Abstract
Irisin is a clinically significant protein playing a valuable role in regulating various diseases. Irisin attenuates synaptic and memory dysfunction, highlighting its importance in Alzheimer’s disease. On the other hand, Microtubule Affinity Regulating Kinase 4 (MARK4) is associated with various cancer types, uncontrolled neuronal migrations, and disrupted microtubule dynamics. In addition, MARK4 has been explored as a potential drug target for cancer and Alzheimer’s disease therapy. Here, we studied the binding and subsequent inhibition of MARK4 by irisin. Irisin binds to MARK4 with an admirable affinity (K = 0.8 × 107 M−1), subsequently inhibiting its activity (IC50 = 2.71 µm). In vitro studies were further validated by docking and simulations. Molecular docking revealed several hydrogen bonds between irisin and MARK4, including critical residues, Lys38, Val40, and Ser134. Furthermore, the molecular dynamic simulation showed that the binding of irisin resulted in enhanced stability of MARK4. This study provides a rationale to use irisin as a therapeutic agent to treat MARK4-associated diseases.
Keywords: protein-protein interaction, kinase inhibitors, cancer therapy, neurodegenerative diseases, molecular dynamics simulation, microtubule dynamics
1. Introduction
Irisin is a recently identified myokine, a cleaved fibronectin type III domain containing 5 (FNDC5) and secreted into blood circulation upon physical exercise. It was found to stimulate adipose tissue browning and regulate thermogenesis [1,2]. Irisin is widely expressed in skeletal muscles, heart, brain, liver, and salivary glands [1,3]. Recent studies have reported various other beneficial effects of irisin, including the regulation of depression [4], insulin sensitivity [5], glucose homeostasis [6], osteoblast proliferation [7], and neurogenesis [8], highlighting the importance of irisin. Instead of these irisin effects, it can be a potent drug target for many diseases ranging from obesity to dementia. According to a recently published piece of literature, irisin stimulates brain-derived neurotrophic factor (BDNF) in the brain’s hippocampal region and plays an important role in brain function [9].
Moreover, irisin was suggested to be involved in the prevention of Alzheimer’s disease (AD). Irisin modulates various AD risk factors and shows protective action against the abnormal expression of synapse-related genes, which can attenuate the synaptic and memory dysfunction in AD models [10]. All these studies imply that irisin is a potential therapeutic agent for neurodegenerative disorders. Hence, pharmacologically, boosting brain irisin levels can be a therapeutic strategy to protect/repair synapse function, thereby preventing cognitive decline in AD.
One of the pathological hallmarks of AD is the accumulation of the microtubule-associated protein tau and hyper-phosphorylation of tau in the brain that is intricately involved in the progression of AD. Overexpression of different kinases is related to multiple cancers and neurodegenerative diseases. MAP (Mitogen-activated protein kinase)/Microtubule Affinity Regulating Kinase 4 (MARK4) is a Ser-Thr kinase that plays a key role in early tau phosphorylation, leading to Alzheimer’s disease [11]. MARK4 has an important role in AD progression. It phosphorylates the tau at the Ser262 site, resulting in the detachment of tau from microtubules, thus allowing it to be phosphorylated by other kinases [12]. Numerous reports have suggested MARK4 as one of the best targets for AD therapy and other neurodegenerative diseases [13,14]. Due to the various important roles of MARK4 in neurodegenerative diseases, MARK4 is considered an attractive drug target; therefore, its inhibition can be a therapeutic strategy for treating neurodegenerative disorders [15,16,17].
For pharmacological profiling, it is important to investigate the interaction mechanisms of therapeutic proteins or drugs with target proteins [18,19,20]. The pharmacokinetic properties of therapeutic proteins or drugs depend upon their interaction with target proteins or tissues [21,22,23,24]. It is essential to study the protein-protein or protein-drug interaction to compete with pharmaceutical industry advancements [25,26,27,28]. Protein-based therapeutics are very successful and have high clinical significance [29,30]. More than 100 genuine and modified therapeutic proteins are approved for clinical use in the European Union and the USA [31]. To date, a vast number of drugs have been designed against the targeted proteins [32,33,34]. Naturally derived and chemically synthesize inhibitors are used effectively against the targeted proteins [35,36,37,38,39,40,41,42,43]. Irisin has a vital role in various metabolic and neurological problems, indicating its high therapeutic value, so using irisin as a therapeutic agent can be a good strategy for pharmaceutical industries.
For the first time, this study gives a rationale for targeting the inhibition of MARK4 with irisin, a protein–protein interaction. This approach might provide us a platform to use irisin as a therapeutic agent in targeting different kinases that are overexpressed in various diseased conditions [44]. In this study, we have expressed and purified irisin and MARK4 using Ni-NTA chromatography. Fluorescence-based binding and isothermal titration calorimetry (ITC) ascertained the actual binding of irisin with MARK4. Molecular docking and simulation were performed to validate the in vitro observations, and these suggested that the binding of irisin increases the stability of MARK4.
2. Materials and Methods
2.1. Materials
The expression construct of irisin (pET15b-His-3C-Irisin) was purchased from Addgene (122612). Unless stated, all the chemicals were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). Other analytical grade reagents were procured from local suppliers.
2.2. Expression and Purification of Irisin and MARK4
Human irisin and MARK4 proteins were expressed and purified as per our published protocol, respectively [3,45]. Kinase assay, SDS-PAGE investigated the quality of the purified recombinant MARK4. The MARK4 protein was further confirmed with Western blot using peptide-specific primary antibodies [46].
2.3. Fluorescence Measurements
For studying the binding affinity of irisin with MARK4, the fluorescence emission spectrum was recorded using the Jasco spectrofluorometer (FP-6200) and analyzed as per previous studies [3,47]. Fluorescence quenching data were fitted into a modified Stern−Volmer equation to estimate the different binding parameters of MARK4–irisin interaction.
2.4. Isothermal Titration Calorimetry
ITC measurements were carried out on a VP-ITC microcalorimeter from MicroCal, Inc (MicroCal, Amherst, MA, USA) as previous reports [30,47]. The first injection was a false one of 2 s with other injections set at 10 s. Before loading, degassing was carried out to remove the bubble hindrance. The sample cell was filled with 15 µM MARK4, and the syringe was loaded with 200 µM irisin. The final figure was obtained with the attached software (MicroCal 8.0), and the data was plotted as a two-site model.
2.5. Kinase Assay
An ATPase assay was conducted to check the inhibitory effect of varying irisin concentrations on MARK4 as previously published protocols [48,49].
2.6. Protein Structure Modeling and Preparation
The high-resolution crystal structures of human MARK4 and irisin were retrieved from the Protein Data Bank [50]. We used atomic coordinates of the PDB code of 5ES1 (https://www.rcsb.org/structure/5es1, accessed on 10 September 2021) for MARK4 (2.80 Å resolution) for molecular docking residue numbers 51–366. The structure was further refined, and all missing residues were added. We used the structure of MARK4 since it mainly consists of kinase and UBA domains. We aimed to see the kinase inhibitory potential of irisin; therefore, our focus was on the kinase domain. We have cloned, expressed, and purified the recombinant MARK4 containing both kinase and UBA domains as described [51,52].
The atomic coordinates of the irisin structure were downloaded from the PDB (https://www.rcsb.org/structure/4LSD, accessed on 10 September 2021). The 2.28 Å resolution structure of irisin contains residues 30–126. Irisin is produced by proteolytic processing of a transmembrane receptor, fibronectin domain-containing protein 5 (FNDC5), which is comprised of an N-terminal 29-residue signal sequence, followed by the irisin domain or fibronectin III (FN III)2 domain, a transmembrane domain, and a 39-residue cytoplasmic segment [1]. Thus, we cloned the mature peptide of irisin consisting of amino acid residues 30–140 only for experimental studies.
The structure of MARK4 was pre-processed, minimized, and refined using the Protein Preparation Wizard implemented in the Schrödinger suite [53]. This correlated by removing crystallographic waters and adding missing side chain or hydrogens atoms, with an accurate charge and protonation state assigned to the enzyme structure persistent to neutral pH (7.0) while recognizing the appropriate ionization states for the basic as well as acidic amino acid residues. The structure was then exposed to energy minimization using the OPLS-2005 force-field [54] with an RMSD cut-off value of 0.30 Å to reduce the steric clashes between the residues due to the addition of hydrogen atoms. A standard proceedure of docking was implicated as described [55,56,57].
2.7. Protein–Protein Docking
Molecular docking was performed using the PIPER program implemented in the BioLuminae module of Schrodinger’s protein–protein docking workflow to identify the best binding scores relevant for the MARK4 enzyme [58]. It is an effective program that removes false-positive poses and executes a comprehensive search with a fast Fourier transform (FFT) approach. Using 100 conformations of input structures, the top 20 clusters were selected with cluster radius 10 Å, which then centralized, and the docking results based on cluster size were assessed. The docked complex out of 5 complexes was studied for molecular dynamics simulation with a large cluster size.
2.8. Molecular Dynamics Simulations
Molecular dynamics (MD) simulation has served as an essential tool for comprehending macromolecules’ structural, functional and dynamic behavior [59]. GPU accelerated simulation engine PMEMD programmed in the Amber 18 package implemented MD simulations on two prepared systems [60]. FF14SB force-field [61] was used to describe the enzyme. Both systems were fully solvated in a TIP3P computer-generated water box [62] inside 10 Å box edge with LEaP component [63] of Amber 18. We have added Na+ and Cl− counter ions for neutralizing the system leading to the minimization phase with the same LEaP component of Amber 18. A two-step—partial and full minimization of 1500 and 1000 steps were achieved with a 500 kcal/mol restraint potential and conjugate gradient method followed by eliminating the restraints. Systems were then heated from 0 K to 300 K for 50 ps. Later, equilibration was accomplished by retaining the 500 ps equilibration step ensuring 300 K firm temperature. To maintain a steady atom numbers of and pressure for each system, NPT (isobaric-isothermal ensemble) was permitted. Berendsen barostat was applied to preserve the pressure at 1 bar on both systems [64]. Finally, a 100 ns MD simulations run was performed on both systems by incorporating the SHAKE algorithm [65] to control the hydrogen bond atoms. Details of MD simulation method has been described elsewhere [66].
2.9. Post-Dynamic Trajectories Analysis
Apo and MARK4-Myricetin complex were stored after every 1 ps, and the CPPTRAJ component [67] incorporated into Amber 18 package was used to analyze the MD trajectory curves. The RMSD We have assessed RMSF Rg, SASA, formation of intramolecular hydrogen bonds, distance correlation matrix with the secondary structure on the final MD trajectories. For scheming and analysis of these MD trajectories, Origin [68] and VMD visualization tools [69] were utilized.
2.10. Dynamic of the Cross-Correlation Matrix
The DCCM was assessed using the CPPTRAJ module employed in Amber 18 and all the matrices schemes using Origin software as described earleir [70]. Factors for i and j cross-correlation Cα atoms are shown underneath:
| (1) |
where Δri,j is the movement of the ith and jth atom average point and the angle braces are specified over the complete curves. All correlated movements are symbolized by Cij = 1; however, Cij = −1 denoted highly anti-correlated movements over time. The deviation of movements from 1 to −1 describes that i and j movements are correlated and anti-correlated, respectively. The DCCM was assessed using the CPPTRAJ module employed in Amber 18 and all the matrices schemes using Origin software.
2.11. Binding Energy Calculation
PRODIGY (PROtein binDIng enerGY) [58] is a freely available web server used to evaluate the binding free energy (ΔG) and to predict the dissociation constant (Kd) of the protein-protein complex. This server implements a highly effective predictive model based on intermolecular contacts and properties derived from the non-interface surface.
3. Result and Discussion
3.1. Fluorescence-Based Binding Analysis
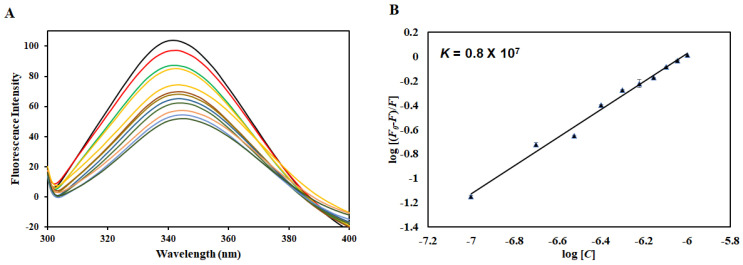
Human irisin and MARK4 proteins were successfully purified as per previously published reports [3,45]. Fluorescence-based binding is a commonly deployed technique retorted to analyze the actual interaction between protein and ligand [71]. With the use of intrinsic fluorescence, changes in the local microenvironment of aromatic amino acid residues are investigated [72,73]. In fluorescence quenching, a decrease in fluorescence intensity is evident with increasing ligand concentration suggesting the formation of the complex [74]. Native MARK4 shows a fluorescence emission spectrum at 344 nm, characteristic of the native protein. However, with increasing irisin concentration (0–1 µM), a decline in the fluorescence intensity was apparent implicative of the complex formation between MARK4 and irisin (Figure 1A). This decrease in the fluorescence was analyzed employing the MSV equation as per previously published reports [48,70].
Figure 1.
(A) Intrinsic fluorescence of MARK4 in the absence and presence of irisin (0–1 µM). (B) Modified Stern–Volmer plot of MARK4–irisin interaction to find binding constant.
Figure 1B shows the experimental fitting obtained according to this equation, with the slope of the plot giving the number of binding sites (n) and the intercept giving the binding constant (K). Irisin binds to MARK4 with an excellent binding affinity evident from the obtained binding constant (K = 0.8 × 107 M−1). This analysis reveals that irisin binds to MARK4 with an excellent affinity and can serve as a potent binding partner of MARK4 that can be implicated in the management of diseases in which MARK4 is overexpressed, contributing to the development of diseases viz. various cancers and neurodegenerative diseases such as AD.
3.2. Isothermal Titration Calorimetry
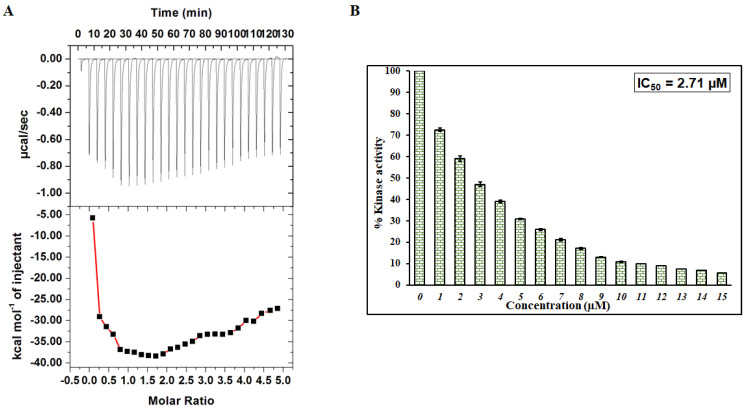
ITC is an affirmative technique deployed to complement the binding studies [30]. Thus, to validate earlier observations, ITC was performed to understand the binding parameters of MARK4-irisin interaction and find its associated thermodynamic parameters [75]. Figure 2A depicts a typical isotherm obtained upon titration of 200 µM irisin with 15 µM MARK4. The obtained isotherm for MARK4-irisin interaction suggests that irisin is binding spontaneously to MARK4 and these observations are in line with earlier results affirming strong binding between MARK4 and irisin. The upper panel shows the raw data owing to consecutive irisin injections into MARK4. In contrast, the bottom panel depicts the binding curves obtained after subtracting the dilution heat of the MARK4 and irisin. Table 1 depicts various thermodynamic parameters obtained for MARK4-irisin interaction, and the results presented were obtained from two-site model fitting.
Figure 2.
(A) ITC profile of MARK4-irisin system. The sample cell was filled with 15 µM MARK4 while the syringe contained 200 µM irisin. (B) Kinase inhibition assay of MARK4 with varying irisin concentration (0−15 µM).
Table 1.
Thermodynamic parameters of MARK4–irisin interaction obtained from ITC.
|
Ka (Association Constant) M−1 |
∆H (Enthalpy Change) cal/mol |
∆S (cal/mol/deg) |
|---|---|---|
| Ka1 = 2.59 × 103 ± 2.10 × 104 | ∆H1 = −1.549 × 106 7617 ± 6.75 × 107 | ∆S1 = −5.18 × 103 |
| Ka2 = 1.19 × 106 ± 9.73 | ∆H2 = 1.898 × 105 ± 1.04 × 106 | ∆S2 = 664 |
3.3. Kinase Assay
In vitro and in silico observations, it is evident that irisin binds to MARK4 with an excellent affinity. Thus, it becomes imperative to evaluate the kinase inhibitory potential of irisin. MARK4 in the absence of irisin was taken as 100% for reference. Figure 2B shows the effect of varying concentrations of irisin on the kinase activity of MARK4, suggesting that when irisin concentration increases, there is a corresponding decrease in the activity of MARK decreases. IC50 is that ligand concentration where 50% inhibitory effect is observed. According to earlier published studies [48,76] it was calculated as 2.71 µM making use of AAT Bioquest calculator. IC50 of such order implies irisin to be a potent MARK4 inhibitor and corroborates with earlier observations.
3.4. Molecular Docking
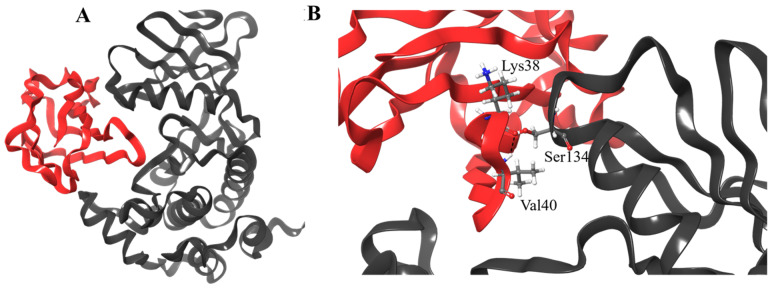
The three-dimensional structure of irisin was retrieved from the PDB database as stated before, to assess the potential interaction within the binding site of MARK4 protein by the molecular docking approach (Figure 3). We achieved an optimized positioning of the irisin chain in the protein’s binding site by minimizing the comprehensive energies of the corresponding complex. The predictable value of the binding affinity of irisin was −10.0 kcal/mol. As shown in Figure 4B, the protein–protein complex of irisin and MARK4 proteins has been formed by multiple hydrogen bonds through the active site residues Lys38, Val40, and Ser134 of the irisin-MARK4 complex. On the other hand, the predicted dissociation constant was reported as 4.8 × 10−8 M.
Figure 3.
(A) Structural representation of MARK4 enzyme (ribbon view—black color) in complex with irisin (ribbon view—red color). (B) Close view of interactions formed between the carbon backbone atoms of MARK4 and irisin.
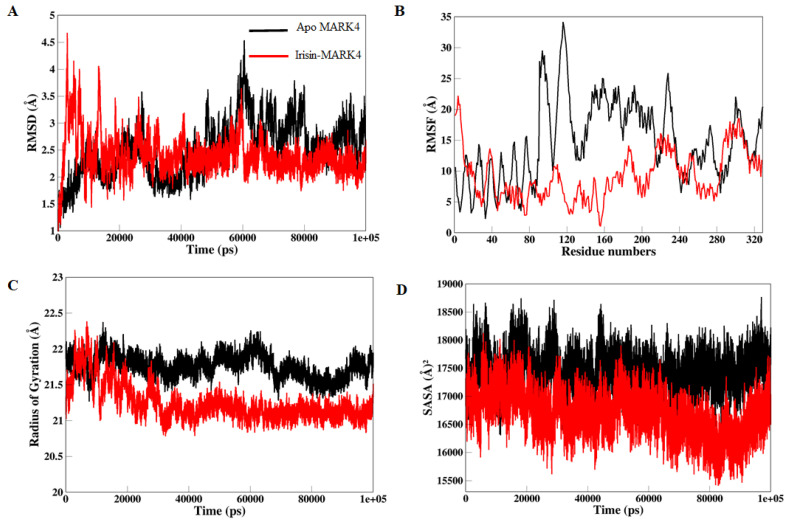
Figure 4.
Structural dynamics of MARK4 apo (black color) and irisin bound MARK4 (black color) enzyme. (A) RMSD, (B) RMSF, (C) Rg values, and (D) SASA values across Cα backbone in Å of apo MARK4 and irisin-MARK4 complex in Å across Cα backbone of both of the two conditions calculated after 100 ns of MD trajectories.
3.5. Post-Dynamics Trajectories Analysis
Protein-protein interactions manage a broad range of biological activities, involving cell-to-cell interfaces, metabolic and developmental influence [77]. Differences within protein structure are explicitly correlated with their genomic behaviors [78]. The inhibitory activity of many enzymes convoluted in various infectious pathways is extremely affected upon the binding of small molecules. Hence there is necessary to consider the conformational changes and structural dynamics correlated with the inhibitory activity of these complexes [79]. To determine the efficiency and consistency of the simulated MARK4 protein in complex with Irisin, we have estimated the time-variability through RMSD of Cα atoms from constructed trajectories. As shown in Figure 4A the RMSD perturbed values uncovered possible symmetrical deviations in protein structure after binding of studied protein. Apo MARK4 and Irisin-MARK4 complex were stabilized and achieved convergence after 40 ns of the simulation time as indicated in the 2D plot displayed in Figure 4A. The least RMSD value of 2.57 Å was observed in the Irisin-MARK4 complex, while apo MARK4 suggested an average of 3.25 Å separately. Apo MARK4 protein revealed high fluctuations in contrast with the bound MARK4 complex. This RMSD assessment suggests that the irisin-MARK4 complex showed the least deviation of Cα backbone atoms, indicating that binding of this employed irisin protein boosted stability on MARK4 protein comparative to apo MARK4. The RMSD evaluation also proposes that any further reports on the determined trajectories for the apo and irisin-MARK4 complex were reliable.
The biological activity of the target protein is supported by the rigidity and flexibility of its amino acid residues and biological activity in several biological pathways [80]. RMSF of Cα atoms were evaluated from the generated trajectories during 100 ns of MD simulations to explain the flexibility and rigidity of overall MARK4 residues upon irisin binding. The Irisin-MARK4 complex suggested the least fluctuations in the amino acid residues with an average RMSF value of 9.93 Å as shown in Figure 4B. Apo MARK4 protein displayed a significantly higher RMSF value of 18.36 Å. The RMSF value of Irisin-MARK4 is notably lower than apo MARK4, disclosing a substantial activity of the Irisin-MARK4 complex against apo MARK4. The residues from 80–240 revealed high fluctuations throughout the MD simulation time as this region contributes to the binding of Irisin to MARK4 protein. The effective binding of this Irisin protein chain in the active site might be connected with the structural inactivation of MARK4 protein and have a more complex structural effect on various parts of the overall MARK4 structure.
Furthermore, to be consistent with our findings, we have assessed radius of gyration (Rg) values, a factor associated with the overall conformational improvements within protein structure after Irisin binding. It also uncovers protein’s structural compactness, folding behavior and stability [66]. We calculated the Rg values to study the compactness of Irisin-MARK4 and apo MARK4 protein. As shown in Figure 4C, a notable increase was seen in the Rg value of apo MARK4 protein, whereas the Irisin-MARK4 complex showed a least Rg value of 21.14 Å. Apo MARK4 protein uncovered the elevated Rg value of 22.24 Å in comparison with the Irisin-MARK4 complex. This estimate suggested improved compactness and increased activity of Irisin upon MARK4 binding. The stability, flexibility, and efficiency of irisin with MARK4 protein indicate positive discoveries associated with its conformational dynamics.
Solvent accessible surface area (SASA) was measured after studying the conformational binding studies to describe the efficiency of hydrophobic and hydrophilic amino acid residues and energies subjected to the solvent throughout the simulation time [81]. The enzyme inhibition usually affects by influences between the hydrophobic intrinsic contacts inside the protein structure. These hydrophobic networks generated between the non-polar amino acid residues justify the protein structure’s stability inside the solution by defending the non-polar residues in the hydrophobic core isolated from an aqueous mixture [82]. For the Irisin-MARK4 complex and apo MARK4 protein, total SASA values were assessed throughout the 100 ns MD simulation production period (Figure 4D). We exposed the Irisin-MARK4 complex and apo MARK4 protein to the solvent and obtained the standard SASA value of 16,517 Å2 in the Irisin-MARK4 complex leading to an increased average SASA value of 17,874 Å2 was noted in apo MARK4 protein as compared to the Irisin-MARK4 complex. An improved SASA value was revealed in Irisin-MARK4 against apo MARK4 protein. The folding and unfolding behavior of the MARK4 protein was marked through the modifications in SASA values for the Irisin-MARK4 complex during this simulation time. All these predictions favored the improved exposure of irisin to solvent and also indicated its enhanced inhibitory activity over apo MARK4 protein.
3.6. Hydrogen Bond Analysis
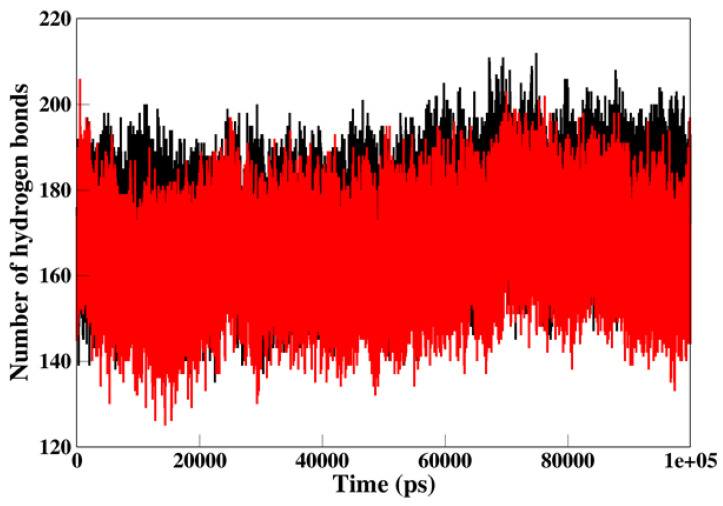
Intramolecular hydrogen bond assessment helps in estimating the stability of the protein-ligand complex (Figure 5). This analysis offers an overall understanding of the mechanism involved in protein-protein binding. The number of intramolecular hydrogen bonds in the Irisin-MARK4 complex was approximately 125–180, whereas 145–190 in the apo MARK4 protein, respectively. Apo MARK4 protein indicated the least number of intramolecular hydrogen bonds formation in comparison to the Irisin-MARK4 complex.
Figure 5.
Hydrogen bond analysis. Intramolecular hydrogen bonds in apo (black color) and irisin-MARK4 complex (red color) were calculated after 100 ns MD simulation.
3.7. Distance Correlation Matrix
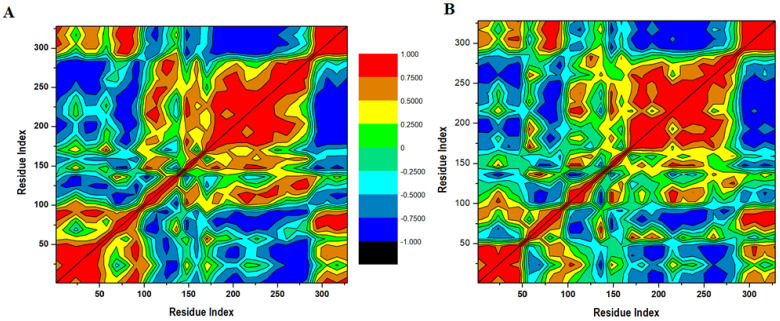
A distance correlation matrix was analyzed for the apo MARK4 and Irisin-MARK4 complex for the correlated and anti-correlated developments of all the amino acid residues, as displayed in Figure 6. Various residual movements were observed and MARK4 protein was scattered into several communities across positive and negative correlations. More negative and less positive residual movements were seen in apo MARK4 protein during the whole 100 ns MD simulation run (Figure 6A). On the other side, the Irisin-MARK4 complex produced a significant positive correlation of motions in the amino acid residues (Figure 6B). The residues between 150–270 in apo MARK protein contributed to the most negative correlation indicative of no inhibition without Irisin (Figure 6A). On the other side, the residues between 1–50 and 160–280 contributed to positive developments in the Irisin-MARK4 complex, indicative of improved Irisin activity to MARK4 protein.
Figure 6.
Dynamics cross-correlation matrix analysis (A) apo MARK4 enzyme and (B) irisin-MARK4 complex calculated after 100 ns of MD trajectories.
3.8. Secondary Structure Analysis
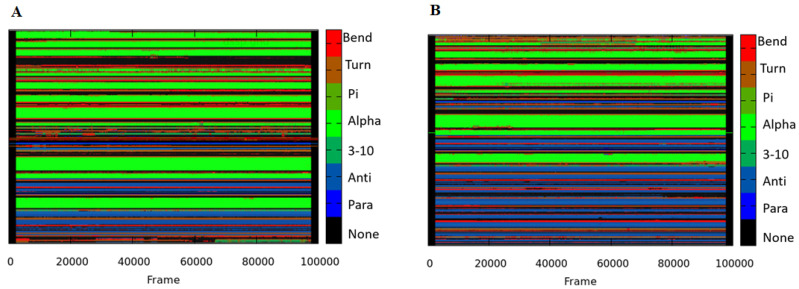
We also measured the secondary structure modifications for the apo and Irisin-MARK4 complex in favor of all the above-mentioned findings. This analysis serves as an important tool to observe the changes in the MARK4 structure in the absence and presence of Irisin. The 2D plots for apo and Irisin-MARK4 complex are displayed in Figure 7. The structural components are higher in the Irisin-MARK4 complex in contract with apo MARK4 protein, as shown in Table 2. The number of α-helix in apo MARK4 protein is slightly increased, whereas the β-strands and 310-helix contributed majorly in the Irisin-MARK4 complex. The strands and helices in the Irisin bound protein indicate improved activity of MARK4 protein after Irisin binding in the active site of this protein. Strong stability and flexibility of Irisin residues to MARK4 protein was revealed through this secondary structure analysis that could indicate Irisin importance to MARK4 protein.
Figure 7.
Secondary structure analysis (A) apo-MARK4 enzyme and (B) irisin-MARK4 complex calculated after 100 ns of MD trajectories.
Table 2.
Percentage of residues contributing to the secondary structure of apo MARK4 enzyme and irisin-MARK4 complex calculated after 100 ns of MD trajectories.
| Protein–Protein Complex | α | β | 310-Helix | Turn | Bend | Other |
|---|---|---|---|---|---|---|
| Apo MARK4 | 28 | 25 | 4 | 13 | 7 | 21 |
| Irisin-MARK4 | 26 | 27 | 6 | 16 | 11 | 23 |
4. Conclusions
The present study establishes irisin as an inhibitor of MARK4, highlighting its role in cancer and AD therapy. This work reveals the therapeutic aspect of irisin via its binding to MARK4 with an excellent affinity and subsequent inhibition of kinase activity. Overexpression of MARK4 is associated with cancers and neurodegenerative diseases. Many studies have reported MARK4 inhibitors that can be retorted in the management of MARK4-associated diseases. For the first time, this study reports the inhibition of MARK4 by irisin, an exercise-induced myokine, opening a new avenue in the therapeutics to the use of irisin as a potential therapeutic agent for targeting different kinases. Thus, pharmacologically or through exercise-induced irisin levels, there can be a therapeutic strategy to treat MARK4–directed cancers and neurodegenerative diseases.
Acknowledgments
This work was supported by Taif University Researchers Supporting Project Number (TURSP-2020/131), Taif University, Taif, Saudi Arabia. The M.I.H. acknowledges the Council of Scientific and Industrial Research for financial support (Project No. 27(0368)/20/EMR-II). The S.K. would like to acknowledge the NRF (National Research Foundation) for providing the financial assistance and the CHPC server based in Cape Town, South Africa, for conducting this research. The authors thank the Department of Science and Technology, Government of India, for the FIST support (FIST program No. SR/FST/LSII/2020/782).
Author Contributions
Conceptualization, R.W., S.A. and M.I.H.; methodology, R.W., S.A., A.S. (Anas Shamsi), D.K.Y., S.K. and A.S. (Anas Shamsi); software, A.S. (Alaa Shafie) and F.A.; validation, A.S. (Anas Shamsi) and M.I.H.; formal analysis, A.S. (Anas Shamsi), A.S. (Alaa Shafie) and S.K.; investigation, S.A., S.K., A.S. (Alaa Shafie) and A.I.; resources; data curation, D.K.Y. and A.I.; writing—original draft preparation, R.W., S.A., A.S. (Alaa Shafie), D.K.Y. and S.K.; writing—review and editing, A.I., F.A. and M.I.H.; visualization, F.A., A.I. and M.I.H.; supervision, M.I.H. and A.I.; project administration, D.K.Y. and A.I.; funding acquisition, F.A. and A.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Taif University Researchers Supporting Project Number (TURSP-2020/131), Taif University, Taif, Saudi Arabia. National Medicinal Plants Board (NMPB), Ministry of AYUSH, Government of India [F. No. Z. 18017/187/CSS/R&D/DL-01/2019-20-NMPB-IVA].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Boström P., Wu J., Jedrychowski M.P., Korde A., Ye L., Lo J.C., Rasbach K.A., Boström E.A., Choi J.H., Long J.Z., et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jedrychowski M.P., Wrann C.D., Paulo J.A., Gerber K.K., Szpyt J., Robinson M.M., Nair K.S., Gygi S.P., Spiegelman B.M. Detection and quantitation of circulating human irisin by tandem mass spectrometry. Cell Metab. 2015;22:734–740. doi: 10.1016/j.cmet.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waseem R., Shamsi A., Mohammad T., Alhumaydhi F.A., Kazim S.N., Hassan M.I., Ahmad F., Islam A. Multispectroscopic and Molecular Docking Insight into Elucidating the Interaction of Irisin with Rivastigmine Tartrate: A Combinational Therapy Approach to Fight Alzheimer’s Disease. ACS Omega. 2021;6:7910–7921. doi: 10.1021/acsomega.1c00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang S., Pan J. Irisin ameliorates depressive-like behaviors in rats by regulating energy metabolism. Biochem. Biophys. Res. Commun. 2016;474:22–28. doi: 10.1016/j.bbrc.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 5.Liu T.-Y., Shi C.-X., Gao R., Sun H.-J., Xiong X.-Q., Ding L., Chen Q., Li Y.-H., Wang J.-J., Kang Y.M., et al. Irisin inhibits hepatic gluconeogenesis and increases glycogen synthesis via the PI3K/Akt pathway in type 2 diabetic mice and hepatocytes. Clin. Sci. 2015;129:839–850. doi: 10.1042/CS20150009. [DOI] [PubMed] [Google Scholar]
- 6.Chen J.-Q., Huang Y.-Y., Gusdon A.M., Qu S. Irisin: A new molecular marker and target in metabolic disorder. Lipids Health Dis. 2015;14:2. doi: 10.1186/1476-511X-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiao X., Nie Y., Ma Y., Chen Y., Cheng R., Yin W., Hu Y., Xu W., Xu L. Irisin promotes osteoblast proliferation and differentiation via activating the MAP kinase signaling pathways. Sci. Rep. 2016;6:18732. doi: 10.1038/srep18732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novelle M.G., Contreras C., Romero-Picó A., López M., Diéguez C. Irisin, two years later. Int. J. Endocrinol. 2013;2013:746281. doi: 10.1155/2013/746281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim O.Y., Song J. The role of irisin in Alzheimer’s disease. J. Clin. Med. 2018;7:407. doi: 10.3390/jcm7110407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lourenco M.V., Frozza R.L., de Freitas G.B., Zhang H., Kincheski G.C., Ribeiro F.C., Gonçalves R.A., Clarke J.R., Beckman D., Staniszewski A., et al. Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer’s models. Nat. Med. 2019;25:165–175. doi: 10.1038/s41591-018-0275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lund H., Gustafsson E., Svensson A., Nilsson M., Berg M., Sunnemark D., von Euler G. MARK4 and MARK3 associate with early tau phosphorylation in Alzheimer’s disease granulovacuolar degeneration bodies. Acta Neuropathol. Commun. 2014;2:22. doi: 10.1186/2051-5960-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandelkow E.-M., Thies E., Trinczek B., Biernat J., Mandelkow E. MARK/PAR1 kinase is a regulator of microtubule-dependent transport in axons. J. Cell Biol. 2004;167:99–110. doi: 10.1083/jcb.200401085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun W., Lee S., Huang X., Liu S., Inayathullah M., Kim K.-M., Tang H., Ashford J.W., Rajadas J. Attenuation of synaptic toxicity and MARK4/PAR1-mediated Tau phosphorylation by methylene blue for Alzheimer’s disease treatment. Sci. Rep. 2016;6:34784. doi: 10.1038/srep34784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenardhanan P., Mannu J., Mathur P.P. The structural analysis of MARK4 and the exploration of specific inhibitors for the MARK family: A computational approach to obstruct the role of MARK4 in prostate cancer progression. Mol. Biosyst. 2014;10:1845–1868. doi: 10.1039/C3MB70591A. [DOI] [PubMed] [Google Scholar]
- 15.Naz F., Khan F.I., Mohammad T., Khan P., Manzoor S., Hasan G.M., Lobb K.A., Luqman S., Islam A., Ahmad F., et al. Investigation of molecular mechanism of recognition between citral and MARK4: A newer therapeutic approach to attenuate cancer cell progression. Int. J. Biol. Macromol. 2018;107:2580–2589. doi: 10.1016/j.ijbiomac.2017.10.143. [DOI] [PubMed] [Google Scholar]
- 16.Naz F., Anjum F., Islam A., Ahmad F., Hassan M.I. Microtubule affinity-regulating kinase 4: Structure, function, and regulation. Cell Biochem. Biophys. 2013;67:485–499. doi: 10.1007/s12013-013-9550-7. [DOI] [PubMed] [Google Scholar]
- 17.Naz F., Shahbaaz M., Bisetty K., Islam A., Ahmad F., Hassan M.I. Designing New Kinase Inhibitor Derivatives as Therapeutics Against Common Complex Diseases: Structural Basis of Microtubule Affinity-Regulating Kinase 4 (MARK4) Inhibition. OMICS. 2015;19:700–711. doi: 10.1089/omi.2015.0111. [DOI] [PubMed] [Google Scholar]
- 18.Tiwari M.K., Coghi P., Agrawal P., Yadav D.K., Yang L.J., Congling Q., Sahal D., Wong V.K.W., Chaudhary S. Novel halogenated arylvinyl-1, 2, 4 trioxanes as potent antiplasmodial as well as anticancer agents: Synthesis, bioevaluation, structure-activity relationship and in-silico studies. Eur. J. Med. Chem. 2021;224:113685. doi: 10.1016/j.ejmech.2021.113685. [DOI] [PubMed] [Google Scholar]
- 19.Kalani K., Yadav D.K., Alam S., Khan F., Kashyap M.P., Srivastava S.K., Pant A.B. In-silico Studies and Wet-Lab Validation of Camptothecin Derivatives for Anti-Cancer Activity Against Liver (HepG2) and Lung (A549) Cancer Cell Lines. Curr. Top. Med. Chem. 2021;21:1–12. doi: 10.2174/1568026621666210426124719. [DOI] [PubMed] [Google Scholar]
- 20.Yadav D.K. Recent Advances on Small Molecule Medicinal Chemistry to Treat Human Diseases-Part II. Curr. Top. Med. Chem. 2021;21:849. doi: 10.2174/156802662110210601102135. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.W., Park S., Kim S.Y., Um S.H., Moon E.Y. Curcumin hampers the antitumor effect of vinblastine via the inhibition of microtubule dynamics and mitochondrial membrane potential in HeLa cervical cancer cells. Phytomedicine. 2016;23:705–713. doi: 10.1016/j.phymed.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 22.Gadhe C.G., Kim M.H. Insights into the binding modes of CC chemokine receptor 4 (CCR4) inhibitors: A combined approach involving homology modelling, docking, and molecular dynamics simulation studies. Mol. Biosyst. 2015;11:618–634. doi: 10.1039/C4MB00568F. [DOI] [PubMed] [Google Scholar]
- 23.Kim J.H., Yi S.H., Lee J., Kim Y.S. Effects of clozapine on heart rate dynamics and their relationship with therapeutic response in treatment-resistant schizophrenia. J. Clin. Psychopharmacol. 2013;33:69–73. doi: 10.1097/JCP.0b013e31827d14e3. [DOI] [PubMed] [Google Scholar]
- 24.Kim J.H., Ann J.H., Lee J., Kim M.H., Han A.Y. Altered heart rate dynamics associated with antipsychotic-induced subjective restlessness in patients with schizophrenia. Neuropsychiatr. Dis. Treat. 2013;9:989–994. doi: 10.2147/NDT.S48701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadav D.K., Kumar S., Teli M.K., Kim M.H. Ligand-based pharmacophore modeling and docking studies on vitamin D receptor inhibitors. J. Cell. Biochem. 2020;121:3570–3583. doi: 10.1002/jcb.29640. [DOI] [PubMed] [Google Scholar]
- 26.Shyamlal B.R.K., Mathur M., Yadav D.K., Chaudhary S. Microwave-assisted modified synthesis of C8-analogues of naturally occurring methylxanthines: Synthesis, biological evaluation and their practical applications. Fitoterapia. 2020;143:104533. doi: 10.1016/j.fitote.2020.104533. [DOI] [PubMed] [Google Scholar]
- 27.Dileep K., Ashok S., Remya C., Dharmendra K., Pérez-Sánchez H., Sadasivan C. Indole fragments for the design of lead molecules against pancreatitis. J. Biomol. Struct. Dyn. 2020;38:263–267. doi: 10.1080/07391102.2019.1567389. [DOI] [PubMed] [Google Scholar]
- 28.Yadav D.K., Kumar S., Choi E.-H., Chaudhary S., Kim M.-H. Computational Modeling on Aquaporin-3 as Skin Cancer Target: A Virtual Screening and Molecular Dynamic Simulation Study. Front. Chem. 2020;8:250. doi: 10.3389/fchem.2020.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav D.K., Saloni S.K., Kim M.-H. Proteomics and Molecular Medicine Bioinformatics. J. Proteom. 2017;10 doi: 10.15761/IMM.1000233. [DOI] [Google Scholar]
- 30.Khan P., Rahman S., Queen A., Manzoor S., Naz F., Hasan G.M., Luqman S., Kim J., Islam A., Ahmad F., et al. Elucidation of Dietary Polyphenolics as Potential Inhibitor of Microtubule Affinity Regulating Kinase 4: In silico and In vitro Studies. Sci. Rep. 2017;7:9470. doi: 10.1038/s41598-017-09941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrov D.S. Therapeutic Proteins. Springer; Berlin/Heidelberg, Germany: 2012. pp. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheon J.H., Kim K.S., Yadav D.K., Kim M., Kim H.S., Yoon S. The JAK2 inhibitors CEP-33779 and NVP-BSK805 have high P-gp inhibitory activity and sensitize drug-resistant cancer cells to vincristine. Biochem. Biophys. Res. Commun. 2017;490:1176–1182. doi: 10.1016/j.bbrc.2017.06.178. [DOI] [PubMed] [Google Scholar]
- 33.Venkanna A., Kwon O.W., Afzal S., Jang C., Cho K.H., Yadav D.K., Kim K., Park H.-g., Chun K.-H., Kim S.Y. Pharmacological use of a novel scaffold, anomeric N, N-diarylamino tetrahydropyran: Molecular similarity search, chemocentric target profiling, and experimental evidence. Sci. Rep. 2017;7:1–17. doi: 10.1038/s41598-017-12082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yadav D.K., Kumar S., Misra S., Yadav L., Teli M., Sharma P., Chaudhary S., Kumar N., Choi E.H., Kim H.S. Molecular insights into the interaction of RONS and Thieno [3, 2-c] pyran analogs with SIRT6/COX-2: A molecular dynamics study. Sci. Rep. 2018;8:1–16. doi: 10.1038/s41598-018-22972-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dwivedi G.R., Maurya A., Yadav D.K., Khan F., Darokar M.P., Srivastava S.K. Drug resistance reversal potential of ursolic acid derivatives against nalidixic acid-and multidrug-resistant Escherichia coli. Chem. Biol. Drug Des. 2015;86:272–283. doi: 10.1111/cbdd.12491. [DOI] [PubMed] [Google Scholar]
- 36.Jaiswal P.K., Sharma V., Kumar S., Mathur M., Swami A.K., Yadav D.K., Chaudhary S. Non-peptide-based new class of platelet aggregation inhibitors: Design, synthesis, bioevaluation, SAR, and in silico studies. Archiv. Der. Pharmazie. 2018;351:1700349. doi: 10.1002/ardp.201700349. [DOI] [PubMed] [Google Scholar]
- 37.Yadav D.K., Khan F., Negi A.S. QSAR and Docking studies of Gallic acid derivatives for anticancer and Immunomodulatory activity. Nat. Chem. 2011:1. doi: 10.1038/npre.2011.6620.1. [DOI] [Google Scholar]
- 38.Lee Y.B., Ah Lee J., Soobin J., Lim Lee H. Herbal medicine (Suoquan) for treating nocturnal enuresis: A protocol for a systematic review of randomized controlled trials. Medicine (Baltimore) 2018;97:e0391. doi: 10.1097/MD.0000000000010391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee K., Cho S.G., Choi Y.K., Choi Y.J., Lee G.R., Jeon C.Y., Ko S.G. Herbal prescription, Danggui-Sayuk-Ga-Osuyu-Senggang-Tang, inhibits TNF-alpha-induced epithelial-mesenchymal transition in HCT116 colorectal cancer cells. Int. J. Mol. Med. 2018;41:373–380. doi: 10.3892/ijmm.2017.3241. [DOI] [PubMed] [Google Scholar]
- 40.Lee H.L., Lee Y.B., Choi J.Y., Lee J.A. Herbal medicine for idiopathic central precocious puberty: A protocol for a systematic review of controlled trials. Medicine (Baltimore) 2018;97:e0267. doi: 10.1097/MD.0000000000010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park S.J., Kim M.O., Kim J.H., Jeong S., Kim M.H., Yang S.J., Lee J., Lee H.J. Antioxidant Activities of Functional Beverage Concentrates Containing Herbal Medicine Extracts. Prev. Nutr. Food. Sci. 2017;22:16–20. doi: 10.3746/pnf.2017.22.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ku S.K., Kim H., Kim J.W., Kang K.S., Lee H.J. Ameliorating effects of herbal formula hemomine on experimental subacute hemorrhagic anemia in rats. J. Ethnopharmacol. 2017;198:205–213. doi: 10.1016/j.jep.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Hussain A., Yadav M.K., Bose S., Wang J.H., Lim D., Song Y.K., Ko S.G., Kim H. Daesiho-Tang Is an Effective Herbal Formulation in Attenuation of Obesity in Mice through Alteration of Gene Expression and Modulation of Intestinal Microbiota. PLoS ONE. 2016;11:e0165483. doi: 10.1371/journal.pone.0165483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kashyap M.P., Singh A.K., Yadav D.K., Siddiqui M.A., Srivastava R.K., Chaturvedi V., Rai N. 4-Hydroxy-trans-2-nonenal (4-HNE) induces neuronal SH-SY5Y cell death via hampering ATP binding at kinase domain of Akt1. Arch. Toxicol. 2015;89:243–258. doi: 10.1007/s00204-014-1260-4. [DOI] [PubMed] [Google Scholar]
- 45.Naz F., Asad M., Malhotra P., Islam A., Ahmad F., Hassan M.I. Cloning, expression, purification and refolding of microtubule affinity-regulating kinase 4 expressed in Escherichia coli. Appl. Biochem. Biotechnol. 2014;172:2838–2848. doi: 10.1007/s12010-014-0733-5. [DOI] [PubMed] [Google Scholar]
- 46.Khan P., Idrees D., Moxley M.A., Corbett J.A., Ahmad F., von Figura G., Sly W.S., Waheed A., Hassan M.I. Luminol-based chemiluminescent signals: Clinical and non-clinical application and future uses. Appl. Biochem. Biotechnol. 2014;173:333–355. doi: 10.1007/s12010-014-0850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shamsi A., Anwar S., Mohammad T., Alajmi M.F., Hussain A., Rehman M., Hasan G.M., Islam A., Hassan M.I. MARK4 inhibited by AChE inhibitors, donepezil and Rivastigmine tartrate: Insights into Alzheimer’s disease therapy. Biomolecules. 2020;10:789. doi: 10.3390/biom10050789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anwar S., Mohammad T., Shamsi A., Queen A., Parveen S., Luqman S., Hasan G.M., Alamry K.A., Azum N., Asiri A.M., et al. Discovery of Hordenine as a potential inhibitor of pyruvate dehydrogenase kinase 3: Implication in lung Cancer therapy. Biomedicines. 2020;8:119. doi: 10.3390/biomedicines8050119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anwar S., Shamsi A., Shahbaaz M., Queen A., Khan P., Hasan G.M., Islam A., Alajmi M.F., Hussain A., Ahmad F., et al. Rosmarinic acid exhibits anticancer effects via MARK4 inhibition. Sci. Rep. 2020;10:10300. doi: 10.1038/s41598-020-65648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naz F., Sami N., Islam A., Ahmad F., Hassan M.I. Ubiquitin-associated domain of MARK4 provides stability at physiological pH. Int. J. Biol. Macromol. 2016;93:1147–1154. doi: 10.1016/j.ijbiomac.2016.09.087. [DOI] [PubMed] [Google Scholar]
- 52.Naz F., Sami N., Naqvi A.T., Islam A., Ahmad F., Imtaiyaz Hassan M. Evaluation of human microtubule affinity-regulating kinase 4 inhibitors: Fluorescence binding studies, enzyme, and cell assays. J. Biomol. Struct. Dyn. 2017;35:3194–3203. doi: 10.1080/07391102.2016.1249958. [DOI] [PubMed] [Google Scholar]
- 53.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 54.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., Wang L., Lupyan D., Dahlgren M.K., Knight J.L., et al. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 55.Sharma V., Jaiswal P.K., Saran M., Yadav D.K., Saloni, Mathur M., Swami A.K., Misra S., Kim M.H., Chaudhary S. Discovery of C-3 Tethered 2-oxo-benzo[1,4]oxazines as Potent Antioxidants: Bio-Inspired Based Design, Synthesis, Biological Evaluation, Cytotoxic, and in Silico Molecular Docking Studies. Front. Chem. 2018;6:56. doi: 10.3389/fchem.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma V., Jaiswal P.K., Kumar S., Mathur M., Swami A.K., Yadav D.K., Chaudhary S. Discovery of Aporphine Analogues as Potential Antiplatelet and Antioxidant Agents: Design, Synthesis, Structure-Activity Relationships, Biological Evaluations, and in silico Molecular Docking Studies. Chem. Med. Chem. 2018;13:1817–1832. doi: 10.1002/cmdc.201800318. [DOI] [PubMed] [Google Scholar]
- 57.Kim H.H., Hyun J.S., Choi J., Choi K.E., Jee J.G., Park S.J. Structural ensemble-based docking simulation and biophysical studies discovered new inhibitors of Hsp90 N-terminal domain. Sci. Rep. 2018;8:368. doi: 10.1038/s41598-017-18332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozakov D., Brenke R., Comeau S.R., Vajda S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins Struct. Funct. Bioinform. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 59.Hospital A., Goñi J.R., Orozco M., Gelpí J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. AABC. 2015;8:37. doi: 10.2147/AABC.S70333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee T.-S., Cerutti D.S., Mermelstein D., Lin C., LeGrand S., Giese T.J., Roitberg A., Case D.A., Walker R.C., York D.M. GPU-accelerated molecular dynamics and free energy methods in Amber18: Performance enhancements and new features. J. Chem. Inf. Modeling. 2018;58:2043–2050. doi: 10.1021/acs.jcim.8b00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez A., MacCallum J.L., Brini E., Simmerling C., Dill K.A. Grid-based backbone correction to the ff12SB protein force field for implicit-solvent simulations. J. Chem. Theory Comput. 2015;11:4770–4779. doi: 10.1021/acs.jctc.5b00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrach M.F., Drossel B. Structure and dynamics of TIP3P, TIP4P, and TIP5P water near smooth and atomistic walls of different hydroaffinity. J. Chem. Phys. 2014;140:174501. doi: 10.1063/1.4872239. [DOI] [PubMed] [Google Scholar]
- 63.Wang J., Wang W., Kollman P.A., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 64.Lin Y., Pan D., Li J., Zhang L., Shao X. Application of Berendsen barostat in dissipative particle dynamics for nonequilibrium dynamic simulation. J. Chem. Phys. 2017;146:124108. doi: 10.1063/1.4978807. [DOI] [PubMed] [Google Scholar]
- 65.Gonnet P. P-SHAKE: A quadratically convergent SHAKE in O (n2) J. Comput. Phys. 2007;220:740–750. doi: 10.1016/j.jcp.2006.05.032. [DOI] [Google Scholar]
- 66.Gupta P., Khan S., Fakhar Z., Hussain A., Rehman M., AlAjmi M.F., Islam A., Ahmad F., Hassan M.I. Identification of potential inhibitors of calcium/calmodulin-dependent protein kinase IV from bioactive phytoconstituents. Oxidative Med. Cell. Longev. 2020;2020:2094635. doi: 10.1155/2020/2094635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 68.Seifert E. OriginPro 9.1: Scientific data analysis and graphing software-software review. J. Chem. Inf. Modeling. 2014;54:1552. doi: 10.1021/ci500161d. [DOI] [PubMed] [Google Scholar]
- 69.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 70.Anwar S., Shamsi A., Kar R.K., Queen A., Islam A., Ahmad F., Hassan M.I. Structural and biochemical investigation of MARK4 inhibitory potential of cholic acid: Towards therapeutic implications in neurodegenerative diseases. Int. J. Biol. Macromol. 2020;161:596–604. doi: 10.1016/j.ijbiomac.2020.06.078. [DOI] [PubMed] [Google Scholar]
- 71.Shamsi A., Ahmed A., Khan M.S., Al Shahwan M., Husain F.M., Bano B. Understanding the binding between Rosmarinic acid and serum albumin: In vitro and in silico insight. J. Mol. Liq. 2020;311:113348. doi: 10.1016/j.molliq.2020.113348. [DOI] [Google Scholar]
- 72.Soares S., Mateus N., De Freitas V. Interaction of different polyphenols with bovine serum albumin (BSA) and human salivary α-amylase (HSA) by fluorescence quenching. J. Agric. Food Chem. 2007;55:6726–6735. doi: 10.1021/jf070905x. [DOI] [PubMed] [Google Scholar]
- 73.Klajnert B., Stanisławska L., Bryszewska M., Pałecz B. Interactions between PAMAM dendrimers and bovine serum albumin. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2003;1648:115–126. doi: 10.1016/S1570-9639(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 74.Shamsi A., Al Shahwan M., Ahamad S., Hassan M.I., Ahmad F., Islam A. Spectroscopic, calorimetric and molecular docking insight into the interaction of Alzheimer’s drug donepezil with human transferrin: Implications of Alzheimer’s drug. J. Biomol. Struct. Dyn. 2020;38:1094–1102. doi: 10.1080/07391102.2019.1595728. [DOI] [PubMed] [Google Scholar]
- 75.Banerjee T., Kishore N. 2, 2, 2-trifluoroethanol-induced molten globule state of concanavalin A and energetics of 8-anilinonaphthalene sulfonate binding: Calorimetric and spectroscopic investigation. J. Phys. Chem. B. 2005;109:22655–22662. doi: 10.1021/jp053757r. [DOI] [PubMed] [Google Scholar]
- 76.Khan N.S., Khan P., Inam A., Ahmad K., Yousuf M., Islam A., Alib S., Azam A., Husain M., Hassan M.I. Discovery of 4-(2-(dimethylamino) ethoxy) benzohydrazide derivatives as prospective microtubule affinity regulating kinase 4 inhibitors. RSC Adv. 2020;10:20129–20137. doi: 10.1039/D0RA00453G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rao V.S., Srinivas K., Sujini G., Kumar G. Protein-protein interaction detection: Methods and analysis. Int. J. Proteom. 2014;2014:47648. doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khan S., Bjij I., Soliman M.E. Selective covalent inhibition of “Allosteric Cys121” distort the binding of PTP1B enzyme: A novel therapeutic approach for cancer treatment. Cell Biochem. Biophys. 2019;77:203–211. doi: 10.1007/s12013-019-00882-5. [DOI] [PubMed] [Google Scholar]
- 79.Khan S., Bjij I., Betz R.M., Soliman M.E. Reversible versus irreversible inhibition modes of ERK2: A comparative analysis for ERK2 protein kinase in cancer therapy. Future Med. Chem. 2018;10:1003–1015. doi: 10.4155/fmc-2017-0275. [DOI] [PubMed] [Google Scholar]
- 80.Bornot A., Etchebest C., De Brevern A.G. Predicting protein flexibility through the prediction of local structures. Proteins Struct. Funct. Bioinform. 2011;79:839–852. doi: 10.1002/prot.22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khan S., Fakhar Z., Hussain A., Ahmad A., Jairajpuri D.S., Alajmi M.F., Hassan M.I. Structure-based identification of potential SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1848634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen H., Panagiotopoulos A.Z. Molecular modeling of surfactant micellization using solvent-accessible surface area. Langmuir. 2019;35:2443–2450. doi: 10.1021/acs.langmuir.8b03440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.