznyakova GY, Cao H, Schiöth HB. BDNF DNA methylation changes as a biomarker of psychiatric disorders: literature review and open access database analysis. Behav Brain Funct. 2016;12:17. doi: 10.1186/s12993-016-0101-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C, Domschke K. Epigenetic signature of MAOA and MAOB genes in mental disorders. J Neural Transm. 2018;125:1581–1588. doi: 10.1007/s00702-018-1929-6. [DOI] [PubMed] [Google Scholar]
- Ziegler C, Richter J, Mahr M, et al. MAOA gene hypomethylation in panic disorder-reversibility of an epigenetic risk pattern by psychotherapy. Transl Psychiatry. 2016;6:e773. doi: 10.1038/tp.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C, Wolf C, Schiele MA, et al. Monoamine oxidase A gene methylation and Its role in posttraumatic stress disorder: first evidence from the south eastern europe (SEE)-PTSD study. Int J Neuropsychopharmacol. 2018;21:423–432. doi: 10.1093/ijnp/pyx111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on reasonable request.