Abstract
Oral squamous cell carcinoma (OSCC) represents 90% of oral malignant neoplasms. The search for specific biomarkers for OSCC is a very active field of research contributing to establishing early diagnostic methods and unraveling underlying pathogenic mechanisms. In this work we investigated the salivary metabolites and the metabolic pathways of OSCC aiming find possible biomarkers. Salivary metabolites samples from 27 OSCC patients and 41 control individuals were compared through a gas chromatography coupled to a mass spectrometer (GC-MS) technique. Our results allowed identification of pathways of the malate-aspartate shuttle, the beta-alanine metabolism, and the Warburg effect. The possible salivary biomarkers were identified using the area under receiver-operating curve (AUC) criterion. Twenty-four metabolites were identified with AUC > 0.8. Using the threshold of AUC = 0.9 we find malic acid, maltose, protocatechuic acid, lactose, 2-ketoadipic, and catechol metabolites expressed. We notice that this is the first report of salivary metabolome in South American oral cancer patients, to the best of our knowledge. Our findings regarding these metabolic changes are important in discovering salivary biomarkers of OSCC patients. However, additional work needs to be performed considering larger populations to validate our results.
Keywords: metabolomics, biomarkers, metabolites, oral squamous cell carcinoma, oral cancer, saliva, mass spectrometry, GC-MS
1. Introduction
Oral cancer refers to the set of malignant neoplasms that affect the lips and other intraoral regions [1]. It represents the 16th most common neoplasm in the world, with 355,000 new diagnoses and 177,000 deaths in 2018 [2]. It is a highly relevant problem for global public health since there is no evidence of significant improvement for fast treatment and prevention in spite of all the progress in current research and therapies [3]. Among oral malignancies squamous cell carcinoma (OSCC) is the most prevalent histological type representing approximately 90% of cases. OSCC is often preceded by the presence of oral potentially malignant disorders. They are clinically identifiable as either white or red patches known as leukoplakia and erythroplakia, respectively. Non-healing ulcers may also be noticed along cancer development [4]. The highest incidence of OSCC occurs in the middle-aged population although the number of young individuals diagnosed with the disease has increased [5,6]. The most common site for OSCC is the tongue followed by the floor of the mouth. Less common sites include the gingiva, buccal mucosa, labial mucosa, and hard palate [4]. OSCC has a survival rate of approximately 80% for individuals detected with early stage disease (stage I) when compared to a rate of 20–30% in patients diagnosed at advanced stages (stages III–IV) [7]. This fact emphasizes the importance of early diagnosis. Unfortunately about 50% of cases are diagnosed in advanced stages (III and IV) [8,9] which implies a worse prognosis, increased costs, and a high mortality rate [10,11].
The predominant etiological factors for oral cancer are well established in the literature and include the use of tobacco and alcohol which act as carcinogenic substances responsible for constituting the so-called “field cancer” [12]. The carcinogenesis process is complex, being influenced by genetic and epigenetic alterations [13,14]. The fact is that the sooner these changes are detected, the earlier the disease will be discovered, contributing to a better prognosis for patients [15]. Conventional biopsy is considered the gold standard for the diagnosis of OSCC. However, it is inconvenient for large population screening and monitoring of patients due to its invasiveness, high cost, and need for trained personnel and equipment [16]. Thus, it is important to investigate biological molecules acting as biomarkers that may provide valuable diagnostic data on OSCC [14].
The search for biomarkers for chronic diseases and malignant neoplasms is a very active field of research worldwide [17]. Metabolomics employ state-of-the-art analytical techniques to recognize and study metabolic alterations in individuals who are undergoing some pathophysiological process or are undergoing pharmacological interventions and genetic modifications [18,19,20].
Biofluids—urine, blood, and saliva—are often used as clinical specimens of patients for metabolomic analysis [15]. Saliva is an oral fluid capable of reflecting the oral and systemic health conditions of individuals [21]. It is a complex and valuable composition that includes proteins, peptides, nucleic acids, enzymes, hormones, antibodies, electrolytes, antimicrobial constituents, growth factors, and other molecules associated with the phenotype and even diseases of individuals [22,23,24,25]. The main functions of saliva are related to digestion, swallowing, tasting, and lubrication of the oral mucosa. However, it is known that in addition to these functions saliva acts as a protective substance against pathogens and toxins due to its specific composition [26].
Previous studies have identified metabolomic biomarkers for OSCC [27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43]. Some of these probed salivary metabolites [27,28,29,30,31,32,33,34,35,36]. The fact that the metabolites profile can be influenced by the sample collection time [31,44], the food intake [28,30,31,45], the general oral health status [46], and even the oral microbiome [38,47] represents a challenge for standardization of salivary studies in order to avoid inconsistencies and reproducibility drawbacks. Ethnicity has also been shown to play an important role in the differentiation of metabolites since populations of distinct ethnicities presented distinct salivary metabolic profiles [31,37]. Most studies involving salivary metabolome in OSCC patients come from Asian individuals [38], showing the importance of studying different ethnic groups [48].
However, despite existing limitations, previous studies have shown consistent changes between OSCC and healthy patients [28], mainly due to the direct contact between saliva and the oral cancer lesion [30].
In the present work we investigated the salivary metabolites profile from a sample of OSCC patients from a South American population. The objectives were to identify possible salivary metabolomic biomarkers and also altered metabolic pathways.
2. Results
2.1. Demographic Data
The main clinical data of patients are summarized in Table 1. Data on sex and age did not show a statistically relevant difference between the groups (p < 0.05).
Table 1.
Demographic data of patients.
| Variable | OSCC 1
(n = 27) |
CONTROL (n = 41) |
p-Value * |
|---|---|---|---|
| Sex 2 | |||
| Female | 8 (29.6%) | 20 (49%) | 0.3326 |
| Male | 19 (70.4%) | 21 (51%) | 0.9131 |
| Age 3 | 57 ± 13.87 | 57.34 ± 11.66 | 0.9131 |
| (28–88) | (31–86) |
1 OSCC, oral squamous cell carcinoma group. 2 Sex was described with their respective means and (%) percentages. 3 Age described as mean ± standard deviation and in parentheses the minimum and maximum age of the patients. n represents the number of patients in each group. * p-values according to the Student’s t-test considering as significant p < 0.05.
Table 2 presents the TNM cancer staging system, smoking habits, and racial ethnicity data of the patients.
Table 2.
Cancer staging system, smoking habits, and racial ethnicity of patients.
| TNM 1 | OSCC (n = 27) |
Control (n = 41) |
|---|---|---|
| T (tumor) | ||
| T1 | 5 (19%) | |
| T2 | 7 (26%) | Not applicable |
| T3 | 6 (22%) | |
| T4 | 9 (33%) | |
| N (node) | ||
| N0 | 14 (52%) | |
| N1 | 4 (15%) | Not applicable |
| N2 | 8 (30%) | |
| N3 | 1 (4%) | |
| M (metastasis) | ||
| M0 | 27 (100%) | Not applicable |
| Stages | ||
| I | 4 (15%) | |
| II | 4 (15%) | Not applicable |
| III | 6 (22%) | |
| IV | 13 (48%) | |
| Smokers | 20 (74%) | 8 (20%) |
| Non smokers | 7 (26%) | 20 (49%) |
| Ex smokers | 0 (0%) | 13 (32%) |
| Racial ethnicity | ||
| Leucoderma | 24 (89%) | 32 (78%) |
| Melanoderm | 1 (4%) | 4 (10%) |
| Pheoderm | 2 (7%) | 4 (10%) |
| Xanthoderm | 0 (0%) | 1 (2%) |
1 TNM—classification of malignant tumors. The TNM system is used to describe the anatomical extension of the disease, where T—the extension of the primary tumor, N—the absence or presence and extension of metastasis in regional lymph nodes, M—the absence or presence of distant metastasis. All data are described with their respective n of each group and their respective (%) percentages.
2.2. Metabolomic Analysis
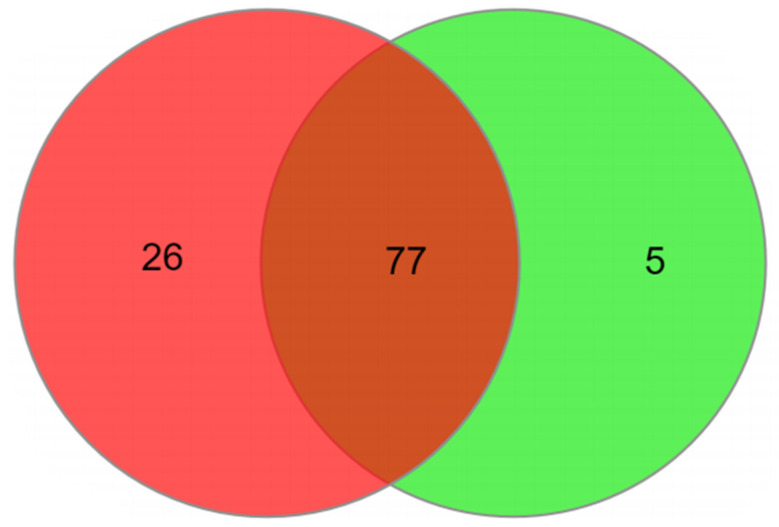
A total of 108 metabolites were identified as relevant for OSCC and control discrimination. All metabolites found in both groups studied were allocated on a Venn diagram to assess their distribution between groups (Figure 1). The analysis showed that the OSCC group has a higher number of specific metabolites (26 metabolites), while the control group had 5 specific metabolites. Seventy-seven metabolites were common for both groups. These metabolites are show on Table 3.
Figure 1.
Venn diagram for salivary metabolites probed on OSCC (red) and control (green) groups.
Table 3.
Exclusive and shared salivary metabolites for OSCC and control groups.
| OSCC | CONTROL | OSCC AND CONTROL |
|---|---|---|
| 2-Hydroxyglutaric acid | 2-Ketoadipic acid | 1,6-Anhydroglucose |
| 2-Ketoglutaric acid | Catechol | 1-Hexadecanol |
| 3-Hydroxypropionic acid | Lactose | 2-Aminoethanol |
| 4-Hydroxyphenyllactic acid | Leucine | 2-Deoxy-glucose |
| Cystamine | Urea | 2-Hydroxyisovaleric acid |
| Dihydroxyacetone phosphate | 3-Aminoglutaric acid | |
| Galacturonic acid | 3-Aminoisobutyric acid | |
| Gluconic acid | 3-Aminopropanoic acid | |
| Hippuric acid | 3-Hydroxyisovaleric acid | |
| Indol-3-acetic acid | 3-Phenyllactic acid | |
| Inosine | 4-Aminobutyric acid | |
| Isocitric acid | 5-Aminovaleric acid | |
| Lactitol | Acetoacetic acid | |
| Lyxose | Adenine | |
| Malic acid | Allose | |
| Maltose | Arabitol | |
| Methionine | Arachidonic acid | |
| O-Phospho-Serine | Arginine | |
| Pantothenic acid | Aspartic acid | |
| Protocatechuic acid | Batyl alcohol | |
| Ribose 5-phosphate | Cadaverine | |
| Sorbose | Caproic acid | |
| Spermidine | Citramalic acid | |
| Thymidine | Citric acid | |
| Uracil | Cysteine | |
| Ureidosuccinic acid | Dopamine | |
| Eicosapentaenoic acid | ||
| Elaidic acid | ||
| Fructose | ||
| Galactosamine | ||
| Galactose | ||
| Glucono-1,5-lactone | ||
| Glucosamine | ||
| Glucose | ||
| Glucuronic acid | ||
| Glutamic acid | ||
| Glycerol | ||
| Glycerol 2-phosphate |
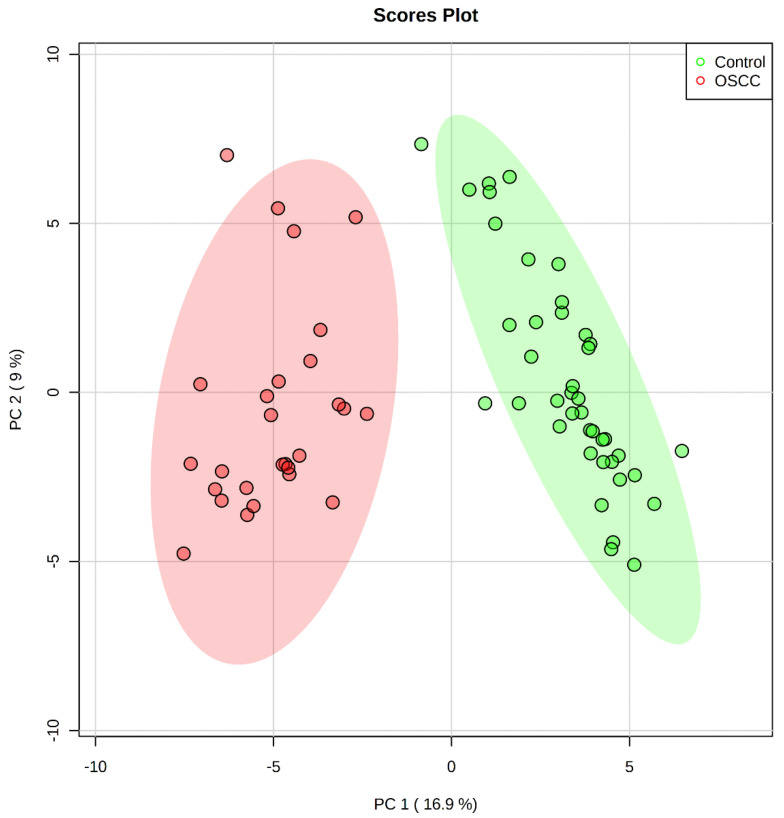
The dispersion score plot PC2 against PC1 (Figure 2) shows a clear separation among groups.
Figure 2.
PCA score plot OSCC (green) and control (red) groups. Ellipses represent the loci of maximum variance of data.
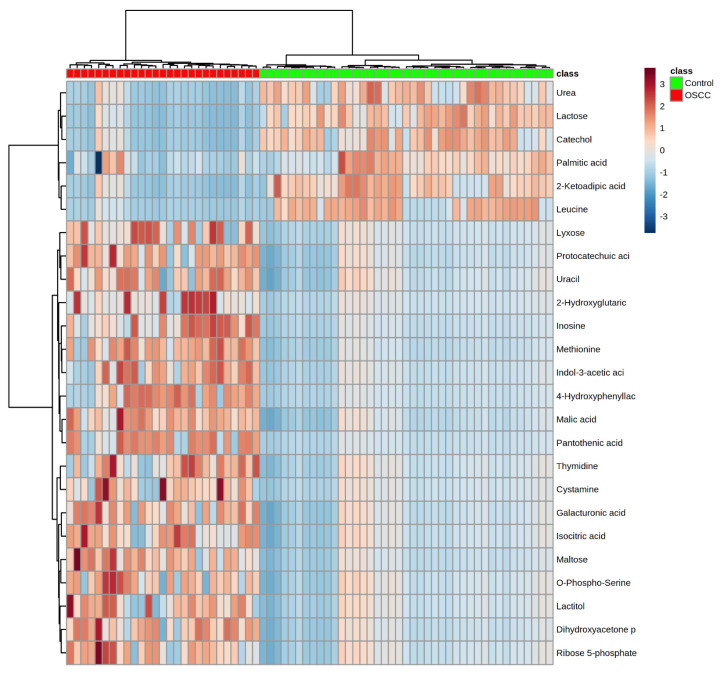
The heatmap showing the clustering of classes against metabolites is shown in Figure 3. Samples in OSCC and control classes clustered in two big groups with 100% discrimination. Metabolites urea, lactose, catechol, palmitic acid, 2-ketoadipic acid and leucine appeared underexpressed in the OSCC group. On the other hand, lyxose, protocatechuic acid, uracil, 2-hydroxyglutaric, inosine, methionine, indol-3-acetic acid, 4-hydroxyphenyllac, malic acid, pantothenic acid, isocitric acid, maltose, O-phospho-serine, lactitol, dihydroxyacetone and ribose 5-phosphate were overexpressed in patients with cancer.
Figure 3.
Heatmap using PCA data for OSCC and control classes.
Table 4 displays the up- and down-regulated metabolites presenting statistical relevance. Twenty metabolites were up-regulated (malic acid, methionine, maltose, protocatechuic acid, inosine, pantothenic acid, dihydroxyacetone phosphate, hydroxyphenylatic acid, galacturonic acid, indole-3-acetic acid, uracil, isocitric acid, ribose-5-phosphate, o-phospho serine, lactitol, gluconic acid, hippuric acid, 3-hydroxypropionic acid and spermidine) and 20 down-regulated (lactose, catechol, 2-ketoadipic acid, leucine, urea, maleic acid, palmitic acid, ornithine, margaric acid, sucrose, octadecanol, threitol, acetoacetic acid, methionine sulfone, phosphoric acid, elaidic acid, mannose, sorbitol, citric acid, 3-aminopropanoic acid) in OSCC samples.
Table 4.
Set of metabolites up- and down-regulated in OSCC samples according to PCA analyses.
| Metabolites | OSCC | Control | p-Value 1 | q-Value (FDR) 2 | FC | Volcano Plot 3 | ||
|---|---|---|---|---|---|---|---|---|
| Mean | Standard Deviation | Mean | Standard Deviation | |||||
| Lactose * | −1.090 | 0.492 | 0.718 | 0.673 | <0.0001 | 3.1755 × 10−16 | 0.015832 | Down |
| Malic acid ** | 0.917 | 0.622 | −0.604 | 0.444 | <0.0001 | 3.7012 × 10−16 | 40.712 | Up |
| Methionine ** | 1.088 | 0.939 | −0.717 | 0.367 | <0.0001 | 3.0633 × 10−15 | 311.66 | Up |
| Catechol * | −0.952 | 0.521 | 0.627 | 0.734 | <0.0001 | 7.1635 × 10−13 | 0.035587 | Down |
| 2-Keto adipic acid * | −0.925 | 0.522 | 0.609 | 0.768 | <0.0001 | 6.363 × 10−12 | 0.029706 | Down |
| Maltose ** | 0.889 | 0.959 | −0.586 | 0.407 | <0.0001 | 2.0868 × 10−11 | 325.18 | Up |
| Protocatechuic acid ** | 0.806 | 0.827 | −0.531 | 0.447 | <0.0001 | 2.7666 × 10−11 | 35.723 | Up |
| Leucine * | −1.177 | 0.394 | 0.775 | 1.173 | <0.0001 | 8.7168 × 10−11 | 8.2595 × 10−4 | Down |
| Inosine ** | 1.070 | 1.317 | −0.704 | 0.330 | <0.0001 | 9.7882 × 10−11 | 2873.0 | Up |
| Pantothenic acid ** | 1.153 | 1.459 | −0.759 | 0.304 | <0.0001 | 1.4172 × 10−10 | 4271.4 | Up |
| Urea * | −0.861 | 0.530 | 0.567 | 0.810 | <0.0001 | 1.687 × 10−10 | 0.037894 | Down |
| Dihydroxyacetone phosphate ** | 0.793 | 0.895 | −0.522 | 0.439 | <0.0001 | 1.687 × 10−10 | 45.791 | Up |
| 4-hydroxyphenylactic acid ** | 1.092 | 1.403 | −0.719 | 0.318 | <0.0001 | 2.1476 × 10−10 | 2173.8 | Up |
| Galacturonic acid ** | 0.725 | 0.831 | −0.477 | 0.467 | <0.0001 | 8.9307 × 10−10 | 19.383 | Up |
| Indole-3-acetic acid ** | 0.906 | 1.242 | −0.597 | 0.365 | <0.0001 | 3.0805 × 10−9 | 341.04 | Up |
| Uracil ** | 0.644 | 0.817 | −0.424 | 0.491 | <0.0001 | 3.04 × 10−8 | 10.819 | Up |
| Isocitric acid ** | 0.665 | 0.885 | −0.438 | 0.472 | <0.0001 | 3.6657 × 10−8 | 20.802 | Up |
| Ribose-5-phosphate ** | 0.647 | 0.969 | −0.469 | 0.461 | <0.0001 | 3.1666 × 10−7 | 41.912 | Up |
| O-Phospho-Serina ** | 0.609 | 0.945 | −0.401 | 0.474 | <0.0001 | 9.548 × 10−7 | 17.64 | Up |
| Lactitol ** | 0.630 | 1.061 | −0.415 | 0.446 | <0.0001 | 2.1547 × 10−6 | 41.538 | Up |
| Gluconic acid ** | 0.609 | 1.101 | −0.401 | 0.443 | <0.0001 | 7.7433 × 10−6 | 183.99 | Up |
| 2-Ketoglutaric acid ** | 0.515 | 0.836 | −0.339 | 0.512 | <0.0001 | 1.3092 × 10−5 | 6.7421 | Up |
| Hipuric acid ** | 0.518 | 0.888 | −0.341 | 0.506 | <0.0001 | 1.4925 × 10−5 | 7.3906 | Up |
| Maleic acid | −0.664 | 1.049 | 0.437 | 0.817 | <0.0001 | 3.294 × 10−5 | 0. 8093 | Down |
| Palmitic acid | −0.430 | 0.657 | 0.283 | 0.551 | <0.0001 | 3.3213 × 10−5 | 0.38165 | Down |
| 3-hydroxypropionic acid ** | 0.608 | 1.265 | −0.400 | 0.411 | 0.0002 | 4.4319 × 10−5 | 202.32 | Up |
| Spermidine ** | 0.481 | 0.887 | −0.317 | 0.514 | 0.0001 | 5.3374 × 10−5 | 10.562 | Up |
| Ornithine | −0.614 | 1.197 | 0.405 | 0.986 | 0.0003 | 0.0010593 | 0.33872 | Down |
| Margaric acid | −0.453 | 1.055 | 0.298 | 0.648 | <0.0001 | 0.0018846 | 0.28057 | Down |
| Sucrose | −0.487 | 1.005 | 0.321 | 0.928 | 0.0002 | 0.0039383 | 0.25406 | Down |
| Octadecanol | −0.310 | 0.666 | 0.204 | 0.628 | 0.0010 | 0.0064518 | 0.56165 | Down |
| Threitol | −0.465 | 1.148 | 0.307 | 0.847 | 0.0012 | 0.0069549 | 0.37775 | Down |
| Acetoacetic acid | −0.373 | 0.732 | 0.246 | 0.826 | 0.0024 | 0.0074047 | 0.25319 | Down |
| Methionine sulfone | −0.306 | 0.767 | 0.202 | 0.582 | 0.0001 | 0.0085698 | 1.123 | Down |
| Phosphoric acid | −0.374 | 0.806 | 0.246 | 0.968 | 0.0103 | 0.022159 | 0.12317 | Down |
| Elaidic acid | −0.254 | 0.578 | 0.167 | 0.722 | 0.0134 | 0.038044 | 0.4826 | Down |
| Mannose | −0.398 | 1.309 | 0.262 | 0.881 | 0.0324 | 0.042273 | 0.51969 | Down |
| Sorbitol | −0.361 | 0.890 | 0.238 | 1.048 | 0.0173 | 0.046325 | 0.11612 | Down |
| Citric acid | −0.416 | 1.200 | 0.274 | 1.111 | 0.0369 | 0.046725 | 0.11946 | Down |
| 3-Aminopropanoic acid | −0.324 | 0.895 | 0.213 | 0.907 | 0.0004 | 0.048905 | 0.39703 | Down |
1 p-value was calculated using the Wilcoxon-Mann-Whitney test (p-value < 0.05). 2 All metabolites shown in the table were statistically significant with a false discovery rate (FDR) of 5%. 3 Volcano plot shows up- and down-regulated metabolites in patients with OSCC. * Metabolites exclusively found in control patients. ** Metabolites exclusively found in OSCC patients.
2.3. Analysis of Altered Metabolic Pathways in the OSCC Group
The precedent analysis enables us to investigate the altered metabolic pathways in OSCC patients and find the role of each metabolite in these pathways.
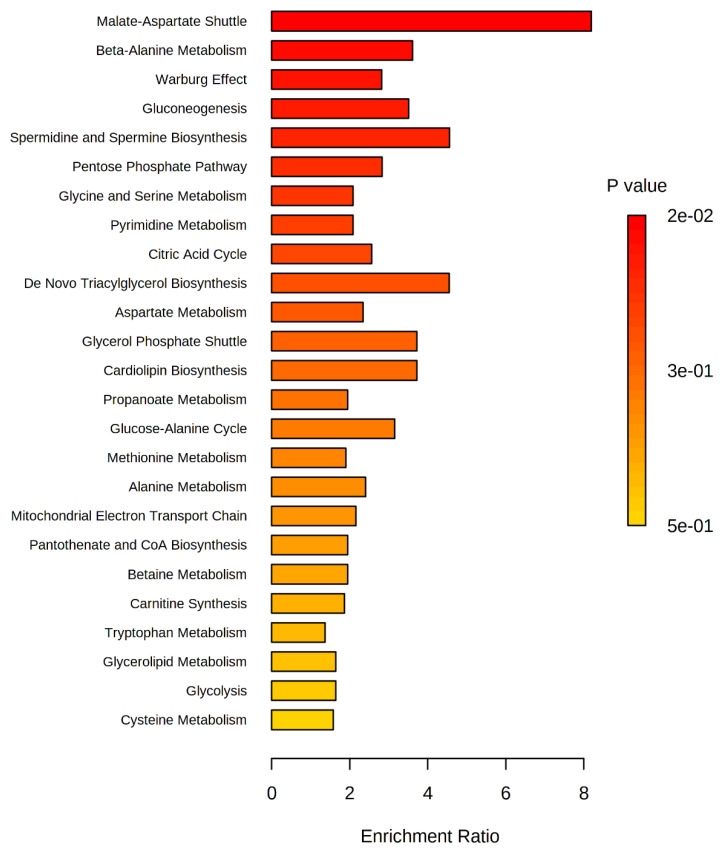
We analyzed 25 metabolites which were found exclusively in the OSCC group. Cystamine was absent from the databases of the chosen metabolomic compound and was excluded from further analysis. Thus, the role of 2-ketoglutaric acid, 2-hydroxyglutaric acid, 3-hydroxypropionic acid, 4-hydroxyphenylatic acid, galacturonic acid, gluconic acid, hippuric acid, indol-3-acetic acid, isocitric acid, malic acid, pantothenic acid, protocatechuic acid, ureidosuccinic acid, spermidine, dihydroxyacetone phosphate, inosine, lactitol, lyxose, maltose, methionine, O-phospho-serine, ribose 5-phosphate, sorbose, thymidine, and uracil in metabolic pathways was investigated. The pathway enrichment analysis is shown in Figure 4.
Figure 4.
Statistically significant (p < 0.05) for OSCC.
A total of 41 metabolic pathways were identified as present in OSCC salivary samples. However, only 25 presented statistical relevance. From these we can mention the malate-aspart (p = 0.0229), beta-alanine metabolism (p = 0.0467), and the Warburg effect (p = 0.048) signaling pathways.
2.4. Analysis of Possible Salivary Biomarkers for the OSCC Group
A receiver operating characteristic (ROC) curve was used to establish promising biomarkers for OSCC. The area under the ROC curve value (AUC) measures the performance of the biomarkers. Thus, an excellent biomarker has an AUC value of 1.0. Good biomarkers have AUC > 0.80. Using this criterion, we list in Table 5 the set of possible good salivary biomarkers for OSCC.
Table 5.
Area Under the Receiving—Operator Curve (AUC) for possible OSCC salivary biomarkers.
| Metabolite | AUC |
|---|---|
| Malic acid | 0.98103 |
| Lactose | 0.96387 |
| Catecol | 0.94670 |
| 2-ketoadipic acid | 0.94128 |
| Maltose | 0.93360 |
| Methionine | 0.92502 |
| Urea | 0.92502 |
| Leucine | 0.92322 |
| Inosine | 0.92186 |
| Protocatechuic acid | 0.91192 |
| Dihydroxyacetone phosphate | 0.89657 |
| Galacturonic acid | 0.88573 |
| Margaric acid | 0.86902 |
| Uracil | 0.86721 |
| Isocitric acid | 0.86585 |
| Ribose 5-phosphate | 0.84146 |
| O-Phospho-Serine | 0.82385 |
| Indole-3-acetic acid | 0.82204 |
| Palmitic acid | 0.82204 |
| 2-ketoglutaric acid | 0.81798 |
| Maleic acid | 0.81030 |
| Pantothenic acid | 0.80307 |
| Spermidine | 0.80217 |
3. Discussion
The relevance of the investigation of the salivary metabolome of OSCC relies on the identification of predominantly altered metabolic pathways which may lead to the discovery of possible biomarkers. This could improve the capacity of early diagnosis and, consequently, the quality of life of patients.
Reports of the salivary metabolome of patients with oral cancer described in the literature are presented in Table 6 and compared to our findings. The present study sought the main altered salivary metabolic pathways in OSCC patients and, additionally, the main metabolites that can be used as future salivary biomarkers for early diagnosis. To the best of our knowledge, this is the first research in this area focusing on Latin American patients.
Table 6.
Main salivary metabolomic studies of patients with OSCC.
| Possible Salivary Metabolic Biomarkers |
Studied Population | Notes | References |
|---|---|---|---|
| Malic acid ↑, Lactose ↓, Catecol ↓, 2-Keto adipic acid ↓, Maltose ↑, Methionine ↑, Urea ↓, Leucine ↓, Inosine ↑, Protocatechuic acid ↑ and others metabolites present in Table 3 | South American | We compared OSCC patients with healthy control | This study |
| Lactic acid ↑, phenylalanine ↓, valine ↓ | Not mentioned in the study | They compared OSCC patients with healthy control and oral leukoplasia | [36] |
| L-phenylalanine ↓, L-leucine ↓, Propionylcholine ↑, Acetylphenylalanine ↓, sphinganine ↓, phytosphingosine ↓, S-carboxymethyl-L-cysteine ↓, Choline ↑, betaine ↑, pipecolinic acid ↑, L-carnitine ↓ |
Chinese | They compared OSCC patients with healthy control | [32,33,34,35] |
| S-adenosylmethionine ↑, pipecolate ↑ | Not mentioned in the study | Two cases from the oral cancer group were oral melanoma | [28] |
| Ornithine ↓, o-hydroxybenzoate ↓, ribose-5-phosphate ↓ | Caucasian, African American, Hispanic, Asian | They compared OSCC patients and oral epithelial dysplasia patients with the healthy control | [29] |
| Alanine ↑, choline ↑, Leucine + isoleucine ↑, glutamic acid ↑, 120.0801 m/z ↑, phenylalanine ↑, alpha-aminobutyric acid ↑, serine ↑ |
Caucasian, Asian, African-American, Hispanic | They compared OSCC patients with healthy control | [31] |
| Indole-3-acetate ↑, ethanolamine phosphate ↑ | Not mentioned in the study | They compared OSCC patients with control patients with oral lichen planus | [27] |
| They studied conductive polymer spray ionization mass spectrometry (CPSI-MS) associated with machine learning (ML) as a viable tool for the diagnosis of OSCC | Chinese | They compared OSCC patients with oral lichen planus and oral leukoplakia controls | [30] |
↑ Up arrow indicates increased metabolites in OSCC patients. ↓ Down arrow indicates decreased metabolites in OSCC patients.
OSCC is mostly diagnosed at late stages, as also evidenced by our study, in which only 15% of patients were diagnosed with early-stage cancer (stage I), revealing that early diagnosis remains a challenge [49]. It is noteworthy that early diagnosis implies greater possibilities of successful treatment, less mutilation of the patient concerning the treatments carried out, decreased mortality rate, and reduced costs [50,51,52].
3.1. The Malate-Aspartate Shuttle Pathway
ATP consumption is higher for cancer cells compared to healthy ones. Thus, high glycolytic rates and mitochondrial oxidative phosphorylation are observed in tumor cells to deliver a greater amount of ATP in a short period of time [53]. Glycolysis is the metabolic pathway chosen by the body in the absence of oxygen, so less energy is generated, although the pathway normally occurs without the presence of oxygen [54]. If the body has plenty of oxygen, the glycolysis process will only be the beginning of the aerobic respiration cycle, in which the lactate generated by glycolysis will be consumed by the tricarboxylic acid cycle, also known as the Krebs cycle [55]. During the Krebs cycle, ATP is not produced directly. NADH and FADH2 are produced, which are essential for the production of ATP during oxidative phosphorylation. Oxidative phosphorylation is the preferred method of generating energy in the presence of oxygen, since the process generates 38 ATPs in contrast to anaerobic glycolysis that generates 2 ATPs [56].
The tricarboxylic acid cycle (Krebs cycle) and oxidative phosphorylation occur within the mitochondria. The malate-aspartate shuttle is responsible for transporting NADH from the cytoplasm to the mitochondrial matrix in the ATP production process [57,58]. In our study, the malate-aspartate launcher is one of the altered pathways and it is directly related to the energy production of tumor cells. The malic acid metabolite was abundant in most patients with OSCC. Malic acid is an intermediate product of the Krebs cycle explaining its higher concentration in patients with OSCC [59]. Based on our results, there is a high energy production in cancer cells that provides a favorable environment for disorderly growth.
3.2. Warburg Effect Pathway
Cancer metabolism has been studied for decades, mainly because cells exhibit rapid growth, proliferation, and survival [60]. These characteristics are inherent in an altered metabolism. In 1920, Otto Warburg observed a common characteristic in the metabolism of tumor cells. This characteristic consisted of increased glucose uptake with high lactate release, signaling that the rate of glycolysis in tumor cells is high even in the presence of oxygen and perfectly functioning mitochondria [61]. This process, known as the Warburg effect, was established as the form of energy generation in tumor cells [60,61,62,63]. Our findings indicate that the Warburg effect was also one of the metabolic pathways activated in OSCC patients.
Although the Warburg effect is established as the way tumor cells acquire energy, some studies have shown that several types of cancer can obtain energy through oxidative phosphorylation in conjunction with glycolysis. A study on breast cancer metabolism reported that 20% of energy production comes from the glycolytic pathway and 80% from oxidative phosphorylation [64]. Furthermore, in a study on hepatoma cells, it was found that the cells obtained energy mainly through the oxidative pathway, in contrast to a small portion via the glycolytic pathway [65].
Xu and Guppy conducted a study with different types of cancers (breast, ovary, lung, uterus, melanoma, various types of hepatomas, and many others) to measure the rate of ATP production through glycolytic and oxidative processes. Among the types of cancer studied, the average contribution of the glycolytic pathway to the production of ATP was 17%. The authors concluded that the vast majority of tumor cells can generate ATP via oxidative phosphorylation, but also through glycolysis, in addition to the fact that some tumors are glycolytic as a result of the hypoxic environment [66]. This corroborates our findings since both the Warburg effect and malate-aspartate pathways contributed to the maintenance of oxidative phosphorylation. Studies involving OSCC show that the Warburg effect is present in cell metabolism [28,30,34,36,37]. However, our study is the pioneer in demonstrating that oxidative phosphorylation is also present in OSCC.
3.3. Beta-Alanine Pathway
Beta-alanine is a non-essential amino acid responsible for reducing fatigue and increasing muscle strength [67,68]. Its metabolism was indirectly involved with uracil and spermidine metabolites [69], both up-regulated in OSCC. A previous study on oral cancer metabolome revealed the beta-alanine metabolite as a possible biomarker for oral cancer [31]. It is related to conditions of hypoxia, hypoglycemia, ischemia, and oxidative stress due to the presence of free radicals [70]. Therefore, this oxidative stress is responsible for damage to neural cells [71]. In this sense, a study on metabolites in breast cancer demonstrated that beta-alanine is one of the metabolites related to high glycolytic activity and associated with aggressiveness of tumor cells [72].
3.4. Biomarkers
We have found that some metabolites such as malic acid, maltose, methionine, and inosine were over-expressed in the saliva of patients with OSCC. Malic acid was reported above to be present in the malate-aspartate pathway. It has been reported that maltose is a possible natural substance with carcinogenic potential [73].
Ishikawa et al. identified the following metabolites over-expressed in the saliva of OSCC patients: hypoxanthine, guanine, guanosine, trimethylamine N-oxide, spermidine, pipecolate, methionine [28]. Of these, methionine and spermidine were also increased in our study, with an AUC of 0.92 and 0.80, respectively. Ishikawa et al. showed that the metabolism of purines was altered since the metabolites hypoxanthine, guanine, and guanosine are part of this pathway. However, in our study, only the inosine metabolite was altered in the purine metabolism pathway, being a potential biomarker for OSCC.
Ohshima et al., also found urea in lower concentrations in OSCC patients and in higher concentrations in control patients, indicating that the urea cycle might be altered in oral cancer. This was the first study to describe urea as a possible biomarker for oral cancer. It was carried out using capillary electrophoresis-mass spectrometry (CE-MS) to evaluate saliva from Japanese patients with OSCC [37]. Liang et al. observed changes in urea concentration when studying the metabolome of patients with gastric cancer [74]. Our study showed a significant change in urea in salivary samples from control patients, corroborating data from the Japanese survey [37]. According to Ohshima et al., OSCC patients may have difficulty eating due to pain and trouble in opening the mouth, which makes it difficult to ingest proteins and, consequently, form urea [37]. In addition, these patients may have a Helicobacter pylori infection, which produces urease, reducing the availability of urea [75].
Another metabolite present in salivary metabolome studies found in our study is leucine. Leucine was present in a study with 24 salivary metabolites that are candidate biomarkers in OSCC [31]. Wei et al. identified leucine, isoleucine, valine, and all intermediate branched-chain amino acids (BCAAs) underexpressed in the saliva of OSCC patients [36]. These metabolites are involved in the Krebs cycle. Thus, the activation of the glycolytic pathway (Warburg effect) decreases the entry of pyruvate into the TCA cycle. Therefore, the aforementioned metabolites are less necessary for the process due to the lack of energy supply via TCA [36].
In summary, we conclude that the whole nature of cellular energy production was altered in the OSCC group. This is the first salivary metabolomic study of a South American population with OSCC. Therefore, carrying out new studies covering larger populations may bring similar results and new insights so that these metabolites can be used as a non-invasive tool in oral cancer screening. Thus, salivary metabolic screening in populations exposed to risk factors, such as smoking and alcohol consumption, can reveal possible salivary biomarkers of oral cancer and improve the early diagnosis of carcinoma.
4. Materials and Methods
This study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Research Ethics Committee of the Institute of Science and Technology of São José dos Campos (ICT-UNESP), as part of the study entitled “Genetic study of the main risk factors in the prognosis of patients with oral squamous cell carcinoma”, protocol number 1.033.312/2015 PH/CEP. Patients were informed about the objectives, propositions, and conditions of this project, and those who agreed to participate signed the Free and Informed Consent Term (FICT). After acceptance, all patients underwent an extra and intraoral physical examination. Patients were divided into OSCC and control groups.
The OSCC group consisted of 27 patients diagnosed with OSCC. Inclusion criteria were patients over 18 years of age concomitant with the diagnosis of OSCC. The exclusion criterion considered patients diagnosed with cancer anywhere on the body that had already undergone some type of treatment, that is, surgery, radiotherapy, and chemotherapy. Cancer staging followed tumor-node-metastasis (TNM) classification according to the 8th edition of the American Joint Committee on Cancer (AJCC) Cancer Staging Manual [76]. The control group was composed of 41 patients from the oral medicine outpatient clinic of the Department of Biosciences and Oral Diagnosis of ICT-UNESP. The inclusion criterion was patients over 18 years of age, who wanted to participate in the research. The exclusion criterion was patients with some type of cancer during their lifetime.
4.1. Collection and Storage of Salivary Samples
Patients were instructed not to ingest pasty or hardened foods for 1 h before collection, as well as not to consume alcoholic beverages for at least 12 h before saliva collection. They could only swallow water and had to brush their teeth at least 2 h before the collection. Patients were instructed to expectorate 3 mL of saliva in the plastic tubes, which were then hermetically closed, immersed in ice, and transported within 1 h to the storage location. Salivary samples were stored in a freezer at −80 °C at the Laboratory of Microbiology and Immunology of the ICT-UNESP.
4.2. Preparation and Metabolomic Analysis of Salivary Samples
The methodology for analyzing the salivary metabolome was adapted from a previous study [77]. First, 300 µL of saliva samples were dried in a vacuum centrifuge (Labconco Centrivap Concentrator, Kansas City, MI, USA). Metabolites were extracted adding 300 µL of methanol containing methionine sulphonate as internal standard and stirred (LCG Vortex Mixer, Taiwan, China) for 2 min, and the supernatant was dried in a vacuum centrifuge. After extraction, derivatization was performed by adding 100 µL of a solution with proportions (1:1) of N-methyl-N-(trimethylsilyl) trifluoroacetamide and a solvent solution: acetonitrile/dichloromethane/cyclohexane (5:4:1) and 5% trimethylamine. The samples were stirred for 30 s and then kept in a thermal bath (Nova Instruments NI 1225, Piracicaba, Brazil) at 60 °C for 1 h. Next, the samples were centrifuged (Eppendorf MiniSpin, Hamburg, Germany) at 12,044× g for 2 min. The supernatant was analyzed via GC-MS. The data obtained were processed using GCMS solution and the metabolites identified using Smart metabolite database version 4.2. GC-MS analysis conditions:
MRM analysis method
running time: 67 min
injection temperature: 280 °C
interface temperature: 280 °C
ionization source temperature: 200 °C
heating rate: from 100 °C to 320 °C in a linear ramp of 4 °C/min, remaining at this temperature for 8 min.
4.3. Statistical Analysis
Clinical data were analyzed from the description of categorical variables with counts and proportions and quantitative variables with normal and asymmetric distribution and described as mean and deviation. For the consideration of normality, visual inspection of histograms or application of a normality test was used when appropriate. For all analyses, we considered the significance level of 5% (p < 0.05). Descriptive statistical analysis of clinical data was performed using GraphPad Prism 5.03 software (GraphPad Software, San Diego, CA, USA). For the salivary metabolomic analysis, principal component analysis (PCA) was performed, which allows the amount of information collected to be reduced. Its results select a subset of n variables capable of describing variability of data. The heatmap cluster was also used as a way to visualize the metabolites and hierarchical grouping of the compounds in each group. To demonstrate the significance of the metabolic data, we used the Wilcoxon-Mann-Whitney test. The p values to assess differences in metabolite concentrations between oral cancer and controls were corrected using the false discover rate (FDR) analysis of Benjamin-Hockberg [78] to consider several independent tests at a value of q < 0.05.
The volcano plot was used to visibly identify and illustrate the metabolites that are significant and most expressed in each study group. The volcano plot presents a diagram showing the set of metabolites in the salivary samples that would be down- and up-regulated. That is, it can indicate whether the compound is present with significance in the control group or in the OSCC group. The volcano plot combines the measure of statistical significance, in this case the q-value (FDR) with the measure of magnitude variation FC (fold change). In order to identify possible salivary biomarkers for OSCC, a ROC (receiver operating characteristic) curve was drawn for each metabolite. For this, the ROC curve uses the parameters of sensitivity and specificity. The area under the ROC curve, also called AUC, allows identification of whether a condition is present or not. That is, an AUC of 0.5 has no discriminating capacity, while an AUC of 1.0 shows an ideal discrimination [79]. For our study, we considered AUCs above 0.8 as the ideal cutoff point. In addition to the statistical tests mentioned above, the averages and standard deviations of the metabolites of each group were also performed. MetaboAnalyst 5.0 software (https://www.metaboanalyst.ca/ accessed on 18 December 2020) was used to analyze the metabolomic data.
All metabolites found were allocated on a Venn diagram to assess their distribution between groups. InteractiVenn (http://www.interactivenn.net/ accessed on 18 December 2020) was used for the analysis of the Venn diagram. The relative quantification of the metabolites for each group was performed from specific peak areas for each metabolite using the MRM analysis method. For the search for metabolites to be effective in the main databases, the Kyoto Encyclopedia of Genes and Genomes (KEGG pathway) and Small Molecule Pathway Database (SMPDB), the initial standardization of compound names was carried out on the MetaboAnalyst platform.
5. Conclusions
In summary, in our study, three important altered metabolic pathways were identified in OSCC for South American patients: the malate-aspartate shuttle, the beta-alanine metabolism pathway and the Warburg effect. These pathways are related to the cellular energy production in carcinogenesis, promoting a favorable environment for high energy consumption and cell survival. It was possible to statistically distinguish the salivary metabolites of control patients compared to patients with oral cancer. These metabolic changes may help in the discovery of salivary biomarkers of oral cancer and stimulate interest for new studies with larger populations to validate our results.
Acknowledgments
The authors are grateful for the technical assistance of Carlos Guedes. In addition to Bruna Fernandes do Carmo Carvalho and Keila Cristina Miranda, Master’s students, who helped us in the data analysis, and Flávio Francisco Godoy Peres, with the collection of samples. The authors would like to express their most sincere gratitude to all patients for their time and effort. In addition, we would like to thank the Universidade Estadual Paulista (Unesp), the University of São Paulo, the Pontifical Catholic University of Campinas Hospital and the Mário Gatti Municipal Hospital.
Author Contributions
J.D.A., M.A.M. and L.A.C.A. conceived and designed the experiments. C.M.B., M.B.N.P. and J.F.S.C. were responsible for collecting saliva from patients with oral cancer. M.d.S.A., N.d.S.R. and M.G.O.A. performed the experiments. M.d.S.A., N.d.S.R., M.G.O.A., G.L.J.T.N., H.d.S.M., M.D. and M.A.M. analyzed the data. M.d.S.A. wrote the manuscript. M.d.S.A., N.d.S.R., M.G.O.A., M.A.M., L.A.C.A., H.d.S.M. and J.D.A. revised the manuscript. All authors contributed to the article and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the São Paulo Research Foundation; (FAPESP) grant #2016/08633-0; the National Council of Technological and Scientific Development (CNPq): scholarship for MSA and grant # 307718/2019-0 for HSM; and Coordination for the Improvement of Higher Education Personnel (CAPES): scholarship for MSA and NR and São Paulo Research Foundation (FAPESP) grant #2021/09168-8.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Research Ethics Committee of the Institute of Science and Technology of São José dos Campos Campus (ICT-Unesp), as part of the study entitled “Genetic study of the main risk factors in the prognosis of patients with oral squamous cell carcinoma” (protocol number 1.033.312/2015 PH/CEP).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to the privacy of the patients assisted in the research.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.MacCarthy D., Flint S.R., Healy C., Stassen L.F. Oral and neck examination for early detection of oral cancer–A practical guide. J. Ir. Dent. Assoc. 2011;57:195–199. [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Rivera C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015;8:11884–11894. [PMC free article] [PubMed] [Google Scholar]
- 4.Neville B.W., Day T.A. Oral cancer and precancerous lesions. CA Cancer J. Clin. 2002;52:195–215. doi: 10.3322/canjclin.52.4.195. [DOI] [PubMed] [Google Scholar]
- 5.Chen J.K., Katz R.V., Krutchkoff D.J. Intraoral squamous cell carcinoma. Epidemiologic patterns in Connecticut from 1935 to 1985. Cancer. 1990;66:1288–1296. doi: 10.1002/1097-0142(19900915)66:6<1288::AID-CNCR2820660632>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 6.Schantz S.P., Yu G.P. Head and neck cancer incidence trends in young Americans, 1973-1997, with a special analysis for tongue cancer. Arch. Otolaryngol. Head Neck Surg. 2002;128:268–274. doi: 10.1001/archotol.128.3.268. [DOI] [PubMed] [Google Scholar]
- 7.Dumache R., Rogobete A.F., Andreescu N., Puiu M. Genetic and Epigenetic Biomarkers of Molecular Alterations in Oral Carcinogenesis. Clin. Lab. 2015;61:1373–1381. doi: 10.7754/Clin.Lab.2015.150327. [DOI] [PubMed] [Google Scholar]
- 8.Gomez I., Seoane J., Varela-Centelles P., Diz P., Takkouche B. Is diagnostic delay related to advanced-stage oral cancer? A meta-analysis. Eur. J. Oral Sci. 2009;117:541–546. doi: 10.1111/j.1600-0722.2009.00672.x. [DOI] [PubMed] [Google Scholar]
- 9.Scott S.E., Grunfeld E.A., McGurk M. The idiosyncratic relationship between diagnostic delay and stage of oral squamous cell carcinoma. Oral Oncol. 2005;41:396–403. doi: 10.1016/j.oraloncology.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Fontes P.C., Correa G.H., Issa J.S., Brandao A.A., Almeida J.D. Comparison of exfoliative pap stain and AgNOR counts of the tongue in smokers and nonsmokers. Head Neck Pathol. 2008;2:157–162. doi: 10.1007/s12105-008-0059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guneri P., Epstein J.B. Late stage diagnosis of oral cancer: Components and possible solutions. Oral Oncol. 2014;50:1131–1136. doi: 10.1016/j.oraloncology.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Sahingur S.E., Yeudall W.A. Chemokine function in periodontal disease and oral cavity cancer. Front. Immunol. 2015;6:214. doi: 10.3389/fimmu.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Califano J., van der Riet P., Westra W., Nawroz H., Clayman G., Piantadosi S., Corio R., Lee D., Greenberg B., Koch W., et al. Genetic progression model for head and neck cancer: Implications for field cancerization. Cancer Res. 1996;56:2488–2492. doi: 10.1016/S0194-5998(96)80631-0. [DOI] [PubMed] [Google Scholar]
- 14.Santosh A.B., Jones T., Harvey J. A review on oral cancer biomarkers: Understanding the past and learning from the present. J. Cancer Res. Ther. 2016;12:486–492. doi: 10.4103/0973-1482.176414. [DOI] [PubMed] [Google Scholar]
- 15.Mikkonen J.J., Singh S.P., Herrala M., Lappalainen R., Myllymaa S., Kullaa A.M. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J. Periodontal Res. 2016;51:431–437. doi: 10.1111/jre.12327. [DOI] [PubMed] [Google Scholar]
- 16.Singh P., Verma J.K., Singh J.K. Validation of Salivary Markers, IL-1beta, IL-8 and Lgals3bp for Detection of Oral Squamous Cell Carcinoma in an Indian Population. Sci. Rep. 2020;10:7365. doi: 10.1038/s41598-020-64494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra A., Verma M. Cancer biomarkers: Are we ready for the prime time? Cancers. 2010;2:190–208. doi: 10.3390/cancers2010190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clayton T.A., Lindon J.C., Cloarec O., Antti H., Charuel C., Hanton G., Provost J.P., Le Net J.L., Baker D., Walley R.J., et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 19.Fiehn O. Metabolomics–The link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- 20.Holmes E., Wilson I.D., Nicholson J.K. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 21.Chicharro J.L., Lucia A., Perez M., Vaquero A.F., Urena R. Saliva composition and exercise. Sports Med. 1998;26:17–27. doi: 10.2165/00007256-199826010-00002. [DOI] [PubMed] [Google Scholar]
- 22.Bigler L.R., Streckfus C.F., Dubinsky W.P. Salivary biomarkers for the detection of malignant tumors that are remote from the oral cavity. Clin. Lab. Med. 2009;29:71–85. doi: 10.1016/j.cll.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Samaranayake L. Saliva as a diagnostic fluid. Int. Dent. J. 2007;57:295–299. doi: 10.1111/j.1875-595X.2007.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 24.Wong D.T. Towards a simple, saliva-based test for the detection of oral cancer ‘oral fluid (saliva), which is the mirror of the body, is a perfect medium to be explored for health and disease surveillance’. Expert Rev. Mol. Diagn. 2006;6:267–272. doi: 10.1586/14737159.6.3.267. [DOI] [PubMed] [Google Scholar]
- 25.Wong D.T. Salivary diagnostics for oral cancer. J. Calif. Dent. Assoc. 2006;34:303–308. [PubMed] [Google Scholar]
- 26.Humphrey S.P., Williamson R.T. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 2001;85:162–169. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa S., Sugimoto M., Kitabatake K., Sugano A., Nakamura M., Kaneko M., Ota S., Hiwatari K., Enomoto A., Soga T., et al. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci. Rep. 2016;6:31520. doi: 10.1038/srep31520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa S., Sugimoto M., Edamatsu K., Sugano A., Kitabatake K., Iino M. Discrimination of oral squamous cell carcinoma from oral lichen planus by salivary metabolomics. Oral Dis. 2020;26:35–42. doi: 10.1111/odi.13209. [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa S., Wong D.T.W., Sugimoto M., Gleber-Netto F.O., Li F., Tu M., Zhang Y., Akin D., Iino M. Identification of salivary metabolites for oral squamous cell carcinoma and oral epithelial dysplasia screening from persistent suspicious oral mucosal lesions. Clin. Oral Investig. 2019;23:3557–3563. doi: 10.1007/s00784-018-2777-3. [DOI] [PubMed] [Google Scholar]
- 30.Song X., Yang X., Narayanan R., Shankar V., Ethiraj S., Wang X., Duan N., Ni Y.H., Hu Q., Zare R.N. Oral squamous cell carcinoma diagnosed from saliva metabolic profiling. Proc. Natl. Acad. Sci. USA. 2020;117:16167–16173. doi: 10.1073/pnas.2001395117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugimoto M., Wong D.T., Hirayama A., Soga T., Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Christison T.T., Misuno K., Lopez L., Huhmer A.F., Huang Y., Hu S. Metabolomic profiling of anionic metabolites in head and neck cancer cells by capillary ion chromatography with Orbitrap mass spectrometry. Anal. Chem. 2014;86:5116–5124. doi: 10.1021/ac500951v. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q., Gao P., Cheng F., Wang X., Duan Y. Measurement of salivary metabolite biomarkers for early monitoring of oral cancer with ultra performance liquid chromatography-mass spectrometry. Talanta. 2014;119:299–305. doi: 10.1016/j.talanta.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q., Gao P., Wang X., Duan Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci. Rep. 2014;4:6802. doi: 10.1038/srep06802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q., Gao P., Wang X., Duan Y. Investigation and identification of potential biomarkers in human saliva for the early diagnosis of oral squamous cell carcinoma. Clin. Chim. Acta. 2014;427:79–85. doi: 10.1016/j.cca.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Wei J., Xie G., Zhou Z., Shi P., Qiu Y., Zheng X., Chen T., Su M., Zhao A., Jia W. Salivary metabolite signatures of oral cancer and leukoplakia. Int. J. Cancer. 2011;129:2207–2217. doi: 10.1002/ijc.25881. [DOI] [PubMed] [Google Scholar]
- 37.Ohshima M., Sugahara K., Kasahara K., Katakura A. Metabolomic analysis of the saliva of Japanese patients with oral squamous cell carcinoma. Oncol. Rep. 2017;37:2727–2734. doi: 10.3892/or.2017.5561. [DOI] [PubMed] [Google Scholar]
- 38.Lohavanichbutr P., Zhang Y., Wang P., Gu H., Nagana Gowda G.A., Djukovic D., Buas M.F., Raftery D., Chen C. Salivary metabolite profiling distinguishes patients with oral cavity squamous cell carcinoma from normal controls. PLoS ONE. 2018;13:e0204249. doi: 10.1371/journal.pone.0204249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sridharan G., Ramani P., Patankar S. Serum metabolomics in oral leukoplakia and oral squamous cell carcinoma. J. Cancer Res. Ther. 2017;13:556–561. doi: 10.4103/jcrt.JCRT_1233_16. [DOI] [PubMed] [Google Scholar]
- 40.Awasthi N. Role of salivary biomarkers in early detection of oral squamous cell carcinoma. Indian J. Pathol. Microbiol. 2017;60:464–468. doi: 10.4103/IJPM.IJPM_140_16. [DOI] [PubMed] [Google Scholar]
- 41.Bel’skaya L.V., Sarf E.A., Solomatin D.V., Kosenok V.K. Diagnostic and Prognostic Value of Salivary Biochemical Markers in Oral Squamous Cell Carcinoma. Diagnostics. 2020;10:818. doi: 10.3390/diagnostics10100818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee P.K., Funchain P., Retuerto M., Jurevic R.J., Fowler N., Burkey B., Eng C., Ghannoum M.A. Metabolomic analysis identifies differentially produced oral metabolites, including the oncometabolite 2-hydroxyglutarate, in patients with head and neck squamous cell carcinoma. BBA Clin. 2017;7:8–15. doi: 10.1016/j.bbacli.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen T.T.H., Sodnom-Ish B., Choi S.W., Jung H.I., Cho J., Hwang I., Kim S.M. Salivary biomarkers in oral squamous cell carcinoma. J. Korean Assoc. Oral Maxillofac. Surg. 2020;46:301–312. doi: 10.5125/jkaoms.2020.46.5.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeda I., Stretch C., Barnaby P., Bhatnager K., Rankin K., Fu H., Weljie A., Jha N., Slupsky C. Understanding the human salivary metabolome. NMR Biomed. 2009;22:577–584. doi: 10.1002/nbm.1369. [DOI] [PubMed] [Google Scholar]
- 45.Kirwan J.A., Brennan L., Broadhurst D., Fiehn O., Cascante M., Dunn W.B., Schmidt M.A., Velagapudi V. Preanalytical Processing and Biobanking Procedures of Biological Samples for Metabolomics Research: A White Paper, Community Perspective (for Precision Medicine and Pharmacometabolomics Task Group—The Metabolomics Society Initiative) Clin. Chem. 2018;64:1158–1182. doi: 10.1373/clinchem.2018.287045. [DOI] [PubMed] [Google Scholar]
- 46.Liebsch C., Pitchika V., Pink C., Samietz S., Kastenmuller G., Artati A., Suhre K., Adamski J., Nauck M., Volzke H., et al. The Saliva Metabolome in Association to Oral Health Status. J. Dent. Res. 2019;98:642–651. doi: 10.1177/0022034519842853. [DOI] [PubMed] [Google Scholar]
- 47.Gardner A., Parkes H.G., So P.W., Carpenter G.H. Determining bacterial and host contributions to the human salivary metabolome. J. Oral Microbiol. 2019;11:1617014. doi: 10.1080/20002297.2019.1617014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vitorio J.G., Duarte-Andrade F.F., Dos Santos Fontes Pereira T., Fonseca F.P., Amorim L.S.D., Martins-Chaves R.R., Gomes C.C., Canuto G.A.B., Gomez R.S. Metabolic landscape of oral squamous cell carcinoma. Metabolomics. 2020;16:105. doi: 10.1007/s11306-020-01727-6. [DOI] [PubMed] [Google Scholar]
- 49.Abati S., Bramati C., Bondi S., Lissoni A., Trimarchi M. Oral Cancer and Precancer: A Narrative Review on the Relevance of Early Diagnosis. Int. J. Environ. Res. Public Health. 2020;17:9160. doi: 10.3390/ijerph17249160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumann E., Koller M., Wiltfang J., Wenz H.J., Moller B., Hertrampf K. Challenges of early detection of oral cancer: Raising awareness as a first step to successful campaigning. Health Educ. Res. 2016;31:136–145. doi: 10.1093/her/cyv099. [DOI] [PubMed] [Google Scholar]
- 51.Gao W., Guo C.B. Factors related to delay in diagnosis of oral squamous cell carcinoma. J. Oral Maxillofac. Surg. 2009;67:1015–1020. doi: 10.1016/j.joms.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 52.Thomas A., Manchella S., Koo K., Tiong A., Nastri A., Wiesenfeld D. The impact of delayed diagnosis on the outcomes of oral cancer patients: A retrospective cohort study. Int. J. Oral Maxillofac. Surg. 2021;50:585–590. doi: 10.1016/j.ijom.2020.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Alam M.M., Lal S., FitzGerald K.E., Zhang L. A holistic view of cancer bioenergetics: Mitochondrial function and respiration play fundamental roles in the development and progression of diverse tumors. Clin. Transl. Med. 2016;5:3. doi: 10.1186/s40169-016-0082-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernie A.R., Carrari F., Sweetlove L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant. Biol. 2004;7:254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 55.Krebs H.A. The citric acid cycle and the Szent-Gyorgyi cycle in pigeon breast muscle. Biochem. J. 1940;34:775–779. doi: 10.1042/bj0340775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boyer P.D., Cross R.L., Momsen W. A new concept for energy coupling in oxidative phosphorylation based on a molecular explanation of the oxygen exchange reactions. Proc. Natl. Acad. Sci. USA. 1973;70:2837–2839. doi: 10.1073/pnas.70.10.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borst P. The aerobic oxidation of reduced diphosphopyridine nucleotide formed by glycolysis in Ehrlich ascites-tumour cells. Biochim. Biophys. Acta. 1962;57:270–282. doi: 10.1016/0006-3002(62)91120-4. [DOI] [PubMed] [Google Scholar]
- 58.Borst P. The malate-aspartate shuttle (Borst cycle): How it started and developed into a major metabolic pathway. IUBMB Life. 2020;72:2241–2259. doi: 10.1002/iub.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J., Li J., Shin H.D., Du G., Chen J., Liu L. Biological production of L-malate: Recent advances and future prospects. World J. Microbiol. Biotechnol. 2017;34:6. doi: 10.1007/s11274-017-2349-8. [DOI] [PubMed] [Google Scholar]
- 60.Liberti M.V., Locasale J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 62.Potter M., Newport E., Morten K.J. The Warburg effect: 80 years on. Biochem. Soc. Trans. 2016;44:1499–1505. doi: 10.1042/BST20160094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaupel P., Schmidberger H., Mayer A. The Warburg effect: Essential part of metabolic reprogramming and central contributor to cancer progression. Int. J. Radiat. Biol. 2019;95:912–919. doi: 10.1080/09553002.2019.1589653. [DOI] [PubMed] [Google Scholar]
- 64.Guppy M., Leedman P., Zu X., Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem. J. 2002;364:309–315. doi: 10.1042/bj3640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodriguez-Enriquez S., Torres-Marquez M.E., Moreno-Sanchez R. Substrate oxidation and ATP supply in AS-30D hepatoma cells. Arch. Biochem. Biophys. 2000;375:21–30. doi: 10.1006/abbi.1999.1582. [DOI] [PubMed] [Google Scholar]
- 66.Zu X.L., Guppy M. Cancer metabolism: Facts, fantasy, and fiction. Biochem. Biophys. Res. Commun. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 67.Caruso J., Charles J., Unruh K., Giebel R., Learmonth L., Potter W. Ergogenic effects of beta-alanine and carnosine: Proposed future research to quantify their efficacy. Nutrients. 2012;4:585–601. doi: 10.3390/nu4070585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jordan T., Lukaszuk J., Misic M., Umoren J. Effect of beta-alanine supplementation on the onset of blood lactate accumulation (OBLA) during treadmill running: Pre/post 2 treatment experimental design. J. Int. Soc. Sports Nutr. 2010;7:20. doi: 10.1186/1550-2783-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parthasarathy A., Savka M.A., Hudson A.O. The Synthesis and Role of beta-Alanine in Plants. Front. Plant Sci. 2019;10:921. doi: 10.3389/fpls.2019.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shetewy A., Shimada-Takaura K., Warner D., Jong C.J., Mehdi A.B., Alexeyev M., Takahashi K., Schaffer S.W. Mitochondrial defects associated with beta-alanine toxicity: Relevance to hyper-beta-alaninemia. Mol. Cell. Biochem. 2016;416:11–22. doi: 10.1007/s11010-016-2688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsuruoka M., Hara J., Hirayama A., Sugimoto M., Soga T., Shankle W.R., Tomita M. Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013;34:2865–2872. doi: 10.1002/elps.201300019. [DOI] [PubMed] [Google Scholar]
- 72.Hutschenreuther A., Birkenmeier G., Bigl M., Krohn K., Birkemeyer C. Glycerophosphoglycerol, Beta-alanine, and pantothenic Acid as metabolic companions of glycolytic activity and cell migration in breast cancer cell lines. Metabolites. 2013;3:1084–1101. doi: 10.3390/metabo3041084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hickey R.J., Clelland R.C., Bowers E.J. Essential hormones as carcinogenic hazards. J. Occup. Med. 1979;21:265–268. [PubMed] [Google Scholar]
- 74.Liang Q., Wang C., Li B. Metabolomic Analysis Using Liquid Chromatography/Mass Spectrometry for Gastric Cancer. Appl. Biochem. Biotechnol. 2015;176:2170–2184. doi: 10.1007/s12010-015-1706-z. [DOI] [PubMed] [Google Scholar]
- 75.Akada J.K., Shirai M., Takeuchi H., Tsuda M., Nakazawa T. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 2000;36:1071–1084. doi: 10.1046/j.1365-2958.2000.01918.x. [DOI] [PubMed] [Google Scholar]
- 76.Huang S.H., O’Sullivan B. Overview of the 8th Edition TNM Classification for Head and Neck Cancer. Curr. Treat. Options Oncol. 2017;18:40. doi: 10.1007/s11864-017-0484-y. [DOI] [PubMed] [Google Scholar]
- 77.Barnes V.M., Ciancio S.G., Shibly O., Xu T., Devizio W., Trivedi H.M., Guo L., Jonsson T.J. Metabolomics reveals elevated macromolecular degradation in periodontal disease. J. Dent. Res. 2011;90:1293–1297. doi: 10.1177/0022034511416240. [DOI] [PubMed] [Google Scholar]
- 78.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 79.Ma H., Bandos A.I., Gur D. On the use of partial area under the ROC curve for comparison of two diagnostic tests. Biom. J. 2015;57:304–320. doi: 10.1002/bimj.201400023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to the privacy of the patients assisted in the research.