Summary
NLR family CARD domain containing protein 4 (NLRC4) inflammasome activation and the associated pyroptosis are critical for protection against infection by bacterial pathogens. This protocol presents a detailed procedure to activate and measure NLRC4 inflammasome activation and pyroptosis upon Salmonella Typhimurium infection. The techniques can be adapted to monitoring the activation of other types of inflammasomes and pathogenic stimuli.
For comprehensive details on the use and execution of this protocol, please refer to Dong et al. (2021).
Subject areas: Cell Biology, Immunology, Microbiology, Microscopy, Molecular Biology
Graphical abstract
Highlights
-
•
Detailed steps to obtain log-phase S. Typhimurium for NLRC4 inflammasome activation
-
•
Reproducible procedures for detection of Caspase-1 and IL-1β from culture supernatant
-
•
Procedures to identify the oligomerization of ASC
NLR family CARD domain containing protein 4 (NLRC4) inflammasome activation and the associated pyroptosis are critical for protection against infection by bacterial pathogens. This protocol presents a detailed procedure to activate and measure NLRC4 inflammasome activation and pyroptosis upon Salmonella Typhimurium infection. The techniques can be adapted to monitoring the activation of other types of inflammasomes and pathogenic stimuli.
Before you begin
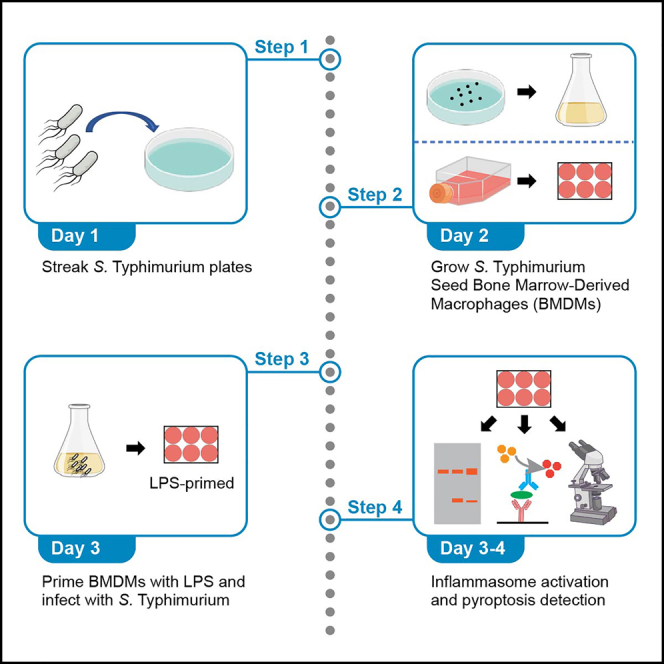
This protocol describes specific steps to monitor the activation of NLRC4 inflammasome and the associated pyroptosis in bone marrow-derived macrophages (BMDMs) upon Salmonella Typhimurium (S. Typhimurium) infection. Therefore, BMDMs and S. Typhimurium strain should be ready before you begin.
BMDMs isolation and culture
Timing: 7 days
-
1.
To differentiate the murine bone marrow cells into BMDMs, we cultured bone marrow cells in DMEM/F12 with 10% FBS, L-Glutamine (2 mM), Penicillin/Streptomycin (1:100), HEPES buffer (10 mM), and 20% L929 conditional medium in sterile plastic Petri dishes. Please refer to Curr. Protoc. Immunol. (Zhang et al., 2008) for the protocol of isolation and culture of murine BMDMs.
Note: We consistently isolated around 50–60 million bone marrow cells from one 8–12 weeks old mouse after red blood cells lysis. We cultured bone marrow cells in 10 cm Petri dishes with 5 million cells per dish. While bone marrow cells or BMDMs can be cryopreserved in liquid nitrogen with 90% FBS and 5% DMSO, we used freshly prepared cells for our experiments.
Note: If L929 conditional medium is not available, 20 ng/ml macrophage colony-stimulating factor (M-CSF) can be used for BMDMs differentiation. However, the yield of macrophages is less when recombinant M-CSF is used.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Polyclonal anti-IL1β | R&D system | AF-401-NA |
| Mouse Monoclonal anti-Caspase-1 (p20) | Adipogen | AG-20B-0042 |
| Rabbit Polyclonal anti-ASC | Adipogen | AG-25B- 0006-C100 |
| Rabbit Monoclonal anti-GSDMD | Abcam | ab209845 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, FITC | Thermo Fisher Scientific | F-2765 |
| Bacterial and virus strains | ||
| Salmonella Typhimurium | ATCC | 14028 |
| Chemicals, peptides, and recombinant proteins | ||
| Lipopolysaccharides from Escherichia coli O111:B4 | Millipore Sigma | L2630 |
| Disuccinimidyl suberate (DSS) | Thermo Fisher Scientific | 21655 |
| M-CSF | PeproTech | 315-02 |
| Pam3csk4 | InvivoGen | tlrl-pms |
| Critical commercial assays | ||
| IL-1β Mouse Uncoated ELISA Kit | Invitrogen | 88-7013-88 |
| Experimental models: Cell lines | ||
| Bone marrow-derived macrophages (BMDMs) | Isolated from C57BL/6J mice | N/A |
| L929 cell line | ATCC | CCL-1 |
| Software and algorithms | ||
| GraphPad prism 9 | GraphPad | https://www.graphpad.com/ |
| Image J | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| Other | ||
| DMEM/F12 | Cell Medium Facility, UIUC | N/A |
| FBS | Sigma | F2442-500ML |
| Penicillin-Streptomycin | Fisher Scientific | MT30002CI |
| HEPES | Fisher Scientific | MT25060CI |
| L-Glutamine | Fisher Scientific | MT25005CI |
| Petri Dishes with Clear Lid | Fisher Scientific | FB0875712 |
| LB broth | Fisher Scientific | BP97235 |
| LB Agar, powder | Thermo Fisher Scientific | 22700025 |
| 2× Laemmli Sample Buffer | Bio-Rad | 1610737 |
| Methanol | Fisher Scientific | A412-500 |
| Chloroform | Millipore Sigma | C2432-500ML |
| Pierce Protease Inhibitor Mini Tablets | Thermo Fisher Scientific | A32953 |
| DAPI Solution | Thermo Fisher Scientific | 62248 |
| Mowiol 4-88 | Millipore Sigma | 475904-100GM-M |
| 1,4-Diazabicyclo[2.2.2]octane | Millipore Sigma | D27802-25G |
| Glycerol | Millipore Sigma | G5516-500ML |
| Tween-20 | Millipore Sigma | P1379-500ML |
| Cover glass | Fisher Scientific | 12-541A |
| Western blot running and transfer system | Bio-Rad Laboratories | 1658001FC |
| Light microscope | Thermo Fisher Scientific | AMF5000 |
| Confocal microscope | Carl Zeiss AG | ZEISS LSM 510 |
| Microcentrifuge (22°C and 4°C) | Thermo Fisher Scientific | 75002432 |
| Cell/bacteria culture incubator | Fisher Scientific | 11-676-600 |
| Water bath | Fisher Scientific | Isotemp 215 |
| Hemocytometer | Fisher Scientific | 0267151B |
| Biosafety cabinet | Fisher Scientific | 1300 Series A2 |
Materials and equipment
TBS Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCL, pH 7.4 (1 M) | 50 mM | 2.5 mL |
| NaCl (2.5 M) | 150 mM | 3 mL |
| Triton X-100 | 0.5% (v/v) | 0.25 mL |
| Protease Inhibitor Mini Tablets | n/a | 5 tablets |
| ddH2O | n/a | 44.75 mL |
| Total | n/a | 50 mL |
Store at 4°C. Add one tablet to 10 mL buffer right before use.
Mounting Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris, pH 8.5 (1 M) | 100 mM | 5 mL |
| DAPI Solution (1 mg/mL) | 0.3 μg/mL | 15 μL |
| Mowiol 4-88 | 10% (w/v) | 5 g |
| 1,4-Diazabicyclo [2.2.2] octane | 1% (w/v) | 0.5 g |
| Glycerol | 25% (v/v) | 12.5 mL |
| ddH2O | n/a | 32.5 mL |
| Total | n/a | 50 mL |
Store at −20°C for 12 months. The medium remains stable for 1 month when stored at 4°C.
Other buffers:
| Name | Composition |
|---|---|
| PBS (1×), 1 L | 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.47 mM KH2PO4, 1 mM CaCl2, 0.5 mM MgCl2, pH adjusted to 7.2 with HCl |
| PBST | PBS with 0.02% Tween-20 (v/v) |
| Immunofluorescence Block buffer | PBST with 5% BSA (w/v) |
Step-by-step method details
Timing: 3 days for step 1
Timing: 24 h for step 2
Timing: 1 h for step 3
Timing: 15 min for step 4
Timing: 24 h for step 5
Timing: 24 h for step 6
-
1.
Prepare S. Typhimurium for infection
This step describes the details to prepare the log-phase S. Typhimurium for activating NLRC4 inflammasome in BMDMs.-
a.On day one, streak S. Typhimurium on a Lennox Broth (LB) plate with aseptic technique and place it in an incubator at 37°C 16 h.
-
b.On day two, seal the LB plate with parafilm in the morning and store it at 4°C to avoid the overgrowth of S. Typhimurium growth on the plate. In the evening, pick one colony into 5 mL sterile LB broth and grow the S. Typhimurium in the incubator at 37°C for 16 h with shaking (200 rpm).
-
c.On day three, sub-culture S. Typhimurium at 1:100 (add 20 μL bacterial culture into 2 mL fresh LB broth) for an additional 4–6 h with shaking (200 rpm) at 37°C.
-
d.Collect S. Typhimurium by centrifuging at 200 g for 3 min at 22°C.
-
e.Remove supernatant and wash the S. Typhimurium pellet twice with sterile PBS.
-
f.Resuspend the S. Typhimurium pellet with 1 mL sterile PBS and measure the OD600.
-
a.
CRITICAL: The sub-culture of S. Typhimurium at 37°C for an additional 4–6 h (Step c) is essential for inflammasome activation. During this period, S. Typhimurium reaches the log-phase of growth for the maximum expression of Salmonella pathogenicity island 1 (SPI-1), which activates NLRC4 inflammasome.
-
2.
Seed BMDMs and LPS priming
This step describes how to seed and prime BMDMs with LPS before S. Typhimurium infection.-
a.On day two, after 7 days of BMDMs differentiation, aspirate off the BMDM culture medium and add 5 mL sterile PBS to the Petri dish.
-
b.Gently detach the BMDMs from the Petri dish by pipetting with a 1 mL pipette.Note: BMDMs can be easily detached from the Petri dishes by pipetting. If tissue culture dishes are used for BMDM differentiation, cell can be collected with cell scraper by pre-treating with EDTA disassociation buffer (10 mM EDTA in PBS) for 10–15 min, followed by trypsin (0.25%) treatment for 2 min. A detailed comparison between these collecting methods is described in J. Immunol Methods (Chen et al., 2015).
-
c.Count the cell numbers with a hemocytometer and seed 1.5×106 BMDMs per well in a 6-well plate in DMEM/F12 with 10% FBS, L-Glutamine (2 mM), Penicillin/Streptomycin (1:100), HEPES buffer (10 mM). Culture BMDMs in a cell culture incubator for 16 h.Note: Adjust the cell numbers accordingly if other cell culture plates are used. We seed 1.0×106 BMDMs per well in a 12-well plate, and 0.5×106 BMDMs per well in a 24-well plate.
-
d.On day three, prime BMDMs with 0.5 μg/mL LPS (dissolved in endotoxin-free water) for 4 h before S. Typhimurium infection.Note: LPS priming for NLRC4 inflammasome activation is not always necessary. S. Typhimurium infection could significantly increase the protein level of IL-1β as early as 30 min, making LPS priming less critical in activating the NLRC4 inflammasome. Nevertheless, we recommend priming BMDMs, especially when your gene of interest could affect Il1b and Il18 expression independent of NLRC4 inflammasome activation during S. Typhimurium infection.
-
e.After LPS priming for 4 h, wash the BMDMs 3 times with sterile PBS. Change to the FBS-free medium before infection.Note: Intracellular LPS can activate non-canonical NLRP3 inflammasome; therefore, PBS washes after LPS priming is necessary. Alternatively, you can prime BMDMs with a TLR2 ligand, such as Pam3csk4.
-
a.
CRITICAL: Changing to FBS-free media before the infection is important since it helps reduce the number of non-specific proteins from FBS when precipitating proteins from cell culture supernatants.
-
3.
Infection of BMDMs with S. Typhimurium
This step describes the final step of infection and how to identify pyroptotic cells after inflammasome activation.-
a.Infect the primed BMDMs with S. Typhimurium at a multiplicity of infection (MOI) of 10. The simple conversion to determine the volume of S. Typhimurium added to one well at a certain Multiplicity of Infection (MOI) is described as:MOI × Cell number/well = OD600 × (1.5×109) × Volume (ml)Note: Log-phase S. Typhimurium expressing SPI-I activates NLRC4 inflammasomes after 30 min of infection at an MOI of 10, and about 80% BMDMs undergo inflammasome-induced pyroptosis within 1 h (Figure 1).
-
a.
-
4.
Cell culture supernatant protein precipitation
This step describes the detailed methanol-chloroform protein precipitation method to detect cleaved caspase-1 and cleaved IL-1β in the cell culture supernatant.
Inflammasome activation features the cleavage of pro-caspase-1, pro-IL-1β, and gasdermin-D (GSDMD) into their functional forms and can be detected by western blot. Western blot is used to detect cleaved caspase-1 (20 kDa) and GSDMD (35 kDa) in the cells, as well as secreted caspase-1 (20 kDa) and IL-1β (17 kDa) in the cell culture supernatant. We provide a reproducible cell culture supernatant protein precipitation method for the detection of secreted cleaved caspase-1 and cleaved IL-1β by western blot. Secreted IL-1β in the supernatant can also be used by ELISA.-
a.After the infection, collect 1 volume of supernatant (e.g., 600 μL) into a 1.5 mL Eppendorf tube. Add ¼ volume (150 μL) of chloroform and 1 volume of methanol (600 μL) to the supernatant and flip the tubes to mix.
-
b.Centrifuge at 13,800 g for 5 min at 22°C.
-
c.Aspirate off the top layer without disrupting the protein layer in the middle and add another 1 volume of methanol (600 μL).
-
d.Centrifuge at 13,800 g for 5 min at 22°C. The protein pellet should be visible at the bottom of the tube.
-
e.Remove the supernatant and air dry the pellet for 5 min.
-
f.Boil the pellet in 1× laemmli buffer before subject to western blot.Note: Since the cleaved caspase-1 and IL-1β have low molecular weights, we recommend using at least 12% SDS-PAGE to make sure that these small proteins don’t run out of the gel. We usually ran SDS-PAGE at a constant 100 V for 60 mins and transfer at a constant 400 A for 60 min.
-
a.
-
5.Detect the oligomerization of apoptosis-associated speck-like protein containing a CARD (ASC).
-
a.After the infection, lyse the cells with 200 ul TBS buffer for 30 min on a rocker at 4°C.
-
b.Collect the cells with cell scrapers and centrifuge at 6,000×g at 4°C for 15 min. Collect the Triton X-100 soluble fraction (lysate) as ASC western blot loading control.
-
c.Wash the Triton X-100 insoluble fraction (pellet) twice with PBS (6,000×g at 4°C for 5 min) and re-suspend the pellet with 300 ul TBS buffer.
-
d.Break the pellet by vigorous pipetting and vortexing.
-
e.Add disuccinimidyl suberate (DSS) to a final concentration of 4 mM and incubate in a water bath at 37°C for 30 min. You will observe precipitation during the crosslinking. Gently vortex the tube periodically.
-
f.After crosslinking, centrifuge at 6,000×g for 15 min at 4°C and dissolve the pellet in 1× laemmli buffer before subject to western blot.
-
a.
-
6.Detect ASC specks by immunofluorescence staining.
-
a.Before seeding BMDMs, autoclave square-shape cover glasses and put them into wells of a 6-well plate. Seed BMDMs at a lower density (5×105/well). Infect the cells with S. Typhimurium as described above.
-
b.After infection, gently move the cover glasses to the wells of a new 6-well plate. Fix and permeabilize cells with 100% methanol (prechilled at −20°C) at 22°C for 5 min.Note: No additional permeabilization step is required when methanol is used to fix the cells since 100% methanol fixes and permeabilizes cells at the same time. Alternatively, cells can be fixed and permeabilized with 4% paraformaldehyde (in PBS) and 0.5% TritonX-100 (in PBS), respectively, for 10 min at 22°C.
-
c.Wash the cells with PBS 3 times and incubate the cells with block buffer for 30 min at 22°C.
-
d.Dilute ASC antibody (2 μg/mL) in PBST and incubate at 4°C 10–16 h without shaking.
-
e.Wash the cells with PBS 3 times and incubate the cells with goat anti-rabbit FITC secondary antibody (2 μg/mL) for 1 h at 22°C in the dark.
-
f.Mount the cells with the mounting medium containing DAPI (0.3 ug/mL) and capture the images with a fluorescent or confocal microscope. We used the Zeiss LSM 510 Meta confocal microscope.Note: Besides FITC, any appropriate fluorophore conjugated goat anti-rabbit antibody can be used for the staining.
-
a.
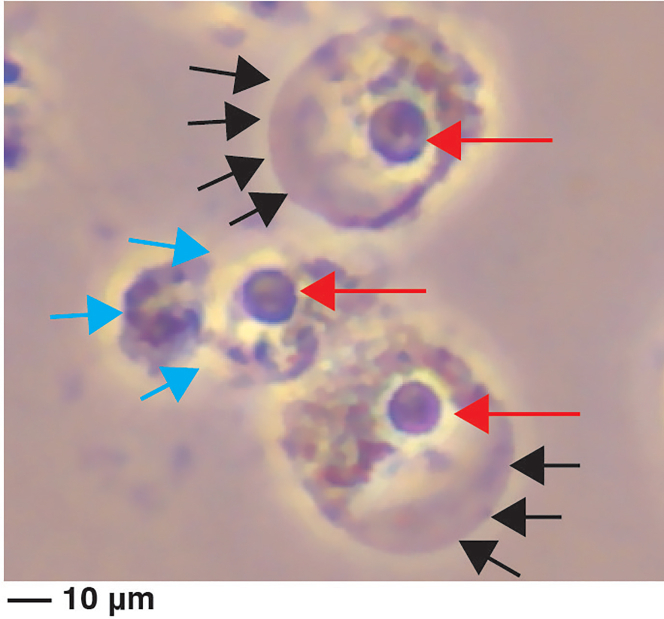
Figure 1.
Morphological characteristics of pyroptotic cells under a light microscope
LPS-primed BMDMs were infected with S. Typhimurium for 1 h (MOI, 10). The pyroptotic cells can be identified with protruding nuclei (red arrow), bubble-like protrusion (black arrow), and cell membrane rupture (cyan arrow). ). Scale bar, 10 μm
Expected outcomes
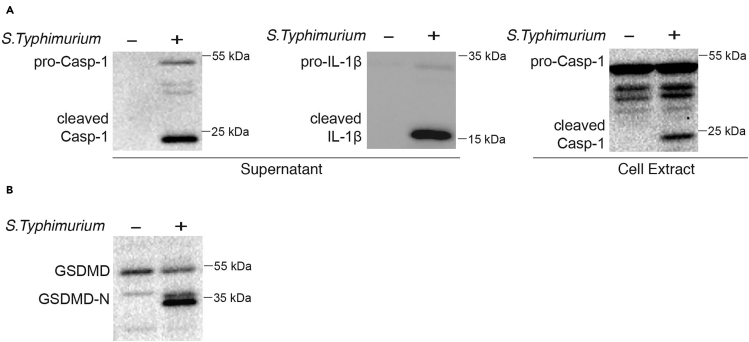
Western blot is the most widely used method to detect inflammasome activation with the cleaved Caspase-1 and IL-1β in cell culture supernatant (Figure 2A). Cleaved form of Caspase-1 could be detected in both cell extracts and supernatant when inflammasome is activated with small amount of pro-caspase-1 in the supernatant (Figure 2A). Cleaved IL-1β could be readily detected in the supernatant with small amount of pro- IL-1β (Figure 2A). Cleaved GSDMD forms pores on cell membrane and is recognized as the executor of pyroptosis. Therefore, cleaved GSDMD in western blot directly indicates pyroptosis and should be readily detected in the cell extracts (Figure 2B).
Figure 2.
Inflammasome activation detected by western blots
(A and B) LPS-primed BMDMs were infected with S. Typhimurium for 1 h (MOI, 10). Cleaved caspase-1, IL-1β in the cell culture supernatant, and cleaved caspase-1 in the cell extract indicates inflammasome activation (A). Cleaved GSDMD in the cell extract implies pyroptosis occurrence (B). Scale bar, 20 μm.
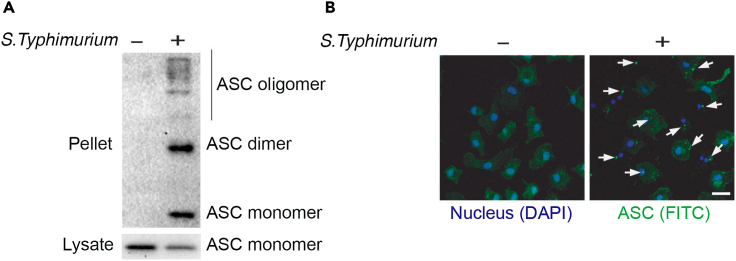
ASC oligomers function as platforms for recruiting pro-caspase-1 during inflammasome activation. Therefore, ASC oligomerization is considered as one of the hallmarks of inflammasome activation and can be detected by western blot (Figure 3A) and immunofluorescence (Figure 3B).
Figure 3.
ASC oligomer formation during inflammasome activation
(A and B) LPS-primed BMDMs were infected with S. Typhimurium for 1 h (MOI, 10). Triton X-100 soluble (lysate) and insoluble (pellet) fractions were immunoblotted with ASC antibody (A). Confocal microscopy of BMDMs infected with S. typhimurium for 1 h (MOI, 10) staining for ASC (FITC) and nucleus (DAPI). ASC specks are marked with arrows (B).
Limitations
Multiple inflammasomes have been identified thus far, including NLRP3, Aim2, Pyrin, and others (Karki and Kanneganti, 2019). Although they share a similar mechanism of activation, they respond differently to distinct stimuli. Therefore, this protocol is limited to NLRC4 inflammasome activation by S. Typhimurium. Other bacteria, such as Pseudomonas aeruginosa or Shigella flexneri, can also activate NLRC4 inflammasome; however, the protocol for bacteria culture and infection may differ from that presented here for S. Typhimurium. Different S. Typhimurium strains are also known to activate NLRC4 inflammasome at different time points via distinct mechanisms. Despite the diversity of stimuli, the preparation of BMDMs and the detection of inflammasome activation described in this protocol can be adapted to detect all other types of inflammasomes.
Troubleshooting
Problem
Failure to observe the characteristics of pyroptotic cells under the microscope (Figure 1).
Potential solution
Failure to activate the NLRC4 inflammasome by the stationary S. Typhimurium, which SPI-1 expression is inhibited, could be one of the reasons. Therefore, it is essential to use the log-phase S. Typhimurium in order to successfully activate NLRC4 inflammasome.
In addition, in the late stage of pyroptosis, the bubble-like protrusions burst eventually. This might lead to the failure of detecting the bubble-like structure for some pyroptotic cells. However, you can still check cleaved Caspase-1, IL-1β, and GSDMD signal by western blot to ensure a successfully inflammasome activation and pyroptosis.
Problem
Weak signals of cleaved Caspase-1 and cleaved IL-1β (Step 4).
Potential solution
During inflammasome activation, BMDMs undergo a hyperactivation state before pyroptosis, where most of the IL-1β secreted through the pores formed by GSDMD-N fragments on the cell membrane. Therefore, the IL-1β level in the cell extract is always low and hard to detect. To our experience, the cleaved caspase-1 levels are relatively even between cell extract and culture supernatant. Therefore, we recommend doing western blots against cleaved caspase-1 and cleaved IL-1β from the cell culture supernatant.
Problem
Non-specific inflammasome activation signal in uninfected control (Step 4).
Potential solution
Intracellular LPS can activate Caspase 11-dependent non-canonical NLRP3 inflammasome. If BMDMs are primed with LPS before infection, it is necessary to wash the cells sufficiently to remove LPS. Otherwise, the signals could be mixed signals from both non-canonical NLRP3 and NLRC4 inflammasomes. This can be evidenced by inflammasome activation in uninfected control cells. To completely eradicate the possibility of non-canonical NLRP3 inflammasome activation, the TLR2 ligand Pam3csk can be used to prime the cells.
Problem
Failure to detect inflammasome activation when using other stimuli.
Potential solution
The time needed to induce the activation of inflammasomes varies significantly with different stimuli. To determine the best time point for inflammasome activation, we recommend checking BMDMs visually for pyroptotic cells every 30 min to ensure that the optimal time points for the detection of inflammasome activation won’t be missed.
LPS priming is another way to secure a successful detection of cleaved caspase-1 and cleaved IL-1β. LPS priming increase pro-caspase-1 and pro-IL-1β drastically, and this provides cells with sufficient pro-caspase-1 and its substrate pro-IL-1β before inflammasome activation.
Problem
No ASC oligomer in western blot (Step 5e).
Potential solution
Failure to detect ASC oligomer is mainly due to the inefficient DSS crosslinking. If DSS crosslinking fails, all the oligomers would break down to monomer during the SDS-PAGE.
The critical step that ensures the detection of ASC oligomers by western blot is to break the Triton X-100 insoluble (pellet) fraction as much as possible before crosslinking. In addition, it is necessary to gently vertex the tube periodically during crosslinking.
Failure to activate the NLRC4 inflammasome could be another reason. We recommend doing a test with immunofluorescence first (Step 6). This will provide a clear visualization of ASC oligomers, an indication for the successful inflammasome activation.
Resource availability
Lead contact
Lin-Feng Chen (lfchen@illinois.edu)
Materials availability
Requests for resources and reagents should be directed to and will be fulfilled by the lead contact.
Acknowledgments
This work is supported in part by funds provided by the University of Illinois at Urbana-Champaign to L.-F.C. (RB18061)
Author contributions
Conceptualization and writing – review & editing, X.D. and L.-F.C.; writing – original draft, X.D.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Xingchen Dong, Email: xdong14@illinois.edu.
Lin-Feng Chen, Email: lfchen@illinois.edu.
Data and code availability
This study did not generate any data sets or code.
References
- Chen S., So E.C., Strome S.E., Zhang X. Impact of detachment methods on M2 macrophage phenotype and function. J. Immunol. Methods. 2015;426:56–61. doi: 10.1016/j.jim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Dong X., Hu X., Bao Y., Li G., Yang X.D., Slauch J.M., Chen L.F. Brd4 regulates NLRC4 inflammasome activation by facilitating IRF8-mediated transcription of Naips. J. Cell Biol. 2021;220:e202005148. doi: 10.1083/jcb.202005148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R., Kanneganti T.D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer. 2019;19:197–214. doi: 10.1038/s41568-019-0123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008;14:Unit 14 1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any data sets or code.