Abstract
Peroxisome proliferator-activated receptor α (PPARα) plays a key role in the transcriptional control of genes encoding mitochondrial fatty acid β-oxidation (FAO) enzymes. In this study we sought to determine whether the recently identified PPAR gamma coactivator 1 (PGC-1) is capable of coactivating PPARα in the transcriptional control of genes encoding FAO enzymes. Mammalian cell cotransfection experiments demonstrated that PGC-1 enhanced PPARα-mediated transcriptional activation of reporter plasmids containing PPARα target elements. PGC-1 also enhanced the transactivation activity of a PPARα-Gal4 DNA binding domain fusion protein. Retroviral vector-mediated expression studies performed in 3T3-L1 cells demonstrated that PPARα and PGC-1 cooperatively induced the expression of PPARα target genes and increased cellular palmitate oxidation rates. Glutathione S-transferase “pulldown” studies revealed that in contrast to the previously reported ligand-independent interaction with PPARγ, PGC-1 binds PPARα in a ligand-influenced manner. Protein-protein interaction studies and mammalian cell hybrid experiments demonstrated that the PGC-1–PPARα interaction involves an LXXLL domain in PGC-1 and the PPARα AF2 region, consistent with the observed ligand influence. Last, the PGC-1 transactivation domain was mapped to within the NH2-terminal 120 amino acids of the PGC-1 molecule, a region distinct from the PPARα interacting domains. These results identify PGC-1 as a coactivator of PPARα in the transcriptional control of mitochondrial FAO capacity, define separable PPARα interaction and transactivation domains within the PGC-1 molecule, and demonstrate that certain features of the PPARα–PGC-1 interaction are distinct from that of PPARγ–PGC-1.
The peroxisome proliferator-activated receptor α (PPARα) is a fatty acid-activated nuclear receptor that plays a key role in the transcriptional regulation of genes involved in cellular lipid and energy metabolism. PPARα together with PPARδ and PPARγ form a subgroup within the nuclear receptor superfamily (12, 17). In contrast to PPARα which is involved in the control of cellular lipid utilization, PPARγ has been shown to be a necessary component of the adipocyte differentiation program (22, 36). The biological function of PPARδ is unknown. A diverse group of compounds can act as activating ligands for PPARα including several prostaglandin derivatives, eicosanoids, and long-chain unsaturated fatty acids (8, 18, 39). To date, the majority of PPARα target genes identified are involved in cellular fatty acid oxidation (FAO) (22). We and others have previously demonstrated that PPARα mediates fatty acid-induced transcriptional control of several nuclear genes encoding mitochondrial FAO enzymes, including medium-chain acyl coenzyme A (acyl-CoA) dehydrogenase (MCAD) (9) and muscle carnitine palmitoyltransferase I (M-CPT I or CPT Iβ) (2, 9, 26, 41). PPARα is enriched in tissues with high oxidative energy demands that depend on mitochondrial FAO as a primary energy source such as heart and liver (17). PPARα is also expressed at high levels in brown adipose tissue (BAT), a specialized tissue in which mitochondrial FAO provides the reducing equivalents necessary for the generation of heat via the uncoupling of oxidative phosphorylation. Consistent with its regulatory role in mitochondrial FAO, the expression of PPARα is much higher in BAT than in white adipose tissue, which is a lipid storage tissue (15, 36). Recent studies of PPARα-null mice have confirmed that PPARα is necessary in vivo for high-level expression of mitochondrial and peroxisomal FAO enzyme genes in heart and liver under basal and stimulated conditions (1, 7, 24).
Evidence has emerged that nuclear receptors regulate transcription, in large part, via interactions with coactivator (e.g., CBP/p300, SRC-1, GRIP1, pCIP) or corepressor (e.g., N-CoR, SMRT) molecules (4, 5, 10, 11, 14, 20). Nuclear receptor interacting proteins regulate transcriptional activity by affecting chromatin structure through changes in the acetylation status of histones. Most coactivators are recruited to nuclear receptors upon ligand binding. Several coactivators such as SRC-1, which possesses intrinsic histone acetylase activity, also serve as adaptor molecules to link nuclear receptors to multiprotein complexes containing larger pleiotropic activator proteins such as CBP or p300 (35, 37, 40). The ligand-mediated activation of PPARs also involves coactivator networks (28, 44). Crystallographic studies have demonstrated that the binding of ligand to PPAR stabilizes the position of an alpha-helical domain (the AF2 helix) forming a “charge clamp” that interacts with an LXXLL motif within coactivator molecules (28). Indeed, SRC-1 has been shown to interact with the PPARs upon ligand binding leading to transcriptional activation (44). However, in vivo disruption of SRC-1 does not appear to impair PPARα's ability to respond to its ligand activators (32). Accordingly, other coactivator molecules must exist to mediate transactivation by PPARα.
Recently the PPAR gamma coactivator 1, or PGC-1, was cloned based on its BAT-enriched expression and ability to bind and coactivate PPARγ (31). In contrast to most nuclear receptor-coactivator interactions, PGC-1 was shown to interact with PPARγ in a ligand-independent manner. Interestingly, PGC-1 exhibits a tissue-enriched expression pattern with abundant levels in tissue types with high capacity for mitochondrial FAO such as BAT and heart (31). In addition, PGC-1 was shown to be markedly induced in BAT upon cold exposure (31), suggesting that it transduces physiologic stimuli to the transcriptional control of genes involved in thermogenesis. Given the key role of mitochondrial FAO in the thermogenic process in BAT and its importance in cardiac energy production, we hypothesized that PGC-1 cooperates with PPARα to regulate mitochondrial FAO enzyme gene expression. We show here that PGC-1 interacts with PPARα to coactivate target genes involved in mitochondrial FAO. Surprisingly, in contrast to the PGC-1–PPARγ interaction, PGC-1 binds PPARα in a ligand-influenced manner and the PPARα binding domains within the PGC-1 molecule are at least partially distinct from that reported for PPARγ. Moreover, we show that PGC-1 domains critical for PPARα interaction and transcriptional activation are distinct and separable. These results establish a role for PGC-1 as a PPARα coactivator in the control of cellular FAO.
MATERIALS AND METHODS
Plasmid constructs. (i) Mammalian expression vectors.
CDMRXRα and CDMPPARα have been described elsewhere (9). A HindIII fragment from pSVSport.PGC-1 (a gift from Bruce Spiegelman, Harvard Medical School) was cloned into pcDNA3.1myc/his (Invitrogen) to give myc/his.PGC-1, which contains amino acids 1 to 794 of PGC-1. PGC-1LXXFF was created by PCR-based mutagenesis and cloned into pcDNA3.1myc/his at the HindIII site to give a construct identical to the wild type except for the LKKLL-to-LKKFF mutation described in Results. The PGC-1 deletion series (PGC338, PGC284, and PGC120) was created by PCR which introduced a BglII site at the start of the coding sequence and a stop sequence at codons 339, 285, and 120. The resultant PCR products were cloned into pCMV-Tag1 (Stratagene) which fuses a FLAG epitope at the 5′ end of the PGC-1 deletions. The PGC-1 deletions were then subcloned into pcDNA3.1 (Invitrogen) and used for subsequent transfection studies. The Gal4-PGC-1 constructs were created by subcloning a BamHI fragment from the pcDNA3.1-PGC-1 plasmids into pCMX-Gal4 (a gift from David D. Moore, Baylor University). PPARαΔAF2 was generated by PCR which introduced a stop codon at codon 445, deleting the carboxy-terminal 18 amino acids. PPARα and PPARαΔAF2 were subcloned into the EcoRI site of pCMX-Gal4 to yield constructs expressing amino acids 25 to 462 and 25 to 444 of PPARα, respectively, fused to the Gal4 DNA binding domain (Gal4DBD).
(ii) Reporter constructs.
MCPTI.Luc.781 has been described elsewhere (2). (PPRE)3TKLuc contains three copies of a known PPARα response element derived from the peroxisomal acyl-CoA oxidase gene promoter (5′-TTCCGAACGTGACCTTTGTCCTGGTCCCCTTTA-3′) cloned into the BamHI site of a construct containing the herpes simplex virus thymidine kinase promoter linked to the luciferase gene (TKLuc). The TKLuc and Gal4TKLuc vectors were gifts from David D. Moore.
(iii) Retroviral expression vectors.
ΔU3nlsLacZ (a gift from Daniel S. Ory, Washington University School of Medicine) has been described previously (29). PPARα and PGC-1 were cloned into the NcoI/BamHI sites of the ΔU3 vector to give ΔU3-PPARα and ΔU3-PGC-1, respectively.
(iv) Bacterial expression vectors.
A BamHI/NotI fragment was isolated from pcDNA-PGC338, -PGC284, and -PGC120 and subcloned into pGex4T-3 (Pharmacia) to give GST-PGC338, GST-PGC284, and GST-PGC120, respectively. An EcoRI fragment from either pcDNA-PGC-1 or pcDNA-PGC-1LXXFF was subcloned into pGEX4T-3 to yield the GST.PGC190 or GST.PGCLXXFF construct, respectively.
Protein-protein interaction studies.
The glutathione S-transferase (GST) fusion proteins were produced in bacteria according to the manufacturer's instructions (Pharmacia). 35S-labeled PPARα, ΔAF2, ΔEF, and ΔDEF were produced in the TNT T7 Quick coupled in vitro transcription/translation system (Promega). For the “pulldown” assays, 50 μl of a 50% slurry of GST fusion protein bound to glutathione beads was resuspended in 500 μl of binding buffer (20 mM Tris [pH 7.5], 100 mM KCl, 0.1 mM EDTA, 0.05% Nonidet P-40, 10% glycerol, bovine serum albumin [BSA] [1 mg/ml], and 0.1 mM phenylmethylsulfonyl fluoride [PMSF]). Ligand was added where indicated. 35S-labeled PPARα or PPARαΔAF2 was added to the resuspended GST fusion proteins and incubated at room temperature for 1 h. The beads were spun down and washed three times in binding buffer alone or binding buffer with ligand. An equal volume of sample reducing buffer was added and boiled for 3 min. The samples were then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Cell culture and transfection studies.
3T3-L1 cells preadipocytes were maintained in Dulbecco's modified Eagle's medium (DME) and 10% calf serum as described previously (3). At confluency, the medium was changed to DME containing 10% fetal calf serum supplemented with 10 μg of insulin per ml, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), and 1 μM dexamethasone for the first 48 h. Thereafter, cells were placed in DME containing 10% fetal calf serum supplemented with 2.5 μg of insulin per ml every 48 h. CV-1 cells were maintained and transfected as previously described (23). Cells were transfected by the calcium phosphate coprecipitation method. Briefly, 4 μg of reporter construct and 500 ng of each expression construct or expression construct without insert was used. The day after transfection, oleic acid complexed to BSA (250 μM), 5,8,11,14-eicosate traynoic acid (ETYA) (10 μM), or vehicle control was added to the cells. The cells were harvested 24 h later in cell lysis buffer (Promega), and luciferase activity was measured as previously described. All transfection data are presented as means (± standard errors of means) of at least three separate transfection experiments done in triplicate.
Retroviral infection.
Production of recombinant Moloney murine leukemia virus (MMLV) was used for retroviral transfer as described elsewhere (29). Expression of the cDNA is driven by a promoter derived from the MMLV long terminal repeat region. Briefly, virus production was carried out in the packaging 293GPG cell line. Retroviral expression constructs were transiently transfected into the cells by use of Lipofectamine (Gibco BRL) per the manufacturer's instructions. Virus produced from this transient transfection was used to infect 293GPG cells to produce a stable population of virus-producing cells. Virus produced from these cells was concentrated by centrifugation at 25,000 × g for 90 min. Concentrated virus was resuspended in 3T3-L1 growth medium. A single 8-h exposure of subconfluent cells to virus resulted in greater than 95% infection efficiency as judged by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining of cells infected with virus producing LacZ. Infected cells were grown to confluency and induced to differentiate as described above. At day 6 after the addition of differentiation media, ETYA or dimethyl sulfoxide (DMSO) vehicle was added to the cells.
RNA blot analyses.
Total RNA was isolated from virus-infected cells 48 h later using the RNAzol (Tel-Test, Inc.) method. Northern blot analysis was performed with QuikHyb (Stratagene) using random-primed 32P-labeled cDNA probes. cDNA probes encoding mouse MCAD, rat long-chain acyl-CoA dehydrogenase (LCAD) (a gift from Bryan Hainline, Indiana University), rat L-CPT I, mouse PPARα, and mouse PGC-1.
[14C]palmitate oxidation studies.
Measurements of palmitate oxidation were performed as described elsewhere (9). Briefly, 3T3-L1 preadipocytes seeded in 25-cm2 flasks were infected as described above with recombinant retroviral particles encoding a LacZ control, PPARα, PGC-1, or PPARα and PGC-1. Seventy-two hours later, [1-14C]palmitate (American Radiolabeled Chemicals, St. Louis, Mo.) was added to a final concentration of 200 nCi/ml. The flasks were sealed and fitted with a center well containing a piece of Whatman no. 1 filter paper (1 in. by 1.5 in.). After 6 h, the 14CO2 was released from the culture medium by acidification with 2 ml of 6 N HCl. The 14CO2 was collected overnight by alkalinization of the filter paper with 250 μl of 2 N NaOH. 14CO2 was then measured by scintillation counting of the filters. The measurements presented are a compilation of three separate experiments performed in duplicate or triplicate. Statistical analysis was performed using analysis of variance coupled with the Scheffe test.
RESULTS
PGC-1 enhances PPARα-mediated transcriptional activation.
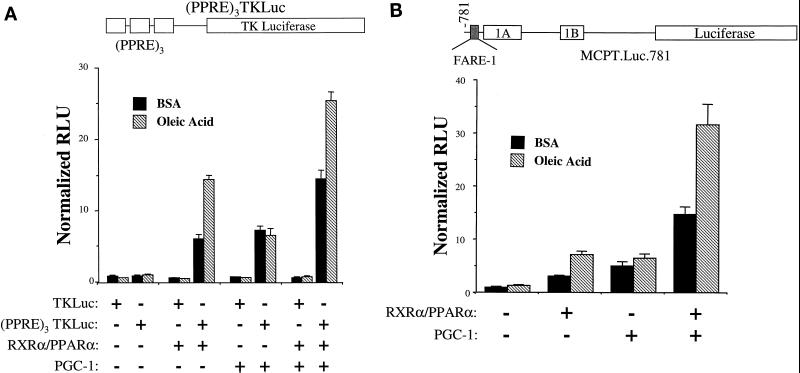
To examine the effect of PGC-1 on PPARα activity, cotransfection studies were performed in the CV-1 cell line. A target reporter plasmid containing a PPARα response element (PPRE) derived from the peroxisomal acyl-CoA oxidase gene promoter multimerized upstream of the thymidine kinase minimal promoter [(PPRE)3TKLuc] was employed in these studies. Cotransfection of expression vectors for RXRα/PPARα or PGC-1 had no effect on a reporter plasmid lacking the PPRE (TKLuc) (Fig. 1). As expected, cotransfection of expression plasmids for RXRα and PPARα activated (PPRE)3TKLuc more than sixfold, an effect that was further enhanced by addition of the PPARα ligand, oleic acid (Fig. 1A). Cotransfection of a PGC-1 expression vector with the RXRα/PPARα expression plasmids increased the level of PPARα-mediated activation of the reporter both in the presence and absence of oleic acid (Fig. 1A). Cotransfection of PGC-1 in the absence of cotransfected RXRα/PPARα also resulted in a reproducible albeit lower magnitude activation of (PPRE)3TKLuc, an effect that may be mediated through interactions with endogenous nuclear receptors other than PPARα because oleic acid had no effect on this activity (Fig. 1A).
FIG. 1.
PGC-1 enhances PPARα-mediated transactivation. The heterologous promoter reporter construct (PPRE)3TKLuc (A) or the homologous promoter reporter MCPT.Luc.781 (B) was transiently transfected into CV-1 cells. Expression constructs encoding RXRα/PPARα and/or PGC-1, were cotransfected in the presence of BSA vehicle or oleic acid (250 μM) complexed to BSA as indicated. Bars represent mean (± standard error) relative luciferase units (RLU) normalized (=1.0) to the activity of (PPRE)3TKLuc (A) or MCPT.Luc.781 (B) cotransfected with expression vector backbone in the absence of ligand. All transfection data represent the means of at least three independent experiments.
The enzyme carnitine palmitoyltransferase I (CPT I) catalyzes the rate-limiting step in the import of long-chain fatty acids into the mitochondrion prior to entering the FAO cycle. We and others have shown that the gene encoding muscle CPT I (M-CPT I) is a PPARα target (2, 26, 41). A reporter construct containing a portion of the human M-CPT I gene promoter (MCPT.Luc.781) containing the known PPARα response element, FARE-1 (2), was employed to further examine the coregulatory effect of PGC-1 with PPARα on a bona fide target gene. As expected, RXRα/PPARα activated the transcription of MCPT.Luc.781, an effect that was enhanced by the addition of oleic acid (Fig. 1B). As with the heterologous PPARα target construct, cotransfection of PGC-1 activated transcription of MCPT.Luc.781 in the absence or presence of exogenous RXRα/PPARα. However, in this case, a clear synergistic effect was seen between PGC-1 and RXRα/PPARα in the presence of PPARα ligand.
PPARα and PGC-1 cooperate to increase mitochondrial FAO enzyme gene expression and cellular FAO rates.
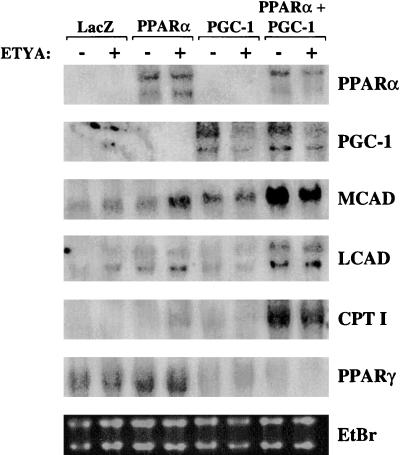
To determine whether PGC-1 coactivates the PPARα-mediated upregulation of mitochondrial FAO enzyme gene expression, PPARα and PGC-1 were ectopically overexpressed alone or together in the 3T3-L1 cell line, using a retroviral expression system. Upon differentiation, 3T3-L1 cells most closely resemble the white adipocyte, a cell with inherent low expression of mitochondrial FAO enzymes. 3T3-L1 preadipocytes were infected with recombinant retroviral particles encoding LacZ (control), PPARα, PGC-1, or PPARα and PGC-1. Each condition was evaluated in the presence or absence of the known PPARα activator, ETYA. Following infection, the cells were switched to differentiation media (see Materials and Methods), and RNA blot analysis was performed to examine the level of expression of PPARα, PGC-1, and several mitochondrial FAO enzyme genes (MCAD, LCAD, and CPT I). The retrovirus-mediated expression of PGC-1 and PPARα mRNA and proteins was documented by RNA blot analysis (Fig. 2) and immunoblotting studies (data not shown), respectively. Independent expression of either PPARα or PGC-1 led to a modest increase in the levels of mRNAs encoding MCAD, LCAD, and CPT I compared to cells infected with the retroviral backbone alone (Fig. 2). The induction conferred by PPARα was greatest with the addition of the exogenous PPARα ligand, ETYA. However, coexpression of both PPARα and PGC-1 led to a marked coordinate increase in the levels of the FAO enzyme mRNAs in the absence or presence of PPARα ligand (Fig. 2). The expression of the PPARγ gene was downregulated in cells overexpressing PGC-1, indicating that the observed effects were not a result of a PGC-1–PPARγ interaction. These results demonstrate that PPARα and PGC-1 cooperatively induce PPARα target genes involved in mitochondrial FAO.
FIG. 2.
PPARα and PGC-1 cooperate to induce PPARα gene target markers of the mitochondrial FAO pathway. Autoradiographs of Northern blot analysis performed with total RNA (15 μg) isolated from 3T3-L1 preadipocytes infected with recombinant retroviral particles encoding LacZ, PPARα, PGC-1, or PPARα and PGC-1 as indicated at the top are shown. Cells were grown to confluence and induced to differentiate as described in Materials and Methods. Six days after addition of differentiation media, ETYA (+) or vehicle control (−) was added. RNA was isolated 48 h after addition of ligand or vehicle. The blot was hybridized with the radiolabeled cDNA probes indicated on the right. The ethidium bromide (EtBr)-stained RNA is included as a control for loading and RNA integrity.
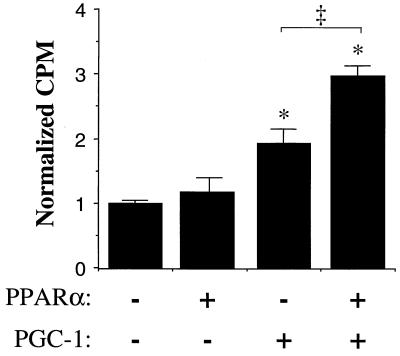
To determine whether cellular FAO rates were increased by the cooperative action of PPARα and PGC-1, palmitate oxidation studies were performed in cells infected with the retroviral vectors described above. Flux through the mitochondrial FAO pathway was determined by measurement of the rate of release of 14CO2 from 3T3-L1 preadipocytes following incubation with [1-14C]palmitate. For these studies, 3T3-L1 preadipocytes were used to avoid dilution of labeled palmitate with intracellular long-chain fatty acid known to be present in lipid droplets within adipocytes. The amount of 14CO2 produced following a 6-h incubation with [1-14C]palmitate was significantly greater in cells overexpressing both PGC-1 and PPARα compared to either alone (Fig. 3). These results are consistent with that of the gene expression studies shown in Fig. 2 and indicate that PPARα and PGC-1 cooperatively increase cellular long-chain FAO capacity.
FIG. 3.
PPARα and PGC-1 increase cellular palmitate oxidation rates. Palmitate oxidation studies were performed on 3T3-L1 preadipocytes in culture infected with retroviral vectors expressing LacZ (control), PPARα, PGC-1, or PPARα and PGC-1 as described in Materials and Methods. Following incubation of the cells with [1-14C]palmitate for 6 h, the amount of 14CO2 liberated from the cells was measured by scintillation counting. The bars represent mean (± standard error) 14CO2 (in counts per minute) normalized (=1.0) to the mean value obtained with the LacZ-infected control cells. An asterisk denotes a significant difference (P < 0.01) compared to the control value. The double dagger denotes a significant difference (P < 0.01) between the values for PPARα without and with PGC-1.
PPARα binds PGC-1 in a ligand-influenced manner.
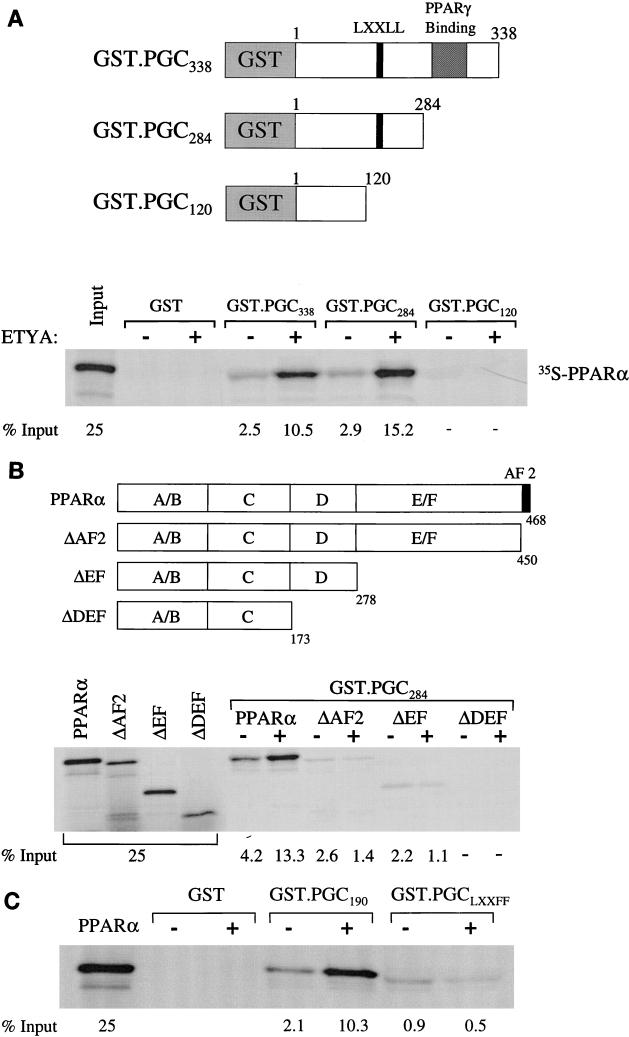
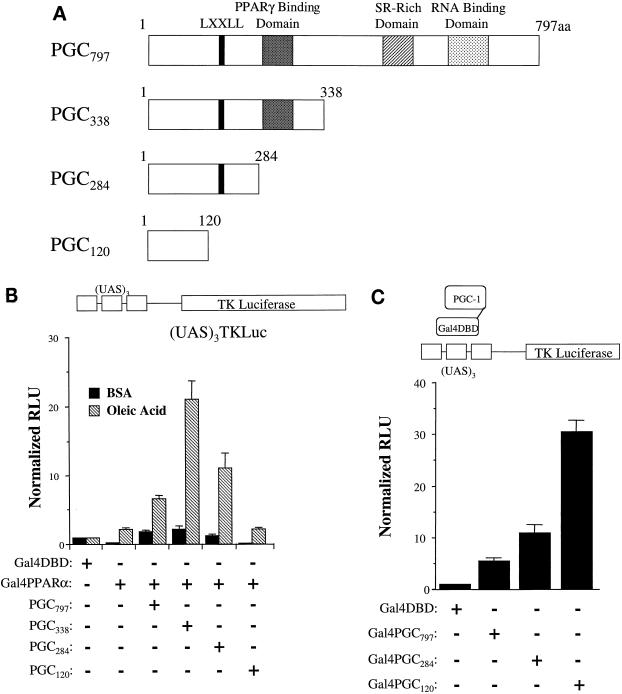
A series of in vitro GST pulldown assays were performed to confirm a direct interaction between PPARα and PGC-1 and to map the corresponding interaction domains. A recent study demonstrated that PGC-1 interacts with PPARγ in a ligand-independent manner via a region located between amino acids 292 and 338 (31). Based on this information, we constructed several GST-PGC-1 fusion proteins (Fig. 4A) to be used in pulldown studies with 35S-methionine-labeled, in vitro-translated PPARα. Initial experiments were performed with a GST-PGC fusion protein containing 338 amino acids of PGC-1 including the PPARγ binding domain (GST.PGC338 [Fig. 4A]). A modest interaction between GST.PGC338 and PPARα was detected in the absence of ligand (Fig. 4A). However, addition of the PPARα ligand ETYA significantly increased the PPARα–GST-PGC338 interaction (Fig. 4A). Surprisingly, strong, ligand-influenced PPARα binding was also observed with a PGC-1 deletion mutant lacking the PPARγ binding domain (GST.PGC284), indicating that in contrast to PPARγ (31), the region between amino acids 284 and 338 does not play a significant role in the interaction between PPARα and PGC-1. However, further deletion from amino acids 284 to 120 (GST.PGC120), abolished the PGC-1–PPARα interaction in the presence or absence of ligand. These results indicate that a critical PPARα binding domain exists in a region located between amino acids 120 and 284 of the PGC-1 molecule. Moreover, these data identify two stark differences between the nature of the interaction between PGC-1 and PPARα compared with that of PPARγ. First, whereas the PGC-1–PPARγ interaction is ligand independent (31), the binding of PPARα by PGC-1 is increased by ligand. Second, the PGC-1 domains required for the interaction with PPARγ and PPARα are distinct.
FIG. 4.
PGC-1 interacts with PPARα. (A) The GST–PGC-1 fusion proteins used for PPARα pulldown assays are shown schematically at the top with numbers corresponding to the amino acids within the PGC-1 molecule (31). The locations of the domain necessary for PPARγ binding (31) and the single LXXLL domain are also shown. Autoradiographs depicting the results of GST pulldown assays performed with 35S-labeled PPARα and several GST–PGC-1 fusion proteins or GST alone in the presence of the DMSO vehicle (−) or the PPARα ligand ETYA (+) are shown at the bottom of each panel. The numbers below each pulldown product shown in the autoradiographs indicate the percent total input as determined by phosphorimager analysis. 25% of the input is shown for comparison. (B) Pulldown studies using 35S-labeled PPARα deletion mutant proteins (shown at the top) and GST-PGC284. (C) The results of pulldown studies performed with the GST.PGC190 fusion protein and the LXXLL mutant, GST.PGCLXXFF.
As demonstrated in Fig. 4A, ligand potentiated the interaction between PPARα and PGC-1. The AF2 domain of nuclear receptors has been shown to mediate ligand-responsive interactions with coactivators through LXXLL motifs present in the latter. PGC-1 contains one such LXXLL sequence located at amino acids 142 to 146 (LKKLL). As shown in Fig. 4A, deletion of a region (amino acids 120 to 184) of the PGC-1 molecule which contains an LKKLL motif abolished the interaction between PPARα and PGC-1. Given these results, we predicted that the PPARα–PGC-1 interaction required the PPARα AF2 domain. To explore this possibility, GST pulldown experiments were performed with a PPARα deletion mutant lacking the AF2 helical domain (ΔAF2). GST-PGC284 was used in these experiments. In contrast to the full-length PPARα, the interaction of GST-PGC-1284 with the ΔAF2 PPARα protein was markedly diminished and was not enhanced by ligand (Fig. 4B). Further removal of the ligand binding domain (ΔEF) of PPARα did not change the PGC-1 interaction binding pattern compared to that of the ΔAF2 protein. However, removal of the D domain (ΔDEF) abolished the residual, ligand-independent PGC-1 binding. Interestingly, the D domain of PPARγ was shown previously to be required for binding to PGC-1 (31). These results demonstrate a key role for the PPARα AF2 domain in the interaction with PGC-1 and identify both ligand-influenced and ligand-independent PGC-1 interaction regions within the PPARα molecule.
To explore further the role of the LXXLL domain of PGC-1 in binding with PPARα, protein-protein interaction studies were repeated with a mutant PGC-1 protein fragment in which the LXXLL motif was mutated. For these experiments, PPARα pulldowns were performed with a PGC-1–GST fusion protein containing the amino-terminal 190 amino acids, including the LXXLL domain (GST.PGC190), or a mutant GST.PGC190 fragment in which LXXLL was changed to LXXFF (GST.PGCLXXFF). As shown in Fig. 4C, the interaction of GST.PGCLXXFF with 35S-labeled PPARα was markedly reduced compared with that of GST.PGC190. Moreover, ligand did not increase the GST.PGCLXXFF-PPARα interaction. These results, which are consistent with the data shown in Fig. 4A and B, demonstrate that the PGC-1 LXXLL motif is a critical participant in the PGC-1–PPAR interaction.
The AF2-LXXLL interaction is necessary for the coactivation of PPARα by PGC-1.
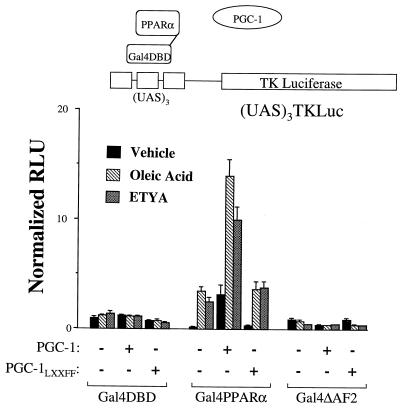
To establish functional correlates of the protein-protein interaction studies shown in Fig. 4, a mammalian cell protein hybrid system was employed. This system also allowed a functional assessment of the PGC-1–PPARα interaction in the absence of the background, PPARα-independent, PGC-1-mediated activation observed in the cotransfection studies shown in Fig. 1. In these experiments we took advantage of the transcriptional activation properties of PGC-1. An expression vector for the full-length PGC-1 was cotransfected with an expression vector encoding PPARα fused, in frame, to the Gal4 DNA binding domain (Gal4-PPARα) or the Gal4 DNA binding domain alone (Gal4DBD), along with a reporter construct containing three copies of the Gal4 binding site upstream of the TK minimal promoter, [(UAS)3TKLuc]. Gal4-PPARα, in the absence of ligand, modestly repressed (UAS)3TKLuc (Fig. 5). Addition of the PPARα ligands (oleic acid or ETYA) activated (UAS)3TKLuc only in the presence of Gal4-PPARα. Addition of PGC-1 markedly increased the transcriptional activation by Gal4-PPARα in the presence or absence of ligand (Fig. 5), whereas PGC-1 had no effect on the Gal4DBD alone.
FIG. 5.
Coactivation of PPARα by PGC-1 requires intact AF2 and LXXLL motifs. To examine functional correlates of the GST pulldown interaction studies, a mammalian cell hybrid system was employed (shown schematically at the top). PPARα or PPARΔAF2 was fused to the Gal4 DNA binding domain (DBD) and cotransfected with an expression plasmid encoding PGC-1 or a mutant PGC-1 in which the LXXLL motif was mutated (PGCLXXFF). A plasmid containing the Gal4 upstream activating sequence (UAS) multimerized upstream of TK luciferase [(UAS)3TKLuc] was used as a reporter in these experiments. Transfections were performed in the presence of the PPARα ligands (oleic acid or ETYA) or vehicle controls. Bars represent mean RLU normalized (=1.0) to the value obtained with Gal4DBD cotransfected with expression plasmid backbone in the presence of vehicle.
To determine whether as predicted by the results of the GST pulldown experiments, the PPARα–PGC-1 interaction was mediated by the AF2 domain of PPARα and the LXXLL motif of PGC-1, the effect of deleting or mutating each of these domains on the PGC-1-mediated coactivation of PPARα was evaluated. As expected, a Gal4-PPARα fusion lacking the AF2 activation domain (Gal4-ΔAF2) was unresponsive to PPARα ligand (Fig. 5) and was unable to confer the coactivation by PGC-1. Conversely, mutation of the LXXLL motif within the full-length PGC-1 (PGC-1LXXFF) prevented the PGC-1-mediated activation of PPARα, either in the presence or absence of ligand (Fig. 5). Taken together with the data shown in Fig. 4, these results demonstrate that the coactivation of PPARα by PGC-1 involves an AF2-LXXLL interaction.
Identification of a potent transactivation domain within the PGC-1 molecule.
In order to delineate the region of PGC-1 critical for its transactivating function, we evaluated the PPARα coactivating function of a series of carboxy-terminal PGC-1 deletion mutants (Fig. 6A). Surprisingly, removal of the C-terminal region (PGC338) actually enhanced the PGC-1-mediated increase in ligand-dependent activation of Gal4-PPARα (Fig. 6B). The portion of the molecule deleted contains a serine-arginine rich (SR) domain and a region which has similarity to RNA binding domains of other proteins. Further carboxy-terminal deletions, which included removal of the region shown previously to be involved in binding to PPARγ (PGC284), also increased PGC-1 activity but not to the same level as PGC338. A vector containing only the NH2-terminal 120 amino acids of PGC-1 (PGC120), however, had no effect on PPARα activity. Parallel immunoblotting experiments performed with an antibody to the FLAG epitope confirmed that each of the PGC protein fragments was expressed (data not shown). These results indicate that a transactivation function is conferred by the NH2-terminal 284 amino acids of the PGC-1 molecule.
FIG. 6.
The NH2-terminal region of PGC-1 is required for transactivation function. (A) Schematic representations of PGC-1 deletion mutants used in the transactivation studies shown in Fig. 6B and 7. aa, amino acids. A region homologous with known RNA binding domains and a serine-arginine (SR)-rich domain are shown. (B) Gal4-PPARα was cotransfected with expression vectors into CV-1 cells for each of the PGC-1 deletion mutants shown in panel A in the presence or absence of oleic acid. Bars represent RLU normalized (=1.0) to the activity of the (UAS)3TKLuc reporter cotransfected with Gal4DBD and empty PGC-1 expression vector. (C) Expression vectors encoding PGC-1 deletion mutants (Fig. 6A), fused to the Gal4DBD, were cotransfected with the (UAS)3TKLuc reporter plasmid (one-hybrid assay). The values represent RLU normalized (=1.0) to that of the Gal4DBD alone.
The data shown in Fig. 6B identify a transactivating function in the NH2-terminal region of PGC-1 but do not distinguish between the action of a distinct transactivation domain and the PPARα binding function which was mapped to this region. Accordingly, to further delineate the PGC-1 transactivation domain, PGC797, PGC284, and PGC120 were fused to the Gal4DBD and were evaluated in a one-hybrid assay in the CV-1 cell line. This strategy allowed us to evaluate the PGC-1 transactivating function independent of the interaction with PPARα. Whereas PGC120 had no activity in the mammalian two-hybrid system (Fig. 6B), Gal4-PGC120 exhibited a potent transcriptional activation function (Fig. 6C), despite the fact that this region lacks the PPARα binding domain. These data demonstrate that the NH2-terminal 120 amino acids of PGC-1 comprises a potent activation region which is distinct and separable from the PPARα interacting domain.
DISCUSSION
Coactivator molecules play a critical role in the transcriptional activation of nuclear receptor target genes. A current challenge in the understanding of nuclear receptor biology is to elucidate the mechanisms involved in the receptor-coactivator interaction. PGC-1 is a coactivator molecule identified recently based on its ability to interact with PPARγ (31). In this report, we extend the role of PGC-1 by demonstrating that it is a bona fide coactivator for PPARα in the transcriptional control of genes involved in mitochondrial FAO. We also demonstrate that the interaction between PPARα and PGC-1 is influenced by ligand and involves domains distinct from that described previously for the PPARγ–PGC-1 interaction. Finally, a potent transactivation domain separable from the PPARα interaction domains has been identified within the PGC-1 molecule.
PGC-1, a new member of the rapidly growing list of nuclear receptor coactivators, has several unique characteristics. First, in contrast to most coactivators reported to date, PGC-1 exhibits a tissue-restricted expression pattern. Second, PGC-1 expression is induced by physiologic stimuli; PGC-1 mRNA levels increase dramatically upon cold exposure in tissues with a role in heat production, namely, BAT and skeletal muscle (31). We have also found that fasting induces PGC-1 gene expression in heart (J. J. Lehman, T. C. Leone, and D. P. Kelly, unpublished data). These observations suggest that PGC-1 transduces extracellular stimuli to the transcriptional control of genes involved in cellular energy metabolism. The observation that PGC-1 expression is induced by fasting and cold exposure, physiologic conditions known to increase cellular lipid utilization, suggested that PGC-1 may function as a regulator of mitochondrial β-oxidation. Accordingly, we explored the possibility that PGC-1 could serve as a coactivator for PPARα, a key factor in the transcriptional control of the mitochondrial FAO pathway (2, 9, 21). Our results indicate that PGC-1 is indeed capable of enhancing PPARα-mediated transactivation based on the following observations. (i) PGC-1 interacts directly with PPARα in GST pulldown assays and mammalian protein hybrid studies. (ii) PGC-1 coactivates PPARα-mediated transactivation of known PPARα target elements in homologous and heterologous promoter contexts. (iii) Ectopic overexpression of PPARα and PGC-1 expression in 3T3-L1 cells, which have an inherently low capacity for FAO, cooperatively induces the expression of mitochondrial FAO enzyme genes and increases cellular palmitate oxidation rates. The coactivating effect of PGC-1 on PPARα in the 3T3-L1 cells was independent of its known interaction with PPARγ given that the expression of the latter was downregulated in the PGC-1 overexpressing cells. The reason for the lower expression of PPARγ in the PGC-1-expressing cells is unclear but could be related to a squelching effect or a biologically relevant feedback inhibition. These results establish PGC-1 as a PPARα coactivator in the control of mitochondrial FAO enzyme gene expression. The high-level expression of PGC-1 in heart and BAT (31), tissues with high expression of mitochondrial FAO enzymes, is consistent with a role for this coactivator in the control of the FAO pathway.
Recently, PGC-1 was shown to induce the expression of genes encoding mitochondrial proteins involved in electron transport, mitochondrial number, and cellular mitochondrial DNA content (31, 38). These results are indicative of mitochondrial biogenesis. In further support of a role for PGC-1 in the control of mitochondrial oxidative capacity, the results of our cellular oxidation studies demonstrated a significant increase in the oxidation of palmitate to CO2 in cells overexpressing both PPARα and PGC-1, consistent with an increase in both FAO and tricarboxylic acid cycle flux, two key mitochondrial pathways. We propose that PGC-1 serves as the elusive link between the gene regulatory pathway involved in the transcriptional control of nuclear genes encoding mitochondrial FAO enzymes and the broad program of mitochondrial biogenesis in tissues with high fatty acid utilization rates such as heart and BAT.
Our results demonstrate several surprising differences in the nature of the interaction of PGC-1 with PPARα compared to that of PPARγ. In contrast to the ligand-independent interaction of PGC-1 with PPARγ (31), we found that ligand influences the PPARα–PGC-1 interaction in GST pulldown studies. Cell cotransfection experiments demonstrated that the AF-2 domain of PPARα and the LXXLL domain of PGC-1 were required for the cooperative PPARα–PGC-1 interaction. We also found that the previously defined PPARγ interaction domain within the PGC-1 molecule was dispensable for the PPARα interaction, consistent with the differences in ligand dependence. However, we did not observe a strong dependence on exogenous PPAR ligand in the cotransfection experiments shown in Fig. 1. This discrepancy may be explained by the presence of endogenous ligand in the cell culture media or in the CV-1 cells. Alternatively, the PGC-1–PPARα interaction could occur via both ligand-independent and ligand-influenced mechanisms.
The primary structure of PGC-1 provides few clues as to how it activates transcription. Many coactivator molecules contain histone acetylase (HAT) activity which is thought to be critical to the transcriptional activation function. However, the PGC-1 molecule does not contain significant amino acid sequence similarity with any known histone acetylase. PGC-1 does contain an SR-rich region juxtaposed to a second region with homology to RNA binding domains of other proteins. These two domains are seen in the SR family of splicing factors as well as a group of proteins which interact with the C-terminal domain of RNA polymerase II (25, 30, 42). However, these latter regions were not necessary for transactivation function in our one-hybrid assay experiments. Rather, a separate domain comprised of the NH2-terminal 120 amino acids of the PGC-1 molecule was responsible for transactivation. The mechanism whereby the transactivation domain of PGC-1 exerts its effects is unknown. There are no similarities between this region of the molecule and any other published protein sequence. It is possible that this region contains HAT activity or binds to a protein with this activity such as CBP/p300. However, other proteins that interact simultaneously with nuclear receptors and other coactivators (e.g., SRC-1) usually contain multiple LXXLL motifs which mediate interactions between both sets of proteins (27). PGC-1 contains only one LXXLL sequence mitigating against the possibility that it interacts with both PPARα and p300/CBP, although interaction with other coactivator proteins via novel domains is possible. Recently, several nuclear receptor coactivators have been found to exist in large preformed complexes containing multiple proteins (13, 33). These complexes are likely involved in mediating activation from a variety of transcription factors in addition to nuclear receptors including p53, VP16, and NF-κB. The delineation of the precise mechanism involved in the PGC-1 transactivation function and its relationship to the recently described transcriptional regulatory complexes are important avenues for future studies.
Recent studies have indicated that PPARα plays a pivotal role in the control of cellular fatty acid utilization pathways in response to diverse physiologic conditions including fasting (16, 19, 23), nutritional alterations (16), and aging (6). The PPARα regulatory pathway has also been implicated in disease states including cardiac hypertrophy (34), obesity (6), and diabetes mellitus (19, 43). The factors involved in the modulation of PPARα activity are largely unknown. Availability of ligand has been considered one potential key regulator of PPARα activity. The identification of a PPARα coactivator molecule that is induced by physiologic stimuli adds a new layer of regulatory complexity for PPARα as well as for the entire nuclear receptor superfamily. We speculate that certain nuclear receptor coactivators, such as PGC-1, serve to transduce physiologic input to changes in gene expression. Our results identify the mitochondrial FAO enzyme genes as one candidate group of targets regulated by such a mechanism.
ACKNOWLEDGMENTS
We thank Bruce Spiegelman for the generous gift of a PGC-1 cDNA and helpful discussions, David D. Moore for the Gal4DBD and (UAS)3TKLuc vectors, Daniel Ory for the ΔU3 vector and helpful advice regarding retroviral particle production, and Kelly Hall for expert secretarial assistance.
This work was supported in part by NIH grant RO1-DK45416. D.P.K. is an Established Investigator of the American Heart Association. R.B.V. received support from an NIH training grant (5-T32-HL07275) during this work.
ADDENDUM IN PROOF
During the final review of this paper, Puigserver et al. (Science 286:1368–1371, 1999) demonstrated that the PGC-1 transactivation domain interacts with the coactivator SRC-1.
REFERENCES
- 1.Aoyama T, Peters J M, Iritani N, Nakajima T, Furihata K, Hashimoto T, Gonzalez F J. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARα) J Biol Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 2.Brandt J, Djouadi F, Kelly D P. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor α. J Biol Chem. 1998;273:23786–23793. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 3.Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 4.Cavailles V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J D, Umesono K, Evans R M. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costet P, Legendre C, Moré J, Edgar A, Galtier P, Pineau T. Peroxisome proliferator-activated receptor α-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. J Biol Chem. 1998;273:29577–29585. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]
- 7.Djouadi F, Weinheimer C, Saffitz J E, Pitchford C, Bastin J, Gonzalez F J, Kelly D P. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator-activated receptor α-deficient mice. J Clin Invest. 1998;102:1083–1091. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman B M, Chen J, Evans R M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulick T, Cresci S, Caira T, Moore D D, Kelly D P. The peroxisome proliferator activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci USA. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 11.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 13.Ito M, Yuan C X, Malik S, Gu W, Fondell L D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 14.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 15.Jones P S, Savory R, Barratt P, Bell A R, Gray T, Jenkins N A, Gilbert D J, Copeland N G, Bell D R. Chromosomal localisation, inducibility, tissue-specific expression and strain differences in three murine peroxisome proliferator-activated receptor genes. Eur J Biochem. 1995;233:219–226. doi: 10.1111/j.1432-1033.1995.219_1.x. [DOI] [PubMed] [Google Scholar]
- 16.Kersten S, Seydoux J, Peters J M, Gonzalez F J, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliewer S A, Sundseth S S, Jones S A, Brown P J, Wisely G B, Koble C S, Devchand P, Wahli W, Willson T M, Lenhard J M, Lehman J M. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroetz D L, Yook P, Costet P, Bianchi P, Pineau T. Peroxisome proliferator-activated receptor α controls the hepatic CYP4A induction adaptive response to starvation and diabetes. J Biol Chem. 1998;273:31581–31589. doi: 10.1074/jbc.273.47.31581. [DOI] [PubMed] [Google Scholar]
- 20.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee S S T, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemberger T, Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: a nuclear receptor signaling pathway in lipid physiology. Annu Rev Cell Dev Biol. 1996;12:335–363. doi: 10.1146/annurev.cellbio.12.1.335. [DOI] [PubMed] [Google Scholar]
- 23.Leone T C, Cresci S, Carter M E, Zhang Z, Lala D S, Strauss A W, Kelly D P. The human medium chain acyl-CoA dehydrogenase gene promoter consists of a complex arrangement of nuclear receptor response elements and Sp1 binding sites. J Biol Chem. 1995;270:16308–16314. doi: 10.1074/jbc.270.27.16308. [DOI] [PubMed] [Google Scholar]
- 24.Leone T C, Weinheimer C J, Kelly D P. A critical role for the peroxisome proliferator-activated receptor alpha (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 26.Mascaró C, Acosta E, Ortiz J A, Marrero P F, Hegardt F G, Haro D. Control of human muscle-type carnitine palmitoyltransferase I gene transcription by peroxisome proliferator-activated receptor. J Biol Chem. 1998;273:8560–8563. doi: 10.1074/jbc.273.15.8560. [DOI] [PubMed] [Google Scholar]
- 27.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T-M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;13:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 29.Ory D S, Neugeboren B A, Mulligan R C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patturajan M, Wei X, Berezney R, Corden J L. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol Cell Biol. 1998;18:2406–2415. doi: 10.1128/mcb.18.4.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 32.Qi C, Zhu Y, Pan J, Yeldandi A V, Rao M S, Maeda N, Subbarao V, Pulikuri S, Hashimoto T, Reddy J K. Mouse steroid receptor coactivator-1 is not essential for peroxisome proliferator-activated receptor α-regulated gene expression. Proc Natl Acad Sci USA. 1999;96:1585–1590. doi: 10.1073/pnas.96.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:825–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 34.Sack M N, Disch D L, Rockman H A, Kelly D P. A role for Sp and nuclear receptor transcription factors in a cardiac hypertrophic growth program. Proc Natl Acad Sci USA. 1997;94:6438–6443. doi: 10.1073/pnas.94.12.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer T E, Jenste G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 36.Tontonoz P, Hu E, Spiegelman B M. Stimulation of adipogenesis in fibroblasts by PPAR γ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 37.Torchia J, Rose D W, Inostroza J, Damei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 38.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, Spiegelman B M. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 39.Xu H E, Lambert M H, Montana V G, Parks D J, Blanchard S G, Brown P J, Sternbach D D, Lehmann J M, Wisely G B, Willson T M, Kliewer S A, Milburn M V. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 40.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu G-S, Lu Y-C, Gulick T. Co-regulation of tissue-specific alternative human carnitine palmitoyltransferase Iβ gene promoters by fatty acid enzyme substrate. J Biol Chem. 1998;273:32901–32909. doi: 10.1074/jbc.273.49.32901. [DOI] [PubMed] [Google Scholar]
- 42.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y T, Shimabukuro M, Wang M Y, Lee Y, Higa M, Milburn J L, Newgard C B, Unger R H. Role of peroxisome proliferator-activated receptor α in disease of pancreatic β cells. Proc Natl Acad Sci USA. 1998;95:8898–8903. doi: 10.1073/pnas.95.15.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, Qi C, Calandra C, Rao M S, Reddy J K. Cloning and identification of mouse steroid receptor coactivator-1 (mSRC-1), as a coactivator of peroxisome proliferator-activated receptor γ. Gene Expr. 1996;6:185–195. [PMC free article] [PubMed] [Google Scholar]