ABSTRACT
Our understanding of the pathogenic yeasts Cryptococcus neoformans and Cryptococcus gattii has been greatly enhanced by use of genome sequencing technologies. Found ubiquitously as saprotrophs in the environment, inhalation of infectious spores from these pathogens can lead to the disease cryptococcosis. Individuals with compromised immune systems are at particular risk, most notably those living with HIV/AIDS. Genome sequencing in combination with laboratory and clinical studies has revealed diverse lineages with important differences in their observed frequency, virulence and clinical outcomes. However, to date, genomic analyses have focused primarily on clinical isolates that represent only a subset of the diversity in the environment. Enhanced genomic surveillance of these yeasts in their native environments is needed in order to understand their ecology, biology and evolution and how these influence the epidemiology and pathophysiology of clinical disease. This is particularly relevant on the African continent from where global cryptococcal diversity may have originated, yet where environmental sampling and sequencing has been sparse despite harbouring the largest population at risk from cryptococcosis. Here, we review what scientifically and clinically relevant insights have been provided by analysis of environmental Cryptococcus isolates to date and argue that with further sampling, particularly in Africa, many more important discoveries await.
Keywords: fungi, genomics, ecology, epidemiology, microbiology, evolutionary biology
We summarise important insights unveiled by environmental sampling and genomics of the pathogenic fungi, Cryptococcus, but argue there is still much to learn with increased focus in this area.
INTRODUCTION
Fungal diseases cause a considerable and underappreciated burden of disease worldwide (Bongomin et al. 2017). A vast diversity of fungi exist as saprotrophs in the environment, some of which can cause opportunistic disease in at-risk humans—these fungi are often known as ‘sapronoses’. With current climate trends, fungal sapronoses are expected to present an increasing risk and burden to human health since they often thrive in warm and wet conditions (Garcia-Solache and Casadevall 2010). Fungal sapronoses are largely made up of species of moulds in the phylum, Ascomycota, however, the sister phylum of Basidiomycota includes the species complexes of Cryptococcus neoformans and Cryptococcus gattii. Unlike the majority of the Basidiomycota which are filamentous, Cryptococcus spp. are yeasts which are cosmopolitan in environments worldwide and can cause the disease cryptococcosis when aerosolised spores and/or desiccated yeast cells are inhaled by a susceptible individual (Velagapudi et al. 2009; Walsh et al. 2019). The resulting infection can affect any organ but often manifests as an acute pneumonia or a highly fatal meningitis (Kronstad et al. 2011). These fungi predominantly affect immunocompromised individuals, particularly those with HIV/AIDS among whom they are estimated to cause 223 100 new cases and more than 181 000 deaths globally per year, three quarters of which (162 500 new cases and 135 900 deaths) are in sub-Saharan Africa (Rajasingham et al. 2017). Other estimates place the incidence of cryptococcal meningitis among the general population in Africa at 4.8 per 100 000 during the years 1990 to 2017 (Nyazika et al. 2019). In comparison, Asia and Pacific region has the second-highest burden of disease with an estimated 43 200 new cases and 39 700 deaths annually, while Europe has just 4400 and 1800 new cases and deaths, respectively, per year (Rajasingham et al. 2017).
As single-celled yeasts, the increasing availability and affordability of genome sequencing has broadened our understanding of these pathogens and, in combination with complementary laboratory and clinical studies, has revealed a genetically diverse set of lineages. Within C. neoformans there are five main molecular types: VNI, VNII and VNB, collectively known as C. neoformans var. grubii; VNIV, also known as C. neoformans var. neoformans; and VNIII, a hybrid of the two varieties. Within C. gattii there are the lineages VGI, VGII, VGIII, VGIV and the more recently discovered and described, VGV (Farrer et al. 2019). Few and rare inter-species hybrids have also been reported (Bovers et al. 2006, 2008; Aminnejad et al. 2012). The diversity between these lineages revealed by whole-genome sequencing (including single nucleotide polymorphisms (SNPs), insertions and deletions (INDELs) and genomic rearrangements (Desjardins et al. 2017; Rhodes et al. 2017b; Vanhove et al. 2017)) has led to proposals to elevate these molecular types to species level (Hagen et al. 2015).
Since these fungi are acquired from nature, environmental sampling and genomic analysis is key to understanding their diversity, biology, ecology and epidemiology. To date, such analysis of environmental Cryptococcus spp. has, however, been limited, particularly in Africa (Cogliati 2013). Yet, it is this region that warrants greater investigation; not only does southern Africa hold the largest population that are at-risk from cryptococcosis due to the high number of HIV/AIDS-infected individuals (Perfect and Bicanic 2015; Oladele et al. 2017; Rajasingham et al. 2017), it is also hypothesised as being home to the ancestral diversity from which more globalised lineages evolved (Litvintseva et al. 2011). While much has already been learned from the limited number of environmental isolates gathered here, we argue that far more can be discovered by focusing more attention on environmental cryptococcal genomics across this region.
GLOBAL SAMPLING AND SEQUENCING EFFORT TO DATE
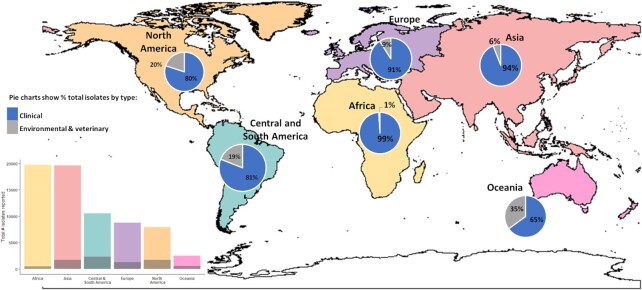
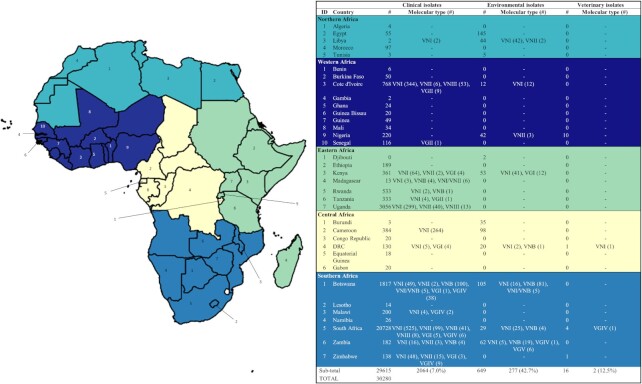
Our understanding of the distribution of cryptococcal genotypes is directly influenced both by the amount of clinical and environmental sampling conducted and by the proportion of sampled isolates that have been molecular typed. Cogliati's 2013 review found there were 69 022 isolates of C. neoformans and C. gattii reported globally, with the vast majority collected from Africa and Asia, and fewest from Oceania (Fig. 1) (Cogliati 2013). Of these isolates, less than 10% were from environmental or veterinary sources (as opposed to clinical), and less than 12% had been examined for molecular type. Of all regions, Africa had the highest volume of clinical isolates collected, as may be expected given this area carries the greatest burden of clinical disease. However, this region also has the lowest proportion of isolates that have been molecular typed. For our current review we updated Cogliati's 2013 study for Africa and found a total of 30 280 isolates reported of which 8% (n = 2343) were examined for molecular type and only 2% (n = 649) were collected from the environment (Fig. 2, Table 1 and Supplementary Table). The proportion of environmental isolates that are molecular-typed is higher at 43% because these are predominantly from research studies, as opposed to most clinical isolates, which are collected in routine diagnoses.
Figure 1.
Reported isolations of C. neoformans and C. gattii across each continental region up to Cogliati's 2013 review. Pie charts show distribution of clinical and environmental/veterinary sources of isolation. Bar chart shows total number of isolates reported with shaded regions as the number that were examined for molecular type. Despite data being from 2013, general patterns and proportions remain true.
Figure 2.
Clinical, environmental and veterinary isolations (published) of C. neoformans and C. gattii by country and region across Africa.
Table 1.
Environmental Cryptococcus neoformans and Cryptococcus gattii isolations from countries in Africa.
| Country (total # isolates) | Region | Environmental source | # isolates recovered | Species, variety, serotype, molecular type (as reported) | Reference |
|---|---|---|---|---|---|
| Libya (44) | Tripoli | Pigeon droppings | 32 | C. neoformans var. grubii, A, VNI | Ellabib et al. 2016 |
| 1 | C. neoformans var. grubii, A, VNI | ||||
| E. camaldulensis | 2 | C. neoformans var. grubii, A, VNII | |||
| Olea europaea | 9 | C. neoformans var. grubii, A, VNI | |||
| Tunisia (5) | Sfax region | E. camaldulensis | 1 | C. neoformans species complex | Mseddi et al. 2011 |
| E. camaldulensis | 2 | C. gattii species complex | |||
| Almond tree (Prunus dulcis) | 2 | C. gattii species complex | |||
| Egypt (145) | Tanta | E. camaldulensis | 1 | C. gattii species complex | Mahmoud 1999 |
| Qutur | E. camaldulensis | 2 | C. gattii species complex | ||
| Gharbia Governatorate | Avian droppings | 95 | C. neoformans species complex | ||
| Nile delta | Pigeon droppings | 30 | C. neoformans/C. gattii | Refai et al. 1983 | |
| Giza | E. camaldulensis | 3 | C. neoformans var. grubii | Elhariri et al. 2016 | |
| Cairo | E. camaldulensis | 2 | C. neoformans var. grubii | ||
| Al-Sharqia | E. camaldulensis | 5 | C. neoformans var. grubii | ||
| Elmenofia | E. camaldulensis | 3 | C. neoformans var. grubii | ||
| Abulnomorous | Ground water | 3 | C. neoformans var. grubii | Elfadaly et al. 2018 | |
| Shabramant | Ground water | 1 | C. neoformans var. grubii | ||
| Kenya (53) | Nairobi | Avian droppings | 23 | C. neoformans var. grubii, VNI | Kangogo et al. 2015 |
| 5 | C. gattii, VGI | ||||
| Tree swabs | 5 | C. neoformans var. grubii, VNI | |||
| 7 | C. gattii, VGI | ||||
| Chicken cages | 5 | C. neoformans var. grubii, VNI | |||
| Garbage dumping | 6 | C. neoformans var. grubii, VNI | |||
| Soil | 2 | C. neoformans var. grubii, VNI | |||
| Djibouti (1) | Djibouti | Pigeon droppings | 2 | C. neoformans/C. gattii | Pal 2015 |
| Cameroon (98) | West region | Pigeon droppings and bat guano | 57 | C. gattii species complex | Dongmo et al. 2016 |
| 41 | C. neoformans species complex | ||||
| Ivory Coast (12) | Adjamé | Pigeon droppings | 12 | C. neoformans var. grubii, A, VNI | Kassi et al. 2018 |
| Nigeria (41) | Southeastern Nigeria | Pigeon droppings | 39 | C. neoformans/C. gattii C. neoformans species complex C. neoformans var. grubii, VNII | Nweze et al. 2015 |
| Jos | Pigeon droppings | 3 | |||
| Nnadi et al. 2016 | |||||
| Democratic Republic of Congo (20) | Zaire | House dust | 1 | C. neoformans var. grubii, A, VNI | Boekhout et al. 2001 |
| Wood | 1 | C. neoformans var. grubii, A, VNI | |||
| Wood | 1 | C. neoformans var. grubii, A, VNB | |||
| Kinshasa | House dust | 2 | C. neoformans var. grubii, A | Varma et al. 1995 | |
| Kinshasa | House dust | 4 | C. neoformans species complex | Swinne et al. 1986 | |
| House air | 2 | C. neoformans species complex | |||
| Chicken droppings | 2 | C. neoformans species complex | |||
| Pigeon droppings | 7 | C. neoformans species complex | |||
| Burundi (35) | Bujumbura | Environment | 15 | C. neoformans species complex | Varma et al. 1995 |
| Bujumbura | Patient's house | 7 | C. neoformans species complex | Swinne et al. 1989 | |
| Bujumbura | House dust | 13 | C. neoformans species complex | Swinne et al. 1991 | |
| Zambia (32) | Zambesi and Miombo woodlands | Trees | 5 | C. neoformans var. grubii, VNI | Vanhove et al. 2017 |
| 19 | C. neoformans var. grubii, VNB | ||||
| 31 | C. gattii species complex | ||||
| Miombo woodlands | Hyrax midden | 4 | C. gattii, VGV | Farrer et al. 2019 | |
| 1 | C. gattii, VGIV | ||||
| Tree hole | 2 | C. gattii, VGV | |||
| Botswana (105) | Gaborone | Pigeon droppings | 3 | C. neoformans var. grubii, A, VNI | Litvintseva et al. 2011 |
| Gaborone | Tree bark | 2 | C. neoformans var. grubii, A, VNI | ||
| Tuli block | Mopane tree | 15 | C. neoformans var. grubii, A, VNB | ||
| Tuki block | Mopane tree | 4 | C. neoformans var. grubii, A, VNI | ||
| Tuli block | Soil | 2 | C. neoformans var. grubii, A, VNI | ||
| Tuli block | Soil | 1 | C. gattii, B | ||
| Tuli block | Baobab | 2 | C. neoformans var. grubii, A, VNB | ||
| Francistown, Gaborone, and Maun | Trees and bird excreta | 5 | C. neoformans var. grubii, VNI | Chen et al. 2015 | |
| 64 | C. neoformans var. grubii, VNB | ||||
| 5 | C. neoformans v. grubii, VNI/VNB | ||||
| 2 | C. gattii species complex | ||||
| South Africa (29) | Durban | Pigeon droppings | 20 | C. neoformans var. grubii, A, VNI | Litvintseva et al. 2011 |
| Johannesburg | Soil | 2 | C. neoformans var. grubii, A, VNI | ||
| Parys | Pigeon droppings | 3 | C. neoformans var. grubii, A, VNI | ||
| Zeerust | Eucalyptus tree | 2 | C. neoformans var. grubii, A, VNB | ||
| Zeerust | Soil | 2 | C. neoformans var. grubii, A, VNB |
GLOBAL DISTRIBUTION OF GENOTYPES AND AFRICA IN CONTEXT
Figure 3 summarises the distribution of molecular types by global region based on Cogliati's 2013 review (Cogliati 2013). Since this review, isolates of the VNB molecular type have also been identified from six clinical cases and one environmental sample in South America and the new C. gattii lineage, VGV, was identified from environmental sources in Africa (Rhodes et al. 2017b; Farrer et al. 2019). However, the overarching patterns within the data documented by Cogliati remain.
Figure 3.
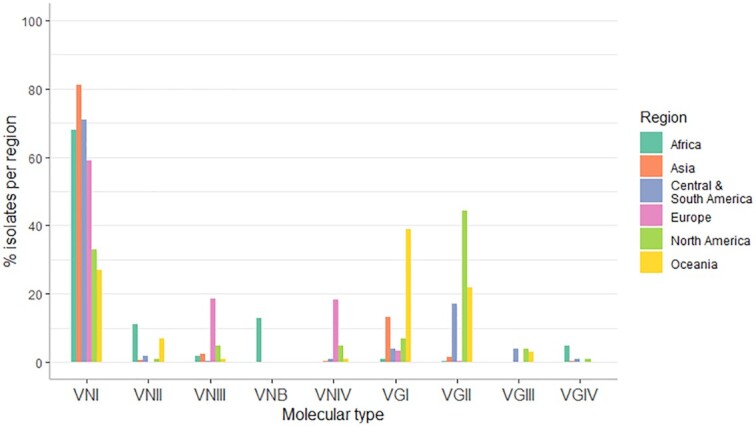
Distribution of the main C. neoformans and C. gattii molecular types identified over different global regions, as reported in Cogliati 2013. Since this review additional molecular types have been identified, including VNB in Central and South America and VGV in Africa, however general distribution patterns remain true.
The VNI molecular type has been isolated from all regions and is the dominant molecular type across all except North America and Oceania where VGI and VGII dominate, respectively (Fig. 3). The majority of global clinical disease is caused by infection with C. neoformans var. grubii which makes up approximately 95% of all cryptococcosis cases worldwide (Maziarz and Perfect 2016). Of these, the vast majority are due to infection with the VNI molecular type. VNI is frequently isolated from the environment where it is associated with trees, pigeon and other bird guano, and with urban sites including churches and dwellings (Table 2) (Litvintseva et al. 2011; Chen et al. 2015; Kangogo et al. 2015; Nweze et al. 2015; Ellabib et al. 2016; Nnadi et al. 2016; Kassi et al. 2018). VNB, on the other hand, has only been isolated from Southern Africa and, more recently, South America, from a small number of clinical isolates and from arboreal tree species.
Table 2.
Sources of environmental and veterinary isolates of C. neoformansand C. gattii in each global region.
| Sources of environmental and veterinary isolates* | ||
|---|---|---|
| Region | C. neoformans | C. gattii |
| Oceania | Environmental: Eucalyptus camaldulensis, pine needles | Environmental: Eucalyptus camaldulensis, Eucalyptus tereticornis, Syncarpia glomulifera, insect frass, olive seedlings, plant debris |
| Veterinary: cat, dog, horse, koala, ferret, Potorous gilbertii | Veterinary: kiwi, cat, dog, horse, sheep, cow, koala, quokka, cockatoo, ferret, Potorous tridactylus, echidna, African grey parrot, dolphin | |
| Asia | Environmental: Mostly from pigeon and other bird excreta, less frequently from trees including Eucalyptus, Tamarindus arjuna, Tamarindus indica, Cassia fistola, Syzygium cumini, and Ficus religiosa; and some fruit and vegetables (tomato, carrot, banana, eggplant, papaya, apple, guava) | Environmental: Trees including Syzgium cumini, Mimusops elengi, Azadirachta indica, Acacia nilotica, Cassia fistola, Manikara hexandra, Polyalthia longifolia, Eucalyptus camaldulensis, Tamarindus indica, Cassia marginata, and Mangifera indica |
| Veterinary: cat, dog, bandicoot | Veterinary: koala | |
| Africa | Environmental: Pigeon and bird excreta, soil, house dust, trees including Eucalyptus camaldulensis, mopane, baobab | Environmental: Soil, Eucalyptus camaldulensis, almond tree |
| Veterinary: N/A | Veterinary: cheetah | |
| Europe | Environmental: Mostly from pigeon, bird and bat guano, and red fox faeces. Few from trees including Eucalyptus camaldulensis and oak tree | Environmental: mostly from trees including Eucalyptus camaldulensis, Douglas tree, carob tree, stone pine |
| Veterinary: cat, dog, magpie, striped grass mouse, degu. | Veterinary: ferret, goat | |
| Central and South America | Environmental: pigeon and bird excreta, soil, dust, contaminated dwellings, Eucalyptus tree, almond tree, kassod tree, pink shower tree, Caesalpinia peltophoroides, Anadenanthera peregrine | Environmental: soil, dust, psittaciformes bird excreta, Eucalyptus camaldulensis, almond tree, kassod tree, pottery tree, jungle tree, Corymbia ficifolia, Cephalocereus royenii |
| Veterinary: insects, bull, sheep | Veterinary: cheetah, goat, psittacine birds | |
| North America | Environmental: Mainly from pigeon droppings, some from fruit and vegetables | Environmental: Soil, trees, air, water |
| Veterinary: ferret | Veterinary: dog, cat, horse, ferret, birds, alpaca, parrots | |
*information taken from Cogliati2013
Cryptococcus gattii rarely causes clinical disease and was previously thought to be restricted to tropical and sub-tropical regions, where it is associated predominantly with arboreal tree species including Eucalyptus, olive trees, and dry-tropical miombo (Brachystegia sp.) (Ellis and Pfeiffer 1990; Pfeiffer and Ellis 1992; Mseddi et al. 2011; Cogliati et al. 2016; Vanhove et al. 2017) (Table 2). However, the molecular type, VGII, has been the cause of recent outbreaks in more temperate and developed areas of the world, such as in PNW, Vancouver and Oregon (Byrnes and Marr 2011) and has become the dominant molecular type reported here owing to intensive clinical and environmental surveillance in regions affected by the outbreak (Bartlett, Kidd and Kronstad 2008; Billmyre et al. 2014; Engelthaler et al. 2014). VGII has been isolated across the Pacific North West region from trees, sea water and marine animals.
In Africa, clinical infection is most commonly associated with VNI infection, although a high diversity of lineages have been identified from clinical cases, especially in Southern Africa (Fig. 2 and Supplementary Table). This region has also uncovered high diversity from the environment, with VNB being most commonly isolated, followed by VNI and several lineages of C. gattii (Fig. 2 and Table 1). Despite the limited sequencing, the diversity of molecular types in southern Africa is one of the factors supporting the ‘out-of-Africa’ hypothesis which postulates that Cryptococcus diversified in Africa prior to subsequent global spread. However, sampling and molecular-typing have been even more limited in regions other than Southern Africa, including Central Africa which shares a large border with the southern region and thus shares some of the ecological habitats which favour cryptococcal growth and harbour diversity (Fig. 2).
WHAT HAVE ENVIRONMENTAL ISOLATES TAUGHT US AND WHAT COULD THEY YET STILL REVEAL?
Despite the limited volume of environmental sampling and genomics analysis conducted to date, the genomes and associated biology of Cryptococcus spp. recovered from environmental sources has provided useful insights into various aspects of Cryptococcus evolution, virulence and epidemiology. The focus on clinical cases is understandable but, we argue, that combining clinical analysis with increased focus on what exists in the environment can help answer some of the key knowledge gaps in understanding the impact of this opportunistic infection. We group these insights and remaining areas of research under four key themes, described here in turn: (i) evolutionary origins, speciation and spread of genotypes, (ii) biology of virulence, (iii) exposure risk and epidemiology and (iv) emergence of drug resistance. We finally discuss some of the challenges in environmental sampling and modelling of Cryptococcus. Although we discuss global research, we highlight where we believe environmental sampling can answer knowledge gaps particularly pertaining to the African context where the highest burden of disease is concentrated and thus the biggest gains are to be made.
EVOLUTIONARY ORIGINS, SPECIATION AND SPREAD OF GENOTYPES
Since the genotypes of C. neoformans and C. gattii that cause clinical infection are a subset of what occurs in the environment, environmental sampling will reveal the true extent of the taxonomic diversity within each species complex. The significance of this was recently demonstrated with the discovery of an entirely new lineage of the C. gattii species complex, VGV, from environmental sampling conducted in Zambia in 2013 (Farrer et al. 2019). This discovery demonstrates there may yet be more diversity to discover given greater surveillance effort in new and more varied ecotypes and ecoregions. Understanding the full taxonomic diversity of Cryptococcus is not only of general biological interest; subsequent phylogenetic and population genomics analyses provide important insights into evolutionary origins, speciation and genotype flow. For example, phylogenetic analyses of loci from environmental genomes in South Africa and Botswana showed a high proportion of C. neoformans isolates from African arboreal trees belong to the genetically diverse and sexual lineage, VNB, which is ancestral to the globalised and asexual VNI and VNII lineages. This finding has been used to propose an ‘out-of-Africa’ hypothesis to account for the current distribution of C. neoformans genotypes (Litvintseva et al. 2011). Conversely, evidence to date suggests that lineages of the C. gattii species complex appear to originate from South America and that the species complexes themselves may have diverged 80–100 million years ago at the time of the breakup of the Pangean supercontinent (Hagen et al. 2013; Casadevall et al. 2017). Although geography can help explain patterns of speciation, closely related genotypes of both C. neoformans and C. gattii have also been found on separate continents, suggesting that relatively recent long-distance dispersal events occur (Ashton et al. 2019). Specifically, the highly virulent VGII lineage is hypothesised to have spread to the North American Pacific Northwest (PNW) 70–90 years ago from Brazil, possibly vectored by trade along shipping routes and assisted by passive dispersal in ocean currents (Engelthaler and Casadevall 2019).
Population genetics comparing environmental and clinical isolates is a powerful approach in not only understanding these long-distance dispersal events, but also the rate at which genotypes move across smaller scales. In Europe, the geographical distribution of clinical and environmental isolates together with analysis of spatial patterns of gene-flow allowed inference of how the main VNI sequence types circulate and highlighted Germany and Italy as the ‘fulcrum’ of diffusion of both endemic and imported genotypes (Cogliati et al. 2019). At finer-scales, genome-sequencing and phylogenetic analysis is now being used to investigate sources of exposure leading to cryptococcosis, for instance in recent attempts to link hospital environments to nosocomial outbreaks of the disease (Farrer et al.2021). Similar increased sampling and sequencing of environmental isolates in Africa would describe the spatial genetic structure of lineages and genotypes throughout the continent. These data would, at last, provide a baseline from which a more nuanced understanding of the epidemiology of exposure and infection for the large at-risk population of people living with HIV/AIDS in Africa could be developed.
BIOLOGY AND EMERGENCE OF VIRULENCE
Comparing the biology of both clinical and environmental cryptococcal isolates lends insight into differences in virulence between isolates and genotypes as well as what genomic mechanisms can generate diversity that may explain the emergence of virulent phenotypes. The ability of Cryptococcus to adapt to selective pressures in the environment is linked to plasticity of its genome which allows changes in ploidy, microevolution and hypermutator states leading to phenotypic switching (Guerrero et al. 2006; Jain and Fries 2008; Magditch et al. 2012; Rhodes et al. 2017a), as well as its ability to recombine it's genome through recombination. The yeast is able to mate both bisexually between two cells of opposing mating types (MAT-a and MAT-α) as well as unisexually between two members of the same mating type, with unisexual reproduction still leading to diverse progeny and biologically important since MAT-a cell types are rare (Nielsen et al. 2003; Ni et al. 2013; Phadke et al. 2014; Fu et al. 2015; Sun et al. 2019). Such genomic mechanisms may also contribute to the emergence and spread of global virulent phenotypes. For example, evidence has implicated both microevolution (via a transient mutator phenotype) and sexual reproduction (either unisexual or bisexual) in the emergence of the virulent VGII strains responsible for the PNW outbreak (Billmyre et al. 2014). Although primarily a haploid organism, changes in ploidy and cell size increases, such as seen in polyploid titan cells, can occur in response to environmental stressors and during human infection this can result in enhanced virulence, dissemination and survival within the host (Gerstein et al. 2015; Hommel et al. 2018; Zhou and Ballou 2018).
Humans are dead-end hosts for C. neoformans and C. gattii; pathogenesis is thus considered to be an ‘accidental’ by-product of traits that have evolved in response to natural selection in the environment rather than selection for virulence within a mammalian host (Casadevall 2008; May et al. 2016). These attributes thus have a ‘dual-use’ survival value that is manifested both in the environment as well as the accidental host (Casadevall, Steenbergen and Nosanchuk 2003). For example, a complex thick-walled polysaccharide capsule protects against desiccation and predation by amoebae in the environment as well as phagocytosis by macrophages in the host; melanin production protects against ultraviolet light and temperature fluctuations in the environment as well as resistance to oxidative stress, body temperature, the immune system and drug treatment pressures in the host; laccase production aids lignin degradation in the environment as well as protecting against oxidative bursts in the host (Williamson 1997; Guerrero et al. 2006; Perfect 2006; Jain and Fries 2008; Magditch et al. 2012; Rhodes et al. 2017a; Casadevall et al. 2019; Zaragoza 2019). The capacity for virulence that is independent of the requirement for animal hosts to aid survival and replication has been termed ‘ready-made’ virulence, as opposed to virulence that is selected for through dependence and/or symbiosis with the host (Casadevall, Steenbergen and Nosanchuk 2003). This hypothesis does not explain the whole story, however, since most species of Cryptococcus (and other environmental fungi) do not appear to infect mammalian hosts yet likely experience similar environmental pressures as C. neoformans/C. gattii (Casadevall, Steenbergen and Nosanchuk 2003).
Although it is accepted that virulent genotypes are acquired from the environment and that virulence factors are largely a result of adaptations to environmental pressures, few studies have specifically compared the virulence of environmental isolates to that of clinical isolates. Since some molecular types are found more frequently in clinical cases than the environment, and vice versa, there must either be biological differences in virulence between molecular types, or differential exposure of the susceptible human population to each molecular type. For example, the division of VNB into two distinct phylogenetic clades, VNBI and VNBII, characterised notable phenotypic differences between these two groups. In Botswana, VNBII was enriched for clinical isolates relative to VNBI which contained a far higher number of environmental isolates (Desjardins et al. 2017). The same trend was seen by a separate study in Zambia where VNBII (which the authors denoted VNB-A) comprised a mix of environmental and clinical isolates while VNBI (denoted VNB-B) was entirely environmental in origin (Vanhove et al. 2017). The comparison is more complex since, although evidence is limited, differences in virulence can occur not only between lineage types but also between environmental and clinical isolates of the same lineage type. Perhaps surprisingly, high-throughput phenotyping showed that VNBI environmental isolates were more resistant to oxidative stress and more heavily melanized that VNBI clinical isolates. Here, lack of melanisation was associated with loss-of-function mutations in the BZP4 transcription factor and likely reflects a greater breadth of selective pressures in the environment than in the human host (Desjardins et al. 2017). This may suggest, then, that the lower incidence of VNBI clinical cases is due to more limited exposure to their infectious propagules rather than a lack of intrinsic ability to infect the human host. However, earlier studies found differential ability of environmental strains of C. neoformans to cause disease in murine models (Da Silva et al. 2006) and lower virulence than clinical isolates (Fromtling, Abruzzo and Ruiz 1989), although these studies did not distinguish molecular type. Litvintseva & Mitchell (Litvintseva and Mitchell 2009) found that only one VNI isolate of 11 environmental isolates of C. neoformans (including 10 VNI and 1 VNII) caused infection in mice up to 60 days post-infection, whereas 7 of 10 clinical isolates were lethal at median times of 19 and 40 days (lethal clinical isolates included 6/7 VNI and 2/3 VNII).
These intriguing findings suggest that genetically encoded mechanisms driving emergence of virulent phenotypes may be complex and it is yet to be conclusively determined what genetic and/or epigenetic factors may play a role. If virulence is a result of adaptation to the yeast's local environment then it may be determined by the micro-ecological niche that each isolate occupies, resulting in differences between apparently similar populations. Further dissection of the eco-evolutionary basis of cryptococcal virulence is certainly warranted and may provide insight into how to better manage infection when it does occur.
HOW ECOLOGY CAN SHAPE CLINICAL EPIDEMIOLOGY AND EXPOSURE
Environmental genomic surveillance also helps explain patterns of clinical disease and risk of human exposure to Cryptococcus. It is hypothesised that growth on bird guano as a key niche may have led to VNI's widening global distribution in concert with bird domestication and association with urban locales (Nielsen, De Obaldia and Heitman 2007). In comparison, VNB environmental isolates have only been isolated from arboreal trees in rural Africa and, once, from Brazil. In turn, VNB infections are rare and restricted to these areas of Africa and South America, suggesting that patients are acquiring VNB infections as a consequence of their exposures to these arboreal reservoirs (Litvintseva et al. 2011; Vanhove et al. 2017; Rhodes et al. 2017b). The exact extent and type of VNB arboreal reservoir in South America remains unknown, however.
How ecological niche has shaped C. gattii distribution is less clear since, although a global infection, C. gattii is also predominantly associated with arboreal tree species. It is hypothesised that C. gattii’s spread to the Pacific Northwest may have been through shipping ballast combined with ocean currents and perhaps aided by extreme events such as Tsunami (Engelthaler and Casadevall 2019), and/or via the plant and seed trade (Roe et al. 2018).
How the biotic and abiotic environment shapes exposure and epidemiology of cryptococcosis at a more local level is yet to be determined. Environmental surveillance in Zambia has suggested an ecological split between C. neoformans which was found mostly in the southern, arid and low altitude Zambezi Mopane ecoregion, and C. gattii in the northern, wet and high altitude Central Miombo ecoregion (Vanhove et al. 2017). This ecological divide could be significant if it affects the distribution of clinical cases and the relative risk of exposure, particularly among populations of HIV-infected individuals that inhabit each part of the country (Maziarz and Perfect 2016). Further environmental sampling and enhanced clinical diagnosis to distinguish, at minimum, the infectious agent at the level of the species complex could disentangle the effect of geographic species distribution on clinical incidence at sub-national levels. This could ultimately affect recommendations given to health service providers on diagnosis and drug stewardship upon presentation of a case of pneumonia or meningitis, as well as the utility of prophylaxis, if the risk of local acquisition of C. neoformans infection is high (Oladele et al. 2017).
It is highly likely that not all environmental niches of Cryptococcus have as yet been identified. This was recently demonstrated by the discovery of VGV from investigative sampling of an entirely new ecological niche, the rock hyrax midden, where it was found to co-exist with other cryptococcal molecular types (Farrer et al. 2019). Hyrax middens are extremely stable and long-lasting structures that can exist in the same place for thousands of years (Chase et al. 2012). Middens have a high nitrogen content which is known to aid cryptococcal growth, which likely results in the development of patchy high-burden hotspots of Cryptococcus. Twinned with their extreme environmental stability, hyrax middens may therefore provide stable long-term evolutionary arenas that are important in generating diversity of Cryptococcus (Staib et al. 1978; Vreulink et al. 2020).
How each identified niche relates to being a reservoir of infection and hence when and where people are exposed remains unclear. In California, USA, isolation of VGIII environmental isolates showed a very close relationship with clinical isolates suggesting a local environmental reservoir of infection (Springer et al. 2014), and similar studies are ongoing in the UK (Farrer et al.). However, as yet the genomic epidemiology to explore these links have not been made in Africa. Since VNI is found frequently around the globe in pigeon faeces from urban locations, it is easy to anticipate how people may be exposed to VNI more often, thereby leading to more frequent infection. Yet many observed (and more diverse) ecological niches are found in very rural locations far from human activity and thus may not pose an immediate clinical threat through exposure. Conversely, some clinically significant molecular types, such as VNII, are rarely found in the environment and thus their infectious reservoir is, as yet, unknown.
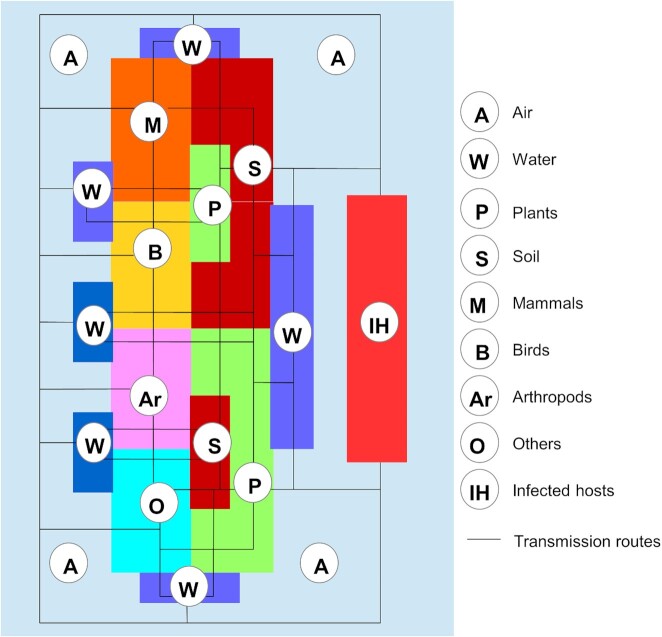
The interaction of Cryptococcus with its environment and susceptible hosts is complex (Fig. 4). Although Cryptococcus spp. are environmental saprotrophs, thriving on decaying wood, soil and animal droppings, they can also be found in water, including ocean saltwater (Emmons 1955; Kidd et al. 2007b; Kandasamy, Alikunhi and Subramanian 2012). Cryptococcus is likely actively dispersed between sites through contamination of a variety of animal species that live or feed on colonised trees or soil, including insects (23). Passive dispersal occurs through the production and aerosolization of desiccated yeast cells or through basidiospores that are produced during sexual reproduction (Zhao et al. 2019; Cogliati et al. 2020). These cells and spores may disperse widely before colonising new habitats, and are thought to represent the principle exposure to susceptible hosts through inhalation (Velagapudi et al. 2009; Rieux et al. 2014).
Figure 4.
Schematic representation of the relationships among biotic and abiotic components (coloured polygons) of the Cryptococcus ecosystem, and dynamic flow of the fungus through the different niches (solid lines). Cryptococcus can circulate in the environment through several vectors (wind, water, animals) and reach the main reservoirs (soil and plants). From its habitat in these reservoirs, Cryptococcus can produce and release aerosolised basidiospores which are able to colonize other niches or infect susceptible hosts.
Since trees have been shown to be one of the main reservoirs for cryptococcal yeasts, understanding the biotic and abiotic components that comprise these tree-scale ecological niches alongside which vectors contribute to the spread of Cryptococcus in the environment could aid understanding of the mechanisms involved in human infection. In a recent study, biotic and abiotic factors affecting the distribution of both C. neoformans var. neoformans and C. neoformans var. grubii found living on the same oak tree were investigated (Cogliati et al. 2020). Ants and other arthropods were shown to contribute to the distribution of the yeasts on the tree as well as to the colonisation of other trees. Microscopy showed how the yeast cells use filamentous protrusions to anchor to the bark, leaving the non-adherent surface free for budding, the resulting spores of which were identified in the surrounding air. These studies may implicate arthropods as important hosts for Cryptococcus, and may in part explain the utility of the wax-moth larvae Galleria mellonella as a model for cryptococcal virulence (Mylonakis et al. 2005).
Although infection is caused by aerosolised infectious propagules, airborne isolations of Cryptococcus are scarce. Most attempts to isolate cryptococcal spores have simply exposed agar Petri dishes to the air, a few of which have been successful, mostly when plates are exposed directly next to pigeon guano sources or when spores have been aerosolized through human intervention (Baroni et al. 2006; Randhawa et al. 2006; Pedroso, Ferreira and Candido 2009). Other attempts have been made with high-throughput air samplers to trap Cryptococcus bioaerosols (Lazera et al. 2000; Kidd et al. 2007b, 2007a). Use of high-throughput air sampling in Canada found that forestry activities led to a higher concentration of C. gattii spores in the air (Kidd et al. 2007a). This may be relevant to exposure risk in southern Africa since mopane trees, which are strongly associated with colonisation by C. neoformans (Litvintseva et al. 2011; Vanhove et al. 2017), form an important part of the local culture and are frequently cut and used for charcoal, traditional medicine, building materials and the cultivation of edible mopane worms (Chidumayo 1993; Woollen et al. 2016; Ziba and Grouwels 2017). Seasonality may also affect the concentration of infectious propagules released into the air, with autumn conditions associated with a greater concentration of airborne cryptococcal propagules observed in the temperate climate of northern Italy (Cogliati et al. 2020).
An added complication in assessing from where and when infection occurs is the hypothesis that infection may occur many months-to-years before symptoms. A study by Beale et al. (Beale et al. 2015) found a lack of geographic clustering between genetic sequences from patients in Cape Town, suggesting against local acquisition of infection, though the study did not attempt to support this with surveillance of the environment. Combining clinical genetic studies such as this with environmental surveillance around people's houses and in line with their travel and activity history (particularly activities related to forestry), may give more insights into from where and when infection is acquired.
EMERGENCE OF ANTIFUNGAL DRUG RESISTANCE
Treatment failure and subsequent relapse of infection can occur as a result of cryptococcal resistance to first-line drug treatment, including azoles and flucytosine (FLC) (Birley et al. 1995; Aller et al. 2000; Musubire 2013; Billmyre et al. 2020). Development of resistance and emergence of heteroresistant colonies is apparent in serially collected isolates from patients and relapse patients, suggesting resistance can develop as a within-host response to drug treatment (Chen et al. 2017; Stone et al. 2019). In some clinical cases, nonsense mutations in the gene encoding DNA mismatch repair proteins (MSH2, MSH5, RAD5 and POL3) are associated with hypermutator phenotypes that can lead to very rapid within-host microevolution (Rhodes et al. 2017a; Boyce et al. 2020). When twinned with drug-pressure, hypermutating genotypes are associated with the emergence of drug-resistance in vitro and present a novel pathway for rapid evolution of resistance to first-line antifungal drugs (Boyce et al. 2017). The relevance of hypermutators in the environmental stages of Cryptococcus has not been established, however. Differing levels of resistance to antifungals have been identified in environmental isolates suggesting that either hypermutator or other, perhaps innate, resistance mechanisms may be ecologically relevant. For instance, sampling in Cameroon found both C. neoformans and C. gattii in pigeon and bat guano with high antifungal resistance (Dongmo et al. 2016). In another region of Africa, both environmental VGIV / VGV strains from Zambia showed unusually high resistance to flucytosine (FLC), and in particular isolates from a specific clade of VGV (VGV-A) (Farrer et al. 2019). The ability of environmental isolates to manifest resistance to first-line drugs could either be the indirect consequence of adaptation to antifungal-like chemicals in the environment or the direct consequence of exposure to fungicides (such as azoles) that are used in agriculture or forestry. Evidence that azole resistance works at least partly through upregulation of ABC transporters which act to remove molecules from cells in a non-specific manner suggests the former may be true (Posteraro et al. 2003; Sanguinetti et al. 2006). Of relevance, there is widespread concern that widespread use of azoles in agriculture and forestry industries is contributing to emerging resistance in other fungi, most notably Aspergillus fumigatus (Snelders et al. 2012; Chowdhary et al. 2013; Kleinkauf et al. 2013; Ren et al. 2017). Surveillance of resistance in environmental cryptococcal populations may well be important to monitor the emergence and spread of resistance and thus the threat to clinical management of disease. Mapping environmental isolations against areas of intensive farming and commercial forestry may also indicate whether there is an effect of azole usage on propagating these genotypes by creating hotspots for the evolution of antifungal resistance.
CHALLENGES IN ENVIRONMENTAL SURVEYING AND MODELLING CRYPTOCOCCAL DISTRIBUTIONS
It is clear there is much to learn from the genomics of environmental cryptococcal populations. However, isolating Cryptococcus spp. from the environment is challenging—it can be difficult to find and, once found, can be problematic to isolate into pure culture due to competition from faster-growing filamentous fungi (Lazera et al. 2000; Pham et al. 2014; de Matos Castro e Silva et al. 2015). Surveying and subsequent culturing can thus be labour and time-intensive and results in limited recovery rates (Vilcins et al. 2002; Kidd et al. 2007b; Litvintseva et al. 2011; Cogliati et al. 2016; Vanhove et al. 2017). Because of this and the propensity to find Cryptococcus spp. in certain ecological niches, targeted sampling should be used in order to generate a larger number of isolates for study. However, targeted sampling leads to issues if using data to conduct environmental niche modelling (ENM) due to positive selection bias (Mak et al. 2010; Cogliati et al. 2017; Vanhove et al. 2017; Alaniz et al. 2020). ENM studies attempt to map the distribution of Cryptococcus spp. across entire countries or continents using climatic variables highly dependent on a small number of sampled collection sites. Models have focused on use of presence-only data since absence of the pathogen from locations that may not have been sampled cannot be assumed and negatively sampled locations may not indicate true absence since the yeast may just not have been recovered successfully in culture. Presence-absence models perform better than presence-only models but models for wide-ranging and tolerant species can be particularly sensitive to absence data, as has been shown in predictions of bird habitats (Brotons et al. 2004; Elith et al. 2006). Use of pseudo-absence data has been proposed as a potential strategy in such situations (Gu and Swihart 2004) (Gu and Swihart 2004) (108) (108) (108) (Zaniewski, Lehmann and Overton 2002; Engler, Guisan and Rechsteiner 2004; Gu and Swihart 2004; Phillips et al. 2009; Lobo, Jiménez-Valverde and Hortal 2010; Senay, Worner and Ikeda 2013). Since different species distribution models also show differences in predictive performance and stability, different algorithms should be compared to give an indication of uncertainty between methods, in a process that is analogous to the use of climatic ensemble models (Ren-Yan et al. 2014).
CONCLUSION
Despite insights into the ecology, biology, evolution and epidemiology that environmental isolates of C. neoformans/C. gattii provide, sampling and subsequent genomic and phenotypic analysis of environmental isolates have, to date, been limited, particularly within the African context. This is despite recent progress stemming from both ecological surveys and genomic epidemiology showing that we are underestimating the scale and clinical importance of cryptococcal diversity. While increased sampling and genomics analysis of Cryptococcus in the southern Africa region would be of benefit since this region appears to be the origin of global diversity and has the highest clinical impact, sampling has been very limited in other regions, particularly Central Africa which may be important given that it borders the southern region. Although here we have focused on Cryptococcus, the significance of the methods and analyses we describe are applicable to other environmental fungi and microbes that pose an increasing threat to human, animal and plant health and biosecurity (Fisher et al. 2012). The integration of data from multiple sources, including environmental, clinical, bioclimatic, molecular and epidemiological, is becoming increasingly important in understanding the complexity of microbial threats. Indeed, the integrative environment-health science frameworks that we describe here are increasingly needed to understand and model future scenarios with the aim of thwarting future outbreaks of infection (Fisher and Murray 2021).
Supplementary Material
ACKNOWLEDGEMENTS
HME and MCF were funded by the UK Medical Research Councilx2 MR/R015600/1; MR/K000373/1, and MCF by the UK Natural Environmental Research Council. MCF is a CIFAR Fellow in the ‘Fungal Kingdoms’ program.
Contributor Information
Hannah M Edwards, MRC Centre for Global Infectious Disease Analysis, Imperial College School of Public Health, Imperial College London, Norfolk Place, London W2 1PG, UK.
Massimo Cogliati, Dip. Scienze Biomediche per la Salute, Università degli Studi di Milano, Via Pascal 36, 20133 Milano, Italy.
Geoffrey Kwenda, Department of Biomedical Sciences, School of Health Sciences, University of Zambia, Ridgeway Campus, PO Box 50110, Lusaka, Zambia.
Matthew C Fisher, MRC Centre for Global Infectious Disease Analysis, Imperial College School of Public Health, Imperial College London, Norfolk Place, London W2 1PG, UK.
Conflicts of interest
None declared.
REFERENCES
- Alaniz AJ, Carvajal JG, Carvajal MA et al. Spatial Quantification of the Population Exposed to Cryptococcus neoformans and Cryptococcus gattii Species Complexes in Europe: estimating the Immunocompetent and HIV/AIDS Patients Under Risk. Risk Anal. 2020;40. DOI: 10.1111/risa.13410. [DOI] [PubMed] [Google Scholar]
- Aller AI, Martin-Mazuelos E, Lozano F et al. Correlation of fluconazole MICs with clinical outcome in cryptococcal infection. Antimicrob Agents Chemother. 2000;44:1544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminnejad M, Diaz M, Arabatzis M et al. Identification of Novel Hybrids Between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia. 2012;173:337–46. [DOI] [PubMed] [Google Scholar]
- Ashton PM, Thanh LT, Trieu PH et al. Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans. Nat Commun. 2019;10:2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni FDA, Paula CR, Da Silva ÉG et al. Cryptococcus neoformans strains isolated from church towers in Rio de Janeiro City, RJ, Brazil. Revista do Instituto de Medicina Tropical de São Paulo. 2006;48:71–5. [DOI] [PubMed] [Google Scholar]
- Bartlett KH, Kidd SE, Kronstad JW. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Current Infectious Disease Reports. 10, 2008. DOI: 10.1007/s11908-008-0011-1. [DOI] [PubMed] [Google Scholar]
- Beale MA, Sabiiti W, Robertson EJ et al. Genotypic diversity is associated with clinical outcome and phenotype in cryptococcal meningitis across Southern Africa. PLoS NeglTrop Dis. 2015;9:e0003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre RB, Applen Clancey S, Li LX et al. 5-fluorocytosine resistance is associated with hypermutation and alterations in capsule biosynthesis in Cryptococcus. Nat Commun. 2020;11:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre RB, Croll D, Li W et al. Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. MBio. 2014;5:e01494–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birley HDL, Johnson EM, Mcdonald P et al. Azole Drug Resistance as a Cause of Clinical Relapse in AIDS Patients with Cryptococcal Meningitis. Int J STD AIDS. 1995;6:353–5. [DOI] [PubMed] [Google Scholar]
- Boekhout T, Theelen B, Diaz M et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. [DOI] [PubMed] [Google Scholar]
- Bongomin F, Gago S, Oladele RO et al. Global and multi-national prevalence of fungal diseases—estimate precision. Journal of Fungi. 2017;3:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Kuramae EE et al. AIDS patient death caused by novel Cryptococcus neoformans x C. gattii hybrid. Emerg Infect Dis. 2008;14:1105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Kuramae EE et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607. [DOI] [PubMed] [Google Scholar]
- Boyce KJ, Cao C, Xue C et al. A spontaneous mutation in DNA polymerase POL3 during in vitro passaging causes a hypermutator phenotype in Cryptococcus species. DNA Repair (Amst). 2020;86:102751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce KJ, Wang Y, Verma S et al. Mismatch Repair of DNA Replication Errors Contributes to Microevolution in the Pathogenic Fungus Cryptococcus neoformans. MBio. 2017;8:e00595–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotons L, Thuiller W, Araújo MB et al. Presence-absence versus presence-only modelling methods for predicting bird habitat suitability. Ecography. 2004;27. DOI: 10.1111/j.0906-7590.2004.03764.x. [Google Scholar]
- Byrnes EJ, Marr KA. The outbreak of Cryptococcus gattii in western North America: epidemiology and clinical issues. Current Infectious Disease Reports. 2011;13:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Coelho C, Cordero RJB et al. The capsule of Cryptococcus neoformans. Virulence. 2019;10:822–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Freij JB, Hann-Soden C et al. Continental Drift and Speciation of the Cryptococcus neoformans and Cryptococcus gattii Species Complexes. mSphere. 2017;2:e00103–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Steenbergen JN, Nosanchuk JD.“Ready made” virulence and “dual use” virulence factors in pathogenic environmental fungi - The Cryptococcus neoformans paradigm. Curr Opin Microbiol. 2003;6:332–7. [DOI] [PubMed] [Google Scholar]
- Casadevall A. Evolution of Intracellular Pathogens. Annu Rev Microbiol. 2008;62:19–33. [DOI] [PubMed] [Google Scholar]
- Chase BM, Scott L, Meadows ME et al. Rock hyrax middens: a palaeoenvironmental archive for southern African drylands. Quat Sci Rev. 2012;56:107–25. [Google Scholar]
- Chen Y, Farrer RA, Giamberardino C et al. Microevolution of Serial Clinical Isolates of Cryptococcus neoformans var. grubii and C. gattii. MBio. 2017;8:e00166–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Litvintseva AP, Frazzitta AE et al. Comparative analyses of clinical and environmental populations of Cryptococcus neoformans in Botswana. Mol Ecol. 2015;24:3559–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidumayo EN. Zambian charcoal production. Energy Policy. 1993;21:586–97. [Google Scholar]
- Chowdhary A, Kathuria S, Xu J et al. Emergence of Azole-Resistant Aspergillus fumigatus Strains due to Agricultural Azole Use Creates an Increasing Threat to Human Health. PLoS Pathog. 2013;9:e1003633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati M, D'Amicis R, Zani A et al. Environmental distribution of Cryptococcus neoformans and C. gattii around the Mediterranean basin. FEMS Yeast Res. 2016;16:fow045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati M, Desnos-Ollivier M, McCormick-Smith I et al. Genotypes and population genetics of Cryptococcus neoformans and Cryptococcus gattii species complexes in Europe and the Mediterranean area. Fungal Genet Biol. 2019;129:16–29. [DOI] [PubMed] [Google Scholar]
- Cogliati M, Patrizia P, Vincenzo C et al. Cryptococcus neoformans species complex isolates living in a tree micro-ecosystem. Fungal Ecology. 2020;44:100889. [Google Scholar]
- Cogliati M, Puccianti E, Montagna MT et al. Fundamental niche prediction of the pathogenic yeasts Cryptococcus neoformans and Cryptococcus gattii in Europe. Environ Microbiol. 2017;19:4318–25. [DOI] [PubMed] [Google Scholar]
- Cogliati M. Global Molecular Epidemiology of Cryptococcus neoformans and Cryptococcus gattii: an Atlas of the Molecular Types. Scientifica (Cairo). 2013;2013:675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva EG, Baroni FDA, Viani FC et al. Virulence profile of strains of Cryptococcus neoformans var. grubii evaluated by experimental infection in BALB/c mice and correlation with exoenzyme activity. J Med Microbiol. 2006;55:139–42. [DOI] [PubMed] [Google Scholar]
- de Matos Castro e Silva D, Santos DCS, Pukinskas SRBS et al. A new culture medium for recovering the agents of cryptococcosis from environmental sources. Brazilian Journal of Microbiology. 2015;46:355–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Giamberardino C, Sykes SM et al. Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 2017;27:1207–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongmo W, Kechia F, Tchuenguem R et al. In Vitro Antifungal Susceptibility of Environmental Isolates of Cryptococcus spp. from the West Region of Cameroon. Ethiopian Journal of Health Sciences. 2016;26:555–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfadaly HA, Hassanain NA, Hassanain MA et al. Evaluation of primitive ground water supplies as a risk factor for the development of major waterborne zoonosis in Egyptian children living in rural areas. J Infect Public Health. 2018;11:203–8. [DOI] [PubMed] [Google Scholar]
- Elhariri M, Hamza D, Elhelw R et al. Eucalyptus Tree: A Potential Source of Cryptococcus neoformans in Egyptian Environment. Int J Microbiol. 2016;2016:4080725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elith J.H., Graham C.P., Anderson R et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29. DOI: 10.1111/j.2006.0906-7590.04596.x. [Google Scholar]
- Ellabib MS, Aboshkiwa MA, Husien WM et al. Isolation, Identification and Molecular Typing of Cryptococcus neoformans from Pigeon Droppings and Other Environmental Sources in Tripoli, Libya. Mycopathologia. 2016;181:603–8. [DOI] [PubMed] [Google Scholar]
- Ellis DH, Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990;28:1642–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons CW. Saprophytic sources of Cryptococcus neoformans associated with the pigeon (Columba livia). Am J Epidemiol. 1955;62:227–32. [DOI] [PubMed] [Google Scholar]
- Engelthaler DM, Casadevall A. On the Emergence of Cryptococcus gattii in the Pacific Northwest: ballast Tanks, Tsunamis, and Black Swans. MBio. 2019;10:e02193–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler DM, Hicks ND, Gillece JD et al. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. MBio. 2014;5. DOI: 10.1128/mBio.01464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler R, Guisan A, Rechsteiner L. An improved approach for predicting the distribution of rare and endangered species from occurrence and pseudo-absence data. J Appl Ecol. 2004;41. DOI: 10.1111/j.0021-8901.2004.00881.x. [Google Scholar]
- Farrer R, Borman A, Inkster T et al. Genomic epidemiology of a Cryptococcus neoformans case cluster in Glasgow, Scotland, 2018. Microb genomics. 2021. DOI 10.1099/mgen.0.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer RA, Chang M, Davis MJ et al. A new lineage of Cryptococcus gattii (VGV) discovered in the central Zambezian Miombo woodlands. MBio. 2019;10:e02306–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Henk DA, Briggs CJ et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Murray KA. Emerging infections and the integrative environment-health sciences: the road ahead. Nat Rev Microbiol. 2021;19. DOI: 10.1038/s41579-021-00510-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromtling RA, Abruzzo GK, Ruiz A. Virulence and antifungal susceptibility of environmental and clinical isolates of Cryptococcus neoformans from Puerto Rico. Mycopathologia. 1989;106:163–6. [DOI] [PubMed] [Google Scholar]
- Fu C, Sun S, Billmyre RB et al. Unisexual versus bisexual mating in Cryptococcus neoformans: consequences and biological impacts. Fungal Genet Biol. 2015;78:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Solache MA, Casadevall A. Hypothesis: global warming will bring new fungal diseases for mammals. MBio. 2010;1:e00061–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Fu MS, Mukaremera L et al. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. MBio. 2015;6:e01340–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Swihart RK. Absent or undetected? Effects of non-detection of species occurrence on wildlife-habitat models. Biol Conserv. 2004;116:195–203. [Google Scholar]
- Guerrero A, Jain N, Goldman DL et al. Phenotypic switching in Cryptococcus neoformans. Microbiology. 2006;152:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Ceresini PC, Polacheck I et al. Ancient Dispersal of the Human Fungal Pathogen Cryptococcus gattii from the Amazon Rainforest. PLoS One. 2013;8:e71148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Khayhan K, Theelen B et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015;78:16–48. [DOI] [PubMed] [Google Scholar]
- Hommel B, Mukaremera L, Cordero RJB et al. Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog. 2018;14:e1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Fries BC. Phenotypic switching of Cryptococcus neoformans and Cryptococcus gattii. Mycopathologia. 2008;166:181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K, Alikunhi NM, Subramanian M. Yeasts in marine and estuarine environments. J Yeast Fungal Res. 2012;3:74–82. [Google Scholar]
- Kangogo M, Bader O, Boga H et al. Molecular types of Cryptococcus gattii/Cryptococcus neoformans species complex from clinical and environmental sources in Nairobi, Kenya. Mycoses. 2015;58:665–70. [DOI] [PubMed] [Google Scholar]
- Kassi FK, Bellet V, Drakulovski P et al. Comparative typing analyses of clinical and environmental strains of the Cryptococcus neoformans/Cryptococcus gattii species complex from Ivory Coast. J Med Microbiol. 2018;67:87–96. [DOI] [PubMed] [Google Scholar]
- Kidd SE, Bach PJ, Hingston AO et al. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg Infect Dis. 2007a;13:51–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd SE, Chow Y, Mak S et al. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl Environ Microbiol. 2007b;73:1433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinkauf N, Verweij PE, Arendrup MC et al. Risk Assessment on the Impact of Environmental Usage of Triazoles on the Development and Spread of Resistance to Medical Triazoles in Aspergillus Species. ECDC Technical Report, 2013. [Google Scholar]
- Kronstad JW, Attarian R, Cadieux B et al. Expanding fungal pathogenesis: c ryptococcus breaks out of the opportunistic box. Nat Rev Microbiol. 2011;9:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazera MS, Cavalcanti MAS, Londero AT et al. Possible primary ecological niche of Cryptococcus neoformans. Med Mycol. 2000;38:379–83. [DOI] [PubMed] [Google Scholar]
- Litvintseva AP, Carbone I, Rossouw J et al. Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS One. 2011;6:e19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Mitchell TG. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect Immun. 2009;77:3188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo JM, Jiménez-Valverde A, Hortal J. The uncertain nature of absences and their importance in species distribution modelling. Ecography. 2010;33. DOI: 10.1111/j.1600-0587.2009.06039.x. [Google Scholar]
- Magditch DA, Liu TB, Xue C et al. DNA Mutations Mediate Microevolution between Host-Adapted Forms of the Pathogenic Fungus Cryptococcus neoformans. PLoS Pathog. 2012;8:e1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud Y. First environmental isolation of Cryptococcus neoformans var. neoformans and var. gatti from the Gharbia Governorate, Egypt. Mycopathologia. 1999;148:83–6. [DOI] [PubMed] [Google Scholar]
- Mak S, Klinkenberg B, Bartlett K et al. Ecological niche modeling of Cryptococcus gattii in British Columbia, Canada. Environ Health Perspect. 2010;118:653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RC, Stone NRH, Wiesner DL et al. Cryptococcus: from environmental saprophyte to global pathogen. Nat Rev Microbiol. 2016;14:106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziarz EK, Perfect JR Cryptococcosis. Infect Dis Clin North Am. 2016;30:179–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mseddi F, Sellami A, Jarboui MA et al. First Environmental Isolations of Cryptococcus neoformans and Cryptococcus gattii in Tunisia and Review of Published Studies on Environmental Isolations in Africa. Mycopathologia. 2011;171:355–60. [DOI] [PubMed] [Google Scholar]
- Musubire AK. Diagnosis and Management of Cryptococcal Relapse. J AIDS Clin Res. 2013;Suppl 3:S3–003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis E, Moreno R, El Khoury JB et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73:3842–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Li W et al. Unisexual and Heterosexual Meiotic Reproduction Generate Aneuploidy and Phenotypic Diversity De Novo in the Yeast Cryptococcus neoformans. PLoS Biol. 2013;11:e1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P et al. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun. 2003;71:4831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, De Obaldia AL, Heitman J. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryotic Cell. 2007;6:949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nnadi NE, Enweani IB, Cogliati M et al. Molecular characterization of environmental Cryptococcus neoformans VNII isolates in Jos, Plateau State, Nigeria. Journal de Mycologie Médicale. 2016;26:306–11. [DOI] [PubMed] [Google Scholar]
- Nweze EI, Kechia FA, Dibua UE et al. Isolation of Cryptococcus neoformans from environmental samples collected in southeastern Nigeria. Revista do Instituto de Medicina Tropical de São Paulo. 2015;57:295–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyazika TK, Kamtchum-Tatuene J, Kenfak-Foguena A et al. Prevalence and mortality of cryptococcal meningitis in Africa from 1950 to 2017 and associated epidemiological mapping of C. neoformans and C. gattii species complexes: a systematic review and meta-analysis. SSRN Electronic Journal. 2019. DOI: 10.2139/ssrn.3393702. [Google Scholar]
- Oladele RO, Bongomin F, Gago S et al. HIV-associated cryptococcal disease in resource-limited settings: a case for “prevention is better than cure”?. Journal of Fungi. 2017;3:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso RS, Ferreira JC, Candido RC. The isolation and characterization of virulence factors of Cryptococcus spp. from saprophytic sources in the city of Ribeirão Preto, São Paulo, Brazil. Microbiol Res. 2009;164:221–7. [DOI] [PubMed] [Google Scholar]
- Perfect JR, Bicanic T. Cryptococcosis diagnosis and treatment: what do we know now. Fungal Genet Biol. 2015;78:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR. Cryptococcus neoformans: the yeast that likes it hot. FEMS Yeast Res. 2006;6:463–8. [DOI] [PubMed] [Google Scholar]
- Pfeiffer TJ, Ellis DH. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. Med Mycol. 1992;34:127–31. [PubMed] [Google Scholar]
- Phadke SS, Feretzaki M, Clancey SA et al. Unisexual reproduction of Cryptococcus gattii. PLoS One. 2014;9:e111089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CD, Ahn S, Turner LA et al. Development and validation of benomyl birdseed agar for the isolation of Cryptococcus neoformans and Cryptococcus gattii from environmental samples. Med Mycol. 2014;52:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SJ, Dudík M, Elith J et al. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol Appl. 2009;19. DOI: 10.1890/07-2153.1. [DOI] [PubMed] [Google Scholar]
- Posteraro B, Sanguinetti M, Sanglard D et al. Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Mol Microbiol. 2003;47:357–71. [DOI] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17:873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa HS, Kowshik T, Preeti Sinha K et al. Distribution of Cryptococcus gattii and Cryptococcus neoformans in decayed trunk wood of Syzygium cumini trees in north-western India. Med Mycol. 2006;44:623–30. [DOI] [PubMed] [Google Scholar]
- Refai M, Taha M, Selim SA et al. Isolation of Cryptococcus neoformans, Candida albicans and other yeasts from pigeon droppings in Egypt. Med Mycol. 1983;21:163–5. [PubMed] [Google Scholar]
- Ren J, Jin X, Zhang Q et al. Fungicides induced triazole-resistance in Aspergillus fumigatus associated with mutations of TR46/Y121F/T289A and its appearance in agricultural fields. J Hazard Mater. 2017;326:54–60. [DOI] [PubMed] [Google Scholar]
- Ren-Yan D, Xiao-Quan K, Min-Yi H et al. The predictive performance and stability of six species distribution models. PLoS One. 2014;9:e112764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Beale MA, Vanhove M et al. A Population Genomics Approach to Assessing the Genetic Basis of Within-Host Microevolution Underlying Recurrent Cryptococcal Meningitis Infection. G3 Genes|Genomes|Genetics. 2017a;7:1165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Desjardins CA, Sykes SM et al. Tracing genetic exchange and biogeography of Cryptococcus neoformans var. grubii at the global population level. Genetics. 2017b;207:327–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieux A, Soubeyrand S, Bonnot F et al. Long-distance wind-dispersal of spores in a fungal plant pathogen: estimation of anisotropic dispersal kernels from an extensive field experiment. PLoS One. 2014;9:e103225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe CC, Bowers J, Oltean H et al. Dating the Cryptococcus gattii Dispersal to the North American Pacific Northwest. mSphere. 2018;3:e00499–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M, Posteraro B, Sorda LaM et al. Role of AFR1, an ABC transporter-encoding gene, in the in vivo response to fluconazole and virulence of Cryptococcus neoformans. Infect Immun. 2006;74:1352–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senay SD, Worner SP, Ikeda T. Novel Three-Step Pseudo-Absence Selection Technique for Improved Species Distribution Modelling. PLoS One. 2013;8:e71218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E, Camps SMT, Karawajczyk A et al. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS One. 2012;7:e31801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer DJ, Billmyre RB, Filler EE et al. Cryptococcus gattii VGIII Isolates Causing Infections in HIV/AIDS Patients in Southern California: identification of the Local Environmental Source as Arboreal. PLoS Pathog. 2014;10:e1004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib F, Grave B, Altmann L et al. Epidemiology of Cryptococcus neoformans. Mycopathologia. 1978;65:73–6. [DOI] [PubMed] [Google Scholar]
- Stone NRH, Rhodes J, Fisher MC et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J Clin Invest. 2019;129:999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Coelho MA, David-Palma M et al. The Evolution of Sexual Reproduction and the Mating-Type Locus: links to Pathogenesis of Cryptococcus Human Pathogenic Fungi. Annu Rev Genet. 2019;53:417–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinne D, Deppner M, Maniratunga S et al. AIDS-associated cryptococcosis in Bujumbura, Burundi: An epidemiological study. J Med Vet Mycol. 1991;29:25–30. [DOI] [PubMed] [Google Scholar]
- Swinne D, Deppnert M, Larochet R et al. Isolation of Cryptococcus neoformans from houses of AIDS-associated cryptococcosis patients in Bujumbura (Burundi). AIDS. 1989;3:389–90. [DOI] [PubMed] [Google Scholar]
- Swinne D, Kayembe K, Niyimi M. Isolation of saprophytic Cryptococcus neoformans var. neoformans in Kinshasa, Zaïre. Ann Soc Belg Med Trop (1920). 1986;66:57–61. [PubMed] [Google Scholar]
- Vanhove M, Beale MA, Rhodes J et al. Genomic epidemiology of Cryptococcus yeasts identifies adaptation to environmental niches underpinning infection across an African HIV/AIDS cohort. Mol Ecol. 2017;26:1991–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Swinne D, Staib F et al. Diversity of DNA fingerprints in Cryptococcus neoformans. J Clin Microbiol. 1995;33:1807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi R, Hsueh YP, Geunes-Boyer S et al. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun. 2009;77:4345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcins I, Krockenberger M, Agus H et al. Environmental sampling for Cryptococcus neoformans var. gattii from the Blue Mountains National Park, Sydney, Australia. Med Mycol. 2002;40:53–60. [DOI] [PubMed] [Google Scholar]
- Vreulink JM, Boekhout T, Vismer H et al. The growth of Cryptococcus gattii MATα and MATa strains is affected by the chemical composition of their woody debris substrate. Fungal Ecology. 2020;47:100943. [Google Scholar]
- Walsh NM, Botts MR, McDermott AJ et al. Infectious particle identity determines dissemination and disease outcome for the inhaled human fungal pathogen Cryptococcus. PLoS Pathog. 2019;15:e1007777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson PR. Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front Biosci. 1997;2:e99–107. [DOI] [PubMed] [Google Scholar]
- Woollen E, Ryan CM, Baumert S et al. Charcoal production in the mopane woodlands of Mozambique: what are the trade-offs with other ecosystem services?. Philosophical Transactions of the Royal Society B: Biological Sciences. 2016;371:20150315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaniewski AE, Lehmann A, Overton JMC. Predicting species spatial distributions using presence-only data: a case study of native New Zealand ferns. Ecol Modell. 2002;157:261–80. [Google Scholar]
- Zaragoza O. Basic principles of the virulence of Cryptococcus. Virulence. 2019;10:490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lin J, Fan Y et al. Life Cycle of Cryptococcus neoformans. Annu Rev Microbiol. 2019;73:17–42. [DOI] [PubMed] [Google Scholar]
- Zhou X, Ballou ER. The Cryptococcus neoformans Titan Cell: from In Vivo Phenomenon to In Vitro Model. Current Clinical Microbiology Reports. 2018;5:252–60. [Google Scholar]
- Ziba V, Grouwels S. Greening Zambia's Charcoal Business for Improved Livelihoods and Forest Management through Strong Producer Groups. Country Case. Rome, 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.