Abstract
This study investigated the complex phenotypic and genetic response of Monterey pine (Pinus radiata) seedlings to co-infections by F. circinatum, the causal agent of pine pitch canker disease, and the oomycetes Phytophthora xcambivora and P. parvispora. Monterey pine seedlings were wound-inoculated with each single pathogen and with the combinations F. circinatum/P. xcambivora and F. circinatum/P. parvispora. Initially, seedlings inoculated only with F. circinatum showed less severe symptoms than seedlings co-inoculated or inoculated only with P. xcambivora or P. parvispora. However, 30 days post-inoculation (dpi), all inoculated seedlings, including those inoculated only with F. circinatum, showed severe symptoms with no significant differences among treatments. The transcriptomic profiles of three genes encoding pathogenesis-related proteins, i.e., chitinase (PR3), thaumatin-like protein (PR5), phenylalanine ammonia-lyase (PAL), and the pyruvate decarboxylase (PDC)-encoding gene were analyzed at various time intervals after inoculation. In seedlings inoculated with single pathogens, F. circinatum stimulated the up-regulation of all genes, while between the two oomycetes, only P. xcambivora induced significant up-regulations. In seedlings co-inoculated with F. circinatum and P. xcambivora or P. parvispora none of the genes showed a significant over-expression 4 dpi. In contrast, at 11 dpi, significant up-regulation was observed for PR5 in the combination F. circinatum/P. xcambivora and PDC in the combination F. circinatum/P. parvispora, thus suggesting a possible synergism of multiple infections in triggering this plant defense mechanism.
Keywords: pitch canker disease, Monterey pine, Phytophthora xcambivora, P. parvispora, plant- oomycetes- fungal interactions, gene expression, housekeeping genes, plant-defense molecular mechanisms, PR3, PR5, PAL
1. Introduction
Pinus radiata D. Don, commonly known as Monterey pine, is native to California (Western United States) and has been introduced into areas with a Mediterranean climate of Australia, Chile, New Zealand, South Africa, Spain, and Uruguay [1,2]. Due to the rapid growth and excellent quality of the wood, this pine species, of great interest for forestry, is mainly used for the production of timber [3]. The growth and productivity of P. radiata can be severely hampered by various parasites and diseases, resulting in significant economic losses. Several diseases of P. radiata have been described; among these, the resinous canker called “pine pitch canker”, caused by the ascomycete Fusarium circinatum, is considered one of the most important diseases of Pinus species worldwide [4,5].
Fusarium circinatum Nelson Nirenberg & O’Donnell, which was formerly named under Gibberella circinata Nirenberg & O’Donnell, its teleomorphic stage, is a necrotrophic fungus. Fusarium circinatum, listed in the EPPO (European and Mediterranean Plant Protection Organization) quarantine pathogens, has been reported on more than 60 Pinus species and on Pseudotsuga menziesii (Mirb.) Franco [6,7,8], albeit P. radiata, is among the most susceptible ones [5,6,8]. The fungus is able to infect pines of all ages and in all seasons, but mostly during the rainy season [4,5,6,7,8,9]. The first report of this pathogen was on Pinus virginiana in the southeastern United States in 1946. Afterward, its presence was reported in other countries, such as Haiti, Japan, South Africa, South Korea, Mexico, Chile, Spain, Italy, Uruguay, Portugal, and Colombia [10,11,12]. The infections originate from lesions that allow the penetration of F. circinatum into the susceptible pine organs, including the main stem, secondary branches, and roots [13]. In nurseries, the main symptom of the pine pitch canker consists of dying off of the seedlings (damping off), while in plantations, the most common symptoms are the drying of the branches and cankers on the stems, with the production of abundant resinous exudate, which leads to the death of the apex of the plant, resulting in stunted growth and reduced crown and stem volume as a consequence of this malformation [4,10,14].
Phytophthora spp. are a group of major pathogens of Pinus species. The genus Phytophthora (Oomycetes, Stramenopila) encompasses at least 200 recognized species, considering they were over 180 by 2018, and most of these species are plant pathogens [15,16,17,18,19,20,21,22,23,24,25]. The majority of Phytophthora spp. are soil-borne and prevalently infect the roots and the basal stem. However, many soil-borne Phytophthora species have adapted, at least temporarily, to an aerial lifestyle and are able to infect leaves, twigs, and fruit [18]. Stem cankers and root and crown rot are the most common symptoms induced by these pathogens on woody hosts, both in nurseries and on mature trees [18,26,27,28,29]. Several Phytophthora species were reported to be associated with damping-off of seedlings, stem cankers, root rot, and decline of Pinus species, including P. austrocedri, P. cactorum, P. xcambivora, P. capsici, P. castaneae, P. cinnamomi sensu lato, P. citricola sensu lato, P. citrophthora, P. cryptogea, P. crassamura, P. drechsleri, P. fallax, P. humicola, P. kernoviae, P. nicotianae, P. obscura, P. pini, P. plurivora, P. syringae, and other unidentified Phytophthora spp. [29,30,31,32,33,34,35]. Pinus plants affected by Phytophthora crown and root rot show growth reduction, chlorosis, and decline [18]. The genus Phytophthora includes species that have a significant impact in European pine forests and are believed to be among major pine pathogens occurring together with F. circinatum. Infections by these Oomycetes are favored by climatic factors, such as high humidity, rain, and warm temperatures, which are known to be conducive also for infections by Fusarium spp. Therefore, interaction, including co-infection and synergy, between species of these two genera of pathogens can be assumed to be very probable [34].
During pathogen attack, different metabolic pathways are activated in the plant, including changes in primary energy-producing metabolism, plant defense mechanisms, and hormone signaling. Defense mechanisms are controlled by a series of genes, individually or synergistically involved in the resistance traits of plants [35,36,37,38]. As in the majority of plants, in conifers, the hypersensitive responses are the most commonly induced defense mechanisms against pathogens. These mechanisms include early lignification of fibers and the production of terpenoids and alkaloids that are phenolic compounds [39]. These responses are activated upon recognition of a pathogen by triggering signal transduction mechanisms that stimulate secondary metabolic pathways resulting in the production of defense compounds, which represents the result of the expression of a diverse range of genes [36,37,38,39,40]. Previous studies have identified a number of genes implicated in response to pathogen infection in coniferous species [41,42]. These encompass genes codifying for pathogenesis-inherent proteins (PR proteins), which represent the most numerous group of plant defense proteins, as well as genes coding for secondary metabolites with a wide range of antifungal activity, like the ones linked to the phenylalanine metabolic pathway [43,44]. Phenylalanine is pivotal in connecting primary and secondary plant metabolism and is being utilized by phenylalanine ammonia-lyase (PAL) for the synthesis of a number of compounds that are critical to the plant under stress situations [45]. In addition to PAL, other major pathogenesis-related (PR) proteins, including PR3 (chitinase) and PR5 (thaumatin-like protein), are commonly involved in resistance to fungal pathogens [46]. Also, plant respiration is usually enhanced after infection of the pathogen. Pyruvate decarboxylase (PDC) is triggered under stressed circumstances and can transform pyruvate into acetaldehyde and CO2 under aerobic fermentation. Acetaldehyde may be converted to ethanol during fermentation or be transformed into acetyl-CoA by pyruvate dehydrogenase and enter the tricarboxylic acid (TCA) cycle [47]. While the expression levels of genes encoding for pathogenesis-related proteins and PDC [46] has been extensively investigated in pathosystems where a single pathogen is involved, little information is available on the relative expression levels of genes that respond to the plant-pathogen interaction in more complex pathosystems where two or more pathogens simultaneously infect the host-plant.
The aim of this study was to investigate: i. the phenotypic response of Monterey pine to co-infections by F. circinatum and Phytophthora species, and ii. the relative expression levels of genes encoding for pathogenesis-related proteins and PDC in seedlings of Monterey pines co-infected by these pathogens.
2. Results
2.1. Symptom Progression
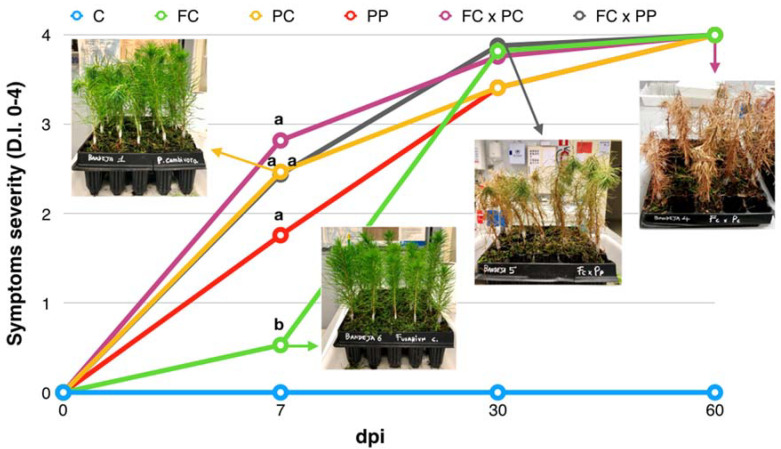
A common symptom on inoculated seedlings was the folding of the apex (damping-off). Other symptoms included restricted resin exudations and necrotic lesions around the wound, which were visible in the advanced stage of the disease. Non-inoculated control seedlings were asymptomatic. In detail, 7 days post-inoculation (dpi), the first symptom on all inoculated seedlings was the damping-off of the apex; all inoculated seedlings showed severe symptoms with disease severity in terms of disease index (D.I.) ranging from 2 to 3, except seedlings inoculated only with F. circinatum (FC) which initially showed significantly less severe symptoms (mean D.I. 0.5) (Figure 1). Thirty dpi, seedlings from all the treatments, including those inoculated only with F. circinatum, showed an advanced state of decay (D.I. ranging from 3 to 4). At this evaluation date, additional symptoms were wilting and browning of needles, indicating the plant was about to die. As a matter of fact, the death of all inoculated seedlings occurred 60 dpi.
Figure 1.
Time course of symptoms progression in Pinus radiata seedlings non-inoculated (C) or inoculated with Fusarium circinatum (FC), P. xcambivora (PC), P. parvispora (PP), F. circinatum ∗ P. xcambivora (FC × PC) and F. circinatum ∗ P. parvispora (FC × PP). Symptom severity was expressed as mean value of disease index (D.I.) at 7, 30, and 60 days post-inoculation (dpi). Images show the type of symptoms at each time interval post-inoculation. At each time interval, values sharing the same letters are not significantly different according to Tukey’s honestly significant difference (HSD) test (p ≤ 0.05).
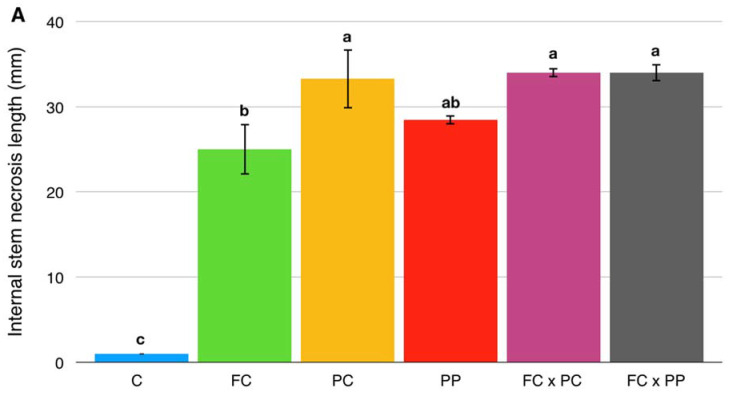
The time course of symptom progression is summarized in Figure 1. All inoculated seedlings, with no exceptions, showed an extended necrotic stem lesion, while non-inoculated control seedlings did not show any symptoms (Figure 1 and Figure 2B). The length of the internal stem lesions was measured at 14 dpi, when more than 50% of the inoculated seedlings in each treatment showed symptoms. The statistically significant differences between the length of stem lesions in seedlings inoculated with single pathogens or their combinations (Figure 1 and Figure 2A) were consistent with the severity of visual symptoms, as evaluated 7 dpi (Figure 1).
Figure 2.
(A) Mean values of the length of internal stem necrosis (mm) in Pinus radiata seedlings non-inoculated (C) or inoculated with Fusarium circinatum (FC), P. xcambivora (PC), P. parvispora (PP), F. circinatum ∗ P. xcambivora (FC × PC) and F. circinatum ∗ P. parvispora (FC × PP), 14 days post-inoculation (dpi), when more than 50% of the inoculated seedlings from each treatment showed disease symptoms. Data are presented as mean ± SE of four biological replicates per treatment. Values sharing the same letters are not statistically different according to Tukey’s honestly significant difference (HSD) test (p ≤ 0.05). (B) Stem internal necrosis from representative samples of Pinus radiata seedlings non-inoculated (C) or inoculated with Fusarium circinatum (FC), P. xcambivora (PC), P. parvispora (PP), F. circinatum ∗ P. xcambivora (FC × PC) and F. circinatum ∗ P. parvispora (FC × PP) observed using a zoom stereomicroscope when more than 50% of the inoculated seedlings of each treatment showed disease symptoms (14 dpi).
2.2. Housekeeping Gene Selection
In order to obtain reliable results, the expression level of target genes was standardized using internal control genes, also referred to as reference or housekeeping genes [47,48]. The expression level of these genes should be quite stable between diverse samples and environmental conditions. Until now, there have been no housekeeping genes that have been validated for the purpose of gene expression on conifers being infected by F. circinatum or similar necrotrophic fungi [46]. The stability of the three selected housekeeping genes was estimated by using the comparative delta Ct method and geNorm software. For both methods, ubiquitin (UBQ) was the more stable housekeeping gene, while actin (ACT) and β-Tubulin (TUB) TUB had the highest delta Ct values and the highest M values when analyzed with the geNorm software. The current study indicated that UBQ was the overall most stable reference gene for P. radiata that was infected with F. circinatum or similar necrotrophic fungi. This evidence was congruent with the results of other research aiming to provide validation of reference genes on plants under biotic and abiotic stress situations [46,49,50,51]. Therefore, UBQ was used for normalizing qRT-PCR results.
2.3. Differential Expression of Candidate Genes
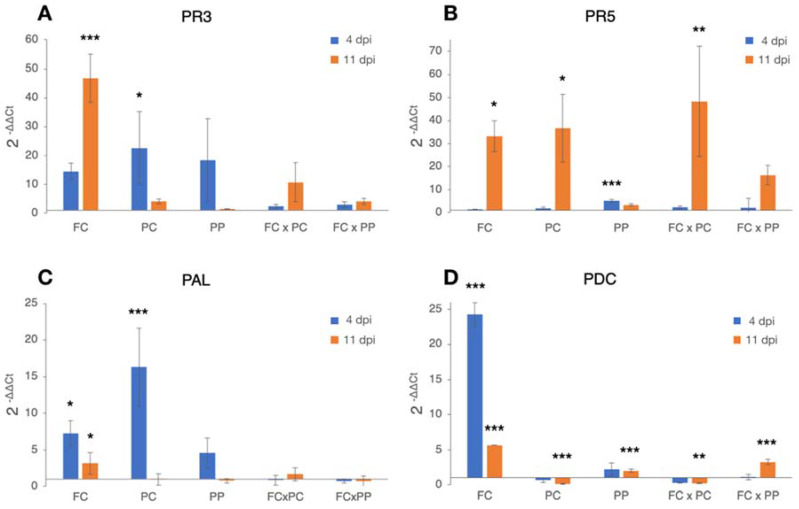
The analysis of the transcriptomic profile of the candidate genes (PR3, PR5, PAL, and PDC) evidenced variable responses depending on both the treatment and the time interval after inoculation. The PR3-encoding gene for chitinase resulted up-regulated in all treatments at both time intervals after inoculation (4 and 11 dpi), with significant differences over the non-inoculated control in seedlings inoculated with P. xcambivora (PC) (4 dpi) and F. circinatum (FC) (11 dpi) (Figure 3A).
Figure 3.
Differences in the expression levels of PR3- (A), PR5- (B), PAL- (C) and PDC- (D) -encoding genes in Pinus radiata seedlings at 4 (blue bars) and 11 (orange bars) days-post-inoculation (dpi) with Fusarium circinatum (FC), P. xcambivora (PC), P. parvispora (PP), F. circinatum ∗ P. xcambivora (FC × PC) and F. circinatum ∗ P. parvispora (FC × PP). Columns with asterisks are statistically different according to Dunnett’s test (* p < 0.05, ** p < 0.01, *** p < 0.001), compared with their calibrator (i.e., wounded and non-inoculated control seedlings).
The PR5-encoding gene for thaumatin-like protein, like the PR3, had a generalized up-regulation pattern, although it was significantly up-regulated mainly at 11 dpi. In particular, at 4 dpi, a weak significant up-regulation was observed only in seedlings inoculated with P. parvispora, while 11 dpi a marked up-regulation was observed in F. circinatum- (FC) and P. xcambivora-(PC) inoculated seedlings as well as in seedlings inoculated with F. circinatum ∗ P. xcambivora (FC × PC), and F. circinatum ∗ P. parvispora (FC × PC) (Figure 3B).
The PAL-encoding gene resulted generally up-regulated 4 dpi only in treatments that received the tested pathogens (FC, PC, PP) singularly, although the up-regulation was significant only in seedlings inoculated with F. circinatum (FC) and P. xcambivora (PC). Fusarium circinatum (FC) was the only significantly up-regulated treatment at 11 dpi (Figure 3C).
Finally, the PDC-encoding gene was significantly modulated in all the treatments, although with opposite trends. The strongest up-regulation was observed in seedlings from treatments with F. circinatum (FC) only at both time intervals (4 and 11 dpi). The gene was also up-regulated at both time intervals in the treatment with P. parvispora (PP), although the modulation was significant only at 11 dpi. Generalized significant down-regulation was recorded in seedlings from treatments with P. xcambivora alone (PC) and with F. circinatum (FC × PC). Finally, in the interaction system F. circinatum ∗ P. parvispora (FC × PP), the gene was significantly up-regulated only at 11 dpi (Figure 3D).
3. Discussion
The complex interactions of plant-multiple microorganisms represent a major topic of modern Plant Pathology [52,53]. Indeed, several studies started to investigate the mechanisms by which microorganisms interact with each other and affect the phenotypic and genetic response of plants as a consequence of their pathogenetic activity [52,53,54,55]. In order to try to elucidate how simultaneous infections by different plant pathogens can affect the progression of a specific plant disease traditionally imputed to a single pathogen, this study addressed the plant-microorganism interaction in the pine pitch canker pathosystem (Pinus radiata-Fusarium circinatum) under the additional influence of two aggressive Phytophthora species that cause stem cankers of pines. The synergism between pathogens is key to understanding the etiology and pathogenesis mechanisms of several complex diseases. Examples of this type of disease involving a Phytophthora species include the Phytophthora-Diaprepes complex and the Huanglongbing syndrome of citrus [56]. However, special attention was paid to the interactions between pathogenic fungi, including Fusarium species [57,58]. A typical example of a complex fungal disease is the esca disease of grapevine, which is present across many regions worldwide and is caused by diverse fungal pathogens present alone or in combination, including Ilyonectria spp., Phaeomoniella chlamydospora, Togninia spp., and Botryosphaeriaceae species [59,60]. A more recent study demonstrated that co-infection of several species of Botryosphaeriaceae and Ilyonectria results in a more severe decline of young grafted grapevines in the field [60]. Similarly, laboratory experiments further confirmed that co-inoculation of Ilyonectria and Botryosphaeriaceae isolates induced an increased disease severity compared to single inoculation of Ilyonectria isolates [61].
In the present study, the experimental approach was aimed at investigating the phenotypic and gene-mediated response of P. radiata plants to an infective process due to the simultaneous infection by two plant pathogens, F. circinatum and P. xcambivora or F. circinatum and P. parvispora. Previous studies reported synergic effects of diverse pathogens in the phenotypic plant response to complex diseases involving Fusarium species and other fungus and oomycete pathogens belonging to different genera. In the case of cassava root rot, the pathogens involved were Fusarium sp., Botryodiplodia theobromae, and Armillaria sp. [61]. In the gray necrosis of hazelnut, the species of fungal pathogens involved were Alternaria sp., Fusarium lateritium, and Phomopsis sp. [62]. Pythium sp., Fusarium sp., Cylindrocarpon sp., and Rhizoctonia sp. were responsible for the root rot of strawberry [63], and Pythium sp., Rhizoctonia sp., and Fusarium sp. were responsible for the root disease of Trifolium vesiculosum [64]. Potential interactions between F. circinatum and other fungal pathogens can also be expected. Indeed, the genera Heterobasidion, Armillaria, and Phytophthora include root and butt rot pathogens with a wide distribution and high impact in pine forests in Europe, and their infections are favored by factors such as high humidity, which are also favorable to Fusarium spp. [65,66]. Therefore, although a co-occurrence of F. circinatum has been only reported so far for Diplodia sapinea [66,67], it is most likely that diverse aggressive fungal species can infect simultaneously with F. circinatum. In the present study, the possible interaction of F. circinatum and aggressive Phytophthora species was investigated for the first time. Results showed the simultaneous infection by these pathogens exacerbated the severity of symptoms caused by F. circinatum alone in the early stages of the infection process, possibly due to more rapid colonization of the stem of pine seedlings by Phytophthora spp. In this respect, the co-infection has produced an additive rather than a synergistic effect. However, it has to be considered that, in this case, it would be difficult to evaluate any synergistic effect only on the basis of phenotypic response as all three tested pathogens alone had lethal effects on pine seedlings.
The study of gene expression was expected to be more effective than symptom severity to get a better insight into the response of the plant to multiple infections. It is well known that in plants, the infection by a pathogen triggers the activation of the main resistance mechanism, the systemic acquired resistance (SAR), which provides long-term resistance throughout the plant to subsequent infection by different pathogens [51]. This kind of resistance is correlated with the synthesis of pathogenesis-related (PR) proteins, which, in turn, is mediated by the up-regulation of genes encoding enzymes involved in the biosynthesis of salicylic acid (SA) [68]. Plant cellular respiration is also usually stimulated by pathogenic infections [45,50,51,52]. In such a kind of oxygen stress condition, it has been observed that plants respond by the synthesis of pyruvate decarboxylase (PDC), which then converts the pyruvate into acetaldehyde and CO2 and makes possible the production of energy [46]. In order to decrypt how the simultaneous infection by F. circinatum and Phytophthora species affects the plant defense response, this study describes the modulation of the SAR and plant cellular respiration by the analysis of the transcriptomic profile of three genes encoding for pathogenesis-related proteins (PR3, PR5, and PAL) as well as that of the pyruvate decarboxylase- (PDC) encoding gene.
The results from the interaction systems (F. circinatum ∗ P. xcambivora, and F. circinatum ∗ P. parvispora) showed that, at the early stages of the infection (4 dpi), none of the studied encoding genes had a significant modulation over the non-inoculated while at 11 dpi, significant up-regulation was exclusively reported for PR5 in the treatment F. circinatum ∗ P. xcambivora and PDC in F. circinatum ∗ P. parvispora. Interestingly, a significantly higher up-regulation of gene coding for PR5 was observed in the F. circinatum ∗ P. xcambivora interaction, suggesting a possible synergism of multiple infections in triggering this plant defense mechanism. Results from inoculations with a single pathogen overall evidenced that F. circinatum markedly stimulated the up-regulation of all the studied genes mainly at the late stages of infection, while between the two Phytophthora species, only P. xcambivora seemed to stimulate significant up-regulations. These effects can be explained assuming that: i. the competition between pathogens for the infection site and substrate could have determined a delay in the onset of the infective process by F. circinatum and consequently of the plant defensive response, and ii. multiple infections might have repressed the expression of defense-related genes, with the only exception of the PR5 encoding gene. In this respect, co-infection by aggressive pathogens might act synergistically by circumventing or silencing plant defense responses, thus exacerbating the final effect of the infection. Considering that this is the first study that investigated the pine response to multiple infections by different pathogens simultaneously, further studies are needed to validate these two hypotheses.
4. Materials and Methods
4.1. Plant Material
Ten-month-old Pinus radiata seedlings (Spanish provenance) were used for the experiment. Seedlings had an average height of 19.84 ± 1.8 cm and an average diameter at the soil level of 0.34 ± 0.03 cm. They were maintained in a greenhouse at 20–22 °C and a photoperiod of 16 h light/8 h darkness and inoculated after two weeks of acclimation.
4.2. Fungal Inoculum and Inoculation Methods
An isolate of F. circinatum from Pinus radiata (Fc 072) sourced in Spain and two isolates of Phytophthora, P. xcambivora from Quercus ilex (PH 14.012) sourced from forest soil in Spain, and P. parvispora recovered from rhizosphere soil of Salix pedicellata in a riparian forest in Sicily [19] were used for plant inoculation. Spore suspensions (F. circinatum microconidia and Phytophthora zoospores) were used for inoculum.
Fusarium circinatum was grown on potato dextrose agar (PDA) at 25 °C in the dark until the mycelium covered at least 90% of the Petri dish. Three to five pieces of mycelium (5 mm diameter) were grown under agitation on potato dextrose broth (PDB) for at least 24 h. Microconidia were then recovered by centrifugation, rinsed, and resuspended in sterile distilled water (s.d.w.). The final conidium concentration was adjusted to 106 conidia/mL using a hemocytometer.
Phytophthoraxcambivora and P. parvispora were first grown on V8 juice agar (V8A) at 20° C in the dark for 7 days. For the zoospore production, mycelium plugs from these colonies were flooded in s.d.w. with the addition of autoclaved soil extract and incubated for 2–3 days at 20–22 °C with a 16/8 h photoperiod. Sporangia formation was monitored during this incubation period, and once mature sporangia were observed, the plates were cold shocked by incubation at 4 °C for 45 min after which they were removed and left at room temperature for 1 h to stimulate zoospore release. The zoospores were removed from the plates, pooled together, and used for the inoculation. The zoospore concentration was determined by using a hemocytometer and standardized to 106 zoospores/mL.
The experimental scheme consisted of the following treatments: i. wounded plants control (C); ii. plants inoculated with F. circinatum (FC); iii. plants inoculated with P. xcambivora (PC); iv. plants inoculated with P. parvispora (PP); v. plants inoculated with F. circinatum ∗ P. xcambivora (FC × PC); vi. plants inoculated with F. circinatum ∗ P. parvispora (FC × PP). For the inoculation, 180 seedlings of Pinus radiata were used, 30 replicates per treatment (i.e., 1 forest tray of 30 cells). The stem surface of plants from all treatments was wounded by two cuts at the same height, at two opposite sides of the stem, using a sterile scalpel; 106 spores (F. circinatum microconidia and Phytophthora zoospores) were then applied at one of the two wounds; the other wound received sterile distilled water in plants from treatments ii., iii. and iv., while each wound received singularly one of the two pathogens in plants from treatments v. and vi. Wounds were then sealed with Parafilm® to prevent desiccation and contamination. Control plants were equally wounded and received an equal volume of s.d.w. After inoculation, controls and inoculated plants were kept in separate climate chambers under controlled conditions at a temperature of 20 ± 2 °C, 40–50% relative humidity, a photoperiod of 16/8 h light/dark for 60 days and irrigated 30 min per day.
4.3. Evaluation of Symptoms and Internal Necrosis Length
The typical symptoms of pine pitch canker disease are tip dieback, wilting, browning of needles, and resin formation and were evaluated and monitored once a week. The seedlings were visually scored for disease symptoms according to a 0–4 Disease Index (D.I.) empirical scale proposed by Correll et al. [69]: 0 = healthy plant or no symptoms; 1 = resin and/or necrosis at the point of inoculation and healthy foliage; 2 = resin and/or necrosis beyond the point of inoculation; 3 = severe wilting and noticeable dieback or damping-off and 4 = dead plant. Symptoms were evaluated at 3-time intervals after inoculation, i.e., 7, 30, and 60 days post-inoculation (dpi). To re-isolate the pathogens, stem cuttings were plated onto PDA or V8A and incubated at 20–22 °C for 7 days. The internal stem lesion length (mm) was also measured in 4 cm longitudinal stem cuts of three biological replicates per treatment when more than 50% of the inoculated plants showed disease symptoms (14 dpi). Images of the necrosis were obtained using a zoom stereomicroscope.
Data were analyzed by using one-way ANOVA followed by Tukey’s HSD test (Honestly Significant Difference) as a post hoc test (R software). Differences at p ≤ 0.05 were considered significant.
4.4. Sample Collection, RNA Extraction, and cDNA Synthesis
Stem fragments of approximately 4 cm in length, 1 cm above the inoculation point, were sampled for RNA extraction at 4 and 11 dpi, using 5 seedlings per treatment at each sampling time interval. Samples were stored at −80 °C until used. Total RNA was extracted by using Spectrum™ Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA) and quantified using a Qubit 4 Fluorometer (Invitrogen, Waltham, MA, USA). The integrity of RNA samples was visualized by loading 5 μL of RNA on a 1.5% agarose gel stained with SYBR Safe. cDNA was synthesized using Revert Aid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA), random hexamers, and 1μg of RNA in 20 µL reactions following the manufacturer’s specifications.
4.5. Selection of Primers and Housekeeping Genes
The primers used to amplify the four candidate genes, namely, PR3, PR5, PAL, and PDC [33,34,35,36,37,38,39], and three housekeeping genes, namely actin (ACT), β-tubulin (TUB), and ubiquitin (UBQ), are shown in Table 1. Primers were selected on the basis of previous studies [45,46,47,48,49,50]. Housekeeping genes’ stability was analyzed using the geNORM v. 3.4 software and the comparative delta-Ct method [70,71,72]. For the latter, the reference gene analysis tool refFinder (http://www.leonxie.com/referencegene.php) was used. After the analysis, the most stable reference gene was used for the normalization of the gene expression data.
Table 1.
Primer sequences used for the amplification of housekeeping and candidate genes studied in the multi-factorial system Pinus radiata-Fusarium circinatum-Phytophthora species.
| Gene | Primer Sequence | GenBank ID | Functions/Putative Functions | References |
|---|---|---|---|---|
| Chitinase (PR3) | F: TGGCAACACGGACGCCCATT | HM219849.1 | Hydrolyzation of chitin. | [45,50,73,74] |
| R: ACCGGCGTCGTTTCTGTGCTT | ||||
| Thaumatin-like protein (PR5) | F: AGGAGCGCGTGTGATGCGTT | JQ015859.1 | Involved in cell wall damage and formation of pores on the plasma membrane. | [45,46,47,48,49,50,75,76] |
| R: TGAAAGTGCTGGTGGCGTCGT | ||||
| Phenylalanine ammonia-lyase (PAL) | F: TGCTGGCCACTGTGAAGCAGA | AY641535.1 | Lignin and phenolic accumulation in plants. Cinnamic acid synthesis | [45,46,47,48,49,50,77,78] |
| R: TCGCAGAAACGGCCTGGCAA | ||||
| Pyruvate decarboxylase (PDC) | F: CCCGCAAACAATGACGTGGGGT | JQ264496.1 | Involved in aerobic fermentation. | [45,46,47,48,49,50,79,80] |
| R: TGCGAGCAGATGGTCCAGCA | ||||
| Actin (ACT) | F: TGGACCTTGCTGGGCGTGATCT | GQ339779.1 | Major component of cytoskeleton microfilaments. |
[45,46,47,48,49,50,81] |
| R: ACAATCTCGCGCTCTGCGGT | ||||
| β-Tubulin (TUB) | F: AAGGGGGTCAGTGTGGCAACCA | KM496536.1 | Structural units of the cytoskeleton microtubes | [45,46,47,48,49,50,82] |
| R: ACAGCCCGCGGAACAAACCT | ||||
| Ubiquitin (UBQ) | F: AGCCCTTATGCCGGAGGGGTTT | AF461687.1 | Participates in protein recognition by the proteasome | [45,47,50] |
| R: AGTGCGGGACTCCACTGTTCCT |
4.6. Relative Expression of Candidate Genes
Amplifications were performed by using the iCycler iQ™ Real-Time PCR Detection System (Biorad). Reactions were performed in a total volume of 20 μL by mixing 10 ng of cDNA with 1 μL of 10 μM of each primer and 10 μL of PowerUp™ SYBR™ Green Master Mix (2X) (Applied Biosystems). qRT-PCR experiments were carried out in triplicate. The thermo-cycling conditions were 2 min at 50 °C (UDG activation), 2 min at 95 °C (Dual-Lock™ DNA polymerase) followed by 40 cycles of two steps: 95 °C for 15 s (denaturation) and 65 °C (annealing/extension) for 1 min. The quantification of gene expression was carried out by using the 2−ΔΔCt method where ΔΔCt = (Ct of target gene-Ct of reference gene) sample-(Ct of target gene-Ct of reference gene) calibrator and Ct is the threshold cycle of each transcript, defined as the point at which the amount of amplified target reaches a fixed threshold above the background fluorescence and using housekeeping genes for data normalization as described by Vandesompele et al. [70,71].
Data on gene expression were analyzed by using one-way ANOVA followed by Dunnett’s multiple comparisons test by using R software. Differences at p ≤ 0.05 were considered significant.
5. Conclusions
This study investigated for the first time the effects of co-infections of pine seedlings by diverse aggressive pathogens, the true fungus Fusarium circinatum, causing pine pitch canker disease, and the oomycetes Phytophthora xcambivora and P. parvispora, causing crown and root rot. According to the aim, it provided a preliminary contribution to the understanding of the genetics of plant defense mechanisms in multiple infections. Based on the development of symptoms and the genetic analysis of the transcriptomic profile of the pyruvate decarboxylase-(PDC)-encoding gene and three genes encoding pathogenesis-related proteins (PR3, PR5, and PAL), two tentative hypotheses were proposed: i. the competition between pathogens could have delayed the infective process by F. circinatum and the plant defense response; ii. co-infection might have repressed the expression of defense-related genes, thus exacerbating the severity of the disease.
Acknowledgments
The authors are grateful to Ann Davies for the English revision of the text.
Author Contributions
Conceptualization, S.O.C., J.M.-G., and J.J.D.; methodology, S.O.C., J.M.-G., J.J.D., and F.A.; software, F.A. and C.Z.-B.; validation, S.O.C., and J.J.D.; formal analysis, F.A. and C.Z.-B.; investigation, F.A. and C.Z.-B.; resources, S.O.C. and J.J.D.; data curation, F.A.; writing—original draft preparation, F.A.; writing—review and editing, S.O.C., J.M.-G., and J.J.D.; visualization, S.O.C.; supervision, S.O.C. and J.J.D.; project administration, S.O.C.; funding acquisition, S.O.C. and J.J.D. All authors have read and agreed to the published version of the manuscript.
Funding
This article is based upon the activity of COST Action FP1406 PINESTRENGTH (Pine Pitch Canker strategies for management of Gibberella circinata in greenhouses and forests) supported by COST (European Cooperation in Science and Technology); this research was funded by the University of Catania, Italy “Investigation of phytopathological problems of the main Sicilian productive contexts and eco-sustainable defense strategies (MEDIT-ECO)” “PiaCeRi-PIAno di inCEntivi per la Ricerca di Ateneo 2020-22 linea 2” “5A722192155”; by the project “Smart and innovative packaging, postharvest rot management and shipping of organic citrus fruit (BiOrangePack)” Partnership for Research and Innovation in the Mediterranea Area (PRIMA)-H2020 (E69C20000130001). F.A. has been granted a Ph.D. fellowship “Scienze Agrarie, Alimentari, Forestali e Ambientali—XXXIII cycle”, University of Palermo.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Row Data can be shared upon a reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rogers D.L. In situ genetic conservation of a naturally restricted and commercially widespread species, Pinus radiata. For. Ecol. Manag. 2004;197:311–322. doi: 10.1016/j.foreco.2004.05.022. [DOI] [Google Scholar]
- 2.Garbelotto M. Molecular analysis to study invasions by forest pathogens: Examples from Mediterranean ecosystems. Phytopath. Medit. 2008;47:183–203. [Google Scholar]
- 3.Guerrero P.C., Bustamante R.O. Can native tree species regenerate in Pinus radiata plantations in Chile? Evidence from field and laboratory experiments. For. Ecol. Manag. 2007;253:97–102. doi: 10.1016/j.foreco.2007.07.006. [DOI] [Google Scholar]
- 4.Wingfield M.J., Hammerbacher A., Ganley R.J., Steenkamp E.T., Gordon T.R., Wingfield B.D., Coutinho T.A. Pitch canker caused by Fusarium circinatum—A growing threat to pine plantations and forests worldwide. Australas. Plant Pathol. 2008;37:319–334. doi: 10.1071/AP08036. [DOI] [Google Scholar]
- 5.Hodge G., Dvorak S. Differential responses of Central American and Mexican pine species and Pinus radiata to infection by the pitch canker fungus. New For. 2000;19:241–258. doi: 10.1023/A:1006613021996. [DOI] [Google Scholar]
- 6.EPPO PM7/91(2) Fusarium circinatum (formerly Giberella circinata) Bull. OEPP/EPPO Bull. 2019;49:228–247. doi: 10.1111/epp.12587. [DOI] [Google Scholar]
- 7.Ioos R., Aloi F., Piškur B., Guinet C., Mullett M., Berbegal M., Bragança H., Cacciola S.O., Oskay F., Cornejo C., et al. Transferability of PCR-based diagnostic protocols: An international collaborative case study assessing protocols targeting the quarantine pine pathogen Fusarium circinatum. Sci. Rep. UK. 2019;9:8195. doi: 10.1038/s41598-019-44672-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon T.R., Wikler R., Clark K.R., Okamoto S.L., Storer D., Bonello A.J. Resistance to pitch canker disease, caused by Fusarium subglutinans f. sp. pini, in Monterey pine (Pinus radiata) Plant Pathol. 1998;47:706–711. [Google Scholar]
- 9.Garbelotto M., Smith T., Schweigkofler W. Variation of spore dispersal of Fusarium circinatum, the causal agent of pine pitch canker, over a 12-month-period at two locations in Northern California. Phytopathology. 2008;98:137–143. doi: 10.1094/PHYTO-98-1-0137. [DOI] [PubMed] [Google Scholar]
- 10.Hepting G.H., Roth E.R. Pitch canker, a new disease of southern pines of forestry. J. For. 1946;44:742–744. [Google Scholar]
- 11.Viljoen A., Wingfield M.J., Marasas W.F.O. First report of Fusarium subglutinans f. sp. pini on seedlings in South Africa. Plant Dis. 1994;78:309–312. doi: 10.1094/PD-78-0309. [DOI] [Google Scholar]
- 12.Lee J.K., Lee S., Yang S., Lee Y. First report of pitch canker disease on Pinus rigida in Korea. Plant Pathol. 2000;16:52–54. [Google Scholar]
- 13.Thoungchaleun V., Kim K.W., Lee D.K., Chang C.S., Park E.W. Pre-infection behavior of the pitch canker fungus Fusarium circinatum on pine stems. Plant Pathol. J. 2008;24:112–117. doi: 10.5423/PPJ.2008.24.2.112. [DOI] [Google Scholar]
- 14.Gordon T.R., Storer A.J., Wood D.L. The pitch canker epidemic in California. Plant Dis. 2001;85:1128–1139. doi: 10.1094/PDIS.2001.85.11.1128. [DOI] [PubMed] [Google Scholar]
- 15.Abad G.Z. The taxonomy of Phytophthora: What is done and what is needed for the correct identification and diagnostics of species in the genus; Proceedings of the 7th International Union of Forest Research Organizations, IUFRO Working Party 7-02-09 Meeting, Phytophthora in Forests and Natural Ecosystems; Esquel, Argentina. 10–14 November 2014. [Google Scholar]
- 16.Ruano-Rosa D., Schena L., Agosteo G.E., Magnano di San Lio G., Cacciola S.O. Phytophthora oleae sp. nov., causing fruit rot of olive in southern Italy. Plant Pathol. 2018;67:1362–1373. doi: 10.1111/ppa.12836. [DOI] [Google Scholar]
- 17.Brasier C.M. Phytophthora Pathogens of Trees: Their Rising Profile in Europe. Forestry Commission; Stockport, UK: 1999. p. 30. [Google Scholar]
- 18.Jung T., Pérez-Sierra A., Duran A., Horta Jung M., Balci Y., Scanu B. Canker and decline diseases caused by soil-and airborne Phytophthora species in forests and woodlands. Persoonia. 2018;40:180–220. doi: 10.3767/persoonia.2018.40.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riolo M., Aloi F., La Spada F., Sciandrello S., Moricca S., Santilli E., Pane A., Cacciola S.O. Diversity of Phytophthora communities across different types of Mediterranean vegetation in a nature reserve area. Forests. 2020;11:853. doi: 10.3390/f11080853. [DOI] [Google Scholar]
- 20.Jung T., Horta Jung M., Cacciola S.O., Cech T., Bakonyi J., Seress D., Mosca S., Schena L., Seddaiu S., Pane A., et al. Multiple new cryptic pathogenic Phytophthora species from Fagaceae forests in Austria, Italy and Portugal. IMA Fungus. 2017;8:219–244. doi: 10.5598/imafungus.2017.08.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garbelotto M., Frankel S., Scanu B. Soil- and waterborne Phytophthora species linked to recent outbreaks in Northern California restoration sites. Calif. Agric. 2018;72:208–216. doi: 10.3733/ca.2018a0033. [DOI] [Google Scholar]
- 22.Scott P., Bader M.K.-F., Burgess T., Hardy G., Williams N. Global biogeography and invasion risk of the plant pathogen genus Phytophthora. Environ. Sci. Policy. 2019;101:175–182. doi: 10.1016/j.envsci.2019.08.020. [DOI] [Google Scholar]
- 23.Jung T., La Spada F., Pane A., Aloi F., Evoli M., Horta Jung M., Scanu B., Faedda R., Rizza C., Puglisi I., et al. Diversity and distribution of Phytophthora species in protected natural areas in Sicily. Forests. 2019;10:259. doi: 10.3390/f10030259. [DOI] [Google Scholar]
- 24.Erwin D.C., Ribeiro O.K. Phytophthora Diseases Worldwide. APS Press; Saint Paul, MN, USA: 1996. p. 562. [Google Scholar]
- 25.Marçais B., Caël O., Delatour C. Interaction between root rot basidiomycetes and Phytophthora species on pedunculate oak. Plant Pathol. 2011;60:296–303. doi: 10.1111/j.1365-3059.2010.02378.x. [DOI] [Google Scholar]
- 26.Migliorini D., Tondini E., Luchi N., Ghelardini L., Capretti P., Santini A. Detection of Phytophthora species on different woody species in nurseries; Proceedings of the 7th International Union of Forest Research Organizations, IUFRO Working Party 7-02-09 Meeting, Phytophthora in Forests and Natural Ecosystems; Esquel, Argentina. 10–14 November 2014. [Google Scholar]
- 27.Jung T., Orlikowski L., Henricot B., Abad-Campos P., Aday Kaya A.G., Aguín Casal O., Bakonyi J., Cacciola S.O., Cech T., Chavarriaga D., et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Pathol. 2016;46:134–146. doi: 10.1111/efp.12239. [DOI] [Google Scholar]
- 28.Santilli E., Riolo M., La Spada F., Pane A., Cacciola S.O. First report of root rot caused by Phytophthora bilorbang on Olea europaea in Italy. Plants. 2020;9:826. doi: 10.3390/plants9070826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chavarriaga D., Bodles W.J.A., Leifert C., Belbahri L., Woodward S. Phytophthora cinnamomi and other fine root pathogens in north temperate pine forests. FEMS Microbiol. Lett. 2007;276:67–74. doi: 10.1111/j.1574-6968.2007.00914.x. [DOI] [PubMed] [Google Scholar]
- 30.Jung T., Burgess T.I. Re-evaluation of Phytophthora citricola isolates from multiple woody hosts in Europe and North America reveals a new species, Phytophthora plurivora sp. nov. Persoonia. 2009;22:95–110. doi: 10.3767/003158509X442612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tkaczyk M., Sikora K., Nowakowska J., Ani´sko E., Oszako T., Belbahri L., Milenković I. Four different Phytophthora species that are able to infect Scots pine seedlings in laboratory conditions. Folia For. Pol. Ser. 2016;58:123–130. doi: 10.1515/ffp-2016-0014. [DOI] [Google Scholar]
- 32.Cleary M., Blomquist M., Vetukuri R.R., Böhlenius H., Witzell J. Susceptibility of common tree species in Sweden to Phytophthora cambivora, P. plurivora and P. cactorum. For. Pathol. 2017;47:e12329. doi: 10.1111/efp.12329. [DOI] [Google Scholar]
- 33.Sechi C., Seddaiu S., Linaldeddu B.T., Franceschini A., Scanu B. Dieback and mortality of Pinus radiata trees in Italy associated with Phytophthora cryptogea. Plant Dis. 2014;98:159. doi: 10.1094/PDIS-05-13-0572-PDN. [DOI] [PubMed] [Google Scholar]
- 34.Elvira-Recuenco M., Cacciola S.O., Sanz-Ros A.V., Garbelotto M., Aguayo J., Solla A., Mullett M., Drenkhan T., Oskay F., Aday Kaya A.G., et al. Potential interactions between invasive Fusarium circinatum and other pine pathogens in Europe. Forests. 2020;11:7. doi: 10.3390/f11010007. [DOI] [Google Scholar]
- 35.Landa B.B., Arias-Giraldo L.F., Henricot B., Montes-Borrego M., Shuttleworth L.A., Pérez-Sierra A. Diversity of Phytophthora species detected in disturbed and undisturbed British soils using high-throughput sequencing targeting ITS rRNA and COI mtDNA regions. Forests. 2021;12:229. doi: 10.3390/f12020229. [DOI] [Google Scholar]
- 36.Eyles A., Bonello P., Ganley R., Mohammed C. Induced resistance to pests and pathogens in trees. New Phytol. 2010;185:893–908. doi: 10.1111/j.1469-8137.2009.03127.x. [DOI] [PubMed] [Google Scholar]
- 37.Fiorilli V., Catoni M., Lanfranco L., Zabet N.R. Interactions of Plants with Bacteria and Fungi: Molecular and Epigenetic Plasticity of the Host. Front. Plant Sci. 2020;11:274. doi: 10.3389/fpls.2020.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yadav V., Wang Z., Wei C., Amo A., Ahmed B., Yang X., Zhang X. Phenylpropanoid pathway engineering: An emerging approach towards plant defense. Pathogens. 2020;9:312. doi: 10.3390/pathogens9040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franceschi V.R., Krokene P., Christiansen E., Krekling T. Anatomical and chemical defenses of conifer bark against bark beetles and other pests. New Phytol. 2005;167:353–375. doi: 10.1111/j.1469-8137.2005.01436.x. [DOI] [PubMed] [Google Scholar]
- 40.Morse A.M., Nelson C.D., Covert S.F., Holliday A.G., Smith K.E., Davis J.M. Pine genes regulated by the necrotrophic pathogen Fusarium circinatum. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2004;109:922–932. doi: 10.1007/s00122-004-1719-4. [DOI] [PubMed] [Google Scholar]
- 41.Quesada T., Gopal V., Cumbie W.P., Eckert A.J., Wegrzyn J.L., Neale D.B., Goldfarb B., Huber D.A., Casella G., Davis J.M. Association mapping of quantitative disease resistance in a natural population of loblolly pine (Pinus taeda L.) Genetics. 2010;186:677–686. doi: 10.1534/genetics.110.117549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Loon L.C., Rep M., Pieterse C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 43.Veluthakkal R., Dasgupta M. Pathogenesis-related genes and proteins in forest tree species. Trees. 2010;24:993–1006. doi: 10.1007/s00468-010-0489-7. [DOI] [Google Scholar]
- 44.Pascual M.B., El-Azaz J., Fernando N., Cañas R.A., Avila C., Cánovas F.M. Biosynthesis and metabolic fate of phenylalanine in conifers. Front. Plant Sci. 2016;7:1030. doi: 10.3389/fpls.2016.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donoso A., Rodriguez V., Carrasco A., Ahumada R., Sanfuentes E., Valenzuela S. Relative expression of seven candidate genes for pathogen resistance on Pinus radiata infected with Fusarium circinatum. Physiol. Mol. Plant Pathol. 2015;92:42–50. doi: 10.1016/j.pmpp.2015.08.009. [DOI] [Google Scholar]
- 46.Bolton M.D. Primary metabolism and plant defense-fuel for the fire. Mol. Plant Microbe Interact. 2009;22:487–497. doi: 10.1094/MPMI-22-5-0487. [DOI] [PubMed] [Google Scholar]
- 47.Long X.Y., Wang J.R., Ouellet T., Rocheleau H., Wei Y.M., Pu Z.E., Jiang Q.T., Lan X.J., Zheng Y.L. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 2010;74:307–311. doi: 10.1007/s11103-010-9666-8. [DOI] [PubMed] [Google Scholar]
- 48.Brunner A.M., Yakovlev I.A., Strauss S.H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004;4:14. doi: 10.1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jarosova J., Kundu J.K. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 2010;10:146. doi: 10.1186/1471-2229-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amaral J., Correia B., António C., Rodrigues A.M., Gómez-Cadenas A., Valledor L., Hancock R.D., Alves A., Pinto G. Pinus susceptibility to pitch canker triggers specific physiological responses in symptomatic plants: An integrated approach. Front. Plant Sci. 2009;10:509. doi: 10.3389/fpls.2019.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.La Spada F., Stracquadanio C., Riolo M., Pane A., Cacciola S.O. Trichoderma counteracts the challenge of Phytophthora nicotianae infections on Tomato by modulating plant defense mechanisms and the expression of crinkler, necrosis-inducing Phytophthora protein 1, and cellulose-binding elicitor lectin pathogenic effectors. Front. Plant Sci. 2020;11:583539. doi: 10.3389/fpls.2020.583539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.La Spada F., Aloi A., Coniglione M., Pane A., Cacciola S.O. Natural biostimulants elicit plant immune system in an integrated management strategy of the postharvest green mold of orange fruits incited by Penicillium digitatum. Front. Plant Sci. 2021;12:684722. doi: 10.3389/fpls.2021.684722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tucci M., Ruocco M., De Masi L., De Palma M., Lorito M. The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 2011;12:341–354. doi: 10.1111/j.1364-3703.2010.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalio R.J.D., Máximo H.J., Oliveira T.S., Azevedo T.M., Felizatti H.L., Campos M.A., Machado M.A. Molecular basis of Citrus sunki susceptibility and Poncirus trifoliata resistance upon Phytophthora parasitica attack. Mol. Plant Microb. Interact. 2018;31:386–398. doi: 10.1094/MPMI-05-17-0112-FI. [DOI] [PubMed] [Google Scholar]
- 55.Panabières F., Ali G.S., Allagui M.B., Dalio R.J.D., Gudmestad N.C., Kuhn M.-L., Gua Roy S., Schena L., Zampounis A. Phytophthora nicotianae diseases worldwide: New knowledge of a long-recognised pathogen. Phytopath. Medit. 2016;55:20–40. [Google Scholar]
- 56.Lamichhane J.R., Venturi V. Synergisms between microbial pathogens in plant disease complexes: A growing trend. Front. Plant Sci. 2015;6:385. doi: 10.3389/fpls.2015.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costa M.M., Silva B.A.A.S., Moreira G.M., Pfenning L.H. Colletotrichum falcatum and Fusarium species induce symptoms of red rot in sugarcane in Brazil. Plant Path. 2021;70:1807–1818. doi: 10.1111/ppa.13423. [DOI] [Google Scholar]
- 58.Mugnai L., Graniti A., Surico G. Esca (black measles) and brown wood-streaking: Two old and elusive diseases of grapevines. Plant Dis. 1999;83:404–418. doi: 10.1094/PDIS.1999.83.5.404. [DOI] [PubMed] [Google Scholar]
- 59.Gramaje D., Armengol J. Fungal trunk pathogens in the grapevine propagation process: Potential inoculum sources, detection, identification, and management strategies. Plant Dis. 2011;95:1040–1055. doi: 10.1094/PDIS-01-11-0025. [DOI] [PubMed] [Google Scholar]
- 60.Whitelaw-Weckert M.A., Sergeeva V., Priest M.J. Botryosphaeria stevensii infection of Pinot Noir grapevines by soil-root transmission. Australas. Plant Path. 2006;35:369–371. doi: 10.1071/AP06034. [DOI] [Google Scholar]
- 61.Bandyopadhyay R., Mwangi M., Aigbe S.O., Leslie J.F. Fusarium species from the cassava root rot complex in west Africa. Phytopathology. 2006;96:673–676. doi: 10.1094/PHYTO-96-0673. [DOI] [PubMed] [Google Scholar]
- 62.Belisario A., Maccaroni M., Coramusi A., Corazza L., Pryor B.M., Figuli P. First report of Alternaria species groups involved in disease complexes of hazelnut and walnut fruit. Plant Dis. 2004;88:426. doi: 10.1094/PDIS.2004.88.4.426A. [DOI] [PubMed] [Google Scholar]
- 63.Wing K.B., Pritts M.P., Wilcox W.F. Strawberry black root rot: A review. Adv. Strawb. Res. 1994;13:13–19. [Google Scholar]
- 64.Pemberton I.J., Smith G.R., Philley G.L., Rouquette F.M., Yuen G.Y. First Report of Pythium ultimum, P. irregulare, Rhizoctonia solani AG 4 and Fusarium proliferatum from Arrowleaf clover (Trifolium vesiculosum): A disease complex. Plant Dis. 1998;82:128. doi: 10.1094/PDIS.1998.82.1.128B. [DOI] [PubMed] [Google Scholar]
- 65.Garbelotto M., Gonthier P. Biology, epidemiology and control of Heterobasidion species worldwide. Annu. Rev. Phytopath. 2013;51:39–59. doi: 10.1146/annurev-phyto-082712-102225. [DOI] [PubMed] [Google Scholar]
- 66.Bezos D., Martínez-Álvarez P., Diez J.J., Fernández M.M. Association levels between Pityophthorus pubescens and Fusarium circinatum in pitch canker disease affected plantations in northern Spain. Entomol. Gen. 2016;36:43–54. doi: 10.1127/entomologia/2016/0314. [DOI] [Google Scholar]
- 67.Bezos D., Martínez-Álvarez P., Fernández-Fernández M.M., Diez J.J. Fungi and insect diversity associated with Pinus radiata in pitch-canker-affected stands. Int. For. Rev. 2014;16:336. [Google Scholar]
- 68.Zhang Y., Xu S., Ding P., Wang D., Cheng Y.T., He J., Gao M., Xu F., Li X., Zhang Y. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA. 2010;107:18220–18225. doi: 10.1073/pnas.1005225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Correll J.C., Gordon T.R., McCain A.H., Fox J.W., Koehler C.S., Wood D.L., Schultz M.E. Pitch canker disease in California–Pathogenicity, distribution, and canker development on Monterey Pine (Pinus radiata) Plant Dis. 1991;75:676–682. doi: 10.1094/PD-75-0676. [DOI] [Google Scholar]
- 70.Vandesompele J., De Preter D., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Silver N., Best S., Jiang J., Thein S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 73.Collinge D.B., Kragh K.M., Mikkelsen J.D., Nielsen K.K., Rasmussen U., Vad K. Plant chitinases. Plant J. 1993;3:31–40. doi: 10.1046/j.1365-313X.1993.t01-1-00999.x. [DOI] [PubMed] [Google Scholar]
- 74.Mauch F., Mauch-Mani B., Boller T. Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and beta-1,3-glucanase. Plant Physiol. 1988;88:936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grenier J., Potvin C., Trudel J., Asselin A. Some thaumatin-like proteins hydrolyse polymeric b-1,3-glucans. Plant J. 1999;19:473–480. doi: 10.1046/j.1365-313X.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- 76.Roberts W.K., Selitrennikoff C.P. Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J. Gen. Microbiol. 1990;136:1771–1778. doi: 10.1099/00221287-136-9-1771. [DOI] [Google Scholar]
- 77.Campbell M.M., Ellis B.E. Fungal elicitor-mediated responses in pine cell cultures: III. Purification and characterization of phenylalanine ammonialyase. Plant Physiol. 1992;98:62–70. doi: 10.1104/pp.98.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang X., Dron M., Cramer C.L., Dixon R.A., Lamb C.J. Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J. Biol. 1989;264:14486–14492. [PubMed] [Google Scholar]
- 79.Mithran M., Paparelli E., Novi G., Perata P., Loreti E. Analysis of the role of the pyruvate decarboxylase gene family in Arabidopsis thaliana under low-oxygen conditions. Plant Biol. (Stutt.) 2013;16:28–34. doi: 10.1111/plb.12005. [DOI] [PubMed] [Google Scholar]
- 80.Kürsteiner O., Dupuis I., Kuhlemeier C. The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol. 2003;132:968–978. doi: 10.1104/pp.102.016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi J.K., Holtzer S., Chacko S.A., Lin Z.X., Hoffman R.K., Holtzer H. Phorbol esters selectively and reversibly inhibit a subset of myofibrillar genes responsible for the ongoing differentiation program of chick skeletal myotubes. Mol. Cell Biol. 1991;11:4473–4482. doi: 10.1128/MCB.11.9.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jeong Y.-M., Mun J.-H., Lee I., Woo J.C., Hong C.B., Kim S.-G. Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol. 2006;140:196–209. doi: 10.1104/pp.105.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Row Data can be shared upon a reasonable request.