Abstract
Striated muscle needs to maintain cellular homeostasis in adaptation to increases in physiological and metabolic demands. Failure to do so can result in rhabdomyolysis. The identification of novel genetic conditions associated with rhabdomyolysis helps to shed light on hitherto unrecognized homeostatic mechanisms. Here we report seven individuals in six families from different ethnic backgrounds with biallelic variants in MLIP, which encodes the muscular lamin A/C-interacting protein, MLIP. Patients presented with a consistent phenotype characterized by mild muscle weakness, exercise-induced muscle pain, variable susceptibility to episodes of rhabdomyolysis, and persistent basal elevated serum creatine kinase levels. The biallelic truncating variants were predicted to result in disruption of the nuclear localizing signal of MLIP. Additionally, reduced overall RNA expression levels of the predominant MLIP isoform were observed in patients’ skeletal muscle. Collectively, our data increase the understanding of the genetic landscape of rhabdomyolysis to now include MLIP as a novel disease gene in humans and solidifies MLIP’s role in normal and diseased skeletal muscle homeostasis.
Keywords: MLIP, myopathy, hyperCKemia, rhabdomyolysis, cardiomyopathy
Lopes Abath Neto et al. identify recessive pathogenic genetic variants in MLIP in six families from different ethnic backgrounds with a myopathy characterized by exercise-induced muscle pain, susceptibility to spontaneous episodes of rhabdomyolysis, and elevated baseline serum creatine kinase levels.
See Ravenscroft and Cabrera-Serrano (doi:10.1093/brain/awab308) for a scientific commentary on this article.
Introduction
Rhabdomyolysis refers to intermittent catastrophic failure of muscle homeostasis and integrity, resulting in muscle breakdown and release of muscle cytosolic content into the circulation. Rhabdomyolysis can give rise to serious complications such as myoglobinuria with acute kidney injury. Clinical symptoms can include severe muscle weakness, myalgia, and muscle swelling with a serum creatine kinase (CK) rising above 1000 U/l or >5 times the upper limit of normal.1 Rhabdomyolysis can result from acquired (trauma, ischaemia, infection, toxin or drug-related) or from genetic causes.
Genetic susceptibility to rhabdomyolysis has been associated with variants in many genes coding for proteins important for muscle energy production. For example, genes encoding proteins involved in glycogen and fatty acid metabolism, mitochondrial complexes, or genes that cause disorders of intramuscular calcium release (RYR1, CACNA1S), have been implicated in rhabdomyolysis.2 Certain muscular dystrophies (DMD, FKRP, ANO5) and miscellaneous neurodegenerative disorders (SIL1, TSEN54, TANGO2) have also been associated with rhabdomyolysis.2-5 Identification of a trigger leading to the episodes (including exercise, temperature, diet, infection, emotional stress, and exposure to drugs or anaesthetics), in addition to ancillary tests, such a muscle biopsy and targeted metabolic testing, can help guide genetic diagnosis towards established conditions.2,6 However, a significant number of patients with recurrent episodes of rhabdomyolysis remain undiagnosed despite extensive clinical and genetic investigations.2,7,8
Muscular lamin A/C-interacting protein (MLIP, encoded by MLIP, MIM *614106) was first identified through its direct interaction with lamins A and C, which are intermediate filament proteins of the nuclear lamin.9 MLIP is a ubiquitously expressed protein with highest expression levels found in the heart, skeletal muscle and brain.9,10 Work on MLIP has focused on its potential role in protecting heart muscle from stress. The murine orthologue Mlip has been shown to be essential for the heart’s capacity to adapt to stress through regulation of the Akt/mTOR pathways.11 Additionally, Mlip-deficient mice show accelerated development of cardiomyopathy11,12 and a 7-fold increase in the prevalence of centralized nuclei in muscle fibres.10 From a clinical perspective, patients with end-stage heart failure due to dilated cardiomyopathy were shown to have reduced cardiac expression of MLIP.12 More recently, an exome-wide association study highlighted a common single nucleotide variant in MLIP as a contributing risk allele for dilated cardiomyopathy.13 Despite these observations suggesting a role in heart protection, the exact function of the protein has yet to be elucidated. Moreover, MLIP has not yet been implicated in causing Mendelian disease in humans.
Here we report seven individuals of different ethnic and geographic backgrounds with biallelic inactivating variants in MLIP, who present with a consistent phenotype characterized by mild muscle weakness, exercise-induced muscle pain, and variable susceptibility to episodes of rhabdomyolysis superimposed on persistent basal elevated serum creatine kinase levels (hyperCKemia). This patient cohort expands the genetic landscape of vulnerability to rhabdomyolysis and proposes a potential metabolic function for MLIP in skeletal muscle.
Materials and methods
Standard protocol approvals and patient consents
The study was approved by the Institutional Review Board (IRB) of the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health (Protocol 12-N-0095), the IRB of the Japanese National Center of Neurology and Psychiatry, and the Ethics Committee of the Instituto de Investigación Hospital 12 de Octubre (i+12). Written informed consent and appropriate assent were obtained by qualified investigators, or the revised Federal Policy for the Protection of Human Subjects from the United States Department of Health and Human Services was followed. Medical history, clinical evaluations, and ancillary exams were performed as part of the standard neurological or genetic evaluations of each institution. Genomic DNA was obtained from blood based on standard procedures.
Muscle biopsies
Muscle biopsies were obtained as part of the regular clinical diagnostic work-up of each institution and were evaluated by standard light microscopy protocols. Electron microscopy was performed on a single patient (Patient P6), whose muscle specimen was fixed with cold glutaraldehyde in phosphate buffer, postfixed in osmium tetroxide, dehydrated in graded alcohol series and embedded in epoxy resin according to the standard procedure. Semithin sections were prepared with an ultramicrotome, stained with toluidine blue, and examined with a light microscope. Thin sections were made with an ultramicrotome and examined under a JEOL JEM-100SX TEM (Jeol Ltd).
Whole exome sequencing
Whole exome sequencing (WES) on genomic DNA extracted from blood was performed on all patients. The protocol details can be found in the Supplementary material.
Immunofluorescence studies
Cold methanol fixed 9-μm muscle cross-sections from biopsy samples of Patients P1, P3 and P4 were incubated with primary antibodies at 4°C overnight [MLIP (Abcam ab131324) and emerin (Novocastra NCL-EMERIN)]. Antibody labelling was detected with secondary antibodies left for 1 h at room temperature (Alexa 568-conjugated goat anti-mouse IgG and Alexa 488-conjugated goat anti-rabbit IgG) (ThermoFisher Scientific). Prepared muscle sections were imaged with a Leica SP5 confocal microscope (Leica).
Immunoblotting studies
Total protein was extracted from frozen skeletal muscle samples of Patients P1–P3 using a 1% NP40 lysis buffer. Nuclear protein fractionation was additionally extracted with a urea buffer. Aliquots of cytosolic and nuclear protein (50 μg) were loaded on 4–12% SDS-polyacrylamide gels (NuPAGETM 4-12% Bis-Tris Protein Gels, ThermoFisher Scientific), transferred to a PVDF membrane with an iBlotTM PVDF Mini transfer stack (ThermoFisher Scientific), and immunostained with polyclonal antibodies against MLIP (Abcam ab131324, 1:200; and a non-commercial antibody developed by Dr Patrick Burgon, University of Ottawa, 1:2009) and a polyclonal antibody against lamin A/C (Leica NCL-LAM-A/C, 1:200).
Droplet digital PCR
RNA was isolated from frozen skeletal muscle specimens of Patients P1, P3 and P4, as well as control muscle, using TRIzol™ manufacturer’s instructions (ThermoFisher Scientific). Complementary DNA was prepared from 500 ng of RNA using SuperScriptIV manufacturer’s instructions (ThermoFisher Scientific). Custom TaqMan primers and FAM-labelled probe assays were designed for each exon-exon junction and synthesized as PrimeTime Assays (Integrated DNA Technologies). Sequences of the primers and probe were as follows: E1-E2 Forward 5′-CCTCTGTGTCTCTCAGTCTTTC-3′, Reverse 5′-AGATCAGAGGTTTGGCTTCAG-3′, Probe 5′-CCACCAGCAGAGACCGTCAGTTTC-3′; E3-E4 Forward 5′-TCCTGAAATGCTTCATGGGAT-3′, Reverse 5′-CAAGCTGCTCAGAGGTAGTG-3′, Probe 5′- AGCATGGGCAGTTAACTTCTTCTCC-3′; E4-E5 Forward 5′-GTGCATTGTCCATGTCAACAG-3′, Reverse 5′-AAGCAATGTGTTTGTAGGGATTG-3′, Probe 5′- ACAAGACCAAGTCAAGCTACAAGGCT-3′. Housekeeping gene β-2-microglobulin (B2M) assay was purchased from ThermoFisher (Assay hs00187842). The droplet digital PCR (ddPCR) assay was performed at the National Cancer Institute/Center for Cancer Research Genomics Core on a C1000 Touch Thermal Cycler (Bio-Rad; Cat. #1851197) after droplet generation with a Bio-Rad QX200 Automated Droplet Generator (Cat. #1864101) and a QX200 Droplet Reader (Cat. #1864003). Probe Supermix (No dUTP; Cat. #1863023) at a 1× final concentration was used with 900 nM final primer concentration, 200 nM probe concentration, and a 1:50 dilution of cDNA in water as template. Droplet generation and thermocycling were completed according to the manufacturer’s instructions (Bio-Rad Laboratories). QuantaSoftTM software was used for droplet analysis. The threshold for each assay was determined with a negative control (RNA without reverse transcriptase). Concentration (copies/µl) values for all assays were normalized by dividing with the concentration obtained from B2M housekeeping assay, for each sample.
Alternative splicing study in Patient P7
Muscle biopsy from Patient P7 and a control was disaggregated in NZYol™ (NZYtech) using a Dounce homogenizer on ice. Total RNA was isolated following the manufacturer’s instructions. Reverse transcription was performed on total RNA from experimental samples using an NZY First-Strand cDNA Synthesis Kit (NZYtech) according to the manufacturer’s protocol. Complementary DNAs were used as templates for reverse transcription PCR (RT-PCR) using BioMix™ Red (Bioline). Thermal cycling conditions were 94°C (2 min), 45 cycles at 94°C (10 s), 60°C (10 s) and 72°C (1 min 30 s), and a final extension at 72°C for 7 min on a 2720 Thermal Cycler (Applied Biosystems). The primers used were: MLIPF3 CCAAAACACAGGGGACTGAT in exon 3, and MLIPR4 TTCAGAGGTCAGTCCAGGCT in exon 4 downstream of the examined variant for MLIP. The PCR products were purified and the variant screening was performed by bidirectional sequencing using the BigDyeTM Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on an ABI 3730 DNA Analyzer (Applied Biosystems).
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
Clinical characterization
We identified seven individuals (henceforth referred to as Patients P1–P7) from six families (Patients P6 and P7 are siblings) with MLIP pathogenic variants. Detailed individual clinical data can be found in the Supplementary material. Clinical, imaging, and genetic findings are summarized in Table 1, with further details provided in Supplementary Table 1.
Table 1.
Clinical, imaging, and molecular data of seven patients with MLIP pathogenic variants
| Patient | P1 | P2 | P3 | P4 | P5 | P6 | P7 |
|---|---|---|---|---|---|---|---|
| Age (y)/sex | 17/Male | 9/Male | 13/Female | 8/Female | 5/Female | 19/Male | 12/Male |
| Age at onset (y) | 3 | 2 | 7 | 2 | 2.5 | 0.75 (8 months) | 1.5 (19 months) |
| Symptoms | Muscle cramping, rhabdomyolysis | Myalgia on exertion, rhabdomyolysis | Muscle cramping and stiffness on exertion, rhabdomyolysis | Recurrent muscle pain and tenderness on exertion | Recurrent thigh pain (quadriceps) relieved by rest | Myalgia and severe episodes of rhabdomyolysis | Myalgia and severe episodes of rhabdomyolysis |
| Triggers | No apparent triggers | No apparent triggers | No apparent triggers | No overt CK spikes | No overt rhabdomyolysis | Mild exercise, muscle overuse | Tantrums, cold, mild exercise |
| Baseline serum CK level (U/l) | 500–4000 | 2000 | 500–3000 | 2000–3000 | 297 | 1287 | 2325 |
| Highest CK level (U/l) | 200 000 | 4810 | 32 000 | 5000 | 9348 | 60 000 | 30 000 |
| Muscle imaging | MRI: subtle STIR hyperintensity on vastus lateralis | MRI: mild hyperintensity in bilateral gastrocnemius muscles | MRI: mild atrophy of quadratus femoris, subtle TIRM hyperintensities in the vastus lateralis | Not performed | Normal | Normal | Normal |
| Variant status | Homozygous | Compound heterozygous | Homozygous | Homozygous | Compound heterozygous | Homozygous | Homozygous |
| Variant (MLIP NM_1281746) | c.1706delA (p.His569ProfsTer7) |
c.1799delT (p.Phe600SerfsTer10) and c.2410delT (p.Ser804LeufsTer49) |
c.595C>T (p.Gln199Ter) | c.2497C>T (p.Arg833Ter) |
c.2461_2464delGTCA (p.V821KfsX31) and c.2556+1G>A (IVS10+1G>A) |
r.[682a>g;613_682del] (p.Thr206IlefsTer17) | r.[682a>g;613_682del] (p.Thr206IlefsTer17) |
For additional details see Supplementary Table 1. STIR = short tau inversion recovery; TIRM = turbo inversion recovery magnitude.
There were four male and three female patients with ages varying from 5 to 19 years. Two patients (Patients P1 and P4) were born from consanguineous parents (first degree cousins), while Patient P3’s parental status is unknown because she was adopted. The onset of symptoms varied from 8 months to 7 years of age. All patients (7/7) had muscle pain on exertion, which was relieved with rest. Five (5/7) showed recurrent episodes of rhabdomyolysis. Of these, three (3/5) did not have apparent consistent triggers, and two (2/5, siblings Patients P6 and P7) had rhabdomyolysis associated with excessive muscle use. Patient P7 potentially had episodes triggered by exposure to cold temperatures. Serum CK levels during episodes of rhabdomyolysis reached spikes of 5000 to 200 000 U/l. All patients (7/7) consistently had elevated baseline serum CK levels ranging from 300 to 4000 U/l.
Physical exam was notable for mild proximal muscle weakness [Medical Research Council (MRC) grade 4], in five patients (5/7), while two had normal strength on examination. Patient P5 also had joint hypermobility in fingers and knees. In the two siblings (Patients P6 and P7), physical exam was unremarkable. Patient P3 was found to have mild left ventricular dysfunction on an echocardiogram, for which an angiotensin converting enzyme (ACE) inhibitor was introduced. None of the other patients had clinically significant involvement of the heart. Incidental structural imaging findings identified in other patients included a small patent foramen ovale in Patient P1 and a mild dilation of the aortic valve in Patient P6.
Muscle MRI was performed in six patients (6/7), with normal findings in three (3/6) and with minimal changes in the remainder (3/6). Among the mild changes observed, there were subtle short tau inversion recovery (STIR) and turbo inversion recovery magnitude (TIRM) hyperintensities on the vastus lateralis in Patients P1 and P3, respectively, and mild hyperintensity in bilateral gastrocnemii in Patient P2. Muscle ultrasound for two patients (2/7, Patients P1 and P3) showed mild symmetrical involvement, predominantly in proximal muscles. In Patient P3, there was additionally moderate involvement of the lateral gastrocnemii.
EMG was performed on only two patients (2/7; Patients P1 and P3), and showed a myopathic process without membrane irritability. Due to the need for patient cooperation and the discomfort associated with needle insertion, in the paediatric population EMG testing frequently is not obtained when the primary clinical concern is for a purely myopathic process, in which case muscle imaging can yield more relevant information.
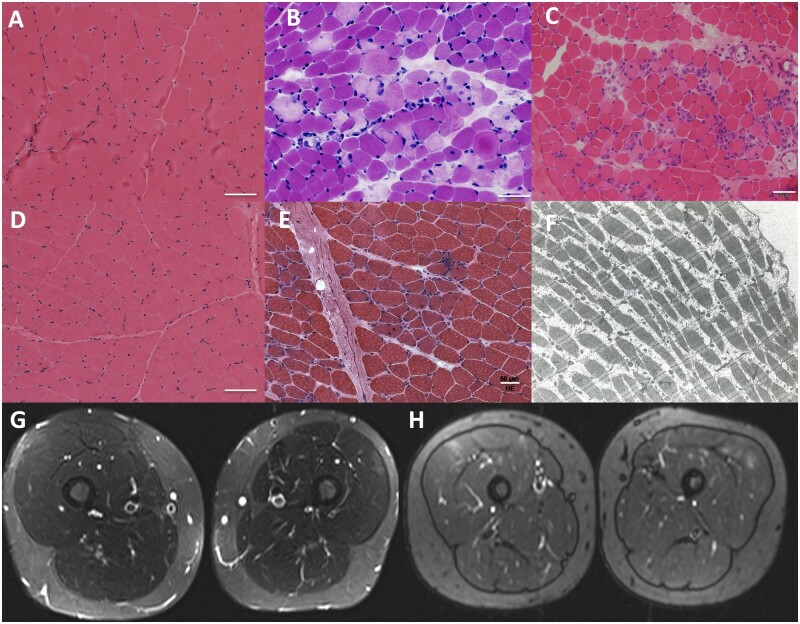
Muscle biopsy was performed on six patients (6/7). Two patients (2/6) showed mild and non-specific myopathic changes, while four (4/6) showed variable degrees of myofibre necrosis and regeneration, ranging from scattered isolated necrotic fibres to overt dystrophic changes (Fig. 1).
Figure 1.
Histological and imaging findings in patients with MLIP pathogenic variants. Muscle biopsies varied from showing normal to minimal findings in Patients P1 and P3 (A and D, respectively; haematoxylin and eosin, ×200) to revealing focal areas of necrosis and regeneration of muscle fibres in Patients P2, P5 and P4 (B, C and E, respectively; haematoxylin and eosin, ×200). Electron microscopy imaging performed on Patient P6 (F) did not reveal ultrastructural abnormalities. MRI of thighs in Patients P1 and P3 show mild signal hyperintensity in the vastus lateralis (G and H, respectively; G: STIR sequence; H: TIRM sequence).
Identification of MLIP pathogenic variants
WES analysis identified variants in the MLIP gene (NM_001281746.2) in all seven patients described in this study, which were predicted to be pathogenic.
Patient P1 was homozygous for the frameshift variant p.His569ProfsTer7 (c.1706delA), located in exon 4, predicted to lead to a downstream premature stop codon at position 575 (Fig. 2). The variant was not identified in the genome aggregation population variant database (gnomAD).14 Parents of Patient P1 were found to be heterozygous for this variant, consistent with autosomal recessive inheritance.
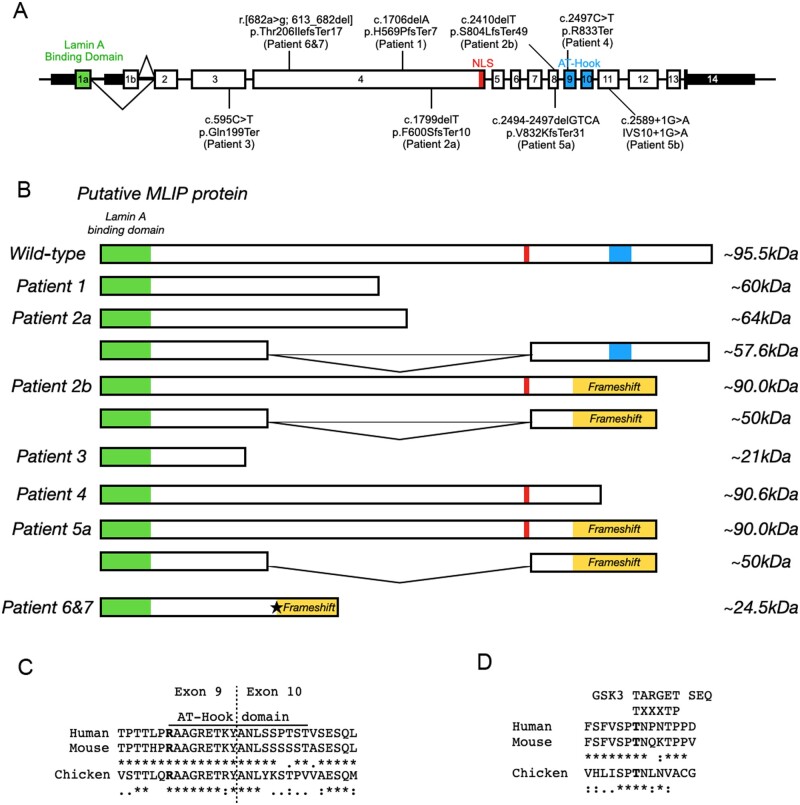
Figure 2.
Localization and effect of MLIP variants. (A) MLIP genomic map with identified position of each pathogenic variant. (B) Expected effects of the MLIP pathogenic variants on the size of MLIP protein for the various patients. Star: Non-canonical 3′ splice site in exon 4. (C and D) Alignment of MLIP between species in the AT-Hook domain (C) and GSK3 phosphorylation motif (D). The consensus sequence for C and D is denoted by: asterisk as identical; a single dot as conserved substitutions; and double dot as semi-conservative substitutions.
Patient P2 was found to be a compound heterozygous for two frameshift variants: a maternally inherited p.Phe600SerfsTer10 (c.1799delT) variant in exon 4, and a paternally inherited p.Ser804LeufsTer49 (c.2410delT) in exon 8. Both variants generate downstream premature stop codons, at positions 609 and 852 (Fig. 2), respectively, and are not reported in gnomAD.
Patient P3 was found to have a stop-gain variant p.Gln199Ter (c.595C>T) in a homozygous state in exon 3 (Fig. 2). The variant is not reported in gnomAD. The patient is adopted, and the biological parents were not available for segregation testing.
Patient P4 was homozygous for stop-gain variant p.Arg833Ter (c.2497C>T) in exon 9 (Fig. 2). The variant has been identified three times in gnomAD but only in a heterozygous state, with an allele frequency of 1.28 × 10−5. Segregation analysis revealed that the parents are heterozygous for this variant, confirming an autosomal recessive inheritance pattern.
Patient P5 was found to carry the compound heterozygous pathogenic variants: p.Val821LysfsTer31 (c.2461_2464delGTCA) in exon 8, inherited from the father, and c.2556+1G>A (IVS10+1G>A), inherited from the mother (Fig. 2). The first variant generates a stop codon at amino acid position 851 and is reported in gnomAD at very low allele frequencies in various populations, the highest of which is in non-Finnish Europeans (0.000328), but never in homozygosity. Likewise, the second variant has a very low allele frequency (0.0000353 in non-Finnish Europeans), while no homozygous individuals are identified.
Siblings Patients P6 and P7 carry the homozygous variant c.682A>G, g.54001582A>G (GRCh37.p13) in exon 4 of MLIP (Fig. 2). We investigated the possibility that this variant affected splicing through the generation of a non-canonical 3′ splice site in exon 4. In vivo splicing analysis revealed the generation of a new splicing site that results in the loss of the first 70 nucleotides of exon 4 (Supplementary Fig. 1). The resultant deletion is predicted to produce a frameshift and a putative truncated protein, p.(Thr206IlefsTer17) (Fig. 2). The variant is not reported in gnomAD. Sanger sequencing on parental DNA revealed both to be heterozygous for the variant.
Thus, all seven patients were found to have biallelic truncating variants (Fig. 2A) in MLIP. MLIP displays a complex tissue-specific pattern of alternative splicing,9,10 resulting in at least 12 protein-coding transcripts, of variable exon constellations and differential expression in various tissues. The largest single exon, exon 4, has a length of 524 amino acids and is exclusively included in two transcript isoforms, which according to the Genotype-Tissue Expression Project (GTEx; https://www.gtexportal.org/, accessed September 2021) data have their highest expression in skeletal muscle [MLIP-213 (ENST00000514921) and MLIP-206 (ENST00000502396)]. Exon 4 concentrates three of the eight variants identified in our cohort (homozygous p.His569ProfsTer7 in Patient P1, homozygous p.Thr206IlefsTer17 in Patients P6 and P7, and one of the heterozygous variants of Patient P2, p.Phe600SerfsTer10) (Fig. 2A). Another potential hotspot for pathogenic variants was identified in a region spanning exons 8 and 9, where four null variants were identified in Patients P2, P4 and P5. Modelling of the identified variants of Patients P1, P2 and P6/P7 predicted a loss of the nuclear localizing signal, which is located towards the 3′ end of exon 4,9,10 indicating a potential shift of MLIP localization away from the nucleus. On the other hand, variants identified in Patients P3, P4 and P5 show premature truncations that result in the disruption or loss of MLIP’s AT-hook DNA binding motif (exon 9 and 10)12 and MLIP’s putative capacity to bind DNA. For Patients P6 and P7, siblings, the identified variant (p.Thr206IlefsTer17) within a highly conserved region of MLIP may result in the loss of a putative glycogen synthase kinase-3 (GSK3) phosphorylation site15 (Fig. 2D).
Immunofluorescence and immunoblotting studies
We attempted to demonstrate the effect of MLIP pathogenic variants at the protein level through immunofluorescence and immunoblotting (western blot) studies. Immunofluorescence with the available antibodies showed similar localization of MLIP in Patients P1, P3 and P4 when compared to control subjects, with mostly nuclear and some cytoplasmic staining (Fig. 3B). Western blot analysis was difficult to interpret due to the complexity of the banding pattern, but patients and control subjects had comparable bands. We are thus unable to conclusively interpret these results, likely due to the limitations of available antibodies and the intricate isoform composition of MLIP.10
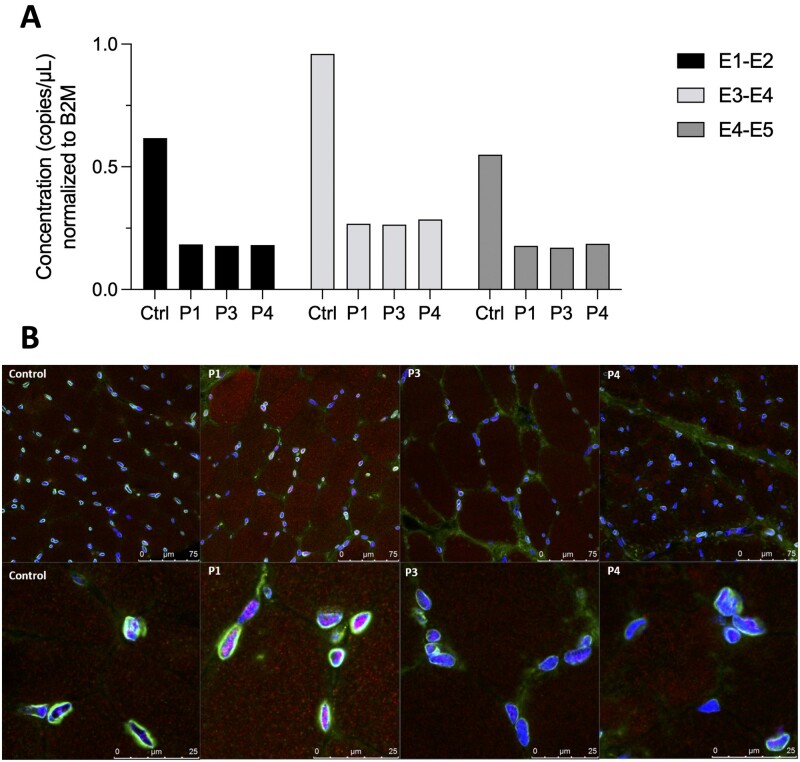
Figure 3.
RNA expression and protein localization experiments for MLIP. (A) Droplet digital PCR performed on muscle samples for Patients P1, P3 and P4, compared to control muscle (Ctrl). Amplicon concentration in copies/µl is shown normalized to B2M. Concentrations are shown for the boundaries between exons 1 and 2 (E1-E2), exons 3 and 4 (E3-E4), and exons 4 and 5 (E4-E5) of the transcript MLIP-213. (B) Immunofluorescence studies on muscle biopsy samples of Patients P1, P3 and P4. Confocal microscope pictures show a similar pattern of staining for MLIP and emerin in the control tissue compared to patient samples. Emerin (green) stains the nuclear envelope, while MLIP (red) is decorated mostly in the nucleus, with some cytoplasmic staining. DAPI (blue) is used for nuclear staining control. Top: ×200 magnification; bottom: ×630 magnification.
RNA expression studies
Next we quantified MLIP mRNA levels in patient (Patients P1, P3 and P4) skeletal muscle compared to control samples using ddPCR technology. To efficiently assay for the muscle relevant transcript MLIP-213, primer pairs were designed to cover exon boundaries 1–2, 3–4 and 4–5. Patients P1, P3 and P4 had comparably decreased cDNA levels for the three regions assayed by the chosen primer pairs, at ∼30% of the concentration in control muscles, after normalization to B2M (Fig. 3A). Additional primer pairs to target exon boundaries 1–2 and 11–12 to specifically address transcript MLIP-206 failed to generate detectable copies, suggesting very low baseline expression for this transcript in skeletal muscle.
Discussion
Here we report seven individuals from six unrelated families of diverse ethnic backgrounds presenting with exercise-induced myalgia, baseline hyperCKemia and susceptibility to episodes of rhabdomyolysis. All seven patients were found to have biallelic pathogenic variants in MLIP, a gene highly expressed in skeletal and cardiac muscle. Population variant frequency data from gnomAD shows only a few truncating variants in MLIP with more than a single heterozygous hit in the database, and a single individual has been reported with a homozygous intronic variant (c.64-37829_64-37828delTG). While this variant involves a splice donor site, the computational prediction is conflicting for pathogenicity, as there are multiple heterozygous hits in the population. In keeping with that observation, variants identified in four of six families from this cohort were not reported in gnomAD. In the two families with listed hits in the population variant database, variants are reported in very low allelic frequencies, and none in a homozygous state, consistent with recessive disease alleles.
Clinical features common for all patients are an onset of symptoms during early childhood, predominantly with muscle pain and cramping. Mild proximal muscle weakness concentrated in the lower limbs was observed in all families, except for the Spanish siblings (Patients P6 and P7), who had normal strength. Disease manifestations were relatively stable and did not impact activities of daily living outside of the episodes of rhabdomyolysis. While myalgia and cramping were induced by non-strenuous or strenuous physical activity, rhabdomyolysis episodes have manifested in response to effort, exercise, or cold, but also without obvious triggers.
Muscle biopsies of various patients showed a range of myopathic changes, varying in severity from mild non-specific to overtly dystrophic, in line with the presence of ongoing degeneration and regeneration. Internalized nuclei were identified in scattered fibres in some of the patient biopsies (Patients P1 and P4), a finding that is commonly seen as a reflection of regeneration following degeneration and thus does not on its own signify a primary structural myopathy. An MLIP mouse model was reported to show nuclear internalization in most muscle fibres, thus consistent with the human biopsies even though this finding was more prominent in the mouse.10 In conjunction with the elevated baseline CK, ranging from 300 IU/ml to 4000 IU/ml, these features suggest the presence of a mild ongoing dystrophic process with superimposed episodes of intermittent muscle breakdown. However, patients have thus far shown no indication of significant progressive permanent muscle degeneration, at least within the age range that our cohort currently represents (5 to 19 years of age), judging from the good functional status that the patients have maintained. This conclusion is also supported by the minimal findings on imaging studies, which showed no evidence of permanent fatty replacement at this point. In future studies, it will be important to identify patients later in adulthood to develop a better sense of the longer-term natural history of this condition.
Cardiac evaluation in four patients has been normal, while Patient P3 was found to have mild left ventricular dysfunction at age 14 years. In addition, two patients have reported mild structural abnormalities, including the presumably unrelated small patent foramen ovale in Patient P1 and a mildly dilated aortic valve in Patient P6. It remains to be established whether patients with MLIP pathogenic variants are at a higher risk for acquiring cardiomyopathy over time, as has been suggested by one association study linking an increased risk of developing dilated cardiomyopathy to a common missense polymorphism in MLIP.13 Careful interval evaluations of cardiac function thus remain advisable for patients with biallelic pathogenic MLIP variants.
The specific roles and physiological functions of MLIP in cardiac and skeletal muscle remain to be fully explored. Mice deficient in cardiac predominant MLIP are vulnerable to the development of cardiomyopathy and heart failure in response to mechanical and/or pathophysiological stimuli,11,12 suggesting a role in adaptation to cardiac stressors, potentially related to an overactivation of Akt/mTOR11 and FOXO112 pathways. While skeletal muscle of these mice shows increased centralized nuclei, consistent ongoing muscle repair and intermittent muscle breakdown has not been reported in this model.10 More recently, studies in mice have suggested that MLIP interacts with chromatin17 and may be required for myoblast commitment.10,17 Furthermore, MLIP may function as an anchoring protein that regulates nuclear positioning in skeletal muscle fibres.16 Preliminary co-immunoprecipitation data (unpublished) suggests that the interactome of MLIP includes proteins from pathways important for energy production (PKM2, PDHX and DLD, involved in glycolysis and gluconeogenesis), thus providing a potential link to pathways that have been associated with rhabdomyolysis.
The tissue-specific splicing pattern of MLIP is highly complex, making a prediction of the consequences of variants and their analysis challenging. Three of the identified variants locate to the large muscle-specific exon 4, while another four nonsense variants locate to exons 8 and 9. The identified variants are predicted to either cause a loss of the nuclear localizing signal at the end of exon 4, or a disruption of MLIP’s AT-hook DNA binding motif (Fig. 2B and C), although we have not yet been able to confirm this experimentally. The sibling patients (Patients P6 and P7) harbour the homozygous variant (r.[682a>g;613_682del]), located in a highly conserved region of MLIP, which we show generates a non-canonical splice site within exon 4. This results in the deletion of the first 70 nucleotides of exon 4 with a frameshift leading to a protein truncation (p.Thr206IlefsTer17). The disruption of this region of MLIP may also result in the loss of a putative GSK3 phosphorylation site (Fig. 2D). GSK3 is a highly conserved ubiquitously expressed eukaryotic serine/threonine-protein kinase that was originally identified as a negative regulator in the hormonal control of glucose homeostasis.15 Therefore, disruption of this putative GSK phosphorylation site may result in a dysregulation of MLIP activity with a subsequent impact on MLIP’s capacity to mitigate metabolic maladaptation in response to physiological stress.
An assessment of the effect of the pathogenic variants on the protein level through immunofluorescence and immunoblotting studies failed to show a clear difference in protein localization or difference in the complex immunostaining banding pattern between a subset of patients and controls. We used the commercial antibody Abcam ab131324, as well as an antibody developed initially for the Mlip knockout mouse model, which probes a region encoded by exon 11 in transcripts MLIP-213 and MLIP-206, but blot interpretations were challenging due to the unclear specificity of the antibodies in skeletal muscle. However, RNA quantification experiments of MLIP mRNA for the muscle predominant isoform using ddPCR revealed convincing reductions of overall expression levels in Patients P1, P3 and P4 compared to control muscle, suggesting engagement of nonsense-mediated decay as a result of the premature termination codons, and supporting the detrimental nature of the variants.
Full understanding of the impact of MLIP sequence variants remains challenging as available antibody tools are limited and not fully characterized. Since none of the variants identified in the patients appear to affect all MLIP transcripts, at least it would not be unexpected that there was no complete loss of various forms of MLIP immunoreactivity in muscle samples from any of the patients.
In summary, our findings in seven individuals establish MLIP as a gene implicated in a novel rhabdomyolysis phenotype. Pathogenic variants in MLIP should be sought in undiagnosed patients with exercise-induced myalgia and episodes of rhabdomyolysis, in particular in those patients with baseline hyperCKemia. It will be of great importance to further characterize MLIP function in skeletal muscle to fully understand how its dysfunction would lead to mild muscular dystrophy with intermittent rhabdomyolysis.
Supplementary Material
Acknowledgements
We thank patients and their families for volunteering to participate in this study. We thank Christopher Mendoza, Christine Jones, Gilberto (“Mike”) Averion and Kia Brooks for their help in clinic.
Funding
The work in C.G.B. laboratory is supported by intramural funds from the NIH National Institute of Neurological Disorders and Stroke. The work in the Instituto de Investigación Hospital 12 de Octubre (i+12) was supported by the Spanish Instituto de Salud Carlos III (ISCIII) and European Regional Development Fund (ERDF) (PI17/00487 to F.M.-A.). F.J.C.-V. was supported by fellowship from the Instituto de Investigación Hospital 12 de Octubre (i+12). M.E.R.-G. was supported by fellowship from ISCIII and ERDF (PI17/00487). The work in P.G.B. laboratory was supported, in whole or in part, by Canadian Institutes of Health Research Grant MOP119470.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- CK
creatine kinase
- gnomAD
genome aggregation database
References
- 1. Stahl K, Rastelli E, Schoser B.. A systematic review on the definition of rhabdomyolysis. J Neurol. 2020;267(4):877–882. [DOI] [PubMed] [Google Scholar]
- 2. Scalco RS, Gardiner AR, Pitceathly RD, et al. Rhabdomyolysis: A genetic perspective. Orphanet J Rare Dis. 2015;10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Muller-Felber W, Zafiriou D, Scheck R, et al. Marinesco Sjogren syndrome with rhabdomyolysis. A new subtype of the disease. Neuropediatrics. 1998;29(2):97–101. [DOI] [PubMed] [Google Scholar]
- 4. Zafeiriou DI, Ververi A, Tsitlakidou A, Anastasiou A, Vargiami E.. Recurrent episodes of rhabdomyolysis in pontocerebellar hypoplasia type 2. Neuromuscul Disord. 2013;23(2):116–119. [DOI] [PubMed] [Google Scholar]
- 5. Kremer LS, Distelmaier F, Alhaddad B, et al. Biallelic truncating mutations in TANGO2 cause infancy-onset recurrent metabolic crises with encephalocardiomyopathy. Am J Hum Genet. 2016;98(2):358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lilleker JB, Keh YS, Roncaroli F, Sharma R, Roberts M.. Metabolic myopathies: A practical approach. Pract Neurol. 2018;18(1):14–26. [DOI] [PubMed] [Google Scholar]
- 7. Vivante A, Ityel H, Pode-Shakked B, et al. Exome sequencing in Jewish and Arab patients with rhabdomyolysis reveals single-gene etiology in 43% of cases. Pediatr Nephrol. 2017;32(12):2273–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu L, Brady L, Shoffner J, Tarnopolsky MA.. Next-generation sequencing to diagnose muscular dystrophy, rhabdomyolysis, and hyperckemia. Can J Neurol Sci. 2018;45(3):262–268. [DOI] [PubMed] [Google Scholar]
- 9. Ahmady E, Deeke SA, Rabaa S, et al. Identification of a novel muscle A-type lamin-interacting protein (MLIP). J Biol Chem. 2011;286(22):19702–19713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cattin ME, Deeke SA, Dick SA, et al. Expression of murine muscle-enriched A-type lamin-interacting protein (MLIP) is regulated by tissue-specific alternative transcription start sites. J Biol Chem. 2018;293(51):19761–19770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cattin ME, Wang J, Weldrick JJ, et al. Deletion of MLIP (muscle-enriched A-type lamin-interacting protein) leads to cardiac hyperactivation of Akt/mammalian target of rapamycin (mTOR) and impaired cardiac adaptation. J Biol Chem. 2015;290(44):26699–26714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang ZP, Kataoka M, Chen J, et al. Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis. J Clin Invest. 2015;125(11):4122–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Esslinger U, Garnier S, Korniat A, et al. Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS One. 2017;12(3):e0172995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karczewski KJ, Francioli LC, Tiao G, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss of function intolerance across human protein-coding genes. bioRxiv. [Preprint] doi:10.1101/531210 [Google Scholar]
- 15. Beurel E, Grieco SF, Jope RS.. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol Ther. 2015;148:114–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu J, Huang ZP, Nie M, et al. Regulation of myonuclear positioning and muscle function by the skeletal muscle-specific CIP protein. Proc Natl Acad Sci U S A. 2020;117(32):19254–19265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahmady E, Blais A, Burgon PG.. Muscle enriched lamin interacting protein (Mlip) binds chromatin and is required for myoblast differentiation. Cells. 2021;10(3):615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.